Abstract
In the biological immune process, the major histocompatibility complex (MHC) plays an indispensable role in the expression of HLA molecules in the human body when viral infection activates the T-cell response to remove the virus. Since the first case of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in 2019, how to address and prevent SARS-CoV-2 has become a common problem facing all mankind. The T-cell immune response activated by MHC peptides is a way to construct a defense line and reduce the transmission and harm of the virus. Presentation of SARS-CoV-2 antigen is associated with different types of HLA phenotypes, and different HLA phenotypes induce different immune responses. The prediction of SARS-CoV-2 mutation information and the design of vaccines based on HLAs can effectively activate autoimmunity and cope with virus mutations, which can provide some references for the prevention and treatment of SARS-CoV-2.
Keywords: MHC functions, The HLA phenotype, SARS-CoV-2, T cell response, Vaccine design
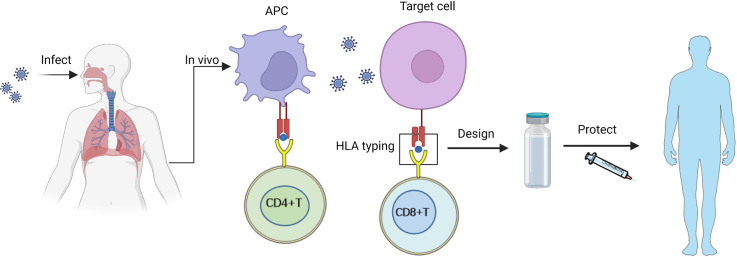
Graphical abstract
1. Introduction
In the process of human immunity, MHC (major histocompatibility complex) plays a crucial role in the rejection reaction; for instance, it participates in the processing of antigens in multiple links, inhibits the interaction between immune cells, and induces the differentiation of T cells [1], [2], [3]. MHC is classified as MHC class I (MHC-I) and MHC class II in organisms (MHC-II). MHC-I is responsible for presenting endogenous antigens to CD8+ T cells, while MHC-II is responsible for presenting exogenous antigens to CD4+ T cells. The human MHC gene, located on the short arm of human chromosome 6, is the most complex genetic system in humans, with molecules known as human leukocyte antigens (HLAs). During the process of virus infection, HLA polymorphism will affect the susceptibility of the virus and severity of the disease, which is caused by the difference in the ability of different individuals to respond to specific antigens, affecting the body to regulate the immune response.
Coronaviruses are a group of viruses that are round or oval in shape, have spike-like protrusions on their surface, and are restricted to infecting vertebrates. Human coronavirus (HCoV) infection primarily causes respiratory diseases [4]. Humans have faced three zoonotic coronaviruses since the beginning of the 21st century. They are severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute SARS-CoV-2; they have emerged in recent years and are rapidly spreading across the globe. Hundreds of millions of cases have been confirmed since the first COVID-19 infection, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in 2019. SARS-CoV-2 has become a major global public health threat. Many of the characteristics of SARS-CoV-2 are derived from its structure's spike proteins, which are the primary target of human antigen production and the virus's primary mutation site [5]. HLA class molecules play a crucial role in SARS-CoV-2 infection. In addition to presenting antigens to T cells, distinct HLA class molecules influence virus susceptibility [6]. Different countries and ethnic groups on different continents have different frequencies of HLA alleles, resulting in varying morbidity, mortality, and severity of SARS-CoV-2 in different populations. Consequently, the study of HLA alleles is crucial for the global prevention and treatment of SARS-CoV-2.
T cells play a crucial role in the SARS-CoV-2 immune response. The presence of short viral peptides of SARS-CoV-2 on the surface of antigen-presenting cells (APCs) by HLA molecules induces the immune response of T cells. CD8+ T cells primarily drive the T-cell response to HLA-I binding peptides. CD4+ T cells recognize HLA-II class-binding peptides [7]. Functionally, CD8+ T cells clear intracellular viral regions, whereas CD4+ T cells have a broader spectrum of functions, including homologous assistance to B and T cells, promotion of memory formation, and cytotoxic activity indirectly (for example, through IFNγ) or directly against target cells expressing MHC-II (10) [8]. Studying the T-cell response to SARS-CoV-2 is essential for developing protective immunity, immune pathogenesis, and COVID-19 vaccines. However, current studies on SARS-CoV-2 vaccines are basically based on neutralizing antibodies combined with the spike protein, which is susceptible to mutations and deletions [9], making it difficult to design long-term effective SARS-CoV-2 vaccines at this time. With a large amount of evidence indicating the crucial role of T cells in combating SARS-CoV-2, research on COVID-19 vaccines that induce a strong T-cell response is also a new direction for vaccine design, and the coordinated response of neutralizing antibodies and T cells has a greater protective effect against COVID-19. One key to designing a SARS-CoV-2 vaccine that elicits a T-cell response is to identify the viral epitopes responsible for activating T cells. SARS-CoV-2 proteins and presents them on the surface of host cells. Consequently, investigation of the epitopes of the HLA-I peptide derived from SARS-CoV-2 can identify the viral epitopes responsible for the activation of cytotoxic T cells and serve as a guide for developing a more effective SARS-CoV-2 vaccine.
This article describes the primary functions of MHC molecules and their roles after SARS-CoV-2 invasion and presents SARS-CoV-2 viral proteins that induce CD4+ T-cell and CD8+ T-cell responses. Because of the importance of T-cell responses to the COVID-19 immune response, this review summarizes the design of vaccines that can induce T-cell responses in the hopes of providing references for SARS-CoV-2 outbreaks.
2. Basic function and structure of MHC
MHC plays a crucial role in the presentation of biological immune antigens, such as dendritic cells that use MHC molecules to present antigens to CD4+ T and CD8+ T cells. In living organisms, there are two classes of MHC: MHC class I (MHC-I) and MHC class II (MHC-II). Often, MHC-I is responsible for presenting endogenous antigens to CD8+ T cells, and MHC-II is responsible for presenting exogenous antigens to CD4+ T cells, but this mode of operation is not fixed. Both MHC-I and MHC-II have exhibited cross-presentation. Human MHC products are commonly known as HLAs (or HLA complexes). MHC-I and MHC-II are both polymorphic molecules that exist in numerous forms; as a result, their ability to bind to viral fragments and present them to T cells varies significantly.
2.1. MHC-I
MHC class I molecules are present on the surface of nucleated cells and present as antigens. During presentation, transporters associated with antigen presentation (TAP) transport peptides to the endoplasmic reticulum (ER), where they approach MHC-I molecules [10]. In general, MHC-I is stabilized intracellularly by endoplasmic reticulum proteins, including ERp57 (also known as PDIA3), protein disulfide isomerase, and tap-binding chaperone (Tapasin). Tapasin interacts with TAP to translocate, couple, and transport peptides to MHC-I. The molecular chaperone is released when the peptide binds to MHC-I molecules, and the fully assembled peptide-MHC-I complex is presented on the cell surface [11]. Peptides delivered to MHC-I are recognized by CD8+ T cells, which ultimately initiate an immune response. Peptides and MHC-I molecules that are unable to bind in the ER are degraded in the cytosol [12], [13].
After the human immune response, APCs bind endogenous antigens to MHC-I and express them on the cell surface. Meanwhile, T-cell antigen receptor scanning of MHC class I molecules on APCs activates naive CD8+ T cells, which then function as effector cytotoxic T lymphocytes. When APCs are not directly infected, they must acquire exogenous antigens from infectious sources and present them via cross-presentation to MHC class I molecules [14]. These MHC ligands that directly stimulate the immune response of T cells are referred to as epitopes. T cells that recognize epitopes can perform immune functions such as producing inflammatory or regulatory cytokines and cytotoxicity and assisting B cells in regulating the maturation and development of antibody responses. Once an epitope is recognized, T cells proliferate to form an effector cell population, which can detect the same epitope on other cells and form a long-lived memory cell population, allowing the host to respond quickly upon subsequent encounters with the same epitope [15]. Therefore, T-cell recognition of the MHC ligand is a crucial step in the formation of adaptive immune [16] responses and memories. In addition to activating CD8+ T cells, MHC-I serves other crucial purposes. For instance, in the 2019 novel coronavirus outbreak (SARS-CoV-2), polymorphisms in the MHC-I protein sequence of human populations significantly affect the binding ability of viral peptides, thereby altering T-cell immunity to the virus [17].
