Abstract
Tripartite motif (TRIM) 31 is a new member of the TRIM family and functions as an E3 ubiquitin ligase. Abnormal TRIM31 expression leads to a variety of pathological conditions, such as cancer, innate immunity diseases, sepsis-induced myocardial dysfunction, cerebral ischemic injury, nonalcoholic fatty liver disease and hypertensive nephropathy. In this review, we comprehensively overview the structure, expression and regulation of TRIM31 in cancer. Moreover, we discuss the dual role of TRIM31 in human cancer, and this dual role may be linked to its involvement in the selective regulation of several pivotal cellular signaling pathways: the p53 tumor suppressor, mTORC1, PI3K-AKT, NF-κB and Wnt/β-catenin pathways. In addition, we also discuss the emerging role of TRIM31 in innate immunity, autophagy and its growing sphere of influence across multiple human pathologies. Finally, a better understanding of the dual role of TRIM31 in cancer may provide new therapeutic strategies aimed at inhibiting the cancer-promoting effects of TRIM31 without affecting its tumor suppressor effects.
Keywords: TRIM31, cancer, oncogene, tumor suppressor, innate immunity
1 Introduction
Ubiquitination is a common posttranslational modification of proteins. It participates in various cellular processes and physiological responses in cancer, inflammatory disorders, infection and other diseases by regulating the degradation and activation of intracellular proteins (1). The ubiquitination process is catalyzed by E1, E2 and E3, among which E3 ubiquitin ligase is mainly involved in the recognition and binding of target proteins (2). E3 ubiquitin ligases can be divided into two major classes: homologous to E6-AP COOH terminal (HECT) E3 ubiquitin ligases and RING finger-containing E3 ubiquitin ligases. Although TRIMs are considered to be RING finger-containing E3 ubiquitin ligases, not all TRIM E3 ubiquitin ligases have a RING domain. To date, there are 9 ring-domain-free TRIM proteins in humans (3). Apart from the RING finger domain, TRIM proteins also contain one or two zinc-binding motifs, named B-boxes, and a coiled-coil domain. According to their domains, TRIM proteins are divided into I to XI subfamilies (4–6). TRIM proteins regulate important cellular processes, such as intracellular signal transduction, innate immunity, transcriptional regulation, autophagy, and carcinogenesis (7, 8). In cancer research, TRIM members act as oncogenes or tumor suppressor genes in ovarian cancer, renal cell carcinoma, gastric cancer, and breast cancer by controlling multiple processes such as transcriptional regulation, DNA repair, cell proliferation, apoptosis, and metastasis (9–13).
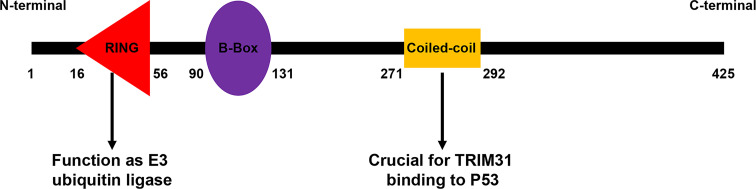
TRIM31 is a member of the TRIM family, and structural analysis found that it contains a RING domain, which makes it an E3 ubiquitin-protein ligase (14). The RING domain is a zinc-binding motif located in amino acids 10-20 of the first methionine in nearly all TRIM protein N-terminal portions (15). General insights into RING domain function are derived from the report that the RING domain contains the CBL protein, which has shown that the RING domain regulates ubiquitination events (16–18). It has been reported that C16, C36, C53, C56, and C58 are the key amino acids of the RING domain, and mutation of these amino acid sites can inhibit the E3 ubiquitin ligase activity of TRIM31 (19–21). Apart from the RING domain, TRIM31 also contains one zinc-finger domain named the B box (type 2 box). B-box domains exist in more than 1500 proteins from a variety of organisms, and they can be divided into two groups, in which the intervals of 7-8 zinc binding residues of type 1 and type 2 B-box domains are different. Type 2 B-box proteins play a role in the ubiquitination process. After the B boxes are the coiled-coil region at the N-terminus, this domain regulates homomeric and heteromeric interactions between TRIM proteins and other proteins, especially self-association. In our research, the coiled-coil region was important for the binding of TRIM31 and p53 (22). TRIM31 has no domain at the C-terminus ( Figure 1 ).
Figure 1.
Domain organization of the TRIM31 protein. The different TRIM31 domains are reported with numbers and indicate the first and last amino acids of each domain.
Recent studies on TRIM31 strongly advocate for the critical role of TRIM31 in cancer, immunity and inflammation. In this review, in addition to accumulating recent corroborations that endorse this dual role of TRIM31 in cancer, we also discuss the emerging role of TRIM31 in innate immunity, inflammation and autophagy and its growing sphere of influence across multiple other human pathologies.
