Abstract
P fimbriae of extraintestinal pathogenic Escherichia coli mediate digalactoside-specific adherence via the tip adhesin molecule PapG, which occurs in three known variants (I to III), which are encoded by the corresponding three alleles of papG. In the present study, newly discovered variants of papG allele I and the respective wild-type source strains were characterized. One of the new papG allele I variants conferred a unique agglutination phenotype that combined the phenotypes associated with papG alleles I, II, and III. Comparative hydrophilicity analysis of predicted PapG peptides revealed regions that might explain the observed phenotypic similarities and differences between the PapG variants. The new papG allele I variants occurred either as the sole papG allele or together with both papG alleles II and III, rather than with only papG allele III, as in archetypal strains J96 and CP9. They also occurred in the absence of the usual F13 papA allele. One of the new papG allele I variants occurred in a serogroup O6 strain that, according to random amplified polymorphic DNA analysis, was phylogenetically distant from the “J96-like” clonal group of E. coli O4:H5, which includes all previously identified examples of papG allele I. Cluster analysis of nucleotide and predicted peptide sequences suggested that papG allele I represents the earliest evolutionary branch from a common papG ancestor. These results demonstrate unexpected diversity within papG allele I and, together with previous findings, suggest that the J96-like clonal group of E. coli O4:H5 may represent the original source of papG within the species.
P fimbriae of extraintestinal pathogenic Escherichia coli (ExPEC) are heteropolymeric proteinaceous fibers that mediate adherence to Gal(α1-4)Gal-containing isoreceptors on host cell surfaces (13). PapG, the P fimbrial tip adhesin molecule, is responsible for digalactoside-specific receptor recognition and binding (10, 13, 38). A thin, flexible fibrillum connects PapG to the main fimbrial shaft (13). The fimbrial shaft in turn is composed of hundreds to thousands of identical PapA structural subunits, linked end-to-end by “donor strand exchange” to form a rigid alpha-helix which is attached to an anchor protein in the outer membrane (13, 48). Adherence mediated by P fimbriae contributes to the pathogenesis of extraintestinal infections such as pyelonephritis (17, 46) and may promote intestinal colonization of the host (59, 60).
PapG occurs in three known molecular variants (I to III), which are encoded by the three alleles of the corresponding gene, papG (32, 40). The three PapG variants exhibit subtly different receptor binding preferences (15, 19, 32–34, 36, 37, 40, 50, 52–55). They also exhibit divergent associations with clinical syndromes, with specific antigenic variants of the major structural subunit PapA, and with phylogenetic groups. PapG variant III (PapG III) requires for binding terminal substitutions on the Gal(α1-4)Gal consensus receptor (34, 36, 50, 53, 54). Thus, it mediates agglutination of sheep erythrocytes, which contain Forssman glycolipid [with its extended Gal(α1-4)Gal-containing oligosaccharide chain] and human erythrocytes (which contain the extended digalactoside-containing glycolipid sialosyl-Gal-globoside), but not Gal(α1-4)Gal-coated latex beads (P beads) or neuraminidase-treated human type O erythrocytes (25, 34, 36, 50, 53). Clinically, PapG III is associated with cystitis in humans and with genitourinary infections in dogs and cats (16, 25, 26). PapG III typically occurs in conjunction with the F12, F13, and F14 PapA variants (22, 31) and is concentrated within E. coli phylogenetic group B2 (22, 40).
PapG variant II (PapG II) binds well to both terminally substituted and nonsubstituted Gal(α1-4)Gal-containing isoreceptors and hence mediates agglutination of sheep erythrocytes, human erythrocytes (irrespective of neuraminidase treatment), and P beads (25, 34, 36, 50, 53). Clinically, it is associated with pyelonephritis and bacteremia in humans (16, 18, 21, 43). It often occurs in conjunction with the F7-2, F10, and F11 PapA variants (22, 31). It is distributed across E. coli phylogenetic groups B2 and D and occurs sporadically in other phylogenetic groups (22, 40).
PapG variant I (PapG I), although able to bind both substituted and nonsubstituted Gal(α1-4)Gal-containing isoreceptors, agglutinates sheep erythrocytes poorly but agglutinates human erythrocytes and P beads well (25, 36, 39, 40, 53, 54). Until recently, PapG I had been encountered only in pyelonephritis isolate J96 (O4:K−:H5) (27), which has two pap operons, one with papG allele I and the other with papG allele III (33, 39); both operons contain the F13 papA allele (31, 39). Recently, however, a disseminated clonal group of “J96-like” strains of E. coli O4:H5 was discovered that includes archetypal extraintestinal pathogenic strains J96 and CP9 (27, 29). The members of this clonal group, like J96 and CP9, typically contain papG alleles I and III plus the F13 papA allele, with or without another papA allele (27, 29). These findings suggested that although papG allele I is not unique to strain J96, it nonetheless may be restricted to the J96-like clonal group and may occur only in conjunction with the F13 papA allele; hence, it may represent a comparatively recent evolutionary development within (or acquisition by) E. coli.
In this report we describe wild-type strains and variants of PapG I that contradict conventional assumptions regarding PapG I. The new findings include molecular variants of PapG I that differ both structurally and functionally from PapG I of strain J96, that occur either together with PapG II and PapG III or alone, that occur in the absence of the F13 papA allele, and that appear in lineages distant from the J96-like clonal group. Evidence also is presented that the PapG I line may actually represent the earliest evolutionary branch from a common PapG ancestor and that the J96-like clonal group may represent the original source of papG within E. coli.
MATERIALS AND METHODS
Strains.
Canine urinary tract infection (UTI) isolates 7U and L31 and canine rectal isolate 7R, which was from the same host as strain 7U (original designations 5557 [7U], 5808 [L31], and 5565 [7R]), were obtained from Gerald Ling. The collection from which 7U and 7R were derived comprised paired pap-positive urine and rectal isolates from eight dogs with UTI, representing serotypes O4, O6, O120, and O149 (25). Isolate L31 was from an unpublished collection of paired urine and rectal isolates from 60 dogs with UTI, representing serotypes O1, O2, O4, O6, O7, O8, and O25, plus 10 other non-UTI-associated O antigens. Strain C367-82, an O4 urine isolate from a human with UTI, was provided by Flemming Scheutz. C367-82 was derived from an unpublished collection comprising approximately 328 isolates of serogroup O4. Archetypal ExPEC strains J96 and CP9 (O4:H5), members of the J96-like clonal group, were used as controls for papG alleles I and III (25, 27). Strain IA2, a non-J96-like O4 isolate, was used as a control for papG allele II (5, 29). Other wild-type comparator strains included BOS080 and BOS060 (papG allele III) (21), CL021 (papG alleles II and III) (23), HU968-298 (sfa: S fimbriae) (25), COR149 (O4, papG allele II; provided by Jorge Blanco), 536 (O6, papG allele III) (9), BOS050 (O6, papG allele III) (21, 41), and R61 (O6, papG allele III). Recombinant comparator strains included P678-54/pRHU845 (papG allele I) (33), HB101/pDC1 (papG allele II) (5), and HB101/pJFK102 (papG allele III) (33).
Detection of pap elements and other putative E. coli virulence genes.
