Abstract
Embryonic morphogenesis is powered by dramatic changes in cell shape and arrangement driven by the cytoskeleton and its connections to adherens junctions. This requires robust linkage allowing morphogenesis without disrupting tissue integrity. The small GTPase Rap1 is a key regulator of cell adhesion, controlling both cadherin-mediated and integrin-mediated processes. We have defined multiple roles in morphogenesis for one Rap1 effector, Canoe/Afadin, which ensures robust junction–cytoskeletal linkage. We now ask what mechanisms regulate Canoe and other junction–cytoskeletal linkers during Drosophila morphogenesis, defining roles for Rap1 and one of its guanine nucleotide exchange factor (GEF) regulators, Dizzy. Rap1 uses Canoe as one effector, regulating junctional planar polarity. However, Rap1 has additional roles in junctional protein localization and balanced apical constriction–in its absence, Bazooka/Par3 localization is fragmented, and cells next to mitotic cells apically constrict and invaginate, disrupting epidermal integrity. In contrast, the GEF Dizzy has phenotypes similar to but slightly less severe than Canoe loss, suggesting that this GEF regulates Rap1 action via Canoe. Taken together, these data reveal that Rap1 is a crucial regulator of morphogenesis, likely acting in parallel via Canoe and other effectors, and that different Rap1 GEFs regulate distinct functions of Rap1.
INTRODUCTION
Small GTPases in the Ras superfamily regulate virtually all aspects of cell biology, ranging from receptor tyrosine kinase signaling to vesicular trafficking to nuclear import to cell adhesion (Cherfils and Zeghouf, 2013). Small GTPases cycle between GTP-bound (ON) states and GDP-bound (OFF) states, with the transitions promoted by guanine nucleotide exchange factors (GEFs), which enhance nucleotide exchange and thus turn GTPases on, and GTPase-activating proteins (GAPs), which stimulate GTPase activity to promote deactivation. Active GTPases bind to and activate diverse effector proteins, stimulating downstream events. The small GTPase Rap1 belongs to the Ras superfamily and is a key regulator of cell adhesion, controlling both cadherin-mediated and integrin-mediated processes (Boettner and Van Aelst, 2009). Our recent work defined key roles for the Rap1 effector Canoe (Cno), homologue of mammalian Afadin, in morphogenesis. We now want to move upstream, defining the roles of Rap1 itself, its upstream regulators, and other potential Rap1 effectors implicated in regulating cell shape change and cell migration during embryonic development.
Rap1 is a conserved GTPase that plays important roles across the animal kingdom. The two mammalian Rap1 proteins regulate diverse processes ranging from neuronal migration to leukocyte trafficking to platelet activation. Rap1 regulation of blood vessel development and homeostasis, promoting angiogenesis and endothelial barrier function, provides an illustrative example (Chrzanowska-Wodnicka, 2017). In this tissue, Rap1 acts downstream of VE-cadherin and the receptor tyrosine kinase VEGFR and upstream of multiple effectors, including Rasip1, Krit-1, and Afadin. In leukocytes and platelets, Rap1 acts via RIAM and Talin to modulate integrin function (Bromberger et al., 2018; Lagarrigue et al., 2020). Mammals have an extensive suite of Rap1 GEFs, which act in different tissues and times—for example, the GEF EPAC activates Rap1 to ensure endothelial barrier function (Pannekoek et al., 2020), while RA-GEF-1 regulates Rap1 to promote callosal axons crossing the midline (Bilasy et al., 2011) and RA-GEF-2 regulates Rap1 during spermatogenesis (Okada et al., 2014).
Drosophila Rap1 was first identified via dominant gain-of-function mutations affecting the development of the stereotyped ommatidial cell arrangements in the developing eye (Hariharan et al., 1991; Asha et al., 1999; Walther et al., 2018). Rap1 was subsequently implicated in regulating diverse cellular events in Drosophila. Rap1 regulates cell adhesion in the larval wing disc (Knox and Brown, 2002) and the germline stem cell niche (Wang et al., 2006) and planar cell polarity and cell shape in the pupal wing (O’keefe et al., 2009). It regulates the migration of somatic border cells in the ovary (Sawant et al., 2018), embryonic macrophages (Huelsmann et al., 2006), germline precursors (Asha et al., 1999), and embryonic mesodermal cells (Asha et al., 1999; McMahon et al., 2010). Rap1 also regulates organ development and function, including synaptogenesis at neuromuscular junctions (Ou et al., 2019), axon guidance (Yang et al., 2016), epidermal muscle attachment (Camp et al., 2018), and nephrocyte function (Dlugos et al., 2019).
Given these diverse roles of Drosophila Rap1, scientists explored which GEFs regulate Rap1 in different contexts. Several GEFs were implicated in different events. For example, C3G mutants have defects in body wall muscle development (Shirinian et al., 2010) and nephrocyte function (Dlugos et al., 2019), while inappropriate C3G activation alters cell patterning in the eye and wing (Ishimaru et al., 1999). The Rap1 GEF EPAC is implicated in Malpighian tubule function (Efetova et al., 2013), in the role of mushroom body Kenyon cell neurons in aversive learning (Richlitzki et al., 2017), and in the response to anthrax toxin, via a role in blocking fusion of recycling endosomes with the plasma membrane (Guichard et al., 2017). While these two GEFs have relatively limited roles, Dizzy (Dzy), also known as PDZ-GEF, has broader roles. Dizzy is the GEF that mediates Rap1’s roles in macrophage (Huelsmann et al., 2006) and border cell migration (Sawant et al., 2018), germline stem cell adhesion (Wang et al., 2006), and synapse development and function at the neuromuscular junction (Heo et al., 2017; Ou et al., 2019), while in the developing eye and wing Dzy acts in parallel with the atypical GEF Sponge (Lee et al., 2002; Eguchi et al., 2013).
Our interest is in the regulation of cell adhesion and the cytoskeleton during embryonic morphogenesis, using Drosophila as a model. These events provide an outstanding place to explore the complexities of the Rap1 pathway and define its regulators and effectors (Figure 1A). We have focused on the Drosophila Rap1 effector Cno. Cno plays important roles in linking cell–cell adherens junctions (AJs) to the cytoskeleton, reinforcing cell junctions under mechanical tension. Through this function, Cno regulates the initial apical positioning of AJs during polarity establishment (Choi et al., 2013; Bonello et al., 2018), apical constriction of mesodermal cells (Sawyer et al., 2009), cell intercalation during germband elongation (Sawyer et al., 2011; Perez-Vale et al., 2021), dorsal closure (Boettner et al., 2003; Choi et al., 2011; Choi et al., 2013), and epithelial integrity (Sawyer et al., 2009; Manning et al., 2019). However, we and others have only begun to explore the roles of Rap1 and its GEFs in these events, to define in which events Rap1 uses Cno as its effector, and to determine which GEFs activate Rap1 in these contexts. Thus far, these efforts focused almost exclusively on Rap1’s initial roles in the first two events of embryonic development: polarity establishment during cellularization and mesoderm invagination. Examination of Rap1 maternal/zygotic mutants (henceforth referred to as M/Z mutants) revealed that Rap1 uses Cno as an effector during polarity establishment, mesoderm invagination, and potentially dorsal fold formation (Asha et al., 1999; Spahn et al., 2012; Choi et al., 2013; Wang et al., 2013; Bonello et al., 2018). The GEF Dzy plays an important role in these events (Spahn et al., 2012; Bonello et al., 2018). However, things are complex, as the GEF Sponge is also involved in apical-basal positioning of AJs (Schmidt et al., 2018), and dzy mutants mediate only a subset of the effects of Rap1 mutants during polarity establishment (Bonello et al., 2018). The use of dominant negative and activated variants supports the idea that Rap1 acts via Cno in collective cell migration during dorsal closure (Boettner et al., 2003), and there has been some examination of the effect of dzy zygotic mutants on dorsal closure (Boettner and Van Aelst, 2007), but beyond this we do not know what roles Rap1 or Dzy plays in the many diverse events of morphogenesis that require Cno, including cell intercalation during germband extension and maintenance of epidermal integrity.
FIGURE 1:
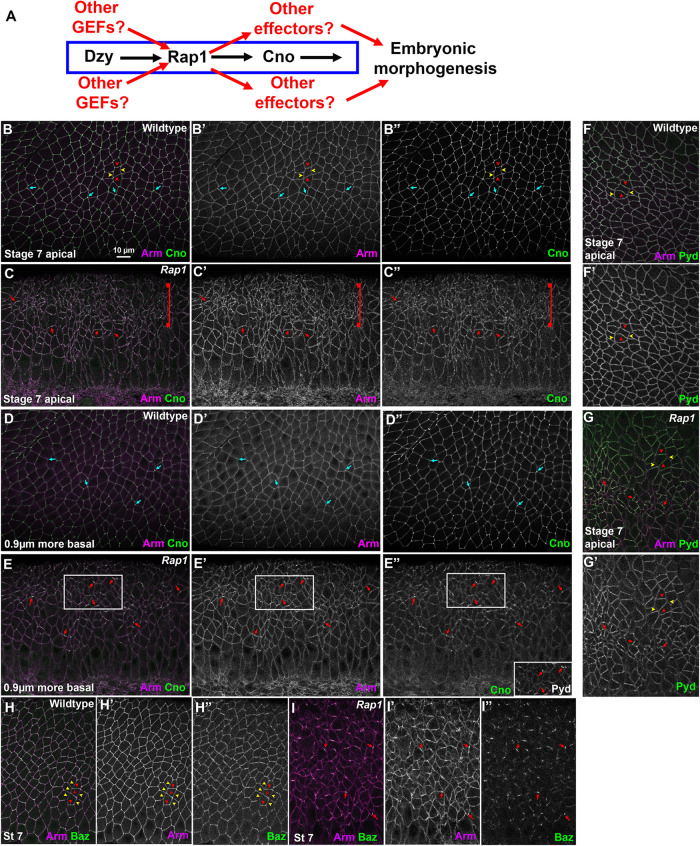
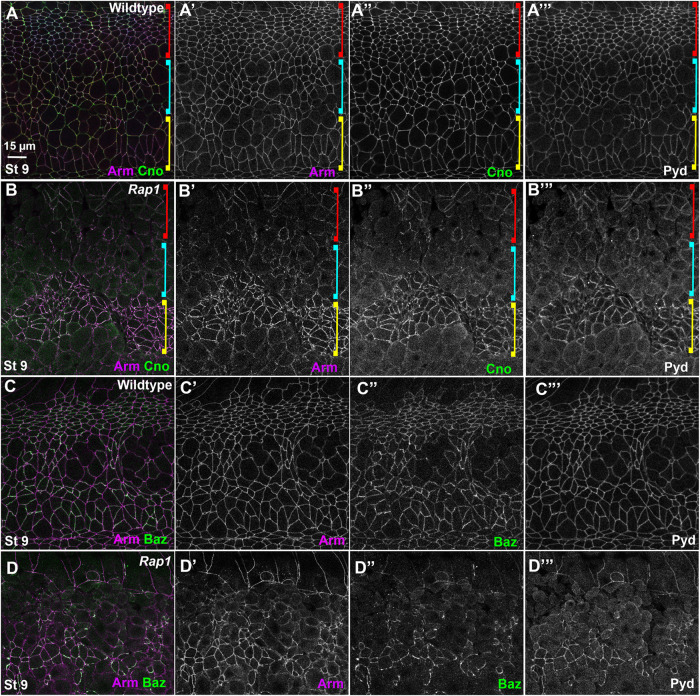
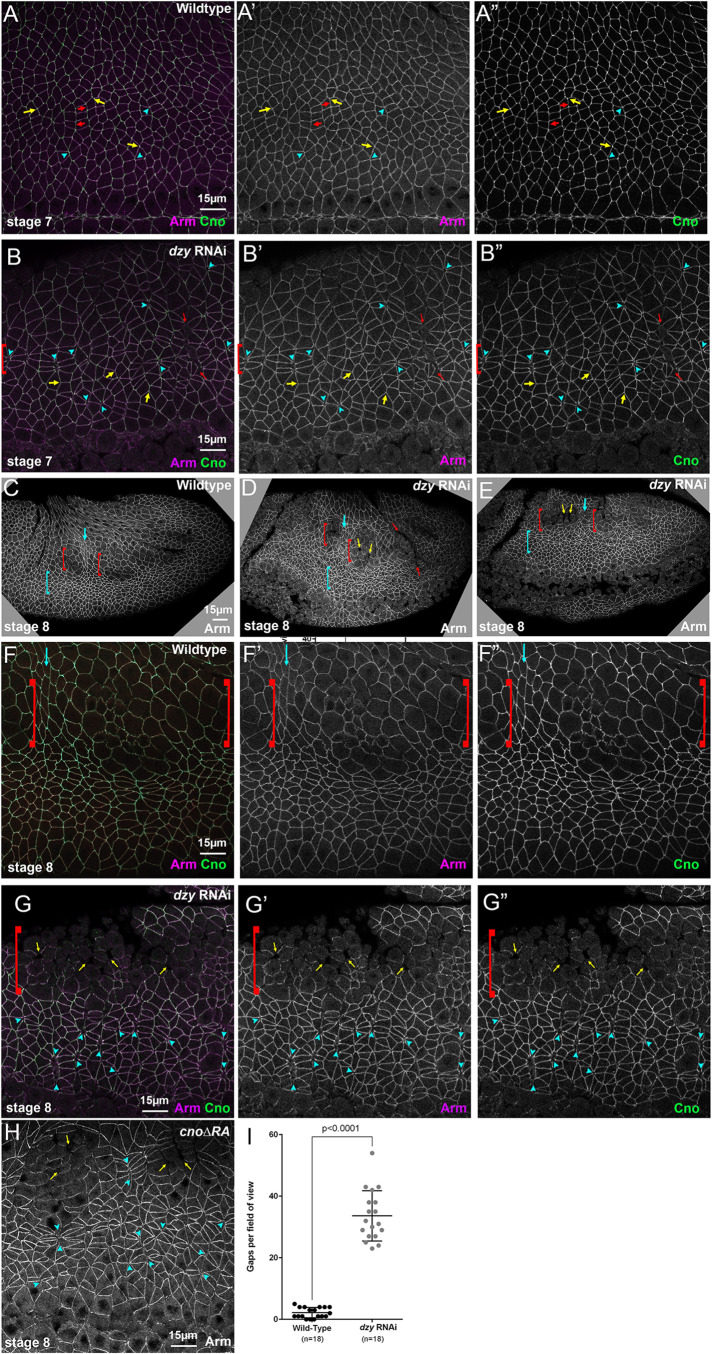
Rap1 RNAi rapidly leads to alterations in apical junctions, variability in apical cell area, and fragmentation of junctional Baz. (A) Diagram of the Rap1 “pathway.” (B–I) Stage 7 embryos, imaged at the level of apical AJs (B, C, F–I) or 0.9 µm more basal (D, E). Unless noted, in this figure and all others, embryos are anterior left and dorsal up. (B, D) WT. Apical cell areas are relatively similar at the two sectional planes. Cno is enriched at TCJs (cyan arrows). (C) Rap1 RNAi. Many cell junctions become difficult to follow in the apicalmost plane (brackets), and at other AJs Arm and Cno staining become more punctate (red arrows). (E) Rap1 RNAi. More basally cell areas remain much more variable than in WT, and junctional accumulation of Arm, Cno and Pyd is not as continuous as in WT (red arrows). (F) WT. Pyd accumulates on all AJs, without strong planar polarity. (G) Rap1 RNAi. Pyd is less continuous at AJs, with places of stronger accumulation (red arrows), and planar polarization to DV borders is accentuated (red vs. yellow arrowheads). (H) WT. Baz is found all around the cell circumference but is planar polarized to DV borders (red arrowheads) as opposed to AP borders (yellow arrowheads). (I) Rap1 RNAi. Baz staining is fragmented, and regions where it remains also accumulate elevated levels of Arm (arrows).