2 Expression of TRIM31 in cancer and its clinical value
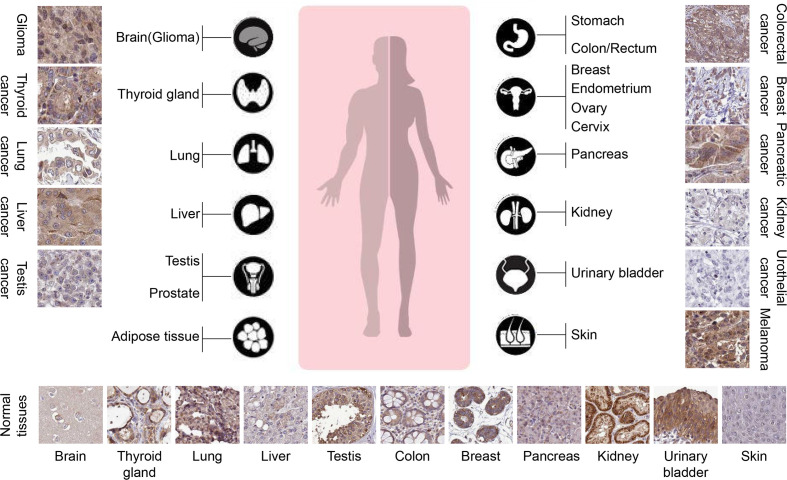
To date, the clinical correlation of TRIM31 in cancer is still elusive. Several reports have revealed that there is a positive correlation between the expression of TRIM31 and cancer prognosis in specific cancer types. TRIM31 is upregulated in gastric adenocarcinoma and may be a potential biomarker of gastric cancer because it is overexpressed in the precancerous stage (23). TRIM31 was markedly upregulated in hepatocellular carcinoma, gallbladder cancer, colorectal cancer, high-grade glioma, pancreatic cancer and acute myeloid leukemia, and the high expression of TRIM31 was also associated with an aggressive phenotype, advanced disease status and poor prognosis (24–29). Multivariate survival analysis demonstrated that TRIM31 was an independent prognostic factor for glioma patients (27). From the Human Protein Atlas data, immunohistochemical analysis found that TRIM31 was more highly expressed in liver, gastric, pancreatic, gallbladder, colorectal tumors and glioma ( Figure 2 , more information please see www.proteinatlas.org). It is suggested that upregulation of TRIM31 is a common feature of many epithelial cancers and predicts a poor prognosis. However, several reports have indicated that TRIM31 is downregulated in cancer; for example, TRIM31 expression is downregulated in lung cancer tissues and cell lines and correlates with clinic-pathological factors (30). Our research showed that TRIM31 expression was decreased in breast cancer tissues and that lower TRIM31 levels were associated with worse survival of breast cancer patients (22). Altogether, the aforementioned studies showed that TRIM31 was upregulated in liver, gastric, pancreatic, gallbladder, colorectal tumors and glioma, and higher levels of TRIM31 are related to the poor prognosis of cancer patients. However, there are also opposite conclusions. TRIM31 was downregulated in lung and breast cancer, and higher expression of the TRIM31 gene is linked to better overall survival of patients.
Figure 2.
TRIM31 is highly expressed in several types of tumors (The Human Protein Atlas data). The rabbit polyclonal antibody HPA046400 (Sigma Aldrich) was used for the immunohistochemistry assay. The cancer tissues of glioma, melanoma, thyroid gland, pancreatic cancer and liver cancer were strongly cytoplasmic stained. Occasional membranous positivity was observed in colorectal cancer. Urothelial cancer and most kidney cancers were negatively stained. Normal tissues showed weak to strong cytoplasmic positivity with HPA046400 antibody, as presented at the bottom. In glioma, thyroid cancer, liver cancer, colorectal cancer, and pancreatic cancer, the expression of TRIM31 in cancer cell cytoplasm is higher than that in adjacent normal tissues. However, in lung cancer, testis cancer, kidney cancer, and urothelial cancer, the expression of TRIM31 was lower than that in adjacent normal tissues.
3 Regulation of TRIM31 expression
The expression of TRIM31 in diverse cancer cells is different, and its expression is tightly controlled by various factors. Recent reports have shown that TRIM31 expression is regulated by retinoid, microRNA and posttranslational modifications. Retinoids, natural or synthetic derivatives of vitamin A, are effective in the treatment of acute promyelocytic leukemia (APL) and are used in the chemoprophylaxis of cancers such as breast, skin, head and neck and liver cancers. Retinoid bound to the promoter of TRIM31 to induce TRIM31 expression and then suppressed the proliferation of breast cancer (31). MicroRNA is a small single-stranded noncoding RNA with a length of 21-23 nt that regulates the transcriptional inhibition, cleavage and degradation of mRNA (32). According to related studies, abnormal miRNA function is closely related to tumor invasion and metastasis (33, 34). In ovarian cancer, microRNA-551b downregulates TRIM31 expression by targeting its 3’ UTR to promote cancer progression (35). In addition, microRNA-29c-3p is abnormally expressed in many cancers, including gastric cancer, colon cancer, pancreatic cancer and hepatocellular carcinoma (36–38). Overexpression of microRNA-29c-3p significantly inhibited the proliferation and migration of hepatocellular carcinoma (HCC) cells in vitro and the growth of HCC tumors in vivo. Mechanistically, microRNA-29c-3p directly bound to the TRIM31 promoter and suppressed TRIM31 expression (39).
Posttranslational modifications including phosphorylation, ubiquitination and acetylation have been shown to modulate various biological functions, such as cell signal conduction, protein–protein interactions, protein transport, cell differentiation and proliferation through regulating the protein conformation, localization, stability and activity (40, 41). The TRIM31 protein is polyubiquitinated in gastric cancer, which leads to its proteasomal degradation. Furthermore, the ubiquitin proteasome-regulated degradation of TRIM31 was confirmed in AsPC-1 pancreatic cancer cells (23). These discoveries suggest that posttranslational modification can control the abundance of endogenous TRIM31.
4 The dual role of TRIM31 in cancer: Oncogene or Tumor Suppressor?
4.1 The tumor suppressor role of TRIM31
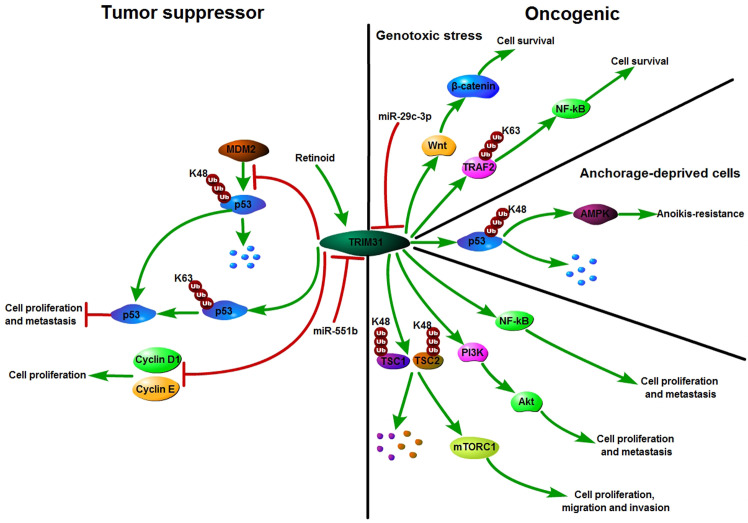
Recently, increasing evidence has shown that TRIM31 plays an important tumor suppressor role in the occurrence and development of various cancers. TRIM31 was first reported in breast cancer in 2002. Retinoid induced proliferation inhibition of breast carcinoma cells by targeting the TRIM31 promoter (31). In our study, we found that TRIM31 directly interacted with p53 and subsequently stabilized and activated p53 by inducing K63-linked ubiquitination as well as inhibiting MDM2-mediated K48-linked ubiquitination of p53 and then suppressing breast cancer progression (22). In addition, TRIM31 plays a potential tumor suppressor role in non-small cell lung cancer. The expression of TRIM31 in lung cancer cell lines was lower than that in the normal bronchial cell line HBE. TRIM31 inhibited the cell proliferation rate and colony formation by reducing the expression of the cell cycle regulators cyclin D1 and cyclin E (30). Moreover, TRIM31 can be recognized as a growth suppressor at the early stage of gastric adenocarcinoma (23). Therefore, TRIM31 may act as a tumor suppressor in the early stage of the tumor. Altogether, these studies have shown that TRIM31 might play a tumor suppressor role in breast cancer, non-small cell lung cancer and the early stage of gastric adenocarcinoma ( Figure 3 ).
