Abstract
Improvements in genome analysis technology using next-generation sequencing have revealed that abnormalities in the composition of the intestinal microbiota are important in numerous diseases. Furthermore, intestinal commensal pathogens that are directly involved in the onset and exacerbation of disease have been identified. Specific control of them is strongly desired. However, antibiotics are not appropriate for the control of intestinal commensal pathogens because they may kill beneficial bacteria as well. The intestinal tract contains many viruses: most are bacteriophages (phages) that infect intestinal bacteria rather than viruses that infect human cells. Phages have very high specificity for their host bacteria. Therefore, phage therapy is considered potentially useful for controlling intestinal commensal pathogens. However, the intestinal tract is a specialized, anaerobic environment, and it is impossible to isolate phages that infect host intestinal bacteria if the bacteria cannot be cultured. Furthermore, genomic analysis methods for intestinal phages have not been well established, so until recently, a complete picture of the intestinal phage has not been clear. In this review, I summarize the importance of next-generation phage therapy based on metagenomic data and describe a novel therapy against Clostridioides difficile developed using such data.
Keywords: bacteriophage, phage therapy, Clostridioides difficile, dysbiosis, microbiome, pathobiont
INTRODUCTION
There is a large body of evidence regarding the impact of the gut microbiota (the bacteriome) on disease. Abnormalities of the human gut microbiota (dysbiosis) have been implicated in allergy, obesity, cancer, inflammatory bowel disease, diabetes mellitus, atherosclerosis, and autoimmune diseases [1,2,3,4], among others. In recent years, research has been conducted worldwide to improve dysbiosis using fecal microbiota transplantation and other therapies to control disease. In addition to dysbiosis, pathobionts—which are directly involved in the pathogenesis of diseases—have been identified, such as in Crohn’s disease [5,6,7,8,9]. Therefore, controlling pathobionts may be useful for disease prevention and treatment. However, antibiotics kill not only pathobionts but also beneficial bacteria, which could promote dysbiosis. Thus, development of pathobiont-specific control strategies is highly desirable.
In recent years, the trans-kingdom interactions between the bacteriome and the viral microbiome (virome) have been shown to be related to the pathogenesis of intestinal bacteria-mediated diseases [10,11,12,13,14,15]. Less well known are the viruses that dominate our intestinal tract—these are not viruses that infect our cells (such as norovirus and rotavirus) but are viruses that infect our intestinal bacteria [16]. They are called bacteriophages (or, more commonly, phages). A phage recognizes a characteristic receptor molecule on the membrane of its host bacterium and delivers the genome stored in its head into the bacterium when infection is established. The phage then uses the bacterial cell to replicate daughter phages in large numbers and destroy the bacterium via membrane-disrupting lytic enzymes, thereby releasing new phage particles.
Historically, Ernest Hankin discovered in 1896 that there was “an antiseptic substance” in the water of the Ganges River that killed certain bacteria, and in 1915, Frederick Twort discovered “a transparent material” that changed the properties of Staphylococcus aureus [17]. In 1917, Félix d’Hérelle named an “invisible microbe” that lysed Shigella a “bacteriophage”. Furthermore, d’Hérelle proposed phage therapy, i.e., the treatment of bacterial infections using phages, and practiced phage therapy in humans and animals, including for Shigella and cholera (which is caused by the bacterium Vibrio cholerae). D’Hérelle also met Giorgi Eliava at the Pasteur Institute, and between 1923 and 1936, they established several institutes in Tbilisi, Georgia, with the support of the Soviet Union. One of them, the Eliava Institute, became a hotbed of phage research and phage therapy, and it continues to operate to this day. With the discovery of the antibacterial drug penicillin in 1928 and the subsequent commercialization of antibiotics [18], phage therapy was abandoned in Western countries. However, during the Cold War, the Soviet Union developed phage therapy as an alternative way to treat infectious diseases because of the lack of supply of the newly developed antibiotic drugs in that country. Even today, phage cocktail solutions are formulated and used as treatments for infectious diseases in Russia, Georgia, and Poland [19]. Meanwhile, the overuse of antibiotics has led to the emergence of a variety of multidrug-resistant bacteria, which has developed into a critical global medical problem. Therefore, phage therapy against multidrug-resistant bacteria has begun to attract renewed attention around the world as a next-generation treatment method.
