ABSTRACT
The amoeba parasite Entamoeba histolytica is the causative agent of human amebiasis, an enteropathic disease affecting millions of people worldwide. This ancient protozoan is an elementary example of how parasites evolve with humans, e.g. taking advantage of multiple mechanisms to evade immune responses, interacting with microbiota for nutritional and protective needs, utilizing host resources for growth, division, and encystation. These skills of E. histolytica perpetuate the species and incidence of infection. However, in 10% of infected cases, the parasite turns into a pathogen; the host-parasite equilibrium is then disorganized, and the simple lifecycle based on two cell forms, trophozoites and cysts, becomes unbalanced. Trophozoites acquire a virulent phenotype which, when non-controlled, leads to intestinal invasion with the onset of amoebiasis symptoms. Virulent E. histolytica must cross mucus, epithelium, connective tissue and possibly blood. This highly mobile parasite faces various stresses and a powerful host immune response, with oxidative stress being a challenge for its survival. New emerging research avenues and omics technologies target gene regulation to determine human or parasitic factors activated upon infection, their role in virulence activation, and in pathogenesis; this research bears in mind that E. histolytica is a resident of the complex intestinal ecosystem. The goal is to eradicate amoebiasis from the planet, but the parasitic life of E. histolytica is ancient and complex and will likely continue to evolve with humans. Advances in these topics are summarized here.
KEYWORDS: Entamoeba, lifecycle, stress responses, virulence, pathogenesis
Introduction
The persistence of a parasite species is the trade-off between the rapid reproduction of the parasite and, in doing so, the measured exploitation of the host’s resources; disruption of this paradigm can result in host damage and hence loss of fitness for the parasite. Virulence is the term used to designate the mechanisms leading to the breakdown of homoeostasis in the parasitized host, whereas the extent of damage, due to the appearance of virulence, is the measure of pathogenicity. Important mechanisms increase parasite survival during pathogenicity, including the presence of different genotypes within a single parasite species leading to rapid growth, the efficient establishment of stress response systems, and the induction of resistant cell forms. These parasitic skills leading to its adaptation, reduce the likelihood that the hosts will kill them; or at least, allow parasites to persist within tolerance limits. A successful parasite, surviving throughout the life of the host without damaging it, should be considered a commensal-like organism, but emerging notions point out that in complex ecosystems, commensals can disrupt homoeostasis. This is the case of the human intestinal ecosystem because the presence of non-virulent parasites can modify the composition of the microbiota [1,2] and consequently, they impact the nutritional and immune status of the host, without any sign of intestinal damage [3,4]. This complexity has led to the idea of naming eukaryotic intestinal microbes as symbionts, thus encompassing mutualists, commensals, and parasites [3] and, therefore, enriching perspectives for the study of host-microbial eukaryote interactions. Under certain circumstances, parasites can become pathogenic, for example when their numbers increase indiscriminately in the absence of an effective host immune response.
Ancient parasites have evolved with humans as in the case of the Entamoeba species, which are endobiotic amoebae colonizing animal species [5,6]. The persistence of at least seven species of Entamoeba depends on their ability to infect humans primarily in the intestinal tract, where they divide and encyst. Most Entamoeba infections in humans appear to be caused by non-pathogenic species (e.g. Entamoeba dispar and Entamoeba coli), the only amoebic species that could become virulent and harm the host is Entamoeba histolytica, the causative agent of amoebiasis, responsible for dysentery and liver abscesses [7]. Asymptomatic infections are most common, while symptoms of invasive amoebiasis develop in about 10% of infected people [8]. Clinical features of symptomatic amoebic infection may include blood and mucus in diarrhoeal stools, which are dysentery phenotypes; in some cases, the parasite invades the liver and forms abscesses that may occur for months or years after travel or residence in a geographic region where amoebiasis is endemic. The exact burden of amoebiasis is difficult to quantify due to limited diagnostic and surveillance capacities in endemic regions, including developing countries in Central and South America, Africa, and Asia. This infectious disease is also present in industrialized countries mainly due to returning travellers or immigrants from endemic countries [9]. Although poorly quantified overall, the impact of amoebiasis remains significant as prevalence estimates are as high as 40% in some populations (e.g. Mexico [10], China [11], India [12], Turkey [13]). No vaccine against amoebiasis currently exists; the drug of choice for therapy is metronidazole, which may cause severe side effects in humans [14] and on anaerobic members of the microbiota [15]. Therefore, understanding what determines the outcome of amoebic infection and the nature of amoebic virulence is a key challenge for Entamoeba research to advance therapeutic and vaccine development.
Here is reviewed our current knowledge of the lifecycle and mechanisms of pathogenic conversion of E. histolytica considering several characteristics by which this parasite survives in the human intestine. Due to the abundant literature in the field, it is important to note several compilations of various aspects of pathogenesis already published [7,16–18].
The lifecycle of Entamoeba histolytica
The occurrence of amoebiasis is largely based on the simple two-stage lifecycle of E. histolytica, with an environmental infectious cyst and a dividing trophozoite residing in the human intestine. Although other non-pathogenic amoeba species share this same lifestyle, E. histolytica has been able to take advantage of gene regulation and activated pathways to adapt to the host responses when the balance between the parasite and the intestinal ecosystem is disturbed. The cyst, which is the contaminating form, ensures the survival of the species because it resists to environmental changes and is easily transmitted. Due to their high resilience, cysts overcome the eradication of the pathogen by the immune system or by antibiotic treatment. Mature cysts are excreted in the host’s stool and are transferred to another human via a faecal-oral route through contaminated food or water or person-to-person contact; thus, amoebic infection occurs when water quality, sanitation, and hygiene practices are inadequate. This finding has been largely confirmed during the COVID-19 pandemic, as good hand hygiene compliance in endemic regions significantly correlated with a reduced rate of E. histolytica and other intestinal parasitic infections [19]. After ingestion by a new host, in the small intestine, the cyst emerges into four disease-causing trophozoite forms that migrates and colonizes the large intestine, where they feed on bacteria and divide or encyst themselves (Figure 1). While most infected people are asymptomatic carriers, they shed millions of cysts daily and can perpetuate amoebic infection.
Figure 1.
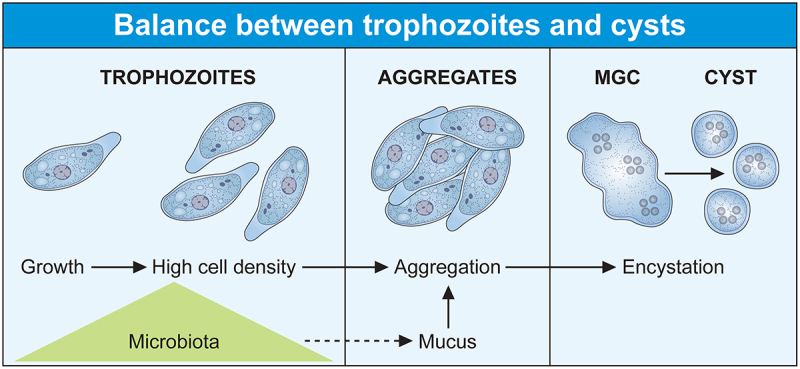
The lifecycle of Entamoeba histolytica is influenced by the microbiota. (Two cellular forms make up the lifecycle of Entamoeba, the contaminating cyst and the trophozoite. Infection occurs directly upon ingestion of the cyst by a new host, without intermediate hosts as a vector. The trophozoite colonizes the large intestine as a commensal-like, where it feeds on bacteria and divides. Trophozoite populations can reach high densities and aggregates due to amoebic surface molecules recognizing galactose ligands (e.g. lectins) that interact with mucus. High trophozoite density is a key parameter of encystation, and aggregation should trigger the transition from exponential growth to encystation following the formation of multicellular giant cells (MGC) where the cyst forms. The MGC phenotype depends on the transcription factor ERM-BP which has no role in the aggregation process.
Formation of cysts in Entamoeba
Encystation in Entamoeba occurs after a slowdown in cellular metabolism following food deprivation or environmental conditions unfavourable to the growth and division of the parasite. The common model to study amoebic encystation in laboratory conditions is Entamoeba invadens (a reptile pathogen), which has a lifecycle similar to E. histolytica. Trophozoites retract their cytoplasmic projections such as pseudopodia, round off, detach from the substrate, and aggregate into multicellular clumps [20,21]. At early stages of encystation, a tetra nucleated intermediate cell (early cyst) forms due to two successive cycles of chromosomal replication without cell division [22,23]. Gradual dehydration induces the decrease in cytoplasmic volume by approximately 80% leading to retraction of the cytoplasm from the cyst wall, glycogen accumulation, and maturation of cysts by addition of the wall elements [22] such as chitin-rich microfibrils that are associated with the lectins Jacob and Jessie. The main biochemical and molecular features of Entamoeba encystation have been reviewed [18,24]. Clearly, changes in the environment such as starvation of the carbon source or osmotic shock induces encystation.
Signalling molecules involved in cyst formation include galactose-terminated ligands, sphingolipids (especially long-chain fatty acids), small-molecule intermediates in signalling pathways as cyclic adenosine monophosphate, calcium, phospholipase D, cholesteryl sulphate, and the metabolic cofactor nicotinamide adenine dinucleotide (NAD+). Encystation is due to the activation of specific amoebic surface receptors including a β1-adrenergic receptor sensitive to catecholamines [25] and lectin receptors sensitive to galactosidase-terminated ligands [26]. Profiling gene transcription in Entamoeba species (both E. histolytica and E. invadens) determined the role of at least three transcription factors involved in encystation: Myb [27], ERM-BP [28], and NF-YC [29,30], these factors have temporary sequential activities according to the cyst stage. Epigenetic modifications also control encystation since treatment of trophozoites with histone acetylation modifiers lead to the downregulation of expression for genes implicated in the synthesis of chitin, cyst wall proteins, polyamines, or gamma-aminobutyric acid pathways [31]. The data suggest that the genetic regulations and entire signalling pathways activating gene transcription necessary for Entamoeba encystation are far from completely identified.
The trophozoite as the vegetative cellular form of Entamoeba histolytica
E. histolytica trophozoites (20 to 40 µm in size) reside in the intestinal lumen, feed on bacteria and divide by binary fusion. The genome of E. histolytica is aneuploid; sometimes larger than tetraploid and contains one or more extra copies of shorter truncated chromosomes [32]. Cytokinesis in E. histolytica is not coupled with nuclear division, therefore, cell division can be asymmetrical [33], delinked from S phase and mitosis [34], leading to daughter cells with zero, one, or several nuclei [33]. Nevertheless, genome analysis determines that E. histolytica has orthologues of all cytokinesis-related proteins, compared to budding yeast, but the fact that amoebae can accumulate polyploid cells suggests that checkpoints regulating the synchrony between genome duplication and cell division are absent [35,36].
The current AT-rich genome assembly of E. histolytica is sized at 20 Mbp with 8,201 predicted genes of which the vast majority (~76%) do not contain introns and encode short open reading frames (on average ~389 amino acids), compared with other unicellular eukaryotes. There are distinctive genomic features during the evolution of Entamoeba species such as the remarkable loss of genes related to adaptation to low oxygen niches (e.g. genes involved in the tricarboxylic acid cycle and oxidative phosphorylation), a fact correlated with the absence of classic mitochondria [37] and as a result, glycolysis is the primary source of energy for cellular functions [35,38], and ethanol is the main end-product of glucose catabolism [39].
Entamoeba species retain a mitochondrion-derived organelle called mitosome devoid of an organellar genome [40,41], it is the seat for the synthesis of sulfolipids [42]. The mitochondrial iron – sulphur assembly system of eukaryotes is replaced in E. histolytica by an unrelated nitrogen fixation system whose coding genes were acquired by lateral gene transfer (LGT) from ε-proteobacteria [43,44]. Recently, another organelle, the peroxisome, has been identified in E. histolytica [45]; this finding breaks the paradigm of peroxisome absence in anaerobic parasites. Amoebic peroxisomes contain myo-IDH, an enzyme that catalyses the conversion of myo-inositol, which is a substrate for the synthesis of phosphatidylinositol and phosphoinositide derivatives. It seems that mitosomes and peroxisomes are involved in the mechanism of lipid synthesis and transfer, but the interplay between components of mitosomes, peroxisomes, and the cytosolic membrane system is still under investigation [46]. In addition to gene loss, Entamoeba genomes also show expansion in some gene families such as those encoding proteins containing leucine-rich repeats (LRRs) homologous to bacterial adhesins (BspA from Bacteroides forsythus) [47], surface antigen Ariel1 and AIG1-like GTPases [6].
An interesting aspect of the biology of E. histolytica is the absence of well-defined morphological structures corresponding to the rough endoplasmic reticulum (rER), Golgi bodies [36] or microtubular spindle [48]. However, there are protein orthologs of established eukaryotic endomembrane systems, and functional ER and Golgi can be predicted [49,50]; in addition, α, β, and γ tubulin have been identified but they differ significantly from other eukaryotic tubulins, sharing only 50%, 58%, and 46% identity, respectively [48]. The actin-rich cytoskeleton plays a crucial role in all life processes of E. histolytica: division, encystation, motility, endocytosis, and in other processes supporting pathogenesis, such as phagocytosis of human cells and parasite resistance to host immune responses. However, E. histolytica has a single actin protein, which differs significantly in structure from its human counterparts [51,52], but actin performs all the various functions of the actin-rich cytoskeleton through many actin-binding proteins, 390 of them have been identified by bioinformatics [53], but their specific functions are only partially determined
Encystation of trophozoites in the presence of the microbiota
The four primary hallmarks of E. histolytica encystation (e.g. acquisition of a spherical shape, resistance to detergents, chitin accumulation at the cell wall, and tetranucleation); these steps are accomplished in the complex environment of the human colon where proliferation of trophozoites occurs simultaneously. Extensive research under laboratory conditions, using E. invadens as an amoebic encystation model, has provided rich information on encystation pathways; nevertheless, the results may not fully reflect the reality of this phenomenon in E. histolytica due to genetic differences between the two species and the need of distinct hosts for their proliferation [54]. Moreover, the intestinal environment complexity is different from the culture medium due to the fact that the human intestinal lumen harbours the microbiota which plays important roles in tissue homoeostasis including protection against pathogens [55], maintenance of mucosal barrier integrity [56], host nutritional elements sustain [57], and factoring the efficiency of the immune response [56]. For instance, some bacteria from the microbiota can degrade complex non-digestible carbohydrates (fibbers) to produce short-chain fatty acids (SCFAs) such as acetate, butyrate, and propionate [58], which are energy providers for colon epithelial cells and parasite growth and SCFAs can inhibit encystation [59]. Levels of glucose in the human colon does not reflect the carbon source deprivation, which is important for in vitro encystation, and SCFAs also stimulate the production of mucin 2 [60], which contains oligosaccharides that might be used as a carbon source. Moreover, the potential impact of eukaryote symbionts on intestinal homoeostasis is not often considered; for example, microbiota composition is modified due to intestinal colonization by Blastocystis spp [61], which can be associated with other protozoa such as non-pathogenic Entamoeba species [62] and these changes are associated with amoebiasis [63,64]. Likewise, more than 140 fungal genera have been discovered as permanent or transient biota in the intestinal tract; many are either beneficial or commensal (e.g. Saccharomyces); as well as viruses and bacteriophages [65].