Figure 3.
Schematic diagram of TRIM31 function in cancer. TRIM31 can act as either a tumor suppressor or an oncogene. TRIM31 inhibits cell growth by reducing the expression of Cyclin D1 and Cyclin E. TRIM31 stabilizes P53 expression to suppress tumor cell proliferation and metastasis. TRIM31 promotes tumor onset and progression by regulating the P53, mTORC1, PI3K-Akt, NF-kB and Wnt/β-catenin pathways. Retinoid induced cell growth arrest by targeting the promoter of TRIM31. miR-551b suppresses the expression of TRIM31 by targeting its 3’-UTR and further promotes cell invasion and drug resistance. miR-29c-3p can inhibit TRIM31 expression by targeting its 3’-UTR. TRIM31 activated Wnt/β-catenin signaling to promote cell survival.
4.2 The oncogene role of TRIM31
Recently, studies have shown that TRIM31 is an oncogene in various cancers. A number of carcinogenic mechanisms have been proposed for TRIM31, such as regulating the P53, mTORC1, PI3K-Akt, NF-kB and Wnt/β-catenin pathways to promote tumor onset and progression (24–30). P53 is one of the most important tumor suppressor proteins. In differentiated cells, the abundance and activity of p53 are strictly regulated. With an increased risk of acquiring mutations, the accumulation and activation of p53 protein are regulated by posttranslational modifications (42). TRIM31 was significantly upregulated in the anchorage-deprived HCC cells compared with their attached counterparts and promoted anoikis resistance. TRIM31 can directly interact with p53, which is an inhibitor of the AMPK pathway, and regulate the K48-linked ubiquitous degradation of p53. It was further confirmed that excessive activation of the AMPK pathway is the cause of TRIM31-mediated HCC cell resistance to anoikis. That is, TRIM31 facilitates anoikis resistance by targeting the degradation of p53 and subsequently overactivating the AMPK pathway (24). AMPK phosphorylates tuberous sclerosis complex 2 (TSC2) and enhances its activity (43). TSC2 is an upstream inhibitor of the mTORC1 pathway. MTOR forms two unique catalytic subunits of the complex, called mTORC1 and mTORC2, and it plays critical roles in a variety of biological processes, such as cell growth, survival, autophagy, metabolism, and immunity (44, 45). TRIM31 facilitates the malignant behaviors of HCC cells by overactivating the target of the mTORC1 pathway. Further studies have shown that TRIM31 plays a carcinogenic role by directly interacting with the TSC1 and TSC2 complexes and facilitating the ubiquitination of K48 and the degradation of the complex (19).
The phosphoinositide 3-kinase (PI3K)–AKT pathway is an important node in controlling cell growth, migration, proliferation, and metabolism in mammalian cells and is the most commonly activated pathway in human cancers (46). In glioma, by silencing or overexpressing TRIM31 expression, the proliferation, invasion and migration of glioma cells could be downregulated or upregulated through the PI3K/Akt signaling pathway (27). Moreover, TRIM31 can activate the PI3K/Akt signaling pathway to enhance chemoresistance in glioblastoma (47). In gallbladder cancer, TRIM31 promotes proliferation and invasion via the PI3K/Akt signaling pathway (25). The PI3K/AKT/IKK alpha pathway regulates the activation of NF kappa B and β-catenin in CRC cell lines (48). Nuclear factor kappa B (NF-κB) is activated in various cancers and not only coordinates with immunity and inflammation but also plays a vital role in the development of cancer (49, 50).. Past research has shown that ubiquitin modification plays an important role in the regulation of NF-κB signaling (51–53). As an E3 ubiquitin ligase, TRIM31 promotes K63-linked polyubiquitination of tumor necrosis factor receptor-associated factor 2 (TRAF2) to upregulate the levels of nuclear p65 and then maintains the activation of NF-κB in pancreatic cancer cells. Furthermore, TRIM31 promotes gemcitabine resistance in pancreatic cancer cells by activating the NF-κB signaling pathway (28). In addition, TRIM31 activates the NF-κB pathway to promote migration and invasion in glioma and colorectal cancer (26, 54). TRIM31 regulates Wnt/β-catenin signaling to promote acute myeloid leukemia progression and sensitivity to daunorubicin (29). Collectively, TRIM31 plays an oncogene role in cancer by regulating the p53, mTORC1, NF-κB, and PI3K-Akt pathways ( Figure 3 ).
