Novel mouse models of Myd88-driven DLBCL serve as preclinical platforms to evaluate combined BTK/BCL2 inhibition and immune-therapeutic approaches.
Abstract
Genomic profiling revealed the identity of at least 5 subtypes of diffuse large B-cell lymphoma (DLBCL), including the MCD/C5 cluster characterized by aberrations in MYD88, BCL2, PRDM1, and/or SPIB. We generated mouse models harboring B cell–specific Prdm1 or Spib aberrations on the background of oncogenic Myd88 and Bcl2 lesions. We deployed whole-exome sequencing, transcriptome, flow-cytometry, and mass cytometry analyses to demonstrate that Prdm1- or Spib-altered lymphomas display molecular features consistent with prememory B cells and light-zone B cells, whereas lymphomas lacking these alterations were enriched for late light-zone and plasmablast-associated gene sets. Consistent with the phenotypic evidence for increased B cell receptor signaling activity in Prdm1-altered lymphomas, we demonstrate that combined BTK/BCL2 inhibition displays therapeutic activity in mice and in five of six relapsed/refractory DLBCL patients. Moreover, Prdm1-altered lymphomas were immunogenic upon transplantation into immuno-competent hosts, displayed an actionable PD-L1 surface expression, and were sensitive to antimurine-CD19-CAR-T cell therapy, in vivo.
Significance:
Relapsed/refractory DLBCL remains a major medical challenge, and most of these patients succumb to their disease. Here, we generated mouse models, faithfully recapitulating the biology of MYD88-driven human DLBCL. These models revealed robust preclinical activity of combined BTK/BCL2 inhibition. We confirmed activity of this regimen in pretreated non–GCB-DLBCL patients.
See related commentary by Leveille et al., p. 8.
This article is highlighted in the In This Issue feature, p. 1
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is a heterogeneous disease and represents the most common lymphoid malignancy in adults (1). DLBCL is subdivided into germinal center B cell–like (GCB) and activated B cell–like (ABC) DLBCL using gene-expression profiling, which separates DLBCL according to the presumed cell of origin (COO; refs. 1–3). GCB-DLBCL was proposed to originate from light-zone germinal center (GC) B cells, whereas ABC-DLBCL are thought to originate from either post-germinal center plasmablasts or memory B cells (3–7). This COO-based classifier distinguishes subentities displaying distinct biological features, pathogenesis, and clinical response to first-line anthracycline-based chemoimmune therapy (3, 8, 9). Recent genomic analyses led to the discovery of partially overlapping genetically defined DLBCL subsets (10, 11). One analysis used a supervised approach, allowing the classification of ∼50% of the cases into four genetically defined DLBCL subtypes (11). This approach enabled the clustering of lymphomas with cooccurring MYD88- and CD79B mutations (MCD), BCL6 rearrangements and NOTCH2 mutations (BN2), EZH2 mutations and BCL2 rearrangements (EZB), as well as NOTCH1 mutations (N1; ref. 11). Another analysis initially defined recurrent genetic driver lesions in DLBCL and then used a clustering approach, allowing to classify 98% of cases into 5 distinct clusters with specific hallmark genetic signatures (10). These clusters were defined by (i) BCL6 structural variants in combination with NOTCH2 aberrations (C1 DLBCL); (ii) biallelic TP53 inactivation (TP53 mutations and 17p copy-number alterations) in combination with haploinsufficiencies of 9p21.13/CDKN2A and 13q14.2/RB1 (C2 DLBCL); (iii) BCL2 mutations together with BCL2 structural variants in combination with EZH2-, CREBBP-, and KMT2D mutations and additional activating alterations of the PI3K pathway (C3 DLBCL); (iv) mutations in histone genes together with aberrations in immune evasion molecules, NF-κB and RAS/JAK/STAT signaling molecules (C4 DLBCL); and (v) near-uniform 18q copy-number gains with concurrent MYD88- and CD79B mutations (C5 DLBCL; ref. 10). Notably, the amplified region on 18q involves the BCL2 gene, which in these cases is not rearranged, as commonly observed in C3/EZB cases, but amplified. Concordantly, the MCD/C5 DLBCL subtype shows the highest BCL2 expression (11). We further note that approximately 40% of MYD88-mutated cases in both the MCD (21 of 51 cases) and C5 clusters (18 of 28) do not harbor an additional mutation in CD79B (10, 11).
Although approximately 60% of DLBCL patients are cured through first-line treatment, relapsed or refractory disease remains a major challenge, as these patients are often difficult to salvage and even high-dose chemotherapy regimens with autologous stem cell support or CAR-T cell therapies frequently do not provide long-term disease control (12–16). Thus, the development and preclinical validation of treatment strategies for relapsed/refractory disease, as well as less toxic treatment options for elderly and frail DLBCL patients are urgently needed.
We previously reported that B cell–specific expression of Myd88p.L252P (the murine orthologue of the human MYD88 p.L265P mutant) cooperates with BCL2 overexpression in ABC-DLBCL lymphomagenesis in mice (17, 18). Although Myd88cond.p.L252P/wt;Rosa26LSL.BCL2.IRES.GFP/wt;Cd19Cre/wt-derived lymphomas (referred to as MBC) displayed transcriptome signatures that were reminiscent of human ABC-DLBCL, these lymphomas were CD138+ and lacked expression of the B-cell marker B220 (18). This constellation is frequently observed in plasmablastic lymphomas (19, 20). Here, we set out to engineer a plasma cell differentiation block into this model through B cell–specific deletion of Prdm1 or Spib overexpression.
RESULTS
Generation of Prdm1- or Spib-Altered Lymphoma Models
Amplifications of SPIB and mutations in PRDM1 and TBL1XR1 are significantly associated with the MCD cluster and 90% (46/51) of the MYD88-mutated MCD cases analyzed carried one or more of these lesions (ref. 11; Supplementary Fig. S1A). Similarly, 79% (19/24) of MYD88-mutant C5 DLBCL carry one or more of these lesions (ref. 10; Supplementary Fig. S1A). Hence, we speculated that engineering B cell–specific Prdm1 deletions or Spib copy-number gains may prevent plasma cell differentiation programs in MBC lymphomas, ultimately resulting in refined autochthonous mouse models of MYD88-mutant MCD/C5 DLBCL. To this end, we pursued two experimental approaches. First, to mimic Spib amplification in vivo, we generated a novel conditional allele, where Spib.IRES.GFP was targeted into the Rosa26 locus. In this setting, Spib expression from the Rosa26 locus is prevented through the insertion of a loxP.STOP.loxP cassette upstream of the translation-initiating codon (Supplementary Fig. S1B). Using this allele, we generated Myd88cond.p.L252P/wt;Rosa26LSL.BCL2.IRES.GFP/LSL.Spib.IRES.GFP;Cd19Cre/wt mice (referred to as SMBC), in which Cd19-driven Cre mediates the excision of the STOP cassettes, leading to BCL2 and Spib overexpression, in addition to Myd88p.L252P expression. As shown in Supplementary Fig. S1C, CMV promoter-driven Cre expression mediated by adenoviral Cre recombinase (Ad-CMV-Cre) in Rosa26LSL.Spib.IRES.GFP/wt murine embryonic fibroblasts (MEF) led to robust Spib expression, which was not detectable in Rosa26LSL.Spib.IRES.GFP/wt MEFs that were not exposed to Ad-CMV-Cre or in wild-type MEFs, irrespective of Ad-CMV-Cre transduction.
In a parallel approach, we generated Prdm1fl/fl; Myd88cond.p.L252P/wt;Rosa26LSL.BCL2.IRES.GFP/wt;Cd19Cre/wt mice (referred to as PPMBC), which, in addition to B cell-specific Myd88p.L252P and BCL2 expression, harbor a biallelic Prdm1 deletion.
To assess the effects of Spib overexpression and Prdm1 deletion, we performed serial MRI scans to monitor spontaneous splenomegaly in wild-type, MBC, SMBC, and PPMBC mice at 10 and 20 weeks of age. As shown in Fig. 1A, both Spib overexpression and Prdm1 deletion significantly accelerated splenomegaly in MBC mice.
Figure 1.
Deletion of Prdm1 and amplification of Spib increase splenomegaly in MBC animals and abolish plasma cell differentiation. A, Representative axial MR images of 10-week-old animals (spleens are outlined by dashed lines) and volume quantifications of 10- and 20-week-old animals are depicted. Wild-type (n ≥ 5), MBC (n = 9), PPMBC (n = 5), and SMBC (n ≥ 6) spleens were quantified from MR images. B, Splenocytes of 10-week-old C (n = 6), MBC, PPMBC, and SMBC animals (n = 7 each) were analyzed by flow cytometry to analyze the frequencies of B, marginal zone B (MZB), and follicular B (FoB) cells, as well as memory B (MB) and germinal center B (GCB) cells (C). D, Quantification of plasmablasts (PB) and plasma cells (PC) in spleens from 10-week-old C (n = 11), MBC (n = 10), SMBC (n = 8), or PPMBC (n = 9) mice. B–D, Representative pseudocolor plots and gatings (left) as well as quantification of populations (right). Full gating strategies are depicted in Supplementary Fig. S12A–S12G. E, Serum immunoglobulin levels of 10-week-old wild-type (n = 8), MBC (n = 7), PPMBC (n = 5), and SMBC (n = 6) animals were measured by ELISA. F, GCs were stained by biotinylated PNA in splenic FFPE sections of 10-week-old wild-type (n = 10), MBC (n = 5), PPMBC (n = 5) and SMBC animals (n = 5) and quantified as GC area per spleen area and mean GC size (right). Additional stainings of these samples are provided in Supplementary Fig. S2A. Representative stainings are depicted (left). Scale bar represents 500 μm. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; Welch unpaired two-tailed t test; error bars, SD.
We next performed flow cytometry to assess the composition of the splenic B-cell compartment in 10-week-old mice. These analyses revealed, that compared with Cd19Cre/wt (termed “C”) controls, MBC, SMBC, and PPMBC mice displayed an increased percentage of splenic B2 cells among the CD45-positive population. A further inspection of this subpopulation demonstrated that MBC and SMBC animals showed a significantly reduced percentage of marginal zone B (MZB) cells and a relative increase in the follicular B (FoB) cell population, compared with control mice. These phenotypes were reversed to the control level in PPMBC mice (Fig. 1B). In line with previously published data indicating reduced surface BCR expression in Myd88-mutant B cells (21), we observed significantly reduced surface IgM levels in MBC- and SMBC-derived naïve B cells, compared with control mice. In contrast, PPMBC-naïve B cells did not show a reduced surface IgM expression, compared with controls (Supplementary Fig. S1D).
An analysis of the B1 cell compartment showed a significant reduction of this population in MBC and SMBC mice, compared with control animals, whereas PPMBC mice showed a B1-cell population that was significantly increased, compared with control animals (Supplementary Fig. S1E). Of note, among the B1 cells, MBC, SMBC, and PPMBC mice showed a similar shift toward decreased B1a and increased B1b populations, relative to control mice (Supplementary Fig. S1E). Moreover, we detected significantly more splenic GC B cells with a DZ/LZ B cell ratio that was significantly shifted toward DZ B cells in MBC, SMBC, and PPMBC animals, compared with control mice (Fig. 1C; Supplementary Fig. S1F). Furthermore, memory B cell (MB) pools were significantly enlarged in MBC, SMBC, and PPMBC, compared with controls (Fig. 1C). As expected, both overexpression of Spib and loss of Prdm1 were associated with significantly reduced early and late plasmablasts and plasma cells in the spleens of SMBC and PPMBC, compared with MBC controls (Fig. 1D). Moreover, PPMBC spleens contained significantly less late plasmablasts and plasma cells than SMBC spleens, potentially indicating that the plasma cell differentiation block imposed by Prdm1 deficiency is more potent than that induced by Spib overexpression (Fig. 1D).
Consistent with the reduced late plasmablast/plasma cell abundance in SMBC and PPMBC mice, IgM, IgA, and IgG levels in the serum of 10-week-old SMBC and PPMBC mice were significantly reduced, compared with age-matched MBC controls (Fig. 1E). Fittingly, we also observed significantly lower levels of IgM, IgA, and IgG in PPMBC, compared with SMBC mice (Fig. 1E). We note that loss of Prdm1 appears to install a more pronounced plasma cell differentiation block than Spib overexpression, as judged by the number of CD138+ splenic cells (early and late plasmablasts and plasma cells), splenic GC number, and serum immunoglobulin levels.
