Abstract
Purpose
MRI is integral to the diagnosis of multiple sclerosis (MS) and is important for clinical prognostication. Quantitative volumetric reporting tools (QReports) can improve the accuracy and objectivity of MRI-based assessments. Several QReports are commercially available; however, validation can be difficult to establish and does not currently follow a common pathway. To aid evidence-based clinical decision-making, we performed a systematic review of commercial QReports for use in MS including technical details and published reports of validation and in-use evaluation.
Methods
We categorized studies into three types of testing: technical validation, for example, comparison to manual segmentation, clinical validation by clinicians or interpretation of results alongside clinician-rated variables, and in-use evaluation, such as health economic assessment.
Results
We identified 10 companies, which provide MS lesion and brain segmentation and volume quantification, and 38 relevant publications. Tools received regulatory approval between 2006 and 2020, contextualize results to normative reference populations, ranging from 620 to 8000 subjects, and require T1- and T2-FLAIR-weighted input sequences for longitudinal assessment of whole-brain volume and lesions. In MS, six QReports provided evidence of technical validation, four companies have conducted clinical validation by correlating results with clinical variables, only one has tested their QReport by clinician end-users, and one has performed a simulated in-use socioeconomic evaluation.
Conclusion
We conclude that there is limited evidence in the literature regarding clinical validation and in-use evaluation of commercial MS QReports with a particular lack of clinician end-user testing. Our systematic review provides clinicians and institutions with the available evidence when considering adopting a quantitative reporting tool for MS.
Keywords: Systematic review, Multiple sclerosis, MRI, Quantitative volumetric reporting tools, Validation
Introduction
Multiple sclerosis (MS) is a chronic inflammatory and neurodegenerative disease of the central nervous system characterized by demyelinating lesions and atrophy [1, 2]. Brain atrophy is accelerated in MS compared to the healthy population [3]. Both lesion evolution and brain volume loss over time correlate with clinical disability [3, 4].
Structural MRI is routinely used in the diagnostic workup of MS and to assess and monitor demyelinating lesions [5]. MRI-based measurement of brain atrophy is becoming increasingly recognized as an important clinical prognostication tool [3, 6, 7]. Brain and lesion volumes measured using image segmentation have become established biomarkers for determining treatment efficacy in research studies and clinical trials [8–11]. Previous research has shown that brain atrophy [12] and lesion volumes [13] significantly predict long-term disability in all MS phenotypes, especially when used in combination [4, 9]. Manual segmentation of the brain and lesions is time-consuming and can also be prone to imprecision and error [14–16]. The development and use of automated and semi-automated brain and lesion segmentation methods, such as in quantitative volumetric reporting tools (QReports), has increased in recent years [11, 17]. These tools aim to improve the objectivity of image interpretation by increasing the sensitivity of MRI analysis [18, 19], the accuracy[20–25] and reproducibility of results [22, 26], and potentially decreasing reporting time [18]. QReports can facilitate cross-sectional diagnosis [20, 26–30], longitudinal assessment [20, 22, 23, 31], and therapy response monitoring [32] via user-friendly graphical displays. QReports may also offer automatic contextualization of an individual patient’s volumetric results against a relevant reference population [33], which could assist clinicians in disease course prognostication and deciding on therapeutic strategies. Various QReports for MS have been developed for use in the clinical setting, and many of these tools are commercially available having received regulatory approval.
Currently, the application of QReports in the clinic is limited [11, 12]. Clinical institutions may not have adequate resources to assess how tools have been tested and validated, despite commercialization for medical use. To encourage evidence-based use and to aid clinicians in deciding how and whether to adopt these tools, the validity of results and the impact on clinical management should be established. Technical and clinical validation and evaluation of quantitative reporting tools do not currently follow standardized methods. The quantitative neuroradiology initiative (QNI) addresses this issue and provides a six-step translational pathway for quantitative reporting tools [34]. The QNI model distinguishes three types of testing: technical validation of tool performance, for example, comparison to manual segmentation or other segmentation techniques; clinical validation by clinicians or by interpretation of results alongside clinician-rated variables; and finally in-use evaluation, such as health economic assessment [34].
Our previous work demonstrated a lack of technical and notably clinical validation of commercial QReports in dementia [35]. In the current paper, we replicated this methodology and performed a systematic review of the literature aiming to validate or evaluate commercial QReports for use in MS. We (1) presented the range of tools, including details of their technical features and characteristics and (2) provided a descriptive synthesis of the evidence published regarding their validation. We assessed the literature according to the QNI framework, categorizing studies into technical and clinical validation and in-use evaluation. The aim is to increase transparency and help clinicians to make informed decisions about whether to adopt commercial QReports into clinical routine for the assessment of patients with MS and provide an overview of the features of each commercially available tool.
Methods
This review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [36–38] and is registered with the Prospective Register of Systematic Reviews (PROSPERO) database under number CRD42021286139.
Vendor and product search
Product inclusion and exclusion criteria
The inclusion criteria for QReports are as follows: (1) FDA or CE clearance; (2) target disorder MS or a population with suspected MS (specified on the company website or in the literature); (3) uses structural-MRI-based input (4) to generate brain and lesion volumetric results; (5) incorporates normative reference data for single-subject comparison; (6) presents results in a structured report format.
Search methodology: FDA-cleared product identification
Keyword screening
The FDA medical device databases were used (last access: 28 January 2022) to find FDA-cleared automated quantitative MRI reporting tools in MS (https://www.fda.gov/medical-devices/device-advice-comprehensive-regulatory-assistance/medical-device-databases). A total of 83,556 premarket 510(k) FDA notification clearances dating from 1996 to present were downloaded in a text file from https://www.fda.gov/medical- devices/510k-clearances/downloadable-510k-files. The text file was searched using the keywords listed below and 821 “medical devices” were identified for further review. Terms with an * use “wild-cards,” covering relevant suffixes of each word stem, for example, “Radiolog*” covers “Radiology,” “Radiologist,” and “Radiological”:
| • Neuro* | • Cortex | • Structur* |
| • Brain | • Dementia | • Segment* |
| • Quant* | • Volume | • Automat* |
| • MRI | • Multiple | • Spinal |
| • Hippocamp* | • Sclerosis | • Cord |
| • Radiolog* | • Lesion | • MS |
| • Atroph* | • Lobar | • Demyelinat* |
| • Cortical | • Lobe |
-
2.
Eligibility screening
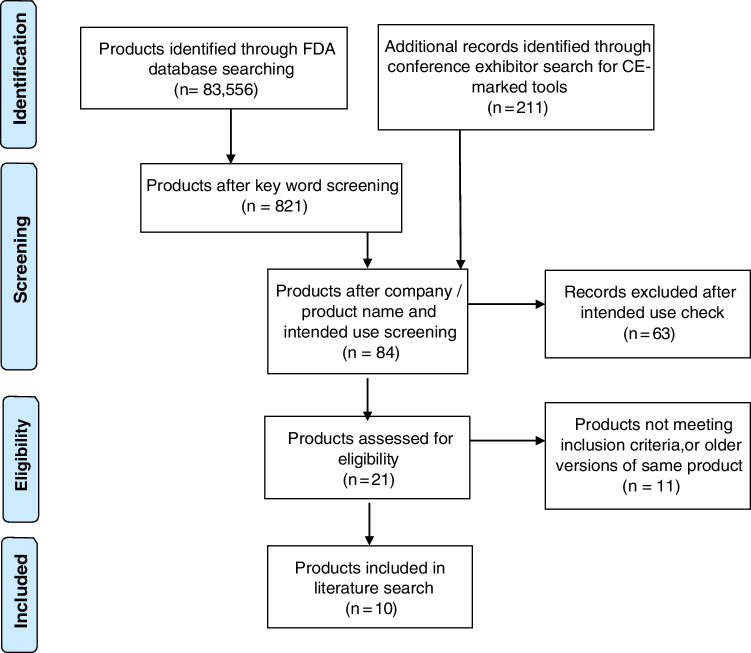
Manual checks were performed to verify the company name, product name, approval date, and description on the FDA database. Tools considered hardware were excluded at this stage. The websites of all remaining companies were searched to further investigate the intended use of their products. Seven companies that had not specified MS as the target disorder were excluded from further review. Two quantitative reporting tools that were acquisition dependent were also excluded at this stage. After manual checks and searching company websites, four companies were identified as meeting our inclusion criteria (see Fig. 1 for PRISMA flowchart outlining search for companies).
Fig. 1.
PRISMA flow diagram outlining a systematic search for CE- and FDA-approved QReports. The FDA medical device databases and lists of all companies that exhibited at relevant medical imaging conferences (ISMRM, ESMRMB, RSNA, ECR, ESR AIX, ASNR, SIIM, and ESNR) were searched and the website https://grand-challenge.org/aiforradiology/ was used to cross-check results
Search methodology: CE-marked product identification
As there was no freely available, searchable database of CE-marked products, the websites of relevant medical imaging conferences (ISMRM, ESMRMB, RSNA, ECR, ESR AIX, ASNR, SIIM, and ESNR) were searched to identify companies that exhibited their products in 2019–2021. The websites of identified companies were searched in detail to find CE-marked quantitative reporting tools. Sixty-three tools were excluded after screening the company and product name and intended use. Results were cross-checked using the following website: https://grand-challenge.org/aiforradiology/. Two companies that had not specified MS as the target disorder were excluded from further review. Finally, six companies with CE-marked tools were identified that fit our inclusion criteria.