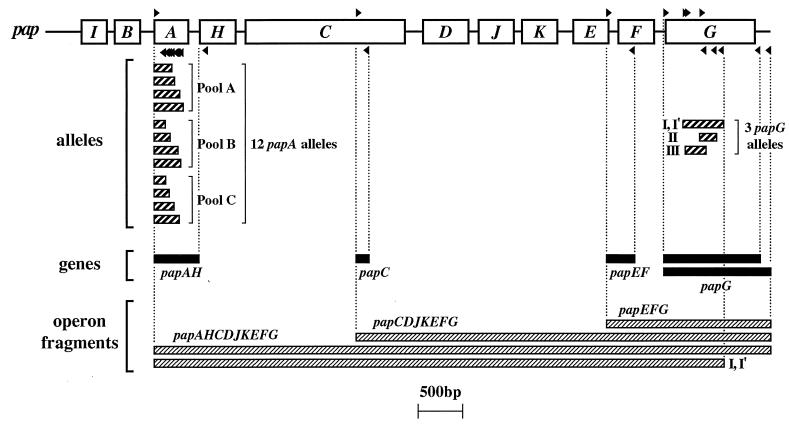
The three papG alleles were detected both by PCR using allele-specific internal primers (Fig. 1) and by dot blot hybridization in which the allele-specific PCR products were digoxigenin labeled and used as probes, as previously described (20, 26). papG also was detected by using flanking primers, as previously described, including a consensus forward primer that detects all three papG alleles and two reverse primers, one specific for the region downstream from papG allele I in J96 and one specific for the consensus region downstream from papG alleles II and III in strains IA2 and J96, respectively (Fig. 1) (25, 30). Other pap regions (papAH, papC, and papEG) were detected by PCR, as previously described (Fig. 1) (30).
FIG. 1.
Map positions of pap primers and corresponding PCR products. Open boxes represent genes within the pap operon. Forward and reverse primers (right-pointing and left-pointing black triangles, respectively, above and below the pap operon) are used in combinations as shown to yield the indicated PCR products (thin rectangles below the pap operon). Heavily striped rectangles indicate papA and papG allele PCR products; solid black rectangles indicate pap gene PCR products; finely striped rectangles indicate long PCR operon fragments (as generated using either flanking or internal allele-specific papG reverse primers, as illustrated for allele I-I′). Adapted from Fig. 1 of reference 25.
The forward primers for papA, papC, and papE also were used in combination with reverse (internal or flanking) allele-specific papG primers to generate long pap amplicons, as a way to assess the integrity and extent of the pap operons in which specific papG copies were situated (Fig. 1). Long-product PCR was done using the LA PCR kit, version 2.1 (Takara Shuzo Biomedical Group, Otsu, Shiga, Japan) as specified by the manufacturer, with annealing temperatures adjusted empirically to achieve optimal amplification.
papA alleles corresponding to the 11 serologically defined P fimbrial F types and the recently described F48 PapA variant were detected by PCR, as previously described (Fig. 1) (31). Twenty-five non-pap putative virulence factor (VF) genes of ExPEC were detected by a combination of PCR and dot blot hybridization, as previously described (28, 30). Throughout, boiled lysates were used as template DNA and PCR products were resolved by size in agarose gels. All assays were done at least in duplicate, with additional determinations used as needed to resolve discrepancies. Appropriate positive and negative controls were included in all assays.
Nucleotide sequencing and sequence analysis.
After column purification, selected full-length papG amplicons underwent direct sequencing of both strands by using a dye terminator method and an automated DNA sequencer, as previously described (25, 31). Newly determined papG sequences were compared with all available papG sequences from the databanks, as previously described (25) and, to provide an outgroup, with the fimH sequence from strain J96 (GenBank accession no. AF089840) and the sfaS sequence from strain 536 (GenBank accession no. X16664). Sequences were edited and assembled, and alignments of predicted mature PapG, FimH, or SfaS peptides (and of the corresponding nucleotide sequences) were made using CLUSTAL-W (57). From these alignments, trees were inferred by the neighbor-joining (NJ) method (47) and the unweighted paired group method with averaging (UPGMA) (51) by using the application MEGA (S. Kumar, K. Tamura, and M. Nei, MEGA: Molecular Evolutionary Genetics Analysis, version 1.01, 1993). Hydrophilicity plots were generated by the method of Kyte and Doolittle (35) for a representative of each of the three allele-specific variants (classes) of PapG and for the newly identified PapG I′ variant from strain 7U.
RAPD analysis.
To assess the phylogenetic context within which the various papG variants occurred, selected wild-type strains were subjected to random amplified polymorphic DNA (RAPD) analysis with (separately) arbitrary decamer primers 1247, 1254, and 1281, as previously described (2, 58), and commercial PCR beads (24, 25). Single-primer RAPD fingerprints were digitally combined head-to-tail to create a “virtual” composite fingerprint for each strain by using a computerized image analysis system (Molecular Analyst; Bio-Rad, Hercules, Calif.). (24, 25). Composite fingerprints were then compared in a pairwise fashion by using Pearson's correlation coefficient to assess similarities between analog densitometric scans of each gel track. From the resulting similarity matrix, a dendrogram was inferred by using UPGMA (24, 25).
Hemagglutination phenotypes.
Strains were tested for mannose-resistant hemagglutination (MRHA) of diverse erythrocyte types, with or without neuraminidase pretreatment of the erythrocytes, in the presence or absence of pigeon egg white (an inhibitor of P fimbriae) or fetuin (an inhibitor of S fimbriae), by using microscope slide assays as previously described (25, 45). MRHA inhibition and neuraminidase sensitivity were defined as a two-level decrease in the intensity of MRHA in the presence of inhibitor or following neuraminidase treatment of erythrocytes, respectively. P-pattern MRHA was defined as MRHA inhibition by pigeon egg white, and S-pattern MRHA was defined as inhibition by fetuin. P beads were from Chembiomed (Alberta, Canada; now defunct). Human A1P1 and OP1 erythrocytes were from the investigators (J.R.J. and T.T.O., respectively). Human Pk2 erythrocytes were provided by Jane Swanson.
Nucleotide sequences accession numbers.
Newly determined papG allele I sequences were deposited in GenBank under accession numbers AF237471 (strain 7U), AF247505 (strain L31), AF237476 (strain C367-82), and AF237473 (CP9).
RESULTS
Discovery of a molecular variant of papG allele I.
Strains 7U and 7R, which are matching urine and rectal E. coli isolates from an individual canine host, were previously shown by allele-specific internal primer PCR to contain papG alleles II and III but not papG allele I (25). However, when amplified using a universal papG forward primer in combination with flanking reverse primers specific for papG alleles II and III versus papG allele I, respectively (Fig. 1), these strains yielded not only the expected ∼1,100-bp PCR product with the papG allele II- and III-specific reverse primer but also (like strains J96 and CP9) a ∼1,200-bp PCR product with the papG allele I-specific reverse primer. The possibility of a chimeric papG region consisting of papG allele II or III recombined with allele I-specific 3′ flanking sequence, although consistent with the initial PCR results, was excluded by reassortment experiments in which each of three individual papG allele-specific internal forward primers was combined (separately) in PCR with the papG allele I-specific flanking reverse primer without producing an amplicon (data not shown).
