Abstract
Selective modification of the N-terminus of peptides and proteins is a promising strategy for single site modification methods. Here we report N-terminal selective modification of peptides and proteins by using 2-ethynylbenzaldehydes (2-EBA) for the production of well-defined bioconjugates. After reaction screening with a series of 2-EBA, excellent N-terminal selectivity is achieved by the reaction in slightly acidic phosphate-buffered saline using 2-EBA with electron-donating substituents. Selective modification of a library of peptides XSKFR (X = either one of 20 natural amino acids) by 2-ethynyl-4-hydroxy-5-methoxybenzaldehyde (2d) results in good-to-excellent N-terminal selectivity in peptides (up to >99:1). Lysozyme, ribonuclease A and a therapeutic recombinant Bacillus caldovelox arginase mutant (BCArg mutant) are N-terminally modified using alkyne- and fluorescein-linked 2-EBA. Alkyne-linked BCArg mutant is further modified by rhodamine azide via copper(I)-catalyzed [3 + 2] cycloaddition indicating that the reaction has high functional group compatibility. Moreover, the BCArg mutant modified by 2-ethynyl-5-methoxybenzaldehyde (2b) exhibits comparable activity in enzymatic and cytotoxic assays with the unmodified one.
Subject terms: Synthetic chemistry methodology, Chemical modification
Selective functionalisation of the N-terminus of peptides and proteins may offer a useful tool for single-site modification. Here a general method for selective N-terminal labeling of peptides is reported, based on cyclisation of ethynylbenzaldehyde to form stable isoquinolinium salts.
Introduction
Site-selective chemical modification of peptides and proteins has become an emerging research field in chemical biology, which allows the production of well-defined bioconjugates for biological studies and drug development1–4. Although a number of bioconjugation reactions for specific amino acid modification have been developed in the past decade, due to the prevalence of multiple targeted residues on protein surface, only a few of them are amenable to give single-site modification5,6. To achieve site-selective modification, current methods mainly focus on labeling of the low abundant free cysteine residue or non-canonical amino acids, which always require sophisticated sequence engineering7–12. Besides, a few examples targeting the C-terminus or specific lysine ε-amino group have also been reported13–16. Despite these advances, it is still of ongoing interest to develop new methods for site-selective protein functionalization.
Targeting the N-terminus of peptides and proteins is a promising strategy to achieve single-site modification as a single-chain protein contains only one N-terminal residue in its sequence and it is mostly solvent exposed for functionalization17,18. On the other hand, recent studies suggest that a change of the charge in the N-terminal region of the signal peptide would disrupt the translocation of the small secretory preproteins19. Thus, development of an efficient N-terminal modification method will not only be an important direction for site-selective protein bioconjugation but also provide a chemical biology approach to study the biological functions of the protein N-terminus.
To achieve site-selective modification on the N-terminus, the important strategy is to perform the modification using pH control. As the N-terminal α-amino group possesses a lower basicity (pKa ≈ 6–8) compared with the lysine ε-amino group (pKa ≈ 10), a reaction medium with well-controlled pH value could favor the modification on the N-terminal α-amino group20. Based on this mechanism, N-terminal azidation, acylation, oxidation, and reductive alkylation have been reported by our group and others21–25. Another strategy is to utilize the specific residues on the N-terminus. Following this strategy, pyridoxal-5-phosphate (PLP) or Rapoport’s salt (RS)-mediated transamination reaction involving tautomerization triggered by the lower pKa α-proton on the N-terminal amino acid26–28, as well as 2-pyridinecarboxyaldehyde (2-PCA)-mediated imidazolidinone formation via cyclization of the imine intermediate with the nearby amide group on the N-terminus29, have been reported by Francis and co-workers. Among those reactions, the one-step modifications using ketenes22,23 or 2-PCA29 without addition of oxidizing or reducing reagent are promising approaches to achieve regioselective modified bioconjugates. However, preparation of their derivatives requires multi-step synthesis and it hampered the further studies on their structure-reactivity relationship, stability of the conjugates, as well as their applications. Thus, it is still of importance to investigate new, efficient, and convenient approaches for site-specific labeling of the N-terminus.
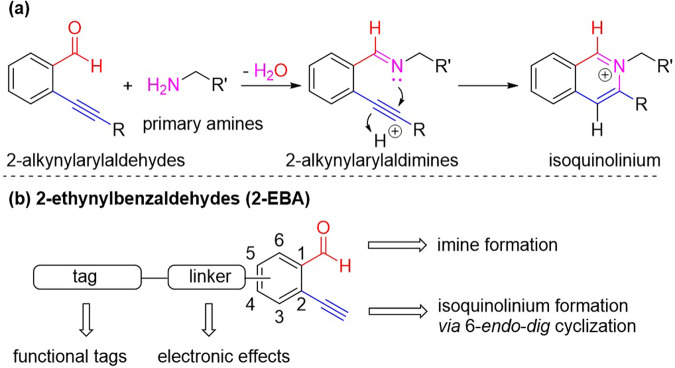
2-Alkynylarylaldehydes are versatile building blocks in organic synthesis30–33. Under transition metal catalysis, the in situ generated 2-alkynylarylaldimines between 2-alkynylarylaldehydes and primary amines could undergo 6-endo-dig cyclizations to give the corresponding isoquinoliniums, which have been demonstrated as key intermediates for syntheses of complex heterocycles31,32. Despite the fact that this transformation has been extensively explored in organic synthesis, studies on its applicability on protein modification remain largely elusive. We hypothesize that the efficient imine formation and tandem cyclization would render 2-alkynylarylaldehydes amenable to selectively modify the N-terminal α-amino group (Fig. 1).
Fig. 1. N-terminus selective modification.
a General mechanism of isoquinolinium formation. b Our design of 2-ethynylbenzaldehydes for N-terminal modification.
In this work, we first report a metal-free one-step N-terminal modification of peptides and proteins using 2-ethynylbenzaldehydes (2-EBA) under mild reaction conditions. The isoquinoliniums formed have been isolated and characterized by a model reaction. After a comprehensive study on the reaction conditions and the structure-reactivity relationship of the reagent, we demonstrate that, apart from the pH control, electronic effects also play important roles in controlling the N-terminal selectivity of the modification. We have also extended this reaction to protein modification, including labeling a therapeutic Bacillus caldovelox arginase mutant (BCArg mutant). The enzymatic and anti-cancer activities of the modified BCArg mutant have also been studied.