Our overall goal is to define the roles of Rap1 during morphogenesis, identify its regulators during these events, and ultimately identify its full suite of effectors. To fully define the role of Rap1 signaling during these critical processes, we need to address two key knowledge gaps: 1) Is Cno the sole Rap1 effector during morphogenesis, or does Rap1 have broader effects through other effectors (Figure 1A)? 2) Is Dzy the relevant Rap1 GEF during these stages, or do other GEFs play roles (Figure 1A)? To address these questions, we set out to define the roles of Rap1 and Dzy after gastrulation, interrogating which events occur via a Rap1-Cno pathway, what roles Rap1 plays that require other effectors, and in which Cno-regulated events Dzy is the GEF involved in Cno activation.
RESULTS
Our first goal was to define the roles of Rap1 during morphogenesis and to determine which of these roles require its effector Cno and which might involve other effectors. To do so, we compared the effects of Rap1 RNA interference (RNAi) to those of Cno loss, which we and others previously characterized. We examined cell shape, cell rearrangements, cell junction stability under tension, junction protein planar polarity, and overall tissue integrity from the onset of gastrulation to the completion of dorsal closure. We envisioned two possibilities: 1) Rap1 RNAi would precisely mimic loss of Cno, suggesting that Cno is its only effector during these events, or 2) Rap1 RNAi would affect additional cell biological events, suggesting the existence of other important effector(s).
Previous analyses of the role of Rap1 in embryos were largely confined to the earliest events of morphogenesis–initial establishment of apical-basal polarity and invagination of the mesoderm, the first event of gastrulation (Sawyer et al., 2009; Spahn et al., 2012; Choi et al., 2013). In these events, Rap1 and Cno loss lead to similar defects, but because Cno is only one of Rap1’s effectors, we suspected that Rap1 might use other effectors to carry out additional roles in junctional integrity as AJs are challenged by cell intercalation, cell division, and neuroblast invagination (Figure 1A). Cuticle analysis was potentially consistent with this hypothesis, as the Rap1 cuticle phenotype is as or more severe than that of most cno mutants (Bonello et al., 2018). We thus examined the effects of Rap1 loss. We used a previously validated RNAi line that, when driven by the matGAL4 driver, reduces Rap1 protein to undetectable levels through the end of dorsal closure (Bonello et al., 2018). The effects of Rap1 RNAi on apical-basal polarity establishment match those of Rap1 maternal/zygotic mutants (Choi et al., 2013; Bonello et al., 2018), and Rap1 RNAi leads to fully penetrant defects in mesoderm invagination (31/31 stage 7 and 8 embryos), another previously characterized Rap1 phenotype (Spahn et al., 2012). Finally, both Rap1M/Z mutants (Sawyer et al., 2009) and Rap1 RNAi (Bonello et al., 2018) can lead to disruption of epidermal integrity as assessed by examining cuticles. Rap1 RNAi thus is a well-validated, on-target reagent to substantially reduce or eliminate Rap1 function.
Rap1 RNAi disrupts germband extension and affects the process even earlier than loss of Cno
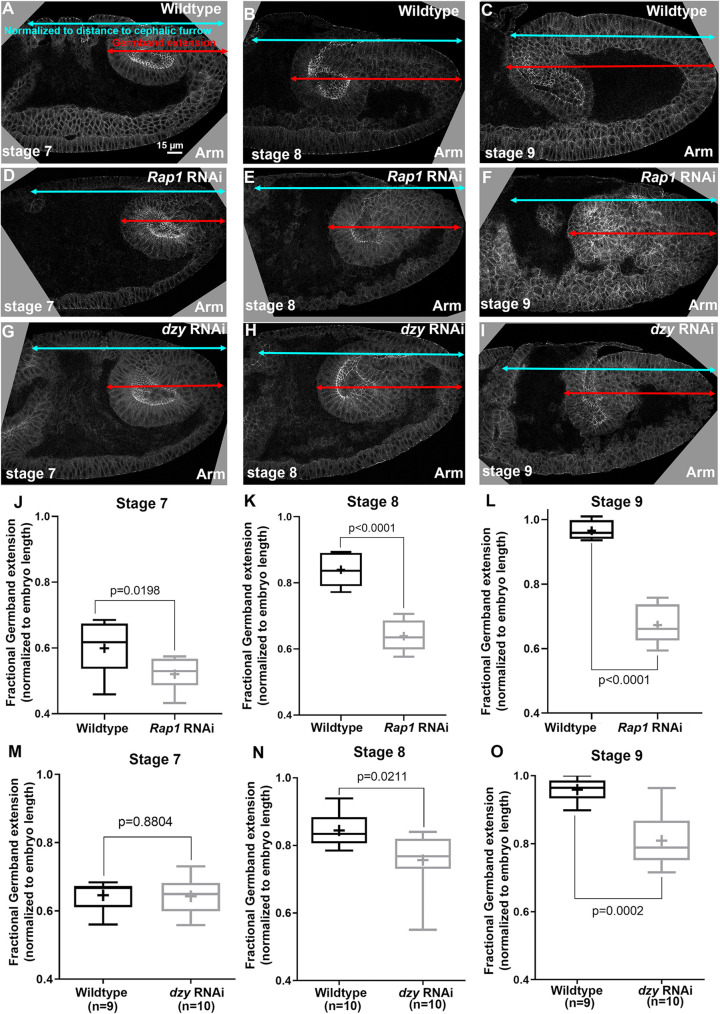
To determine whether Rap1 primarily acts via Cno or has additional effectors during embryonic morphogenesis, we compared the effects of Rap1 RNAi with those of Cno loss. We first examined germband extension, an embryonic event in which cells in the ectoderm undergo cell intercalation, which in turn helps narrow and extend the germband (Kong et al., 2017). In our earlier work, we found that the first phase of germband extension occurs normally in cnoM/Z mutants but then germband extension slows substantially (Sawyer et al., 2011). We thus first examined how Rap1 RNAi affects germband extension. To stage embryos, we used a “timer” likely to be independent of the cell movements we wanted to assess: the pattern of cell divisions in mitotic domains (Foe, 1989). We then measured the progress of the germband, normalizing it to the distance between the posterior end of the embryo and the cephalic furrow. Rap1 RNAi slowed germband extension at all three stages (Figure 2, A–C vs. D–F, quantified in J–L), thus suggesting that Rap1 loss leads to additional defects relative to those caused by Cno loss. The early effect may reflect the “twisted gastrulation” phenotype that we previously observed in Rap1M/Z mutants (Supplemental Figure S1, A vs. B; Sawyer et al., 2009).
FIGURE 2:
Both Rap1 and dzy RNAi delay germband extension, but effects of Rap1 RNAi occur earlier and are more severe. (A–I) Cross-sections of representative WT (A–C), Rap1 RNAi (D–F), and dzy RNAi (G–I) embryos, revealing extension of posterior end of the gut (red arrows) vs. the full distance to the cephalic furrow (cyan arrows). (J–O) Fractional germband extension at the indicated stages. (J–L) Rap1 RNAi substantially reduces germband extension at all stages. (M–O) dzy RNAi does not delay early germband extension (stage 7, G) but does reduce germband extension at later stages (H, I).
While Rap1 RNAi mimics the effect of Cno loss on AJ planar polarity, it also dramatically alters apical AJs and destabilizes Bazooka/Par3 localization
Germband extension is driven in part by reciprocal planar polarization of actin and myosin to anterior/posterior (AP) cell borders and AJ proteins to dorsal/ventral (DV) cell borders. Myosin drives constriction of AP borders, creating T1 transition or rosettes, which then rearrange to complete intercalation. Cno is enriched at the sites where AJs are under elevated molecular tension (Sawyer et al., 2009, 2011; Yu and Zallen, 2020)–with subtle enrichment at AP borders (Figure 1B, yellow vs. red arrowheads) and tricellular junctions (TCJs) (Figure 1B, cyan arrows). Cno is important to reinforce these AJs, and in Cno’s absence junctions separate at these sites (Sawyer et al., 2011). The fly ZO-1 homologue Polychaetoid (Pyd) is enriched at DV borders with AJ proteins and acts in parallel with Cno to maintain epithelial integrity (Manning et al., 2019).
We began our exploration of the mechanisms by which Rap1 regulates morphogenesis by examining whether it regulates AJ protein localization and stability. In wild-type (WT) embryos, the fly β-catenin homologue Armadillo (Arm) and Cno rapidly transition from the spot AJs seen during cellularization to a more-uniform localization along apical junctions as germband extension accelerates during stage 7 (Figure 1B). Cno is enriched at TCJs (Figure 1B, cyan arrows) and at AP borders (Figure 1B, yellow vs. red arrowheads; quantified in Figure 3C), while Arm and Pyd are more uniformly localized between AP and horizonal borders (Figure 1B; quantified in Figure 3, A and B). Bazooka (fly Par3; Baz) is obviously planar polarized, with enrichment at DV borders at this stage (Figure 1H; quantified in Figure 3D), but remains present on both AP and DV cell borders. Previous work revealed that, in cno null mutants or after strong cno RNAi, planar polarization of Arm, Pyd, and especially Baz is enhanced by strongly reducing their accumulation on AP borders (Sawyer et al., 2011; Manning et al., 2019). This weakens AJ–cytoskeletal connections at these locations, leading to apical gaps at AP borders and TCJs (Sawyer et al., 2011; Manning et al., 2019). However, core AJ proteins remain localized to AJs in cno mutants (Sawyer et al., 2011; Manning et al., 2019).
FIGURE 3:
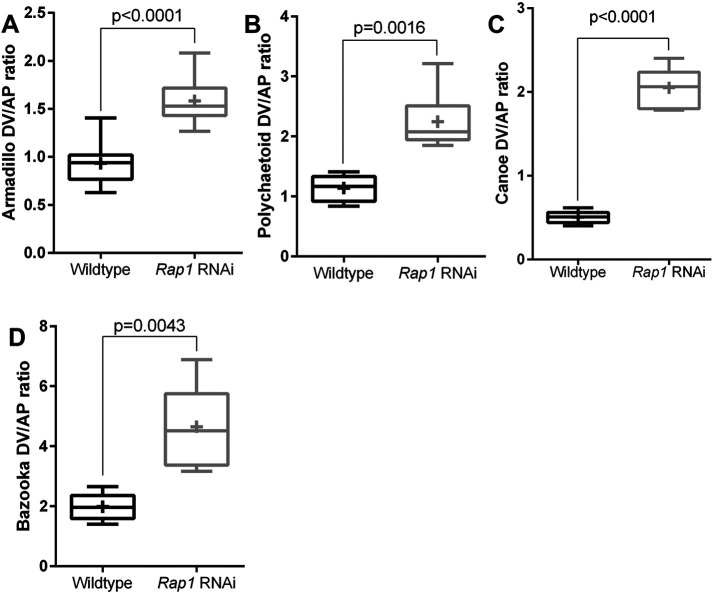
Rap1 RNAi alters junctional protein planar polarity in ways similar to loss of Cno when assessed in cells where shape is not drastically altered. (A–D) Planar polarity quantification. Rap1 RNAi enhances planar polarity of Arm (A), Pyd (B), and Baz (D) and flips the planar polarity of Cno (C). The numbers of cells analyzed are in Supplemental Table S1.
AJ protein localization is altered early after Rap1 RNAi. While Arm and Cno are either basally mislocalized (i.e., Arm) or lost from the membrane (i.e., Cno) during cellularization in Rap1 RNAi embryos, both relocalize to apical AJs as gastrulation begins (Bonello et al., 2018). However, we found that their junctional localization was abnormal in Rap1 RNAi embryos. This was most striking apically, where in Rap1 RNAi embryos both Arm and Cno localization to cell borders became more punctate (Figure 1C). Their localization was less continuous along less-affected cell junctions (Figure 1C, arrows), while their localization to other cell junctions was sufficiently disrupted to make cell borders hard to define at the most apical end of many cells (Figure 1C, bracket; observed in 16/16 stage 7 embryos). Cell shapes were more-regular 0.9 µm basally, but AJ localization of Arm was less continuous than in the WT, and this was even more evident for Cno (Figure 1, D vs. E, arrows).
To quantify the effect of Rap1 RNAi on junctional Arm and Cno enrichment at AJs, we stained WT embryos (marked with Histone:RFP) and Rap1 RNAi embryos together in the same tube, visualized them using the same confocal microscope settings, and assessed levels at bicellular AJs, subtracting the cytoplasmic background and normalizing levels to those in WT. Levels of Arm were modestly reduced at AJs (Supplemental Figure S1, H′, J′ vs. I′, and K′, quantified in L; mean 73% of WT; 12 bicellular borders per embryo in five stage 7 and 8 embryos). Levels of Cno were more substantially reduced (Supplemental Figure S1, H′′, J′′ vs. I′′, and K′′, quantified in L; mean = 33% of WT). We also examined the localization of Arm and Cno along the apical-basal axis after Rap1 RNAi. Because Rap1 regulates initial apical poisoning of AJs during cellularization (Choi et al., 2013), we examined whether these effects continue after gastrulation. During stage 7, Arm continued to localize all along the lateral membrane with enrichment in the apical AJs, and Cno remained enriched in apical AJs, both in lateral (Supplemental Figure S1, C vs. D, arrows) and dorsal ectodermal cells (Supplemental Figure S1, E vs. F, arrows). Both were also enriched in apical AJs of invaginating posterior midgut cells (Supplemental Figure S1F, arrowhead). AJ enrichment was maintained at stage 8 (Supplemental Figure S1G, arrows).
We also examined Pyd and Baz localization. Apically, Pyd was less continuous at AJs after RNAi (Figure 1, F vs. G, arrows), and Pyd planar polarization to DV borders was strongly enhanced (Figure 1, F vs. G, red vs. yellow arrowheads). Sites where junctional Cno was more intense were also places where both Arm and Pyd were enriched (Figure 1, E–E′′, box, and E inset). The effect of Rap1 RNAi on Baz localization was even more substantial. Baz localization already was highly fragmented at stage 7 in Rap1 RNAi embryos, with just small regions of AJs retaining Baz (Figure 1I, arrows)–again, these often coincided with the regions where Arm localization was most intense. The strong disruption of apical junctions in many cells and the fragmentation of junctional Baz were all phenotypes absent in cno null mutants (Sawyer et al., 2011; Manning et al., 2019).
We next quantified the effect of Rap1 RNAi on AJ protein planar polarity, choosing the subset of stage 8 cells that were less affected by the more-drastic cell shape changes described above. Rap1 RNAi mimicked the effect of Cno loss on Arm and Pyd localization (Sawyer et al., 2011; Manning et al., 2019), enhancing planar polarization of Arm (Figure 3A) and Pyd (Figure 3B) by reducing their enrichment on AP borders. Rap1 RNAi also reversed the planar polarity of Cno (Figure 3C), altering mild enrichment on AP borders to strong enrichment on DV borders. In each case, these alterations appeared to reflect the fact that AJ protein localization to AP borders was reduced. In the subset of cells where we could measure Baz, its planar polarity was also strongly enhanced (Figure 3D). Together, these data are consistent with Cno being one of Rap1’s effectors during these stages, as their effects on junctional planar polarity were similar in direction. However, the dramatically more-severe effects of Rap1 loss on apical cell junctions and Baz junctional integrity strongly suggest that Cno is not its sole effector.