Direct nucleotide sequencing of the putative papG allele I PCR product from strain 7U yielded a sequence 90.9% identical to papG from J96, the only version of papG allele I for which the sequence was available prior to the present study, and the closest match in the sequence databases to the new papG sequence (Fig. 2). Polymorphisms in this new papG variant at the predicted binding sites for both the published forward and reverse papG allele I-specific internal primers (20), in comparison with papG allele I from J96 (five and two nucleotides, respectively) (40), were sufficient to explain the failure of the published allele I primers to react with the new papG variant (data not shown). The new papG allele I variant from strains 7U and 7R was designated papG allele I′. Dot blot hybridization using allele-specific papG probes (26) showed that strains 7U and 7R hybridized with all three probes, consistent with presence of all three papG alleles, as suggested by the PCR results (data not shown).
FIG. 2.
Sequence comparisons of five papG allele I or I′ variants and corresponding predicted PapG peptides, identified as to strain name and allele. Alignments were by CLUSTAL-W (57). Positions in the gene, as numbered from the putative start codon, or in predicted mature peptides, as numbered from the putative amino-terminal residue (31), are shown above polymorphisms. Only positions at which at least one variant differs from strain J96 are included. Dashes indicate identity to J96; asterisks indicate a gap. In the peptide alignment, symbols below the polymorphism data signify the folowing: colon, highly similar amino acids; period, somewhat similar amino acids; blank, different amino acids. The single-letter amino acid code is used.
Alternative primers were designed to recognize approximately the same sites in the new papG variant as recognized by the published allele I-specific primers in papG allele I from J96. When substituted for the published papG allele I-specific internal primers, the new primers (AlleleI′-f: 5′-CTACTATAGTTCATGCTCAGGTC-3′, bases 183 to 205; AlleleI′-r: 5′-CTGACATCCTCCAACATTATCGA-3′, bases 657 to 635) yielded the predicted ∼470-bp PCR product with strains 7U and 7R but not with strain J96 or any of the papG allele I-, II-, or III-containing positive control strains tested (data not shown). When the new primers were added to the allele-specific multiplex papG PCR primer pool along with the three published allele-specific primer pairs (20), control strains for papG alleles I, II, and III, as well as strain 7U and 7R, yielded combinations of PCR products of the sizes expected for each strain's known papG allele content, with no spurious products (data not shown).
Discovery of additional examples of papG allele I′.
The modified allele-specific papG primer pool, now containing the new allele I′-specific primers, was used to screen a collection of approximately 60 canine UTI isolates of diverse serotypes and a collection of approximately 300 clinical isolates of serogroup O4 from animals and humans. Strains that were positive for an allele I-specific product were tested with the allele I and allele I′ primers separately. Within each collection, a single strain was identified that yielded an appropriately sized PCR product (∼470 bp) with the new allele I′ primers but not the standard allele I primers (Table 1).
TABLE 1.
Characteristics of E. coli isolates containing new variants of papG allele I (i.e., I′) versus comparator strains
| Serogroup and strain(s)a | This study | Hostb | Serotypec | Presence ofd:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
pap
|
sfa/foc | sfaS | focG | iha | fim | hly | cnf1 | cdt | fyuA | iroN | iutA |
kpsMT
|
rfc | traT | malX | |||||||||
| F type | A | C | EF | allele | II | III | ||||||||||||||||||
| Serogroup O4 | ||||||||||||||||||||||||
| J96 | H | O4:K-:H5 | 13 | + | + | + | I, III | + | − | + | − | + | + | + | − | + | + | − | − | + | + | + | + | |
| CP9 | H | O4:54:H5e | 13, 14 | + | + | + | I, III | + | − | + | − | + | + | + | − | + | + | − | − | + | + | − | + | |
| C367-82 | + | H | O4:H5 | 7–2, 10 | + | + | + | I, II, III | + | − | + | − | + | + | + | − | + | + | − | − | + | + | + | + |
| 7U and 7R | + | D | O4:H5 | 10 | + | + | + | I′, II, III | + | − | − | − | + | + | + | − | + | + | − | + | − | + | − | + |
| IA2 | H | O4:K12:H− | 11, 16 | + | + | + | II | + | − | − | − | + | + | − | − | + | − | + | + | − | + | + | + | |
| COR149 | H | O4:H5 | 10, 15 | + | + | + | II | + | − | + | + | + | + | − | − | + | + | − | + | − | − | − | + | |
| Serogroup O6 | ||||||||||||||||||||||||
| 536 | H | O6:K15:H31 | “536” | + | + | + | III | + | + | − | − | + | + | + | − | + | + | − | − | − | − | − | + | |
| BOS050 | H | O6:HU | 48 | + | + | + | III | + | + | − | − | + | + | + | − | + | + | − | + | − | − | + | + | |
| L31 | + | D | O6 | 12, 15 | + | + | + | I′ | + | + | − | − | + | + | + | − | + | + | − | + | − | − | + | + |
| R61 | H | O6:H31 | 12 | + | + | + | III | + | + | − | − | + | + | + | − | + | − | − | + | − | − | − | + | |
Strains C367-82, 7U and 7R, and L31 (“This study”) contained variants of papG allele I that were not detected by published primers based on papG allele I from J96. Reference strains J96 and CP9, which are archetypal members of the J96-like clonal group of serogroup O4, and strain IA2, which is an archetypal non-J96-like O4 strain, were included for comparison with O4 isolates C367-82 and 7U-7R. Reference strain COR149 was included for comparison with strain L31 because of its F15 papA allele content and O4 antigen status (unpublished data). Reference strains 536, BOS050, and R61 were included since they resembled strain L31 with respect to virulence gene profile and O6 antigen expression.
H, human; D, dog.
Serotypes shown include only antigens for which the particular strains were tested. Hu, H untypeable.
F type, papA allele (as determined by PCR). Allele, papG allele. (All strains also were positive for papG per se with flanking primers.) sfa/foc, central consensus regions of S fimbrial and F1C fimbrial operons; sfaS, S fimbrial adhesin; focG, F1C fimbrial adhesin; iha, novel putative adhesin-siderophore gene; fim, type 1 fimbriae; hly, hemolysin; cnf1, cytotoxic necrotizing factor; fyuA, yersiniabactin (siderophore); iroN, novel putative siderophore; iutA, aerobactin receptor; kpsMT, capsule synthesis (group II or III); rfc, O4 lipopolysaccharide synthesis; traT, serum resistance-associated gene; malX, marker for pathogenicity-associated island from strain CFT073 (28, 30). All strains were negative for afa/dra (Dr-binding adhesins), bmaE (M adhesins), gafD (G fimbriae), nfaE (nonfimbrial adhesins), cdtB (cytolethal distending toxin), the K1 variant of kpsMT II, and ibeA (invasion of brain endothelium) (30).
Strain CP9 is serologically positive for K10, K54, and K96 antigens.
One of the two newly detected putative allele I′-positive strains, C367–82, was a human UTI isolates of serotype O4:H5, which, like canine isolates 7U and 7R, also contained papG alleles II and III and the F10 papA allele. When amplified with papG-flanking primers, this strain yielded both the ∼1,200-bp papG allele I-I′ PCR product and the ∼1,100-bp papG allele II-III product. Like strains 7U and 7R, this strain also hybridized with all three allele-specific papG probes (data not shown).
