Abstract
Establishing structure–activity relationships is crucial to understand and optimize the activity of peptide-based inhibitors of protein–protein interactions. Single alanine substitutions provide limited information on the residues that tolerate simultaneous modifications with retention of biological activity. To guide optimization of peptide binders, we use combinatorial peptide libraries of over 4,000 variants—in which each position is varied with either the wild-type residue or alanine—with a label-free affinity selection platform to study protein–ligand interactions. Applying this platform to a peptide binder to the oncogenic protein MDM2, several multi-alanine-substituted analogs with picomolar binding affinity were discovered. We reveal a non-additive substitution pattern in the selected sequences. The alanine substitution tolerances for peptide ligands of the 12ca5 antibody and 14-3-3 regulatory protein are also characterized, demonstrating the general applicability of this new platform. We envision that binary combinatorial alanine scanning will be a powerful tool for investigating structure–activity relationships.
Subject terms: Combinatorial libraries, Drug screening, Peptides
Alanine substitution in peptides is crucial for studying peptide-based inhibitors of protein–protein interactions, but limited information is obtained from single alanine substitution. Here, the authors develop a label-free combinatorial alanine affinity selection platform to establish multi-alanine mutational tolerance and provide structure-activity relationships.
Introduction
Protein–protein interactions (PPIs) drive many aspects of biological function and are heavily involved in disease progression. The extensive (1000–5000 Å2), shallow and flat PPI interface challenges the development of PPI modulators using small molecules1–3. Peptides, on the other hand, can mimic the native binding epitope to recognize the PPI interface with high binding affinity and specificity. Recent advances in affinity selections and biological display methods have accelerated the generation of peptide-based PPI inhibitors1,4. To improve biophysical and pharmacological properties, iterative optimization is necessary. This process involves various structural modifications, for example, side chain modifications or macrocyclization to develop better analogs5,6. Modifying peptides while maintaining their binding affinity is crucial for hit-to-lead drug development, thus calling for a deep understanding of structure–activity relationships (SAR)7.
Alanine scanning informs SAR of peptides by systematically substituting each residue with alanine. This approach characterizes alanine tolerable residues and irreplaceable ‘hotspot’ residues essential for activity. Hotspots are identified by point alanine mutations that give rise to inactive mutants8. The alanine tolerable residues are often subjected to structure modifications without impairing the bioactivity. When multiple modifications happen simultaneously, non-additive combination effects emerge9–13, leading to unforeseen boosts or disruptions in activity. Complementary to single-point alanine scanning, shotgun alanine scanning is widely employed in protein mutagenesis to interrogate the pairwise and higher order combination effect of multi-point mutations14–18. As an example, the phage-displayed γ-receptor protein library was constructed by varying eleven residues to wild type (WT) or alanine19,20. The library was subjected to bioactivity assays to select for active strains, which presented a specific ratio of wild type to alanine at each residue. Pairwise analysis showed the frequency of most double alanine mutations followed a normal distribution when the two mutated residues were located in discontiguous regions14,21. However, for peptides an alanine mutation at one residue may affect the tolerance of an alanine mutation at a neighbouring residue. An analysis of the combined effects of multi-site modifications is thus critical at revealing the comprehensive peptide SAR landscape.
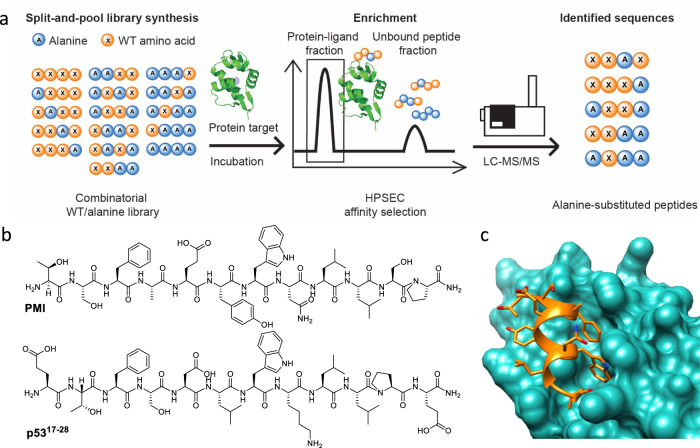
We established a label-free combinatorial alanine library affinity selection (Fig. 1a) platform to rapidly identify multiple sites in peptide-based lead compounds that tolerate modification while maintaining bioactivity. Peptide libraries were synthesized by the split-and-pool solid-phase peptide synthesis (SPPS) method and enriched by affinity selection according to an established protocol22. The peptide library was incubated with the target protein and subjected to high-performance size-exclusion chromatography (HPSEC) to separate bound and unbound ligands. The bound variants were decoded by liquid chromatography-tandem mass spectrometry (LC-MS/MS)23. Synthetic peptide libraries avoid possible loss of sequences due to expression limitation or proteolysis characteristic of biological methods4,24. This in-solution affinity selection is a powerful alternative to on-bead screening and provides fine control over the selection conditions.
Fig. 1. Combinatorial alanine scanning enables identification of alanine-containing peptide binders to proteins of interest.
a A chemically synthesized combinatorial alanine library was generated by split-and-pool solid phase peptide synthesis. The library was cleaved, deprotected, solid phase extracted, and incubated with protein targets. The peptide–protein complex eluate was separated from unbound peptides by size-exclusion chromatography. Bound peptide binders were dissociated from the protein and then sequenced by Q-TOF liquid chromatography-tandem mass spectrometry (LC-MS/MS). b Chemical structure of p5317–28 peptide (ETFSDLWKLLPE-OH) and PMI (TSFAEYWNLLSP-NH2). c Crystal structure of the PMI–MDM2 complex (TSFAEYWNLLS-NH2, PDB entry 3LNZ)23.
The strategy was applied to the PMI peptide inhibitor of oncoprotein mouse double minute 2 homolog (MDM2). The PMI sequence (TSFAEYWNLLSP-NH2) with high affinity (dissociation constant, KD = 7.7 ± 4.5 nM) against MDM2 was discovered by phage display25,26 (Fig. 1b, c). MDM2 is an E3 ubiquitin ligase of the transcription factor p53 that responds to stress by promoting DNA repair, cell cycle arrest, senescence and apoptosis27. MDM2 recognizes the N-terminal p53 transactivation domain (p5317–28, see Fig. 1b for amino acid sequence) and promotes the ubiquitin-mediated p53 degradation. Disrupting p53–MDM2 binding is a strategy to restore p53 activity and promote apoptosis of cancer cells28. Aided by the combinatorial alanine scanning technique developed here, we identified several PMI analogs with simultaneous multi-alanine substitutions that maintained high affinity for MDM2. In some cases, we found the multi-alanine PMI variants lead to active cysteine (Cys)-substituted peptide macrocycles. To expand the application scope, we applied the strategy on peptide binders of antibody 12ca5 and the signalling protein 14-3-3σ, and identified their alanine tolerance.
Results and discussion
The combinatorial alanine scanning platform was developed to identify alanine-containing peptide binders of the target protein (Fig. 1a). A PMI-derived combinatorial alanine library was prepared by split-and-pool SPPS. During synthesis, each position was evenly pooled to give either the wild-type amino acid or alanine, resulting in a library of 4,096 peptide variants. A C-terminal lysine is included to improve de novo LC-MS/MS sequencing. Prior to affinity selection, the library solution was incubated with MDM2 in Tris buffer (pH = 7.5) to reach equilibrium. The peptide–protein complexes were enriched by HPSEC where the early protein fraction eluted. Bound peptides were dissociated from the protein, sequenced by LC-MS/MS and analyzed with the PEAKS software suite23. Identified sequences were filtered based on the library design23. Quality control of the peptide library confirmed the recovery of sequences and even distribution prior to screening. To identify non-specific binders in the PMI-based library, we screened in parallel against the 12ca5 clone of antihemagglutinin antibody, for which no sequences were enriched.