Recently, the development of analytical technology using next-generation sequencing has made it possible to obtain whole genome sequences of intestinal phages as well as their host intestinal bacteria, which can be technically difficult to cultivate [20]. However, phage research was originally developed based on morphology using electron microscopy, and the development of reference genomes for phages has not progressed worldwide. Therefore, when metagenomic sequencing reads have been subjected to homology analysis using known phage genome databases, most of them have been assigned to unknown phages. The difficulty of analyzing phages has led them to be described as “viral dark matter” [21].
IMPENDING THREAT OF DRUG-RESISTANT BACTERIA
While we are currently confronting an unprecedented pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), we should not forget that the threat of drug-resistant bacteria is also close at hand. If no action is taken, it is predicted that infections caused by drug-resistant bacteria will be the leading cause of death by 2050 [22, 23]. The problem of drug-resistant bacteria is becoming more serious worldwide. In Japan, methicillin-resistant Staphylococcus aureus (MRSA) and drug-resistant Escherichia coli (fluoroquinolone-resistant E. coli and third-generation cephalosporin-resistant E. coli) are the most frequently occurring drug-resistant bacteria [24]. Given that S. aureus is one of the species that makes up the indigenous microbiota of our skin, preventing the spread of MRSA is very important. Until about 20 to 30 years ago, detection of drug-resistant bacteria was rare, even in large hospitals in Japan, but in recent years, their detection has become increasingly common, even in medium and small hospitals. Considering that these drug-resistant bacteria have been detected not only in infections within hospitals but also in outpatients and newly admitted patients, it is natural to assume that they have already spread outside hospitals. Therefore, countermeasures outside of medical institutions are extremely important, and even the general public now need to know how to prevent the spread of drug-resistant bacteria (such as infection control measures like hand washing and proper use of antimicrobial agents).
Carbapenem-resistant Enterobacteriaceae (CRE) are less frequently detected than the aforementioned drug-resistant bacteria, but they are resistant to carbapenems such as meropenem and broad-spectrum β-lactams. These bacteria are considered an urgent public health threat by the U.S. Centers for Disease Control and Prevention [25]. The rate of carbapenem resistance among the major Enterobacteriaceae, such as E. coli, Klebsiella, and Enterobacter, has reportedly increased over the past 10 years, and countermeasures have already begun in many countries [26]. However, there are still no CRE-specific and effective treatments. Deaths due to diseases caused by these drug-resistant organisms, which are normally treatable, have already begun to occur. The current problem of drug-resistant bacteria requires a “One Health” approach that includes not only human health but also animals and the environment, and the use of antibiotics must be reviewed in cooperation with various sectors, including agriculture, fisheries, and livestock production.
PHAGE THERAPY
Phage therapy is considered a trump card in the fight against drug-resistant bacteria [27]. In Japan, the National Action Plan (Research and Development and Drug Discovery) calls for “research on drug resistance and promotion of research and development to ensure preventive, diagnostic, and therapeutic measures against drug-resistant microorganisms”. Hence, phage therapy, a treatment method that does not use antimicrobial agents, has gradually been attracting attention. However, it must be emphasized that there are still no clinical cases or preparations of phage therapy in Japan.
In the United States, Intralytix has commercialized a phage spray using phages as a preventive measure against Listeria food poisoning, and this phage preparation has been approved by the Food and Drug Administration as a food additive for spraying on the surfaces of meat during shipment. In France, a phage cocktail preparation for burns (PhagoBurn) is being developed, and a European Union-led multicenter clinical trial is underway [28]. Other phage therapies are being developed against S. aureus and Pseudomonas aeruginosa, and phage therapy using modified phage technology is being tested [29, 30].
DEVELOPMENT OF NOVEL PHAGE THERAPIES FOR INTESTINAL BACTERIA-MEDIATED DISEASES
With the improvement of genome analysis technology using next-generation sequencers, it has become important to understand in detail how the intestinal microbiota affects the host and its involvement in various pathological mechanisms. In particular, how to improve dysbiosis and how to control pathobionts are very important for disease prevention and treatment.
Therefore, my colleagues and I considered the use of intestinal phages to control intestinal bacteria, because intestinal phages are abundant in the intestinal tract. First, we worked on clarifying the complete picture of an intestinal phage (i.e., visualizing the viral dark matter). Unlike bacteria and fungi, viruses do not have universal genome signatures (e.g., 16S rRNA genes for bacteria, internal transcribed spacers for fungi) [31]. Therefore, targeted sequencing analysis (such as 16S rRNA gene analysis) cannot be performed.