Each company was directly contacted to verify the product name, date of approval, description, and intended use of the product. The companies were informed of their inclusion in the review and given the opportunity to add to and correct information gathered from company websites and the literature. Excluded companies are summarized in the “Results” section.
Company and product features
The following technical details of the 10 tools included in the review were extracted from company websites, publications identified in the literature search, and by direct vendor contact:
FDA/CE approval
Date of approval
Target disorder
Input sequences
Brain and lesion segmentation and volumetry method
Lesion filling
Brain atrophy data
Cross-sectional or longitudinal analysis available
Details of normative reference populations
Provision of segmentation overlays
Strategies to account for inter-scanner variability
Image quality control method(s)
Report deployment procedure
Literature search on technical and clinical validation of identified products
A literature search was conducted independently by two authors according to PRISMA guidelines [36–38]. The results were checked and verified by a third author; any inclusion or exclusion discrepancies were settled by consensus. The 10 company names and their associated product names were used as search terms. Both company and product names were used to ensure the identification of studies published before product branding. Searches were simultaneously conducted in PubMed, Ovid Medline “All fields” and Scopus (latest search: 29 March 2022). Company and product names comprising several words are bracketed to indicate single search terms. Companies were directly contacted to verify company and product names. The search terms were as follows:
AIRAmed OR (AIRAscore)
Combinostics OR (cNeuro cMRI)
(CorTechs Labs) OR (NeuroQuant MS) OR LesionQuant
Icometrix OR MSmetrix OR (icobrain ms)
(Jung diagnostics) OR Biometrica
mediaire OR mdbrain
Pixyl OR Pixyl.Neuro.MS OR Pixyl.Neuro.BV
Quibim OR (Quibim Precision)
Qubiotech OR (Neurocloud VOL)
Qynapse OR QyScore
References in identified publications, papers listed under “similar articles” in PubMed, and all publications listed on company websites were also searched to identify additional relevant validation studies. Companies were given the opportunity to provide further relevant studies.
Study inclusion criteria
The study inclusion criteria used in this review are based on the QNI framework for the translation of quantitative reporting tools into the clinic. Studies were included in the review on the basis that they met the following inclusion criteria: (1) published in English as original research in academic peer-reviewed journals or conference proceedings (conference abstracts and posters excluded), (2) which involve automated lesion, or brain and lesion, segmentation and volumetry computed from structural MR images (3) in an MS population and/or healthy controls, and (4) fit either:
Technical validation
Papers validating the technical performance of lesion or brain and lesion segmentation methods. For example, test-retest studies or comparison to manual segmentation and/or other state-of-the-art brain volumetry tools, such as FreeSurfer [39], SPM (www.fil.ion.ucl.ac.uk/spm), SIENA(X) [40], and lesion segmentation tools, for example, LST [41, 42], nicMSlesions [43], and samseg [44], and testing for robustness to different input data. Papers focusing only on brain segmentation were not included unless conducted in an MS population.
Clinical validation
Testing of the tool by clinicians on an MS population focusing on one of more of the following: (a) determining diagnostic accuracy, confidence, and differential diagnoses vs. “ground truth” clinician-rated diagnoses, i.e., using receiver operating characteristics; (b) assessing the tool’s effect on clinical management (usability, prognostic value); (c) inter-rater reliability metrics or percent agreement.
Clinical trials in MS using the tool’s results as an outcome measurement.
Interpretation of results alongside clinician-rated variables, such as the Expanded Disability Status Scale (EDSS) and Symbol Digit Modalities Test (SDMT), as measures of physical and cognitive disability in MS, respectively.
In-use evaluation
Studies assessing any of (1) benefit to patients, (2) the effect on reporting time in the context of normal clinical routine, (3) clinical and population perception, or (4) socioeconomic impact of using QReports in the clinic.
Data extraction
Two raters independently categorized all studies that met our inclusion criteria into technical validation, clinical validation, or in-use evaluation. A third researcher checked the results, and a consensus was reached on any discrepancies.
Results
Company and product search
Following the systematic search outlined above, 10 companies were identified that produce tools meeting our inclusion criteria; see Fig. 1 for a research flow diagram summarizing the search for relevant products.
Excluded tools
According to PRISMA guidelines, inclusion criteria were decided on in advance (see “Methods” section). The results of the eligibility screening are presented below.
MS brain and lesion segmentation and volumetry tools were excluded if they were not FDA or CE approved, such as SegPlus by Neurophet (https://www.neurophet.com), which has conducted technical validation [45], and TensorMedical (https://www.tensormedical.ai) that developed and uses the nicMSlesions software and has evidence of technical validation in MS [43]. Research tools that did not have FDA or CE approval, such as FreeSurfer [39], SPM (www.fil.ion.ucl.ac.uk/spm) or SIENA(X) [40] and LST [41, 42], samseg [44], or BaMoS [46], were excluded.
QReports that, according to the literature and the company websites, did not conduct brain and MS lesion segmentation, including Childmetrix by Icometrix (a pediatric non-MS-related QReport) (http://icometrix.com), Quantib ND by Quantib (https://www.quantib.com), neuroreader by Brainreader (https://brainreader.net), THINQ by Corticometrics (https://www.corticometrics.com), tools by JLK Inc (https://www.jlkgroup.com), and Corinsights MRI by ADM diagnostics (https://admdx.com), VUNO Med-DeepBrain by Vuno (https://www.vuno.co), AI-Rad Companion Brain by Siemens Health (https://www.siemens-healthineers.com), AQUA by Neurophet (https://www.neurophet.com), and DIADEM by Brainminer (https://www.brainminer.co.uk), were excluded. Our research group has conducted a systematic review including several of these tools for other indications [35].
Acquisition-dependent quantitative neuroimaging tools were also excluded, including SyMRI Neuro by SyntheticMR (https://syntheticmr.com/) and STAGE by SpinTechMRI (https://spintechmri.com), which include dedicated quantitative MRI-based reporting tools that can be used in the setting of MS. SyMRI Neuro is an FDA- and CE-approved tool providing brain and myelin segmentation and has been technically and clinically validated in MS populations [47–50]. STAGE (strategically acquired gradient echo) is an FDA-approved quantitative MRI-based reporting tool providing atrophy and MS lesion characterization using susceptibility-weighted images. STAGE has been validated on healthy subjects and several MS cases [51, 52].
Included tools
The 10 companies and their QReports identified using the search strategy described in the “Methods” section and illustrated in Fig. 1 are presented in Table 1 along with key technical details.
Table 1.
A database of the QReports and their key technical details presented in alphabetical order of vendor name
| Vendor | Product name | CE/FDA status | Approval received | Target disorder(s) | Input sequences | Brain/lesion segmentation/volumetry method | Lesion filling | Brain atrophy data | Cross sectional + longitudinal brain volumetry analysis | Normative reference database | Segmentation Overlays | Methods to account for inter-scanner variability | Image quality control | Deployment procedure |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
AIRAmed |
AIRAscore | CE – class I | Aug-2020 | All neurodegenerative diseases with brain volume reduction, e.g., dementia and MS | 3D T1, 3D T2-FLAIR | In-house and CNN-based | Not required | ml, % TIV and normative percentile | Both, indirect longitudinal comparison | 8000 + healthy subjects from private and public datasets, mix of field strength and scanner vendors | Brain and lesion segmentation overlays | Mix of field strength and scanner vendors in the normative reference data. For longitudinal comparisons, it is recommended to only use images from the same scanner sequence combination | Automatic checks for technical image parameters in the DICOM header, image QC for movement artifacts should be performed by the reading physician | Cloud-based PACS integration |
|
Combinostics |
cNeuro cMRI |
CE – class IIa FDA – 510(k) cleared, class II |
Sep-2016 | Dementia, MS and other diseases where analysis of atrophy or WMHs are of interest | 3D T1 and 2D or 3D T2-FLAIR | In-house | Did not disclose | ml, normative percentile for cross-sectional data + annualized atrophy rate and its percentile for longitudinal data from T1 | Both, direct longitudinal comparison | ~ 2000 subjects from private and public US/Europe datasets, 18–94 y, mix of field strength and scanner vendors | Brain and lesion segmentation overlays | Mix of field strength and scanner vendors in the normative reference data | Automated QC for CNR, abnormal signal intensities and acquisition parameter checks | Cloud-based PACS integration |
|
CorTechs.ai |
NeuroQuant MS (also known as LesionQuant) | CE – class IIa, FDA – 510(k) cleared, class II | Aug-2006 | Multiple sclerosis | 3D T1 and 2D or 3D T2-FLAIR | In-house | Using neighborhood information | ml, % change, % TIV and normative percentile | Both, direct longitudinal comparison | ~ 5000 subjects from private and public datasets—age range 3–100 y, equal male/female ratio, acquired using Siemens, GE, and Philips MRI scanners with both 1.5 T and 3 T field strength | Lesion and brain segmentation overlays | Scanner-specific 3D gradient field distortion correction; voxel intensity normalization; custom dynamic atlas–based contrast adjustment by anatomical region to correct the measured contrast variability in patients | Automated QC checks for acquisition parameter, atlas fit, atlas tissue class contrast, scan noise estimation and image quality | PACS integration via local hardware, local virtualization or cloud based |
|
Icometrix |
