SUMMARY
Epstein–Barr virus (EBV) is nearly ubiquitous in adults. EBV causes infectious mononucleosis, and is associated with B cell lymphomas, epithelial cell malignancies and multiple sclerosis. The EBV gH/gL glycoprotein complex facilitates fusion of virus membrane with host cells, and is a target of neutralizing antibodies. Here we examined the sites of vulnerability for viral neutralization and fusion-inhibition within EBV gH/gL. We developed a panel of human monoclonal antibodies (mAbs) that targeted five distinct antigenic sites on EBV gH/gL and prevented infection of epithelial and B cells. Structural analyses using X-ray crystallography and electron microscopy revealed multiple sites of vulnerability and defined the antigenic landscape of EBV gH/gL. One mAb provided near-complete protection against viremia and lymphoma in a humanized mouse EBV-challenge model. Our findings provide structural and antigenic knowledge on the viral fusion machinery, yield a potential therapeutic antibody to prevent EBV disease and emphasize gH/gL as a target for herpesvirus vaccines and therapeutics.
Keywords: Herpes virus, Epstein-Barr virus, antibody therapeutics, sites of vulnerability, glycoprotein H, fusion machinery, gH/gL, vaccine
Graphical Abstract
In Brief
EBV is associated with several malignancies. Chen and colleagues identify six human monoclonal antibodies (mAb) that target five distinct sites on EBV gH/gL, neutralize virus infection, and inhibit virus-cell fusion. mAb 769B10 reduces viremia and prevents lymphoma in mice challenged with EBV and may be useful as a therapeutic.
INTRODUCTION
Epstein-Barr virus (EBV), a human γ-herpesvirus, is the major cause of infectious mononucleosis, and is associated with B cell lymphomas such as Burkitt and Hodgkin lymphoma and epithelial cell carcinomas including nasopharyngeal and gastric carcinoma (Cohen 2010). EBV also causes lymphomas in immunocompromised organ and bone marrow transplant recipients, and in patients with certain primary immunodeficiencies. EBV is a trigger for multiple sclerosis (Bjornevik et al. 2022; Lanz et al. 2022). There is no licensed prophylactic or therapeutic vaccine to protect against the virus. Intravenous immunoglobulin (IVIG) or antiviral therapy is used to reduce infection in EBV seronegative immunocompromised patients, and for the related human cytomegalovirus (Seemayer et al., 1995; Snydman et al., 1984), but breakthrough infections and disease still occur. Immunocompromised patients may not elicit sufficient protective responses by vaccination, and the use of passive immunoprophylaxis with monoclonal antibodies (mAbs), to prevent EBV infection and/or disease may be required.
EBV infection requires fusion of the viral membrane with the host cell plasma or endosomal membrane. The core fusion machinery for EBV includes a set of glycoproteins, gH/gL or gH/gL/gp42, that activate gB the viral fusogen (Kirschner et al., 2007; McShane and Longnecker, 2004; Wu and Hutt-Fletcher, 2007). In addition to cell fusion, EBV gH/gL binds to ephrin receptor A2 (EphA2) and integrins on epithelial cells (Chen et al., 2018; Chesnokova et al., 2009; Zhang et al., 2018), while gp42 binds to MHC class II molecules on B cells (Spriggs et al., 1996).
The EBV gH/gL heterodimer molecule is made up of four tightly packed domains on gH, with large interdomain contacts that form an extended structure (Matsuura et al., 2010). The gL molecule is a globular domain that intertwines between the N-terminus of gH between the first two domains (D-I and D-II) (Matsuura et al., 2010). EBV gp42 associates with gH/gL to form a heterotrimer, with the N-terminal region of gp42 having an elongated linear interaction with gH, while the larger gp42 globular domain locates on the gH surface located between D-II and D-III (Sathiyamoorthy et al., 2016).
Antibodies against herpesvirus gH/gL molecules potently inhibit virus infection. Structural and functional studies of antibodies binding to gH/gL have facilitated understanding of the mechanism of herpesvirus entry machinery and pathways. More recently, interest has focused on gH/gL as a potential vaccine or therapeutic target. EBV gH/gL mAbs E1D1, CL40, and CL59 block epithelial cell infection, but not B cell infection (Molesworth et al., 2000), while mAb AMMO1 blocks infection of both cell types and protects against EBV infection in both murine and nonhuman primate studies (Singh et al., 2020). The structures of mAbs E1D1, CL40, CL59, and AMMO1, in complex with gH/gL or gH/gL/gp42 provide insights into their neutralizing activities (Sathiyamoorthy et al., 2016; Sathiyamoorthy et al., 2017; Snijder et al., 2018) while gH/gL antibodies in human plasma inhibit EBV infection of both epithelial and B cells (Bu et al., 2019).
To understand the molecular determinants of EBV neutralization, and to further characterize the antigenic sites of vulnerability on EBV gH/gL, we isolated a set of human mAbs that target EBV gH/gL. Six gH/gL human mAbs (769A7, 769B10, 769C2, 769C5, 770F7, 770F8) neutralized virus infection in both B cells and epithelial cells. X-ray crystallography and negative stain electron microscopy defined the neutralizing epitopes on gH/gL. We found that multiple epitopes on gH/gL are sites of vulnerability for virus neutralization and inhibition of fusion. These findings enabled us to define similarities to recognition by other herpesvirus gH/gL neutralizing antibodies. The most promising antibody 769B10 showed protective efficacy against viremia and EBV lymphoma in a humanized mouse EBV-challenge model. These data support EBV gH/gL as a highly immunogenic antigen whose neutralizing antibody epitopes share similar structural features with other human herpesviruses and identify a target for a highly effective antibody than can reduce viremia and prevent lymphoma in an animal model
RESULTS
Isolation and characterization of gH/gL monoclonal Abs from healthy donors
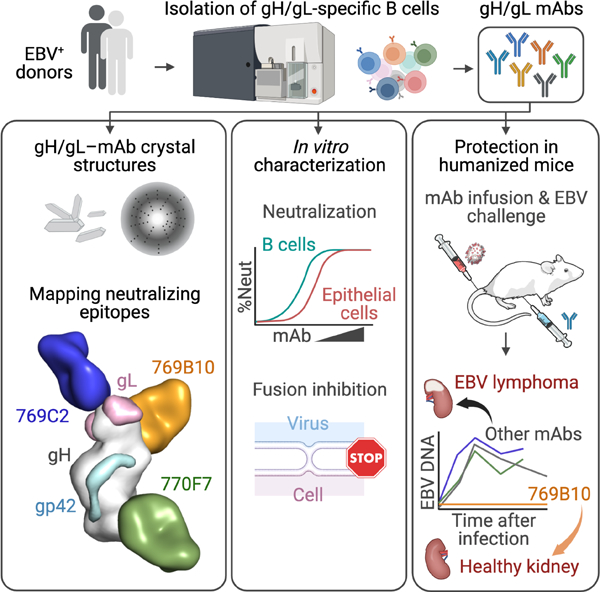
We used fluorescent labeled gH/gL probes to isolate EBV gH/gL-specific B cells from healthy donors with high B cell and epithelial cell EBV neutralization titers. We sequenced the immunoglobulin genes from these sorted B cells and produced six mAbs that bound specifically to gH/gL and gH/gL/gp42 (Figures 1A, S1 and S2). We determined if the gH/gL mAbs were able to neutralize EBV infection in B cells and epithelial cells using virus expressing GFP (Bu et al., 2019). All six mAbs were highly effective in neutralizing EBV infection in both B cells and epithelial cells (Figures 1B and S1B). Comparison of these 6 gH/gL mAbs showed that mAb 769B10, which was previously reported (Bu et al., 2019), was the most potent for neutralizing B cell infection followed by mAbs 769C2 and 769A7 (Figure 1B left panel, Figure S1B left panel, Figure S1C), while mAb 769B10 was most effective to neutralize epithelial cell infection followed by mAbs E1D1, 770F7 and 769C5 (Figure 1B right panel, Figure S1B right panel, Figure S1C).
Figure 1. Isolation and characterization of EBV gH/gL-specific mAbs.
(A) EBV gH/gL-specific B cells were isolated from two convalescent donor samples. Immunoglobulin gene assignments, and antibody characteristics are shown for isolated antibodies, and previously described EBV gH/gL-targeting antibodies for reference.
(B) Antibody neutralization of EBV infection of Raji B cells and SVKCR2 epithelial cells.
(C) Inhibition of B cell and epithelial cell fusion by isolated mAbs.
(D) Octet Biolayer interferometry antibody-antigen affinity measurements using EBV gH/gL (left) and gH/gL/gp42 (right) antigens. The association, dissociation rates, and KD values are shown for antibodies. 769B10 values were previously described (Bu et al., 2019).
(E) Antibody competition profile using previously described antibodies (mAbs E1D1, CL40, 769B10) as competitors. Antibodies noted in the left column were first bound to gH/gL, followed by binding to a second antibody noted in the top row, with percent competition indicated.
Since gH/gL together with gB comprise the EBV core-fusion machinery, we measured fusion-inhibitory activities of the six gH/gL EBV-neutralizing mAbs using a luciferase-based cell-cell fusion assay (Bu et al., 2019). mAbs 769C2, 769B10, and 769C5 were the most effective in blocking fusion with B cells (Figure 1C left panel, Figure S1D left panel, Figure S1E), while mAbs 769B10, 769C5, and 770F7 were the most potent for inhibiting fusion in epithelial cells (Figure 1C right panel, Figure S1D right panel, Figure S1E). Thus, within this set of antibodies, gH/gL-targeting mAb 769B10 is in the top tier and potently blocks EBV glycoprotein-mediated fusion in both B cells and epithelial cells.
We assessed the isolated gH/gL-targeting antibodies by biolayer interferometry to determine the affinity and to provide initial characterization of the antibody epitopes (Figure 1D and 1E). Binding affinity of each Fab to either gH/gL and gH/gL/gp42 was largely the same, except for 769B10 which has a significantly lower affinity to gH/gL/gp42 compared to gH/gL. Among the Fabs tested, 769B10 and 770F7 had the highest overall affinity to gH/gL and gH/gL/gp42 with KD less than 1 nM (Figure 1D). We next performed an antibody competition assay to determine the epitopes targeted by the mAbs by measuring antibody binding to gH/gL-antibody complexes (Figure 1E). Using the newly identified antibodies, combined with previously described antibodies including CL40 (Sathiyamoorthy et al., 2017), 769B10 (Bu et al., 2019), and E1D1 (Sathiyamoorthy et al., 2016), we defined six distinct antibody groups with 769B10 competing with CL40 as previously described (Bu et al., 2019), and mAb 769C2 competing with mAb 770F8. The other mAbs 769A, 769C5, and 770F7 did not show competition with any of the tested antibodies, indicating that these mAbs bind to novel epitopes. Interestingly, the known gL-binding mAb E1D1 also did not compete with binding to any of the tested antibodies.
Crystal structure of mAb 769B10 in complex with EBV gH/gL/gp42peptide
To understand the detailed recognition of neutralizing antibodies to EBV gH/gL/gp42 glycoproteins, we determined two crystal structures of EBV gH/gL/gp42peptide (gp42p) ternary antibody complexes containing mAbs 769B10 and 769C2 at a resolution of 3.0 Å (Figure 2 and 3, respectively, and S3, and Table S1), and mAbs 769C2 and 770F7 at a resolution of 3.3 Å (Figure 3 and 4, and S3, and Table S1).
