Abstract
Introduction of new covalent bonds in proteins is affording novel avenues for protein research and applications, yet it remains difficult to generate covalent linkages at all possible sites and across diverse protein classes. Herein, we genetically encoded meta-fluorosulfate-L-tyrosine (mFSY) to selectively react with lysine, tyrosine, and histidine via proximity-enabled SuFEx reaction. mFSY was able to target residues elusive to previous Uaas, and permitted engineering of various proteins including affibody, nanobody, and Fab into covalent binders that irreversibly cross-linked EGFR and HER2 receptors. mFSY is thus valuable for developing covalent proteins for biological research, synthetic biology, and biotherapeutics.
Adding new covalent bonding capability to proteins would offer diverse properties unattainable with natural proteins, and would enable novel avenues for researching and engineering proteins and protein-involved biological processes.1 In recent years, latent bioreactive unnatural amino acids (Uaas) have been designed and site-specifically incorporated into proteins through genetic code expansion.2–5 These latent bioreactive Uaas react with natural amino acid residues through proximity-enabled reactivity, generating covalent linkages within or between proteins specifically. The resultant covalent linkages have been harnessed to enhance protein properties, to probe protein interactions, and to develop covalent protein drugs. 6–11 The most challenging aspect of developing a latent bioreactive Uaa is to finely balance its biocompatibility and reactivity.12 The Uaa should not react with any biomolecules inside cells to avoid off-target reactions and cytotoxicity, while simultaneously being able to react with the target residue in high efficiency under mild cellular conditions.
In fulfilling these demanding requirements, aryl fluorosulfate has emerged as one of the most attractive warheads to use in latent bioreactive Uaas.13 Aryl fluorosulfates are quite stable and inert in cells, but become reactive toward weak nucleophiles only when brought into close proximity, proceeding via the sulfur fluoride exchange (SuFEx) click reaction in water, at physiological pH, and without any catalyst or additive needed.14–16 We previously genetically encoded the first aryl fluorosulfate-containing Uaa, fluorosulfate-L-tyrosine (FSY), demonstrating its ability to selectively crosslink proteins in vitro and in cells through SuFEx reaction with proximal Lys, His, or Tyr residues.17 FSY has been subsequently used to study protein-protein interactions in cells and to develop covalent protein therapeutics.9,18 Most recently, we further developed and genetically encoded fluorosulfonyloxybenzoyl-L-lysine (FSK), which has a longer side chain than FSY to achieve a larger radius of reactivity.19 Nonetheless, we discovered many situations where FSK was too long to be accommodated and FSY’s rigid para-pointing warhead did not orient toward the target residue for reaction despite the close proximity. To date, only a few proteins have been covalently engineered with FSY and FSK;1 the general applicability of a SuFEx warhead to diverse proteins still awaits demonstration.
To expand the proximity-enabled SuFEx reactivity to a broader range of proteins with greater site diversity, here we developed another aryl fluorosulfate Uaa, meta-fluorosulfate-L-tyrosine (mFSY). A new orthogonal tRNA/synthetase pair was generated to genetically encode mFSY into proteins in E. coli and mammalian cells. Through the incorporation of mFSY, we engineered affibody and nanobody proteins into covalent binders for HER2 and EGFR. We further showcased the first example of generating a covalent Fab irreversibly crosslinked to HER2. Moreover, owing to the meta positioning of the fluorosulfate, mFSY was able to react with residues elusive to FSY. These results collectively indicate that mFSY can be generally incorporated into various proteins to expand the covalent engineering of proteins.
We designed mFSY to complement FSY and FSK and further expand covalent targeting abilities (Fig. 1a). While FSK has a long and flexible side chain to reach target residues at further distances, FSY is similar to tyrosine in structure and side chain length. Thus, FSK is suitable for use at sites peripheral to a protein-protein binding interface, whereas FSY can be used inside the interface without disrupting the protein interaction. However, the rigid side chain of FSY and the often compact protein binding interface can make FSY unable to react with the target residue when its para-fluorosulfate warhead is not oriented toward the target residue despite their close proximity. We reasoned that putting fluorosulfate at the meta position, as in mFSY, would allow for the targeting of residues that are unable to react with FSY due to orientation misalignment. In addition, the phenyl ring of mFSY or FSY could rotate around the Cβ-Cγ bond, which would increase the reaction area of fluorosulfate when installed at the meta than the para position. Using the same warhead, mFSY should have similar reactivity as FSY to target multiple nucleophilic residues via proximity-enabled SuFEx reaction.