Altogether, these diverse microbes and their derivative metabolic products, suggest their potential specific roles in the control of intestinal homoeostasis and therefore in amoebic encystation. Thus, the major question that arises is how E. histolytica maintains the balance between vegetative growth and the onset of encystation? The answer to this question is largely unknown due to a significant gap in the development of experimental models mimicking the human colon, which are needed to study the amoebic lifecycle and amoebiasis [66,67]. Significant efforts have been made to this end; for example, 30% of cyst-like structures were obtained after treatment of trophozoites with hydrogen peroxide and metals, but these structures are not tetranucleate and may reflect a transient trophozoite survival response when subjected to a stressful environmental state [68,69]. An encouraging result was obtained using media conditioned by Escherichia coli and Enterococcus faecalis and an alkaline pH, the cyst-like cells obtained were multinucleated, but were not viable for excystation [70]. Recently, a promising model for encystation studies was obtained using two key parameters: high trophozoite density and glucose limitation in the culture medium [71]. The cysts are viable and exhibit all four characteristics of the cyst phenotype: round shape, chitinous cell wall, four nuclei, and detergent resistance. These cysts are viable for excystation generating the growing trophozoite form. A remarkable difference compared with E. invadens is that encystation commitment of E. histolytica was enhanced by SCFAs in this model, with acetate having the strongest effect [59]. Moreover, treatment with SCFAs induced minimal transcriptional changes in E. histolytica [72], all data support the recently proposed working hypothesis that epigenetic regulations leading to encystation of E. histolytica could occur under the influence of microbiota metabolites, such as SCFAs [73] and reinforce the idea that the encystation signals of E. histolytica only partially overlap those of E. invadens.
High trophozoite density in glucose-free medium allows cyst formation in E. histolytica in the above cited model. This characteristic is already known for E. invadens, the cells aggregate and merge to give multinuclear giant cells (MGC) where cysts form [26,74]. The signal molecules and pathways inducing trophozoite aggregation are not known, but the addition of galactose (Gal) inhibits aggregation and hence encystation [75], a fact indicating that surface molecules sensitive to Gal are involved in amoebae interactions necessary for aggregation. Gal-inhibitable cell surface lectins have been proposed as an important Gal-responsive component bridging trophozoites [26] or allowing their interaction with the extracellular medium rich in Gal-terminated ligands including mucin [26,76]. Several lectins from E. histolytica are potential candidates for trophozoite aggregation, including a protein recognizing oligosaccharides found in ovalbumin [77], β trefoil lectin [78], and galactose/N-acetyl galactosamine lectin (Gal/GalNAc lectin) which recognizes Gal and N-acetyl galactosamine [79,80]. The molecular mechanism and signalling pathways implicating these lectins in encystation are far from determined; in particular, the Gal/GalNAc lectin is a versatile receptor activating multiple overlapping signalling pathways depending on the cellular function to be performed [18]. Nevertheless, encystation signalling pathways may occur through changes in the actin-rich cytoskeleton including reduction in actin levels that lead to rounded quiescent cells [81] or disruption of actin polymerization dynamics [82]; moreover, the carboxyl terminal domain of the Gal/GalNAc lectin heavy chain interacts with the cytoskeleton [18] triggering the activation of the above-described transcription factors involved in encystation. Recent data reinforce this hypothesis; aggregation occurs as an independent step leading eventually to encystation since the ERM-BP transcription factor participates in MGC formation in E. invadens, but silencing ERM-BP has no effect on the aggregation process [29]; indicating that trophozoite aggregation follows specific molecular regulation independent of MGC formation. Furthermore, ERM-BP is an important factor for MGC induction in E. histolytica following heat stress under conditions preceding cell death [29], these observations correlate stress responses and encystation. The MGC phenotype depends on the ERM-BP transcription factor which uses NAD+ as a cofactor, the level of NAD+ increases during both phenomena [29] and genes involved in cyst wall formation (e.g. chitinase and Jacob) are upregulated in both conditions [83]. These observations are reinforced by a detailed view of E. histolytica transcriptome, which confirm that the gene encoding the cyst wall-specific glycoprotein Jacob 1 (EHI_028930) is upregulated during Oxidative Stress (OS) [84] and HS [85]; the gene encoding chitinase 1 is upregulated during HS, and ERM-BP is upregulated during HS (8 times) but downregulated in OS. However, orthologous genes, including cyst wall – specific glycoprotein Jacob 2, chitinase Jessie 1–2, −3, and −4 are not significantly altered during HS, indicating the existence of selective regulation for encystation-related genes.
Trophozoites, oxidative stress, and the microbiota
The epithelium provides a physical separation between the immune cells and the microbiota present in the intestinal lumen and the smooth mucus. In tissue homoeostasis, epithelial cells participate in immune defence by secreting mucus and releasing reactive oxygen species (ROS), antimicrobial peptides, chemokines, and cytokines. ROS derive from the reduction of oxygen to form: (i) superoxide (O2−) a reaction involving NADPH oxidase and dual oxidase; (ii) hydrogen peroxide (H2O2) generated by dismutation of two molecules of O2− by the enzyme superoxide dismutase (SOD), (iii) hydroxyl radical (HO) produced by either oxidation of water or hydroxide ions. Catalase or glutathione (GSH) reduce H2O2 [86]. Some bacteria from the microbiota contribute to ROS levels in the intestinal lumen [87], bacteria as well as other intestinal microorganisms accommodate to live with ROS levels related to homoeostasis. During the onset and establishment of intestinal diseases ROS level may increase triggering oxidative stress (OS), which damage lipids, proteins, and DNA and lead to cell death.
E. histolytica as a single-celled microaerophilic organism lives in low oxygen tension conditions, this parasite faces ROS during their growth and undergoes OS upon contact with bacteria and upon activation of immune responses from the host. E. histolytica lacks known important components of antioxidant defence mechanisms (e.g. catalase, peroxidase, glutathione, and the glutathione recycling enzymes glutathione peroxidase and glutathione reductase) and uses L-cysteine, instead of glutathione, as the main thiol. Nevertheless, this parasite has several anti-oxidative enzymes, including peroxiredoxins, SOD, dismutase, flavoprotein A, ferredoxin, thioredoxin, and thioredoxin reductase. Additionally, other scavenging proteins have been identified, such as rubrerythrin and hybrid-cluster protein. ROS cause oxidation of amoebic proteins and trigger OS responses [88], including transcription changes for genes whose protein products are involved in mRNA translation, signalling regulatory cascades, DNA repair processes, energy metabolism, stress response, and solute transport [84,89,90]. A stress-responsive transcription factor (HRM-BP) in E. histolytica has been identified [91].
Commensal or pathogenic enterobacteria [84] and their derived metabolites (e.g. oxaloacetate [92], queuine [93]) protect trophozoites against OS. The molecular mechanisms of such bacterial protection on trophozoites are being studied, and it has already been determined that: (i) Enterobacteriaceae modify the transcription of numerous amoebic genes (approximately 31% of the coding sequences), including genes involved in protein synthesis and homoeostasis (ribosomal proteins and translation-related factors), nutrition (hydrolases, peptidases and trafficking), cell survival, encystation factors, as well as genes for several types of antioxidant enzymes (e.g. peroxidases and thioredoxins) [84]; (ii) oxaloacetate, as a non-enzymatic antioxidant H2O2 scavenger, reduces the amount of oxidized proteins [92], and (iii) queuine, as a hypermodified nucleobase, leads to hypermethylation of certain tRNAs affecting the protein synthesis [93] and, by unknown mechanism induce the upregulation of genes encoding anti-stress proteins, including heat shock protein 70, antioxidant enzymes and enzymes involved in DNA repair [93]. The protective effect of Enterobacteriaceae on E. histolytica is not universal since the production of H2O2 by Lactobacillus acidophilus (a probiotic) modifies the amoebic transcriptome differently compared to other Enterobacteriaceae [84]. L. acidophilus causes oxidation of vital proteins of E. histolytica (e.g. Gal/GalNAc lectin heavy chain, thioredoxin, cysteine protease A5, oxidoreductases, and signalling molecules), thus triggering parasite death [94].
In summary, the success of the parasitic lifestyle, due to the balance of the two cell forms of Entamoeba (trophozoite and cyst), is strongly influenced by the microbiota which, on the one hand, provides the bacteria and its derived nutrients promoting the growth of trophozoites and, on the other hand, acts as a motor for the survival of the parasite in stressful situations. Thus, the microbiota appears as the natural and adapted fuel to increase the population density of trophozoites. Their interaction with the extracellular environment, and their number, allow trophozoites aggregation; then, initiating the process of encystation after resources limitation. The cyst spreads in nature with the human stools, infects a new host and perpetuates the parasitic species (Figure 1). In some intestinal conditions (explained later), the lifecycle of E. histolytica can be disrupted switching trophozoites to a virulent phenotype.
Virulence of E. histolytica strains isolated from asymptomatic or symptomatic patients, genetic diversity
The outcomes of E. histolytica infection are mostly asymptomatic, only a fraction of those infected develop dysentery and, in rare cases, liver abscesses. In areas where amoebic infections are endemic, the incidence rate of E. histolytica has been found to be higher than that of E. dispar, while the population does not suffer from amoebiasis [95]. Molecular diagnosis among asymptomatic carriers determined a significant number of cases of mixed infections with Entamoeba [95,96] or in association with another protozoan [2]. However, a possible synergy in the mixed species population favouring the pathogenicity of E. histolytica has not yet been considered; mainly because it is not possible to conduct field experiments with these parasites strictly infecting humans. Nevertheless, this hypothesis is plausible because, as mentioned above, the presence of different protozoa in the same individual leads to changes in the microbiota [2] and potentially, interactions between amoebic species could modify the parameters of the lifecycle; for example, by inducing efficient aggregation of trophozoites because many molecules on the amoebic surface are common to various species [80]. Relevant knowledge on amoebic virulence has been acquired by studying strains of E. histolytica isolated in different geographical areas: while HM1: IMSS strain, isolated in Mexico from a patient suffering from amoebiasis is virulent; the Rahman strain, isolated in England from an asymptomatic carrier, is naturally attenuated and shows reduced virulence in experimental models [97,98]. Isolates of E. histolytica with various virulence patterns have also been reported from other countries [99]. To decipher the mechanisms supporting these varying levels of virulence among strains, parasite genotypes have been compared to determine changes in genetic polymorphism (e.g. using polymorphic markers linked to tRNA or interspersed nuclear retrotransposable elements) [100–103]; data suggest a high genetic diversity in strains of E. histolytica isolated worldwide [104]; but their link to the outcome of infection is still elusive.
Genome sequencing of trophozoites isolated worldwide has established [105] single nucleotide polymorphisms (SNPs) in these strains. Although their divergence from the reference genome is small (between 0.312 and 0.857 SNPs per kilobase), the SNPs indicate genomic plasticity and suggest genetic recombination in Entamoeba [105]. To test a potential association between SNPs and disease outcome, samples of human stool and liver abscess aspirates were collected from areas endemic for amebiasis. After short-term xenic cultures of parasites, extensive DNA sequencing and bioinformatics analysis identified SNPs from 21 loci [106]. Two genes (EHI_065250 and EHI_080100) have SNPs with a significant link to parasites isolated from liver abscesses. Recently, SNPs at non-repetitive loci, which should be genetically stable, were identified in 49 isolates of E. histolytica from human cases with different disease outcomes (22 were from diarrhoeal, 7 from asymptomatic, and 20 with liver abscess) [107]. A total of 26 SNPs were present in three coding DNA sequences (lysine and glutamic acid – rich protein gene, kerp1, EHI_098210; glutamic acid and lysine – rich protein 2 gene, kerp2, EHI5A_120210; and amoebapore C gene, apc, X76903), and in two non-coding sequences (intron AY956435, intergenic region AY956439). Analysis of the genetic distribution within the kerp2 gene shows that 10 SNPs are associated with disease outcome clusters (7 asymptomatic, 2 liver abscess). These SNP-disease correlations, in addition to potential recombination events and significant population distinction between pathogenic and non-pathogenic strains of E. histolytica, indicate that SNPs might be under constant selection pressure from the host and may be a potential determinant of disease outcome regarding amoebiasis [107]. SNPs of the kerp2 gene are the first to be suggested as potential prognostic markers linked to a high risk of developing liver abscesses in people with persistent asymptomatic E. histolytica infection. Overall, these encouraging data, implying an impact of genetic diversity of field isolates of E. histolytica on the outcome of the disease, call for more studies, with a large sampling, in different geographical areas where amoebiasis is endemic, to obtain genetic markers relevant for detection of E. histolytica strains capable of invading the intestinal parenchyma.