Hence, we purified viral fractions from human fecal samples derived from 101 healthy subjects, extracted the viral genomic DNA, and performed shotgun sequencing [13]. Although a large number of viral genome sequences were obtained, it was not possible to classify the phage species based on the sequencing reads because the reference genome database for intestinal phages was insufficient (as mentioned above). Therefore, we decided to create contigs by assembling the obtained sequence reads and tried to classify phage species based on the sequence information.
First, we attempted to classify the obtained contigs by referring to existing databases, but only about 0.5% of the contigs could be classified. Next, we added the open reading frames (ORFs) encoding proteins to the analyses and classified the phage species based on ORF information. Because it is known that crAss-like phages have terminases and polymerases characteristic of crAss-like phages, we classified crAss-like phages by using these sequences as landmarks. Next, Caudovirales were classified according to tail protein sequences, followed by Microviridae according to capsid protein sequences. Phage species that could not be determined by the above classifications were classified using a combination of multiple ORFs in the contigs. Using this method, >90% of the obtained contigs could be classified [13].
Next, host searches of the obtained phage sequences were performed. For host searching, we considered it important to obtain intestinal bacterial genomes from the same feces samples used for the viral analysis. Thus, we purified bacterial fractions from the human fecal samples of the aforementioned 101 healthy subjects, extracted bacterial genomic DNA, and performed shotgun sequencing. As with the phage-derived sequencing reads, contigs were generated from the resulting bacteria-derived sequencing reads.
Phages have two life cycles: the lytic cycle and the lysogenic cycle. Both lytic and lysogenic cycles can infect host bacteria, but lytic phages have been considered suitable for phage therapy because they lyse their host bacteria after infecting and replicating in them. Lysogenic phages incorporate their phage genomes into the chromosomes of their host bacteria as prophages after infection and repeatedly multiply with their host bacteria. Because lysogenic phages are incorporated into the genomes of their host bacteria, they are considered unsuitable for phage therapy using phages themselves.
We used two analytical methods to identify associations between intestinal host bacteria and intestinal phages based on genomic information. In the first method, we identified prophage regions in the bacterial contigs and performed homology analysis with the phage-derived contigs. This method can be used to identify the host bacteria–phage associations of lysogenic phages. In the second method, clustered regularly interspaced short palindromic repeat (CRISPR) sequences in the bacterial contigs were identified, and a homology analysis was performed with the phage-derived contigs to identify host bacteria–phage associations. This method can be used to identify the host bacteria–phage associations of lytic phages. Previously, host bacteria–phage associations were generally clarified by experimentally isolating phages that lyse host bacteria, but by using large-scale metagenomic information, it is now possible to identify phages that infect pathobionts [13].
The next step was to develop a phage therapy to control a pathobiont. However, the intestinal lumen contains many difficult-to-culture bacteria, making it very difficult to isolate the host-specific phage itself. In this regard, it may not be realistic to apply phage therapy using phages themselves for all pathobionts. Therefore, we tried to develop a method of controlling host bacteria based on lysogenic phages by using the genomic information on host bacteria–phage associations that we had established so far.
Clostridioides difficile, a Gram-positive, spore-forming, anaerobic bacterium, is endemic in the intestinal tract of healthy people. It is a representative cause of nosocomial diarrhea following antibiotic treatment. In Japan, improvement is often observed after administration of antibiotics to which C. difficile is susceptible, such as vancomycin and metronidazole, but in Western countries, there have been cases of unsuccessful treatment and relapse due to the emergence of highly virulent strains of C. difficile and resistance to antimicrobial agents [32].