Given the increased abundance of GC B cells in MBC, SMBC, and PPMBC, compared with control mice, we next performed IHC on spleens derived from the different genotypes. Consistent with the relative expansion of GC B cells in our flow cytometry analysis, we observed significantly more and larger germinal centers in MBC, SMBC, and PPMBC mice at 10 weeks of age, compared with age-matched wild-type animals (Fig. 1F; Supplementary Fig. S2A). Although the average GC size was similar between PPMBC, SMBC, and MBC samples, we found a significantly increased number of GCs/spleen area in PPMBC animals, compared with MBC mice. This observation is fully in line with the increased relative number of GC B cells in PPMBC, compared with MBC mice (Fig. 1C). No significant differences in the GC area/mm2 spleen area or the number of GCs/spleen area were observed, when we compared MBC and SMBC tissues, whereas PPMBC mice displayed a significantly increased number of GCs/spleen area and a significantly increased GC area/mm2 spleen area, compared with MBC controls.
We next analyzed the BCR repertoire of nonmalignant GC B cells for a possible V-segment restriction, to ask whether a limited set of (auto-)antigens might drive the spontaneous GC expansion observed in MBC, SMBC, and PPMBC mice, compared with wild-type controls. To this end, we performed RNA-based full-length BCR sequencing on PNA+ cells from wild-type, MBC, SMBC, and PPMBC animals (Supplementary Fig. S2B). In these experiments, we did not detect any significant differences in VH usage, neither on the family, gene, or allele level (Supplementary Fig. S2B; Supplementary Table S1), indicating that a diverse set of antigens might drive the GC hyperplasia. We further observed significantly increased class-switching to IgG in MBC- and SMBC-derived GC B cells, compared with the control setting, whereas loss of Prdm1 on the Myd88/BCL2-mutant background repressed class-switching (Supplementary Fig. S2C). When we analyzed the rate of somatic hypermutation (SHM) in class-switched reads, we observed significantly reduced SHM in MBC- and SMBC-derived GC B cells, compared with controls (Supplementary Fig. S2D). In agreement with a more efficient GC retention of B cells induced by Prdm1-deficiency, compared with Spib overexpression, this reduction of SHM was abolished in PPMBC, but not in SMBC samples (Supplementary Fig. S2D). Altogether, the increased SHM frequency in conjunction with reduced class-switching in PPMBC, compared with MBC GC B cells, might suggest that Prdm1 deletion retains PPMBC GC B cells in the GC reaction at a stage prior to class-switch and GC exit.
Deletion of Prdm1 and Spib Overexpression Accelerate Lymphomagenesis
We next aimed to determine the effect of B cell–specific Prdm1 deletion or Spib overexpression in MBC animals on overall survival. We previously reported that MBC animals succumb to (oligo-)clonal B cell lymphomas, which display transcriptome signatures akin to human ABC-DLBCL (17, 18). However, these lymphomas were CD138+ and lacked expression of the B cell marker B220—a surface marker profile that is also consistent with plasmablastic lymphoma (17–20). Here, we performed a direct comparison of wild-type, MBC, SMBC, and PPMBC animals. MBC animals displayed a median overall survival of 49 weeks (Fig. 2A; refs. 17, 18). Importantly, the overall survival of SMBC and PPMBC mice was significantly reduced, compared with MBC animals with a median of 37.1 and 34.1 weeks, respectively (Fig. 2A).
Figure 2.
PPMBC and SMBC animals develop ABC-DLBCL–like tumors. A, Survival curves of wild-type (n = 6), MBC (n = 45, median 49 weeks), SMBC (n = 28, median 37.1 weeks), and PPMBC animals (n = 26, median 34.1 weeks). B, Representative H&E and IHC stainings of MBC, SMBC, and PPMBC tumors. Scale bars represent 100 μm (low magnification) and 10 μm (high magnification H&E). C, Quantification of the terminal phenotype of MBC, SMBC, and PPMBC animals shown in B. D, Clonality plots generated from BCR sequencing data. Each circle represents a specific V(D)J sequence with the circle size being proportional to the respective read count. Sequences with one mismatch are connected to clones. Clones with a size > 10% of reads are colorized, smaller clusters are in gray. E, Maximum clone sizes in MBC (n = 18), SMBC (n = 9), and PPMBC (n = 18) lesions compared with the polyclonal scenario observed in wild-type spleens (n = 3). F, Representative axial MR images of preterminal MBC (n = 16), SMBC (n = 8), and PPMBC (n = 12) animals. Lymphomas and spleens are outlined (“L” and “S,” respectively). G, Quantification of the number of lymph node regions infiltrated by lymphoma of moribund MBC (n = 14), SMBC (n = 10) and PPMBC (n = 15) animals at necropsy. H, Representative H&E and B220 stainings of infiltrated liver and lungs of terminal PPMBC animals and quantification of animals with observed extranodal manifestation. Scale bars represent 500 μm. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; Welch unpaired two-tailed t test; error bars represent SD.
MBC mice developed life-limiting (oligo-)clonal lymphoma with 85% penetrance, as we previously reported (Fig. 2B and C; refs. 17, 18). These lymphomas were exclusively B220−/CD138+ (Fig. 2B and C). SMBC and PPMBC mice developed lymphoma in 73% and 97%, respectively (Fig. 2B and C). IHC-based profiling revealed that PPMBC-derived lymphomas were exclusively B220+/CD138− (Fig. 2B and C; Supplementary Fig. S3). In contrast, we observed B220−/CD138+, B220+/CD138−, as well as B220+/CD138+ lymphomas in SMBC mice (in 5, 4, and 2 of 11 cases, respectively; Fig. 2B and C; Supplementary Fig. S4). All three genotypes displayed similar Ki67 indices (Fig. 2B; Supplementary Fig. S5A). We note that lymphomas of all three genotypes were transplantable in Rag1−/− recipient animals (Supplementary Fig. S5B). Moreover, PPMBC lymphomas were also transplantable into syngeneic recipients, albeit at a significantly lower engraftment rate (2 of 8 individual primary tumors engrafted; Supplementary Fig. S5C), suggesting immunogenicity of this model, which is corroborated by significantly higher Pd-l1 and Pd1 mRNA expression in primary PPMBC, compared with primary MBC lymphomas (Supplementary Fig. S5D). We further note that syngeneically transplanted PPMBC lymphomas did not display any detectable difference in B220, CD3, or Ki67 staining, compared with their respective primary tumors (Supplementary Fig. S5C).
To determine whether the autochthonous lesions that were histologically classified as lymphoma indeed represent clonal expansions, we next performed RNA-based BCR sequencing (22). As expected, the BCR repertoire in wild-type spleens was highly polyclonal (Fig 2D and E). In sharp contrast, MBC, SMBC, and PPMBC lesions displayed a strongly reduced variation with the majority of cases being dominated by a single or two parallel large clones (Fig 2D and E).
When analyzing the intraclonal diversity of the dominant clones, we observed significantly smaller Gini coefficients in the PPMBC-derived malignant clones, compared with the MBC setting, fitting with ongoing SHM activity following the transformation step in those lesions (Supplementary Fig. S5E; Tables S2 and S3). In agreement with these data, we also observed a significantly increased CDR (complementarity-determining region) mutation frequency in PPMBC, compared with MBC lymphomas. Of note, all analyzed PPMBC lymphomas harbored CDR mutations leading to amino acid alterations, whereas only two out of four MBC cases displayed amino acid-altering mutations (Supplementary Table S2). No differences in the mutation frequency within the framework region were detected between the lymphoma genotypes (Supplementary Table S2). The increased SHM activity in PPMBC lymphomas is further corroborated by a significantly higher Aicda expression in PPMBC, compared with MBC and SMBC lesions (Supplementary Fig. S5F). BCR sequencing data did not reveal an obvious bias in VH usage within the malignant clones, arguing against a restricted set of (auto-)antigens provoking lymphomagenesis (Supplementary Fig. S5G; Supplementary Table S2). However, the limited number of cases analyzed might not allow the detection of a subtle VH segment preference.
We next asked whether we could detect preferential Ig isotype expression in MBC, SMBC, or PPMBC-derived lymphomas. This analysis showed that the majority of these lymphomas were class-switched to IgG or IgA (Supplementary Fig. S5H; Supplementary Table S2). Altogether, BCR sequencing clearly demonstrated that lymphomas in all three genotypes were of clonal origin, were largely class-switched, and that SHM activity was highest in PPMBC lymphomas.
Next to IHC-based surface marker profiling and clonality analyses, we also performed MRI scans of preterminal MBC, SMBC, and PPMBC mice, as well as extensive necropsies, to assess the degree of lymphadenopathy and extranodal infiltration among the three genotypes. These analyses revealed that the malignant lesions observed in MBC mice were largely confined to mesenteric lymph nodes (Fig. 2F and G; Supplementary Fig. S6A). In marked contrast, preterminal PPMBC and SMBC mice displayed lymphadenopathy in significantly more lymph node regions (Fig. 2F and G; Supplementary Fig. S6A) and significantly more extranodal infiltrations (Fig. 2H), compared with preterminal MBC animals.
The long latency of lymphoma development in our models suggests that in addition to the engineered mutations, further genomic lesions might be required for full-blown lymphoma manifestation. To directly address this hypothesis, we also performed whole-exome sequencing (WES) on 17 MBC and 12 PPMBC lymphomas derived from individual mice. We retrieved recurrent aberrations in Irf2bp2, Pim1, Pou2f2, Stat3, and Hist1h1e, which have all been previously shown to be recurrently mutated in human DLBCL (refs. 10, 11, 23, 24; Supplementary Fig. S6B and S6C; Supplementary Table S4). These WES data provide further cross-species validation of our models. Thus, in summary, B cell–specific Spib overexpression or Prdm1 deletion results in (oligo-)clonal, largely B220+/CD138− lymphomas, which display a more aggressive infiltration pattern and disease course, compared with MBC controls.
Transcriptomic Profiling of MBC, SMBC, and PPMBC Lymphomas
To map the biology of MBC, SMBC, and PPMBC lymphomas onto physiologic B cell developmental stages, we next performed mass cytometry analyses (CyTOF). In brief, we defined individual cell clusters, using UMAP dimensionality reduction on the expression levels of several established lineage markers (Fig. 3A and B; Supplementary Fig. S7A). No significant differences in the relative size of the identified clusters were observed between the three models (Fig. 3C; Supplementary Fig. S7B). Within the cluster containing normal and malignant B cells (B/lymphoma), we observed a significantly increased expression of the GC markers (BCL6, FAS, and GL7) in the PPMBC samples, compared with MBC lymphomas (Fig. 3D). We note that no significant differences were observed in IRF4 expression between the three genotypes (Fig. 3D). These mass cytometry data were validated using IHC, which demonstrated that MBC lymphomas were consistently BCL6−/IRF4+, whereas SMBC and PPMBC lymphomas displayed 50% and 75% BCL6 positivity, respectively, in conjunction with 100% IRF4 positivity (Fig. 3E).
Figure 3.
PPMBC and SMBC lymphomas display transcriptomic and surface marker features associated with germinal center–derived lymphomas. A, Cell suspensions from MBC (n = 5), SMBC (n = 7), and PPMBC (n = 8) tumors were analyzed by mass cytometry. Dimensionality reduction of the data set was performed by UMAP and clusters were gated manually. B, Heat map of lineage markers used for cluster identification. C, Cluster sizes observed in MBC, SMBC, and PPMBC samples. D, Expression of BCL6, FAS, GL7, and IRF4 in the “B/lymphoma” cluster. E, Representative images (left) and quantification (right) of IHC BCL6 and IRF4 stainings in MBC, SMBC, and PPMBC lymphomas. Scale bar represents 20 μm. F, Gene set enrichment plots comparing the transcriptional profiles of MBC (n = 13), SMBC (n = 11), and PPMBC (n = 13) lymphomas to the KBC model (n = 6) as reference. The NES is illustrated as an approximation of the distance between the genotypes. G, Bulk transcription profiles of MBC, SMBC, and PPMBC lymphomas were clustered in an unsupervised manner by their expression levels of published gene sets derived from single-cell transcriptomic analyses of human GC B cells (26). DZ, dark zone; Int, intermediate; LZ, light zone; PreM, prememory B; PBL, plasmablast. H, Representative flow cytometry plots and gating strategy of splenic MB, GCB, and DZ/LZ B cells from a 10-week-old healthy C mouse (top) and lymphoma cells from a PPMBC lesion (middle). Bottom, an overlay of both samples. A total of 10 PPMBC lymphomas was analyzed; additional samples are illustrated in Supplementary Fig. S8, full gating strategy is depicted in Supplementary Fig. S12A. H–I, *, P ≤ 0.05, Welch unpaired two-tailed t test.
