Abstract
More than 30% of eukaryotic proteins contain domains that must translocate across or integrate into the endoplasmic reticulum (ER) membrane. With few exceptions, protein translocation and transmembrane domain integration at the ER require the conserved Sec61 translocon. Decades of studies have established a clear mechanistic model for how the Sec61 translocon functions. The biosynthesis of distinct subsets of proteins at the ER also involves accessory factors that interact with the Sec61 translocon and translocating nascent proteins. However, assigning specific functions to many translocon accessory factors has been a persistent challenge in the field. This Perspective discusses recent insights into mechanisms that promote protein biosynthesis at the ER through accessory factors that directly regulate the Sec61 translocon or chaperone nascent proteins within the ER membrane. These translocon accessory factor functions, and more still to be discovered, are essential for producing a diverse and high-fidelity proteome at the ER.
THE SEC TRANSLOCON FACILITATES PROTEIN BIOSYNTHESIS AT THE ER
Protein translocation across or integration into a lipid bilayer are fundamental cellular processes facilitated by the conserved “Sec translocon” at the eukaryotic endoplasmic reticulum (ER) or the prokaryotic periplasmic membrane (Komarudin and Driessen, 2019; Itskanov and Park, 2022). The Sec translocon provides proteins access to the exterior of the cell, either directly in prokaryotes or through vesicular trafficking in eukaryotes. The core translocon contains a central subunit, Sec61α (SecY in prokaryotes), in complex with Sec61β and Sec61γ (SecG and SecE, respectively, in prokaryotes). Sec61α forms a membrane-spanning channel that allows for the translocation of unfolded hydrophilic polypeptides from the cytosol into the ER lumen (van den Berg et al., 2004). The Sec61α channel also has a lateral gate that provides polypeptides access from the channel to the lipid bilayer (Figure 1A). The lateral gate engages and is opened by hydrophobic α-helices in the form of cleavable N-terminal signal sequences that initiate protein translocation or transmembrane helices that partition into the membrane.
FIGURE 1:
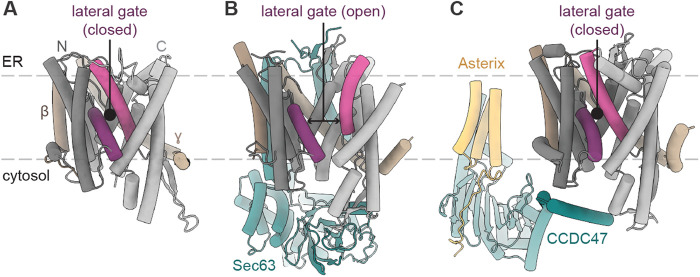
Accessory factors modulate translocon conformations. (A) The isolated Sec translocon (Protein Data Bank [PDB] 1RH5). Lateral gate helices (purple) and the N-terminal (dark gray) and the C-terminal (light gray) halves of the Sec61α homologue are indicated along with the Sec61β and Sec61γ homologues. (B) The yeast posttranslational translocon (PDB 6N3Q). Sec63 (teal) acts as a brace holding the Sec61α lateral gate open; Sec62 and the yeast-specific Sec72 and Sec73 subunits are not shown. (C) The mammalian translocon associated with the PAT complex (PDB 7TM3) comprising Asterix (yellow) and CCDC47 (teal). The translocon-bound ribosome and the GEL and BOS complexes are not shown.
The Sec translocon functions with several partners. The most common translocon partners are ribosomes, which bind the cytosolic side of Sec61α and drive cotranslational translocation of nascent polypeptides through the channel. While cotranslational protein import into the ER is dominant in metazoans, posttranslational translocation is common in yeast and prokaryotes. In yeast, posttranslational translocation requires the Sec61 translocon in complex with Sec62/Sec63/Sec71/Sec72, which recruits the ER-resident Hsp70 protein BiP through a J domain on Sec63. BiP binds translocating nascent proteins in the ER lumen, thereby preventing the proteins from sliding back into the cytosol. In prokaryotes, the cytosolic ATPase SecA binds and facilitates nascent protein movement through the translocon through cycles of ATP hydrolysis (Komarudin and Driessen, 2019). Extensive biochemical and structural studies on the mechanism of the Sec translocon in cooperation with these partners are well-reviewed elsewhere (Komarudin and Driessen, 2019; Hegde and Keenan, 2022; Itskanov and Park, 2022). This Perspective discusses recent mechanistic insights into eukaryotic translocon accessory factors.