Fig. 1. mFSY incorporation into proteins in E. coli and mammalian cells via genetic code expansion.
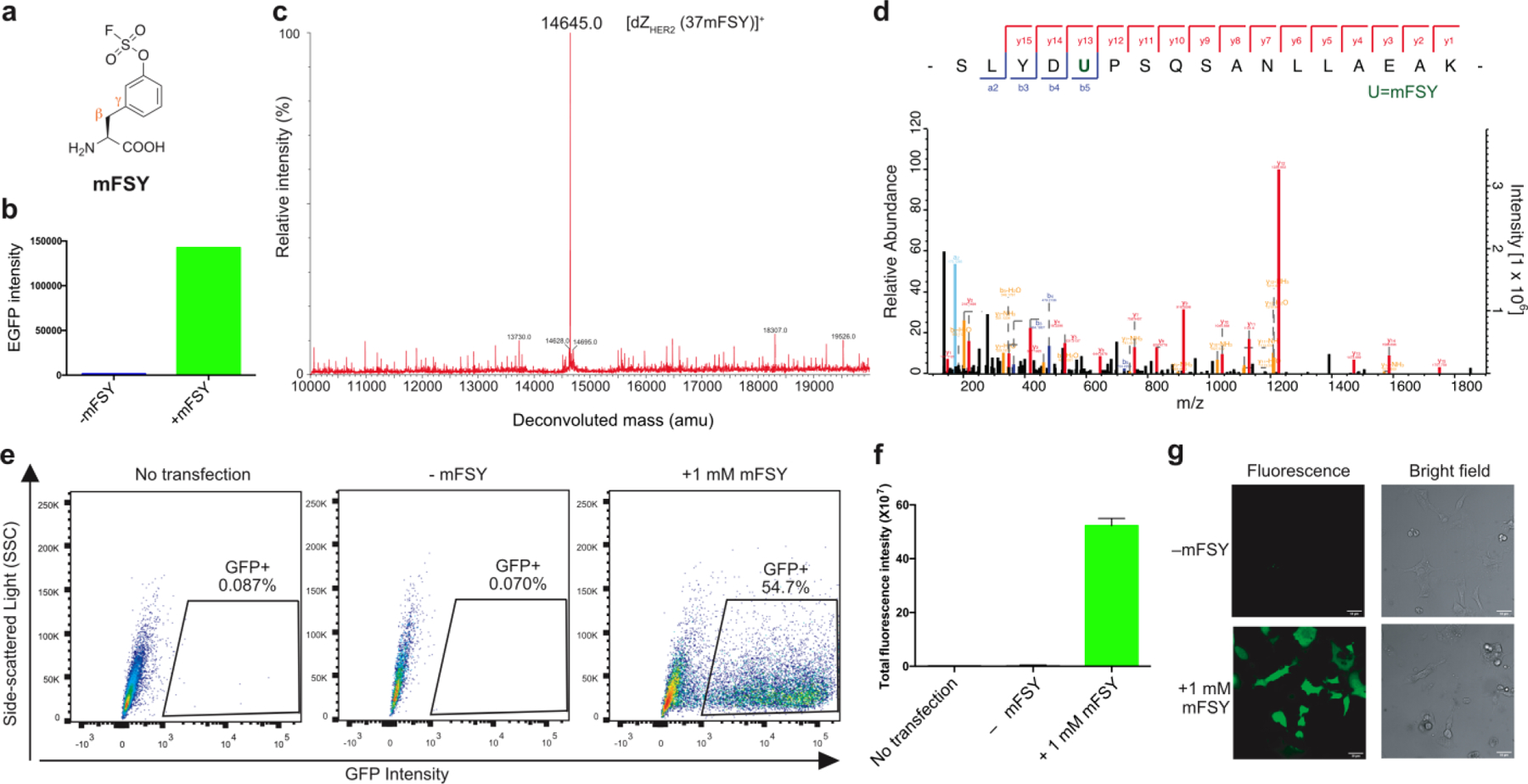
(a) Structure of mFSY. (b) Fluorescence intensity of E. coli cells expressing tRNAPyl/mFSYRS and EGFP(182TAG) in the absence or presence of 1 mM mFSY. Same number of cells were compared. (c) Mass spectrum of the intact dZHER2 (37mFSY) protein. (d) Tandem mass spectrometric analysis showing the dZHER2 peptide with mFSY clearly incorporated at position 37 (indicated by the green U). (e) Flow cytometric analysis of mFSY incorporation into HeLa-GFP(182TAG) reporter cells. Data is representative of three biological replicates. (f) Total GFP fluorescence intensity from flow cytometric analysis of the same number of HeLa-GFP(182TAG) reporter cells. Error bar: s.d., n = 3. (g) Fluorescence and brightfield microscopic images of HeLa-GFP(182TAG) reporter cells with or without 1 mM mFSY added in cell culture.