The other putative new papG allele I′-positive strain, canine UTI isolate L31, was of serogroup O6, which was surprising since papG allele I had previously been described only in strains of serogroup O4. Although strain L31 was positive for the F12 and F15 papA alleles, which suggested the presence of two copies of the pap operon, unlike strains 7U, 7R, and C367-82, it was negative by internal primer PCR for both papG alleles II and III; i.e., it had allele I′ as its only papG allele. This was confirmed by papG-flanking primer PCR, which yielded only the ∼1,200-bp putative allele I-I′-specific product, and by dot blot hybridization of genomic DNA using allele-specific probes for all three papG alleles (data not shown). These findings also were surprising, since papG allele I previously had been encountered only in combination with papG allele III and in conjunction with the F13 papA allele (27, 29, 31).
Analysis of predicted PapG peptides.
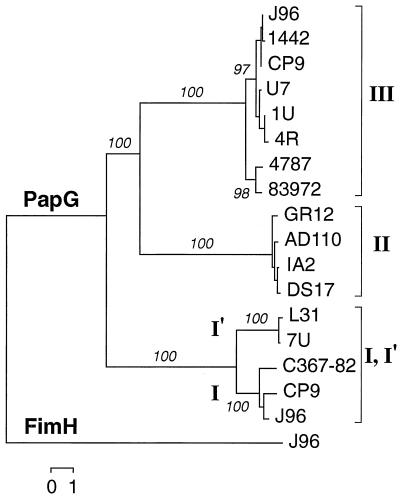
The papG allele I sequence was determined for putative allele I′-containing strains L31 and C367–82 and for strain CP9, which was used as a second example (in addition to papG from J96) of the “orthodox” version of papG allele I (27). The four corresponding predicted PapG I peptides, and PapG I from strain 7U, were compared with all other available versions of PapG from the databases (25). In a similarity dendrogram that was rooted by using the type I fimbrial adhesin molecule FimH as an outgroup, the five predicted PapG I (I′) peptides formed a cluster that was quite distant from the cluster containing representatives of PapG II and PapG III (Fig. 3). In both the NJ dendrogram (Fig. 3) and an UPGMA dendrogram (not shown), the PapG I (I′) group was the first to branch off from an ancestral PapG trunk, suggesting that it represented the most basal or primitive version of PapG. Similar conformation trees were obtained when SfaS was used as the outgroup and when nucleotide data were used instead of peptide data (results not shown).
FIG. 3.
Dendrogram of predicted PapG and FimH peptides. Alignment of predicted mature peptides was done by CLUSTAL-W (57). Tree construction was done by the NJ method (47). PapG variants are identified as to class (brackets: I, I′, II, or III) and individually according to strain name. FimH from strain J96 (shown) and SfaS (data not shown) were used to provide outgroups to root the tree. Similar tree conformations were obtained by UPGMA and the NJ method, whether nucleotide or peptide sequences were analyzed (data not shown). Numbers (in percentages) indicate reproducibility of nodes in bootstrap analysis.
In these dendrograms, the PapG I cluster was split into two subclusters, one of which contained the PapG variants from (canine) isolates 7U and L31 and one of which contained the PapG variants from (human) isolates J96, CP9, and C367-82 (Fig. 3). Thus, although papG from strain C367-82 was recognized only by the allele I′ internal primers, the gene and the corresponding PapG peptide (Fig. 2) actually represented variant I rather than variant I′. As with strain 7U, nucleotide sequence polymorphisms were identified within papG from strains C367-82 and L31 that predictably would interfere with binding of both the forward and reverse standard allele I internal primers (data not shown).
A comparison of peptide sequences between the I and I′ PapA variants (Fig. 2) showed that PapA I from strains CP9 and C367-82 were highly similar to PapA I from strain J96 throughout the amino-terminal half of the peptide, which putatively contains the receptor binding region (12). Polymorphisms were largely confined to the carboxy-terminal portion of the molecule, which putatively is responsible for subunit-subunit interactions (12). In contrast, PapA I′ peptides from L31 and 7U, which were highly similar to one another, differed from PapA I from J96 primarily within the amino-terminal (receptor binding) half of the molecule. However, they were no more divergent from PapA from J96 within the carboxy-terminal (subunit-subunit interactions) half of the molecule than were the PapA I variants from CP9 and C367-82 (Fig. 2).
Long-product PCR.
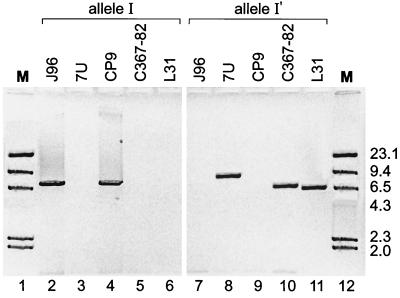
We next sought to determine whether the newly identified papG allele I variants were integrated into intact pap operons, which should allow for expression, or instead occurred as isolated operon fragments, in which case expression would not be expected. Long-product PCR “operon walking” was done by using (flanking or internal) papG allele I- and I′-specific reverse primers, in combination (individually) with forward primers for papA, papC, and papE (Fig. 1). From each of the controls strains (J96 and CP9) and from strains 7U, C367-82, and L31, suboperonic amplicons representing papEFG, papCDJKEFG, and papAHCDJKEFG were obtained, in each instance only with the reverse primers corresponding to the variant of papG allele I known to be present in the particular strain (Fig. 4). This demonstrated that each of the papG allele I variants was contiguous with a seemingly intact pap operon. However, whereas strains C367-82 and L31 yielded papAHCDJKEFG amplicons of approximately the same size as that obtained from strain J96, strain 7U yielded an amplicon that was approximately 1.4 kb longer than those obtained from strains J96 and CP9 (Fig. 4). This suggested the possibility of an insertion or partial duplication within the allele I′ pap operon of strains 7U that might account for the previously noted MRHA negativity of 7U (25). As predicted, long PCR products (i.e., papAHCDJKEFG and/or papCDJKEFG) also were obtained with the reverse papG allele II and III primers with strains 7U, 7R, and C367-82 but not with strain L31 (data not shown).
FIG. 4.
Long PCR products from strains containing papG allele I or I′. Amplification was done using the universal papA forward primer combined with the allele-specific papG reverse primer for either allele I (left, lanes 2 to 6) or allele I′ (right, lanes 7 to 11). With the published papG allele I reverse primer, bands of the size expected for the papAHCDJKEFG amplicon from strain J96 (∼7,540 bp [40]) were obtained with strains J96 (lane 2, ∼7,430 bp) and CP9 (lane 4, ∼7,540 bp) whereas no amplicon was obtained with strains 7U (lane 3), C367-82 (lane 5), or L31 (lane 6). In contrast, with the new papG allele I′ reverse primer, comparable bands were obtained with strains 7U (lane 8, ∼8,950 bp), C367-82 (lane 10, ∼7,280 bp), and L31 (lane 11, ∼6,920 bp), whereas no amplicon was obtained with strains J96 (lane 7) or CP9 (lane 10). Lanes 1 and 12, molecular weight marker (M, HindIII-restricted lambda DNA).
Agglutination phenotypes.