TRANSLOCON ACCESSORY FACTORS MEDIATE DISTINCT ASPECTS OF PROTEIN BIOSYNTHESIS
Protein import into the ER has been reconstituted with purified components comprising the Sec61 complex, a functional partner, and targeting factors (Görlich and Rapoport, 1993; Panzner et al., 1995; Matlack et al., 1999). However, these components are insufficient for the successful biogenesis of many proteins at the ER. Particularly in metazoan cells, various “accessory” factors identified through interactions with Sec61 and/or translocating nascent proteins are thought to aid protein biogenesis at the ER (Wiedmann et al., 1987; Görlich et al., 1992; Görlich and Rapoport, 1993; McGilvray et al., 2020; Hegde and Keenan, 2022; Itskanov and Park, 2022). Unlike the translocon partners described above, accessory factors are not required for the basic process of protein translocation through Sec61 but instead facilitate biosynthetic processes specific to distinct subsets of substrates. A few translocon accessory factors, such as the OST (oligosaccharyltransferase) complex, which mediates N-linked glycosylation in the ER, or the signal peptidase complex, which cleaves off signal sequences, have clear functions (Itskanov and Park, 2022). Other accessory factors, such as TRAP (translocon-associated protein complex; also known as the signal sequence receptor, SSR) and TRAM (translocating chain-associating membrane protein), interact with and enhance the biogenesis of numerous substrates (Wiedmann et al., 1987; Görlich et al., 1992; Görlich and Rapoport, 1993; Voigt et al., 1996; Hegde et al., 1998; Meacock et al., 2002; Fons et al., 2003; Sommer et al., 2013; Nguyen et al., 2018; Kriegler et al., 2020; Huang et al., 2021) but have poorly defined mechanisms. Only recently have more precise mechanistic insights into certain translocon accessory factors emerged.
SEC62/SEC63: A TRANSLOCON MODIFIER
Discovered more than three decades ago (Rothblatt et al., 1989), Sec62/Sec63 is now well appreciated to be physically and functionally linked with the Sec61 translocon. In yeast, Sec62/Sec63/Sec71/Sec72 mediates the posttranslational translocation of proteins that generally have less hydrophobic signal sequences than cotranslationally translocated proteins (Ng et al., 1996). Although posttranslational translocation is uncommon in mammals, Sec62/Sec63 (but not Sec71 or Sec72) is conserved and implicated in an accessory role during the cotranslational biogenesis of certain substrates (Conti et al., 2015; Schorr et al., 2020). Mammalian substrates that rely on Sec62/Sec63 are reported to have less hydrophobic signal sequences and may be especially dependent on BiP recruitment to fold (Conti et al., 2015; Schorr et al., 2020).
A significant advance in understanding how Sec62/Sec63 regulates translocon activity came from recent cryogenic electron microscopy (cryo-EM) structures of the yeast posttranslational translocon complex (Itskanov and Park, 2019; Wu et al., 2019; Itskanov et al., 2021; Weng et al., 2021). In the structures, Sec62/Sec63 acts as a dynamic brace capable of fully opening the lateral gate of the channel (Figure 1B). The effect of Sec62/Sec63 on the conformation of Sec61α is distinct from how translocon binding partners slightly crack the lateral gate to “prime” the translocon for signal sequence engagement (Itskanov and Park, 2022). Full lateral gate opening by Sec62/Sec63 may lower the energetic barrier for nonoptimal signal sequences to initiate protein translocation. Once engaged at the lateral gate, signal sequences may be further stabilized by interaction with Sec62 (Lyman and Schekman, 1997; Plath et al., 1998; Itskanov et al., 2021; Weng et al., 2021). Thus, these structures establish Sec62/Sec63 as an accessory complex that directly regulates the conformation and function of the translocon in addition to its role in recruiting BiP. How Sec62/Sec63 operates cotranslationally, when a ribosome bound to Sec61α is predicted to clash with cytosolic portions of the complex (Itskanov and Park, 2019; Wu et al., 2019), remains to be determined.
