Abstract
Factor inhibiting hypoxia-inducible factor (FIH) is a 2-oxoglutarate-dependent protein hydroxylase that catalyses C3 hydroxylations of protein residues. We report FIH can accept (D)- and (L)-residues for hydroxylation. The substrate selectivity of FIH differs for (D) and (L) epimers, e.g., (D)- but not (L)-allylglycine, and conversely (L)- but not (D)-aspartate, undergo monohydroxylation, in the tested sequence context. The (L)-Leu-containing substrate undergoes FIH-catalysed monohydroxylation, whereas (D)-Leu unexpectedly undergoes dihydroxylation. Crystallographic, mass spectrometric, and DFT studies provide insights into the selectivity of FIH towards (L)- and (D)-residues. The results of this work expand the potential range of known substrates hydroxylated by isolated FIH and imply that it will be possible to generate FIH variants with altered selectivities.
Subject terms: Biocatalysis, X-ray crystallography
Hypoxia-inducible factor (FIH) is an oxygenase which post-translationally hydroxylates proteins and is implicated in a range of biological processes. Here a wide substrate tolerance for FIH is demonstrated, including for d-amino acids, where double hydroxylation of d-leucine is observed.
Introduction
Biopolymer hydroxylation as catalysed by Fe(II) and 2-oxoglutarate (2OG)-dependent oxygenases1,2 is important in biological processes including oxygen sensing3–7, chromatin regulation, carnitine biosynthesis, lipid metabolism, collagen biosynthesis, and DNA/RNA modification/repair2,7–10. The 2OG oxygenase factor inhibiting hypoxia-inducible factor (FIH) is of particular interest from an enzymology selectivity perspective, because it catalyses the posttranslational hydroxylation of multiple protein residues11–15. FIH was first identified as an asparaginyl C3 hydroxylase acting on hypoxia-inducible factor α-subunits; such modification reduces the transcriptional activity of HIF in a manner relevant to the hypoxic response in animals16–20. FIH also catalyses C3 hydroxylation of residues on multiple human members of the ubiquitous ankyrin repeat domain (ARD) protein family12–15,21–23. In the case of the ARD proteins, FIH catalyses hydroxylation not only of Asn-residues but also of Asp- and His-residues12,14,15. More recent work has shown that FIH can catalyse the hydroxylation of residues with hydrophobic side chains, such as leucine, to form C3 hydroxylated products with the same relative stereoselectivity as in the hydroxylation of residues with polar side chains, i.e., to form the (3S)-hydroxylated product (Fig. 1)12.
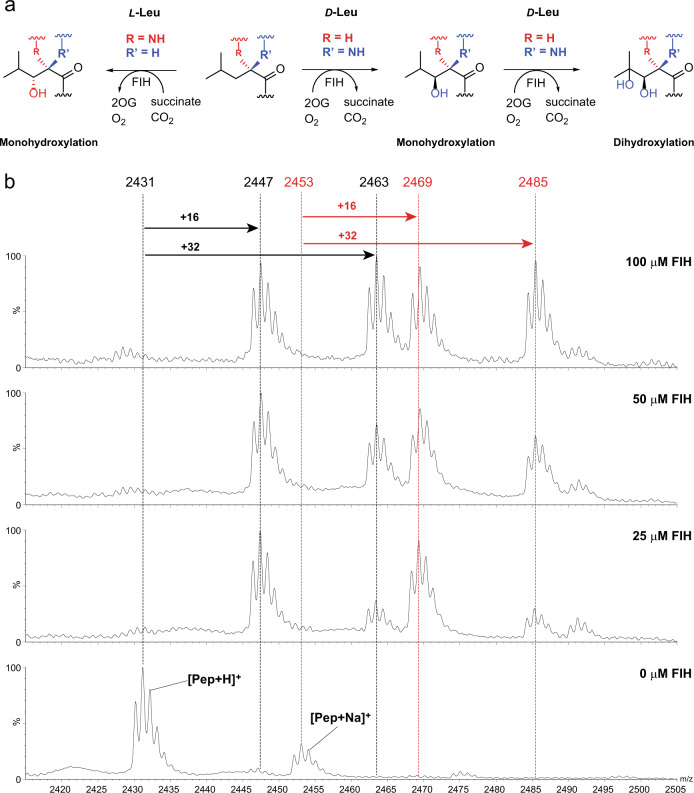
Fig. 1. Reactions catalysed by FIH.
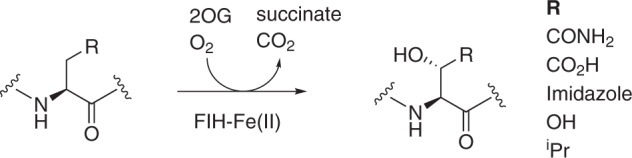
FIH is a 2OG oxygenase catalyzing the posttranslational C3 hydroxylation of (L)-residue substrates including the hypoxia-inducible factor α-subunits (HIFα; R = CONH2) and ankyrin repeat domain proteins (R = CONH2, COOH, OH, imidazole and isopropyl) to give mono-hydroxylated products. Note all studied (L)/(2S)-substrates give products with the same relative stereochemistry at C3, i.e., (2S,3S) for R = CONH2, COOH, Imidazole, OH or (2S,3R) for R = iPr.
Here we report that FIH can catalyse the hydroxylation of (D)-residues, interestingly with altered substrate and product selectivities compared with the analogous (L)-residues. Unexpectedly, in the case of the (D)-Leu residue substrate, a dihydroxylated product is observed.
