Co-infection with multiple SARS-CoV-2 lineages can result in recombination of the viral genomes and the emergence of novel, recombinant SARS-CoV-2 lineages.1, 2, 3, 4 In January, 2022, the recombinant SARS-CoV-2 XBB lineage was first detected in India and incidence is increasing in Asia and Europe.5 The XBB lineage is the result of recombination of two omicron variant sublineages, BJ.1 and BM.1.1.1, and the breakpoint is located in the gene for the spike protein (appendix p 10),6 which is responsible for host cell entry and constitutes the target of neutralising antibodies. Five major XBB sublineages (XBB.1 to XBB.5) have evolved so far, and sublineage XBB.1 accounts for most cases.5
We report an initial assessment of the ability of the SARS-CoV-2 XBB.1 lineage to enter host cells and to evade antibody-mediated neutralisation. For this, we used spike-protein-carrying pseudovirus particles (pp) that represent a suitable model to study host-cell entry of SARS-CoV-2 and its neutralisation.7 Particles pseudotyped with the spike protein of the ancestral B.1 (B.1pp) or the currently dominating omicron BA.5 (BA.5pp) lineage were used for comparison. Compared with B.1pp, BA.5pp entered Vero cells (kidney cells of the African green monkey) with 2·2 times higher efficiency and 293T cells (human kidney cells) with 5·3 times higher efficiency, whereas entry into Calu-3 cells (human lung cells) was 1·9 times less efficient compared with B.1pp, as expected (appendix p 10).8 Particles carrying XBB.1 spike protein (XBB.1pp) showed significantly reduced efficiency of cell entry compared with BA.5pp for all cell lines analysed (1·7–3·9 times reduced) and compared with B.1pp for Calu-3 cells (3·4 times reduced), whereas entry efficiency of XBB.1pp and B.1pp was similar for 293T and Vero cells (1·3–1·4 times increased efficiency of XBB.1pp; appendix p 10).
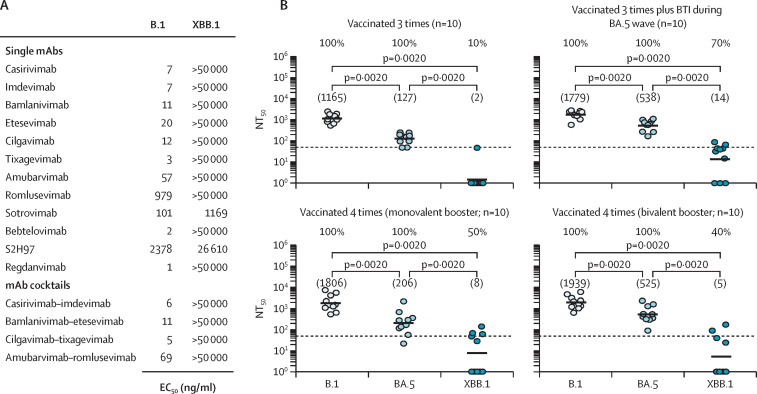
We analysed the sensitivity of XBB.1pp to neutralisation by monoclonal antibodies (mAbs) and mAb cocktails that are in clinical use (or for which clinical use has been stopped) or in development for COVID-19 prophylaxis and therapy (figure A ). All tested mAbs and mAb cocktails efficiently neutralised B.1pp (effective concentration 50 [EC50] 1–2378 ng/mL), whereas for XBB.1pp, only sotrovimab (EC50 1169 ng/mL) and S2H97 (EC50 26 610 ng/mL) were able to neutralise, and efficiency of neutralisation was reduced by more than 10 times compared with the neutralisation of B.1pp.
Figure.
Sensitivity of the SARS-CoV-2 XBB.1 lineage to neutralisation by monoclonal antibodies, antibodies induced by vaccination, and antibodies induced by vaccination plus breakthrough infection
(A) Sensitivity of the SARS-CoV-2 XBB.1 lineage to neutralisation by monoclonal antibodies. Pseudoviruses were preincubated with different concentrations of individual mAb or mAb cocktails before being inoculated onto Vero cells. At 16–18 h after inoculation, pseudovirus entry was analysed, normalised against samples without mAb (0% inhibition), and the EC50, which indicates the mAb concentration required for half-maximal inhibition, was calculated. Data represent the mean of three biological replicates (done with four technical replicates; appendix pp 3–6 and 11–12 for details on the experimental set-up and additional data). (B) Sensitivity of the SARS-CoV-2 XBB.1 lineage to neutralisation by antibodies induced by vaccination or vaccination plus breakthrough infection. Pseudoviruses were preincubated with serially diluted plasma and subsequently inoculated onto Vero cells. At 16–18 h after inoculation, pseudovirus entry was analysed and the NT50 was calculated. Data represent geometric mean NT50 values from a single biological replicate (done with four technical replicates). Numbers in brackets indicate NT50 values and percentages above the graphs represent responder rates (ie, proportion of samples with detectable neutralising activity). Dashed lines indicate the lowest plasma dilution tested. We used the Wilcoxon matched pairs signed rank test to analyse the statistical significance of the effects observed (appendix pp 13–16 for individual data). BTI=breakthrough infection. EC50=effective concentration 50. mAb=monoclonal antibodies. NT50=neutralising titre 50.