PAT-10/ASTERIX: AN INTRAMEMBRANE PROTEIN CHAPERONE
Many translocon accessory factors probably act directly on nascent proteins. A recently clarified example is the eukaryotic protein Asterix. Early site-specific cross-linking studies identified an interaction between PAT-10 (protein associated with the ER translocon of 10 kDa) and the first transmembrane helix of the multispanning membrane protein rhodopsin (Meacock et al., 2002). This interaction was specific, as PAT-10 cross-links were not observed with other transmembrane helices of the protein (Meacock et al., 2002; Ismail et al., 2006). PAT-10 continues to interact with the first transmembrane helix even after that helix ceases to interact with Sec61α and dissociates only after complete synthesis of the entire protein. These findings implicated PAT-10, whose sequence was unknown at the time, in the biogenesis of multispanning membrane proteins. The probable identity of PAT-10 would not be revealed until nearly 20 years later. With improved mass spectrometry approaches facilitating protein identification, affinity purifications identified the ∼10 kDa nascent protein interactor as Asterix, which forms the obligate PAT complex with CCDC47 (Chitwood and Hegde, 2020). Asterix interacts more strongly with transmembrane helices containing hydrophilic amino acids, which may be critical for membrane protein folding and function but are energetically unfavorable within the lipid bilayer. Depleting either CCDC47 or Asterix destabilizes the other PAT complex partner and impairs the biosynthesis of numerous mutispanning membrane proteins (Chitwood and Hegde, 2020; Sundaram et al., 2022), thereby functionally linking the interaction between Asterix and nonoptimal transmembrane helices to protein biosynthesis.
Assignment of Asterix and CCDC47 as translocon accessory factors is further validated by cryo-EM structures (McGilvray et al., 2020; Smalinskaitė et al., 2022). The structures showed ribosome–Sec61 complexes associated not only with 1) the PAT complex but also with 2) the GEL (Get and EMC-like) complex comprising the putative protein insertase TMCO1 (Anghel et al., 2017) and OPTI (obligate partner of TMCO1 insertase; also known as C20orf24 or RAB5IF) and 3) the BOS (back of Sec61) complex comprising TMEM147, Nicalin, and NOMO. The positions of these complexes sterically exclude the OST glycosylation complex from associating with the translocon. The structures additionally revealed that CCDC47 makes multiple interactions with the translocon-bound ribosome (McGilvray et al., 2020) and with the cytosolic side of the N-terminal half of Sec61α, which may keep the Sec61α lateral gate closed (Figure 1C) (Smalinskaitė et al., 2022). The effect of CCDC47 on Sec61α, the relatively large distance between Asterix and Sec61 (Figure 1C), and the observation of a nascent transmembrane helix outside of Sec61α on the side opposite the lateral gate raised the intriguing hypothesis that some transmembrane helices of multispanning proteins insert into the ER membrane independently of the Sec61α lateral gate (Smalinskaitė et al., 2022). How this may occur awaits future investigations. As PAT complex subunits are present in most eukaryotes, the chaperoning function of Asterix and the potential regulation of Sec61 by CCDC47 appear to be broadly used. In contrast, GEL and BOS components occur primarily in metazoans, and their specific roles in protein biosynthesis at the ER remain to be defined.
