Abstract
Cytosine methylation at the C5-position – generating 5-methylcytosine (5mC) – is a DNA modification found in many eukaryotic organisms, including fungi, plants, invertebrates and vertebrates, albeit its levels vary greatly in different organisms. In mammals, cytosine methylation occurs predominantly in the context of CpG dinucleotides, with the majority (60-80%) of CpG sites in their genomes being methylated. DNA methylation plays crucial roles in the regulation of chromatin structure and gene expression and is essential for mammalian development. Aberrant changes in DNA methylation and genetic alterations in enzymes and regulators involved in DNA methylation are associated with various human diseases, including cancer and developmental disorders. In mammals, DNA methylation is mediated by two families of DNA methyltransferases (Dnmts), namely Dnmt1 and Dnmt3 proteins. Over the last three decades, genetic manipulations of these enzymes, as well as their regulators, in mice have greatly contributed to our understanding of the biological functions of DNA methylation in mammals. In this chapter, we discuss genetic studies on mammalian Dnmts, focusing on their roles in embryogenesis, cellular differentiation, genomic imprinting, and human diseases.
Keywords: DNA methylation, Dnmt1, Dnmt3a, Dnmt3b, Dnmt3c, Dnmt3L, Uhrf1, Genomic imprinting
1. Distinct roles of Dnmt1 and Dnmt3 families in DNA methylation
In 1975, long before the identification of any mammalian DNA methyltransferase, Holliday and Pugh and Riggs independently proposed a theory that DNA methylation could serve as a heritable epigenetic mark for cellular memory. Recognizing that the CpG dinucleotide is self-complementary, they postulated that methylated and unmethylated CpG sites could be copied when cells divide so that DNA methylation patterns would be replicated semi-conservatively like the base sequence of DNA itself (Holliday and Pugh, 1975; Riggs, 1975). A prediction of the theory was the existence of two DNA methyltransferase activities: de novo methyltransferase(s) would methylate unmodified DNA and establish DNA methylation patterns, and maintenance methyltransferase(s) would recognize hemi-methylated sites and “copy” the methylation patterns from the parental strand onto the daughter strand at each round of DNA replication.
1.1. Dnmt1 — the maintenance DNA methyltransferase
The first mammalian DNA methyltransferase gene, Dnmt1 was cloned from murine cells (Bestor et al., 1988). The Dnmt1 locus has several transcription start sites and produces two major protein products (Mertineit et al., 1998; Rouleau et al., 1992). Transcription initiation within a somatic cell-specific exon (exon 1s) results in the Dnmt1s isoform (generally referred to as Dnmt1), which consists of 1620 amino acids (mouse). Transcription initiation within an oocyte-specific exon (exon 1o) produces a transcript that utilizes a downstream AUG as the translation initiation codon. As a result, the oocyte-specific isoform, Dnmt1o, lacks the N-terminal 118 amino acids of Dnmt1 (Fig. 5.1). Genetic evidence suggests no functional differences between the two isoforms, although Dnmt1o appears to be more stable (Ding and Chaillet, 2002). Human DNMT1, consisting of 1616 amino acids, is 80% identical to mouse Dnmt1 at the amino acid level.
Fig. 5.1. DNMTs and major regulators.
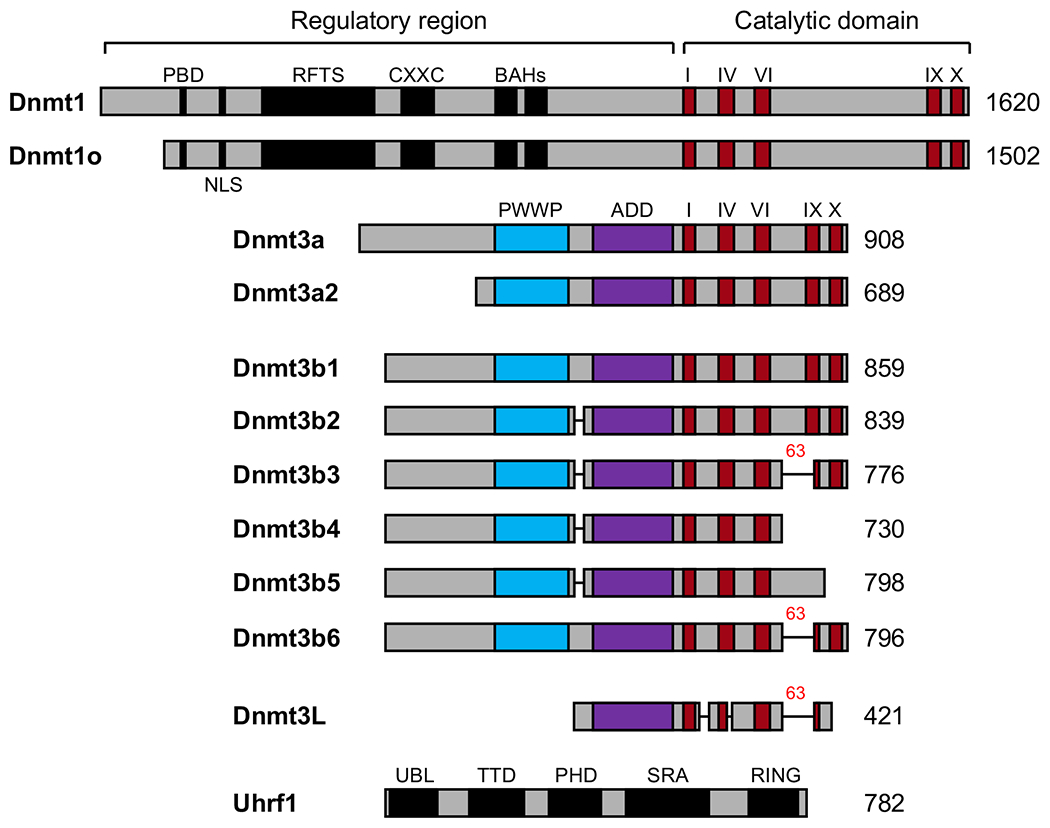
Shown are schematic diagrams of major Dnmt1, Dnmt3a and Dnmt3b isoforms, as well as Dnmt3L and Uhrf1, in mouse. The C-terminal catalytic domains of the Dnmt1 and Dnmt3 families are conserved (the highly conserved signature motifs I, IV, VI, IX, and X are shown in red), but their N-terminal regulatory regions are distinct. Note that Dnmt3L, Dnmt3b3 and Dnmt3b6 have the same 63-residue deletion in their C-terminal region. PBD, PCNA-binding domain; NLS, nuclear localization signal; RFTS, replication foci-targeting sequence; CXXC, a cysteine-rich domain implicated in binding CpG-containing DNA sequences; BAHs, bromo-adjacent homology domains; PWWP, proline-tryptophan-tryptophan-proline domain; ADD, ATRX-Dnmt3-Dnmt3L domain; UBL, ubiquitin-like domain; TTD, tandem tudor domain; PHD, plant homeodomain; SRA, SET and RING associated domain; RING, Really Interesting New Gene domain.
Dnmt1 contains a C-terminal catalytic domain with special sequence motifs that are homologous to bacterial DNA methyltransferases, and an N-terminal regulatory region that is not present in bacterial enzymes (Bestor et al., 1988). The N-terminal regulatory region contains several functional domains, including a proliferating cell nuclear antigen (PCNA) binding domain (PBD) responsible for the interaction with the DNA replication machinery, a nuclear localization signal (NLS), a replication foci-targeting sequence (RFTS) that mediates the association with late replicating heterochromatin, a zinc finger CXXC domain that recognizes unmethylated CpG-containing DNA, and a pair of bromo-adjacent homology (BAH) domains (Fig. 5.1). Structural data revealed that the RFTS domain binds to the catalytic domain and blocks the catalytic center, suggesting an autoinhibitory role in the regulation of enzymatic activity (Takeshita et al., 2011).
In vitro biochemical assays revealed that, although Dnmt1 is capable of methylating both unmethylated and hemi-methylated CpG dinucleotides, its activity toward hemi-methylated substrates is far more efficient (Pradhan et al., 1999). Dnmt1 is ubiquitously expressed through development, with high levels in proliferating cells. Dnmt1 associates with the DNA replication machinery at S phase and with heterochromatin at late S and G2 phases (Chuang et al., 1997; Easwaran et al., 2004; Leonhardt et al., 1992; Schneider et al., 2013), suggesting that Dnmt1-mediated methylation is coupled to DNA replication. These findings supported the notion that Dnmt1 mainly functions as a maintenance enzyme (Fig. 5.2). However, because Dnmt1, the only known DNA methyltransferase at the time, also had de novo methylation activity in vitro, it was initially debated whether de novo methylation and maintenance methylation are carried out by Dnmt1 alone or by two or more distinct enzymes.
Fig. 5.2. De novo and maintenance methylation.
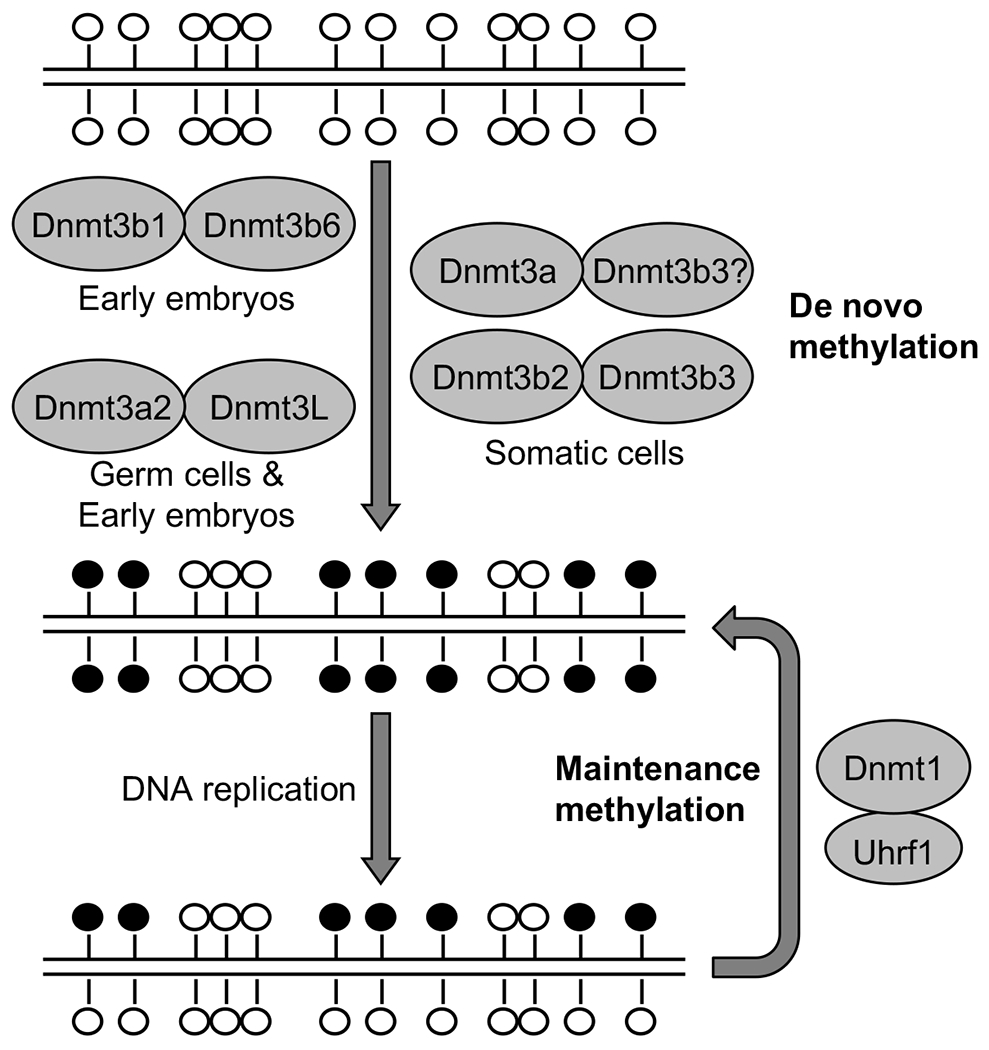
The de novo methyltransferases Dnmt3a and Dnmt3b, in complex with their accessory factors Dnmt3L, Dnmt3b3 or Dnmt3b6, methylate unmodified DNA and establish methylation patterns. The major Dnmt3 isoforms in early embryos, germ cells and somatic cells are shown. At each round of DNA replication, the maintenance methyltransferase Dnmt1, aided by its accessory factor Uhrf1, “copies” the methylation pattern from the parental strand onto the daughter strand. Open circles represent unmethylated CpG dinucleotides, and filled circles represent methylated CpG dinucleotides.