2.2. MHC-II
MHC class II molecules are predominantly found on APCs, such as macrophages and dendritic cells, as opposed to MHC class I molecules (DCs). In the ER of APCs, the α- and β-chains of MHC class II molecules form the Li-MHC-II complex with an invariant chain (li). The complex is transported to the MIIC compartment via the Golgi apparatus or the plasma membrane (MHC class II compartment). Exogenous proteins and li are degraded into small peptides by proteases in MIIC, and li is continuously degraded into the Class II-associated II peptide (CLIP), which is retained in the peptide-binding tank of the MHC-II dimer. Subsequently, it is exchanged with antigenic peptides from the endosomal pathway via the action of HLA-DM [18]. MHC class II molecules are transported to the membrane of APCs following the exchange, thereby providing antigenic peptides to CD4+ T cells and activating CD4+ T cells.
Additionally, three polymorphic genes code for MHC class II molecules. Human MHC class II molecules encoded by HLA-DR, HLA-DQ, and HLA-DP bind to various peptides. The α and β chains are two noncovalently linked polypeptide chains found in class II MHC molecules. Both α and β chains extend into the cytoplasm from the cell membrane. Outside of the two chains are two Ig-like functional regions, called α1, α2, β1, and β2. α1 and β1 bind to antigenic peptides, while α2 and β2 bind to CD4 molecules. The peptide-binding II groove of MHC-II is more inclusive than that of MHC-I, allowing peptides to extend beyond the MHC-II structure. Therefore, MHC-II can bind not only long peptides but also to unfolded proteins and native proteins with different conformations [19], [20].
In most APCs and a small number of cells that can express MHC class II molecules, such as mesenchymal stromal cells, fibroblasts, and endothelial cells [19], [20], MHC class II transactivators regulate the expression of MHC class II molecules (CIITA). Exogenous or bacterial recombinant CIITA can bind to numerous histone acetyltransferases (HATs), including cyclic adenosine monophosphate response element-binding protein and P300, P300/CAF-related factor (pCAF), and steroid receptor coactivator (SRC) [21]. This mode of action enhances MHC-II gene activation. In addition, it has been discovered that CIITA is involved in host–virus defense through the upregulation of the P41 isoform of CD74. This isoform prevents the division of viral glycoproteins mediated by cathepsin, thereby preventing viral fusion and inducing viral resistance. This antiviral activity protects macrophages and dendritic cells from a wide variety of cathepsin-dependent viruses, including filoviruses and coronaviruses such as Ebola virus and SARS-like coronaviruses [22].
Human HLAs are an essential immunomodulatory component of MHC gene encoding and one of the causes of infection susceptibility. The diversity of the HLA genes is one of their defining characteristics. There are thousands of published accounts of HLA gene polymorphisms. Individually distinct immune responses to pathogens are a result of HLA gene-based differences. Numerous infectious diseases caused by RNA viruses, such as the current epidemic coronavirus SARS-CoV-2, influenza, HIV, hepatitis C, and rabies [23], are associated with HLA gene polymorphism. Class HLA-I molecules are essential for initiating the specific immune response to SARS-CoV-2. Numerous correlations between HLA genotypes and susceptibility to SARS-CoV-2 have been reported. In COVID-19 cases, the HLA-B*07:03 [24], HLA-B*46:01 [25], and HLA-C*08:01 [26] alleles are associated with severe symptoms, whereas the HLA-C*15:02 allele is associated with mild symptoms [27]. In addition, numerous studies have examined the impact of HLA gene polymorphisms on SARS susceptibility, pathogenesis, and outcome. Several HLA class I polymorphisms (including HLA-B*46:01, HLA-B*07:03.76, and HLA-CW*08:01.77) are significantly associated with the susceptibility to or severity of SARS in various populations [24], [25], [26]. Similarly, HLA class II gene polymorphisms (such as HLA-DRB4*01 and HLA-DRB1*12:02) have been demonstrated to be significantly linked to susceptibility to SARS infection. In addition to these HLA genotypes, specific HLA gene polymorphisms are described in greater detail in the following sections. Determining a patient's HLA gene profile is essential for understanding the fundamental mechanisms that protect innate and adaptive immunity and may lead to the development of genetic markers as protective factors (Fig. 1).
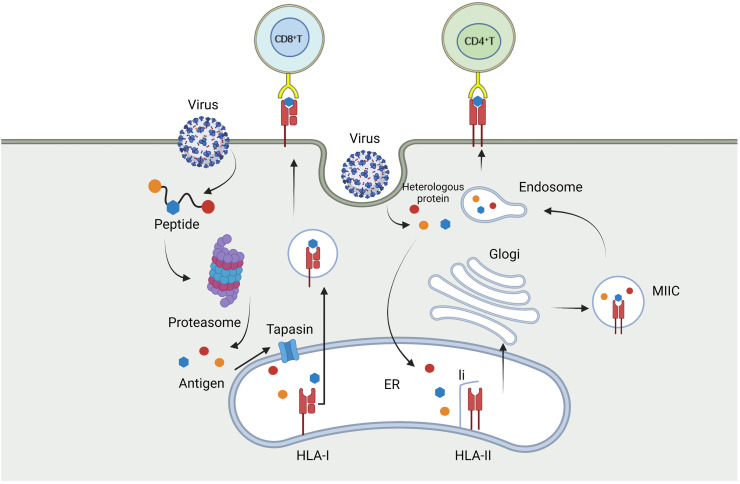
Fig. 1.
After the virus infects host cells, the endogenous antigen is degraded by proteasome, and the resulting peptide approaches HLA-I molecules through tapasin on the endoplasmic reticulum and binds to them to form a complex. Then the complex leaves the endoplasmic reticulum to reach the surface of the infected cells, and transmits the antigen information to CD8+ T cells through TCR. Exogenous antigen presentation pathway is more complex. Exogenous antigens form li-HLA-II complexes with li and HLA-II in the endoplasmic reticulum and are transported via the Golgi apparatus to the MIIC compartment, where they are degraded into small molecules, while li is continuously decomposed to generate CLIP, which is exchanged with antigenic peptides from the endosomal pathway. After exchange, class HLA-II molecules are transported to the cell membrane of infection, providing antigenic peptides to CD4+T cells and activating CD4+T cells.
3. SARS-CoV-2 is associated with HLA
3.1. Structure and properties of SARS-CoV-2
Coronaviruses are RNA viruses with widespread single plus-stranded genomes and possess the largest genome size of all known RNA viruses. SARS-CoV-2 is the seventh known human-infecting coronavirus. It primarily targets the human respiratory system and causes pneumonia and lymphocytopenia in patients. SARS-CoV-2 is comparable to previous epidemics of SARS and MERS [23]. One of its significant features is the ability to cross the species barrier, and this particular ability is closely related to its particular structure. MERS-CoV had been transmitted in camels for approximately 30 years before the first known human coronavirus case appeared [28], while SARS-CoV-2 probably originated in bat hosts [29]. SARS-CoV-2 infects alveolar epithelial cells through receptor-mediated endocytosis [30]. This ability is primarily derived from its structurally unique spike protein (S), a 1200–1400 amino acid spike protein encoded by the S gene downstream of ORF 1ab, consisting of two subunits, S1 and S2.
Compared with SARS-CoV-2 nucleocapsid, envelope (E) and membrane (M) structural proteins, the S protein has more important biological and pathogenic functions. The S protein is the primary target of neutralizing antibodies and a major component of existing vaccines. It impedes virus entry into the target cell (50, 66, 67) [31], [32], [33]. By identifying T-cell responses to six different HLA alleles in samples from COVID-19 cases, the S protein was found to be the potential target of 23.2 % of CD8+ T-cell responses to SARS-CoV-2 [34], while the viral N protein and other viral structures [35] were identified as the target of 2/3 CD4+ T cells. Other SARS-CoV-2 proteins, in addition to spike proteins, are targets of CD8+ T-cell responses. It has been reported that the S protein induces 50 % of CD8+ T-cell-mediated responses [36], while the N protein is involved in over 35 % of CD8+ T-cell-mediated responses. To date, the number of CD8 T-cell epitopes identified in the N protein is approximately one-third of that found in the S protein, but the S protein is approximately three times longer than the N protein [37], [38], [39], so the N protein also deserves careful study in areas such as vaccine design. Additionally, the variability of the N protein is less than that of the S protein. To date, only seven VoC/VoI mutations have occurred in the N protein, compared to 34 mutations in the S protein, indicating that the N protein is more conserved. Most of the key residual mutations of SARS-CoV-2 also occur in the S protein, which can evade the immune system and reduce the immunogenicity and efficacy of vaccines, thus posing a significant barrier to the effectiveness of current prevention and control strategies [40]. Therefore, it is particularly important to find a target to address SARS-CoV-2 mutations in the autoimmune process of SARS-CoV-2 patients, and HLA alleles may be helpful to find such a target (Fig. 2).