Single position SAR analysis
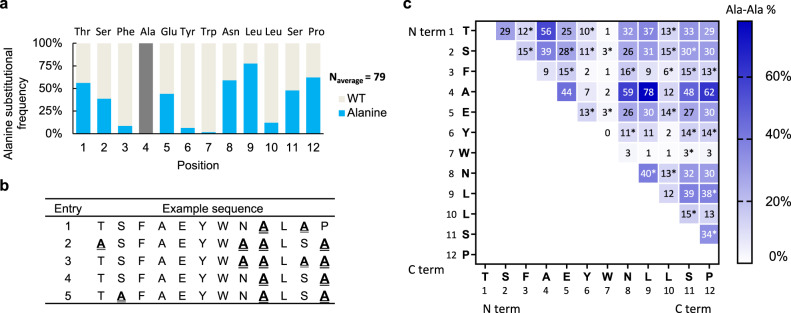
Positional alanine substitution frequency (Fig. 2a) can be used to differentiate binding hotspots from non-essential residues. Unique alanine mutant peptides were recovered from the affinity selection (Fig. 2b). For each position, the alanine frequency was determined by dividing the number of alanine mutations to the total number of identified sequences, averaging by three replicates (Naverage = 79). Four positions Phe3, Tyr6, Trp7 and Leu10 are of low alanine frequency (Ala%<10%), consistent with hotspots determined by point alanine mutagenesis26.
Fig. 2. Combinatorial alanine scanning of peptide PMI combined with affinity selection mass spectrometry identifies multi-alanine substituted variants.
a 1-D single position alanine substitution frequency. The alanine tolerance was indicated by the single position alanine substitution frequency on the y-axis. Residues displaying low alanine substitution frequency (Phe3, Tyr6, Trp7 and Leu10) correspond to the hotspot residues. The x-axis indexes the PMI sequence from the N- to the C-terminus. Ala4 is excluded. b A subset of identified sequences from the affinity selections. c The pairwise alanine tolerance was indicated by the 2-D pairwise alanine substitution frequency matrix. Each box represents the pairwise alanine substitution frequency (Ala-Ala%) of two residues, calculated as the ratio of number of observed simultaneous pairwise alanine substitutions to the total number of identified sequences, expressed as Ala-Ala% = [(nAla,Ala)/ntotal] × 100%. At 95% confidence level, 25 out of 55 total possible pairwise substitution frequencies are non-additive of single alanine frequencies. Non-additive pairs are marked with asterisks (*). Ala4 is excluded from the analysis.
To provide quantitative data on residue-specific contributions to binding affinity, positional alanine frequencies from the combinatorial scanning were converted to changes in Gibbs free binding energy (∆∆Gscanning). This calculation assumes that the ratio of WT to Ala for each position (nWT/nAla) approximates the ratio of equilibrium association constants KA,WT to KA,Ala, such that ∆∆Gscanning is given by: ∆∆GAla-WT = RT ln(KA,WT/KA,Ala) = RT ln(nWT/nAla)19. By comparing the ∆∆GAla-WT values calculated from combinatorial scanning reported here (∆∆Gscanning) to the ∆∆Gbinding values previously measured by point alanine mutagenesis26, we found the two correlated linearly (R2 = 0.88, Supplementary Fig. 1 and Supplementary Note 1). While ∆∆Gscanning for each position derived from the combinatorial scanning correlated well with the previously reported ∆∆Gbinding values, the slope was found to be 0.36. This significant deviation from 1.0 suggests a numerical discrepancy between the ∆∆Gscanning and ∆∆Gbinding and indicates that combinatorial alanine scanning tends to underestimate the effect of individual alanine point mutations. In particular, the ∆∆Gscanning values of the four hotspot residues (Phe3, Tyr6, Trp7 and Leu10) were all >1.0 kcal/mol, consistent with the conventional definition of hotspot8. Therefore, the combinatorial alanine scanning informs on SAR at the single position level.
Pairwise SAR analysis reveals non-additive Ala-substituted pairs
To identify pairs of nonadditive Ala substitutions that might contribute to binding, a pairwise alanine substitution frequency matrix was generated9,14 (Fig. 2c). The pairwise alanine substitution frequency (Ala-Ala%) was computed by dividing the number of simultaneous pairwise alanine substitutions by the total number of identified sequences for each pair. Each box in the matrix presents a distinct pair of residues. For example, the residue pair (Thr1, Ser2) located in the first row and the second column of the matrix shows that 29% of the decoded sequences contained simultaneous (T1A, S2A) substitutions.
A statistical test showed that a number of Ala-Ala%’s were not a mere product of two single Ala%’s but statistically different from such a simple combination. A moderate non-additive combination effect was revealed by comparing the observed pairwise alanine substitution frequency (Ala-Ala%)Observed to the product of single alanine frequencies (Ala-Ala%)Additive. The theoretical additive double-mutant frequencies were computed from a large number of sets (1,000 sets) of randomly-generated and independent alanine-substituted sequences, in which the randomization of each position is weighted by its positional alanine substitution frequency14. Each set contains hypothetical peptide sequences that follow the positional SAR. If a non-additive combination effect is present, (Ala-Ala%)Observed would not be equal to (Ala-Ala%)Additive. To assess the statistical significance of the deviation, we compared the observed (Ala-Ala%)Observed to the theoretical (Ala-Ala%)Additive. The difference (Ala-Ala%)Observed–(Ala-Ala%)Additive was compared to the standard deviation (σ) of the theoretical additive values calculated from the random sets of sequences and assessed by the z-test (Supplementary Fig. 2). To normalize for randomness, the statistical test was averaged by three replicate selections.
At 95% confidence (|z score | >1.96σ), 25 out of 55 (44%) pairwise alanine substitution probabilities are statistically distinct from a simple combination of the corresponding two single alanine substitution probabilities (marked with asterisks in Fig. 2c). Among these 25 non-additive pairs, 23 pairs show positive non-additivity, and 2 pairs show negative non-additivity (Supplementary Fig. 2).
Pairwise SAR validation
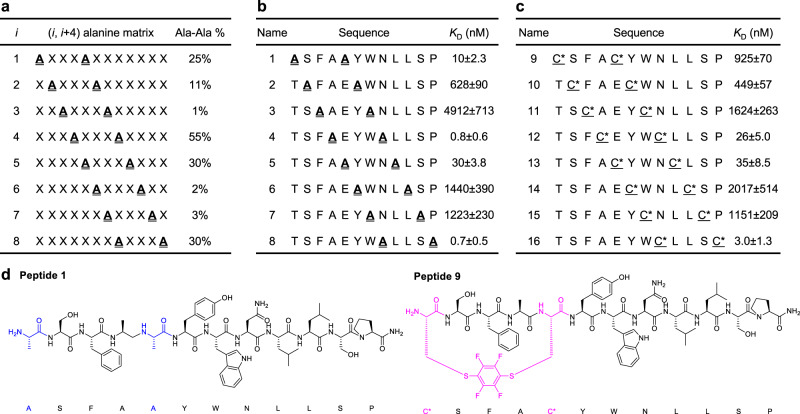
We envisioned the positions displaying high pairwise substitution frequencies in our combinatorial alanine library (Fig. 3a, extracted from Figs. 2c and 3d) would tolerate double mutations. To validate the correlation between pairwise substitution frequencies and binding affinity of double-mutants, a series of (i, i + 4) pairwise alanine substituted peptides were prepared by automated fast-flow peptide synthesis29. (i, i + 4) Positions are chosen for subsequent macrocyclization30. The binding constant was determined by performing a competition assay by biolayer interferometry (BLI). Replacing the above average-frequency (i, i + 4) alanine-substituted pairs to alanine has minor impact on the binding affinity (Fig. 3b; PMI, KD = 7.7 ± 4.5 nM).