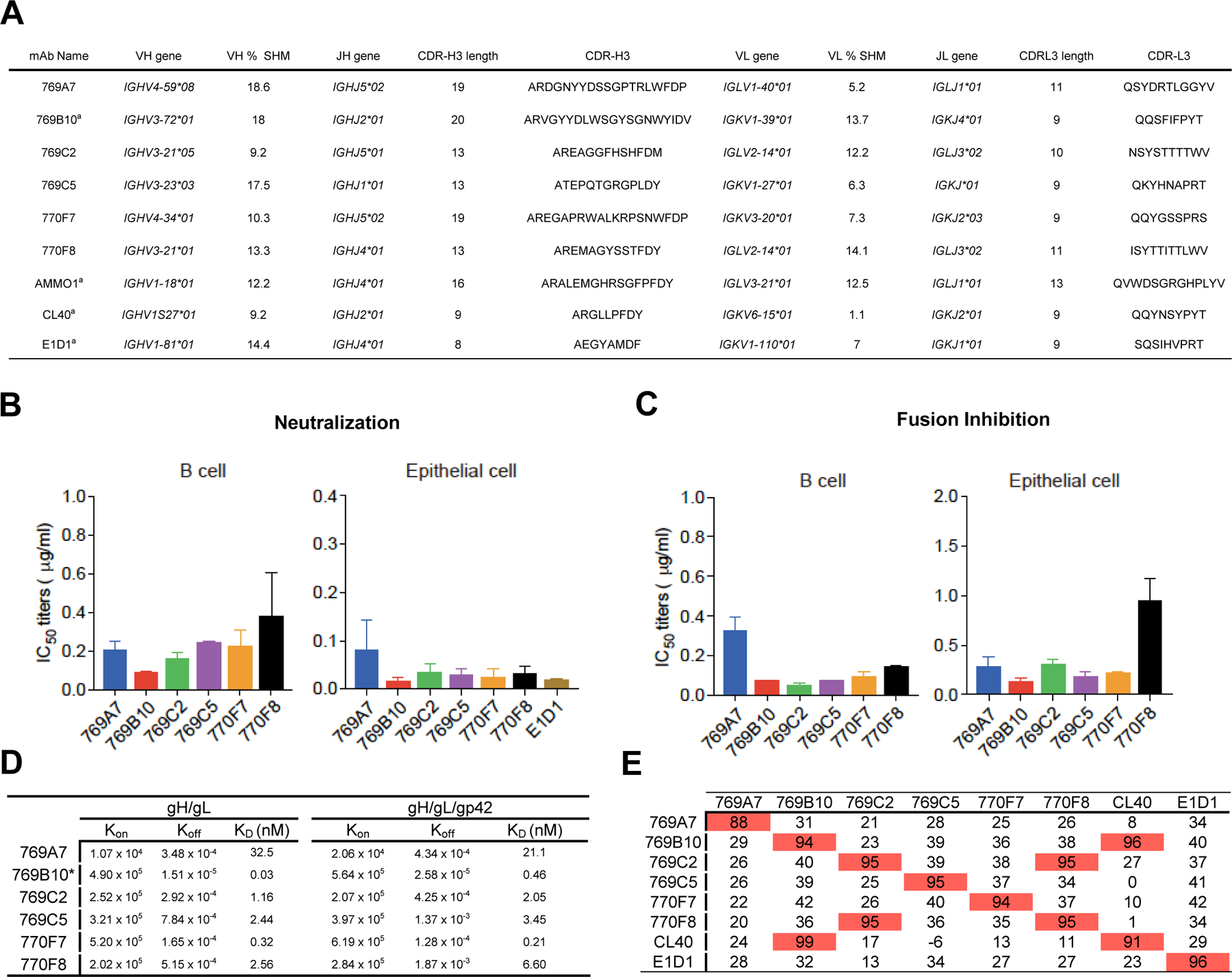
Figure 2. mAb 769B10 binds to a gH/gL antigenic supersite on EBV gH/gL.
(A) Crystal structure of 769B10 Fab in complex with EBV gH/gL/gp42peptide. EBV glycoproteins gH/gL/gp42p are shown in surface representation; gH, gL, and gp42p (colored in light grey, light pink, and pale cyan respectively) bound to mAb 769B10 (cartoon representation, heavy chain and light chain are colored with bright orange and light orange respectively). The integrin tripeptide motif ‘KGD’ is indicated by a red circle.
(B) Major antibody loops involved in gH/gL recognition are shown in ribbon representation with gH/gL in surface representation. Antibody residue numbering and CDR loops are designated using the Kabat numbering system.
(C) CDR-gH/gL epitope BSA percentage is shown in pie-chart representation. The 769B10 epitope is made up of 73% gH and 27% gL (inner circle), with both heavy and light chain recognition of gH, and predominantly heavy chain recognition of gL (middle), with multiple CDR loops used to recognize the gH/gL (outer).
(D) mAb 769B10 contact residues are shown in stick representations based on (1) CDRs H2, and H3; (2) CDR H2; (3) FR H3 antibody contacting regions; (4) CDR L1 and L3.
(E) Negative-stain EM maps of 769B10 (top), or 769A7 (bottom) in complex with EBV gH/gL. The EBV gH/gL/gp42p structure is fitted into the electron density for reference.
(F) Definition of the antigenic supersite. Structure-based alignment of CL40, AMMO1, and 769B10 (ribbon representation) in complex with EBV gH/gL (surface representation) are shown.
(G) Comparison of the 769B10 epitope to CL40 and AMMO1 epitopes. The 769B10 (orange), AMMO1 (green), and CL40 (blue) epitopes are outlined on the gH/gL molecule individually, and overlaid (right panel). The equivalent % BSA between the 769B10 and either AMMO1, or CL40 epitopes are indicated at the base of the two figures, and shown based on total, gH-only, or gL-only overlap.
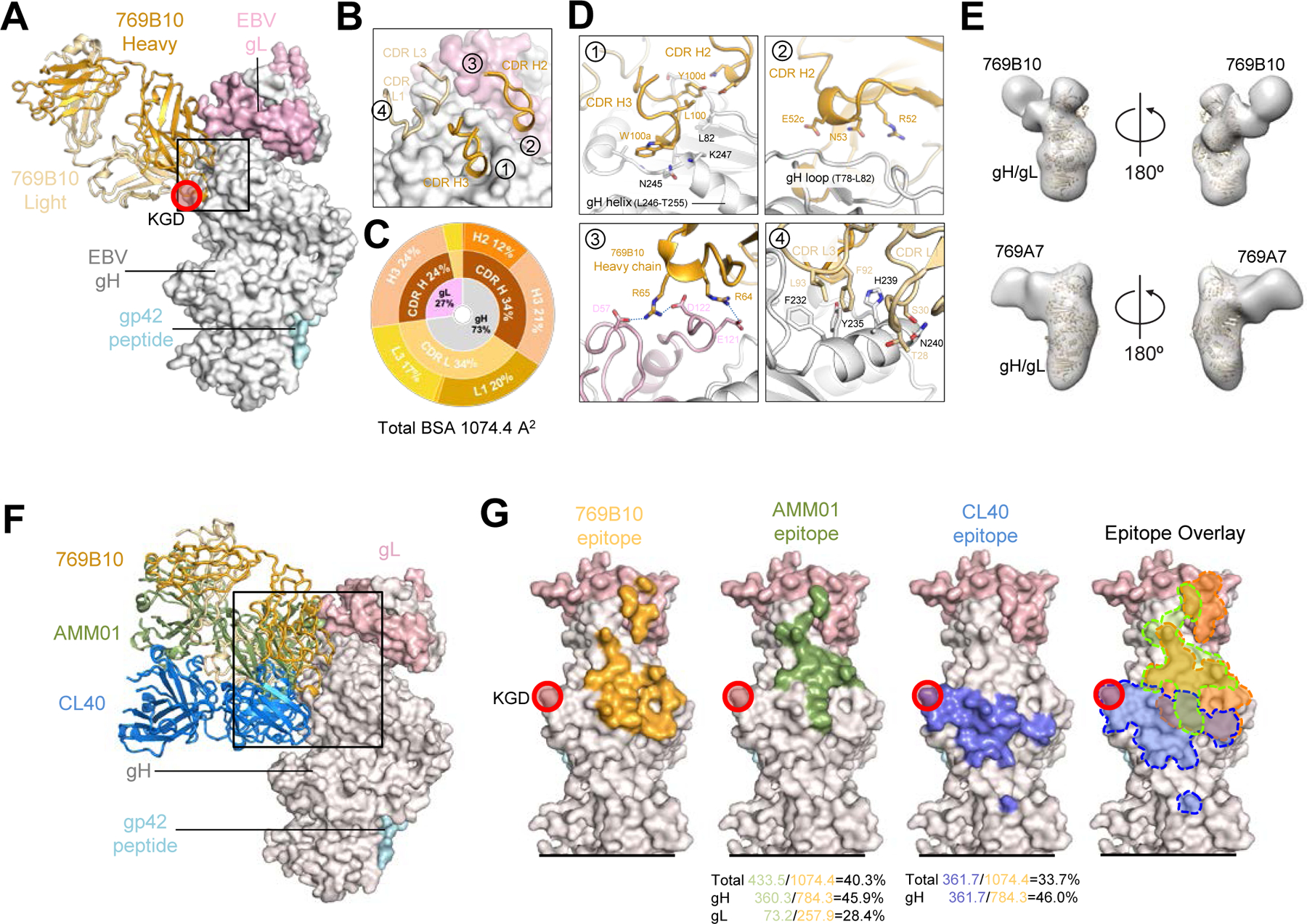
Figure 3. mAb 769C2 defines a site of vulnerability with antibody contacts on both gH and gL molecules.
(A) Structure of 769C2 Fab (ribbon representation, heavy chain and light chain are colored with sky blue and light blue respectively) in complex with EBV gH/gL/gp42peptide (surface representation; gH, gL, and gp42p are colored in light grey, light pink, and pale cyan respectively) are shown.
(B) Major antibody loops involved in gH/gL recognition are shown in ribbon representation with gH/gL in surface representation.
(C) CDR-glycoprotein epitope BSA % are shown represented in pie-chart format. The 769C2 epitope is made up of 72% gH and 28% gL (inner circle), with both heavy and light chain recognition of gH and gL (middle), with multiple CDR loops used to recognize the gH/gL molecule (outer).
(D) mAb 769C2 contact residues are shown in stick representations based on (1, 2) CDR H1, H2 and H3; (3) CDR L1 and H3; (4) CDR L1 and L3.
(E) Negative-stain EM maps of 769C2 (top) and competing mAb 769F8 (bottom) in complex with EBV gH/gL. The EBV gH/gL/gp42p structure is fitted into the electron density for reference.
(F) Structural comparison of gH/gL/gp42p (surface representation) in complex with 769C2 or EphA2 (left) or E1D1 (right) respectively.
(G) Comparison of the 769C2 epitope (blue, upper left) to mAb E1D1 epitope (purple, lower left) or the EphA2 (bright green, top right) binding site with all shown overlaid (lower right). The % BSA that is identical between the EphA2 binding site (top) and the antibody epitopes is indicated at the base of the figure.
Figure 4. mAb 770F7 defines a novel epitope on EBV gH D-IV.
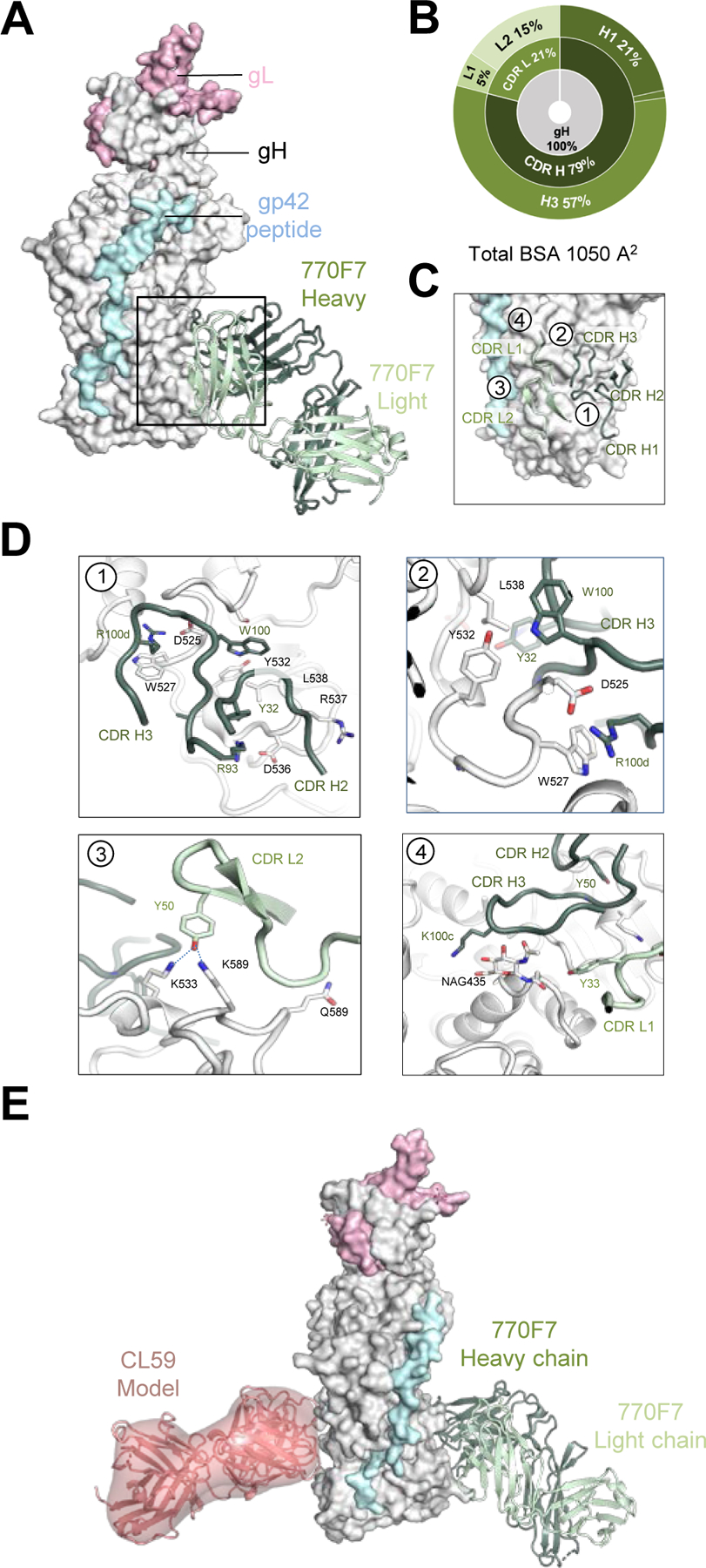
(A) Structure of 770F7 Fab in complex with EBV gH/gL/gp42peptide (surface representation; gH, gL, and gp42p are colored in light grey, light pink, and pale cyan respectively) are shown bound to mAb 770F7 (ribbon representation, heavy chain and light chain are colored with forest green and pale green respectively).
