Summary:
In this issue of Blood Cancer Discovery, Kodgule, Goldman, Monovichet al. cleverly analyzed the transcription regulatory elements to investigate why the second copy of ETV6 is often lost in ETV6::RUNX1-translocated in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). It turns out that ETV6 suppresses the enhancer activity of GGAA microsatellite repeats, preventing ERG from subverting them to activate aberrant oncogene transcription.
See related article by Kodgule, Goldman, Monovich et al., p. 34 (5).
The E Twenty-Six (ETS) family of transcription factors is encoded by 27 genes in humans, making it one of the largest families of transcription factors. They all bind the core DNA sequence [5′-GGA(A/T)-3′] through the ETS domain with moderate diversity in flanking sequences, suggesting competition for similar binding sites. They, however, differ in expression profile and interacting partners. Their role in the transcriptional response to intracellular signaling is known. Their involvement in cancer processes is extremely diverse, with either activating and inactivating consequences, recurrent gene fusions in both solid and hematopoietic tumors as well as gene dosage effects. The effect of ETS mutations includes predisposition, tumorigenesis, and cancer maintenance (1).
ETV6 is an ETS transcriptional repressor, which activity relies on the strong polymerization property of the pointed (PNT) domain. ETV6 is frequently mutated in human leukemia through gene fusions with transcription factors [e.g., ETV6::RUNX1 in B-cell precursor acute lymphoblastic leukemia (BCP-ALL)] or kinases (e.g., ETV6::JAK2) in which the PNT domain is maintained and the DNA binding domain of ETV6 is lost generally (2). Somatic inactivating mutations are also observed along with germline single-nucleotide changes in ETV6, leading to a decreased transcriptional repression, that are associated with thrombocytopenia and a predisposition to leukemia (3).
In ETV6::RUNX1 leukemic cells, the fusion protein represses RUNX1 targets affecting genes essential for maturing lymphoid progenitor cells and contributing to the differentiation alteration (4). Notably, secondary inactivating ETV6 mutations are recurrently acquired, underscoring the importance of loss of the ETV6 activity for transformation. However, although these observations suggested that the derepression of ETV6 targets may also contribute to disease development, their identification was still elusive.
Here, Kodgule and colleagues cleverly thought to investigate the transcription factor motifs under open chromatin regions (intersect between ATAC-seq and H3K27ac peaks) in 26 cell lines derived from human B-cell malignancies, including 13 BCP-ALL cell lines (5). Regions with at least 3 repeats of a GGAA motif were particularly enriched in samples with both ETV6::RUNX1 and loss of the second ETV6 copy. Reanalysis of primary BCP-ALL patients’ chromatin immunoprecipitation (ChIP) and ATAC-seq data confirmed higher accessibility of these regions in ETV6::RUNX1 samples. Importantly, GGAA repeats were not enriched upon ETV6::RUNX1 ChIP-seq in REH and UoCB cell lines, which lack the second ETV6 copy, but were enriched upon ETV6 ChIP-seq in ETV6-intact cells or in ETV6::RUNX1 cells ectopically expressing ETV6, supporting that GGAA repeats are bound by wild-type ETV6 but not by ETV6::RUNX1.
ETV6::RUNX1-activated human genes were located significantly close to the GGAA repeats, supporting their regulatory role. Notably, such proximity was not observed in a mouse genome and because of the poor conservation of GGAA repeat locations during evolution, these data highlight the importance of developing human cell-based models of leukemogenesis. Functional involvement of ETV6 was obtained by the identification of a list of 40 genes showing both binding by ETV6 and a decreased H3K27ac mark upon ectopic ETV6 expression. This list could be used as a classifier to identify ETV6::RUNX1 leukemia samples among other BCP-ALL samples. Then, functional involvement of the GGAA repeats in the regulation of ETV6::RUNX1-activated genes was obtained through CRISPR interference or deletion targeting the GGAA repeats of 6 genes (including the EPOR gene) showing that these repeats positively control their transcription. These data clearly define the associated GGAA repeats as regulatory elements, hence named microsatellite enhancers, that appear otherwise quiescent in normal cells.
Guided by motif analysis and DepMap data (https://depmap.org/portal/) in ETV6::RUNX1 B-ALL, the authors then investigated the role of the other ETS factor ERG in the transcriptional activity of GGAA repeats. ChIP-seq analyses revealed that ERG directly binds more intensely at 6× GGAA repeats in ETV6-deficient cells than in ETV6-intact cells and that CRISPRi-mediated ERG knockdown significantly decreased transcript levels for 36/40 ETV6-controlled genes.
