Abstract
Background:
Anterior cruciate ligament tear (ACLT) leads to protracted quadriceps atrophy. Protein turnover largely dictates muscle size and is highly responsive to injury and loading. Regulation of quadriceps molecular protein synthetic machinery following ACLT has largely been unexplored, limiting development of targeted therapies.
Purpose:
To define the effect of ACLT on 1) activation of protein synthetic and catabolic signaling within quadriceps biopsies from human participants, and 2) the time course of alterations to protein synthesis and its molecular regulation in a mouse ACL injury model.
Study Design:
Descriptive laboratory study
Methods:
Muscle biopsies were obtained from the ACL-injured and non-injured vastus lateralis of young adults following an overnight fast (n=21, 19±5 years). Mice had their limbs assigned to ACLT or control, and whole quadriceps were collected 6 hours, 1, 3, or 7 days post-injury with puromycin injected before tissue collection for assessment of relative protein synthesis. Muscle fiber size and expression and phosphorylation of protein anabolic and catabolic signaling proteins were assessed at the protein and transcript level (RNA-sequencing).
Results:
Human quadriceps showed reduced phosphorylation of ribosomal protein S6 (−41%) in the ACL-injured limb (P = .007), in addition to elevated phosphorylation of eukaryotic initiation factor 2α (+98%, P = .006), indicative of depressed protein anabolic signaling in the injured limb. No differences in E3 ubiquitin ligase expression were noted. Protein synthesis was lower at 1 (P = .01 vs. control limb) and 3 days post-ACLT in mice (P = .002 vs. control limb). Pathway analyses revealed shared molecular alterations between human and mouse quadriceps following ACLT.
Conclusions:
1) Global protein synthesis and anabolic signaling deficits occur in the quadriceps in response to ACL injury, without notable changes in measured markers of muscle protein catabolism. 2) Importantly, these deficits occur prior to the onset of significant atrophy, underscoring the need for early intervention.
Clinical Relevance:
These findings suggest blunted protein anabolism as opposed to increased catabolism likely mediates quadriceps atrophy following ACL injury. Thus, future interventions should aim to restore muscle protein anabolism rapidly following ACLT.
Keywords: skeletal muscle, anterior cruciate ligament, protein metabolism, E3 ubiquitin ligase
Introduction
Protracted quadriceps weakness is a debilitating consequence of anterior cruciate ligament tears (ACLT) 1, 25, 35. Strength deficits persist following reconstructive surgery and rehabilitation when patients return to sports or activities involving greater muscle demands 26, 44, 53, 54, 61. Recent research emphasizes derangements in quadriceps quality and morphology that significantly contribute to strength deficits 23, 37, 38. Quadriceps cross-sectional area (CSA) is a strong predictor of muscle strength several months after surgical reconstruction, underscoring the contribution of muscle size to strength deficits 36, 41, 43, 64, 71. Following ACL injury there is sizeable quadriceps fiber atrophy that is obstinate to surgical reconstruction and physical therapy 17, 50, 64, 65, necessitating the need for innovative strategies to enhance recovery of quadriceps muscle.
The maintenance of skeletal muscle mass is largely dependent on the balance between protein synthesis and breakdown. Skeletal muscle atrophy can be resultant from deficits in protein synthesis and/or increases in protein degradation 3, 68. However, whether blunted anabolism or elevated catabolism contributes more substantially to muscle atrophy remains unknown 57, 59. The molecular regulation of muscle protein metabolism following ACL injury has largely been unexplored with the exception of elevated expression of protein catabolic factors in pre-clinical models 12, 28. Importantly, mitigation of atrogene expression (muscle ring finger 1 [MuRF1]/Tripartite Motif Containing 63 [TRIM63] and Atrogin-1/muscle atrophy F-box [MAFbx]/F-Box Protein 32 [FBXO32]) was unable to offset quadriceps atrophy12, suggesting that protein catabolism is not the dominant mechanism causing ACL injury-induced muscle atrophy. Limited and underpowered data 27 from human subjects has impeded translation of pre-clinical findings as well as the development of targeted therapies to attenuate atrophy.
Molecular effectors of quadriceps atrophy following ACL injury need to be defined in order to develop targeted nutritional, rehabilitative and/or pharmacologic-based therapies. Additionally, the rapidity of molecular derangements is unclear, limiting evidence to inform the timing of prospective interventions. Thus, investigation of early molecular effectors of atrophy following ACLT is imperative to fully define the anabolic and catabolic response to the injury. Our purpose in the current study is to define the effect of ACLT on 1) activation of protein synthetic and catabolic signaling within quadriceps biopsies from human participants, and 2) the time course of alterations to protein synthesis and its molecular regulation in a mouse model of ACL injury. Therefore, we hypothesize deficits in protein synthetic and/or elevated catabolic signaling contributes to and precedes quadriceps atrophy resultant from ACL injury.
Methods
Ethical Approval
This study was approved by the Institutional Review Board (protocol #43046) at the University of Kentucky and performed in accordance with the ethical standards of the 1964 Declaration of Helsinki. Informed written consent was obtained from each patient before participation in this study. Additionally, all minors in this study assented to participation. Animal care and experimentation was administered under the University of Kentucky Institutional Animal Care and Use Committee (protocol #2019-3241). All mice were housed in a temperature-controlled room on a 14:10 light:dark cycle and provided chow and water ad libitum.
Study Design
Human
To be considered for inclusion, subjects had to be between 15-40 years of age, with their ACL tear occurring no more than 6 months prior to enrollment and register at least a 5/10 on the Tegner Activity Scale, or have torn their ACL during an athletic activity. Exclusion criteria included complete prior ACL injury/reconstruction, total knee dislocation, spinal fusion, taking anti-coagulant medication, recent inflammation or infection of the lower limb(s), diabetes, pregnancy, diminished capacity to provide informed consent, or inability to attend physical therapy or study visits. The diagnosis of the ACL injury was made by a single orthopedic surgeon (DLJ) via magnetic resonance imaging (MRI) and physical examination. All subjects were confirmed to have a complete tear of the ACL; 16 of 21 subjects also had a meniscus tear. Subject demographic information can be found in Table 1.
Table 1.
Patient demographics
| Mean ± SD | Range | Median | |
|---|---|---|---|
| Combined | |||
| N | 21 | ||
| Age | 18.5 ± 4.9 | 15 - 35 | 17 |
| BMI | 26.0 ± 3.5 | 21.4 - 34.0 | 25.4 |
| Days post-injury | 37 ± 36 | 9 - 149 | 21 |
| Males | |||
| N | 9 | ||
| Age | 16.8 ± 1.0 | 16 - 18 | 16 |
| BMI | 27.2 ± 3.9 | 22.2 - 34.0 | 25.8 |
| Days post-injury | 33 ± 33 | 9 - 116 | 21 |
| Females | |||
| N | 12 | ||
| Age | 19.8 ± 6.2 | 15 - 35 | 17 |
| BMI | 25.1 ± 3.0 | 21.4 - 30.6 | 24.5 |
| Days post-injury | 40 ± 40 | 11 - 149 | 20.5 |
No statistical differences are noted between male and female patients (all P > .05). BMI, body mass index.
Mouse
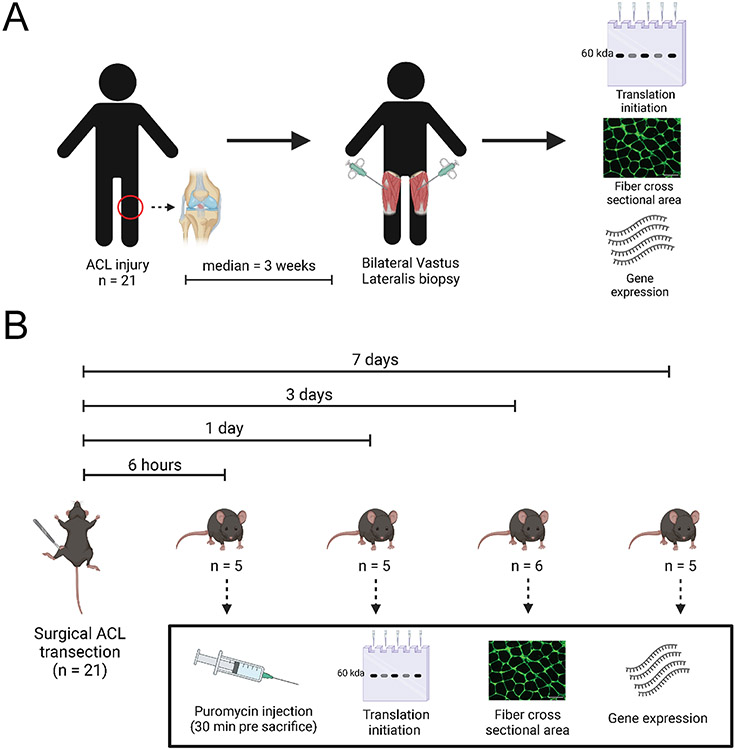
C57BL/6J mice (4-6 months of age) had one limb assigned to ACL surgical transection under general anesthesia with the contralateral limb serving as a healthy, un-injured control. An additional cohort of mice received a sham surgery (described below). At 6 hours (6h; n=5), 1 day (1d; n=5), 3 days (3d; n=6), and 7 days (7d; n=5) following ACL transection, mice were euthanized by cervical dislocation under deep anesthesia and quadriceps muscle was rapidly collected. Sample size estimates were based on published literature using murine models of limb immobilization to study muscle protein synthesis 40. We performed a power analysis on the puromycin protein synthesis differences at the 1 and 3 day post-ACLT time point. The difference in mean puromycin protein synthesis (AU) at the 1 day post-ACLT time point was 1.309 (AU) with a standard deviation of 0.520 (AU). With n=5 paired limbs per group, we had an observed power of 0.935. The difference in mean puromycin protein synthesis 3 day post-ACLT time point was 1.869 (AU) with a standard deviation of 0.817 (AU). With n=6 paired limbs per group, we had an observed power of 0.992. ACL sham surgery mice were euthanized under anesthesia and quadriceps muscle collected at 3 days (n=4) and 7 days (n=4) following sham surgery. Whole quadriceps muscles were collected following an overnight fast. A portion of quadriceps muscle from each limb was immediately frozen in liquid nitrogen for molecular analyses, and the remaining portion (corresponding to the vastus lateralis) was mounted in optimal cutting temperature mounting media (OCT, Tissue Tek) on cork and frozen in liquid nitrogen-cooled 2-methylbutane for immunohistochemical analyses. All samples were stored at −80°C. A graphical representation of the study design can be seen in Figure 1.