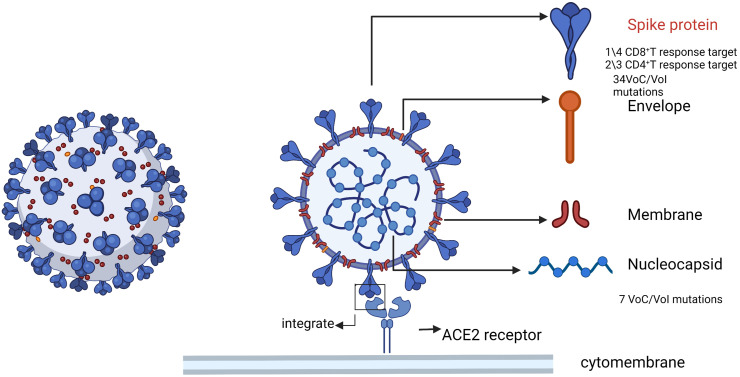
Fig. 2.
Schematic diagram of SARS-CoV-2 structure. The main components of the virus are spike, nucleocapsid, Envelope and membrane, as well as the single-stranded RNA of the virus. Spike protein is the key to SARS-CoV-2 infection. SARS-CoV-2 is also the main antigen that triggers T cell response. Most of the mutation sites of SARS-CoV-2 also occur on spike protein, while a small number of mutations occur on nucleocapsid protein.
3.2. HLA allele with COVID-19
To identify the target genes for susceptibility to and severity of SARS-CoV-2, it is necessary to understand the interaction between viral components and human proteins and the resulting immune mechanisms against infection. The S and N proteins trigger an immune response from the host to destroy the virus, which can be recognized by B cells as viral antigens and presented to T lymphocytes via MHC complexes. MHC gene polymorphisms promote the presentation of certain T-cell epitopes more effectively than other genes, so the severity of coronavirus is related to the expression level of MHC alleles. According to some functional studies, HLA-A*0201 T-cell epitopes are derived from SARS-CoV N and S proteins [41], [42]. Recent studies have determined that the HLA-A*32 allele is more frequent in healthy people than in SARS-CoV-2 patients, whereas the HLA-B*39 and HLA-C*16 alleles are more frequent in patients with SARS-CoV-2 than in healthy individuals [43]. This result indicated that MHC gene polymorphism (HLA gene polymorphism) affected the prevalence of SARS-CoV-2. Changes in the theoretical ability to bind SARS-CoV-2 peptides were also proposed in one paper to explain the connection between HLAs and clinical heterogeneity of the disease [44]. Thus, considering the role of HLA molecules in regulating the immune response to SARS-CoV-2, variability in this locus could explain the risk susceptibility of various populations to identify risk and develop personalized therapies [45].
Studying the HLA-I peptide sequence from SARS-CoV-2 can identify the epitopes [46] responsible for activating cytotoxic T cells, and the HLA epitopes are also associated with the expression of immune cells after SARS-CoV-2 infection. In a study of 28 COVID-19 patients with severe respiratory failure, extremely low expression of HLA-DR was observed, along with significant decreases in CD4 lymphocytes, CD19 lymphocytes, and natural killer cells [47]. The HLA-A, HLA-B, and HLA-C genes may contribute to the susceptibility to and severity of SARS-CoV-2 infection according to another in silico study of genetic variability in HLA class I genes [6]. In fact, susceptibility to infectious diseases such as tuberculosis, leprosy, AIDS, hepatitis B, and influenza has been linked to specific HLA haplotypes, and MHC class II haplotypes have also been associated with influenza susceptibility in certain mice [48]. Researchers computed the binding affinity between MHC class I molecules and viral peptides for all 145 HLA-A, -B, and -C genotypes of SARS-CoV-2. It has been observed that the HLA-B *46:01 allele increases susceptibility to COVID-19, as it has the least predictive binding peptide for SARS-CoV-2. At the haplotype level, HLA-A*02:02, HLA-B*15:03, and HLA-C*12:03 exhibited the greatest ability, whereas HLA-A*25:01, HLA-B*46:01, and HLA-C*01:02 exhibited the least ability. Some SARS-CoV-2 epitopes [49] are associated with five different HLA alleles, including HLA-A*02:01, HLA-B*40:01, HLA-DRA*01:01, HLA-DRB1*07:01, and HLA-DRB1*04:01 [50]. Another bioinformatics prediction and molecular modeling study identified a highly immunogenic SARS-CoV-2 epitope and its corresponding HLA allele. This model predicts that HLA-A*02:03 and A*31:01 are effective antigen carriers of SARS-CoV-2, meaning that human expression of these antigens will reduce the risk of SARS-CoV-2 infection, while HLA-A*03:02 is an A-type risk allele. Subsequently, a second in silico study revealed that the SARS-CoV polypeptide had a high propensity to bind to HLA-A*02:01 [51]. There are also a number of studies on the HLA association with SARS-CoV-1, but the results of each study are contradictory. Some studies reported that some HLA alleles increase the risk of SARS-CoV-1 infection, including HLA-B*46:01, HLA-B*07:03, HLA-C*08:01, and HLA-DRB1*1202, while other HLA alleles offer protection, including HLA-DRB1*03:01.22 HLA-C*15:02, and HLA-DRB1*03:01 (29, 31, 34) [25], [27], [52].
As a key regulator of the immune response to viral infection, HLAs play an important role in susceptibility to SARS-CoV-2 infection, but there may also be associations between HLA genetic diversity and the geographic distribution patterns, risk, severity, or outcome of SARS-CoV-2 infection. The identification of HLA alleles in different populations is one of the most important factors for protective immunity against SARS-CoV-2, which can make some individuals and populations resistant or susceptible to COVID-19. There are several molecular subtypes of HLA-A2 that specifically target Caucasians, Africans, Middle Easterners, and Asians [53]. In a recent study, certain HLA-A*02 alleles, including A*02:01, *02:03, *02:05, *02:06, *02:07, and *02:11 [54], were discovered to be prevalent in North and Central Indian populations but not in Caucasian or Middle Eastern populations. The HLA-B*46:01 allele is also believed to increase susceptibility to COVID-19. Similar to the distribution of HLA-A2, HLA-B*46:01 is present in Southeast Asia, where it is widely distributed, but completely absent in Indian and African populations and rarely present in European populations [55]. A functional study revealed that HLA-B *46:01 was expressed at a low level on the cell surface and that the diversity of the peptide group was also low, which may be a result of its long-term association with HLA-specific chaperones and intracellular retention [56], [57]. Epidemiological studies have shown that HLA-B*46:01 carriers are also more susceptible to tuberculosis, malaria, HIV, and SARS coronaviruses [58]. Similarly, the protective allele HLA-B*15:03 is completely absent from the East Asian gene pool, while the susceptibility allele HLA-C*12:03 is the most common allele in African populations and the most common allele in individuals of European descent. This suggests that susceptibility to viruses based on HLA loci appears to vary among different ethnic groups. In a study involving 99 subjects, the alleles HLA-DRB1 *15:01, -DQB1*06:02, and -B*27:07 were associated with sensitivity to COVID-19 in Italians [59]. Another study demonstrated that HLA-C*01 and B*44 were responsible for the increase in SARS-CoV-2 infections in Italy [60]. In China, the HLA-A *11:01, -B*51:01, and -C*14:02 alleles have been linked to severe cases of COVID-19 [61]. Three HLA alleles (HLA-A*11, HLA-C*01, and HLA-DQB1*04) were associated with higher mortality in a study involving 72 Spanish individuals with COVID-19 when controlling for Sequential Organ Failure Assessment and Acute Physiological and Chronic Health Assessment II (APACHE II) scores [43]. This allele does not bind any SARS-CoV-2 peptide with high affinity [62], as determined by peptide binding predictive analysis. In an ecological study, the HLA-C*05 allele was also related to COVID-19 mortality [63]. Regional frequency studies of common HLA haplotypes in Italy indicate that HLA-A*01:01-B*08:01-C*07:01-DRB1*03:01 and HLA-A*02:01-B*18:01-C*07:01-DRB1*11:04 are associated with morbidity and mortality of COVID-19, indicating risk- and protection-related haplotypes, respectively [64]. In an Italian Sardinian population association study, the HLA-A*30:02-B*14:02-C*08:02 triad haplotype was more prevalent in patients with COVID-19 [65]. In addition, HLA-B*46:01 was identified as the least predicted HLA allele for the SARS-CoV-2 binding peptide and was predicted to present the least SARS-CoV peptide, which is consistent with previous clinical data indicating that this allele is associated with severe disease [49]. However, the HLA frequency data in the study's database only represent a small portion of the population and thus may not reflect the actual gene pool of the larger population [25], [49]. To better distinguish the impact of HLA genes on the risk, severity, and outcome of SARS-CoV-2, it is necessary to collect comprehensive HLA genotyping data from the world's major populations [66]. In some cases, the HLA allele associated with a disease was found to be shared by multiple populations, and HLA-A*11 was associated with samples from Chinese and Spanish populations but not with patients from Italy. Therefore, ethnic differences in HLA allele frequency should be considered when determining genetic markers for COVID-19. Additionally, the effect of SARS-CoV-2 genomic variation on the relevant host alleles must be evaluated, as the efficiency with which HLA molecules present antigens varies based on where the virus mutates [67], [68]. This mechanism may provide a biological explanation for the severity of SARS-CoV-2 in individuals who have been exposed to other coronaviruses [69].