Strain L31, which had PapG I′ as its sole PapG variant and was MRHA positive, provided the only opportunity to examine the phenotype correlates of PapG I′ in the absence of confounding from a coexpressed alternative PapG variant. The agglutination phenotype of L31 represented a unique combination of reaction patterns that included agglutination of P beads (characteristic of PapG I and II) and of sheep erythrocytes (characteristic of PapG II and III), but also NS-pattern MRHA of human OP1 but not AP1 erythrocytes (suggestive of PapG III only, according to previous data [25]; specific to PapG I′ in the present study) (Table 2). Confounding by coexpression of S fimbriae, which was possible given the presence of sfa/focDE and sfaS in strain L31 (Table 1), was excluded by MRHA inhibition assays that showed P-pattern MRHA with L31 (i.e., inhibition of MRHA by pigeon egg white but not by fetuin) and S-pattern MRHA with the S fimbria control (i.e., inhibition of MRHA by fetuin but not by pigeon egg white) (Table 2).
TABLE 2.
Agglutination phenotypes associated with PapG variants and S fimbriae
| Adhesina | Agglutination intensity
|
HA inhibition (A1P1 RBC)c
|
Neuraminidase sensitivity of HAc
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| A1P1, OP1, or pig RBCb | Sheep RBC | Pk1 RBC | P beads | Pigeon egg white | Fetuin | A1P1 RBC | OP1 RBC | Sheep RBC | |
| PapG III | +++ | ++++ | − | − | Yes | No | No | Vard | No |
| PapG II | ++++ | +++ | +++ | +++ | Yes | No | No | Var | No |
| PapG I + III | ++++ | ++++ | +++ | ++++ | Yes | No | No | Var | No |
| PapG I | +++ | + | ++ | +++ | Yes | No | No | Var | NAe |
| PapG I′ | +++ | ++++ | ++ | +++ | Yes | No | No | Yes | No |
| S fimbriae | +++ | +++ | ++ | − | No | Yes | Yes | Nof | Yes |
Strains used included BOS080, BOS060, and P678-54/pJFK102 (PapG III), IA2 and HB101/pDC1 (PapG II), J96 (PapG I + III), P678-54/pRHU845 (PapG I), L31 (PapG I′), and HU968-298 (S fimbriae).
RBC, erythrocytes. A1P1, OP1, and Pk1 RBC are from human donors.
HA, hemagglutination. Inhibition and sensitivity defined as at least a two-level decrease in agglutination intensity (strongest to weakest, ++++ to −) in the presence of inhibitor or after neuraminidase digestion, respectively.
var., variable between experiments.
NA, not applicable (no HA of sheep RBC).
HA intensity decreased by one intensity level with neuraminidase digestion in both of two trials.
Hydrophilicity analysis.
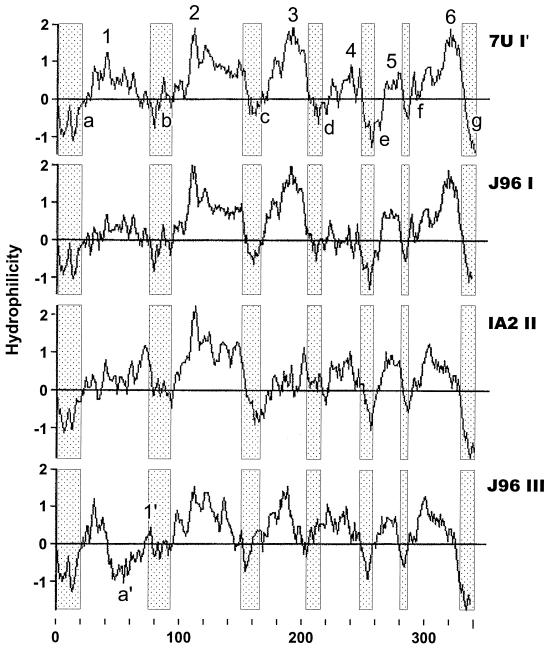
To explore the possible structural basis for the phenotypic differences noted between PapG I′ from 7U and conventional PapG I, II, and III (Table 2), hydrophilicity plots for these predicted PapG peptides were generated by the method of Kyte and Doolittle (35) and were aligned for comparative analysis (Fig. 5). The four PapG variants exhibited a consensus hydrophilicity profile characterized by six hydrophilic maxima (peaks 1 to 6) flanked by seven hydrophilic minima (valleys a to g), which included the amino- and carboxy-terminal regions of the peptides (Fig. 5). The only significant deviations from this pattern were exhibited by PapG III, which exhibited a secondary hydrophilicity minimum (designated a′) and a secondary hydrophilicity peak (designated I′) between minima a and b (Fig. 5). The hydrophilicity profiles of PapG I′ and PapG I were similar, except that in PapG I′ peaks 1 and 4 were higher and peak 5 was narrower (Fig. 5). Comparisons according to agglutination phenotypes revealed that the PapG I′, II, and III peptides, which were associated with MRHA of sheep erythrocytes, exhibited higher maximal hydrophilicity values for peaks 1, 3, and 4 than did the PapG I peptide, which was not associated with this phenotype. Similarly, the PapG I′, I, and II peptides, which conferred agglutination of human Pk2 erythrocytes and P beads, exhibited higher hydrophilicity values for peak 2 than did the PapG III peptide, which was not associated with these phenotypes.
FIG. 5.
Hydrophilicity profiles of PapG peptides I′, I, II, and III. Hydrophilicity was assessed by the method of Kyte and Doolittle (35), based on predicted mature PapG peptides from strains 7U (PapG I′) (top), J96 (PapG I) (second from top), IA2 (PapG II) (second from bottom), and J96 (PapG III) (bottom). The horizontal axis shows residue number in the mature peptide; the vertical axis shows the hydrophilicity index. Shaded regions identify conserved hydrophilicity minima (a to g, present in all variants; a′, unique to PapG III); intervening regions are hydrophilicity maxima (1 to 6, present in all variants; 1′, unique to PapG III).
Clonal associations.
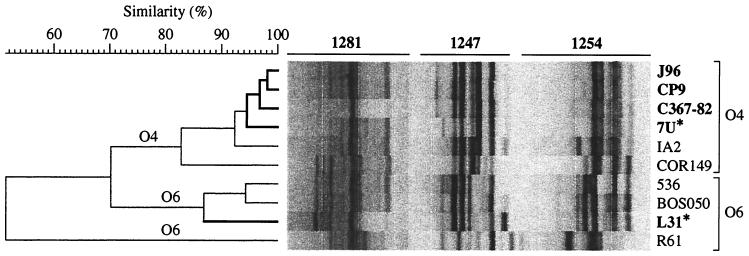
The unusual occurrence in a serogroup O6 isolate (strain L31) of a version of papG allele I, which previously had been identified only within serogroup O4, suggested the possibility of horizontal transfer of O antigen determinants from an O6 donor into a papG allele I′-containing O4 recipient or, alternatively, transfer of papG allele I′ from an O4 donor into an O6 recipient. To differentiate between these two hypotheses, strains 7U, C367-82, and L31 were compared with a panel of control O4 and O6 strains, which were selected on the basis of their status as archetypal representatives of the J96-like versus non-J96-like clonal groups within serogroup O4 (strains J96, CP9, and IA2, respectively) or their similarities to strain L31 with respect to O antigen, VF profiles, and/or papA allele content (Table 1). In a UPGMA-derived similarity dendrogram based on three-primer composite RAPD fingerprints, the papG allele I-containing O4 strains clustered closely within the larger O4 cluster (Fig. 6). Consistent with its O6 antigen status, strain L31 was placed among the O6 controls, well removed from the O4 cluster and the other papG allele I-containing strains, including the F15 papA allele-positive O4 strain COR149 (Fig. 6). This supported the hypothesis of horizontal transfer of papG allele I′ from an O4 ancestor into a recipient within an O6 clonal group.