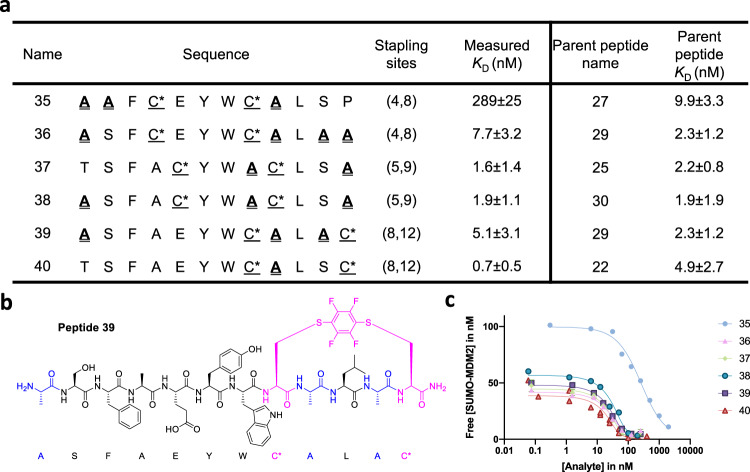
Fig. 3. High-frequency alanine pairs observed in the affinity selection show high tolerance for double mutations.
a (i, i + 4) pairwise alanine substitution frequency extracted from Fig. 2c. b Binding affinity of (i, i + 4) pairwise alanine-substituted peptides. c Binding affinity of (i, i + 4) perfluoroaryl stapled peptides. The three most potent stapled peptides correlate with the three highest frequency pairwise alanine substitutions. Therefore, the (i, i + 4) pairwise alanine substitution frequencies accurately indicate retention of high affinity in peptide binders substituted at the corresponding positions. Binding dissociation constants (KD) were determined by a competition assay using BLI. d Representative chemical structures of an (i, i + 4) pairwise alanine-substituted peptide (1) and an (i, i + 4) perfluoroaryl-stapled peptide (9).
Pairwise alanine-tolerated positions prompt sites amenable to further modifications, i.e., stapling. Side chain stapling is used for reinforcing helical structure and improving peptide stability toward proteases31–33. To leverage the pharmacological properties of the identified MDM2 binders, a hexafluorobenzene-mediated cysteine arylation reaction was employed to generate (i, i + 4) stapled PMI analogs (peptides 9-16)34. BLI competition assays showed that the three highest frequency (i, i + 4) pairwise alanine substitutions (peptides 4, 5 and 8 in Fig. 3c) gave rise to the three highest affinity (i, i + 4) perfluoroaryl-stapled peptide binders at the corresponding positions. Peptides 12 and 13 showed slightly attenuated binding, and 16 exhibited a comparable binding to the parent PMI inhibitor (KD = 7.7 ± 4.5 nM). This result indicates that peptides stapled at high (i, i + 4) alanine frequency positions can maintain low nanomolar binding affinity to MDM2.
Multiple alanine-substituted peptides retain potent binding affinity
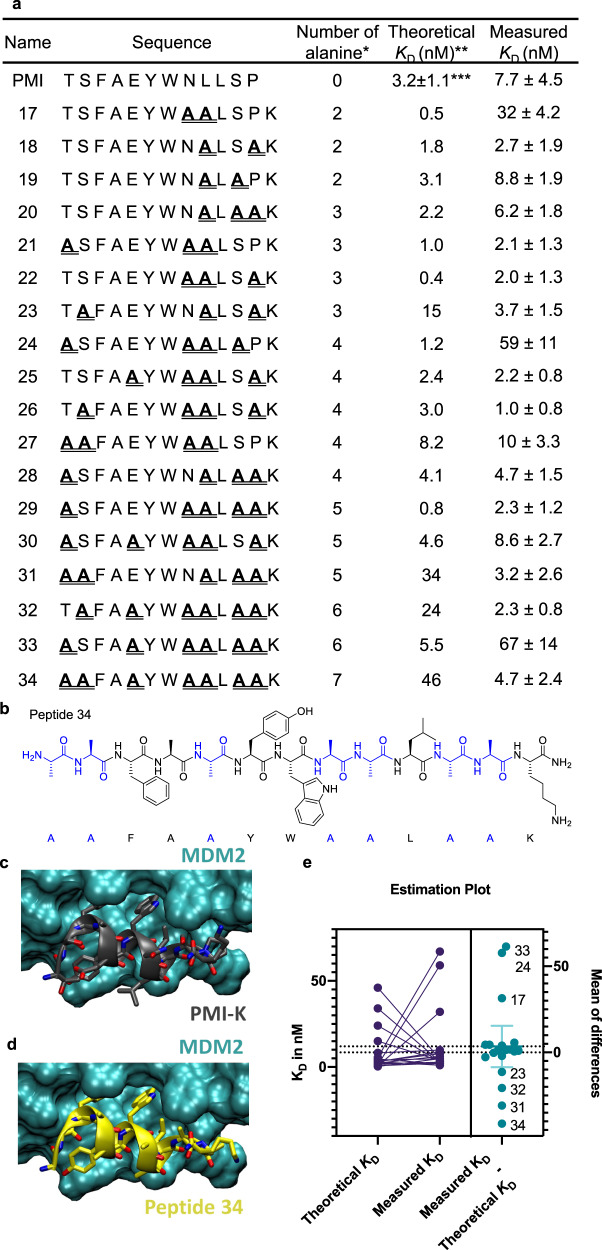
By increasing the selection stringency, a small number of potent multiple alanine-substituted binders were identified (Fig. 4a and Supplementary Method 2). Using a smaller amount of the library increased the ligand–protein binding threshold, and consequently reduced the number of identified peptides. We used the same algorithm shown in Supplementary Figs. 2 and 3 to generate a pairwise matrix of the multiple alanine-substituted peptides (Supplementary Fig. 3). As shown by the matrix, high alanine percentage (Ala% > 45%) was observed with the substitution pairs (T1, L9), (N8, L9), (L9, S11), and (L9, P12). Peptides with different alanine content were individually synthesized, and their binding affinity was validated by a competition assay using BLI. Several peptides exhibited low nanomolar binding affinity. Triple alanine sequences frequently occurred in the most stringent selection (Supplementary Fig. 4). They retained or even enhanced the protein–ligand binding interaction (Fig. 4a).
Fig. 4. Multiple alanine-substituted peptides exhibit nanomolar binding affinity.
a A subset of alanine-substituted peptides were identified and resynthesized and validated after increasing the selection stringency. Dissociation constants (KD) were determined by a competition assay using BLI. *Ala4 is not considered. **Theoretical KD (KD’) was calculated by adding the residue-specific energy contributions of individual alanine mutations (detailed calculation described in Supplementary Fig. 4). ***The reported KD of PMI25. b A 13-mer peptide 34 with seven alanine substitutions was identified (KD = 4.7 ± 2.5 nM). Molecular docking results of (c) PMI-K (gray) and (d) peptide 34 (yellow) bound to MDM2 (cyan). e Paired t-test of theoretical KD’ and measured KD. The left plot shows the KD and KD’ values. The right plot denotes the mean of difference calculated from the paired t-test. Error bar denotes 95% confidence level.
To study the non-additive effect of multi-alanine substituted variants, we calculated the theoretical binding affinity (KD’) by adding ∆∆Gbinding, the residue-specific energy contribution of alanine mutations, of all substituted positions for a given peptide (Supplementary Fig. 5) and using the reported affinity of PMI (KD = 3.2 ± 1.1 nM26). The theoretical binding affinity KD’ falls under the additivity assumption. Then we compared the theoretical KD’ to the measured KD of alanine substituted variants by running a paired t-test (Fig. 4c). At 95% confidence, 7 out of 19 (37%) variants showed significant difference in the theoretical KD’ and measured KD. Three variants showed negative non-additivity while four variants showed positive non-additivity.
To analyze the non-additivity observed in the multi-alanine variants with a complex substitution pattern, we analyzed the 7 variants. Comparing the theoretical and measured binding affinities, peptides 17, 22 and 24 displayed ten-fold higher experimental KD values, while peptides 31, 32, 33 and 34 exhibited ten-fold lower experimental KD values relative to the calculated KD’ (Fig. 4a). Within the three negative non-additive variants, they all share N8A and L9A substitutions. The pairwise matrix indicated that the (N8, L9) pair is negatively non-additive, therefore, this pair may contribute to the non-additive decrease in binding affinity of peptide 17, even though it only contains a smaller number of alanine substitutions. On the other hand, the four positive non-additive variants all contain the (S11, P12) Ala substitution pair, which may contribute to the non-additive increase in the binding affinity of peptides 31, 32, and 34. The theoretical KD’ given by simple addition of positional ∆∆Gbinding can thus deviate significantly from the measured KD, and may not accurately predict high-affinity multi-alanine substituted PMI variants.