In addition to being associated with susceptibility to COVID-19, HLA genes may also influence the infectious phenotype of COVID-19. A European study demonstrated that half of COVID-19 patients developed anosmia, and studies in the United States showed that almost all COVID-19 patients exhibited reduced olfactory function. This difference may be due to olfactory receptor (OR) genes and MHC and OR polymorphisms contributing to the expansion of HLA/OR [70], [71] haplotypes. However, olfactory problems with COVID-19 may be related to the geographic distribution and genotype of the population. For instance, patients in Wuhan were more likely to experience fever and dyspnea (91.7 % and 21.1 %, respectively) than patients in other regions of China (78.1 % and 3.80 %) [72]. In addition, loss of smell, or anosmia, appears to be a common symptom among European and American patients but is uncommon in Asian patients (Fig. 3 ). Differences in population, culture and diet may account for this variation, but genetic variation occurs worldwide [73], [74], [75].
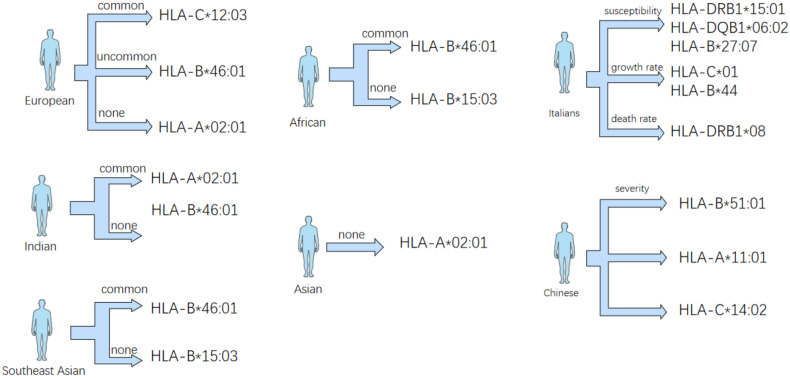
Fig. 3.
Different HLA phenotypes were expressed differently in different regions, while Italians and Chinese had relationship with the survival rate, mortality and symptoms of SARS-CoV-2 patients.
4. T cell response and HLA induced by SARS-CoV-2
SARS-CoV-2 infection causes extensive activation of innate and adaptive immunity [43], [76], [77], [78], with neutralizing antibodies and cytotoxic CD8+ T lymphocytes (CTLs [79]) serving as the primary protective factors. In contrast, virus-specific cytotoxic T lymphocytes influence viral infection and provide immune memory, allowing the body to develop long-term protection. CD8 (cytotoxic, lethal) T cells are activated upon recognition of viral peptides presented by HLA-I class molecules following SARS-CoV-2 infection, allowing them to recognize and destroy SARS-CoV-2-infected cells [80]. Similarly, CD4 T lymphocyte receptors bind to complexes formed by viral peptides and HLA-II class molecules, activating T cells [81]. The primary functions of activated CD4 (helper) T cells include regulating the immune system, stimulating B cells to produce antibodies, and enhancing CD8 T-cell responses [82]. CD4+ helper T cells coordinate the immune response and stimulate the production of antibodies by B cells, whereas CD8+ cytotoxic T cells eliminate virus-infected cells [7]. Increasing evidence also shows that the SARS-CoV-2-specific T-cell response plays a key role in regulating the pathogenesis of COVID-19. SARS-CoV-2 infection induces an extensive T-cell response in most patients, and the CD4+ T-cell response is more important than the CD8+ T-cell response [83]. Therefore, recognizing viral antigens presented on HLAs as short peptides is essential for both types of T cells.
In a study of selected SARS-CoV-2-derived T-cell epitopes, CD8+ T cells drove the T-cell response to HLA class I binding peptides, while CD4+ T cells recognized HLA-DR binding peptides [7]. More HLA-DR-dominated T-cell epitopes were also found in SARS-CoV-2 donors than class HLA-A [7] epitopes, suggesting that CD4+ T cells play a greater role in the body after viral infection. In measuring SARS-CoV-2-specific T cells, the ability of HLA alleles to bind to various viral peptides is also crucial. Three T-cell epitopes, for instance, were found to express HLA-I restriction antigens strongly in nucleocapsid polypeptide combinations, whereas only 2.6 % of S protein-derived T-cell epitopes had the ability to bind class HLA-I molecules [84]. Moreover, it was discovered that the faster COVID-19 recovers, the more viral peptides with high affinity bind to HLA-I class molecules [85]. This suggests that in addition to the patient's age and underlying disease as the cause of death, HLA genotype is also a significant factor, as age-related reduction of T-cell receptor complexes can negatively affect the prognosis of COVID-19 [86]. Consequently, it is of great significance to study the HLA-induced T-cell response in COVID-19.
Multiple stages of lung infection are present in severe COVID-19 cases. Some COVID-19 strains also cause severe respiratory failure, accompanied by deep depletion of CD4 lymphocytes, CD19 lymphocytes and natural killer cells due to macrophage activation syndrome or extremely low expression of HLA-DR [87], [88], [89]. The researchers also found an interesting similarity between COVID-19-induced immune dysregulation and a reduction in the number of HLA-DR molecules in septicemic CD14 monocytes. Tocilizumab partially restored HLA-DR expression in SARS-CoV-2 patients whose plasma inhibited HLA-DR expression. Thus, severe immune dysregulation of COVID-19 is characterized by IL-6-mediated low HLA-DR expression and lymphocytopenia as well as persistent cytokine production and excessive inflammation [47].
For CD8+ T lymphocyte activation, SARS-CoV-2 RNA induces viral protein translation after virus entry into host cells. During this step, the protein enters the proteasome of the infected cell and is cleaved into an 8- to 12-residue peptide that binds to HLA-I class receptors [85]. The complex of HLA-I class molecules and peptides is transferred to the surface of the infected cell upon binding, where it can interact with and activate the T-cell receptors of CD8+ T lymphocytes. After activation, CD8+ T lymphocytes began to divide; within 5–7 days, a population of virus-specific cytotoxic CD8+ T lymphocytes capable of destroying infected cells with perforin and serine proteases [90] was formed. In a group of patients with milder symptoms, researchers investigated the critical role of long-term activation of CD8+ T cells in the immune response to COVID-19 and discovered that activated CD8+ T cells expressed higher levels of HLA-DR and CD38 on their surface [91]. In addition to HLA-DR, some HLA alleles contain more SARS-CoV-2 epitopes, which may also influence disease severity [92].