FIG. 6.
Phylogenetic analysis of PapG I′-containing strains and comparator strains. Pearson correlation coefficient analysis of composite RAPD fingerprints from primers 1281, 1247, and 1254 was used to establish pairwise similarity relationships between strains, which were then used construct a dendrogram by UPGMA (51). Strains that contain papG allele I or I′ (asterisk) are shown in boldface. Strains of serogroups O4 (top) and O6 (bottom) are bracketed. Note that (papG allele I-containing) strain L31, which serologically is an O6 strain, is placed with the O6 comparator strains according to RAPD analysis. Strains J96, CP9, C367-82, and 7U constitute a “J96-like” subcluster within the larger O4 cluster. (Strains are shown in the same sequence in Table 1 as in this RAPD-based dendrogram.)
DISCUSSION
In the present study we used molecular and phenotypic methods to characterize novel variants of papG allele I and the corresponding wild-type E. coli strains. The new papG allele I variants exhibited unconventional agglutination phenotypes, papG and papA allele environments, and clonal associations. Comparative sequence analysis suggested that the papG allele I family may represent the earliest evolutionary branch from a common papG ancestor. In conjunction with previous findings, the data support the hypothesis that the J96-like clonal group of E. coli O4:H5 may represent the origin of papG within the species.
Our serendipitous discovery of the first known molecular variant of papG allele I led to the demonstration that genetic diversity is actually greater among representatives of papG allele I than among the known representatives of papG alleles II or III. We found that the various versions of papG allele I (or of the PapG I peptide) segregated into two clusters. One cluster included PapG I from strains J96, CP9, and C367-82, despite the reactivity of the latter strain with the alternate papG allele I′ primers rather than with standard papG allele I primers. The other cluster included PapG I from canine isolates L31 and 7U. We termed this variant PapG I′ and the corresponding gene papG allele I′. The novel agglutination phenotype of strain L31, which expressed papG allele I′ in the absence of another papG allele, suggested that PapG I′ confers a combination of the agglutination phenotypes associated with PapG I or II and those associated with PapG III. Comparison of the distinctive hydrophilicity profile of PapG I′ with the hydrophilicity profiles of the three traditional PapG variants identified regions that could be responsible for the observed phenotypic similarities and differences between these four adhesin molecules. Ideally, the adherence phenotype conferred by papG allele I′ should also be assessed using a recombinant strain to avoid artifacts from other adhesins and surface structures expressed by wild-type strains such as L31. However, recombinant strains may be associated with their own artifacts (11, 19).
In each strain analyzed, each copy of papG appeared to be part of an intact pap operon, as suggested by long-product PCR, which was done by using reverse primers specific for each allele of papG in combination with forward primers for upstream genes within the pap operon, as far back as papA. A possible explanation for the apparent nonexpression of papG allele I′ by strains 7U and 7R was provided by the unusually long papAHCDJKEFG PCR products that were obtained from these strains with the papG allele I′ reverse primer. These extra-long amplicons suggested the presence of insertions or partial duplications somewhere within the corresponding pap operons in these strains compared with strains J96 or CP9 (40).
The findings of the present study show that in contrast to what has been thought previously on the basis of available evidence, papG allele I (or I′) can coexist with papG allele II; can occur alone, i.e., in the absence of another papG allele; and can appear in evolutionary lineages distant from the J96-like O4:H5 clonal group (21, 27, 29). Moreover, rather than being a recent evolutionary development within (or acquisition by) E. coli, the papG allele I family appears to represent the most primitive branch of the papG tree. These observations present a series of seeming paradoxes. If papG allele I (I′) is ancestral within E. coli, is compatible with each of the other papG alleles, and is horizontally mobile between lineages, what can account for its comparative rarity (particularly papG allele I′) within the E. coli population, its near-total confinement to the J96-like clonal group, and its consistent association with papG allele III, almost to the exclusion of papG allele II (18, 21, 26, 27, 29, 40)?
A possible scenario would be that the J96-like clonal group, which uniquely contains representatives of all three papG alleles (both collectively and within individual strains, as demonstrated here for the first time) (18), may be the ancestral home of papG within E. coli. papG allele I, putatively the most primitive form of papG, by itself may confer only a minor fitness advantage. However, it may synergize with (presumably later-evolving) papG allele III, but not with (presumably even later-evolving) papG allele II, to confer a greater fitness advantage than does papG allele III alone. The presumably minimal selective advantage conferred by papG allele I itself thus would limit the driving force for horizontal spread of this papG allele. In contrast, papG allele III, which presumably confers a greater fitness advantage than does papG allele I, has spread to (and has been maintained within) multiple lineages within phylogenetic group B2 (21, 30, 40). (The logistic difficulties of coordinate horizontal transfer of both papG alleles I and III, which occur on separate pathogenicity islands, may account for the limited diffusion of this papG allele combination, despite its presumably greater selective advantage than that of either papG allele I or III individually.) Finally, papG allele II, which hypothetically confers an even greater fitness advantage than does papG allele III, has been propelled by selective pressure horizontally from the J96-like clonal group not only throughout phylogenetic group B2 but also into phylogenetic group D and beyond (21, 30, 40). This scenario represents a substantial departure from the hypothesis that papG allele I is the most recently evolved (or acquired) papG variant (21, 27, 29).
The occurrence in strain L31 of papG allele I′ by itself, and in a lineage other than the J96-like clonal group, suggests that this version of papG allele I may confer a greater selective advantage than does papG allele I from strain J96. If so, the even greater rarity of papG allele I′ than of papG allele I per se presents another paradox to be explained.
In the present study, both examples of papG allele I′ were from canine UTI isolates (strains 7U and L31). In contrast, a papG allele I variant that was initially detected only by the allele I′ primers but that according to full-length DNA sequence actually represented “orthodox” papG allele I was from a human UTI isolate (strain C367-82). These data might be interpreted as suggesting that papG allele I′ is specific to dogs. However, the limited sample size precludes firm conclusions. It must be remembered that for some time after its discovery, papG allele III was presumed to be specific to dogs (3, 6, 40, 53), whereas it is now known to be common also among human isolates from diverse clinical syndromes (1, 14, 16, 21, 26, 42). Additional investigation is needed to define the epidemiological associations and host range of papG allele I′.
The varied papA alleles encountered in the present study in conjunction with papG alleles I and I′ provide additional evidence that horizontal transfer and recombination involving papA can proceed independently of papG. Thus, horizontal transfer of virulence genes can be envisioned as involving entire pathogenicity islands or large fragments thereof (i.e., of multiple virulence operons) (7, 8, 49, 56), single operons such as pap (4, 40, 44), or individual genes from within these operons, such as papA or papG (28, 31, 40). Possible driving forces for reassortment of papA and papG alleles through horizontal transfer may include the antigenic diversity provided by the various PapA structural subunit variants (31) and the diversity of receptor recognition capabilities provided by the various PapG adhesion variants (15, 19, 32, 36, 37, 53, 54).
Summary.