(B) CDR-glycoprotein epitope BSA % are shown represented in pie-chart format. The 770F7 epitope is on the gH molecule (inner circle), with both heavy and light chain recognition of gH (middle), with predominant recognition by the CDR H2 and H3 loops (outer).
(C) Major antibody loops involved in gH/gL recognition are shown in ribbon representation with gH/gL in surface representation.
(D) mAb 770F7 contact residues are shown in stick representations based on (1) CDR H2, and H3; (2) CDR H3; (3) CDR L2; (4) CDR H3 and CDR L1.
(E) Structure model of 770F7 and CL59 in complex with gH/gL/gp42p (surface representation); 770F7 Fab is shown in ribbon representation colored as in A, and CL59 Fab is shown as both cartoon and surface representation in salmon color.
mAb 769B10 binds to EBV gH/gL/gp42p protein with a total buried surface area (BSA) of 1074 Å2 engaging both antibody heavy (691 Å2) and light chain (383 Å2) to recognize both gH (846 Å2) and gL (228 Å2) (Figure 2A and Table S2 and S3). All antibody residue numbering and CDR loops are designated using the Kabat numbering system. mAb 769B10 recognition of gH is through CDR H2, H3, L1 and L3, while recognition of gL is through CDRs H3 and L3 (Figure 2B and 2C). The CDR H3 residues 100a-100d bind to gH residues 243–247 region through main chain hydrogen bonds (Figure 2D, insert 1). The residues L106 and Y110 also form hydrophobic interaction with gH residues G79-L82. 769B10 CDR H2 residues R52 and N53 engage additional gH main chain S79 and G80 contacts (Figure 2D, insert 2). The Framework H3 also binds to gL through two somatically mutated residues, Arg64 and Arg65, that form salt bridges with gL-D57, D122 and E121 and stabilize the flexible gL C-terminus (Figure 2D, insert 3). The light chain CDRs interact with the gH D-II α-helix 4 (residues 232–240). The CDR L3 utilizes aromatic and hydrophobic amino acid residues (Phe92, Leu93, Phe94, and Tyr96) to engage gH Tyr76 and a recessed α-helical region. This α-helix also forms extensive aromatic residue interactions with the CDR L1 loop (Asn27-Phe32) by interacting with the outermost surface of the gH D-II α-helix 4 (Figure 2D, insert 4).
Negative-stain EM mapping of mAbs 769B10 and 769A7 indicates these antibodies approach gH/gL from similar angles and bind adjacent to each other on the gH/gL molecule (Figure 2E, and S4). In agreement with the competition studies (Figure 1E), these antibodies have distinct and non-overlapping epitopes, but the mapping of the mAb 769A7 epitope is in the gH DI-DII region and gL.
Previously described neutralizing mAbs CL40 (which binds solely to gH) and AMMO1 have overlapping epitopes with mAb 769B10 (Figure 2F). Both mAbs AMMO1 and 769B10 interact with gH and gL, and share ~50% of contact residues. Both mAbs 769B10 and AMMO1 bind proximal to the ‘KGD’ integrin binding motif (Figure 2G), and while not directly interacting with the tripeptide motif, there would be significant steric hindrance to prevent gH/gL-integrin binding.
Crystal structure of mAb 769C2 in complex with EBV gH/gL/gp42p
We determined the structure of mAb 769C2 bound to gH/gL/gp42 in two heterologous crystal forms (Figure S3 and Table S1). The overall structures of the 769C2-gH/gL/gp42p complex in both crystal forms are very similar. Structural comparison of the antibody Fv domains resulted in a root mean square deviation (RMSD) of 0.58 Å for equivalent 184 C-α atoms, with minor differences in flexible loop regions (Figure S3B). The figures are based on the structure of mAb 769C2–770F7-gH/gL/gp42p complex determined at 3.0 Å (Figure 3A, and S3).
mAb 769C2 binds to both gH D-I (674 Å2) and gL (267 Å2) with a total BSA of 941 Å2 using both antibody heavy (558 Å2) and light chains (383 Å2) (Figure 3A, and Table S2 and S4). mAb 769C2 uses CDR H1-H3, CDR L1 and L3 to bind to gH and CDRH1, H2 and CDR L1 and L3 to interact with gL (Figure 3B and 3C). The major antibody contact region is with the typically unstructured H27-S34 region of D-I, where the antibody CDR H2 and H3 bind to both sides of the hairpin-like motif, with the CDR H1 binding to the tip of the hairpin (Figure 3D, insert 1 and 2). A collection of hydrogen-bonding interactions between 769C2 CDRs and gH and gL residues including CDR L1 D33 with gL K68 provide critical contacts (Figure 3D, insert 3 and 4).
Negative-stain EM mapping of mAb 769F8 with gH/gL showed that this antibody recognizes gH/gL in a similar manner as mAb 769C2, both with a comparable angle of approach (Figure 3E, and S4) and with overlapping epitopes which is in agreement with the antibody competition assay (Figure 1E). Sequence comparison between these two mAbs showed high sequence similarity, while having genetic precursor differences indicating that they are not clonally related.
EphA2 was previously described as an EBV gH/gL receptor involved in epithelial cell entry (Chen et al., 2018). Recently, the structure of EphA2 ligand-binding domain (LBD) in complex with gH/gL was determined (Su et al., 2020). Comparison of the structures of gH D-I/gL targeting mAb 769C2 and gL-targeting E1D1 to the EphA2-bound structure (Figure 3F) showed that both antibody epitopes overlap with the EphA2-binding site with > 500 Å2 BSA shared. mAbs 769C2 and E1D1 have distinct epitopes adjacent to each other but are not overlapping (Figure 3G), in agreement with the antibody-competition assays (Figure 1). This would explain how both mAbs block EBV infection of epithelial cells, while the ability of mAb 769C2 to block B cell infection may be due to the antibody angle of approach, steric hindrance, or another mechanism to prevent viral fusion.
Crystal structure of mAb 770F7 in complex with EBV gH/gL/gp42p
mAb 770F7 binds solely to EBV gH using both antibody heavy (855 Å2) and light chain (195 Å2) recognition, with total BSA of 1,050 Å2 (Figure 4A and 4B, Table S5). The site of recognition is a novel epitope located in the gH D-III and D-IV regions distal from most previously described gH/gL antibody epitopes. mAb 770F7 recognition of gH protein is largely through the CDR H1 and H3, with minor contacts from CDR L1 and L2 (Figure 4B-4D). The 770F7 CDR H3, residues 97–115 in the unbound Fab structure adopts a different structural conformation to that seen in the gH/gL co-crystal structure, due to crystal contacts, which indicates that the CDR H3 has some flexibility in recognition (Figure S3C-S3D). mAb 770F7 CDR H2 engages the main chain atoms of gH residues Arg537 and Lys533, while CDR H3 Trp100 and Arg100d form extensive gH contacts (Figure 4D, insert 1 and 2). Trp100 protrudes outwards and interacts with Tyr532 and Ser519 side chains while also inserting into an opening between two gH D-III loops at positions 536–540 and 517–522. Arg100d forms a salt bridge with gH Asp525 while also interacting with Trp527 through a cation-π interaction. Additional hydrogen bonds are formed by CDR L2 Tyr50 and the gH Lys533 and Lys589 residues (Figure 4D, insert 2 and 3). CDR H3 and L1 also form interactions with the N-glycan attached to Asn435, and Tyr50 forming a hydrogen bond network with D-III Lys533 and D-IV Lys586 (Figure 4D, insert 4). The previously reported low resolution negative-stain map of the EBV neutralizing antibody CL59-gH/gL complex shows that mAb CL59 binds to the gH D-III/D-IV region (Figure 4E). However, mAbs CL59 and 769F7 bind on opposite faces of gH, with no epitope overlap. Antibody targeting of D-IV may prevent efficient viral-cell fusion as residues in D-IV have been identified as critical to enable efficient EBV cell infection.
EBV gH/gL sites of vulnerability
By combining the previously described gH/gL-targeting antibody structures and those determined in this study, we assessed the overall antigenic sites of vulnerability on the EBV gH/gL molecule. The herpesvirus gH/gL molecules is typically divided into four domains with the N-terminal domain identified as Domain I, and the C-terminal domain designated as Domain IV, with Domain II and III located between these domains (Chowdary et al., 2010; Matsuura et al., 2010)). All four domains of gH and gL are targeted to neutralize EBV infection of B cells or epithelial cells. The identification of antibodies from naturally infected humans, which target each of the gH D-I-IV, and gL and utilize multiple immunoglobulin germline gene precursors without unusually long CDR3s (Figure S2) indicates that elicitation of these types of human antibodies is not constrained by immunogenetic recombination compositions, and may be readily elicited by immunogenic gH/gL- or gH/gL/gp42-based vaccines.
To date, there are seven distinct antigenic sites on EBV gH/gL, recognized by multiple antibodies. These include two sites in D-I (Figure 5A), the 769C2–770F8 site (Epitope 1) which is associated with both B cell and epithelial cell neutralization, and the E1D1 site (Epitope 2) which is associated with only epithelial cell neutralization. Within the DI-II region (Figure 5B), there are two sites, the site recognized by mAbs AMMO1, 769B10, and CL40 that have considerable overlap in their epitopes (Epitope 3). Distinct from these gH/gL junctional epitopes is the site (Epitope 4) bound by mAb 769A7 which also can provide fusion-blocking and viral neutralization in both cell types. In the DII-III region (Figure 5C), mAb 769C5 and the recently described mAb 1D8 bind to a similar location (Epitope 5) with partially overlapping epitopes, and both mAbs can also protect both cell types against EBV infection. In the DIII-IV region (Figure 5D), mAbs CL59 and 770F7 recognize distinct epitopes (Epitope 6 and 7, respectively) located on opposite sides of the gH/gL molecule.
Figure 5. Antigenic sites of vulnerability on the EBV gH/gL and gH/gL/gp42 fusion machinery.