Rap1 RNAi leads to major defects in balanced apical contractility and cell shape regulation
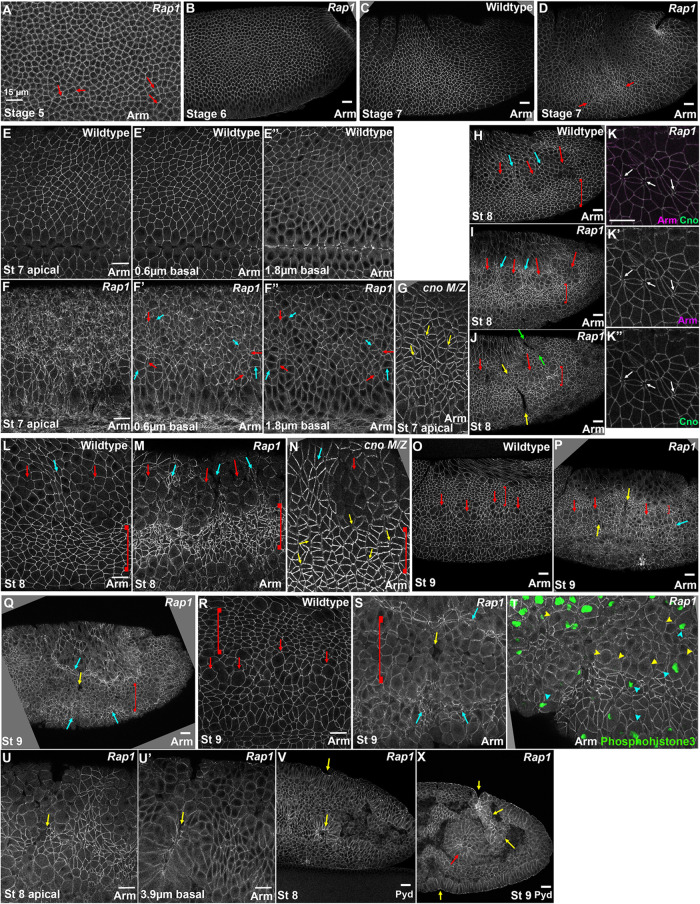
We next explored more broadly the effects of Rap1 RNAi in cell shape change and AJ integrity as morphogenesis continued. Until the onset of germband extension, Rap1 RNAi embryos were virtually normal (Figure 4, A and B), like Rap1M/Z mutants, with only modest alterations in the regularity of apical cell area during cellularization (Figure 4A, arrows; Choi et al., 2013). However, as germband extension accelerated during stage 7, Rap1 RNAi embryos began to exhibit striking defects. During stage 7, all cells assemble both a junctional and an apical contractile actomyosin cytoskeleton. In WT, contractility is relatively balanced. While some cell stretching is seen in the WT next to the invaginated mesoderm, cell shapes remain largely regular (Figure 4, C and E), with relatively consistent apical areas even as individual cell borders contract to mediate cell intercalation. In contrast, in Rap1 RNAi embryos apical cell shapes became highly abnormal (Figure 4, C vs. D). In WT, cell shapes are consistent from the apical ends of the cells to more-basal slices (Figure 4, E–E′′). In Rap1 RNAi embryos the alteration in cell shape was most apparent in the apicalmost region of the cells, where cell borders became difficult to define (Figure 4, F–F′′). Cell borders were more clearly visible just 0.6 µm more-basal in Rap1 RNAi embryos, but cell cross-sectional areas were highly variable, with apically constricted cells (Figure 4, F and F′, red arrows) adjacent to cells with much larger apical areas (Figure 4, F and F′, cyan arrows). In a subset of embryos, apically constricted cells were aligned along the anterior–posterior axis, potentially presaging the deeper folds seen at stage 8 (9/12 stage 7 embryos had at least one such incipient fold). In contrast, while cno null mutants do exhibit abnormal cell shapes and AJ gaps at AP borders and TCJs (Figure 4G, arrows; Sawyer et al., 2011), they do not share the extreme alterations in the apicalmost AJs seen after Rap1 RNAi.
FIGURE 4:
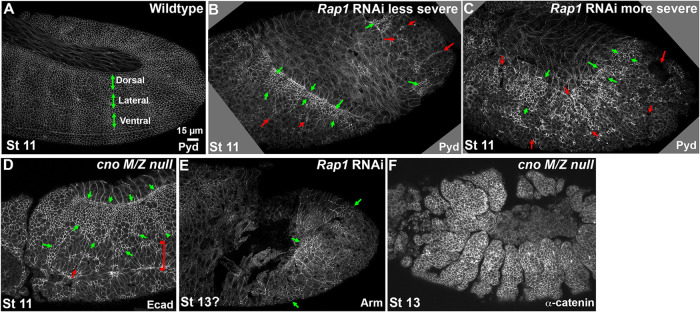
After Rap1 RNAi apical cell contractility becomes progressively unbalanced. (A, B) At stages 5 and 6, apical cell areas in Rap1 RNAi embryos remain relatively uniform with only modest variation. (C, D) At stage 7, while apical area remains relatively uniform in WT (C), in Rap1 RNAi embryos groups of cells begin to apically constrict relative to their neighbors (D, arrows). (E–G) Stage 7 closeups. In the most-apical sections, junctions are difficult to trace in Rap1 RNAi embryos (E vs. F). More basally, junctions are more apparent but apical cell areas are quite variable relative to WT, with small (F′, F′′, red arrows) and large (cyan arrows) cross-sectional areas. (G) cnoM/Z null mutants do not exhibit the dramatic defects in AJs in the apical plane. (H–N) Stage 8. In WT, cells round up to divide in mitotic domain 11 (H, L, red arrows) and cells between or adjacent undergo some apical constriction (H, L, cyan arrows and brackets). (K) Rap1 RNAi. In less-affected regions gaps in AJs open at TCJs and AP borders (arrows). (I, J, M) In Rap1 RNAi embryos, cells between and adjacent to mitotic domains are hyperconstricted (I, J, M, cyan arrows and brackets) and at times constriction leads to deep folds (J, yellow and green arrows). (N) cnoM/Z null mutants have gaps between cells at AJs under tension (yellow arrows) but do not exhibit the extreme accentuation of apical constriction of cells ventral to mitotic domain 11 (bracket). (O–S) Stage 9. In WT (O, R), dorsal ectodermal cells that divided in mitotic domain 11 resume columnar architecture (brackets) while cells in mitotic domain N round up for division (arrows). In milder Rap1 RNAi embryos (P), both dorsal ectodermal cells (bracket) and cells ventral to the mitotic cells (cyan arrow) are hyperconstricted, and folds remain (yellow arrows). Other Rap1 RNAi embryos have more severe defects. (Q, S). Some mitotic domain 11 and domain N cells remain rounded up (bracket), and flanking cells are very hyperconstricted (cyan arrows) and are folding inward (yellow arrows). (T) Use of the mitotic marker phosphohistone-3 confirms that a subset of rounded-up cells are no longer in mitosis (yellow arrowheads). (U–X) Examples of deep epithelial folds after Rap1 RNAi (yellow arrows) descending to the level of the posterior midgut (red arrow).
The cell shape defects after Rap1 RNAi became even more pronounced in embryonic stage 8. Drosophila cells divide in programmed groups called mitotic domains. In WT during stage 8, mitotic domain 11 becomes prominent in the thoracic and abdominal regions as cells in this mitotic domain round up to divide (Figure 4, H and L, red arrows). In the WT, cells between (Figure 4, H and L, cyan arrows) or more-ventral to (Figure 4, H and L, brackets) the mitotic domains apically constrict slightly, presumably due to reduced pulling forces from mitotic neighbors. In Rap1 RNAi embryos these differences were strongly exaggerated, with cells between (Figure 4, I and M, cyan arrows) or more-ventral to (Figure 4, I and M, brackets) the mitotic domains becoming irregular in shape and highly apically constricted. This phenotype was fully penetrant (17/17 stage 8 embryos). The folds observed during stage 7 also became more prevalent and deeper, extending across the ectoderm (Figure 4J, yellow arrows) and sometimes extending into the amnioserosa (Figure 4J, green arrows; 17/18 stage 8 embryos had folds). In less-affected regions of the ectoderm, gaps formed at TCJs and AP cell borders (Figure 4K), similar to those seen in cno mutants (Sawyer et al., 2011; Perez-Vale et al., 2021). While cno null mutants continue to exhibit altered cells shapes and gaps in apical AJs during stage 8 (Figure 4N; Sawyer et al., 2011; Manning et al., 2019), they lack the strongly unbalanced apical contractility seen after Rap1 RNAi.
In WT embryos during stage 9, dorsal ectodermal cells in mitotic domain 11 resume columnar shape (Figure 4, O and R, brackets) while cells in mitotic domain N begin to round up to divide (Figure 4, O and R, arrows). In Rap1 RNAi embryos epithelial defects continued or worsened at this stage. In less-severe embryos (14/23 scored), dorsal ectodermal cells were hyperconstricted (Figure 4, O vs. P, brackets) and grooves remained (Figure 4P, yellow arrows). In more-severe embryos (9/23 scored), some ectodermal cells failed to resume columnar architecture after their divisions (Figure 4, Q and R vs. S, brackets). We verified that some of these rounded-up cells had completed mitosis by staining for the mitotic marker phosphohistone3, which stained only some of the rounded-up cells (Figure 4T, yellow vs. cyan arrowheads). Adjacent cells became more apically constricted (Figure 4, Q and S, cyan arrows) or folded into the embryo (Figure 4, Q and S, yellow arrows). These infolding cells formed epithelial balls or folded sheets (Figure 4, U, U′, and V, arrows), which could become quite extensive (Figure 4X, yellow arrows), matching the posterior gut in depth and extent (Figure 4X, red arrow). Once again, this deep infolding seen after Rap1 RNAi was not observed after Cno loss (Manning et al., 2019).
The alterations in AJ protein localization observed after Rap1 RNAi at stages 7 and 8 continued during stage 9. At this stage in WT, Arm and Cno AJ localization is continuous both in the dorsal ectodermal cells that have regained columnar architecture after having divided (Figure 5A, red bracket) and in the most ventral cells that have yet to divide (Figure 5A, yellow bracket). It is also continuous in cells in mitotic domain N that are rounding up to divide (Figure 5A, cyan bracket), though at slightly lower levels. In Rap1 RNAi embryos, Arm, Cno, and Pyd junctional localization was reduced at AJs in dorsal and mitotic cells (Figure 5B, red and cyan brackets). In the more-ventral, highly apically constricted cells, Arm, Cno, and Pyd retained strong junctional localization but they appeared somewhat less uniform (Figure 5B, yellow bracket). Junctional Baz remained very fragmented, even in those cells that retained more columnar architecture (Figure 5, C vs. D). Thus, Rap1 is essential for maintaining uniform localization of Arm, Cno, and Pyd along AJs, and for continued Baz localization to AJs.
FIGURE 5:
While core AJ proteins remain at cell junctions at stage 9 after Rap1 RNAi, their cortical localization becomes less continuous and defects in junctional Baz localization continue or intensify. (A, B) In WT (A) Arm, Cno, and Pyd remain at junctions in both mitotic and columnar cells. After Rap1 RNAi (B) junctional Arm, Cno, and Pyd become less continuous in both cell populations. (C, D) In WT (C) Baz is strong in nonmitotic cells and weaker in mitotic cells. After Rap1 RNAi (D), Baz localization to junctions is reduced and fragmented.
To confirm that the cell shape and epidermal integrity defects we observed were due to knockdown of Rap1 rather than off-target effects, we examined embryos expressing a GDP-locked Rap1 mutant, Rap1S17A, as an alternate way of disrupting Rap1 activity. In our earlier work we found that expressing Rap1S17A mimics the effects of Rap1M/Z mutants and of Rap1 RNAi on Cno localization and initial AJ positioning during cellularization (Bonello et al., 2018). We thus expressed Rap1S17A maternally and zygotically using the GAL4/UAS system and the matGAL4 drivers. Cuticle analysis revealed that Rap1S17A led to the same strong disruption of epidermal integrity seen in Rap1M/Z mutants (Sawyer et al., 2009) and after Rap1 RNAi (Supplemental Figure S2, A, C vs. B, and D). We then examined the effects of expressing Rap1S17A on morphogenesis, comparing it to Rap1 RNAi. Rap1S17A expression also blocked mesoderm invagination (Supplemental Figure S2E, red arrow). At stage 7, we observed the same dramatically altered apical cell shapes (Supplemental Figure S2, E–E′′) that we had observed after Rap1 RNAi (Figures 1C and 4F). Cno localization was more disrupted than that of Arm (Supplemental Figure S2F), but places where Cno was retained also retained elevated levels of Arm (Supplemental Figure S2F, arrows), as observed after Rap1 RNAi (Figure 1, C and E). By stage 8, Rap1S17A expression led to unbalanced contractility, with hyperconstricted cells ventral to (Supplemental Figure S2, G–I, brackets) or between (Supplemental Figure S2, G–I, cyan arrows) the mitotic domains, epithelial folds (Supplemental Figure S2, G and J yellow arrows), and gaps appearing at AP borders and rosettes (Supplemental Figure S2J, red arrows), all defects that matched those seen after Rap1 RNAi (Figure 4). Thus, two different methods of reducing Rap1 activity both lead to major defects in balanced apical contractility and cell shape regulation.
Rap1 RNAi leads to major disruption of epidermal integrity that is more severe than that after Cno loss
These cell shape and junctional defects after Rap1 RNAi ultimately had striking effects on epidermal integrity. By stage 11, phenotypes diverged somewhat, with more and less severely affected embryos, likely due to differences in the zygotic copy number of the RNAi construct (0–2 copies). However, all Rap1 RNAi embryos had defects in epithelial integrity. At this stage, the WT epidermis is continuous from the amnioserosa dorsally to the ventral midline (Figure 6A). In the more severely affected Rap1 RNAi embryos, epidermal integrity was largely lost by stage 11, with the ventral and lateral epidermis completely disrupted, and only patches of dorsal epidermis remaining intact (Figure 6C, green arrows). Laterally and ventrally one could see the apical ends of cells that have folded into the interior (Figure 6B, red arrows). Less severely affected embryos had similar defects, but the amount of remaining dorsal epidermis was more extensive (Figure 6B). cnoM/Z null mutant embryos also have defects in the ventral epidermis, leading to eventual holes in the ventral cuticle, but more epidermis remains (Figure 6D; Sawyer et al., 2009, 2011; Manning et al., 2019; Perez-Vale et al., 2021). After stage 11, many Rap1 RNAi embryos had such substantial disruptions of the epidermis that they were difficult to stage precisely, because only patches of epidermis remained intact (Figure 6E, arrows). In contrast, while cnoM/Z null mutant embryos have defects in head involution, dorsal closure, and ventral epidermal integrity at stages 13 and 14, their lateral and dorsal epidermis remains intact (Figure 6F; Sawyer et al., 2009, 2011; Manning et al., 2019; Perez-Vale et al., 2021). Embryos expressing Rap1S17A had a similarly disrupted epidermis (Supplemental Figure S2, K and L, vs. Figure 6, B and C), with only patches of intact epidermis, consistent with their cuticle defects (Supplemental Figure S2, A–D). Thus, Rap1 is essential for maintaining epithelial integrity during morphogenesis.
