Abstract
The activation of innate immune responses by genomic DNA from bacteria and several nonvertebrate organisms represents a novel mechanism of pathogen recognition. We recently demonstrated the CpG-dependent mitogenic activity of DNA from the protozoan parasite Babesia bovis for bovine B lymphocytes (W. C. Brown, D. M. Estes, S. E. Chantler, K. A. Kegerreis, and C. E. Suarez, Infect. Immun. 66:5423–5432, 1998). However, activation of macrophages by DNA from protozoan parasites has not been demonstrated. The present study was therefore conducted to determine whether DNA from the protozan parasites B. bovis, Trypanosoma cruzi, and T. brucei activates macrophages to secrete inflammatory mediators associated with protective immunity. DNA from Escherichia coli and all three parasites stimulated B-lymphocyte proliferation and increased macrophage production of interleukin-12 (IL-12), tumor necrosis factor alpha (TNF-α), and nitric oxide (NO). Regulation of IL-12 and NO production occurred at the level of transcription. The amounts of IL-12, TNF-α, and NO induced by E. coli and protozoal DNA were strongly correlated (r2 > 0.9) with the frequency of CG dinucleotides in the genome, and immunostimulation by DNA occurred in the order E. coli ≥ T. cruzi > T. brucei > B. bovis. Induction of inflammatory mediators by E. coli, T. brucei, and B. bovis DNA was dependent on the presence of unmethylated CpG dinucleotides. However, at high concentrations, E. coli and T. cruzi DNA-mediated macrophage activation was not inhibited following methylation. The recognition of protozoal DNA by B lymphocytes and macrophages may provide an important innate defense mechanism to control parasite replication and promote persistent infection.
It is well established that genomic DNA from bacteria, but not mammals, is mitogenic for B cells and stimulates macrophages and dendritic cells to produce proinflammatory cytokines and nitric oxide (NO) (reviewed in references 18 and 27). The immunostimulatory properties of bacterial DNA are attributed to the high frequency of unmethylated CG dinucleotides (18, 27 28). These motifs are largely absent from mammalian DNA because CG dinucleotides have a reduced frequency of occurrence, are generally methylated, and are often flanked by bases that constitute immunosuppressive motifs (27). The B-lymphocyte pattern recognition of unmethylated CpG motifs in bacterial DNA was found to extend to other nonvertebrate genomes, including insects, mollusks, nematodes, yeasts, and protozoa (5, 42). Recognition of nonmammalian DNA by the innate immune system is a newly discovered host immune response to infectious organisms that is distinct from protein or carbohydrate immune recognition (17).
In mice, bacterial DNA generates a type 1 immune response, marked by enhanced cytotoxic T-lymphocyte and antibody responses and by production of interleukin-1β (IL-1β), IL-6, IL-12, IL-18, gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) (reviewed in references 18 and 27). Induction of a T-helper 1 (Th1)-dominated immune response by bacterial DNA has provided an explanation for the remarkable efficacy of “naked” DNA vaccines and indicated the basis for new vaccine strategies (46). Bacterial and invertebrate genomic DNA have been used in mice as potent adjuvants for immunization with soluble or particulate antigens (18, 26–28, 44, 46). Furthermore, through the addition of immunostimulatory CpG motifs, plasmid DNA or synthetic oligodeoxynucleotides can be engineered to provide a Th1-promoting adjuvant function (18, 26, 27, 46). Several reports have demonstrated similar effects of Escherichia coli DNA and defined oligodeoxynucleotides on leukocytes of nonhuman primate, human, and bovine leukocytes (27).
Recognition of CpG motifs of microbial origin as a “danger signal” (32) represents a novel innate immune defense mechanism to enable the discrimination of pathogen from host and to trigger a selective immune response at the site of infection (17, 34). Activation of innate immune responses by DNA released from dying parasites could contribute to host survival and encourage persistent parasitic infection. In fact, protozoan parasite infections of humans, such as malaria, African sleeping sickness, and Chagas' disease, as well as those of cattle, such as theileriosis, babesiosis, and trypanosomiasis, result in parasite persistence, thereby ensuring parasite survival by providing a reservoir for subsequent arthropod vectored transmission. It is also possible that immunostimulation by protozoal DNA could provoke hyperactivation of B cells and macrophages, with pathological consequences.
We recently reported that DNA from the protozoan parasite Babesia bovis induced CpG-dependent proliferation of bovine B cells and enhanced immunoglobulin G (IgG) secretion (5), indicating that DNA is a mitogenic component of the parasite with the potential to stimulate innate defenses against the foreign pathogen. We have also demonstrated that B. bovis-infected erythrocytes stimulated NO and inflammatory cytokine production by bovine macrophages (37, 41). However, with the exception of a parasite-derived lipid fraction that stimulated the induction of NO but not cytokines, the immunostimulatory components in the parasite were not defined. The present study was designed to determine if DNA from this parasite and others that cause persistent infection, namely, Trypanosoma cruzi, and T. brucei, is capable of activating macrophages. We demonstrate that protozoal DNA induces IL-12, TNF-α, and NO, which are thought to be important for the resolution of infection and/or increased pathologic changes caused by these parasites (4, 13, 22, 23, 25, 31, 37, 51). In addition, we show that, like B. bovis DNA, DNA from T. cruzi and T. brucei is mitogenic for B lymphocytes.
MATERIALS AND METHODS
Macrophage isolation.
Monocyte-derived macrophages were isolated from peripheral blood mononuclear cells (PBMC) of six noninfected cattle by plastic adherence and culture as previously described (37). After 6 to 7 days of culture, macrophages were harvested with Ca2+- and Mg2+-free Hanks balanced salt solution containing 0.5 mM EDTA. The procedure regularly yielded greater than 80% CD14-expressing cells by fluorescence-activated cell sorting with monoclonal antibody (MAb) CAM36A. Unless indicated otherwise, all MAbs were purchased from the Washington State University Monoclonal Antibody Center, Pullman, Wash.
B-cell isolation.
B cells were isolated from bovine PBMC by positive selection as previously described (5). Briefly, PBMC were incubated with a MAb (GB25a) to bovine CD21 at 4°C for 40 min with gentle agitation. Bead-bound B cells were isolated using goat anti-mouse IgG-coated magnetic beads (Dynabead M-450; Dynal Inc., Lake Success, N.Y.) as specified by the manufacturer. The procedure routinely yielded >90% of cells that expressed surface immunoglobulin. The purified cell population was negative for T cells and monocytes as determined by fluorescence-activated cell sorter analysis using MAb specific for bovine CD14 (CAM36A), CD2 (MAb MUC2A), CD3 (MAb MM1A), CD4 (MAb CACT 138A), CD8 (MAbs CACT 80C and BAT 82B), and the δ chain of the γ/δ T-cell receptor (MAb CACT 61A).
DNA preparation.
DNA from E. coli, B. bovis, and T. cruzi was prepared essentially as described previously (5). Briefly, lyophilized E. coli (strain B, ATCC 11303) (Sigma) was resuspended in TE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 25 mM EDTA [pH 8.0]) and treated with lysozyme, proteinase K, and sodium dodecyl sulfate (SDS). E. coli DNA was successively extracted with phenol, phenol-chloroform-isoamyl alcohol, and chloroform-isoamyl alcohol. The DNA was then precipitated with 3 M sodium acetate–absolute ethanol, washed with 70% ethanol, and air dried. B. bovis Mexico strain merozoites were maintained in continuous culture in bovine erythrocytes and purified as described previously (6). T. cruzi DNA was prepared from the RA strain (15). Parasites were washed in phosphate-buffered saline (PBS) and incubated in a digestion buffer containing NaCl, Tris-HCl, EDTA, SDS, and proteinase K. RNase A was added to a final concentration of 100 μg per ml, and the mixture was incubated at 37°C for 2 h. B. bovis and T. cruzi DNA was extracted as described above. Pelleted DNA was resuspended in phosphate-buffered saline or Tris-buffered saline at 37°C. T. brucei bloodstream trypomastigotes were isolated from rats by cardiac puncture and purified over a DE52 column, and DNA was prepared as described previously (52). Trypomastigotes were lysed by the addition of 0.5% SDS and then incubated sequentially with 100 μg of RNase A per ml and 1 mg of pronase per ml. DNA was extracted with phenol, extensively dialyzed, precipitated with absolute ethanol, and washed with 75% ethanol. Pelleted DNA was lyophilized and resuspended in TE buffer. Bovine (Bos taurus) DNA was prepared from bovine buffy coats obtained from 10 ml of anticoagulated blood from normal adult Holstein cattle (33). DNA concentrations were determined by spectrophotometric analysis.