Several recent studies have confirmed the crucial role of the T-cell response in the severity and long-term immunity of COVID-19 [87], [93], [94]. Patients with a milder form of SARS-CoV-2 exhibit a greater proportion of CD8+ T-cell responses than those with a severe form of the virus. Different epitope-specific CD8+ T-cell responses may also result in distinct clinical outcomes for COVID-19 [95], whereas CD4+ T cells play a crucial role in the natural progression of infection [7], but CD4+ T-cell responses are more important than CD8+ T-cell responses in patients [83]. These phenomena contribute to understanding the pathogenesis of SARS-CoV-2 infection.
The immunity of preexisting T cells to CCC may affect the immunity of SARS-CoV-2 and the clinical outcome of COVID-19 patients; this result is a consequence of T-cell cross-reaction formed in response to more than one different peptide-MHC ligand. The T-cell receptor (TCR) is also expressed by peptide-MHC ligand-bound cell surface markers CD154 and CD69. Therefore, CD4+ memory T cells reactive to SARS-CoV-2 can be isolated and screened [96]. In this way, Low et al. identified many T-cell clones that respond extensively to SARS-CoV-2 and other coronavirus S proteins [96], [97]. This finding provides evidence that infection with SARS-CoV-2 affects preexisting cross-reactive memory T cells. Preexisting SARS-CoV-2 cross-reactive T cells may influence various COVID-19 symptoms, and SARS-CoV-2 cross-reactive T cells may have been acquired during a previous infection with an endemic human coronavirus. Cross-reactive T cells have been found in 20 %–50 % of different populations worldwide who have not been exposed to SARS-CoV-2 [98].
It is worth mentioning that cross-reactive T-cell studies are also relevant to developing a SARS-CoV-2 vaccine. According to an analysis of the pathogenic components of SARS-CoV-2, the S protein and nucleocapsid elicit a robust response from CD4+ T cells. The reason for this is that the RBD of CD4+ T cells contains a conserved immunodominant region and many T-cell epitopes, allowing the identification of cross-reactive T cells targeting the S protein site, which could inform the development of new vaccines against novel SARS-CoV-2 variants [99].
In conclusion, T cells play a crucial role in the immune response against SARS-CoV-2 and mediate long-term protection against the virus. Understanding the response and differentiation of CD4+ and CD8+ T cells in COVID-19 through HLAs and other epitopes is of great significance for the diagnosis, treatment and enhancement of COVID-19.
5. T cell responses and HLA can be involved in SARS-CoV-2 vaccine design
Currently, countries lack an effective method for controlling COVID-19. Typically, traditional public health measures are used to control the spread of SARS-CoV-2, such as wearing masks in public and isolating people in close proximity to COVID-19 to prevent and reduce the virus's spread. Traditional methods have been ineffective in reducing the spread of COVID-19 and the number of cases; therefore, developing a vaccine that is effective against SARS-CoV-2 is one way to reduce the global spread and infection of the virus. The majority of SARS-CoV-2 vaccine strains that are currently in development were created using traditional and modern techniques with attenuated or inactivated viruses. Traditional vaccines have been effective against infectious diseases caused by viruses, including polio, measles, rubella and chickenpox. However, using traditional techniques to develop vaccines against HIV, hepatitis C and dengue has not been successful, and the same is true regarding vaccines against coronaviruses. The use of modern vaccine techniques, such as viral peptides or DNA/RNA, represents a promising new direction. Most SARS-CoV-2 vaccines in phase II or III clinical trials are based on modern vaccine [9] techniques. These techniques introduce specific virus components or viral genes into the body to induce a targeted immune response. The vaccine induces a cellular and humoral response mediated by T cells and antibodies. Most current COVID-19 vaccine candidates concentrate on spike proteins [100], which are also prime neutralizing antibody targets [101], [102].
The T-cell response is critical for vaccine effectiveness [36], [103]. As described in the previous narrative, a robust T-cell response facilitates recovery from COVID-19 and produces durable immunity in the body [104]. In addition to antiviral approaches that rely solely on T-cell responses, coordinated responses of neutralizing antibodies and T cells provide better protection against COVID-19 [9], [105]. One direction of COVID-19 vaccine development is to mimic this process by stimulating immune cells that specialize in recognizing SARS-CoV-2. T cells are activated by recognizing SARS-CoV-2 peptides (short linear amino acid sequences) that are presented on the surface of infected cells via HLA molecules. This prepares the immune system to combat the virus should a natural infection occur. While different HLA haplotypes are associated with different disease sensitivities, HLA typing can provide information about disease sensitivity for prevention, treatment, vaccination, and clinical strategies. For example, HLA-I is associated with H1N1 infection in humans, HLA-A*11, HLA-B*35, and HLA-DRB1*10 confer susceptibility to (PDM09) influenza A (H1N1) infection [106], and the previously mentioned MHC Class II haplotype found in mice is also associated with influenza infection [48]. Identifying the association of the virus with these specific HLA loci allows the detection of alleles that demonstrate induction of protective immunity, which can be used to assess the effectiveness of vaccination in the entire population. Understanding the potent T-cell response induced by HLAs in COVID-19 vaccines may therefore aid in developing and evaluating such vaccines. The following are some of the most recent discoveries in this area.
In the SARS-CoV-2 vaccine currently under development, neutralizing antibodies are an important part of the vaccine, but this may not be sufficient to address the current spread of the virus. Viral variants of SARS-CoV-2, such as B.1.1.7 alpha, B.1.351 Beta, P.1 Gamma, and B.1.617.2 Delta have increased transmissible ability and ability to evade convalescence and vaccine-induced antibody responses [107], [108], [109], [110], [111]. Unlike the sharp restriction neutralization antibody response, CD8+ T cells can target the entire SARS-CoV-2 proteome region, enabling them to target the restriction mutation site [112] in the SARS-CoV-2 mutant. Therefore, inducing SARS-CoV-2-specific CD8+ T cells can significantly improve the protective ability of the vaccine. Increased CD8+ T-cell clonal expansion was found in bronchoalveolar lavage fluid in individuals with mild symptoms of COVID-19 [113], indicating strong responsiveness of CD8+ T cells to SARS-CoV-2 epitopes and rapid viral clearance mediated by CD8+ T cells [114], [115], [116]. Similarly, Shabak virus-induced CD8+ T cells showed persistent immune memory against SARS-CoV-1 infection [117], and vaccine-induced CD8+ T cells protected mice from the fatal symptoms of SARS-CoV-1. Thus, inducing CD8+ T cells to respond is the key to vaccine design, and it is necessary to search for inducing factors.
One entry point is that SARS-CoV-2 can evade cellular immunity by mutating class HLA-I-restricted epitopes [118]. Anusha Nathan et al. recently demonstrated that CD8+ T-cell epitopes identified by class HLA-I restrictive epitopes are confined by mutations in highly mutated HIV. For SARS-CoV-2, targeting of CD8+ T cells in vivo was observed by predicting whether the highly networked SARS-CoV-2 epitopes could bind and stabilize 18 HLA alleles. The results showed that at least 50 % of the HIV epitopes were deemed promising SARS-CoV-2 T-cell immunogens relative to HLA class I [119]. This induced CD8+ T-cell response to highly networked epitopes may offer enhanced protection against emerging SARS-CoV-2 variants in humans. Thus, highly networked T-cell vaccines could complement current neutralizing antibody-based vaccines to provide stronger and broader immunity to the global population to prevent the emergence of new SARS-like coronaviruses and the continued mutation of SARS-CoV-2.