icobrain ms (previously MSmetrix) | CE – class I, FDA – 510(k) cleared, class II | Jul-2015 | Multiple sclerosis | 3D T1 and 2D or 3D T2-FLAIR | In-house | Gaussian smoothed mean WM values | ml, % change, normative percentile | Both, direct longitudinal comparison | 1903 healthy subjects (1069 female and 834 male subjects) available from several public collections, 6–96 y, mix of field strength and scanner vendors | Lesion and brain segmentation overlays | Cross-sectional report has been tested across multiple scanners. Longitudinal comparisons require same scanner and acquisition protocol for accuracy | Automated flagging for manual QC: incomplete head coverage, insufficient CNR or distortions between sequences or time points | Cloud-based PACS integration, optional EMR integration |
|
jung diagnostics GmbH |
Biometrica | CE – class I | Jun-2009 | Multiple sclerosis, dementia, neurodegenerative diseases |
2D/3D MPRAGE and 2D/3D T2-FLAIR and 2D/3D Gd enhancing T1 |
In-house, CNN-based and SIENA for longitudinal comparison | Using neighborhood information | z-score using residuals method | Both, direct longitudinal comparison | ~ 2000 subjects from a proprietary dataset using a single scanner and protocol, 18–99 y | Lesion and brain segmentation overlays | Site qualification process—protocol set-up using 30 healthy scans for each individual scanner using the service. Manual checks for global offset between each site and proprietary normative database | Site qualification process and expert manual QC/QA checks by vendor | Cloud-based PACS integration |
|
mediaire |
mdbrain | CE – class I | Jan-2019 | Multiple sclerosis, dementia, NPH, neurodegenerative diseases, vascular dementia, brain aneurysm, brain tumor | 2D or 3D T1 and T2-FLAIR | In-house | Not required | ml and normative percentile (in %) | Both, direct longitudinal comparison | ~ 8000 scans from mainly private datasets, 18–93 y, mix of field strength and scanner vendors | Lesion and brain segmentation overlays | Mixture of field strength and scanner vendors in the segmentation algorithm training data | Automated checks for acquisition parameters and artifact | PACS integrated via local hardware or local virtualization |
|
Pixyl |
Pixyl.Neuro.MS Pixyl.Neuro.BV |
CE – class IIa | Nov-2019 | Multiple sclerosis, neurodegenerative diseases, dementias, Alzheimer’s disease, Parkinson’s disease |
Pixyl.Neuro.MS: 2D or 3D T2 FLAIR; 3D T1 and 3D T1 with contrast optional Pixyl.Neuro.BV: 3D T1 |
In-house, deep learning-based | T1 white matter hypo-intensity segmentation is incorporated as white matter labels | Pixyl.Neuro.BV: ml and in % | Both, indirect longitudinal comparison |
Pixyl.Neuro.BV: ~ 3000 subjects from private and public datasets, 18–97 y, multi-center and multi-country (mix of field strength and scanner vendors) |
Lesions labeled according to their individual evolution | Data augmentation using proprietary 3DT1 library with clinically relevant MRI and patient variability (noise, contrast, artifacts, distortions, style transfer) | Automated QC based on voxel size, DICOM headers, and acquisition parameters. The user is warned when insufficient input image quality could impact quality of results | PACS integration via local hardware or local virtualization. Stand-alone web based; Integration via AI marketplace and distribution platforms |
|
Quibim |
Quibim Precision: (white matter lesions; atrophy screening) | CE – class IIa | Dec-2018 | Neurodegenerative diseases, multiple sclerosis, dementia Parkinson’s disease, epilepsy, amyotrophic lateral sclerosis (ALS) | White matter lesions: 2D or 3D FLAIR; atrophy screening: 3D T1 | In-house, lesions: CNN-based; atrophy: atlas-based | Not done or not applied | Atrophy screening: ml, %TIV, normative percentile | Both, indirect longitudinal comparison | 620 Caucasians from private and public datasets, 20–86 y, mix of field strength and scanner vendors | Lesion and brain segmentation overlays | Onboarding with sample of new site data, amendments to data pre-processing/acquisition protocols where necessary |
Automated QC checks on required acquisition parameter ranges |
PACS integrated via local hardware, local virtualization or cloud-based |
|
Qubiotech |
Neurocloud VOL | CE – class I | Jan-2019 | Multiple sclerosis, epilepsy, dementia, Alzheimer's disease, Parkinson's disease |
Cross-sectional: 3D T1 recommended (3D T2-FLAIR also possible) Longitudinal: multi-sequence only (3D T1 and T2 FLAIR) analyses |
In-house and SPM-based | Using neighborhood information | ml, %TIV and normative percentile | Both, direct longitudinal comparison | ~ 852 subjects from private dataset, age 19–94 y (375 male, 477 female), mix of scanner vendors and field strength | Lesion and brain segmentation overlays | Mixture of field strength and scanner vendors in the normative reference, training, and validation data | Automated QC checks on required acquisition parameters, DICOM headers, and cross-correlation metrics to evaluate image co-registration results | Locally virtualized or cloud-based PACS integration |
|
Qynapse |
QyScore | CE – class IIa, FDA – 510(k) cleared, class II |
CE 2017 FDA 2019 |
All central nervous system diseases | 3D T1 and 2D or 3D T2 FLAIR | Lesion: in-house; atrophy: open source | Not done or not applied | ml, %ICV, z-score, normative percentile | Both, indirect longitudinal comparison | 20–90 y, mix of field strength and scanner vendors | Lesion and brain segmentation overlays | Specific MRI parameters are required which have been tested to provide good QyScore results. Early manual QC checks by vendor for each site | Automated QC checks on required acquisition parameters, SNR, CNR | PACS integrated via local hardware or cloud-based |
Information gathered from the literature and direct contact with vendors can be assessed according to individual needs. All information was checked and confirmed with vendors before publication. It was not possible for the authors to independently verify technical details without access to commercial software packages
MS, multiple sclerosis; CE, Conformitè Europëenne; FDA, Food and Drug Administration; FLAIR, fluid-attenuated inversion recovery; SIENA, Structural Image Evaluation, using Normalisation, of Atrophy; SPM, statistical parametric mapping; CNN, convolutional neural network; WMH, white matter hyperintensity; SNR, signal-to-noise ratio; CNR, contrast-to-noise ratio; QC, quality control; PACS, picture archiving and communication system; TIV, intracranial volume; CSF, cerebrospinal fluid; GM, grey matter; WM, white matter; DICOM, Digital Imaging and Communications in Medicine; QA, quality assurance
Company and product features
Table 1 is a structured database of the technical features and characteristics of the QReports. Company and product features are summarized below. Report processing times were not included, as measurement and comparison should be conducted independently by the authors using the same cases and resources, which was not possible without access to the software packages.
CE/FDA approval status
All 10 companies have obtained either CE class I/II or FDA 510(k) clearance, as “software as a medical device.”
Date of approval
CorTechs.ai was the first company to receive FDA clearance in 2006. The most recent of the 10 companies to receive CE or FDA regulatory approval was AIRAmed in 2020.
Target disorder
All companies produced a report for the assessment of MS lesions and brain atrophy. For some tools, the MS QReport was an extension of a previously established brain volume quantification tool.
Input sequences
Most companies required 3D T1 and 2D or 3D T2-FLAIR input sequences for brain and lesion segmentation. Two tools also provided the option to use a 2D or 3D gadolinium-enhancing T1-weighted sequence for contrast-enhancing T1 lesion detection.
Brain/lesion segmentation/volumetry method
All companies used proprietary methods developed “in house,” of which four claimed to use deep learning. Two companies reported using modified versions of research methods, including SIENA(X)[40] and SPM (www.fil.ion.ucl.ac.uk/spm). Brain and lesion segmentation software was commercialized as a single package or as two different tools (which is the case for two companies, see Table 1).
Lesion data
All tools reported longitudinal lesion volume. Nine out of ten tools reported longitudinal lesion count. Nine out of ten QReports provided the spatial distribution of lesions according to the McDonald criteria [53] categorized into periventricular, juxtacortical, deep white matter, and infratentorial. Companies that have not yet included lesion count and spatial distribution of lesions claimed to be working to provide this information in upcoming updates and releases of the tools.
Lesion filling
Lesion filling is commonly used to accurately compute brain volumes in MS [54]. Eight out of ten companies used either automatic lesion filling or deep learning approaches, which did not require lesion filling. The approaches used for lesion filling are outlined in Table 1.
Brain atrophy data
Brain atrophy was reported in milliliters, as a percentage of the total intracranial volume (TIV), as a normative percentile, or as a z-score.
Cross-sectional and longitudinal brain volumetry analysis
All 10 companies provided both cross-sectional and longitudinal analyses of lesions and whole brain atrophy. Longitudinal analysis approaches were indirect for four QReports, i.e., the difference in volume/percentile per structure between two visits and direct for six QReports, such as using SIENA [40].
Details of a normative reference population
The normative reference populations of all tools comprised a large age range, typically from 20 to 90 years with a sex balance, and were compiled from public and/or private datasets. Nine out of ten companies used datasets with a range of scanner types and field strength. The size of the datasets varied between 620 and ~8000 subjects.
Segmentation/atrophy visual overlays
All QReports provided visual lesion and brain segmentation overlays.
Image quality control method
All tools used image quality control (QC) processes. The methods used varied and were mainly automatic, including checks for artifacts and acquisition parameters, computing of standard measures of image quality, such as signal-to- noise ratio (SNR) (comparing the level of the target signal to background noise), and automatic flagging of the need for manual QC.
Strategies to account for inter-scanner variability
All companies claimed to use strategies to account for diverse input data, including a mix of scanner type and field strength in the normative reference population, algorithm training, using independent validation datasets, accounting for vendor-specific acquisition parameters, implementing AI-based augmentation to anticipate the variability between images, and using site qualification procedures.
