Abstract
Cancer cells need to evade the immune system for their progression. In this issue of Blood Cancer Discovery, Gargiulo and colleagues report that in a mouse model of chronic lymphocytic leukemia, small extracellular vesicles inhibit antitumor immunity by altering CD8 T-cell transcriptome, proteome, and metabolome.
Chronic lymphocytic leukemia (CLL) is the most common type of leukemia in adults in Western countries (1). An immunosuppressive tumor microenvironment (TME) is required for CLL progression. Small extracellular vesicles (sEV) have recently been established as important constituents of the TME that affect tumor progression (2). sEVs generally refer to small vesicles (30–150 nm in diameter) released from cells. A major fraction of the sEVs are exosomes. Exosomes are generated when the limiting membrane of the endosomes invaginate to form intraluminal vesicles, which are released to the extracellular milieu when the multivesicular endosomes (MVE) fuse with the plasma membrane. Proteins such as Rab GTPases and endosomal sorting complexes required for transport (ESCRT) mediate the biogenesis and secretion of the exosomes (2). For solid tumors, sEVs, mostly in the form of exosomes, systemically inhibit antitumor immunity through immune-checkpoint molecules such as programmed death-ligand 1 (PD-L1; refs. 3–7). In blood cancers such as CLL, tumor cells secrete sEVs to educate their surrounding cells to enhance tumor cell proliferation, invasion, and immune escape (8). Previous studies of sEVs in blood cancers were mostly based on in vitro assays. Considering the complexity of the leukemia microenvironment (LME), studying sEVs isolated from CLL tissues is important for the understanding of CLL progression.
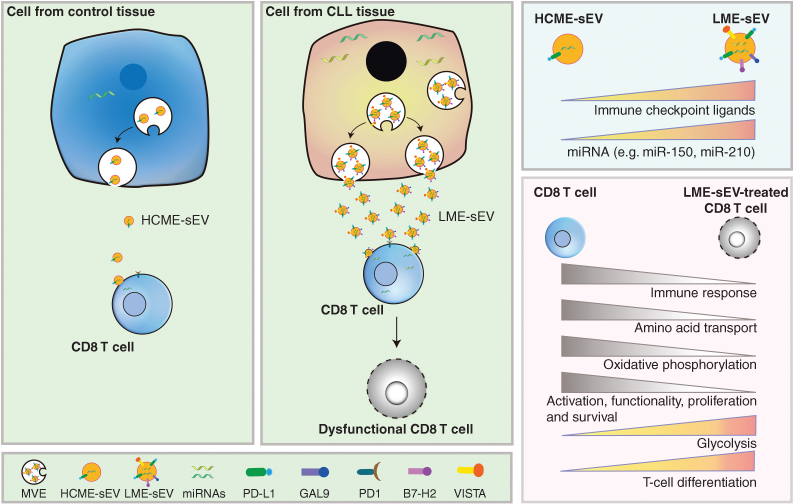
Gargiulo and colleagues established a protocol to purify sEVs from the spleen of a murine CLL model [leukemia microenvironment-derived small extracellular vesicle (LME-sEV; ref. 9)]. They show a 10-fold enrichment of LME-sEVs compared with the control tissue, and the level of circulating sEVs in the peripheral blood of the CLL mice was significantly increased (Fig. 1). Using CLL patient information in the database and their murine model, the authors find that a set of genes related to exosome biogenesis, such as ALIX, RAB10, RA35, are expressed at higher levels in CLL cells compared with the normal B cells. The authors then examined the composition of the isolated sEVs. Proteomics analysis shows that proteins such as those related to protein translation, RNA splicing and binding, and metabolic processes are upregulated in LME-sEVs, whereas proteins involved in intracellular transport and activation of immune processes are decreased. They further confirmed that LME-sEVs carry immune-checkpoint proteins, such as PD-L1, GAL9, B7-H2, and VISTA (Fig. 1). Single-sEV analysis combined with hierarchical stochastic neighbor embedding clustering confirmed that PD-L1 and GAL9 are often coexpressed on CD20+ MHC-II+ vesicles, suggesting LME-sEVs may act as functional units with immunosuppressive capabilities. The authors also fingerprinted several miRNAs enriched both in CLL patients and their murine model (Fig. 1). A high level of miR-210, which is closely associated with hypoxia, was identified in LME-sEVs but not sEVs from MEC-1 culture supernatant, suggesting the influence from the TME.
Figure 1.
Cells from CLL tissues secrete a higher level of sEVs (LME-sEVs) compared with cells from control tissue (HCME-sEVs). With immune-checkpoint ligands (e.g., PD-L1, GAL9, and VISTA) and miRNAs (e.g., miR-150, miR-210), LME-sEVs suppress CD8 T-cell function by changing their transcriptome, proteome, and metabolome. HCME-sEV, healthy control tissue-derived small extracellular vesicle; GAL9, galectin-9; PD-1, programmed death-1; B7-H2, programmed death-ligand 2; VISTA, V-domain Ig suppressor of T-cell activation.