Results and discussion
2-Ethynylbenzaldehydes as N-terminal selective reagents
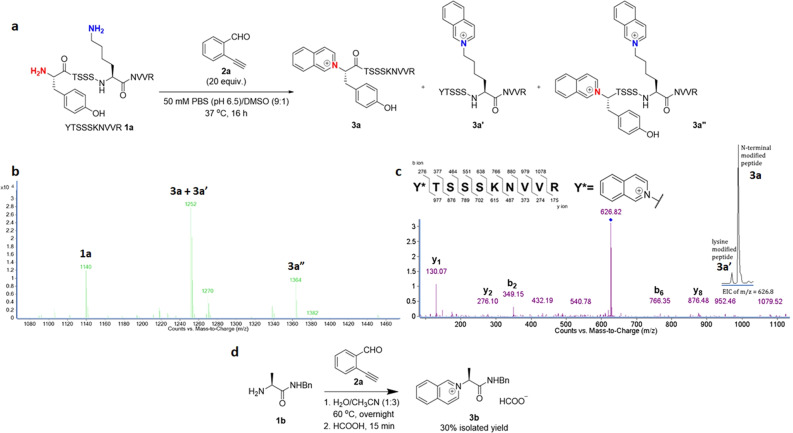
To begin our study, peptide YTSSSKNVVR 1a (molecular mass of 1140 Da, 0.1 mM) was treated with 20 equivalents of 2-ethynylbenzaldehyde 2a (2-EBA, 130 Da) in 50 mM phosphate-buffered saline (PBS)/DMSO (9:1) at pH 6.5 for 16 h (Fig. 2a). After the reaction, we found that peptide 1a was modified to give mono-modified peptides (N-terminally modified peptide 3a and lysine-modified peptide 3a′) in 64% conversion and di-modified peptide 3a″ in 8% conversion (Fig. 2b). An increase of the molecular mass by 112 Da indicated that 2-EBA 2a was incorporated on the peptide 1a with loss of a H2O molecule, which was presumably ascribed to the formation of a quinolinium conjugate after the modification. As N-terminal selectivity23–25 is calculated based on the ratio of the mono-modified peptide at N-terminal α-amino group to lysine ε-amino group as determined by extracted ion chromatogram (EIC) of LC-MS analysis, we achieved N-terminal selectivity (3a:3a′) of 96:4 in the mono-modified peptides, with the corresponding MS/MS spectrum of N-terminally modified peptide 3a as the major product (Fig. 2c). Therefore, the conversions of N-terminally modified peptide 3a and lysine-modified peptide 3a′ were calculated as 61 and 3% respectively. To give an understanding on the proportion of N-terminally modified peptide in overall modified products, we included another method for determining the efficiency of N-terminal modification, referring to the ratio of the conversion of N-terminally modified peptide over the conversion of all modified peptides (i.e. 3a/(3a + 3a′+3aʺ)). Thus, the efficiency of N-terminal modification of YTSSSKNVVR 1a with 2-EBA 2a was 0.85.
Fig. 2. N-terminal modification of a model peptide.
a N-terminal modification of YTSSSKNVVR 1a with 2-ethynylbenzaldehyde 2a. b Deconvoluted mass spectrum of the reaction mixture. c MS/MS spectrum of N-terminally modified peptide 3a (inset: extracted ion chromatogram of mono-modified peptide). d Model reaction of isoquinolinium salt 3b from l-alanine β-benzylamide 1b and 2a.
To investigate the structure of N-terminally modified peptide 3a, we conducted a model study by treatment of L-alanine β-benzylamide 1b with 2-EBA 2a (1.1 equiv.) in H2O/CH3CN (1:3) at 60 °C overnight, followed by addition of formic acid (2 equivalents) for 15 min (Fig. 2d). After the reaction, the corresponding isoquinolinium 3b was isolated in 30% yield. The formation of 3b suggested that the imine was first generated by reaction of 1b and 2a to give 2-ethynylbenzaldimine as a key intermediate. Then, the 2-ethynylbenzaldimine intermediate underwent subsequent intramolecular 6-endo-dig cyclization to give isoquinolinium 3b as the product. Remarkably, the formation of isoquinolinium salts by reaction of 2-EBA and primary amines under a metal-free condition in aqueous media has not yet been reported previously30–33.
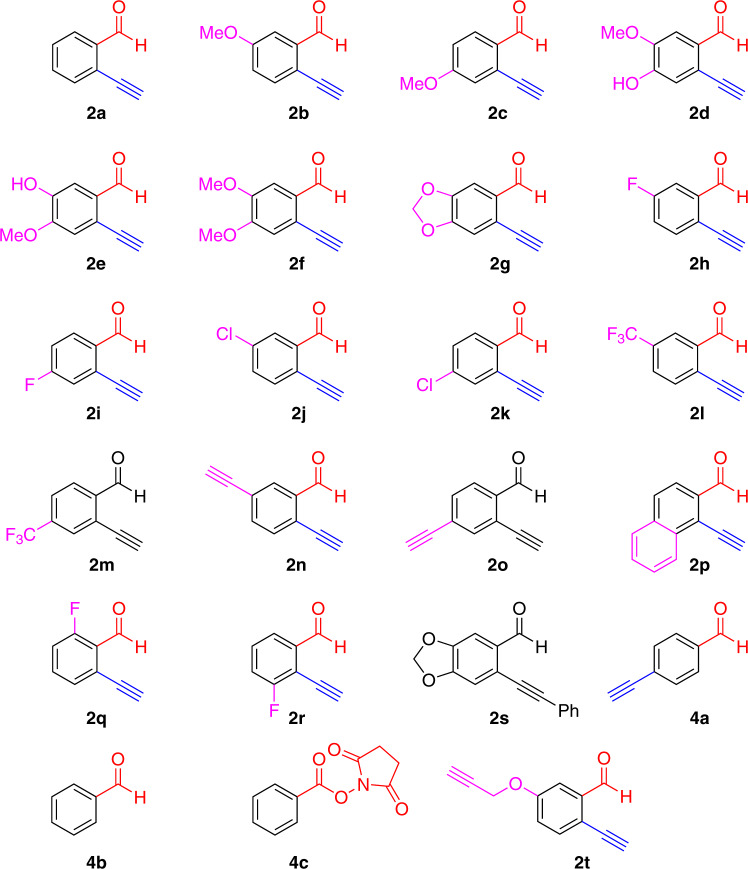
With these promising findings, we next moved on to screen the peptide modification using a series of 2-EBA to improve the N-terminal selectivity of the modification and to study the structure-reactivity relationship of the 2-EBA (Table 1 and Fig. 3). In all, 2-EBA 2a was commercially available and the others 2b–r were easily prepared by Sonogashira coupling reaction of the commercially available aromatic halides with trimethylsilylacetylenes, followed by desilylation34. Screening reactions in 50 mM PBS at pH 6.5 indicated that the reaction could be conducted with good to high conversions (up to 86%) with excellent N-terminal selectivity (up to > 99:1). 