Figure 1. Study design.
(A) Human subjects study design; (B) Mouse study design.
Quadriceps Muscle Biopsies and Tissue Processing
Muscle biopsy specimens from the ACL-injured (ACL-inj) and contralateral Healthy vastus lateralis were collected immediately prior to ACL surgical reconstruction. Biopsies were obtained with the patients reporting to the operating room following an overnight fast. Patients were completely weight bearing at the time of biopsy collection. In addition, patients were given a home exercise program to follow and were encouraged to perform range of motion and strengthening exercises. Percutaneous muscle biopsy specimens (~100 mg) from the vastus lateralis were obtained. Samples were dissected free of fat and connective tissue; approximately 50 mg each were 1) immediately flash frozen in liquid nitrogen for protein and RNA analyses or 2) mounted in OCT on cork and flash frozen in liquid nitrogen–cooled 2-methylbutane. Samples were stored at −80°C until processing. Human muscle outcomes: protein expression, transcript read counts, fiber cross-sectional area.
Mouse Surgical ACL Transection
Surgical ACL transection was performed on a single limb for all mice, with the contralateral hind limb serving as an internal healthy control, similar to other studies19, 32. Aseptic surgical techniques were use at all times. Mice were anesthetized with 2.5% isoflurane in 1.0 L/min oxygen in a closed chamber and the surgical limb was carefully shaved and prepared with povidone-iodide scrub. The mice were then transferred to a headed, sterile surgical platform. A 3 mm longitudinal incision was made with a #11 blade over the distal patella to proximal tibial plateau. The joint capsule was opened by incision. The infrapatellar fat pad over the intercondylar area was dissected and the patellar tendon gently dislocated laterally to provide greater exposure of the femoral-tibial joint and allow visualization of the ACL in the intercondylar region. The ACL was completely transected using the #11 blade under visualization with a stereo dissecting microscope, and damage to the posterior cruciate ligament was avoided. The incision was closed with a 7-0 suture, and the mouse was placed in a clean cage over a heat pad and monitored for recovery. Transection was confirmed with a positive anterior drawer. Mice were monitored regularly throughout the post-surgical time course for signs of pain and inflammation and to confirm proper healing of the incision site. To control for the effects of surgery on study outcomes, we performed sham surgery on a subset of mice (n=4 each at 3 and 7 days post sham surgery). Sham surgery consisted of the same pre-operative care, longitudinal incision and opening of the joint capsule with no disturbance of the ACL, followed by the same suture protocol to close the incision.
Puromycin dosage and tissue collection
Puromycin is an aminonucleoside antibiotic and structural analogue of aminoacyl-transfer RNA that is incorporated into nascent polypeptide chains during elongation 49. At a low concentration insufficient to terminate the elongation phase of translation, incorporation of puromycin into peptides serves as an indicator of protein synthesis 60. The SUnSET (SUrface Sensing of Translation) technique utilizes an anti-puromycin antibody to detect peptides labeled by puromycin 60 and accurately measures in vivo whole muscle protein synthesis20.
Mice were given a single intraperitoneal injection of puromycin (0.04 μmol/g body mass) in 100 μl sterile phosphate buffered saline (PBS) exactly 30 minutes prior to tissue collection, as previously reported 20. Mice were humanely euthanized by cervical dislocation under deep anesthesia with isoflurane at 6 hour, 1 day, 3 days, and 7 days after ACL-transection. Quadriceps muscles were rapidly dissected from the ACLT and control hind limbs exactly 30 minutes following puromycin administration and processed for downstream analyses as described above. Mouse muscle outcomes: puromycin protein synthesis, protein expression, gene expression, fiber type and cross-sectional area.
Immunohistochemistry
Human
Frozen tissue was sectioned (7μm thickness) on a cryostat and air dried for one hour. Laminin immunohistochemical staining was performed with sections fixed in acetone for 3 minutes at −20°C, brought to room temperature, washed in PBS and then incubated overnight in and rabbit polyclonal anti-laminin (1:100, #L9393, Sigma, St. Louis, MO) in 1% bovine serum albumin 45. The following day, slides were incubated in goat anti rabbit AF350 (#A21068, Life Technologies) and then cover slipped.
Mouse
Frozen tissue was sectioned (7μm thickness) on a cryostat and air dried for one hour. Puromycin immunohistochemical staining was performed in accordance with a previously published method 20. In brief, sections were fixed in acetone for 10 minutes at −20°C, brought to room temperature, then blocked and permeabilized for one hour in PBS solution containing 0.5% triton, 0.5% TSA and 10% mouse on mouse blocking reagent (#MKB2213, Vector). After a brief wash, slides were incubated overnight 1:1 in Bovine IgG1 myosin heavy chain type 2a antibody (#SC-71, Developmental Studies Hybridoma Bank) and Bovine monoclonal IgM myosin heavy chain type 2b (#BF-F3, Developmental Studies Hybridoma Bank) antibody solution. Mouse monoclonal IgG2a anti-Puromycin (#MABE343, EMD Millipore) was diluted 1:1000 and rabbit polyclonal anti laminin antibody (#L9393, Sigma) was diluted 1:200 in the same fiber type solution. After another wash, slides were incubated for one hour in a PBS solution containing the following secondary antibodies diluted 1:500: Goat anti mouse IgG2a AF488 (#A21131, Life Technologies), goat anti mouse IgM AF555 (#S21426, Life Technologies), and goat anti rabbit AF350 (#A21068, Life Technologies).
For identification of green fluorescent protein (GFP) + myonuclei, unfixed sections were blocked in 1% bovine serum albumin and 10% mouse on mouse blocking reagent, and then incubated overnight in pericentriolar material 1 (PCM1, 1:100, # HPA023370, Sigma) and dystrophin (#VPD505, 1:50, Vector Labs) primary antibodies. The next day, slides were incubated for one hour in a PBS solution containing the following secondary antibodies diluted 1:500: goat anti mouse IgG1 AF647 (#A21240, Life Technologies) and goat anti rabbit AF555 (#A21428, Life Technologies). Lastly, sections were incubated for 10 min in 4’,6-diamidino-2-phenylindole (DAPI; 10 nM, LifeTechnologies), washed in PBS, and mounted using Vectashield fluorescence mounting media (Vector Laboratories).
Image acquisition and analysis
Human
Images were captured at 100x magnification with a Zeiss upright microscope (AxioImager M2; Zeiss, Oberkochen, Germany). Cross-sectional area of both the ACL-inj and Healthy limbs were performed in a blinded manner using MyoVision software 70.
Mouse
Images were captured at 200x magnification with a Zeiss upright microscope (AxioImager M2; Zeiss, Oberkochen, Germany). Image analysis was performed by a single assessor (ARK) in a blinded manner (blinded to group allocation and injury status of the muscle) using Zen Blue software for manual tracing of fibers for both cross-sectional area and puromycin intensity. Manual analysis of fiber-type specific cross-sectional area was performed 46. Briefly, fibers were classified as type 2a or type 2b positive, and cross-sectional area was determined for both fiber types, in addition to pooled fiber size irrespective to fiber type. Puromycin intensity analyses were performed according to methods published by Goodman et al. 22. Briefly, Zen Blue was used for manual analysis of puromycin intensity across the entire area of each individual fiber analyzed. A minimum of 30 fibers of each type (2a and 2b, 60 fibers total) were analyzed for cross-sectional area and puromycin intensity, which has been shown to produce reliable outcomes 21, 22. All puromycin image analysis was performed by a single assessor (ARK) in a blinded manner to the limb injury status.
RNA Isolation (human and mouse)
Quadriceps muscle tissue (~25-30mg) was homogenized in Tri-reagent using Zirconium Oxide beads (2.0 mm, RNase-free, Next Advance, Inc. Troy, NY). The homogenate was used to isolate total RNA in accordance with manufacturer guidelines (Direct-zol RNA Miniprep Kit, Zymo) 6. RNA content, purity, and integrity was quantified using the 2100 Bioanalyzer (Agilent) (RIN > 8.1) and the NanoDrop 2000 (Thermo Fisher) at the University of Kentucky Genomics Core.
RNA-sequencing (RNA-seq; human)
Six hundred nanograms of total RNA was sent to Novogene Corporation (Chula Vista, CA) for library construction and sequencing on an Illumina HiSeq 4000 system using a paired-end 150 bp dual-indexing protocol. Raw FASTQ files underwent pre-alignment quality control, then were aligned using STAR aligner (reference genome: GRCh38/hg38) using Partek Flow, and analyzed to obtain differential gene expression using DESeq2 (minimum read cutoff of 50) 6. Differential gene expression was calculated comparing ACL-inj and Healthy. Raw p-values were adjusted for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) step-up method. Pathway over-representation analysis was performed using gProfiler58 with non-ordered query and up-or-downregulated genes with FDR < .05. RNA-sequencing data are deposited in Gene Expression Omnibus: GSE211681.