Genetic studies in mouse models and murine cells helped settling the debate. Several Dnmt1 mutant alleles were generated by gene targeting. The Dnmt1n allele (n stands for N-terminal disruption) was reported in 1992 (Li et al., 1992). This allele, in which a genomic region coding for 60 amino acids near the N-terminal end was replaced by a neomycin resistance cassette, is a partial loss-of-function (hypomorphic) mutation. Dnmt1n/n embryos have a ~70% reduction in global DNA methylation and show mid-gestation lethality (Li et al., 1992). Subsequently, the Dnmt1s allele (s stands for SalI site) was reported, which had a neomycin resistance cassette inserted into a SalI site in exon 17, disrupting the RFTS (Li et al., 1993). The Dnmt1s allele is functionally more severe than the Dnmt1n allele, as Dnmt1s/s embryos show lower levels of DNA methylation and earlier lethality (Lei et al., 1996). However, it was unclear whether the Dnmt1s allele was a null mutation, because the C-terminal catalytic domain was intact. To completely inactivate Dnmt1, Lei et al. generated the Dnmt1c allele (c stands for C-terminal disruption) by disrupting the catalytic domain, including the highly conserved PC and ENV motifs that are essential for enzymatic activity (Lei et al., 1996). The development of Dnmt1c/c embryos is arrested prior to the 8-somite stage, significantly earlier than the developmental phenotype of Dnmt1n/n embryos, while the viability and proliferation of Dnmt1 null embryonic stem cells (mESCs) is not affected. DNA methylation analyses revealed that Dnmt1 null embryos and mESCs contain low but stable levels of 5-methylcytosine (5mC) and methyltransferase activity. Moreover, the de novo methylation activity is not impaired by Dnmt1 loss, as integrated provirus DNA in MoMuLV-infected Dnmt1 null mESCs becomes methylated at a similar rate as in wild-type (WT) mESCs (Lei et al., 1996). Taken together, these early studies provided compelling evidence for the existence of one or more DNA methyltransferases other than Dnmt1 that are important for de novo methylation.
1.2. Dnmt3 family — key components of de novo methylation machinery
Results from genetic studies of Dnmt1 prompted the search for more DNA methyltransferase genes. In 1998, several groups reported the identification of a second putative DNA methyltransferase gene, named Dnmt2, which encodes a protein of 391 amino acids in human or 415 amino acids in mouse (Okano et al., 1998b; Van den Wyngaert et al., 1998; Yoder and Bestor, 1998). Despite the presence of all the conserved motifs shared by known prokaryotic and eukaryotic DNA cytosine methyltransferases, Dnmt2 has no detectable DNA methyltransferase activity in standard in vitro assays. Furthermore, inactivation of Dnmt2 in mESCs by gene targeting has no effect on preexisting genomic methylation patterns or on the ability to methylate newly integrated retrovirus DNA de novo (Okano el al., 1998b). Indeed, a subsequent study demonstrated that Dnmt2 is a tRNA methyltransferase, specific for cytosine 38 in the anticodon loop of aspartic acid tRNA and has been renamed tRNA aspartic acid (D) methyltransferase 1 (Trdmt1) (Goll et al., 2006).
By searching an expressed sequence tag (EST) database using full-length bacterial type II cytosine-5 methyltransferase sequences as queries, Okano et al. identified two additional homologous genes, Dnmt3a and Dnmt3b, in both mouse and human. Their protein products contain the highly conserved DNA methyltransferase motifs in their C-terminal regions, but their N-terminal regulatory regions show little sequence similarity to that of Dnmt1 (Okano et al., 1998a). The N-terminal regions of Dnmt3a and Dnmt3b contain a variable region and two conserved domains, the proline-tryptophan-tryptophan-proline (PWWP) domain and the ATRX-Dnmt3-Dnmt3L (ADD) domain (Fig. 5.1). Both domains are implicated in chromatin binding. The PWWP domain is required for heterochromatin localization and binds di- and tri-methylated histone H3 lysine 36 (H3K36me2/3) (Baubec et al., 2015; Chen et al., 2004; Dhayalan et al., 2010; Weinberg et al., 2019), and the ADD domain interacts with the N-terminal tail of histone H3 when H3K4 is unmethylated (Otani et al., 2009).
Dnmt3a produces two major isoforms, Dnmt3a and Dnmt3a2, driven by different promoters (Chen et al., 2002). The full-length Dnmt3a protein, consisting of 908 amino acids in mouse and 912 amino acids in human, is expressed ubiquitously at relatively low levels. The Dnmt3a2 transcript is driven by a downstream intronic promoter and encodes a protein that lacks the N-terminal 219 (in mouse) or 223 (in human) amino acids of Dnmt3a. Dnmt3a2 is the predominant form in mESCs, early embryos, and developing germ cells, as well as human embryonal carcinoma cells, and is also detectable in spleen and thymus (Chen et al., 2002). While both Dnmt3a and Dnmt3a2 are catalytically active (Chen et al., 2002), recent studies suggest functional differences between them. Zeng et al., showed that, in mESCs, Dnmt3a2 requires Dnmt3L (Dnmt3-like), an accessory factor (see below), for de novo methylation, whereas Dnmt3a has substantial activity by itself (Zeng et al., 2020). Moreover, Weinberg et al., showed that the very N terminus of Dnmt3a, which is absent in Dnmt3a2, contains a putative ubiquitin-dependent recruitment (UDR) domain that interacts with nucleosomes modified by monoubiquitylation of histone H2A lysine 119 (H2AK119ub) (Weinberg et al., 2021).
The Dnmt3b gene produces multiple alternatively spliced isoforms (>30 have been reported), many of which encode catalytically inactive protein products. The longest isoform, Dnmt3b1, consists of 859 amino acids in mouse and 853 amino acids in human, respectively. Active and inactive Dnmt3b isoforms appear to co-express in most, if not all, cell types. For example, Dnmt3b1, an active form, and Dnmt3b6, an inactive form, are the predominant forms in early embryos and mESCs, whereas Dnmt3b2, an active form, and Dnmt3b3, an inactive form, are expressed in differentiated somatic cells (Chen et al., 2002). There is evidence that catalytically inactive Dnmt3b protein products play regulatory roles in DNA methylation. For example, overexpression of human DNMT3B7, a truncated isoform frequently found in cancer cells, leads to higher levels of total genomic methylation and altered gene expression in both transgenic mice and human cancer cells (Ostler et al., 2012; Shah et al., 2010). Several catalytically inactive Dnmt3b isoforms were shown to facilitate DNA methylation in gene bodies in differentiated cells (Duymich et al., 2016). Interestingly, two Dnmt3b isoforms, Dnmt3b3 and Dnmt3b6, have the exact 63-residue deletion corresponding to that of Dnmt3L in the catalytic domain (Fig. 5.1). A recent study demonstrated that Dnmt3b3 plays a similar role as Dnmt3L but preferentially enhances Dnmt3b-mediated DNA methylation (Zeng et al., 2020).
Several lines of evidence suggest that Dnmt3a and Dnmt3b primarily function as de novo methyltransferases (Fig. 5.2). First, Dnmt3a and Dnmt3b are highly expressed in early embryos (and mESCs) and developing germ cells, where active de novo methylation takes place, but are downregulated in somatic tissues and when mESCs are induced to differentiate (Okano et al., 1998a). Second, recombinant Dnmt3a and Dnmt3b proteins methylate unmethylated and hemi-methylated DNA with equal efficiency (Okano et al., 1998a). Genetic studies provided definitive evidence that Dnmt3a and Dnmt3b were the long-sought de novo methyltransferases. Inactivation of both Dnmt3a and Dnmt3b by gene targeting blocks de novo methylation in mESCs and early embryos but has no effect on maintenance of imprinted methylation patterns (Okano et al., 1999).
Dnmt3a deficiency also leads to failure to establish DNA methylation imprints in developing germ cells (Kaneda et al., 2004). Recently, the de novo methyltransferase activity of Dnmt3a was utilized for targeted genome de novo methylation by CRISPR-based epigenome editing (Nunez et al., 2021; Stepper et al., 2017) Previously, different DNMTs including DNMT3A have been used for epigenome editing after fusion to zinc fingers (see Chapter 17 of this book). Based on the expression patterns of various Dnmt3 proteins and developmental phenotypes of knockout (KO) mice (Bourc’his and Bestor, 2004; Bourc’his et al., 2001; Hata et al., 2002; Kaneda et al., 2004; Okano et al., 1999; Zeng et al., 2020), Dnmt3a2 and its accessory factor Dnmt3L are responsible for de novo methylation in developing germ cells, Dnmt3b1 and Dnmt3b6 (an inactive isoform similar to Dnmt3L), as well as Dnmt3a2 and Dnmt3L, likely play important roles in de novo methylation during early embryogenesis, and Dnmt3b2 and Dnmt3b3 (an inactive isoform similar to Dnmt3L), as well as Dnmt3a (either alone or complexed with Dnmt3b3), are likely involved in methylation during later stages of development and in somatic cells (Fig. 5.2).
It is worth noting that the de novo DNA methyltransferase activity of Dnmt3a and Dnmt3b is not only essential for the establishment of new methylation patterns, but also important for the faithful maintenance of these patterns. In culture, Dnmt3a/3b double KO (DKO) mESCs exhibit gradual loss of global DNA methylation and, after multiple passages, show severe hypomethylation (Chen et al., 2003), suggesting that Dnmt1 and Dnmt3 enzymes have distinct and non-redundant functions but act cooperatively in the maintenance of global DNA methylation. Recent evidence suggests competition between de novo methylation by Dnmt3 proteins and demethylation initiated by the ten-eleven translocation (TET) family of 5mC dioxygenases at many loci in mESCs (Charlton et al., 2020; Wang et al., 2020). There is also evidence that Dnmt1 has de novo methylation activity. For example, oocytes deficient for Dnmt3a and Dppa3, which encodes a maternal factor DPPA3 (also known as PGC7 and Stella), exhibit higher methylation levels than WT oocytes, suggesting a robust de novo methylation activity of Dnmt1 in mouse oocytes that is normally suppressed (Li et al., 2018). Dnmt1 has also been shown to be involved in de novo methylation of specific retrotransposons, such as intracisternal A particles (Haggerty et al., 2021).
A third member of the Dnmt3 family, Dnmt3L, was originally discovered by database analysis of the human genome sequence (Aapola et al., 2000). Its murine homolog was subsequently identified (Aapola et al., 2001; Hata et al., 2002). The human and mouse Dnmt3L proteins consist of 387 and 421 amino acids, respectively. Dnmt3L contains an ADD domain, but lacks a PWWP domain, in the N-terminal region. Its C-terminal region is highly related to the catalytic domains of Dnmt3a and Dnmt3b but lacks some motifs and regions essential for enzymatic activity (Aapola et al., 2000; Aapola et al., 2001; Hata et al., 2002) (Fig. 5.1). Therefore, Dnmt3L has no methyltransferase activity. However, Dnmt3L has been shown to interact with Dnmt3a and Dnmt3b, stimulate their enzymatic activities, and target them to chromatin (Hata et al., 2002; Jia et al., 2007; Ooi et al., 2007; Suetake et al., 2004). The expression pattern of Dnmt3L during development is similar to that of Dnmt3a and Dnmt3b, including high expression in developing germ cells, early embryos, and mESCs (Hata et al., 2002). These findings indicate that Dnmt3L may regulate de novo DNA methylation (Fig. 5.2). However, genetic studies suggest that Dnmt3L functions mainly as an accessory factor for Dnmt3a (mainly Dnmt3a2) in the germline. Dnmt3L null mice are viable and grossly normal, but both male and female mice are infertile (Bourc’his et al., 2001; Hata et al., 2002). Male mice show activation of retrotransposons in spermatogonia and spermatocytes, due to hypomethylation, and are azoospermic (Bourc’his and Bestor, 2004). Female mice fail to establish maternal methylation imprints in oocytes and, as a result, embryos derived from these oocytes cannot survive beyond mid-gestation (Bourc’his et al., 2001; Hata et al., 2002). The phenotype is indistinguishable from that of mice with conditional Dnmt3a deletion in germ cells (Kaneda et al., 2004). Contrary to a previous report that Dnmt3L positively and negatively regulates DNA methylation depending on genomic regions (Neri et al., 2013), recent genome-wide methylation analysis revealed loss of methylation at many Dnmt3a target regions, but no gain of methylation at any loci in Dnmt3L-deficient mESCs, consistent with the widely accepted view that Dnmt3L is a positive regulator of DNA methylation (Veland et al., 2019). Although Dnmt3L interacts with both Dnmt3a and Dnmt3b, its deficiency results in degradation of Dnmt3a (especially Dnmt3a2), but not Dnmt3b, in mESCs, which explains, at least in part, its functional specificity in vivo (Veland et al., 2019).