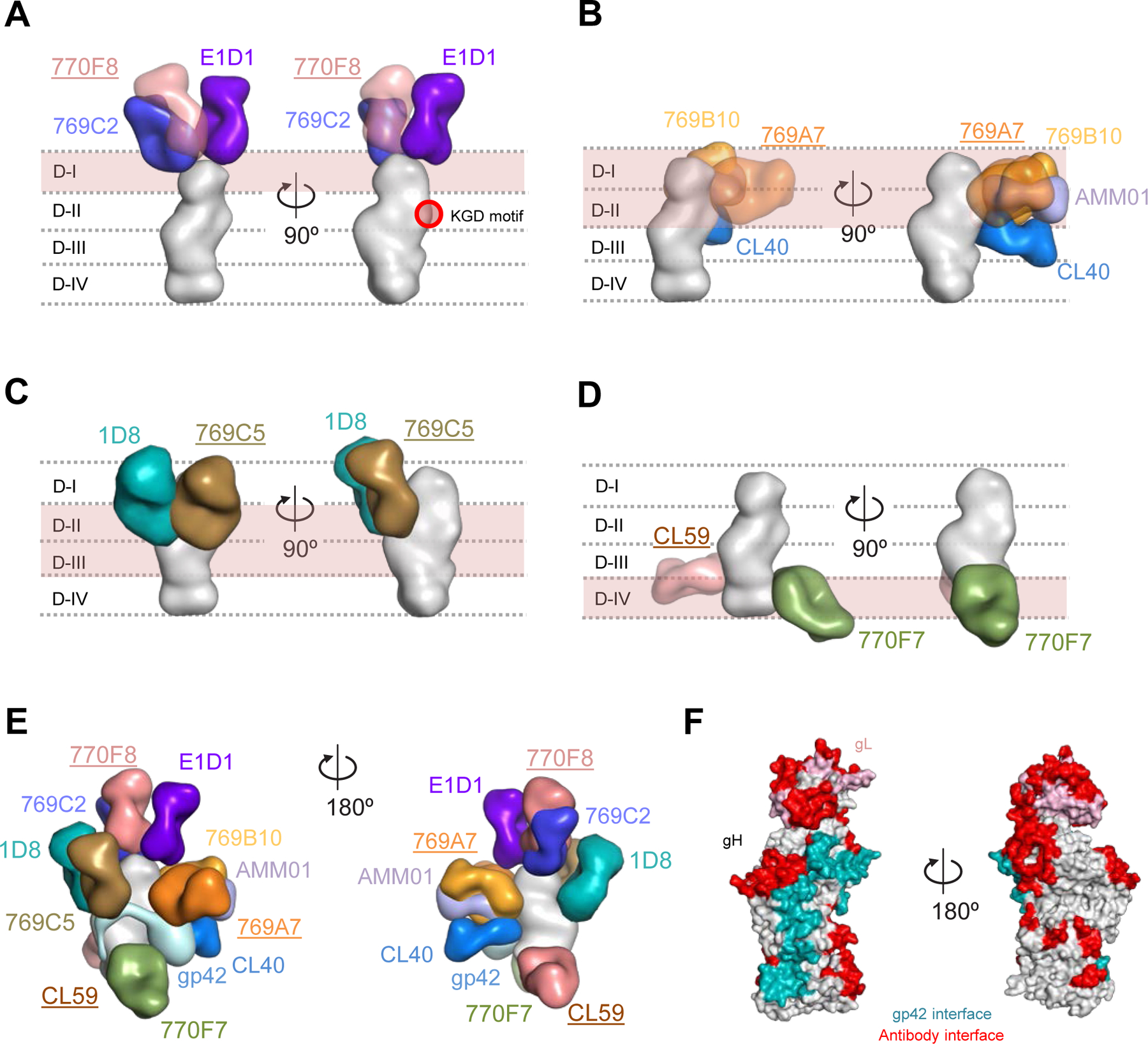
Structure-based composite model of EBV gH/gL and gH/gL/gp42 with neutralizing antibodies. EBV gH domains are indicated by dotted lines for (A-D). Antibody names that are underlined indicate that the structural information is provided by negative-stain EM maps and antibody docking. Antibodies and gH/gL molecules are shown in surface representation.
(A) mAbs 769C2 (blue), 770F8 (rose), and E1D1 (purple) targeting the gH/gL D-I region.
(B) mAbs 769B10 (orange), 769A7 (gold), AMMO1 (periwinkle blue), and CL40 (blue) targeting gH/gL D-I and D-II.
(C) mAbs 1D8 (teal), and 769C5 (brown) targeting the gH/gL D-II and D-III region.
(D) mAbs CL59 (salmon), and 770F7 (green), targeting the gH/gL D-IV region.
(E) EBV glycoproteins gH/gL/gp42 (surface representation) are shown bound to eleven mAbs. The antibody-antigen model is shown in two orientations to allow visibility of the antibodies.
(F) Antibody and gp42 interfaces are shown on the gH/gL/gp42p molecules (surface representation).
See also Figures S4-S5.
Based on the mapped antibody epitopes, more than 40% of the surface of gH/gL is targeted (Figure 5E, 5F and Figure S5). There are two regions of the gH/gL molecule where no known blocking or neutralizing antibodies have been identified to date. These include the region adjacent to the globular domain of gp42 and above the 770F7 epitope, and a second large surface area located between the CL59 epitope and the 769C2 epitope on the opposite side (Figure 5F, right).
Comparison of herpesvirus gH/gL antigenic sites of vulnerability
Structural analysis of other herpesvirus-neutralizing antibodies that target gH/gL molecules showed that two varicella-zoster virus (VZV) neutralizing mAbs VZV RC and VZV 94 with overlapping epitopes (Xing et al., 2015) target a remarkably similar epitope as the 769B10 or CL40 epitope on their respective gH/gL molecules (Figure 6A). Despite the differences between the structures of the different herpesvirus gH/gL molecules whose DI-DII domains are rotated compared to each other, the VZV neutralizing mAb epitopes are clearly related to that seen for EBV. The human cytomegalovirus (HCMV) mAb 13H11 (Gerna et al., 2016; Kabanova et al., 2016; Kschonsak et al., 2021) (Figure 6A and B) binds to an epitope also located on D-II and D-III and overlaps with the equivalent CL40 epitope while extending towards the base of the gH molecule, with the majority of the epitope located on D-III. The related nature of these epitopes indicates that targeting this region on multiple herpesvirus gH/gL fusion machinery may represent a common site of vulnerability, that is broadly applicable to other herpesviruses. An additional antibody which targets HCMV is mAb 3G16 (Gerna et al., 2016; Kschonsak et al., 2021). Structural overlay of the HCMV gH/gL-antibody negative-stain EM maps showed that mAb 3G16 binds in a location equivalent to the EBV CL59 epitope, while the mAb 13H11 epitope is located above mAb 770F7 in a location equivalent to the CL40 EBV gH/gL epitope (Figure 6B). In addition, a recently described HCMV-neutralizing mAb 1–32 targets the HCMV gH/gL region at a position analogous to mAb 769C2 with both antibodies having a similar angle of approach to the corresponding herpesvirus gH/gL and interact with multiple chains of the fusion machinery (Wrapp et al., 2022) (Figure 6C). The EBV mAbs that bind to gH/gL are EBV-specific; VZV RC, CMV 3G16, 13 H11, 1–32 did not bind to EBV gH/gL (unpublished data). However, since the target epitopes are conserved in location, but not in sequence across the herpesvirus family, these results are informative for development of herpesvirus vaccines and therapeutic antibodies.
Figure 6. Comparison of herpesvirus antigenic sites of vulnerability on the gH/gL fusion machinery.
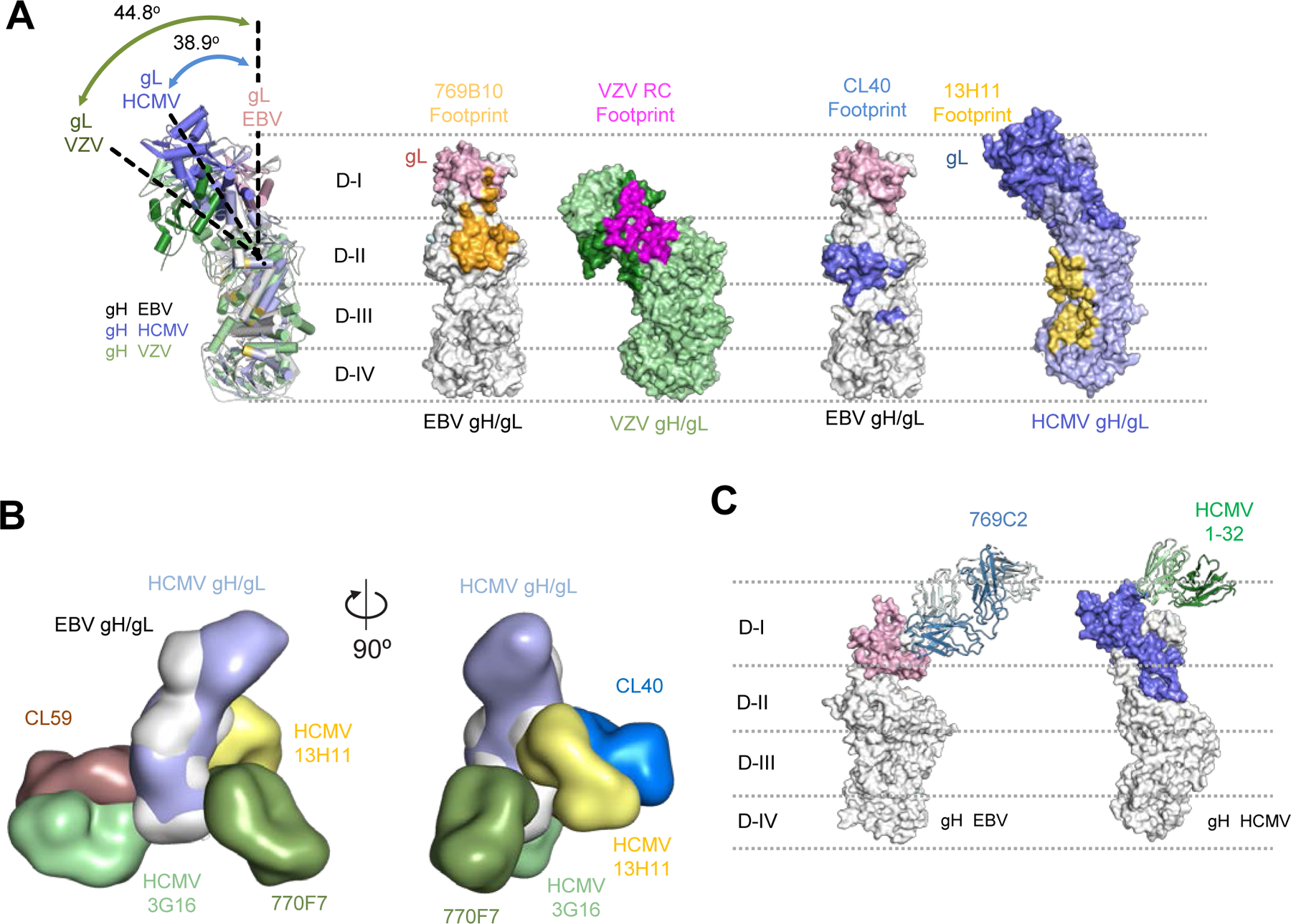
(A) Comparisons of the 796B10 epitope to VZV RC (purple), EBV CL40 (blue), and HCMV 13H11 (yellow) antibody epitopes are mapped on the respective viral gH/gL molecules, shown in surface representation. The EBV gH/gL structural overlap (left) with VZV and HCMV gH/gL molecules indicates the different orientation or tilt of the gH D-I/gL domains in each molecule relative to each other. The approximate location of the gH DI-DIV domains are indicated by horizontal dotted lines.
(B) Overlay of HCMV and EBV gH/gL molecules with HCMV mAbs 3G16, 13H11 and EBV mAbs 770F7, CL40, and CL59 shown in surface representation with two orientations.
(C) Comparison of mAb 796C2-gH/gL recognition to HCMV gH/gL-targeting mAb 1–32 (green) shown in ribbon representation, with herpesvirus gH/gL molecules shown in surface representation. The approximate location of the gH DI-DIV domains are indicated by horizontal dotted lines.
See also Figure S5.
Protection studies of gH/gL antibodies in a humanized mouse model
In order to understand how targeting of different epitopes could protect against a stringent EBV challenge, we assessed mAbs 769B10, 769C2, and 770F7 for their ability to mitigate weight loss, increase survival, reduce the amount of virus in blood and tissues, and prevent lymphomas in humanized mice. The humanized mice used in the study were irradiated NOG mice engrafted with human CD34+ hematopoietic stem cells that develop human lymphocytes. Groups of 5 humanized mice were inoculated with different mAbs and challenged with 1 × 106 Green Raji Units of EBV B95–8/F (Figure 7A). Four out of 5 mice receiving PBS, mAbs 769C2 or 770F7 lost ≥10% of their weight after EBV challenge, while only 2 out of 5 mice receiving mAb 769B10 lost ≥10% weight (Figure S6A). Mice in the PBS control group or those receiving gH/gL mAb 770F7 all died or were euthanized by 11 weeks or 10 weeks after EBV challenge, respectively. Mice that received gH/gL mAb 769B10 had the highest survival rate (60%) after challenge with EBV, followed by animals that received mAb 769C2 (20% survival) at the end of the study (Figure 7B). A repeat experiment using a different set of humanized mice showed that mAbs 769B10 and 770F7 had the highest survival rates (Figure S7A)
Figure 7. Passive transfer of gH/gL mAbs in humanized mice and challenge with EBV.