Results
FIH hydroxylation assays with D-residue containing peptides
The promiscuity of some 2OG oxygenase protein hydroxylases is precedented by earlier work on 2OG oxygenases acting on small molecules, including those involved in β-lactam biosynthesis24,25. In some cases, unsaturated substrate analogues were found to undergo rearrangements, in some examples likely via radical intermediates26,27. Inspired by these studies, we tested whether FIH can catalyse hydroxylation of an allylglycine residue; anticipating hydroxylation of allylglycine might be more efficient than for leucine due to its relatively weak C3 allylic C–H bond, or that 1,3-allylic rearrangement or epoxide formation may occur, as precedented for 2OG oxygenases catalyzing small molecule oxidations9. A consensus ARD peptide substrate22,23 for FIH with an allylglycine residue at the potential hydroxylation position was therefore prepared. For synthetic efficiency reasons, the peptide was initially produced with an epimeric (D/L) mixture of allylglycine at the anticipated potential hydroxylation position. When incubated with FIH under standard conditions22,23, the (D/L) epimeric mixture reproducibly manifested ~50% conversion to a hydroxylated product; as evidenced by a +16 Da mass increment observed by matrix-assisted laser desorption ionization time-of-flight (MALDI-ToF) mass spectrometry (MS) analysis. To investigate further, the corresponding peptide with an (L)-allylglycine residue at the hydroxylation position was made. Surprisingly, given the extensive evidence for FIH-catalysed (L)-residue hydroxylation, this peptide did not undergo hydroxylation (<5%) upon incubation with FIH. By contrast, the subsequently synthesized (D)-allylglycine peptide underwent relatively efficient hydroxylation, demonstrating that FIH can, and at least in this case prefers to, catalyse hydroxylation of (D)-residues (Supplementary Fig. 1g).
To investigate the scope of FIH-catalysed hydroxylation of (D)-residue containing substrates, systematic substitutions at the hydroxylation position of the consensus ankyrin (CA) scaffold peptide22,23 (i.e., HLEVVKLLLEHGADVXAQDK where X is the substituted residue) were then made. The results (Supplementary Fig. 1 and Supplementary Table 1) show that almost half of the D-residue containing peptides tested (9/19) manifested apparent oxidation in the presence of FIH. An approximate order of efficiency of hydroxylation under standard conditions are (D)-Leu ≈ (L)-Asn > (D)-Ile > (D)-Asn ≈ (D)-AllylGly > (D)-His ≈ (D)-Phe ≈ (D)-Try > (D)-Phe > (D)-Trp (Supplementary Table 1b). (D)-Residue substrates with amide, basic, nucleophilic, aromatic, and hydrophobic side chains, were identified indicating FIH may accept an even wider range of (D)- than (L)-residues12. Differences in the selectivity for (D)-residues were apparent with (D)-Phe and (D)-Tyr being apparently hydroxylated by FIH, whereas the (L) version of these peptides were not hydroxylated. Further, the (D)-Leu, (D)-Ile, and (D)-Trp-containing peptides were apparently oxidized more efficiently than their (L)-counterparts12. By contrast, the (D)-Asp CA peptide was not hydroxylated, whereas the (L)-Asp peptide was12. The results also suggested (D)-Cys- and (D)-Met-containing CA sequences may be substrates, although these observations require further work due to the readily oxidized sulfur of these substrates.
Most unexpectedly, the (D)-Leu CA peptide appeared to undergo a double hydroxylation, with both +16 Da and +32 Da peaks being significant in the MS spectrum after incubation, albeit with high levels of FIH (Fig. 2a). In the case of the (L)-Leu CA peptide substrate, only a +16 Da mass shift was observed, as reported12, suggesting a single hydroxylation. To investigate whether the apparent dihydroxylation observed with (D)-Leu CA peptide reflects 1,2 or 1,3 diol formation, the high-performance liquid chromatography (HPLC)-purified product peptide was treated with NaIO4, which reacts with 1,2-diols (Supplementary Fig. 2)28. Partial cleavage of the apparent dihydroxylated product (2462.8 Da) to give a product of mass 2402.7 Da was observed, consistent with 1,2- and not 1,3- oxidation to give a C3 aldehyde (Supplementary Fig. 2). Addition of biotin hydrazide to the reaction mixture gave a new product with a +212 Da increment, again consistent with formation of a C3 aldehyde.
Fig. 2. FIH can catalyse dihydroxylation reactions.
a FIH catalyses single- and double-hydroxylations of (L)- and (D)-leucine-containing residues, respectively. b Mass spectrometric analysis of FIH-catalysed hydroxylation of a (D)-leucine residue in a consensus ankyrin (CA) (HLEVVKLLLEHGADV((D)-L)AQDK) substrate. The CA (D)-Leu peptide (50 μM) was incubated with 0 μM, 25 μM, 50 μM and 100 μM FIH. Sub-stoichiometric quantities of FIH result in predominantly mono-hydroxylated product, whereas at higher FIH concentrations the dihydroxylated product is produced. Note the different C3 hydroxylation stereochemistries for the (L)-Leu and (D)-Leu substrates. See text for discussion.
To investigate whether a mono-hydroxylated (D)-Leu peptide can undergo further hydroxylation to form the overall +32 Da product, a synthetic CA peptide containing the (2R,3S) 3-hydroxyleucine residue was assayed. This peptide was observed by MS to be hydroxylated to only a small extent, if at all (much less than the (D)-Leu CA peptide) (Supplementary Fig. 1g).
Calculations on FIH-catalysed dihydroxylation
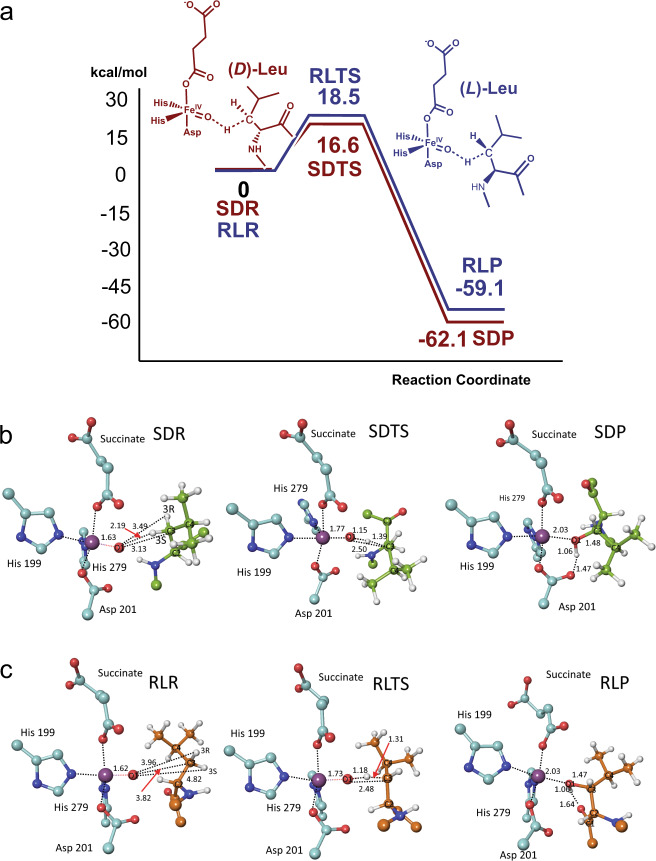
To investigate the differences in hydroxylation of (D)- and (L)-leucine-containing CA substrates, we investigated the reactions using density functional calculations (B3LYP/6–31 G** level), using simplified model systems at the ferryl intermediate stage of catalysis (Supplementary Fig. 3). The modelling studies were based on crystallographic analyses on (D)- and (L)- residue containing FIH substrates as described below.