Purified DNA (150 to 200 μg/ml) was digested with 1 mg of DNase I (Sigma) per ml for 2 h at 37°C (5) and then stored at −20°C until use. Complete digestion of the DNA was confirmed by agarose gel electrophoresis. Prior to use in proliferation assays, the DNA concentration was determined again by spectrophotometric analysis.
DNA was methylated using CpG methylase (SssI methylase; New England Biolabs, Beverly, Mass.) as specified by the manufacturer and as described previously (5, 43). DNA methylation was complete after 24 h, as determined by measuring the resistance of treated DNA samples to cleavage by HpaII (New England Biolabs). Methylated DNA was extracted, precipitated, and quantified by spectrophotometry.
Limulus amebocyte lysate assay.
Cell culture reagents and all DNA preparations were tested for the presence of endotoxin by using the Limulus amebocyte lysate assay (Whittaker M. A. Bioproducts, Walkersville, Md.) The sensitivity of the assay is 0.06 EU of endotoxin per ml (6 pg per ml). All cell culture reagents and protozoal and bovine DNA samples had undetectable levels of endotoxin (<6.0 pg in 25-μg/ml DNA samples). E. coli DNA contained 6.0 pg of endotoxin in a 25-μg/ml sample. Elsewhere, we have shown that 10 μg of polymyxin B sulfate per ml is sufficient to block up to 10 ng per ml of lipopolysaccharide-induced cytokine or inducible nitric oxide synthase (iNOS) transcription (36a). All leukocyte and DNA cultures contained 10 μg of polymyxin B sulfate per ml.
B-lymphocyte proliferation assays.
Purified B cells (2 × 106 cells per ml) were incubated at 37°C for 72 h in duplicate or triplicate cultures of 100 μl in complete RPMI 1640 medium with 1 μg of pokeweed mitogen (Sigma) per ml, 1 μg of concanavalin A (Sigma) per ml, or 1.0 to 12.5 μg of DNA per ml. In the final 18 h of culture, the cells were radiolabeled with 0.25 μCi of [3H]uridine (New England Nuclear, Boston, Mass.). The cells were harvested and counted in a liquid scintillation counter. Results are presented as the mean cpm of replicate cultures ± 1 standard deviation (SD) or as the stimulation index (SI), calculated as the mean cpm of cells cultured with DNA/mean cpm of cells cultured with medium. SI > 3.0 is considered significant.
Stimulation of macrophages for cytokine and NO production.
Macrophages (5 × 105) were cultured for 6 h (for RNA extraction), 24 h (for secreted cytokines), or 48 h (for secreted NO) in 24-well plates in 400-μl volumes with 25 μg of protozoal DNA, E. coli DNA (used as a positive control), or bovine DNA (used as a negative control) per ml plus 50 U of IFN-γ (generously provided by Lorne Babiuk, Veterinary Infectious Disease Organization (VIDO), Saskatoon, Saskatchewan, Canada) per ml and 10 μg of polymyxin B sulfate per ml. In some experiments, 0.01 to 25 μg of E. coli DNA per ml was used. For assays measuring IL-12 p40, serum-free Iscove's medium supplemented with 25 mM HEPES (Mediatech, Herndon, Va.), 2 mM l-glutamine (Mediatech), 5 × 10−5 M 2-mercaptoethanol (Sigma), and 50 μg of gentamicin sulfate per ml was used. Supernatants were collected and stored at −70°C until analysis.
IL-12 p40 detection by dot blot assay.
IL-12 was detected using MAb 17827 specific for the bovine IL-12 p40 subunit (Serotec, Raleigh, N.C.). Macrophage supernatants and recombinant human IL-12 (rHuIL-12; kindly provided by Genetics Institute, Cambridge, Mass.) were serially diluted and applied to a nitrocellulose membrane using a HybriDot manifold (GIBCO BRL, Rockville, Md.). Bound antibody was identified using the Western-Star chemiluminescent detection system (Tropix, Inc, Bedford, Mass.). Briefly, the membrane was incubated on a rocker for 1 h at room temperature or overnight at 4°C with I-block blocking solution and then incubated with IL-12 p40 MAb at a final concentration of 1 μg per ml in I-block for 1 h. After six 15-min washes with TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween 20 [pH 7.6]), goat anti-mouse IgG-plus-IgM alkaline phosphatase conjugate diluted 1:15,000 in I-block was added for 1 h. The membrane was washed, incubated twice for 2 min in 1× assay buffer (20 mM Tris [pH 9.8], 1 mM MgCl2), and transferred to a Western-Star development folder. The substrate solution, composed of 3 ml of CDPStar-RTU and 150 μl of NitroBlock, was spread evenly over the membrane and allowed to bind for 5 min. Excess solution was smoothed out of the development folder, and the blot was exposed to autoradiography film.
IL-12 bioassay.
The presence of the biologically active IL-12 heterodimer was determined as described (37, 38). Briefly, macrophage supernatants (200 μl) were incubated with 2 × 106 PBMC and phytohemagglutinin PHA (1 μg/ml) in a total volume of 400 μl in a 48-well plate for 2 days. PHA-stimulated PBMC were also incubated with rHuIL-12 (1 to 1,000 pg per ml) as a standard. PBMC supernatants were collected and assayed for bovine IFN-γ using a commercially available enzyme-linked immunosorbent assay (ELISA) as specified by the manufacturer instructions (CSL Ltd., Parkeville, Victoria, Australia). IFN-γ activity was determined from a standard curve derived with serial dilutions of a T-cell supernatant, estimated by the vesicular stomatitus virus cytopathic effect reduction assay to contain 440 U of IFN-γ per ml. By comparison with recombinant bovine IFN-γ, 1 U of IFN-γ activity represents approximately 1.7 ng. Macrophage supernatants were also evaluated for residual exogenous IFN-γ.
Detection of TNF-α by ELISA.
Samples were serially diluted and analyzed for TNF-α by ELISA as described previously (12, 37). Briefly, Immulon II ELISA plates (Dynax Technologies, Chantilly, Va.) were coated with anti-bovine TNF-α MAb 1D11-13, kindly provided by Dale Godson (VIDO). After the plates were washed with TBST, samples diluted in TBST-g (TBST containing 0.5% gelatin) were added to the plates and incubated for 2 h at room temperature or overnight at 4°C. Bound TNF-α was detected with a rabbit anti-TNF-α serum (VIDO), followed by biotinylated goat-anti rabbit IgG (heavy plus light chains; Zymed Laboratories, San Francisco, Calif.), streptavidin-alkaline phosphatase (GIBCO BRL), and p-nitrophenylphosphate di(Tris)-salt crystalline (pNPP). The reaction was stopped by addition of 0.3 M EDTA (pH 8.0), and the optical density at 405 nm was determined with an ELISA plate reader. Samples were quantified by comparison against a standard curve generated with recombinant bovine TNF-α (VIDO) diluted from 0.02 to 10 ng per ml.
Detection of nitrite by the Griess assay.
Nitrite (NO2−) present in macrophage supernatants was tested in a Griess assay (37). Macrophages were cultured for 48 h at a concentration of 0.5 × 105 to 1 × 105 cells per well in 96-well flat-bottom plates with 25 μg of DNA per ml, 50 U of IFN-γ per ml, and 10 μg of polymyxin B sulfate per ml. Culture supernatants were transferred (50 μl per well) to new 96-well, flat-bottom plates, a 50 μl of Griess reagents per well was added to the supernatants, and the absorbance at 540 nm was compared to a NaNO2 standard curve. Results are presented as the mean (micromolar) concentration of NO2− in triplicate cultures ± 1 SD.