Noticeably, peptide 34 has all of its non-hotspot residues substituted to alanine, while still maintaining a binding affinity comparable to PMI (Fig. 4b). Single alanine mutagenesis predicts the KD of peptide 34 to be 10-fold weaker than PMI, while peptide 34, shows a KD of 4.7 ± 2.4 nM. Rigid molecular docking of peptide 34 to MDM2 using AutoDock Vina (except the side chains of the peptide, which were kept flexible) also shows that the calculated binding affinity was improved upon Ala substitutions at all seven non-hotspot residues, with a ∆∆G of –3.4 kcal/mol for PMI and –2.5 kcal/mol for PMI-Lys (Fig. 4c, d, Supplementary Table 1 and Supplementary Method 3). Even though it is generally accepted that multiple alanine substitutions are detrimental to binding affinity35, in our hands multi-alanine-substituted MDM2 binding peptides could be identified in the enriched sequences.
Multi-alanine substituted variants tolerate stapling
To test the tolerance of multi-alanine-substituted peptides for subsequent modifications, we applied the (i, i + 4) perfluoroaryl stapling strategy to alanine-substituted sites of peptides 22, 25, 27, 29 and 30 (Fig. 5)34. Three stapling positions, (4, 8), (5, 9) and (8, 12), were chosen based on the tolerance for pairwise substitutions. The highest affinity (KD = 0.7 ± 0.5 nM) was found with the (8,12) perfluoroaryl-stapled peptide 40. Except stapled peptide 35, stapled peptides 36, 37, 38 and 39 showed comparable binding affinity to PMI.
Fig. 5. Perfluoroaryl-stapled and alanine-substituted peptides demonstrate high-affinity binding.
a Perfluoroaryl-stapled and alanine-substituted peptides 35–40 with low nanomolar binding affinity were generated at tolerable (i, i + 4) stapling sites (Fig. 3c). b Structure of peptide 39, alanine is colored in purple and stapling sites are colored in blue. c Example competition binding curves of peptides. C* = cysteine stapled with hexafluorobenzene.
To assess the stability of stapled peptides, we performed a serum stability assay (Supplementary Fig. 6)31,36,37. Three stapled peptides were selected for assessment. Compared with the unmodified PMI-NH2 peptide (t1/2 = 2.2 min), linear peptide 32 (t1/2 = 3.9 min) and 34 (t1/2 = 5.1 min), the half-life of stapled peptides 36, 38 and 39 increased to 26, 27 and 25 min, respectively (Supplementary Fig. 6).
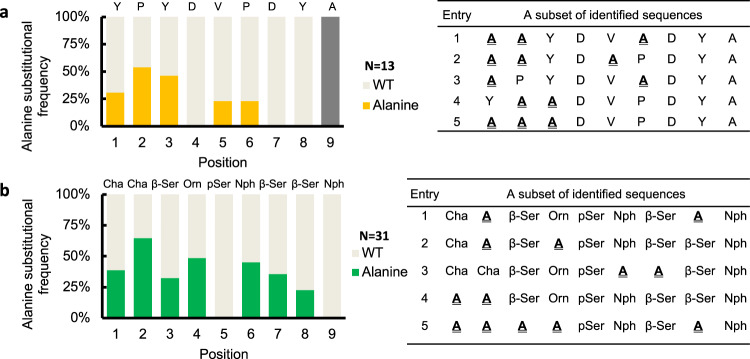
To demonstrate a more general applicability of this platform, in addition to MDM2, the combinatorial alanine scanning was also applied to peptide binders of antibody 12ca5 and the signalling protein 14-3-3σ. The epitope used for 12ca5 has the sequence YPYDVPDYA; the previously characterized protein 14-3-3σ binder 14-3-3.6 was used for 14-3-3σ and its sequence can be found in Fig. 6b. A beta-alanine spacer was used between the library construct and the C-terminal lysine. The combinatorial alanine scanning of YPYDVPDYA (KD = 4 nM) showed no alanine substitution at Asp4, Asp7 and Tyr8, which were previously reported as the hotspot residues of the epitope (Fig. 6a)26. The combinatorial alanine scanning of peptide 14-3-3.6 (KD = 3 nM) showed no alanine replacement at phosphoserine 5 and nitro-phenylalanine 9 (Fig. 6b), consistent with the close interactions these two residues have with 14-3-3σ, as seen in the co-crystal structure38. Remarkably, multiple alanine-substituted binders were identified under the most stringent condition as shown in the subset of sequences (Fig. 6).
Fig. 6. Combinatorial alanine scanning is applicable to peptide binders for antibody 12ca5 and regulatory protein 14-3-3σ.
a Alanine substitution frequency at each position of the 12ca5 binder HA epitope (sequence: YPYDVPDYA). b Alanine substitution frequency at each position of the protein 14-3-3σ binder 14-3-3.6 construct (sequence: Cha-Cha-β-Ser-Orn-pSer-Nph-β-Ser-β-Ser-Nph); abbreviations: Cha cyclohexyl alanine, β-Ser beta-homoserine, Orn ornithine, pSer phosphoserine, Nph 4-nitro phenylalanine. Wild-type alanine is excluded from the bar graph.
Conclusions
We developed a label-free combinatorial alanine affinity selection platform to establish mutational tolerance, inform structure–activity relationships, and facilitate the optimization of peptide-based PPI modulators. Using various statistical analyses and peptide modifications, several sequence activity relationships were inferred. At the single substitution level, the determined alanine substitution frequencies differentiated between hotspot and non-hotspot residues of the MDM2 peptide binder PMI. At the double substitution level, statistical analyses of the pairwise alanine substitution frequencies identified a moderate but significant non-additive combination effect. We can identify suitable sites that can tolerate simultaneous mutations and stapling based on high pairwise alanine substitution percentages, as validated by binding experiments with (i, i + 4) doubly-alanine-substituted peptides and (i, i + 4) stapled peptides.
A handful of multiple alanine-substituted binders were found to maintain their binding affinity. This finding occurred when alanine was combinatorically introduced to peptide libraries that underwent affinity selections. The multi-substituted peptides can further tolerate cysteine-based stapling and still retain nanomolar binding affinity, despite the fact that stapling can potentially change the binding affinity of peptides30. This study shows the potential of the combinatorial alanine scanning platform to identify multiple positions within a sequence that can simultaneously accommodate further modifications. These high affinity stapled peptides may further be modified at the remaining alanine residues and provide a starting point in the development of the next generation of therapeutics targeting the oncogenic p53–MDM2 interaction.
Our study demonstrates a broad alanine tolerance landscape of peptide-based binders, provides insights into the non-additivity of combinatorial substitutions, and identifies multi-alanine-substituted MDM2-binding peptides. These variants can guide peptide ligand optimization through chemical stapling and improve the throughput of hit-to-lead optimization processes. Albeit these peptide variants are synthetically accessible, their sequencing may be limited by solubility and ionization efficiency, to the extent that library quality control studies are necessary. We envision that combinatorial alanine scanning can be a valuable tool for determining the feasible structural modifications of potentially therapeutic peptide leads toward enhancing their biophysical properties, such as binding affinity, serum stability, rigidity, and lipophilicity.