mFSY was efficiently synthesized using [4-(acetylamino)phenyl] imidodisulfuryl difluoride (AISF) in two steps.20 To genetically encode mFSY, we evolved an orthogonal tRNAPyl/synthetase pair for mFSY incorporation in response to the amber stop codon. Using the small-intelligent mutagenesis approach,21 residues Ala302, Leu305, Tyr306, Leu309, Ile322, Asn346, Cys348, Tyr384, Val401, and Trp417 of the Methanosarcina mazei PylRS were mutated to create a PylRS mutant library, which was subjected to selection as described.22,23 A clone showing an mFSY-dependent phenotype was identified (Fig. S1), and the synthetase was found to contain the following mutations L305M/I322T/N346G, herein called mFSYRS.
To evaluate mFSY incorporation into proteins, we expressed tRNAPyl/mFSYRS together with the EGFP gene containing a TAG codon at the permissive site 182 in E. coli. In the absence of mFSY in the growth media, only background EGFP fluorescence was detected; when 1 mM of mFSY was added, strong EGFP fluorescence was measured with intensity increased over 100-fold (Fig. 1b), suggesting mFSY incorporation into EGFP. Using the tRNAPyl/mFSYRS pair, we also incorporated mFSY into a dimeric affibody dZHER2 at site 37 in E. coli. Full-length dZHER2 protein was obtained only when mFSY was added to growth media. The purified intact dZHER2(37mFSY) protein was analyzed by electrospray ionization high-resolution mass spectrometry (Fig. 1c). A peak observed at 14645.0 Da corresponds to intact dZHER2 containing mFSY at site 37 (expected 14645.0 Da). Notably, no peaks corresponding to dZHER2 containing other amino acids at site 37 were observed. The dZHER2(37mFSY) protein was further digested and analyzed by tandem MS (Fig. 1d). A series of b and y ions clearly indicated that mFSY was incorporated at site 37 specified by the TAG codon. These results showed that the evolved tRNAPyl/mFSYRS pair was able to incorporated mFSY into proteins with high efficiency and specificity in E. coli.
We further assessed the incorporation of mFSY in mammalian cells (Fig. 1e). mFSYRS and tRNAPyl were cloned into a mammalian expression vector then transfected into HeLa-GFP(182TAG) cells, a stable cell line expressing genome-integrated GFP gene with a TAG codon at permissive site 182.24 When 1 mM of mFSY was added to the cell culture, flow cytometric analysis showed that 54.70 % of cells became green fluorescent (Fig. 1e). Cells cultured without mFSY had negligible background fluorescence, whereas cells cultured with 1 mM mFSY showed 200-fold increase in fluorescence intensity (Fig. 1f). Fluorescence microscopic images further confirmed that fluorescent full-length GFP was produced in transfected HeLa-GFP(182TAG) cells only when mFSY was added to growth media (Fig. 1g). These results indicate that the tRNAPyl/mFSYRS pair was able to efficiently incorporate mFSY into GFP in mammalian cells.
To evaluate mFSY as a new latent bioreactive Uaa for creating covalent linkages in proteins, we incorporated it into various protein binders and tested the resultant protein binders’ ability to crosslink their targets. First, we incorporated mFSY into an affibody ZHER2 specific for the tyrosine kinase receptor HER2. Affibodies are derived from staphylococcal protein A as antibody mimetics and can be evolved to bind different proteins. On the basis of the crystal structure of ZHER2 in complex with HER2,25 two sites, D36 and D37, were chosen for mFSY incorporation to target the proximal nucleophilic residue H490 on the HER2 receptor (Fig. 2a). The dimeric form of ZHER2 (dZHER2) was used to increase the binding affinity (KD = 6 pM),26 and mFSY was incorporated in the N-terminal ZHER2 monomer. mFSY-incorporated dZHER2 proteins were expressed and purified from E. coli, and incubated with HER2 extracellular domain (ECD) in PBS buffer to allow cross-linking for different time durations, followed by Western blot analysis (Fig. 2b, c). FSY was similarly incorporated into dZHER2 for comparison. At both sites, mFSY was shown to efficiently crosslink with the HER2 receptor in a time-dependent manner (Fig. 2b, c). Cross-linking could be detected at 0.5 h. When compared with dZHER2 mutants incorporating FSY at the same site, mFSY mutants crosslinked HER2 with a similar efficiency, suggesting that mFSY was as capable as FSY for certain protein cross-linking purposes. When WT-dZHER2 was added to compete, the cross-linking efficiency of dZHER2(37mFSY) with HER2 ECD decreased with the increasing concentration of WT-dZHER2 (Fig. 2d), indicating that the cross-linking was dependent on dZHER2/HER2 interaction. These results demonstrate that mFSY could be used to engineer affibody proteins into covalent binders.