PACS integration/report deployment procedure
All companies claimed to provide PACS integration either using a cloud-based solution and/or local virtualization and/or local hardware.
Peer-reviewed technical and clinical validation
All companies had conducted internal validation processes, including the necessary steps for CE and/or FDA clearance. Also, all companies claimed to be carrying out further peer-reviewed validation studies. It is of note that several companies had conducted studies validating their tools in other disease areas [55–64]. These papers have only been included if lesion, or brain and lesion, quantification techniques were under investigation in MS and if the tool is commercialized for use in MS either on the company website or in publications.
The number and category of studies identified in the literature search are presented in Fig. 2 and described below in the “Literature search” section.
Fig. 2.
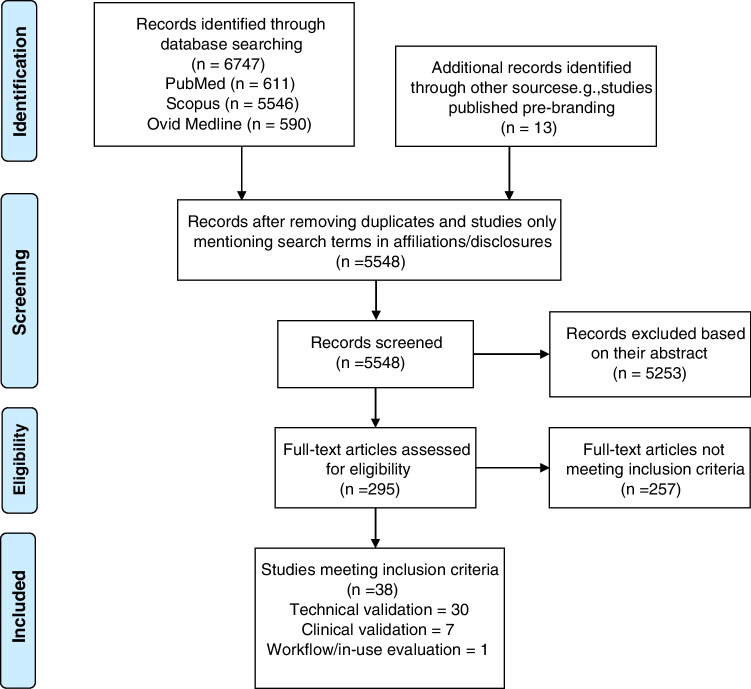
PRISMA flow diagram outlining the search and selection for publications included in the review
Literature search
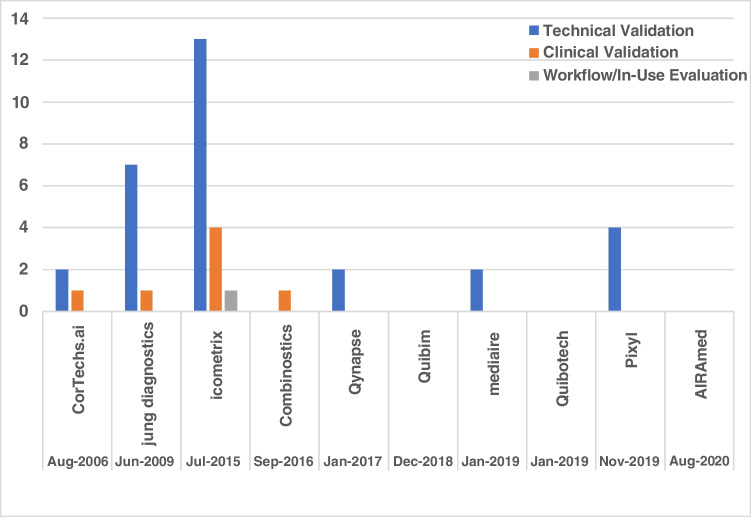
The results of the literature search are outlined in the PRISMA workflow diagram in Fig. 2 and documented further below. A total of 38 peer-reviewed publications covering technical (n = 30), clinical (n = 7) validation, or in-use evaluation (n = 1) were identified. In total, 6 companies have conducted technical validation, 4 have published clinical validation, 1 has conducted an in-use evaluation, and 3 have not published studies meeting our inclusion criteria. The distribution of studies identified is presented in Fig. 3.
Fig. 3.
The distribution of publications included in the review for each company identified. The vendors are listed chronologically according to the date of their first FDA/CE approval
Validation studies identified
To remain unbiased, a narrative synthesis of the studies identified for each company is provided and referenced below (in alphabetical order). All publications were conducted in MS and/or healthy control populations. In summary, technical validation mainly consisted of comparison with manual lesion counting, manual segmentation, or state-of-the-art automated brain volumetry and lesion segmentation tools, including SIENA(X) [40], Freesurfer [39], SPM (www.fil.ion.ucl.ac.uk/spm), FIRST [65], Cascade [66], LST [41, 42], Lesion-TOADS [67], lesionbrain [68], BIANCA [69], and nicMSlesions [43]. Technical validation was also conducted as repeatability studies and by testing different acquisition protocols. Clinical validation mainly comprised correlation of the tool’s results with clinical variables, such as EDSS scores. Only one clinical validation study incorporates clinician end-user testing [18]. Validation studies conducted by each company are summarized below.
Combinostics
Clinical: The cNeuro cMRI results were correlated with EDSS scores [70].
CorTechs Labs
Technical: NeuroQuant MS has been tested for longitudinal consistency [20] and compared to visual radiological assessment [20], icobrain ms [28], and established brain and lesion segmentation methods (FIRST [71], LST [71], SIENA(X) [28], FreeSurfer [20], and Cascade [20]).
Clinical: NeuroQuant MS results were correlated with clinical variables, including EDSS and timed 25-foot walk test (T25FT) and 9-Hole Peg Test (9HPT) scores as measures of MS-related physical disability [20].
Icometrix
Technical: icobrain ms has been tested for robustness to different input data [26, 31, 33, 72–74], reproducibility [26, 31, 72, 74], repeatability [73, 74], and consistency over time [19, 73]. The tool has been compared to manual segmentation [19, 21, 26, 31, 74, 75], LesionQuant [28], and automated established brain and lesion segmentation and atrophy quantification methods, such as SIENA(X) [28, 72–74, 76, 77], LST [26, 31], Lesion-TOADS [26], FreeSurfer [77], and SPM [74, 77] and has been included in a longitudinal MS lesion segmentation challenge [21]. Earlier or other versions of the tool have been compared to the current version [31, 75]. An automated method to decrease the effect of inter-scanner variability on results has been tested [78, 79].
Clinical: The tool has been tested by clinicians as end-users investigating the impact on intra- and inter-rater variability, reporting time (outside of their normal clinical routine), and detection of disease activity in comparison to visual radiological assessment [18]. The ability of the tool to differentiate MS clinical phenotypes has been investigated [18]. The tool’s results were correlated with EDSS [18, 76, 80, 81] and SDMT scores [81] and the number of relapses [80].
In-use evaluation: Icometrix has investigated the health-economic impact of icobrain ms in a microsimulation study with a decision analytical model based on a hypothetical cohort of MS patients testing for disease detection, treatment decision-making, patient quality of life, and costs using the tool in comparison to clinical and visual radiological assessment [32].
Jung diagnostics
Technical: Jung diagnostics have compared the current method to earlier versions of the tool (which included LST) [22, 27, 82, 83] and to manual segmentation [22, 27, 82]. The tool has been tested for repeatability [27, 84, 85], reproducibility [22, 27], and robustness to different input data [22, 27, 86]. Jung diagnostics has compared two methods for brain atrophy data adjustment for head size and age [86].
Clinical: Biometrica results were correlated with clinical variables, including EDSS and SDMT scores, disease duration, and MS phenotypes [87].
Mediaire
Technical: The tool has been tested for robustness to different input data [88] and was compared to manual segmentation and other lesion segmentation tools in a longitudinal MS lesion segmentation challenge [23].
Pixyl
Pixyl.Neuro.MS is an MS lesion segmentation tool and Pixyl.Neuro.BV can be used for brain volumetry. Technical: The lesion segmentation method has been compared to manual segmentation [24, 29, 89] and to older established automated methods [24, 29, 89], including in an MS lesion segmentation challenge [24]. Newer improved versions of the tool have been compared to previous versions [90] and to manual segmentation [90].
Qynapse
Technical: Qynapse has compared the current method for lesion segmentation to a previous method, to state-of-the-art lesion segmentation methods (including LST, Lesion-TOADS, lesionBrain, BIANCA, and nicMSlesions), and to manual segmentation [25, 30]. QyScore has been tested for robustness to different input data [30].
Discussion
This systematic review identified 10 companies currently offering FDA- and CE-cleared QReports for use in MS. Most tools identified in this review have obtained regulatory approval in the last 5 years. By reviewing commercial QReports in MS and previously in dementia [35], we aimed to provide the information needed by clinicians to navigate the rapidly developing market for quantitative reporting tools. Studies identified in this review have been categorized according to the QNI model framework to encourage the adoption of a common translational pathway with rigorous and structured testing. We have identified 38 relevant validation and evaluation studies: 30 technical validation studies, 7 clinical validation studies, and 1 in-use evaluation. In total, 6 QReports have evidence of technical validation, 4 companies have conducted clinical validation, and 1 has conducted in-use evaluation. The date of approval of tools did not always correlate with the number of validation studies identified. For example, CorTechs.ai, which received FDA approval in 2006, began developing and validating their tools in MS after validation in other diseases, such as dementia. Clinical validation studies were more prevalent for companies that had received regulatory approval earlier. All companies claimed to be conducting (further) validation studies.