2-EBA bearing electron-donating groups at 5- or 4-positions (2b–g) gave the highest conversions (up to 86%) with excellent N-terminal selectivity (up to >99:1) (Table 1, entries 2–7). Comparable conversions (up to 78%) and high N-terminal selectivity (up to >99:1) were obtained when using 2-EBA with weakly electron-withdrawing groups (fluoro or chloro) at 5- or 4- positions (2h–k, entries 8–11). Employment of 2-EBA with strongly electron-withdrawing groups (trifluoromethyl) at 5- or 4- positions (2l and 2m) lead to moderate conversions (65 and 72%) and lower N-terminal selectivity (95:5 and 93:7). Incorporation of an alkyne moiety on the 2-EBA (2n and 2o) also gave 41–64% conversions with up to 96:4 of N-terminal selectivity, indicating that the present reaction has high compatibility with unsaturated C-C bond (Entries 14–15). Modification using 1-ethynyl-2-naphthaldehyde (2p) gave poor conversion (5%), which was probably attributed to the poor solubility of the compound (Entry 16). Incorporation of fluoro group at 6-position (2q) resulted in high N-terminal selectivity (>99:1) but the high proportion of the di-modified products (74%) hampered its application (Entry 17). Moreover, introduction of fluoro group at 3-position (2r) lead to a lower N-terminal selectivity (93:7) (Entry 18). These findings revealed that 2-EBA were promising reagents for selective modification of the peptide N-terminus and incorporation of electron-donating groups or weakly electron-withdrawing groups would give high N-terminal selectivity of the modification.
Table 1.
Screening of 2-EBA 2a–t and compounds 4a–c for modification of YTSSSKNVVR 1aa.
| Entry | Reagent | Conversion (%)b | N-terminal selectivity of mono-modified peptided | Efficiency of N-terminal modificatione | |||
|---|---|---|---|---|---|---|---|
| Mono-modifiedc | Di-modified | Total | |||||
| N-terminus | Lysine | ||||||
| 1 | 2a | 61 | 3 | 8 | 72 | 96:4 | 0.85 |
| 2 | 2b | 64 | 2 | 6 | 71 | 98:2 | 0.90 |
| 3 | 2c | 60 | 3 | 5 | 68 | 96:4 | 0.88 |
| 4 | 2d | 73 | - | 13 | 86 | >99:1 | 0.85 |
| 5 | 2e | 58 | 2 | 7 | 67 | 96:4 | 0.87 |
| 6 | 2f | 27 | 1 | - | 28 | 97:3 | 0.96 |
| 7 | 2g | 39 | 1 | 1 | 41 | 98:2 | 0.95 |
| 8 | 2h | 64 | - | 4 | 68 | >99:1 | 0.94 |
| 9 | 2i | 69 | 2 | 7 | 78 | 98:2 | 0.88 |
| 10 | 2j | 46 | 1 | 2 | 49 | 97:3 | 0.94 |
| 11 | 2k | 53 | 2 | 3 | 58 | 97:3 | 0.91 |
| 12 | 2l | 64 | 3 | 5 | 72 | 95:5 | 0.89 |
| 13 | 2m | 58 | 4 | 3 | 65 | 93:7 | 0.89 |
| 14 | 2n | 52 | 2 | 3 | 57 | 96:4 | 0.91 |
| 15 | 2o | 56 | 3 | 5 | 64 | 95:5 | 0.88 |
| 16 | 2p | 4.7 | 0.4 | – | 5 | 93:7 | 0.94 |
| 17 | 2q | 26 | – | 74 | 100 | >99:1 | 0.26 |
| 18 | 2r | 65 | 5 | 25 | 95 | 93:7 | 0.68 |
| 19 | 2s | – | – | – | – | – | – |
| 20 | 4a | – | – | – | – | – | – |
| 21 | 4b | – | – | – | – | – | – |
| 22f | 4c | 55 | 20 | 25 | 100 | 73:28 | 0.55 |
| 23 | 2t | 70 | 1 | 9 | 80 | 98:2 | 0.88 |
aConditions: YTSSSKNVVR 1a (0.1 mM) and reagent (2 mM) in 50 mM PBS (pH 6.5)/DMSO (9:1) solution (100 μL), 37 °C, 16 h.
bDetermined by total ion count (TIC) of LC–MS analysis.
cConversion of N-terminal modified peptide (or lysine-modified peptide) is determined by the conversion of mono-modified peptide and N-terminal selectivity of mono-modified peptide.
dN-terminal selectivity is obtained by ratio of mono-modified peptide at N-terminal α-amino group to lysine ε-amino group as determined by extracted ion chromatogram (EIC) of LC-MS analysis.
eEfficiency of N-terminal modification is equal to the conversion of N-terminal modified peptide over the total conversion.
fModification was conducted at room temperature for 15 min.
Fig. 3. Substrate scope.
Structures of 2-EBA 2a–t and compounds 4a–c.
We have also conducted some control experiments (Table 1 and Fig. 3). No reaction was observed when 2-EBA 2s bearing an internal alkyne was used which was ascribed to the lower reactivity of the internal alkyne for the intramolecular cyclization (Table 1, entry 19). Using 4-ethynylbenzaldehyde (4a) and benzaldehyde (4b) gave no peptide conversion suggesting that a terminal ethynyl group located at the ortho-position of benzaldehyde played a key role for isoquinolinium formation to give the resulting conjugate (Entries 20–21). We have also compared our reagents with the N-(benzoyloxy)succinimide (4c) which was widely used for amine modification (Entry 22). It was found that N-(benzoyloxy)succinimide (4c) was highly reactive towards the amine groups on the peptide and poor N-terminal selectivity (73:28) was achieved even though we conducted the modification in a slightly acidic condition (pH 6.5), indicating that 2-EBA displayed unique properties towards N-terminal modification.