In the present study we used molecular and phenotypic methods to characterize novel variants of papG allele I and the corresponding wild-type E. coli strains. The new papG allele I variants exhibited novel agglutination phenotypes, papG and papA allele environments, and clonal associations. Comparative sequence analysis suggested that the papG allele I family may represent the earliest evolutionary branch from a common papG ancestor. Together with previous findings, these data support the hypothesis that the J96-like clonal group of E. coli O4:H5 may represent the original source of papG within the species.
ACKNOWLEDGMENTS
Strains were generously provided by Jorge Blanco (COR 149), Gabriele Blum-Oehler (536), Steve Clegg (IA2 and HB101/pCD1), Sheila Hull (HB101/pJFK102, P678-54/pRHU845, and HU968-298), Candice Johnson (CL021), Gerald Ling (7U, 7R, and L31), Joel Maslow (BOS080 and BOS060), Flemming Scheutz (C367-82), and Ann Stapleton (R61). Dave Prentiss helped prepare the figures. Ann Emery assisted with manuscript preparation.
This work was supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.); National Institutes of Health grant DK-47504 (J.R.J.); and National Research Initiative Competitive Grants Program/USDA grant 00-35212-9408 (J.R.J.).
REFERENCES
- 1.Andreu A, Stapleton A E, Fennell C, Lockman H A, Xercavins M, Fernandez F, Stamm W E. Urovirulence determinants in Escherichia coli strains causing prostatitis. J Infect Dis. 1997;176:464–469. doi: 10.1086/514065. [DOI] [PubMed] [Google Scholar]
- 2.Berg D E, Akopyants N S, Kersulyte D. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol Cell Biol. 1994;5:13–24. [Google Scholar]
- 3.Beutin L. Escherichia coli as a pathogen in dogs and cats. Vet Res. 1999;30:285–298. [PubMed] [Google Scholar]
- 4.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clegg S. Cloning of genes determining the production of mannose-resistant fimbriae in a uropathogenic strain of Escherichia coli belonging to serogroup O6. Infect Immun. 1982;38:739–744. doi: 10.1128/iai.38.2.739-744.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia E, Bergmans H E N, van den Bosch J F, Orskov I, van der Zeijst B A M, Gaastra W. Isolation and characterization of dog uropathogenic Escherichia coli strains and their fimbriae. Antonie Leeuwenhoek. 1988;54:149–163. doi: 10.1007/BF00419202. [DOI] [PubMed] [Google Scholar]
- 7.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 8.Guyer D M, Kao J-S, Mobley H L T. Genomic analysis of a pathogenecity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect Immun. 1998;66:4411–4417. doi: 10.1128/iai.66.9.4411-4417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hacker J, Bender L, Ott M, Wingender J, Lund B, Marre R, Goebel W. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog. 1990;8:213–225. doi: 10.1016/0882-4010(90)90048-u. [DOI] [PubMed] [Google Scholar]
- 10.Hoschützky H, Lottspeich F, Jann K. Isolation and characterization of the α-galactosyl-1,4β-galactosyl-specific adhesin (P adhesin) from fimbriated Escherichia coli. Infect Immun. 1989;57:76–81. doi: 10.1128/iai.57.1.76-81.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hull R A, Nowicki B, Kaul A, Runyan R, Svanborg C, Hull S I. Effect of pap copy number and receptor specificity on virulence of fimbriated Escherichia coli in a murine urinary tract colonization model. Microb Pathog. 1994;17:79–86. doi: 10.1006/mpat.1994.1054. [DOI] [PubMed] [Google Scholar]
- 12.Hultgren S J, Abraham S, Caparon M, Falk P, St. Germe J W, Normark S. Pilus and non-pilus bacterial adhesins: assembly and function in cell recognition. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 13.Hultgren S J, Jones C H, Normark S. Bacterial adhesins and their assembly. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 14.Jantunen M E, Siitonen A, Koskimies O, Wikstrom S, Karkkainen U M, Salo I, Saxen H. Predominance of class II papG allele of Escherichia coli in pyelonephritis in infants with normal urinary tract anatomy. J Infect Dis. 2000;181:1822–1824. doi: 10.1086/315446. [DOI] [PubMed] [Google Scholar]
- 15.Johanson I, Lindstedt R, Svanborg C. Roles of the pap- and prs-encoded adhesins in Escherichia coli adherence to human uroepithelial cells. Infect Immun. 1992;60:3416–3422. doi: 10.1128/iai.60.8.3416-3422.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johanson I-M, Plos K, Marklund B-I, Svanborg C. pap, papG and prsG DNA sequences in Escherichia coli from the fecal flora and the urinary tract. Microb Pathog. 1993;15:121–129. doi: 10.1006/mpat.1993.1062. [DOI] [PubMed] [Google Scholar]
- 17.Johnson J R. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson J R. papG alleles among Escherichia coli strains causing urosepsis: associations with other bacterial characteristics and host compromise. Infect Immun. 1998;66:4568–4571. doi: 10.1128/iai.66.9.4568-4571.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson J R, Ahmed P, Brown J J. Diversity of hemagglutination phenotypes among P fimbriated wild-type strains of Escherichia coli according to papG repertoire. Clin Diagn Lab Immunol. 1998;5:160–170. doi: 10.1128/cdli.5.2.160-170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson J R, Brown J J. A novel multiply-primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(α1-4)Gal-binding PapG adhesins of Escherichia coli. J Infect Dis. 1996;173:920–926. doi: 10.1093/infdis/173.4.920. [DOI] [PubMed] [Google Scholar]
- 21.Johnson J R, Brown J J, Maslow J N. Clonal distribution of the three alleles of the Gal(α1-4)Gal-specific adhesin gene papG among Escherichia coli strains from patients with bacteremia. J Infect Dis. 1998;177:651–661. doi: 10.1086/514230. [DOI] [PubMed] [Google Scholar]
- 22.Johnson J R, Delavari P, Kuskowski M, Stell A L. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis. 2001;183:154–159. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- 23.Johnson J R, Johnson C E, Maslow J N. Clinical and bacteriologic correlates of the papG alleles among Escherichia coli strains from children with acute cystitis. Pediatr Infect Dis J. 1999;18:446–451. doi: 10.1097/00006454-199905000-00009. [DOI] [PubMed] [Google Scholar]
- 24.Johnson J R, O'Bryan T T. Improved repetitive element-(rep-) polymerase chain reaction (rep-PCR) fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin Diagn Lab Immunol. 2000;7:265–273. doi: 10.1128/cdli.7.2.265-273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson J R, O'Bryan T T, Low D A, Ling G, Delavari P, Fasching C, Russo T A, Carlino U, Stell A L. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli that express papG allele III. Infect Immun. 2000;68:3327–3336. doi: 10.1128/iai.68.6.3327-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson J R, Russo T A, Brown J J, Stapleton A. papG alleles of Escherichia coli strains causing first episode or recurrent acute cystitis in adult women. J Infect Dis. 1998;177:97–101. doi: 10.1086/513824. [DOI] [PubMed] [Google Scholar]
- 27.Johnson J R, Russo T A, Scheutz F, Brown J J, Zhang L, Palin K, Rode C, Bloch C, Marrs C F, Foxman B. Discovery of disseminated J96-like strains of uropathogenic Escherichia coli O4:H5 containing genes for both PapGJ96 (“class I”) and PrsGJ96 (“class III”) Gal(α1-4)Gal-binding adhesins. J Infect Dis. 1997;175:983–988. doi: 10.1086/514006. [DOI] [PubMed] [Google Scholar]