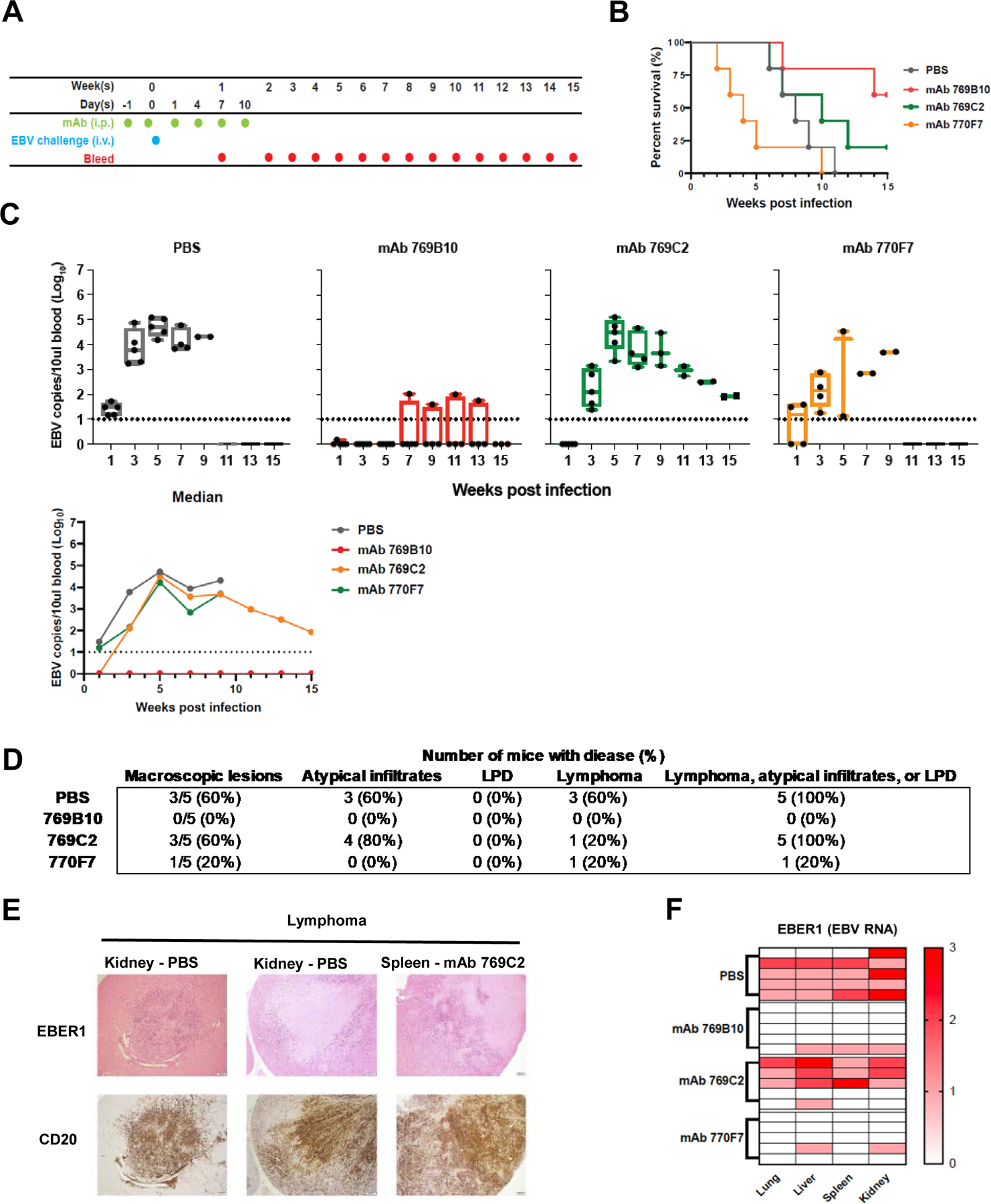
(A) Mice were given six doses of gH/gL mAbs 769C2, 770F7, 769B10, or PBS intraperitoneally and challenged intravenously with EBV after the second dose of mAb.
(B) Survival rate for each group of mice after challenge.
(C) EBV DNA copies in the blood was quantified by real-time qPCR. Each dot represents one mouse and the dotted line indicates the detection limit of the assay. Whiskers indicates minimum and maximum data points and box represents upper and lower quartiles; the horizontal line in the box is the median. Median EBV DNA copies in the blood were calculated.
(D) Number and percent of animals in each treatment group with macroscopic lesions on the surface of liver, kidney, spleen, or lung, and with microscopic lesions in these organs showing lymphoma, atypical B cell infiltrates, or lymphoproliferative lesions that do not meet the criteria for lymphoma.
(E) Representative images of lymphoma in the kidneys, spleen, and lungs of the mice. Treatment groups are indicated. In situ hybridization was performed for EBV EBER1 with eosin counterstain (upper panel) and CD20 staining for human B cells using 3,3’ diaminobenzidine as a chromogen (lower panel).
(F) Heatmap of EBER1 expression in lung, liver, spleen, and kidney in mouse. A score of 0 indicates no EBER1 expression, 1 represents scattered EBER+ cells, 2 indicates a moderate number of EBER+ cells, 3 represents intense infiltration of the tissue with EBER+ cells.
See also Figures S6-S7.
All mice in the PBS control group and those that received gH/gL mAbs 769C2 and 770F7 had EBV DNA detected in the blood after challenge with EBV with peak levels at 5 weeks after infection (Figure 7C). An area under the curve analysis showed that the viral loads were lower in the animals that received mAb than those receiving PBS (p = 0.006 for 769C2 vs. PBS and p=0.018 for 770F7 vs. PBS). In contrast, only 20% of mice that received gH/gL mAb 769B10 had EBV detectable in blood samples following challenge. Mice that received gH/gL mAb 769B10 also had low numbers of EBV DNA copies that were significantly reduced compared to the PBS, mAb 769C2, or mAb 770F7 groups (p <0.001 for 769B10 vs. each of the other three groups). A repeat experiment using another set of humanized mice showed that mice receiving mAb 769B10 had no detectable viremia, while those receiving PBS, mAb 769C2, or 770F7 had viremia (Figure S7B). Thus, gH/gL mAb 769B10 was the most effective mAb at reducing EBV DNA copy numbers in the blood of humanized mice after EBV challenge.
Levels of gH/gL mAbs in the plasma of humanized mice were measured by ELISA. Mice that received gH/gL mAb 769C2 or 769B10 had higher peak antibody levels than those receiving mAb 770F7, with a more gradual decrease of antibody level with mAb detected for at least 9 weeks after infection (Figure S6B). Because the tested mAbs were produced as recombinant human IgG1 antibodies, the half-life of gH/gL mAbs 769C2, 770F7, and 769B10 was also examined in human FcRn transgenic mice after injecting a single dose of each mAb, determining the concentration of the mAb in the serum by ELISA to gH/gL (Figure S6C), and calculating the half-life using a non-compartmental model. mAbs 769C2 and 769B10 had similar half-lives of 19.7 and 17.3 days, respectively, and similar clearance rates of 5.2 mL/day/kg and 6.6 mL/day/kg in the mice (Figure S6D). mAb 770F7 had a shorter half-life of 10.7 days and a faster clearance rate of 10.5 mL/day/kg than mAbs 769C2 and 769B10. An identical IgG1 constant region backbone plasmid was used to produce the antibodies, indicating that the difference in half-lives is dues to the antibody Fv region.
Liver, lungs, spleen, and kidneys were collected from each animal and examined; 60% of mice in the PBS control and gH/gL mAb 769C2 groups had macroscopic lesions visible on the surface of at least one organ (Figure 7D, Figure S6E). In contrast, 1 out of 5 mice that received gH/gL mAb 770F7 and none of the animals that received mAb 769B10 had observable lesions on the organs. Mice treated with 769B10 had no lymphomas in the four organs examined, while ≥2 out of 5 mice receiving mAb769C2 or 770F7 had lymphomas (Figure 7D, and 7E). Atypical lymphoid infiltrates were detected in animals receiving mAb 769C2 or PBS.
In situ hybridization was performed using an EBV-encoded RNA (EBER) probe to determine which organs were infiltrated with EBV-infected cells. Tissues were scored based on the number of EBER-positive cells (0: no EBER+ cells; 1+ to 3+ based on the intensity of tissue infiltration with EBER+ cells) by a pathologist blinded to the study groups (Figure S6F). 80% of mice in the PBS group and 60% of mice in the mAb 769C2 group had scores of 2-to-3 in at least one organ, while only 20% of mice in the mAb 770F7 and 769B10 groups had a score of 1 in at least one organ and the other animals in these latter two groups had scores of 0 in all the other organs (Figure 7F). One caveat is that the majority of mice in the mAb 770F7 group were euthanized prior to week 5 post infection when viremia is normally at its peak. Therefore, the cause of early death in this group of mice was unlikely to be due to EBV infection; thus, EBER scoring of this group may not represent true efficacy of gH/gL mAb 770F7. In summary, mice that received gH/gL mAb 769B10 or 770F7 had the lowest levels of EBV in tissues, and animals that received gH/gL mAb 769B10 had the lowest levels of EBV in the blood and no lymphomas after EBV challenge.
DISCUSSION
Differences in virus cell tropism, multiple receptors for virus entry, and varying virus entry and fusion mechanisms have made vaccine and antibody therapeutic development challenging for herpesviruses. Herpesvirus fusion with host cell membranes involves multiple proteins including gH and gL often in complex with additional glycoproteins depending on the virus (e.g. gp42 for EBV, and gO or UL128, UL130 and UL131 for HCMV) to enable activation of the gB fusogen molecule. Given that the human immune response targets multiple components of the fusion machinery during infection (Bu et al., 2019; Macagno et al., 2010), therapeutic antibody and vaccine development has focused on these components and results have been promising. For example, the demonstration that mAbs to the HCMV pentameric complex (gH/gL/UL128/UL130/UL131) can potently neutralize endothelial, epithelial, and myeloid cell infection (Macagno et al., 2010; Wang et al., 2011) has led to development of several candidate HCMV vaccines focused on these glycoproteins as immunogens (Kabanova et al., 2014; Li et al., 2021).
EBV-infected humans have significant neutralizing antibody responses targeted to gH/gL that can block EBV infection of both B cells and epithelial cells (Bu et al., 2019). This is also seen in studies where animals vaccinated with gH/gL immunogens produce neutralizing antibodies that prevent both B cell and epithelial cell infection (Bu et al., 2019; Cui et al., 2016). To better understand how antibodies targeting gH/gL neutralize infection and block fusion, and to define antigenic sites of EBV vulnerability, we identified and structurally characterized gH/gL antibodies that potently neutralize virus infection including five distinct non-overlapping epitopes that are important for antibody mediated neutralization of EBV infection of B cells and epithelial cells.
Epitope-binning studies indicated that several of the antibodies we isolated bound to neutralizing epitopes on gH/gL. The epitope recognized by mAb 770F7 is located in gH D-IV, on the face opposite to that recognized by mAb CL59 (Sathiyamoorthy et al., 2017). mAb 770C5 binds to gH D-II at an epitope that overlaps with that recognized by mAb 1D8 which has been reported to block EBV infection in both B cells and epithelial cells and can protect humanized mice from EBV infection (Zhu et al., 2021). mAbs 769C2 and 769F8 prevent both epithelial cell and B cell infection with epitopes located in gH D-I. The epitopes of these antibodies overlap with the gH/gL-EphA2 receptor binding site. The 769C2 epitope is adjacent to, but does not overlap with the previously described E1D1 epitope (Sathiyamoorthy et al., 2016). The E1D1 mAb binds to gH/gL with a different angle of approach compared to mAbs 769C2 and 769F8. mAb E1D1 exerts EphA2-blocking activity and prevents epithelial cell infection but not B cell infection.