4.3 The possible mechanism of TRIM31 in promoting or suppressing cancer
TRIM31 is a critical factor that is able to perform multiple functions in cancer, and the complex function of TRIM31 makes it difficult to identify TRIM31 as an oncogene or tumor suppressor. Here, we will discuss why TRIM31 can promote or inhibit cancer from the following aspects. First, TRIM genes are usually expressed in a variety of splicing forms (55). There are three isoforms of TRIM31 (TRIM31α, TRIM31β and TRIM31γ). TRIM31α was the most common splicing form and has been registered in the public database. TRIM31 β is a truncated form of TRIM31 at the C-terminus. TRIM31 γ is a mutant protein of TRIM31 truncated at the C-terminus. Studies have shown that the TRIM31 isoforms have different biological roles in cancer. Therefore, the differential expression of TRIM31 isoforms may lead to the different roles of TRIM31 in cancer. Second, proteins containing a RING finger domain can serve as E3 ubiquitin ligases (17), and Sugiura proved that TRIM31 has autoubiquitylating activity in vitro. It has been demonstrated that the autoubiquitination activity of TRIM31 regulates the intracellular abundance of TRIM31 (23). The strict regulation of the TRIM31 protein level may be linked to its seemingly contradictory behaviors in the cancer process. Third, several cancer-associated proteins that can be posttranslationally regulated by TRIM31 have been reported, such as TRAF2, TSC1/TSC2 and P53. The different target proteins of TRIM31 may decide the tumor promoter or tumor suppressor of TRIM31 in cancer. Therefore, discovering new target proteins of TRIM31 is necessary to further understand the role of TRIM31 in cancer.
5 TRIM31: Growing influence in innate immunity and autophagy
5.1 The emerging role of TRIM31 in innate immunity
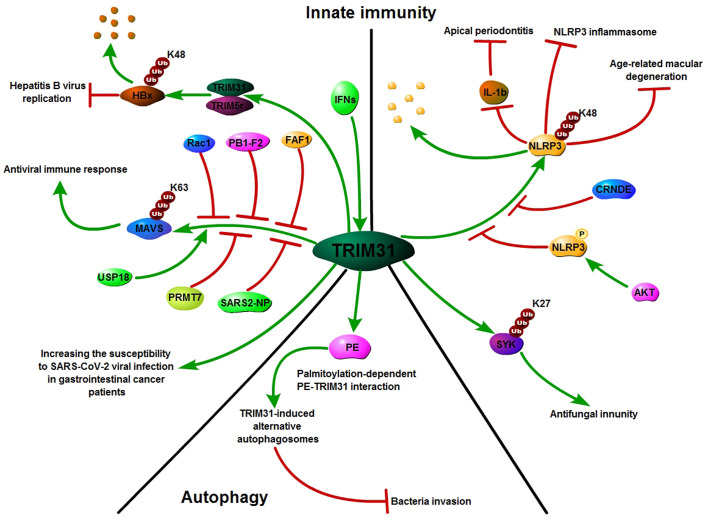
Innate immunity provides the first line of defense against invading pathogens. Activation of innate immunity requires the recognition of pathogen-associated molecular patterns through pattern-recognition receptors (56, 57). As a regulator of Mitochondrial antiviral signaling protein (MAVS) aggregation, TRIM31 can be recruited to mitochondria after viral infection and specifically regulate antiviral signaling mediated by RIG-I-like receptor (RLR) pattern-recognition receptors. Further study showed that TRIM31 interacted with MAVS and catalyzed the Lys63 (K63)-linked polyubiquitination of MAVS by Lys10, Lys311 and Lys461. This modification promoted the formation of prion-like aggregates of MAVS after viral infection (20). Moreover, USP18 interacts with TRIM31 to promote the K63-linked polyubiquitination of MAVS and then positively regulates innate antiviral immunity (58). PB1-F2, Rac1, PRMT7 and FAF1 disrupt TRIM31 interaction with MAVS to inhibit MAVS activation and negatively regulate innate antiviral immune responses (59–62). HBV (hepatitis B virus) infection was reported to induce type III IFNs, and TRIM31 was found to be a type III IFN-stimulated gene. IFN-induced TRIM5γ recruits TRIM31 to degrade HBx, resulting in suppression of hepatitis B virus replication (63, 64). In addition to its crucial role in antiviral processes, TRIM31 also has an important role in promoting viral infection. COVID-19 caused by the novel severe acute respiratory syndrome (SARS) coronavirus 2 (SARS-CoV-2) is rapidly emerging and spreading worldwide. Wang et al. reported that the dimeric domain protein (SARS2-NP) of the SARS-CoV-2 nucleocapsid is required for liquid-liquid phase separation of SARS2-NP and RNA, which suppresses Lys63-linked polyubiquitination and aggregation of MAVS by reducing TRIM31 binding to MAVS, thus inhibiting the innate antiviral immune response (65). Moreover, Temena et al. also found that TRIM31 is positively correlated with SARS−CoV−2 associated genes TMPRSS2−TMPRSS4 and knockdown of TRIM31 significantly altered viral replication and viral processes in gastrointestinal cancer samples. This result suggests that TRIM31 may play a role in increasing the susceptibility to SARS-CoV-2 viral infection in patients with gastrointestinal cancers (66).
The NLRP3 inflammasome is a multiprotein platform that comprises NLRP3, ASC, and caspase-1 and plays crucial roles in host defense against pathogens. The NLRP3 inflammasome is involved in many kinds of diseases, such as cancer, gout, autoimmune disorders, atherosclerosis, type 2 diabetes and obesity (67–70). TRIM31 has been reported to be a feedback suppressor of the NLRP3 inflammasome. TRIM31 directly binds to NLRP3 and promotes K48-linked polyubiquitination and proteasomal degradation of NLRP3. Furthermore, TRIM31 deficiency attenuates the severity of dextran sodium sulfate (DSS)-induced colitis, an inflammatory bowel disease model in which NLRP3 exerts a protective effect (21). Moreover, AKT bound to NLRP3 and phosphorylated it on S5. This phosphorylation event also stabilized NLRP3 by reducing its ubiquitination on lysine 496, which inhibits its proteasome-mediated degradation by TRIM31 (71). In addition, TRIM31 promoted the ubiquitination of NLRP3 to alleviate IL−1ß secretion and diminished the development of apical periodontitis (72). TRIM31 also inhibited the NLRP3 inflammasome and pyroptosis through ubiquitination of NLRP3 in retinal pigment epithelial cells (73). CRNDE interacted with NLRP3 and decreased TRIM31-mediated NLRP3 ubiquitination to activate the NLRP3 inflammasome and exacerbate IgA nephropathy progression (74). In addition to the NLRP3 inflammasome, TRIM31 also plays a crucial role in fungal infections. TRIM31 regulates antifungal immunity by facilitating K27-linked polyubiquitination of SYK (75).