Previous reviews of MS QReports compare both the methodologies used in research and commercially available tools without naming vendors—mainly due to publication prior to their branding [11, 17, 91–95]. In this paper, we review all identified commercial MS QReports that offer a combination of lesion and brain segmentation and volumetry. We aimed to remain unbiased by synthesizing and categorizing papers avoiding direct comparison and evaluation. There is little scope and evidence to recommend one commercial MS QReport over another, as the needs of purchasers may vary and tools have mainly not been tested under the same conditions using the same database (other than in one study identified in this review, which directly compares the performance of two commercial MS QReports) [28].
Our review has highlighted a lack of clinical validation of MS QReports and in particular testing of tools by clinicians. Only four out of ten vendors had conducted clinical validation in an MS population and three of these companies correlated QReport results with clinical variables without directly involving clinicians in the use of the tool. Correlation with clinical variables, such as EDSS, is a first clinical exploration only and can be successful without demonstrating clinical utility. Only one company has tested the tool by clinician end-users investigating reporting time (outside of normal clinical routine), diagnostic accuracy, and intra- and inter-observer variability [18]. We have demonstrated that testing by clinicians in a clinical context is extremely scarce. In 2021, Pemberton et al also demonstrated a lack of clinical validation of dementia QReports [35]. Clinical validation is part of step 4 of the QNI model framework, which encourages studying the impact of QReports on intra- and inter-rater reliability, diagnostic confidence and accuracy, and clinical management, such as reporting time within the context of normal clinical routine, to promote user-confidence and evidence-based care [34]. The collaboration of clinicians and vendors is key for refining these tools, increasing their clinical uptake, and aiding future developments.
This review has demonstrated a lack of in-use evaluation, which is set out as step 6 of the QNI framework [34]. Only one in-use evaluation study was identified, which is a microsimulation investigating the health economic impact of a QReport in a hypothetical cohort of MS patients [32]. Socioeconomic validation may encourage clinical translation, as the added value for stakeholders such as insurers should be demonstrated to encourage reimbursement for widespread clinical use. The effect of an MS QReport on treatment choice and escalation was explored in the simulated in-use evaluation study; however, this should be further explored in a real-life clinical context [32]. The patient perspective on their digital MS care pathway has been investigated by the same company [18]. Patient-reported outcome measures (PROMs) could be incorporated into in-use evaluation, especially if patients have access to results. Several companies claimed that patient access can be provided in the form of a simplified patient-oriented report and icometrix has developed the icompanion patient app, which provides access to their MRI scans. It is of note that seven out of ten companies have received regulatory approval in the last 5 years and clinical use of commercial QReports is still limited; therefore, in-use evaluation may become more prevalent over time. Presenting the evidence as we have done in this review is important for informed implementation in clinical settings, which in turn may facilitate an increase in opportunities for in-use evaluation.
Conducting clinical validation and in-use evaluation could help companies optimize their tools for application in different clinical settings using diverse input data. Step 5 of the QNI framework focuses on workflow integration, including overcoming barriers to generalizability [34]. All companies provided some form of PACS integration and DICOM standard data format. QReports should be tested for robustness to different scanners and field strengths and the normative reference data provided for contextualization of results should be generalizable, as input data can vary. Vendors had mostly compiled large datasets of normative reference values; however, only one company had compared their control population dataset intercontinentally [33]. The tools identified typically rely on 3D MRI input sequences, which are becoming increasingly available (as recommended by imaging guidelines [96, 97]) but may not yet be used in many clinical settings. Furthermore, there is a discrepancy between the MRI sequences used in standard clinical routine and in commercial QReports for use in MS, as most identified QReports rely on both T1-weighted and T2-FLAIR sequences; however, non-contrast T1-weighted images are not routinely included in the imaging guidelines for MS [5]. Companies should continue to be transparent about the generalizability of their tools and clinical usability should be studied to address translational barriers. Table 1, the database of technical features and characteristics, demonstrates the variation in generalizability measures and can help clinicians select the most appropriate tool for a specific clinical setting.
Structured validation and evaluation procedures could facilitate comparison between tools and their improvement. The QNI framework can provide a structure and guidelines for future studies, especially by highlighting the need for the testing of tools by clinicians [34]. In January 2021, the FDA published a regulatory framework action plan for artificial intelligence/machine learning as a medical device [98] and in October 2021, the FDA, Health Canada, and the UK’s Healthcare products Regulatory Agency (MHRA) defined 10 guidelines for Good Machine Learning Practices (GMLP) [99], which reference testing in a clinical setting and validation of robustness and generalizability. The EU has recently introduced new clinical evaluation requirements for regulatory-approved medical devices [100, 101] and in April 2021, the European Commission published the Artificial Intelligence Act to stimulate the development of AI and ensure its trustworthiness focusing on investment and policy [102]. Furthermore, in March 2022, Icometrix received the first Medtech Innovation Briefing in MS by NICE, which provides advice on use and a summary of the evidence (https://www.nice.org.uk/advice/mib291/chapter/summary). By addressing the testing of QReports and providing guidance for use, regulatory bodies could support transparency and encourage structured validation and evaluation procedures.
Limitations
Different search strategies were required to identify FDA- and CE-cleared tools. Without a fully searchable database of CE-marked tools, it is possible that tools could have been missed. It is possible that a tool may have been granted FDA or CE regulatory approval or a company published relevant studies during the publication process of this review. The conclusion remains unchanged that there is a lack of clinical validation and in-use evaluation of MS QReports. Some technical information on tools was provided by the companies and could not be independently verified by the authors without access to the software packages.
Conclusion
This review has identified 10 commercially available MS QReports. We have summarized validation and evaluation studies and provided a database of technical details of the tools to increase transparency and aid evidence-based decision-making in the clinic. We used the QNI framework to classify validation and evaluation studies to promote a common, structured pathway for clinical translation. We revealed an evidence gap in the clinical validation and in-
use evaluation of QReports for use in MS, especially in studies testing the use of the tool by clinicians. In total, 10 companies producing commercial MS QReports were identified, of which 4 have conducted clinical validation (only one study involving clinician end-user testing), and 1 in-use evaluation. With this review, we aim to encourage rigorous, structured testing of QReports to elucidate how these tools can be integrated into clinical workflow for the assessment of MS.
Acknowledgements
The authors would like to thank all vendors for their participation and provision of product details.
Abbreviations
- MRI
Magnetic resonance imaging
- MS
Multiple sclerosis
- QReport
Quantitative volumetric reporting tool
- FLAIR
Fluid-attenuated inversion recovery
- QNI
Quantitative Neuroradiology Initiative
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- PROSPERO
Prospective Register of Systematic Reviews
- FDA
Food and Drug Administration
- CE
Conformité Européenne (French for “European conformity”)
- EDSS
Expanded Disability Status Scale
- SDMT
Symbol Digit Modalities Test
- WMH
White matter hyperintensity
- SPM
Statistical parametric mapping
- LST
Lesion Segmentation Tool
- samseg
Sequence Adaptive Multimodal SEGmentation
- SIENA(X)
Structural Image Evaluation, using Normalisation, of Atrophy
- BaMoS
Bayesian Model Selection
- Lesion-TOADS
Lesion TOpology-preserving Anatomical Segmentation
- BIANCA
Brain Intensity AbNormality Classification Algorithm
- T25FT
Timed 25-foot walk test
- 9HPT
9-Hole Peg Test
- NICE
National Institute for Health and Care Excellence
Funding
F.B. and F.P. are supported by the NIHR biomedical research center at UCLH.
Data availability
This review paper covers public data and data provided by the companies.
Code availability
The script for keyword searching in the FDA database is available at https://github.com/hughp1/FDA_KeyWordSearch/tree/main.
Declarations
Conflict of interest
Z.M., J.G., O.G., M.S., and J.N. declare no relationships with any companies whose products or services are featured in the subject matter of the article and no relevant financial or non-financial interests to disclose. H.P. is a full-time employee of GE Healthcare. F.P.C. received a Guarantors of Brain fellowship 2017–2020. F.B. is a steering committee and iDMC member for Biogen, Merck, Roche, and EISAI; consultant for Roche, Biogen, Merck, IXICO, Jansen, and Combinostics; and has research agreements with Novartis, Merck, Biogen, GE, and Roche. F.B. is a co-founder and shareholder of Queen Square Analytics Ltd.
Ethics approval/informed consent/consent to participate/consent to publish
Ethics approval, informed consent, consent to participate, and consent to publish were not necessary since this review paper covers public data and data provided by the companies with no human or animal data collection.
Footnotes
Key points
• A PROSPERO-registered systematic review identified 10 FDA- and/or CE-approved quantitative volumetric reporting tools (QReports) for the assessment of multiple sclerosis (MS).
• All tools identified provide longitudinal assessment of whole-brain volume and lesions with contextualization of results to normative reference populations.
• A total of 38 peer-reviewed studies covered technical (30 papers) and clinical validation (7 papers) and in-use evaluation (1 paper) of QReports in MS.
• Only one company has conducted a clinical validation study in which their QReport is tested by clinicians as end-users.