Fig. 2. mFSY facilitates cross-linking between affibody dimer dZHER2 and HER2 receptor.
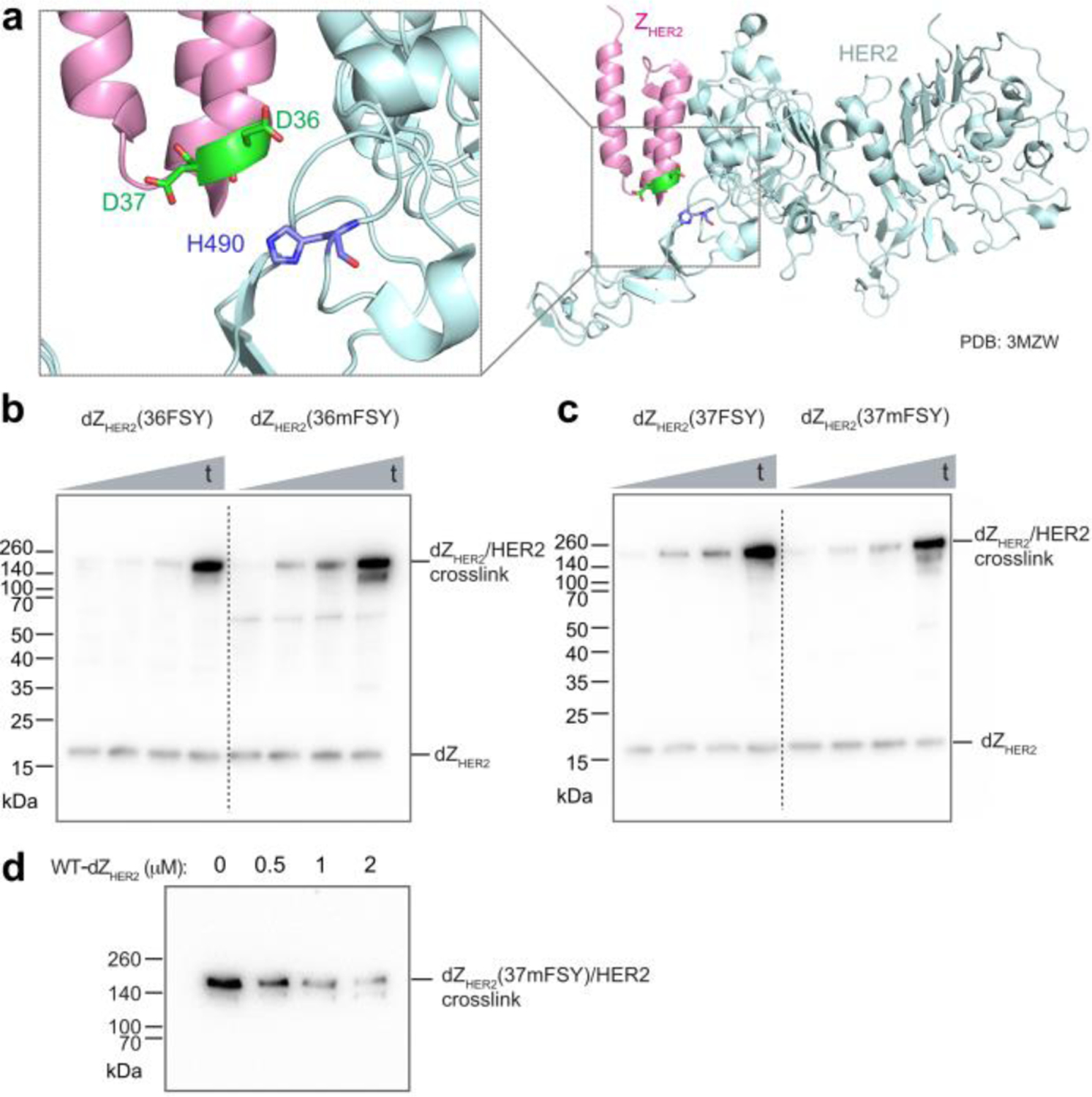
(a) Structure of affibody ZHER2 in complex with the ECD of HER2, showing D36 and D37 on the affibody in proximity to H490 of HER2. (b) Western blot analysis of in vitro cross-linking between HER2 ECD and dZHER2-36FSY or -36mFSY mutant. t: 0.5, 2, 4, and 24 h. (c) Western blot analysis of in vitro cross-linking between HER2 ECD and dZHER2-37FSY or -37mFSY mutant. t: 0.5, 2, 4, and 24 h. (d) Western blot analysis showing that increasing WT-dZHER2 decreased cross-linking between dZHER2(37mFSY) (1 μM) and HER2 ECD (1 μM).