Another set of antibodies target the gH D-II region, including mAb 769B10 which identifies an antigenic site recognized by mAbs CL40 and AMMO1 (Snijder et al., 2018). Structural studies revealed that the epitopes of these antibodies substantially overlap and define an antigenic supersite, which is located proximal to the ‘KGD’ integrin-binding motif. In addition to these high-resolution structures, mAb 769A7 appears to bind closely to the supersite, but does not compete with mAb 769B10 for binding to gH/gL, indicating that an extended area across the gH D-II is a neutralization-sensitive site.
Despite significant sequence divergence in the gH/gL molecules across herpesviruses and different cell tropisms, it was striking that antibodies that neutralized highly divergent herpesviruses including VZV and HCMV showed similar areas of recognition on their respective gH/gL molecules. This included epitopes in all four domains of gH and gL, with the neutralizing antibodies for these divergent herpesviruses also having similar angles of approach, as observed for EBV-neutralizing antibodies. It is likely that additional neutralizing epitopes on the gH/gL molecules of other herpesviruses are yet to be identified and could have therapeutic potential. The conserved location on the gH/gL molecule for these sites of vulnerability indicates conserved viral fusion mechanics and indicates a potential common therapeutic strategy for targeting herpesviruses.
mAb 769B10, like AMMO1 (Singh et al., 2020), protected humanized mice from challenge with EBV. These animals are engrafted with human lymphocytes, but still have murine epithelial cells; the former, but not the latter support EBV infection. Despite the differences in the experimental design between our study and that of Singh et al., (2020), following passive transfer of mAb 769B10 in our study, EBV was undetectable in mouse blood at all timepoints with the exception of one mouse, which had very low levels of EBV in the blood. In contrast, higher levels of EBV were detected in the blood of mice following AMMO1 passive transfer (Singh et al., 2020). We also showed that mAb 769B10 prevented lymphomas and death of mice challenged with EBV, unlike the AMMO1 study.
Limitations of our humanized mouse study include the absence of epithelial cell infection in mice and therefore the need to challenge the animals intravenously. Since humans are typically infected via the oropharynx, the humanized mouse model cannot fully recapitulate the natural route of infection. While mAb 769B10 prevented EBV lymphoproliferative disease, atypical infiltrates, and lymphoma in all the mice after EBV challenge, one of 10 mice had viremia, although the level of EBV DNA in the blood was low relative to mice that received the other mAbs.
Our studies demonstrate that there are multiple sites of vulnerability extending across a broad surface area of the gH/gL fusion machinery. The mAbs described here could be used alone or in combination for prophylactic or therapeutic treatment of transplant recipients or patients with certain immunodeficiencies who are highly susceptible to developing EBV-related disease. In addition, the observation that multiple genetically diverse and potent neutralizing human antibodies can readily be identified indicates that gH/gL is a viable vaccine target. With the numerous epitopes recognized by gH/gL neutralizing mAbs, a polyclonal immune response could provide a multi-faceted approach to neutralize EBV, by blocking EphA2 receptor binding, integrin binding, and by inhibiting viral fusion. Thus, EBV gH/gL- and gH/gL/gp42-based vaccines either alone (Bu et al., 2019) or in combination with gB- or gp350-based vaccines (Kanekiyo et al., 2015; Wei et al., 2022) have the potential to protect against infection and disease reducing the rates of infectious mononucleosis, or preventing EBV-associated B cell, epithelial cell malignancies, or multiple sclerosis. Furthermore, the observation that EBV gH/gL epitopes show similarities with both β- and γ-herpesviruses is intriguing and may suggest common principles for developing herpesvirus vaccines or therapeutics targeting the complex, yet vulnerable viral fusion machinery.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey I. Cohen (jcohen@niaid.nih.gov).
Materials Availability
All reagents will be made available on request after completion of a Materials Transfer Agreement.
Data Availability
X-ray coordinates and structure factors have been deposited in the RCSB Protein Data Bank under accession codes PDBs: 7S07, 7S1B, 7S08, 7S0J. Antibody heavy and light chain sequences have been deposited in GenBank under accession numbers OP453123-OP453132. Additional data supporting the findings of this study are found within the paper and its Supplementary Information and are available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Specimens
PBMCs from EBV-seropositive blood donors were obtained from de-identified healthy blood bank donors age 18 or older. All donors provided written informed consent on a National Cancer Center Institutional Review Board approved protocol at the National Institutes of Health.
Mice
Female HSCCB-NOG-F (NOD.Cg-Prkdcscid Il2rgtm1Sug/ JicTac) mice that were 19 to 20 weeks post engraftment with human CD34+ hematopoietic stem cells were purchased from Taconic and female and male C57BL/6, B6.mFcRn−/− hFcRn Tg32 mice were purchased from Jackson Laboratory. Animals were housed in sterile microisolator cages (5 animals per cage) and were specific pathogen-free for multiple viruses, bacteria, and parasites. All animal experiments were performed in accordance with federal regulations and NIH guidelines and approved by the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases.
METHOD DETAILS
Flow Cytometry, Single-Cell Sorting, Ig Gene Amplification, and Antibody Production
Cryopreserved peripheral blood mononuclear cells from healthy blood bank donors with high serum EBV neutralization titers were stained with anti-human mAbs against surface proteins to identify IgG+ memory B cells and an EBV gH/gL probe was used to stain specific B cells which were sorted into 96-well plates as described previously (Bu et al., 2019). Isolation of Ig light and heavy chains antibody genes, cloning into IgG1 and kappa expression vectors, and production of antibodies were performed as reported previously (Bu et al., 2019). EBV mAbs E1D1 and CL40 were purified from the media of hybridoma cells by protein G chromatography.
Recombinant Protein Production
EBV Glycoprotein Production
Mammalian 293F cells (Life Technologies) were co-transfected with plasmids to express soluble gH with 6x His-tag and gL in the presence of glycosidase inhibitor, kifunensine to allow correct folding of gH/gL proteins, but leave them sensitive to Endoglycosidase H (Kong et al., 2010). His-tagged gH/gL proteins were purified with by Ni-NTA agarose (Thermo Fisher Scientific) followed by size exclusion chromatography using a Superdex 200 10/300 GL column. The gH/gL/gp42p complexes were made by incubation of deglycosylated gH/gL proteins with gp42 peptide (residues 47–81) synthesized by Genscript, and further purified by size exclusion chromatography.
EBV gH-AviHis/gL proteins were purified from the supernatant of Expi293F cells co-transfected with plasmids expressing gH-AviHis and gL by Ni-NTA agarose (Thermo Fisher Scientific) followed by size exclusion chromatography using a Superdex 200 10/300 GL column. Biotinylation of gH-AviHis/gL proteins was described previously (Bu et al. 2009)
Fab Production
Endoproteinase LysC (20 μg; New England BioLabs) in 0.5 mL PBS was added to 9.5 mL freshly prepared PBS containing 20 mg of each of the purified 769B10, 769C2, or 770F7 mAbs (IgGs) for a final ratio of 1:1000. The reaction was allowed to proceed for 4 – 6 hr in a shaker incubator at 37°C temperature and 100 rpm shaking speed. Digestion was analyzed by SDS-PAGE and upon completion, the reaction mixture was passed through protein-A beads (0.5–1 ml beads) 3 times and the final flow through was assessed by SDS-PAGE for purity.
Protein Deglycosylation
EBV gH/gL in the supernatant of cell culture was purified as described above, buffer exchanged to 10 mM HEPES, 150 mM NaCl pH 7.4, and deglycosylated with Endo HF (New England Biolabs, Hitchin, England) at room temperature for 3 h. The deglycosylated gH/gL was passed over a Concanavalin A-affinity chromatography column (1 mL beads) to ensure complete deglycosylation, followed by size-exclusion chromatography using a Superdex 200 10/300GL gel filtration column.
Cell lines, mAbs, and virus
SVKCR2 (human male) epithelial cells stably express gp350 receptor CR2 (Li et al., 1992) and were cultured in DMEM and F-12 (1:1) medium supplemented with 400 mg/ mL G418 and 10 ng/ mL cholera toxin (Li et al., 1992). 293T14 cells that express T7 polymerase were cultured in DMEM medium supplemented with 100 mg/mL zeocin. Raji cells (a human male EBV-positive Burkitt lymphoma cell line) were propagated in RPMI 1640 medium. Akata BX1 (human female) cells (Molesworth et al., 2000) that contain the Akata EBV genome and express GFP were cultured in RPMI 1640 medium supplemented 500 mg/ mL G418. Daudi (human male) B cells that express T7 polymerase were cultured in RPMI 1640 medium supplemented with 1 mg/ mL G418. Expi293F (human female) cells were cultured in Expi293 expression medium (Thermo Fisher Scientific). All cell cultures were supplemented with 10% fetal bovine serum (FBS), 100 U/ mL penicillin, and 100 mg/ mL streptomycin. mAbs 769B10 (Bu et al., 2019), AMM01 (Snijder et al., 2018), CL40 (Moleworth et al., 2000, and E1D1 (Sathiyamoorthy et al., 2016) were reported previously.
293/2089 cells, which contain the EBV B95–8/F genome which expresses GFP, were cultured in DMEM with 100 µg/ml hygromycin (Delecluse et al., 1998). EBV was produced from 293/2089 cells as previously reported (Bu et al., 2019)
EBV neutralization and fusion inhibition assays
Neutralization and fusion inhibition assays of EBV B95–8/F infection in B cells and epithelial cells were performed as previously reported (Bu et al. 2019).
Octet Biolayer Interferometry
Real-time interactions between purified gH/gL or gH/gL/gp42 proteins and Fabs were monitored on an Octet RED96 instrument (FortéBio). His-tagged recombinant gH/gL or gH/gL/gp42 proteins were diluted in kinetics buffer (0.1% [w/v] bovine serum albumin [BSA], 0.02% [v/v] Tween-20 in PBS; FortéBio) and immobilized on Anti-Penta-HIS (HIS1K) biosensors (FortéBio) at ~50% of the sensor maximum binding capacity. Baseline was established in kinetics buffer. To measure Fab affinities, loaded biosensors were dipped into wells containing serial dilutions of the antibody Fab fragments for 300 s. gH/gL:Fab or gH/gL/gp42:Fab complexes were then allowed to dissociate for 900 s in buffer. After reference subtraction, binding kinetic constants were determined using at least 4 concentrations of Fab, fitting the curves to a 1:1 Langmuir binding model using the Data analysis software 9.0 (FortéBio). Antibody cross-competition was performed as described previously (Bu et al., 2019).
Negative-stain Electron Microscopy Data Collection and Single Particle Analysis
EBV gH/gLgp42-Fab complexes were diluted to approximately 0.025 mg/ml with buffer containing 10 mM HEPES, pH 7, and 150 mM NaCl. A 4.7-µl drop of a diluted sample was applied to a glow-discharged carbon-coated copper grid for 15 s and then removed with filter paper. The grid was washed with three drops of the same buffer and negatively stained by applying consecutively three drops of 0.75% uranyl formate. After removing excess negative stain, the grid was allowed to dry. Datasets for the gH/gL/gp42–769B10, gH/gL/gp42–769C2, and gH/gL/gp42–770F7 binary complexes and the gH/gL/gp42–769B10–769C2–770-F7 quaternary complex were collected on an FEI T20 electron microscope equipped with a LaB6 thermionic electron source and an Eagle 2048×2048-pixel CCD camera and operated at 200 kV. The nominal magnification was 100,000x, corresponding to a pixel size of 2.2 Å, and SerialEM was used for data collection (Mastronarde, 2005). Datasets for the gH/gLgp42–769A7, gH/gLgp42–769C5, and gH/gL/gp42–769F8 binary complexes were collected using a ThermoScientific Talos F200C EM equipped with a Ceta 4096×4096-pixel CCD and operated at 200kV. The nominal magnification was 57,000x, corresponding to a pixel size of 2.53 Å, and EPU software was used for data collection.