In the rodent genome, a Dnmt3b duplicated gene, initially annotated as a pseudogene (Lees-Murdock et al., 2004), was subsequently identified as a new functional member of the Dnmt3 family. This gene, renamed Dnmt3c, encodes a protein of 709 amino acids which is similar to Dnmt3b but lacks the PWWP domain in the N-terminal regulatory region. It is specifically expressed in male germ cells. Genetic studies in mice demonstrated that Dnmt3c is not required for mouse development but is essential for normal spermatogenesis by methylating evolutionally young retrotransposons in the male germline (Barau et al., 2016; Jain et al., 2017).
1.3. Uhrf1 — a major regulator of maintenance DNA methylation
A number of DNA methylation regulators have been identified, including the multi-domain protein Uhrf1 (ubiquitin-like with PHD and RING finger domains 1), also known as NP95 (mouse) and ICBP90 (human) (Fig. 5.1). Genetic studies demonstrated an essential role for Uhrf1 in maintaining DNA methylation (Fig. 5.2). Uhrf1 deficiency leads to embryonic lethality and global DNA hypomethylation (Bostick et al., 2007; Muto et al., 2002; Sharif et al., 2007), resembling the phenotype of Dnmt1 deficiency. Cellular and biochemical data indicate physical and functional interactions between Uhrf1 and Dnmt1. Uhrf1 co-localizes with Dnmt1 at DNA replication foci and heterochromatin, and Dnmt1 fails to enrich at these regions in the absence of Uhrf1 (Bostick et al., 2007; Liu et al., 2013; Sharif et al., 2007). These findings suggest that Uhrf1 is a key accessory factor of Dnmt1.
Uhrf1 harbors five known functional domains: a ubiquitin-like domain (UBL) at the N terminus, followed by a tandem tudor domain (TTD), a plant homeodomain (PHD), a SET and RING associated (SRA) domain, and a Really Interesting New Gene (RING) domain (Fig. 5.1). All the domains have been shown to be important for Dnmt1 subnuclear localization and maintenance of DNA methylation. Biochemical and structural evidence revealed that the SRA domain preferentially binds hemi-methylated DNA and is thought to play an important role in loading Dnmt1 onto newly synthesized DNA substrates (Arita et al., 2008; Avvakumov et al., 2008; Bostick et al., 2007; Hashimoto et al., 2008; Sharif et al., 2007). The association of Uhrf1 with heterochromatin is mediated by TTD, which contains an aromatic cage for binding of the heterochromatic H3K9me3 mark. The PHD acts in combination with TTD to read the H3K9me3 mark and, additionally, interacts with the unmethylated arginine 2 of histone H3 tail (H3R2me0) (Cheng et al., 2013; Liu et al., 2013; Rothbart et al., 2013; Rothbart et al., 2012; Rottach et al., 2010). There is also evidence that Uhrf1, via the E3 ligase activity of its RING domain, mediates ubiquitylation of histone H3 at several lysine residues on the N-terminal tail, creating binding sites for Dnmt1 (Ishiyama et al., 2017; Nishiyama et al., 2013; Qin et al., 2015). Recently, the UBL domain was shown to interact with the E2 ubiquitin conjugating enzyme UBE2D to facilitate H3 monoubiquitylation (DaRosa et al., 2018; Foster et al., 2018). It is worth noting that Uhrf1 also controls the ubiquitylation and stability of itself and Dnmt1 (Dan et al., 2017; Du et al., 2010; Qin et al., 2011). Indeed, transgenic Uhrf1 overexpression in zebrafish hepatocytes drives DNA hypomethylation by destabilizing Dnmt1 (Mudbhary et al., 2014). Thus, the role of Uhrf1 in the regulation of DNA methylation is complex.
Recent studies indicate that Uhrf1 is a major point of regulation in modulating DNA methylation. Methylation of DNA ligase 1 (LIG1) by GLP/G9a (also known as EHMT1/EHMT2) enhances the recruitment of Uhrf1 to replicating DNA (Ferry et al., 2017), while Uhrf1 acetylation by PCAF at K490 disrupts Uhrf1 binding to hemi-methylated DNA (Hahm et al., 2020). The DNA helicase HELLS (also known as LSH) facilitates Uhrf1 chromatin association and Uhrf1-catalyzed histone H3 ubiquitylation, which in turn promotes Dnmt1 recruitment to replication forks and DNA methylation (Han et al., 2020). The protein arginine methyltransferase PRMT6 is overexpressed in multiple types of cancer (Yang and Bedford, 2013). It has been reported that PRMT6-mediated asymmetric dimethylation of H3R2 (H3R2me2a) disrupts Uhrf1 association with chromatin and contributes to global DNA hypomethylation in cancer cells (Veland et al., 2017). In culture, pluripotent mESCs sporadically convert to a transient totipotent state, known as two cell embryo-like ESCs (2CLCs), to extend shortened telomeres and repair damaged DNA (Macfarlan et al., 2012; Zalzman et al., 2010). Zscan4, a zinc finger protein specifically and highly expressed in 2CLCs, has been shown to facilitate telomere elongation by inducing global DNA demethylation due to Uhrf1-dependent ubiquitylation and degradation of Uhrf1 itself and Dnmt1 (Dan et al., 2017).
2. Dnmts in embryonic development and cellular differentiation
2.1. Roles of Dnmts in embryonic development
DNA methylation is relatively stable in somatic tissues but exhibits dynamic changes in early embryos. During preimplantation development, both the maternal and paternal genomes undergo global DNA demethylation, albeit the mechanisms involved are distinct. Demethylation of the paternal genome involves both active and passive mechanisms. Shortly after fertilization and before the first cell division, the 5mC dioxygenase Tet3, which is highly expressed in oocytes and abundant in zygotes, converts the majority of 5mC in the male pronucleus to 5-hydroxymethylcytosine (5hmC) (Gu et al, 2011; Wossidlo et al, 2011). 5hmC can be further oxidized to 5-formylcytosone (5fC) and 5-carboxylcytosine (5caC), which can be excised by thymine DNA glycosylase (TDG) and replaced by unmodified cytosine (He et al., 2011; Ito et al., 2011). The oxidized derivatives of 5mC can also persist in the paternal genome and undergo passive dilution during cleavage divisions (Inoue et al., 2011; Inoue and Zhang, 2011). Even though exposed to an identical environment in the zygote as the paternal genome, the maternal genome is protected from Tet3-mediated active demethylation. DPPA3, a maternal factor, protects the maternal genome (as well as imprinted loci in the paternal genome) from Tet3-mediated oxidation of 5mC by binding to chromatin containing H3K9 methylation (Nakamura et al., 2007). Deletion of the H3K9me2 methyltransferase G9a or the H3K9me3 methyltransferase Setdb1 (also known as ESET and KMT1E) in growing oocytes results in significant reductions of the asymmetry of global 5mC oxidation (Zeng et al., 2019). The maternal genome is demethylated mainly through DNA replication-dependent passive dilution because of deficient maintenance methylation, presumably due to the exclusion of Dnmt1 from the nucleus (Hirasawa et al., 2008; Howell et al., 2001). As a result of active and passive demethylation, DNA methylation marks inherited from gametes are largely erased by the blastocyst stage in mice, with the exception of imprinting control regions (ICRs) and some retrotransposons, which resist this wave of global demethylation. Around the time of implantation, a wave of de novo methylation takes place to establish the embryonic methylation patterns, which are then maintained in a lineage-specific manner. In contrast to DNA demethylation in preimplantation embryos in mice, rhesus monkey embryos undergo remethylation at 8-cell stage, resulting in an increase in global DNA methylation, highlighting the difference in DNA methylation reprogramming during embryogenesis among different species (Gao et al., 2017).
Most of our knowledge about the significance of DNA methylation in mammalian development came from genetic manipulations of Dnmt genes in mice. Results from characterization of Dnmt mutant mice demonstrated that the establishment of embryonic methylation patterns requires both de novo and maintenance Dnmts, and that maintaining genomic methylation above a threshold level is essential for embryonic development (Lei et al., 1996; Li et al., 1992; Okano et al., 1999). Complete inactivation of Dnmt1 results in the arrest of embryonic development between presomite and 8-somite stage around E9.5 (Lei et al., 1996). DNA methylation analysis showed that embryos deficient for Dnmt1 undergo dramatic loss of global DNA methylation (Lei et al., 1996; Li et al., 1992), in agreement with its role in maintenance methylation. Disruption of Dnmt3b also leads to embryonic lethality after E12.5, with multiple defects, including growth impairment and rostral neural tube defects (Okano et al., 1999). In contrast, Dnmt3a-deficient mice develop to term and appear normal at birth but die around 4 weeks of age (Okano et al., 1999). Consistent with the developmental phenotypes, DNA methylation analysis of E9.5 embryos revealed that germline-specific genes, pluripotency genes, hematopoietic genes, and eye genes are severely hypomethylated in the absence of Dnmt3b, but not Dnmt3a (Borgel et al., 2010). This suggests that Dnmt3b is the main enzyme responsible for de novo methylation during embryogenesis. Dnmt3b shows a dynamic expression change during pre- and early post-implantation development, with preferential expression in the trophectoderm at the mid blastocyst stage and subsequent transition of expression in the embryonic lineage (Hirasawa and Sasaki, 2009). Notably, DNA methylation at certain genes such as Brdt, Dpep3, Cytip, and Crygd is only partially reduced in Dnmt3b KO embryos (Borgel et al., 2010), suggesting that Dnmt3a cooperates with Dnmt3b to methylate some loci. Indeed, Dnmt3a/3b DKO embryos exhibit more severe defects than Dnmt3b KO embryos. Specifically, DKO embryos show smaller size, lack somites, do not undergo embryonic turning, and die before E11.5, indicating that their growth and morphogenesis are arrested shortly after gastrulation (Okano et al., 1999).
DNA methylation plays critical roles in gene repression and silencing of transposable elements (TEs). Thus, aberrant gene expression and reactivation of TEs likely contribute to the developmental defects observed in Dnmt mutant mice. Dnmt1, Dnmt3a, and Dnmt3b are all highly expressed in pluripotent mESCs, but disruption of these genes individually, both Dnmt3a and Dnmt3b, or even all three Dnmts has no deleterious effects on mESCs in the undifferentiated state (Lei et al., 1996; Li et al., 1992; Okano et al., 1999; Tsumura et al., 2006). However, Dnmt1 KO and Dnmt3a/3b DKO mESCs die upon induction of differentiation (Chen et al., 2003; Lei et al., 1996; Tucker et al., 1996). In contrast to mESCs, undifferentiated human ESCs (hESCs) require DNMT1, but not DNMT3A and DNMT3B, for survival (Liao et al., 2015). mESCs and hESCs represent different pluripotent states, with hESCs resembling the more mature epiblast state (Tesar et al., 2007), which could explain the sensitivity of hESCs to severe loss of DNA methylation.