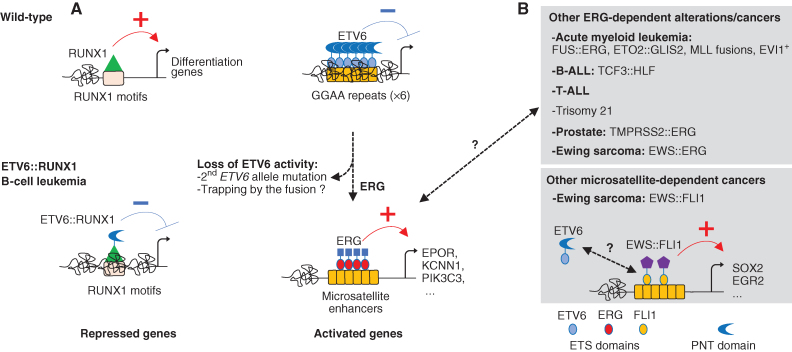
This study provides important novel insights by implicating ETV6 as a safeguard against aberrant activation of GGAA repeats by ERG in acute leukemia (Fig. 1) and by providing some keys for the understanding of how the genomic features/landscape is leading to locus-specific transcriptional control (i.e., number of TF motifs and their organization).
Figure 1.
Relative ETS factor activities control transcriptional gene expression nearby microsatellite enhancers. A, Schematic representation of the transcriptional alterations ETV6::RUNX1 leukemia. Previous studies demonstrated that RUNX1-dependent transcription is repressed by ETV6::RUNX1. Here, Kodgule et al show that upon loss of ETV6, which normal function is to suppress the enhancer activity at GGAA microsatellite repeats, ERG can bind these repeats and subvert them to activate aberrant oncogene transcription. B, Other ERG-dependent alterations/cancers or microsatellite-dependent cancers have been described. The unknown functional contribution of ETV6 activity in these cancers is indicated by a question mark.
Ewing's sarcoma, another ETS fusion oncogene-driven pediatric cancer, has previously been associated with GGAA microsatellite enhancer-dependent aberrant gene expression (6). Ewing's sarcoma generally presents with fusion between EWS, containing a prion-like domain, and one of five members of the ETS family (FLI1, ERG, FEV, ETV1, and E1AF), the EWS::FLI1 fusion representing 85% to 90% of cases. EWS::FLI1 was shown to directly bind GGAA microsatellite enhancers inducing aberrant expression of several oncogenes through the recruitment of the BAF chromatin-remodeling complex. This mechanism is also hijacked by genetic changes as a noncoding genetic susceptibility variant was shown to convert an interspaced GGAT motif into a GGAA motif at the EGR2 locus, resulting in an increased number of consecutive GGAA repeats and a predisposition to develop Ewing's sarcoma (7). Such genetic alterations, either germline or acquired secondarily to ETV6::RUNX1, could theoretically also represent a basis for ETV6::RUNX1 BCP-ALL leukemia predisposition or disease progression. Reciprocally, studying the contribution of ETV6 to the activity of EWS::FLI1-dependent GGAA microsatellite enhancers may reveal a functional relevance for ETV6 in Ewing's sarcoma (Fig. 1). Comparative analyses would also establish whether the GGAA repeats involved in two different cancers originating from different cell types are overlapping or dependent on cell-context specificities.
This study further highlights that the functional balance between ETS factors, revealed by genetic studies in Drosophila (8) involving the closest ETV6-related factor Yan, is relevant for human leukemia. The antagonistic relationship between ETV6 and ERG uncovered in this study raises the question of whether the involvement of GGAA microsatellite repeats is limited to leukemia presenting biallelic targeting of ETV6 or whether this functional antagonism between ETV6 and ERG may in part control the gene-expression program in other leukemia. Indeed, ERG is a known oncogene located on chromosome 21 and presenting functional relevance in several acute myeloid leukemia (AML) either as a direct target (e.g., Trisomy 21, FUS::ERG fusion) or as a dependency in several molecular subgroups (e.g., ETO2::GLIS2, EVI1; Fig. 1). Other ETS fusions, including the TMPRSS2::ERG fusion found in prostate cancer, could behave as the EWS::FLI1 in Ewing's sarcoma. Here, the comparison of ERG binding between human BCP-ALL cells and mouse pre–pro-B cells by Kodgule and colleagues suggests that the ETV6 versus ERG functional antagonism is not observed at evolutionarily conserved ERG binding sites (e.g., MYB gene), supporting that the relative affinity and number of GGAA motifs likely favor these specific microsatellite enhancers. Whether GGAA repeat aberrant activity is involved in other ERG-associated leukemia or other cancers and whether the gene loci controlled by an ETS imbalance depend on the relative expression of ETV6 and ERG imposed by the different oncogenes will require further analyses.
These data raise interesting questions that relate to the interplay between this ETV6 versus ERG imbalance and other cellular or molecular-context specificities and that could highlight mechanisms common to other hematopoietic malignancies or cancers.