Collective recruitment of the PAT, GEL, and BOS complexes to ribosome–Sec61 complexes correlates with two nascent protein features: a hydrophilic transmembrane helix that favors Asterix interaction and the presence of multiple transmembrane helices indicative of a multipass membrane protein (Smalinskaitė et al., 2022; Sundaram et al., 2022). These features were identified through pull downs of stable mimics of cotranslational translocation intermediates assembled in a cell-free system using nonstop mRNAs truncated at defined points to stall ribosomes after the synthesis of specific lengths of model membrane proteins. Such pull downs also indicated that the PAT, GEL, and BOS complexes can be exchanged with the OST complex based on changing substrate requirements (Sundaram et al., 2022). Whether accessory factor exchanges occur at similar points during ongoing protein synthesis occurring at ∼5–10 amino acids per second remains unclear. Altogether, these studies establish that Asterix acts within a larger translocon accessory complex to chaperone early hydrophilic transmembrane helices during multispanning membrane protein biogenesis at the ER (Meacock et al., 2002; Chitwood and Hegde, 2020; Smalinskaitė et al., 2022; Sundaram et al., 2022).
PROSPECTS FOR STUDYING TRANSLOCON ACCESSORY FACTORS
The many years needed to gain coherent mechanistic insights into Sec62/Sec63 and Asterix highlight several general lessons and challenges in studying translocon accessory factors. First, knowing precise substrate features that confer reliance on an accessory factor is essential. Current insights into such substrate features are primarily limited to the N-terminal regions of ER-targeted substrates. Identifying specific elements in other parts of multispanning membrane proteins that may require multiple accessory factor functions for biosynthesis becomes exponentially more complicated. Failure to correctly generate any part of such proteins often leads to the degradation of the entire protein. Loss of accessory factor functions also may induce cellular responses that indirectly affect the nascent proteome. Thus, endpoint and steady-state assays, even on a proteome-wide scale, may be insufficient to assign specific substrate elements to a given accessory factor. Moreover, cataloguing substrate features is critical but often still insufficient to generate a clear mechanistic model. For example, both TRAP and TRAM are linked to the successful production of certain substrates, but how they facilitate the biosynthesis of those proteins is still ambiguous. Thus, much work remains to identify and uncouple translocon accessory functions on different nascent protein elements.
Second, although the growing accessibility of cryo-EM will generate more structures of translocon accessory factors in the coming years, gleaning a clear mechanistic understanding from such structures requires sufficient context. Prior knowledge of substrates that use Sec62/Sec63 and of how other binding factors alter the translocon was critical to interpreting how the conformational changes that Sec62/Sec63 induce on Sec61 confer a specific function. By comparison, structures of TRAP have revealed previously unknown ribosomal interactions and the organization of an ER lumenal domain that probably interacts with translocating proteins but are nonetheless linked to various mechanistic interpretations (Pfeffer et al., 2017; Jaskolowski et al., 2022; Karki et al., 2022; Pauwels et al., 2022). Similarly, the structures of the PAT, GEL, and BOS complexes (McGilvray et al., 2020; Smalinskaitė et al., 2022) have generated many new and unexpected questions about translocon mechanisms. Future structural breakthroughs will rely on new approaches for identifying and isolating functional translocon intermediates to visualize how accessory factors engage and handle specific substrate elements.
Finally, the context for understanding translocon accessory factors continually evolves with new knowledge of mechanisms that influence protein biosynthesis at the ER. Notable examples include the identification of putative protein insertases, such as the EMC (ER membrane protein complex) (Guna et al., 2018) and TMCO1 (Anghel et al., 2017), that may carry out functions historically assumed to be handled by Sec61. In addition, a recently reported protein dislocation activity mediated by the ER-resident P5A-ATPase is required for the correct topogenesis of some Sec61 substrates (McKenna et al., 2020; McKenna et al., 2022). These factors are proposed to act on various cotranslational translocation substrates (Chitwood et al., 2018; Feng et al., 2020; McGilvray et al., 2020; McKenna et al., 2020, 2022; Li et al., 2021) and must therefore coordinate their activities with the translocon through currently unknown mechanisms. Some translocon accessory factors may help manage these different activities and facilitate accurate substrate transfer between Sec61 and other insertases or protein quality control factors. Hence, further understanding of translocon accessory factors awaits the discovery of more tractable model substrates and technical innovations to assay specific steps of complex protein biosynthesis pathways.