Studies of SARS-CoV-2-derived HLA-I peptides can identify the viral epitopes responsible for the activation of cytotoxic T cells, but HLA-I binding alone is insufficient to predict the recognition of antigens. First, there are limitations. Antigen processing and presentation is a complex, multistep biological pathway [18] that involves the degradation of viral proteins by proteasomes, the cleavage of peptides by aminopeptidases, the transfer to the endoplasmic reticulum and the binding of HLA-I, and the presentation of antigens on the cell surface by HLA-I. Many computational predictors now account for some of the steps in the two-point presentation process, but the average positive predictive value of testing for different HLA alleles is still approximately 64 % [120], indicating that a portion of the antigen remains undetected. In addition, the prediction method did not account for the possibility that the virus might affect the cells' ability to express antigens during the infection process. For instance, the virus can inhibit the host's protein translation, cut the proteasome mechanism, and interfere with the expression of HLA-I. These changes may not induce a complete and effective immune response, and HLA-I monitoring is also [121], [122] incomplete. Third, predictive models fail to account for the dynamic evolution of viral proteins during infection. Kinetic studies of vaccines and influenza viruses have demonstrated that HLA-I presentation of virus epitopes reaches a maximum between 3.5 and 9.5 h (HPI) after infection [123], [124]. Fourth, the virus can also inhibit and interfere with HLA-I presentation, so there are more viral proteins present in the early phase of viral infection, whereas two-point proteins expressed later in the viral life cycle cannot be expressed more effectively. Infection with SARS-CoV-2 substantially decreased the expression of protease POMP and ubiquitination pathway proteins. By influencing ubiquitin-mediated proteasome degradation and immune signaling proteins, SARS-CoV-2 may reduce downstream processing and HLA-I presentation precursors and alter immune responses [46]. Additionally, inhibition of translation by NSP1 and degradation of host transcripts attenuated HLA-I expression and antigen presentation [125]. The ORF8 protein also interferes with HLA-I antigen presentation and influences CTLs' ability to recognize and eliminate virus-infected cells [126].
Cross-reactivity of T cells following infection with other coronavirus-infected cells can hinder the effectiveness of COVID-19. Consequently, the identification of genetically similar epitopes of SARS-CoV-2 and other coronavirus-like viruses (e.g., SARS-CoV, MERS-CoV, and human common cold coronavirus-like viruses) could aid in the development of coronavirus-like vaccines. To prevent current human coronaviruses and future coronaviruses that may jump from other species to humans [127], [128], silicon-based T-cell epitope prediction using SARS-CoV-2 could aid in developing such vaccines [129]. At present, the detection of epitopes of SARS-CoV-2 virus is mainly predicted by bioinformatics of HLA-I and HLA-II binding affinity as well as biochemical binding tests and reactivity tests of SARS-CoV-2 peptides [7], [36], [130]. However, this prediction has limitations, and it is not certain that all peptides predicted by SARS-CoV-2 bioinformatics can be processed and presented by APCs or infected cells during SARS infection. Adi Nagler et al. therefore combined bioinformatics tools with HLA peptides to further identify HLA-I and HLA-II viral antigens through viral gene transduction of selected cells or infection with SARS-CoV-2 [131].
Moreover, SARS-CoV-2 mutant vaccines can be engineered to induce neutralizing antibody responses to adaptive vaccines via cross-reactive T helper cell function against conserved SARS-CoV-2 sites. CD4+ T cells respond primarily to the spike protein via the RBD, which is also the primary target of neutralizing antibodies [132], [133]. RBD is highly immunogenic in vivo and contains a large number of T-cell epitopes as well as a conserved immunodominant region. RBD- and S346–S365-specific T-cell clones were identified in distinct memory subsets of COVID-19 survivors, and these clones were also isolated from individuals inoculated with the SARS-CoV-2 mRNA vaccine [97]. The availability of a large number of cross-reactive T-cell clones identified by RBD facilitates the identification of targets in associated pathogens, which may aid in developing vaccines against SARS-CoV-2 variants.
6. Discussion
In this review, we discuss the role of certain HLA molecules in the presentation of the SARS-CoV-2 protein and summarize the research on the susceptibility of different HLA genotypes to SARS-CoV-2 and the postinfection symptoms of different populations in different regions and countries. Regarding the association of human complex HLA genes with SARS-CoV-2, such as the association of HLA-I genotypes with COVID-19 severity and mortality and the association of HLAs with SARS-CoV-2 mutations, more research is needed. Individual HLA haplotypes and whole-gene variants may also influence the capacity to mount an immune response against SARS-CoV-2. We also summarized some characteristics of SARS-CoV-2, including the virus spike protein. Recent studies have described the structure and function of the virus, as well as the crucial role of the spike protein, as an antigen in stimulating the T-cell response.
As of May 2022, there have been over 500 million confirmed cases of SARS-CoV-2 worldwide. SARS-CoV-2 has gradually become an unavoidable part of contemporary human life. Furthermore, confronting and resolving SARS-CoV-2 is a challenging issue for every country today. As SARS-CoV-2 spreads and the virus develops new mutations, the best solution for now may lie in the body's own immune response, one of which is T cells. Therefore, vaccines that can induce a durable and effective T-cell response are one of the most important solutions. The prediction of T-cell epitopes presented by SARS-CoV-2 antigens presented by HLA class molecules can serve as a benchmark. However, there are still some flaws in this strategy. Moreover, the effectiveness of the T-cell response to SARS-CoV-2 infection remains unknown. By studying the induced T-cell response, it is possible to design vaccines based on the ability of HLA class molecules to present antigens, which provides the opportunity to prevent SARS-CoV-2 and slow the spread of the epidemic.
Funding sources
This work was supported by National Natural Science Foundation of China, SGC's Rapid Response Funding for COVID-19 (No. C-0002), the National Natural Science Foundation of China (No. 81970008, 82000020), the Graduate Research and Innovation Foundation of Chongqing, China (No. CYS22080), the Fundamental Research Funds for the Central Universities (No. 2021CDJZYJH-002, 2019CDYGZD009 and 2020CDJYGRH-1005), Natural Science Foundation of Chongqing, China (No. cstc2020jcyj-msxmX0460) and Chongqing Talents: Exceptional Young Talents Project (No. cstc2021ycjh-bgzxm0099). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CRediT authorship contribution statement
Feng Lin, Xiaoyuan Lin, Beibei Fu, Yan Xiong, Mohamed Y. Zaky.
All the authors conceived the article together. Feng Lin wrote the manuscript; the figures were prepared by Feng Lin; Haibo Wu, Beibei Fu, Yan Xiong, Mohamed Y. Zaky reviewed the literature; Haibo Wu and Xiaoyuan Lin jointly supervised and revised the manuscript together. All the authors approved the final manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Data availability
Data will be made available on request.
References
- 1.Trowsdale J. Genomic structure and function in the MHC. Trends Genet. 1993;9(4):117–122. doi: 10.1016/0168-9525(93)90205-v. [DOI] [PubMed] [Google Scholar]
- 2.Germain R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002;2(5):309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 3.Klein L., Kyewski B., Allen P.M., Hogquist K.A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see) Nat. Rev. Immunol. 2014;14(6):377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 5.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debnath M., Banerjee M., Berk M. Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. 2020;34(7):8787–8795. doi: 10.1096/fj.202001115R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22(1):74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 8.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595(7868):572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 9.Le T.T., Cramer J.P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(10):667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 10.Wieczorek M., Abualrous E.T., Sticht J., Alvaro-Benito M., Stolzenberg S., Noe F., et al. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falk K., Rotzschke O., Stevanovic S., Jung G., Rammensee H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 12.Paul S., Kolla R.V., Sidney J., Weiskopf D., Fleri W., Kim Y., et al. Evaluating the immunogenicity of protein drugs by applying in vitro MHC binding data and the immune epitope database and analysis resource. Clin Dev Immunol. 2013;2013 doi: 10.1155/2013/467852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyngaa R., Pedersen N.W., Schrama D., Thrue C.A., Ibrani D., Met O., et al. T-cell responses to oncogenic merkel cell polyomavirus proteins distinguish patients with merkel cell carcinoma from healthy donors. Clin. Cancer Res. 2014;20(7):1768–1778. doi: 10.1158/1078-0432.CCR-13-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joffre O.P., Segura E., Savina A., Amigorena S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012;12(8):557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 15.Bommareddy P.K., Shettigar M., Kaufman H.L. Integrating oncolytic viruses in combination cancer immunotherapy. Nat. Rev. Immunol. 2018;18(8):498–513. doi: 10.1038/s41577-018-0014-6. [DOI] [PubMed] [Google Scholar]