Myonuclear-specific low-input RNA-seq (mouse)
Male and female HSA-rtTA+/−-;H2B-GFP+/− (HSA-GFP) mice were generated as previously described 11, 30, 69 and aged until 4-6 months. Nine mice (7M; 2F) were treated with low-dose doxycycline (0.5 mg/ml doxycycline in drinking water with 2% sucrose) for 1 week prior to undergoing ACLT surgery. This treatment strategy resulted in myonuclear labeling immediately prior to ACL injury. Mouse underwent a 1 week washout, and then were subjected to ACLT injury. Three days after the ACLT injury and following an overnight fast, myonuclei from both the ACLT and healthy contralateral quadriceps muscles were processed via Dounce homogenization in a physiological buffer (5 mM PIPES, 85 mM KCl, 1 mM CaCl2, 5% Sucrose, 0.5x HALT Protease Inhibitor [ThermoFisher], 0.5% NP-40) following an overnight fast, purified via fluorescent activated myonuclear sorting (GFP+ and had incorporated DAPI)67, then analyzed using myonucleus-specific RNA-sequencing. Immediately prior to fluorescent activated myonuclear sorting, samples were filtered through a 40μm filter (Fisher Scientific), DAPI was added as a viability marker (4μg/ml), then myonuclei were sorted directly into TRIzol LS Reagent (ThermoFisher), omitting any centrifugation steps to maximize myonuclear yield. Myonuclear RNA was extracted using the Zymo Direct-zol RNA Microprep Kit (Irvine, CA) in accordance with manufacturer guidelines. RNA content and integrity were quantified using the 2100 Bioanalyzer (Agilent) at the University of Kentucky Genomics Core. RNA samples were pooled (n=3 mouse muscles/pooled sample for both the ACLT and Healthy limb) to maximize RNA yield from isolated myonuclei as described previously48. This resulted in n=3 ACLT and n=3 Healthy distinct pooled samples that were sequenced. Prior work has shown that gene expression from RNA pools are similar to averages of individuals that comprise the pool 33. Three hundred nanograms of total RNA per pooled sample was sent to Novogene Corporation (Chula Vista, CA) for low-input (rRNA removal) library construction and sequencing on an Illumina HiSeq 4000 system using a paired-end 150 bp dual-indexing protocol. Raw FASTQ files underwent pre-alignment quality control, then were aligned using STAR aligner (reference genome: GRCm38/mm10) in Partek Flow, and analyzed to obtain differential gene expression using DESeq2 (minimum read cutoff of 50) 6, 11. Differential gene expression was calculated comparing ACLT and Healthy. Raw p-values were adjusted for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) step-up method. Pathway over-representation analysis was performed using gProfiler58 with non-ordered query and up-or-downregulated genes with FDR < .05. RNA-sequencing data are deposited in Gene Expression Omnibus: GSE211682.
qRT-PCR (mouse)
One microgram of RNA was reverse transcribed using iScript reaction mix (#170-8890, BioRad) and a MasterCycler Gradient thermocycler (EP-MC, Marshall Scientific). cDNA was then diluted 1:7 in nuclease free water and pipetted into a 96 well plate with mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH), MuRF-1, and Atrogin-1 primer probes (GAPDH: 4352339E, MuRF-1: 4331182, Atrogin-1: 4331182), and TaqMan fast advanced master mix (#4444556, ThermoFisher). The qPCR reaction was performed on the QuantStudio3 Real Time PCR system (Bio-rad) and data were analyzed using the 2− ΔΔCT method with GAPDH as a reference gene 42 following verification that GAPDH was stable across time points and between limbs in mouse quadriceps.
Statistics
Data are presented as mean ± SD with individual data points overlaid on bar charts. No data points were excluded from any human or murine sample, and there were no missing data. Human: Paired t-tests were performed between ACL-inj and Healthy limbs to compare each dependent variable, with significance set at P < .05. Mouse: Paired t-tests were performed between ACL-inj and Healthy limbs to compare each dependent variable, with significance set at P < .05. Comparisons between time points were not performed as our primary purpose was to define limb-specific differences that occur following ACLT. Both human and murine data met assumptions for parametric statistical testing: approximate normal distribution and equal variance. All analyses were performed with GraphPad Prism 9.0.2.
Results
Human quadriceps anabolic signaling
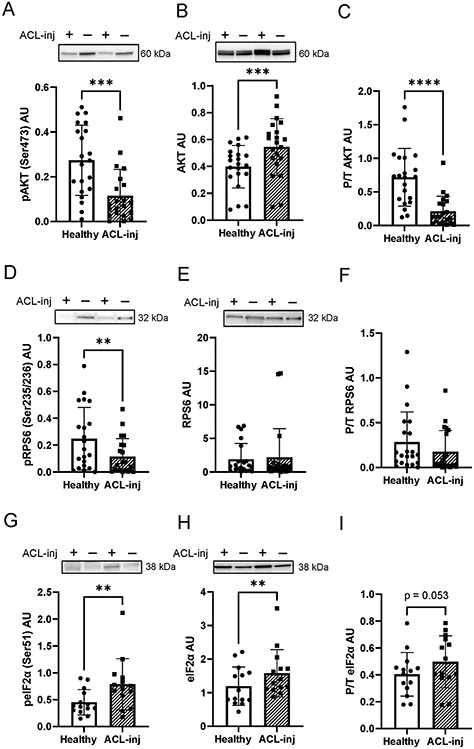
Phosphorylation status of hallmark anabolic signaling factors was significantly lower in human quadriceps following ACLT. We observed a 44% decrease in phospho-AKTSer473 (Figure 2A, P < .001) when comparing ACL-inj to Healthy. Total AKT was 46% higher in the ACL-inj limb (Figure 2B, P < .001), leading to a 64% decrease in phospho/total AKT (Figure 2C, P < .001). This difference was expounded with downstream effectors directly involved in translation initiation. Phospho-RPS6Ser235/236 decreased by 41% (Figure 2D, P = .0078) in the ACL-inj limb; however, no change was observed in total (Figure 3E, P = .72) or phospho/total RPS6 (Figure 2F, P = .14). A significant 98% increase was observed in phospho-eIF2αSer51 in the ACL-inj limb (Figure 2G, P =.006) with concomitant increase in total (Figure 2H, 45% increase, P =.0098) and phospho/total eIF2α (Figure 2I, 27% increase, P = .053) in the ACL-inj limb.
Figure 2. ACL injury decreases translational signaling in human quadriceps skeletal muscle.
Human quadriceps muscle protein lysates were probed with (A) phospho-AKTSer473, (B) total AKT, (C) phospho/total AKT, (D) phospho-RPS6Ser235/236, (E) total RPS6, (F) phospho/total RPS6, (G) phospho eIF2αSer51, and (H) total eIF2α, and (I) phospho/total eIFα. ** P < .01 vs Healthy limb, *** P <.005 vs Healthy limb, **** P < .0001 vs Healthy limb via paired t-tests. Values are presented as mean ± SD with individual data points overlaid. n=21 subjects in panels A-F and n=14 subjects in panels G-I. “+” denotes ACL injured limb, “-“ denotes healthy contralateral limb.
Human quadriceps catabolic signaling
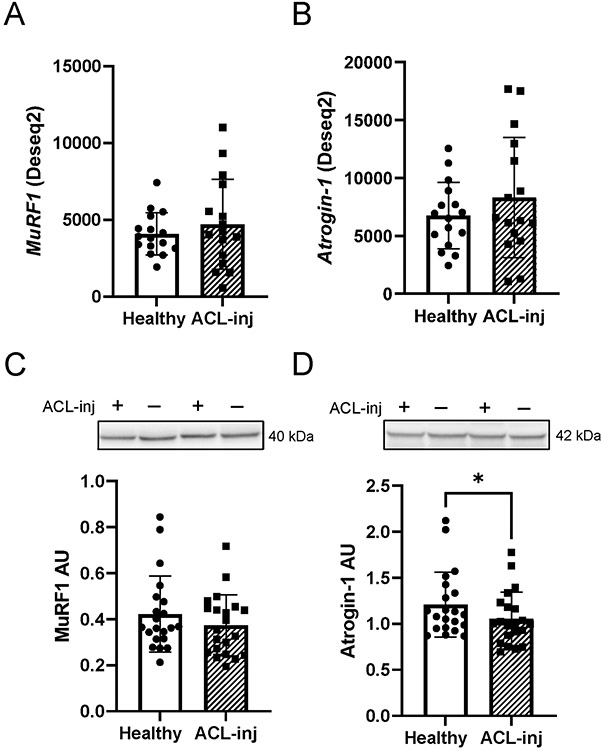
We observed clear deficits in indices of protein synthetic machinery in response to ACLT, and we sought next to define the effects on important molecular effectors of muscle protein catabolism. Specifically, we assessed expression of Atrogin-1/FBXO32/MAFbx and MuRF1/TRIM63 at the transcript and protein level in human quadriceps. MuRF-1 and Atrogin-1 transcript abundances were not different in the ACL-inj limb as compared to Healthy (Figure 3A: MuRF1 FDR-adjusted P = .99; Figure 3B: Atrogin-1 FDR-adjusted P = .99). Additionally, we observed no differences in MuRF1 protein expression (Figure 3C, P = .30). Atrogin-1 protein expression was significantly lower in the ACL-inj limb (Figure 3D, 11% decrease, P = .02).