FIGURE 6:
Rap1 RNAi leads to widespread disruption of epidermal integrity, more severe than that seen after Cno loss. (A–D) Stage 11. (A) The WT epidermis remains intact despite segmental cell shape change and tracheal pit invagination. Dorsal, lateral, and ventral epidermal regions are indicated. (B) In less severe Rap1 RNAi embryos, most of the ventral epidermis is lost and infolding cells are seen there (red arrows). The dorsal epidermis remains more intact (green arrows). (C) In more severe Rap1 RNAi embryos only small regions of intact dorsal epidermis remain near the amnioserosa (green arrows). The lateral and ventral epidermis are totally disrupted, with the apical ends of infolded cells seen in some places (red arrows). (D) In cnoM/Z null mutants the ventralmost epidermis is disrupted (bracket) and occasional infolding is seen (red arrow) but the region of remaining intact epidermis is more substantial (green arrows). (E) Later stage Rap1 RNAi embryo. Staging becomes difficult as only small regions of intact epidermis remain (green arrows). (F) In contrast, stage 13 cnoM/Z null mutants retain intact dorsal and lateral epidermis but do fail during dorsal closure and have the deep segmental groove phenotype.
Putting all these analyses together, Rap1 and Cno loss share some features, including parallel defects in junctional protein planar polarity, consistent with Cno being one of Rap1’s effectors. However, the multiple defects seen after Rap1 RNAi that are not shared by cno mutants, including dramatic destabilization of Baz localization, very unbalanced apical contractility, and extreme loss of epidermal integrity, suggest that Rap1 relies on additional effectors during this phase of morphogenesis.
Rap1 RNAi disrupts AJ–cytoskeletal connections but does not eliminate aPKC localization
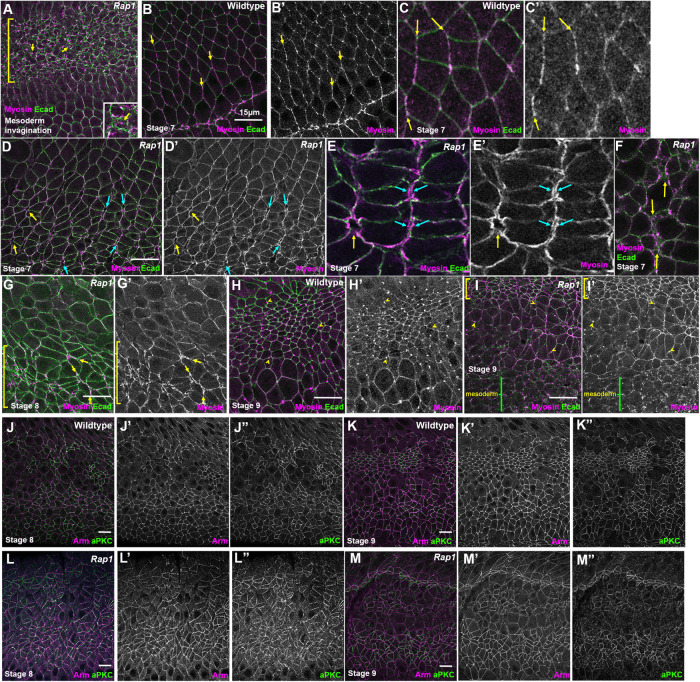
We completed our analysis of the effects of Rap1 knockdown by exploring two potential mechanistic explanations for the unbalanced contractility we observed after Rap1 RNAi. Cno loss weakens AJ–cytoskeletal linkage (Sawyer et al., 2009, 2011), helping explain its morphogenetic effects. Thus, the normally tight localization of myosin to AJs is disrupted in cnoM/Z mutants, with myosin localizing to two bands flanking the AJ. We thus asked how Rap1 affects myosin localization, exploring whether alterations in AJ–cytoskeletal linkage might explain the defects we observed after Rap1 RNAi. To do so, we imaged embryos expressing an mCherry-tagged myosin and a GFP-tagged E-cadherin (Ecad). Consistent with the idea that Rap1 RNAi mimics Rap1M/Z mutants, the apical myosin network that drives mesoderm apical constriction disconnected from AJs and continued to constrict (Figure 7A, bracket, inset), as previously described in Rap1M/Z mutants (Sawyer et al., 2009).
FIGURE 7:
Rap1 RNAi leads to detachment of myosin from many AJs under elevated tension but not to loss of cortical myosin and leads to less-continuous cortical localization of aPKC. (A–I) Embryos expressing Ecad-GFP and mCherry-Myosin. (A) Stage 6. Rap1 RNAi blocks mesoderm invagination. In mesoderm cells (bracket) the myosin network continues to contract but detaches from the AJs (arrows), as we previously observed in Rap1 and cnoM/Z null mutants. (B–F) Stage 7. (B, C) In WT, myosin is enriched at AP borders and is tightly associated with AJs (arrows). (D, E) After Rap1 RNAi, myosin often formed two lines at AP cell junctions (blue arrows) and also appeared to detach from junctions at the center of rosettes (yellow arrows). (F) Tight cortical myosin localization at places where folds were forming was often disrupted. (G) At stage 8, we observed myosin detachment from junctions (yellow arrows) and loss of tight cortical localization in hyperconstricted cells (bracket). (H, I) Stage 9. (H) In WT, myosin is both cortical and strongly enriched at the persistent midbodies (yellow arrows) in dorsal cells that have competed division and is also cortical in more-ventral rounded-up mitotic cells. (I) After Rap1 RNAi, cell shapes are very distorted but myosin remains both cortical and enriched in midbodies. (J–M) aPKC localization. Maximum-Intensity-Projections (MIPs) as aPKC is apical to the AJs. (J, K) WT. Like Arm, aPKC is cortically localized. (L, M) aPKC remains cortical after Rap1 RNAi, but junctional localization appeared less continuous.
We then examined myosin localization during germband elongation. In WT, junctional myosin is enriched at AP borders and tightly localized to the AJ (Figure 7, B and C, arrows). In contrast, after Rap1 RNAi junctional myosin along AP borders was often separated into two lines, suggesting disconnection from AJs (Figure 7, D and E, cyan arrows). Similarly, myosin localization encircled TCJs rather than being concentrated at vertices (Figure 7, D and E, yellow arrows). Both were phenotypes we observed in cno mutants (Sawyer et al., 2009). This myosin disruption was prominent at places where AJ gaps were forming (Figure 7F, arrows). During stage 8, in regions of unbalanced contractility, cells with smaller apical areas had strong but disorganized cortical myosin (Figure 7G, bracket) and myosin detachment from AJs continued (Figure 7G, arrows). In WT stage 9 embryos, myosin is cortical, both in cells that had already completed divisions (as indicated by the myosin in midbodies; Figure 7H, arrowheads) and in cells that had rounded up for division. Many cells in similar stage Rap1 RNAi embryos also retained cortical myosin (Figure 7I, bracket), although the epithelium was more disorganized. Some cells with midbodies had not resumed columnar architecture after dividing (Figure 7I, arrowheads). Thus, Rap1 RNAi disrupts tight linkage of myosin to AJs but does not eliminate cortical myosin. More analysis is needed to define how Rap1 regulates myosin localization.
We also examined another potential mechanism by which Rap1 might regulate balanced contractility. Work from the Morais-de-Sa lab (Osswald et al., 2022), published while our manuscript was being revised, revealed that reducing the activity of the apical polarity regulator aPKC led to unbalanced contractility in the Drosophila follicle cell epithelium, with hyperconstriction of cells neighboring those bordering mitotic cells and subsequent gaps in the epithelium. This was reminiscent of what we observed in Rap1 RNAi embryos. We thus examined whether Rap1 RNAi led to loss of cortical aPKC. During stages 8 and 9 in WT embryos, aPKC is cortical in nonmitotic cells (Figure 7, J and K). Rap1 RNAi did not eliminate cortical aPKC at these stages, though both Arm and aPKC localization became less continuous, likely reflecting AJ gaps (Figure 7, L and M). Thus, Rap1 RNAi does not lead to total loss of cortical aPKC, but future work is needed to determine whether aPKC localization or activity is reduced.
Developing and validating tools to strongly reduce Dzy function to define its role in morphogenesis
We also want to know which GEFs activate Rap1 during morphogenesis. Dzy is one of several Rap1 GEFs. During cellularization, Dzy loss mimics the effects of loss of Cno on junctional polarization (Bonello et al., 2018), and both Cno and Dzy are required for effective apical constriction during mesoderm invagination (Spahn et al., 2012), the first event of gastrulation. Thus, Dzy is an important regulator of Rap1/Cno during these early stages. However, Dzy’s roles in other events of embryonic morphogenesis remain essentially unexplored.
We and others previously used the FLP/FRT/DFS technique to generate maternal/zygotic dzy mutants (Boettner and Van Aelst, 2007; Spahn et al., 2012; Bonello et al., 2018), but females lay few eggs, suggesting possible roles in oogenesis. To circumvent this, we sought to create an effective short hairpin RNA (shRNA) reagent that we could drive maternally and thus use to deplete both maternal and zygotic Dzy (Staller et al., 2013). We and others have used this approach very effectively, in our case to reduce maternal and zygotic expression of Rap1 and Cno (Bonello et al., 2018). We generated such an shRNA reagent, using the pWalium22 vector to create an shRNA targeting dzy’s 5th exon (the oligos used to target dzy are listed in Materials and Methods). We inserted the shRNA plasmid into the right arm of the 3rd chromosome using phiC31/attP2 site integration. We drove this UAS-dzy shRNA construct in the female germline with a strong two-component maternal germline driver line, matGAL4 (Staller et al., 2013). Using this GAL4 driver to drive shRNAs has worked well for us in the past to greatly reduce or eliminate maternal contribution of both Cno and Rap1 (Bonello et al., 2018).
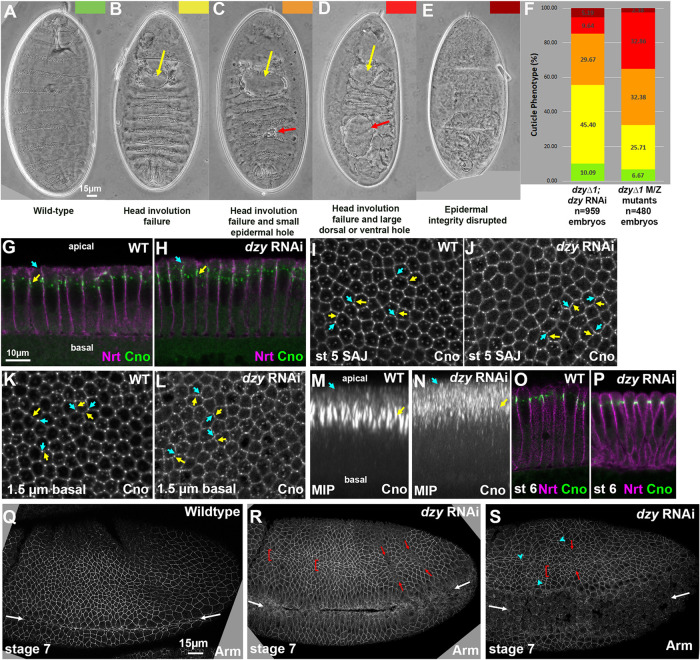
To further ensure reduction of both maternal and zygotic Dzy function, we crossed females that carried the matGAL4 drivers, the dzy shRNA construct, and that were heterozygous for a null allele of dzy (dzy∆1) to males carrying the dzy shRNA construct and were also heterozygous for dzy∆1 (Supplemental Figure S3). Thus, all progeny should have strong maternal dzy RNAi knockdown and one quarter of progeny will also be zygotically null for dzy, preventing any recovery of Dzy function. We assessed the effectiveness by examining the cuticle phenotype, which allows us to assess effects on major morphogenetic movements and epithelial integrity. WT embryos have an intact cuticle, secreted by the embryonic epidermis, and have completed major morphogenetic movements like germband retraction, head involution, and dorsal closure (Figure 8A). Our previous cuticle analysis of dzy∆1 maternal/zygotic null mutants revealed that head involution failed in essentially all embryos (Figure 8, B–D, yellow arrows, quantified in F), and, in most embryos, this was accompanied by small or large holes in the epidermis (Figure 8, C and D, red arrows), with a small fraction of embryos exhibiting more severe disruption of epidermal integrity (Figure 8E). Intriguingly, this is less severe than full maternal/zygotic loss of Cno (Sawyer et al., 2009); instead, it is more similar to the phenotype of Cno∆RA, which lacks the Ras-associated (RA) domains (Perez-Vale et al., 2021). Embryos from the dzy∆1 dzy RNAi cross exhibited a similar range of phenotypes (Figure 8F), with nearly universal defects in head involution, and with many embryos having holes in the cuticle. The distribution of phenotypes was somewhat less severe in the progeny of the dzy RNAi cross than among dzy∆1 maternal/zygotic null mutants (p < 0.0001, Chi-square contingency test based on the number of embryos in each phenotypic category), but given that only 25% of the dzy∆1 dzy RNAi embryos would be predicted to also be zygotically dzy null mutant, the overlap in cuticle phenotype suggests that our RNAi reagent is on-target and that we achieved strong reduction of maternal and zygotic Dzy function in this subset of embryos. To further assess whether these effects on morphogenesis and epidermal integrity were truly due to reduction in Dzy function and not off-target effects, we compared our dzy RNAi approach with or without inclusion of the dzy∆1 mutant. Including the dzy∆1 mutant increased the fraction of embryos that have the more-severe epidermal disruption phenotypes (in addition to defects in head involution) from 36 to 45% (n ≥ 270 embryos of each genotype). This supports the idea that the defects we see are due to reduction in Dzy expression.
FIGURE 8:
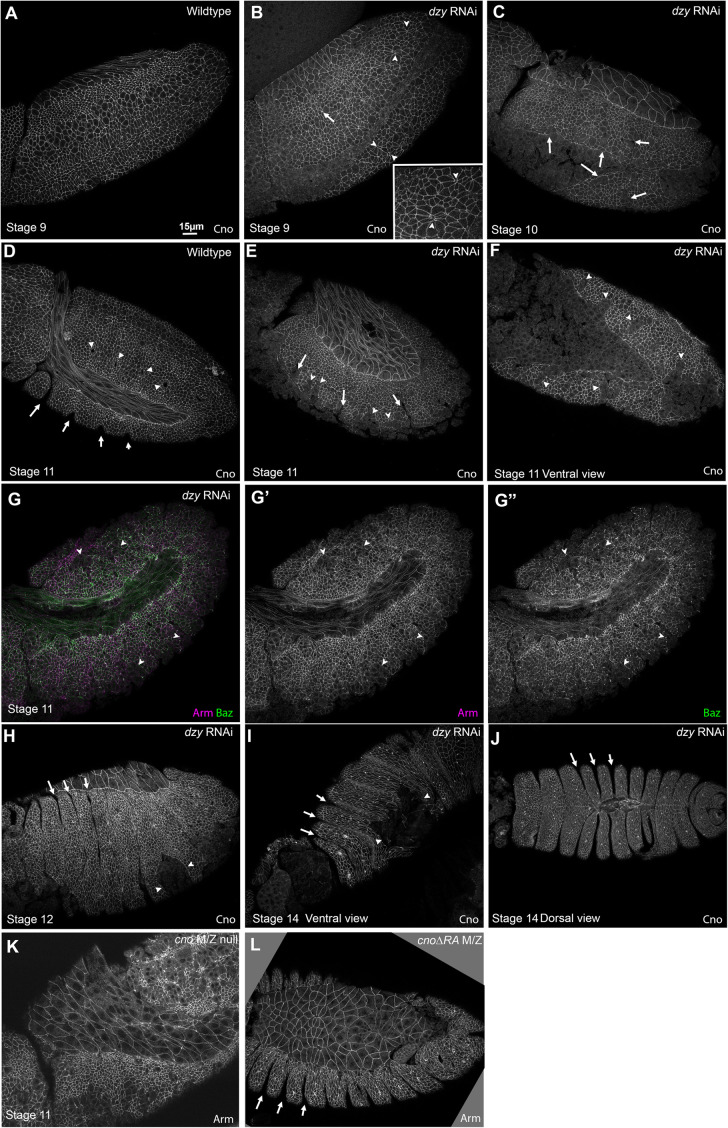
dzy RNAi largely mimics total maternal/zygotic loss of Dzy. (A–E) Representative embryonic cuticles illustrating different phenotypic categories. (F) Cuticle defects in lethal embryonic progeny from the cross in Supplemental Figure S3 (dzy∆1; dzy RNAi) vs. those in maternal/zygotic dzy∆1 mutants. (G, H) Apical-basal cross-sections. Neurotactin (Nrt) marks the plasma membrane. Cno remains apically enriched at nascent SAJs after dzy RNAi (yellow arrow), but some Cno moves more apically (cyan arrow). (I–L) En face sections at the level of the SAJs (I, J) or 1.5 µm more basal (K, L). In WT, Cno localizes to all SAJs but is somewhat enriched at TCJs relative to bicellular junctions (I, cyan vs. yellow arrows) and is strongly enriched at TCJs 1.5 µm basal (K). dzy RNAi leads to loss of TCJ enrichment at both the SAJ level (J) and more basally (L). (M, N) MIPs of apical-basal cross-sections. In WT, TCJ Cno forms cables (M, yellow arrows) and is largely excluded apically (M, cyan arrows). dzy RNAi (N) disrupts TCJ cables and increases levels apical to the SAJs. (O, P) Apical-basal cross-sections. At gastrulation onset (stage 6) Cno localization returns to its normal apical position after dzy RNAi. (Q–S) Embryos, stage 7. (Q) WT stage 7. Mesoderm invagination is complete (white arrows). (R, S) dzy RNAi. Mild (R) or strong (S) defects in mesoderm invagination are seen in virtually all embryos (white arrows). Some ectodermal cells are abnormally elongated along the AP axis (brackets), ectopic grooves appear (red arrows), and gaps appear at TCJs and AP borders (cyan arrowheads).