2.2. Roles of Dnmts in cellular differentiation and maintenance of cell identity
The effects of DNA methylation deficiency become apparent during or after gastrulation, when the embryo differentiates to form the three germ layers (Lei et al., 1996; Li et al., 1992; Okano et al., 1999). Conditional inactivation of Dnmt1 in mouse embryonic fibroblasts (MEFs) leads to severe hypomethylation and cell death, while Dnmt3b-deficient MEFs show modest hypomethylation, chromosomal instability, and abnormal cell proliferation (Dodge et al., 2005; Jackson-Grusby et al., 2001). Furthermore, although a hypomorphic mutation affecting the N-terminal region of human DNMT1 has no effect on the survival and proliferation of the colon cancer cell line HCT116 (Rhee et al., 2000), disruption of the DNMT1 catalytic domain in HCT116 leads to mitotic catastrophe and cell death (Chen et al., 2007). Conditional KO studies in mice have also demonstrated that DNA methylation is essential in various organs and tissues. For example, Dnmt1 is important in the regulation of cell survival and neuronal maturation in the central nervous system and for T cell development, function, and survival (Hutnick et al., 2009; Lee et al., 2001). These results suggest crucial roles for DNA methylation in cellular differentiation and in the viability and proper functioning of differentiated cells.
2.3. DNMT mutations in human diseases
Consistent with the role of DNA methylation in cellular differentiation and maintenance of differentiated cell state, DNMT mutations have been identified in cancer and developmental disorders. DNMT1 mutations in RFTS are reported in two related neurodegenerative diseases, hereditary sensory and autonomic neuropathy with dementia and hearing loss type IE (HSAN IE) and autosomal dominant cerebellar ataxia, deafness and narcolepsy (ADCA-DN) (Klein et al., 2011; Winkelmann et al., 2012). Somatic DNMT3A mutations are frequently observed in patients with hematologic malignancies including acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and T cell acute lymphoblastic leukemia (T-ALL), with almost all mutations being heterozygous (Yang et al., 2015). The DNMT3A R882 hotspot mutation, which are found in ~30% of normal karyotype AML, has been shown to dominantly inhibit WT DNMT3A and alter the flanking sequence preferences of DNMT3A, resulting in hypomethylation at specific CpGs throughout the genome (Emperle et al., 2019; Emperle et al., 2018; Kim et al., 2013; Norvil et al., 2020; Russler-Germain et al., 2014). Changes in DNA methylation results in inhibition of hematopoietic cell differentiation, facilitating leukemogenesis. Germline mutations in DNMT3A are associated with Tatton-Brown-Rahman syndrome, a congenital disorder characterized by overgrowth, macrocephaly, and intellectual disability (Tatton-Brown et al., 2014). A recent study identified heterozygous gain-of-function missense mutations (W330R and D333N) in the PWWP domain of DNMT3A in patients with microcephalic dwarfism, who exhibit hypermethylation of Polycomb-regulated regions (Heyn et al., 2019). DNMT3B mutations account for ~50% of cases with the immunodeficiency, centromeric instability, and facial anomalies (ICF) syndrome (Hansen et al., 1999; Okano et al., 1999; Xu et al., 1999), and the other ~50% of cases are caused by mutations in ZBTB24, CDCA7, HELLS and one or more unknown genes (de Greef et al., 2011; Thijssen et al., 2015). Recent evidence indicates that the four known ICF-associated genes are functionally connected in regulating the specificity of DNA methylation. Specifically, ZBTB24 directly activates CDCA7 transcription, and CDCA7 recruits the chromatin remodeler HELLS to (peri)centromeric heterochromatin to facilitate the chromatin accessibility to the DNA methylation machinery, including DNMT3B (Hardikar et al., 2020; Jenness et al., 2018; Ren et al., 2019; Thompson et al., 2018).
In recent years, progress has been made in modeling some of these diseases in mice. Mice heterozygous for W326R or D329A substitution (corresponding to human W330 and D333, respectively) in the Dnmt3a PWWP domain recapitulate the dwarfism phenotype observed in human patients. The Dnmt3a PWWP domain is important for the normal DNA methylation landscape, as the point mutations result in gain of DNA methylation in some hypomethylated regions, including those marked by H3K27me3 and H3K4me3/H3K27me3 (bivalent) chromatin (Heyn et al., 2019; Kibe et al., 2021; Sendzikaite et al., 2019). A recent study suggests that, when the Dnmt3a PWWP domain is mutated, the UDR domain in the Dnmt3a N terminus, which recognizes H2AK119ub, may target Dnmt3a to Polycomb-regulated regions (Weinberg et al., 2021). Mice carrying Dnmt3a R878H mutation (corresponding to the hotspot DNMT3A R882H mutation in AML patients) initiate leukemogenesis due to activation of mTOR and loss of DNA methylation at specific genomic regions. Indeed, the mTOR inhibitor rapamycin elicits a significant therapeutic response in these mice (Dai et al., 2017). Dnmt3b mutations in mice recapitulate some aspects of ICF syndrome (e.g. facial anomalies), but not antibody deficiency, the major cause of infection and death occurring in patients with ICF syndrome (Ueda et al., 2006). While germline deletion of Zbtb24 leads to early embryonic lethality (Wu et al., 2016), conditional ablation of Zbtb24 in B cells impairs nonhomologous end-joining and class-switch recombination (Helfricht et al., 2020). Hells KO mice display defects in B lymphocyte development and immunoglobulin class switch recombination (He et al., 2020). The defects observed in Zbtb24 KO and Hells KO mice could contribute to antibody deficiency observed in ICF syndrome.
3. Dnmts in genomic imprinting
In the 1980s, elegant nuclear transplantation experiments using pronuclear stage embryos showed that mouse embryos constructed to contain only maternal or paternal diploid genome complements fail to develop beyond mid-gestation. This suggested that the parental genomes are functionally non-equivalent and marked or “imprinted” differently during male and female gametogenesis (Barton et al., 1984; McGrath and Solter, 1984; Surani et al., 1984). Separate experiments using chromosome translocations in mice showed that specific chromosome segments function differently depending on the parental origin (Cattanach and Kirk, 1985). In the early 1990s, the first murine imprinted genes, Igf2r, Igf2 and H19, were discovered, which are expressed only from one parental allele (Barlow et al., 1991; Bartolomei et al., 1991; DeChiara et al., 1991). To date, hundreds of imprinted genes, which exhibit mono-allelic expression strictly according to the parental origin, have been identified in mouse and human, although only some of them are imprinted in both species. Imprinted genes are involved in diverse biological processes, including embryonic development, placental formation, fetal and postnatal growth, metabolism and cognitive behaviors (Tucci et al., 2019). In human, altered expression of imprinted genes, due to genetic and epigenetic changes, is linked to infertility, molar pregnancy, and various congenital disorders such as Prader-Willi syndrome, Angelman syndrome, Beckwith-Wiedemann syndrome, and Silver-Russell syndrome (Tomizawa and Sasaki, 2012). Loss of imprinting (i.e. either biallelic expression or complete silencing of imprinted genes) is frequently observed in cancer (Jelinic and Shaw, 2007).
The majority of imprinted genes are arranged in chromosomal clusters, which usually span hundreds to thousands of kilobases. Each of the imprinting clusters contains an imprinting control region (ICR), an essential regulatory sequence that contains one or more differentially methylated regions (DMRs) – i.e. regions marked by DNA methylation only on one of the two parental alleles. Thus, allele-specific DNA methylation is considered the primary epigenetic mark controlling the mono-allelic expression of most imprinted genes. Studies in recent years have also identified DNA methylation-independent (noncanonical) imprinting that occurs at a small number of loci (see below).
3.1. Establishment of DNA methylation imprints during gametogenesis
The life cycle of DNA methylation imprints consists of three major steps: establishment, maintenance, and erasure (Fig. 5.3). DNA methylation imprints are acquired in the germline, with the majority being established during oogenesis (maternally imprinted) and only four known loci (H19, Dlk1-Gtl2, Rasgrf1, and Zdbf2) being established during spermatogenesis (paternally imprinted). Conditional deletion of Dnmt3a in primordial germ cells (PGCs) disrupts both maternal and paternal imprinting. Embryos from crosses between conditional Dnmt3a mutant females and WT males die around E10.5, and conditional Dnmt3a mutant males are sterile due to impaired spermatogenesis (Kaneda et al., 2004). Dnmt3L KO mice show an identical phenotype, with the exception of one paternally methylated locus, Dlk1-Gtl2, which does not require Dnmt3L for methylation (Bourc’his et al., 2001; Hata et al., 2002; Kaneda et al., 2004). In contrast, conditional deletion of Dnmt3b in PGCs shows no apparent phenotype (Kaneda et al., 2004). These results provide compelling genetic evidence that Dnmt3a (mainly Dnmt3a2) is responsible for the establishment of germline imprints and Dnmt3L is an essential cofactor for Dnmt3a in this process.
Fig. 5.3. Life cycle of DNA methylation imprints.
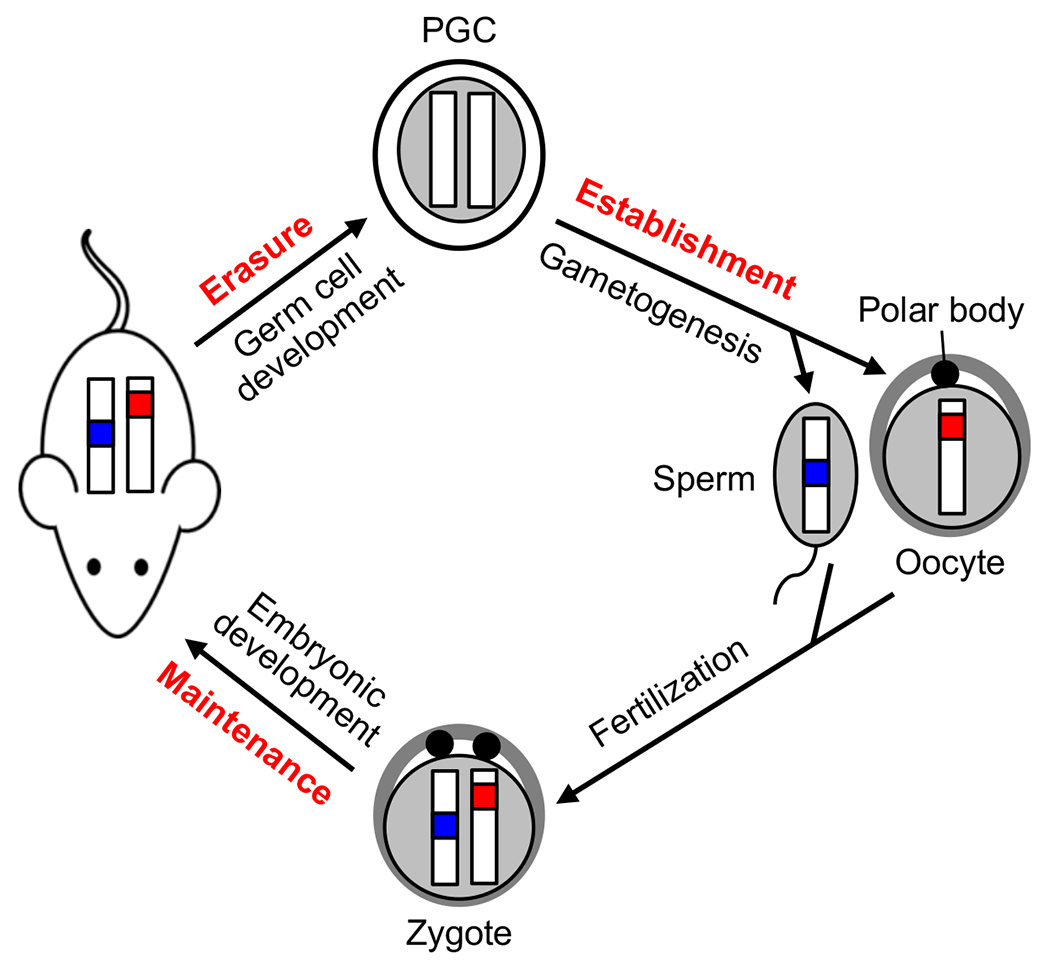
The paternal (blue) and maternal (red) methylation imprints are established during gametogenesis and transmitted to the offspring through fertilization. These marks are maintained and control mono-allelic expression of imprinted genes during embryogenesis and in somatic cells throughout adult life. However, they are erased in primordial germ cells (PGCs) before sex-specific methylation imprints are re-established in later stages of germ cell development.