Statistical analyses.
B-cell proliferation and cytokine and iNOS production were analyzed for statistical significance using the Student's one-tailed t test.
Analysis of CG dinucleotide content in genomic DNA.
Genomic DNA sequences constituting at least 20,000 bases from E. coli, T. cruzi, T. brucei, and B. bovis were obtained from the GenBank database and analyzed for the presence of CG dinucleotides with the Genetics Computer Group (version 10.0) package (11). Genbank accession numbers are as follows: for E. coli, 111721.em_ba, e02087.gb_pat, af016587.gb_sy, m93424.em_ba, 148948.em_ba, a00047.gb_pat, a01447.gb_pat, e01484.gb_pat, a22409.gb_pat, e03599.gb_patz21706.gb_sy, k02969.em_ba; for T. cruzi, AF032907.GB_in, AF004423, AB010287, AB005063, AF044732, AF061250, AQ445526, AQ445530, AQ445531, AQ445537, AQ445541; for T. brucei, TBR132925, TBR012199; and for B. bovis, BBU18792, AF027149. The results of the analysis are presented in Table 1.
TABLE 1.
Frequency of CG dinucleotides in genomic DNA of E. coli and protozoan parasites used in this study
| Organism | % CG | No. of bases analyzed |
|---|---|---|
| E. coli | 7.7 | 32,362 |
| T. cruzi | 4.6 | 20,489 |
| T. brucei | 3.9 | 36,189 |
| B. bovis | 3.4 | 20,333 |
RT-PCR.
The induction of cytokine and iNOS mRNA was determined by reverse transcription-PCR (RT-PCR) as previously described (37). Briefly, RNA was isolated using TRIzol reagent (GIBCO BRL), treated with DNase (Ambion, Inc., Austin, Tex.), and analyzed for cytokine and iNOS transcripts by RT-PCR. The primers for bovine IL-12 p40, IL-12 p35, TNF-α, IL-1β, iNOS, IL-10, and β-actin and the optimal PCR conditions were described recently (37). Additional primers used to amplify IFN-α were as follows: forward, 5′-CGACAACTGAGGAGGGTCTC-3′; and reverse, 5′-AGGCTCTCATGACTTGTGCTC-3′. The cycle number chosen for each primer set was empirically determined for each set of samples, based on the positive control, and was selected to fall within the linear range of amplification. Samples were compared by normalizing the target signal to the β-actin signal from each sample and comparing the normalized values.
RESULTS
Stimulation of B-cell proliferation by T. cruzi and T. brucei DNA.
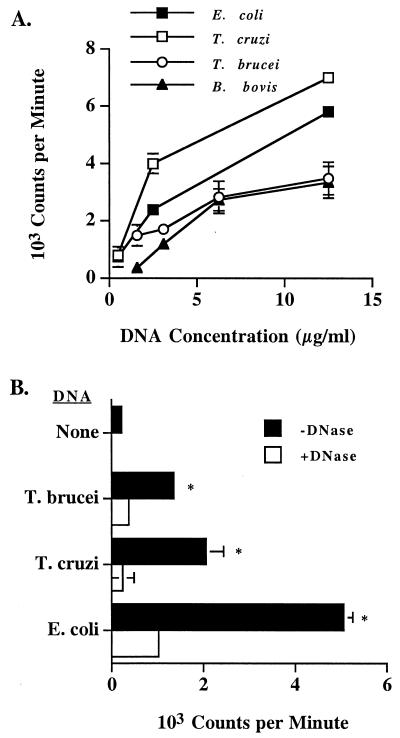
In previous work, we demonstrated that B lymphocytes were stimulated to proliferate in response to both E. coli and B. bovis DNA but not bovine DNA (5). We have now extended these studies to two additional protozoan parasites, T. cruzi and T. brucei. Purified B cells proliferated in response to pokeweed mitogen but not concanavalin A (data not shown). DNA from both parasites stimulated dose-dependent B-cell proliferation, and in general the response to E. coli and T. cruzi DNA was higher than the response to T. brucei or B. bovis DNA (Fig. 1A) (5). Interestingly, E. coli and T. cruzi DNA, which have the highest CG dinucleotide content (Table 1), stimulated more proliferation. As shown previously for B. bovis DNA (5), treatment of the E. coli or trypanosome DNA preparations with DNase resulted in a significant (P < 0.02) reduction in the B-lymphocyte proliferative response (Fig. 1B), indicating that the mitogenic activity was not due to contaminants in the preparation.
FIG. 1.
Trypanosome DNA stimulates dose-dependent B-lymphocyte proliferation. (A) Bovine B cells were cultured at 2 × 105 cells per well with 0.5 to 12.5 μg of E. coli, T. cruzi, T. brucei, or B. bovis DNA per ml and 10 μg of polymyxin B sulfate per ml. Results are presented as the mean of duplicate (E. coli and T. cruzi DNA) or triplicate (T. brucei and B. bovis DNA) cultures and 1 SD. These data are representative of at least two independent experiments performed with different cattle. (B) B cells were cultured as described for panel A with 12.5 μg of DNA per ml that was untreated or treated with DNase. Results are presented as the mean cpm and 1 SD of duplicate cultures and are representative of two experiments. DNase treatment significantly reduced the mitogenic activity of all DNAs (P < 0.02, indicated by asterisk).
Trypanosome DNA contains unmethylated CpG motifs and stimulates B-cell proliferation by a CpG-dependent mechanism.
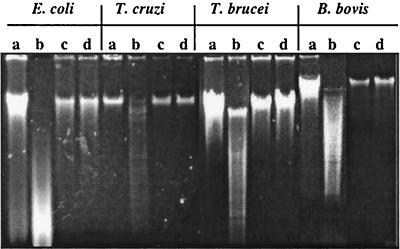
The mitogenic effects of DNA from E. coli, Drosophila, yeast, and B. bovis were reduced or abrogated following methylation with CpG methylase (5, 27, 42). To determine whether trypanosome DNA is unmethylated and to determine the requirement for unmethylated CpG dinucleotides in stimulating B-lymphocyte proliferation, DNA was methylated with CpG methylase, examined for HpaII sensitivity, and tested for mitogenicity. Although this method of analysis is not quantitative, DNA from B. bovis, T. cruzi, and T. brucei appears to be somewhat less sensitive than E. coli DNA to HpaII digestion (Fig. 2, lanes a and b), suggesting that the parasite DNA may contain fewer unmethylated CG dinucleotides than the E. coli DNA does. However, all were resistant to HpaII cleavage following methylation (lanes c and d). Similar results were reported for DNA from other nonvertebrate organisms (42).
FIG. 2.
Unmethylated DNA is susceptible but methylated DNA is resistant to digestion with HpaII. DNAs from E. coli, T. cruzi, T. brucei, and B. bovis were treated with SssI methylase and tested for sensitivity to HpaII. DNA was visualized following electrophoresis on ethidium bromide-agarose gels. Lanes: a and b, unmethylated DNA; c and d, methylated DNA. DNA in lanes a and c was not subjected to HpaII digestion, whereas DNA in lanes b and d was subjected to HpaII digestion.
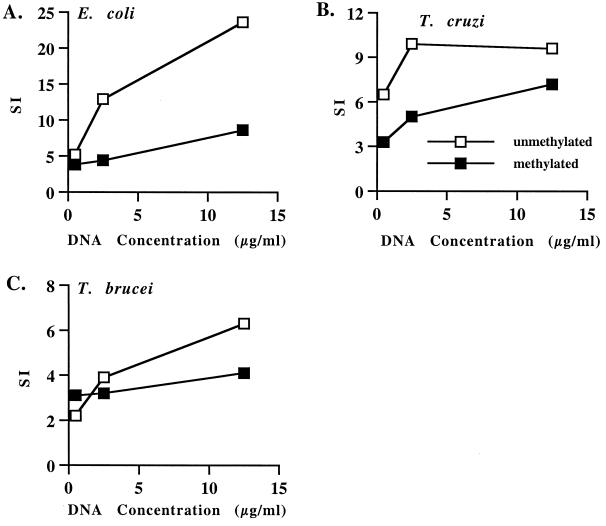
T. brucei and T. cruzi DNA stimulated dose-dependent proliferation of B lymphocytes, and methylation reduced but did not eliminate the stimulatory activity of each DNA (Fig. 3). These results are similar to what was previously observed with B. bovis DNA (5) and yeast DNA (42), where methylation did not completely eliminate mitogenicity for B lymphocytes. The effect of methylation was significant (P < 0.05) for E. coli DNA with 2.5 and 12.5 μg of DNA per ml and for T. cruzi DNA with 2.5 μg DNA per ml. These data indicate that unmethylated CpG sequences are important for the mitogenic property of protozoal DNA.