Acknowledgments
I thank M. McKenna, G. Nelson, J. Coelho, and M. Rale for critical reading of the manuscript and Shao lab members for useful discussions.
Abbreviations used:
- ER
endoplasmic reticulum
- Hsp70
70 kDa heat shock protein.
Footnotes
REFERENCES
- Anghel SA, McGilvray PT, Hegde RS, Keenan RJ (2017). Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep 21, 3708–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood PJ, Hegde RS (2020). An intramembrane chaperone complex facilitates membrane protein biogenesis. Nature 584, 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood PJ, Juszkiewicz S, Guna A, Shao S, Hegde RS (2018). EMC is required to initiate accurate membrane protein topogenesis. Cell 175, 1507–1519.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti BJ, Devaraneni PK, Yang Z, David LL, Skach WR (2015). Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol Cell 58, 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhao Y, Li T, Nie W, Yang X, Wang X, Wu J, Liao J, Zou Y (2020). CATP-8/P5A ATPase regulates ER processing of the DMA-1 receptor for dendritic branching. Cell Rep 32, 108101. [DOI] [PubMed] [Google Scholar]
- Fons RD, Bogert BA, Hegde RS (2003). Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol 160, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Hartmann E, Prehn S, Rapoport TA (1992). A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 357, 47–52. [DOI] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA (1993). Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75, 615–630. [DOI] [PubMed] [Google Scholar]
- Guna A, Volkmar N, Christianson JC, Hegde RS (2018). The ER membrane protein complex is a transmembrane domain insertase. Science 359, 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Keenan RJ (2022). The mechanisms of integral membrane protein biogenesis. Nat Rev Mol Cell Biol 23, 107–124. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Voigt S, Rapoport TA, Lingappa VR (1998). TRAM regulates the exposure of nascent secretory proteins to the cytosol during translocation into the endoplasmic reticulum. Cell 92, 621–631. [DOI] [PubMed] [Google Scholar]
- Huang Y, Xu X, Arvan P, Liu M (2021). Deficient endoplasmic reticulum translocon-associated protein complex limits the biosynthesis of proinsulin and insulin. FASEB J 35, e21515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N, Crawshaw SG, High S (2006). Active and passive displacement of transmembrane domains both occur during opsin biogenesis at the Sec61 translocon. J Cell Sci 119, 2826–2836. [DOI] [PubMed] [Google Scholar]
- Itskanov S, Kuo KM, Gumbart JC, Park E (2021). Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62. Nat Struct Mol Biol 28, 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskanov S, Park E (2019). Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science 363, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskanov S, Park E (2022). Mechanism of protein translocation by the Sec61 translocon complex. Cold Spring Harb Perspect Biol 2022, a041250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolowski M, Jomaa A, Gamerdinger M, Shrestha S, Leibundgut M, Deuerling E, Ban N (2022). Molecular basis of the TRAP complex function in ER protein biogenesis. bioRxiv 10.1101/2022.10.04.510795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Javanainen M, Tranter D, Rehan S, Huiskonen JT, Happonen L, Paavilainen VO (2022). Molecular view of ER membrane remodeling by the Sec61/TRAP translocon. bioRxiv 10.1101/2022.09.30.510141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarudin AG, Driessen AJM (2019). SecA-mediated protein translocation through the SecYEG channel. Microbiol Spectr 7, doi: 10.1128/microbiolspec.PSIB-0028-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegler T, Kiburg G, Hessa T (2020). Translocon-associated protein complex (TRAP) is crucial for co-translational translocation of pre-proinsulin. J Mol Biol 432, 166694. [DOI] [PubMed] [Google Scholar]
- Li T, Yang X, Feng Z, Nie W, Fang Z, Zou Y (2021). P5A ATPase controls ER translocation of Wnt in neuronal migration. Cell Rep 37, 109901. [DOI] [PubMed] [Google Scholar]