Figure 3. ACL injury does not induce expression of muscle E3 ubiquitin ligases at the transcript or protein level.
(A) MuRF1 and (B) Atrogin-1 transcript read count following DESeq2 differential gene expression analysis. (C) MuRF1 and (D) Atrogin-1 protein expression. Values are presented as mean ± SD. * P < .05 vs Healthy limb. n=16 for panels A-B and n=21 for panels C-D. “+” denotes ACL injured limb, “-“ denotes healthy contralateral limb.
Human quadriceps muscle fiber size
We observed significant quadriceps atrophy in the ACL-inj limb; quadriceps mean fiber cross-sectional area (CSA) was 8% lower in the ACL-inj limb (Figure 4, P =.03).
Figure 4. Quadriceps fiber atrophy following ACL-injury.
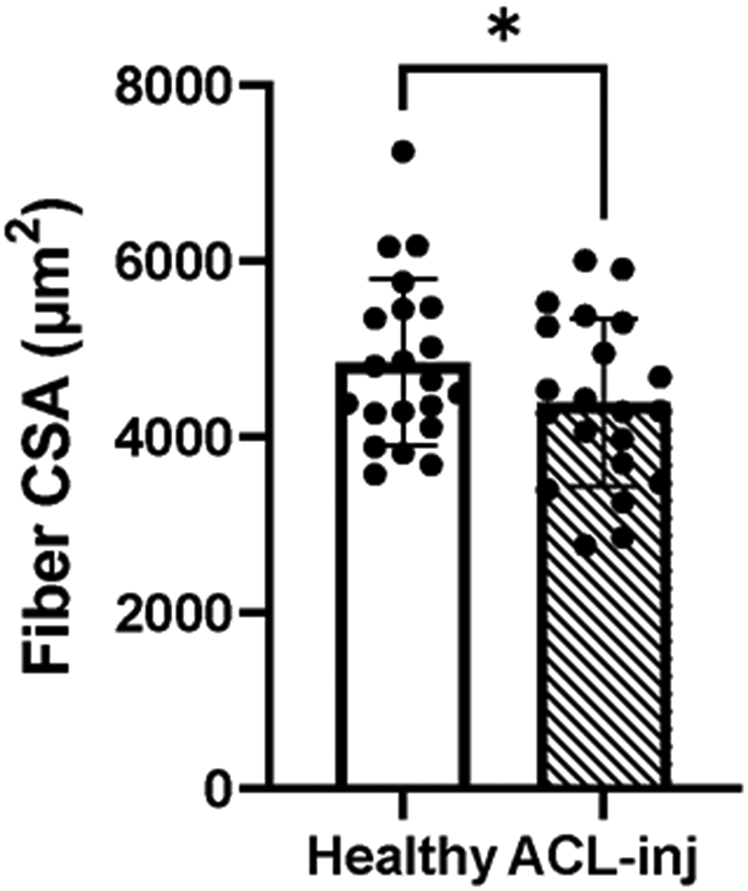
Skeletal muscle fiber cross-sectional area (CSA) in the ACL injured and Healthy limbs of human participants. Values are presented as mean ± SD. * P < .05 vs Healthy limb. n=21.
Murine quadriceps protein synthesis
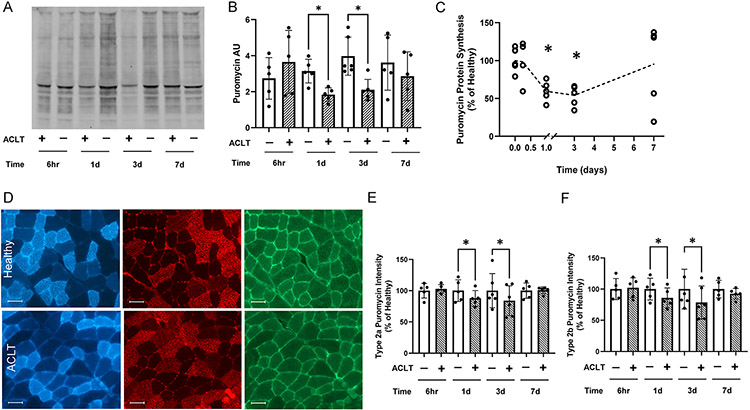
To elucidate the onset of perturbed anabolic signaling post ACLT, murine ACLs were transected, and quadriceps muscle was analyzed 6 hours, 1, 3, and 7 days post transection. Protein synthesis, as assessed by puromycin incorporation into nascent polypeptides, was significantly decreased at 1 (−41%, P = .012) and 3 (−49%, P = .003) days post-injury (Figure 5A-C). Puromycin incorporation was then secondarily assessed via immunohistochemistry to further interrogate protein synthetic deficits in a fiber type specific manner. Decreased puromycin incorporation was observed at 1 and 3 days post ACLT in both fiber types 2a (Figure 5D-E, 12% decrease at 1 day, P = .02, 16% decrease at 3 days, P = .04) and 2b (Figure 5F, 15% decrease at 1 day, P = .015, 22% decrease at 3 days, P = .03), coinciding with deficits in protein synthesis assayed via immunoblotting (Figure 5A-C).
Figure 5. Quadriceps protein synthesis is depressed following ACL transection in mouse quadriceps.
(A) Representative image of whole murine quadriceps muscle homogenate showing puromycin-labeled peptides. (B) Quantification of puromycin band intensity in mouse quadriceps. (C) Normalized presentation of puromycin protein synthesis in the quadriceps of the ACL transected limb. (D) Representative images of type 2a fibers (blue), type 2b fibers (red) and intensity of puromycin-labeled peptides (green). (E) Normalized puromycin intensity of type 2a fibers in the quadriceps. (F) Normalized puromycin intensity of type 2b fibers in the quadriceps. Values are presented as mean ± SD with individual points overlaid. * P < .05 vs control limb. n=5-6 mice per time point. “+” denotes ACL transected (ACLT) limb, “-“ denotes healthy contralateral limb.
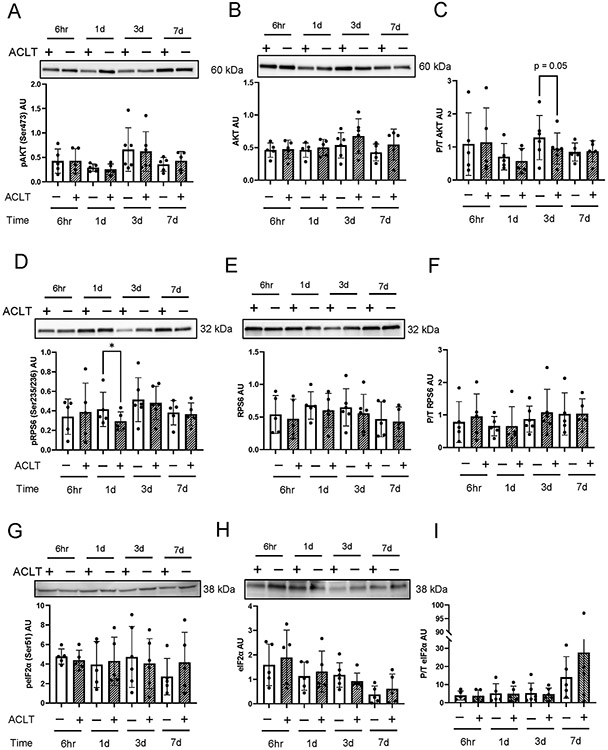
Murine quadriceps anabolic signaling
We observed subtle declines in phosphorylated states of hallmark proteins in translation signaling cascades post ACLT. Reduced phospho/total AKT (Figure 6A-C) was observed 3 days post injury (−24%, P = .05). Downstream, phospho RPS6 (Figure 6D-F) was 28% lower 1 day post injury (P = .04). No significant differences in eIF2α were observed following ACLT in murine quadriceps (Figure 6G-I).
Figure 6. ACL transection does not lead to robust alterations in murine quadriceps protein anabolic signaling.
ACL transected murine quadriceps muscle homogenates were assessed 6 hours, 1 day, 3 days, and 7 days post-ACLT injury. Membranes were probed with (A) phospho-AKTSer473, (B) total AKT, (C) phospho/total AKT, (D) phospho-RPS6Ser235/236, (E) total RPS6, (F) phospho/total RPS6, (G) phospho eIF2αSer51, and (H) total eIF2α, and (I) phospho/total eIFα. Values are presented as mean ± SD with individual points overlaid. * P < .05 vs control limb. n=5-6 mice per time point. “+” denotes ACL transected (ACLT) limb, “-“ denotes healthy contralateral limb.