EVs affect the biological behaviors of their recipient cells through two different modes. The first involves internalization of EVs by the recipient cells, resulting in a horizontal transfer of bioactive molecules (e.g., signaling proteins and miRNA). The second is through ligand–receptor interactions on the cell surface (3–7). Using fluorescence-labeled LME-sEVs, the authors detected 5% to 10% of the T cells and 40% of B cells uptake sEVs. The study suggests that LME-sEVs influence their target cells through both internalization and ligand–receptor interactions on the cell surface. Authors find that CD8+ T cells, but not Treg, CD4+ Tconv, and CD19+ B cells, have decreased expression of genes associated with immune response and amino acid transport, and increased expression of genes involved in the inhibition of immunity and T-cell differentiation (Fig. 1). In an ex vivo experiment with LME-sEV treatment, decrease of the costimulatory molecules is observed, whereas expression of negative regulators for T-cell activation, migration, and adhesion, is significantly increased in CD8 T cells. In contrast, no significant changes were detected in Tregs, CD4+ Tconv, and CD19+ B cells, again suggesting that the immunosuppressive function of LME-sEV is cell-type selective. The authors further profiled gene expression and protein contents of sorted CD8+ T lymphocytes treated with LME-sEVs. The genes involved in CD8+ T-cell activation, survival, proliferation, and immune activation are significantly downregulated, whereas genes negatively associated with the above processes were increased in these cells (Fig. 1). In addition, LME-sEV treatment inhibited oxidative phosphorylation and leads to an unfavorable metabolic profile in CD8 T cells. Proteomics analysis with a miRNA target prediction algorithm implicates that miRNA transferred by LME-sEVs to CD8+ T cells may contribute to these proteome changes.
The small GTPases Rab27a and Rab27b control the secretion of exosomes (10). To confirm the role of sEVs in vivo, a CLL murine model deficient in exosome secretion was generated by knocking out RAB27A and RAB27B genes (TCL1-RAB27DKO). A significant delay in CLL progression was observed in these mice. In addition to regulating exosome secretion, Rab27a and Rab27b are also involved in regulating the secretion from other lysosome-related organelles. To verify the role of LME-sEVs, the authors intravenously infused purified LME-sEVs, which rescued the growth of tumors generated from CLL cells isolated from spleens of TCL1-RAB27DKO mice. The data suggest that LME-sEVs are important in the inhibition of the antitumor immune response in immune-competent mice. Finally, the authors find a correlation between sEV-related gene expression and the clinical parameters (prognostic markers and survival) in CLL patients.
This is a comprehensive study profiling the inhibitory function of tumor tissue–derived sEVs on CD8 T-cell transcriptome, proteome, and metabolome. The biogenesis, cargo-sorting, and secretion of exosomes are not only regulated by tumor-intrinsic oncogenic signaling, but also likely influenced by factors in the TME. As such, sEVs isolated from tumor tissues would better reflect the pathologic context of CLL. On the other hand, it is important to realize that sEVs isolated from tumor tissues are not solely originated from tumor cells. Other cell types such as macrophages and fibroblasts also generate sEVs that very likely contribute to the observed immune-suppressive effect. Further studies along this line will provide a more comprehensive understanding of the mechanisms by which different cell types in the tumor tissue generate a suppressive TME that functionally exhaust CD8 T cells and/or spatially block T-cell infiltration. These studies will not only help develop more effective therapeutic strategies to treat cancer but also help identify diagnostic biomarkers to predict patient response to immune therapies.
Authors’ Disclosures
No disclosures were reported.
References
- 1. Burger JA. Treatment of chronic lymphocytic leukemia. N Engl J Med 2020;383:460–73. [DOI] [PubMed] [Google Scholar]
- 2. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020;367:eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560:382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monypenny J, Milewicz H, Flores-Borja F, Weitsman G, Cheung A, Chowdhury R, et al. ALIX regulates tumor-mediated immunosuppression by controlling EGFR activity and PD-L1 presentation. Cell Rep 2018;24:630–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 2019;177:414–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ricklefs FL, Alayo Q, Krenzlin H, Mahmoud AB, Speranza MC, Nakashima H, et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv 2018;4:eaar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res 2018;28:862–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gargiulo E, Paggetti J, Moussay E. Hematological malignancy-derived small extracellular vesicles and tumor microenvironment: the art of turning foes into friends. Cells 2019;8:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gargiulo E, Viry E, Morande PE, Largeot A, Gonder S, Xian F, et al. Extracellular vesicle secretion by leukemia cells in vivo promotes CLL progression by hampering antitumor T-cell responses. Blood Cancer Discov 2023;4:54–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 2010;12:19–30; sup 1–13. [DOI] [PubMed] [Google Scholar]