• There is limited evidence regarding clinical validation and in-use evaluation of commercial MS QReports, particularly involving clinician end-user testing.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.McGinley MP, Goldschmidt CH, Rae-Grant AD. Diagnosis and treatment of multiple sclerosis. JAMA. 2021;325:765. doi: 10.1001/jama.2020.26858. [DOI] [PubMed] [Google Scholar]
- 3.De Stefano N, Airas L, Grigoriadis N, Mattle HP, O’Riordan J, Oreja-Guevara C, et al. Clinical relevance of brain volume measures in multiple sclerosis. CNS Drugs. 2014;28:147–156. doi: 10.1007/s40263-014-0140-z. [DOI] [PubMed] [Google Scholar]
- 4.Popescu V, Agosta F, Hulst HE, Sluimer IC, Knol DL, Sormani MP, et al. Brain atrophy and lesion load predict long term disability in multiple sclerosis. J Neurol Neurosurg Psychiatry. 2013;84:1082–1091. doi: 10.1136/jnnp-2012-304094. [DOI] [PubMed] [Google Scholar]
- 5.Wattjes MP, Ciccarelli O, Reich DS, Banwell B, de Stefano N, Enzinger C, et al. 2021 MAGNIMS–CMSC–NAIMS consensus recommendations on the use of MRI in patients with multiple sclerosis. Lancet Neurol. 2021;20:653–670. doi: 10.1016/S1474-4422(21)00095-8. [DOI] [PubMed] [Google Scholar]
- 6.Bermel RA, Bakshi R. The measurement and clinical relevance of brain atrophy in multiple sclerosis. Lancet Neurol. 2006;5:158–170. doi: 10.1016/S1474-4422(06)70349-0. [DOI] [PubMed] [Google Scholar]
- 7.Giorgio A, De Stefano N. Clinical use of brain volumetry. J Magn Reson Imaging. 2013;37:1–14. doi: 10.1002/jmri.23671. [DOI] [PubMed] [Google Scholar]
- 8.Barkhof F, Calabresi PA, Miller DH, Reingold SC. Imaging outcomes for neuroprotection and repair in multiple sclerosis trials. Nat Rev Neurol. 2009;5:256–266. doi: 10.1038/nrneurol.2009.41. [DOI] [PubMed] [Google Scholar]
- 9.Sormani MP, Arnold DL, De Stefano N. Treatment effect on brain atrophy correlates with treatment effect on disability in multiple sclerosis. Ann Neurol. 2014;75:43–49. doi: 10.1002/ana.24018. [DOI] [PubMed] [Google Scholar]
- 10.Khaleeli Z, Ciccarelli O, Manfredonia F, Barkhof F, Brochet B, Cercignani M, et al. Predicting progression in primary progressive multiple sclerosis: a 10-year multicenter study. Ann Neurol. 2008;63:790–793. doi: 10.1002/ana.21375. [DOI] [PubMed] [Google Scholar]
- 11.Danelakis A, Theoharis T, Verganelakis DA. Survey of automated multiple sclerosis lesion segmentation techniques on magnetic resonance imaging. Comput Med Imaging Graph. 2018;70:83–100. doi: 10.1016/j.compmedimag.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Sastre-Garriga J, Pareto D, Rovira À. Brain atrophy in multiple sclerosis. Neuroimaging Clin N Am. 2017;27:289–300. doi: 10.1016/j.nic.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 13.van Walderveen MAA, Barkhof F, Hommes OR, Polman CH, Tobi H, Frequin STFM, et al. Correlating MRI and clinical disease activity in multiple sclerosis: relevance of hypointense lesions on short-TR/short- TE (T1-weighted) spin-echo images. Neurol. 1995;45:1684–1690. doi: 10.1212/WNL.45.9.1684. [DOI] [PubMed] [Google Scholar]
- 14.Filippi M, Horsfield MA, Bressi S, Martinelli V, Baratti C, Reganati P, et al. Intra- and inter-observer agreement of brain MRI lesion volume measurements in multiple sclerosis. Brain. 1995;118:1593–1600. doi: 10.1093/brain/118.6.1593. [DOI] [PubMed] [Google Scholar]
- 15.Ashton EA, Takahashi C, Berg MJ, Goodman A, Totterman S, Ekholm S. Accuracy and reproducibility of manual and semiautomated quantification of MS lesions by MRI. J Magn Reson Imaging. 2003;17:300–308. doi: 10.1002/jmri.10258. [DOI] [PubMed] [Google Scholar]
- 16.Miller DH. Measurement of atrophy in multiple sclerosis: pathological basis, methodological aspects and clinical relevance. Brain. 2002;125:1676–1695. doi: 10.1093/brain/awf177. [DOI] [PubMed] [Google Scholar]
- 17.Zeng C, Gu L, Liu Z, Zhao S (2020) Review of deep learning approaches for the segmentation of multiple sclerosis lesions on brain MRI. Front Neuroinform 1414:610967. 10.3389/fninf.2020.610967 [DOI] [PMC free article] [PubMed]
- 18.Van Hecke W, Costers L, Descamps A, Ribbens A, Nagels G, Smeets D, et al. A novel digital care management platform to monitor clinical and subclinical disease activity in multiple sclerosis. Brain Sci. 2021;11:1171. doi: 10.3390/brainsci11091171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain S, Ribbens A, Sima DM, Van Huffel S, Maes F, Smeets D (2017) Unsupervised framework for consistent longitudinal MS lesion segmentation. Medical computer vision and bayesian and graphical models for biomedical imaging. BAMBI MCV 2016 2016. Lect Notes Comput Sci 10081.208–19 10.1007/978-3-319-61188-4_19
- 20.Brune S, Høgestøl EA, Cengija V, Berg-Hansen P, Sowa P, Nygaard GO et al (2020) LesionQuant for assessment of MRI in multiple sclerosis—a promising supplement to the visual scan inspection. Front Neurol 11:546744. 10.3389/fneur.2020.546744 [DOI] [PMC free article] [PubMed]
- 21.Carass A, Roy S, Jog A, Cuzzocreo JL, Magrath E, Gherman A, et al. Longitudinal multiple sclerosis lesion segmentation: resource and challenge. Neuroimage. 2017;148:77–102. doi: 10.1016/j.neuroimage.2016.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krüger J, Opfer R, Gessert N, Ostwaldt A-C, Manogaran P, Kitzler HH, et al. Fully automated longitudinal segmentation of new or enlarged multiple sclerosis lesions using 3D convolutional neural networks. NeuroImage Clin. 2020;28:102445. doi: 10.1016/j.nicl.2020.102445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Commowick O, Cervenansky F, Cotton F, Dojat M (2021) MSSEG-2 challenge proceedings: multiple sclerosis new lesions segmentation challenge using a data management and processing infrastructure. MICCAI 2021 - 24th International Conference on Medical Image Computing and Computer Assisted Intervention, Strasbourg, France pp 126, 2021. hal-03358968v3f
- 24.Commowick O, Istace A, Kain M, Laurent B, Leray F, Simon M, et al. Objective evaluation of multiple sclerosis lesion segmentation using a data management and processing infrastructure. Sci Rep. 2018;8:13650. doi: 10.1038/s41598-018-31911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran P, Thoprakarn U, Gourieux E, dos Santos CL, Cavedo E, Guizard N, et al. Automatic segmentation of white matter hyperintensities: validation and comparison with state-of-the-art methods on both Multiple Sclerosis and elderly subjects. NeuroImage Clin. 2022;33:102940. doi: 10.1016/j.nicl.2022.102940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain S, Sima DM, Ribbens A, Cambron M, Maertens A, Van Hecke W, et al. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage Clin. 2015;8:367–375. doi: 10.1016/j.nicl.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krüger J, Ostwaldt AC, Spies L, Geisler B, Schlaefer A, Kitzler HH, et al. Infratentorial lesions in multiple sclerosis patients: intra- and inter-rater variability in comparison to a fully automated segmentation using 3D convolutional neural networks. Eur Radiol. 2021;32(4):2798–2809. doi: 10.1007/s00330-021-08329-3. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Beadnall HN, Hatton SN, Bader G, Tomic D, Silva DG, et al. Automated brain volumetrics in multiple sclerosis: a step closer to clinical application. J Neurol Neurosurg Psychiatry. 2016;87:754–757. doi: 10.1136/jnnp-2015-312304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forbes Florence, Doyle Senan, García-Lorenzo Daniel, Christian Barillot MD. A weighted multi-sequence Markov model for brain lesion segmentation. AISTATS. 2010;2010:225–32. [Google Scholar]
- 30.Cavedo E, Tran P, Thoprakarn U, Martini JB, Movschin A, Delmaire C, et al. Validation of an automatic tool for the rapid measurement of brain atrophy and white matter hyperintensity: QyScore®. Eur Radiol. 2022;32(5):2949–2961. doi: 10.1007/s00330-021-08385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain S, Ribbens A, Sima DM, Cambron M, De Keyser J, Wang C et al (2016) Two time point MS lesion segmentation in brain MRI: an expectation-maximization framework. Front Neurosci 10:576. 10.3389/fnins.2016.00576 [DOI] [PMC free article] [PubMed]