MCD/C5 DLBCL, which harbors recurrent aberrations in MYD88, BCL2, and PRDM1/SPIB, is enriched for ABC-DLBCL cases (10). As transcriptome profiling remains the gold standard for differentiation of GCB- and ABC-DLBCL (2), we next generated 3′-RNA-seq reference data from GC B and activated blood B (AB) cells, isolated from wild-type animals. From these data sets, we derived gene sets of upregulated transcripts enriched in each of the two normal B-cell populations (ABup and GCBup). Using these reference gene sets, we compared and contrasted MBC, SMBC, PPMBC, as well as Kmt2dfl/fl;VavP-Bcl2;Cɣ1Cre/wt (KBC)-derived lymphomas, which served as an established reference akin to GCB-DLBCL (ref. 25; Fig. 3F). Using our MBC samples as an internal reference, we observed that KBC lymphomas were most strongly enriched for the GCBup gene set, while displaying a negative normalized enrichment score (NES) for ABup (Supplementary Fig. S7C). Of note, SMBC and PPMBC lymphomas displayed a similar enrichment pattern, albeit NES were lower for these lymphoma genotypes (Supplementary Fig. S7C). Furthermore, transcriptomes derived from MBC lymphomas displayed the strongest positive and negative enrichment for the ABup and GCBup gene sets, respectively, when compared with KBC lymphomas (Fig. 3F). When we used the GC-derived KBC model as a reference, MBC, SMBC, and PPMBC lymphomas showed a positive enrichment for the ABup gene set, while being negatively enriched for GCBup (Fig. 3F). Altogether, these transcriptome data indicate that SMBC and PPMBC lymphomas are developmentally closely related to GCB cells than their MBC counterparts. This more distant relationship of MBC lymphomas with GCB cells is also reflected in the CD138 positivity that we had observed in MBC, but not in PPMBC, and to a lesser extent in SMBC lymphomas (Fig. 2B and C). It is also important to reiterate that, compared with KBC lymphomas, which represent a canonical GCB-DLBCL model, SMBC and PPMBC lymphomas display a clear and significant AB cell transcriptome phenotype (Fig. 3F).
To further carve out the precursor population of our MBC, SMBC, and PPMBC lymphomas, we next mapped transcriptomic data from these three lymphoma subtypes onto gene sets derived from a recently published single-cell transcriptome data set that was generated to characterize the distinct human GC B cell populations (26). Unsupervised clustering of these data showed that particularly MBC and PPMBC lymphomas cluster distinctly, whereas SMBC cases were interspersed between these genotypes (Fig. 3G). This analysis further revealed that MBC lymphomas showed significantly higher ssGSEA scores for gene sets associated with late light-zone and plasmablast stages (Supplementary Fig. S8A). In contrast, PPMBC lymphomas were enriched for early light zone and intermediate stages (Supplementary Fig. S8A). Moreover, PPMBC lymphomas were significantly enriched for the prememory-B-cell gene signature (Supplementary Fig. S8A). The combined presence of light-zone GC and memory B cell features in the absence of plasmablast/plasma cell features is consistent with the malignant transformation of PPMBC cells occurring prior to plasmablast differentiation at an intermediate stage between GC exit and memory B cell differentiation. These data are further supported by flow cytometry experiments, indicating that 6 of 10 PPMBC lymphomas were CD38+/FAS−/IgD−, in line with a memory B cell phenotype (Fig. 3H; Supplementary Fig. S8B). At the same time, these PPMBC lymphomas were largely CXCR4−/CD86+, consistent with a light-zone surface marker profile (Fig. 3H; Supplementary Fig. S8C). Of note, consistent with the expansion of a CD38+/FAS+ population also in the premalignant setting (Fig. 1C), a fraction of PPMBC and SMBC lymphomas fell into CD38+/FAS+ gate, suggesting that this population might also constitute a possible precursor population. We believe that our data are consistent with a model in which transformed cells are not arrested at one precisely defined B cell developmental stage, but rather retain the ability to dynamically switch between different stages without passing beyond a certain developmental stage. For instance, PPMBC-derived lymphomas clearly display transcriptomic features of prememory B cells, as well as light-zone B cells (Fig. 3G; Supplementary Fig. S8A), while not showing any plasmablastic differentiation features (CD138 negativity, Figs. 2B–C and 3G; Supplementary Fig. S8A). In contrast, MBC cases clearly demonstrate CD138 positivity as an indicator for plasmablastic differentiation, while simultaneously displaying transcriptomic features still mapping to light-zone B cells (Figs. 2B–C and 3G; Supplementary Fig. S8A).
The PPMBC Model as a Platform to Assess Immunotherapeutic Approaches
To evaluate immune-therapeutic approaches in our PPMBC model, we next established a murine syngeneic anti-CD19 chimeric antigen receptor (CAR)-T cell platform. For that purpose, we transduced splenic T cells with a construct driving the expression of an antimurine-CD19 CAR (derived from ref. 27; Fig. 4A). We next cocultured these CAR-T cells, as well as mock-transduced T cells, with a CD19-expressing PPMBC lymphoma cell line at various effector:target ratios (Fig. 4B). After 48 hours, the experiment was terminated and population sizes of T cells and lymphoma cells were assessed by flow cytometry. These experiments demonstrated that even at an effector:target ratio of 0.25:1, antimurine-CD19 CAR-transduced cells efficiently eradicated PPMBC lymphoma cells (Fig. 4B), compared with the control condition.
Figure 4.
Murine PPMBC lymphomas are susceptible to immune-therapeutic approaches. A, Schematic depiction of the anti-mCD19scFv-CD28-CD3z CAR construct (27). B, Population sizes of mock- and CAR-transduced T cells isolated from C57BL6/J wild-type spleens and a CD19+ PPMBC lymphoma cell line at the indicated effector:target ratios after 48 hours of coculture after gating for live cells. C, PPMBC mice with a low or high lymphoma-burden [spleen volume 500–700 μL (n = 3) or >700 μL (n = 3), respectively] were treated with a single injection of 2 × 106 anti-mCD19 CAR-T cells. Tumor volume was monitored weekly via MRI and is visualized as fold change from baseline. D, Kaplan–Meier curve showing the overall survival of CAR-T–treated mice and untreated controls. The CAR-T–treated cohort is depicted in total, as well as separated into high and low lymphoma-burden animals. E, MRI-determined tumor volume of PPMBC animals treated with an anti–PD-L1 (n = 7), displayed as fold change from baseline. Untreated animals (n = 8) served as controls. F, Kaplan–Meier curve showing the overall survival of anti–PD-L1-treated mice (n = 7) and untreated controls (n = 8). *, P ≤ 0.05; **, P ≤ 0.01; log-rank test.
We next assessed the therapeutic efficacy of these CAR-T cells for the treatment of lymphoma-bearing PPMBC mice. Animals with MRI-verified lymphomas were infused with a single dose of 2 × 106 anti-mCD19 CAR-T cells or were left untreated. We further stratified animals according to their tumor volume (spleen volume between 500 and 699 μL or above 700 μL, as an indicator for low vs. high tumor volume). Lymphoma responses were monitored by weekly MRI scans. We observed substantial tumor shrinkage and a significantly prolonged overall survival in CAR-T cell-treated animals, compared with untreated controls (Fig. 4C and D). Moreover, animals with a spleen volume below 700 μL survived significantly longer after CAR-T infusion than animals with a spleen volume >700 μL, indicating that lymphoma burden prior to CAR-T infusion might be a predictor for response, similar to what has been observed in human DLBCL (28).
We and others have recently shown that ABC-DLBCL cases frequently display PD-L1 overexpression, compared with GCB-DLBCL cases (18, 23). Thus, we next asked whether intercepting the PD1/PD-L1 immune checkpoint might display therapeutic efficacy in our PPMBC model. For this purpose, we treated lymphoma-bearing PPMBC mice with a PD-L1 antibody (29). We observed a stabilization of tumor growth in the treatment group (Fig. 4E), which resulted in a significantly prolonged survival in these animals, compared with untreated controls (Fig. 4F). In these experiments, PD-L1 blockade significantly repressed lymphoma growth and led to a significantly increased overall survival, compared with untreated animals. Altogether, these experiments indicate that the PPMBC model is a suitable tool to study immune-therapeutic approaches in an immune-competent in vivo model.
Combined BTK/BCL2 Inhibition Displays Efficacy in PPMBC Lymphomas
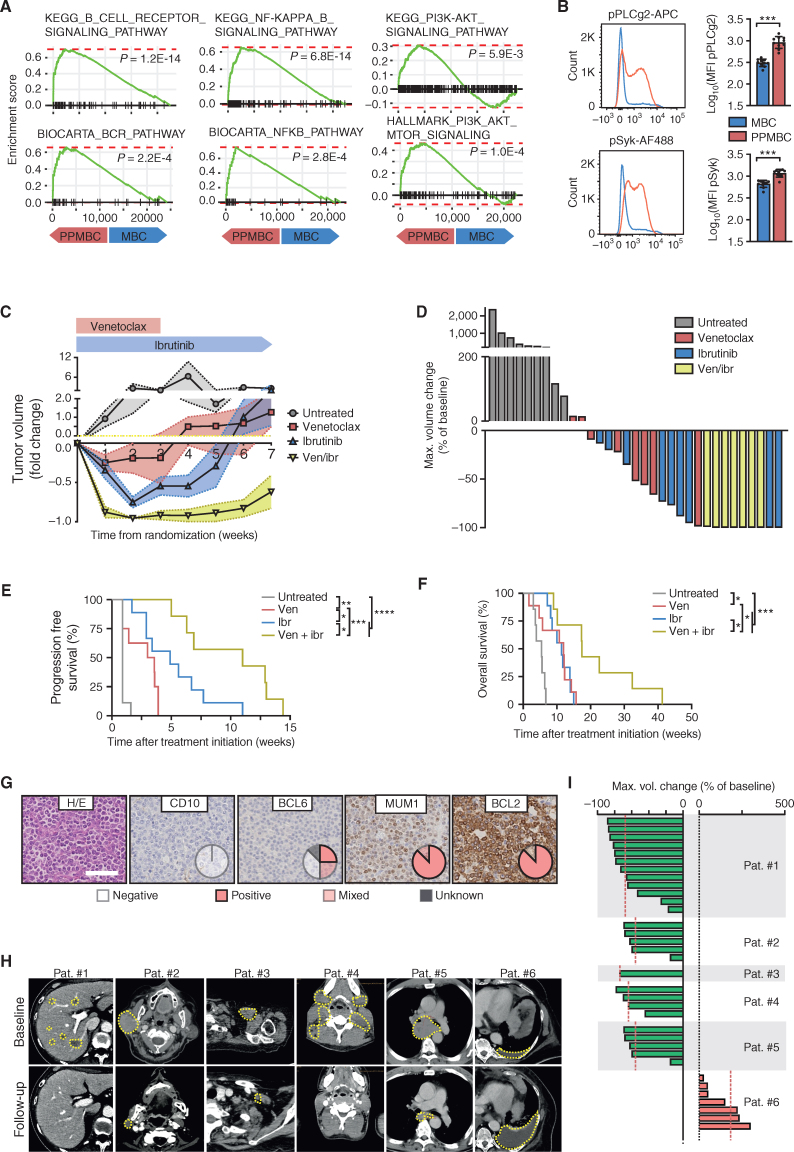
We next aimed to use our PPMBC model as a preclinical tool to assess the efficacy of BTK and/or BCL2 inhibition in vivo. In this regard, we note that BCL2 has emerged as a potential therapeutic target in DLBCL during the last years. Particularly, the BCL2 inhibitor venetoclax produces response rates of ∼18% in relapsed/refractory DLBCL (30). Similarly, in a phase I/II clinical trial involving 80 patients with relapsed/refractory DLBCL, the BTK inhibitor ibrutinib induced complete or partial remissions in 37% of ABC-DLBCL patients, but in only 5% GCB-DLBCL patients (31). Moreover, comparing PPMBC and MBC lymphomas using GSEA, we find that PPMBC cases were significantly enriched for B-cell receptor pathway–associated gene sets (Fig. 5A; Supplementary Table S5). In line with this, gene sets associated with the BCR downstream pathways NFκB- and PI3K signaling were also enriched in PPMBC lymphomas (Fig. 5A; Supplementary Table S5). Furthermore, phospho-flow cytometry experiments demonstrated increased p-PLCγ2 and p-SYK levels in PPMBC lymphomas, compared with MBC lesions (Fig. 5B).