We next incorporated mFSY into nanobodies, which are single-domain antibodies able to bind different antigens. In one example, we incorporated mFSY into nanobody 7D12, which specifically binds to the epidermal growth factor receptor (EGFR). The structure of 7D12-EGFR complex suggests that E44 of 7D12 is in close proximity to K443 of EGFR (Fig. 3a).27 7D12(44mFSY) mutant protein was purified in E. coli, and showed robust cross-linking with EGFR in a higher efficiency (47.3%) than the 7D12(44FSY) mutant (28.7%) and the 7D12(44FSK) mutant (22.3%) (Fig. 3b). In another example, we incorporated mFSY into nanobody 2Rs15d, which is specific for the HER2 receptor. On the basis of the crystal structure of 2Rs15d-HER2 complex, within the binding interface, Tyr37 of 2Rs15d has its meta position oriented toward the hydroxyl of Tyr112 of HER2, while its para hydroxyl is pointing away (Fig. 3c).28 We reasoned that mFSY, but not FSY, incorporated at site 37 should be able to crosslink Tyr112 of HER2. We characterized the purified intact 2Rs15d(37mFSY) protein with mass spectrometry, which showed major monomeric and minor non-cross-linked dimeric 2Rs15d(37mFSY) species (Fig. S3). As expected, 2Rs15d(37mFSY) led to crosslinking of the HER2 receptor, whereas 2Rs15d(37FSY) did not (Fig. 3d), demonstrating the mFSY could complement FSY in crosslinking side chains of different orientation as designed.
Fig. 3. mFSY-containing nanobodies crosslink their target proteins.
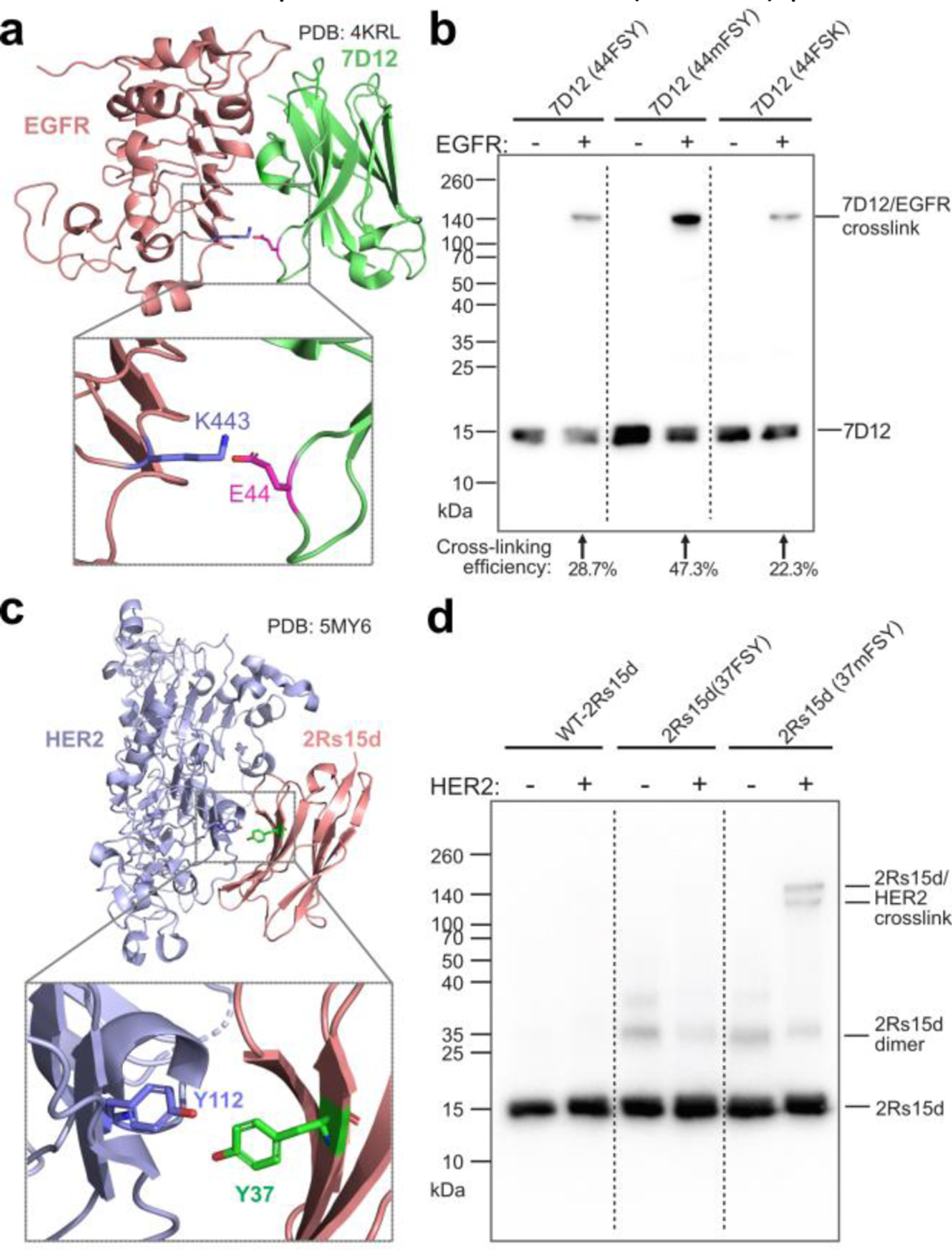