Because we could not find a Dzy antibody that worked in immunoblotting, we also verified the effectiveness of depleting maternal Dzy by comparing the effects of dzy RNAi on cellularization and mesoderm invagination with those previously characterized for maternal/zygotic dzy null mutants. We first compared the effects of dzy RNAi on AJ polarization during cellularization, a process likely driven exclusively by maternal contribution, comparing it to our previous analysis of dzy null maternal mutants (Bonello et al., 2018). During cellularization, Cno localizes to nascent apical spot AJs (SAJs; Figure 8, G, yellow arrow, and I). At the apical position of SAJs, Cno localizes to bicellular junctions and is also enriched at TCJs (Figure 8I, yellow vs. cyan arrows). At TCJs, strong Cno localization extends more basally (Figure 8K, yellow vs. cyan arrows), forming cable-like structures visible in maximum-intensity projections (MIPs; Figure 8M). While loss of Rap1 leads to complete loss of Cno from the plasma membrane (Sawyer et al., 2009), dzy null maternal mutants have less-severe effects, suggesting that additional Rap1 GEFs act at this stage (Bonello et al., 2018). In dzy maternal mutants, while Cno remains enriched in spot AJs and roughly localized to the apicolateral region, its localization spreads more apically and Cno enrichment at TCJs and formation of cable-like structures is lost (Bonello et al., 2018). dzy RNAi replicated the effect of dzy null maternal mutants on Cno localization during cellularization. While Cno remained roughly localized apically (Figure 8H) and still accumulated in bicellular SAJs (Figure 8, H and J, yellow arrows), Cno localization expanded more apically (Figure 8, H and N, cyan arrows) and enrichment at TCJs was lost, at both the levels of normal nascent spot AJs (Figure 8J, yellow vs. cyan arrows) and 1.5 µm basally (Figure 8L, yellow vs. cyan arrows). As a result, in MIPs, Cno cables were fragmented (Figure 8N). Thus, dzy RNAi replicates all the cellularization stage phenotypes of dzy null maternal mutants (Bonello et al., 2018), consistent with our RNAi effectively knocking down maternal Dzy. As we had observed with dzy null maternal mutants (Bonello et al., 2018), apical junctional enrichment of Cno was largely restored as gastrulation began in stage 6 (Figure 8, O vs. P). As a second test of the effectiveness of maternal Dzy depletion by dzy RNAi, we assessed its effects on mesoderm invagination. In dzy null maternal mutants, mesoderm invagination fails (Spahn et al., 2012). Dzy RNAi also had an extremely penetrant effect on this process, with partial (Figure 8, Q vs. R, white arrows) or complete (Figure 8S, white arrows) failure of mesoderm invagination in 52 of 53 embryos (as assessed at stage 7). Together these data suggest that our shRNA is effective at very strongly reducing or eliminating the Dzy maternal contribution, suggesting that our strategy provided a reagent for defining the role of Dzy during morphogenesis. We note that we cannot absolutely rule out off-target effects, but the correspondences we reveal below between the effects of dzy RNAi and those of Cno loss also support the idea that the effects observed are due to reduction in Dzy function. For all remaining experiments, we used the aforementioned dzy∆1 dzy RNAi cross, referred to for simplicity below as “dzy RNAi embryos.”
dzy RNAi slows germband extension, with defects similar to those of Cno loss and less severe than those seen after Rap1 RNAi
Dzy is one of multiple Rap1 GEFs in the Drosophila genome. Our goal was to determine which of Cno’s many roles during morphogenesis require Dzy-mediated Rap1 activation and which may rely on other Rap1 GEFs. We envisioned three broad possibilities. First, given the multiple roles of Rap1 revealed above, if Dzy is the predominant GEF regulating Rap1 during morphogenesis, Dzy loss would lead to Rap1-like phenotypes and thus be more severe in its effects than Cno loss. Second, it was possible that Dzy activates Rap1 to mediate its action via Cno, but other GEFs regulate Rap1’s Cno independent roles. In this case, Dzy loss would mimic loss of Cno. Finally, if Dzy was not an important Rap1 GEF during morphogenesis, dzy RNAi might not have later morphogenesis phenotypes.
We thus used the cross outlined above (Supplemental Figure S3) to strongly reduce maternal and zygotic Dzy and distinguish between these possibilities, beginning by exploring Dzy’s role during convergent elongation during germband extension, using our dzy RNAi approach. In cnoM/Z mutants, germband elongation is significantly slowed (Sawyer et al., 2011). We thus assessed the effect of dzy RNAi on this process and compared them to cnoM/Z mutants and Rap1 RNAi embryos. While the degree of germband extension was similar between WT and dzy RNAi embryos at stage 7 (Figure 2, A vs. G, and M), at both stage 8 (Figure 2, B vs. H, and N) and stage 9 (Figure 2, C vs. I, and O) germband extension in dzy RNAi embryos slowed substantially relative to WT. This delay resembles that seen after cno loss, which also preferentially affects the later stages of germband extension (Sawyer et al., 2011), and differs from the earlier and more-severe reduction in germband extension we observed after Rap1 RNAi above (Figure 2, J–L vs. M–O).
Like Cno, Dzy is required to reinforce junctions under tension during germband extension
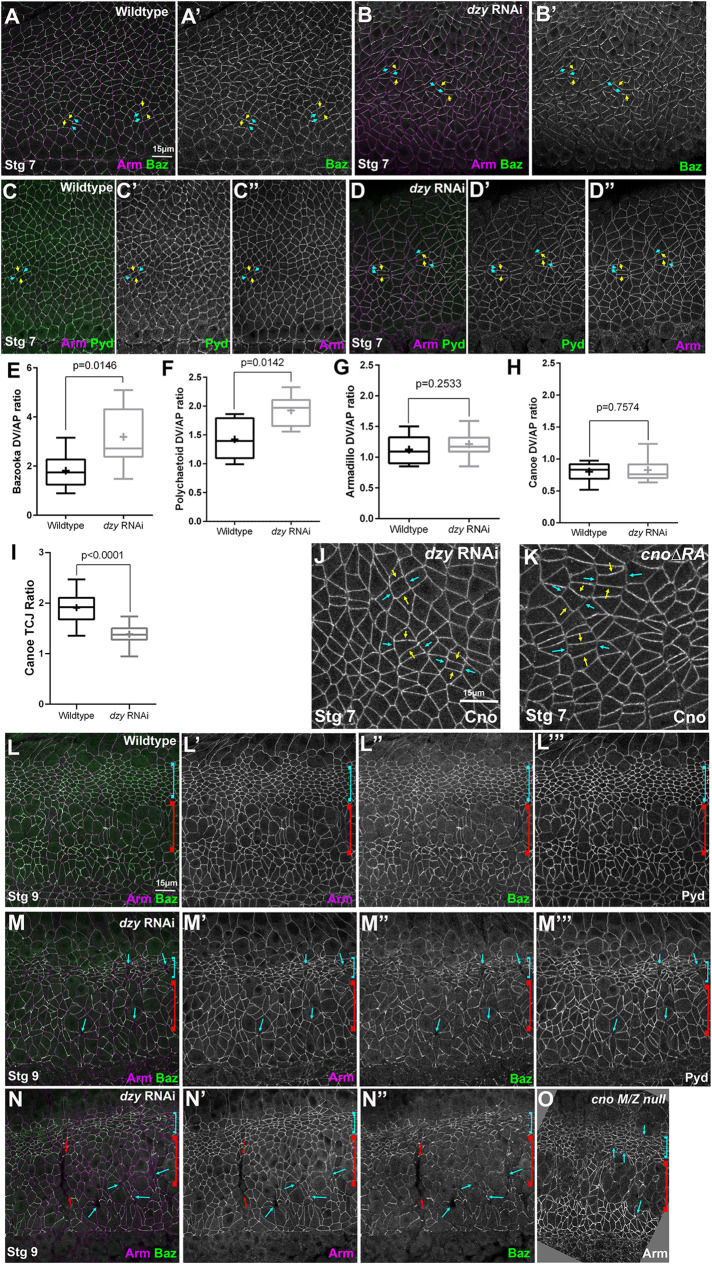
We next examined the effects of dzy RNAi on the orchestrated cell shape changes and AJ integrity during cell intercalation and germband extension. While loss of Dzy affects initial assembly of AJ and Cno proteins into apical AJs during cellularization, this is largely rescued as gastrulation begins (Bonello et al., 2018). Consistent with this, in dzy RNAi embryos the overall localization of both Arm (Figures 8, Q vs. R, and S, and 9, A′ vs. B′) and Cno (Figure 9, A′′ vs. B′′) to AJs appeared relatively normal during gastrulation. However, as the germband extended in stage 7, we observed the first defects in dzy RNAi embryos: stacks of cells became abnormally elongated along the AP axis (Figures 8, L and M, and 9B, brackets). The dorsal folds also extended farther ventrally than normal and ectopic folds were observed (Figures 8, R and S, and 9B, red arrows; 14/21 stage 7 embryo had folds extending into the ectoderm). In WT embryos, Arm and Cno were continuous around the cell circumference, both along aligned AP borders (Figure 9A, red arrows) and at TCJs and short multicellular junctions (Figure 9A, cyan arrowheads). In contrast, in dzy RNAi embryos gaps opened in apical AJs at many TCJs and AP borders (Figure 9B, cyan arrowheads). All of these defects closely resembled cno mutant phenotypes (Sawyer et al., 2011; Manning et al., 2019; Perez-Vale et al., 2021).
FIGURE 9:
dzy RNAi leads to defects in cell shapes and at AJs under elevated tension similar to those seen in cno mutants. Embryos, stages 7 (A, B) and 8 (C–H). (A) WT closeup. Arm and Cno remain strong all around cells, including those at aligned AP borders (red arrows), those at TCJs (yellow arrows), where Cno is enriched, and at the centers of rosettes (cyan arrowheads). (B) dzy RNAi. Gaps appear at aligned AP borders and rosette centers (cyan arrowheads), especially in regions where cells are abnormally elongated along the AP axis (brackets). Ectopic grooves are present (red arrows). Cno enrichment at TCJs is reduced (yellow arrows). (C, F) At stage 8 in WT, cells in mitotic domain 11 round up to divide (red brackets) and cells between the mitotic domains apically constrict as their mitotic neighbors reduce apical contractility (cyan arrows). (D, E, G) dzy RNAi. Note ectopic fold (red arrows), gaps between mitotic cells (yellow arrows), elongated cells in the ventral ectoderm (cyan brackets), and gaps at TCJs and aligned AP borders (cyan arrowheads). (H) Similar gaps are seen in cno∆RA mutants. (I) Quantification of gaps.
In addition to the challenges of cell shape change and rearrangement, AJs also must be remodeled as cells round up for cell division. During stage 8, as cells in mitotic domain 11 round up to divide (Figure 9, C and F, red brackets), intervening cells apically constrict (Figure 9, C and F, cyan arrows), perhaps due to reduced pulling force from mitotic neighbors (Ko et al., 2020). However, in WT embryos AJs remain intact around the circumference of mitotic cells, although protein localization per unit membrane is reduced (Figure 9F, brackets). More-ventral cells, which are yet to divide, continue intercalation and retain intact apical AJs throughout the process. In dzy RNAi embryos, mitotic domains divided on schedule (Figure 9, D and E, red brackets), but gaps appeared between rounded-up mitotic cells (Figure 9, D, E, and G, yellow arrows). More ventrally, gaps remained in apical AJs at many TCJs and AP borders (Figure 9G, arrowheads). Most embryos retained ectopic folds (Figure 9D, red arrow; 11/15 stage 8 embryos). However, Arm and Cno continued to localize to AJs in places where AJs retained their integrity (Figure 9G). All of these defects were quite similar to those we previously observed in cno∆RA embryos lacking Cno’s Rap1-binding RA domains (Figure 9H; Perez-Vale et al., 2021). To quantitatively compare the effects on junctional stability of Cno and Dzy loss, we quantified gaps in WT versus dzy RNAi embryos (Figure 9I). While WT embryos had only occasional gaps in apical junctions (2.2 gaps per 133 × 133 μm field of cells; n = 18 stage 7 and 8 embryos), gaps were much more frequent in dzy RNAi embryos (33.6 gaps per 133 × 133 μm field of cells; n = 18 stage 7 and 8 embryos). This was comparable to the average number of gaps we previously observed in both cno∆RA embryos and cno maternal/zygotic null mutants (22 and 30 gaps per field, respectively; Perez-Vale et al., 2021). Together these data suggest that Dzy function is dispensable for Cno and Arm localization to AJs but is important to reinforce AJs under elevated tension, like TCJs and constricting AP borders, as reflected by the gaps that appear at these locations after dzy RNAi (Figure 9, G and I). This is consistent with Dzy being a major GEF involved in Rap1 regulation of Cno during these events. However, dzy RNAi embryos lacked the unbalanced apical contractility and fragmentation of junctional Baz seen after Rap1 RNAi. Thus Dzy is clearly not the only GEF responsible for Rap1 activation during embryonic morphogenesis.
Dzy is important for maintaining most but not all aspects of AJ planar polarity and for Cno recruitment to TCJs
As noted above, planar polarization of AJ proteins and the actomyosin cytoskeleton are essential for cell intercalation during germband extension (Perez-Vale and Peifer, 2020). In WT embryos, Baz is enriched on DV borders (Figure 10A, yellow vs. cyan arrows; enrichment is approximately twofold, Figure 10E), where it plays an important role in driving intercalation (Zallen and Wieschaus, 2004). Cno is important for maintaining balanced planar polarity. In Cno’s absence, Ecad, Arm, and Pyd are reduced on AP borders, while Baz is essentially lost there, thus strongly enhancing its planar polarization to DV borders (Sawyer et al., 2011; Perez-Vale et al., 2021). Given the parallel effects of loss of Cno and loss of Dzy on AJs under tension, we anticipated that effects on planar polarity of junctional proteins would be identical. However, when we measured junctional planar polarity the results proved to be more complex. Like cno mutants, dzy RNAi embryos had significantly enhanced Baz planar polarity, with Baz strongly reduced or eliminated on AP borders (Figure 10B, yellow vs. cyan arrows, quantified in E). In WT, Pyd is also more subtly planar polarized, with enrichment on DV borders (Figure 10C, yellow vs. cyan arrows, quantified in F). Pyd planar polarity was also further enhanced by dzy RNAi (Figure 10D, yellow vs. cyan arrows, quantified in F), thus resembling the effect of cno mutants (Manning et al., 2019). However, in contrast to cno mutants (Sawyer et al., 2011), the planar polarity of Arm was not significantly enhanced in dzy RNAi embryos when borders with obvious apical gaps were excluded (Figure 10G).