Entamoeba histolytica and humans: from asymptomatic to symptomatic phenotype
The complex balance between “commensal-like” and pathogenic phenotypes for E. histolytica has allowed this parasite to become one of the most common human pathogens causing life-threatening infections. As a pathogenic parasite, E. histolytica leads to intestinal invasion in the 10% of infected people. For reasons still poorly understood, trophozoites begin to destroy the muco-epithelial barrier, thereby inducing the overproduction of mucus, killing host cells, and causing inflammation and dysentery. Trophozoites may disseminate through the portal vein system and invade the liver where it forms abscesses [108]. These stages of amoebiasis, illustrating amoebic virulence, depend on the capacity of the parasite to adapt rapidly to the new environments it encounters (mucus, epithelium, blood). An effective immune response to the parasiticinvasion leads to trophozoites death since in nature they cannot encyst outside the intestinal lumen. Invasive trophozoites trigger pathogenicity that is associated with several parasitic factors, including markers for: adhesion to mammalian cells, motility, extracellular matrix (ECM) degradation, cytotoxicity and engulfment of human cells, induction of host cell death and inflammation. Due to extensive literature and complexity of amoebic infection; in this topic we will focus on some newly identified features correlating to the very early virulent stages of E. histolytica, the reader is referred to excellent overviews elsewhere on the pathogenic process [7,17,109].
Entamoeba histolytica virulent trophozoites and the heat-shock proteins
Amoebic virulence is generally attributed to the ability of the parasite to produce liver abscesses in susceptible laboratory animals (e.g. gerbils and hamsters); however, long-term culture of E. histolytica HM-1: IMSS strain drifts towards an irreversible loss of the virulent phenotype [110,111]. The reasons for the attenuation of trophozoite virulence should be multiple [112]. To date, several studies have highlighted the correlation between attenuation and the lack of amoebae ability to maintain an intracellular hypoxic environment due to reduced performance of their OS responses [113,114]. Moreover, glycolytic flux is irreversibly decreased in attenuated trophozoites exposed to O2 [115] reinforcing the idea of a link between virulence and survival mechanisms under host immune attack.
Gene expression analysis of virulent trophozoites cultured under OS indicates massive upregulation of genes encoding heat shock proteins (e.g. ClpB, HSP70, HSP90, and HSP101); in contrast, this response is absent in cultured attenuated trophozoites [113]. Careful differential expression analysis of transcriptome data obtained by RNASeq reinforces and clarifies these observations [116]: in highly virulent amoebae, 31 hsp genes are exclusively upregulated (e.g. hsp70), 14 hsp genes are upregulated in both strains (e.g. hsp101), and only 1 hsp gene is exclusively upregulated in attenuated trophozoites; moreover, 42 hsp genes are expressed under OS at comparable levels in the two types of trophozoites (Figure 2).
Figure 2.
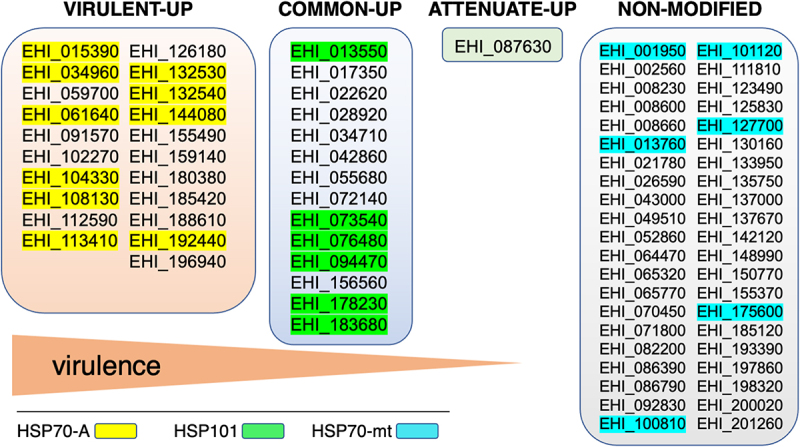
The stress response of virulent trophozoites involves heat shock proteins of the Hsp70 family. In the genome of E. histolytica HM1:IMSS strain, 86 genes encoding HSP proteins have been identified; of these, 78 genes have a transcriptional response in trophozoites undergoing OS. Gene expression analysis of stressed trophozoites indicates that the gene family encoding Hsp70 is preferentially upregulated (fold changes >3) in highly virulent amoebae; the Hsp101 encoding genes appear preferentially upregulated in virulent and attenuated amoebae, whereas only a gene encoding one HSP transcription factors is upregulated in attenuated trophozoites. In addition, 42 genes encoding HSPs are expressed at comparable mRNA levels in both types of trophozoites subjected to OS. The EHI_ prefix followed by numbers matches the gene identity in AmoebaDB database (https://amoebadb.Org).
Hsps are molecular chaperones that allow cells to survive during stress by maintaining proper protein folding limiting their denaturation and aggregation, some Hsps also have a role in maintaining genomic stability [117,118]. These proteins are classified according to their molecular mass: Hsp100(−101, ClpB), Hsp90, Hsp70, Hsp60, Hsp40, and Hsp20. In E. histolytica, Hsp70-A (EHI_113410), belonging to the Hsp70 family (13 members), is cytosolic [119], the coding gene is upregulated following O2, H2O2, or NO treatments and during the formation of hepatic abscesses [89,113,116,120]; therefore, inhibition of Hsp70-A blocks virulence [113]. Hsp101 is highly overexpressed when trophozoites are treated with NO [120], and the only gene exclusively overexpressed in attenuated parasites is EHI_087630, which encodes an isoform of the Hsp transcription factor, the other 5 isoforms are expressed at the same level in all trophozoites. Overall, the data support the conclusion that the attenuation of virulence in E. histolytica involves the inhibition of the stress response through Hsps. To date, efforts to reverse the virulence-attenuated parasite phenotype have been unsuccessful and, consistent with these recent data, this objective call for experiments devoted to reverse the Hsp gene expression in attenuated parasites.
Virulent E. histolytica invade and destroy the intestinal mucus, the role of glycosidases
Human intestinal mucus is composed of highly glycosylated mucins, mainly mucin-2 (MUC2) [66,121]. O-glycans make up about 80% of the MUC2 mucin mass, these have a sialic acid on the GalNAc component attached to the protein core [121]. Mucus also contains several other molecules, such as IgGFc binding protein and calcium-activated chloride channel 1 [122]; these components are produced and secreted by goblet cells [66]. Some members of the microbiota, pathogenic bacteria, and parasites are mucus degraders [123–125], these are known to harbour glycosyl hydrolases that cleave specific glycan bonds in mucin. Degradation of the intestinal mucus layer by E. histolytica is crucial for colonic invasion and this activity induces rapid hypersecretion of new mucus from goblet cells [126]. Thanks to the human colon explant model of amoebiasis [127], the early pathophysiological pathways of intestinal invasion by virulent E. histolytica HM-1: IMSS show mucus degradation in 2 hours [127]. Mucus breakdown by E. histolytica was recently visualized using the in vitro 3D scaffold model of the human colon containing goblet cells [67]. Immunofluorescence with specific anti-MUC2 antibodies and two-photon microscopy locate mucin fragments either in the mucosal environment or inside trophozoites. The amoebic signalling pathways activated by ingestion of MUC2 remain to be determined.
E.histolytica poses various glycosidases, such as N-acetylgalactosamidase, N-acetyl-glucosaminidase, β-galactosidase, β-N-acetyl-hexosaminidase, and sialidases [128]; some of these enzymes can cleave polysaccharides present in MUC2, as they can be secreted [125] or be a component of cytoplasmic granules released by exocytosis after amoeba interaction with collagen [129]. E. histolytica meeting the colon rapidly changes its transcriptomic profile [116]. The increased expression of genes related to carbohydrate and glycosylated residue metabolisms of virulent amoebae compared with attenuated amoebae correlates with gene expression patterns obtained by comparing virulent HM-1:IMSS and non-virulent Rhaman strains, in the same colon explant model [130]. The glycosidases encoding genes highly upregulated include: β-galactosidase which catalyzes the hydrolysis of β-galactosides into monosaccharides, as well as α- and β-amylases acting on starch, glycogen, and related polysaccharides and oligosaccharides [131]. Knock down of genes encoding β-amylases leads to trophozoites that are unable to degrade the mucus layer and therefore, with limitations to invade the human colon [130].
In agreement with the pathophysiological analysis and the transcriptomic profile of virulent trophozoites, it can be hypothesized that the degradation of saccharides within the mucus layer is an important source of energy promoting the viability of amoebas during mucosal invasion. Mucus-derived components can compensate for the deprivation of carbon sources due to the decrease in the bacterial population characteristic of the deep layer of mucus. Moreover, this expected nutritional need could be accompanied by the stress response of amoebae imposed by the passage from the lumen to the mucosal environment. This hypothesis correlates the upregulation in E. histolytica of genes encoding β-amylases during intestine invasion, heat shock, and OS [85]; in contrast, β-amylase genes are downregulated in trophozoites subjected to OS in the presence of bacteria [84], a fact that reinforces the microbiota role in parasite survival. Links between nutritional requirements and stress responses for β-amylase genes have been observed in plants where they respond positively to growth, development, and abiotic stress [132,133]. The importance of these β-amylase gene behaviours during amoebic pathogenesis is still under investigation; however, humans lack β-amylase and the fact that blocking the amoebic enzyme suppresses pathogenesis makes this enzyme a highly relevant candidate target for new anti-amoebiasis drugs.
Virulent E. histolytica invade and destroy the intestinal mucosa, the role of cysteine protease A5
Cysteine proteases (CP) play a key role in the pathogenesis of E. histolytica [134]; among them, cysteine protease A5 (CP-A5, EHI_168240) is present only in E. histolytica [135]; it is an important amoebic factor for mucus depletion because it targets peptides in glycosylated poor regions of MUC2 [135,136]. This enzyme localizes to the surface of the trophozoite [137], contains an Arg-Gly-Asp (RGD) integrin-binding motif that has also been described in higher eukaryotic cathepsin X [138]. CP-A5 binds via its RGD motif to the αVβ3 integrin on enterocyte-like cells and stimulates inflammatory responses [139]. The interaction of amoebic CP-A5 with integrin αVβ3 triggers in goblet cells the outside-inside signalling that result in mucin secretion with activation of several kinases (e.g. kinases of the SRC family, the focal adhesion kinase FAK, PI3K, PKCδ, and MARCKS) [140]. An E. histolytica cyclooxygenase-like regulates CPA5 activity through a protein–protein interaction that stabilizes CP-A5 and increases virulence [141]. Recent data indicate the role of CP-A5 in the degradation of bacterial biofilms [142].
To enter the tissues, trophozoites disorganize the loose collagen scaffold to escape into the mucosa [127]. Epigenetically silenced trophozoites for CP-A5, which are negative for its proteinase activity, fail to trigger the tissue inflammatory response [127] or to induce collagen remodelling necessary for lamina propria invasion [143]. In the explant colon model, enzymatic activities of human matrix metalloproteinases (MMP-1 and −3) have been determined to be responsible for the alteration of collagen fibrillar structures during intestinal invasion by E. histolytica [144]. Clearly, CP-A5 activity convert pro-MMP-3 into active MMP-3 enzyme, which in turn activates pro-MMP-1, leading to collagen degradation [144]. In addition to the role in intestinal amoebiasis, CP-A5 activity is also required to induce the formation of liver lesions in hamsters [145] and, is implicated in the loss of the actin cytoskeleton and the organization of focal adhesion structure triggering endothelial cell death [146].
Overall, we can conclude that CP-A5 is a multifunctional enzyme that plays a critical role in the pathogenesis of E. histolytica in the human colon: by degrading mucus and bacterial biofilms, binding to αvβ3 integrin receptors on goblet cells to induce pro-inflammatory responses, by activating the remodelling of the intestinal fibrillar collagen structure and in addition, by playing a role in the formation of liver abscess.
Interplay of virulent E. histolytica and human intestine at early stages of amoebiasis
To be successful during intestinal invasion, E. histolytica must be motile and the actin-rich cytoskeleton is central to the complex invasive process [147]. In the early stages of intestinal invasion [148], once the protective mucus layer has been destroyed, virulent trophozoites penetrate the intestinal parenchyma, adhere and destroy the extracellular matrix (ECM) through the action of amoebic surface receptors recognizing ECM components and proteinase activities [149]. The trophozoite evolves in the colon epithelium by attaching to the mammalian cell by means of surface glycosylated components like the Gal-GalNAc lectin and the glycocalyx-bound lipopeptidophosphoglycan (LPPG) [18]; or by direct amoeba-cell contact involving proteins like KERP1, KERP2 [150]; serine, threonine-, and isoleucine-rich proteins STIRPs [151] and the adhesin ADH112 [152]. Cell-cell junctions of epithelial cells are disrupted by proteinases [153], mammalian cell killing, and phagocytosis occurs; alternatively, after attaching to host cells, amoebae “chew off” and ingest host cell fragments by trogocytosis [154,155]. Onset of host immune responses begins with neutrophil infiltration to sites of parasitic invasion, followed by activation of mast cells, macrophages, natural killer T cells, and subsequent secretion of pro-inflammatory cytokines [148]. By studying the pro-inflammatory response and mucus secretion due to E. histolytica, two recent data indicate that: (i) pro-inflammatory responses correlate with the downregulation of the cell differentiation-related transcription factor Math1 (homologous mouse atonal 1) [156], the inhibition of which leads, during tissue development, to a reduction in the population of goblet cells [157]; (ii) microbiota restore normal responses for mucus secretion and pro-inflammatory cytokine activation, in a mouse model of antibiotic-induced dysbiosis infected with E. histolytica [158]. These results suggest firstly a direct effect of trophozoites on the regulation of goblet cells persistence and secondly, a role of the microbiota in the control of pro-inflammatory responses, which acts in addition to the nutritional and protective role of amoebae mentioned above. Research on the nature, activity, and importance (in terms of pathogenicity) of amoebic “virulence factors” and signalling supporting the intestinal invasive process has been intensive and are the subject of detailed reviews [7,159]. Several aspects will be summarized below aimed at further enriching these publications.