The mechanisms by which Dnmt3L facilitates Dnmt3a function are complex. Dnmt3L, via its C-terminal domain, forms a tetrameric complex with Dnmt3a (Jia et al., 2007). In vitro experiments revealed that Dnmt3L stimulates the enzymatic activity of Dnmt3a (Hata et al., 2002; Suetake et al., 2004). Via its ADD domain, Dnmt3L interacts with the N-terminal tail of histone H3 when H3K4 is unmethylated (Ooi et al., 2007). Indeed, mice homozygous for an engineered point mutation (D124A) in the Dnmt3L ADD domain exhibit defects in DNA methylation and spermatogenesis (Vlachogiannis et al., 2015), supporting a critical role of the ADD domain in Dnmt3L function in the male germline. It remains to be determined whether female mice homozygous for the D1124A mutation show defects in the establishment of maternal imprints. A recent study showed that Dnmt3L also plays a role in maintaining Dnmt3a (especially Dnmt3a2) stability (Veland et al., 2019).
The observation that H3K4 methylation disrupts the interaction between the Dnmt3a/3L complex and histone H3 (Ooi et al., 2007; Otani et al., 2009) suggests that chromatin organization may be an important determinant of the sites of de novo DNA methylation in the germ line. Indeed, genetic evidence indicated that the H3K4 demethylase KDM1B (also known as LSD2 and AOF1) is essential for the establishment of a subset of maternal imprints (Ciccone et al., 2009). KDM1B is highly expressed in growing oocytes, where maternal imprints are acquired, but shows little expression in most somatic tissues. Kdm1b KO mice are viable and show no defects in spermatogenesis and oogenesis, and male mice are fertile. However, oocytes from KDM1B-deficient females exhibit global accumulation of H3K4me2 and fail to establish DNA methylation imprints at a subset of imprinted loci. Consequently, embryos derived from these oocytes die around mid-gestation (Ciccone et al., 2009), similar to embryos derived from Dnmt3L- or Dnmt3a-deficient female mice (Bourc’his et al., 2001; Hata et al., 2002; Kaneda et al., 2004). These results strongly suggest that removal of H3K4 methylation is a prerequisite for de novo DNA methylation at imprinted loci.
Transcription is another requirement for DNA methylation, at least at some imprinted loci. In the mouse Gnas locus, transcription initiated at the promoter of Nesp55, a gene upstream of the DMRs of the Gnas locus, occurs in growing oocytes, placing a large genomic region, including the DMRs, within an active transcription unit. Deletion of the Nesp55 promoter or insertion of a transcription termination cassette downstream of Nesp55 to ablate transcription results in failure to establish DNA methylation at the ICR of the Gnas locus (Chotalia et al., 2009; Frohlich et al., 2010; Williamson et al., 2011). Methylation of the DMR at the Snrpn locus also depends upon transcription (Smith et al., 2011). The proposed model for transcription-dependent genomic imprinting is that transcription elongation causes an enrichment of H3K36me2/3 at the transcribed regions, which recruit the Dnmt3a/Dnmt3L complex to establish DNA methylation in oocytes. In support of this model, depletion of the H3K36me3 methyltransferase Setd2 in oocytes causes genome-wide loss of H3K36me3, aberrant acquisition of H3K4me3 instead of DNA methylation at ICR of maternally imprinted loci (Xu et al., 2019). In contrast, Setd2 is dispensable for de novo DNA methylation in male germline. Instead, the H3K36me2 methyltransferase Nsd1 plays a critical role in de novo DNA methylation in prospermatogonia, including at imprinted genes (Shirane et al., 2020).
3.2. Maintenance of DNA methylation imprints during development
The second step of the life cycle of genomic imprints is the maintenance of mono-allelic DNA methylation marks in the offspring. The paternal and maternal imprints are transmitted to the zygote through fertilization and, despite extensive demethylation during preimplantation development, parental allele-specific DNA methylation imprints are faithfully maintained through development and adult life. Notably, genome-wide DNA methylation analyses revealed far more differentially methylated loci in oocytes and sperm than the number of imprinted genes. Therefore, the previous notion that imprinted loci are determined by distinct methylation patterns in gametes has been revised to the current view that genomic imprinting results from selective maintenance of germline-derived allele-specific methylation. Genetic studies using conditional KO mice demonstrated that Dnmt1, but not Dnmt3a or Dnmt3b, is responsible for maintaining methylation marks at imprinted loci during preimplantation development (Hirasawa et al., 2008). The oocyte-specific variant, Dnmt1o, is the predominant Dnmt1 isoform in preimplantation embryos (Hirasawa et al., 2008; Kurihara et al., 2008). However, offspring of females lacking Dnmt1o exhibit only a ~50% reduction of methylation at certain imprinted loci (Howell et al., 2001). While initial evidence suggested that the somatic form, Dnmt1, is not expressed until the blastocyst stage (Ratnam et al., 2002), subsequent work showed that Dnmt1 is present at very low levels in the nucleus of oocytes and preimplantation embryos (Hirasawa et al., 2008; Kurihara et al., 2008). Conditional deletion of Dnmt1 (both Dnmt1o and Dnmt1) in growing oocytes leads to a partial loss of methylation imprints in the offspring (Hirasawa et al., 2008), resembling the effect of Dnmt1o loss (Howell et al., 2001). However, ablation of both maternal and zygotic Dnmt1 results in a complete loss of methylation at paternally and maternally methylated DMRs in embryos (Hirasawa et al., 2008). Therefore, both maternal and zygotic Dnmt1 proteins are necessary for the maintenance of methylation imprints in preimplantation embryos. Dnmt1 is also responsible for the maintenance of methylation imprints in postimplantation embryos (Li et al., 1993) and likely in adult somatic cells as well.
It is not well understood what confers the specificity of Dnmt1, such that methylation is maintained at imprinted genes, but not at other sequences in preimplantation embryos. Genetic and epigenetic features may distinguish imprinted loci from other regions. Several factors have been shown to be essential for the maintenance of DNA methylation imprints. DPPA3, a DNA-binding protein, is highly expressed in oocytes and persists in preimplantation embryos. Genetic evidence suggests that, in early embryos, maternal DPPA3 plays a crucial role in protecting the maternal genome against DNA demethylation. DPPA3 also protects the paternally imprinted H19 and Rasgrf1 against demethylation (Nakamura et al., 2007). While the mechanisms involved remain to be determined, DPPA3 has been shown to play a role in chromatin condensation during oogenesis and to protect the maternal genome against Tet3-mediated conversion of 5mC to 5hmC in early embryos (Bian and Yu, 2014; Liu et al., 2012; Nakamura et al., 2012). The Krüppel associated box (KRAB)-containing zinc-finger proteins ZFP57 and ZFP445 are also involved in the maintenance of genomic imprints (Li et al., 2008; Takahashi et al., 2019), as mouse embryos lacking ZFP57 or ZFP445 fail to maintain DNA methylation at most ICRs. Human ZFP57 mutations are associated with hypomethylation at multiple imprinted loci in patients with transient neonatal diabetes (Mackay et al., 2008), suggesting conserved roles of ZFP57 in evolution. Mechanistic studies indicate that ZFP57 and ZFP445 act together to specifically bind the methylated allele of ICRs, recognizing a hexanucleotide sequence (TGCmCGC) shared by all murine ICRs and most human putative ICRs (Quenneville et al., 2011; Takahashi et al., 2019). ZFP57 and ZFP445 recruit KAP1 (also known as Trim28) to ICRs, which acts as a scaffold protein for various heterochromatin proteins, including heterochromatin protein 1 (HP1), the histone H3K9me3 methyltransferase Setdb1, the nuclear remodeling and histone deacetylation (NuRD) complex, and the DNA methylation machinery (Nielsen et al., 1999; Quenneville et al., 2011; Ryan et al., 1999; Schultz et al., 2002; Schultz et al., 2001; Zuo et al., 2012). Ablation of either maternal or zygotic KAP1 causes partial loss of DNA methylation imprints, and ablation of both maternal and zygotic KAP1 leads to a complete loss of imprinting (Lorthongpanich et al., 2013; Messerschmidt et al., 2012; Quenneville et al., 2011). Depletion of the NuRD components MBD3 or MTA2 also results in reduction of methylation at some imprinted loci in preimplantation embryos (Ma et al., 2010; Reese et al., 2007). N-α-acetyltransferase 10 protein (Naa10p) is another protein that has been shown to facilitate Dnmt1 binding to ICRs of imprinted loci during S phase (Lee et al., 2017). In addition to Dnmt1-mediated maintenance of DNA methylation marks at ICRs, it is equally important to prevent ICRs from being demethylated. DPPA3 has been shown to prevent erasure of DNA methylation marks at maternally imprinted genes, as well as two paternally imprinted genes, H19 and Rasgrf1, presumably by binding to H3K9me2/3 and blocking the recruitment of Tet3 (Nakamura et al., 2007; Zeng et al., 2019).
3.3. Erasure of DNA methylation imprints in primordial germ cells
The last step of the imprint life cycle is the erasure of methylation imprints in PGCs, which ensures the establishment of sex-specific imprints in later stages of germ cell development. In mice, PGCs are specified around E7.25 in the epiblast of the developing embryo. Shortly afterwards, PGCs begin migrating along the embryonic-extraembryonic interface and eventually arrive at the genital ridge by E12.5. Genome-wide DNA methylation analyses reveal that PGCs undergo demethylation in two major phases (Guibert et al., 2012; Kobayashi et al., 2013; Popp et al., 2010; Seisenberger et al., 2012). The first phase takes place during PGC expansion and migration from ~E8.5, which leads to a global demethylation affecting almost all genomic regions. Passive demethylation likely plays a major role in this phase, as a result of repression of Uhrf1, as well as Dnmt3a and Dnmt3b, in PGCs (Kagiwada et al., 2013; Kurimoto et al., 2008). The second phase occurs from E9.5 to E13.5 and affects specific loci including ICRs, germline-specific genes, and CpG islands on the X chromosome (Guibert et al., 2012; Hackett et al., 2013; Popp et al., 2010; Seisenberger et al., 2012; Yamaguchi et al., 2013a). Tet1- and Tet2-mediated 5mC oxidation is important in the second phase of demethylation (Zeng and Chen, 2019; Zhao and Chen, 2013). Genetic studies in mouse indicate that Tet1 deficiency leads to aberrant DNA hypomethylation at a subset of ICRs in germ cells and somatic tissues and results in activation of germline genes involved in gametogenesis and meiosis (Hill et al., 2018; SanMiguel et al., 2018; Yamaguchi et al., 2012; Yamaguchi et al., 2013b). Thus, both passive and active demethylation pathways are involved in the erasure of parental imprints in PGCs.
3.4. Noncanonical genomic imprinting
In addition to DNA methylation-dependent “canonical” imprinting, recent studies in mice have shown that maternal H3K27me3 can lead to DNA methylation-independent “noncanonical” imprinting, which occurs at several placenta-specific genes (e.g. Gob1, Sfmbt2, Slc38a4, and Phf17) (Inoue et al., 2017a; Okae et al., 2012). These genes, which are not associated with germline DMRs, are paternally expressed in preimplantation embryos. After implantation, they become biallelically expressed or repressed in the epiblast (which gives rise to the embryonic lineages), while retaining imprinted expression in the extraembryonic lineages and placenta (Inoue et al., 2017a). Loss of H3K27me3-mediated imprinting in mice cloned by somatic cell nuclear transfer (SCNT) could contribute to placental hyperplasia and postnatal developmental defects (Matoba et al., 2018; Okae et al., 2014), highlighting the importance of noncanonical imprinting. Oocyte-derived H3K27me3 also serves as a maternal imprint for the long non-coding RNA Xist, leading to paternal X chromosome inactivation (XCI) in female preimplantation embryos and extraembryonic tissues (i.e. imprinted XCI). The H3K27me3 imprinting mark is established during oocyte growth. Oocyte-specific deletion of Eed, which encodes an essential component of Polycomb repressive complex 2 (PRC2), abrogates noncanonical imprinting and imprinted XCI (Inoue et al., 2018; Inoue et al., 2017b). In contrast, embryos derived from DNA methylation-deficient oocytes show intact noncanonical imprinting (Chen et al., 2019; Hanna et al., 2019). Unlike DNA methylation-dependent canonical imprinting, which is maintained throughout development, H3K27me3-mediated noncanonical imprinting is transient in preimplantation embryos and absent after implantation (Chen et al., 2019; Hanna et al., 2019; Inoue et al., 2017a). Instead, maintenance of noncanonical imprinting in extraembryonic cells requires de novo DNA methylation of the maternal allele (i.e. acquisition of somatic DMRs) at implantation, which is mediated by zygotic Dnmt3a/3b (Chen et al., 2019; Hanna et al., 2019). Recent evidence suggests that the H3K9me2 methyltransferase G9a is also critical in the maintenance of noncanonical imprinting, likely by regulating the acquisition of somatic DMRs (Andergassen et al., 2021; Zeng et al., 2021).