FIGURE 10:
dzy RNAi alters junctional protein planar polarity in ways similar but not identical to loss of Cno. (A–D, J, K) Stage 7 embryos. (A) WT. Baz is planar polarized to DV borders (yellow vs. cyan arrows). (B) dzy RNAi. Baz planar polarization is enhanced. (C) WT. Pyd is mildly enhanced at DV borders (yellow vs. cyan arrows). (D) dzy RNAi. Pyd planar polarization is enhanced. (E–H) Planar polarity quantification. (I) dzy RNAi reduces Cno enrichment at TCJs. (J, K) dzy RNAi does not alter Cno planar polarity (J), unlike cno∆RA (K). (L–N) Stage 9. (L) WT. Dorsal ectodermal cells in mitotic domain 11 resume columnar shape (cyan bracket), while neuroectodermal cells in mitotic domain N round up to divide (red bracket). (M, N) dzy RNAi. Dorsal ectodermal cells are hyperconstricted (cyan brackets). Gaps (cyan arrows) and folds (red arrows) persist. (O) cnoM/Z null mutant for comparison. The numbers of cells analyzed for planar polarity are in Supplemental Table S1.
Cno itself is not uniformly localized to AJs. Cno is enriched at TCJs (Sawyer et al., 2009) in a tension-sensitive manner (Yu and Zallen, 2020; Figure 9A, yellow arrows; quantified in Figure 10I) and also subtly planar polarized, with enrichment on AP borders in the WT, opposite that of Baz (Sawyer et al., 2011; Figure 10H). TCJs and AP borders are locations where tension is thought to be elevated (Fernandez-Gonzalez et al., 2009; Yu and Zallen, 2020; Perez-Vale et al., 2021). Cno enrichment at both TCJs and AP borders requires Cno’s Rap1-binding RA domain, as in cno∆RA mutants Cno enrichment at TCJs is strongly reduced and planar polarity is flipped, as loss at AP borders means that it is now enriched at DV borders (Perez-Vale et al., 2021; Figure 10K, yellow vs. cyan arrows). Our data above confirm that this also requires Rap1. On the basis of this, we anticipated that Dzy would be required for both proper Cno TCJ enrichment and planar polarization. However, once again things were more complicated. Cno TCJ enrichment was strongly reduced after dzy RNAi (Figure 9, A vs. B, yellow arrows; quantified in Figure 10I). Intriguingly, however, Cno planar polarity was unchanged in dzy RNAi embryos (Figure 10J, quantified in H), unlike what we observed in cno∆RA mutants (Figure 10, J vs. K). Together, these data suggest that regulation by Dzy is important for most but not all aspects of AJ protein localization, thereby paralleling the complexity we saw in the regulation of Cno by Rap1 versus Dzy during cellularization (Bonello et al., 2018).
dzy RNAi embryos largely maintain epithelial integrity, with some defects ventrally
We finished this analysis by examining how loss of Dzy affected the maintenance of epithelial integrity as morphogenesis proceeded. Despite the defects in junctional integrity at AJs exposed to elevated tension in dzy RNAi embryos, the epidermal epithelium remained relatively intact during germband extension (Figure 9, C vs. D, and E). In WT embryos at stages 9 and 10, dorsal ectodermal cells that divided in domain 11 have resumed columnar architecture (Figure 10L, cyan bracket). During these stages, AJs are further challenged as cells in mitotic domain N (Figure 10L, red bracket) and mitotic domain M round up to divide, and a subset of cells invaginate as neuroblasts. In WT embryos, Arm and Pyd staining remain strong in both mitotic and nonmitotic cells while Baz is continuous in nonmitotic cells and reduced but still generally present at junctions of mitotic cells (Figure 10L′′). In dzy RNAi embryos, while the ectoderm remained largely intact, multiple defects in cell shapes and AJ integrity persisted. Dorsal ectodermal cells were somewhat hyperconstricted along the AP axis (Figure 10, L vs. M, and N, cyan brackets). However, this contrasted with the earlier onset and much stronger hyperconstriction observed after Rap1 RNAi, leading to tissue infolding–instead, the dzy RNAi defect was more similar to what we observed in cno∆RA mutants (Manning et al., 2019; Perez-Vale et al., 2021). Gaps remained at TCJs and AP borders (Figure 10, M and N, cyan arrows; Figure 11B, arrowheads; 17/22 stage 9 and 10 embryos), and occasional ectopic grooves were present (Figure 10N, red arrows; Figure 11B, arrow; 7/22 stage 9 and 10 embryos). Arm and Pyd staining remained largely continuous around cells, except at gaps (Figure 10, L′, L′′′ vs. M′, M′′′, and N′), and Baz staining, while less continuous (Figure 10, L′′ vs. M′′, and N′′), was not fragmented as we observed in Rap1 RNAi embryos. At this stage, defects were similar to those seen in cnoM/Z null mutants (Figure 10O).
FIGURE 11:
dzy RNAi leads to defects in the ventral epidermis and persistent deep segmental grooves. (A, B) Stage 9. (A) WT. (B) dzy RNAi. Note remaining fold (arrow) and gaps at some AP borders and TCJs (arrowheads, inset). (C) dzy RNAi, stage 10. Gaps in the ventral epidermis are evident (arrows). (D–G) Stage 11. (D) WT. Segment grooves become apparent (arrows), and tracheal pits invaginate (arrowheads). (E–G) dzy RNAi. Note deep folds (arrows) and gaps in the ventral epidermis (arrowheads). (H–J) dzy RNAi, stages 12 and 14. Note abnormally deep segmental grooves (arrows) and holes in the ventral epidermis (arrowheads). (K) Epidermal defects are more frequent and widespread in cnoM/Z null mutants. (L) cno∆RA mutants share the deep segmental groove phenotype.
By stage 10, more severely affected embryos exhibited defects in epithelial integrity (Figure 11C, arrows), perhaps where cells failed to regain columnar architecture following division. During stage 11, cell junctions in WT embryos are challenged by the formation of segmental grooves (Figure 11D, arrows) and invagination of cells to form tracheal pits (Figure 11D, arrowheads). At this stage, dzy RNAi embryo phenotypes become more variable, perhaps because RNAi knockdown is diminished in the 75% of embryos that are not dzy zygotic mutants. However, while epidermal integrity was broadly maintained, defects were observed in the ventral epidermis (Figure 11, E–G, arrowheads; 14/16 stage 11 and 12 embryos), often coinciding with the location of segmental grooves (Figure 11E, arrows), which are also known to be sites of increased tension (Mulinari et al., 2008). These defects were less severe than those we observed in cnoR2M/Z null mutants (Figure 11K; Sawyer et al., 2009); instead they were more similar in severity to those observed in cno∆RA mutants (Figure 11L; Perez-Vale et al., 2021). As in cno mutants (Manning et al., 2019), Baz localization to junctions was more affected than that of Arm (Figure 11G), but we did not see the complete junctional fragmentation that we saw after Rap1 RNAi (above) or when we simultaneously reduced both Cno and Pyd (Manning et al., 2019). During dorsal closure, epidermal integrity remained largely intact, but many dzy RNAi embryos had ventral holes in the epidermis (Figure 11, H and I, arrowheads), consistent with what we observed in the dzy RNAi cuticles. They also exhibited deep persistent segmental grooves (Figure 11, H and J, arrows), another characteristic of cno mutants (Figure 11L; Manning et al., 2019; Perez-Vale et al., 2021). However, consistent with their respective cuticle phenotypes, the effects of dzy RNAi on cuticle integrity and dorsal closure were less severe than those seen in cnoR2M/Z null mutants (e.g., Figure 11, G vs. K).
Putting all of these observations together, the defects in morphogenesis seen in dzy RNAi embryos are similar to those of cno mutants, suggesting that Dzy is the major GEF acting via Rap1 to regulate Cno in these events. It is intriguing that in many ways, especially the effect on overall epidermal integrity, dzy RNAi more closely resembles the phenotype of cno∆RA rather than the even more severe effects of complete loss of Cno, suggesting that Dzy may mediate Rap1 action via that direct interaction, with other GEFs regulating Rap1 and affecting Cno via less-direct interactions. Thus, subtle differences in the effect on planar polarity of dzy RNAi and cno∆RA, along with the reduced loss of epidermal integrity, may suggest that other GEFs act through Rap1 to regulate Cno. Finally, the substantially less-severe phenotype of dzy RNAi relative to Rap1 RNAi clearly suggests that Rap1 utilizes additional GEFs to regulate morphogenesis.
DISCUSSION
The diverse cell shape changes and arrangements during embryonic development require robust connections between cell–cell AJs and the contractile actomyosin cytoskeleton. AJs need to withstand the forces generated without disrupting tissue integrity. Drosophila Cno and its mammalian homologue Afadin are both essential for these events. One key challenge now is to define the mechanisms by which Cno is regulated. The small GTPase Rap1 regulates many events involving cell–cell and cell–matrix adhesion, using diverse effectors, and Rap1, in turn, is activated by numerous GEFs, acting at different times and places. Thus far, analyses of Rap1’s roles in Drosophila embryonic development have been largely confined to the first two events. Rap1 uses Cno as an effector during cellularization and mesoderm invagination, and during these stages Rap1 is regulated by PDZ-GEF/Dzy. We sought to define Rap1’s roles in the many complex events of later morphogenesis, asking whether its roles match those of Cno, or whether it might play additional roles using other effectors. In parallel, we asked whether Dzy remained the predominant GEF involved in Rap1 regulation. Our results suggest that Rap1 has both Cno-dependent and Cno-independent roles in embryonic morphogenesis, allowing it to ensure tissue integrity. They further suggest that Dzy is the main GEF involved in Cno regulation during these events, but that Rap1’s Cno-independent roles must involve another GEF.
Rap1 regulates morphogenesis by restraining apical contractility via Cno-dependent and Cno-independent effects
Given the parallel effects of loss of Cno and loss of Rap1 on cellularization and mesoderm apical constriction, we initially expected Rap1 loss to precisely mimic loss of Cno. However, the phenotypes we observed suggest that Rap1 plays additional roles (Table 1). Some early effects of Rap1 loss parallel those of loss of Cno: gaps appear at many TCJs and AP borders, and AJ proteins and Baz planar polarity are altered in similar ways. However, quite rapidly additional defects appear in Rap1 RNAi embryos that are not present in cno mutants. As gastrulation begins, apical cell shapes are strongly altered, with some cell apical areas much smaller and others much larger. This suggests unbalanced apical contractility. These differences are accentuated as subsets of cells round up to divide in mitotic domains, thus reducing their junctional contractility. Adjacent cells hyperconstrict, sometimes doing so to such an extent that groups of cells fold inward to form epithelial folds or balls. This may be due to reduced pulling forces across the tissue, both from the mitotic cells and from the open ventral furrow. It also could be that rounded-up cells push on their neighbors. Many cells that rounded up to divide in stages 8 and 9 fail to restore columnar cell shape. As a result of these two defects, infolding of some cells and failure of others to resume columnar shape, tissue integrity is dramatically disrupted by stage 11. While loss of Cno or Dzy leads to some unbalanced contractility and occasional tissue folds, these defects are much more severe and pervasive after Rap1 loss, providing strong evidence that Rap1 has additional effectors during morphogenesis.
TABLE 1:
Comparison of the effects of different mutations of perturbations on multiple aspects of morphogenesis.
| cno null(Sawyer et al. 2009; 2011; Manning et al., 2019) | Rap1 RNAi (This work) | cno∆RA (Perez-Vale et al. 2021) | dzy RNAi(This work) | |
| Overall morphogenesis as assessed by cuticles | Head involution and dorsal closure failure Moderate-Strong defects in overall epidermal integrity | Head involution and dorsal closure failureModerate-Strong defects in overall epidermal integrity | Complete failure of both head involution and dorsal closure | Failure of head involution. Dorsally closed but frequent dorsal or ventral holes |
| Cell shape defects and gaps at AJs under tension | Gaps common (20–30 gaps/field of view) but with only modest and late hyperconstriction | Widespread unbalanced apical constriction and epithelial infolding; gaps present in less affected areas | Gaps common (20–30 gaps/field of view) but with only modest and late hyperconstriction | Gaps common (20–30 gaps/field of view) but with only modest and late hyperconstriction |
| Planar cell polarity | ||||
| Baz | Planar polarity enhanced | AJ localization strongly disrupted | Planar polarity enhanced | Planar polarity enhanced |
| Cno | NA | Planar polarity enhanced and more punctate | Planar polarity enhanced | Planar polarity unaffected |
| Arm | Planar polarity enhanced | Planar polarity enhanced and more punctate | Planar polarity enhanced | Planar polarity unaffected |
| AJ integrity | Frequent large holes in the ventral epidermis | Only dorsal epidermis remains intact | Small holes in the ventral epidermis | Small holes in the ventral epidermis |
Rap1’s effects on AJ protein localization provide some clues. Cno loss accentuates the planar polarity of AJ proteins and Baz, but they remain localized to AJs. In contrast, after Rap1 RNAi Baz localization is affected much earlier and much more substantially. While Baz returned to apical junctions after the end of cellularization after Rap1 RNAi (Bonello et al., 2018), as germband extension began, Baz localization in Rap1 RNAi embryos rapidly became discontinuous. This fragmentation persisted and was accentuated by stages 8 and 9. In contrast, Arm, Cno, and Pyd remain localized to junctions, though their accumulation around the cell circumference is less regular than is true in WT–all are enriched at places where Baz remains. This suggests that effects of Rap1 loss on Baz are not simply the consequence of alterations in AJs but, instead, that Rap1 may have more direct effects on Baz. However, Rap1 loss does not fully replicate the effects of total maternal/zygotic Baz loss, which leads to complete fragmentation of the epithelium and the cuticle it secretes (Müller and Wieschaus, 1996; Harris and Peifer, 2004). One speculative possibility is that the initial apical relocalization of Baz to AJs allows their assembly, but after Baz fragmentation, AJs lose the ability to balance contractility among neighbors. Consistent with Rap1 and Baz operating together or in parallel in balanced contractility, both Rap1 and Baz loss lead to increased variability of apical cell shape at cellularization (Choi et al., 2013). Rap1 has also been demonstrated to regulate AJ accumulation of both Arm and Baz in the developing adult photoreceptors (Walther et al., 2018). However, in that tissue Cno appears to be the relevant Rap1 effector involved in regulating Baz.
One important task going forward will be to identify additional Rap1 effectors. At this point, we can only speculate about possibilities. Combined reduction of Cno and loss of the ZO-1 homologue Pyd also leads to dramatic defects in epithelial integrity and infolding of epidermal cells, effects that are much more severe than loss/reduction of either alone (Manning et al., 2019) but are more similar to the effects of Rap1 loss. Could Pyd be another Rap1 effector in the Drosophila ectoderm? In mammalian cultured epithelial cells and in the developing mouse lens epithelium, Rap1 can regulate ZO-1 localization (Maddala et al., 2015). However, while the combined reduction of Cno and loss of Pyd disrupts Baz in a way similar to Rap1 loss, the effect on Arm localization is stronger, as both Arm and Baz localization to AJs are fragmented after combined reduction of Cno and loss of Pyd (Manning et al., 2019).