- 16.Peters B., Nielsen M., Sette A. T cell epitope predictions. Annu. Rev. Immunol. 2020;38:123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson E.A., Hirneise G., Singharoy A., Anderson K.S. Total predicted MHC-I epitope load is inversely associated with population mortality from SARS-CoV-2. Cell Rep. Med. 2021;2(3) doi: 10.1016/j.xcrm.2021.100221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neefjes J., Jongsma M.L., Paul P., Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011;11(12):823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 19.Sercarz E.E., Maverakis E. Mhc-guided processing: binding of large antigen fragments. Nat. Rev. Immunol. 2003;3(8):621–629. doi: 10.1038/nri1149. [DOI] [PubMed] [Google Scholar]
- 20.Castellino F., Zappacosta F., Coligan J.E., Germain R.N. Large protein fragments as substrates for endocytic antigen capture by MHC class II molecules. J. Immunol. 1998;161(8):4048–4057. [PubMed] [Google Scholar]
- 21.Zika E., Ting J.P. Epigenetic control of MHC-II: interplay between CIITA and histone-modifying enzymes. Curr. Opin. Immunol. 2005;17(1):58–64. doi: 10.1016/j.coi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Bruchez A., Sha K., Johnson J., Chen L., Stefani C., McConnell H., et al. MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science. 2020;370(6513):241–247. doi: 10.1126/science.abb3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dendrou C.A., Petersen J., Rossjohn J., Fugger L. HLA variation and disease. Nat. Rev. Immunol. 2018;18(5):325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 24.Ng M.H., Lau K.M., Li L., Cheng S.H., Chan W.Y., Hui P.K., et al. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J. Infect. Dis. 2004;190(3):515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin M., Tseng H.K., Trejaut J.A., Lee H.L., Loo J.H., Chu C.C., et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y.M., Liang S.Y., Shih Y.P., Chen C.Y., Lee Y.M., Chang L., et al. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J. Clin. Microbiol. 2006;44(2):359–365. doi: 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S.F., Chen K.H., Chen M., Li W.Y., Chen Y.J., Tsao C.H., et al. Human-leukocyte antigen class I cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol. 2011;24(5):421–426. doi: 10.1089/vim.2011.0024. [DOI] [PubMed] [Google Scholar]
- 28.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fricke-Galindo I., Falfan-Valencia R. Genetics insight for COVID-19 susceptibility and severity: a review. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.622176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arya R., Kumari S., Pandey B., Mistry H., Bihani S.C., Das A., et al. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021;433(2) doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371(6529) doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131(5) doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–501 e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2(2) doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat. Med. 2021;27(1):78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 39.Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., et al. Unbiased screens show CD8(+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53(5):1095–107 e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C., et al. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem. Biophys. Res. Commun. 2006;344(1):63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang B., Chen H., Jiang X., Zhang M., Wan T., Li N., et al. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104(1):200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorente L., Martin M.M., Franco A., Barrios Y., Caceres J.J., Sole-Violan J., et al. HLA genetic polymorphisms and prognosis of patients with COVID-19. Med. Intens. (Engl. Ed.) 2021;45(2):96–103. doi: 10.1016/j.medin.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iturrieta-Zuazo I., Rita C.G., Garcia-Soidan A., de Malet Pintos-Fonseca A., Alonso-Alarcon N., Pariente-Rodriguez R., et al. Possible role of HLA class-I genotype in SARS-CoV-2 infection and progression: a pilot study in a cohort of Covid-19 Spanish patients. Clin. Immunol. 2020;219:108572. doi: 10.1016/j.clim.2020.108572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavasolian F., Rashidi M., Hatam G.R., Jeddi M., Hosseini A.Z., Mosawi S.H., et al. HLA, immune response, and susceptibility to COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.601886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weingarten-Gabbay S., Klaeger S., Sarkizova S., Pearlman L.R., Chen D.Y., Gallagher K.M.E., et al. Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell. 2021;184(15):3962–80 e17. doi: 10.1016/j.cell.2021.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94(13) doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee C.H., Koohy H. In silico identification of vaccine targets for 2019-nCoV. F1000Research. 2020;9:145. doi: 10.12688/f1000research.22507.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keicho N., Itoyama S., Kashiwase K., Phi N.C., Long H.T., Ha L.D., et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum. Immunol. 2009;70(7):527–531. doi: 10.1016/j.humimm.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Middleton D., Williams F., Meenagh A., Daar A.S., Gorodezky C., Hammond M., et al. Analysis of the distribution of HLA-A alleles in populations from five continents. Hum. Immunol. 2000;61(10):1048–1052. doi: 10.1016/s0198-8859(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 54.Saxena A., Sharma G., Tyagi S., Mourya M., Coshic P., Tiwari P.K., et al. HLA-A*02 repertoires in three defined population groups from North and Central India: Punjabi Khatries, Kashmiri Brahmins and Sahariya tribe. HLA. 2019;93(1):16–23. doi: 10.1111/tan.13447. [DOI] [PubMed] [Google Scholar]
- 55.Gonzalez-Galarza F.F., Takeshita L.Y., Santos E.J., Kempson F., Maia M.H., da Silva A.L., et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(Database issue):D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sibilio L., Martayan A., Setini A., Monaco E.L., Tremante E., Butler R.H., et al. A single bottleneck in HLA-C assembly. J. Biol. Chem. 2008;283(3):1267–1274. doi: 10.1074/jbc.M708068200. [DOI] [PubMed] [Google Scholar]
- 57.Hilton H.G., McMurtrey C.P., Han A.S., Djaoud Z., Guethlein L.A., Blokhuis J.H., et al. The intergenic recombinant HLA-B *46:01 has a distinctive peptidome that includes KIR2DL3 ligands. Cell Rep. 2017;19(7):1394–1405. doi: 10.1016/j.celrep.2017.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L.M., Kimura A., Satoh M., Mineshita S. HLA linked with leprosy in southern China: HLA-linked resistance alleles to leprosy. Int. J. Lepr. Other Mycobact. Dis. 1999;67(4):403–408. [PubMed] [Google Scholar]
- 59.Novelli A., Andreani M., Biancolella M., Liberatoscioli L., Passarelli C., Colona V.L., et al. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA. 2020;96(5):610–614. doi: 10.1111/tan.14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Correale P., Mutti L., Pentimalli F., Baglio G., Saladino R.E., Sileri P., et al. HLA-B*44 and C*01 prevalence correlates with Covid19 spreading across Italy. Int. J. Mol. Sci. 2020;21(15) doi: 10.3390/ijms21155205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F., Huang S., Gao R., Zhou Y., Lai C., Li Z., et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020;6(1):83. doi: 10.1038/s41421-020-00231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amoroso A., Magistroni P., Vespasiano F., Bella A., Bellino S., Puoti F., et al. HLA and AB0 polymorphisms may influence SARS-CoV-2 infection and COVID-19 severity. Transplantation. 2021;105(1):193–200. doi: 10.1097/TP.0000000000003507. [DOI] [PubMed] [Google Scholar]
- 63.Sakuraba A., Haider H., Sato T. Population difference in allele frequency of HLA-C*05 and its correlation with COVID-19 mortality. Viruses. 2020;12(11) doi: 10.3390/v12111333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pisanti S., Deelen J., Gallina A.M., Caputo M., Citro M., Abate M., et al. Correlation of the two most frequent HLA haplotypes in the italian population to the differential regional incidence of Covid-19. J. Transl. Med. 2020;18(1):352. doi: 10.1186/s12967-020-02515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Littera R., Campagna M., Deidda S., Angioni G., Cipri S., Melis M., et al. Human leukocyte antigen complex and other immunogenetic and clinical factors influence susceptibility or protection to SARS-CoV-2 infection and severity of the disease course. The Sardinian experience. Front. Immunol. 2020;11:605688. doi: 10.3389/fimmu.2020.605688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020;94(5) doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Sousa E., Ligeiro D., Lerias J.R., Zhang C., Agrati C., Osman M., et al. Mortality in COVID-19 disease patients: correlating the association of major histocompatibility complex (MHC) with severe acute respiratory syndrome 2 (SARS-CoV-2) variants. Int. J. Infect. Dis. 2020;98:454–459. doi: 10.1016/j.ijid.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pretti M.A.M., Galvani R.G., Vieira G.F., Bonomo A., Bonamino M.H., Boroni M. Class I HLA allele predicted restricted antigenic coverages for spike and nucleocapsid proteins are associated with deaths related to COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.565730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22(2):72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Younger R.M., Amadou C., Bethel G., Ehlers A., Lindahl K.F., Forbes S., et al. Characterization of clustered MHC-linked olfactory receptor genes in human and mouse. Genome Res. 2001;11(4):519–530. doi: 10.1101/gr.160301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ehlers A., Beck S., Forbes S.A., Trowsdale J., Volz A., Younger R., et al. MHC-linked olfactory receptor loci exhibit polymorphism and contribute to extended HLA/OR-haplotypes. Genome Res. 2000;10(12):1968–1978. doi: 10.1101/gr.10.12.