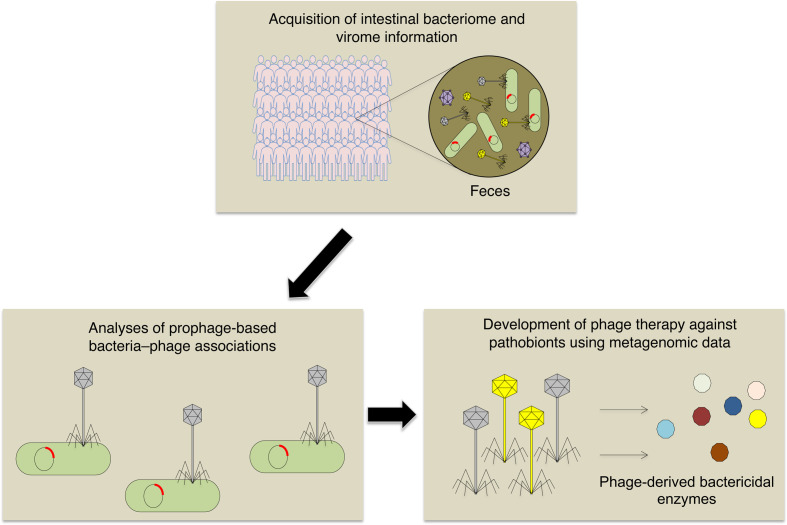
By using sequencing data obtained from healthy subjects and clinical isolates of C. difficile strains, we developed a new phage therapy that is specific for C. difficile. We searched for novel phage-derived bacteriolysis enzymes specific for C. difficile using the obtained sequencing data. Using our phage genome analysis pipeline, we were able to identify several novel sequences of endolysins—bacteriolytic enzymes used when phages are released from bacteria after growth inside the bacteria—from C. difficile prophage sequences. These endolysins were synthesized and shown to have bacteriolytic activity in vitro; they were also found to be effective in a mouse model of C. difficile infection. This is a practical example of a next-generation phage therapy based on metagenomic information, and its strategy can be applied to various targets in the future [13] (Fig. 1).
Fig. 1.
Generation of a next-generation phage therapy based on metagenomic information.
To detect phage-derived antibacterial enzymes that can specifically control pathobionts, intestinal bacterial and viral metagenomic information is acquired from human fecal samples. Phage-derived bactericidal enzymes can kill host bacteria specifically.
CONCLUSION
Analysis of the gut microbiota has advanced dramatically, and relationships between the gut microbiota and diseases have gradually become clear. We can now also analyze intestinal phages, which was difficult in the past; this will not only be a very powerful analytical tool for the future practice of phage therapy but will also lead to various industrial applications of phages. The integration of phage science with a wide range of fields, including medicine, microbiology, bioinformatics, and synthetic biology, will be promoted in the near future.
The number of genes possessed by intestinal bacteria is 100 to 1,000 times greater than the number possessed by humans, and the commensal microbiota, in which each bacterium functions in concert, may be thought of as an organ. Designing and managing the intestinal microbiota will be necessary for maintaining our health and improving diseases in the future. I would like to continue to confront intractable diseases from multiple perspectives and promote new medical developments.
CONFERENCE PRESENTATION
The contents of this article received a 2021 Research Encouragement Award and were presented at the 26th Annual Meeting of Intestinal Microbiology held on July 7th, 2022.
FUNDING
This study was supported by the Japan Agency for Medical Research and Development (AMED; 21ae0121048h0001).
Acknowledgments
I thank K. Ogawa, M. Maeda, and K. Suetsugu for administrative assistance. I thank Edanz Group (https://en-author-services.edanz.com/ac) for editing a draft of this manuscript.
REFERENCES
- 1.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13: 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157: 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch SV, Pedersen O. 2016. The human intestinal microbiome in health and disease. N Engl J Med 375: 2369–2379. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. 2018. Current understanding of the human microbiome. Nat Med 24: 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrios-Villa E, Martínez de la Peña CF, Lozano-Zaraín P, Cevallos MA, Torres C, Torres AG, Rocha-Gracia RDC. 2020. Comparative genomics of a subset of Adherent/Invasive Escherichia coli strains isolated from individuals without inflammatory bowel disease. Genomics 112: 1813–1820. [DOI] [PubMed] [Google Scholar]
- 6.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498: 99–103. [DOI] [PubMed] [Google Scholar]
- 7.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD, Pedersen O, MetaHIT consortium. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- 8.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR. 2013. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2: e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujimoto K, Kawaguchi Y, Shimohigoshi M, Gotoh Y, Nakano Y, Usui Y, Hayashi T, Kimura Y, Uematsu M, Yamamoto T, Akeda Y, Rhee JH, Yuki Y, Ishii KJ, Crowe SE, Ernst PB, Kiyono H, Uematsu S. 2019. Antigen-specific mucosal immunity regulates development of intestinal bacteria-mediated diseases. Gastroenterology 157: 1530–1543.e4. [DOI] [PubMed] [Google Scholar]