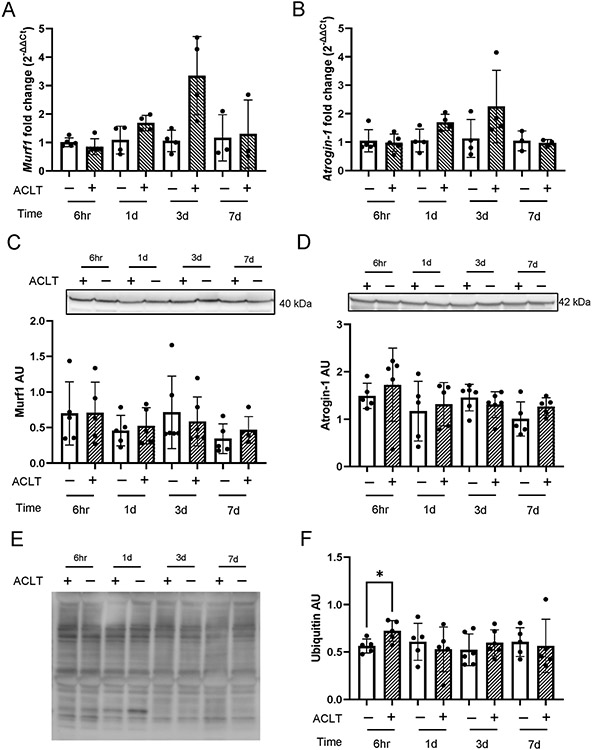
Murine quadriceps catabolic signaling
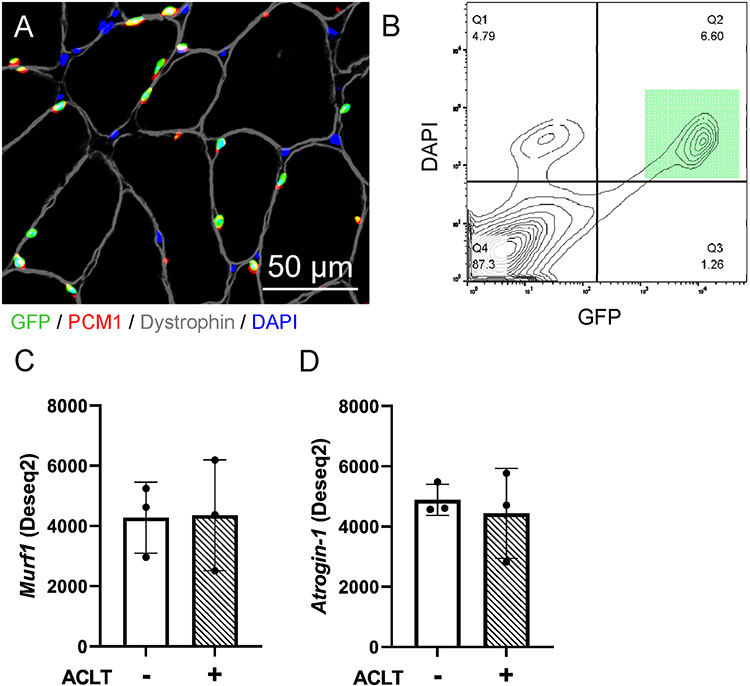
Similar to human quadriceps muscle, ACLT did not substantially alter expression of E3 ubiquitin ligases MuRF1 or Atrogin-1 at either the gene or protein level (Figure 7A-D). We assessed total ubiquitinated protein abundance in murine quadriceps muscle and we found a transient significant elevation in ubiquitinated protein at 6hr post-ACLT (Figure 7E-F, 28% increase, P = .013). To provide information on transcriptional adaptation occurring following ACLT within the quadriceps muscle fibers, we conducted low input RNA sequencing on quadriceps myonuclei from Healthy (n = 3 pooled samples) and ACLT-injured limbs (n = 3 pooled samples) using the HSA-GFP model (Figure 8A-B), as previously described11, 30, 69. HSA-GFP mice underwent ACLT injury on 1 limb, and quadriceps muscles were analyzed at 3 days post ACLT. Three days was chosen as it is when we observed the greatest relative deficit in protein synthesis. Similar to whole quadriceps muscle analysis, ACLT injury did not substantially alter expression of MuRF1 or Atrogin-1 at the transcript level (Figure 8C-D).
Figure 7. ACL transection does not lead to robust alterations in murine quadriceps protein catabolic signaling.
Ubiquitin ligase gene and protein expression in ACL transected murine quadriceps muscle was determined. (A) MuRF-1 and (B) Atrogin-1 gene and (C) MuRF-1 and (D) Atrogin-1 protein expression were assayed 6 hours, 1 day, 3 days, and 7 days post injury. (E) Representative image of murine quadriceps muscle homogenate showing ubiquitinated proteins via western blot. (F) Ubiquitinated protein abundance in murine quadriceps tissue. Values are presented as mean ± SD with individual points overlaid. * P < .05 vs control limb. n=5-6 mice per time point. “+” denotes ACL transected (ACLT) limb, “-“ denotes healthy contralateral limb.
Figure 8. ACL transection does not lead to robust alterations in murine myonuclear E3 ubiquitin ligase expression.
A) Representative fluorescent image showing GFP-labeled myonuclei (green), that co-express PCM1 (red) in addition to being DAPI (blue) positive and residing within the dystrophin border (gray). Scale bar=50μm. B) Flow cytometry plot illustrating GFP+ nuclei co-labeled with DAPI (green square), which was used to identify and purify myonuclei via FACS. C) MuRF1 and (D) Atrogin-1 transcript read count following DESeq2 differential gene expression analysis of isolated quadriceps myonuclei. n=3 pooled biological replicates for panels C-D. “+” denotes ACL transected (ACLT) limb, “-“ denotes healthy contralateral limb.
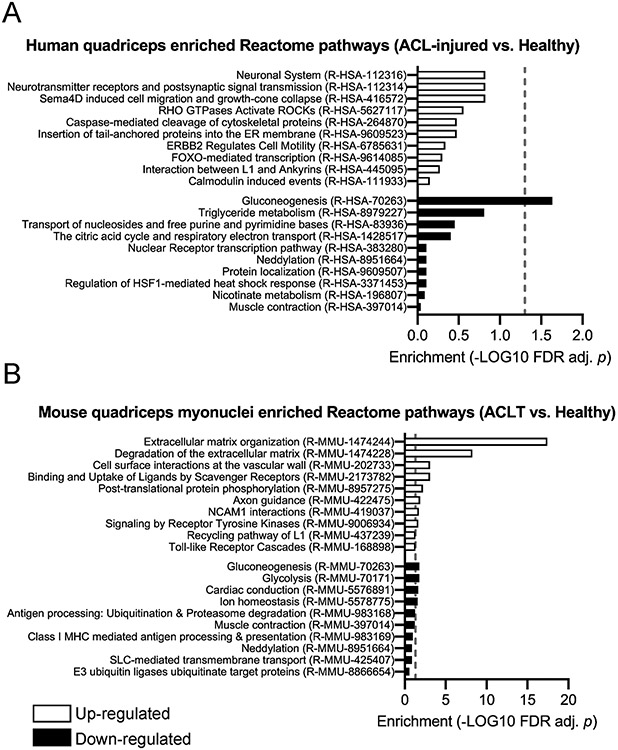
Similarities in transcriptional features of murine quadriceps myonuclei and human whole quadriceps following ACLT
We performed RNA-seq on murine quadriceps myonuclei isolated 3 days following ACLT and compared these results to RNA-seq analyses of human quadriceps muscle following ACLT. At the pathway level, ACLT was associated with down-regulation of genes related to gluconeogenesis, neddylation and muscle contraction respective to Healthy limb controls in both murine quadriceps myonuclei and human quadriceps (Figure 9). In murine quadriceps myonuclei, genes in pathways related to ubiquitination and proteasome degradation, ion homeostasis and glycolysis were down-regulated respective to the respective Healthy limb quadriceps myonuclei (Figure 9). Only gluconeogenesis-related genes were significantly lower (FDR-adjusted P < .05) in human quadriceps pathway analyses; however we observed non-significant down-regulation of genes involved in oxidative metabolism (e.g. triglyceride metabolism and the citric acid cycle) (Figure 9). We observed a similar response to ACLT at the pathway level with up-regulation of genes in pathways associated with neuromuscular plasticity (e.g. neurotransmitter receptors, postsynaptic signaling, axon guidance, neural cell adhesion molecule [NCAM] interactions) respective to Healthy limb controls in both murine quadriceps myonuclei and human quadriceps (Figure 9). Unique to the murine quadriceps myonuclei was an up-regulation of genes in pathways associated with extracellular matrix organization and degradation in the ACLT limb (FDR-adjusted P < .05; Figure 9).
Figure 9. Quadriceps gene expression changes in human whole muscle and murine myonuclei in response to ACL injury.
Over-representation and under-representation analysis of pathways with up-regulated and down-regulated genes following ACL injury in (A) human quadriceps and (B) murine quadriceps myonuclei. Blue bars = up-regulated pathways, red bars = down-regulated pathways. (A) n=16 human subjects; (B) n=3 pooled biological replicates of murine quadriceps myonuclei. Dashed line represents FDR adjusted P < .05.
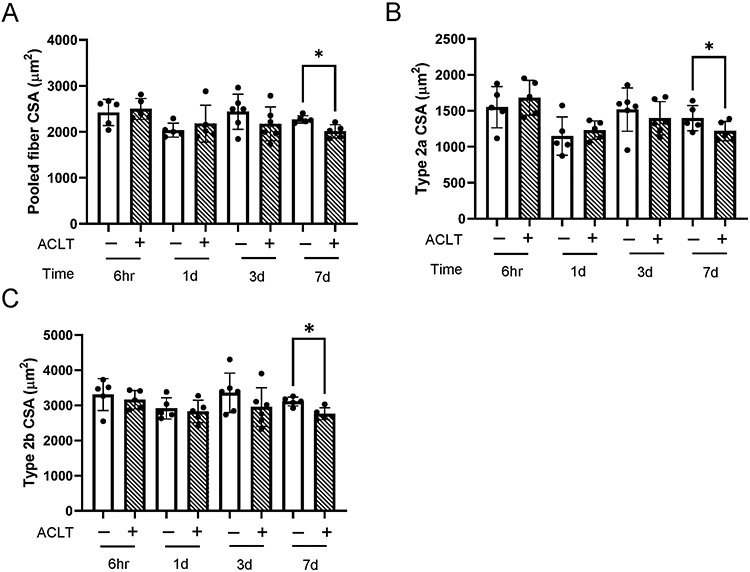
Murine quadriceps fiber size
Protein synthetic and anabolic signaling deficits precede reductions in muscle fiber cross sectional area (CSA) in murine tissue. Relative to the non-injured limb, mean fiber CSA steadily declines in the injured limb from 3 to 7 days post injury, with statistically significant atrophy occurring at 7 days post-ACLT (Figure 10A, 12% atrophy, P = .004). We note a similar degree of murine quadriceps atrophy at 7 days post-ACLT (12% atrophy) to human quadriceps atrophy (8%) following ACL injury (Figure 4. Fiber type specific differences in murine quadriceps support findings in pooled fiber CSA. Specifically, 7 days following ACLT, we found atrophy in type 2a fibers (Figure 10B, 13% atrophy, P = .01) and type 2b fibers (Figure 10C, 11% atrophy, P = .03).