Another speculative possibility is that aPKC might be a Rap1 effector. In cultured mammalian cells, Rap1 can act through the adapter Shank, which binds aPKC and regulates junctional integrity (Sasaki et al., 2020). There are interesting similarities between the phenotype of aPKC M/Z mutants and those of Rap1 RNAi, including disruption of ventral furrow invagination and variable cell apical areas at stage 8 (Harris and Peifer, 2007). However, complete loss of aPKC has earlier and more severe consequences in ectodermal integrity in Drosophila, with complete loss of epithelial integrity by stage 9 (Harris and Peifer, 2007). We also examined aPKC localization after Rap1 RNAi—it remained cortical though both it and Arm appeared less continuous at the cortex. One intriguing possibility, inspired by recent work from the Morais-de-Sá lab (Osswald et al., 2022), is that loss of Rap1 reduces but does not eliminate aPKC function. They examined a different Drosophila epithelium–the ovarian follicle cells. Reducing aPKC activity led to phenotypes quite reminiscent of those we saw in the embryo after Rap1 RNAi–epithelial contractility became unbalanced, with cells next to mitotic cells undergoing unrestrained apical constriction. aPKC is known to regulate apical contractility in other contexts, such as the Drosophila amnioserosa (David et al., 2010). It will be important in the future to assess whether Rap1 RNAi reduces rather than eliminating aPKC localization or function. Another way Rap1 could regulate apical contractility is via effects on Rho, the activity of which it is known to regulate in endothelial cells (Pannekoek et al., 2014). Intriguingly, a recent preprint that investigated the roles of Rap1 in Drosophila wound closure suggested that in that process Rap1 acts in parallel via Cno and Rho (Rothenberg et al., 2022). It will be of interest to explore these and other speculative possibilities.
However, this speculation about additional effectors should not distract from the fact that Cno is one key effector. In our earlier work we made an observation that emphasizes this. Simultaneous overexpression of WT Cno can substantially alleviate the defects in morphogenesis caused by Rap1 RNAi, as assessed by cuticle phenotype, although it does not fully rescue either morphogenesis or viability (Bonello et al., 2018). Examining whether the roles we observed for Rap1 in Drosophila morphogenesis are conserved in other animals is an important future direction. We were excited to see a paper published during review that suggests that mammalian Rap1 is required for epithelial morphogenesis and epiblast tricellular junction formation during embryo implantation, using Talin and likely other effectors (Kim et al., 2022).
In the future, it will also be important to more directly examine the effects of Rap1 RNAi on the balance of apical contractility, using approaches like laser cutting to directly measure tension on different borders. In our work on the cno∆RA mutant (Perez-Vale et al., 2021), we explored whether reducing tension by treatment with a ROCK inhibitor to reduce myosin activation reduced or reversed the junctional disruption in the mutant. Surprisingly, this accentuated the junctional gaps seen, rather than reducing them. We speculated that this was due to the fact that reducing tension also reduces the tension-dependent strengthening of AJ–cytoskeletal connections. Similar analyses could be done after-Rap1 RNAi.
Dzy is the predominant GEF involved in Cno regulation during morphogenesis, but Rap1 must use additional GEFs during these events
Rap1 has many known GEFs that—in both flies and mammals—play roles at different times and in different tissues. Defining the relevant GEFs that regulate junction–cytoskeletal linkage during embryonic morphogenesis remains an important task. Previous work examined the two earliest stages of Drosophila development, cellularization and mesoderm invagination, and the answer was complex. During cellularization, we know of two GEFs that act sequentially: the atypical GEF Sponge regulates the initial placement of Cno and AJ proteins before membrane invagination (Schmidt et al., 2018), while Dzy regulates the correct assembly of nascent AJs (Bonello et al., 2018). However, loss of Dzy does not affect either Cno localization or AJ placement as dramatically as loss of Rap1. Another complexity emerges from the fact that Rap1 is essential for Cno recruitment to the plasma membrane, while Cno remains localized to the membrane in dzy mutants, but its precise apical positioning and enrichment at TCJs is disrupted. The effect of Dzy loss on AJ protein localization is also somewhat milder than the effect of loss of Rap1. Intriguingly, the phenotype of Dzy loss mimics that of the loss of Cno’s Rap1-binding RA domains. These data suggested that Rap1 affects Cno and AJ positioning during cellularization via both RA-dependent and RA-independent mechanisms, and that Dzy mediates the RA-domain–dependent effects, while other GEFs act in parallel to regulate Rap1 and mediate its RA-domain–independent regulation of Cno, likely via other intermediate effectors. In contrast, losses of Dzy, Cno, and Rap1 have similar effects on mesoderm invagination (Sawyer et al., 2009; Spahn et al., 2012).
Our current work suggests an even greater level of complexity of Rap1 regulation and action during germband extension and the maintenance of epithelial integrity. Loss of Dzy has effects similar to the loss of Cno, in terms of altered cell rearrangements, stability of AJs under elevated tension, and planar polarization of junctional proteins (Table 1). However, some effects of Dzy loss do not fully replicate the effects of complete loss of Cno. These include a lack of effect on Arm planar polarity and differences in the degree of loss of epithelial integrity. While some of these differences may be due to a failure to completely eliminate Dzy using our knockdown/zygotic mutant approach, we think that it is more likely that Cno retains some residual activity in Dzy’s absence. Intriguingly, the somewhat less-severe effects of dzy RNAi on epithelial integrity are shared by cno∆RA mutants. Thus, Dzy may be required for the “direct” Cno activation mediated by active Rap1 bound to the RA domains, but Rap1 may also activate Cno to a lesser extent via RA-domain and Dzy-independent mechanisms, presumably when a different GEF activates it. The latter regulation may be indirect, via the other speculative effectors discussed above. These data also raise another intriguing possibility: that different pools of Rap1 at different subcellular locations carry our Rap1’s Cno-dependent and Cno-independent roles and that different GEFs localize to and act at these different locations. In future studies, it will be important to identify those GEFs and, as discussed above, to identify Rap1’s additional effectors regulating morphogenesis.
MATERIALS AND METHODS
Fly stocks and generation of the dzy shRNA construct
We generated an UAS-driven 21 nucleotide siRNA targeting a region of the 5th exon of dizzy. We used the Designer of Small Interfering RNA (DSIR: http://biodev.extra.cea.fr/DSIR/DSIR.html) tool to design the 21 nucleotide target. The target sequence was input into the Predicted Off-Target Free Sequence Regions (https://www.flyrnai.org/RNAi_find_frag_free.html). The targeted sequence is cgttctatccgatcgtcaaga (see additional oligo design information below). We followed the TRiP protocols (Perkins et al., 2015), cloning the hairpins into the WALIUM20 vector. The UAS-driven siRNA was inserted by BestGene into the 3rd chromosome attP docking site for phiC31 integrase-mediated transformation (position 68A4; Bloomington Drosophila Stock Center stock #8622 (genotype: y1 w67c23; P{CaryP}attP2).
Oligo design is done as follows:
Top strand oligo: ctagcagt–insert sense oligo–tagttatattcaagcata–insert anti-sense oligo–gcg
Bottom strand oligo: aattcgc–sense oligo–tatgcttgaatataacta–anti-sense oligo–actg
For targeting dzy, this involved the following oligonucleotides:
Top strand oligo: ctagcagtcgttctatccgatcgtcaagatagttatattcaagcatatcttgacgatcggatagaacggcg
Bottom strand oligo: aattcgccgttctatccgatcgtcaagatatgcttgaatataactatcttgacgatcggatagaacgactg
We used yellow white flies as our control. These are referred to as WT in figures and text. All experiments involving dzy RNAi and the associated WT embryos were conducted at 27°C to enhance GAL4 activity. The other experiments were performed at 25°C. The fly stocks used and their sources are given in Supplemental Table S2. Rap1 knockdown by shRNA was completed by crossing double-copy mat-tub-GAL4 females to UAS.Rap1 RNAi v20/TM3, Sb males (Supplemental Figure S3).
Embryo fixation and immunofluorescence
Flies were crossed in cages over apple juice agar plates with yeast paste and left to lay eggs for 4–18 h before collection. Our method of embryo collection, embryo fixation, and embryo staining was previously described by Bonello et al. (2018). Briefly, for heat fixation: We removed the chorion membrane by nutating in 50% bleach. Afterward, the embryos were washed three times in 0.03% Triton X-100 with 68 mM NaCl, and then fixed in 95°C Triton salt solution (0.03% Triton X-100 with 68 mM NaCl and 8 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid [EGTA]) for 10 s. We fast-cooled samples by adding ice-cold Triton salt solution and placing on ice for at least 30 min. We removed the vitelline membrane by vigorous shaking in 1:1 heptane/methanol solution. The embryos were again washed thrice with 95% methanol/5% EGTA and stored in 95% methanol/5% EGTA for up to 24 h at −20°C before staining. Before staining, the heat-fixed embryos were washed three times with 0.01% Triton X-100 in phosphate-buffered saline (PBS) (PBS-T). Embryos were then blocked in 1% normal goat serum in PBS-T (PNT) for 1 h. The embryos were then nutated in the primary antibodies overnight at 4°C, washed three times with PNT, and nutated in secondary antibodies overnight at 4°C. We used PNT to dilute the primary and secondary antibodies. After incubation in the secondary antibody, the embryos were washed three times with PNT and stored in 50% glycerol at −20°C. Later, we used Aquapolymount (Polysciences) to mount onto glass slides. The antibodies used and their sources are given in Supplemental Table S3. To optimize visualization of endogenous myosin fluorescence, we employed a modified formaldehyde fixation protocol: After removing chorion membranes by nutating for 4 min in 50% bleach, the embryos were washed three times in 0.03% Triton X-100 with 68 mM NaCl and then fixed in 1:1 n-heptane/4% formaldehyde (prepared fresh by diluting paraformaldehyde powder in 1× PBS, pH 7.2) for 25 min on nutator at room temperature. Following removal of fixative solution, embryos were washed once with n-heptane, devitellinized by vigorous shaking for 10 s in 1:1 n-heptane/cold methanol solution (95% methanol/5% EGTA, stored at –20°C), and washed once in cold methanol solution (with minimal incubation). Finally, embryos were washed thrice in 0.03% Triton X-100 in PBS, incubated in 1% bovine serum albumin/0.1% saponin in PBS on nutator for 1 h, and then mounted onto glass slides with a homemade Gelvatol solution (recipe from the University of Pittsburgh’s Center for Biological Imaging).
Image acquisition and analysis
All images were obtained from fixed embryos. We imaged on a Carl Zeiss LSM 880 confocal laser-scanning microscope. Images were captured on the 40×/1.3 NA Plan-Apochromat oil objective. Brightness and contrast were fine-tuned using both ZEN 2009 software and after image processing in Adobe Photoshop. To capture the enrichment of proteins at the adherens junctions, we created MIPs with tools from ImageJ (National Institutes of Health). MIPs for Cno TCJ enrichment, planar-cell-polarity, and antigen intensity analysis were created from Z-stacks (1024 × 1024 pixels) through the ventrolateral ectoderm of the embryo at stages 7 and 8. The Z-stack images were captured using a digital zoom of 1.6 and a step size of 0.3 µm.
Cno TCJ enrichment analysis
We obtained TCJ enrichment data from MIPs. First, we determined the length of the AJs through the embryo, which was on average seven slices from the most apical portion of the junction to the most basal, spanning 1.5–2.4 µm of the apical region of stage 7 epithelial cells. We captured the mean intensity of Cno in ImageJ software by measuring the pixel intensity along the lines of the cell borders using the line tool at a width of 3 pixels. We next created a short line at the TCJ, careful to avoid overlapping the bicellular border lines. For each TCJ or multicellular junction we analyzed, the connecting three or four bicellular junctions were measured for comparison. We quantified 10 cells per embryo measured, and a total of four (dzy RNAi) or five (Rap1 RNAi) embryos were assessed from three independent experiments. We calculated the Cno TCJ ratio by dividing the mean pixel intensity of the TCJ by the average intensity of the bicellular junctions. Statistical analysis was calculated by GraphPad Prism 9. The significance was determined by Welch’s corrected unpaired t test. We did not assume equal SD. We generated box-and-whisker plots using GraphPad Prism software, where the box represents the 25th to 75th percentile, the whiskers represent 5th to 95th percentiles, the median is displayed by a horizontal line within the box, and the mean is represented by a plus sign (+).
Antigen intensity comparison
We used MIPs spanning a 2.4 µm apical section of AJs from embryos at stages 7 and 8 to compare antigen intensity in WT embryos versus Rap1RNAi embryos. For this experiment, our WT expressed Histone:RFP, allowing us to identify those embryos. We heat-fixed, stained, and mounted WT and Rap1 RNAi embryos together on the same slide to eliminate any imaging discrepancies and receive comparable fluorescence from each sample, differentiating between the two genotypes by the endogenous Histone:RFP marker. A six-panel grid was generated over the area of interest in ImageJ to ensure randomized selection of cell borders. Using the Line tool with a width of 4 pixels, we selected the two brightest borders in each panel (measured by the mean intensity) for a total of 12 cell measurements per embryo. We also drew a solid line across the cytoplasm of two cells per grid panel for a total of 12 background measurements to subtract from the average mean intensity. The mean intensity of Arm and Cno in Rap1 RNAi embryos was normalized to the WT levels. In Figure S1L we used GraphPad Prism 9 to generate the scatter-plot where the horizontal bold line represents the mean, and smaller vertical lines represent the minimum and maximum range.
Gap analysis
Data for gaps per field of view were collected visually at the level of apical AJs. A field of view measured 133 × 133 µm, collected from Z-stacks at the ventrolateral epidermis of each embryo. Data were collected from embryos at stages 7 and 8. Gaps were visualized using the Arm channel. Gaps formed at TCJs, rosette centers, or along AP borders spanning several cells (exceeding ∼1 µm). Along AP borders, gaps that transversed the length of about four cells (likely the location of a failed rosette) were considered one gap. Graphs and statistical analysis were executed in GraphPad Prism software using Welch’s unpaired t test.
Germband extension analysis
Cross-section and apical view images of embryos were used to determine the rate of germband extension as compared with the embryo stage. Three time points of germband extension were chosen: stages 7, 8, and 9. These stages were matched across mutant and WT embryos using the mitotic domains as described by Foe (1989). The length of the embryo was normalized as the distance between the posterior end and the cephalic furrow on the dorsal side of the embryo. The degree of germband extension at stage 7, 8, or 9 was quantified as the percent of the length of the embryo that the anteriormost columnar gut cells had reached at the time of fixation. This was measured by creating vertical lines in ImageJ at the posterior end of the embryo, at the cephalic fold, and at the anteriormost gut cells. We then created perpendicular lines to measure the pixel distance between each specified point for the length of the embryo and the length of germband extension. The graph was plotted as average length of germband extension divided by average normalized length of embryo. Data statistical analysis was done using GraphPad Prism 9. The statistical significance was calculated by Welch’s unpaired t test.