Imprinted XCI is absent in humans (Looijenga et al., 1999; Migeon and Do, 1979), and the Xist locus is devoid of H3K27me3 in human preimplantation embryos (Xia et al., 2019). Whether noncanonical imprinting by H3K27me3 exists in humans is controversial, although candidate genes have been reported (Hanna and Kelsey, 2021).
4. Concluding remarks
DNA methylation, a relatively stable epigenetic mark, acts in concert with other epigenetic mechanisms such as histone modifications to stably maintain gene silencing and chromatin structure. Aberrant DNA methylation patterns are associated with various human diseases, including cancer and developmental disorders. Since the discovery of mammalian Dnmts (Bestor et al., 1988; Okano et al., 1998a), great progress has been made in understanding the biological functions of DNA methylation in mammals. Genetic studies using Dnmt mutant mice and murine cells have provided important insights into the roles of DNA methylation in various biological processes and the mechanisms by which mutations in DNMTs contribute to disease phenotypes (Tables 1 and 2). Nevertheless, some key questions remain to be answered. For example, what retains Dnmt1 in the cytoplasm in oocytes and early embryos that presumably leads to passive demethylation during preimplantation development? How are genes selected for the establishment of canonical or noncanonical imprinting during gametogenesis? In the coming years, we expect to see the generation of mouse models that better recapitulate the major features of human diseases associated with DNMT mutations. This becomes more feasible with the development of new technologies such as CRISPR/Cas9-mediated gene editing. Genomic, epigenomic, transcriptomic and proteomic analyses of these models will be powerful approaches for defining the molecular mechanisms and pathways involved in pathogenesis. These studies will likely lead to novel therapeutic strategies, and the genetically engineered mouse models would be valuable tools for testing therapeutics.
Table 1.
Phenotypes of Dnmt mutant mice
| Gene | Mutations | Major Phenotypes | References |
|---|---|---|---|
|
| |||
| Dnmt1 | Dnmt1 n/n | ~70% reduction in global DNA methylation, embryonic lethality at E12.5-15.5 | (Li et al., 1992) |
| Dnmt1c/c | ~90% reduction in global DNA methylation, embryonic lethality at ~E9.5. Unstable XCI | (Lei et al., 1996; Sado et al., 2000) | |
| Dnmt1s/s | Similar to Dnmt1c/c | (Beard et al., 1995; Lei et al., 1996)Lei, 1996 #10} | |
| Dnmt1o −/− | Maternal-effect phenotype: partial loss of DNA methylation imprints, defects in imprinted XCI, embryonic lethality at mid-gestation | (Howell et al., 2001; McGraw et al., 2013) | |
| Both maternal and zygotic Dnmt1−/− | Complete loss of DNA methylation imprints, embryonic lethality at mid-gestation | (Hirasawa et al., 2008) | |
|
| |||
| Dnmt3a | Dnmt3a −/− | Gut malfunction, spermatogenesis defects, death at ~4 weeks of age | (Okano et al., 1999) |
| Dnmt3a−/− in PGCs | Failure to establish DNA methylation imprints, spermatogenesis defects | (Kaneda et al., 2004) | |
| Heterozygous W326R or D329A KI in PWWP domain (corresponding to human W330 and D333) | Hypermethylation of regions marked by H3K27me3 and H3K4me3/H3K27me3 (bivalent) chromatin, Dwarfism | (Heyn et al., 2019; Kibe et al., 2021; Sendzikaite et al., 2019) | |
|
| |||
| Dnmt3b | Dnmt3b −/− | Hypomethylation of some regions, including minor satellite DNA, neural tube defects, embryonic lethality at E14.5-18.5 | (Okano et al., 1999) |
|
| |||
| Dnmt3a & 3b | Dnmt3a−/−, Dnmt3b−/− | Failure to initiate de novo methylation after implantation, embryonic lethality before E11.5. | (Okano et al., 1999) |
|
| |||
| Dnmt3L | Dnmt3L −/− | Failure to establish DNA methylation imprints, spermatogenesis defects | (Bourc’his et al., 2001; Hata et al., 2002) |
| Homozygous D124A KI in ADD domain | Hypomethylation in male germ cells, spermatogenesis defects | (Vlachogiannis et al., 2015) | |
|
| |||
| Dnmt3c | Dnmt3c −/− | Failure to silence evolutionally young retrotransposons, spermatogenesis defects | (Barau et al., 2016) |
Table 2.
Human diseases caused by DNMT mutations
| Gene | Mutations | Diseases | References |
|---|---|---|---|
|
| |||
| DNMT1 | Heterozygous missense mutations (D490E, P491Y, Y495C) in RFTS | Hereditary sensory and autonomic neuropathy with dementia and hearing loss type IE (HSAN IE) | (Klein et al., 2011) |
| Heterozygous missense mutations (A570V, G605A, V606F) in RFTS | Autosomal dominant cerebellar ataxia, deafness and narcolepsy (ADCA-DN) | (Winkelmann et al., 2012) | |
|
| |||
| DNMT3A | Somatic heterozygous mutations (multiple), with R882 as hotspot | Acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), T cell acute lymphoblastic leukemia (T-ALL) | (Yang et al., 2015) |
| De novo heterozygous mutations (multiple) | Tatton-Brown-Rahman syndrome | (Tatton-Brown et al., 2014) | |
| Gain-of-function heterozygous mutations (W330R, D333N) in PWWP domain | Microcephalic dwarfism | (Heyn et al., 2019) | |
|
| |||
| DNMT3B | Hypomorphic compound heterozygous or homozygous mutations (multiple) | Immunodeficiency, centromeric instability, and facial anomalies (ICF) syndrome | (Hansen et al., 1999; Okano et al., 1999; Xu et al., 1999) |
Acknowledgments
Work in the Chen laboratory is supported by grants from the National Institutes of Health (R01DK106418 and R01AI12140301). Work in the Dan laboratory is supported by grants from the Natural Science Foundation of Yunnan Province (202001BC070001 and 202102AA100053).
References
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Minoshima S, Krohn K, Antonarakis SE, et al. (2000). Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 65, 293–298. 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- Aapola U, Lyle R, Krohn K, Antonarakis SE, and Peterson P (2001). Isolation and initial characterization of the mouse Dnmt31 gene. Cytogenet Cell Genet 92, 122–126. 56881. [DOI] [PubMed] [Google Scholar]
- Andergassen D, Smith ZD, Kretzmer H, Rinn JL, and Meissner A (2021). Diverse epigenetic mechanisms maintain parental imprints within the embryonic and extraembryonic lineages. Dev Cell 56, 2995–3005 e2994. 10.1016/j.devcel.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, and Shirakawa M (2008). Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821. 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, and Dhe-Paganon S (2008). Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822–825. 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Herault Y, Guillou F, and Bourc’his D (2016). The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354, 909–912. 10.1126/science.aah5143. [DOI] [PubMed] [Google Scholar]
- Barlow DP, Stoger R, Herrmann BG, Saito K, and Schweifer N (1991). The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349, 84–87. 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, and Tilghman SM (1991). Parental imprinting of the mouse H19 gene. Nature 351, 153–155. 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Barton SC, Surani MA, and Norris ML (1984). Role of paternal and maternal genomes in mouse development. Nature 311, 374–376. 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, and Schubeler D (2015). Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247. 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- Beard C, Li E, and Jaenisch R (1995). Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev 9, 2325–2334. 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, and Ingram V (1988). Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 203, 971–983. [DOI] [PubMed] [Google Scholar]
- Bian C, and Yu X (2014). PGC7 suppresses TET3 for protecting DNA methylation. Nucleic Acids Res 42, 2893–2905. 10.1093/nar/gkt1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, and Weber M (2010). Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42, 1093–1100. 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, and Jacobsen SE (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764. 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, and Bestor TH (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99. 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Xu GL, Lin CS, Bollman B, and Bestor TH (2001). Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539. 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Cattanach BM, and Kirk M (1985). Differential activity of maternally and paternally derived chromosome regions in mice. Nature 315, 496–498. 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- Charlton J, Jung EJ, Mattei AL, Bailly N, Liao J, Martin EJ, Giesselmann P, Brandl B, Stamenova EK, Muller FJ, et al. (2020). TETs compete with DNMT3 activity in pluripotent cells at thousands of methylated somatic enhancers. Nat Genet 52, 819–827. 10.1038/s41588-020-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, and Li E (2007). Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet 39, 391–396. 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- Chen T, Tsujimoto N, and Li E (2004). The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol 24, 9048–9058. 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, and Li E (2003). Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 23, 5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, and Li E (2002). A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277, 38746–38754. 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yin Q, Inoue A, Zhang C, and Zhang Y (2019). Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci Adv 5, eaay7246. 10.1126/sciadv.aay7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Yang Y, Fang J, Xiao J, Zhu T, Chen F, Wang P, Li Z, Yang H, and Xu Y (2013). Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem 288, 1329–1339. 10.1074/jbc.M112.415398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, and Kelsey G (2009). Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev 23, 105–117. 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, and Li BF (1997). Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277, 1996–2000. 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, and Chen T (2009). KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418. 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Dai YJ, Wang YY, Huang JY, Xia L, Shi XD, Xu J, Lu J, Su XB, Yang Y, Zhang WN, et al. (2017). Conditional knockin of Dnmt3a R878H initiates acute myeloid leukemia with mTOR pathway involvement. Proc Natl Acad Sci USA 114, 5237–5242. 10.1073/pnas.1703476114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Rousseau P, Hardikar S, Veland N, Wong J, Autexier C, and Chen T (2017). Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells. Cell Rep 20, 1936–1949. 10.1016/j.celrep.2017.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa PA, Harrison JS, Zelter A, Davis TN, Brzovic P, Kuhlman B, and Klevit RE (2018). A bifunctional role for the UHRF1 UBL domain in the control of hemi-methylated DNA-dependent histone ubiquitylation. Mol Cell 72, 753–765 e756. 10.1016/j.molcel.2018.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef JC, Wang J, Balog J, den Dunnen JT, Frants RR, Straasheijm KR, Aytekin C, van der Burg M, Duprez L, Ferster A, et al. (2011). Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet 88, 796–804. 10.1016/j.ajhg.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, and Efstratiadis A (1991). Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859. 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, and Jeltsch A (2010). The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285, 26114–26120. 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, and Chaillet JR (2002). In vivo stabilization of the Dnmt1 (cytosine-5)-methyltransferase protein. Proc Natl Acad Sci USA 99, 14861–14866. 10.1073/pnas.232565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Okano M, Dick F, Tsujimoto N, Chen T, Wang S, Ueda Y, Dyson N, and Li E (2005). Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem 280, 17986–17991. 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. (2010). DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal 3, ra80. 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duymich CE, Charlet J, Yang X, Jones PA, and Liang G (2016). DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat Commun 7, 11453. 10.1038/ncomms11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran HP, Schermelleh L, Leonhardt H, and Cardoso MC (2004). Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep 5, 1181–1186. 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emperle M, Adam S, Kunert S, Dukatz M, Baude A, Plass C, Rathert P, Bashtrykov P, and Jeltsch A (2019). Mutations of R882 change flanking sequence preferences of the DNA methyltransferase DNMT3A and cellular methylation patterns. Nucleic Acids Res 47, 11355–11367. 10.1093/nar/gkz911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emperle M, Rajavelu A, Kunert S, Arimondo PB, Reinhardt R, Jurkowska RZ, and Jeltsch A (2018). The DNMT3A R882H mutant displays altered flanking sequence preferences. Nucleic Acids Res 46, 3130–3139. 10.1093/nar/gky168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S, Kirsh O, Amouroux R, Dohmae N, Suzuki T, et al. (2017). Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to Replicating DNA and Regulates DNA Methylation. Mol Cell 67, 550–565 e555. 10.1016/j.molcel.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Foster BM, Stolz P, Mulholland CB, Montoya A, Kramer H, Bultmann S, and Bartke T (2018). Critical role of the UBL domain in stimulating the E3 ubiquitin ligase activity of UHRF1 toward chromatin. Mol Cell 72, 739–752 e739. 10.1016/j.molcel.2018.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich LF, Mrakovcic M, Steinborn R, Chung UI, Bastepe M, and Juppner H (2010). Targeted deletion of the Nesp55 DMR defines another Gnas imprinting control region and provides a mouse model of autosomal dominant PHP-Ib. Proc Natl Acad Sci USA 107, 9275–9280. 10.1073/pnas.0910224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Niu Y, Sun YE, Lu H, Chen Y, Li S, Kang Y, Luo Y, Si C, Yu J, et al. (2017). De novo DNA methylation during monkey pre-implantation embryogenesis. Cell Res 27, 526–539. 10.1038/cr.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, and Bestor TH (2006). Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398. 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. (2011). The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610. 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guibert S, Forne T, and Weber M (2012). Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22, 633–641. 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, and Surani MA (2013). Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339, 448–452. 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty C, Kretzmer H, Riemenschneider C, Kumar AS, Mattei AL, Bailly N, Gottfreund J, Giesselmann P, Weigert R, Brandl B, et al. (2021). Dnmt1 has de novo activity targeted to transposable elements. Nat Struct Mol Biol 28, 594–603. 10.1038/s41594-021-00603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm JY, Park JW, Kang JY, Park J, Kim CH, Kim JY, Ha NC, Kim JW, and Seo SB (2020). Acetylation of UHRF1 Regulates Hemi-methylated DNA Binding and Maintenance of Genome-wide DNA Methylation. Cell Rep 32, 107958. 10.1016/j.celrep.2020.107958. [DOI] [PubMed] [Google Scholar]