- 10.Yutin N, Makarova KS, Gussow AB, Krupovic M, Segall A, Edwards RA, Koonin EV. 2018. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat Microbiol 3: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, McDonnell SA, Khokhlova EV, Draper LA, Forde A, Guerin E, Velayudhan V, Ross RP, Hill C. 2019. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26: 527–541.e5. [DOI] [PubMed] [Google Scholar]
- 12.Shkoporov AN, Hill C. 2019. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25: 195–209. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto K, Kimura Y, Shimohigoshi M, Satoh T, Sato S, Tremmel G, Uematsu M, Kawaguchi Y, Usui Y, Nakano Y, Hayashi T, Kashima K, Yuki Y, Yamaguchi K, Furukawa Y, Kakuta M, Akiyama Y, Yamaguchi R, Crowe SE, Ernst PB, Miyano S, Kiyono H, Imoto S, Uematsu S. 2020. Metagenome data on intestinal phage-bacteria associations aids the development of phage therapy against pathobionts. Cell Host Microbe 28: 380–389.e9. [DOI] [PubMed] [Google Scholar]
- 14.Bajaj JS, Sikaroodi M, Shamsaddini A, Henseler Z, Santiago-Rodriguez T, Acharya C, Fagan A, Hylemon PB, Fuchs M, Gavis E, Ward T, Knights D, Gillevet PM. 2021. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 70: 1162–1173. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto K, Kimura Y, Allegretti JR, Yamamoto M, Zhang YZ, Katayama K, Tremmel G, Kawaguchi Y, Shimohigoshi M, Hayashi T, Uematsu M, Yamaguchi K, Furukawa Y, Akiyama Y, Yamaguchi R, Crowe SE, Ernst PB, Miyano S, Kiyono H, Imoto S, Uematsu S. 2021. Functional restoration of bacteriomes and viromes by fecal microbiota transplantation. Gastroenterology 160: 2089–2102.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. 2016. Healthy human gut phageome. Proc Natl Acad Sci USA 113: 10400–10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abedon ST, Thomas-Abedon C, Thomas A, Mazure H. 2011. Bacteriophage prehistory: is or is not Hankin, 1896, a phage reference? Bacteriophage 1: 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchings MI, Truman AW, Wilkinson B. 2019. Antibiotics: past, present and future. Curr Opin Microbiol 51: 72–80. [DOI] [PubMed] [Google Scholar]
- 19.Abedon ST, García P, Mullany P, Aminov R. 2017. Editorial: phage therapy: past, present and future. Front Microbiol 8: 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat Protoc 4: 470–483. [DOI] [PubMed] [Google Scholar]
- 21.Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, Ryan FJ, Draper LA, Plevy SE, Ross RP, Hill C. 2019. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26: 764–778.e5. [DOI] [PubMed] [Google Scholar]
- 22.de Kraker ME, Stewardson AJ, Harbarth S. 2016. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med 13: e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abat C, Fournier PE, Jimeno MT, Rolain JM, Raoult D. 2018. Extremely and pandrug-resistant bacteria extra-deaths: myth or reality? Eur J Clin Microbiol Infect Dis 37: 1687–1697. [DOI] [PubMed] [Google Scholar]
- 24.Ohmagari N. 2019. National action plan on antimicrobial resistance (AMR) 2016–2020 and relevant activities in Japan. Glob Health Med 1: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomon SL, Oliver KB. 2014. Antibiotic resistance threats in the United States: stepping back from the brink. Am Fam Physician 89: 938–941. [PubMed] [Google Scholar]
- 26.Codjoe FS, Donkor ES. 2017. Carbapenem resistance: a review. Med Sci (Basel) 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kortright KE, Chan BK, Koff JL, Turner PE. 2019. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25: 219–232. [DOI] [PubMed] [Google Scholar]
- 28.Jault P, Leclerc T, Jennes S, Pirnay JP, Que YA, Resch G, Rousseau AF, Ravat F, Carsin H, Le Floch R, Schaal JV, Soler C, Fevre C, Arnaud I, Bretaudeau L, Gabard J. 2019. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis 19: 35–45. [DOI] [PubMed] [Google Scholar]
- 29.Nobrega FL, Costa AR, Kluskens LD, Azeredo J. 2015. Revisiting phage therapy: new applications for old resources. Trends Microbiol 23: 185–191. [DOI] [PubMed] [Google Scholar]
- 30.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25: 730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motooka D, Fujimoto K, Tanaka R, Yaguchi T, Gotoh K, Maeda Y, Furuta Y, Kurakawa T, Goto N, Yasunaga T, Narazaki M, Kumanogoh A, Horii T, Iida T, Takeda K, Nakamura S. 2017. Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals. Front Microbiol 8: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372: 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]