- 32.Sima DM, Esposito G, Van Hecke W, Ribbens A, Nagels G, Smeets D. Health economic impact of software-assisted brain MRI on therapeutic decision-making and outcomes of relapsing-remitting multiple sclerosis patients—a microsimulation study. Brain Sci. 2021;11:1570. doi: 10.3390/brainsci11121570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finkelsztejn A, Fragoso YD, Bastos EA, Duarte JA, Varela JS, Houbrechts R, et al. Intercontinental validation of brain volume measurements using MSmetrix. Neuroradiol J. 2018;31:147–149. doi: 10.1177/1971400917721521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodkin O, Pemberton H, Vos SB, Prados F, Sudre CH, Moggridge J, et al. The quantitative neuroradiology initiative framework: application to dementia. Br J Radiol. 2019;92:20190365. doi: 10.1259/bjr.20190365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pemberton HG, Zaki LAM, Goodkin O, Das RK, Steketee RME, Barkhof F, et al. Technical and clinical validation of commercial automated volumetric MRI tools for dementia diagnosis—a systematic review. Neuroradiol. 2021;63:1773–1789. doi: 10.1007/s00234-021-02746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700–b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4:1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta- analyses: the PRISMA statement. BMJ. 2009;339:b2535–b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews PM, Federico A, et al. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage. 2002;17:479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt P, Gaser C, Arsic M, Buck D, Förschler A, Berthele A, et al. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage. 2012;59:3774–3783. doi: 10.1016/j.neuroimage.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt P, Pongratz V, Küster P, Meier D, Wuerfel J, Lukas C, et al. Automated segmentation of changes in FLAIR-hyperintense white matter lesions in multiple sclerosis on serial magnetic resonance imaging. NeuroImage Clin. 2019;23:101849. doi: 10.1016/j.nicl.2019.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valverde S, Salem M, Cabezas M, Pareto D, Vilanova JC, Ramió-Torrentà L, et al. One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks. NeuroImage Clin. 2019;21:101638. doi: 10.1016/j.nicl.2018.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerri S, Puonti O, Meier DS, Wuerfel J, Mühlau M, Siebner HR, et al. A contrast-adaptive method for simultaneous whole-brain and lesion segmentation in multiple sclerosis. Neuroimage. 2021;225:117471. doi: 10.1016/j.neuroimage.2020.117471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rieu Z, Kim J, Kim RE, Lee M, Lee MK, Oh SW, et al. Semi-supervised learning in medical MRI segmentation: brain tissue with white matter hyperintensity segmentation using FLAIR MRI. Brain Sci. 2021;11:720. doi: 10.3390/brainsci11060720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudre CH, Cardoso MJ, Bouvy WH, Biessels GJ, Barnes J, Ourselin S. Bayesian model selection for pathological neuroimaging data applied to white matter lesion segmentation. IEEE Trans Med Imaging. 2015;34:2079–2102. doi: 10.1109/TMI.2015.2419072. [DOI] [PubMed] [Google Scholar]
- 47.Granberg T, Uppman M, Hashim F, Cananau C, Nordin LE, Shams S, et al. Clinical feasibility of synthetic MRI in multiple sclerosis: a diagnostic and volumetric validation study. Am J Neuroradiol. 2016;37:1023–1029. doi: 10.3174/ajnr.A4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vågberg M, Lindqvist T, Ambarki K, Warntjes JBM, Sundström P, Birgander R, et al. Automated determination of brain parenchymal fraction in multiple sclerosis. Am J Neuroradiol. 2013;34:498–504. doi: 10.3174/ajnr.A3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salzer J, Svenningsson R, Alping P, Novakova L, Björck A, Fink K, et al. Rituximab in multiple sclerosis. Neurol. 2016;87:2074–2081. doi: 10.1212/WNL.0000000000003331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagiwara A, Hori M, Kamagata K, Warntjes M, Matsuyoshi D, Nakazawa M, et al. Myelin measurement: comparison between simultaneous tissue relaxometry, magnetization transfer saturation index, and T1w/T2w ratio methods. Sci Rep. 2018;8:10554. doi: 10.1038/s41598-018-28852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Liu S, Wang Y, Kang Y, Haacke EM. STrategically Acquired Gradient Echo (STAGE) imaging, part I: creating enhanced T1 contrast and standardized susceptibility weighted imaging and quantitative susceptibility mapping. Magn Reson Imaging. 2018;46:130–139. doi: 10.1016/j.mri.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Haacke EM, Chen Y, Utriainen D, Wu B, Wang Y, Xia S, et al. STrategically Acquired Gradient Echo (STAGE) imaging, part III: technical advances and clinical applications of a rapid multi-contrast multi-parametric brain imaging method. Magn Reson Imaging. 2020;65:15–26. doi: 10.1016/j.mri.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 53.Thompson AJ, Banwell BL, Barkhof F, Carroll WM, Coetzee T, Comi G, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17:162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- 54.Valverde S, Oliver A, Roura E, Pareto D, Vilanova JC, Ramió-Torrentà L, et al. Quantifying brain tissue volume in multiple sclerosis with automated lesion segmentation and filling. NeuroImage Clin. 2015;9:640–647. doi: 10.1016/j.nicl.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaki LAM, Vernooij MW, Smits M, Tolman C, Papma JM, Visser JJ et al (2022) Comparing two artificial intelligence software packages for normative brain volumetry in memory clinic imaging. Neuroradiol 64(7):1359–1366 10.1007/s00234-022-02898-w [DOI] [PMC free article] [PubMed]
- 56.Jokinen H, Koikkalainen J, Laakso HM, Melkas S, Nieminen T, Brander A, et al. Global burden of small vessel disease–related brain changes on MRI predicts cognitive and functional decline. Stroke. 2020;51:170–178. doi: 10.1161/STROKEAHA.119.026170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lötjönen J, Wolz R, Koikkalainen J, Julkunen V, Thurfjell L, Lundqvist R, et al. Fast and robust extraction of hippocampus from MR images for diagnostics of Alzheimer’s disease. Neuroimage. 2011;56:185–196. doi: 10.1016/j.neuroimage.2011.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ochs AL, Ross DE, Zannoni MD, Abildskov TJ, Bigler ED. Comparison of automated brain volume measures obtained with NeuroQuant® and FreeSurfer. J Neuroimaging. 2015;25:721–727. doi: 10.1111/jon.12229. [DOI] [PubMed] [Google Scholar]
- 59.Relkin NR, Thomas RG, Rissman RA, Brewer JB, Rafii MS, van Dyck CH, et al. A phase 3 trial of IV immunoglobulin for Alzheimer disease. Neurol. 2017;88:1768–1775. doi: 10.1212/WNL.0000000000003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niemantsverdriet E, Ribbens A, Bastin C, Benoit F, Bergmans B, Bier J-C, et al. A retrospective Belgian multi-center MRI biomarker study in Alzheimer’s disease (REMEMBER) J Alzheimer’s Dis. 2018;63:1509–1522. doi: 10.3233/JAD-171140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hedderich DM, Dieckmeyer M, Andrisan T, Ortner M, Grundl L, Schön S, et al. Normative brain volume reports may improve differential diagnosis of dementing neurodegenerative diseases in clinical practice. Eur Radiol. 2020;30:2821–2829. doi: 10.1007/s00330-019-06602-0. [DOI] [PubMed] [Google Scholar]
- 62.Opfer R, Suppa P, Kepp T, Spies L, Schippling S, Huppertz H-J. Atlas based brain volumetry: how to distinguish regional volume changes due to biological or physiological effects from inherent noise of the methodology. Magn Reson Imaging. 2016;34:455–461. doi: 10.1016/j.mri.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 63.Menze BH, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) IEEE Trans Med Imaging. 2015;34(10):1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chupin M, Gérardin E, Cuingnet R, Boutet C, Lemieux L, Lehéricy S, et al. Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009;19:579–587. doi: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Damangir S, Manzouri A, Oppedal K, Carlsson S, Firbank MJ, Sonnesyn H, et al. Multispectral MRI segmentation of age related white matter changes using a cascade of support vector machines. J Neurol Sci. 2012;322:211–216. doi: 10.1016/j.jns.2012.07.064. [DOI] [PubMed] [Google Scholar]
- 67.Shiee N, Bazin P-L, Ozturk A, Reich DS, Calabresi PA, Pham DL. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage. 2010;49:1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coupé P, Tourdias T, Linck P, Romero JV, Manjon JV (2018) Lesion brain: an online tool for white matter lesion segmentation. international workshop on patch-based techniques in medical imaging (MICCAI) p 95 - 103, hal-01918438
- 69.Griffanti L, Zamboni G, Khan A, Li L, Bonifacio G, Sundaresan V, et al. BIANCA (Brain Intensity AbNormality Classification Algorithm): a new tool for automated segmentation of white matter hyperintensities. Neuroimage. 2016;141:191–205. doi: 10.1016/j.neuroimage.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hänninen K, Viitala M, Paavilainen T, Karhu JO, Rinne J, Koikkalainen J et al (2020) Thalamic atrophy predicts 5-year disability progression in multiple sclerosis. Front Neurol 11:606. 10.3389/fneur.2020.00606 [DOI] [PMC free article] [PubMed]
- 71.Pareto D, Sastre-Garriga J, Alberich M, Auger C, Tintoré M, Montalban X, et al. Brain regional volume estimations with NeuroQuant and FIRST: a study in patients with a clinically isolated syndrome. Neuroradiol. 2019;61:667–674. doi: 10.1007/s00234-019-02191-3. [DOI] [PubMed] [Google Scholar]
- 72.Lysandropoulos AP, Absil J, Metens T, Mavroudakis N, Guisset F, Van Vlierberghe E et al (2016) Quantifying brain volumes for multiple sclerosis patients follow‐up in clinical practice – comparison of 1.5 and 3 Tesla magnetic resonance imaging. Brain Behav 6(2):e00422. 10.1002/brb3.422 [DOI] [PMC free article] [PubMed]