(a) Structure of nanobody 7D12 in complex with EGFR, showing E44 on 7D12 in proximity to K443 of EGFR. (b) Western blot analysis of 7D12(44FSY), 7D12(44mFSY), and 7D12(44FSK) incubated with or without EGFR receptor. Results for WT-7D12 control are shown in Fig. S2. (c) Structure of nanobody 2Rs15d in complex with HER2 ECD. Residue Y37 of 2Rs15d is shown in proximity to residue Y112 of HER2. (d) Western blot analysis of WT and mutant 2Rs15d cross-linking with HER2 ECD in vitro. Two cross-linking bands were detected for 2Rs15d(37mFSY), corresponding to its monomeric and dimeric form.
We further incorporated mFSY into the fragment antigen-binding (Fab) region of an antibody to generate covalent Fabs. Fab consists of one constant and one variable domain of each of the heavy and light chain and is a common small binding fragment of monoclonal antibodies designated for diagnostic and therapeutic use. Trastuzumab is a well-known HER2-specific antibody for treating breast and stomach cancer. We decided to incorporate mFSY into the Trastuzumab Fab, TrasFab. Guided by the structure of TrasFab-HER2 complex,29 we incorporated mFSY into sites Ser50 and Tyr92 on the light chain of TrasFab, aiming to target Lys593 and Lys569 of HER2, respectively (Fig. 4a). Through simultaneously expressing the heavy and light chains, we were able to produce and purify the mutant Fab proteins from E. coli cells. When incubated with the HER2 ECD, TrasFab(50mFSY) showed detectable cross-linking, while TrasFab(92mFSY) showed more robust cross-linking (Fig. 4b), showcasing that mFSY-mediated cross-linking was site-dependent. Similar time-dependent strong cross-linking was also seen for the TrasFab(92FSY) mutant. These results represent the first example of a Fab that has been engineered to crosslink its target receptor via latent bioreactive Uaas.
Fig. 4. Incorporation of mFSY into TrasFab enables first shown instance of Fab-receptor cross-linking with HER2.
