Abstract
Serum antibodies IgM and IgG are elevated during Coronavirus Disease 2019 (COVID-19) to defend against viral attacks. Atypical results such as negative and abnormally high antibody expression were frequently observed whereas the underlying molecular mechanisms are elusive. In our cohort of 144 COVID-19 patients, 3.5% were both IgM and IgG negative, whereas 29.2% remained only IgM negative. The remaining patients exhibited positive IgM and IgG expression, with 9.3% of them exhibiting over 20-fold higher titers of IgM than the others at their plateau. IgG titers in all of them were significantly boosted after vaccination in the second year. To investigate the underlying molecular mechanisms, we classed the patients into four groups with diverse serological patterns and analyzed their 2-year clinical indicators. Additionally, we collected 111 serum samples for TMTpro-based longitudinal proteomic profiling and characterized 1494 proteins in total. We found that the continuously negative IgM and IgG expression during COVID-19 were associated with mild inflammatory reactions and high T cell responses. Low levels of serum IgD, inferior complement 1 activation of complement cascades, and insufficient cellular immune responses might collectively lead to compensatory serological responses, causing overexpression of IgM. Serum CD163 was positively correlated with antibody titers during seroconversion. This study suggests that patients with negative serology still developed cellular immunity for viral defense and that high titers of IgM might not be favorable to COVID-19 recovery.
Keywords: COVID-19, serology, proteomics, inflammation, cellular immunity
Abbreviations: C1, complement 1; CLIA, chemiluminescence immunoassay; COVID-19, Coronavirus Disease 2019; DEP, differentially expressed protein; EHR, electronic hospital record; HDL-C, high-density lipoprotein cholesterol; IQR, interquartile range; LDL-C, low-density lipoprotein cholesterol; NAb, neutralizing antibody; nonVac, nonvaccinated; PCA, principal component analysis; R1, 1-year follow-up; R2, 2-year follow-up; RT-PCR, reverse-transcriptase polymerase-chain-reaction; TG, triglyceride; TMT, tandem mass tag; Vac, vaccinated
Graphical Abstract
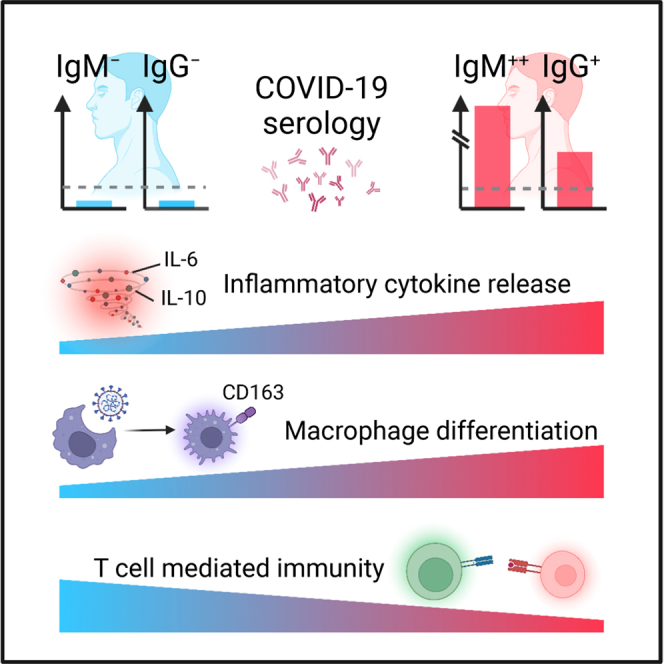
Highlights
-
•
Two-year IgM and IgG manifestation of 144 COVID-19 patients.
-
•
Longitudinal serum proteomics characterization of four serological patterns is done.
-
•
Negative serology was associated with mild inflammation and enhanced T cell immunity.
-
•
Overexpressed IgM was related to dysregulated complement and cellular immunity.
-
•
IgG expression was boosted in the COVID-19 survivors after vaccination.
In Brief
Unexpected serological patterns, such as continuous negative IgM and IgG expression, or exceptionally high titers of IgM were observed in a cohort of 144 COVID-19 patients. To understand the host responses behind the diverse serology, we applied 2-year clinical manifestation and longitudinal serum proteomics analysis. Our findings suggest that COVID-19 patients who do not express antibodies developed cellular immunity for viral defense and that high titers of IgM might not be favorable to COVID-19 recovery.
Coronavirus Disease 2019 (COVID-19) remains a threat to global health. The production of serum antibodies in the human body is a major defensive mechanism to neutralize SARS-CoV-2. Within them, IgM is initiated during the acute phase for early defense, whereas IgG is secreted afterward with a higher affinity for SARS-CoV-2 (1). Typically, COVID-19 patients underwent seroconversion (from negative to positive) of IgM and IgG within 20 days (2). The IgM and IgG expression kept elevating before reaching the plateau, with IgG plateau titers higher and long-lasting than IgM plateau titers (3). The timespans of seroreversion (from positive to negative) were around 3 to 6 months since disease onset for IgM (4, 5), whereas hardly observed for IgG in 1 year (6). After vaccination, convalescent COVID-19 patients exhibited higher titers of IgM and IgG compared to healthy individuals (7).
Several atypical serological patterns were documented in the literature. 3.2% to 6.9% of the COVID-19 patients remained low expression or seronegative for both IgM and IgG throughout the disease stage (1, 2). It has also been reported that less than 10% of the patients exhibited 10- to 20-fold higher antibody titers than the average values when reaching the plateau (1, 8). These unexpected serological patterns indicate heterogeneous host responses during COVID-19, with unclear molecular mechanisms.
This study was designed to investigate the diverse expression patterns of IgM and IgG from a single-center cohort across 2 years of monitoring and to explore the molecular evidence associated with atypical antibody expression via longitudinal proteomic profiling.
Experimental Procedures
Patient Information
One hundred forty-four COVID-19 patients who were admitted to Taizhou Public Health Medical Center, Taizhou Hospital, from January 17, 2020 to April 2, 2020 were recruited in this study. Within them, 73 patients participated in the 1-year follow-up (R1) between day 363 and 397 (interquartile range [IQR], 10) since disease onset, and 58 patients participated in the 2-year follow-up (R2) between day 728 and 763 (IQR, 7) since disease onset.
All enrolled patients were confirmed to be infected with SARS-CoV-2 by use of real-time reverse transcriptase—polymerase chain reaction (RT-PCR) assay on the viral RNA extracted from nasopharyngeal or sputum specimens, and the classification of their disease severity was based on Diagnostic and Treatment Protocol for COVID-19 (Trial Version 5) issued by National Health Commission of the People’s Republic of China (9), unless otherwise mentioned. The onset date was defined as the day when any symptoms were noticed by the patients. None of the recruited patients had self-reported reinfection of SARS-CoV-2.
Removal of Identifying Information
All information that would allow the patient/study participant or their family, friends, or neighbors to identify them (e.g., age, past medical history, etc.) has been removed. Patient IDs from hospital records have been replaced with identifiers that cannot reveal the identity of the study subjects (e.g., R001, R002, etc.). The correspondence between identifiers and patient IDs was not known to anyone outside the research group.
Experimental Design and Statistical Rationale
A flowchart of the study design is illustrated in Fig. 1. This study has been approved by the Ethical/Institutional Review Board of Westlake University and Taizhou Hospital (approval notice: K20210218). The studies in this work abide by the Declaration of Helsinki principles. Since archived specimens were used, informed consent from the patients was waived by the boards.