FIG. 3.
Trypanosome DNA stimulates B-cell proliferation in a CpG-dependent manner. Positively selected B cells were cultured at 2 × 105 cells per well with 0.5 to 12.5 μg of unmethylated or methylated E. coli (A), T. cruzi (B) or T. brucei (C) DNA per ml and 10 μg of polymyxin B sulfate per ml. Results are presented as the SI of duplicate cultures of B cells cultured with DNA compared with B cells cultured with medium. These data are representative of at least two experiments performed with different cattle.
Induction of IL-12, TNF-α, and NO production by protozoal DNA.
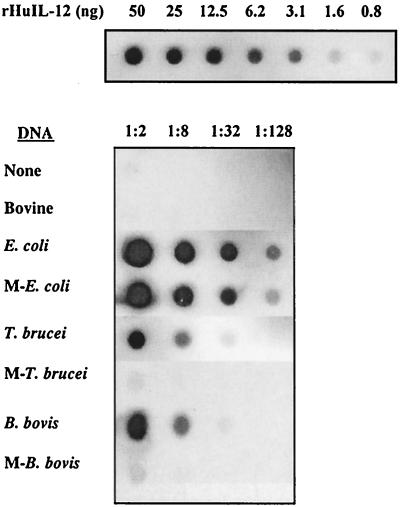
In addition to activating B cells, bacterial DNA stimulates monocytes and macrophages to increase the expression of inflammatory mediators and costimulatory molecules (3, 9, 16, 40). However, to our knowledge, there have been no reports on the ability of protozoal DNA to activate macrophages. Therefore, bovine macrophages were stimulated with untreated or methylated protozoal DNA and induction of TNF-α, IL-12, and NO was determined. The presence of IL-12 in macrophage supernatants was initially evaluated by a dot blot assay using a MAb specific for the bovine IL-12 p40 subunit. E. coli, T. brucei, and B. bovis DNA all induced IL-12 p40 expression (Fig. 4). (T. cruzi DNA was not evaluated due to the limited amount of sample available.) E. coli DNA had a more potent effect than either T. brucei or B. bovis DNA, and while methylation had little to no effect on the induction of IL-12 p40 by E. coli DNA, it completely prevented the induction of IL-12 p40 by T. brucei and B. bovis DNA. Untreated macrophages and macrophages treated with bovine DNA produced little or no IL-12.
FIG. 4.
Protozoal DNA stimulates the production of IL-12 p40. Macrophages were cultured in serum-free medium with 25 μg of DNA per ml, 50 U of IFN-γ per ml and 10 μg of polymyxin B sulfate per ml. After 24 h, supernatants were collected, serially diluted, and transferred to nitrocellulose membranes by using a dot blot apparatus. The upper blot contains serial dilutions (0.8 to 50 ng) of rHuIL-12. The lower blot contain serial dilutions (1:2 to 1:128) of supernatants from macrophages cultured with the indicated untreated or methylated (M) DNA. IL-12 was detected with bovine IL-12p40-specific MAb and visualized using an alkaline phosphatase conjugate detection system. Autoradiograph films are shown. These data are representative of three experiments.
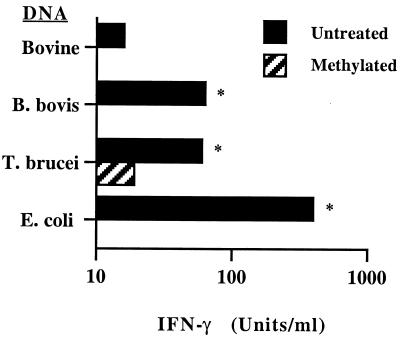
Because the dot blot assay detects only p40 expression, a bioassay which measures the induction of IFN-γ by bovine PBMC stimulated with functional heterodimeric IL-12 was also used to measure IL-12-like bioactivity (37, 38). Supernatants from macrophages treated with E. coli, T. brucei, or B. bovis DNA induced significant IFN-γ production by PBMC compared with those from macrophages treated with bovine DNA (Fig. 5). A strong correlation of the level of IFN-γ produced and the CG dinucleotide content (Table 1) was revealed when DNA from the different organisms was compared (r2 = 0.993). Methylated T. brucei or B. bovis DNA induced significantly less downstream IFN-γ production than did untreated DNA. Together, these data indicate that bacterial and protozoal DNA induce bovine macrophage expression of biologically active IL-12.
FIG. 5.
Protozoal DNA induces IL-12-like activity. Macrophages were cultured for 24 h with 25 μg of bovine DNA (negative control), E. coli DNA (positive control), or untreated or methylated B. bovis or T. brucei DNA per ml, IFN-γ, and polymyxin B sulfate. Supernatants were assayed by ELISA for induction of IFN-γ by PHA-costimulated PBMC. Results are presented as the mean and 1 SD of duplicate determinations and are representative of two experiments. Significantly more IFN-γ was induced by T. brucei and B. bovis DNA than by bovine DNA or the corresponding methylated DNA (P < 0.01, indicated by asterisk).
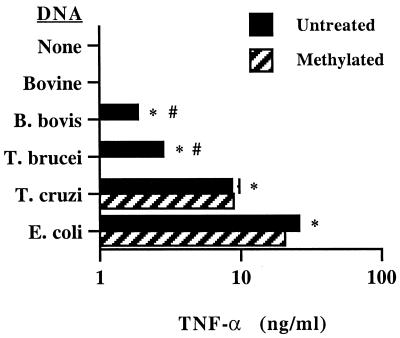
E. coli, T. cruzi, T. brucei, and B. bovis DNA all induced significant amounts of TNF-α production by macrophages (Fig. 6). E. coli DNA was clearly the most potent activator, while T. cruzi DNA was more potent than T. brucei or B. bovis DNA. The amount of secreted TNF-α was highly correlated with CG dinucleotide frequency in DNA of the four organisms (Table 1) (r2 = 0.997). Methylated E. coli DNA and T. cruzi DNA retained their ability to induce TNF-α production, whereas methylation abolished the TNF-α-inducing activity of T. brucei and B. bovis DNA. Untreated macrophages and macrophages treated with bovine DNA did not produce TNF-α.
FIG. 6.
Protozoal DNA induces TNF-α. Macrophages were cultured with 25 μg of the indicated DNA per ml, IFN-γ, and polymyxin B sulfate for 24 h, and supernatants were assayed for TNF-α by ELISA. Results are presented as the mean and 1 SD of duplicate determinations and are representative of at least two independent assays. Significantly more TNF-α was induced by protozoal DNA than by medium or bovine DNA (P < 0.02, indicated by asterisk) or the corresponding methylated DNA (P < 0.02, indicated by octothorp).