Figure 10. ACL transection induces murine quadriceps fiber atrophy 7 days following injury.
Fiber type specific and pooled fiber cross-sectional area (CSA) were determined via immunohistochemistry. (A) Pooled muscle fiber CSA, (B) Type 2a muscle fiber CSA, and (C) Type 2b muscle fiber CSA. Values are presented as mean ± SD with individual points overlaid. * P < .05 vs control limb. n=5-6 mice per time point. “+” denotes ACL transected (ACLT) limb, “-“ denotes healthy contralateral limb.
Sham ACL surgery does not induce deficits in quadriceps protein synthesis or fiber size
To rule out an effect of surgery, we performed sham surgery on mice. Three days following sham surgery, we observed no difference in protein synthesis via puromycin incorporation (Appendix Figure A2A and A2B, available in the online version of this article), and 7 days following sham surgery we observe no difference in quadriceps fiber CSA (Figure A2AC). These time points were selected as they showed the greatest deficits in protein synthesis (3d post ACLT, Figure 5A-C) and fiber size (7 days post-ACLT, Figure 10) in mice that underwent ACL transection.
Discussion
Our findings demonstrate deficits in protein anabolic signaling and protein synthesis following ACL injury in human and murine quadriceps that is associated with atrophy. These deficits occur early following ACLT, highlighting rapid dysregulation of critical molecular effectors of muscle growth/size maintenance that could represent early therapeutic targets to offset muscle loss post-injury. Importantly, there was no induction in the expression of key regulators of muscle protein catabolism through the ubiquitin ligase system, namely the E3 muscle ubiquitin ligases MuRF1 and Atrogin-1. Our findings following ACL injury stand in contrast to other etiologies of muscle atrophy in murine and human muscle tissue including immobilization3, 9 , spinal cord injury66, and bed rest52, in which E3 ligase-mediated ubiquitination plays an important role in unloading-induced muscle atrophy. Collectively, our data provide evidence for a greater contribution of altered muscle protein anabolism versus catabolism in the etiology of quadriceps atrophy following ACL injury.
We provide the first direct evidence for dysregulation of signaling proteins critical for translation initiation within the quadriceps following ACL injury in human subjects. Bilateral biopsies of the healthy and ACL-injured limbs in human participants showed stark decrements in the phosphorylation of AKT and RPS6, upstream and downstream factors in the mechanistic (mammalian) target of rapamycin complex 1 (mTORC1) signaling pathway. The mTORC1 kinase phosphorylates and activates several effectors that regulate translation and cell growth 8, 24, 62, and AKT regulates protein synthesis via mTORC1 to govern muscle adaptation 31. Additionally, ACL-injury induced phosphorylation of eIF2α in human quadriceps muscle; eIF2α downregulates protein synthesis when activated during times of cellular stress 10. These findings are supported by our observations in a preclinical murine model of ACL injury that induced decrements in quadriceps protein synthesis as assessed by puromycin incorporation. Intriguingly, we did not detect increases in the expression of muscle E3 ubiquitin ligases MuRF1 and Atrogin-1, key effectors of muscle protein catabolism 3. Two prior studies in rodent models of ACL injury have shown elevated quadriceps expression of MuRF1 and Atrogin-1 12, 28; importantly however, mitigation of MuRF1 and Atrogin-1 expression via electrical stimulation was insufficient to offset quadriceps atrophy12. These findings, in conjunction with our data showing no elevation in E3 ligase expression in human quadriceps, strongly suggest that decreased protein anabolism, and not increased protein catabolism, is the dominant mechanism causing ACL injury-induced muscle atrophy 57, 59.
Deficits in protein synthesis and dysregulation of factors regulating translation occurred rapidly following injury; relative rates of protein synthesis declined ~40% and ~50% in the murine ACL-injured quadriceps 1 and 3 days following injury, respectively. The rapidity and magnitude of the decline in protein synthesis is similar to unilateral limb immobilization 40, a striking model of atrophy. The deficits in translation regulatory factors and protein synthesis likely contribute to our observed quadriceps atrophy in both human (~8%) and murine (~12%) quadriceps in the current study. While the degree of atrophy in human quadriceps is less than previously reported 50, it is similar to quadriceps atrophy reported via computed tomography occurring prior to reconstruction 43. We note the rapid time to biopsy following ACL injury in our current participants. Prior work has reported quadriceps fiber atrophy of ~18% prior to surgery; however, biopsies from that study were collected at a mean time post-injury of 60 days 50. To the best of our knowledge, the median time post-injury of 21 days to biopsy collection represents the earliest assessment of human quadriceps fiber atrophy, capturing atrophic processes that occur quickly following injury. Our collective murine and human data, in conjunction with work from other groups 12, 28, 43 indicate perturbations in protein metabolism and subsequent atrophy occur rapidly post ACL tear. A recent pilot study showed that provision of a protein supplement did not rescue deficits in quadriceps protein synthesis following ACL injury 27. It appears that the addition of protein alone is insufficient to rescue the intrinsic deficits in translation signaling that we find in the ACL-injured quadriceps. Adjunctive therapies to stimulate muscle protein synthesis, including pharmacological 63, electrical stimulation 65, mesenchymal/pluripotent stem cell derived treatment 14, 34, 72, and/or medical nutrition therapy 2, following injury may better preserve quadriceps mass and function following reconstruction, compared to traditional rehabilitation alone.
Intriguingly, in murine quadriceps the decline in protein synthesis was not accompanied by robust changes in the abundance or phosphorylation of proteins implicated in the regulation of translation. While we observed effects of ACL injury at certain time points with reduced phosphorylation of AKT-mTORC1 signaling proteins, our data do not underscore a systematic and robust alteration in abundance or phosphorylation state AKT-mTORC1 signaling proteins. This is in line with recent work leveraging proteomic and phospho-proteomic approaches highlighting that immobilization-induced decreases in protein synthesis are not mediated by changes in the phosphorylation state of translational signaling proteins 40. These findings may represent a disconnect between translation signaling and protein synthesis that occurs in murine skeletal muscle. Recent literature also shows an uncoupling between transcript and protein expression of E3 ubiquitin ligases in various models of atrophy in mice 29. This disconnect could be unique to murine skeletal muscle, as prior work from our group and others has shown strong associations between phosphorylation of AKT-mTORC1 signaling proteins and rates of protein synthesis in human muscle 15, 27, 39. In addition, our current data show agreement between E3 ubiquitin ligase transcript and protein expression data in human skeletal muscle. Our current data offer intriguing insight into comparative differences between human and murine muscle in the regulation of protein metabolism.
Transcriptome analyses reveal several shared molecular alterations between human and murine muscle following ACL injury. At the pathway level, ACL injury was associated with down-regulation of genes related to gluconeogenesis and neddylation respective to Healthy limb controls in both murine quadriceps myonuclei and human quadriceps. Neddylation is the process by which the ubiquitin-like protein NEDD8 is conjugated to a target protein, analogous to ubiquitination. Neddylation has emerged as a novel ubiquitin signaling and proteome remodeling agent in muscle that is exercise-responsive 55. In conjunction with our data showing no elevation in the expression of canonical muscle E3 ubiquitin ligases, the down-regulation of genes involved in neddylation provides further evidence against protein catabolism as a dominant mechanism contributing to ACL injury-induced quadriceps atrophy. We observed further similarities between species in response to ACLT at the pathway level with up-regulation of genes in pathways associated with neuromuscular plasticity respective to Healthy limb controls. Prior evidence exists showing quadriceps muscle fiber denervation following ACL injury17, and our current findings further highlight neuromuscular remodeling that occurs in both murine and human quadriceps following ACL injury. Murine quadriceps myonuclei showed significant upregulation of pathways enriched for extracellular matrix organization and degradation in the ACLT limb. Recent work underscores collagen and extracellular matrix remodeling within the quadriceps following ACL injury 17, 50, 51, 56, and our current data show that murine myonuclei may contribute to extracellular matrix turnover in the ACL-injured quadriceps. Muscle fibers have an underappreciated role in extracellular matrix remodeling7, 47, but our results suggest that murine quadriceps myonuclei are active participants in the remodeling of their extracellular niche following ACL injury.