Methods
Materials
Unless otherwise noted, all commercial chemicals and reagents were used without further purification. H-Rink Amide-ChemMatrix resin was purchased from PCAS BioMatrix Inc. TentaGel S NH2 (130 μm, 0.2-0.35 mmol/g) was purchased from Rapp Polymere GmbH (Tuebingen, Germany). Linkers: 4-[(R,S)-α-[1-(9H-Fluoren-9-yl)-methoxyformamido]-2,4-dimethoxybenzyl]-phenoxyacetic acid (Fmoc-Rink amide linker) was purchased from Chem-Impex International (Wood Dale, IL). Amino acids: Fmoc-Ala-OH, Fmoc-β-Ala-OH, Fmoc-Asn(Trt)-OH, Fmoc-Asp(tBu)-OH, Fmoc-Cys(Trt)-OH, Fmoc-Glu(tBu)-OH, Fmoc-Leu-OH, Fmoc-Lys(Boc)-OH, Fmoc-Phe-OH, Fmoc-Pro-OH, Fmoc-Ser(tBu)-OH, Fmoc-Thr(tBu)-OH, Fmoc-Trp(Boc)-OH, Fmoc-Tyr(tBu)-OH, and Fmoc-Val-OH were purchased from Novabiochem (Billerica, MA). Noncanonical amino acids: Fmoc-β-cyclohexyl-L-alanine (Cha), Fmoc-O-tert-butyl-L-β-homoserine (β-Ser), Fmoc-L-Orn (Boc)-OH (Orn), Fmoc-Ser(PO(OBzl)OH)-OH (pSer) and Fmoc-4-nitro-L-phenylalanine (Nph) were purchased from Chem-Impex International (Wood Dale, IL). N-α-Fmoc-O-benzyl-L-phosphoserine was purchased from MilliporeSigma (St. Louis, MO). Reagents used in solid phase peptide synthesis: Piperidine (ReagentPlus; 99%), formic acid (≥98%) were purchased from Sigma-Aldrich (St. Louis, MO). Diisopropylethylamine (DIEA; biotech. grade; 99.5%) was purchased from MilliporeSigma and purified by a Seca Solvent Purification system from Pure Process Technology (Nashua, NH). Reagents for peptide cleavage: Trifluoroacetic acid (TFA; for HPLC, ≥ 99%), triisopropylsilane (TIPS; 98%), 1,2-ethanedithiol (EDT; ≥ 98%), phenol (≥99%), and thioanisole (≥99%) were purchased from Sigma-Aldrich (St. Louis, MO). Reagents used in peptide post-synthesis modifications: hexafluorobenzene (≥99%) was purchased from TCI (Portland, OR), 2-Amino-2-(hydroxymethyl)propane-1,3-diol Tris (base) was purchased from J.T. Baker (Center Valley, PA). Acetic anhydride (≥98%) was purchased from Sigma-Aldrich (St. Louis, MO). Solvents: Deionized water was obtained from a Milli-Q Reference water purification system (Millipore). N,N-Dimethylformamide (DMF; peptide synthesis grade), dichloromethane (DCM), diethyl ether (Et2O), and acetonitrile (MeCN; HPLC grade) were purchased from VWR International (Philadelphia, PA). Proteins: Mouse anti-hemagglutinin (HA) monoclonal antibody clone 12ca5 (anti-HA mAb 12ca5) was purchased from Columbia Biosciences (Frederick, MD). Bovine serum albumin (BSA; RIA grade) was purchased from Amresco (Solon, OH). 14-3-3sigma was kindly provided by Professor Christian Ottmann38. Buffers and other reagents: Phosphate-buffered saline 10x (10x PBS, Molecular biology grade) was purchased from Corning (Manassas, VA). Tween 20 (reagent grade) was purchased from Amresco (Solon, OH). Trichloroacetic acid (ACS grade, TCA) was purchased from MilliporeSigma (St. Louis, MO). Bradford assay kit purchased from Sigma-Aldrich (St. Louis, MO). Human serum was purchased from MilliporeSigma (St. Louis, MO).
SUMO-25-109MDM2 expression
The plasmid construct and E. coli expression of SUMO-25-109MDM2 were achieved39. In brief, the 25-109MDM2 gene was purchased from Addgene (pGEX-4T MDM2 wild type (WT), 16237). The SUMO tag was incorporated using the Champion™ pET SUMO Expression System (Invitrogen, CA). SUMO-25-109MDM2 was expressed in Rosetta (DE3) pLysS cells. The bacteria were inoculated and cultured for 6 hours to reach OD600 = 0.5 under ambient temperature, induced with 0.4 mM IPTG for 4 hours, and pelleted. Approximately 1 L broth produced 10 g cell pellet, which was resuspended in 30 mL of 50 mM Tris-HCl, 150 mM NaCl, pH 7.5 buffer containing 40 mg lysozyme, 1 mg Roche DNAase I, and one tablet of Roche protease inhibitor cocktail, and then sonicated for three times for 20 s. The suspension was then centrifuged at 30,000 rcf for 30 min to clarify the lysate. The supernatant was loaded onto a 5 mL HisTrap FF crude Ni-NTA columns (GE Healthcare, UK), and washed sequentially with 30 mL of 20 mM Tris-HCl, 150 mM NaCl, pH 8.5 and then 30 mL 20 mM Tris-HCl 150 mM NaCl, 80 mM imidazole pH 8.5. The protein was eluted with 10 mL 20 mM Tris-HCl, 500 mM imidazole, 500 mM NaCl, pH 8.5. The eluted protein was buffer exchanged into 20 mM Tris-HCl, 50 mM NaCl, pH 8.5 using a HiPrep 26/10 Desalting column (GE Healthcare, UK). The resulting protein solution was purified the same day using a 5 mL HiTrap Q HP (GE Healthcare, UK) anion exchange columns with a linear NaCl gradient (B% graded from 5% to 25%, B = 20 mM Tris-HCl, 500 mM NaCl, pH 8.5). Fractions containing pure SUMO-25-109MDM2, as determined by SDS-PAGE, were collected and concentrated to 0.5 mg/mL using a 3,000 Da nominal molecular weight limit Amicon Ultra-15 Centrifuge Filter Unit (EMD Millipore), and immediately flash frozen and stored at −80 °C.
Peptide synthesis
H-Rink Amide-ChemMatrix resin (200 mg, 0.49 mmol/g, 0.10 mmol) was used to prepare peptides. Peptides containing noncanonical amino acids were prepared by manual solid-phase peptide synthesis. Peptides without noncanonical amino acids were prepared by fully automated SPPS29. Upon completion, resins were washed with DCM (3x) and dried under reduced pressure.
Solid-phase peptide synthesis
200 mg of H-Rink Amide-ChemMatrix resin (0.10 mmol) was placed in a 5 mL Torviq fritted syringe. After swelling in DMF for 10 min, the resin was then washed with DMF (3x). Each Fmoc-protected amino acid (0.50 mmol, 5.0 eq) was dissolved in 0.38 M HATU solution (DMF as solvent, 1.3 mL, 0.50 mmol). Immediately before the coupling, DIEA (130 μL, 0.72 mmol, 6.9 eq) was added to the mixture. The resulting mixture was sonicated briefly, and then transferred to the fritted syringe containing the resin. Coupling was performed for 20 min. The resin was washed with DMF (3x). Fmoc deprotection was done by treating the resin with 20% piperidine in DMF (3 mL) for 5 min and this step was repeated twice. The resulting resin was then washed with DMF (3x). The synthetic cycle was repeated to completion of the peptide sequence.
Cleavage of peptides without cysteine
Synthesized peptide was cleaved from resin and globally deprotected by treating the peptidyl resin with a cleavage cocktail containing 94% TFA, 2.5% EDT, 2.5% water, and 1.0% TIPS (v/v), for 1 h at ambient temperature. TFA was removed under a gentle stream of nitrogen gas, and crude peptide was precipitated by the addition of cold Et2O (−80 °C). After centrifugation at 3220 rcf for 3 min, supernatant was removed, and precipitated peptide was triturated three times with cold Et2O. The resulting material was dissolved in 50% MeCN in water with 0.1% TFA, and lyophilized.
Cleavage of cysteine-containing peptides
Synthesized peptide was cleaved from resin and globally deprotected by treating the peptidyl resin with reagent K containing 82.5% TFA, 5.0% phenol, 5.0% water, 5.0% thioanisole, and 2.5% EDT (v/v), for 1 h at ambient temperature. TFA was removed under a gentle stream of nitrogen gas, and crude peptide was precipitated by the addition of cold Et2O (−80 °C). After centrifugation at 3220 rcf for 3 min, supernatant was removed, and precipitated peptide was triturated three times with cold Et2O. The resulting material was dissolved in 50% MeCN in water (v/v) with 0.1% TFA, and lyophilized.
Purification of crude peptide
Crude peptides were purified by a Biotage Selekt flash purification system. Water with 0.1% TFA (solvent A) and MeCN with 0.1% TFA (solvent B) were utilized as mobile phases for purifications. The crude peptide was dissolved in minimal amount of 10% MeCN in water with 0.1% TFA, and then loaded onto a 10 g Biotage SNAP Bio C18 20 μm column. The purification was performed using a gradient as following: 10% B for 3 column volume (CV), linear ramp from 10% B to 70% B for 30 CV, 25 mL/min flow rate.