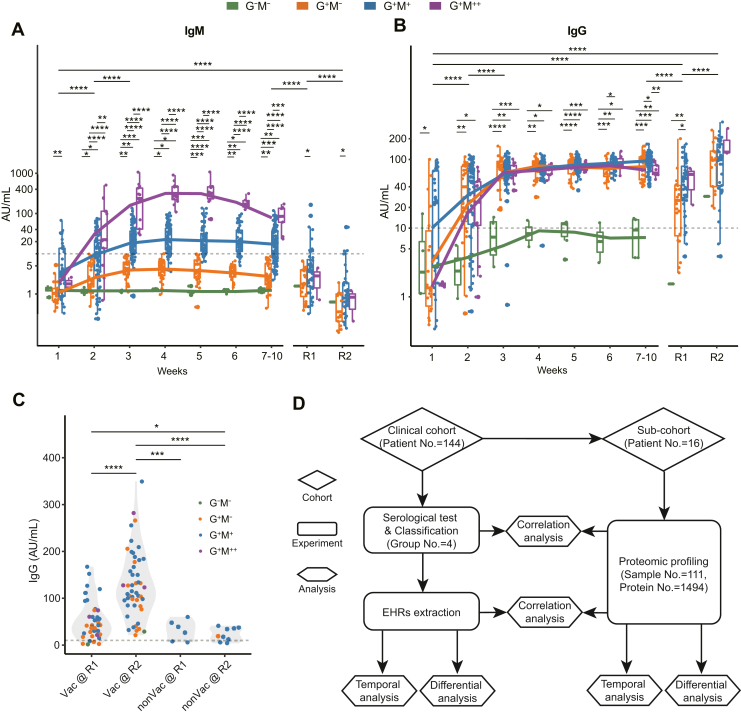
Fig. 1.
Overview of antibody expression in COVID-19.A and B, classification and 2-year monitoring of SARS-CoV-2-specific IgM and IgG expression in COVID-19 patients. The y-axis denotes log-transformed antibody titers. The statistical significance was calculated within and across each timepoint. C, comparison of IgG expression before and after vaccination. D, study design for clinical and proteomic analyses of diverse serology in COVID-19. p values were calculated by two-sided Wilcoxon rank-sum tests. Significances: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005; ∗∗∗∗p < 0.001. EHR, electronic hospital record; nonVac, nonvaccinated; R1, 1-year follow-up; R2, 2-year follow-up; Vac, vaccinated.
Information about demographics, epidemiological history, clinical symptoms, and laboratory data were collected through an electronic hospital record (EHR) system. One hundred seventy eight laboratory indexes in total were retrieved from EHR for laboratory data analysis. For IgM and IgG antibody assays, 790, 72, and 58 tests were conducted during the first 10 weeks since disease onset, R1 and R2, respectively. For proteomics analysis, 111 serum samples from 16 COVID-19 patients were collected from weeks 1 to 10 since disease onset. Nine technical replicates were designed in the MS experiment. Because this study analyzes the diverse host responses associated with COVID-19-specific IgM and IgG, no control individuals were recruited.
For demographical and epidemiological data, categorical variables were described as frequency and percentage, and continuous variables were shown as mean and standard deviation or median and IQR values as appropriate. p values for the categorical variables were calculated using Chi-square analysis. p values for the continuous variables were calculated using the Kruskal–Wallis statistics. For laboratory data, p values were calculated by two-sided Wilcoxon rank-sum tests. For proteomics data, the adjusted p values were calculated by one-way ANOVA and corrected with Benjamini & Hochberg test. Differentially expressed proteins (DEPs) at each week were calculated with an adjusted p value of ≤0.05. Because four groups were involved, we did not set a fold-change cut-off for the DEPs. The p values for the pairwise comparisons of the DEPs in the boxplots (Figs. 3, F and H, and 4B) were calculated by two-sided Student's t test. Technical replicates were not included in the statistical analysis. Significances: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.005; ∗∗∗∗, p < 0.001. Principal component analysis (PCA) was applied to display patient and sample distribution, and the ellipses were shown at a confidence interval of 95%. Missing values in PCA (Figs. 3C, S2, B and C, and S4, B, and C) were imputed with a software tool missMDA (10). Unsupervised hierarchical clustering was applied to display the DEPs as heatmap. Locally weighted scatterplot smoothing model was applied for fitting analysis. Spearman correlation coefficients were calculated for the correlation of clinical data and proteomic data. Statistical analyses were performed with R software (version 3.6.0).
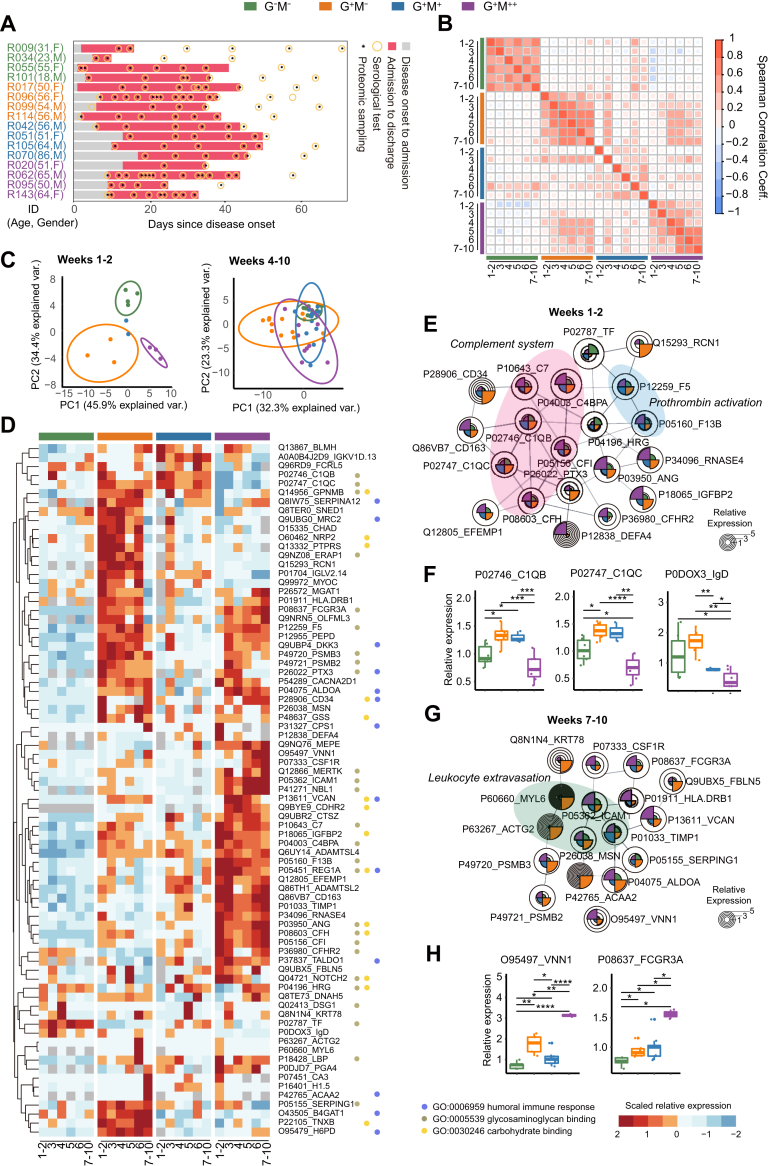
Fig. 3.