As several ETS factors are generally expressed in a cell, do other ETS factors contribute to the relative repression or activation of microsatellite enhancers and how do ETS factors precisely compete for the binding to these regions? Here, ectopic expression of ETV6 in the ETV6:RUNX1 REH cell line allowed ETV6 binding to microsatellite enhancers and repress their transcriptional activity, supporting that ETV6 successfully competes with ERG. The precise mechanism underlying ETV6 replacement by ERG and its reversibility, including stoichiometry requirements, warrants further investigations. As it relates to the involvement of other ETS factors, the authors excluded here the contribution of the FLI1, the closest ERG-related ETS factor, in the activation of ETV6:RUNX1 microsatellites. However, they also report that other ETS factors (e.g., ETV1, ELK3, and SPI1) showed some degree of correlation of expression with ETV6, although to a lower extent than ERG. Intriguingly, a recent report showing an interplay between TET2 status, methylation, and the SPI1 gene regulatory network suggests a relevance for SPI1 in ETV6::RUNX1 leukemia (9).
Also, what is the functional relationship between ETS factors and chromatin regulators or in the control of microsatellite transcriptional activity? It is becoming more and more evident that fusion transcription factors impose constraints that may not be all favorable for transformation. In the context of aggressive forms of Ewing's sarcoma, it was recently shown that the associated STAG2 mutations result in a downmodulation of the EWS::FLI1 transcriptional activity. Therefore, chromatin and chromatin organizers including cohesin/CTCF complexes, which represent mechanisms to fine-tune the transcription activity at important genomic loci, could be of importance for ETV6::RUNX1 BCP-ALL leukemogenesis or in response to treatments. GGAA enhancers–controlled expression of genes located generally less than 50 kb away in the present study and sometimes >400 kb away in Ewing's sarcoma. Together, identifying chromatin compaction and 3D chromatin looping regulators involved would likely help understand the transcriptional output upon microsatellite enhancer activation and define whether a single gene or multiple genes are deregulated in the vicinity of an enhancer.
Finally, how are these ETV6 versus ERG processes also regulated by signaling pathways activated by extrinsic cues? Indeed, ETS factors’ activity was genetically linked to intracellular signaling pathway activation in Drosophila, including the MAPK pathway (8), which was also reported to control ETS factor phosphorylation and protein conformation. In addition, SRC-mediated intracellular signaling regulates the nucleo-cytoplasmic localization of ETV6 (10). Here, Kodgule and colleagues showed that the high EPOR expression associated with ETV6::RUNX1 B-ALL cells results from the activation of a nearby microsatellite enhancer that also controlled responsiveness to EPO stimulation. Together, whether EPOR through the MAPK pathway, or other signaling pathways, may control the relative activity of ERG and ETV6 at microsatellite satellite enhancers and contribute to early-stage disease development should be another interesting area of investigation.
Acknowledgments
The author thanks Dr. Olivier A. Bernard for suggestions and apologizes for not citing some important related studies due to size constraints.
Authors’ Disclosures
No disclosures were reported.
References
- 1. Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer 2017;17:337–51. [DOI] [PubMed] [Google Scholar]
- 2. Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol 2005;15:162–74. [DOI] [PubMed] [Google Scholar]
- 3. Noetzli L, Lo RW, Lee-Sherick AB, Callaghan M, Noris P, Savoia A, et al. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat Genet 2015;47:535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zelent A, Greaves M, Enver T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene 2004;23:4275–83. [DOI] [PubMed] [Google Scholar]
- 5. Kodgule R, Goldman J, Monovich A, Saari T, Aguilar A, Hall C, et al. ETV6 deficiency unlocks ERG-dependent microsatellite enhancers to drive aberrant gene activation in B-lymphoblastic leukemia. Blood Cancer Discov 2023;4:34–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Riggi N, Suvà ML, Stamenkovic I. Ewing's sarcoma. N Engl J Med 2021;384:154–64. [DOI] [PubMed] [Google Scholar]
- 7. Grünewald TGP, Bernard V, Gilardi-Hebenstreit P, Raynal V, Surdez D, Aynaud M-M, et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet 2015;47:1073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 1994;78:137–47. [DOI] [PubMed] [Google Scholar]
- 9. Aivalioti MM, Bartholdy BA, Pradhan K, Bhagat TD, Zintiridou A, Jeong JJ, et al. PU.1-dependent enhancer inhibition separates Tet2-deficient hematopoiesis from malignant transformation. Blood Cancer Discov 2022;3:444–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lopez RG, Carron C, Ghysdael J. v-SRC specifically regulates the nucleo-cytoplasmic delocalization of the major isoform of TEL (ETV6). J Biol Chem 2003;278:41316–25. [DOI] [PubMed] [Google Scholar]