Disruption of epithelial cells junctions by E. histolytica
The epithelium facing the intestinal lumen is composed of a single layer of polarized cells (enterocytes, goblet, enteroendocrine, Paneth, microfold, cup and chemosensory) constituting the physicochemical and immunological barrier against luminal antigens and enteric microbiota while allowing the absorption of nutrients and water [66]. Sealing cells together needs of tight junctions (TJs), adherens junctions (AJs), and desmosomes (DSMs) [160]; particularly, TJs regulate paracellular traffic of macromolecules and ions across the epithelium. TJ-specific families of integral membrane proteins include occludin, claudins, and junctional adhesion molecules (JAMs) that are responsible for maintaining the ion barrier of the paracellular space; claudins interact with scaffolding proteins like zona occludens (ZO-1 and ZO-2), which link them to other cytoplasmic proteins and actin microfilaments [161]. Occludins regulate the flux of macromolecules, while claudins provide control of ionic flux and limit the diffusion of proteins and lipids acting as a membrane fence [162] and, in combination with JAMs, regulate epithelial polarity [163].
The cysteine proteinase CP112 from E. histolytica is involved in the degradation of the cell-cell junction [164]. CP112 has been described with a canonical catalytic domain and an RGD motif, this enzyme degrades type I collagen, fibronectin, and haemoglobin. The proteinase CP112, together with the adhesin ADH, forms the virulence complex EhCPADH [165]. Trophozoites overexpressing CP112 or the purified enzyme cause a drop in transepithelial electrical resistance of cells and disrupt the fence function of TJs in enterocyte-like cells (Caco-2 cells); in contrast, CP112 does not affect the paracellular permeability of macromolecules [164]. By means of high-resolution confocal microscopy and immunoprecipitation essay, the authors demonstrated that purified CP112 affects claudin-1 and claudin-2 integrity; no effect was found on claudin-4, occludin, ZO-1, or ZO-2; treatment of mice with CP112 protease confirmed these data. Additional findings suggest that trophozoites could also affect AJs and DSMs and indicate a role for CP112 in the multifactorial virulence event of E. histolytica [166]. CP112 is not the only factor disrupting TJs; for example, trophozoites secretes the inflammatory molecule prostaglandin E2 (PGE2), which, in mammalian cells, activates secretion of chloride and breaks TJs ion barrier acting on claudins with the increased Na+ permeability; PGE2 from E. histolytica dissociates claudin-4 from mammalian cell membrane affecting TJs ion barrier [167]. Other additional proteases participate in alterations of TJ proteins: CP-A5 (tested by amoebic infection in Muc2−/− mice) [168] and serine proteases participating in transepithelial electrical resistance dropping of epithelial cells [169].
Due to the primary function of cell junctions as regulators of ion transport in the intestine, it is tempting to correlate the activities described above, concerning TJ disruption by amoebic proteases, with two important sets of data regarding the early stages of amoebic infection. Based on the cytotoxic properties of E. histolytica leading to mammalian cells death, the authors screened a library of human interfering RNAs (RNAi) to select enterocyte-like cells resistant to die in the presence of virulent trophozoite [170]. The identification of genes significantly enriched, whose silencing by RNAi increase cell survival upon contact with E. histolytica, highlight ion transporters channels for Ca2±dependent K+ (5 genes); Cl− (4 genes), Na+ (3 genes) and Ca2+ (2 genes). Further drug inhibition of K+ efflux prevented the death of intestinal epithelial cells and macrophages, indicating direct activation of K+ efflux as an early marker of tissue responses following intestinal invasion by E. histolytica [170]. The second dataset corresponds to the transcriptome and proteome analysis of human factors significantly modulated at the early stages of the interaction between virulent E. histolytica and the 3D scaffold model of the human colon [67]. Upregulated genes for receptors acting as ion channels have been identified: the epsilon subunit of the gamma-aminobutyric acid receptor; the CL channel involved in ulcerative colitis [171], and the epsilon subunit of the acetylcholine receptor that can open ion channels [172]. The long non-coding SLC9A3 antisense RNA 1 [173] targeting the SLC9A3 gene that encodes the Na+/H+ exchanger NHE3 associated with congenital diarrhoea [174] was also upregulated. At the protein level, abundantly secreted are components of desmosomes and factors linking them to the cytoskeleton (desmoplakin, plakophilin-3, and plectin-1); these are important for the maintenance of tissue integrity [175].
Taken together, the data suggest that disruption of structures responsible for cell-cell junctions combined with disruption of ion channels, are major lesions occurring at the early stages of contact between E. histolytica and the intestinal epithelium, they both lead to the collapse of tissue architecture. Therefore, mucus secretagogue activity, tissue leakage, and epithelial collapse, are responsible for the early symptoms of amoebic dysentery. They are the premises for the expected death of host cells, although how epithelial cells die because of interaction with E. histolytica remains an open question.
Mammalian cell death upon contact with E. histolytica
Following the entry of E. histolytica in the intestinal mucosa, the cells die. The data demonstrates that the destruction of mammalian cells requires contact with trophozoite that trigger calcium influx. Adhesion of trophozoites to enterocytes occurs via various factors on the amoebic surface, including LPPG, Gal-GalNAc lectin, KERP1, STIRP, and the CPAHD112 complex [18]; the contact led to the death of human cells in large proportions; approximately 40% of cultured enterocyte-like cells are killed within 30 minutes of interaction [150,176]. Dead cells can be ingested by phagocytosis, which is an important process for clearance of pathogens, necrotic or apoptotic cells [177]. In E. histolytica, phagocytosis is a way to acquire nutrients as this parasite is a professional phagocytic cell ingesting bacteria [178] and diverse mammalian cell types [179]. However, the interaction of living cells (e.g. epithelial, erythrocytes or lymphocytes) and E. histolytica involves complex steps. Amoebae that encounter mammalian cells rapidly induce trogocytosis, a phenomenon where amoebae bite and ingest distinct host cell fragments, contributing to cell killing [180]. Uptake and display of mammalian membrane proteins on the surface of the amoeba protect the parasite from lysis by inhibiting the complement cascade, indicating that trogocytosis contributes to amoebic defence strategies for immune evasion [181,182]. Trogocytosis and phagocytosis occur by different mechanisms, although many of the early steps and molecules involved may be common [183,184].
Contact-dependent cell killing by E. histolytica triggers apoptosis of T cells and neutrophils through activation of host cell caspase 3 [185], but caspase-independent death mechanisms in T cells have also been described; these include protein tyrosine dephosphorylation and ROS generation and are associated with the amoebic kinases, PI3K and PKC [186]. Human enterocytic cell lines can undergo apoptosis (associated with caspases-3 and −9 activation) upon interaction with E. histolytica [187]. In search of the molecular mechanisms inducing the apoptotic process of enterocytes, the authors investigated non-coding RNAs, such as microRNAs (miRNAs), which are negative regulators of gene expression. They revealed a set of six miRNAs with complex miRNA-mRNA co-regulatory networks in which the miRNAs target the anti-apoptotic genes XIAP, API5, BCL2, and AKT1 [187]. One of them, miR-643 exerts post-transcriptional downregulation of XIAP by targeting its mRNA 3’-UTR. The use of antagonists of miR-643 leads to the restoration of XIAP levels and the suppression of apoptosis and activation of caspases 3 and 9. The authors conclude that there is a functional role for the miR-643/XIAP axis in the apoptosis of enterocytes activated by E. histolytica [187]. The role of the other miRNAs discovered in this work remains ongoing experimentation; as well as the role of upregulated long non-coding RNAs discovered in the 3D scaffold model of the human colon [67].
LncRNAs are involved in many cellular processes and can interact with miRNAs to modulate their roles, including in epigenetic modifications regulating gene expression [188]. This area of research is terra incognita with respect to amoebic infection and progression of amoebiasis; however, interesting advances have been made in the study of intestinal host-bacteria interactions. Induction of miRNA has been correlated with the relative abundance of several microbiota taxa [189]; suggesting that the microbiota modulates miRNA induction mainly through metabolites [190]. Conversely, miRNAs secreted by intestinal epithelial cells modify the composition of the microbiota [190]. Certainly, new avenues of research concerning the control by human cells of the amoebic cytotoxic effect call for studies determining the involvement of non-coding human RNAs in the regulation of enterocyte death; in return, it is also important to understand the role of specific amoebic natural antisense RNAs [85] appearing during the invasion of the human colon in the control of virulence factors.
Human host defences at the epithelium and cell-mediated innate immunity responses against E. histolytica
Antimicrobial peptides followed by cell-mediated innate immunity corresponds to the second level of intestine defence against invasion by E. histolytica; these include defensins, regenerating islet-derived proteins REGs and alarmins. Defensins are small peptides that integrate into the membranes and form pores devoted to kill the invading microbe. E. histolytica induce secretion by enterocytic-like cells of α and β defensins [191], which, when purified, can kill trophozoites in vitro [192]; but at the same time, the genes encoding β defensins are downregulated in the 3D-scaffold model of infection [67] and in the human xenograft model [193], probably as an amoebic defence mechanism that requires more investigations. The antimicrobial peptide REG1A (anti-apoptotic) and REG1B mRNA increase during acute amoebic infection in humans [194], and REG4 (anti-inflammatory) is secreted by cells in the 3D-intestinal scaffold model [67]. Alarmins are endogenous molecules released to signal cell or tissue stress or damage. The ubiquitous nuclear DNA-binding protein call high-mobility group box 1 protein (HMGB1) is an alarmin activating innate immunity, it is released by necrotic cells or secreted by macrophages, natural killer cells, and dendritic cells. HMGB1 is activated and released from macrophages in response to E. histolytica invasion [195]. HMGB1 secretion depends on the microbiota and this molecule shapes the magnitude of the pro-inflammatory response during amoebiasis.
A very broad view of intestinal immune responses emerges when considering the activities of the microbiota (for example, the production of metabolites) and the diversity of responses of the immune system due to the presence of enteric pathogens [196]. A brief summary of these responses highlights several factors. The Toll-like receptors (TLR) on enterocytes surface, which recognizes microbial-derived compounds and trigger signalling cascades ending with nuclear translocation of NF-κB transcription factor and secretion of cytokines and chemokines, (e.g. TNF, IL-6, IL-8, IL-18, and CCL20). These molecules activate immune cells underlying the enterocytes layer, which migrate to the infection site (e.g. dendritic cells, monocytes, neutrophils, and T lymphocytes). Antimicrobial factors, including β-defensins and the inducible nitric oxide synthase generating nitric oxide, are also induced following TLR activation. The pathogen and the inflammation damage the enterocytes, these can physically expel themselves from the intestinal epithelium by a mechanism that involves the activation of inflammatory caspases (caspase-1, −4, and −11) as along with caspase-8. Activated caspases induce the macromolecular signalling complex called “inflammasome,” triggering the processing and secretion of the pro-inflammatory cytokine IL-18, as well as the induction of a specialized form of inflammatory cell death called pyroptosis [197]. The last step of pyroptosis requires the cleavage of the pore-forming protein gasdermin D by caspase-1; the N-terminus end of gasdermin D forms a transmembrane pore that disturbs ion and water regulation, and the dying cells release cytokines such as IL-1β, IL-18 and the prostaglandin PGE2, resulting in subsequent interferon-γ (IFN-γ) production and neutrophil recruitment [198].
Many of these components marking immune responses are evoked during invasion of the colonic mucosa by E. histolytica, which triggers a complex inflammatory process with production of interleukins (IL-1β, IL-6, IL-8), IFN-γ, and Tumour Necrosis Factor (TNF) [17]. These factors activate immune cells, including neutrophils, lymphocytes, and macrophages, which are further expected to kill trophozoites through the action of ROS and NO.
Regarding macrophages, the LPPG of E. histolytica is recognized by TLR 2 and TLR4, with activation of NF-kB and release of IL-8, IL-10, IL-12p40, and TNF [199]. Caspase-1 (processed by CP-A5) activates the NLRP3-like inflammasome [200] and cleaves pro-IL-1β into the active form IL-1β, the secretion of which also depends on the activity of caspase-4 and gasdermin D [201].
Neutrophil infiltration is intense in the human colon due to the density of E. histolytica [202], and they kill E. histolytica in the presence of TNF and IFN-γ primarily via ROS [203,204]. Decreased microbiota diversity in humans or in the mouse model increases the severity of E. histolytica infection, likely due to decreased neutrophil recruitment to the infection site [205]; however, the mechanisms of amoeba−neutrophil interaction, and consequent parasite and cell death, are far from determined. Exposure of human neutrophils to E. histolytica, or purified LPPG, induced the release of neutrophil extracellular traps (NET) in a time- and dose-dependent manner [206,207], but it is not clear that these NETs are able to kill E. histolytica. Mechanisms of NETosis involve translocation of elastase to the nucleus due to generation of ROS by NADPH oxidase (NOX2) or by activation of peptidyl arginine deiminase 4 (PAD4); both mechanisms result in DNA decondensation. Amoeba-induced NETosis is independent of ROS and PAD4 activity; however, it depends on the presence of extracellular calcium and serine protease activity [207], the signalling pathway leading to NETosis involves the activation of Raf/MEK/ERK and NF-κB [208]. Besides oxidative and nitrosative stresses, some cytokines (e.g. TNF) can act as a chemoattractant for E. histolytica by increasing trophozoite motility and directionality [209], the amoebic surface protein CSP has homologies with human TNF receptor, even a blockage of CSP abolishes the chemotaxis towards TNF and blocks the parasitic invasion of explants of the human colon [210].