5.2 TRIM31 and autophagy
Autophagy is one of the major intracellular degradation systems in addition to the ubiquitin–proteasome system. A primary role of autophagy is to maintain cellular homoeostasis by degrading intracytoplasmic proteins and organelles through starvation and by recycling multiple sources (76–78). Recent studies have shown that several TRIM proteins regulate cancer progression via autophagy. TRIM59 inhibits p62 selective autophagy degradation of PDCD10 to promote the motility of breast cancer (79). In A549/DDP cells, knockdown of TRIM65 can inhibit autophagy and cisplatin resistance by regulating miR-138-5P/ATG7 (80). TRIM31, an intestine-specific protein localized in mitochondria, is essential for promoting lipopolysaccharide-induced Atg5/Atg7-independent autophagy. TRIM31 directly interacts with phosphatidylethanolamine in a palmitoylation-dependent manner, leading to the induction of autolysosome formation (81). Altogether, TRIM31 plays an important role in innate immunity, autophagy and other human pathologies ( Figure 4 ).
Figure 4.
TRIM31: Growing influence in innate immunity and autophagy. TRIM31 plays an important role in innate immunity; TRIM31 interacts with MAVS and catalyzes the Lys63 (K63)-linked polyubiquitination of MAVS to promote the formation of prion-like MAVS aggregates after viral infection. USP18 promotes TRIM31-mediated K63-linked MAVS polyubiquitination, while PB1-F2, Rac1, PRMT7, SARS2-NP and FAF1 inhibit TRIM31-mediated K63-linked MAVS polyubiquitination. TRIM31 directly binds to NLRP3 and promotes K48-linked polyubiquitination and proteasomal degradation of NLRP3. AKT and CRNDE decreased TRIM31-mediated NLRP3 ubiquitination. Alongside its emerging role in innate immunity, TRIM31 is also known to be involved in autophagy.
6 Conclusion and future perspectives
In conclusion, although the function of TRIM31 has been studied for many years, there is still much to be clarified regarding the role of TRIM31 in cancer. A number of studies suggest that TRIM31 may serve as an oncogene when it is highly expressed and may facilitate cancer progression, metastasis and drug resistance by inducing the mTORC1 pathway (19), activating the NF-κB and AKT signaling pathways (25–28), and downregulating the activity of p53 (24). However, some studies have shown that TRIM31 can act as a tumor suppressor, and its high expression inhibits the proliferation and metastasis of cancer by stabilizing p53 or decreasing the expression of cyclin D1 and cyclin E. Although TRIM31 has been extensively studied, some questions should also be considered. First, the regulation of TRIM31 in cancer should be further researched. Recent research has shown only that TRIM31 can be regulated by posttranslational modification, and whether there are other regulatory mechanisms is still unclear. Second, the mechanism by which TRIM31 promotes and suppresses cancer needs further study. For example, whether TRIM31 regulates cancer progression via autophagy or innate immunity is unknown. Third, as an E3 ubiquitin ligase, TRIM31 can target many proteins ( Table 1 ). Therefore, it is very important to find new target proteins for studying the function of TRIM31 in cancer. Although there are still many questions to be addressed, we believe that in-depth understanding of the TRIM31 in carcinogenesis may help to answer whether TRIM31 possesses the potential to become a new anticancer target.
Table 1.
The TRIM31 targets or interacting proteins.
| Target or interacting proteins | Modification | Effect | Outcome | Reference |
|---|---|---|---|---|
| P53 | K48-linked poly-ubiquitination | Degradation of p53 | Promoting the anoikis-resistance | (24) |
| TSC1/TSC2 | K48-linked poly-ubiquitination | Degradation of TSC1/TSC2 | Promoting HCC progression | (19) |
| TRAF2 | K63-linked poly-ubiquitination | Activation of NF-κB | Promoting the gemcitabine resistance | (28) |
| P53 | K63-linked poly-ubiquitination | Activation of p53 | Suppressing the proliferation and migration of breast cancer cell |
(22) |
| MAVS | K63-linked poly-ubiquitination | Promoting the formation of prion-like aggregates | Activation of antiviral immunity | (20) |
| USP18 | No | Promoting the K63-linked polyubiquitination of MAVS | Activation of antiviral immunity | (58) |
| HBx | K48-linked poly-ubiquitination | Degradation of HBx | Inhibiting HBV Replication | (62) |
| NLRP3 | K48-linked poly-ubiquitination | Degradation of NLRP3 | Attenuating NLRP3 inflammasome activation |
(70) |
| SYK | K27-linked poly-ubiquitination | Activation of SYK | Promoting antifungal immunity | (74) |
Currently, pharmaceutical companies have entered the era of E3 ubiquitin ligase-targeted therapy, and targeting the E3 ligase is gradually becoming a considerable cancer treatment option (82). Proteolysis-targeting chimera (PROTAC) has been developed as a useful protein-targeted degradation technique. A bifunctional PROTAC molecule is composed of a ligand of the protein of interest (POI) and a covalently linked ligand of an E3 ubiquitin ligase (E3). Upon binding to POI, PROTAC can recruit E3 for POI ubiquitination, which is subject to proteasome-mediated degradation (83). However, the PROTAC technique has not been applied to the TRIM31 protein, and future studies may focus on the application of PROTAC to TRIM31 protein.