With the aforementioned findings, we designed and synthesized 2-EBA 2t for N-terminal modification of the peptides and proteins. The propargyl ether structure would provide an electron-donating effect towards the 2-EBA core structure to improve the N-terminal selectivity, while the free alkyne moiety allows attachment of versatile functional tags after the modification. Treatment of 2t with peptide 1a gave the high conversion (80%) with good N-terminal selectivity (98:2), suggesting that it would be a promising reagent for N-terminal modification of peptides and proteins.
Optimization of reaction conditions
We next sought to examine the effects of the reagent amount, temperature, and pH values of the media on the N-terminal selectivity of the modification. With the highest N-terminal selectivity (Table 1, entry 5), 2-EBA 2d was selected for further screening reactions. Different concentrations of 2d were employed in the present reaction at 37 °C (Supplementary Table 2). Good conversion (86%) was observed using 20 equivalents of 2d with excellent N-terminal selectivity (>99:1). When the reaction temperature was reduced to 25 °C and 4 °C respectively, lower conversions were observed. Noticeably, poor N-terminal selectivity (60:40) was found at 4 °C, suggesting that the low temperature would favor the reaction of the 2-EBA with the less hindered lysine ε-amino group.
We conducted time course experiments to test the effect of pH values on the modification. Bioconjugation reactions of peptide YTSSSKNVVR 1a with 2d (20 equiv.) in different pH values of 50 mM PBS and DMSO (9:1) were studied (Fig. 4a). At pH 6.5, excellent N-terminal selectivity (>99:1) was observed. As pH increased from 7.4 to 9, an increasing amount of mono-internal lysine-modified peptide and di-modified peptide as well as lower N-terminal selectivity were found at higher pH, indicating that the N-terminal selectivity of the modification was strongly influenced by the pH effects. Besides, the present reaction was optimized at 16 h to reach the highest conversion during the time course experiments.
Fig. 4. Effect of pH and substrate substituents.
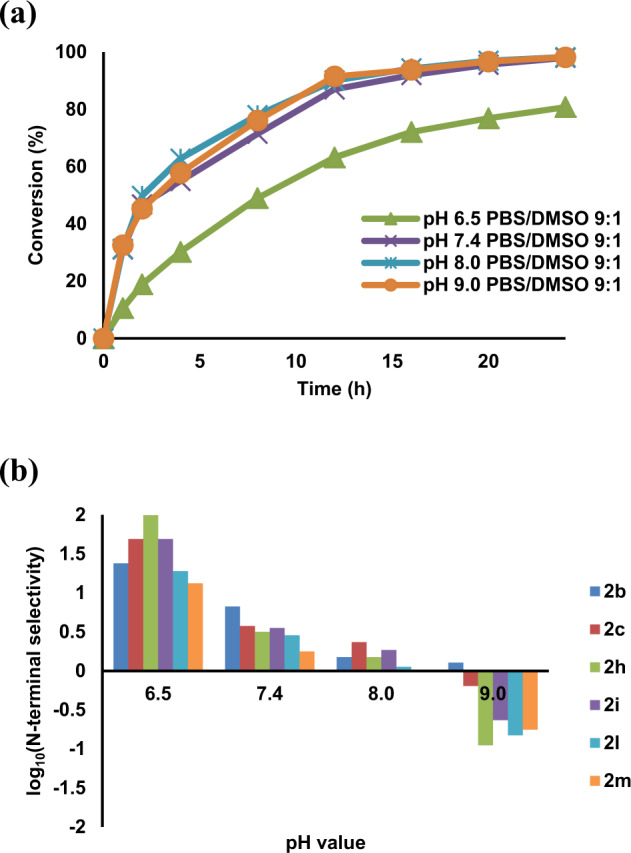
a Time course of N-terminal modification of YTSSSKNVVR 1a using 2d at pH 6.5–9.0. b Electronic effects of the substituents on 2-EBA towards the N-terminal modification of 1a at pH 6.5–9.0.
To provide more insights on electronic effects of the substituents on the phenyl moieties of the 2-EBA towards the N-terminal selectivity of the modification, we performed the modification in different pH values of the media using 2-EBA with strongly electron-donating methoxy group (2b and 2c), weakly electron-withdrawing fluoro group (2h and 2i) as well as strongly electron-withdrawing trifluoromethyl group (2l and 2m). As shown in Fig. 4b, by increasing the pH of the medium, the N-terminal selectivity of the modification decreased. Surprisingly, 2-EBA bearing electron-donating substituents still gave moderately N-terminal selective modification (2b) or weakly lysine selective modification (2c) at pH 9.0 PBS medium, while 2-EBA bearing electron-withdrawing substituents (2h, 2i, 2l, and 2m) changed to give lysine selective modification. For example, modification of peptide 1a with 2-EBA 2h in pH 9.0 PBS/DMSO medium afforded mono-modified peptide with 9:81 of N-terminal selectivity, indicating the labeling was highly selective to the lysine ε-amino group. The above findings implicated that the site-selectivity of the modification was not only controlled by the pH effects but also by the electronic effects of the substituents on 2-EBA.
After screening of the effects towards the N-terminal modification, the stability of the bioconjugates was then studied by incubating the 2d-modified YTSSSKNVVR with excess amount of reducing or oxidizing reagents (glutathione (GSH), homocysteine, l-cysteine, dl-dithiothreitol (DTT), 2-mercaptoethanol, tris(2-carboxyethyl)phosphine (TCEP), ascorbic acid, and hydrogen peroxide) in 50 mM PBS (pH 6.5)/DMSO (9:1) at 37 °C for 2 h (Supplementary Fig. 49). LC-MS/MS analysis revealed that the 2d-modified YTSSSKNVVR was stable towards the additives with no significant decomposition or scrambling product.
Screening of a peptide library
We next studied the applicability of this reaction on modification of a library of 20 unprotected peptides, XSKFR (X = either one of 20 natural amino acids). The peptide sequences with nucleophilic Ser and Lys were chosen for examination of the N-terminal selectivity of this reaction. As shown in Table 2, peptides with N-terminal Ala, Cys, Asp, Glu, Gly, His, Lys, Asn, Gln, Ser, or Tyr gave excellent N-terminal selectivity (>99:1) (Table 2, entries 1–11). Moderate-to-high N-terminal selectivities (86:14 to 98:2) were obtained for the N-terminal Ile, Leu, Trp, Phe, Val, Met, Thr, and Arg peptides (Entries 12–19). However, a low N-terminal selectivity of 46:54 was observed for PSKFR having N-terminal proline residue (Entry 20), which is presumably due to the iminium intermediate formed between proline and 2-EBA cannot undergo subsequent cyclization with the proximal alkyne group.