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park J.H., Jang W., Kim S.W., Lee J., Lim Y.S., Cho C.G., et al. The clinical manifestations and chest computed tomography findings of coronavirus disease 2019 (COVID-19) patients in China: a proportion meta-analysis. Clin. Exp. Otorhinolaryngol. 2020;13(2):95–105. doi: 10.21053/ceo.2020.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A., et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter european study. Eur. Arch. Otorhinolaryngol. 2020;277(8):2251–2261. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan C.H., Faraji F., Prajapati D.P., Boone C.E., DeConde A.S. Association of chemosensory dysfunction and COVID-19 in patients presenting with influenza-like symptoms. Int. Forum Allergy Rhinol. 2020;10(7):806–813. doi: 10.1002/alr.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah V.K., Firmal P., Alam A., Ganguly D., Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., et al. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 78.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508) doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 81.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590(7847):630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nersisyan S., Zhiyanov A., Shkurnikov M., Tonevitsky A. T-CoV: a comprehensive portal of HLA-peptide interactions affected by SARS-CoV-2 mutations. Nucleic Acids Res. 2022;50(D1):D883–D887. doi: 10.1093/nar/gkab701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci. Immunol. 2020;5(49) doi: 10.1126/sciimmunol.abd6160. [DOI] [PubMed] [Google Scholar]
- 84.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–27 e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shkurnikov M., Nersisyan S., Jankevic T., Galatenko A., Gordeev I., Vechorko V., et al. Association of HLA class I genotypes with severity of coronavirus Disease-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.641900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gutierrez L., Beckford J., Alachkar H. Deciphering the TCR repertoire to solve the COVID-19 mystery. Trends Pharmacol. Sci. 2020;41(8):518–530. doi: 10.1016/j.tips.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol. 2012;12(2):136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosendahl Huber S., van Beek J., de Jonge J., Luytjes W., van Baarle D. T cell responses to viral infections - opportunities for peptide vaccination. Front. Immunol. 2014;5:171. doi: 10.3389/fimmu.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khan N., Best D., Bruton R., Nayak L., Rickinson A.B., Moss P.A. T cell recognition patterns of immunodominant cytomegalovirus antigens in primary and persistent infection. J. Immunol. 2007;178(7):4455–4465. doi: 10.4049/jimmunol.178.7.4455. [DOI] [PubMed] [Google Scholar]
- 90.Wherry E.J., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78(11):5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kratzer B., Trapin D., Ettel P., Kormoczi U., Rottal A., Tuppy F., et al. Immunological imprint of COVID-19 on human peripheral blood leukocyte populations. Allergy. 2021;76(3):751–765. doi: 10.1111/all.14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shomuradova A.S., Vagida M.S., Sheetikov S.A., Zornikova K.V., Kiryukhin D., Titov A., et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53(6):1245–57 e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Z., Yang X., Zhou Y., Sun J., Liu X., Zhang J., et al. COVID-19 severity correlates with weaker T-cell immunity, hypercytokinemia, and lung epithelium injury. Am. J. Respir. Crit. Care Med. 2020;202(4):606–610. doi: 10.1164/rccm.202005-1701LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bange E.M., Han N.A., Wileyto P., Kim J.Y., Gouma S., Robinson J., et al. CD8(+) T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27(7):1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mallajosyula V., Ganjavi C., Chakraborty S., McSween A.M., Pavlovitch-Bedzyk A.J., Wilhelmy J., et al. CD8(+) T cells specific for conserved coronavirus epitopes correlate with milder disease in COVID-19 patients. Sci. Immunol. 2021;6(61) doi: 10.1126/sciimmunol.abg5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meckiff B.J., Ramirez-Suastegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell. 2020;183(5):1340–53 e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Low J.S., Vaqueirinho D., Mele F., Foglierini M., Jerak J., Perotti M., et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372(6548):1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Riou C., du Bruyn E., Stek C., Daroowala R., Goliath R.T., Abrahams F., et al. Relationship of SARS-CoV-2-specific CD4 response to COVID-19 severity and impact of HIV-1 and tuberculosis coinfection. J. Clin. Invest. 2021;131(12) doi: 10.1172/JCI149125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.von Massow G., Oh S., Lam A., Gustafsson K. Gamma Delta T cells and their involvement in COVID-19 virus infections. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.741218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee E., Sandgren K., Duette G., Stylianou V.V., Khanna R., Eden J.S., et al. Identification of SARS-CoV-2 nucleocapsid and spike T-cell epitopes for assessing T-cell immunity. J. Virol. 2021;95(6) doi: 10.1128/JVI.02002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yong S.E.F., Anderson D.E., Wei W.E., Pang J., Chia W.N., Tan C.W., et al. Connecting clusters of COVID-19: an epidemiological and serological investigation. Lancet Infect. Dis. 2020;20(7):809–815. doi: 10.1016/S1473-3099(20)30273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zohar T., Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 2020;20(7):392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Snyder T.M., Gittelman R.M., Klinger M., May D.H., Osborne E.J., Taniguchi R., et al. medRxiv; 2020. Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. [Google Scholar]
- 105.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012 e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dutta M., Dutta P., Medhi S., Borkakoty B., Biswas D. Polymorphism of HLA class I and class II alleles in influenza A(H1N1)pdm09 virus infected population of Assam, Northeast India. J. Med. Virol. 2018;90(5):854–860. doi: 10.1002/jmv.25018. [DOI] [PubMed] [Google Scholar]
- 107.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–83 e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hoffmann M., Hofmann-Winkler H., Kruger N., Kempf A., Nehlmeier I., Graichen L., et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36(3) doi: 10.1016/j.celrep.2021.109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., et al. bioRxiv; 2021. mRNA Vaccine-elicited Antibodies to SARS-CoV-2 and Circulating Variants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., et al. SARS-CoV-2 501Y.V2 escapes neutralization by south african COVID-19 donor plasma. Nat. Med. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 112.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88(19):11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 114.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Stralin K., Gorin J.B., Olsson A., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–68 e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34(6) doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34(17):2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Agerer B., Koblischke M., Gudipati V., Montano-Gutierrez L.F., Smyth M., Popa A., et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8(+) T cell responses. Sci Immunol. 2021;6(57) doi: 10.1126/sciimmunol.abg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nathan A., Rossin E.J., Kaseke C., Park R.J., Khatri A., Koundakjian D., et al. Structure-guided T cell vaccine design for SARS-CoV-2 variants and sarbecoviruses. Cell. 2021;184(17):4401–13 e10. doi: 10.1016/j.cell.2021.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sarkizova S., Klaeger S., Le P.M., Li L.W., Oliveira G., Keshishian H., et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 2020;38(2):199–209. doi: 10.1038/s41587-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hansen T.H., Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nat Rev Immunol. 2009;9(7):503–513. doi: 10.1038/nri2575. [DOI] [PubMed] [Google Scholar]
- 122.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Croft N.P., Smith S.A., Wong Y.C., Tan C.T., Dudek N.L., Flesch I.E., et al. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog. 2013;9(1) doi: 10.1371/journal.ppat.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu T., Guan J., Handel A., Tscharke D.C., Sidney J., Sette A., et al. Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses. Nat. Commun. 2019;10(1):2846. doi: 10.1038/s41467-019-10661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.A., et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27(10):959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 126.Park M.D. Immune evasion via SARS-CoV-2 ORF8 protein? Nat Rev Immunol. 2020;20(7):408. doi: 10.1038/s41577-020-0360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., et al. bioRxiv; 2020. Origin and Cross-species Transmission of Bat Coronaviruses in China. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 128.Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sohail M.S., Ahmed S.F., Quadeer A.A., McKay M.R. In silico T cell epitope identification for SARS-CoV-2: Progress and perspectives. Adv. Drug Deliv. Rev. 2021;171:29–47. doi: 10.1016/j.addr.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang F., Gan R., Zhen Z., Hu X., Li X., Zhou F., et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal. Transduct. Target Ther. 2020;5(1):156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nagler A., Kalaora S., Barbolin C., Gangaev A., Ketelaars S.L.C., Alon M., et al. Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA peptidomics. Cell Rep. 2021;35(13) doi: 10.1016/j.celrep.2021.109305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183(4):1024–42 e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57 e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.