To improve angular coverage, datasets for the gH/gLgp42–769B10, gH/gL/gp42–769C2, and gH/gL/gp42–770F7 binary complexes included micrographs collected with the stage tilted to 0°, 15°, 30°, and 45°, whereas for the 769A7-gH/gL/gp42, 769C5-gH/gL/gp42, and 769F8-gH/gL/gp42 complexes the angles were 0°, 30°, and 45°. Micrographs collected at a tilt were divided into slices for more accurate contrast transfer function parameters estimation. Particle detection was performed automatically with in-house written software (Y.T., unpublished). Particles were extracted from micrographs into 144×144 (gH/gL/gp42–769B10), 136×136 (gH/gL/gp42–769C2, gH/gL/gp42–770F7), and 128×128 (gH/gL/gp42–769A7, gH/gL/gp42–769C5, gH/gL/gp42–769F8) pixel boxes. Particle stacks were subjected to 2D classification using SPIDER (Shaikh et al., 2008), or Relion (Scheres, 2012), and high-quality 2D classes were selected for further analysis. The final stacks contained 18,974 (gH/gL/gp42–769B10), 13,845 (gH/gL/gp42–769C2), 20,704 (gH/gL/gp42–770F7), 20,432 (gH/gL/gp42–769A7), 13,240 (gH/gL/gp42–769C5), and 26,773 (gH/gL/gp42–769F8) particles. Initial three-dimensional references were generated with EMAN2 (Tang et al., 2007) from selected 2D classes and used as starting models for three-dimensional reconstruction and refinement using reference projections in SPIDER (Shaikh et al., 2008). The resulting maps were then further refined with FREALIGN (Grigorieff, 2007). The final resolutions, determined using the 0.5 Fourier shell correlation (FSC) threshold with FREALIGN, were 24 Å (gH/gL/gp42–769B10), 24 Å (gH/gL/gp42–769C2), 23 Å (gH/gL/gp42–770F7), 30 Å (gH/gL/gp42–769A7), 30 Å (gH/gL/gp42–769C5), and 27 Å (gH/gL/gp42–769F8).
The negative stain-EM map of the gH/gL/gp42–769B10–769C2–770-F7 quaternary complex was obtained using the random conical tilt (RCT) approach (Radermacher et al., 1987) as described in (Lee et al., 2016). Briefly, tobacco mosaic virus was added to the sample to simplify alignment of untilted and tilted images. Micrograph pairs at 0° and 45° tilt were collected using the T20 electron microscope using the settings described above. Particle tilt pairs were picked manually using JWeb (Frank et al., 1996) and extracted into 180×180 pixel boxes. RCT reconstruction followed by refinement with limited angular search was performed with SPIDER (Shaikh et al., 2008). The resulting map, obtained using 5,647 particles, was subjected to refinement in FREALIGN (Grigorieff, 2007), and the final resolution (FSC 0.5 threshold) was 31 Å.
X-ray crystallography.
Protein Crystallization
All proteins were crystallized by hanging-drop vapor diffusion at 293 K. 769B10 Fab (8.5 mg/ml), 770F7 Fab (7.8 mg ml), gH/gL/gp42p-769B10–769C2 complex (8.0 mg/ ml), and gH/gL/gp42p-769C2–770F7 complex (8.5 mg/ml) were screened for crystallization conditions using an Art Robbins Gryphon crystallization robot, 0.2 µl drops, and a set of 1200 conditions. Crystal drops were observed daily using a Jan Scientific UVEX-PS hotel with automated UV and brightfield drop imaging. Initial crystallization conditions were optimized manually in larger 1 µl drops, and crystals used for data collection grew in the following crystallization conditions: 769B10 Fab: 0.1M Tris-HCl pH8.0, 18% PEG5000 MME, 0.2M NaCl, 2.5% Glycerol; 770F7 Fab: 0.1M HEPES PH7.5, 0.1M CaCl2, 25% PEG3350, 4% isopropanol; gH/gL/gp42p-769B10–769C2 complex: 0.1M Bis-Tris pH6.5, 20% PEG3350, 7.5% Glycerol, 0.7% Dioxane; gH/gL/gp42p-769C2–770F7 complex: 0.1M HEPES pH7.4, 24% PEG3350, 7.5% Glycerol.
Diffraction Data Collection and Processing
Single crystals were transferred to mother liquor containing 20–25% glycerol, and cryo-cooled in liquid nitrogen prior to data collection. Diffraction data were collected at Advanced Photon Source (APS), Argonne National Laboratory beamlines. Diffraction data for the 769B10–769C2-gH/gL/gp42p and 769C2–770F7-gH/gL/gp42p crystals and 770F7 crystals were collected at APS 24-ID-E beamline and measured using a Dectris Eiger 16M PIXEL detector to a final resolution of 3.3 Å, 3.0 Å and 2.4 Å respectively. 769B10 diffraction data to 2.2 Å were collected on SER-CAT ID-22. Diffraction data indexing, integration, and scaling were carried out using the HKL2000 suite (Otwinowski and Minor, 1997). Data collection statistics are reported in Table S1.
Structure Solution and Refinement
Phenix.xtriage was used to analyze all the scaled diffraction data output from HKL2000 and XDS. Primarily, data was analyzed for measurement value significance, completeness, asymmetric unit volume, and possible twinning and/or pseudotranslational pathologies. All crystal structures described in this study were solved by molecular replacement using the program Phaser. Refinement for all structure models was carried out using Phenix refine with positional, global isotropic B-factor refinement and defined TLS groups. Manual model building was performed in Coot. All structure figures were generated using PyMOL (The PyMOL Molecular Graphics System, Schrodinger).
Mouse experiments
Female HSCCB-NOG-F (NOD.Cg-Prkdcscid Il2rgtm1Sug/ JicTac) mice that were 19 to 20 weeks post-engraftment, purchased from Taconic, were used for gH/gL mAb passive transfer and EBV challenge studies. Due to limit on the number of animals that could be engrafted with cells from a single human donor, mice from two different donors were equally distributed among the different groups. Mice were injected with 500 µg of mAbs 769C2, 770F7, 769B10, or 300 µl of PBS by intraperitoneal injection on days −1, 0, 1, 4, 7, and 10 relative to day of challenge of animals with 1 × 106 Green Raji Units of EBV B95–8/F by intravenous tail injection on day 0. Mice were weighed and blood was collected weekly for 15 weeks after infection. Mice were euthanized when they had 30% of weight loss, were unable to obtain food or water, were moribund, or at after 15 weeks. Tissues were fixed in 10% neutral-buffered formalin, and sections on slides were stained with hematoxylin and eosin (H&E), CD20, and in situ hybridization was performed using a probe for EBV EBER1. Area under the curve analyses were performed using data for weeks 1 to 9 after infection; for animals that died, the last EBV copy number obtained while alive was used for subsequent values after death. Only p values <0.001 for this analysis were considered to be significant.
Female and male C57BL/6, B6.mFcRn−/− hFCRN Tg32 mice, purchased from Jackson Laboratory, were used to measure mAb levels to determine the half-life of the antibody. Mice were injected with 5 mg/kg of mAb intravenously by tail injection on day 0. Serum was collected before injection and on days 1, 2, 5, 7, 9, 14, 21, 28, and 35 after injection. The concentration of each mAb was quantified by ELISA assay. Half-life, area under the curve, and clearance rate were calculated using WinNonLin software (Certara).
Quantification of EBV DNA copies in the blood by qPCR
EBV DNA was isolated from mouse blood and EBV DNA copy numbers were quantified as described previously (Kim et al., 2021). Welch two sample t-test was used to calculate p-values.
Quantification of gH/gL mAb concentration by ELISA
96-well ELISA plates were coated with 200 ng/well of EBV gH/gL in ELISA coating buffer (BioLegend, San Diego, CA) and incubated overnight at 4°C. Plates were washed 6 times with PBS with 0.05% Tween-20 (washing buffer) and blocked with PBS containing 0.05% Tween-20, 5% skim milk, and 2% BSA (blocking buffer) for 1 hr at room temperature. Plates were washed again with washing buffer. Known concentrations of purified gH/gL mAb was used as a standard. Control wells were incubated with blocking buffer without any serum or mAb. Plates were incubated with mouse sera, mouse plasma, or control mAb for 1 hr at room temperature and washed 6 times with washing buffer. Following washing, a 1:8000 dilution of goat anti-human IgG-HRP (Southern Biotech, Birmingham, AL) in blocking buffer was added to each well, including mAb control wells, and incubated at room temperature for 1 hr. Plates were washed with washing buffer and 100 µl of SureBlue Reserve TMB Microwell Peroxidase substrate. 100 µl of 1 N sulfuric acid was added to each well after 3 min and the absorbance of yellow color was read at A450. All assays were performed in duplicate and samples were corrected to subtract the background level, averaged A450 values of control wells, and the standard curve was generated by plotting corrected averaged A450 values to known concentration of gH/gL mAb by fitting data to sigmoidal dose-response curve using GraphPad PRISM software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Biolayer interferometry measurements were taken over 2 independent experiments, and after reference subtraction, binding kinetic constants were determined using at least 4 concentrations of Fab, fitting the curves to a 1:1 Langmuir binding model using the Data analysis software 9.0 (FortéBio).
Throughout the structure refinement processes, a cross validation (Rfree) test set consisting of 5% of the data was used. Structure validations were performed periodically during the model building/refinement process with MolProbity. Data collection statistics and final refinement statistics are summarized in Table S1.
To compare viral loads over time in mice, an area-under-the curve analysis was performed. The mean value for the level of EBV DNA in the blood for each animal was calculated over weeks 1, 3, 5, 7, and 9 and then the group means were compared using t-statistics. When mice died during the experiment, the last data obtained before death was used for subsequent (missing) time points, based on the last observation carried forward method. To account for multiple comparisons, P <0.001 was used to define differences that were statistically significant. The statistical software R package was used in the analysis.
Supplementary Material
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 488 goat-anti mouse IgG (H+L chain) | ThermoFisher Scientific | Cat# A11001 |
| Anti-human CD3 BV510 | BioLegend | Cat# 317332 |
| Anti-human CD56 BV510 | BioLegend | Cat# 318340 |
| Anti-human CD14 BV510 | BioLegend | Cat# 301842 |
| Anti-human CD27 BV605 | BioLegend | Cat# 302830 |
| Anti-human CD38 AF700 | BioLegend | Cat# 303524 |
| Anti-human CD19 ECD | Beckman Coulter | Cat# A07770 |
| IgG BV421 | BD Biosciences | Cat# 562581 |
| IgM PerCP-Cy5.5 | BD Biosciences | Cat# 561285 |
| E1D1 | (Sathiyamoorthy et al., 2016) | N/A |
| 769B10 | (Bu et al., 2019) | N/A |
| 769C2 | This study | N/A |
| 769C5 | This study | N/A |
| 769A7 | This study | N/A |
| 770F7 | This study | N/A |
| 770F8 | This study | N/A |
| Bacterial and Virus Strains | ||
| AKATA EBV GFP | (Molesworth et al., 2000) | N/A |
| B95–8/F | (Delecluse et al., 1998) | N/A |
| Biological Samples | ||
| PBMC from Healthy blood bank donors | National Institutes of Health Warren G. Magnuson Clinical Center | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Streptavidin PE | ThermoFisher Scientific | Cat# S21388 |
| Streptavidin APC | ThermoFisher Scientific | Cat# S32362 |
| gp42 peptide (residues 47–81) | This paper | GenScript Biotech Corp |
| Critical Commercial Assays | ||
| In Vitro Biotin Ligase kit | Avidity | Cat# BirA-500 |
| Superscript® III Reverse Transcriptase | ThermoFisher Scientific | Cat# 18080093 |
| Random Hexmers | Gene Link | Cat# CCL-86 |
| Igepal CA-630 | Sigma-Aldrich | Cat# I8896 |
| RNaseOUT | ThermoFisher Scientific | Cat# 10777019 |
| DNA Ligation Kits, Quick | Takara | Cat# 6021 |
| Britelite plus Reporter Gene Assay System | PerkinElmer | Cat# 6066766 |
| Expi293 Expression System Kit | ThermoFischer Scientific | Cat#A39250 |
| Deposited Data | ||
| Crystal structure gH/gL/gp42p-769C2–769B10 | This Paper | PDB code :7S07 |
| Crystal structure gH/gL/gp42p-769C2–770F7 | This Paper | PDB code: 7S1B |
| Crystal structure 769B10 Fab | This paper | PDB code: 7S08 |
| Crystal structure 770F7 Fab | This paper | PDB code: 7S0J |
| EM gH/gL-769B10 | This Paper | |
| EM gH/gL-769A7 | This Paper | |
| EM gH/gL-769C2 | This Paper | |
| EM gH/gL-769F8 | This Paper | |
| EM gH/gL-769C5 | This Paper | |
| 769A7 | This Paper | OP453131/ OP453132 |
| 769C2 | This Paper | OP453123/ OP453124 |
| 769C5 | This Paper | OP453129/ OP453130 |
| 769F7 | This Paper | OP453125/ OP453126 |
| 769F8 | This Paper | OP453127/ OP453128 |
| Experimental Models: Cell Lines | ||
| 293T | ATCC | Cat# ACS-4500 |
| CHO-K1 | ATCC | Cat# CCL-61 |
| Raji | ATCC | Cat# CCL-86 |
| Expi293F | ThermoFisher Scientific | Cat# A14527 |
| Daudi | (Silva et al., 2004) | N/A |
| 293T-14 | (Omerović et al., 2005) | N/A |
| SVKCR2 | (Li et al., 1992) | N/A |
| 293/2089 | (Delecluse et al., 1998) | N/A |
| Akata BX1 | (Molesworth et al., 2000) | N/A |
| Experimental Models: Organisms/Strains | ||
| Female HSCCB-NOG-F (NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac) mice | Taconic | N/A |
| female and male C57BL/6, B6. mFcRn−/− hFcRn Tg32 mice | Jackson Laboratory | N/A |
| Recombinant DNA | ||
| pCAGGS EBV gH | (Silva et al., 2004) | N/A |
| pCAGGS EBV gL | (Silva et al., 2004) | N/A |
| pCAGGS EBV gB | (Silva et al., 2004) | N/A |
| pCAGGS EBV gp42 | (Silva et al., 2004) | N/A |
| pCMV/R-gH-AviHis | (Bu et al., 2019) | N/A |
| pCMV/R-gL | (Bu et al., 2019) | N/A |
| Software and Algorithms | ||
| Flowjo | Flowjo | http://www.flowjo.com |
| Prism | Graphpad | http://www.graphpadcom |
| Biorender | Biorender | http://www.biorender.com |
| Pymol | Schrödinger, Inc. | https://pymol.org/2/ |
| phenix.refine | Phenix software suite | https://phenix-online.org/ |
| Coot | MRC Laboratory of Molecular Biology in Cambridge | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Other | ||
| Captia EBV VCA (P-18) IgG Kit | Trinity Biotech | Cat# 2324700 |
| Ni-NTA agarose | ThermoFisher Scientific | Cat# R901–15 |
| Tecnai™ T20 | FEI Company | N/A |
| Streptavidin biosensors | FortéBio | Cat# 18–5019 |
| Octet HTX instrument | FortéBio | N/A |
Highlights.