Finally, we assessed the sensitivity of XBB.1pp to neutralisation by antibodies induced by vaccination or vaccination plus breakthrough infection (figure B; appendix pp 1–2). Plasma of triple vaccinated individuals had almost no detectable neutralising activity against XBB.1pp (neutralising titre 50 [NT50] 2), whereas the neutralising activity against B.1pp was high (NT50 1165) and against BA.5pp was moderate (NT50 127). Next, we measured the plasma of triple vaccinated individuals with breakthrough infection during the BA.5 wave in Germany (June to November, 2022). The plasma samples showed high neutralising activity against B.1pp (NT50 1779), moderate neutralising activity against BA.5pp (NT50 538), and low neutralising activity against XBB.1pp (NT50 14). Similar findings were made for plasma from triple vaccinated individuals who received either monovalent or bivalent (ie, B.1 or B.1 plus BA.5) booster vaccination: B.1pp NT50 1806 for B.1 or 1939 for B.1 plus BA.5; BA.5pp NT50 206 for B.1 or 525 for B.1 plus BA.5; and XBB.1pp NT50 8 for B.1 or 5 for B.1 plus BA.5.
Collectively, our data suggest that the SARS-CoV-2 XBB.1 lineage exhibits an extraordinarily strong ability for antibody evasion, which makes XBB.1 similar to BQ.1 and BQ.1.1;9 two highly neutralisation-resistant sublineages of omicron that are currently increasing in incidence in several countries worldwide. The finding that most mAbs do not neutralise XBB.1pp highlights that novel mAbs are needed for the treatment of COVID-19 and that other or additional treatment options (eg, paxlovid, molnupiravir, or remdesivir) should be considered in areas with high incidence of the XBB sublineages. The observation that host-cell entry of XBB.1pp is reduced as compared with BA.5pp suggests that the increased ability of XBB.1 to evade antibody-mediated neutralisation might have come at the cost of a moderately reduced efficiency of host-cell entry.
SP and MH do contract research on the testing of vaccinee serum samples for neutralising activity against SARS-CoV-2 for Valneva, unrelated to this work. GMNB served as an advisor for Moderna and SP served as an advisor for BioNTech, unrelated to this work. All other authors declare no competing interests. SP acknowledges funding for this project by the German Federal Ministry of Education and Research (01KI2006D), the EU project UNDINE (grant agreement number 101057100), the Ministry for Science and Culture of Lower Saxony (14-76103-184, MWK HZI COVID-19), and the German Research Foundation (PO 716/11-1 and PO 716/14-1). H-MJ received funding from the German Federal Ministry of Education and Research (01KI2043, NaFoUniMedCovid19-COVIM 01KX2021), Bavarian State Ministry for Science and the Arts; and DFG through the research training groups RTG1660 and TRR130, the Bayerische Forschungsstiftung (Project CORAd), and the Kastner Foundation. GMNB acknowledges funding by the German Center for Infection Research (grant number 80018019238) and a European Regional Development Fund (Defeat Corona, ZW7-8515131). The funding sources had no role in study design, data collection, data analysis, data interpretation, writing of the Correspondence, or the decision to submit the manuscript for publication. We did not receive payment by a pharmaceutical company or other agency to write this Correspondence. We were not precluded from accessing data in the study and we accept responsibility to submit for publication.
Supplementary Material
References
- 1.Focosi D, Maggi F. Recombination in coronaviruses, with a focus on SARS-CoV-2. Viruses. 2022;14 doi: 10.3390/v14061239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taghizadeh P, Salehi S, Heshmati A, et al. Study on SARS-CoV-2 strains in Iran reveals potential contribution of co-infection with and recombination between different strains to the emergence of new strains. Virology. 2021;562:63–73. doi: 10.1016/j.virol.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turakhia Y, Thornlow B, Hinrichs A, et al. Pandemic-scale phylogenomics reveals the SARS-CoV-2 recombination landscape. Nature. 2022;609:994–997. doi: 10.1038/s41586-022-05189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wertheim JO, Wang JC, Leelawong M, et al. Detection of SARS-CoV-2 intra-host recombination during superinfection with alpha and epsilon variants in New York city. Nat Commun. 2022;13 doi: 10.1038/s41467-022-31247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Nadeau S, Yared M, et al. CoV-spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics. 2021;38:1735–1737. doi: 10.1093/bioinformatics/btab856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO Tracking SARS-CoV-2 variants. https://www.who.int/activities/tracking-SARS-CoV-2-variants
- 7.Schmidt F, Weisblum Y, Muecksch F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med. 2020;217 doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora P, Zhang L, Nehlmeier I, et al. The effect of cilgavimab and neutralisation by vaccine-induced antibodies in emerging SARS-CoV-2 BA.4 and BA.5 sublineages. Lancet Infect Dis. 2022;22:1665–1666. doi: 10.1016/S1473-3099(22)00693-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu P, Evans JP, Faraone J, et al. Enhanced neutralization resistance of SARS-CoV-2 omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.11.012. published online Nov 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.