Figure 3b shows calculated structures for FIH complexed with a ‘truncated’ (D)-leucine substrate model (i.e., with an N-methylated amino and C-terminal methyl ketone, referred to as (D)-Leu) (SDR), the mono-hydroxylated product model (SDP), and the calculated transition state (SDTS)). In SDR, the reactive iron-bound oxygen (O1) is proximate to (D)-Leu C3, being ∼2.2 Å from the pro-S hydrogen. By contrast, the pro-R (D)-Leu C3 hydrogen projects away from O1 (∼3.5 Å), suggesting formation of the (2R,3S) mono-hydroxylated product (as observed—see below).
Fig. 3. Mechanistic insights into FIH-catalysed C3 hydroxylation of (D)- and (L)-leucine.
a Potential energy profile diagrams along the modelled reaction coordinates for the FIH-catalysed hydroxylation of (D)-leucine (brown) or (L)-leucine (blue)-derived substrate models. The potential energies of stationary-state structures for the (D)- and (L)-leucine residue substrate hydroxylations are measured from the SDR or RLR intermediate structures, respectively. b Optimized structures of energy minima and transition state for C3 hydroxylation of the (D)-leucine substrate. c Optimized structures of the energy minima and the transition state for C3 hydroxylation of the (L)-leucine substrate. Colours: Fe (blue purple), N (blue), O (red), C (turquoise), substrate (D)-leucine (yellow-green), (L)-leucine (orange), H (white). Selected distances are in Å.
When the O1···C3 distance contracts from 3.1 Å (in SDR) to 2.5 Å, a transition state (SDTS) manifests, wherein the pro-S hydrogen is partially transferred to O1. The O···H distance decreases from 2.2 Å in SDR to 1.2 Å in SDTS, corresponding to >90% formation of alcohol product (SDP). Despite investigations, there was no evidence for carbanionic or radical intermediates, suggesting an efficient radical rebound type mechanism (Fig. 3a), as proposed for other 2OG oxygenases29–33. In SDP, the newly formed alcohol points towards the Asp201 carboxylate, forming a hydrogen bond (1.5 Å).
With (L)-Leu, the model implies C3 pro-R hydroxylation; because the distance between O1 and C3 is increased from 3.1 Å in SDR to 3.8 Å in RLR, a higher activation barrier is predicted for (L)- compared with (D)-Leu hydroxylation, consistent with the experimental observations. The calculations imply hydroxylation of both (D)- and (L)-Leu is thermodynamically favourable; SDP and RLP are both ∼60 kcal/mol lower in potential energy than SDR and RLR (Fig. 3). The calculated activation barrier for hydroxylation of (D)-Leu is somewhat lower than that of (L)-Leu by 1.9 kcal/mol. It is noteworthy that the role of hydrogen bond acceptor with respect to the product alcohol changes from Asp201 in SDP (Fig. 3b) to the backbone carbonyl oxygen of Leu803 in RLP (Fig. 3c).
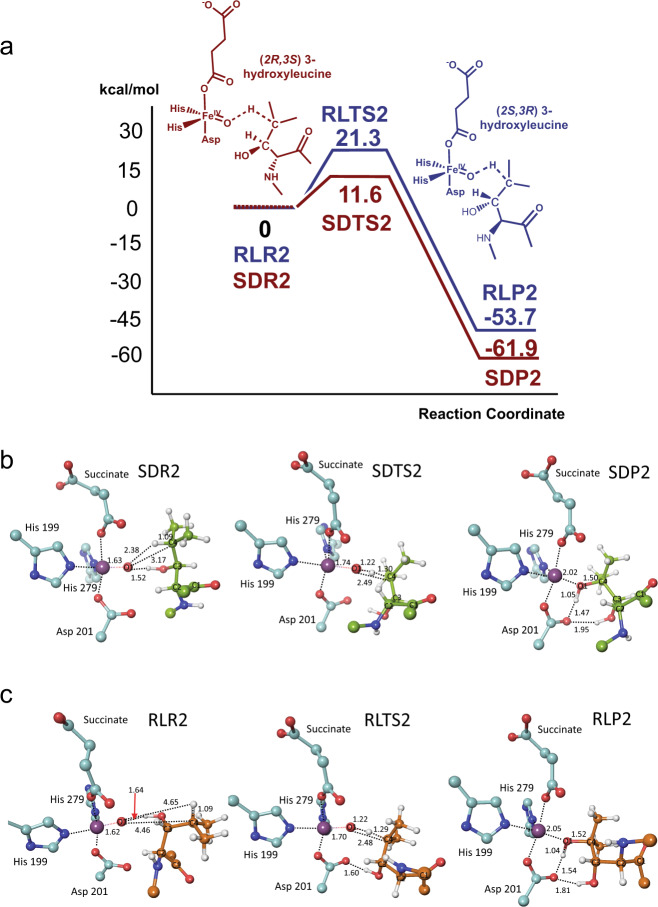
We then modelled the (potential) second hydroxylation of the mono-hydroxylated (D)- and (L)-Leu products (B3LYP/6–31 G** level). The model for the proposed reactive (2R,3S) 3-hydroxyleucine complex (SDR2) is shown in Fig. 4b, with the dihydroxylated product (SDP2) and transition state (SDTS2). In SDR2, the C4 hydrogen points towards the iron-bound O1, consistent with the experimental results implying C3/C4 vicinal diol formation, although O1 is similarly close to C3 (3.09 Å for C3 vs. 3.17 Å for C4). A second hydroxylation at C3 would give an unstable gem diol, for which there is precedent in 2OG oxygenase catalysis34; however, the C3 hydrogen is directed away from O1 in SDR2. Thus, these observations are consistent with the second hydroxylation occurring at C4.