- 73.Smeets D, Ribbens A, Sima DM, Cambron M, Horakova D, Jain S et al (2016) Reliable measurements of brain atrophy in individual patients with multiple sclerosis. Brain Behav 6(9):e00518. 10.1002/brb3.518 [DOI] [PMC free article] [PubMed]
- 74.Storelli L, Rocca MA, Pagani E, Van Hecke W, Horsfield MA, De Stefano N, et al. Measurement of whole-brain and gray matter atrophy in multiple sclerosis: assessment with MR imaging. Radiol. 2018;288:554–564. doi: 10.1148/radiol.2018172468. [DOI] [PubMed] [Google Scholar]
- 75.Rakić M, Vercruyssen S, Van Eyndhoven S, de la Rosa E, Jain S, Van Huffel S, et al. icobrain ms 5.1: Combining unsupervised and supervised approaches for improving the detection of multiple sclerosis lesions. NeuroImage Clin. 2021;31:102707. doi: 10.1016/j.nicl.2021.102707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beadnall HN, Wang C, Van Hecke W, Ribbens A, Billiet T, Barnett MH. Comparing longitudinal brain atrophy measurement techniques in a real-world multiple sclerosis clinical practice cohort: towards clinical integration? Ther Adv Neurol Disord. 2019;12:175628641882346. doi: 10.1177/1756286418823462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steenwijk MD, Amiri H, Schoonheim MM, de Sitter A, Barkhof F, Pouwels PJW, et al. Agreement of MSmetrix with established methods for measuring cross-sectional and longitudinal brain atrophy. NeuroImage Clin. 2017;15:843–853. doi: 10.1016/j.nicl.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Billast M, Meyer MI, Sima DM, Robben D (2020) Improved Inter-scanner MS Lesion Segmentation by Adversarial Training on Longitudinal Data. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes, (2019) Lect Notes Comput Sci 11992. Springer, Cham. 10.1007/978-3-030-46640-4_10
- 79.Meyer MI, de la Rosa E, Pedrosa de Barros N, Paolella R, Van Leemput K, Sima DM (2021) A Contrast Augmentation Approach to Improve Multi-Scanner Generalization in MRI. Front Neurosci. 15:708196. 10.3389/fnins.2021.708196 [DOI] [PMC free article] [PubMed]
- 80.Fragoso YD, Wille PR, Abreu M, Brooks JBB, Dias RM, Duarte JA, et al. Correlation of clinical findings and brain volume data in multiple sclerosis. J Clin Neurosci. 2017;44:155–157. doi: 10.1016/j.jocn.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 81.D’hooghe MB, Gielen J, Van Remoortel A, D’haeseleer M, Peeters E, Cambron M, et al. Single MRI-based volumetric assessment in clinical practice is associated with MS-related disability. J Magn Reson Imaging. 2019;49:1312–21. doi: 10.1002/jmri.26303. [DOI] [PubMed] [Google Scholar]
- 82.Gessert N, Krüger J, Opfer R, Ostwaldt A-C, Manogaran P, Kitzler HH, et al. Multiple sclerosis lesion activity segmentation with attention-guided two-path CNNs. Comput Med Imaging Graph. 2020;84:101772. doi: 10.1016/j.compmedimag.2020.101772. [DOI] [PubMed] [Google Scholar]
- 83.Opfer R, Ostwaldt A-C, Sormani MP, Gocke C, Walker-Egger C, Manogaran P, et al. Estimates of age-dependent cutoffs for pathological brain volume loss using SIENA/FSL—a longitudinal brain volumetry study in healthy adults. Neurobiol Aging. 2018;65:1–6. doi: 10.1016/j.neurobiolaging.2017.12.024. [DOI] [PubMed] [Google Scholar]
- 84.Opfer R, Ostwaldt A-C, Walker-Egger C, Manogaran P, Sormani MP, De Stefano N, et al. Within-patient fluctuation of brain volume estimates from short-term repeated MRI measurements using SIENA/FSL. J Neurol. 2018;265:1158–1165. doi: 10.1007/s00415-018-8825-8. [DOI] [PubMed] [Google Scholar]
- 85.Opfer R, Krüger J, Spies L, Hamann M, Wicki CA, Kitzler HH, et al. Age-dependent cut-offs for pathological deep gray matter and thalamic volume loss using Jacobian integration. NeuroImage Clin. 2020;28:102478. doi: 10.1016/j.nicl.2020.102478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Opfer R, Krüger J, Spies L, Kitzler HH, Schippling S, Buchert R (2022) Single-subject analysis of regional brain volumetric measures can be strongly influenced by the method for head size adjustment. Neuroradiol 64(10):2001–2009 10.1007/s00234-022-02961-6 [DOI] [PMC free article] [PubMed]
- 87.Raji A, Ostwaldt A-C, Opfer R, Suppa P, Spies L, Winkler G (2018) MRI-based brain volumetry at a single time point complements clinical evaluation of patients with multiple sclerosis in an outpatient setting. Front Neurol 9:545. 10.3389/fneur.2018.00545 [DOI] [PMC free article] [PubMed]
- 88.Dieckmeyer M, Roy AG, Senapati J, Wachinger C, Grundl L, Döpfert J, et al. Effect of MRI acquisition acceleration via compressed sensing and parallel imaging on brain volumetry. Magn Reson Mater Physics, Biol Med. 2021;34:487–497. doi: 10.1007/s10334-020-00906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Forbes F, Doyle S, Garcia-Lorenzo D, Barillot C, Dojat M (2010) Adaptive weighted fusion of multiple MR sequences for brain lesion segmentation. IEEE Int Symp Biomed Imaging From Nano to Macro 69–72. 10.1109/ISBI.2010.5490413
- 90.Lambert B, Louis M, Doyle S, Forbes F, Dojat M, AT (2021) Leveraging 3D information in unsupervised Brain MRI segmentation. ISBI - 18th International Symposium on Biomedical Imaging, pp1–4 ⟨hal-03273953⟩
- 91.Mortazavi D, Kouzani AZ, Soltanian-Zadeh H. Segmentation of multiple sclerosis lesions in MR images: a review. Neuroradiol. 2012;54:299–320. doi: 10.1007/s00234-011-0886-7. [DOI] [PubMed] [Google Scholar]
- 92.García-Lorenzo D, Francis S, Narayanan S, Arnold DL, Collins DL. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal. 2013;17:1–18. doi: 10.1016/j.media.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 93.Pagnozzi AM, Fripp J, Rose SE. Quantifying deep grey matter atrophy using automated segmentation approaches: a systematic review of structural MRI studies. Neuroimage. 2019;201:116018. doi: 10.1016/j.neuroimage.2019.116018. [DOI] [PubMed] [Google Scholar]
- 94.Kaur A, Kaur L, Singh A. State-of-the-art segmentation techniques and future directions for multiple sclerosis brain lesions. Arch Comput Methods Eng. 2021;28:951–977. doi: 10.1007/s11831-020-09403-7. [DOI] [Google Scholar]
- 95.Shanmuganathan M, Almutairi S, Aborokbah MM, Ganesan S, Ramachandran V. Review of advanced computational approaches on multiple sclerosis segmentation and classification. IET Signal Process. 2020;14:333–341. doi: 10.1049/iet-spr.2019.0543. [DOI] [Google Scholar]
- 96.Traboulsee A, Simon JH, Stone L, Fisher E, Jones DE, Malhotra A, et al. Revised recommendations of the Consortium of MS Centers Task Force for a Standardized MRI Protocol and Clinical Guidelines for the Diagnosis and Follow-Up of Multiple Sclerosis. Am J Neuroradiol. 2016;37:394–401. doi: 10.3174/ajnr.A4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu XY, Rajendran L, Lapointe E, Tam R, Li D, Traboulsee A, et al. Three-dimensional MRI sequences in MS diagnosis and research. Mult Scler J. 2019;25:1700–1709. doi: 10.1177/1352458519848100. [DOI] [PubMed] [Google Scholar]
- 98.US Food and Drug Administration (2021) Artificial intelligence/machine learning (ai/ml)-based software as a medical device (SaMD) Action Plan. FDA U.S. food & drug administration center for devices & radiological health. https://www.fda.gov/media/145022/download. Accessed 21 Apr 2022
- 99.U.S. Food and Drug Administration (FDA), Health Canada and the UKM and H products RA (MHRA). Good Machine Learning Practice for Medical Device Development: Guiding Principles 2021. https://www.fda.gov/medical-devices/software-medical-device-samd/good-machine-learning-practice- medical-device-development-guiding-principles (accessed April 21, 2022).
- 100.European Parliament and Council of the European Union (2017) Regulation (EU) 2017/745 of the European parliament and of the councilof 5 April 2017 on medical devices, amending Directive 2001/83/EC, Regulation (EC) No 178/2002 and Regulation (EC) No 1223/2009 and repealing Council Directives 90/385/EEC and 93/42/EEC. Official Journal of the European Union. https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32017R0745. Accessed 21 Apr 2022
- 101.European Parliament and the Council of the European Union (2020) Regulation (EU) 2020/561 of the European parliament and of the council of 23 April 2020 amending Regulation (EU) 2017/745 on medical devices, as regards the dates of application of certain of its provisions. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex:32020R0561. Accessed 21 Apr 2022
- 102.European Commission (2021) Communication from the commission to the European parliament, the council, the European economic and social committee and the committee of the regions: fostering a European approach to artificial intelligence. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=COM%3A2021%3A205%3AFIN. Accessed 21 Apr 2022
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This review paper covers public data and data provided by the companies.
The script for keyword searching in the FDA database is available at https://github.com/hughp1/FDA_KeyWordSearch/tree/main.