Intestinal epithelial cells normally produce interleukin-25 (IL-25), but this is decreased in humans with amoebic colitis [211]; this cytokine plays a role in maintaining intestinal barrier function and inhibit TNF production. IL-25-mediated protection is accompanied by a decrease in TNF levels in experimental models of amoebiasis [211]. IL-33 (nuclear alarmin) activates innate lymphoid cell inducers of type 2 immune responses; the IL-33 encoding gene is upregulated in humans with colonic amebiasis, suggesting that IL-33 is involved in the induction of barrier repair mechanisms to protect intestinal tissue from E. histolytica [212]. Overall, this rapid overview of the data confirms the complexity of innate immune responses against E. histolytica considering the diversity of activated cells, secreted products and their consequences on tissue integrity (Figure 3).
Figure 3.
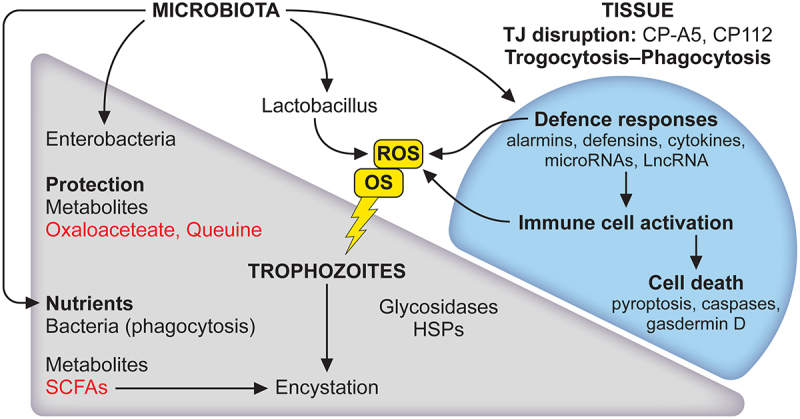
The microbiota has a role to play in the lifecycle of E. histolytica and tissue responses. The balance between trophozoite and cyst is strongly influenced by the microbiota. First, amoebae phagocytose bacteria and derived nutrients promote the growth of trophozoites. Secreted bacterial products such as short-chain fatty acids (SCFAs) are expected to provide energy for parasite growth. During carbon source deprivation, SCFAs stimulate the production of mucin, which could be used as a carbon source. Acetate is a SCFA that induces encystation in vitro in E. histolytica. Second, an important activity of bacteria is to act as a parasite survival aid in stressful situations like oxidative stress (OS); it regulates the expression of amoebic genes, metabolites such as oxalacetate and queuine, derived from Enterobacteriaceae, protect E. histolytica from OS. This response is not universal since Lactobacillus acidophilus (a probiotic) modifies the amoebic transcriptome differently and causes oxidation of vital E. histolytica proteins leading to the death of trophozoites. Major lesions occurring at the early stages of contact between E. histolytica and the intestinal epithelium led to disruption of tight junctions combined with disruption of ion channels tissue architecture collapse. Mammalian cells die by trogocytosis, apoptosis and phagocytosis. The inflammatory response occurs leading to cell death, the microbiota influence alarmins activity. For references see the main text.
Multiple other factors of interest implicated in the intestinal defence against E. histolytica, or having an impact on the development of amoebiasis, include genetic [213,214], social and nutritional aspects [215–217]; they will not be discussed in this review. For the implications of virulence-related factors as potential targets for novel therapies for amoebiasis, the following publications should be considered [218–221].
Conclusion
Research on the pathways of intestinal amoebiasis and its prevention remains a real challenge due to the complex and multifaceted relationship between virulent E. histolytica and the host during the different stages of amoebiasis. The confrontation of trophozoites with microbiota, human immunity and their stress-inducing products, activate regulatory pathways necessary for the genetic, transcriptomic, and epigenetic phenomena that maintain fitness of E. histolytica and ensure its survival in the human body. Although insufficiently considered, the microbiota plays a major role in the relationship of E. histolytica with the intestine; remarkably, it can influence lifecycle, stress response, and pathogenicity. The simplicity of the amoebic lifecycle guarantees the dissemination of the parasite among humans and therefore the persistence of Entamoeba species as commensal-like (or symbiotic) microbes. Even in the future, when virulence and pathogenicity will be controlled by new pharmacological treatments or vaccination, E. histolytica, as a parasite, will probably continue to evolve with humans like other species of Entamoeba.
Acknowledgements
The author apologizes to the researchers who could not be cited due to text length limitations. Thanks to Sherri Smith for her critical revision of this manuscript.
This article is a tribute to our colleague Dr. Alfonso Olivos Garcia (UNAM, Mexico) who died in May 2020 from COVID-19, in the fullness of his youth. I acknowledge here his important contribution to our understanding of virulence attenuation in E. histolytica.
Funding Statement
The author received funding from the European ERA-NET Infect-ERA program AMOEBAC (ANR-14-IFEC-0001-02) and the support of the French Parasitology consortium Labex ParaFrap (grant ANR-11-LABX0024). The funders had no role in the study design, data collection, or in the analysis, decision to publish, or preparation of the manuscript.ORCID ID: https://orcid.org/0000-0001-6551-9012
Data sharing
Data sharing not applicable – no new data generated.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Di Cristanziano V, Farowski F, Berrilli F, et al. Analysis of human gut microbiota composition associated to the presence of commensal and pathogen microorganisms in Côte d’Ivoire. Microorganisms. 2021. Aug 18;9(8):1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Even G, Lokmer A, Rodrigues J, et al. Changes in the human gut microbiota associated with colonization by Blastocystis sp. and Entamoeba spp. in non-industrialized populations. Front Cell Infect Microbiol. 2021;11:533528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lukes J, Stensvold CR, Jirku-Pomajbikova K, et al. Are human intestinal eukaryotes beneficial or commensals? PLOS Pathogens. 2015. Aug;11(8):e1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chiaranunt P, Burrows K, Ngai L, et al. NLRP1B and NLRP3 Control the Host Response following Colonization with the Commensal Protist Tritrichomonas musculis. J Immunol. 2022. Apr 1;208(7):1782–22. [DOI] [PubMed] [Google Scholar]
- [5].Le Bailly M, Maicher C, Dufour B.. Archaeological occurrences and historical review of the human amoeba, Entamoeba histolytica, over the past 6000years. Infect Genet Evol. 2016. Aug;42:34–40. [DOI] [PubMed] [Google Scholar]
- [6].Wilson IW, Weedall GD, Lorenzi H, et al. Genetic diversity and gene family expansions in members of the genus Entamoeba. Genome Biol Evol. 2019. Jan 21;11(3):688–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marie C, Petri WA Jr.. Regulation of virulence of Entamoeba histolytica. Annu Rev Microbiol. 2014;68:493–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Haque R, Duggal P, Ali IM, et al. Innate and acquired resistance to amebiasis in bangladeshi children. J Infect Dis. 2002. Aug 15;186(4):547–552. [DOI] [PubMed] [Google Scholar]
- [9].Shirley DT, Farr L, Watanabe K, et al. A review of the global burden, new diagnostics, and current therapeutics for amebiasis. Open Forum Infect Dis. 2018. Jul;5(7):ofy161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Alvarado-Esquivel C, Hernandez-Tinoco J, Francisco Sanchez-Anguiano L, et al. Serosurvey of Entamoeba histolytica exposure among Tepehuanos population in Durango, Mexico. Int J Biomed Sci. 2015. Jun;11(2):61–66. [PMC free article] [PubMed] [Google Scholar]
- [11].Zhou F, Li M, Li X, et al. Seroprevalence of Entamoeba histolytica infection among Chinese men who have sex with men. PLoS negl trop dis. 2013;7(5):e2232. DOI: 10.1371/journal.pntd.0002232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nath J, Ghosh SK, Singha B, et al. Molecular epidemiology of amoebiasis: a cross-sectional study among North East Indian population. PLoS negl trop dis. 2015. Dec;9(12):e0004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ozin Y, Kilic MZ, Nadir I, et al. Presence and diagnosis of amebic infestation in Turkish patients with active ulcerative colitis. Eur J Intern Med. 2009. Sep;20(5):545–547. [DOI] [PubMed] [Google Scholar]
- [14].Shirley DT, Watanabe K, Moonah S. Significance of amebiasis: 10 reasons why neglecting amebiasis might come back to bite us in the gut. PLoS negl trop dis. 2019. Nov;13(11):e0007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Weir CB, Le JK. Metronidazole. Treasure Island (FL): StatPearls; 2022. [Google Scholar]
- [16].Marie C, Petri WA Jr.. Amoebic dysentery. BMJ Clin Evid. 2013. Aug 30;2013;1:16. [PMC free article] [PubMed] [Google Scholar]
- [17].Nakada-Tsukui K, Nozaki T. Immune response of amebiasis and immune evasion by Entamoeba histolytica. Front Immunol. 2016;7:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Guillen N. Signals and signal transduction pathways in Entamoeba histolytica during the life cycle and when interacting with bacteria or human cells. Mol Microbiol. 2021. May;115(5):901–915. [DOI] [PubMed] [Google Scholar]
- [19].Seid M, Yohanes T, Goshu Y, et al. The effect of compliance to Hand hygiene during COVID-19 on intestinal parasitic infection and intensity of soil transmitted helminthes, among patients attending general hospital, southern Ethiopia: observational study. PloS One. 2022;17(6):e0270378. DOI: 10.1371/journal.pone.0270378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Krishnan D, Ghosh SK. Morphological and motility features of the stable bleb-driven monopodial form of entamoeba and its importance in encystation. Infect Immun. 2020. Jul 21;88(8). DOI: 10.1128/IAI.00903-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mi-Ichi F, Tsugawa H, Arita M, et al. Pleiotropic roles of cholesteryl sulfate during Entamoeba Encystation: involvement in cell rounding and development of membrane impermeability. mSphere. 2022. Aug 31;7(4):e0029922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mousa EAA, Sakaguchi M, Nakamura R, et al. The dynamics of ultrastructural changes during Entamoeba invadens encystation. Parasitology. 2020. Oct;147(12):1305–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Singh N, Naiyer S, Bhattacharya S. Ultra-structural analysis and morphological changes during the differentiation of trophozoite to cyst in Entamoeba invadens. Mol Biochem Parasitol. 2021. Mar;242:111363. [DOI] [PubMed] [Google Scholar]
- [24].Manna D, Ehrenkaufer GM, Lozano-Amado D, et al. Entamoeba stage conversion: progress and new insights. Curr Opin Microbiol. 2020. Dec;58:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Coppi A, Merali S, Eichinger D. The enteric parasite Entamoeba uses an autocrine catecholamine system during differentiation into the infectious cyst stage. J Biol Chem. 2002. Mar 8;277(10):8083–8090. [DOI] [PubMed] [Google Scholar]
- [26].Eichinger D. A role for a galactose lectin and its ligands during encystment of Entamoeba. J Eukaryot Microbiol. 2001. Jan-Feb;48(1):17–21. [DOI] [PubMed] [Google Scholar]
- [27].Ehrenkaufer GM, Hackney JA, Singh U. A developmentally regulated Myb domain protein regulates expression of a subset of stage-specific genes in Entamoeba histolytica. Cell Microbiol. 2009. Jun;11(6):898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Manna D, Lentz CS, Ehrenkaufer GM, et al. An NAD(+)-dependent novel transcription factor controls stage conversion in Entamoeba. Elife. 2018. Oct 30;7. DOI: 10.7554/eLife.37912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Manna D, Lozano-Amado D, Ehrenkaufer G, et al. The NAD(+) responsive transcription factor ERM-BP functions downstream of cellular aggregation and is an early regulator of development and heat shock response in Entamoeba. Front Cell Infect Microbiol. 2020;10:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Manna D, Singh U. Nuclear Factor Y (NF-Y) modulates Encystation in Entamoeba via stage-specific expression of the NF-YB and NF-YC Subunits. MBio. 2019. Jun 18;10(3). DOI: 10.1128/mBio.00737-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lozano-Amado D, Avila-Lopez PA, Hernandez-Montes G, et al. A class I histone deacetylase is implicated in the encystation of Entamoeba invadens. Int J Parasitol. 2020. Oct;50(12):1011–1022. [DOI] [PubMed] [Google Scholar]
- [32].Kawano-Sugaya T, Izumiyama S, Yanagawa Y, et al. Near-chromosome level genome assembly reveals ploidy diversity and plasticity in the intestinal protozoan parasite Entamoeba histolytica. BMC Genomics. 2020. Nov 23;21(1):813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mukherjee C, Majumder S, Lohia A. Inter-cellular variation in DNA content of Entamoeba histolytica originates from temporal and spatial uncoupling of cytokinesis from the nuclear cycle. PLoS negl trop dis. 2009;3(4):e409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Das S, Lohia A. Delinking of S phase and cytokinesis in the protozoan parasite Entamoeba histolytica. Cell Microbiol. 2002. Jan;4(1):55–60. [DOI] [PubMed] [Google Scholar]
- [35].Clark CG, Alsmark UC, Tazreiter M, et al. Structure and content of the Entamoeba histolytica genome. Adv Parasitol. 2007;65:51–190. [DOI] [PubMed] [Google Scholar]
- [36].Lohia A. The cell cycle of Entamoeba histolytica. Mol Cell Biochem. 2003. Nov;253(1–2):217–222. [DOI] [PubMed] [Google Scholar]
- [37].Clark CG, Roger AJ. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc Natl Acad Sci USA. 1995. Jul 3;92(14):6518–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ali V, Nozaki T. Current therapeutics, their problems, and sulfur-containing-amino-acid metabolism as a novel target against infections by “amitochondriate” protozoan parasites. Clinical Microbiology Reviews. 2007. Jan;20(1):164–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Saavedra E, Encalada R, Vazquez C, et al. Control and regulation of the pyrophosphate-dependent glucose metabolism in Entamoeba histolytica. Mol Biochem Parasitol. 2019. Apr;229:75–87. [DOI] [PubMed] [Google Scholar]
- [40].Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999. Jun;32(5):1013–1021. [DOI] [PubMed] [Google Scholar]