Longitudinal serum proteomics of the 16 selected patients.A, temporal serum sampling for the 16 characteristic patients. B, Spearman correlation of the serum proteins for the four groups across 10 weeks. C, principal component analysis (PCA) of serum samples based on the differentially expressed proteins (DEPs) at weeks 1 to 2 and weeks 4 to 10, respectively, and stratified by the four groups. Protein expression with multiple serum samples from the same patient at each week were averaged. The ellipses are shown at a confidence interval of 95%. D, heatmap and functional annotation of 77 DEPs across 10 weeks of proteomics profiling. DEPs are determined with an adjusted p value of ≤0.05. The adjusted p values were calculated by one-way analysis of variance (ANOVA) and corrected with Benjamini & Hochberg test. E, interaction network of selected DEPs at weeks 1 to 2. F, Relative expression of DEPs C1QB, C1QC, and IgD at weeks 1 to 2. The p values between each of the two groups were calculated by two-sided Student's t test. G, interaction network of selected DEPs at weeks 7 to 10. H, relative expression of DEPs VNN1 and FCGR3A at weeks 7 to 10. The p values were calculated in the same way as Fig. 3E. Significances: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005; ∗∗∗∗p < 0.001.
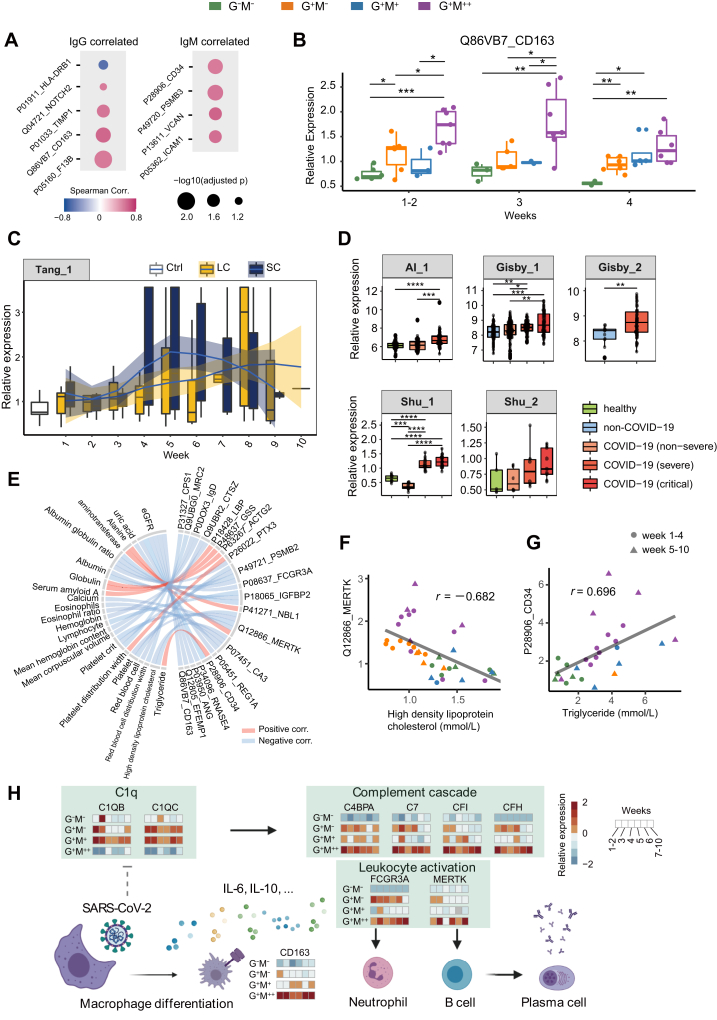
Fig. 4.
Correlations of DEPs with antibody and clinical indicators expression.A, nine DEPs that were correlated with antibody expression during weeks 1 to 4 (absolute values of Spearman correlation coefficient >0.5). B, temporal expression of CD163 during weeks 1 to four in our dataset. The p values between each of the two groups were calculated by two-sided Student's t test. C, temporal expression of CD163 during weeks 1 to 10 since disease onset, in a plasma proteomics dataset by Tang et al. (29). SC, shorter SARS-CoV-2 RNA shedding course; LC, longer SARS-CoV-2 RNA shedding course. Ctrl, non-COVID-19 patients. D, boxplots of CD163 expression in five additional plasma/serum proteomics datasets by Al-Nesf et al. (Al_1) (32), Gisby et al. (Gisby_1 and Gisby_2) (31), and Shu et al. (Shu_1 and Shu_2) (30). The identities of these datasets could be referred to in COVIDpro (https://www.guomics.com/covidPro/). E, correlations of clinical indicators with DEPs during weeks 1 to 10 (absolute values of Spearman correlation coefficient >0.65). F–G, scatterplots of two sets of correlations: MERTK∼HDL-C and CD34∼TG, respectively. r denotes the Spearman correlation coefficient. H, a putative working model for the host responses behind diverse COVID-19 serology. DEPs, differentially expressed proteins. Significances: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005; ∗∗∗∗p < 0.001.
Laboratory Characteristic Tests
Nasopharyngeal or sputum specimens were collected to extract SARS-CoV-2 RNA, using a nucleic acid extractor (EX3600, Shanghai Zhijiang) and a virus nucleic acid extraction kit (P20200201, Shanghai Zhijiang). For nucleic acid detection, fluorescence quantitative PCR (ABI 7500, Thermo) coupled to a SARS- CoV-2 nucleic acid detection kit (P20200203, Shanghai Zhijiang) was used, which applies a one-step RT-PCR combined with Taqman technology to detect RdRp, E, and N genes. “Positive” was concluded when the test for RdRp was positive (threshold cycle <43) and one of the tests for E or N was positive (threshold cycle <43), or when two sequential tests of RdRp were positive whereas the tests for E and N were negative. Other laboratory characteristic tests were conducted according to the manufacturers’ protocols.
Antibody Analyses
The IgM and IgG antibodies against SARS-CoV-2 in serum samples were measured with the chemiluminescence immunoassay (CLIA) kits (iFLASH3000, Shenzhen YHLO). CLIAs were conducted based on two-step indirect immunization according to the manufacturer’s instructions. The recombinant nucleocapsid protein (N) and spike protein (S) antigens of SARS-CoV-2 were enveloped on magnetic beads, and an acridine ester–labeled mouse anti-human IgM/IgG antibody was used as the detection antibody. The IgM/IgG antibody concentrations were positively correlated with the relative luminescence unit. The cut-off to determine positivity was set at 10 AU/ml.
The neutralizing antibodies (NAbs) against SARS-CoV-2 in serum samples were measured using CLIA kits (Caris 200, Wantai). CLIAs for NAb detection were based on competition immunization according to the manufacturer’s instructions. The NAb in the sample and the biotinylated SARS-CoV-2 specific antibody compete with acridine ester–labeled S protein. Next, streptavidin-coated magnetic particles were added. Through the interaction of biotin and streptavidin, a complex consisting of the magnetic particles coated by streptavidin, biotinylated SARS-CoV-2–specific antibody, and acridine ester S protein was formed. After washing and removing the substances that do not bind to the magnetic particles, the NAb concentration in the sample was inversely proportional to the instrumentally detected relative luminescence unit. The cut-off value to determine NAb positivity was set at 0.1 μg/ml.
Patient Classification Based on Antibody Titers
The classification of COVID-19 patients was based on the maximum expression levels of IgM and IgG when reaching the plateau. The cut-off value, as determined by the detection kit, was 10 AU/ml. A total of 47 patients had IgM titers below the cut-off value (IgM−), and three had IgG titers below the cut-off value (IgG−). They were classified as seronegative for IgM and IgG, respectively.