Peptide acetylation
A 100 mg portion of peptidyl resin was placed into a 5 mL Torviq fritted syringe and subsequently swelled in DMF. After removing DMF, a solution of Ac2O, DIEA, and DMF (2 mL, 85:315:1600, v/v) was added to the peptidyl resin. The resulting mixture was occasionally agitated for 45 min. After draining the solution, the remaining resin was washed with DMF (3x), DCM (3x) and dried under reduced pressure.
Peptide stapling
Crude cysteine-containing peptide (~14 mg, 0.010 mmol) was placed in a falcon tube, followed by the addition of DMF (1 mL). 200 mM hexafluorobenzene (DMF as solvent, 2.5 mL) and 150 mM Tris base (DMF as solvent, 1.5 mL) were added to the falcon tube. The tube was sealed and mixed on a nutating mixer for 4 h. The mixture was quenched by adding water with 0.1% TFA (20 mL, v/v), and then purified by a Biotage Selekt flash purification system with the gradient: 25% B for 3 CV, linear ramp from 25% B to 75% B for 30 CV; 25 mL/min flow rate.
LC-MS characterization
LC-MS characterizations were carried out using an Agilent 6520 quadrupole time-of-flight LC-MS or an Agilent 6550 quadrupole time-of-flight LC-MS. The Agilent 6520 ESI-QTOF mass spectrometer was placed in-line with an Agilent 1290 Infinity liquid chromatography system, whereas the Agilent 6550 ESI-QTOF mass spectrometer was placed in-line with an Agilent 1290 Infinity LC system. Total ion current (TIC) chromatograms were plotted. Mass spectra were integrated over the principal TIC peaks. High-performance liquid chromatography was done by following methods: (solvent A: water with 0.1% formic acid; solvent B: MeCN with 0.1% formic acid). Method A: Column: Phenomenex Luna C18(2) column (0.5 ×150 mm, 3 μm particle size, 100 Å pore size). Gradient: 1% B (A-B min), linearly ramp from 1% B to 61% B (B-C min), 61% B to 95% B (C-D min). Flow rate is 50 μL/min. MS acquisition is from 4 to 14 min. Method B: Column: Agilent Zorbax 300SB C18 column (2.1 ×150 mm, 5 μm particle size, 300 Å pore size). Gradient: 1% B (0-2 min), linearly ramp from 1% B to 61% B (2-11 min), 61% B to 95% B (11-12 min). Flow rate is 0.6 mL/min. MS acquisition is from 4 to 14 min. Method C: Column: Phenomenex Jupiter C4 column (1.0 ×150 mm, 5 μm particle size, 300 Å pore size). Gradient: 1% B (0-2 min), linearly ramp from 1% B to 91% B (2-18 min), 61% B to 91% B (18-21 min). Flow rate is 100 μL/min. MS acquisition is from 4 to 18 min. Method D: Column: Phenomenex Jupiter C4 column (1.0 ×150 mm, 5 μm particle size, 300 Å pore size). Gradient: 1% B (0-2 min), linearly ramp from 1% B to 61% B (2-12 min), 61% B to 95% B (11-16 min). Flow rate is 100 μL/min. MS acquisition is from 4 to 12 min. Method E: Column: Agilent Zorbax 300SB C3 column (2.1 ×150 mm, 5 μm particle size, 300 Å pore size). Gradient: 1% B (0-2 min), linearly ramp from 1% B to 91% B (2-12 min), 91% B to 91% B (12-13 min). Flow rate is 500 μL/min. MS acquisition is from 4 to 12 min. LC-MS characterization data are shown in Supplementary Note 2.
Representative protocol for split-and-pool solid phase peptide synthesis
Coupling
400 mg of 130 μm TentaGel resin (0.26 mmol/g, 0.10 mmol, 782770 beads/g, 3.13 ×105 beads) was placed in a 10 mL polypropylene syringe containing a porous polypropylene disc (Torviq). After swelling in DMF for 10 min, the resin was washed with DMF (3x). Fmoc-Rink amide linker (560 mg, 1.0 mmol, 10.0 equiv) was dissolved in 0.38 M HATU solution in DMF (2.4 mL, 0.49 mmol, 9.0 equiv), activated with DIEA (544 μL, 1.56 mmol, 30.0 equiv). The mixture was sonicated briefly, transferred to the fritted syringe containing the resin, and allowed to stir for 20 minutes. After the coupling solution was drained, and the resin was washed with DMF (3x). Fmoc protection group was achieved by soaking the resin with 20% piperidine in DMF (6 mL) for 5 min for twice, and then washed with DMF (3x). A sequential Fmoc-Lys(Boc)-OH coupling, Fmoc removal and DMF washes were performed in the same manner.
Portioning
The peptidyl resin was suspended in DMF (6 mL), homogenized, then evenly divided among two 10 mL Torviq fritted syringes. After draining DMF from the syringes, the couplings were carried out as follows: Fmoc-protected WT amino acid and Fmoc-Ala-OH (0.26 mmol) were separately dissolved in 0.38 M HATU (0.66 mL, 0.26 mmol), activated with DIEA (136 μL, 0.36 mmol). Each of the activated amino acids was added to the individual resin-containing syringes (~200 mg, 0.052 mmol). After coupling for 20 min, the coupling solution was removed, and the resin was washed with DMF (3x). Then, all portions of peptidyl resin were combined and washed with DMF. Fmoc deprotection was achieved by treating the resin with 20% piperidine in DMF (6 mL, 5 min batch treatment), and this step was repeated twice. The resulting resin was then washed with DMF (3x). Twelve cycles of split-and-pool synthesis were performed using this procedure.
Cleavage from resin and global side chain deprotection
The library was cleaved from resin and globally deprotected by treating the peptidyl resin with a cleavage cocktail containing 94% TFA, 2.5% EDT, 2.5% water, and 1.0% TIPS (v/v), for 1 h at ambient temperature. TFA was removed under a gentle stream of nitrogen gas, and the crude peptide was precipitated using cold Et2O (−80 °C). After centrifugation at 3220 rcf for 3 min, the supernatant was removed, and the precipitated peptide was triturated three times with cold Et2O. The resulting material was dissolved in 50% MeCN in water with 0.1% TFA, and lyophilized.
Solid phase extraction
The library (60 mg) was dissolved in 6.0 mL of 5% MeCN in water with 0.1% TFA. 6 mL Bond Elut C18 cartridge (Agilent, P/N 12256130) was used for the solid phase extraction. A cartridge was conditioned with MeCN with 0.1% TFA (~10 mL), and then equilibrated with 1% MeCN in water with 0.1% TFA (~10 mL). Afterward, the library solution was loaded, and the cartridge was washed with 1% MeCN in water with 0.1% TFA (~10 mL). Sample elution was achieved by passing 70% MeCN in water with 0.1% TFA (~10 mL) through the cartridge. The final eluate was collected separately and lyophilized.
HPSEC-based in-solution affinity selection
High-performance size exclusion chromatography (HPSEC) was carried out using an Agilent 1260 Infinity II LC System with the Agilent BIOSEC-3 HPLC column (7.8 × 150 mm, 3 μm particle size, 100 Å pore size). HPSEC samples, such as proteins, libraries, or protein-library mixtures (30 min incubation at 4 °C), were prepared in 100 μL buffer, and then eluted in buffered mobile phase at 1 mL/min flow rate for 15 min. While performing affinity selection experiments, the protein-binder complex fraction was detected by UV (214 and 280 nm) and collected. Prior to the LC-MS/MS-based de novo peptide sequencing, the protein-binder fraction was lyophilized. After the HPSEC affinity selection experiments, SEC column was cleaned with an IPA/water/MeCN/MeOH (1:1:1:1, v/v) mixture containing 0.1% formic acid (FA).
Method of adjusting affinity selection conditions
Stringency of affinity selection is adjusted by the ratio of protein and peptide library. Using our HPSEC method, a less stringent selection condition is achieved with a molar ratio of peptide:protein = 1:10, e.g., protein concentration 10 pM, peptide library per member concentration 1 pM, volume 100 µL. For a stringent selection condition, molar ratio of peptide:protein = 1:1000, e.g., protein concentration 10 pM, peptide library per member concretion 10 fM, volume 100 µL.