- 28.Johnson J R, Russo T A, Tarr P I, Carlino U, Bilge S S, Vary J C J, Stell A L. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect Immun. 2000;68:3040–3047. doi: 10.1128/iai.68.5.3040-3047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson J R, Stapleton A E, Russo T A, Scheutz F S, Brown J J, Maslow J N. Characteristics and prevalence within serogroup O4 of a “J96-like” clonal group of uropathogenic Escherichia coli O4:H5 containing the “class I” and “class III” alleles of papG. Infect Immun. 1997;65:2153–2159. doi: 10.1128/iai.65.6.2153-2159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson J R, Stell A L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 31.Johnson J R, Stell A L, Scheutz F, O'Bryan T T, Russo T A, Carlino U B, Fasching C C, Kavle J, van Dijk L, Gaastra W. Analysis of F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex polymerase chain reactions-based assay. Infect Immun. 2000;68:1587–1599. doi: 10.1128/iai.68.3.1587-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson J R, Swanson J L, Barela T J, Brown J J. Receptor specificities of variant Gal(α1-4)Gal-binding PapG adhesins of uropathogenic Escherichia coli as assessed by hemagglutination phenotypes. J Infect Dis. 1997;175:373–381. doi: 10.1093/infdis/175.2.373. [DOI] [PubMed] [Google Scholar]
- 33.Karr J F, Nowicki B, Truong L D, Hull R A, Hull S I. Purified P fimbriae from two cloned gene clusters of a single pyelonephritogenic strain adhere to unique structures in the human kidney. Infect Immun. 1989;57:3594–3600. doi: 10.1128/iai.57.11.3594-3600.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karr J F, Nowicki B J, Truong L D, Hull R A, Moulds J J, Hull S I. Pap-2-encoded fimbriae adhere to the P blood group-related glycosphingolipid stage-specific embryonic antigen 4 in the human kidney. Infect Immun. 1990;58:4055–4062. doi: 10.1128/iai.58.12.4055-4062.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 36.Lindstedt R, Baker N, Falk P, Hull R, Hull S, Karr J, Leffler H, Svanborg Eden C, Larson G. Binding specificities of wild-type and cloned Escherichia coli strains that recognize Globo-A. Infect Immun. 1989;57:3389–3394. doi: 10.1128/iai.57.11.3389-3394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindstedt R, Larson G, Falk P, Jodal U, Leffler H, Svanborg Eden C. The receptor repertoire defines the host range for attaching Escherichia coli strains that recognize Globo-A. Infect Immun. 1991;59:1086–1092. doi: 10.1128/iai.59.3.1086-1092.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lund B, Lindberg F, Marklund B I, Normark S. The PapG protein is the α-d-galactopyranosyl-(1-4)-β-d-galactopyranose-binding adhesin of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1987;84:5898–5902. doi: 10.1073/pnas.84.16.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund B, Marklund B, Strömberg N, Lindberg F, Karlsson K, Normark S. Uropathogenic Escherichia coli can express serologically identical pili of different receptor binding specificities. Mol Microbiol. 1988;2:255–263. doi: 10.1111/j.1365-2958.1988.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 40.Marklund B I, Tennent J M, Garcia E, Hamers A, Baga M, Lindberg F, Gaastra W, Normark S. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 41.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitsumori K, Terai A, Yamamoto S, Ishitoya S, Yoshida O. Virulence characteristics of Escherichia coli in acute bacterial prostatitis. J Infect Dis. 1999;180:1378–1381. doi: 10.1086/314976. [DOI] [PubMed] [Google Scholar]
- 43.Otto G, Sandberg T, Marklund B I, Ullery P, Svanborg Eden C. Virulence factors and pap genotype in Escherichia coli isolates from women with acute pyelonephritis, with or without bacteremia. Clin Infect Dis. 1993;17:448–456. doi: 10.1093/clinids/17.3.448. [DOI] [PubMed] [Google Scholar]
- 44.Plos K, Hull S I, Hull R A, Levin B R, Orskov I, Orskov F, Svanborg-Edén C. Distribution of the p-associated-pilus (pap) region among Escherichia coli from natural sources: evidence for horizontal gene transfer. Infect Immun. 1989;57:1604–1611. doi: 10.1128/iai.57.5.1604-1611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prats G, Navarro F, Mirelis B, Dalmau D, Margall N, Coll P, Stell A, Johnson J R. Escherichia coli serotype O15:K52:H1 as a uropathogenic clone. J Clin Microbiol. 2000;38:201–209. doi: 10.1128/jcm.38.1.201-209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts J A, Marklund B, Ilver D, Haslam D, Kaack M B, Baskin G, Louis M, Möllby R, Winberg J, Normark S. The Gal(α1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc Natl Acad Sci USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Sauer F G, Futterer K, Pinkner J S, Dodson K W, Hultgren S J, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 49.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senior D, Baker N, Cedergren B, Falk P, Larson G, Lindstedt R, C. S E. Globo-A—a new receptor specificity for attaching Escherichia coli. FEBS Lett. 1988;237:123–127. doi: 10.1016/0014-5793(88)80184-4. [DOI] [PubMed] [Google Scholar]
- 51.Sokal R R, Sneath P H A. Principles of numerical taxonomy. W. H. San Francisco, Calif: Freeman & Co.; 1963. [Google Scholar]
- 52.Stapleton A E, Stroud M R, Hakomori S I, Stamm W E. The globoseries glycosphingolipid sialosyl galactosyl globoside is found in urinary tract tissues and is a preferred binding receptor in vitro for uropathogenic Escherichia coli expressing pap-encoded adhesins. Infect Immun. 1998;66:3856–3861. doi: 10.1128/iai.66.8.3856-3861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strömberg M, Marklund B I, Lund B, Ilver D, Hamers A, Gaastra W, Karlsson K A, Normark S. Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Galα1-4Gal-containing isoreceptors. EMBO J. 1990;9:2001–2010. doi: 10.1002/j.1460-2075.1990.tb08328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strömberg N, Nyholm P G, Pascher I, Normark S. Saccharide orientation at the cell surface affects glycolipid receptor function. Proc Natl Acad Sci USA. 1991;88:9340–9344. doi: 10.1073/pnas.88.20.9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stroud M R, Stapleton A E, Levery S B. The P histo-blood group-related glycosphingolipid sialosyl galactosyl globoside as a preferred binding receptor for uropathogenic Escherichia coli: isolation and structural characterization from human kidney. Biochemistry. 1998;37:17420–17428. doi: 10.1021/bi9814639. [DOI] [PubMed] [Google Scholar]
- 56.Swenson D L, Bukanov N O, Berg D E, Welch R A. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang G, Whittam T S, Berg C M, Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wold A E, Caugant D A, Lidin-Janson G, de Man P, Svanborg C. Resident colonic Escherichia coli strains frequently display uropathogenic characteristics. J Infect Dis. 1992;165:46–52. doi: 10.1093/infdis/165.1.46. [DOI] [PubMed] [Google Scholar]
- 60.Wold A E, Thorssen M, Hull S, Svanborg-Eden C. Attachment of Escherichia coli via mannose- or Galα1→4Galβ-containing receptors to human colonic epithelial cells. Infect Immun. 1988;56:2531–2537. doi: 10.1128/iai.56.10.2531-2537.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]