Two patients had IgG titers above but close to the cut-off value. Specifically, the IgG plateau was reached on the second and the 18th day after the symptoms’ onset for patients R055 (16.11 AU/ml) and R101 (13.80 AU/ml), respectively. These were the “outliers” of the patients with IgG plateau titers above the cut-off value, accordingly to Tukey's test. Clinicians classified them as seronegative for IgG (IgG−). Similarly, nine patients with very high IgM expression were the “outliers” in the patients with IgM plateau titers above the cut-off value, accordingly to Tukey's test. Clinicians classed them as patients with abnormally high IgM expression (IgM++).
Patient Selection for Proteomics Analysis
Although 920 serum antibody data points were recorded in the EHRs, some serum samples were not archived and not available for proteomics analysis. From the remaining serum samples, 4 G+M− and 4 G+M+ patients were selected randomly, whereas 4 G−M− and 4 G+M++ patients with the lowest and highest IgM plateau levels were selected respectively, as characteristic patients that may better depict the antibody diversity. Their plateau antibody titers are shown in supplemental Fig. S1A in the shape of triangles. All available samples for these 16 patients were used for proteomics profiling.
Clinical representativeness of the 16 selected patients was assessed based on their expression of the highlighted clinical indicators on admission and across weeks 1 to 10, as elaborated in supplemental Fig. S2, B and C. All of the serum samples from these patients fell within the confidence intervals (95%) both on-admission and temporally, except for one datapoint from patient R099 collected at weeks 7 to 10. We therefore considered the selected patients to be characteristic and representative in general.
Proteomic Analysis
The sample preparation procedures for proteomics profiling were conducted as described previously (11) with some minor modifications. Specifically, archived serum specimens in 2 ml cryogenic tubes were taken from −80 °C storage and heated to 56 °C for 60 min for inactivation. No additives were added during the process. Thereafter, 10 μl serum from each specimen was extracted and immediately loaded onto High Select Top14 Abundant Protein Depletion Mini Spin Columns (Thermo Scientific) for high abundance protein depletion. Eluates were concentrated using Pierce Protein Concentrators PES, 3K MWCO (Thermo Scientific), and denatured with 50 μl lysis buffer (6 M urea and 2 M thiourea in 0.1 M triethylammonium bicarbonate) at 31 °C for 30 min. The extracts were reduced with 10 mM tris (2-carboxyethyl) phosphine (Damas-beta) at 31 °C for 40 min and alkylated with 40 mM iodoacetamide (Sigma) at 25 °C for 40 min in darkness. The samples were diluted with 0.1 M triethylammonium bicarbonate buffer till the final concentration of urea was below 1.6 M and trypsinized (Hualishi Technology) in double-step with an enzyme-to-substrate ratio of 1:20 at 31 °C for 60 min and 120 min, respectively. Trypsinization was stopped by adding trifluoroacetic acid (Damas-beta) till the final concentration of 1%, and digests were desalted with SOLAμ (Thermo Fisher Scientific), following the manufacturer's instructions. Clean peptides were labeled with TMTpro 16plex labeling reagent sets (Thermo Scientific) according to the labeling scheme (supplemental Table S3). One hundred eleven samples with nine technical replicates were distributed into eight batches. In each batch, the first channel (126) was set as pool, and the remaining 15 channels were allocated with labeled peptides from different samples. Labeling efficiency was tested using pool samples to ensure an incorporation ratio of over 95%. Afterward, samples in the batch were combined and fractionated in previously described settings (11). Sixty fractions were derived and consolidated into 26 combined fractions. The fractions were dried and re-dissolved in 30 μl 2% acetonitrile (ACN)/0.1% formic acid (FA). For nanoLC-MS/MS analysis, an EASY-nLC 1200 system (Thermo Fisher Scientific) coupled with Q Exactive HF-X hybrid Quadrupole-Orbitrap (Thermo Fisher Scientific) was applied, and data-dependent acquisition mode was used throughout the analysis. For MS data acquisition of each fraction, peptides were loaded onto a precolumn (3 μm, 100 Å, 20 mm × 75 μm i.d.) at a flow rate of 6 μl/min for 4 min. Thereafter, a 60 min LC gradient (from 5% to 28% buffer B) at a flow rate of 300 nl/min (analytical column, 1.9 μm, 120 Å, 150 mm × 75 μm i.d.) was applied to analyze the peptides. 2% ACN, 98% H2O containing 0.1% FA was set as buffer A, and 98% ACN in water containing 0.1% FA was set as buffer B. The m/z range of MS1 was set to 350 to 1800 m/z with a resolution power of 60,000 at 200 m/z, AGC target was set to 3e6, and maximum ion injection time (max IT) was set to 50 ms. Top 15 precursors were selected for MS/MS experiment, with a resolution power of 45,000 at 200 m/z, AGC target of 2e5, and max IT of 120 ms. The isolation window of selected precursor was set to 0.7 m/z. The resultant data were analyzed with Proteome Discoverer (Version 2.4.1.15). A manually annotated and reviewed protein Homo sapiens fasta downloaded from UniprotKB on 14 Apr 2020 containing 20,435 protein entries was used as protein database. The enzyme digestion was set to full-specific trypsin with maximally two missed cleavages. Fixed modifications were set to carbamidomethylation (+57.021464 Da) of cysteine, TMTpro (+304.207145 Da) of lysine residues and peptides’ N termini, while variable modifications were set to oxidation (+15.994915 Da) of methionine and acetylation (+42.010565 Da) of peptides’ N termini. Precursor ion mass tolerance was set to 20 ppm, and product ion mass tolerance was set to 0.06 Da. The peptide spectrum match allowed 1% target false discovery rate (strict) and 5% target false discovery rate (relaxed). Other parameters were set as default. The protein abundance ratios of the target samples to the pooled sample in each batch were used as the relative abundance for data analysis. Only master proteins (protein with largest # Protein Unique Peptides value and smallest value in the Coverage value in a protein group) were considered in protein groups. No further adjustment of the generated protein matrix was performed.
Pathway Analysis
Metascape (12), String (13), and Ingenuine Pathway Analysis were applied for pathway analyses in this study.
Results
Negative and Exceptionally High IgM and IgG Expression in COVID-19
We procured a cohort of 144 COVID-19 patients and used CLIAs to assess their expression levels of IgM and IgG (Methods). 790, 72, and 58 CLIA tests were conducted during the first 10 weeks since disease onset (weeks 1–10), R1, and R2, respectively. 84.5% of the revisited patients received vaccination between R1 and R2 (supplemental Table S1). Based on their plateau titers during weeks 1 to 10, the antibodies’ expression patterns were classified as follows: − for negative results or very low expression, + for positive results, and ++ for exceptionally high expression (Methods). Accordingly, patients were classified into four groups: IgG−IgM− (G−M−, N = 5), IgG+IgM− (G+M−, N = 42), IgG+IgM+ (G+M+, N = 88), and IgG+IgM++ (G+M++, N = 9) (supplemental Fig. S1, A and B).
We first assessed the IgM expression dynamics in the four groups. G+M+ and G+M++ patients underwent IgM seroconversion between weeks 1 and 2 (Fig. 1A). However, the IgM titers in the G+M++ group were significantly higher since week 2 and were over 20-fold in expression when reaching the plateau at week 4, compared to that in the G+M+ group. The IgM titers in the G+M++ group remained over 10-fold higher than the G+M+ group at weeks 7 to 10. Comparatively, none of the patients in the G+M− or the G−M− group underwent IgM seroconversion during weeks 1 to 10. The overall IgM titers of the four groups had a significant decrease from weeks 1 to 10 to R1, which were further decreased at R2. Only 13.9% (N = 10) and 5.2% (N = 3) of the revisited patients were IgM seropositive at R1 and R2, respectively. The statistical differences in IgM titers between the G+M− and the G+M+ groups persisted at R1 and R2, suggesting a long-term effect of COVID-19.