Author contributions
Conception and design: YG and PL. Initial manuscript writing: YG. Confirmation of Manuscript: PL, YH and CW. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (No. 81971457) and the Science-technology Support Plan Projects in Sichuan province (No. 2022YFS0240).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med (2014) 20(11):1242–53. doi: 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- 2. Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. resolution, affinity purification, and role in protein breakdown. J Biol Chem (1983) 258:8206–14. doi: 10.1016/S0021-9258(20)82050-X [DOI] [PubMed] [Google Scholar]
- 3. Hatakeyama S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci (2017) 42(4):297–311. doi: 10.1016/j.tibs.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 4. Short KM, Cox TC. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem (2006) 281(13):8970–80. doi: 10.1074/jbc.M512755200 [DOI] [PubMed] [Google Scholar]
- 5. Ozato K, Shin DM, Chang TH, Morse HC. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol (2008) 8(11):849–60. doi: 10.1038/nri2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol (2011) 23(1):46–56. doi: 10.1016/j.coi.2010.10.021 [DOI] [PubMed] [Google Scholar]
- 7. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer (2011) 11(11):792–804. doi: 10.1038/nrc3139 [DOI] [PubMed] [Google Scholar]
- 8. Napolitano LM, Meroni G. TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life (2012) 64(1):64–71. doi: 10.1002/iub.580 [DOI] [PubMed] [Google Scholar]
- 9. Cambiaghi V, Giuliani V, Lombardi S, Marinelli C, Toffalorio F, Pelicci PG. TRIM proteins in cancer. Adv Exp Med Biol (2012) 770:77–91. doi: 10.1007/978-1-4614-5398-7_6 [DOI] [PubMed] [Google Scholar]
- 10. Qiao HY, Zhang Q, Wang JM, Jiang JY, Huyan LY, Yan J, et al. TRIM29 regulates the SETBP1/SET/PP2A axis via transcription factor VEZF1 to promote progression of ovarian cancer. Cancer Lett (2022) 529:85–99. doi: 10.1016/j.canlet.2021.12.029 [DOI] [PubMed] [Google Scholar]
- 11. Miao CK, Liang C, Li P, Liu BJ, Qin C, Yuan H, et al. TRIM37 orchestrates renal cell carcinoma progression via histone H2A ubiquitination-dependent manner. J Exp Clin Cancer Res (2021) 40(1):195. doi: 10.1186/s13046-021-01980-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li RZ, Xu P, Jin XS, Shi ZC, Zhang QQ, Ye FP, et al. TRIM50 inhibits proliferation and metastasis of gastric cancer via promoting β-catenin degradation. J Oncol (2022) 2022:5936753. doi: 10.1155/2022/5936753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang R, Huang KL, Xing LX. TRIM35 functions as a novel tumor suppressor in breast cancer by inducing cell apoptosis through ubiquitination of PDK1. Neoplasma (2022) 69(2):370–82. doi: 10.4149/neo_2021_210823N1205 [DOI] [PubMed] [Google Scholar]
- 14. Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J (2001) 20(9):2140–51. doi: 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Torok M, Etkin LD. Two b or not two b? overview of the rapidly expanding b-box family of proteins. Differentiation (2001) 67(3):63–71. doi: 10.1046/j.1432-0436.2001.067003063.x [DOI] [PubMed] [Google Scholar]
- 16. Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science (1999) 286(5438):309–12. doi: 10.1126/science.286.5438.309 [DOI] [PubMed] [Google Scholar]
- 17. Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A (1999) 96(20):11364–9. doi: 10.1073/pnas.96.20.11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-cbl mediates desensitization of the epidermal growth factor receptor. J Biol Chem (1999) 274(32):22151–4. doi: 10.1074/jbc.274.32.22151 [DOI] [PubMed] [Google Scholar]
- 19. Guo P, Ma X, Zhao W, Huai W, Li T, Qiu Y, et al. TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1-TSC2 complex. Oncogene (2018) 37(4):478–88. doi: 10.1038/onc.2017.349 [DOI] [PubMed] [Google Scholar]
- 20. Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol (2017) 18(2):214–24. doi: 10.1038/ni.3641 [DOI] [PubMed] [Google Scholar]
- 21. Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun (2016) 7:13727. doi: 10.1038/ncomms13727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo YF, Li Q, Zhao G, Zhang J, Yuan H, Feng TY, et al. Loss of TRIM31 promotes breast cancer progression through regulating K48- and K63-linked ubiquitination of p53. Cell Death Dis (2021) 12(10):945. doi: 10.1038/s41419-021-04208-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sugiura T, Miyamoto K. Characterization of TRIM31, upregulated in gastric adenocarcinoma, as a novel RBCC protein. J Cell Biochem (2008) 105(4):1081–91. doi: 10.1002/jcb.21908 [DOI] [PubMed] [Google Scholar]
- 24. Guo PB, Qiu YM, Ma XM, Li T, Ma XX, Zhu LH, et al. Tripartite motif 31 promotes resistance to anoikis of hepatocarcinoma cells through regulation of p53-AMPK axis. Exp Cell Res (2018) 368(1):59–66. doi: 10.1016/j.yexcr.2018.04.013 [DOI] [PubMed] [Google Scholar]
- 25. Li H, Zhang Y, Hai J, Wang JX, Zhao B, Du LX, et al. Knockdown of TRIM31 suppresses proliferation and invasion of gallbladdercancer cells by down-regulating MMP2/9 through the PI3K/Akt signaling pathway. BioMed Pharmacother (2018) 103:1272–8. doi: 10.1016/j.biopha.2018.04.120 [DOI] [PubMed] [Google Scholar]
- 26. Wang HY, Yao L, Gong YD, Zhang B. TRIM31 regulates chronic inflammation via NF-κB signal pathway to promote invasion and metastasis in colorectal cancer. Am J Transl Res (2018) 10(4):1247–59. [PMC free article] [PubMed] [Google Scholar]
- 27. Shi G, Lv C, Yang Z, Qin T, Sun L, Pan P, et al. TRIM31 promotes proliferation, invasion and migration of glioma cells through akt signaling pathway. Neoplasma (2019) 66(5):727–35. doi: 10.4149/neo_2019_190106N21 [DOI] [PubMed] [Google Scholar]