- [41].Santos HJ, Nozaki T. The mitosome of the anaerobic parasitic protist Entamoeba histolytica: a peculiar and minimalist mitochondrion-related organelle. J Eukaryot Microbiol. 2022. May 19;69:e12923. DOI: 10.1111/jeu.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mi-Ichi F, Miyamoto T, Takao S, et al. Entamoeba mitosomes play an important role in encystation by association with cholesteryl sulfate synthesis. Proc Natl Acad Sci USA. 2015. Jun 2;112(22):E2884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Alsmark UC, Sicheritz-Ponten T, Foster PG, et al. Horizontal gene transfer in eukaryotic parasites: a case study of Entamoeba histolytica and Trichomonas vaginalis. Methods Mol Biol. 2009;532:489–500. [DOI] [PubMed] [Google Scholar]
- [44].Grant JR, Katz LA. Phylogenomic study indicates widespread lateral gene transfer in Entamoeba and suggests a past intimate relationship with parabasalids. Genome Biol Evol. 2014. Sep;6(9):2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Verner Z, Zarsky V, Le T, et al. Anaerobic peroxisomes in Entamoeba histolytica metabolize myo-inositol. PLOS Pathogens. 2021. Nov;17(11):e1010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Santos HJ, Hanadate Y, Imai K, et al. Entamoeba histolytica EHD1 is involved in mitosome-endosome contact. MBio. 2022. Apr 26;13(2):e0384921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Davis PH, Zhang Z, Chen M, et al. Identification of a family of BspA like surface proteins of Entamoeba histolytica with novel leucine rich repeats. Mol Biochem Parasitol. 2006. Jan;145(1):111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vayssie L, Vargas M, Weber C, et al. Double-stranded RNA mediates homology-dependent gene silencing of gamma-tubulin in the human parasite Entamoeba histolytica. Mol Biochem Parasitol. 2004. Nov;138(1):21–28. [DOI] [PubMed] [Google Scholar]
- [49].Perdomo D, Ait-Ammar N, Syan S, et al. Cellular and proteomics analysis of the endomembrane system from the unicellular Entamoeba histolytica. J Proteomics. 2015. Jan 1;112:125–140. [DOI] [PubMed] [Google Scholar]
- [50].Vaithilingam A, Teixeira JE, Huston CD. Endoplasmic reticulum continuity in the protozoan parasite Entamoeba histolytica: evolutionary implications and a cautionary note. Commun Integr Biol. 2008;1(2):172–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Manich M, Hernandez-Cuevas N, Ospina-Villa JD, et al. Morphodynamics of the actin-rich cytoskeleton in Entamoeba histolytica. Front Cell Infect Microbiol. 2018;8:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hernandez-Cuevas NA, Jhingan GD, Petropolis D, et al. Acetylation is the most abundant actin modification in Entamoeba histolytica and modifications of actin’s amino-terminal domain change cytoskeleton activities. Cell Microbiol. 2019. Apr;21(4):e12983. [DOI] [PubMed] [Google Scholar]
- [53].Rath PP, Gourinath S. The actin cytoskeleton orchestra in Entamoeba histolytica. Proteins. 2020. Oct;88(10):1361–1375. [DOI] [PubMed] [Google Scholar]
- [54].Mornico D, Hon CC, Koutero M, et al. RNA Sequencing reveals widespread transcription of natural antisense RNAs in Entamoeba species. Microorganisms. 2022. Feb 8;10(2):396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rolhion N, Chassaing B. When pathogenic bacteria meet the intestinal microbiota. Philos Trans R Soc Lond B Biol Sci. 2016. Nov 5;371(1707):20150504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. 2020. Apr 26;38:23–48. DOI: 10.1146/annurev-immunol-070119-115104. [DOI] [PubMed] [Google Scholar]
- [57].Magnusdottir S, Ravcheev D, de Crecy-Lagard V, et al. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Burgess SL, Oka A, Liu B, et al. Intestinal parasitic infection alters bone marrow derived dendritic cell inflammatory cytokine production in response to bacterial endotoxin in a diet-dependent manner. PLoS negl trop dis. 2019. Jul;13(7):e0007515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Byers J, Faigle W, Eichinger D. Colonic short-chain fatty acids inhibit encystation of Entamoeba invadens. Cell Microbiol. 2005. Feb;7(2):269–279. [DOI] [PubMed] [Google Scholar]
- [60].Burger-van Paassen N, Vincent A, Puiman PJ, et al. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009. May 13;420(2):211–219. [DOI] [PubMed] [Google Scholar]
- [61].Lokmer A, Cian A, Froment A, et al. Use of shotgun metagenomics for the identification of protozoa in the gut microbiota of healthy individuals from worldwide populations with various industrialization levels. PLoS ONE. 2019;14(2):e0211139. DOI: 10.1371/journal.pone.0211139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Iebba V, Santangelo F, Totino V, et al. Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Cote d’Ivoire. J Infect Dev Ctries. 2016. Sep 30;10(9):1035–1041. [DOI] [PubMed] [Google Scholar]
- [63].Verma AK, Verma R, Ahuja V, et al. Real-time analysis of gut flora in Entamoeba histolytica infected patients of Northern India. BMC Microbiol. 2012. Aug 22;12:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ankri S. Entamoeba histolytica—gut microbiota interaction: more than meets the eye. Microorganisms. 2021. Mar 12;9(3):581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zarate S, Taboada B, Yocupicio-Monroy M, et al. Human Virome. Arch Med Res. 2017. Nov;48(8):701–716. [DOI] [PubMed] [Google Scholar]
- [66].Aguilar-Rojas A, Olivo-Marin JC, Guillen N. Human intestinal models to study interactions between intestine and microbes. Open Biol. 2020. Oct;10(10):200199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Aguilar-Rojas A, Castellanos-Castro S, Matondo M, et al. Insights into amebiasis using a human 3D-intestinal model. Cell Microbiol. 2020. Aug;22(8):e13203. [DOI] [PubMed] [Google Scholar]
- [68].Aguilar-Diaz H, Diaz-Gallardo M, Laclette JP, et al. In vitro induction of Entamoeba histolytica cyst-like structures from trophozoites. PLoS negl trop dis. 2010. Feb 16;4(2):e607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Luna-Nacar M, Navarrete-Perea J, Moguel B, et al. Proteomic study of Entamoeba histolytica trophozoites, cysts, and cyst-like structures. PLoS ONE. 2016;11(5):e0156018. DOI: 10.1371/journal.pone.0156018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Barron-Gonzalez MP, Villarreal-Trevino L, Resendez-Perez D, et al. Entamoeba histolytica: cyst-like structures in vitro induction. Exp Parasitol. 2008. Apr;118(4):600–603. [DOI] [PubMed] [Google Scholar]
- [71].Wesel J, Shuman J, Bastuzel I, et al. Encystation of Entamoeba histolytica in axenic culture. Microorganisms. 2021. Apr 18;9(4). doi : 10.3390/microorganisms9040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ehrenkaufer GM, Eichinger DJ, Singh U. Trichostatin a effects on gene expression in the protozoan parasite Entamoeba histolytica. BMC Genomics. 2007. Jul 5;8:216. DOI: 10.1186/1471-2164-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sarid L, Ankri S. Are metabolites from the gut microbiota capable of regulating epigenetic mechanisms in the human parasite Entamoeba histolytica? Front Cell Dev Biol. 2022;10:841586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Krishnan D, Ghosh SK. Cellular events of multinucleated giant cells formation during the Encystation of Entamoeba invadens. Front Cell Infect Microbiol. 2018;8:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Turner NA, Eichinger D. Entamoeba invadens: the requirement for galactose ligands during encystment. Exp Parasitol. 2007. Aug;116(4):467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mi-Ichi F, Yoshida H, Hamano S. Entamoeba Encystation: new targets to prevent the transmission of Amebiasis. PLOS Pathogens. 2016. Oct;12(10):e1005845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Heron BT, Sateriale A, Teixeira JE, et al. Evidence for a novel Entamoeba histolytica lectin activity that recognises carbohydrates present on ovalbumin. Int J Parasitol. 2011. Feb;41(2):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Khan F, Kurre D, Suguna K. Crystal structures of a beta-trefoil lectin from Entamoeba histolytica in monomeric and a novel disulfide bond-mediated dimeric forms. Glycobiology. 2020. Jul 20;30(7):474–488. [DOI] [PubMed] [Google Scholar]
- [79].Petri WA Jr., Smith RD, Schlesinger PH, et al. Isolation of the galactose-binding lectin that mediates the in vitro adherence of Entamoeba histolytica. J Clin Invest. 1987. Nov;80(5):1238–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Petri WA Jr., Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu Rev Microbiol. 2002;56:39–64. [DOI] [PubMed] [Google Scholar]
- [81].Manning-Cela R, Meraz MA, Hernandez JM, et al. Actin mRNA levels and actin synthesis during the encystation of Entamoeba invadens. J Eukaryot Microbiol. 1994. Jul-Aug;41(4):360–365. [DOI] [PubMed] [Google Scholar]
- [82].Makioka A, Kumagai M, Ohtomo H, et al. Entamoeba invadens: enhancement of excystation and metacystic development by cytochalasin D. Exp Parasitol. 2001. Jul;98(3):145–151. [DOI] [PubMed] [Google Scholar]
- [83].Field J, Van Dellen K, Ghosh SK, et al. Responses of Entamoeba invadens to heat shock and encystation are related. J Eukaryot Microbiol. 2000. Sep-Oct;47(5):511–514. [DOI] [PubMed] [Google Scholar]
- [84].Varet H, Shaulov Y, Sismeiro O, et al. Enteric bacteria boost defences against oxidative stress in Entamoeba histolytica. Sci Rep. 2018. Jun 13;8(1):9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mornico D, Hon CC, Koutero M, et al. Genomic determinants for initiation and length of natural antisense transcripts in Entamoeba histolytica. Sci Rep. 2020. Nov 19;10(1):20190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lambeth JD, Neish AS. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu Rev Pathol. 2014;9:119–145. [DOI] [PubMed] [Google Scholar]
- [87].Singh V, Ahlawat S, Mohan H, et al. Balancing reactive oxygen species generation by rebooting gut microbiota. J Appl Microbiol. 2022. Jun;132(6):4112–4129. [DOI] [PubMed] [Google Scholar]
- [88].Shahi P, Trebicz-Geffen M, Nagaraja S, et al. Proteomic identification of oxidized proteins in Entamoeba histolytica by resin-assisted capture: insights into the role of arginase in resistance to oxidative stress. PLoS negl trop dis. 2016. Jan;10(1):e0004340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Vicente JB, Ehrenkaufer GM, Saraiva LM, et al. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cell Microbiol. 2009. Jan;11(1):51–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Rastew E, Vicente JB, Singh U. Oxidative stress resistance genes contribute to the pathogenic potential of the anaerobic protozoan parasite, Entamoeba histolytica. Int J Parasitol. 2012. Oct;42(11):1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Pearson RJ, Morf L, Singh U. Regulation of H2O2 stress-responsive genes through a novel transcription factor in the protozoan pathogen Entamoeba histolytica. J Biol Chem. 2013. Feb 8;288(6):4462–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Shaulov Y, Shimokawa C, Trebicz-Geffen M, et al. Escherichia coli mediated resistance of Entamoeba histolytica to oxidative stress is triggered by oxaloacetate. PLOS Pathogens. 2018. Oct;14(10):e1007295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Nagaraja S, Cai MW, Sun J, et al. Queuine is a nutritional regulator of Entamoeba histolytica response to oxidative stress and a virulence attenuator. MBio. 2021. Mar 9;12(2). doi : 10.1128/mBio.03549-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sarid L, Zanditenas E, Ye J, et al. Insights into the mechanisms of Lactobacillus acidophilus activity against Entamoeba histolytica by using thiol redox proteomics. Antioxidants (Basel). 2022. Apr 22;11(5):814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ramos F, Moran P, Gonzalez E, et al. Entamoeba histolytica and Entamoeba dispar: prevalence infection in a rural Mexican community. Exp Parasitol. 2005. Jul;110(3):327–330. [DOI] [PubMed] [Google Scholar]
- [96].Faqe Mahmood SA, Mustafa HB. Molecular identification and prevalence of Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii in Erbil City, Northern Iraq. Pol J Microbiol. 2020;69:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Davis PH, Schulze J, Stanley SL Jr.. Transcriptomic comparison of two Entamoeba histolytica strains with defined virulence phenotypes identifies new virulence factor candidates and key differences in the expression patterns of cysteine proteases, lectin light chains, and calmodulin. Mol Biochem Parasitol. 2007. Jan;151(1):118–128. [DOI] [PubMed] [Google Scholar]
- [98].Davis PH, Zhang X, Guo J, et al. Comparative proteomic analysis of two Entamoeba histolytica strains with different virulence phenotypes identifies peroxiredoxin as an important component of amoebic virulence. Mol Microbiol. 2006. Sep;61(6):1523–1532. [DOI] [PubMed] [Google Scholar]
- [99].Nakada-Tsukui K, Sekizuka T, Sato-Ebine E, et al. AIG1 affects in vitro and in vivo virulence in clinical isolates of Entamoeba histolytica. PLOS Pathogens. 2018. Mar;14(3):e1006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kumari V, Iyer LR, Roy R, et al. Genomic distribution of SINEs in Entamoeba histolytica strains: implication for genotyping. BMC Genomics. 2013. Jul 1;14:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ali IK, Mondal U, Roy S, et al. Evidence for a link between parasite genotype and outcome of infection with Entamoeba histolytica. J Clin Microbiol. 2007. Feb;45(2):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Ayeh-Kumi PF, Ali IM, Lockhart LA, et al. Entamoeba histolytica: genetic diversity of clinical isolates from Bangladesh as demonstrated by polymorphisms in the serine-rich gene. Exp Parasitol. 2001. Oct;99(2):80–88. [DOI] [PubMed] [Google Scholar]