As for the IgG expression dynamics, all the groups except the G−M− group underwent seroconversion at weeks 1 to 2 and reached the plateau at week 3 (Fig. 1B). Their IgG titers remained at plateau levels during weeks 3 to 6. Thereafter, the IgG titers exhibited a mild decrease in the G+M++ group at weeks 7 to 10. By contrast, IgG seroconversion was hardly observed in the G−M− group, and most of the IgG titers in the G−M− group were below the positivity cut-off value during weeks 1 to 10. The overall IgG titers in the four groups exhibited a significant decrease from weeks 1–10 to R1. Statistical differences in IgG titers between G+M− and G+M+ patients were also observed at R1. The IgG expression of all of the groups was significantly enhanced from R1 to R2, which might be attributed to vaccination (supplemental Table S1).
Vaccination Boosts IgG Expression in COVID-19
To evaluate the effect of vaccination on IgG expression, we classed the convalescent patients at R2 into nonvaccinated (N = 9) and vaccinated (Vac, N = 49) groups and compared their IgG titers (Fig. 1C). The Vac group exhibited significantly elevated IgG titers from R1 to R2. All of the Vac patients were seropositive at R2, including 1 G−M− patient R009. Comparatively, IgG titers in the nonvaccinated group were equivalent between R1 and R2. These observations showed that vaccination can significantly boost IgG expression for COVID-19 convalescents. We also conducted 255 CLIA tests of NAb expression in 85 patients from the cohort throughout weeks 1 to 10 and R1 (supplemental Table S1). The NAb titers were relatively higher in the G+M+ and G+M++ groups (supplemental Fig. S1C) compared to others, suggestive of their stronger neutralizing abilities. NAb and IgG were positively correlated in all of the groups except for G−M− (supplemental Fig. S1C), whereas NAb and IgM were not correlated (supplemental Fig. S1D).
Our observation showed that COVID-19 patients underwent highly diverse antibody expression, especially IgM, after viral infection. IgM turned negative while IgG persisted with a significant decrease 1 year after COVID-19. IgG titers were significantly boosted after vaccination.
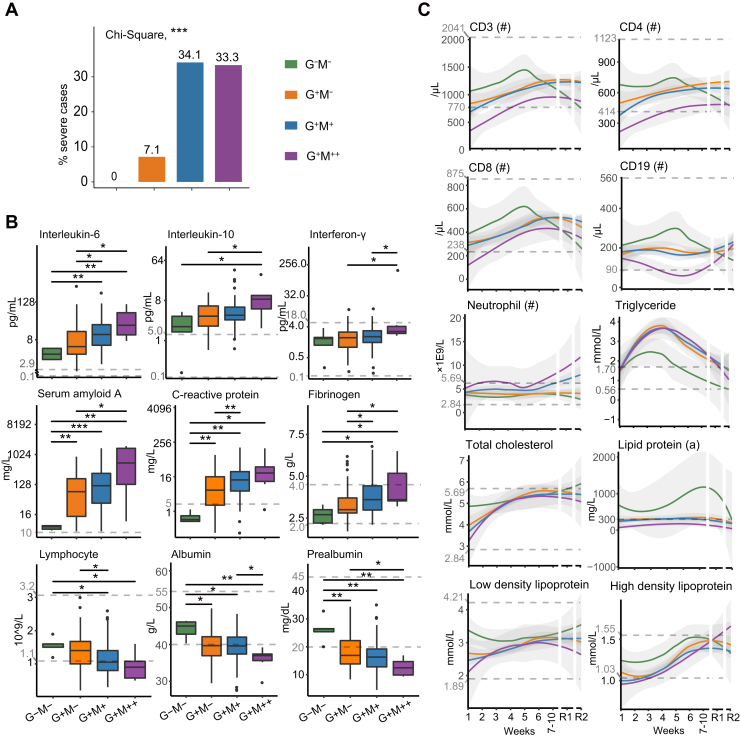
COVID-19 Severity and On-Admission Inflammation Were Positively Correlated With Antibody Expression
We assessed the EHRs of the enrolled patients (Fig. 2 and supplemental Table S2). None of the G−M− patients were severe cases, whereas 34.1% of the G+M+ patients and 33.3% of the G+M++ patients had severe symptoms, respectively (Fig. 2A). Supportively, the G+M+ and G+M++ groups had significantly higher amounts of infected lung lobes and received more drug therapies, including immunoglobulin and methylprednisolone, compared to other groups (supplemental Table S2).
Fig. 2.
Clinical characteristics of the COVID-19 patients with diverse serology.A, distribution of severe cases in the four groups. The y-axis denotes the percentages of severe cases in each group. p value was calculated by chi-square test. B, expression of nine highlighted clinical indicators in the four groups on admission: interleukin-6 (IL-6), interleukin-10, interferon-γ, serum amyloid A (SAA), C-reactive protein (CRP), fibrinogen, lymphocyte, albumin, and prealbumin. p values were calculated by two-sided Wilcoxon rank-sum tests. C, 2-year temporal expression of 10 highlighted clinical indicators in the four groups: CD3, CD4, CD8, CD19, neutrophil, triglyceride, total cholesterol, lipid protein (a), low density lipoprotein, and high density lipoprotein. LOESS model was applied for fitting analysis. The dashed lines denote the upper and lower limits of normal for the indicators. For SAA, CRP, and lipid protein (a), only the upper limits of normal were shown. Significances: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005; ∗∗∗∗p < 0.001.
To investigate the difference in the basic physiological status among the four groups, we compared their clinical indicators on admission (Fig. 2B). Notably, risk factors, such as serum amyloid A and C-reactive protein, and inflammation factors, such as IL-6, IL-10, and IFN-γ, were positively correlated with IgM and IgG expression, suggesting that patients with higher antibody titers also had stronger inflammatory responses. Coagulation factor fibrinogen remained at a normal range in the G−M− group but overexpressed in some patients from the other groups, suggesting a tendency of coagulopathy in these patients. The lymphocyte amounts were negatively correlated with antibody titers and below the lower limit of normal ranges in most of the G+M++ patients, suggesting that their baseline immunological status was inferior to the other patients. A series of nutritional factors such as albumin and pre-albumin, which have been reported as potential indicators for adverse outcomes in COVID-19 (14, 15), were negatively correlated with antibody expression titers.
To sum up, the severity and inflammation status of COVID-19 patients on admission were positively correlated with their antibody expression titers.
Enhanced Cellular Immune Responses are Associated With Negative IgM and IgG Expression
We monitored the 2-year temporal changes of serum clinical indicators in our cohort (Fig. 2C). In the G+M++ group, the expression of CD3, CD4, and CD8, three T lymphocyte markers, was below the lower limit of the reference ranges during weeks 1 to 3 and was significantly lower than the other groups during weeks 1 to 10. CD19, a B lymphocyte marker, was lower in the G+M++ group than the other groups throughout weeks 1 to 10 and fell below the lower limit of the reference range since week 4. Comparatively, these CD markers were highly expressed in the G−M− group until week 5. The intergroup differences of these CD markers decreased after week 7. These observations suggested that the G−M− patients exhibited stronger cellular immune responses compared to the other groups. Neutrophil counts during weeks 1 to 4 increased in the G+M++ group whereas remained equivalent in the other groups. Notably, starting from week 6, neutrophil counts in the G+M++ and G+M+ groups increased beyond the upper limit of the reference range. The decrease of B and T lymphocytes and the increase of neutrophils during hospitalization have been previously observed in severe (16) and deceased (17) COVID-19 patients.