- 28. Yu C, Chen SY, Guo YT, Sun CY. Oncogenic TRIM31 confers gemcitabine resistance in pancreatic cancer via activating the NF-κB signaling pathway. Theranostics (2018) 8(12):3224–36. doi: 10.7150/thno.23259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiao Y, Deng TR, Ming X, Xu JH. TRIM31 promotes acute myeloid leukemia progression and sensitivity to daunorubicin through the wnt/β-catenin signaling. Biosci Rep (2020) 40(4):BSR20194334. doi: 10.1042/BSR20194334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Zhang Y, Zhang Y, Bai X, Peng Y, He P. TRIM31 is downregulated in non-small cell lung cancer and serves as a potential tumor suppressor. Tumour Biol (2014) 35(6):5747–52. doi: 10.1007/s13277-014-1763-x [DOI] [PubMed] [Google Scholar]
- 31. Dokmanovic M, Chang BD, Fang J, Roninson IB. Retinoid-induced growth arrest of breast carcinoma cells involves co-activation of multiple growth-inhibitory genes. Cancer Biol Ther (2002) 1(1):24–7. doi: 10.4161/cbt.1.1.35 [DOI] [PubMed] [Google Scholar]
- 32. Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov (2014) 13(8):622–38. doi: 10.1038/nrd4359 [DOI] [PubMed] [Google Scholar]
- 33. Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res (2006) 66(15):7390–4. doi: 10.1158/0008-5472.CAN-06-0800 [DOI] [PubMed] [Google Scholar]
- 34. Hayes CN, Chayama K. MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. Int J Mol Sci (2016) 17(3):280. doi: 10.3390/ijms17030280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei Z, Liu Y, Wang Y, Zhang Y, Luo Q, Man X, et al. Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med Oncol (2016) 33(11):126. doi: 10.1007/s12032-016-0842-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dong XZ, Song Y, Lu YP, Hu Y, Liu P, Zhang L. Sanguinarine inhibits the proliferation of BGC-823 gastric cancer cells via regulating miR-96-5p/miR-29c-3p and the MAPK/JNK signaling pathway. J Nat Med (2019) 73(4):777–88. doi: 10.1007/s11418-019-01330-7 [DOI] [PubMed] [Google Scholar]
- 37. Chen G, Zhou T, Li Y, Yu Z, Sun L. p53 target miR-29c-3p suppresses colon cancer cell invasion and migration through inhibition of PHLDB2. Biochem Biophys Res Commun (2017) 487(1):90–5. doi: 10.1016/j.bbrc.2017.04.023 [DOI] [PubMed] [Google Scholar]
- 38. Lu Y, Tang L, Zhang Z, Li S, Liang S, Ji L, et al. Long noncoding RNA TUG1/miR-29c axis affects cell proliferation, invasion, and migration in human pancreatic cancer. Dis Markers (2018) 2018:6857042. doi: 10.1155/2018/6857042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lv T, Jiang L, Kong L, Yang J. MicroRNA−29c−3p acts as a tumor suppressor gene and inhibits tumor progression in hepatocellular carcinoma by targeting TRIM31. Oncol Rep (2020) 43(3):953–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marcelli S, Corbo M, Iannuzzi F, Negri L, Blandini F, Nistico R, et al. The involvement of post-translational modifications in alzheimer’s disease. Curr Alzheimer Res (2018) 15(4):313–35. doi: 10.2174/1567205014666170505095109 [DOI] [PubMed] [Google Scholar]
- 41. Wu Z, Huang R, Yuan L. Crosstalk of intracellular post-translational modifications in cancer. Arch Biochem Biophys (2019) 676:108138. doi: 10.1016/j.abb.2019.108138 [DOI] [PubMed] [Google Scholar]
- 42. Boehme KA, Blattner C. Regulation of p53–insights into a complex process. Crit Rev Biochem Mol Biol (2009) 44(6):367–92. doi: 10.3109/10409230903401507 [DOI] [PubMed] [Google Scholar]
- 43. Inoki K, Zhu TQ, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell (2003) 115(5):577–90. doi: 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- 44. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell (2017) 168(6):960–76. doi: 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harwood FC, Klein Geltink RI, O’Hara BP, Cardone M, Janke L, Finkelstein D, et al. ETV7 is an essential component of a rapamycin-insensitive mTOR complex in cancer. Sci Adv (2018) 4(9):eaar3938. doi: 10.1126/sciadv.aar3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature (2014) 505(7484):495–501. doi: 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fan MD, Zhao XY, Qi JN, Jiang Y, Liu BY, Dun ZP, et al. TRIM31 enhances chemoresistance in glioblastoma through activation of the PI3K/Akt signaling pathway. Exp Ther Med (2020) 20(2):802–9. doi: 10.3892/etm.2020.8782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Agarwal A, Das KH, Lerner NL, Sathe S, Cicek M, Casey G, et al. The AKT/I kappa b kinase pathway promotes angiogenic/metastatic gene expression in colorectal cancer by activating nuclear factor-kappa b and β-catenin. Oncogene (2005) 24(6):1021–31. doi: 10.1038/sj.onc.1208296 [DOI] [PubMed] [Google Scholar]
- 49. DiDonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol Rev (2012) 246(1):379–400. doi: 10.1111/j.1600-065X.2012.01099.x [DOI] [PubMed] [Google Scholar]
- 50. Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol (2009) 1(5):a000141. doi: 10.1101/cshperspect.a000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen ZJ, Fuchs SY. Ubiquitin-dependent activation of NF-kappaB: K63-linked ubiquitin chains: a link to cancer? Cancer Biol Ther (2004) 3(3):286–8. doi: 10.4161/cbt.3.3.798 [DOI] [PubMed] [Google Scholar]
- 52. Lang V, Rodríguez MS. Innate link between NF-kappaB activity and ubiquitin-like modifiers. Biochem Soc Trans (2008) 36(Pt 5):853–7. doi: 10.1042/BST0360853 [DOI] [PubMed] [Google Scholar]
- 53. Haas AL. Linear polyubiquitylation: the missing link in NF-kappaB signalling. Nat Cell Biol (2009) 11(2):116–8. doi: 10.1038/ncb0209-116 [DOI] [PubMed] [Google Scholar]
- 54. Zhou L, Deng ZZ, Li HY, Jiang N, Wei ZS, Hong MF, et al. TRIM31 promotes glioma proliferation and invasion through activating NF-κB pathway. Onco Targets Ther (2019) 12:2289–97. doi: 10.2147/OTT.S183625 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55. Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. BioEssays: news and reviews in molecular, cellular and developmental biology. Bioessays (2005) 27(11):1147–57. doi: 10.1002/bies.20304 [DOI] [PubMed] [Google Scholar]