Method for de novo peptide sequencing
The lyophilized sample was dissolved in 50 μL water containing 0.2% FA and 10% trichloroacetic acid (TCA), and then subjected to LC-MS/MS using an Agilent 6550 quadrupole time-of-flight LC-MS with an Agilent Zorbax 300SB C3 column (2.1 × 150 mm, 5 μm particle size, 300 Å pore size). Mobile phases were water with 0.1% FA (solvent A) and MeCN with 0.1% FA (solvent B). A linear gradient of 1% B to 61% B (34 min, flow rate: 0.5 mL/min) was used to perform liquid chromatography. Absolute MS/MS threshold was typically set to 1500 counts and selected precursor ions had 2+ and 3+ charges. MS/MS spectra were imported and analyzed using PEAKS Studio software from Bioinformatics Solutions. Quality control of the peptide library was conducted prior to screening (Supplementary Fig. 7 and Supplementary Table 3).
Method of expected value calculation
The expected pairwise alanine percentage (Ala-Ala%)Additive is calculated as the product of single alanine frequencies (Ala-Ala%)Additive. For example, the (Thr1, Ser2) pair has an expected (Ala-Ala%)Additive = 0.56 × 0.39 = 0.22 (22%). The calculation is repeated for the other possible pairs of residues in the peptide sequence.
Method of calculating standard deviation
To compute the potential error distribution in the expected pairwise alanine frequencies, we generated large sets (1000 sets) of randomized sequences. Each set contains several random sequences. The total number of sequences in each set is equal to the total number of sequences enriched from the experiment. The sequences were generated by a random generator weighted by the positional alanine substitutional frequencies. The theoretical additive double-mutant frequencies were computed from the 1000 sets of randomly-generated and independent alanine-substituted sequences, in which the randomization of each position is weighted by its positional alanine substitution frequency14. From the 1000 output matrices, we calculated the standard deviation (σ) for alanine-substituted pairs and the mean value of the pairwise substitutional frequencies. These pairwise substitutional frequencies represent the expected values that are numerically equal to the product of positional alanine frequencies.
With the experimentally observed sequences, we computed the observed pairwise alanine frequencies, then divided the simultaneous alanine substitutional frequencies to the total number of sequences, (Ala-Ala%)Observed. To evaluate the difference between the expected pairwise alanine substitutional frequencies and the observed alanine substitutional frequencies, we divided the difference by the standard deviation values obtained in the randomization process. This difference, or (Ala-Ala%)Observed–(Ala-Ala%)Additive was compared to the standard deviation (σ) of the theoretical additive values calculated from the random sets of sequences and assessed by the z-test.
Method of statistical model
To provide quantitative data on residue-specific contributions to binding affinity, positional alanine frequencies from the combinatorial scanning were converted to changes in Gibbs free binding energy (∆∆Gscanning). This calculation assumes that the ratio of WT to Ala for each position (nWT/nAla) approximates the ratio of equilibrium association constants KA,WT to KA,Ala, such that ∆∆Gscanning is given by: ∆∆GAla-WT = RT ln(KA,WT/KA,Ala) = RT ln(nWT/nAla)19. By comparing the ∆∆GAla-WT values calculated from combinatorial scanning reported here (∆∆Gscanning) to the ∆∆Gbinding values previously measured by point alanine mutagenesis26, we found the two correlated linearly (R2 = 0.88, Supplementary Fig. 1).
Theoretical KD analysis
The theoretical KD’ was computed by linear addition of the KD values of single alanine mutants. Assuming substitution follows additivity, we calculated the theoretical KD’ by adding up the binding free energy of single alanine substituted peptides. Therefore, theoretical KD’ is given by the equation: RT ln(KD’/KD,PMI) = ∑ (∆∆Gbinding of single alanine mutations) = RT ln(Π KD ratio). KD,PMI = 3.2 ± 1.1 nM3, as reported (Supplementary Table 2). For example, peptide 17 (TSFAEYWAALSPK) has a theoretical KD’ given by the equation: RT ln(KD’/KD,PMI) = RT ln(0.2 0.8), such that the theoretical KD’ = 0.2 0.8 3.2 nM = 0.5 nM.
In-solution competition assay
A competition binding assay was performed as described below using ForteBio Octet RED96 BLI system (Octet RED96) to estimate the binding affinity of peptides.
Calibration curve
Streptavidin sensors were soaked in blocking buffer (PBS supplemented with 0.05% Tween-20 and 1 mg/mL bovine serum albumin) for 5 min. After immobilizing the PEG4-biotinylated15–29 p53 peptide (100 nM of Biotin-PEG4-SQETFSDLWKLLPEN) onto streptavidin sensors, serial dilutions of SUMO-25-109MDM2 in blocking buffer were analyzed for binding. The response was recorded at equilibrium after 2 min. A curve of sensor response (nm) vs. MDM2 concentration (nM) was generated to calibrate the free MDM2 concentration in solution observed in the competition assay. The curve was generated using Prism 8.
Competition assay
Various concentrations of peptides were incubated in wells with a constant concentration of SUMO-25-109MDM2 protein (either ~50 nM or ~100 nM, as described for each peptide in the Supplementary Method 2) in the blocking buffer for 30 min. The PEG4-biotinylated p5315–29 peptide was immobilized onto streptavidin sensors and dipped into preincubated sample wells. The association events were measured at 30 °C, 1,000 rpm. Response at equilibrium after 2 min was recorded. Based on the binding response (nm) values, the concentration of ‘free’ SUMO-MDM2 (Y) was interpolated for each sample using the calibration curve (Supplementary Method 4). Nonlinear regression analysis was performed using the GraphPad Prism 8 software to estimate the KD value based on the equation:
, where Y is ‘free’ [SUMO-MDM2] in nM determined as described above, X is the total [peptide] in nM, KD is the dissociation constant in nM, and b is the maximum [SUMO-MDM2] value fitted by the equation (Ymax), equivalent to the total SUMO-MDM2 concentration provided in the assay. Reorganizing the quadratic equation for Y we obtain: Y2 + (KD + X − b) × Y − b × KD = 0. Solving the equation for the positive Y value, we arrived at the following equation used to generate the fitted curves and calculate KD: Y = 0.5 × [b − KD − X + ((KD + X − b)2 + 4b × KD)0.5]. In fitting this equation, the parameters KD and b (Ymax, the maximum possible protein concentration) were fitted by the software. The fitted Ymax value was compared in each case to the nominal total protein concentration used in the assay. After the regression analysis, we set the following criteria for the fit to be considered acceptable: (1) the fitted Ymax value should be within 10% of the nominal total SUMO-MDM2 concentration, (2) the fitted curve should reach a plateau at the lowest peptide concentration(s) used in the fit. The Ymax value was not fixed in any case. If either of the two criteria were not met for a given fit, the binding study was repeated with fresh solutions of the peptides and protein.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Financial support for this work was provided by Calico Life Sciences (to B.L.P.). Y.-C.L. gratefully acknowledges support from a Deutsche Forschungsgemeinschaft postdoctoral fellowship (DFG, LE 4224/1-1). We thank Bente A. Somsen and Christian Ottmann for generously providing the 14-3-3 protein, Anthony J. Quartararo for helpful discussions and suggestions, and Andrei Loas for valuable comments during manuscript preparation. We acknowledge the Biophysical Instrumentation Facility at MIT for providing access to the Octet Red96 Bio-Layer Interferometry System (NIH S10OD016326). Y.L., J.C.M., and Y.-S.L. thank the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health for financial support under award R01-GM124160 (to Y.-S.L.), the support of the Tufts IRACDA Program (NIGMS award K12-GM133314) for J.C.M, and the support of Laidlaw Undergraduate Research and Leadership Program at Tufts University for Y.L.
Author contributions
X.Y., Y.-C.L., Z.P.G., Y.-S.L. and B.L.P. designed research; X.Y., Y.-C.L., Z.P.G., Y.L., J.C.M. and F.-S.Y. performed the experiments; X.Y., Y.-C.L., Z.P.G., F.-S.Y., Y.-S.L. and B.L.P. analyzed data; X.Y., Y.-C.L., Y.L., J.C.M. and F.-S.Y. generated the figures; X.Y. and Y.-C.L. wrote the manuscript with input from all authors.
Peer review
Peer review information
Communications Chemistry thanks Jian Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
All data generated during this study are available either in the main text or supplementary materials.