The temporal expression of several blood lipids and lipoproteins was different between the G−M− group and the others (Fig. 2C). Triglyceride (TG) increased during weeks 1 to 3 followed by a gradual decrease in the four groups, but only the G−M− group went back to reference ranges by week 5. The G−M− group also exhibited relatively higher expression of total cholesterol, lipoprotein(a), low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C) during weeks 1 to 10. The combination of low HDL-C and high TG, termed atherogenic dyslipidemia, was considered a risk factor for severe COVID-19 (18). These observations suggest different levels of systematic dyslipidemia during COVID-19, which were the mildest in the G−M− group.
Taken together, patients who remained both IgM and IgG seronegative during disease had enhanced cellular immune responses whereas less perturbation of lipid metabolism, compared to the other patients.
Serum Proteomics of 16 Selected Patients
To understand the differences in host responses behind the diverse antibody expression, we collected 111 serum samples from 16 selected COVID-19 patients (supplemental Fig. S2A) spanning weeks 1 to 10 since disease onset (Fig. 3A). Four patients from each group were selected in a fashion that may represent the diverse antibody characteristics, as elaborated in Methods and supplemental Fig. S2. A total of 1494 proteins were characterized using TMTpro 16plex technology (supplemental Fig. S2D and supplemental Table S3). The median value of the protein coefficient of variation for the pooled samples is 0.189 (supplemental Fig. S2E), indicating high quality of our data.
We firstly assessed the Spearman correlation coefficient of the full protein dataset (Fig. 3B). The overall protein expression in the G−M− and G+M− patients are significantly correlated across weeks 1 to 10, suggesting that their alternation of serum host responses during disease was less prominent compared to other groups. Proteins associated with leukocyte-mediated immunity and lipid metabolic process, two dysregulated functions according to our clinical observations, were clustered across different groups (supplemental Fig. S3). Two protein clusters in leukocyte-mediated immunity were consistently high (Cluster 1) and low (Cluster 2) in expression in the G−M− group compared to others, respectively (supplemental Fig. S3A). They were mainly enriched in signaling pathways of different interleukins according to Ingenuine Pathway Analysis (supplemental Fig. S3B). A protein cluster in lipid metabolic process had diverse expression patterns in the four groups (Cluster 3, supplemental Fig. S3C), which is mostly associated with inositol metabolism, and interleukin signaling as well (supplemental Fig. S3D). These functions are known to mediate COVID-19 host responses according to previous COVID-19 studies (19, 20), which prompted us to assess the expression levels of reported COVID-19–related pathways stratified by their antibody patterns in our further analyses.
Differential Analyses of Serum Proteomics
To explore the proteomic differences among the four groups, ANOVA was used to assess DEPs (adjusted p value <0.05) at each week. Seventy-seven DEPs in total were identified, 61.0% (n = 47) of which were from weeks 1 to 2 (supplemental Fig. S4A). Only one of these 47 proteins was overlapped with the DEPs from weeks 4 to 10, suggesting a distinct difference in the host responses during and after seroconversion. Accordingly, we found that the different patient groups could be stratified by the DEPs during weeks 1 to 2 but not weeks 4 to 10 via PCA (Fig. 3C). The DEPs during weeks 4 to 10 also did not have a distinct stratification of intragroup patients nor week intervals (supplemental Fig. S4, B and C). Additionally, the intragroup correlations at each week were significantly higher at weeks 1 to 2 compared to weeks 4 to 10 for each group of patients (supplemental Fig. S4D).
We next analyzed the key functions of the DEPs. The three most enriched pathways throughout weeks 1 to 10 are humoral immune responses, glycosaminoglycan binding, and carbohydrate binding (Fig. 3D). Within them, humoral immune responses include multiple established risk factors of COVID-19, such as complement and fibrinogen factors (11, 21). The enrichment of glycosaminoglycan binding might be attributed to either cytokine recognition or SARS-CoV-2 host entry (22). We then clustered the DEPs at the initial (weeks 1–2) and late (weeks 7–10) stages of COVID-19 via String (13). Complement system and prothrombin activation were the main enriched functions during weeks 1 to 2 (Fig. 3E). Notably, the expression of C1QB and C1QC, subunits of complement 1 (C1), were significantly lower in the G−M− group compared to the G+M−and G+M+ groups and further decreased in the G+M++ group (Fig. 3F). Comparatively, a list of downstream proteins of complement cascade, including C4BPA, C7, CFI, and CFH, were significantly upregulated in the G+M++ group (Fig. 3E). This observation suggested that complement system was strongly activated in the G+M++ group but not via the classical C1-mediated pathway. P0DOX3, an IgD heavy chain residue, was negatively correlated with IgM expression in the four groups (Fig. 3F), suggesting that IgD may compensate for the lack of IgM in the G−M− and G+M− groups (23). At weeks 7 to 10, leukocyte extravasation signaling was enriched (Fig. 3G). Within them, VNN1, a proposed HDL regulator (24), was significantly upregulated in the G+M++ group (Fig. 3H). FCGR3A was also overexpressed in the G+M++ group, which might enhance the production of proinflammatory cytokines and the activities of cytotoxic effector cells (25). Notably, our pathway findings are also in high accordance with two previous studies that compared the serum proteome (11) and monocyte transcriptome (26) differences between COVID-19 patients and non-COVID-19 individuals, including the enrichment of complement and coagulation cascades at weeks 1 to 2 and the degranulation of neutrophils and platelets at weeks 4 to 10 (supplemental Fig. S4E).
In summary, the proteomics differences in the four groups were the most prominent during weeks 1 to 2, which were associated with complement cascades. The main host response differences during the late stage of COVID-19 relate to the leukocyte activities.
Correlation Between Serum Proteomes and Clinical Indicators
Beyond the functional analyses, we explored the correlation between the DEPs and the antibody titers during weeks 1 to 4 since disease onset. Five and four DEPs were significantly correlated with IgM and IgG expression titers (absolute value of Spearman correlation coefficient is over 0.5), respectively (Fig. 4A). Within them, TIMP1, ICAM1, CD163, NOTCH2, and HLD-DRB1 are associated with inflammatory response, corroborating the important role of inflammation in serology. CD163, a marker for monocytic macrophages (27), is the most correlated protein with IgG among these regulators. As exemplified in Fig. 4B, the expression of serum CD163 was significantly higher in the G+M++ group than the others throughout weeks 1 to 4. We explored CD163 in COVIDpro, a web server that integrates over 40 COVID-19 proteome datasets (28). CD163 was comparatively higher expressed in a group of patients with shorter SARS-CoV-2 RNA shedding course (SC) than those with longer course (LC, Fig. 4C) (29). Notably, SC patients were observed to exhibit higher IgG titers than LC in the first 4 weeks of disease, followed by a fast decrease (29), suggesting that a strongly regulated and short-lived humoral immunity during COVID-19 could efficiently defend against SARS-CoV-2 attacks. CD163 was also upregulated in severe and critical patients compared to the mild ones in five additional serum/plasma proteomics datasets (Fig. 4D) (30, 31, 32). These studies also reported activation of alveolar macrophages and neutrophils in severe and critical cases. Taken together, CD163 upregulation suggests a series of leukocyte activities which may strongly modulate antibody overexpression in COVID-19.