Figure 5.
Murine PPMBC lymphomas and human non-GCB lymphomas are sensitive to combined treatment with venetoclax and ibrutinib. A, Gene set enrichment analysis plots for BCR-related gene signatures from the KEGG, Biocarta, and Hallmark collections, comparing PPMBC and MBC lymphomas (n = 13 each). B, Flow-cytometric analysis of phospho-PLCg2 and phospho-Syk levels in IgDneg cells from MBC (n = 10) and PPMBC (n = 10) lymphomas. C, Normalized tumor volumes after tumor detection of untreated (n = 7), venetoclax-treated (n = 8), ibrutinib-treated (n = 9), or combination-treated (n = 7) PPMBC animals. D, Best tumor volume change of untreated, venetoclax-treated, ibrutinib-treated and combination-treated PPMBC animals. E, PFS and (F) overall survival of untreated, venetoclax-treated, ibrutinib-treated and combination-treated PPMBC animals. G, Six patients were stratified as non–GCB-DLBCL by the Hans algorithm. One representative case is visualized; pie charts illustrate the distribution of staining patterns across the cohort (n = 6). Scale bar represents 50 μm. H, CT-images of patients #1 to #6. Target lesions are outlined. I, Best tumor volume change of individual target lesions of each patient. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; B, Welch unpaired two-tailed t test; E and F, log-rank test; error bars represent SD.
We next asked whether ibrutinib as a single agent or combined with venetoclax might display preclinical activity in PPMBC lymphomas. For that purpose, we generated a cohort of PPMBC animals, in which lymphoma development was surveilled through weekly MRI scans. Upon lymphoma manifestation, animals were randomized in a 1:1:1:1 fashion to receive either ibrutinib (30 mg/kg, p.o., q.d. days 1–21, followed by application via drinking water until death), venetoclax (200 mg/kg, p.o., q.d., days 1–21), or combined ibrutinib plus venetoclax. Untreated mice served as controls. Ibrutinib and venetoclax produced transient remissions in the PPMBC model (Fig. 5C–F; Supplementary Fig. S9A). Both ibrutinib and venetoclax resulted in a significantly prolonged median progression-free survival (PFS, assessed by MRI scans), compared with untreated control (3.29 and 4.9 vs. 0.9 weeks, respectively). Moreover, the median PFS in animals treated with the combination regimen was significantly longer than that observed in the single-agent treatment arms (11.0 weeks; Fig. 5E). Similarly, the median overall survival of animals treated with the combination regimen (17.43 weeks) significantly exceeded that observed in untreated mice (5.4 weeks), ibrutinib (11.4 weeks), or venetoclax (12.07 weeks; Fig. 5F). Altogether, these in vivo experiments indicate that combined BTK and BCL2 blockade may represent a viable treatment strategy for a molecularly defined subset of DLBCL cases, harboring MYD88, BCL2, and PRDM1 aberrations.
Combined BTK/BCL2 Inhibition Displays Efficacy in Non–GCB-DLBCL Patients
Given the preclinical activity of combined ibrutinib and venetoclax in our PPMBC model, we also treated six relapsed/refractory non–GCB-DLBCL patients with this regimen in an off-label scenario, after receiving written informed consent by the patients or their legally authorized representatives. Patients in this cohort had a median age of 68 years. Between 2018 and 2021, these patients were treated with ibrutinib (560 mg, q.d., p.o.) and venetoclax (400 mg, q.d., p.o.) at the University Hospitals Cologne and Essen. They had received a median of 3 prior lines of therapy, including autologous stem cell transplantation and anti-CD19 CAR-T cell therapy (Supplementary Fig. S9B; Supplementary Table S6A). All patients had progressive non–GCB-DLBCL (classified by Hans algorithm; Fig. 5G) at the time of treatment initiation. For four of these six patients, we had biopsy material available on which we performed exome sequencing after receiving written informed consent from the patients. While none of these four cases was assigned to the MCD cluster by the LymphGen classification algorithm (ref. 24; Supplementary Table S6B), lymphomas of all 4 sequenced patients harbored distinct genomic aberrations associated with the MCD- or C5 clusters of DLBCL (10, 11), including mutations in PRDM1 (1 of 4 cases), PIM1 (1 of 4 cases), BTG2 (2 of 4 cases), HLA-A (2 of 4 cases) and HLA-C, TAP1, VMP1, IL16, KLHL42, and TNRC18 (1 of 4 cases each; Supplementary Fig. S9C; Supplementary Table S6C–S6F). We further found chromosomal aberrations associated with cluster C5, namely, gains and amplifications of 18p, 18q (harboring BCL2), 3q and 19q (harboring SPIB; Supplementary Fig. S10; Supplementary Table S6G–S6J). Moreover, our analyses retrieved a deletion of the chromosomal location of PRDM1 (6q) in two cases, as well as several additional aberrations highly recurrent in DLBCL, but not significantly associated with any specific cluster (gains and amplifications of 1q, 7p, 7q, 12p, 12q; Supplementary Fig. S10; Supplementary Table S6G–S6J). Lymphoma burden was quantified by contrast-enhanced CT scans or PET-CT prior to treatment initiation and during follow-up (between weeks 2 and 7.5, median 5.5 weeks; Fig. 5H and I; Supplementary Fig. S11). Five of six patients displayed tumor regressions, whereas one patient experienced progressive disease (Fig. 5H and I; Supplementary Fig. S11). Intriguingly, the nonresponding lymphoma lacked detectable BCL2 expression on IHC analysis, whereas the remaining five lymphomas all showed robust BCL2 positivity on IHC (Fig. 5G; Supplementary Fig. S9C). In four of the six patients, combined venetoclax and ibrutinib served as a bridge to CAR-T cell therapy (Supplementary Fig. S9B). We did not observe any dose-limiting toxicities in these six patients. Altogether, our clinical data corroborate our preclinical observations and suggest that combined ibrutinib and venetoclax may display clinical activity in a subset of relapsed/refractory non–GCB-DLBCL.
DISCUSSION
Here, we describe murine models of Myd88 and BCL2-driven DLBCL, which were engineered to harbor B cell–specific Spib overexpression or Prdm1 deletion. It is important to note that we used a Cd19Cre allele for the generation of these models, which induces recombination of loxP sites already at the pro-B cell stage and thus substantially prior to the GC reaction, where the transformation of human ABC-DLBCL presumably occurs (32). While Spib is expressed in pre-B cells, Prdm1 expression is only initiated during the GC reaction (33, 34). Thus, in the Cd19Cre background, we are unlikely to interfere with early B cell development through Prdm1 deletion in the PPMBC model. We observed that B cell–specific loss of Prdm1 installs a plasma cell differentiation defect in MBC-derived, nontransformed B cells. Intriguingly, this plasma cell differentiation block appears to be more robust than that observed when we overexpressed Spib in MBC B cells, at least when judged by the abundance of late plasmablasts, as well as plasma cells by flow cytometry and by assessment of serum immunoglobulin levels. We further observed reduced surface IgM levels in MBC- and SMBC-derived naïve B cells, compared with controls, whereas naïve PPMBC B cells did not show a reduced surface IgM expression. As these naïve B cells have not passed through the GC reaction and are thus likely not expressing substantial PRDM1 levels, this observation warrants further investigation outside the context of this work. One possible explanation might be the contamination of the B220+/CD19+/IgM+/IgD+ population with IgM+/IgD+ memory- or marginal zone B cells. These differences between SMBC and PPMBC mice might be at least partially explained by the fact that Prdm1 deletion in our model prevents any protein expression, whereas Spib overexpression from the Rosa26 locus might be counterregulated by mechanisms leading to SPIB degradation or through partial silencing of the Rosa26 locus.
Next to mutational and epigenetic inactivation, PRDM1 is also transcriptionally repressed by BCL6 (35, 36), which itself is a target of PRDM1-mediated repression, thus constituting a negative feedback loop (37). In addition to its role in blocking terminal plasma cell differentiation via PRDM1 repression, SPIB also plays an important role for proper BCR signaling in mature B cells. In contrast to Spib-proficient B cells, Spib-deficient B cells display reduced proliferation in response to BCR cross-linking and an impaired ability to form GCs (38, 39). The oncogenic role of SPIB in ABC-DLBCL is further underscored by the observation that the ABC-DLBCL cell line OCI-Ly3 harbors a rearrangement, which juxtaposes SPIB to Ig enhancers leading to enforced SPIB expression (40). Altogether, it is conceivable that the similar lymphomagenesis-enhancing effects of Spib overexpression and Prdm1 deletion in the MBC model are the result of different molecular mechanisms, namely, a potent plasma cell differentiation block in the Prdm1-deficient setting and a slightly less pronounced block in terminal differentiation together with augmented BCR signaling in the Spib-overexpressing scenario.
One central disadvantage of the original MBC model is the CD138 positivity, which is not a common feature of human DLBCL, but rather a hallmark of plasmablastic lymphoma (17–20, 41). The PPMBC model described here overcomes this limitation and develops clonal CD138−/B220+ DLBCL. Fitting with the robust CD138 and IRF4 expression of MBC lymphomas (17), as well as the retained B220 expression together with lack of CD138 expression in PPMBC lymphomas, transcriptome analyses of KBC, MBC, SMBC, and PPMBC lymphomas revealed that SMBC and PPMBC lymphomas are more closely related to GC B cells than MBC-derived lymphomas. Nevertheless, SMBC and PPMBC lymphomas displayed a significant enrichment of AB cell transcriptome features, compared with KBC lymphomas, which constitute a canonical GCB-DLBCL model. Thus, altogether, the SMBC and PPMBC models bear a closer resemblance to human ABC-DLBCL cases than the original MBC model. Moreover, these models harbor genetic lesions that constitute key features of MCD/C5 DLBCL, thus nominating the SMBC and PPMBC models as preclinical avatars for this particularly hard-to-treat subentity.
While transcriptomic, genomic, and surface marker profiling revealed that the PPMBC model shares central characteristics with human C5/MCD cases, long-range BCR sequencing revealed that the majority of PPMBC-derived lymphomas were class-switched to IgG. This phenotype differs from what has been reported for most human MCD DLBCL cases, which are predominately nonclass-switched. However, it is important to note that in the MCD DLBCL case series reported by Wright and colleagues, approximately 10% of the MCD cases were indeed class-switched to IgG and to a lesser extent to IgA (24). Thus, while our model might not fully capture this part of MCD biology, we believe that it still reflects a substantial aspect of this disease.