Competing interests
B.L.P. is a co-founder and/or member of the scientific advisory board of several companies focusing on the development of protein and peptide therapeutics. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiyun Ye, Yen-Chun Lee.
Supplementary information
The online version contains supplementary material available at 10.1038/s42004-022-00737-w.
References
- 1.Nevola L, Giralt E. Modulating Protein-Protein Interactions: The Potential of Peptides. Chem. Commun. 2015;51:3302–3315. doi: 10.1039/C4CC08565E. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham AD, Qvit N, Mochly-Rosen D. Peptides and Peptidomimetics as Regulators of Protein–Protein Interactions. Curr. Opin. Struct. Biol. 2017;44:59–66. doi: 10.1016/j.sbi.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee ACL, Harris JL, Khanna KK, Hong JH. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019;20:2383. doi: 10.3390/ijms20102383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sohrabi C, Foster A, Tavassoli A. Methods for Generating and Screening Libraries of Genetically Encoded Cyclic Peptides in Drug Discovery. Nat. Rev. Chem. 2020;11:801–807. doi: 10.1038/s41570-019-0159-2. [DOI] [PubMed] [Google Scholar]
- 5.Derda R, Jafari MR. Synthetic Cross-Linking of Peptides: Molecular Linchpins for Peptide Cyclization. Protein Pept. Lett. 2018;25:1051–1075. doi: 10.2174/0929866525666181120090650. [DOI] [PubMed] [Google Scholar]
- 6.Bhat A, Roberts LR, Dwyer JJ. Lead Discovery and Optimization Strategies for Peptide Macrocycles. Eur. J. Med. Chem. 2015;94:471–479. doi: 10.1016/j.ejmech.2014.07.083. [DOI] [PubMed] [Google Scholar]
- 7.Rogers JM, Passioura T, Suga H. Nonproteinogenic Deep Mutational Scanning of Linear and Cyclic Peptides. Proc. Natl Acad. Sci. 2018;115:10959–10964. doi: 10.1073/pnas.1809901115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.London N, Movshovitz-Attias D, Schueler-Furman O. The Structural Basis of Peptide-Protein Binding Strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Istomin AY, Gromiha MM, Vorov OK, Jacobs DJ, Livesay DR. New Insight into Long-Range Nonadditivity within Protein Double-Mutant Cycles. Proteins Struct. Funct. Genet. 2008;70:915–924. doi: 10.1002/prot.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowler DM, et al. High-Resolution Mapping of Protein Sequence-Function Relationships. Nat. Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler DM, Fields S. Deep Mutational Scanning: A New Style of Protein Science. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung I, Dekel A, Shifman JM, Sidhu SS. Saturation Scanning of Ubiquitin Variants Reveals a Common Hot Spot for Binding to USP2 and USP21. Proc. Natl Acad. Sci. USA. 2016;113:8705–8710. doi: 10.1073/pnas.1524648113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Lalic J, Baeza-Centurion P, Dhar R, Lehner B. Changes in Gene Expression Predictably Shift and Switch Genetic Interactions. Nat. Commun. 2019;10:3886. doi: 10.1038/s41467-019-11735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregoret LM, Sauer RT. Additivity of Mutant Effects Assessed by Binomial Mutagenesis. Proc. Natl Acad. Sci. USA. 1993;90:4246–4250. doi: 10.1073/pnas.90.9.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin L, Pinker RJ, Phillips GN, Kallenbach NR. Stabilization of Myoglobin by Multiple Alanine Substitutions in Helical Positions. Protein Sci. 1994;3:1430–1435. doi: 10.1002/pro.5560030909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouadio JLK, Horn JR, Pal G, Kossiakoff AA. Shotgun Alanine Scanning Shows That Growth Hormone Can Bind Productively to Its Receptor through a Drastically Minimized Interface. J. Biol. Chem. 2005;280:25524–25532. doi: 10.1074/jbc.M502167200. [DOI] [PubMed] [Google Scholar]
- 17.Morrison KL, Weiss GA. Combinatorial Alanine-Scanning. Curr. Opin. Chem. Biol. 2001;5:302–307. doi: 10.1016/S1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 18.Olson CA, Wu NC, Sun R. A Comprehensive Biophysical Description of Pairwise Epistasis throughout an Entire Protein Domain. Curr. Biol. 2014;24:2643–2651. doi: 10.1016/j.cub.2014.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid Mapping of Protein Functional Epitopes by Combinatorial Alanine Scanning. Proc. Natl Acad. Sci. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pál G, Fong S-Y, Kossiakoff AA, Sidhu SS. Alternative Views of Functional Protein Binding Epitopes Obtained by Combinatorial Shotgun Scanning Mutagenesis. Protein Sci. 2005;14:2405–2413. doi: 10.1110/ps.051519805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells JA. Additivity of Mutational Effects in Proteins. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- 22.Touti F, et al. In-Solution Enrichment Identifies Peptide Inhibitors of Protein–Protein Interactions. Nat. Chem. Biol. 2019;15:410–418. doi: 10.1038/s41589-019-0245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinogradov AA, et al. Library Design-Facilitated High-Throughput Sequencing of Synthetic Peptide Libraries. ACS Comb. Sci. 2017;19:694–701. doi: 10.1021/acscombsci.7b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y. Wiedmann, M. M. & Suga, H. RNA Display Methods for the Discovery of Bioactive Macrocycles. Chemical Reviews. American Chemical Society September 11, 2019, pp 10360–10391. [DOI] [PubMed]
- 25.Li J, et al. Structural Basis for High-Affinity Peptide Inhibition of P53 Interactions with MDM2 and MDMX. Proc. Natl Acad. Sci. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, et al. Systematic Mutational Analysis of Peptide Inhibition of the P53-MDM2/MDMX Interactions. J. Mol. Biol. 2010;398:200–213. doi: 10.1016/j.jmb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoe KK, Verma CS, Lane DP. Drugging the P53 Pathway: Understanding the Route to Clinical Efficacy. Nat. Rev. Drug Discov. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 28.Chang YS, et al. Stapled Α−helical Peptide Drug Development: A Potent Dual Inhibitor of MDM2 and MDMX for P53-Dependent Cancer Therapy. Proc. Natl Acad. Sci. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mijalis AJ, et al. A Fully Automated Flow-Based Approach for Accelerated Peptide Synthesis. Nat. Chem. Biol. 2017;13:464–466. doi: 10.1038/nchembio.2318. [DOI] [PubMed] [Google Scholar]
- 30.Walensky LD, Bird GH. Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress. J. Med. Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, et al. Dithiocarbamate-Inspired Side Chain Stapling Chemistry for Peptide Drug Design. Chem. Sci. 2019;10:1522–1530. doi: 10.1039/C8SC03275K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the P53 Tumor Suppressor Pathway by a Stapled P53 Peptide. J. Am. Chem. Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of α-Helix-Mediated Protein-Protein Interactions Using Designed Molecules. Nat. Chem. 2013;5:161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- 34.Spokoyny AM, et al. A Perfluoroaryl-Cysteine SNAr Chemistry Approach to Unprotected Peptide Stapling. J. Am. Chem. Soc. 2013;135:5946–5949. doi: 10.1021/ja400119t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heyne M, Papo N, Shifman JM. Generating Quantitative Binding Landscapes through Fractional Binding Selections Combined with Deep Sequencing and Data Normalization. Nat. Commun. 2020;11:297. doi: 10.1038/s41467-019-13895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhan C, et al. An Ultrahigh Affinity D-Peptide Antagonist of MDM2. J. Med. Chem. 2012;55:6237–6241. doi: 10.1021/jm3005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe JM, et al. Perfluoroaryl Bicyclic Cell-Penetrating Peptides for Delivery of Antisense Oligonucleotides. Angew. Chem. Int. Ed. 2018;57:4756–4759. doi: 10.1002/anie.201801167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quartararo AJ, et al. Ultra-Large Chemical Libraries for the Discovery of High-Affinity Peptide Binders. Nat. Commun. 2020;11:3183. doi: 10.1038/s41467-020-16920-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabideau AE, Liao X, Pentelute BL. Delivery of Mirror Image Polypeptides into Cells. Chem. Sci. 2015;6:648–653. doi: 10.1039/C4SC02078B. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are available either in the main text or supplementary materials.