Six human monoclonal antibodies (mAbs) target five distinct sites on EBV gH/gL
Some of these antigenic sites match sites of vulnerability on other herpesviruses
Each mAb neutralizes EBV infection and blocks virus-cell fusion
mAb 769B10 blocks viremia and prevents lymphoma in an EBV challenge animal model
ACKNOWLEDGMENTS
We thank Lindsey Hutt-Fletcher for mAbs E1D1, and CL40, and Akata BX1 and SVKCR2 cells, Henri-Jacques Delecluse and Bill Sugden for 293/2089 cells, Tong-Ming Fu for CMV mAbs, and Ravi Mahalingam for VZV Ab RC, Richard Longnecker for plasmids encoding EBV gH, gL, gB, gp42, and T7 luciferase and for 293-T14 and Daudi cells, Ed Berger, Stephen Dollery, and Agnes Hajduczki for the KSHV reagents, David Ambrozak and the VRC Flow Cytometry Core for help with B cell sorting, and Jing Qin for help with statistics. This research was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases and in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. Research was conducted under an approved animal use protocol in an AAALAC International-accredited facility in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on the 24-ID-E beam line is funded by a NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. Data were also collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. SER-CAT is supported by its member institutions, and equipment grants (S10_RR25528, S10_RR028976 and S10_OD027000) from the National Institutes of Health. The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army or the Department of Defense or HJF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at...
DECLARATION OF INTERESTS
W.B., Y.T., M.K., M.G.J. and J.I.C. are named as inventors on patent applications describing the data presented in this paper, which have been filed by the Department of Health and Human Services and the Henry M. Jackson Foundation.
REFERENCES
- Bu W, Joyce MG, Nguyen H, Banh DV, Aguilar F, Tariq Z, Yap ML, Tsujimura Y, Gillespie RA, Tsybovsky Y, et al. (2019). Immunization with Components of the Viral Fusion Apparatus Elicits Antibodies That Neutralize Epstein-Barr Virus in B Cells and Epithelial Cells. Immunity 50, 1305–1316 e1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sathiyamoorthy K, Zhang X, Schaller S, Perez White BE, Jardetzky TS, and Longnecker R (2018). Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol 3, 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova LS, Nishimura SL, and Hutt-Fletcher LM (2009). Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc Natl Acad Sci U S A 106, 20464–20469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, and Heldwein EE (2010). Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol 17, 882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Cao Z, Chen Q, Arjunaraja S, Snow AL, and Snapper CM (2016). Rabbits immunized with Epstein-Barr virus gH/gL or gB recombinant proteins elicit higher serum virus neutralizing activity than gp350. Vaccine 34, 4050–4055. [DOI] [PubMed] [Google Scholar]
- Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, and Hammerschmidt W (1998). Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95, 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, and Leith A (1996). SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Gerna G, Percivalle E, Perez L, Lanzavecchia A, and Lilleri D (2016). Monoclonal Antibodies to Different Components of the Human Cytomegalovirus (HCMV) Pentamer gH/gL/pUL128L and Trimer gH/gL/gO as well as Antibodies Elicited during Primary HCMV Infection Prevent Epithelial Cell Syncytium Formation. J Virol 90, 6216–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N (2007). FREALIGN: high-resolution refinement of single particle structures. J Struct Biol 157, 117–125. [DOI] [PubMed] [Google Scholar]
- Kabanova A, Marcandalli J, Zhou T, Bianchi S, Baxa U, Tsybovsky Y, Lilleri D, Silacci-Fregni C, Foglierini M, Fernandez-Rodriguez BM, et al. (2016). Platelet-derived growth factor-alpha receptor is the cellular receptor for human cytomegalovirus gHgLgO trimer. Nat Microbiol 1, 16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabanova A, Perez L, Lilleri D, Marcandalli J, Agatic G, Becattini S, Preite S, Fuschillo D, Percivalle E, Sallusto F, et al. (2014). Antibody-driven design of a human cytomegalovirus gHgLpUL128L subunit vaccine that selectively elicits potent neutralizing antibodies. Proc Natl Acad Sci U S A 111, 17965–17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M, Bu W, Joyce MG, Meng G, Whittle JR, Baxa U, Yamamoto T, Narpala S, Todd JP, Rao SS, et al. (2015). Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 162, 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bu W, Mine S, Tariq Z, Nguyen H, Wang Y, Tolman C, Mond J, and Cohen JI (2021). Epstein-Barr virus (EBV) hyperimmune globulin isolated from donors with high gp350 antibody titers protect humanized mice from challenge with EBV. Virology 561, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner AN, Lowrey AS, Longnecker R, and Jardetzky TS (2007). Binding-site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion. J Virol 81, 9216–9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Sheppard NC, Stewart-Jones GBE, Robson CL, Chen H, Xu X, Krashias G, Bonomelli C, Scanlan CN, Kwong PD, et al. (2010). Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J Mol Biol 403, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kschonsak M, Rouge L, Arthur CP, Hoangdung H, Patel N, Kim I, Johnson MC, Kraft E, Rohou AL, Gill A, et al. (2021). Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry. Cell 184, 1232–1244 e1216. [DOI] [PubMed] [Google Scholar]
- Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee CH, Lavinder JJ, et al. (2016). Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med 22, 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Freed DC, Liu Y, Li F, Barrett DF, Xiong W, Ye X, Adler SP, Rupp RE, Wang D, et al. (2021). A conditionally replication-defective cytomegalovirus vaccine elicits potent and diverse functional monoclonal antibodies in a phase I clinical trial. NPJ Vaccines 6, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, Gerna G, Sallusto F, and Lanzavecchia A (2010). Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128–131A complex. J Virol 84, 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Matsuura H, Kirschner AN, Longnecker R, and Jardetzky TS (2010). Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc Natl Acad Sci U S A 107, 22641–22646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane MP, and Longnecker R (2004). Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc Natl Acad Sci U S A 101, 17474–17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molesworth SJ, Lake CM, Borza CM, Turk SM, and Hutt-Fletcher LM (2000). Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J Virol 74, 6324–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, and Frank J (1987). Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc 146, 113–136. [DOI] [PubMed] [Google Scholar]
- Sathiyamoorthy K, Hu YX, Mohl BS, Chen J, Longnecker R, and Jardetzky TS (2016). Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat Commun 7, 13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathiyamoorthy K, Jiang J, Mohl BS, Chen J, Zhou ZH, Longnecker R, and Jardetzky TS (2017). Inhibition of EBV-mediated membrane fusion by anti-gHgL antibodies. Proc Natl Acad Sci U S A 114, E8703–E8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemayer TA, Gross TG, Egeler RM, Pirruccello SJ, Davis JR, Kelly CM, Okano M, Lanyi A, and Sumegi J (1995). X-linked lymphoproliferative disease: twenty-five years after the discovery. Pediatr Res 38, 471–478. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, and Frank J (2008). SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3, 1941–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Homad LJ, Akins NR, Stoffers CM, Lackhar S, Malhi H, Wan YH, Rawlings DJ, and McGuire AT (2020). Neutralizing Antibodies Protect against Oral Transmission of Lymphocryptovirus. Cell Rep Med 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J, Ortego MS, Weidle C, Stuart AB, Gray MD, McElrath MJ, Pancera M, Veesler D, and McGuire AT (2018). An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity 48, 799–811 e799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snydman DR, McIver J, Leszczynski J, Cho SI, Werner BG, Berardi VP, LoGerfo F, Heinze-Lacey B, and Grady GF (1984). A pilot trial of a novel cytomegalovirus immune globulin in renal transplant recipients. Transplantation 38, 553–557. [DOI] [PubMed] [Google Scholar]
- Spriggs MK, Armitage RJ, Comeau MR, Strockbine L, Farrah T, Macduff B, Ulrich D, Alderson MR, Mullberg J, and Cohen JI (1996). The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR beta chain and inhibits antigen presentation. J Virol 70, 5557–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Wu L, Chai Y, Qi J, Tan S, Gao GF, Song H, and Yan J (2020). Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses. Nat Commun 11, 5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Wang D, Li F, Freed DC, Finnefrock AC, Tang A, Grimes SN, Casimiro DR, and Fu TM (2011). Quantitative analysis of neutralizing antibody response to human cytomegalovirus in natural infection. Vaccine 29, 9075–9080. [DOI] [PubMed] [Google Scholar]
- Wei CJ, Bu W, Nguyen LA, Batchelor JD, Kim J, Pittaluga S, Fuller JR, Nguyen H, Chou TH, Cohen JI, et al. (2022). A bivalent Epstein-Barr virus vaccine induces neutralizing antibodies that block infection and confer immunity in humanized mice. Sci Transl Med 14, eabf3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Ye X, Ku Z, Su H, Jones HG, Wang N, Mishra AK, Freed DC, Li F, Tang A, et al. (2022). Structural basis for HCMV Pentamer recognition by neuropilin 2 and neutralizing antibodies. Sci Adv 8, eabm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, and Hutt-Fletcher LM (2007). Point mutations in EBV gH that abrogate or differentially affect B cell and epithelial cell fusion. Virology 363, 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Oliver SL, Nguyen T, Ciferri C, Nandi A, Hickman J, Giovani C, Yang E, Palladino G, Grose C, et al. (2015). A site of varicella-zoster virus vulnerability identified by structural studies of neutralizing antibodies bound to the glycoprotein complex gHgL. Proc Natl Acad Sci U S A 112, 6056–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Y, Wang HB, Zhang A, Chen ML, Fang ZX, Dong XD, Li SB, Du Y, Xiong D, et al. (2018). Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol 3, 1–8. [DOI] [PubMed] [Google Scholar]
- Zhu QY, Shan S, Yu J, Peng SY, Sun C, Zuo Y, Zhong LY, Yan SM, Zhang X, Yang Z, et al. (2021). A potent and protective human neutralizing antibody targeting a novel vulnerable site of Epstein-Barr virus. Nat Commun 12, 6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
X-ray coordinates and structure factors have been deposited in the RCSB Protein Data Bank under accession codes PDBs: 7S07, 7S1B, 7S08, 7S0J. Antibody heavy and light chain sequences have been deposited in GenBank under accession numbers OP453123-OP453132. Additional data supporting the findings of this study are found within the paper and its Supplementary Information and are available from the Lead Contact upon request.