Our flow cytometry and transcriptome profiling experiments indicate PPMBC lymphomas displayed gene-expression signatures associated with early light zone and intermediate stages, as well as prememory B cell stages. The presence of both light zone GC and memory B cell features in the absence of plasmablastic differentiation markers is coherent with a precursor population at an intermediate stage between GC exit and memory B-cell differentiation. Similar results were recently published by Pindzola and colleagues, using a AicdaCre/wt; Prdm1fl/fl; Myd88cond.p.L252P/wt; Rosa26LSL.BCL2.IRES.GFP/wt; Cd79bcond.p.Y195H model to demonstrate aberrant expansion of spontaneous GCs and a lack of plasma cell differentiation in these mice (42). The nonmalignant GCB cells in this model, which differs from the PPMBC model by the inclusion of an inducible oncogenic Cd79b mutation, displayed a dark zone phenotype and a neglectable memory B-cell expansion (42). Unfortunately, lymphomas arising in the AicdaCre/wt; Prdm1fl/fl; Myd88cond.p.L252P/wt;Rosa26LSL.BCL2.IRES.GFP/wt;Cd79bcond.p.Y195H model were not extensively characterized with regard to their transcriptomic profiles, thus preventing a proper comparison between lymphoma phenotypes in these two models. Nevertheless, in both the PPMBC and the AicdaCre/wt; Prdm1fl/fl; Myd88cond.p.L252P/wt; Rosa26LSL.BCL2.IRES.GFP/wt; Cd79bcond.p.Y195H models an expansion of the GCB pools with a DZ/LZ B cell ratio shifted toward DZ B cells was observed (42). However, the enlarged memory B cell pool that we observed in PPMBC mice was not described in the AicdaCre/wt; Prdm1fl/fl; Myd88cond.p.L252P/wt; Rosa26LSL.BCL2.IRES.GFP/wt; Cd79bcond.p.Y195H model (42). Collectively, these data might indicate that different, genetically defined DLBCL subtypes originate from distinct precursor cell populations.
In the clinical setting, relapsed/refractory DLBCL still remains a major therapeutic challenge. Approximately 50% of patients who are treated with salvage chemoimmune therapy, followed by high-dose chemotherapy and autologous stem cell support, will subsequently progress after transplantation and achieve a median overall survival of approximately 10 months (43, 44). Patients with disease that is refractory to primary or salvage therapy, as well as patients who relapse within the first 12 months after autologous transplantation, display an extraordinarily poor prognosis with a median overall survival of only 6.3 months (45). The advent of anti-CD19 CAR-T cells has recently revolutionized the treatment landscape for relapsed/refractory DLBCL patients, in which two lines of therapy have failed, as well as transplant-eligible patients who experience relapse within 12 months after first-line therapy or refractory disease (46–48). However, recent analyses focusing on the identification of factors that may predict CAR-T cell treatment failure retrieved Eastern Cooperative Oncology Group Performance Status (ECOG PS) ≥2, Ann Arbor stage III/IV disease, involvement of ≥2 extranodal sites, elevated serum lactate dehydrogenase, increased serum C-reactive protein, high international prognostic index at the time of treatment decision and at the time of treatment, as well as increased CRP, bulky lymphoma mass, and high total metabolic tumor volume at the time of treatment, as risk factors for poor response to CAR-T cell therapy (49). These data, particularly the association of bulky disease at the time of treatment with poor outcome, may suggest that remission-inducing bridging therapy between treatment decision and time of CAR-T infusion may be desirable. In light of these clinical circumstances, our observation that combined ibrutinib/venetoclax induces remissions in lymphoma-bearing PPMBC mice, as well as extensively pretreated non–GCB-DLBCL patients, recommends this combination regimen as a potential bridge to CAR-T cell therapy in non–GCB-DLBCL patients. In addition to the data reported here, there is substantial evidence indicating that ibrutinib may display clinical efficacy in selected DLBCL patients. For instance, in a recently reported prospective trial, single-agent ibrutinib produced complete or partial responses in 37% (14/38) of ABC-DLBCL patients, whereas only 5% (1/20) of GCB-DLBCL patients achieved complete or partial responses (31). Moreover, a recently published case series involving 13 relapsed/refractory non–GCB-DLBCL patients described an overall response rate after two cycles of ibrutinib/venetoclax of 61.5%, with 23% of patients achieving a complete remission (50). The median PFS and overall survival was 5.6 and 11.3 months, respectively (50). Nevertheless, these data suggest that combined ibrutinib/venetoclax may be a useful regimen as a bridge to consolidating cellular immune therapy.
METHODS
Experimental Mice
The generation of the Cd19Cre, Myd88cond.p.L252P, and Rosa26LSL.BCL2-IRES-GFP alleles has been described previously (18). The Prdm1flox allele was purchased from the Jackson Laboratory on a C57BL/6J background (Stock No: 008100; ref. 51). To generate the Rosa26LSL.Spib.IRES.GFP allele, murine Spib cDNA was cloned into a Rosa26 locus-targeting vector, in which a CAGGS (cytomegalovirus early enhancer/chicken β actin) promoter drives expression of the transgene (Spib) as well as an internal ribosomal entry site–driven green fluorescent protein (IRES-GFP). Expression of these genes is prevented by a LoxP-flanked STOP cassette. This targeting vector was electroporated into BRUCE4 embryonic stem cells, which were then screened for correct integration by standard Southern blot methods. Correctly targeted ES cells were used to generate chimeras, which were backcrossed onto a pure C57BL/6J background and examined for germline transmission.
For survival analyses, animals were recorded as events that succumbed to disease or that had to be sacrificed due to predefined termination criteria. Animals that died due to genotype-unrelated reasons (appendicitis, abnormal teeth, injuries inflicted by cage mates) were censored. In treatment studies, onset of lymphoma was defined by a lesion larger than 150 μL or splenomegaly larger than 500 μL detectable by MRI, with a robust volume increase in two consecutive scans. Venetoclax (MedChem Tronica) was administered as a suspension in 0.4% methylcellulose by oral gavage at 200 mg/kg daily. Ibrutinib (MedChem Tronica) was administered as a suspension in 0.4% methylcellulose by oral gavage at 30 mg/kg daily for 3 weeks as a single-agent or in combination with venetoclax, then via drinking water until death. For the preparation of drinking water, ibrutinib was resolved in 5% HP-cyclodextrin (PanReac Applichem) at 0.16 mg/mL. Mice were allowed to drink liberally with no other source of water. Mice receiving ibrutinib drinking water received approximately 30 mg/kg ibrutinib per day (52). Upon relapse or progression, animals were rechallenged with venetoclax for 3 weeks, with or without concurrent treatment with ibrutinib via drinking water. To assess the efficacy of immune-checkpoint blockade in our PPMBC model, mice were injected twice weekly with 250 μg of a PD-L1 antibody (clone 80; ref. 29; Astra Zeneca, intraperitoneally) for 8 weeks.
For anti-CD19 CAR-T cell experiments, T cells were isolated from spleens of C57BL/6 wild-type mice using the Mouse CD3 T cell isolation kit (BioLegend; cat. #480031), cultured and activated as previously described (27). In brief, T cells were activated in vitro by antimouse CD3e (200 ng/mL, BioLegend, clone 145-2C11), murine IL2 (100 U/mL, Novartis, 17152.00.00), and anti-mouse CD28 (100 ng/mL, BioLegend, clone 37.51), and subsequently transduced with CAR-retrovirus. Retrovirus for transduction of T cells was produced by HEK 293T cells cotransfected with the retroviral Moloney murine leukemia virus (MMLV)–based helper plasmids coding for the gag, pol, and env genes together with the CAR. After transduction, T cells were stained with a polyclonal goat anti-rat IgG antibody (Biotin, Jackson Immuno, cat. #112-066-072), counterstained with streptavidin (APC, BioLegend, cat. #405207) and anti-mouse CD3 (APC, BioLegend, clone 145-2C11) to analyze transduction efficacy. To treat CD19+ lymphomas with antigen-specific T cell therapy, 2 × 106 engineered CAR-T cells were intravenously injected into lymphoma-bearing C57BL/6 PPMBC mice (identified by MRI). Tumor response was subsequently monitored by weekly MRI and overall survival was recorded.
For transplantation experiments, Rag1−/− mice (C57BL6/J background from Jackson Laboratories 002216) or syngeneic C57BL/6 mice (Cre-negative Prdm1fl/fl; Myd88cond.p.L252P/cond.p.L252P; Rosa26LSL.BCL2-IRES-GFP/LSL.BCL2-IRES-GFP breeder animals) were used as recipients. 5 × 106 cells suspended in PBS were transplanted intraperitoneally (Rag1−/−) or intravenously (syngeneic mice). All animals were housed in a specific pathogen-free facility and animal breedings and experiments were approved by the local animal care committee and the relevant authorities (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, AZ: 84-02.04.2014.A146, 84-02.04.2017.A131, 81-02.04.2019.A009, 81-02.04.2020.A395).
SPIB Detection by Immunoblotting in MEFs
Rosa26 LSL.Spib.IRES.GFP/wt and Rosa26wt/wt MEFs were generated according to standard protocol (53). MEFs were then exposed to Ad-CMV-Cre (University of Iowa) overnight at an MOI of 100. Cells were subsequently lysed in 4% SDS, protein concentration was measured by BCA Assay (Thermo Fisher, 23225), and equivalent amounts of protein (60 μg in Laemmli buffer) were separated on 12.5% SDS-PAGE and blotted on PVDF membranes (Immobilon-P, Millipore, 0.45 μm). Membranes were blocked (5% BSA in TBS-T), incubated with specific antibodies against SPIB (Cell Signaling Technology, D4V9S) and β-Actin (Sigma, A2228) and subsequently washed and incubated with secondary HRP-coupled antibody. Proteins were detected using the ECL solution (Amersham).
MR Imaging
Mouse MR imaging was performed as described previously (18). In brief, mice were anesthetized with 2.5% isoflurane and scanned on a 3.0T MRI system (Igenia, Philips) with a small rodent solenoid coil (diameter 40 mm, Philips Research Europe). Axial T2-weighted images of the abdomen were acquired (TSE factor: 10, TR: 2674 ms, TE: 65 ms, slice thickness: 1.0 mm. Images were exported in DICOM format and spleen and tumor volumes were measured by segmentation using the Horos software.
IHC
Mouse tissue was formalin-fixed and paraffin-embedded (FFPE). Sections (4 μm) were stained for B220 (BD, clone RA3-6B2), CD138 (BD, cat. #553712), Ki67 (Cell Marque), CD3 (Thermo Fisher, clone RM-9107), biotinylated PNA (Vector Laboratories, B-1075-5), BCL6 (CST clone D65C10), and MUM1/IRF4 (Proteintech, cat. #11247-2-AP). GCs were quantified from PNA stainings using the software ImageJ. Ki67+ cells per total cell count was quantified using the ImmunoRatio plugin for ImageJ.
Human FFPE tissue was cut into 4 μm sections and stained for CD10 (NCL-L-CD10–279, Novocastra), MUM-1 (Dako, cat. #M7259), BCL6 (Dako, cat. #M7211), and BCL2 (Dako, cat. #M0877).
Flow Cytometry
Flow cytometry was conducted on a Gallios flow cytometer (Beckman Coulter) or LSRFortessa cell analyzer (BD Biosciences), and data were analyzed with the FlowJo software (version 10.6.1; BD Biosciences). Single-cell suspensions from mouse spleen or lymphomas were stained with the following fluorescent-labeled antibodies: BD Biosciences: CD45 (APC-Cy7 and Alexa Fluor700, clone 30-F11), B220 (Pacific-Blue and BV786, clone RA3-6B2), CD95/Fas (BV510, clone Jo2), CD5 (BUV496, clone 53-7.3), IgD (BUV496, clone 217-170), IgA (BV650, clone C10-1), IgM (BV711, clone II/41), CD19 (BUV395, clone 1D3), CXCR4 (PE, clone 2B11), CD38 (BUV395, clone clone 90/CD38), IgG2b (BV650, clone R12-3), IgG3 (BV650, clone R40-82), IgG1 (BV650, clone RMG1-1), BioLegend: CD3 (PerCP-Cy5.5, clone 281-2), CD138 (PE/Cy7 and PerCP-Cy5.5, clone 281.2), MHCII (AF700, clone M5-1142), CD21/CD35 (PE/Cy7, clone 7E9), CD23 (BV510, clone B3B4), CD86 (PE/Cy7, clone GL1), PLCg2 (PE, biorbyt, cat. #orb504888), Invitrogen: CD93 (PE, clone AA4.1), GL-7 (eFluor660, clone GL7), pPLCg2 (APC, clone 4NPRN4). Cell Signaling Technology: pSyk (AF488, clone C87C1), Syk (AF488, clone D3Z1E). Biozol: IgG2c (AF647, cat. #SBA-1079-31). A Zombie NIR Fixable Viability Kit (BioLegend, cat. #423105) was used for exclusion of dead cells. Intracellular stains were performed as previously described (7). The gating strategies are depicted in Supplementary Fig. S12A–S12I.