- 56. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol (2010) 11(5):373–84. doi: 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 57. Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol (2016) 16(1):35–50. doi: 10.1038/nri.2015.8 [DOI] [PubMed] [Google Scholar]
- 58. Hou JX, Han LL, Zhao Z, Liu HQ, Zhang L, Ma CH, et al. USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. Nat Commun (2021) 12(1):2970. doi: 10.1038/s41467-021-23219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cheung PH, Lee TT, Kew C, Chen HL, Yuen KY, Chan CP, et al. Virus subtype-specific suppression of MAVS aggregation and activation by PB1-F2 protein of influenza a (H7N9) virus. PloS Pathog (2020) 16(6):e1008611. doi: 10.1371/journal.ppat.1008611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang SG, Harding AT, Sweeney C, Miao D, Swan G, Zhou C, et al. Control of antiviral innate immune response by protein geranylgeranylation. Sci Adv (2019) 5(5):eaav7999. doi: 10.1126/sciadv.aav7999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu JJ, Li X, Cai XL, Zha HY, Zhou ZW, Sun XY, et al. Arginine monomethylation by PRMT7 controls MAVS-mediated antiviral innate immunity. Mol Cell (2021) 81(15):3171–86.e8. doi: 10.1016/j.molcel.2021.06.004 [DOI] [PubMed] [Google Scholar]
- 62. Dai T, Wu LM, Wang SA, Wang J, Xie F, Zhang ZK, et al. FAF1 regulates antiviral immunity by inhibiting MAVS but is antagonized by phosphorylation upon viral infection. Cell Host Microbe (2018) 24(6):776–90.e5. doi: 10.1016/j.chom.2018.10.006 [DOI] [PubMed] [Google Scholar]
- 63. Tan GG, Yi ZH, Song HX, Xu FC, Li F, Aliyari R, et al. Type-I-IFN-Stimulated gene TRIM5γ inhibits HBV replication by promoting HBx degradation. Cell Rep (2019) 29(11):3551–63.e3. doi: 10.1016/j.celrep.2019.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu FC, Song HX, Xiao QF, Wei Q, Pang XL, Gao YL, et al. Type-III interferon stimulated gene TRIM31 mutation in an HBV patient blocks its ability in promoting HBx degradation. Virus Res (2022) 308:198650. doi: 10.1016/j.virusres.2021.198650 [DOI] [PubMed] [Google Scholar]
- 65. Wang S, Dai T, Qin ZR, Pan T, Chu F, Lou LF, et al. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat Cell Biol (2021) 23(7):718–32. doi: 10.1038/s41556-021-00710-0 [DOI] [PubMed] [Google Scholar]
- 66. Temena MA, Acar A. Increased TRIM31 gene expression is positively correlated with SARS-CoV-2 associated genes TMPRSS2 and TMPRSS4 in gastrointestinal cancers. Sci Rep (2022) 12(1):11763. doi: 10.1038/s41598-022-15911-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature (2012) 481(7381):278–86. doi: 10.1038/nature10759 [DOI] [PubMed] [Google Scholar]
- 68. Wen HT, Ting JPY, O’Neill LAJ. A role for the NLRP3 inflammasome in metabolic diseases–did warburg miss inflammation? Nat Immunol (2012) 13(4):352–7. doi: 10.1038/ni.2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Choi AJ, Ryter SW. Inflammasomes: molecular regulation and implications for metabolic and cognitive diseases. Mol Cells (2014) 37(6):441–8. doi: 10.14348/molcells.2014.0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Broderick L, Nardo DD, Franklin BS, Hoffman HM, Latz E. The inflammasomes and autoinflammatory syndromes. Annu Rev Pathol (2015) 10:395–424. doi: 10.1146/annurev-pathol-012414-040431 [DOI] [PubMed] [Google Scholar]
- 71. Zhao W, Shi CS, Harrison K, Hwang IY, Nabar NR, Wang M. AKT regulates NLRP3 inflammasome activation by phosphorylating NLRP3 serine 5. J Immunol (2020) 205(8):2255–64. doi: 10.4049/jimmunol.2000649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu XY, Lu MM, Ding S, Zhong Q. Tripartite motif 31 alleviates IL-1ß secretion via promoting the ubiquitination of pyrin domain domains-containing protein 3 in human periodontal ligament fibroblasts. Odontology (2020) 108(3):424–32. doi: 10.1007/s10266-020-00519-7 [DOI] [PubMed] [Google Scholar]
- 73. Huang PR, Liu WJ, Chen JQ, Hu YF, Wang YW, Sun JR. TRIM31 inhibits NLRP3 inflammasome and pyroptosis of retinal pigment epithelial cells through ubiquitination of NLRP3. Cell Biol Int (2020) 44(11):2213–9. doi: 10.1002/cbin.11429 [DOI] [PubMed] [Google Scholar]
- 74. Shen M, Pan XY, Gao YJ, Ye HY, Zhang J, Chen Y. LncRNA CRNDE exacerbates IgA nephropathy progression by promoting NLRP3 inflammasome activation in macrophages. Immunol Invest (2022) 51 (5) 1515–27. doi: 10.1080/08820139.2021.1989461 [DOI] [PubMed] [Google Scholar]
- 75. Wang XE, Zhang HH, Shao ZG, Zhuang WX, Sui C, Liu F, et al. TRIM31 facilitates K27-linked polyubiquitination of SYK to regulate antifungal immunity. Signal Transduct Target Ther (2021) 6(1):298. doi: 10.1038/s41392-021-00711-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature (2011) 469(7330):323–35. doi: 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annu Rev Cell Dev Biol (2011) 27:107–32. doi: 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 78. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature (2008) 451(7182):1069–75. doi: 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tan P, Ye YQ, He L, Xie JS, Jing J, Ma GL. TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PloS Biol (2018) 16(11):e3000051. doi: 10.1371/journal.pbio.3000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pan X, Chen Y, Shen Y, Tantai J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis (2019) 10(6):429. doi: 10.1038/s41419-019-1660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ra EA, Lee TA, Won Kim S, Park A, Choi HJ, Jang I, et al. TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat Commun (2016) 7:11726. doi: 10.1038/ncomms11726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bhaduri U, Merla G. Rise of TRIM8: A molecule of duality. Mol Ther Nucleic Acids (2020) 22:434–44. doi: 10.1016/j.omtn.2020.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li X, Song YC. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J Hematol Oncol (2020) 13(1):50. doi: 10.1186/s13045-020-00885-3 [DOI] [PMC free article] [PubMed] [Google Scholar]