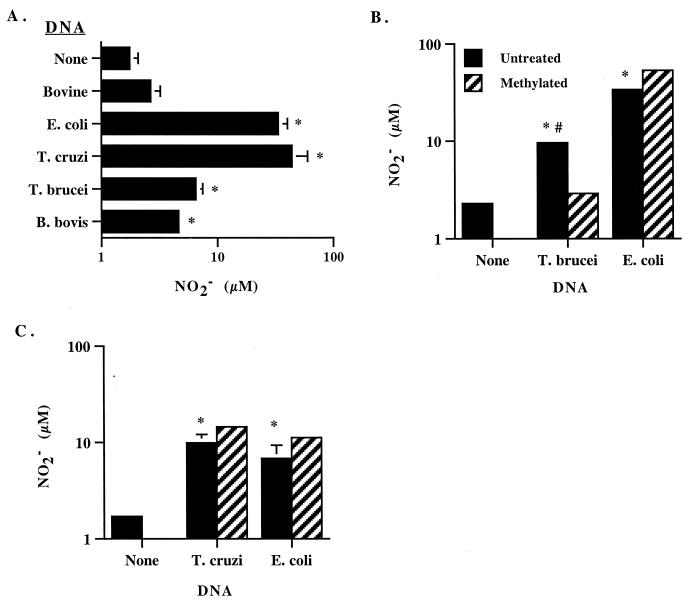
When NO production was determined by the Griess assay, B. bovis, T. brucei, T. cruzi, and E. coli DNA, but not bovine DNA, all stimulated significant amounts of NO2− (Fig. 7A). B. bovis DNA was the weakest stimulus, while DNA from E. coli and T. cruzi DNA were more potent. The correlation of NO2− production and CG dinucleotide frequency was again very high when DNA from E. coli, T. brucei, and B. bovis was compared (r2 = 0.990) or when DNA from all three parasites were compared (r2 = 0.927). The amounts of NO2− induced by T. cruzi DNA and E.coli DNA were comparable within a given experiment that was repeated three times, although these levels varied from experiment to experiment (Fig. 7A and C). Whereas methylation of T. brucei DNA significantly reduced NO2− production (Fig. 7B), there was no negative effect of methylating either T. cruzi or E. coli DNA (Fig. 7C).
FIG. 7.
Protozoal DNA stimulates NO production. Macrophages were cultured with the indicated DNA, IFN-γ, and polymyxin B sulfate for 48 h, and supernatants were assayed for NO2− production by the Griess assay. Results are presented as the mean and 1 SD of triplicate determinations. (A to C) Significantly more NO2− was induced by parasite or E. coli DNA than no DNA (none) or bovine DNA (P < 0.02, indicated by asterisk). (B) Significantly more NO2− was induced by untreated T. brucei DNA than by the corresponding methylated DNA (P < 0.01, indicated by octothrop).
Protozoal DNA activates the expression of macrophage mRNA for inflammatory cytokines and iNOS.
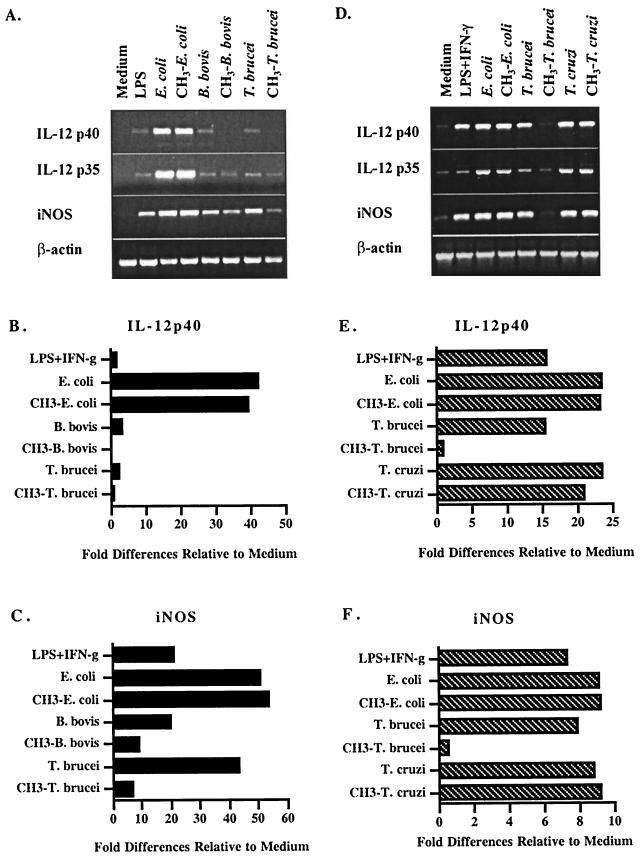
To further examine the effects of protozoal DNA on cytokine and NO induction, steady-state mRNA levels were compared in nonactivated and activated macrophages (Fig. 8). Bovine DNA did not upregulate the expression of any cytokine or iNOS (data not shown). DNA from E. coli and T. cruzi was most effective at stimulating enhanced steady-state levels of IL-12 p40, IL-12 p35, and iNOS mRNA expression, which was unaffected by methylation (Fig. 8A and D). T. brucei and B. bovis DNA also induced increased steady-state levels of expression of IL-12 and iNOS, although they were relatively lower than for E. coli or T. cruzi DNA, and methylation effectively reduced their activity (Fig. 8A and D). The steady-state levels of TNF-α and IL-1β transcript expression were high in unstimulated macrophages and not reproducibly elevated in response to DNA (data not shown). Furthermore, in several experiments, parasite DNA stimulated an increase in IFN-α transcript levels, but did not induce IL-10 mRNA (data not shown). The graphical representations of DNA-mediated increases in steady-state levels of IL-12 p40 and iNOS mRNA relative to medium are shown in Fig. 8B and C and Fig. 8E and F, respectively. The relative levels of expression of cytokine and iNOS mRNA mirrored the CG dinucleotide frequency in genomic DNA.
FIG. 8.
Protozoal DNA stimulates enhanced steady-state levels of transcripts for inflammatory cytokines and iNOS in macrophages. RNA was collected from macrophages stimulated with 1 ng of lipopolysaccharide (LPS) per ml without or with 50 U of IFN-γ per ml or 25 μg of unmethylated or methylated E. coli, T. cruzi, T. brucei, or B. bovis DNA per ml, IFN-γ and 10 μg of polymyxin B sulfate per ml. Following RNA isolation and DNase treatment, RNA was analyzed by RT-PCR. Panels A to C and panels D to F represent two independent experiments. PCR products were semiquantified by nonsaturating densitometry and normalized to actin, and each sample was evaluated relative to the medium control. (B) IL-12 p40 analysis of samples in panel A; (C) iNOS analysis of samples in panel A; (E) IL-12 p40 analysis of samples in panel D; (F) iNOS analysis of samples in panel D. A third experiment gave a similar pattern of results.
Methylation of E. coli DNA effectively inhibits immunostimulatory activity at lower concentrations.
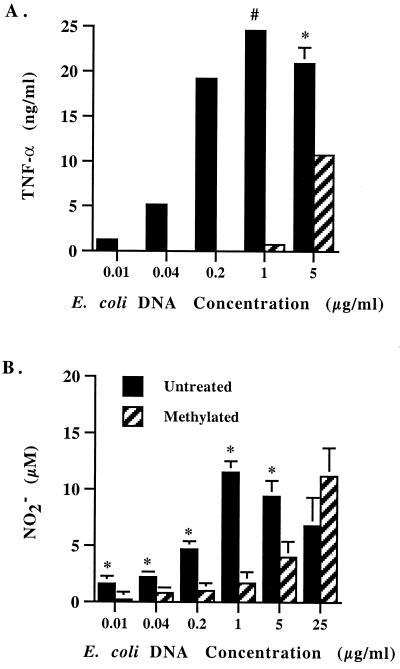
One possible explanation for the ability of methylated E. coli or T. cruzi DNA to activate macrophages is that these DNAs are incompletely methylated, so that at high concentrations, such as the 25 μg per ml used in these studies, sufficient numbers of unmethylated CpG dinucleotides remained. This would be more likely to occur with E. coli and T. cruzi DNA, which have a higher frequency of CG dinucleotides than T. brucei and B. bovis DNA. At lower DNA concentrations, methylation should abrogate the macrophage response to E. coli and T. cruzi DNA. To test this possibility, we measured TNF-α and NO2− production by macrophages treated with different concentrations of methylated or untreated E. coli DNA in the presence of polymyxin B sulfate (Fig. 9). A dose-dependent response to untreated DNA was observed, which was maximal at 1 μg DNA per ml. Interestingly, methylation significantly inhibited the induction of TNF-α and NO when 5 μg of DNA per ml or less was used. When 1 μg of DNA per ml or less was used, methylation nearly or completely abrogated the stimulatory activity.