Serum Ig Analysis
Serum was collected from 10-week-old mice and subjected to measurement of IgA, IgM, or whole IgG via ELISA, which was performed according to the manufacturer’s instructions (Thermo Fisher, cat. #88-50450-22, 88-50470-88, and 88-50400-22)
Mass Cytometry
Tumor single-cell suspensions and stainings with metal-labeled antibodies were performed as previously described (18). In brief, cells were barcoded using a commercial kit following the manufacturer’s instructions (Fluidigm, 201060). Stainings were performed according to standard Fluidigm protocol. Samples were measured on a Helios device (Fluidigm). After the acquisition, the samples were preprocessed as follows: (i) bead normalization (CATALYST R package), (ii) flow rate cleanup (flowAI R package), (iii) compensation (CATALYST), (iv) debarcoding (CATALYST), (v) automatic gating for intact cells (flowDensity R package). 50,000 events per sample were used for the subsequent analysis, which included the following steps: (i) arcsinh transformation (diffcyt); (ii) gating on DNA+/CD45+ cells; (iii) UMAP (umap-learn python package) dimensionality reduction for the markers B220, CD4, CD19, CD8A, CD23, CD3E, NKG2A, CD21, NK1.1, GR1, CD11B, CD11C, FOXP3; and (iv) manual clustering. Analysis and graphical visualization were then performed using custom R scripts.
3′-RNA-Sequencing Analysis
For bulk transcriptomic analysis of lymphomas, RNA was isolated from cryo-frozen lymphoma tissue using a commercial kit (Qiagen). Tissue samples (nodal or splenic) were collected from moribund animals and classified as malignant by histology. Libraries were prepared with the Lexogen QuantSeq kit according to the standard protocol. After validation (2200 TapeStation; Agilent Technologies) and quantification (Qubit System; Invitrogen) pools of cDNA libraries were generated. The pools were quantified using the KAPA Library Quantification kit (Peqlab) and the 7900HT Sequence Detection System (Applied Biosystems) and subsequently sequenced on an Illumina HiSeq4000 sequencer using a 1 × 50 bp protocol. Reads were mapped to the murine genome (mm10) and quantified using Salmon. Data were normalized, and statistics were calculated using DESeq2.
To generate transcriptional profiles of activated murine blood B cells, blood was drawn from wild-type animals and untouched B cells were purified by CD43 depletion with a commercial kit (Miltenyi Biotec, 130-090-862). Cells were then treated with 10 μg/mL anti-CD40 antibody (FGK45.5, Miltenyi), 10 μg/mL anti-IgM antibody (Jackson ImmunoResearch), and 40 ng/mL IL4 (130–094–061, Miltenyi) for 24 hours. For the generation of murine GC B cell transcriptomes, PNA+ cells were purified from wild-type spleens by a commercial kit (130-110-479, Miltenyi). For both cell populations, RNA isolation, library preparation, sequencing, and raw data processing were performed as described above. To generate gene sets for blood-activated B and GC B cells, differentially expressed genes between the two groups were identified. Fold change ≥ 2 and P ≤ 0.01 were used as thresholds.
To map the bulk lymphoma transcriptomes to published gene sets (26) of human GC B-cell developmental stages, their murine homologs were identified. The murine transcriptomes were subsequently clustered in an unsupervised fashion with the heatmap.2 function of the gplots R package and ssGSEA scores were calculated for each sample and gene set.
Gene set analyses were performed using the FGSEA package (R) and ssGSEA scores were calculated with the corto R package throughout the manuscript.
BCR Sequencing
PNA+ cells were isolated by MACS-sorting [Germinal Center B Cell (PNA) MicroBead Kit, mouse, Miltenyi Biotec, 130-110-479]. The BCR sequencing of PNA+ cells was performed using a commercial kit (NEB, NEBNext immune sequencing kit mouse). To assemble the raw sequencing data into unique sequences (unique combinations of barcodes and BCR sequence), the pRESTO package (54) was used, according to the kit manufacturer’s recommendations. Individual deduplicated sequences were then processed using the IgBlast software (55) and the output was analyzed for V, D, J gene usage as well as somatic hypermutation rate. The isotype of each read is given in the pRESTO output and determined by aligning the C gene–specific primer sequences used during library preparation.
The library preparation for the BCR receptor sequencing data in Fig. 2D and E and Supplementary Fig. S5E and S5G was performed as described previously (18). Sample demultiplexing and assembly were performed using the published MIGEC pipeline. To assess clone sizes, the MIGEC output was further processed using a custom R script. Clones were assembled by connecting subclonal sequences that differ by only a single mismatch. The resulting networks were visualized with the software Gephi. Gini coefficients were calculated for the dominant clones of each sample with the R package “reldist.”
For SHM and isotype analysis in lymphoma samples, a commercial kit for full-length Ig sequencing was used (NEBNext immune sequencing kit mouse). The reads were processed using the pRESTO package. The assembled and deduplicated reads were processed with a custom R script to define individual clones by connecting subclonal sequences with a distance of 1 bp. The dominant clone sequences were analyzed using IgBlast to determine nucleotide and amino acid changes from germline.
WES
DNA was isolated from cryo-preserved PPMBC, SMBC, and MBC tumors, as well as human FFPE samples, after written informed consent was provided by the patients. WES was performed on an Illumina NovaSeq6000 according to previously published protocols (18, 56). By means of the BWA mem aligner, we aligned raw sequencing reads of the mouse samples to the reference genome mm10 and the human samples to the reference genome hg19. Identical read pairs that represent possible PCR duplicates were masked out after alignment. All overlapping regions between the read pairs are considered only once in the analysis. Due to a lack of matched normal for some tumor specimens, we generated a representative nontumor sample by combining normals matching to some tumor samples. Our in-house cancer genome analysis pipeline is used for mutation and copy-number alteration calling (57, 58). To correct for genotypes that are not captured by representative normal, we filtered out called mutations that were exactly the same in more than one tumor sample.
CAR-T Coculture Experiments
For coculture experiments, anti-CD19 CAR-T cells were cocultured with a CD19+ PPMBC lymphoma cell line for 48 hours. Subsequently, cell counts were analyzed by flow cytometry after staining for CD3 (PE, BioLegend, clone 145-2C11) and CD19 (V500, BD Biosciences, Clone 1D3) after gating for Zombie-negative live cells.
CT Imaging
Contrast-enhanced CT scans were performed at baseline and after every treatment cycle. Imaging was evaluated according to the Lugano criteria. Objective response was classified based on CT measurements, using the mintLesion software package (MINT; ref. 59).
Patients
For the off-label use of venetoclax and ibrutinib, we treated six patients with relapsed/refractory non–GCB-DLBCL after receiving written informed consent from the patients or their legal representatives, as described in the main text. Patients were between 32 and 71 years of age (median 66.5 years) at treatment initiation. Patient data including age, sex, and number of previous therapies are depicted in Supplementary Table S6A. All patients were Caucasian. All patients were treated in accordance with ethical standards as described in the Declaration of Helsinki. Inclusion criteria were: non–GCB-DLBCL as defined by Hans algorithm, no other available or medically indicated approved line of therapy. Exclusion criteria were cardiac arrhythmia or bleeding complications in the last 2 months, neutropenia below 500/μL, thrombopenia below 50,000/μL, impaired renal function with a glomerular filtration rate < 50 mL/minute, pregnancy, ECOG > 2 and age below 18 years. For four out of these six patients, biopsy material was available for exome sequencing, which we conducted after receiving written informed consent for this procedure from the patients or their legal representatives.
Data Availability
Transcriptome, exome and BCR sequencing data generated in this study are publicly available at the Sequence Read Archive (SRA) under the BioProject ID PRJNA876929 and PRJNA668334.
Supplementary Material
Table S1 contains the p values of comparisons of the V gene usage in non-malignant GC B cells between the individual genotypes. These comparisons were done on the family, gene and allele level. V usage was compared for all BCR sequences as well as for isotype subsets.
Table S2 contains a summary of the B cell receptor sequencing data of lymphoma samples. It includes 1) an overview of the investigated samples, 2) a list of characteristics of the dominant clone of each lymphoma sample, 3) a list comparing the frequencies of V family usage in WT samples to the lymphoma frequency, 4) the frequencies of FR and CDR mutations in samples analyzed by full-length BCR sequencing.
Table S3 contains B cell receptor repertoire sequencing data and lists the individual sublcones of the dominant clones of MBC, SMBC and PPMBC lymphomas.
Table S4 contains the individual mutations of PPMBC and MBC lymphomas.
Table S5 contains statistical values of gene set enrichment analyses of the KEGG, BIOCARTA and HALLMARK gene set collections.
Table S6 contains clinical parameters and sequencing results of human DLBCL samples.
This file contains the Supplementary Figures S1 to S12.
Acknowledgments
We are indebted to our patients, who provided primary material. We thank Alexandra Florin and Marion Müller from the Institute of Pathology, University Hospital Cologne, for their outstanding technical support. Flow cytometry analyses were partially performed in the FACS and Imaging Core Facility at the Max Planck Institute for Biology of Ageing, Cologne. We thank the Cologne Center for Genomics and the Genomics and Transcriptomics Facility Essen for their sequencing support. We are indebted to Scott A. Hammond and Xiaoyoun Wu (Astra Zeneca) for generously providing the anti–PD-L1 antibody “clone 80′.” We thank Laura Pasqualucci for providing Kmt2dfl/fl; VavP-Bcl2;Cɣ1Cre/wt lymphoma samples. This work was funded through the German–Israeli Foundation for Research and Development (I-65-412.20-2016 to H.C. Reinhardt), the Deutsche Forschungsgemeinschaft (RE 2246/13-1, SFB1399-A01, SFB1430-A09, SFB-Geschäftszeichen 455784452 to H.C. Reinhardt), the Deutsche Jose Carreras Leukämie Stiftung (R12/08 to H.C. Reinhardt, Promotionsstipendium to P. Nguyen), the Else Kröner-Fresenius Stiftung (EKFS-2014-A06 to H.C. Reinhardt, 2016_Kolleg.19 to H.C. Reinhardt, R.D. Jachimowicz), the Deutsche Krebshilfe (1117240 and 70113041 to H.C. Reinhardt) and a Mildred Scheel Nachwuchszentrum Grant (Grant number 70113307), as well as the German Ministry of Education and Research (BMBF e:Med 01ZX1303A to H.C. Reinhardt). J. Hansen is a member of the Cologne Graduate School of Ageing Research.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
R.K. Thomas reports other support from NEO New Oncology, PearlRiver Bio, Centessa, DISCO Pharmaceuticals, and grants from Roche outside the submitted work. R. Büttner reports lectures and advisory boards for AbbVie, Amgen, AstraZeneca, Bayer, BMS, Boehringer-Ingelheim, Illumina, Janssen, Lilly, Merck-Serono, MSD, Novartis, Qiagen, Pfizer, Roche, Cofounder and CSO of Targos MP Inc. (until May 2021), Gnothis Inc/Stockholm (Member of Board of Directors and Coowner). D.P. Calado reports other support from Cancer Research UK (CC2078), Wellcome Trust (CC2078), and UK Medical Research Council (CC2078) during the conduct of the study. B. Chapuy reports grants from the German Research Foundation, Jose Carreras Leukemia Foundation, Gerdes Foundation, German Ministery of Science (BMBF), other support from Roche, Incyte, grants, nonfinancial support, and other support from Gilead, BMS, Astra Zenecca, Sandoz, AbbVie, Regeneron, and ADC outside the submitted work; in addition, B. Chapuy has a patent on molecular subtyping of DLBCL pending. B. von Tresckow reports personal fees from Allogene, BMS/Celgene, Cerus, Incyte, Miltenyi, Novartis, Pentixafarm, Roche, Amgen, Pfizer, Takeda, Merck Sharp & Dohme, Gilead Kite, AstraZeneca, grants from Novartis, Merck Sharp & Dohme, Takeda, nonfinancial support from AbbVie, AstraZeneca, Kite-Gilead, Merck Sharp & Dohme, Takeda, and Novartis outside the submitted work. A.M. Melnick reports grants from Janssen and grants and personal fees from Astra Zeneca during the conduct of the study; grants and personal fees from Epizyme and grants from Treeline Biosciences outside the submitted work. H.C. Reinhardt reports grants from German-Israeli Foundation for Research and Development, grants from Deutsche Forschungsgemeinschaft, Deutsche Jose Carreras Leukämie Stiftung, Else Kröner-Fresenius Stiftung, Deutsche Krebshilfe, and German Ministry of Education and Research during the conduct of the study; personal fees and other support from Roche, Novartis, BMS, AbbVie, grants and other support from AstraZeneca, personal fees from Vertex, Merck, grants from Gilead, and nonfinancial support and other support from CDL Therapeutics GmbH outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
R. Flümann: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration. J. Hansen: Investigation. B.W. Pelzer: Investigation. P. Nieper: Conceptualization, data curation, formal analysis, investigation, visualization, methodology. T. Lohmann: Resources, software, formal analysis, visualization. I. Kisis: Investigation. T. Riet: Data curation, formal analysis, investigation, visualization. V. Kohlhas: Investigation, methodology. P.-H. Nguyen: Methodology. M. Peifer: Software, investigation. N. Abedpour: Software, investigation. G. Bosco: Resources, methodology. R.K. Thomas: Resources. M. Kochanek: Investigation. J. Knüfer: Investigation. L. Jonigkeit: Investigation. F. Beleggia: Software. A. Holzem: Investigation, methodology. R. Büttner: Resources, methodology. P. Lohneis: Investigation. J. Meinel: Investigation. M. Ortmann: Investigation. T. Persigehl: Resources, methodology. M. Hallek: Resources. D.P. Calado: Resources. M. Chmielewski: Conceptualization, resources, supervision. S. Klein: Software, formal analysis, visualization, methodology. J.R. Göthert: Resources. B. Chapuy: Resources, software. B. Zevnik: Resources, methodology. F.T. Wunderlich: Resources, methodology. B. von Tresckow: Resources. R.D. Jachimowicz: Resources. A.M. Melnick: Resources, supervision, methodology. H.C. Reinhardt: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration. G. Knittel: Conceptualization, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration.