- Han M, Li J, Cao Y, Huang Y, Li W, Zhu H, Zhao Q, Han JJ, Wu Q, Li J, et al. (2020). A role for LSH in facilitating DNA methylation by DNMT1 through enhancing UHRF1 chromatin association. Nucleic Acids Res 48, 12116–12134. 10.1093/nar/gkaa1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, and Kelsey G (2021). Features and mechanisms of canonical and noncanonical genomic imprinting. Genes Dev 35, 821–834. 10.1101/gad.348422.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Perez-Palacios R, Gahurova L, Schubert M, Krueger F, Biggins L, Andrews S, Colome-Tatche M, Bourc’his D, Dean W, and Kelsey G (2019). Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol 20, 225. 10.1186/s13059-019-1833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, and Gartler SM (1999). The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA 96, 14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardikar S, Ying Z, Zeng Y, Zhao H, Liu B, Veland N, McBride K, Cheng X, and Chen T (2020). The ZBTB24-CDCA7 axis regulates HELLS enrichment at centromeric satellite repeats to facilitate DNA methylation. Protein Cell 11, 214–218. 10.1007/s13238-019-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, and Cheng X (2008). The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826–829. 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, and Li E (2002). Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993. [DOI] [PubMed] [Google Scholar]
- He Y, Ren J, Xu X, Ni K, Schwader A, Finney R, Wang C, Sun L, Klarmann K, Keller J, et al. (2020). Lsh/HELLS is required for B lymphocyte development and immunoglobulin class switch recombination. Proc Natl Acad Sci USA 117, 20100–20108. 10.1073/pnas.2004112117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfricht A, Thijssen PE, Rother MB, Shah RG, Du L, Takada S, Rogier M, Moritz J, H IJ, Stoepker C, et al. (2020). Loss of ZBTB24 impairs nonhomologous end-joining and class-switch recombination in patients with ICF syndrome. J Exp Med 217. 10.1084/jem.20191688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn P, Logan CV, Fluteau A, Challis RC, Auchynnikava T, Martin CA, Marsh JA, Taglini F, Kilanowski F, Parry DA, et al. (2019). Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat Genet 51, 96–105. 10.1038/s41588-018-0274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PWS, Leitch HG, Requena CE, Sun Z, Amouroux R, Roman-Trufero M, Borkowska M, Terragni J, Vaisvila R, Linnett S, et al. (2018). Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 555, 392–396. 10.1038/nature25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, and Sasaki H (2008). Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 22, 1607–1616. 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, and Sasaki H (2009). Dynamic transition of Dnmt3b expression in mouse pre- and early post-implantation embryos. Gene Expr Patterns 9, 27–30. 10.1016/j.gep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Holliday R, and Pugh JE (1975). DNA modification mechanisms and gene activity during development. Science 187, 226–232. [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, and Chaillet JR (2001). Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104, 829–838. [DOI] [PubMed] [Google Scholar]
- Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, Silva AJ, Schweizer FE, and Fan G (2009). DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet 18, 2875–2888. 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Chen Z, Yin Q, and Zhang Y (2018). Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev 32, 1525–1536. 10.1101/gad.318675.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, and Zhang Y (2017a). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424. 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, and Zhang Y (2017b). Genomic imprinting of Xist by maternal H3K27me3. Genes Dev 31, 1927–1932. 10.1101/gad.304113.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, and Zhang Y (2011). Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21, 1670–1676. 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, and Zhang Y (2011). Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194. 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y, et al. (2017). Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance. Mol Cell 68, 350–360 e357. 10.1016/j.molcel.2017.09.037. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, and Zhang Y (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, and Jaenisch R (2001). Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27, 31–39. 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jain D, Meydan C, Lange J, Claeys Bouuaert C, Lailler N, Mason CE, Anderson KV, and Keeney S (2017). rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLoS Genet 13, e1006964. 10.1371/journal.pgen.1006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, and Shaw P (2007). Loss of imprinting and cancer. J Pathol 211, 261–268. 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- Jenness C, Giunta S, Muller MM, Kimura H, Muir TW, and Funabiki H (2018). HELLS and CDCA7 comprise a bipartite nucleosome remodeling complex defective in ICF syndrome. Proc Natl Acad Sci USA 115, E876–E885. 10.1073/pnas.1717509115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, and Cheng X (2007). Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251. 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, and Saitou M (2013). Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 32, 340–353. 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, and Sasaki H (2004). Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903. 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kibe K, Shirane K, Ohishi H, Uemura S, Toh H, and Sasaki H (2021). The DNMT3A PWWP domain is essential for the normal DNA methylation landscape in mouse somatic cells and oocytes. PLoS Genet 17, e1009570. 10.1371/journal.pgen.1009570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, and Chen T (2013). A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 122, 4086–4089. 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, Hojo K, Yamanishi H, Karpf AR, Wallace DC, et al. (2011). Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet 43, 595–600. 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, Sakashita A, Wakai T, Suzuki Y, Ito T, et al. (2013). High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res 23, 616–627. 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Kawamura Y, Uchijima Y, Amamo T, Kobayashi H, Asano T, and Kurihara H (2008). Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev Biol 313, 335–346. 10.1016/j.ydbio.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, and Saitou M (2008). Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev 22, 1617–1635. 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Peng SH, Shen L, Lee CF, Du TH, Kang ML, Xu GL, Upadhyay AK, Cheng X, Yan YT, et al. (2017). The Role of N-alpha-acetyltransferase 10 Protein in DNA Methylation and Genomic Imprinting. Mol Cell 68, 89–103 e107. 10.1016/j.molcel.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. (2001). A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774. 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Lees-Murdock DJ, McLoughlin GA, McDaid JR, Quinn LM, O’Doherty A, Hiripi L, Hack CJ, and Walsh CP (2004). Identification of 11 pseudogenes in the DNA methyltransferase gene family in rodents and humans and implications for the functional loci. Genomics 84, 193–204. 10.1016/j.ygeno.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, and Li E (1996). De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, and Bestor TH (1992). A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71, 865–873. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, and Jaenisch R (1993). Role for DNA methylation in genomic imprinting. Nature 366, 362–365. 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, and Jaenisch R (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, and Ferguson-Smith AC (2008). A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15, 547–557. 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Chen J, Liu W, Lai W, Liu B, Li X, Liu L, Xu S, Dong Q, et al. (2018). Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature 564, 136–140. 10.1038/s41586-018-0751-5. [DOI] [PubMed] [Google Scholar]
- Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, et al. (2015). Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet 47, 469–478. 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, and Wong J (2013). UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun 4, 1563. 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Nakamura T, and Nakano T (2012). Essential role of DPPA3 for chromatin condensation in mouse oocytogenesis. Biol Reprod 86, 40. 10.1095/biolreprod.111.095018. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Verkerk AJ, van Putten WL, and Oosterhuis JW (1999). Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am J Hum Genet 64, 1445–1452. 10.1086/302382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C, Cheow LF, Balu S, Quake SR, Knowles BB, Burkholder WF, Solter D, and Messerschmidt DM (2013). Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 341, 1110–1112. 10.1126/science.1240617. [DOI] [PubMed] [Google Scholar]
- Ma P, Lin S, Bartolomei MS, and Schultz RM (2010). Metastasis tumor antigen 2 (MTA2) is involved in proper imprinted expression of H19 and Peg3 during mouse preimplantation development. Biol Reprod 83, 1027–1035. 10.1095/biolreprod.110.086397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, and Pfaff SL (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63. 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. (2008). Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 40, 949–951. 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X, Inoue K, Yang L, Press W, Lee JT, et al. (2018). Loss of H3K27me3 Imprinting in Somatic Cell Nuclear Transfer Embryos Disrupts Post-Implantation Development. Cell Stem Cell 23, 343–354 e345. 10.1016/j.stem.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, and Solter D (1984). Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183. 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- McGraw S, Oakes CC, Martel J, Cirio MC, de Zeeuw P, Mak W, Plass C, Bartolomei MS, Chaillet JR, and Trasler JM (2013). Loss of DNMT1o disrupts imprinted X chromosome inactivation and accentuates placental defects in females. PLoS Genet 9, e1003873. 10.1371/journal.pgen.1003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, and Bestor TH (1998). Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development 125, 889–897. [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, and Knowles BB (2012). Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335, 1499–1502. 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Migeon BR, and Do TT (1979). In search of non-random X inactivation: studies of fetal membranes heterozygous for glucose-6-phosphate dehydrogenase. Am J Hum Genet 31, 581–585. [PMC free article] [PubMed] [Google Scholar]
- Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, et al. (2014). UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 25, 196–209. 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, and Tatsumi K (2002). Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J Biol Chem 277, 34549–34555. 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. (2007). PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9, 64–71. 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, and Nakano T (2012). PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486, 415–419. 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, Matarese F, Stunnenberg HG, and Oliviero S (2013). Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell 155, 121–134. 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, and Losson R (1999). Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J 18, 6385–6395. 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, et al. (2013). Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502, 249–253. 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- Norvil AB, AlAbdi L, Liu B, Tu YH, Forstoffer NE, Michie AR, Chen T, and Gowher H (2020). The acute myeloid leukemia variant DNMT3A Arg882His is a DNMT3B-like enzyme. Nucleic Acids Res. 10.1093/nar/gkaa139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Chen J, Pommier GC, Cogan JZ, Replogle JM, Adriaens C, Ramadoss GN, Shi Q, Hung KL, Samelson AJ, et al. (2021). Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 184, 2503–2519 e2517. 10.1016/j.cell.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okae H, Hiura H, Nishida Y, Funayama R, Tanaka S, Chiba H, Yaegashi N, Nakayama K, Sasaki H, and Arima T (2012). Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum Mol Genet 21, 548–558. 10.1093/hmg/ddr488. [DOI] [PubMed] [Google Scholar]