FIG. 9.
Methylation inhibits the induction of TNF-α and NO by low concentrations of E. coli DNA. Macrophages were cultured with untreated or methylated E. coli DNA at concentrations ranging from 0.01 to 25.0 μg per ml. Supernatants were collected and tested for the presence of TNF-α and NO as described in the legend to Fig. 6 and 7. (A) Unmethylated DNA induced significantly more TNF-α production than methylated DNA (P < 0.05, indicated by asterisk; P < 0.005, indicated by octothorp). TNF-α was undetectable (<0.08 ng/ml) when <1 μg of methylated DNA per ml was used. (B) Unmethylated DNA induced significantly more NO2− production than did methylated DNA for concentrations of 0.01 to 5 μg of DNA per ml (P < 0.05, indicated by asterisk).
DISCUSSION
The data presented here extend earlier studies demonstrating the mitogenic activity of B. bovis DNA for B lymphocytes to two medically important trypanosome species, the human pathogen T. cruzi and the bovine pathogen T. brucei. Reduced mitogenicity of DNA following either DNase treatment or CpG methylation demonstrates the importance of nonmethylated CG dinucleotides in activation of B-lymphocyte proliferation. Importantly, this study is the first demonstration that protozoal DNA activates macrophages. The production of TNF-α, IL-12, and NO by bovine macrophages in response to protozoal and E. coli DNA was qualitatively similar to what has been reported for murine macrophages and human monocytes (3, 16, 40). We also demonstrate that regulation of the expression of NO and IL-12 occurs at the level of transcription. As observed in previous studies with bovine macrophages (37, 41), some variation occurred from experiment to experiment in the relative amount of transcript or its encoded product that was expressed (Fig. 7 and 9). We attribute this variation to different states of activation by the primary macrophage cultures.
Interestingly, differences in the stimulation of macrophages by methylated DNA from different organisms were observed. In many experiments, methylation of E. coli and T. cruzi DNA had little to no effect on activity whereas methylation of T. brucei and B. bovis DNA routinely reduced or abolished the activity. Contaminating endotoxin does not explain our results, since endotoxin was not detected in any of the parasite DNA samples and since all experiments were conducted in the presence of saturating amounts of polymyxin B sulfate. One possible explanation for the ability of high concentrations of methylated E. coli or T. cruzi DNA to activate macrophages is that these DNAs may contain as yet unidentified immunostimulatory nucleotide motifs independent of CpG motifs that may be present at a higher frequency in these organisms. Nucleotide sequences other than CpG motifs, including poly(G) sequences, can stimulate B-cell proliferation, and dG runs facilitate the uptake by macrophages of oligodeoxynucleotides, thereby enhancing their immunostimulation (29, 34). In this regard, it is of interest that both T. brucei and T. cruzi DNA contain a modified base J, β-d-glucosyl-hydroxymethyluracil (14, 49), although the role of this modified base in macrophage activation has not been determined. An alternative explanation is that the CG dinucleotides were not completely methylated in E. coli, and perhaps T. cruzi, DNA. In support of this possibility, we found that methylating E. coli DNA did inhibit the induction of TNF-α and NO production when concentrations of ≤5 μg per ml were used. Similarly, methylated DNA used at concentrations of ≤12.5 μg per ml had reduced mitogenic activity for B lymphocytes (Fig. 3).
Activation of macrophage cytokines by protozoan parasites including B. bovis, T. brucei, and T. cruzi is well documented (13, 25, 30, 37). Among the parasite-derived molecules known to activate macrophages are membrane-derived glycolipids such as glycosylphosphatidylinositol (GPI) moieties (13, 45). GPI molecules from Plasmodium falciparum, T. brucei, and T. cruzi induced the production of inflammatory molecules, including IL-12, TNF-α, and NO, by murine macrophages (reviewed in reference 13). Lipids extracted from B. bovis-infected erythrocytes induced iNOS and NO production by bovine macrophages but did not induce detectable cytokines (37). Recognition of protozoal DNA by macrophages as an infectious stimulus may be another route by which a host immune response is triggered. Most experiments performed with bacterial DNA indicate that leukocyte activation requires internalization of the DNA (27, 28) although toll-like receptor 9 is required for CpG DNA-mediated activation (19). Thus, for parasites like T. cruzi, which reside within macrophages, DNA released intracellularly could induce signaling events. For parasites such as T. brucei and B. bovis, which are extracellular or reside within nonphagocytes, DNA released following phagocytosis and killing of the parasites or DNA taken up from the extracellular environment could stimulate leukocyte activation.
The mediators IL-12, TNF-α, and NO stimulated by parasite DNA and GPI molecules are known to contribute to the control of infection with many pathogenic protozoa. IL-12, induced by B. bovis, T. brucei and T. cruzi parasites (13, 30, 37), is a key type 1 cytokine that stimulates IFN-γ production by NK cells and T cells and results in macrophage activation and production of molecules, such as NO, that are directly microbicidal (47). Several reports indicated that type 1 immune responses involving IL-12, TNF-α, and NO are protective in experimental babesiosis and trypanosomiasis. In Babesia-immune cattle, antigen-specific CD4+ T lymphocytes produced IFN-γ (7, 8, 36), which was further enhanced by exogenous IL-12 (48). B. bovis- and IFN-γ-stimulated macrophages produced IL-12, TNF-α, and NO, and NO inhibited parasite growth (24, 37, 41). Neutralization of IL-12 in mice infected with T. cruzi resulted in increased parasitemia and mortality, suggesting that the induction of IL-12 was key to resistance (1). T. brucei-resistant strains of mice expressed higher levels of IL-12 and TNF-α than did susceptible mice (25), and in separate studies resistance was dependent on TNF-α (31) and IFN-γ (20). NO killed T. cruzi parasites in vitro (50) but did not play a role in IFN-γ-mediated resistance in mice to T. brucei infection (21). Thus, during an acute infection, the production of inflammatory mediators such as IL-12, TNF-α, and NO in response to DNA and GPI-associated lipid molecules released from dying parasites could serve to amplify an immune response that would promote host survival. The relative contributions of parasite-derived DNA and GPI molecules to macrophage activation during infection cannot be evaluated, but since bacterial lipid-associated molecules and CpG DNA appear to activate macrophages via distinct mechanisms involving different toll-like receptors (16, 19, 45), parasite GPI moieties and DNA may work in concert to stimulate innate immune responses.
It has been argued that if pathogens are under selective pressure to dampen their ability to stimulate a host inflammatory reaction, a reduced frequency of CpG motifs might occur in pathogens that establish persistent infection (42). T. cruzi, T. brucei, and B. bovis all establish chronic infections and all exhibit CG dinucleotide frequencies lower than that predicted by random association, which is 1/16 (6.25%). Furthermore, the mitogenic activities of the different protozoal DNA and E. coli DNA correlated with their CG dinucleotide frequency, and stimulation ranked in the order E. coli ≥ T. cruzi > T. brucei > B. bovis. A reduced frequency of CpG motifs and immunostimulatory activity of protozoal DNA, relative to E. coli DNA, is consistent with a selective pressure to minimize deleterious host inflammatory reactions and enhance parasite survival.
It is also possible that the release of parasite DNA might contribute to increased clinical disease through polyclonal B-cell activation and overproduction of inflammatory mediators. For example, polyclonal B-cell activation is remarkable during acute T. cruzi infection (10), and IFN-γ production during chronic human T. cruzi infection has been implicated in cardiac disease (2). Similarly, overproduction of TNF-α and NO is believed to exacerbate cerebral babesiosis (51). In mice, bacterial DNA sensitizes the animals for septic shock induced by TNF-α (40) and CpG oligonucleotides cause splenomegaly and polyclonal B-cell activation (34, 39).