We also assessed the correlation between clinical indicators and DEPs during weeks 1 to 10 (Fig. 4E). HDL-C was significantly correlated with MERTK (Fig. 4F), a transmembrane kinase that contributes to the B lymphocyte activation (33). TG was positively correlated with serum CD34 (Fig. 4G), a human hematopoietic stem cell marker that also has a role in facilitating inflammatory cell trafficking. These evidences further suggested lipid involvement in the immunological activities during COVID-19.
A Putative Working Model for Diverse Serology in COVID-19
Based on our data, we propose a putative working model regarding the diverse COVID-19 serology (Fig. 4H). Complement cascade is initiated upon disease onset, mediating the secretion of cytokines such as IL-6 and IL-10. This process could trigger macrophage polarization, as exemplified by the upregulation of CD163, which would further modulate local inflammation. The subsequently activated neutrophils and T lymphocytes could initiate B cell differentiation, leading to the activation of humoral immune responses to secret antibodies. These processes might be barely activated in the G−M− group due to a prior cellular immune responses that efficiently confront the invasion of SARS-CoV-2 upon disease onset. On the contrary, the cellular immunity might not be sufficient in the G+M++ group for viral defense. Meanwhile, the activation of complement 1 might be disturbed in the G+M++ group. The high expression of inflammatory factors and rapidly ascending IgM titers might be a complementary process to defend against viral attacks.
Discussion
In this study, we extracted 2-year EHRs and applied TMTpro 16plex-based longitudinal proteomics to investigate the host responses of patients with diverse serology in COVID-19. We found that patients with negative IgM and IgG expression still developed strong T cell immunity for viral defense and that the overexpression of IgM was associated with perturbed complement cascades and insufficient cellular immune responses.
Multiple studies have reported the association between antibody expression and COVID-19 severity. For example, Long et al. observed higher titers of IgM and IgG in severe patients than in mild patients since seroconversion (1). Garcia-Beltran et al. found that severe COVID-19 patients that required intubation or were passed away had the highest levels of IgG and IgA antibodies compared to others (34). However, none of them have studied the mechanisms underlying the differentiated antibody expression. Our manifestation that antibody expression was associated with on-admission inflammatory responses supports previous speculation that severe disease might be caused by hyperinflammation, which induces antibody overproduction (34). Conversely, high antibody titers might be involved in the antibody-dependent enhancement of viral entry, which further induces the expression of inflammatory factors (35).
Our observation that a list of CD molecules (CD3, CD4, CD8, and CD19) were highly expressed in the G−M− patients whereas decreased in the G+M++ patients during COVID-19 suggested their differentially regulated lymphocytes to confront viral attacks. The low expression of T and B lymphocyte markers, namely lymphopenia, has been established as a severity hallmark in previous studies (16, 36), further supporting our data. Notably, intragroup differences of these molecules were not as significant after week 5, which might result from medical treatment (37). Therefore, the evaluation of lymphopenia on COVID-19 severity is recommended within 1 month since disease onset. Comparatively, CD163 as shown in our serum proteomics data was continuously upregulated during seroconversion in the G+M++ group. The high expression of CD163 has also been detected in the autopsy samples of six organs in the deceased COVID-19 patients (38), the peripheral blood mononuclear cells during COVID-19, and the THP-1 cell line after 48 h of SARS-CoV-2 infection (39). A recent study reported that overexpression of pulmonary CD163 might lead to idiopathic pulmonary fibrosis in COVID-19 (40). These observations collectively suggest that overly activated macrophage functions might not be favorable to COVID-19 recovery.
Although lipid metabolism dysregulation is typical in COVID-19 (11), our data showed that seronegative patients had the mildest symptoms of dyslipidemia compared to others. This might be attributed to their relatively milder inflammation, which mediates cytokine secretion that alters lipid homeostasis (41). We also found clues that HDL-C and TG might be associated with leukocyte activation during COVID-19. Mechanistic studies are needed in the future to explore their causality.
Our observation that SARS-CoV-2-specific IgG declined significantly 1 year after COVID-19 was in line with previous reports (42), suggesting a transition into immune memories. This also underlined the necessity of vaccinating recovered populations, which could significantly enhance IgG titers 2 years after COVID-19, according to our data. Notably, 85.7% (N = 42) of the vaccinated patients in our cohort received at least two doses of inactivated vaccines as of R2. Previous studies have shown that one to two doses of adenoviral vector or mRNA vaccines were also viable to boost IgG and NAb titers in the recovered patients (7, 43). A longer-term serological monitoring of the same cohort is expected to understand the long-term humoral immunity in the vaccinated population with previous SARS-CoV-2 infection.
Our study has several limitations. Firstly, the single-center study with a relatively small patient cohort was possibly subject to demographic and experimental biases. Especially, the 16 selected patients for our proteomics profiling, although characteristic in antibody and clinical indicator expression may not be representative enough regarding the biases from the distribution of sample acquisition timepoints. Also, the sampling time points failed to cover the seroreversion stages for IgG. Additionally, the COVID-19 patients in our study were all infected by the original SARS-CoV-2 strain but not its variants. With their diverse virulence, their temporal antibody dynamics may exhibit different results from what we observed, which remains to be studied in the future. Furthermore, we did not validate specific markers in this study due to lack of more samples in our country.
Data availability
Patient information and serology data are available in the supplementary material. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD039722.
Supplemental data
This article contains supplemental material Table S1-S3 and Figures S1-S2.
Conflict of interest
Q. Z. and X. Y. are employees of Westlake Omics Inc. Y. Z. and T. G. are shareholders of Westlake Omics Inc. The remaining authors declare no competing interests.
Acknowledgments
We thank Westlake University Supercomputer Center for assistance in data storage and computation
Funding and additional information
This work was supported by grants from National Key R&D Program of China (2021YFA1301602, 2020YFE0202200), National Natural Science Foundation of China (81972492, 21904107, 82072333), Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (LR19C050001), Hangzhou Agriculture and Society Advancement Program (20190101A04), Medical Science and Technology Project of Zhejiang Province (2021KY394), Westlake Education Foundation, and Scientific Research Foundation of Taizhou Enze Medical Center (Group) (21EZZX01).
Author contributions
Y. Z., B. S., and T. G. methodology; Y. Z., J. W., K. Z., D. W., X. L., S. L., X. Y., and G. Z. investigation; X. L., J. W., K. Z., J. L, S. C., M. L., J. P., J. X., H. Z., and G. Z. resources; X. L., M. L., Y. S., Y. X., and Q. Z. formal analysis; X. L., Y. X., and Q. Z. visualization, X. L. and R. S. writing-original draft; Y. Z., B. S., Y. Z., and T. G. supervision.
Contributor Information
Yufen Zheng, Email: zhengyf@enzemed.com.
Bo Shen, Email: shenb@enzemed.com.
Tiannan Guo, Email: guotiannan@westlake.edu.cn.