References
- 1. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016;127:2375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–11. [DOI] [PubMed] [Google Scholar]
- 3. Shaffer AL 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol 2012;30:565–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Basso K, Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol 2015;15:172–84. [DOI] [PubMed] [Google Scholar]
- 5. Grondona P, Bucher P, Schulze-Osthoff K, Hailfinger S, Schmitt A. NF-kappaB activation in lymphoid malignancies: genetics, signaling, and targeted therapy. Biomedicines 2018;6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Venturutti L, Teater M, Zhai A, Chadburn A, Babiker L, Kim D, et al. TBL1XR1 mutations drive extranodal lymphoma by inducing a pro-tumorigenic memory fate. Cell 2020;182:297–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knittel G, Liedgens P, Korovkina D, Pallasch CP, Reinhardt HC. Rewired NFkappaB signaling as a potentially actionable feature of activated B-cell-like diffuse large B-cell lymphoma. Eur J Haematol 2016;97:499–510. [DOI] [PubMed] [Google Scholar]
- 8. Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008;359:2313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002;346:1937–47. [DOI] [PubMed] [Google Scholar]
- 10. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 2018;378:1396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gisselbrecht C, Glass B, Mounier N, Singh Gill D, Linch DC, Trneny M, et al. Salvage regimens with autologous transplantation for relapsed large B-cell lymphoma in the rituximab era. J Clin Oncol 2010;28:4184–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horwitz SM, Negrin RS, Blume KG, Breslin S, Stuart MJ, Stockerl-Goldstein KE, et al. Rituximab as adjuvant to high-dose therapy and autologous hematopoietic cell transplantation for aggressive non-Hodgkin lymphoma. Blood 2004;103:777–83. [DOI] [PubMed] [Google Scholar]
- 14. Kewalramani T, Zelenetz AD, Nimer SD, Portlock C, Straus D, Noy A, et al. Rituximab and ICE as second-line therapy before autologous stem cell transplantation for relapsed or primary refractory diffuse large B-cell lymphoma. Blood 2004;103:3684–8. [DOI] [PubMed] [Google Scholar]
- 15. Tilly H, Gomes da Silva M, Vitolo U, Jack A, Meignan M, Lopez-Guillermo A, et al. Diffuse large B-cell lymphoma (DLBCL): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2015;26Suppl 5:v116–25. [DOI] [PubMed] [Google Scholar]
- 16. Chow VA, Shadman M, Gopal AK. Translating anti-CD19 CAR T-cell therapy into clinical practice for relapsed/refractory diffuse large B-cell lymphoma. Blood 2018;132:777–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knittel G, Liedgens P, Korovkina D, Seeger JM, Al-Baldawi Y, Al-Maarri M, et al. B cell-specific conditional expression of Myd88p.L252P leads to the development of diffuse large B cell lymphoma in mice. Blood 2016;127:2732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flumann R, Rehkamper T, Nieper P, Pfeiffer P, Holzem A, Klein S, et al. An autochthonous mouse model of Myd88- and BCL2-driven diffuse large B-cell lymphoma reveals actionable molecular vulnerabilities. Blood Cancer Discov 2021;2:70–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koizumi Y, Uehira T, Ota Y, Ogawa Y, Yajima K, Tanuma J, et al. Clinical and pathological aspects of human immunodeficiency virus-associated plasmablastic lymphoma: analysis of 24 cases. Int J Hematol 2016;104:669–81. [DOI] [PubMed] [Google Scholar]
- 20. Rudresha AH, Lakshmaiah KC, Agarwal A, Babu KG, Loknatha D, Jacob LA, et al. Plasmablastic lymphoma in immunocompetent and in immunocompromised patients: Experience at a regional cancer centre in India. South Asian J Cancer 2017;6:69–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang JQ, Jeelall YS, Humburg P, Batchelor EL, Kaya SM, Yoo HM, et al. Synergistic cooperation and crosstalk between MYD88(L265P) and mutations that dysregulate CD79B and surface IgM. J Exp Med 2017;214:2759–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR, et al. Towards error-free profiling of immune repertoires. Nat Methods 2014;11:653–5. [DOI] [PubMed] [Google Scholar]
- 23. Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 2020;37:551–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 2015;21:1190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holmes AB, Corinaldesi C, Shen Q, Kumar R, Compagno N, Wang Z, et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J Exp Med 2020;217:e20200483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kochenderfer JN, Yu Z, Frasheri D, Restifo NP, Rosenberg SA. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood 2010;116:3875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Locke FL, Rossi JM, Neelapu SS, Jacobson CA, Miklos DB, Ghobadi A, et al. Tumor burden, inflammation, and product attributes determine outcomes of axicabtagene ciloleucel in large B-cell lymphoma. Blood Adv 2020;4:4898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schofield DJ, Percival-Alwyn J, Rytelewski M, Hood J, Rothstein R, Wetzel L, et al. Activity of murine surrogate antibodies for durvalumab and tremelimumab lacking effector function and the ability to deplete regulatory T cells in mouse models of cancer. mAbs 2021;13:1857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davids MS, Roberts AW, Seymour JF, Pagel JM, Kahl BS, Wierda WG, et al. Phase I first-in-human study of venetoclax in patients with relapsed or refractory non-hodgkin lymphoma. J Clin Oncol 2017;35:826–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med 2015;21:922–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt-Supprian M, Wunderlich FT, Rajewsky K. Excision of the Frt-flanked neo (R) cassette from the CD19cre knock-in transgene reduces Cre-mediated recombination. Transgenic Res 2007;16:657–60. [DOI] [PubMed] [Google Scholar]
- 33. Su GH, Ip HS, Cobb BS, Lu MM, Chen HM, Simon MC. The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. J Exp Med 1996;184:203–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turner CA Jr., Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 1994;77:297–306. [DOI] [PubMed] [Google Scholar]
- 35. Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 2000;13:199–212. [DOI] [PubMed] [Google Scholar]
- 36. Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol 2004;173:1158–65. [DOI] [PubMed] [Google Scholar]
- 37. Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 2002;17:51–62. [DOI] [PubMed] [Google Scholar]
- 38. Su GH, Chen HM, Muthusamy N, Garrett-Sinha LA, Baunoch D, Tenen DG, et al. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 1997;16:7118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garrett-Sinha LA, Su GH, Rao S, Kabak S, Hao Z, Clark MR, et al. PU.1 and Spi-B are required for normal B cell receptor-mediated signal transduction. Immunity 1999;10:399–408. [DOI] [PubMed] [Google Scholar]
- 40. Lenz G, Nagel I, Siebert R, Roschke AV, Sanger W, Wright GW, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med 2007;204:633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lopez A, Abrisqueta P. Plasmablastic lymphoma: current perspectives. Blood Lymphat Cancer 2018;8:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pindzola GM, Razzaghi R, Tavory RN, Nguyen HT, Morris VM, Li M, et al. Aberrant expansion of spontaneous splenic germinal centers induced by hallmark genetic lesions of aggressive lymphoma. Blood 2022;140:1119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gisselbrecht C, Schmitz N, Mounier N, Singh Gill D, Linch DC, Trneny M, et al. Rituximab maintenance therapy after autologous stem-cell transplantation in patients with relapsed CD20(+) diffuse large B-cell lymphoma: final analysis of the collaborative trial in relapsed aggressive lymphoma. J Clin Oncol 2012;30:4462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Epperla N, Badar T, Szabo A, Vaughn J, Borson S, Saini NY, et al. Postrelapse survival in diffuse large B-cell lymphoma after therapy failure following autologous transplantation. Blood Adv 2019;3:1661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Crump M, Neelapu SS, Farooq U, Van Den Neste E, Kuruvilla J, Westin J, et al. Outcomes in refractory diffuse large B-cell lymphoma: results from the international SCHOLAR-1 study. Blood 2017;130:1800–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Locke FL, Miklos DB, Jacobson CA, Perales MA, Kersten MJ, Oluwole OO, et al. Axicabtagene ciloleucel as second-line therapy for large B-cell lymphoma. N Engl J Med 2022;386:640–54. [DOI] [PubMed] [Google Scholar]
- 47. Kamdar M, Solomon SR, Arnason J, Johnston PB, Glass B, Bachanova V, et al. Lisocabtagene maraleucel versus standard of care with salvage chemotherapy followed by autologous stem cell transplantation as second-line treatment in patients with relapsed or refractory large B-cell lymphoma (TRANSFORM): results from an interim analysis of an open-label, randomised, phase 3 trial. Lancet 2022;399:2294–308. [DOI] [PubMed] [Google Scholar]
- 48. Kersten MJ, Spanjaart AM, Thieblemont C. CD19-directed CAR T-cell therapy in B-cell NHL. Curr Opin Oncol 2020;32:408–17. [DOI] [PubMed] [Google Scholar]
- 49. Vercellino L, Di Blasi R, Kanoun S, Tessoulin B, Rossi C, D’Aveni-Piney M, et al. Predictive factors of early progression after CAR T-cell therapy in relapsed/refractory diffuse large B-cell lymphoma. Blood Adv 2020;4:5607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou Z, Zhang L, Wang X, Li X, Li L, Fu X, et al. Ibrutinib combined with venetoclax for the treatment of relapsed/refractory diffuse large B cell lymphoma. Ann Hematol 2021;100:1509–16. [DOI] [PubMed] [Google Scholar]
- 51. Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 2003;19:607–20. [DOI] [PubMed] [Google Scholar]
- 52. Woyach JA, Bojnik E, Ruppert AS, Stefanovski MR, Goettl VM, Smucker KA, et al. Bruton’s tyrosine kinase (BTK) function is important to the development and expansion of chronic lymphocytic leukemia (CLL). Blood 2014;123:1207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Torgovnick A, Heger JM, Liaki V, Isensee J, Schmitt A, Knittel G, et al. The Cdkn1a(SUPER) mouse as a tool to study p53-mediated tumor suppression. Cell Rep 2018;25:1027–39. [DOI] [PubMed] [Google Scholar]
- 54. Vander Heiden JA, Yaari G, Uduman M, Stern JN, O’Connor KC, Hafler DA, et al. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 2014;30:1930–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 2013;41:W34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herling CD, Abedpour N, Weiss J, Schmitt A, Jachimowicz RD, Merkel O, et al. Clonal dynamics towards the development of venetoclax resistance in chronic lymphocytic leukemia. Nat Commun 2018;9:727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cun Y, Yang TP, Achter V, Lang U, Peifer M. Copy-number analysis and inference of subclonal populations in cancer genomes using Sclust. Nat Protoc 2018;13:1488–501. [DOI] [PubMed] [Google Scholar]
- 59. Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol 2014;32:3059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 contains the p values of comparisons of the V gene usage in non-malignant GC B cells between the individual genotypes. These comparisons were done on the family, gene and allele level. V usage was compared for all BCR sequences as well as for isotype subsets.