In summary, DNA from parasitic protozoa has immunomodulatory effects on both B lymphocytes and macrophages of cattle that are similar to the widely studied effects of bacterial DNA and defined CpG oligonucleotides on murine B lymphocytes, macrophages, and dendritic cells. While the effects of protozoal DNA on the outcome of natural infection are unknown, our results are consistent with the potential for DNA to contribute to the stimulatory effects of parasite extracts on innate immune responses (5). Protozoal DNA could act independently or synergistically with parasite-derived lipid or protein molecules that also activate the production of proinflammatory cytokines and NO (13, 35, 37, 45). Furthermore, the strong activation of bovine macrophages by E. coli DNA supports the potential use of E. coli DNA as an adjuvant or of plasmid DNA as a vector in the design of vaccines against hemoprotozoan parasites of cattle.
ACKNOWLEDGMENTS
We thank Debby Alperin and Nissa Gese for excellent technical assistance.
This research was supported by NIAID NIH grant R01-AI30136 and USDA NRICGP grants 98-35204-6462, 98-35204-6737, and 99-35204-8368. R.S.C. is a member of the Research Career Program from the National Research Council (CONICET, Buenos Aires, Argentina).
REFERENCES
- 1.Aliberti J C S, Cardoso M A G, Martins G A, Gazzinelli R T, Vieira L Q, Silva J S. Interleukin-12 mediates resistance to Trypanosoma cruzi in mice and is produced by murine macrophages in response to live trypomastigotes. Infect Immun. 1996;64:1961–1967. doi: 10.1128/iai.64.6.1961-1967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahia-Oliveira I M, Gomes J A, Rocha M O, Moreira M C, Lemos E M, Luz Z M, Pereira M E, Coffman R L, Dias J C, Cancado J R, Gazzinelli G, Correa-Oliveira R. IFN-gamma in human Chagas' disease: protection or pathology? Braz J Med Biol Res. 1998;31:127–131. doi: 10.1590/s0100-879x1998000100017. [DOI] [PubMed] [Google Scholar]
- 3.Bauer M, Heeg K, Wagner H, Lipford G B. DNA activates human immune cells through a CpG sequence-dependent manner. Immunology. 1999;97:699–705. doi: 10.1046/j.1365-2567.1999.00811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brener Z, Gazzinelli R T. Immunological control of Trypanosoma cruzi infection and pathogenesis of Chagas' disease. Int Arch Allergy Immunol. 1997;114:103–110. doi: 10.1159/000237653. [DOI] [PubMed] [Google Scholar]
- 5.Brown W C, Estes D M, Chantler S E, Kegerreis K A, Suarez C E. DNA and a CpG oligonucleotide derived from Babesia bovis are mitogenic for bovine B cells. Infect Immun. 1998;66:5423–5432. doi: 10.1128/iai.66.11.5423-5432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown W C, Logan K S, Wagner G G, Tetzlaff C L. Cell-mediated immune responses to Babesia bovis merozoite antigens in cattle following infection with tick-derived or cultured parasites. Infect Immun. 1991;59:2418–2426. doi: 10.1128/iai.59.7.2418-2426.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown W C, Palmer G H. Designing blood-stage vaccines against Babesia bovis and B. bigemina. Parasitol Today. 1999;15:275–281. doi: 10.1016/s0169-4758(99)01471-4. [DOI] [PubMed] [Google Scholar]
- 8.Brown W C, Woods V M, Dobbelaere D A E, Logan K S. Heterogeneity in cytokine profiles of Babesia bovis-specific bovine CD4+ T cell clones activated in vitro. Infect Immun. 1993;61:3273–3281. doi: 10.1128/iai.61.8.3273-3281.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chace J H, Hooker N A, Mildenstein K L, Krieg A M, Cowdery J S. Bacterial DNA-induced NK cell IFN-γ production is dependent on macrophage secretion of IL-12. Clin Immunol Immunopathol. 1997;84:185–193. doi: 10.1006/clin.1997.4380. [DOI] [PubMed] [Google Scholar]
- 10.Cheikh M C, Hontebeyrie-Joskowicz M, Minoprio P. CD5 B cells. Potential role in the (auto) immune responses to Trypanosoma cruzi infection. Ann N Y Acad Sci. 1992;651:557–563. doi: 10.1111/j.1749-6632.1992.tb24662.x. [DOI] [PubMed] [Google Scholar]
- 11.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis J A, Godson D, Campos M, Sileghem M, Babiuk L A. Capture immunoassay for ruminant tumor necrosis factor-α: comparison with bioassay. Vet Immunol Immunopathol. 1993;35:289–300. doi: 10.1016/0165-2427(93)90040-b. [DOI] [PubMed] [Google Scholar]
- 13.Gazzinelli R T, Camargo M M, Almeida I C, Morita Y S, Giraldo M, Acosta-Serrano A, Hieny S, Englund P T, Ferguson M A J, Travassos L R, Sher A. Identification and characterization of protozoan products that trigger the synthesis of IL-12 by inflammatory macrophages. Chem Immunol. 1997;68:136–152. doi: 10.1159/000058689. [DOI] [PubMed] [Google Scholar]
- 14.Gommers-Ampt J H, Teixeira A J, van de Werken G, van Dijk W J, Borst P. The identification of hydroxymethyluracil in DNA of Trypanosoma brucei. Nucleic Acids Res. 1993;21:2039–2043. doi: 10.1093/nar/21.9.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez Cappa S M, Bijovsky A T, Freilij H, Muller L, Katzin A M. Aislamiento de una cepa de Trypanosoma cruzi a predominio de formas delgadas, en Argentina. Medicina. 1981;41:118–120. [PubMed] [Google Scholar]
- 16.Hartmann G, Krieg A M. CpG DNA and LPS induce distinct patterns of activation in human monocytes. Gene Ther. 1999;6:893–903. doi: 10.1038/sj.gt.3300880. [DOI] [PubMed] [Google Scholar]
- 17.Heeg K, Sparwasser T, Lipford G B, Häcker H, Zimmerman S, Wagner H. Bacterial DNA as an evolutionary conserved ligand signaling danger of infection to immune cells. Eur J Clin Microbiol Infect Dis. 1998;17:464–469. doi: 10.1007/BF01691128. [DOI] [PubMed] [Google Scholar]
- 18.Heeg K, Zimmermann S. CpG DNA as a Th1 trigger. Int Arch Allergy Immunol. 2000;121:87–97. doi: 10.1159/000024303. [DOI] [PubMed] [Google Scholar]
- 19.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 20.Hertz C J, Filutowicz H, Mansfield J M. Resistance to the African trypanosomes is IFN-γ dependent. J Immunol. 1998;161:6775–6783. [PubMed] [Google Scholar]
- 21.Hertz C J, Mansfield J M. IFN-γ-dependent nitric oxide production is not linked to resistance in experimental African trypanosomiasis. Cell Immunol. 1999;192:24–32. doi: 10.1006/cimm.1998.1429. [DOI] [PubMed] [Google Scholar]
- 22.Hölscher C, Köhler G, Muller G, U, Mossmann H, Schaub G A, Brombacher F. Defective nitric oxide effector functions lead to extreme susceptibility of Trypanosoma cruzi-infected mice deficient in gamma interferon receptor or inducible nitric oxide synthase. Infect Immun. 1998;66:1208–1215. doi: 10.1128/iai.66.3.1208-1215.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hölscher C, Mohrs M, Dai W J, Köhler G, Ryffel B, Schaub G A, Mossman H, Brombacher F. Tumor necrosis factor alpha-mediated toxic shock in Trypanosoma cruzi-infected interleukin 10-deficient mice. Infect Immun. 2000;68:4075–4083. doi: 10.1128/iai.68.7.4075-4083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson W C, Cluff C W, Goff W L, Wyatt C R. Reactive oxygen and nitrogen intermediates and products from polyamine degradation are babesiacidal in vitro. Ann N Y Acad Sci. 1996;791:136–147. doi: 10.1111/j.1749-6632.1996.tb53520.x. [DOI] [PubMed] [Google Scholar]