Supplemental Data
References
- 1.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z., et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int. J. Infect. Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Hartog G., Vos E.R.A., van den Hoogen L.L., van Boven M., Schepp R.M., Smits G., et al. Persistence of antibodies to severe acute respiratory syndrome coronavirus 2 in relation to symptoms in a nationwide prospective study. Clin. Infect. Dis. 2021;73:2155–2162. doi: 10.1093/cid/ciab172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masiá M., Fernández-González M., Telenti G., Agulló V., García J.A., Padilla S., et al. Durable antibody response one year after hospitalization for COVID-19: a longitudinal cohort study. J. Autoimmun. 2021;123:102703. doi: 10.1016/j.jaut.2021.102703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali H., Alahmad B., Al-Shammari A.A., Alterki A., Hammad M., Cherian P., et al. Previous COVID-19 infection and antibody levels after vaccination. Front. Public Health. 2021;9:778243. doi: 10.3389/fpubh.2021.778243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geyer P.E., Arend F.M., Doll S., Louiset M.-L., Virreira Winter S., Müller-Reif J.B., et al. High-resolution serum proteome trajectories in COVID-19 reveal patient-specific seroconversion. EMBO Mol. Med. 2021;13:e14167. doi: 10.15252/emmm.202114167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suhre K., McCarthy M.I., Schwenk J.M. Genetics meets proteomics: Perspectives for large population-based studies. Nat. Rev. Genet. 2020;22:19–37. doi: 10.1038/s41576-020-0268-2. [DOI] [PubMed] [Google Scholar]
- 10.Josse J., Husson F. missMDA: a package for handling missing values in multivariate data analysis. J. Stat. Softw. 2016;70:1–31. [Google Scholar]
- 11.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl. Acids Res. 2019;47:D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aziz M., Fatima R., Lee-Smith W., Assaly R. The association of low serum albumin level with severe COVID-19: A systematic review and meta-analysis. Crit. Care. 2020;24:255. doi: 10.1186/s13054-020-02995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Y., Xue Y., Mao L., Yuan X., Lin Q., Tang G., et al. Prealbumin as a predictor of prognosis in patients with coronavirus disease 2019. Front. Med. (Lausanne) 2020;7:374. doi: 10.3389/fmed.2020.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akbari H., Tabrizi R., Lankarani K.B., Aria H., Vakili S., Asadian F., et al. The role of cytokine profile and lymphocyte subsets in the severity of coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. Life Sci. 2020;258 doi: 10.1016/j.lfs.2020.118167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y., Nie H.X., Hu K., Wu X.J., Zhang Y.T., Wang M.M., et al. Abnormal immunity of non-survivors with COVID-19: predictors for mortality. Infect. Dis. Poverty. 2020;9:108. doi: 10.1186/s40249-020-00723-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masana L., Correig E., Ibarretxe D., Anoro E., Arroyo J.A., Jericó C., et al. Low HDL and high triglycerides predict COVID-19 severity. Sci. Rep. 2021;11:7217. doi: 10.1038/s41598-021-86747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng S., Zou Q., Zhang D., Yu F., Bao J., Lou B., et al. Serum level of testosterone predicts disease severity of male COVID-19 patients and is related to T-cell immune modulation by transcriptome analysis. Clin. Chim. Acta. 2022;524:132–138. doi: 10.1016/j.cca.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Alessandro A., Thomas T., Dzieciatkowska M., Hill R.C., Francis R.O., Hudson K.E., et al. Serum proteomics in COVID-19 patients: altered coagulation and complement status as a function of IL-6 level. J. proteome Res. 2020;19:4417–4427. doi: 10.1021/acs.jproteome.0c00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132.e114. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutz C., Ledermann B., Kosco-Vilbois M.H., Ochsenbein A.F., Zinkernagel R.M., Köhler G., et al. IgD can largely substitute for loss of IgM function in B cells. Nature. 1998;393:797–801. doi: 10.1038/31716. [DOI] [PubMed] [Google Scholar]
- 24.Holleboom A.G., Vergeer M., Hovingh G.K., Kastelein J.J., Kuivenhoven J.A. The value of HDL genetics. Curr. Opin. Lipidol. 2008;19:385–394. doi: 10.1097/MOL.0b013e328306a043. [DOI] [PubMed] [Google Scholar]
- 25.Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Etzerodt A., Moestrup S.K. CD163 and inflammation: biological, diagnostic, and therapeutic aspects. Antioxid. Redox Signal. 2013;18:2352–2363. doi: 10.1089/ars.2012.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang F., Luna A., Tan T., Chen Y., Sander C., Guo T. COVIDpro: database for mining protein dysregulation in patients with COVID-19. bioRxiv. 2022 doi: 10.1101/2022.09.27.509819. [preprint] [DOI] [PubMed] [Google Scholar]
- 29.Tang X., Sun R., Ge W., Mao T., Qian L., Huang C., et al. Enhanced inflammation and suppressed adaptive immunity in COVID-19 with prolonged RNA shedding. Cell Discov. 2022;8:70. doi: 10.1038/s41421-022-00441-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu T., Ning W., Wu D., Xu J., Han Q., Huang M., et al. Plasma proteomics identify biomarkers and pathogenesis of COVID-19. Immunity. 2020;53:1108–1122.e1105. doi: 10.1016/j.immuni.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gisby J., Clarke C.L., Medjeral-Thomas N., Malik T.H., Papadaki A., Mortimer P.M., et al. Longitudinal proteomic profiling of dialysis patients with COVID-19 reveals markers of severity and predictors of death. Elife. 2021;10 doi: 10.7554/eLife.64827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Nesf M.A.Y., Abdesselem H.B., Bensmail I., Ibrahim S., Saeed W.A.H., Mohammed S.S.I., et al. Prognostic tools and candidate drugs based on plasma proteomics of patients with severe COVID-19 complications. Nat. Commun. 2022;13:946. doi: 10.1038/s41467-022-28639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao W.H., Zhen Y., Finkelman F.D., Cohen P.L. The Mertk receptor tyrosine kinase promotes T-B interaction stimulated by IgD B-cell receptor cross-linking. J. Autoimmun. 2014;53:78–84. doi: 10.1016/j.jaut.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–488.e411. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen J., Cheng Y., Ling R., Dai Y., Huang B., Huang W., et al. Antibody-dependent enhancement of coronavirus. Int. J. Infect. Dis. 2020;100:483–489. doi: 10.1016/j.ijid.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K., Yang T., Peng X.F., Lv S.M., Ye X.L., Zhao T.S., et al. A systematic meta-analysis of immune signatures in patients with COVID-19. Rev. Med. Virol. 2021;31 doi: 10.1002/rmv.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He R., Lu Z., Zhang L., Fan T., Xiong R., Shen X., et al. The clinical course and its correlated immune status in COVID-19 pneumonia. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie X., Qian L., Sun R., Huang B., Dong X., Xiao Q., et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184:775–791.e714. doi: 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boumaza A., Gay L., Mezouar S., Bestion E., Diallo A.B., Michel M., et al. Monocytes and macrophages, targets of severe acute respiratory syndrome coronavirus 2: the clue for coronavirus disease 2019 immunoparalysis. J. Infect. Dis. 2021;224:395–406. doi: 10.1093/infdis/jiab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wendisch D., Dietrich O., Mari T., von Stillfried S., Ibarra I.L., Mittermaier M., et al. SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis. Cell. 2021;184:6243–6261.e6227. doi: 10.1016/j.cell.2021.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Zhang Y., Lu R., Dai M., Shen M., Zhang J., et al. Lipid metabolism changes in patients with severe COVID-19. Clin. Chim. Acta. 2021;517:66–73. doi: 10.1016/j.cca.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., Lin H., Ye B., Zhao M., Zhan J., Dong S., et al. One-year sustained cellular and humoral immunities of COVID-19 convalescents. Clin. Infect. Dis. 2021;75:e1072–e1081. doi: 10.1093/cid/ciab884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Favresse J., Gillot C., Di Chiaro L., Eucher C., Elsen M., Van Eeckhoudt S., et al. Neutralizing antibodies in COVID-19 patients and vaccine recipients after two doses of BNT162b2. Viruses. 2021;13:1364. doi: 10.3390/v13071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Patient information and serology data are available in the supplementary material. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD039722.