- Lyman SK, Schekman R (1997). Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell 88, 85–96. [DOI] [PubMed] [Google Scholar]
- Matlack KES, Misselwitz B, Plath K, Rapoport TA (1999). BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97, 553–564. [DOI] [PubMed] [Google Scholar]
- McGilvray PT, Anghel SA, Sundaram A, Zhong F, Trnka MJ, Fuller JR, Hu H, Burlingame AL, Keenan RJ (2020). An ER translocon for multi-pass membrane protein biogenesis. eLife 9, e56889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna MJ, Adams BM, Chu V, Paulo JA, Shao S (2022). ATP13A1 prevents ERAD of folding-competent mislocalized and misoriented proteins. Mol Cell 10.1016/j.molcel.2022.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna MJ, Sim SI, Ordureau A, Wei L, Harper JW, Shao S, Park E (2020). The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science 369, eabc5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacock SL, Lecomte FJL, Crawshaw SG, High S (2002). Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Mol Biol Cell 13, 4114–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P (1996). Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol 134, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Stutz R, Schorr S, Lang S, Pfeffer S, Freeze HH, Förster F, Helms V, Dudek J, Zimmermann R (2018). Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat Commun 9, 3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA (1995). Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81, 561–570. [DOI] [PubMed] [Google Scholar]
- Pauwels E, Shewakramani NR, De Wijngaert B, Camps A, Provinciael B, Stroobants J, Kalies K, Hartmann E, Maes P, Vermeire K, Das K (2022). Structural insights into TRAP association with ribosome-Sec61 complex, and translocon inhibition by a CADA derivative. bioRxiv 10.1101/2022.09.28.509949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Schaffer M, Ng BG, Albert S, Plitzko JM, Baumeister W, Zimmermann R, Freeze HH, Engel BD, Förster F (2017). Dissecting the molecular organization of the translocon-associated protein complex. Nat Commun 8, 14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA (1998). Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94, 795–807. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R (1989). Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol 109, 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr S, Nguyen D, Haßdenteufel S, Nagaraj N, Cavalié A, Greiner M, Weissgerber P, Loi M, Paton AW, Paton JC, et al. (2020). Identification of signal peptide features for substrate specificity in human Sec62/Sec63-dependent ER protein import. FEBS J 287, 4612–4640. [DOI] [PubMed] [Google Scholar]
- Smalinskaitė L, Kim MK, Lewis AJO, Keenan RJ, Hegde RS (2022). Mechanism of an intramembrane chaperone for multipass membrane proteins. Nature 611, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer N, Junne T, Kalies K-U, Spiess M, Hartmann E (2013). TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim Biophys Acta 1833, 3104–3111. [DOI] [PubMed] [Google Scholar]
- Sundaram A, Yamsek M, Zhong F, Hooda Y, Hegde RS, Keenan RJ (2022). Substrate-driven assembly of a translocon for multipass membrane proteins. Nature 611, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004). X-ray structure of a protein-conducting channel. Nature 427, 36–44. [DOI] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA (1996). Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol 134, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng T, Steinchen W, Beatrix B, Berninghausen O, Becker T, Bange G, Cheng J, Beckmann R (2021). Architecture of the active post-translational Sec translocon. EMBO J 40, e105643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M, Kurzchalia TV, Hartmann E, Rapoport TA (1987). A signal sequence receptor in the endoplasmic reticulum membrane. Nature 328, 830–833. [DOI] [PubMed] [Google Scholar]
- Wu X, Cabanos C, Rapoport TA (2019). Structure of the post-translational protein translocation machinery of the ER membrane. Nature 566, 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]