Fig. 4. Mechanistic insights into the second FIH-catalysed hydroxylation of (D)-leucine.
a Potential energy profiles for the ‘second’ hydroxylation of (2R,3S) C3 hydroxyleucine (brown) and (2S,3R) C3 hydroxyleucine (blue) substrates. The potential energies of stationary-state structures on the reaction coordinates are measured from the respective reactant complexes (SDR2 and RLR2). b Optimized structures of energy minima and transition states for the ‘second’ hydroxylation reaction of the (2R,3S) C3 hydroxyleucine substrate. c Optimized structures of the energy minima and the transition state for the ‘second’ hydroxylation reaction of the (2S,3R) C3 hydroxyleucine substrate. Colours: Fe (blue purple), N (blue), O (red), C (turquoise), substrate (D)-leucine (yellow-green), (L)-leucine (orange), H (white). Selected distances are in Å.
As the reaction proceeds, the hydrogen-bond acceptor with respect to the substrate C3 alcohol changes from O1 to the Asp201 carboxylate. Contributions to the single imaginary frequency in SDTS2 come from the proton transfer from C4 to O1 and from the O1−C4 bond-forming motion. This implies that hydrogen abstraction and O1−C4 product bond formation occur efficiently. The O1···H distance contracts from 2.38 to 1.22 Å on going from SDR2 to SDTS2, corresponding to ∼85% completion of O1−H bond formation. By contrast, O1 approaches C4 by only 0.69 Å during formation of SDTS2, with ∼40% progress towards the β,γ-dihydroxy-(D)-leucine product.
Consistent with the lack of its experimentally observed second hydroxylation, the modelling results for (2S,3R) 3-hydroxyleucine differ from those for (2R,3S) 3-hydroxyleucine (Fig. 4c). In RLR2, C4 is relatively distant from O1 compared with SDR2; the C4···O1 distance is 4.5 Å in RLR2 compared with 3.2 Å in SDR2 (Fig. 4). In both SDR2 and RLR2 (Fig. 4), the mono-hydroxylated substrate C3 alcohol can hydrogen bond with O1. The O–H···O hydrogen bond in SDR2 contributes to sustaining a reactive conformation. By contrast, the corresponding hydrogen bond in RLR2 apparently serves as an obstacle to reaction by stabilizing a non-productive conformation.
Despite the difference in activation barriers and reaction energies, most structural features of SDTS2 and SDP2 are similar to those of RLTS2 and RLP2. Differences in distances involving O1 are small and RLTS2 is similar to SDTS2 in forming a hydrogen bond between the substrate alcohol and Asp201 (Figs. 3 and 4). The C3 hydroxyl is a hydrogen bond donor to O1 in both complexes. In SDR2, this contributes to sustaining a reactive conformation of (2R,3S) 3-hydroxyleucine. Although also present in RLR2, the substrate cannot adopt such a favourable conformation due to the effect of the (2S)-sterochemistry on side chain conformation.
The dihydroxy-(D)-leucine product (SDP2) is produced with a calculated potential energy of ~62 kcal/mol below the reactant complex (SDR2), indicating that a second hydroxylation reaction with the (2R,3S) 3-hydroxyleucine substrate is thermodynamically favourable (Fig. 4a); the calculated activation barrier to form the product via SDTS2 is 11.6 kcal/mol, which is 9.7 kcal/mol lower than for (2S,3R) 3-hydroxyleucine. Thus, on the basis of kinetic and thermodynamic grounds and consistent with the experimental observations, the calculations imply the second hydroxylation is likely more favourable for (2R,3S)-hydroxyleucine compared with (2S,3R)-hydroxyleucine.
FIH crystal structures with D-residue containing peptides
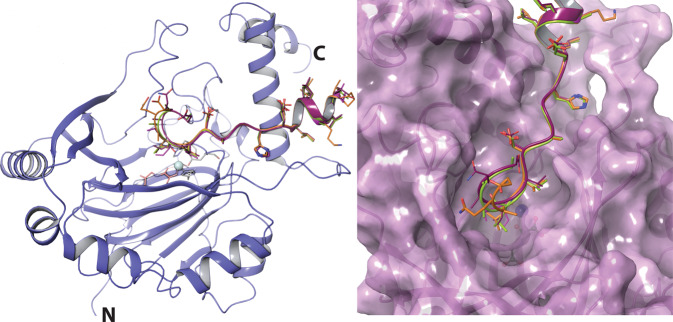
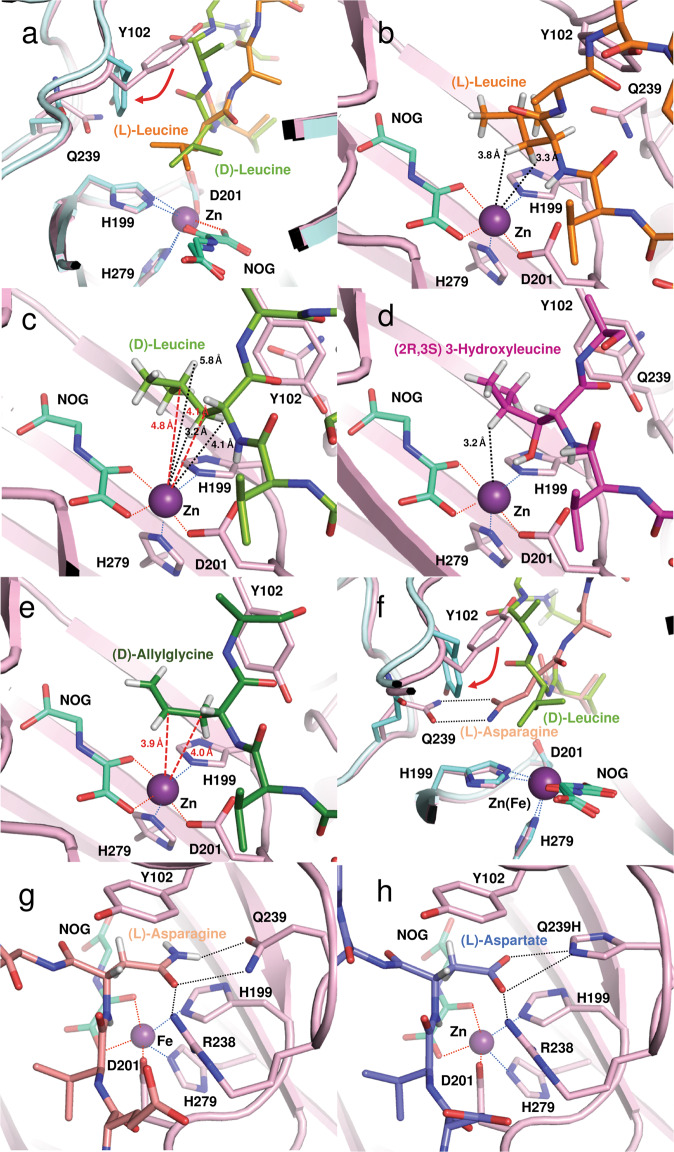
To investigate the structural basis for the differential selectivities of (D)- over (L)-residues reported here, we attempted X-ray crystallographic analysis of FIH with (D)-substrates, to compare with the analogous (L)-substrates. Structures of FIH complexed with (L)-Leu, (D)-Leu, (2R,3S) 3-hydroxyleucine and (D)-allylglycine CA peptides were obtained (Fig. 5, Supplementary Table 2 and Supplementary Fig. 4). We were unable to obtain a structure with (L)-allylglycine CA, which was not observed to be a substrate. The general binding modes of the (D)-substrates were similar to those reported for other (L)-FIH substrates12,14,15,20,21,23. However, based on the available crystal structures and modelling studies, there are differences in the binding modes of the (L)- and (D)-substrates in the active site region close to the iron. Compared with (L)-residue substrate binding, the (D)-residues adopt a distinctive orientation that appears to involve rotation about the Cα-Cβ bond of FIH Tyr102 (Fig. 6a–e).