Our study is not without limitations; our human biopsy data are cross-sectional – a time course post-injury in humans would more clearly elucidate the rapidity of translation signaling deficits, but the invasive and demanding nature of multiple quadriceps biopsies is a challenge. We also note that all muscle tissue collection (both human and murine) were performed in a fasted state given that muscle protein metabolism is highly sensitive to nutrient status 18. It is unknown how the effects of feeding modify the effect of ACLT we observe in the current study, and future research should ascertain whether ACL injury mitigates nutrient-induced stimulation of muscle protein anabolism 27. While anabolic deficits in murine tissue are robust post ACLT, future work would benefit from functional analyses. Assessment of quadriceps strength or gait alteration(s) would add functional depth linking protein metabolic perturbations to weakness. Prior work has shown ACLT results in robust deficits in quadriceps torque in mice4, and future studies should seek to determine whether early reductions in muscle protein anabolism predict functional deficits. Additionally, our murine injury model involves surgical transection of the ACL. This is an invasive procedure, and in an attempt to control for the effects of anesthesia and disruption of the joint capsule, we performed sham surgery in a subset of mice. We show in sham-operated mice no deficits in protein synthesis at 3 days post-sham surgery or quadriceps fiber size 7 days post-sham surgery, both time points where greatest deficits were observed in ACLT-injured mice. Further, we also show a similar degree of quadriceps atrophy at 7 days post-ACLT compared to other surgical and non-invasive rodent models of ACL injury 12, 28. Direct comparison between time post-injury between human and murine species is challenging. We note that mean time post-ACL injury in human participants was 37 days. A recent review article sought to define a relationship between mice age and human age, establishing that across their respective lifespans, 40 human days is equivalent to 1 mouse day 13. With our mean time post-ACL injury in human participants being 37 days, extrapolating the equivalent time post-injury in a mouse equates to approximately 1 day 13. We study a time course of recovery post-ACL injury in mice from 6 hours to 7 days, which encompasses the same ‘relative’ time post-injury as displayed by our human subjects.
The strengths of this study include the identification of putative molecular targets that likely result in ACL tear-resultant quadriceps atrophy. Blunted anabolic signaling was pervasive in human participants’ pre-reconstruction surgery, suggesting an ACL injury-intrinsic mechanism potentiating quadriceps atrophy. Further, our time course analysis of murine tissue reveals rapid deficits in protein synthesis following ACL injury. Together, these data underscore the need for pre-surgical reconstruction interventions to mitigate quadriceps atrophy.
Conclusions
In summary, our results provide compelling evidence for ACLT-mediated deficits in translation signaling and protein synthesis that contribute to quadriceps atrophy. These deficits occur early following ACLT, highlighting rapid dysregulation of critical molecular effectors of muscle growth/size maintenance that could represent early therapeutic targets to offset muscle loss post-injury. Our data provide evidence for a greater contribution of altered muscle protein anabolism versus catabolism in the etiology of quadriceps atrophy following ACL injury.
Supplementary Material
What is known about the subject:
Protracted quadriceps atrophy and weakness occur in response to an anterior cruciate ligament (ACL) tear. These morphological and functional deficits are often obstinate to surgical reconstruction and physical therapy, limiting return to sport and promoting long-term functional deficits. Maintenance of skeletal muscle mass is largely dependent on the balance between protein synthesis and breakdown. Skeletal muscle atrophy can be resultant from deficits in protein synthesis and/or increases in protein degradation. However, whether blunted anabolism or elevated catabolism contributes more substantially to quadriceps atrophy following an ACL tear remains unknown, limiting the development of targeted strategies to enhance recovery.
What this study adds to existing knowledge:
Our research investigates specific anabolic and catabolic mechanisms in the context of ACL resultant quadriceps atrophy. In bilateral human quadriceps biopsies (both ACL-involved and healthy uninvolved limbs) collected prior to reconstructive surgery, we find blunted activation of cellular factors responsible for protein synthesis and translation initiation with no change in catabolic signals. Our data provide evidence for a greater contribution of altered muscle protein anabolism versus catabolism in the etiology of quadriceps atrophy following ACL injury. To determine the onset of this signaling deficit, we utilize a murine ACL injury model in a time-course analysis. We report decrements in protein synthesis and translational signaling that precede muscle atrophy in murine tissue, suggesting these mechanisms underpin observed quadriceps muscle atrophy following ACL injury.
Competing interests
One or more of the authors has declared the following potential conflict of interest or source of funding. Research reported in this publication was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number R01 AR072061 to CSF. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. DLJ has received travel expenses from Linvatec; compensation for services other than consulting, royalty or license, in addition to consulting fees from Smith + Nephew; and consulting fees from Xiros Inc. The myosin heavy chain fiber type 2a (#SC-71) and 2b (#BF-F3) antibodies were developed by S. Schiaffino and obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242.
Data Availability
RNA-sequencing data were deposited in Gene Expression Omnibus: GSE211681 (human) and GSE211682 (murine).
References
- 1.Angelozzi M, Madama M, Corsica C, et al. Rate of force development as an adjunctive outcome measure for return-to-sport decisions after anterior cruciate ligament reconstruction. J Orthop Sports Phys Ther. 2012;42(9):772–780. [DOI] [PubMed] [Google Scholar]
- 2.Arentson-Lantz EJ, Fiebig KN, Anderson-Catania KJ, et al. Countering disuse atrophy in older adults with low-volume leucine supplementation. J Appl Physiol (1985). 2020;128(4):967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. [DOI] [PubMed] [Google Scholar]
- 4.Brightwell CR, Graber TG, Brightwell BD, Borkowski M, Noehren B, Fry CS. In vivo Measurement of Knee Extensor Muscle Function in Mice. JoVE. 2021(169):e62211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brightwell CR, Hanson ME, El Ayadi A, et al. Thermal injury initiates pervasive fibrogenesis in skeletal muscle. Am J Physiol Cell Physiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brightwell CR, Kulkarni AS, Paredes W, et al. Muscle fibrosis and maladaptation occur progressively in CKD and are rescued by dialysis. JCI Insight. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brightwell CR, Latham CM, Thomas NT, Keeble AR, Murach KA, Fry CS. A Glitch in the Matrix: The Pivotal Role for Extracellular Matrix Remodeling During Muscle Hypertrophy. Am J Physiol Cell Physiol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95(4):1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YW, Gregory CM, Scarborough MT, Shi R, Walter GA, Vandenborne K. Transcriptional pathways associated with skeletal muscle disuse atrophy in humans. Physiol Genomics. 2007;31(3):510–520. [DOI] [PubMed] [Google Scholar]
- 10.de Haro C, Méndez R, Santoyo J. The eIF-2alpha kinases and the control of protein synthesis. FASEB J. 1996;10(12):1378–1387. [DOI] [PubMed] [Google Scholar]
- 11.Dungan CM, Brightwell CR, Wen Y, et al. Muscle-Specific Cellular and Molecular Adaptations to Late-Life Voluntary Concurrent Exercise. Function (Oxf). 2022;3(4):zqac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durigan JL, Delfino GB, Peviani SM, et al. Neuromuscular electrical stimulation alters gene expression and delays quadriceps muscle atrophy of rats after anterior cruciate ligament transection. Muscle Nerve. 2014;49(1):120–128. [DOI] [PubMed] [Google Scholar]
- 13.Dutta S, Sengupta P. Men and mice: Relating their ages. Life Sci. 2016;152:244–248. [DOI] [PubMed] [Google Scholar]
- 14.Fix DK, Mahmassani ZS, Petrocelli JJ, et al. Reversal of deficits in aged skeletal muscle during disuse and recovery in response to treatment with a secrotome product derived from partially differentiated human pluripotent stem cells. Geroscience. 2021;43(6):2635–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fry CS, Drummond MJ, Glynn EL, et al. Aging impairs contraction-induced human skeletal muscle mTORC1 signaling and protein synthesis %U http://www.skeletalmusclejournal.com/content/1/1/11. Skeletal Muscle. 2011;1(1 %M doi: 10.1186/2044-5040-1-11):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry CS, Glynn EL, Drummond MJ, et al. Blood flow restriction exercise stimulates mTORC1 signaling and muscle protein synthesis in older men. Journal of Applied Physiology. 2010;108(5):1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry CS, Johnson DL, Ireland ML, Noehren B. ACL injury reduces satellite cell abundance and promotes fibrogenic cell expansion within skeletal muscle. J Orthop Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita S, Dreyer HC, Drummond MJ, et al. Nutrient signalling in the regulation of human muscle protein synthesis. Journal of Physiology-London. 2007;582(2):813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15(9):1061–1069. [DOI] [PubMed] [Google Scholar]
- 20.Goodman CA, Hornberger TA. Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exerc Sport Sci Rev. 2013;41(2):107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman CA, Kotecki JA, Jacobs BL, Hornberger TA. Muscle fiber type-dependent differences in the regulation of protein synthesis. PLoS One. 2012;7(5):e37890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman CA, Mabrey DM, Frey JW, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25(3):1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grapar Zargi T, Drobnic M, Vauhnik R, Koder J, Kacin A. Factors predicting quadriceps femoris muscle atrophy during the first 12weeks following anterior cruciate ligament reconstruction. Knee. 2017;24(2):319–328. [DOI] [PubMed] [Google Scholar]
- 24.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945. [DOI] [PubMed] [Google Scholar]