- 25.Kanshik R S, Uzonna J E, Zhang Y, Gordon J R, Tabel H. Innate resistance to experimental African trypanosomiasis: differences in cytokine (TNF-α, IL-6, IL-10, and IL-12) production by bone marrow-derived macrophages from resistant and susceptible mice. Cytokine. 2000;12:1024–1034. doi: 10.1006/cyto.2000.0685. [DOI] [PubMed] [Google Scholar]
- 26.Klinman D M, Barnhart K M, Conover J. CpG motifs as immune adjuvants. Vaccine. 1999;17:19–25. doi: 10.1016/s0264-410x(98)00151-0. [DOI] [PubMed] [Google Scholar]
- 27.Krieg A M. The role of CpG motifs in innate immunity. Curr Opin Immunol. 2000;12:35–43. doi: 10.1016/s0952-7915(99)00048-5. [DOI] [PubMed] [Google Scholar]
- 28.Krieg A M, Wagner H. Causing a commotion in the blood: immunotherapy progresses from bacteria to bacterial DNA. Immunol Today. 2000;21:521–526. doi: 10.1016/s0167-5699(00)01719-9. [DOI] [PubMed] [Google Scholar]
- 29.Liang H, Lipsky P E. Responses of human B cells to DNA and phosphorothioate oligodeoxynucleotides. Springer Semin Immunopathol. 2000;22:63–75. doi: 10.1007/s002810000017. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Ragua E, Li Z, Nuortio L, Mustafa A, Balchiet M. Interferon-gamma and interleukin 12 genes are preferentially expressed during early experimental African trypanosomiasis and suppressed by denervation of the spleen. Scand J Immunol. 1999;50:485–491. doi: 10.1046/j.1365-3083.1999.00639.x. [DOI] [PubMed] [Google Scholar]
- 31.Magez S, Radwanska M, Beschin A, Sekikawa K, de Baetselier P. Tumor necrosis factor alpha is a key mediator in the regulation of experimental Trypanosoma brucei infections. Infect Immun. 1999;67:3128–3132. doi: 10.1128/iai.67.6.3128-3132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matzinger P. An innate sense of danger. Semin Immunol. 1998;10:399–415. doi: 10.1006/smim.1998.0143. [DOI] [PubMed] [Google Scholar]
- 33.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pisetsky D S. Immune activation by bacterial DNA: a new genetic code. Immunity. 1996;5:303–310. doi: 10.1016/s1074-7613(00)80256-3. [DOI] [PubMed] [Google Scholar]
- 35.Probst P, Skeiky Y A, Steeves M, Gervassi A, Grabstein K H, Reed S G. A Leishmania protein that modulates interleukin (IL)-12, IL-10, and tumor necrosis factor-alpha production and expression of B7–1 in human monocyte-derived antigen-presenting cells. Eur J Immunol. 1997;27:2634–2642. doi: 10.1002/eji.1830271024. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez S D, Palmer G H, McElwain T F, McGuire T C, Ruef B J, Chitko-McKown C G, Brown W C. CD4+ T-helper lymphocyte responses against Babesia bigemina rhoptry-associated protein I. Infect Immun. 1996;64:2079–2087. doi: 10.1128/iai.64.6.2079-2087.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Shoda, L. K. M., K. A. Kegerreis, C. E. Suarez, W. Mwangi, D. P. Knowles, and W. C. Brown. Inclusion of an immunostimulatory CpG oligodeoxynucleotide sequence in plasmid DNA enhances plasimid DNA-induced IL-12, TNF-α, and NO production by bovine monocyte-derived macrophages. J. Leukoc. Biol., in press. [PubMed]
- 37.Shoda L K M, Palmer G H, Florin-Christensen J, Florin-Christensen M, Godson D L, Brown W C. Babesia bovis-stimulated macrophages express interleukin-1β, interleukin-12, tumor necrosis factor alpha, and nitric oxide and inhibit parasite replication in vitro. Infect Immun. 2000;68:5139–5145. doi: 10.1128/iai.68.9.5139-5145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoda L K M, Zarlenga D S, Hirano A, Brown W C. Cloning of a cDNA encoding bovine interleukin-18 and analysis of IL-18 expression in macrophages and its IFN-γ-inducing activity. J Interferon Cytokine Res. 1999;19:1169–1177. doi: 10.1089/107999099313118. [DOI] [PubMed] [Google Scholar]
- 39.Sparwasser T, Hultner L, Koch E S, Luz A, Lipford G B, Wagner H. Immunostimulatory CpG-oligodeoxynucleotides cause extramedulary murine hemopoiesis. J Immunol. 1999;162:2368–2374. [PubMed] [Google Scholar]
- 40.Sparwasser T, Miethke T, Lipford G, Erdmann A, Häcker H, Heeg K, Wagner H. Macrophages sense pathogens via DNA motifs: induction of tumor necrosis factor-α-mediated shock. Eur J Immunol. 1997;27:1671–1679. doi: 10.1002/eji.1830270712. [DOI] [PubMed] [Google Scholar]
- 41.Stich R W, Shoda L K M, Dreewes M, Adler B, Jungi T W, Brown W C. Stimulation of nitric oxide production in macrophages by Babesia bovis. Infect Immun. 1998;66:4130–4136. doi: 10.1128/iai.66.9.4130-4136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun S, Beard C, Jaenisch R, Jones P, Sprent J. Mitogenicity of DNA from different organisms for murine B cells. J Immunol. 1997;159:3119–3125. [PubMed] [Google Scholar]
- 43.Sun S, Cai Z, Langlade-Demoyen P, Kosaka H, Brunmark A, Jackson M R, Peterson P A, Sprent J. Dual function of Drosophila cells as APCs for naive CD8+ T cells: implications for tumor immunity. Immunity. 1996;4:555–564. doi: 10.1016/s1074-7613(00)80482-3. [DOI] [PubMed] [Google Scholar]
- 44.Sun S, Kishimoto H, Sprent J. DNA as an adjuvant: capacity of insect DNA and synthetic oligodeoxynucleotides to augment T cell responses to specific antigen. J Exp Med. 1998;187:1145–1150. doi: 10.1084/jem.187.7.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teixeira M M, Correa-Oliveira R, Gazzinelli R T. Immunoregulation in parasitic infection: insights for therapeutic intervention. Immunol Today. 2000;21:536–538. [PubMed] [Google Scholar]
- 46.Tighe H, Corr M, Roman M, Raz E. Gene vaccination: plasmid DNA is more than just a blueprint. Immunol Today. 1998;19:89–97. doi: 10.1016/s0167-5699(97)01201-2. [DOI] [PubMed] [Google Scholar]
- 47.Trinchieri G. Interleukin-12: a cytokine at the interface of inflammation and immunity. Adv Immunol. 1998;70:83–243. doi: 10.1016/s0065-2776(08)60387-9. [DOI] [PubMed] [Google Scholar]
- 48.Tuo W, Estes D M, Brown W C. Comparative effects of interleukin-12 and interleukin-4 on cytokine responses by antigen-stimulated memory CD4+ T cells of cattle: IL-12 enhances IFN-γ production, whereas IL-4 has marginal effects on cytokine expression. J Interferon Cytokine Res. 1999;19:741–749. doi: 10.1089/107999099313587. [DOI] [PubMed] [Google Scholar]
- 49.van Leeuwen F, Taylor M C, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. beta-d-Glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc Natl Acad Sci USA. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vespa G N, Cunha F Q, Silva J S. Nitric oxide is involved in control of Trypanosoma cruzi-induced parasitemia and directly kills the parasite in vitro. Infect Immun. 1994;62:5177–5182. doi: 10.1128/iai.62.11.5177-5182.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright I G, Goodger B V, Clark I A. Immunopathophysiology of Babesia bovis and Plasmodium falciparum infections. Parasitol Today. 1988;4:214–218. doi: 10.1016/0169-4758(88)90161-5. [DOI] [PubMed] [Google Scholar]
- 52.Young J R, Donelson J E, Majiwa P A, Shapiro S Z, Williams R O. Analysis of genomic rearrangements associated with two variable antigen genes of Trypanosoma brucei. Nucleic Acids Res. 1982;10:803–819. doi: 10.1093/nar/10.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]