Fig. 5. FIH binds peptides with different substrate residues in a similar overall manner.
Ribbons representation of superimposed peptide substrates when bound to FIH as observed by X-ray crystallography (violet); orange, (L)-leucine (PDB: 4B7E); yellow-green, (D)-leucine CA substrate (PDB: 4JAA); maroon, (2R,3S) C3 hydroxyleucine CA substrate (PDB: 6RUJ); purple, (D)-allylglycine CA substrate (PDB: 4NR1). The active site metal is a turquoise sphere. Note only one monomer of the FIH dimer is shown.
Fig. 6. Superposition of views from crystal structures of FIH in complex with consensus ankyrin sequence peptides.
Superimposed structures of FIH with: a (L)-leucine (PDB: 4B7E; orange) and (D)-leucine (PDB: 4JAA; yellow-green) residues at the hydroxylation position. FIH atoms close to the substrates are in pink and cyan, for the (L)- and (D)-substrate views, respectively; b (L)-leucine; c (D)-leucine; d (2R,3S) 3-hydroxyleucine (PDB: 6RUJ; purple); e (D)-allylglycine (PDB: 4NR1; dark-green); f (L)-asparagine (salmon) and (D)-leucine (yellow-green); g (L)-asparagine (salmon) and h (L)-aspartate (PDB: 2XUM); slate-blue). Note that hydrogens are shown on the side chains of the substrate residues in the non-superimposed structures.
In the case of the (D)-allylglycine substrate structure, the side chain does not appear tightly bound, although C3 hydroxylation is likely, C4 is closer than C3 to the active site iron (3.9 Å) (Fig. 6e). Modelling of the (L)-allylglycine substrate complex based on this structure implies its allyl group will project away from the metal, consistent with its observed lack of reaction. By contrast, the side chains of substrates with polar side chains (both (L) and (D)) are positioned to form hydrogen bonds/polar interactions with the side chains of Arg238 and Gln239, as observed in prior FIH-substrate complexes12,14,15,20 (Fig. 6g, h). The (L)-Leu substrate structure implies that conformational movement of the (L)-Leu isopropyl group may be relatively restricted because of close contacts with the Tyr102, His199 and Gln239 side chains of FIH (Fig. 6b). By contrast with the (L)-Leu substrate, with (D)-Leu its isopropyl group projects further away from the protein interior (Fig. 6a). The (D)-Leu backbone carbonyl oxygen and the neighbouring Ala of the (D)-Leu CA peptide substrate appear to push the Tyr102 side chain away from the active site (Fig. 6c). Collectively, these observations suggest more flexibility in binding of (D)-Leu compared with (L)-Leu, consistent with the observed second hydroxylation of (D)-Leu but not (L)-Leu (Fig. 6a–d). The apparently more restricted binding of (L)-Leu may also relate to the lack of activity observed with the (L)- compared with the (D)-allylglycine CA peptide.
Consistent with the calculated SDP structure (Fig. 3), in the (2R,3S)-hydroxyleucine complex crystal structure, the C3 alcohol of the mono-hydroxylated (2R,3S) product/intermediate is positioned to hydrogen bond with the FIH Asp201 iron-binding carboxylate (Fig. 6d). This interaction may influence hydroxylation to give the subsequently formed dihydroxylated product.
Stereochemistry of hydroxylation
The crystallographic analyses lead to the prediction that the product of monohydroxylation of (L)-Leu will be (2S,3R) 3-hydroxyleucine (note this has the same relative stereochemistry as the (2S,3S)-stereochemistry of (L)-Asn hydroxylation12,20), because the pro-3R hydrogen is closer to the iron (3.4 Å) than the pro-3S hydrogen (3.9 Å) consistent with the calculations. However, the relative proximity (as observed by crystallography) of the pro-3R and pro-3S hydrogens to the iron is “swapped” in the (D)-Leu substrate, being 4.1 and 3.2 Å from the iron, respectively (Fig. 6c), in support of the calculations predicting that mono-hydroxylated (D)-Leu has the (2R,3S)-stereochemistry. To test this, we synthesized racemic mixtures of (2S,3R)/(2R,3S) and (2R,3R)/(2S,3S) 3-hydroxyleucines by established procedures (see Supplementary Methods for details). These were used as standards in amino acid analyses of 3-hydroxyleucine from an ankyrin repeat peptide made synthetically and a peptide product after incubation with FIH (Supplementary Fig. 5). The results from the amino acid analyses imply that (2R,3S) mono-hydroxylated leucine is produced after incubation of the (D)-Leu substrate, in agreement with the computational predictions and crystallography. It is noteworthy that this stereochemical outcome is the opposite to that observed for studied FIH-catalysed C3 hydroxylations with (2S)-substrates12,20.