- [103].Zaki M, Reddy SG, Jackson TF, et al. Genotyping of Entamoeba species in South Africa: diversity, stability, and transmission patterns within families. J Infect Dis. 2003. Jun 15;187(12):1860–1869. [DOI] [PubMed] [Google Scholar]
- [104].Blessmann J, Ali IK, Nu PA, et al. Longitudinal study of intestinal Entamoeba histolytica infections in asymptomatic adult carriers. J Clin Microbiol. 2003. Oct;41(10):4745–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Weedall GD, Clark CG, Koldkjaer P, et al. Genomic diversity of the human intestinal parasite Entamoeba histolytica. Genome bio. 2012. May 25;13(5):R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Gilchrist CA, Ali IK, Kabir M, et al. A Multilocus Sequence Typing System (MLST) reveals a high level of diversity and a genetic component to Entamoeba histolytica virulence. BMC Microbiol. 2012. Jul 27;12:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Das K, Sardar SK, Ghosal A, et al. Multilocus sequence typing (MLST) of Entamoeba histolytica identifies kerp2 as a genetic marker associated with disease outcomes. Parasitol Int. 2021. Aug;83:102370. [DOI] [PubMed] [Google Scholar]
- [108].Santi-Rocca J, Rigothier MC, Guillen N. Host-microbe interactions and defense mechanisms in the development of amoebic liver abscesses. Clinical Microbiology Reviews. 2009. Jan;22(1):65–75. DOI: 10.1128/CMR.00029-08 Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Carrero JC, Reyes-Lopez M, Serrano-Luna J, et al. Intestinal amoebiasis: 160 years of its first detection and still remains as a health problem in developing countries. Int J Med Microbiol. 2020. Jan;310(1):151358. [DOI] [PubMed] [Google Scholar]
- [110].Olivos A, Ramos E, Nequiz M, et al. Entamoeba histolytica: mechanism of decrease of virulence of axenic cultures maintained for prolonged periods. Exp Parasitol. 2005. Jul;110(3):309–312. [DOI] [PubMed] [Google Scholar]
- [111].Olivos-Garcia A, Saavedra E, Nequiz M, et al. The oxygen reduction pathway and heat shock stress response are both required for Entamoeba histolytica pathogenicity. Curr Genet. 2016. May;62(2):295–300. [DOI] [PubMed] [Google Scholar]
- [112].Olivos-Garcia A, Saavedra E, Ramos-Martinez E, et al. Molecular nature of virulence in Entamoeba histolytica. Infect Genet Evol. 2009. Dec;9(6):1033–1037. [DOI] [PubMed] [Google Scholar]
- [113].Santos F, Nequiz M, Hernandez-Cuevas NA, et al. Maintenance of intracellular hypoxia and adequate heat shock response are essential requirements for pathogenicity and virulence of Entamoeba histolytica. Cell Microbiol. 2015. Jul;17(7):1037–1051. [DOI] [PubMed] [Google Scholar]
- [114].Pineda E, Perdomo D. Entamoeba histolytica under oxidative stress: what countermeasure mechanisms are in place? Cells. 2017. Nov 21;6(4). DOI: 10.3390/cells6040044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ramos-Martinez E, Olivos-Garcia A, Saavedra E, et al. Entamoeba histolytica: oxygen resistance and virulence. Int J Parasitol. 2009. May;39(6):693–702. [DOI] [PubMed] [Google Scholar]
- [116].Weber C, Koutero M, Dillies MA, et al. Extensive transcriptome analysis correlates the plasticity of Entamoeba histolytica pathogenesis to rapid phenotype changes depending on the environment. Sci Rep. 2016. Oct 21;6:35852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Sottile ML, Nadin SB. Heat shock proteins and DNA repair mechanisms: an updated overview. Cell Stress Chaperones. 2018. May;23(3):303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Dubrez L, Causse S, Borges Bonan N, et al. Heat-shock proteins: chaperoning DNA repair. Oncogene. 2020. Jan;39(3):516–529. [DOI] [PubMed] [Google Scholar]
- [119].Santos F, Marcial-Quino J, Gomez-Manzo S, et al. Functional characterization and subcellular distribution of two recombinant cytosolic HSP70 isoforms from Entamoeba histolytica under normal and stress conditions. Parasitol Res. 2020. Apr;119(4):1337–1351. [DOI] [PubMed] [Google Scholar]
- [120].Santi-Rocca J, Smith S, Weber C, et al. Endoplasmic reticulum stress-sensing mechanism is activated in Entamoeba histolytica upon treatment with nitric oxide. PLoS ONE. 2012;7(2):e31777. DOI: 10.1371/journal.pone.0031777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proc Natl Acad Sci USA. 2011. Mar 15;Suppl 108(supplement_1):4659–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ehrencrona E, van der Post S, Gallego P, et al. The IgGfc-binding protein FCGBP is secreted with all GDPH sequences cleaved but maintained by interfragment disulfide bonds. J Biol Chem. 2021. Jul;297(1):100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Glover JS, Ticer TD, Engevik MA. Characterizing the mucin-degrading capacity of the human gut microbiota. Sci Rep. 2022. May 19;12(1):8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Sicard JF, Le Bihan G, Vogeleer P, et al. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol. 2017;7:387. DOI: 10.3389/fcimb.2017.00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Moncada D, Keller K, Chadee K. Entamoeba histolytica-secreted products degrade colonic mucin oligosaccharides. Infect Immun. 2005. Jun;73(6):3790–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Chadee K, Keller K, Forstner J, et al. Mucin and nonmucin secretagogue activity of Entamoeba histolytica and cholera toxin in rat colon. Gastroenterology. 1991. Apr;100(4):986–997. [DOI] [PubMed] [Google Scholar]
- [127].Bansal D, Ave P, Kerneis S, et al. An ex-vivo human intestinal model to study Entamoeba histolytica pathogenesis. PLoS negl trop dis. 2009. Nov 17;3(11):e551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Martinez-Ocana J, Maravilla P, Olivo-Diaz A. Interaction between human mucins and parasite glycoproteins: the role of lectins and glycosidases in colonization by intestinal protozoa. Rev Inst Med Trop Sao Paulo. 2020. Sep 4;62:e64. DOI: 10.1590/s1678-9946202062064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Riekenberg S, Flockenhaus B, Vahrmann A, et al. The beta-N-acetylhexosaminidase of Entamoeba histolytica is composed of two homologous chains and has been localized to cytoplasmic granules. Mol Biochem Parasitol. 2004. Dec;138(2):217–225. [DOI] [PubMed] [Google Scholar]
- [130].Thibeaux R, Weber C, Hon CC, et al. Identification of the virulence landscape essential for Entamoeba histolytica invasion of the human colon. PLOS Pathogens. 2013;9(12):e1003824. DOI: 10.1371/journal.ppat.1003824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Nag M, Lahiri D, Garai S, et al. Regulation of beta-amylase synthesis: a brief overview. Mol Biol Rep. 2021. Sep;48(9):6503–6511. [DOI] [PubMed] [Google Scholar]
- [132].Monroe JD, Storm AR, Badley EM, et al. Beta-Amylase1 and beta-amylase3 are plastidic starch hydrolases in Arabidopsis that seem to be adapted for different thermal, pH, and stress conditions. Plant Physiol. 2014. Dec;166(4):1748–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ma Y, Han Y, Feng X, et al. Genome-wide identification of BAM (beta-amylase) gene family in jujube (Ziziphus jujuba Mill.) and expression in response to abiotic stress. BMC Genomics. 2022. Jun 13;23(1):438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Siqueira-Neto JL, Debnath A, McCall LI, et al. Cysteine proteases in protozoan parasites. PLoS negl trop dis. 2018. Aug;12(8):e0006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Moncada D, Keller K, Chadee K. Entamoeba histolytica cysteine proteinases disrupt the polymeric structure of colonic mucin and alter its protective function. Infect Immun. 2003. Feb;71(2):838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lidell ME, Moncada DM, Chadee K, et al. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc Natl Acad Sci USA. 2006. Jun 13;103(24):9298–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Jacobs T, Bruchhaus I, Dandekar T, et al. Isolation and molecular characterization of a surface-bound proteinase of Entamoeba histolytica. Mol Microbiol. 1998. Jan;27(2):269–276. [DOI] [PubMed] [Google Scholar]
- [138].Lechner AM, Assfalg-Machleidt I, Zahler S, et al. RGD-dependent binding of procathepsin X to integrin alphavbeta3 mediates cell-adhesive properties. J Biol Chem. 2006. Dec 22;281(51):39588–39597. [DOI] [PubMed] [Google Scholar]
- [139].Hou Y, Mortimer L, Chadee K. Entamoeba histolytica cysteine proteinase 5 binds integrin on colonic cells and stimulates NFkappaB-mediated pro-inflammatory responses. J Biol Chem. 2010. Nov 12;285(46):35497–35504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Cornick S, Moreau F, Chadee K. Entamoeba histolytica Cysteine Proteinase 5 Evokes Mucin Exocytosis from Colonic Goblet Cells via alphavbeta3 Integrin. PLOS Pathogens. 2016. Apr;12(4):e1005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Shahi P, Moreau F, Chadee K. Entamoeba histolytica Cyclooxygenase-Like protein regulates cysteine protease expression and virulence. Front Cell Infect Microbiol. 2018;8:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Zanditenas E, Trebicz-Geffen M, Domínguez-García L, et al. Digestive exophagy of bacterial biofilms by an amoeba predator is mediated by specific biofilm recognition. bioRxiv. 2022. 2022.09.24.509356. [Google Scholar]
- [143].Thibeaux R, Dufour A, Roux P, et al. Newly visualized fibrillar collagen scaffolds dictate Entamoeba histolytica invasion route in the human colon. Cell Microbiol. 2012. May;14(5):609–621. [DOI] [PubMed] [Google Scholar]
- [144].Thibeaux R, Ave P, Bernier M, et al. The parasite Entamoeba histolytica exploits the activities of human matrix metalloproteinases to invade colonic tissue. Nat Commun. 2014. Oct 7;5:5142. [DOI] [PubMed] [Google Scholar]
- [145].Ankri S, Stolarsky T, Mirelman D. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol Microbiol. 1998. May;28(4):777–785. [DOI] [PubMed] [Google Scholar]
- [146].Faust DM, Marquay Markiewicz J, Danckaert A, et al. Human liver sinusoidal endothelial cells respond to interaction with Entamoeba histolytica by changes in morphology, integrin signalling and cell death. Cell Microbiol. 2011. Jul;13(7):1091–1106. [DOI] [PubMed] [Google Scholar]
- [147].Aguilar-Rojas A, Olivo-Marin JC, Guillen N. The motility of Entamoeba histolytica: finding ways to understand intestinal amoebiasis. Curr Opin Microbiol. 2016. Dec;34:24–30. [DOI] [PubMed] [Google Scholar]
- [148].Ghosh S, Padalia J, Moonah S. Tissue destruction caused by Entamoeba histolytica parasite: cell death, inflammation, invasion, and the gut microbiome. Curr Clin Microbiol Rep. 2019;6(1):51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Faust DM, Guillen N. Virulence and virulence factors in Entamoeba histolytica, the agent of human amoebiasis. Microbes Infect. 2012. Dec;14(15):1428–1441. [DOI] [PubMed] [Google Scholar]
- [150].Seigneur M, Mounier J, Prevost MC, et al. A lysine- and glutamic acid-rich protein, KERP1, from Entamoeba histolytica binds to human enterocytes. Cell Microbiol. 2005. Apr;7(4):569–579. [DOI] [PubMed] [Google Scholar]
- [151].MacFarlane RC, Singh U. Identification of an Entamoeba histolytica serine-, threonine-, and isoleucine-rich protein with roles in adhesion and cytotoxicity. Eukaryot Cell. 2007. Nov;6(11):2139–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Garcia-Rivera G, Rodriguez MA, Ocadiz R, et al. Entamoeba histolytica : a novel cysteine protease and an adhesin form the 112 kDa surface protein. Mol Microbiol. 1999. Aug;33(3):556–568. [DOI] [PubMed] [Google Scholar]
- [153].Betanzos A, Banuelos C, Orozco E. Host invasion by pathogenic amoebae: epithelial disruption by parasite proteins. Genes (Basel). 2019. Aug 14;10(8). DOI: 10.3390/genes10080618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Ralston KS. Taking a bite: amoebic trogocytosis in Entamoeba histolytica and beyond. Curr Opin Microbiol. 2015. Dec;28:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Nakada-Tsukui K, Nozaki T. Trogocytosis in unicellular eukaryotes. Cells. 2021. Nov 1;10(11). DOI: 10.3390/cells10112975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Leon-Coria A, Kumar M, Moreau F, et al. Defining cooperative roles for colonic microbiota and Muc2 mucin in mediating innate host defense against Entamoeba histolytica. PLOS Pathogens. 2018. Nov;14(11):e1007466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].VanDussen KL, Samuelson LC. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev Biol. 2010. Oct 15;346(2):215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Leon-Coria A, Kumar M, Workentine M, et al. Muc2 mucin and nonmucin microbiota confer distinct innate host defense in disease susceptibility and colonic injury. Cell Mol Gastroenterol Hepatol. 2021;11(1):77–98. DOI: 10.1016/j.jcmgh.2020.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Watanabe N, Nakada-Tsukui K, Nozaki T. Diversity of phosphoinositide binding proteins in Entamoeba histolytica. Parasitol Int. 2021. Aug;83:102367. [DOI] [PubMed] [Google Scholar]
- [160].Paradis T, Begue H, Basmaciyan L, et al. Tight junctions as a key for pathogens invasion in intestinal epithelial cells. Int J Mol Sci. 2021. Mar 2;22(5):2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Lu Z, Ding L, Lu Q, et al. Claudins in intestines: distribution and functional significance in health and diseases. Tissue Barriers. 2013. Jul 1;1(3):e24978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Lingaraju A, Long TM, Wang Y, et al. Conceptual barriers to understanding physical barriers. Semin Cell Dev Biol. 2015. Jun;42:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Otani T, Nguyen TP, Tokuda S, et al. Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity. J cell Biol. 2019. Oct 7;218(10):3372–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Cuellar P, Hernandez-Nava E, Garcia-Rivera G, et al. Entamoeba histolytica EhCP112 dislocates and degrades Claudin-1 and Claudin-2 at tight junctions of the intestinal epithelium. Front Cell Infect Microbiol. 2017;7:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Ocadiz-Ruiz R, Fonseca W, Linford AS, et al. The knockdown of each component of the cysteine proteinase-adhesin complex of Entamoeba histolytica (EhCPADH) affects the expression of the other complex element as well as the in vitro and in vivo virulence. Parasitology. 2016. Jan;143(1):50–59. [DOI] [PubMed] [Google Scholar]