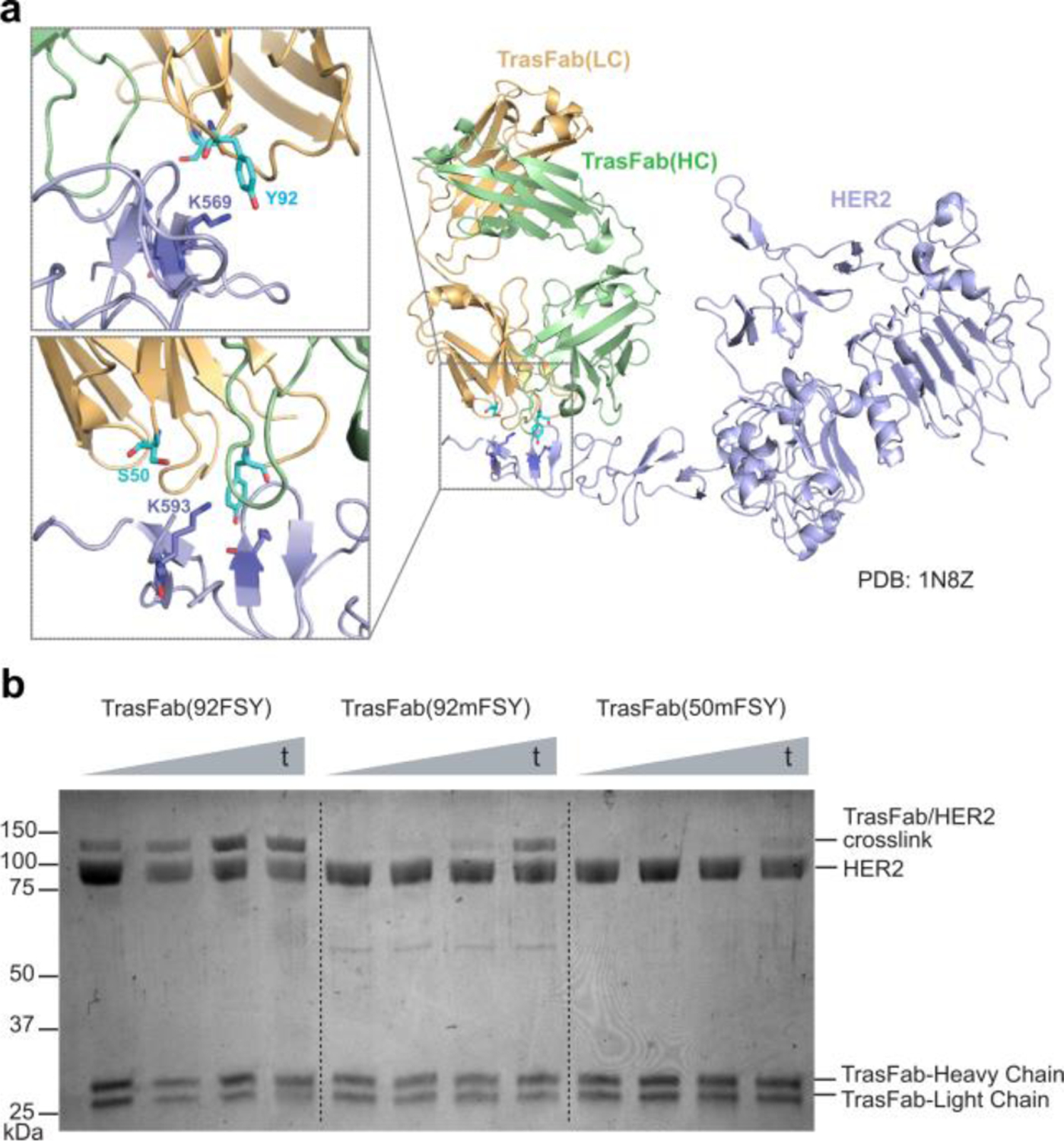
(a) Structure of Trastuzumab Fab (TrasFab, gold and mint) in complex with HER2 ECD. Residues S50 and Y92 of the TrasFab light chain are shown in proximity to targeted residue K593 and K569 on HER2, respectively. (b) SDS-PAGE analysis of in vitro cross-linking between TrasFab mutants and HER2 ECD. t: 0.5, 2, 4, and 24 h.
In summary, we genetically encoded a new latent bioreactive Uaa mFSY into proteins in E. coli and mammalian cells. mFSY was efficiently incorporated into various protein binders including an affibody, nanobodies, and most notably the first case of a Fab. These mFSY-engineered proteins crosslinked with their target receptors upon binding through mFSY reacting with Lys, His, or Tyr residues via proximity-enabled SuFEx reaction. While mFSY generally has comparable cross-linking efficiency with FSY for interchangeable use at some sites, mFSY achieved efficient cross-linking at certain incorporation sites where FSY did not crosslink efficiently. Therefore, mFSY complements FSY in accommodating different target side chain orientations, expanding the proteins covalently targetable by latent bioreactive Uaas. The unique ability of covalent proteins is emerging as recently demonstrated in protein therapeutics9,30 and in studying protein-protein interactions.19, 31 Possessing the biocompatible multi-targeting fluorosulfate warhead with an expanding reaction orientation, mFSY will be a powerful addition to the arsenal of latent bioreactive Uaas for developing covalent proteins for biological research, synthetic biology, and biotherapeutics.