Planar polarity quantification
We collected data for planar polarity from MIPs of a 2.4 µm apical section of AJs from embryos at stage 8. The MIPs were generated with the Stacks tool in ImageJ after determining the length of the AJs from Z-stacks through the embryo. Then we used the line tool at a width of 3 pixels to measure the mean intensity of protein fluorescence at both the AP and DV borders of a selected cell, careful to avoid overlapping the TCJ/multicellular junctions. We distinguished each border by its angle. Borders angled from 60 to 90° were considered vertical or AP borders, while borders angled from 0 to 29° were considered horizontal or DV borders. We then used the Circle tool in the cytoplasmic region of 13 cells to gather the mean background intensity. We subtracted the background intensity for each antigen from the intensity at the bicellular borders to get our results for Cno, Baz, Arm, and Pyd planar polarity. For dzy RNAi, a total of 21 embryos were assessed from at least six different experiments. For Rap1 RNAi, the total of 12 embryos were assessed from at least seven different experiments. The number of cells assessed per antigen can be found in Supplemental Table S1. We normalized our proteins of interest to the AP borders and expressed our results as a DV/AP ratio. We generated box-and-whisker plots to present statistical data using GraphPad Prism software, where the box represents the 25th to 75th percentile, the whiskers represent 5th to 95th percentiles, the median is displayed by a horizontal line within the box, and the mean is represented by a plus sign (+).
Cuticle analysis
Cuticle preparation was performed according to Wieschaus and Nüsslein-Volhard (1986). In brief, embryos were aligned on agar plates and allowed to develop fully. Unhatched embryos were nutated in 50% bleach to remove the chorion membrane. After nutation, the embryos were washed in 0.1% Triton X-100 and mounted on glass slides in 1:1 Hoyer’s medium/lactic acid. The glass slides were incubated at 60°C for 48 h and then stored at room temperature.
To compare the cuticle phenotypes of dzy RNAi dzy∆1 embryos and those of dzy∆1 maternal/zygotic null mutants, statistical comparison was performed in GraphPad Prism 9 using a Chi-square contingency test based on the number of embryos in each phenotypic category (p < 0.0001).
Supplementary Material
Acknowledgments
We are grateful to John Poulton for advice and assistance with statistical analysis, to Teresa Bonello for generating an important double balanced stock and not throwing it out after using it, to Peifer lab members for helpful advice and discussions, to Rodrigo Fernandez-Gonzalez, Katheryn Rothenberg, and especially Kevin Slep for important feedback on the manuscript, to the reviewers for careful reading and thoughtful suggestions, and to the Bloomington Drosophila Stock Center for fly stocks. We thank Tony Perdue of the Biology Imaging Center for confocal imaging advice and support. This work was supported by National Institutes of Health (NIH) R35 GM118096 to M.P. K.Z.P.-V. was supported by NIH F31 GM131521 and by a Graduate Diversity Enrichment Program Award from the Burroughs Wellcome Fund.
Abbreviations used:
- AJs
adherens junctions
- AP
anterior/posterior
- Arm
armadillo
- Baz
Bazooka
- Cno
Canoe
- DV
dorsal/ventral
- Dzy
dizzy
- Ecad
E-cadherin
- GAPs
GTPase-activating proteins
- GEFs
guanine nucleotide exchange factors
- M/Z
mutants, maternal/zygotic mutants
- MIP
maximum-intensity projection
- Pyd
Polychaetoid
- RA
Ras-associated
- RNAi
RNA interference
- shRNA
short hairpin RNA
- TCJs
tricellular junctions
- WT
wild-type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-05-0176) on October 26, 2022.
REFERENCES
- Asha H, de Ruiter ND, Wang MG, Hariharan IK (1999). The Rap1 GTPase functions as a regulator of morphogenesis in vivo. EMBO J 18, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilasy SE, Satoh T, Terashima T, Kataoka T (2011). RA-GEF-1 (Rapgef2) is essential for proper development of the midline commissures. Neurosci Res 71, 200–209. [DOI] [PubMed] [Google Scholar]
- Boettner B, Harjes P, Ishimaru S, Heke M, Fan HQ, Qin Y, Van Aelst L, Gaul U (2003). The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics 165, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Van Aelst L (2007). The Rap GTPase activator Drosophila PDZ-GEF regulates cell shape in epithelial migration and morphogenesis. Mol Cell Biol 27, 7966–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Van Aelst L (2009). Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol 21, 684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello TT, Perez-Vale KZ, Sumigray KD, Peifer M (2018). Rap1 acts via multiple mechanisms to position Canoe and adherens junctions and mediate apical-basal polarity establishment. Development 145, dev157941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberger T, Klapproth S, Rohwedder I, Zhu L, Mittmann L, Reichel CA, Sperandio M, Qin J, Moser M (2018). Direct Rap1/Talin1 interaction regulates platelet and neutrophil integrin activity in mice. Blood 132, 2754–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp D, Haage A, Solianova V, Castle WM, Xu QA, Lostchuck E, Goult BT, Tanentzapf G (2018). Direct binding of Talin to Rap1 is required for cell-ECM adhesion in Drosophila. J Cell Sci 131, jcs225144. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M (2013). Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93, 269–309. [DOI] [PubMed] [Google Scholar]
- Choi W, Harris NJ, Sumigray KD, Peifer M (2013). Rap1 and Canoe/afadin are essential for establishment of apical-basal polarity in the Drosophila embryo. Mol Biol Cell 24, 945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Jung KC, Nelson KS, Bhat MA, Beitel GJ, Peifer M, Fanning AS (2011). The single Drosophila ZO-1 protein Polychaetoid regulates embryonic morphogenesis in coordination with Canoe/Afadin and Enabled. Mol Biol Cell 22, 2010–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M (2017). Rap1 in endothelial biology. Curr Opin Hematol 24, 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DJ, Tishkina A, Harris TJ (2010). The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development 137, 1645–1655. [DOI] [PubMed] [Google Scholar]
- Dlugos CP, Picciotto C, Lepa C, Krakow M, Stober A, Eddy ML, Weide T, Jeibmann AMPK, Van Marck V, Klingauf J, et al. (2019). Nephrin signaling results in integrin beta1 activation. J Am Soc Nephrol 30, 1006–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efetova M, Petereit L, Rosiewicz K, Overend G, Haussig F, Hovemann BT, Cabrero P, Dow JA, Schwarzel M (2013). Separate roles of PKA and EPAC in renal function unraveled by the optogenetic control of cAMP levels in vivo. J Cell Sci 126, 778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi K, Yoshioka Y, Yoshida H, Morishita K, Miyata S, Hiai H, Yamaguchi M (2013). The Drosophila DOCK family protein sponge is involved in differentiation of R7 photoreceptor cells. Exp Cell Res 319, 2179–2195. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Simoes Sde M, Roper JC, Eaton S, Zallen JA (2009). Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell 17, 736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE (1989). Mitotic domains reveal early commitment of cells in Drosophila embryos. Development 107, 1–22. [PubMed] [Google Scholar]
- Guichard A, Jain P, Moayeri M, Schwartz R, Chin S, Zhu L, Cruz-Moreno B, Liu JZ, Aguilar B, Hollands A, et al. (2017). Anthrax edema toxin disrupts distinct steps in Rab11-dependent junctional transport. PLoS Pathog 13, e1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan IK, Carthew RW, Rubin GM (1991). The Drosophila roughened mutation: activation of a rap homolog disrupts eye development and interferes with cell determination. Cell 67, 717–722. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Peifer M (2004). Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol 167, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M (2007). aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Dev Cell 12, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo K, Nahm M, Lee MJ, Kim YE, Ki CS, Kim SH, Lee S (2017). The Rap activator Gef26 regulates synaptic growth and neuronal survival via inhibition of BMP signaling. Mol Brain 10, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann S, Hepper C, Marchese D, Knoll C, Reuter R (2006). The PDZ-GEF dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development 133, 2915–2924. [DOI] [PubMed] [Google Scholar]
- Ishimaru S, Williams R, Clark E, Hanafusa H, Gaul U (1999). Activation of the Drosophila C3G leads to cell fate changes and overproliferation during development, mediated by the RAS-MAPK pathway and RAP1. EMBO J 18, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Fan R, Lith SC, Dicke AK, Drexler HCA, Kremer L, Kuempel-Rink N, Hekking L, Stehling M, Bedzhov I (2022). Rap1 controls epiblast morphogenesis in sync with the pluripotency states transition. Dev Cell 57, 1937–1956.e1938. [DOI] [PubMed] [Google Scholar]
- Knox AL, Brown NH (2002). Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295, 1285–1288. [DOI] [PubMed] [Google Scholar]
- Ko CS, Kalakuntla P, Martin AC (2020). Apical constriction reversal upon mitotic entry underlies different morphogenetic outcomes of cell division. Mol Biol Cell 31, 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Wolf F, Grosshans J (2017). Forces directing germ-band extension in Drosophila embryos. Mech Dev 144, 11–22. [DOI] [PubMed] [Google Scholar]
- Lagarrigue F, Paul DS, Gingras AR, Valadez AJ, Sun H, Lin J, Cuevas MN, Ablack JN, Lopez-Ramirez MA, Bergmeier W, Ginsberg MH (2020). Talin-1 is the principal platelet Rap1 effector of integrin activation. Blood 136, 1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cho KS, Lee J, Kim D, Lee SB, Yoo J, Cha GH, Chung J (2002). Drosophila PDZ-GEF, a guanine nucleotide exchange factor for Rap1 GTPase, reveals a novel upstream regulatory mechanism in the mitogen-activated protein kinase signaling pathway. Mol Cell Biol 22, 7658–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddala R, Nagendran T, Lang RA, Morozov A, Rao PV (2015). Rap1 GTPase is required for mouse lens epithelial maintenance and morphogenesis. Dev Biol 406, 74–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning LA, Perez-Vale KZ, Schaefer KN, Sewell MT, Peifer M (2019). The Drosophila Afadin and ZO-1 homologues Canoe and Polychaetoid act in parallel to maintain epithelial integrity when challenged by adherens junction remodeling. Mol Biol Cell 30, 1938–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A, Reeves GT, Supatto W, Stathopoulos A (2010). Mesoderm migration in Drosophila is a multi-step process requiring FGF signaling and integrin activity. Development 137, 2167–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulinari S, Barmchi MP, Hacker U (2008). DRhoGEF2 and diaphanous regulate contractile force during segmental groove morphogenesis in the Drosophila embryo. Mol Biol Cell 19, 1883–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H-AJ, Wieschaus E (1996). armadillo, bazooka, and stardust are critical for formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J Cell Biol 134, 149–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’keefe DD, Gonzalez-Nino E, Burnett M, Dylla L, Lambeth SM, Licon E, Amesoli C, Edgar BA, Curtiss J (2009). Rap1 maintains adhesion between cells to affect Egfr signaling and planar cell polarity in Drosophila. Dev Biol 333, 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Miyake H, Yamaguchi K, Chiba K, Maeta K, Bilasy SE, Edamatsu H, Kataoka T, Fujisawa M (2014). Critical function of RA-GEF-2/Rapgef6, a guanine nucleotide exchange factor for Rap1, in mouse spermatogenesis. Biochem Biophys Res Commun 445, 89–94. [DOI] [PubMed] [Google Scholar]
- Osswald M, Barros-Carvalho A, Carmo AM, Loyer N, Gracio PC, Sunkel CE, Homem CCF, Januschke J, Morais-de-Sá E (2022). aPKC regulates apical constriction to prevent tissue rupture in the Drosophila follicular epithelium. Curr Biol 32, 4411–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou M, Wang S, Sun M, An J, Lv H, Zeng X, Hou SX, Xie W (2019). The PDZ-GEF Gef26 regulates synapse development and function via FasII and Rap1 at the Drosophila neuromuscular junction. Exp Cell Res 374, 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannekoek WJ, Post A, Bos JL (2014). Rap1 signaling in endothelial barrier control. Cell Adh Migr 8, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannekoek WJ, Vliem MJ, Bos JL (2020). Multiple Rap1 effectors control Epac1-mediated tightening of endothelial junctions. Small GTPases 11, 346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vale KZ, Peifer M (2020). Orchestrating morphogenesis: building the body plan by cell shape changes and movements. Development 147, dev191049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vale KZ, Yow KD, Johnson RI, Byrnes AE, Finegan TM, Slep KC, Peifer M (2021). Multivalent interactions make adherens junction-cytoskeletal linkage robust during morphogenesis. J Cell Biol 220, e202104087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim HS, et al. (2015). The Transgenic RNAi Project at Harvard Medical School: resources and validation. Genetics 201, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richlitzki A, Latour P, Schwarzel M (2017). Null EPAC mutants reveal a sequential order of versatile cAMP effects during Drosophila aversive odor learning. Learn Mem 24, 210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg KE, Chen Y, McDonald J, Fernandez-Gonzalez R (2022). Rap1 coordinates cell-cell adhesion and cytoskeletal reorganization to drive collective cell migration in vivo. BioRxiv, doi: 10.1101/2022.10.07.511328. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kojitani N, Hirose H, Yoshihama Y, Suzuki H, Shimada M, Takayanagi A, Yamashita A, Nakaya MA, Hirano H, et al. (2020). Shank2 b inds to aPKC and controls tight junction formation with Rap1 signaling during establishment of epithelial cell polarity. Cell Rep 31, 107407. [DOI] [PubMed] [Google Scholar]
- Sawant K, Chen Y, Kotian N, Preuss KM, McDonald JA (2018). Rap1 GTPase promotes coordinated collective cell migration in vivo. Mol Biol Cell 29, 2656–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JK, Choi W, Jung KC, He L, Harris NJ, Peifer M (2011). A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol Biol Cell 22, 2491–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JK, Harris NJ, Slep KC, Gaul U, Peifer M (2009). The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J Cell Biol 186, 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Lv Z, Grosshans J (2018). ELMO and Sponge specify subapical restriction of Canoe and formation of the subapical domain in early Drosophila embryos. Development 145, dev157909. [DOI] [PubMed] [Google Scholar]
- Shirinian M, Popovic M, Grabbe C, Varshney G, Hugosson F, Bos H, Rehmann H, Palmer RH (2010). The Rap1 guanine nucleotide exchange factor C3G is required for preservation of larval muscle integrity in Drosophila melanogaster. PLoS One 5, e9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn P, Ott A, Reuter R (2012). The PDZ-GEF Dizzy regulates the establishment of adherens junctions required for ventral furrow formation in Drosophila. J Cell Sci 125, 3801–3812. [DOI] [PubMed] [Google Scholar]
- Staller MV, Yan D, Randklev S, Bragdon MD, Wunderlich ZB, Tao R, Perkins LA, Depace AH, Perrimon N (2013). Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics 193, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther RF, Burki M, Pinal N, Rogerson C, Pichaud F (2018). Rap1, Canoe and Mbt cooperate with Bazooka to promote zonula adherens assembly in the fly photoreceptor. J Cell Sci 131, jcs207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Singh SR, Zheng Z, Oh SW, Chen X, Edwards K, Hou SX (2006). Rap-GEF signaling controls stem cell anchoring to their niche through regulating DE-cadherin-mediated cell adhesion in the Drosophila testis. Dev Cell 10, 117–126. [DOI] [PubMed] [Google Scholar]
- Wang YC, Khan Z, Wieschaus EF (2013). Distinct Rap1 activity states control the extent of epithelial invagination via alpha-catenin. Dev Cell 25, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C (1986). Looking at embryos. In: Drosophila, A Practical Approach, ed. Roberts DB, Oxford, England: IRL Press, 199–228. [Google Scholar]
- Yang DS, Roh S, Jeong S (2016). The axon guidance function of Rap1 small GTPase is independent of PlexA RasGAP activity in Drosophila. Dev Biol 418, 258–267. [DOI] [PubMed] [Google Scholar]
- Yu HH, Zallen JA (2020). Abl and Canoe/Afadin mediate mechanotransduction at tricellular junctions. Science 370, eaba5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Wieschaus E (2004). Patterned gene expression directs bipolar planar polarity in Drosophila. Dev Cell 6, 343–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.