Discussion
Along with the cytochrome 450 and radical SAM enzymes, the 2OG oxygenases form one of the most flexible of all oxidizing enzyme superfamilies7,10,35. In animals, the identified reactions catalysed by 2OG oxygenases, are to date substantially limited to single hydroxylations and N-methyl demethylations proceeding via hydroxylation (as well as related alcohol and aldehyde oxidations)2,9. In plants and microbes, 2OG oxygenases catalyse a much wider range of reactions, including di-hydroxylations of the same residue2,9. Thus, in yeasts, a ribosomal protein (RPS23) prolyl hydroxylase catalyses C3 and C4 prolyl hydroxylation, whereas the homologous enzyme in animals apparently catalyses only C3 prolyl hydroxylation36–38. There is also evidence for dihydroxylation of lysine residues in diatoms39, although the enzyme(s) catalyzing these reactions are unknown. Although its biological/cellular relevance is unclear, if any, the observation that a human 2OG oxygenase, FIH, can catalyse dihydroxylation at different carbons of the same protein residue is thus of interest, because it suggests that the selectivity of human 2OG oxygenases may extend beyond what is presently perceived likely.
In some cases, 2OG oxygenase selectivity is governed by targeting domains in addition to the catalytic domain40, whereas in others including FIH, the catalytic domain appears to be, at least principally, responsible for enabling substrate and product selectivity18,20,41. Some human 2OG oxygenases acting on proteins appear to be highly selective for specific substrates, whereas others appear much more promiscuous, notably JMJD6 and FIH12,14,15,21,42,43. Both of these belong to the JmjC family of 2OG oxygenases, which extends from bacteria to humans18. Some JmjC oxygenases catalyse N-methyl demethylations, whereas others catalyse formation of stable alcohol products via unmodified proteinogenic residue side chain oxidations9,41. JMJD6 catalyses the (3S)-hydroxylation of multiple lysine residues in arginine–serine-rich regions of splicing regulatory proteins and other substrates43,44. FIH was originally identified as interacting with HIF45, then shown to catalyse C3 hydroxylation of an asparagine-residue in the C-terminal transcriptional activation domain of HIF-α isoforms, a modification that hinders binding of HIF to the CBP/p300 histone acetyl transferases16–18. FIH also interacts with, and hydroxylates many ARD proteins, with hydroxylated residues including not only asparagines but also aspartates and histidines, also at the C3 position (and at least with studied polar side chain residues, the reaction proceeds to give the (3S)-stereochemistry products, as with HIF-α hydroxylation)13–15,21.
Studies on the residue selectivity of isolated FIH using ARD sequences have revealed, albeit at a high enzyme:substrate ratio (as used in our work), that FIH can accept even substrates with hydrophobic side chains and has potential for catalyzing desaturation and hydroxylation12,15. These studies were all carried out with (L)/(2S)-substrates. We are interested in exploring (D)/(2R)-substrates for 2OG oxygenases, in part because of the increased prevalence of (D)-residues in aged cells46. Work with JmjC histone demethylases has implied that, at least in some cases, these enzymes are specific for the (L)-substrate stereochemistry47. By contrast, the results presented here clearly demonstrate that isolated FIH can accept (D)-substrates, further expanding the scope of catalysis by isolated FIH.
The substrate/residue selectivity of FIH differs for (L)- and (D)-substrates, as shown by differences in the rank order of conversion of different C-2 epimeric residues. The product selectivity can differ for the same residue with epimeric C-2 stereochemistry. This was most dramatically illustrated in the case of the epimeric C-2 leucine substrates, which we investigated by both experimental and computational studies. In the case of the (L)-leucine substrate, a single hydroxylation is observed, as is typical for FIH12. However, with (D)-leucine we observed a C3/C4 dihydroxylated product. This work echoes observations on VioC, which is naturally an (L)/(2S)-arginine 3-hydroxylase, but which in the case of the unnatural (D)/(2R)-arginine substrate analogue efficiently catalyses an alternative reaction, i.e., oxidative deamintion to give a 2-oxo acid product48.
Crystallographic analyses of FIH in complex with various (D)-CA peptides implies that the C-2 leucine stereochemistry has a deterministic influence on whether or not hydroxylation occurs (including both first and the second hydroxylations with (D)-Leu), due to changes in the side chain binding modes. Using ab initio density functional calculations at the B3LYP/6–31 G** level of theory, we investigated structural and energetic features of the (L)- and (D)-leucine hydroxylation using a simplified model. The results imply stereoselective formation of (2S,3R) 3-hydroxyleucine and (2R,3S) 3-hydroxyleucine from (L)- and (D)-leucine-containing substrates, respectively, consistent with the experimental observations. The observation that (D)-leucine, but not (L)-leucine, undergoes two two-electron oxidations to give C3/C4 di-hydroxyleucine appears to reflect increased conformational flexibility in binding of (D)-leucine (Fig. 6). Notably, we did not observe significant reaction on incubation of the (2R,3S) mono-hydroxylated (D)-Leu peptide with FIH (Supplementary Figure 1g). The structural and modelling studies suggest it is possible that the C3 alcohol of the mono-hydroxylated (D)-Leu product may form a hydrogen bond with the carboxylate of the iron-binding Asp201, thus hindering productive catalysis at the iron-binding site (Fig. 6d); however, this does not explain why we saw more dihydroxylated product from the unhydroxylated (D)-Leu substrate. Thus, factors other than those identified here must also be relevant; indeed, although some of the factors determining the catalytic efficiency of FIH with different substrates have been identified (including residue, local sequence context and ARD fold stability), there is little information on the necessary conformational changes required for the canonical ARD structure to bind productively at the FIH active site13,21–23.