Table S2 contains a summary of the B cell receptor sequencing data of lymphoma samples. It includes 1) an overview of the investigated samples, 2) a list of characteristics of the dominant clone of each lymphoma sample, 3) a list comparing the frequencies of V family usage in WT samples to the lymphoma frequency, 4) the frequencies of FR and CDR mutations in samples analyzed by full-length BCR sequencing.
Table S3 contains B cell receptor repertoire sequencing data and lists the individual sublcones of the dominant clones of MBC, SMBC and PPMBC lymphomas.
Table S4 contains the individual mutations of PPMBC and MBC lymphomas.
Table S5 contains statistical values of gene set enrichment analyses of the KEGG, BIOCARTA and HALLMARK gene set collections.
Table S6 contains clinical parameters and sequencing results of human DLBCL samples.
This file contains the Supplementary Figures S1 to S12.
Data Availability Statement
Transcriptome, exome and BCR sequencing data generated in this study are publicly available at the Sequence Read Archive (SRA) under the BioProject ID PRJNA876929 and PRJNA668334.



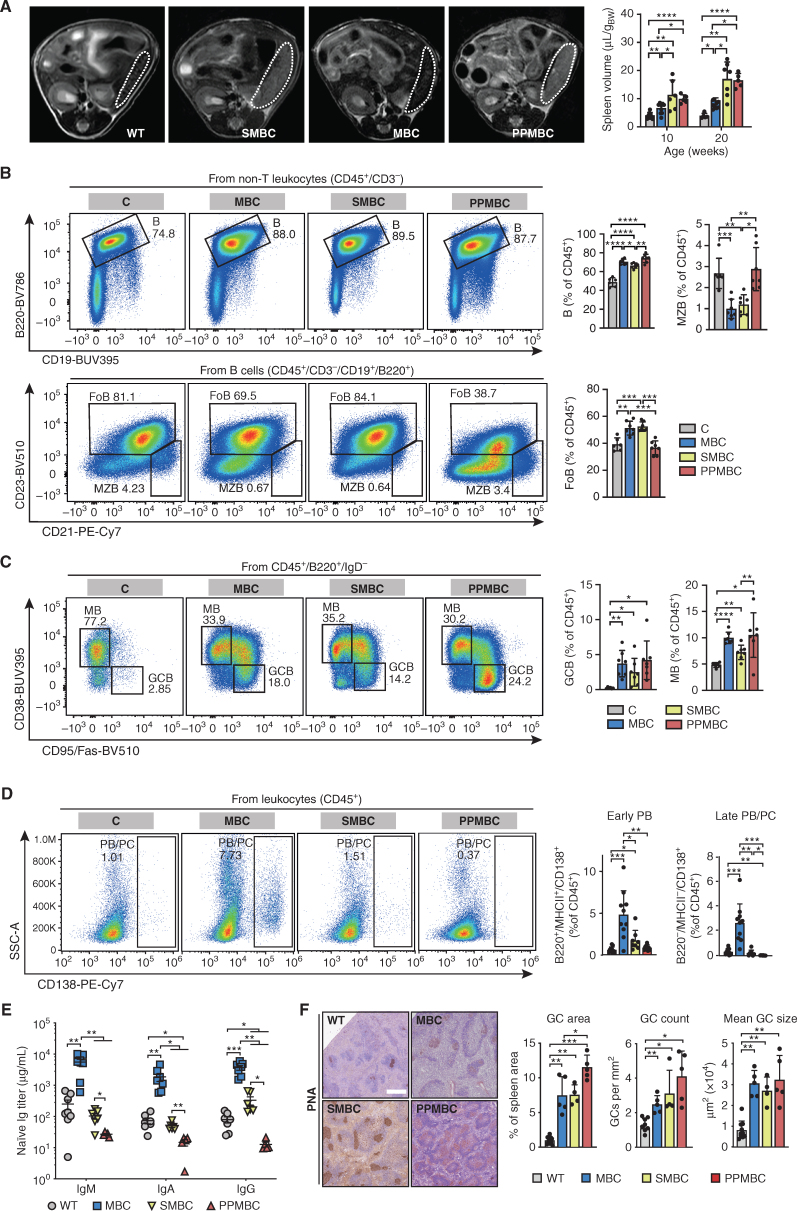
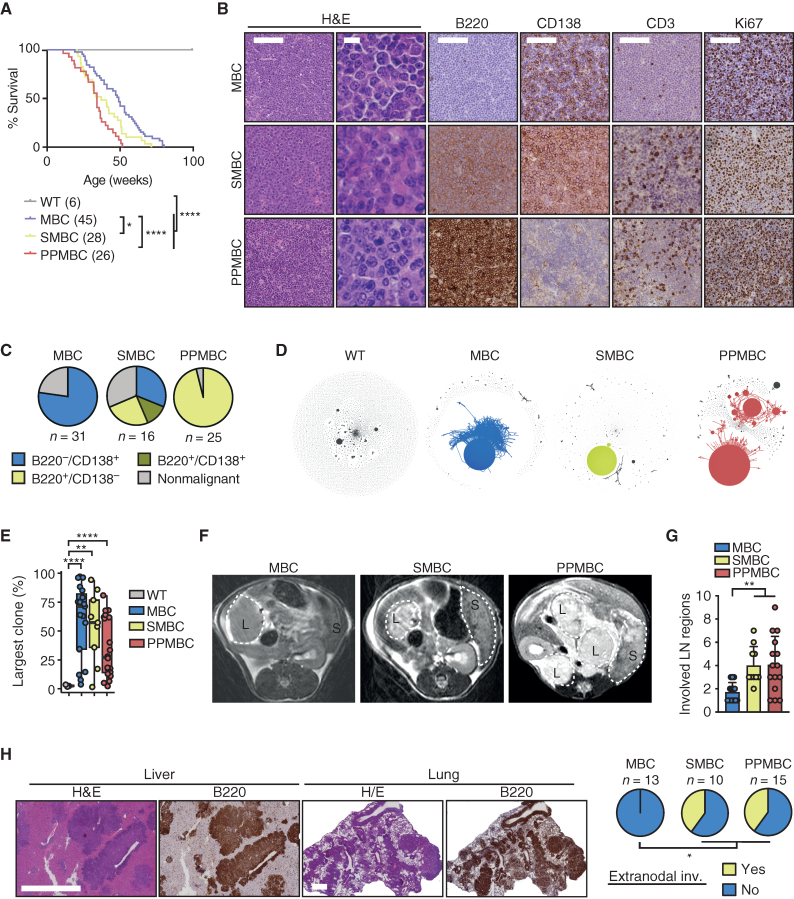
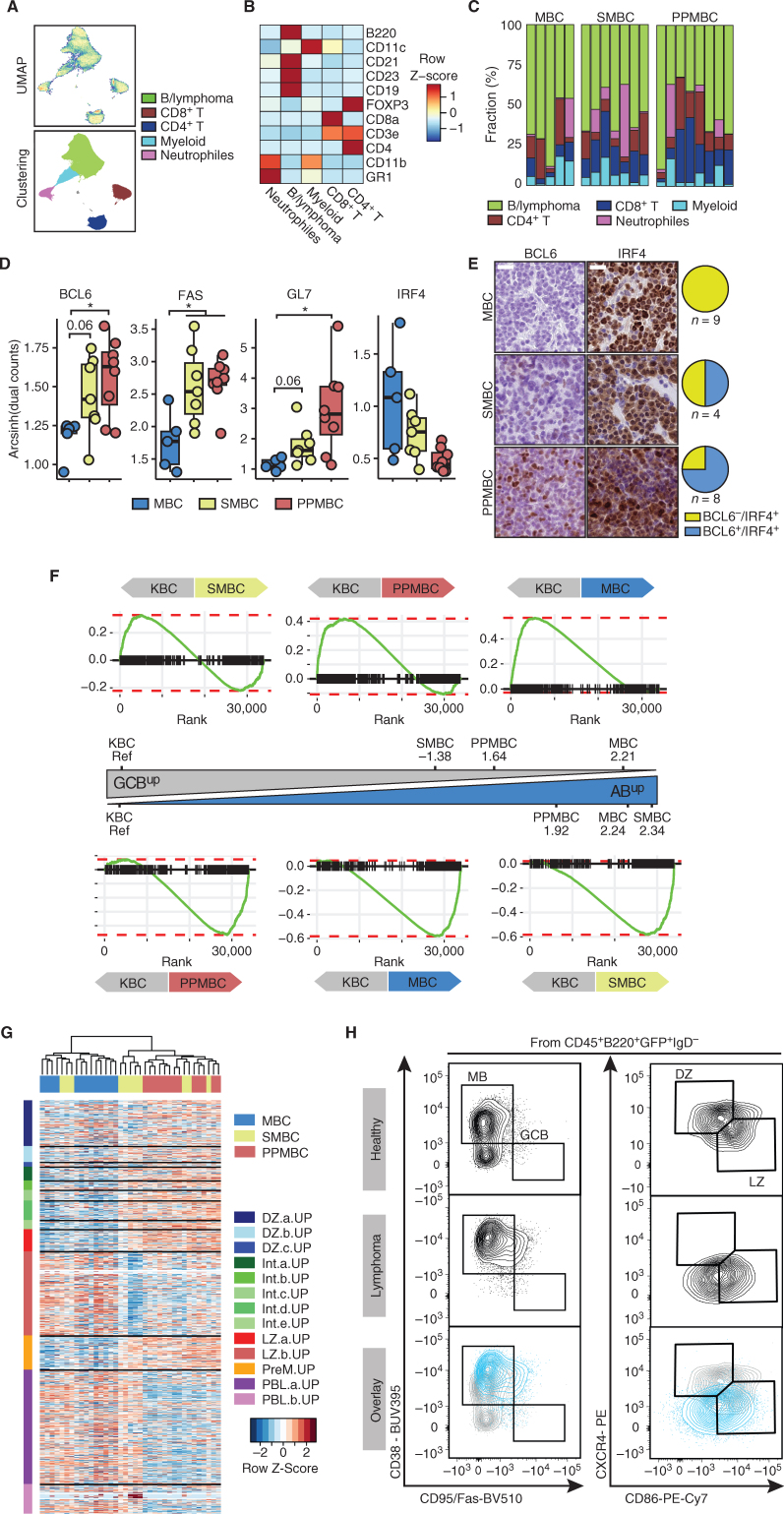
![Figure 4. Murine PPMBC lymphomas are susceptible to immune-therapeutic approaches. A, Schematic depiction of the anti-mCD19scFv-CD28-CD3z CAR construct (27). B, Population sizes of mock- and CAR-transduced T cells isolated from C57BL6/J wild-type spleens and a CD19+ PPMBC lymphoma cell line at the indicated effector:target ratios after 48 hours of coculture after gating for live cells. C, PPMBC mice with a low or high lymphoma-burden [spleen volume 500–700 μL (n = 3) or >700 μL (n = 3), respectively] were treated with a single injection of 2 × 106 anti-mCD19 CAR-T cells. Tumor volume was monitored weekly via MRI and is visualized as fold change from baseline. D, Kaplan–Meier curve showing the overall survival of CAR-T–treated mice and untreated controls. The CAR-T–treated cohort is depicted in total, as well as separated into high- and low lymphoma-burden animals. E, MRI-determined tumor volume of PPMBC animals treated with an anti–PD-L1 (n = 7), displayed as fold change from baseline. Untreated animals (n = 8) served as controls. F, Kaplan–Meier curve showing the overall survival of anti–PD-L1-treated mice (n = 7) and untreated controls (n = 8). *, P ≤ 0.05; **, P ≤ 0.01; log-rank test.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/7d2d/9816818/d980d9b6fc64/78fig4.jpg)