Table 2.
Modification of a peptide library XSKFR using 2-EBA 2da.
| Entry | Peptide | Conversion (%)b | N-terminal selectivity of mono-modified peptided | Efficiency of N-terminal modificatione | |||
|---|---|---|---|---|---|---|---|
| Mono-modifiedc | Di-modified | Total | |||||
| N-terminus | Lysine | ||||||
| 1f | CSKFR | 73 | – | 0 | 73 | >99:1 | 1.0 |
| 2 | ASKFR | 84 | – | 3 | 87 | >99:1 | 0.97 |
| 3 | YSKFR | 56 | – | 2 | 58 | >99:1 | 0.97 |
| 4 | GSKFR | 50 | – | 2 | 52 | >99:1 | 0.96 |
| 5 | HSKFR | 70 | – | 4 | 74 | >99:1 | 0.95 |
| 6 | DSKFR | 39 | – | 3 | 42 | >99:1 | 0.93 |
| 7 | ESKFR | 32 | – | 3 | 35 | >99:1 | 0.91 |
| 8 | NSKFR | 62 | – | 6 | 68 | >99:1 | 0.91 |
| 9 | SSKFR | 42 | – | 6 | 48 | >99:1 | 0.88 |
| 10 | QSKFR | 79 | – | 15 | 94 | >99:1 | 0.84 |
| 11 | KSKFR | 54 | – | 13 | 67 | >99:1 | 0.81 |
| 12 | LSKFR | 51 | 1 | 3 | 55 | 98:2 | 0.93 |
| 13 | ISKFR | 59 | 1 | 10 | 70 | 98:2 | 0.84 |
| 14 | WSKFR | 38 | 1 | 1 | 40 | 97:3 | 0.95 |
| 15 | FSKFR | 58 | 2 | 7 | 67 | 96:4 | 0.87 |
| 16 | VSKFR | 61 | 3 | 2 | 66 | 95:5 | 0.92 |
| 17 | MSKFR | 60 | 4 | 2 | 66 | 94:6 | 0.91 |
| 18 | TSKFR | 51 | 6 | 7 | 64 | 90:10 | 0.80 |
| 19 | RSKFR | 56 | 9 | 5 | 70 | 86:14 | 0.80 |
| 20 | PSKFR | 10 | 12 | 1 | 23 | 46:54 | 0.43 |
aConditions: XSKFR 1a (0.1 mM) and reagent 2d (2 mM) in 50 mM PBS (pH 6.5)/DMSO (9:1) solution (100 μL), 37 °C, 16 h.
bDetermined by total ion count (TIC) of LC–MS analysis.
cConversion of N-terminal modified peptide (or lysine-modified peptide) is determined by the conversion of mono-modified peptide and N-terminal selectivity of mono-modified peptide.
dN-terminal selectivity is obtained by ratio of mono-modified peptide at N-terminal α-amino group to lysine ε-amino group as determined by extracted ion chromatogram (EIC) of LC–MS analysis.
eEfficiency of N-terminal modification is equal to the conversion of N-terminal modified peptide over the total conversion.
fTris(2-carboxyethyl)phosphine (TCEP, 0.5 mM) was added to prevent the formation of disulfide linkage.
To further study the selectivity of the present bioconjugation reaction, we used peptides with cysteine at different positions (ASCGTN, AYEMWCFHQR, and KSTFC). Exclusive N-terminal modification with 70%, 27 and 15% conversions, respectively, was found with the cysteine residue remaining intact (Supplementary Figs. 77–79). In addition, sole modification at the internal lysine in an N-terminally acetylated peptide Ac-YTSSSKNVVR with 19% conversion was observed, indicating that the present bioconjugation reaction was highly chemoselective to the amino group of peptides as only the amino group of lysine was modified when the N-terminus is acetylated (Supplementary Fig. 80). For a peptide containing a second proline residue (YPSSSKNVVR) which has no reactivity towards 2-PCA29, it was found that the bioconjugation proceeded smoothly with 2d to afford 54% conversion with excellent N-terminal selectivity (>99:1) (Supplementary Fig. 81).
Protein modification using 2-ethynylbenzaldehydes
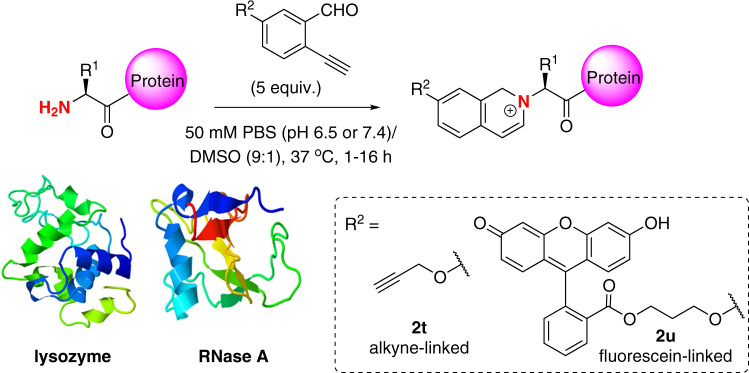
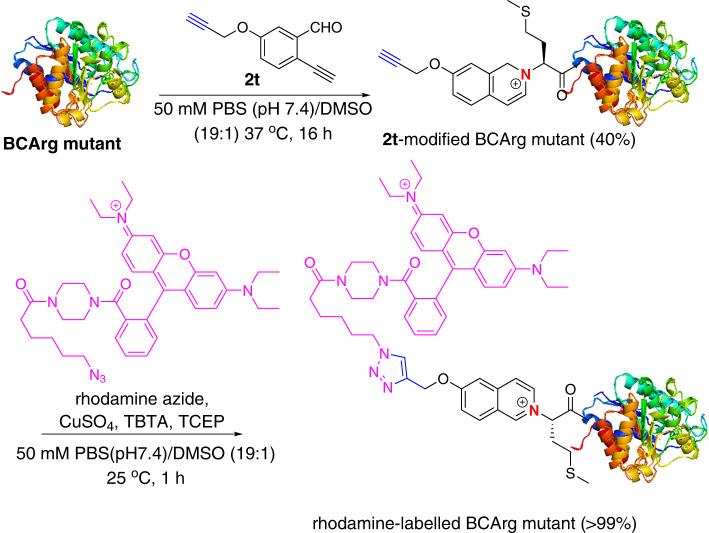
After studying the N-terminal peptide modification, we further explored the present reaction for protein modification (Fig. 5) employing alkyne-linked and fluorescein-linked 2-EBA (2t and 2u, respectively). The presence of the ether linkage was to improve the N-terminal selectivity of the reagent, suggested by the aforementioned findings. In total, 0.1 mM of lysozyme (PDB ID: 1DPX) was treated with 2t (0.5 mM, 5 equivalents) in 50 mM PBS (pH 6.5) at 37 °C for 16 h, giving the 2t-modified lysozyme with 52% conversion (Supplementary Fig. 82a). LC–MS analysis of the reaction mixtures of lysozyme showed peaks at 14470 Da and 14489 Da, which were assigned to the mono-modified lysozyme. Upon trypsin digestion, the modification by 2t was found to selectively occur at the N-terminus, as depicted by LC–MS/MS analysis of the tryptic peptide fragment of KVFGR (Supplementary Fig. 83). Increasing the pH to 7.4 in the N-terminal modification of lysozyme with 2t generally resulted in excellent conversion (92%) (Supplementary Fig. 82b) with mono- and di-modified lysozyme products observed. Note that the N-terminal selectively is still considerably high as depicted in LC-MS/MS analysis. The reaction of RNase A (PDB ID: 3DH5) was performed at pH 7.4 to afford up to 44% protein conversion (Supplementary Figs. 84–85). Modification of lysozyme and RNase A with fluorescein-linked 2-EBA 2u in 50 mM PBS (pH 7.4) gave 34 and 10% conversions with high N-terminal selectivity, respectively (Supplementary Figs. 86–87). As the imidazolidinone conjugate of the 2-PCA-modified RNase A was found to have partially (20–30%) decomposition at 37 °C after 12 h, we also tested the stability of the 2t-modified RNase A by treatment of the modified RNase A in PBS with different pH values (pH 3–11) at 37 °C (Supplementary Fig. 88). Noticeably, no decomposition was found by LC-MS analysis, indicating that the quinolinium conjugate formed was highly stable and this modification would be amenable for preparation of bioconjugates for drug development.
Fig. 5. N-terminal protein modification.