- Okae H, Matoba S, Nagashima T, Mizutani E, Inoue K, Ogonuki N, Chiba H, Funayama R, Tanaka S, Yaegashi N, et al. (2014). RNA sequencing-based identification of aberrant imprinting in cloned mice. Hum Mol Genet 23, 992–1001. 10.1093/hmg/ddt495. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, and Li E (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, and Li E (1998a). Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19, 219–220. 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, and Li E (1998b). Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res 26, 2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. (2007). DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717. 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostler KR, Yang Q, Looney TJ, Zhang L, Vasanthakumar A, Tian Y, Kocherginsky M, Raimondi SL, DeMaio JG, Salwen HR, et al. (2012). Truncated DNMT3B isoform DNMT3B7 suppresses growth, induces differentiation, and alters DNA methylation in human neuroblastoma. Cancer Res 72, 4714–4723. 10.1158/0008-5472.CAN-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, and Shirakawa M (2009). Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep 10, 1235–1241. 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, and Reik W (2010). Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463, 1101–1105. 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Bacolla A, Wells RD, and Roberts RJ (1999). Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem 274, 33002–33010. [DOI] [PubMed] [Google Scholar]
- Qin W, Leonhardt H, and Spada F (2011). Usp7 and Uhrfl control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem 112, 439–444. 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forne I, Pichler G, Horl D, Fellinger K, et al. (2015). DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res 25, 911–929. 10.1038/cr.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, and Trono D (2011). In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44, 361–372. 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnam S, Mertineit C, Ding F, Howell CY, Clarke HJ, Bestor TH, Chaillet JR, and Trasler JM (2002). Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev Biol 245, 304–314. 10.1006/dbio.2002.0628. [DOI] [PubMed] [Google Scholar]
- Reese KJ, Lin S, Verona RI, Schultz RM, and Bartolomei MS (2007). Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genet 3, e137. 10.1371/journal.pgen.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Hardikar S, Horton JR, Lu Y, Zeng Y, Singh AK, Lin K, Coletta LD, Shen J, Lin Kong CS, et al. (2019). Structural basis of specific DNA binding by the transcription factor ZBTB24. Nucleic Acids Res 47, 8388–8398. 10.1093/nar/gkz557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, and Schuebel KE (2000). CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404, 1003–1007. 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- Riggs AD (1975). X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 14, 9–25. [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, and Strahl BD (2013). Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev 27, 1288–1298. 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. (2012). Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 19, 1155–1160. 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, and Leonhardt H (2010). The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res 38, 1796–1804. 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau J, Tanigawa G, and Szyf M (1992). The mouse DNA methyltransferase 5′-region. A unique housekeeping gene promoter. J Biol Chem 267, 7368–7377. [PubMed] [Google Scholar]
- Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, and Ley TJ (2014). The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25, 442–454. 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, and Rauscher FJ 3rd (1999). KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol 19, 4366–4378. 10.1128/MCB.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, and Li E (2000). X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 225, 294–303. 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- SanMiguel JM, Abramowitz LK, and Bartolomei MS (2018). Imprinted gene dysregulation in a Tet1 null mouse model is stochastic and variable in the germline and offspring. Development 145. 10.1242/dev.160622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Fuchs C, Dobay A, Rottach A, Qin W, Wolf P, Alvarez-Castro JM, Nalaskowski MM, Kremmer E, Schmid V, et al. (2013). Dissection of cell cycle-dependent dynamics of Dnmt1 by FRAP and diffusion-coupled modeling. Nucleic Acids Res 41, 4860–4876. 10.1093/nar/gkt191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, and Rauscher FJ 3rd (2002). SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16, 919–932. 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, and Rauscher FJ 3rd (2001). Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 15, 428–443. 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, and Reik W (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48, 849–862. 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendzikaite G, Hanna CW, Stewart-Morgan KR, Ivanova E, and Kelsey G (2019). A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat Commun 10, 1884. 10.1038/s41467-019-09713-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MY, Vasanthakumar A, Barnes NY, Figueroa ME, Kamp A, Hendrick C, Ostler KR, Davis EM, Lin S, Anastasi J, et al. (2010). DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer Res 70, 5840–5850. 10.1158/0008-5472.CAN-10-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shirane K, Miura F, Ito T, and Lorincz MC (2020). NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat Genet 52, 1088–1098. 10.1038/s41588-020-0689-z. [DOI] [PubMed] [Google Scholar]
- Smith EY, Futtner CR, Chamberlain SJ, Johnstone KA, and Resnick JL (2011). Transcription is required to establish maternal imprinting at the Prader-Willi syndrome and Angelman syndrome locus. PLoS Genet 7, e1002422. 10.1371/journal.pgen.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper P, Kungulovski G, Jurkowska RZ, Chandra T, Krueger F, Reinhardt R, Reik W, Jeltsch A, and Jurkowski TP (2017). Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res 45, 1703–1713. 10.1093/nar/gkw1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, and Tajima S (2004). DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem 279, 27816–27823. 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- Surani MA, Barton SC, and Norris ML (1984). Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550. 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Coluccio A, Thorball CW, Planet E, Shi H, Offner S, Turelli P, Imbeault M, Ferguson-Smith AC, and Trono D (2019). ZNF445 is a primary regulator of genomic imprinting. Genes Dev 33, 49–54. 10.1101/gad.320069.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita K, Suetake I, Yamashita E, Suga M, Narita H, Nakagawa A, and Tajima S (2011). Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc Natl Acad Sci USA 108, 9055–9059. 10.1073/pnas.1019629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, Zachariou A, Hanks S, O’Brien E, Aksglaede L, et al. (2014). Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet 46, 385–388. 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, and McKay RD (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199. 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thijssen PE, Ito Y, Grillo G, Wang J, Velasco G, Nitta H, Unoki M, Yoshihara M, Suyama M, Sun Y, et al. (2015). Mutations in CDCA7 and HELLS cause immunodeficiency-centromeric instability-facial anomalies syndrome. Nat Commun 6, 7870. 10.1038/ncomms8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JJ, Kaur R, Sosa CP, Lee JH, Kashiwagi K, Zhou D, and Robertson KD (2018). ZBTB24 is a transcriptional regulator that coordinates with DNMT3B to control DNA methylation. Nucleic Acids Res 46, 10034–10051. 10.1093/nar/gky682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, and Sasaki H (2012). Genomic imprinting and its relevance to congenital disease, infertility, molar pregnancy and induced pluripotent stem cell. J Hum Genet 57, 84–91. 10.1038/jhg.2011.151. [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. (2006). Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814. 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, and Erice Imprinting G (2019). Genomic Imprinting and Physiological Processes in Mammals. Cell 176, 952–965. 10.1016/j.cell.2019.01.043. [DOI] [PubMed] [Google Scholar]
- Tucker KL, Talbot D, Lee MA, Leonhardt H, and Jaenisch R (1996). Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc Natl Acad Sci USA 93, 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Okano M, Williams C, Chen T, Georgopoulos K, and Li E (2006). Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development 133, 1183–1192. 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- Van den Wyngaert I, Sprengel J, Kass SU, and Luyten WH (1998). Cloning and analysis of a novel human putative DNA methyltransferase. FEBS Lett 426, 283–289. 10.1016/s0014-5793(98)00362-7. [DOI] [PubMed] [Google Scholar]
- Veland N, Hardikar S, Zhong Y, Gayatri S, Dan J, Strahl BD, Rothbart SB, Bedford MT, and Chen T (2017). The arginine methyltransferase PRMT6 regulates DNA methylation and contributes to global DNA hypomethylation in cancer. Cell Rep 21, 3390–3397. 10.1016/j.celrep.2017.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veland N, Lu Y, Hardikar S, Gaddis S, Zeng Y, Liu B, Estecio MR, Takata Y, Lin K, Tomida MW, et al. (2019). DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res 47, 152–167. 10.1093/nar/gky947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G, Niederhuth CE, Tuna S, Stathopoulou A, Viiri K, de Rooij DG, Jenner RG, Schmitz RJ, and Ooi SK (2015). The Dnmt3L ADD Domain Controls Cytosine Methylation Establishment during Spermatogenesis. Cell Rep. 10.1016/j.celrep.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yu G, Ming X, Xia W, Xu X, Zhang Y, Zhang W, Li Y, Huang C, Xie H, et al. (2020). Imprecise DNMT1 activity coupled with neighbor-guided correction enables robust yet flexible epigenetic inheritance. Nat Genet 52, 828–839. 10.1038/s41588-020-0661-y. [DOI] [PubMed] [Google Scholar]
- Weinberg DN, Papillon-Cavanagh S, Chen H, Yue Y, Chen X, Rajagopalan KN, Horth C, McGuire JT, Xu X, Nikbakht H, et al. (2019). The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286. 10.1038/s41586-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DN, Rosenbaum P, Chen X, Barrows D, Horth C, Marunde MR, Popova IK, Gillespie ZB, Keogh MC, Lu C, et al. (2021). Two competing mechanisms of DNMT3A recruitment regulate the dynamics of de novo DNA methylation at PRC1-targeted CpG islands. Nat Genet 53, 794–800. 10.1038/s41588-021-00856-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CM, Ball ST, Dawson C, Mehta S, Beechey CV, Fray M, Teboul L, Dear TN, Kelsey G, and Peters J (2011). Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet 7, e1001347. 10.1371/journal.pgen.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G, Melberg A, Cornelio F, Urban AE, Pizza F, et al. (2012). Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet 21, 2205–2210. 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, and Walter J (2011). 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2, 241. 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, Thijssen PE, de Klerk E, Vonk KK, Wang J, den Hamer B, Aytekin C, van der Maarel SM, and Daxinger L (2016). Converging disease genes in ICF syndrome: ZBTB24 controls expression of CDCA7 in mammals. Hum Mol Genet 25, 4041–4051. 10.1093/hmg/ddw243. [DOI] [PubMed] [Google Scholar]
- Xia W, Xu J, Yu G, Yao G, Xu K, Ma X, Zhang N, Liu B, Li T, Lin Z, et al. (2019). Resetting histone modifications during human parental-to-zygotic transition. Science 365, 353–360. 10.1126/science.aaw5118. [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, and Viegas-Pequignot E (1999). Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191. 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Xu Q, Xiang Y, Wang Q, Wang L, Brind’Amour J, Bogutz AB, Zhang Y, Zhang B, Yu G, Xia W, et al. (2019). SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat Genet 51, 844–856. 10.1038/s41588-019-0398-7. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K, and Zhang Y (2013a). Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res 23, 329–339. 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, and Zhang Y (2012). Tet1 controls meiosis by regulating meiotic gene expression. Nature 492, 443–447. 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, and Zhang Y (2013b). Role of Tet1 in erasure of genomic imprinting. Nature 504, 460–464. 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Rau R, and Goodell MA (2015). DNMT3A in haematological malignancies. Nat Rev Cancer 15, 152–165. 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, and Bedford MT (2013). Protein arginine methyltransferases and cancer. Nat Rev Cancer 13, 37–50. 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- Yoder JA, and Bestor TH (1998). A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum Mol Genet 7, 279–284. 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. (2010). Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863. 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng TB, Han L, Pierce N, Pfeifer GP, and Szabo PE (2019). EHMT2 and SETDB1 protect the maternal pronucleus from 5mC oxidation. Proc Natl Acad Sci USA 116, 10834–10841. 10.1073/pnas.1819946116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng TB, Pierce N, Liao J, and Szabo PE (2021). H3K9 methyltransferase EHMT2/G9a controls ERVK-driven noncanonical imprinted genes. Epigenomics 13, 1299–1314. 10.2217/epi-2021-0168. [DOI] [PubMed] [Google Scholar]
- Zeng Y, and Chen T (2019). DNA methylation reprogramming during mammalian development. Genes (Basel) 10, 257. 10.3390/genes10040257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Ren R, Kaur G, Hardikar S, Ying Z, Babcock L, Gupta E, Zhang X, Chen T, and Cheng X (2020). The inactive Dnmt3b3 isoform preferentially enhances Dnmt3b-mediated DNA methylation. Genes Dev 34, 1546–1558. 10.1101/gad.341925.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, and Chen T (2013). Tet family of 5-methylcytosine dioxygenases in mammalian development. J Hum Genet 58, 421–427. 10.1038/jhg.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, and Li X (2012). Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem 287, 2107–2118. 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]