- [166].Hernandez-Nava E, Cuellar P, Nava P, et al. Adherens junctions and desmosomes are damaged by Entamoeba histolytica: participation of EhCPADH complex and EhCP112 protease. Cell Microbiol. 2017. Nov;19(11):e12761. [DOI] [PubMed] [Google Scholar]
- [167].Lejeune M, Moreau F, Chadee K. Prostaglandin E2 produced by Entamoeba histolytica signals via EP4 receptor and alters claudin-4 to increase ion permeability of tight junctions. Am J Pathol. 2011. Aug;179(2):807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Kissoon-Singh V, Moreau F, Trusevych E, et al. Entamoeba histolytica exacerbates epithelial tight junction permeability and proinflammatory responses in Muc2(-/-) mice. Am J Pathol. 2013. Mar;182(3):852–865. [DOI] [PubMed] [Google Scholar]
- [169].Lauwaet T, Oliveira MJ, Callewaert B, et al. Proteinase inhibitors TPCK and TLCK prevent Entamoeba histolytica induced disturbance of tight junctions and microvilli in enteric cell layers in vitro. Int J Parasitol. 2004. Jun;34(7):785–794. [DOI] [PubMed] [Google Scholar]
- [170].Marie C, Verkerke HP, Theodorescu D, et al. A whole-genome RNAi screen uncovers a novel role for human potassium channels in cell killing by the parasite Entamoeba histolytica. Sci Rep. 2015. Sep 8;5:13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Aggarwal S, Ahuja V, Paul J. Attenuated GABAergic signaling in intestinal epithelium contributes to pathogenesis of ulcerative colitis. Dig Dis Sci. 2017. Oct;62(10):2768–2779. [DOI] [PubMed] [Google Scholar]
- [172].Shen XM, Okuno T, Milone M, et al. Mutations causing slow-channel myasthenia reveal that a valine ring in the channel pore of muscle AChR is optimized for stabilizing channel gating. Human mutation. 2016. Oct;37(10):1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Cheng Z, Bai Y, Wang P, et al. Identification of long noncoding RNAs for the detection of early stage lung squamous cell carcinoma by microarray analysis. Oncotarget. 2017. Feb 21;8(8):13329–13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Low END, Mokhtar NM, Wong Z, et al. Colonic mucosal transcriptomic changes in patients with long-duration ulcerative colitis revealed colitis-associated cancer pathways. J Crohn’s Colitis. 2019. May 27;13(6):755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Bouameur JE, Favre B, Borradori L. Plakins, a versatile family of cytolinkers: roles in skin integrity and in human diseases. J Invest Dermatol. 2014. Apr;134(4):885–894. [DOI] [PubMed] [Google Scholar]
- [176].Arhets P, Olivo JC, Gounon P, et al. Virulence and functions of myosin II are inhibited by overexpression of light meromyosin in Entamoeba histolytica. ?Mol Biol Cell. 1998. Jun;9(6):1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Jaganathan D, Bruscia EM, Kopp BT. Emerging concepts in defective macrophage phagocytosis in cystic fibrosis. Int J Mol Sci. 2022. Jul 13;23(14):7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Iyer LR, Verma AK, Paul J, et al. Phagocytosis of gut bacteria by Entamoeba histolytica. Front Cell Infect Microbiol. 2019;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Marion S, Guillen N. Genomic and proteomic approaches highlight phagocytosis of living and apoptotic human cells by the parasite Entamoeba histolytica. Int J Parasitol. 2006. Feb;36(2):131–139. [DOI] [PubMed] [Google Scholar]
- [180].Ralston KS, Solga MD, Mackey-Lawrence NM, et al. Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature. 2014. Apr 24;508(7497):526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Miller HW, Suleiman RL, Ralston KS. Trogocytosis by Entamoeba histolytica mediates acquisition and display of human cell membrane proteins and evasion of lysis by human serum. MBio. 2019. Apr 30;10(2). DOI: 10.1128/mBio.00068-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Miller HW, Tam TSY, Ralston KS. Entamoeba histolytica develops resistance to complement deposition and lysis after acquisition of human complement-regulatory proteins through trogocytosis. MBio. 2022. Apr 26;13(2):e0316321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Somlata NTK, Nozaki T. AGC family kinase 1 participates in trogocytosis but not in phagocytosis in Entamoeba histolytica. Nat Commun. 2017. Jul 17;8(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Bharadwaj R, Kushwaha T, Ahmad A, et al. An atypical EhGEF regulates phagocytosis in Entamoeba histolytica through EhRho1. PLOS Pathogens. 2021. Nov;17(11):e1010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Huston CD, Houpt ER, Mann BJ, et al. Caspase 3-dependent killing of host cells by the parasite Entamoeba histolytica. Cell Microbiol. 2000. Dec;2(6):617–625. [DOI] [PubMed] [Google Scholar]
- [186].Lee YA, Kim KA, Min A, et al. Amoebic PI3K and PKC is required for Jurkat T cell death induced by Entamoeba histolytica. Korean J Parasitol. 2014. Aug;52(4):355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Lopez-Rosas I, Lopez-Camarillo C, Salinas-Vera YM, et al. Entamoeba histolytica up-regulates MicroRNA-643 to promote apoptosis by targeting XIAP in human epithelial colon cells. Front Cell Infect Microbiol. 2018;8:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Maia BM, Rocha RM, Calin GA. Clinical significance of the interaction between non-coding RNAs and the epigenetics machinery: challenges and opportunities in oncology. Epigenetics. 2014. Jan;9(1):75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Yuan C, Burns MB, Subramanian S, et al. Interaction between host MicroRnas and the gut microbiota in colorectal cancer. mSystems. 2018. May-Jun;3(3). DOI: 10.1128/mSystems.00205-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Liu S, Rezende RM, Moreira TG, et al. Oral administration of miR-30d from Feces of MS patients suppresses MS-like symptoms in mice by expanding Akkermansia muciniphila. Cell Host Microbe. 2019. Dec 11;26(6):779–794 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Ayala-Sumuano JT, Tellez-Lopez VM, Dominguez-Robles Mdel C, et al. Toll-like receptor signaling activation by Entamoeba histolytica induces beta defensin 2 in human colonic epithelial cells: its possible role as an element of the innate immune response. PLoS negl trop dis. 2013;7(2):e2083. DOI: 10.1371/journal.pntd.0002083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Preet S, Bharati S, Shukla G, et al. Evaluation of amoebicidal potential of Paneth cell cryptdin-2 against Entamoeba histolytica. PLoS negl trop dis. 2011. Dec;5(12):e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Seydel KB, Li E, Swanson PE, et al. Human intestinal epithelial cells produce proinflammatory cytokines in response to infection in a SCID mouse-human intestinal xenograft model of amebiasis. Infect Immun. 1997. May;65(5):1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Peterson KM, Guo X, Elkahloun AG, et al. The expression of REG 1A and REG 1B is increased during acute amebic colitis. Parasitol Int. 2011. Sep;60(3):296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Begum S, Moreau F, Leon Coria A, et al. Entamoeba histolytica stimulates the alarmin molecule HMGB1 from macrophages to amplify innate host defenses. Mucosal Immunol. 2020. Mar;13(2):344–356. [DOI] [PubMed] [Google Scholar]
- [196].Allaire JM, Crowley SM, Law HT, et al. The Intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 2018. Sep;39(9):677–696. [DOI] [PubMed] [Google Scholar]
- [197].Allaire JM, Crowley SM, Law HT, et al. The Intestinal epithelium: central coordinator of mucosal immunity: (Trends in Immunology 39, 677-696, 2018). Trends Immunol. 2019. Feb;40(2):174. [DOI] [PubMed] [Google Scholar]
- [198].Burdette BE, Esparza AN, Zhu H, et al. Gasdermin D in pyroptosis. Acta Pharm Sin B. 2021. Sep;11(9):2768–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Maldonado-Bernal C, Kirschning CJ, Rosenstein Y, et al. The innate immune response to Entamoeba histolytica lipopeptidophosphoglycan is mediated by toll-like receptors 2 and 4. Parasite Immunol. 2005. Apr;27(4):127–137. [DOI] [PubMed] [Google Scholar]
- [200].Mortimer L, Moreau F, Cornick S, et al. The NLRP3 inflammasome is a pathogen sensor for invasive Entamoeba histolytica via activation of alpha5beta1 Integrin at the macrophage-amebae intercellular junction. PLOS Pathogens. 2015. May;11(5):e1004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Quach J, Moreau F, Sandall C, et al. Entamoeba histolytica-induced IL-1beta secretion is dependent on caspase-4 and gasdermin D. Mucosal Immunol. 2019. Mar;12(2):323–339. [DOI] [PubMed] [Google Scholar]
- [202].Dickson-Gonzalez SM, de Uribe ML, Rodriguez-Morales AJ. Polymorphonuclear neutrophil infiltration intensity as consequence of Entamoeba histolytica density in amebic colitis. Surg Infect (Larchmt). 2009. Apr;10(2):91–97. [DOI] [PubMed] [Google Scholar]
- [203].Denis M, Chadee K. Human neutrophils activated by interferon-gamma and tumour necrosis factor-alpha kill Entamoeba histolytica trophozoites in vitro. J Leukoc Biol. 1989. Sep;46(3):270–274. [DOI] [PubMed] [Google Scholar]
- [204].Sim S, Yong TS, Park SJ, et al. NADPH oxidase-derived reactive oxygen species-mediated activation of ERK1/2 is required for apoptosis of human neutrophils induced by Entamoeba histolytica. J Immunol. 2005. Apr 1;174(7):4279–4288. [DOI] [PubMed] [Google Scholar]
- [205].Watanabe K, Gilchrist CA, Uddin MJ, et al. Microbiome-mediated neutrophil recruitment via CXCR2 and protection from amebic colitis. PLOS Pathogens. 2017. Aug;13(8):e1006513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Avila EE, Salaiza N, Pulido J, et al. Entamoeba histolytica Trophozoites and Lipopeptidophosphoglycan trigger human neutrophil extracellular traps. PLoS ONE. 2016;11(7):e0158979. DOI: 10.1371/journal.pone.0158979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Diaz-Godinez C, Fonseca Z, Nequiz M, et al. Entamoeba histolytica Trophozoites induce a rapid non-classical NETosis mechanism independent of NOX2-derived reactive oxygen species and PAD4 activity. Front Cell Infect Microbiol. 2018;8:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Fonseca Z, Diaz-Godinez C, Mora N, et al. Entamoeba histolytica induce signaling via Raf/MEK/ERK for Neutrophil Extracellular Trap (NET) formation. Front Cell Infect Microbiol. 2018;8:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Blazquez S, Zimmer C, Guigon G, et al. Human tumor necrosis factor is a chemoattractant for the parasite Entamoeba histolytica. Infect Immun. 2006. Feb;74(2):1407–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Silvestre A, Plaze A, Berthon P, et al. In Entamoeba histolytica, a BspA family protein is required for chemotaxis toward tumour necrosis factor. Microb Cell. 2015. Jul 6;2(7):235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Noor Z, Watanabe K, Abhyankar MM, et al. Role of eosinophils and tumor necrosis factor alpha in Interleukin-25-mediated protection from Amebic Colitis. MBio. 2017. Feb 28;8(1). doi : 10.1128/mBio.02329-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Uddin MJ, Leslie JL, Burgess SL, et al. The IL-33-ILC2 pathway protects from amebic colitis. Mucosal Immunol. 2022. Jan;15(1):165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Duggal P, Haque R, Roy S, et al. Influence of human leukocyte antigen class II alleles on susceptibility to Entamoeba histolytica infection in Bangladeshi children. J Infect Dis. 2004. Feb 1;189(3):520–526. [DOI] [PubMed] [Google Scholar]
- [214].Wojcik GL, Marie C, Abhyankar MM, et al. Genome-wide association study reveals genetic link between diarrhea-associated Entamoeba histolytica infection and Inflammatory Bowel disease. MBio. 2018. Sep 18;9(5). doi : 10.1128/mBio.01668-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Verkerke HP, Petri WA Jr., Marie CS. The dynamic interdependence of amebiasis, innate immunity, and undernutrition. Semin Immunopathol. 2012. Nov;34(6):771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Duggal P, Guo X, Haque R, et al. A mutation in the leptin receptor is associated with Entamoeba histolytica infection in children. J Clin Invest. 2011. Mar;121(3):1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Petri WA Jr., Mondal D, Peterson KM, et al. Association of malnutrition with amebiasis. Nutr Rev. 2009. Nov;67 Suppl 2:S207–15. [DOI] [PubMed] [Google Scholar]
- [218].Nagaraja S, Ankri S. Target identification and intervention strategies against amebiasis. Drug Resist Updat. 2019. May;44:1–14. [DOI] [PubMed] [Google Scholar]
- [219].Juarez-Saldivar A, Campillo NE, Ortiz-Perez E, et al. In Silico analysis of potential drug targets for Protozoan infections. Med Chem. 2022. Aug 16. DOI: 10.2174/1573406418666220816121912. [DOI] [PubMed] [Google Scholar]
- [220].Shrivastav MT, Malik Z. Revisiting drug development against the neglected tropical disease, Amebiasis. Front Cell Infect Microbiol. 2020;10:628257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Kangussu-Marcolino MM, Singh U. Ponatinib, Lestaurtinib, and mTOR/PI3K Inhibitors are promising repurposing candidates against Entamoeba histolytica. Antimicrob Agents Chemother. 2022. Feb 15;66(2):e0120721. DOI: 10.1128/AAC.01207-21 [DOI] [PMC free article] [PubMed] [Google Scholar]