Supplementary Material
Acknowledgments
L.W. acknowledges the support of Mary Anne Koda-Kimble Seed Award and NIH (R01CA258300).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Cao L, Wang L, Protein Sci, 2022, 31, 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Brock A, Herberich B, Schultz PG, Science, 2001, 292, 498–500. [DOI] [PubMed] [Google Scholar]
- 3.Xiang Z, Ren H, Hu YS, Coin I, Wei J, Cang H, Wang L, Nat. Methods, 2013, 10, 885–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiang Z, Lacey VK, Ren H, Xu J, Burban DJ, Jennings PA, Wang L, Angew. Chem. Int. Ed. Engl, 2014, 53, 2190–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xuan W, Shao S, Schultz PG, Angew. Chem. Int. Ed. Engl, 2017, 56, 5096–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coin I, Katritch V, Sun T, Xiang Z, Siu FY, Beyermann M, Stevens RC, Wang L, Cell, 2013, 155, 1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoppmann C, Maslennikov I, Choe S, Wang L, J. Am. Chem. Soc, 2015, 137, 11218–11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang B, Tang S, Ma C, Li ST, Shao GC, Dang B, DeGrado WF, Dong MQ, Wang PG, Ding S, et al. , Nat. Commun, 2017, 8, 2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Chen Q, Klauser PC, Li M, Zheng F, Wang N, Li X, Zhang Q, Fu X, Wang Q, et al. , Cell, 2020, 182, 85–97.e16. [DOI] [PubMed] [Google Scholar]
- 10.Berdan VY, Klauser PC, Wang L, Bioorg. Med. Chem, 2021, 29, 115896. [DOI] [PubMed] [Google Scholar]
- 11.Tian Z, Wu L, Yu C, Chen Y, Xu Z, et al. , Sci. Adv, 2021, 7, DOI 10.1126/sciadv.abf2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Biotechnol N., 2017, 38, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong J, Krasnova L, Finn MG, Sharpless KB, Angew. Chem. Int. Ed. Engl, 2014, 53, 9430–9448. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Dong J, Plate L, Mortenson DE, Brighty GJ, Li S, Liu Y, Galmozzi A, Lee PS, Hulce JJ, et al. , J. Am. Chem. Soc, 2016, 138, 7353–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Li J, Li S, Li G, Sharpless KB, Wu P, J. Am. Chem., Soc 2018, 140, 2919–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan A, Jones LH, Chem. Sci, 2015, 6, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang N, Yang B, Fu C, Zhu H, Zheng F, Kobayashi T, Liu J, Li S, Ma C, Wang PG, et al. , J. Am. Chem. Soc, 2018, 140, 4995–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang N, Wang L, Curr. Opin. Chem. Biol, 2022, 66, 102106. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Cao L, Klauser PC, Cheng R, Berdan VY, Sun W, Wang N, Ghelichkhani F, Yu B, Rozovsky S, et al. , J. Am. Chem. Soc, 2021, 143, 10341–10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Mukherjee P, Liu R, Evrard E, Wang D, Humphrey JM, Butler TW, Hoth LR, Sperry JB, Sakata SK, et al. , Org. Lett, 2018, 20, 812–815. [DOI] [PubMed] [Google Scholar]
- 21.Lacey VK, Louie GV, Noel JP, Wang L, ChemBioChem, 2013, 14, 2100–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takimoto JK, Dellas N, Noel JP, Wang L, ACS Chem. Biol, 2011, 6, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi T, Hoppmann C, Yang B, Wang L, J. Am. Chem. Soc, 2016, 138, 14832–14835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee K-F, Slesinger PA, Wang L, Nat. Neurosci, 2007, 10, 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eigenbrot C, Ultsch M, Dubnovitsky A, Abrahmsen L, Hard T, Proc. Natl. Acad. Sci. U. S. A, 2010, 107, 15039–15044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekerljung L, Lennartsson J, Gedda L, PloS one, 2012, 7, e49579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz KR, Bagchi A, Roovers RC, van PMP en Henegouwen Bergen, Ferguson KM, Structure, 2013, 21, 1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Huyvetter M, De Vos J, Xavier C, Pruszynski M, Sterckx YGJ, et al. , Clin. Cancer Res, 2017, 23, 6616–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DWJ, Leahy DJ, Nature, 2003, 421, 756–760. [DOI] [PubMed] [Google Scholar]
- 30.Yu B, Li S, Tabata T, Wang N, Kumar GR, et al. 10.1101/2022.03.11.483867 [DOI] [Google Scholar]
- 31.Liu C, Wu T, Shu X, Li S-T, Wang DR, et al. , Adv. Biol, 2021, 5, e2000308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