The results presented here and elsewhere12–15,21,23 demonstrate flexibility in FIH catalysis and suggest FIH is a good target for mutagenesis aimed at producing posttranslational protein modifications of choice. Thus, FIH-catalysed hydroxylation or desaturation could, e.g., be used to increase proteolytic thermodynamic stability, induce conformational changes or enable further posttranslational modifications. Finally, the results raise the possibility that oxygenase related (L) to (D) C-2 residue epimerization could regulate protein oxygenase function, perhaps even in an oxygen/hypoxia-regulated capacity. Posttranslational hydroxylation has not been studied in the context of protein ageing but it has the potential to impact on aging via altering stability or promoting/hindering repair. The prevalence of (D)-residues is reported to increase in protiens with aging46. Given that FIH differently acts on (L)- and (D)-residues, it (or other oxygenases acting on proteins) has the potential to be involved in the regulation of protein aging. Given the promiscuity of FIH, there are various ways this could be envisaged to occur. Isoaspartyl residues are formed spontaneously from aspartyl and asparaginyl residues, and are a mark of protein ageing49,50. Thus, of particular interest, given the difference in selectivity of FIH for (L)- over (D)-Asp residues is a role for it in regulating isoaspartyl residue formation/repair.
Methods
FIH incubation assays
Small-scale assays used 0–100 μM FIH, 200 μM Fe(II), 50 μM peptide, 2 mM ascorbate and 1 mM 2OG in 100 mM sodium HEPES/150 mM NaCl pH 7.6 buffer12. Assays were incubated at 37 °C for 2 h and quenched with 5% aqueous (v/v) formic acid. The reaction mixture was then co-crystallized with an equal quantity of saturated α-cyano-4-hydroxycinnamic acid matrix solution before analysis with MALDI-ToF-MS.
Reactions with NaIO4 and biotin hydrazide
Ten microlitres of NaIO4 (5 mM in Milli-Q purified H2O) was added to an HPLC-purified solution of dihydroxylated (D)-Leu peptide (∼1 mM in Milli-Q purified H2O) in a 100 µL PCR tube, and the reaction mixture was left at room temperature for 2 h28. Biotin hydrazide (1 mM in Milli-Q purified H2O) was added, and the reaction mixture was incubated at 37 °C for 2 h before analysis by MALDI-TOF-MS.
MALDI-ToF-MS
One microlitre of saturated α-cyano-4-hydroxycinnamic acid matrix was mixed with 1 µL reaction mixture on a sample plate. The plate was air-dried prior to analysis on a Waters MALDI micro MXTM mass spectrometer in the positive ion reflectron mode. Parameters: Laser energy: 175–250 V, Pulse: 2050 V, Detector: 2600 V, Suppression: 125. Data were analysed using MassLynx 4.0 and spectra obtained were background subtracted (polynomial order = 16, % below curve = 10, tolerance = 0.01).
Amino acid analysis
Amino acid analyses were carried out using the acid hydrolysis method as described in Mantri et al.44.
Computational, crystallographic and synthetic methods
Supplementary information
Acknowledgements
We thank the Wellcome Trust, Cancer Research UK and the Institute for Basic Science, South Korea (Grant: IBS-R10-A2) for funding.
Author contributions
H.C. conducted modelling studies and wrote the paper. A.P.H. carried out assays, prepared proteins, carried out the amino acid analyses and wrote the paper. T.M.L., R.C., Y.N., J.S.S. and M.A.M. carried out crystallography. D.K., S.H., and J.L. synthesized peptides and contributed to the modelling. M.M. synthesized the amino acid standards. W.G. and H.T. made initial substrate discoveries. P.A.G. conducted the NaIO4 experiments. H.P. provided supervision of synthetic aspects of the work. C.J.S. conceived the work, provided supervision and wrote the paper.
Data availability
The main data supporting the findings of this study are available within the paper and its Supplementary Information file. The PDB accession codes for novel crystal structures are 4JAA, 6RUJ and 4NR1. Other relevant data are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hwangseo Park, Email: hspark@sejong.ac.kr.
Christopher J. Schofield, Email: christopher.schofield@chem.ox.ac.uk
Supplementary information
Supplementary information is available for this paper at 10.1038/s42004-020-0290-5.
References
- 1.Schofield CJ, Zhang Z. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol. 1999;9:722–731. doi: 10.1016/S0959-440X(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 2.Schofield, C. J. & Hausinger, R. P. (eds) 2-Oxoglutarate-Dependent Oxygenases (Royal Society of Chemistry, 2015).
- 3.Loenarz C, et al. The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 2011;12:63–70. doi: 10.1038/embor.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001;13:167–171. doi: 10.1016/S0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 5.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 6.Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 8.Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Islam MS, Leissing TM, Chowdhury R, Hopkinson RJ, Schofield CJ. 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 2018;87:585–620. doi: 10.1146/annurev-biochem-061516-044724. [DOI] [PubMed] [Google Scholar]
- 10.Hausinger RP. Fe(II)/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 11.Linke S, et al. Substrate requirements of the oxygen-sensing asparaginyl hydroxylase factor-inhibiting hypoxia-inducible factor. J. Biol. Chem. 2004;279:14391–14397. doi: 10.1074/jbc.M313614200. [DOI] [PubMed] [Google Scholar]
- 12.Yang M, et al. Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor. Angew. Chem. Int Ed. Engl. 2013;52:1700–1704. doi: 10.1002/anie.201208046. [DOI] [PubMed] [Google Scholar]
- 13.Cockman ME, et al. Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH) Proc. Natl Acad. Sci. USA. 2006;103:14767–14772. doi: 10.1073/pnas.0606877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, et al. Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains. FEBS J. 2011;278:1086–1097. doi: 10.1111/j.1742-4658.2011.08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang M, et al. Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor. J. Biol. Chem. 2011;286:7648–7660. doi: 10.1074/jbc.M110.193540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 17.Lando D, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hewitson KS, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 19.Dann CE, 3rd, Bruick RK, Deisenhofer J. Structure of factor-inhibiting hypoxia-inducible factor 1: an asparaginyl hydroxylase involved in the hypoxic response pathway. Proc. Natl Acad. Sci. USA. 2002;99:15351–15356. doi: 10.1073/pnas.202614999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins JM, et al. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 alpha. J. Biol. Chem. 2003;278:1802–1806. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- 21.Coleman ML, et al. Asparaginyl hydroxylation of the notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J. Biol. Chem. 2007;282:24027–24038. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- 22.Hardy AP, Prokes I, Kelly L, Campbell ID, Schofield CJ. Asparaginyl beta-hydroxylation of proteins containing ankyrin repeat domains influences their stability and function. J. Mol. Biol. 2009;392:994–1006. doi: 10.1016/j.jmb.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 23.Kelly L, McDonough MA, Coleman ML, Ratcliffe PJ, Schofield CJ. Asparagine beta-hydroxylation stabilizes the ankyrin repeat domain fold. Mol. Biosyst. 2009;5:52–58. doi: 10.1039/B815271C. [DOI] [PubMed] [Google Scholar]
- 24.Hamed RB, et al. The enzymes of beta-lactam biosynthesis. Nat. Prod. Rep. 2013;30:21–107. doi: 10.1039/C2NP20065A. [DOI] [PubMed] [Google Scholar]
- 25.Rabe P, Kamps JJAG, Schofield CJ, Lohans CT. Roles of 2-oxoglutarate oxygenases and isopenicillin N synthase in beta-lactam biosynthesis. Nat. Prod. Rep. 2018;35:735–756. doi: 10.1039/C8NP00002F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldwin JE, Bradley M. Isopenicillin-N synthase - mechanistic studies. Chem. Rev. 1990;90:1079–1088. doi: 10.1021/cr00105a001. [DOI] [Google Scholar]
- 27.Townsend CA. Convergent biosynthetic pathways to beta-lactam antibiotics. Curr. Opin. Chem. Biol. 2016;35:97–108. doi: 10.1016/j.cbpa.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buist, G. J. & Bunton, C. A. The mechanism of oxidation of alpha-glycols by periodic acid. 1. Ethylene glycol. J. Chem. Soc. 1406–1413, 10.1039/Jr9540001406 (1954).