Modification of lysozyme and RNase A with functionalized 2-EBA 2t and 2u.
Human arginase, which is a manganese-dependent enzyme that degrades arginine into urea, has been reported to treat advanced hepatocellular carcinoma and metastatic melanoma where prior immunotherapy failed in early phase of clinical trial35. PEGylated human arginase I was developed as the first generation of therapeutic proteins with long half-life, which is now undergoing phase II clinical trials36. However, current PEGylation usually involved non-specific lysine/cysteine modification via NHS/maleimide chemistry. The second-generation therapeutic protein, Bacillus caldovelox arginase mutant (BCArg mutant), has been reported to induce a sustained complete remission in a patient with immunotherapy-resistant cancer37,38. In addition to lysozyme and RNase A, we also extended this newly developed N-terminal modification to modify the BCArg mutant. BCArg mutant (0.1 mM) was treated with alkyne- and fluorescein-linked 2-EBA (2t and 2u, 10 equivalent) in PBS (pH 7.4)/DMSO (9:1) at 37 °C for 16 h to give the corresponding 2t- and 2u-modified BCArg mutant in 40 and 19% conversions, respectively (Supplementary Fig. 89). High N-terminal selectivity as revealed by the tryptic peptide fragments MKPISIIGVPMDLGQTR of 2t- and 2u-modified BCArg mutants were observed by LC-MS/MS analysis (Supplementary Fig. 90).
As depicted in Fig. 6, the 2t-modified BCArg mutant containing an alkyne handle (32615 Da) could be smoothly modified with a rhodamine-azide via copper(I)-catalyzed [3 + 2] cycloaddition reaction to give the rhodamine-labeled BCArg mutant (33268Da and 33296Da) in >99% conversion (Supplementary Fig. 91). SDS-PAGE analysis revealed that the rhodamine-labeled BCArg mutant gave a strongly green fluorescent signal while the 2t-modified BCArg mutant had no fluorescent signal at UV 365 nm (Supplementary Fig. 92). Coomassie blue staining on the same gel gave deep blue color signals of unmodified, alkyne-linked as well as rhodamine-labeled proteins, indicating that the fluorescent tag has been successfully labeled on the proteins using the N-terminal selective alkyne-linked 2-EBA 2t and a sequential azide-alkyne click reaction. These results indicated that the present reaction has high compatibility with click chemistry.
Fig. 6. N-terminal protein modification with an alkyne.
Modification of the BCArg mutant with alkyne-functionalized 2-EBA 2t and sequential copper(I)-catalyzed [3 + 2] cycloaddition reaction with rhodamine azide.
Biological studies of N-terminally modified BCArg mutants
To study the influence of the quinolinium conjugates on the biological properties of the therapeutic protein, we compared the enzyme activities and anti-cancer properties of the modified BCArg mutant with the unmodified analogue (Table 3). We prepared 2b-modified BCArg mutant (32%) by bioconjugation with 2-EBA 2b in 50 mM PBS (pH 7.4) at 37 °C for 16 h (Supplementary Figs. 93–94). The enzymatic properties of the 2b-modified BCArg mutant was slightly lower than that of the unmodified BCArg mutant. The anti-cancer properties of the unmodified and 2b-modified BCArg mutants were then examined using breast cancer cell lines MDA-MB-231 and MDA-MB-468. Experimental IC50 values indicated that the antitumor efficacy of the 2b-modified BCArg mutant was comparable to that of the unmodified one. These findings indicated that the 2b-modified BCArg mutant retained its biological activities after the bioconjugation.
Table 3.
Enzymatic activities and IC50 values of BCArg mutant and 2b-modified BCArg mutant.
| Sample | BCArg mutant | 2b-modified BCArg mutant |
|---|---|---|
| Specific activity (U/mg) | 443.96 ± 27.50 | 313.68 ± 4.01 |
| IC50 values for MDA-MB-231 (U/mL) | 4.608 ± 0.995 | 5.491 ± 1.564 |
| IC50 values for MDA-MB-468 (U/mL) | 4.772 ± 1.137 | 5.631 ± 2.410 |
In summary, we have discovered that 2-ethynylbenzaldehydes (2-EBA) are a useful reagent for N-terminal modification of peptides and proteins via isoquinolinium formation with the N-terminal α-amino group. After a comprehensive screening of the reaction conditions and the structure-reactivity relationship of the 2-EBA, we have found that apart from the pH control, the electronic properties of the substituents on the 2-EBA could also strongly affect the N-terminal selectivity of the modification. Under slightly acidic condition (pH 6.5) and employing 2-EBA with electron-donating and weakly electron-withdrawing groups, the modification has achieved excellent N-terminal selectivity. Conducting the reaction in basic medium (pH 9) and using 2-EBA with electron-withdrawing groups can switch the modification to become lysine selective. To help other researchers who are interested in using this bioconjugation reaction, the reaction conditions, substituent effects, and functional group tolerance for various N-terminal residues are summarized in Table 4. The present method is successfully applied on selective modification of proteins, including the therapeutic protein (BCArg mutant). The resulting modified bioconjugates are of good stability and the biological properties of the modified BCArg mutant are comparable to those of the unmodified one.