- 25.Hiemstra LA, Webber S, MacDonald PB, Kriellaars DJ. Knee strength deficits after hamstring tendon and patellar tendon anterior cruciate ligament reconstruction. Medicine Science Sports Exercise. 2000;32(8):1472–1479. [DOI] [PubMed] [Google Scholar]
- 26.Hiemstra LA, Webber S, MacDonald PB, Kriellaars DJ. Knee strength deficits after hamstring tendon and patellar tendon anterior cruciate ligament reconstruction. Med Sci Sports Exerc. 2000;32(8):1472–1479. [DOI] [PubMed] [Google Scholar]
- 27.Howard EE, Margolis LM, Fussell MA, et al. Effect of High-Protein Diets on Integrated Myofibrillar Protein Synthesis before Anterior Cruciate Ligament Reconstruction: A Randomized Controlled Pilot Study. Nutrients. 2022;14(3):563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunt ER, Davi SM, Parise CN, et al. Temporal disruption of neuromuscular communication and muscle atrophy following noninvasive ACL injury in rats. J Appl Physiol (1985). 2022;132(1):46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt LC, Graca FA, Pagala V, et al. Integrated genomic and proteomic analyses identify stimulus-dependent molecular changes associated with distinct modes of skeletal muscle atrophy. Cell Rep. 2021;37(6):109971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwata M, Englund DA, Wen Y, et al. A novel tetracycline-responsive transgenic mouse strain for skeletal muscle-specific gene expression. Skelet Muscle. 2018;8(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal N, Gavin M, Loro E, et al. AKT controls protein synthesis and oxidative metabolism via combined mTORC1 and FOXO1 signalling to govern muscle physiology. J Cachexia Sarcopenia Muscle. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeon OH, Kim C, Laberge RM, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 2017;23(6):775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4(3):465–477. [DOI] [PubMed] [Google Scholar]
- 34.Kim MJ, Kim ZH, Kim SM, Choi YS. Conditioned medium derived from umbilical cord mesenchymal stem cells regenerates atrophied muscles. Tissue Cell. 2016;48(5):533–543. [DOI] [PubMed] [Google Scholar]
- 35.Kline PW, Morgan KD, Johnson DL, Ireland ML, Noehren B. Impaired Quadriceps Rate of Torque Development and Knee Mechanics After Anterior Cruciate Ligament Reconstruction With Patellar Tendon Autograft. Am J Sports Med. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konishi Y, Ikeda K, Nishino A, Sunaga M, Aihara Y, Fukubayashi T. Relationship between quadriceps femoris muscle volume and muscle torque after anterior cruciate ligament repair. Scand J Med Sci Sports. 2007;17(6):656–661. [DOI] [PubMed] [Google Scholar]
- 37.Krishnan C, Williams GN. Factors explaining chronic knee extensor strength deficits after ACL reconstruction. J Orthop Res. 2011;29(5):633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuenze CM, Blemker SS, Hart JM. Quadriceps function relates to muscle size following ACL reconstruction. J Orthop Res. 2016;34(9):1656–1662. [DOI] [PubMed] [Google Scholar]
- 39.Kumar V, Selby A, Rankin D, et al. Age-related differences in the dose-response relationship of muscle protein synthesis to resistance exercise in young and old men. Journal of Physiology-London. 2009;587(1):211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin KH, Wilson GM, Blanco R, et al. A deep analysis of the proteomic and phosphoproteomic alterations that occur in skeletal muscle after the onset of immobilization. J Physiol. 2021;599(11):2887–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindström M, Strandberg S, Wredmark T, Felländer-Tsai L, Henriksson M. Functional and muscle morphometric effects of ACL reconstruction. A prospective CT study with 1 year follow-up. Scand J Med Sci Sports. 2013;23(4):431–442. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 43.Lorentzon R, Elmqvist LG, Sjöström M, Fagerlund M, Fuglmeyer AR. Thigh musculature in relation to chronic anterior cruciate ligament tear: muscle size, morphology, and mechanical output before reconstruction. Am J Sports Med. 1989;17(3):423–429. [DOI] [PubMed] [Google Scholar]
- 44.Maletis GB, Cameron SL, Tengan JJ, Burchette RJ. A prospective randomized study of anterior cruciate ligament reconstruction: a comparison of patellar tendon and quadruple-strand semitendinosus/gracilis tendons fixed with bioabsorbable interference screws. Am J Sports Med. 2007;35(3):384–394. [DOI] [PubMed] [Google Scholar]
- 45.Moro T, Brightwell CR, Phalen DE, et al. Low skeletal muscle capillarization limits muscle adaptation to resistance exercise training in older adults. Exp Gerontol. 2019;127:110723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moro T, Brightwell CR, Volpi E, Rasmussen BB, Fry CS. Resistance exercise training promotes fiber type-specific myonuclear adaptations in older adults. J Appl Physiol (1985). 2020;128(4):795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murach K, Vechetti I, Van Pelt D, et al. Fusion-Independent Satellite Cell Communication to Muscle Fibers During Load-Induced Hypertrophy. Function. 2020;1(1):zqaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murach KA, Dungan CM, von Walden F, Wen Y. Epigenetic evidence for distinct contributions of resident and acquired myonuclei during long-term exercise adaptation using timed in vivo myonuclear labeling. Am J Physiol Cell Physiol. 2022;322(1):C86–C93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.NATHANS D PUROMYCIN INHIBITION OF PROTEIN SYNTHESIS: INCORPORATION OF PUROMYCIN INTO PEPTIDE CHAINS. Proc Natl Acad Sci U S A. 1964;51:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noehren B, Andersen A, Hardy P, et al. Cellular and Morphological Alterations in the Vastus Lateralis Muscle as the Result of ACL Injury and Reconstruction. J Bone Joint Surg Am. 2016;98(18):1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noehren B, Hardy PA, Andersen A, et al. T1ρ imaging as a non-invasive assessment of collagen remodelling and organization in human skeletal muscle after ligamentous injury. J Physiol. 2021;599(23):5229–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogawa T, Furochi H, Mameoka M, et al. Ubiquitin ligase gene expression in healthy volunteers with 20-day bedrest. Muscle Nerve. 2006;34(4):463–469. [DOI] [PubMed] [Google Scholar]
- 53.Palmieri-Smith RM, Lepley LK. Quadriceps Strength Asymmetry After Anterior Cruciate Ligament Reconstruction Alters Knee Joint Biomechanics and Functional Performance at Time of Return to Activity. Am J Sports Med. 2015;43(7):1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmieri-Smith RM, Thomas AC, Wojtys EM. Maximizing quadriceps strength after ACL reconstruction. Clin Sports Med. 2008;27(3):405–424, vii-ix. [DOI] [PubMed] [Google Scholar]
- 55.Parker BL, Kiens B, Wojtaszewski JFP, Richter EA, James DE. Quantification of exercise-regulated ubiquitin signaling in human skeletal muscle identifies protein modification cross talk via NEDDylation. FASEB J. 2020;34(4):5906–5916. [DOI] [PubMed] [Google Scholar]
- 56.Peck BD, Brightwell CR, Johnson DL, Ireland ML, Noehren B, Fry CS. Anterior Cruciate Ligament Tear Promotes Skeletal Muscle Myostatin Expression, Fibrogenic Cell Expansion, and a Decline in Muscle Quality. Am J Sports Med. 2019;47(6):1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phillips SM, McGlory C. CrossTalk proposal: The dominant mechanism causing disuse muscle atrophy is decreased protein synthesis. J Physiol. 2014;592(24):5341–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raudvere U, Kolberg L, Kuzmin I, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019;47(W1):W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reid MB, Judge AR, Bodine SC. CrossTalk opposing view: The dominant mechanism causing disuse muscle atrophy is proteolysis. J Physiol. 2014;592(24):5345–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6(4):275–277. [DOI] [PubMed] [Google Scholar]
- 61.Schmitt LC, Paterno MV, Hewett TE. The impact of quadriceps femoris strength asymmetry on functional performance at return to sport following anterior cruciate ligament reconstruction. J Orthop Sports Phys Ther. 2012;42(9):750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Showkat M, Beigh MA, Andrabi KI. mTOR Signaling in Protein Translation Regulation: Implications in Cancer Genesis and Therapeutic Interventions. Mol Biol Int. 2014;2014:686984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinha-Hikim I, Artaza J, Woodhouse L, et al. Testosterone-induced increase in muscle size in healthy young men is associated with muscle fiber hypertrophy. Am J Physiol Endocrinol Metab. 2002;283(1):E154–164. [DOI] [PubMed] [Google Scholar]
- 64.Thomas AC, Wojtys EM, Brandon C, Palmieri-Smith RM. Muscle atrophy contributes to quadriceps weakness after anterior cruciate ligament reconstruction. J Sci Med Sport. 2016;19(1):7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toth MJ, Tourville TW, Voigt TB, et al. Utility of Neuromuscular Electrical Stimulation to Preserve Quadriceps Muscle Fiber Size and Contractility After Anterior Cruciate Ligament Injuries and Reconstruction: A Randomized, Sham-Controlled, Blinded Trial. Am J Sports Med. 2020;48(10):2429–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urso ML, Chen YW, Scrimgeour AG, Lee PC, Lee KF, Clarkson PM. Alterations in mRNA expression and protein products following spinal cord injury in humans. J Physiol. 2007;579(Pt 3):877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Von Walden F, Rea M, Mobley CB, et al. The myonuclear DNA methylome in response to an acute hypertrophic stimulus. Epigenetics. 2020;15(11):1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wackerhage H, Ratkevicius A. Signal transduction pathways that regulate muscle growth. Essays Biochem. 2008;44:99–108. [DOI] [PubMed] [Google Scholar]
- 69.Wen Y, Englund DA, Peck BD, Murach KA, McCarthy JJ, Peterson CA. Myonuclear transcriptional dynamics in response to exercise following satellite cell depletion. iScience. 2021;24(8):102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wen Y, Murach KA, Vechetti IJ Jr., et al. MyoVision: software for automated high-content analysis of skeletal muscle immunohistochemistry. J Appl Physiol (1985). 2018;124(1):40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams GN, Buchanan TS, Barrance PJ, Axe MJ, Snyder-Mackler L. Quadriceps weakness, atrophy, and activation failure in predicted noncopers after anterior cruciate ligament injury. Am J Sports Med. 2005;33(3):402–407. [DOI] [PubMed] [Google Scholar]
- 72.Wu YF, De La Toba EA, Dvoretskiy S, et al. Development of a cell-free strategy to recover aged skeletal muscle after disuse. J Physiol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data were deposited in Gene Expression Omnibus: GSE211681 (human) and GSE211682 (murine).