- 29.Martinez S, Hausinger RP. Catalytic mechanisms of Fe(II)-and 2-Oxoglutarate-dependent oxygenases. J. Biol. Chem. 2015;290:20702–20711. doi: 10.1074/jbc.R115.648691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grzyska PK, Appelman EH, Hausinger RP, Proshlyakov DA. Insight into the mechanism of an iron dioxygenase by resolution of steps following the Fe-IV=O species. Proc. Natl Acad. Sci. USA. 2010;107:3982–3987. doi: 10.1073/pnas.0911565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price JC, Barr EW, Glass TE, Krebs C, Bollinger JM. Evidence for hydrogen abstraction from C1 of taurine by the high-spin Fe(IV) intermediate detected during oxygen activation by taurine:alpha-ketoglutarate dioxygenase (TauD) J. Am. Chem. Soc. 2003;125:13008–13009. doi: 10.1021/ja037400h. [DOI] [PubMed] [Google Scholar]
- 32.Cho KB, Hirao H, Shaik S, Nam W. To rebound or dissociate? This is the mechanistic question in C-H hydroxylation by heme and nonheme metal-oxo complexes. Chem. Soc. Rev. 2016;45:1197–1210. doi: 10.1039/C5CS00566C. [DOI] [PubMed] [Google Scholar]
- 33.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Krebs C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc. Natl Acad. Sci. USA. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbull JJ, et al. Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3beta-hydroxylase. J. Biol. Chem. 2004;279:1206–1216. doi: 10.1074/jbc.M309228200. [DOI] [PubMed] [Google Scholar]
- 35.Landgraf BJ, McCarthy EL, Booker SJ. Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 2016;85:485–514. doi: 10.1146/annurev-biochem-060713-035504. [DOI] [PubMed] [Google Scholar]
- 36.Loenarz C, et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl Acad. Sci. USA. 2014;111:4019–4024. doi: 10.1073/pnas.1311750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singleton RS, et al. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc. Natl Acad. Sci. USA. 2014;111:4031–4036. doi: 10.1073/pnas.1314482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katz MJ, et al. Sudestada1, a Drosophila ribosomal prolyl-hydroxylase required for mRNA translation, cell homeostasis, and organ growth. Proc. Natl Acad. Sci. USA. 2014;111:4025–4030. doi: 10.1073/pnas.1314485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valegard K, et al. Structural and functional analyses of Rubisco from arctic diatom species reveal unusual posttranslational modifications. J. Biol. Chem. 2018;293:13033–13043. doi: 10.1074/jbc.RA118.003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aik W, McDonough MA, Thalhammer A, Chowdhury R, Schofield CJ. Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases. Curr. Opin. Struc Biol. 2012;22:691–700. doi: 10.1016/j.sbi.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Markolovic S, et al. Structure-function relationships of human JmjC oxygenases - demethylases versus hydroxylases. Curr. Opin. Struc Biol. 2016;41:62–72. doi: 10.1016/j.sbi.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 42.Unoki M, et al. Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6) J. Biol. Chem. 2013;288:6053–6062. doi: 10.1074/jbc.M112.433284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webby CJ, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 44.Mantri M, et al. The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues. ChemBioChem. 2011;12:531–534. doi: 10.1002/cbic.201000641. [DOI] [PubMed] [Google Scholar]
- 45.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev. Pathol.-Mech. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 46.Fujii N. D-amino acid in elderly tissues. Biol. Pharm. Bull. 2005;28:1585–1589. doi: 10.1248/bpb.28.1585. [DOI] [PubMed] [Google Scholar]
- 47.Belle R, et al. Investigating D-lysine stereochemistry for epigenetic methylation, demethylation and recognition. Chem. Commun. 2017;53:13264–13267. doi: 10.1039/C7CC08028J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunham NP, et al. α-amine desaturation of D-arginine by the Iron(II)- and 2-(Oxo)glutarate-dependent L-arginine 3-hydroxylase, VioC. Biochemistry. 2018;57:6479–6488. doi: 10.1021/acs.biochem.8b00901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aswad DW, Paranandi MV, Schurter BT. Isoaspartate in peptides and proteins: formation, significance, and analysis. J. Pharm. Biomed. Anal. 2000;21:1129–1136. doi: 10.1016/S0731-7085(99)00230-7. [DOI] [PubMed] [Google Scholar]
- 50.Chondrogianni N, et al. Protein damage, repair and proteolysis. Mol. Asp. Med. 2014;35:1–71. doi: 10.1016/j.mam.2012.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the findings of this study are available within the paper and its Supplementary Information file. The PDB accession codes for novel crystal structures are 4JAA, 6RUJ and 4NR1. Other relevant data are available from the corresponding author upon reasonable request.