Table 4.
Guide for using the 2-ethynylbenzaldehydes for N-terminal selective modification.
| Reaction conditions | Peptides | Proteins |
|---|---|---|
| Equivalent of 2-EBA | 20 equivalents | ≤10 equivalents |
| pH | 6.5 | 6.5–7.4 |
| Reaction temperature | 37 °C | |
| Reaction time | ≤16 h | |
| Substituent effects of 2-EBA | N-terminal selectivity is excellent (up to >99:1) when 2-EBA with electron-donating group at 5- or 4-positions. | |
| Functional group tolerance | Highly compatible with 19 of the 20 natural amino acids, except for N-terminal proline. | |
Methods
Synthesis and characterization
The synthetic procedures and characterization for compounds as well as chromatography and mass spectrometry data are presented in in Supplementary Methods, Supplementary Figs. 1–48, and Supplementary Figs. 50–76.
General procedure for modification of peptides using 2-ethynylbenzaldehydes
To an eppendorf tube (1.5 mL) with 80 µL of 50 mM PBS buffer pH 6.5, 10 µL of YTSSSKNVR (1a, 1 mM in Milli-Q® water) was added to the buffer, followed by 10 µL of 2-ethynylbenzaldehyde (2a–2t, 20 mM in DMSO). The reactive mixture was allowed to react in a 37 °C water bath for 16 h. 10 µL of the mixture was drawn, diluted with 10 µL of Milli-Q® water and subjected to LC/MS–MS analysis.
Unless otherwise specified, all peptides were treated as same as the above procedure.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are grateful for the financial support from Hong Kong Research Grants Council (PolyU 153001/17P), the State Key Laboratory of Chemical Biology and Drug Discovery, and The Hong Kong Polytechnic University (G-YBM8).
Author contributions
M.-K.W., Y.-C.L., and K.K.-Y.K. conceptualized and supervised the study. J.-R.D., N.C.-H.L., and B.Y. performed the organic synthesis in this work. J.-R.D., N.C.-H.L., and K.K.-Y.K. performed the bioconjugation experiments and mass spectroscopy analysis of this work. K.K.-Y.K. performed the SDS-PAGE analysis. S.-F.C., A.S.-L.L., and M.-C.C. purified and measured the biological activities of the anticancer proteins. M.-K.W., Y.-C.L., K.K.-Y. K., J.-R.D., and N.C.-H.L. prepared this paper.
Data availability
All principal data with detailed experimental procedure and characterization of this work are included in this article, and its Supplementary Information or are available from the corresponding author upon reasonable request.
Competing interests
The authors declare the following competing interests: M.-K.W. has filed a patent application on the use of 2-ethynylbenzaldehydes for bioconjugation (WO2019219002) and all other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jie-Ren Deng, Nathanael Chun-Him Lai.
Contributor Information
Karen Ka-Yan Kung, Email: karen.kung@polyu.edu.hk.
Yun-Chung Leung, Email: thomas.yun-chung.leung@polyu.edu.hk.
Man-Kin Wong, Email: mankin.wong@polyu.edu.hk.
Supplementary information
Supplementary information is available for this paper at 10.1038/s42004-020-0309-y.
References
- 1.Stephanopoulos N, Francis MB. Choosing an effective protein bioconjugation strategy. Nat. Chem. Biol. 2011;7:876–884. doi: 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
- 2.Spicer CD, Davis BG. Selective chemical protein modification. Nat. Commun. 2014;5:4740. doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 3.Boutureira O, Bernardes GJL. Advances in chemical protein modification. Chem. Rev. 2015;115:2174–2195. doi: 10.1021/cr500399p. [DOI] [PubMed] [Google Scholar]
- 4.Koniev O, Wagner A. Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem. Soc. Rev. 2015;44:5495–5551. doi: 10.1039/C5CS00048C. [DOI] [PubMed] [Google Scholar]
- 5.Krall N, da Cruz FP, Boutureira O, Bernardes GJL. Site-selective protein-modification chemistry for basic biology and drug development. Nat. Chem. 2016;8:103–113. doi: 10.1038/nchem.2393. [DOI] [PubMed] [Google Scholar]
- 6.Hoyt EA, Cal PMSD, Oliveira BL, Bernardes GJL. Contemporary approaches to site-selective protein modification. Nat. Rev. Chem. 2019;3:147–171. doi: 10.1038/s41570-019-0079-1. [DOI] [Google Scholar]
- 7.Chalker JM, Bernardes GJL, Lin YA, Davis BG. Chemical modification of proteins at cysteine: opportunities in chemistry and biology. Chem. Asian J. 2009;4:630–640. doi: 10.1002/asia.200800427. [DOI] [PubMed] [Google Scholar]
- 8.Gunnoo SB, Madder A. Chemical protein modification through cysteine. ChemBioChem. 2016;17:529–553. doi: 10.1002/cbic.201500667. [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, et al. π-Clamp-mediated cysteine conjugation. Nat. Chem. 2016;8:120–128. doi: 10.1038/nchem.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng J-R, et al. Chemoselective and photocleavable cysteine modification of peptides and proteins using isoxazoliniums. Commun. Chem. 2019;2:93. doi: 10.1038/s42004-019-0193-5. [DOI] [Google Scholar]
- 11.Xie J, Schultz PG. A chemical toolkit for proteins–an expanded genetic code. Nat. Rev. Mol. Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 12.Lang K, Chin JW. Cellular Incorporation of Unnatural amino acids and bioorthogonal babeling of Proteins. Chem. Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 13.Bloom S, et al. Decarboxylative alkylation for site-selective bioconjugation of native proteins via oxidation potentials. Nat. Chem. 2018;10:205–211. doi: 10.1038/nchem.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matos MJ, et al. Chemo- and regioselective lysine modification on native proteins. J. Am. Chem. Soc. 2018;140:4004–4017. doi: 10.1021/jacs.7b12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsubokura K, et al. In vivo gold complex catalysis within live mice. Angew. Chem. Int. Ed. 2017;56:3579–3584. doi: 10.1002/anie.201610273. [DOI] [PubMed] [Google Scholar]
- 16.Fujiki, K. & Tanaka, K. RIKEN click reagent for protein labelling. In Encyclopedia of Reagents for Organic Synthesis10.1002/047084289x.rn02050 (2020).
- 17.Rosen CB, Francis MB. Targeting the N terminus for site-selective protein modification. Nat. Chem. Biol. 2017;13:697–705. doi: 10.1038/nchembio.2416. [DOI] [PubMed] [Google Scholar]
- 18.Jacob E, Unger R. A tale of two tails: why are terminal residues of proteins exposed? Bioinformatics. 2007;23:e225–e230. doi: 10.1093/bioinformatics/btl318. [DOI] [PubMed] [Google Scholar]
- 19.Guo H, et al. Positive charge in the n-region of the signal peptide contributes to efficient post-translational translocation of small secretory preproteins. J. Biol. Chem. 2018;293:1899–1907. doi: 10.1074/jbc.RA117.000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sereda TJ, Mant CT, Quinn AM, Hodges RS. Effect of the α-amino group on peptide retention behaviour in reversed-phase chromatography determination of the pKa values of the α-amino group of 19 different N-terminal amino acid residues. J. Chromatogr. A. 1993;646:17–30. doi: 10.1016/S0021-9673(99)87003-4. [DOI] [PubMed] [Google Scholar]
- 21.Schoffelen S, et al. Metal-free and pH-controlled introduction of azides in proteins. Chem. Sci. 2011;2:701–705. doi: 10.1039/c0sc00562b. [DOI] [Google Scholar]
- 22.Chan W-K, Ho C-M, Wong M-K, Che C-M. Oxidative amide synthesis and N-terminal α-amino group ligation of peptides in aqueous medium. J. Am. Chem. Soc. 2006;128:14796–14797. doi: 10.1021/ja064479s. [DOI] [PubMed] [Google Scholar]
- 23.Chan AO-Y, et al. Modification of N-terminal α-amino groups of peptides and proteins using ketenes. J. Am. Chem. Soc. 2012;134:2589–2598. doi: 10.1021/ja208009r. [DOI] [PubMed] [Google Scholar]
- 24.Kung KK-Y, Wong K-F, Leung Y-C, Wong M-K. N-terminal α-amino group modification of peptides by an oxime formation–exchange reaction sequence. Chem. Commun. 2013;49:6888–6890. doi: 10.1039/c3cc42261e. [DOI] [PubMed] [Google Scholar]
- 25.Chen D, Disotuar MM, Xiong X, Wang Y, Chou DH-C. Selective N-terminal functionalization of native peptides and proteins. Chem. Sci. 2017;8:2717–2722. doi: 10.1039/C6SC04744K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. N-terminal protein modification through a biomimetic transamination reaction. Angew. Chem. Int. Ed. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 27.Scheck RA, Dedeo MT, Iavarone AT, Francis MB. Optimization of a biomimetic transamination reaction. J. Am. Chem. Soc. 2008;130:11762–11770. doi: 10.1021/ja802495w. [DOI] [PubMed] [Google Scholar]
- 28.Witus LS, et al. Site-specific protein transamination using N-methylpyridinium-4-carboxaldehyde. J. Am. Chem. Soc. 2013;135:17223–17229. doi: 10.1021/ja408868a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald JI, Munch HK, Moore T, Francis MB. One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nat. Chem. Biol. 2015;11:326–331. doi: 10.1038/nchembio.1792. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Kuang Y, Wu J. 2‐Alkynylbenzaldehyde: a versatile building block for the generation of cyclic compounds. Asian J. Org. Chem. 2012;1:302–312. doi: 10.1002/ajoc.201200100. [DOI] [Google Scholar]
- 31.Li L, Huang D, Shi C, Yan G. 2‐Alkynylaryl aldehydes (enynals) in organic synthesis. Adv. Synth. Catal. 2019;361:1958–1984. doi: 10.1002/adsc.201801472. [DOI] [Google Scholar]
- 32.Kumar S, Saunthwal RK, Saini KM, Verma AK. Dual function of carbon tetrachloride: synthesis of chlorinated heterocycles. Chem. Commun. 2019;55:10721–10724. doi: 10.1039/C9CC04746H. [DOI] [PubMed] [Google Scholar]
- 33.Cui J-F, et al. Metal‐free cyclocarboamination of ortho‐formyl phenylacetylenes with secondary amines: Access to 1,3‐diamino‐1H‐indenes and 3‐amino‐1‐indanones. Adv. Synth. Catal. 2019;361:569–577. doi: 10.1002/adsc.201801318. [DOI] [Google Scholar]
- 34.Ohta Y, et al. Rapid access to 3-(aminomethyl)isoquinoline-fused polycyclic compounds by copper-catalyzed four-component coupling, cascade cyclization, and oxidation. J. Org. Chem. 2009;74:6299–6302. doi: 10.1021/jo901090u. [DOI] [PubMed] [Google Scholar]
- 35.García D, Uribe E, Salgado M, Martínez MP, Carvajal N. Mutagenic and kinetic support for an allosteric site in arginase from the extreme thermophile Bacillus caldovelox, which allows activation by arginine. Biochimie. 2015;108:8–12. doi: 10.1016/j.biochi.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 36.Cheng PN-M, et al. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 2007;67:309–317. doi: 10.1158/0008-5472.CAN-06-1945. [DOI] [PubMed] [Google Scholar]
- 37.Yau T, et al. Preliminary efficacy, safety, pharmacokinetics, pharmacodynamics and quality of life study of pegylated recombinant human arginase 1 in patients with advanced hepatocellular carcinoma. Invest. New Drugs. 2015;33:496–504. doi: 10.1007/s10637-014-0200-8. [DOI] [PubMed] [Google Scholar]
- 38.Leung, Y.-C. & Lo, W.-H. Site-directed pegylation of arginases and the use thereof as anti-cancer and anti-viral agents. US 8507245B2 (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All principal data with detailed experimental procedure and characterization of this work are included in this article, and its Supplementary Information or are available from the corresponding author upon reasonable request.