Abstract
The control of gene expression in eukaryotes relies on how transcription factors and RNA polymerases manipulate the structure of chromatin. These interactions are especially important in development as gene expression programs change. Chromatin generally limits the accessibility of DNA, and thus exposing sequences at regulatory elements is critical for gene expression. However, it is challenging to understand how transcription factors manipulate chromatin structure and the sequence of regulatory events. The Drosophila embryo has provided a powerful setting to directly observe the establishment and elaboration of chromatin features and experimentally test the causality of transcriptional events that are shared among many metazoans. The large embryo is tractable by live imaging, and a variety of well-developed tools allow the manipulation of factors during early development. The early embryo develops as a syncytium with rapid nuclear divisions and no zygotic transcription, with largely featureless chromatin. Thus, studies in this system have revealed the progression of genome activation triggered by pioneer factors that initiate DNA exposure at regulatory elements and the establishment of chromatin domains, including heterochromatin, the nucleolus, and nuclear bodies. The de novo emergence of nuclear structures in the early embryo reveals features of chromatin dynamics that are likely to be central to transcriptional regulation in all cells.
Keywords: Chromatin, Pioneer factors, Gene regulation
Introduction
Development in multicellular organisms relies on differential gene expression to direct cell proliferation and differentiation. The identification of conserved transcription factors in genetic screens for embryonic defects in Drosophila in the 1980s reformulated our concept of development1,2. Later studies in other organisms revealed that the genetic control of development is conserved across eukaryotes, with common regulatory principles. A key issue is how transcription factors interact with the chromatin packaging of the genome for differential gene expression, even though the genome in different cell types is identical. Just as exceptional features of Drosophila embryogenesis provided a productive setting for identifying developmental regulators, studies in early embryos are now unraveling long-standing issues of fundamental mechanisms of the chromatin control of transcription.
The onset of chromatin organization in Drosophila embryos
The challenge in understanding transcriptional mechanisms is ordering molecular events and dependencies in living cells. Chromatin features have been correlated to gene expression and repression in many cell types, but it is more difficult to distinguish which features cause others and which actually regulate gene activity. Features of Drosophila embryogenesis are now being exploited to address this gap.
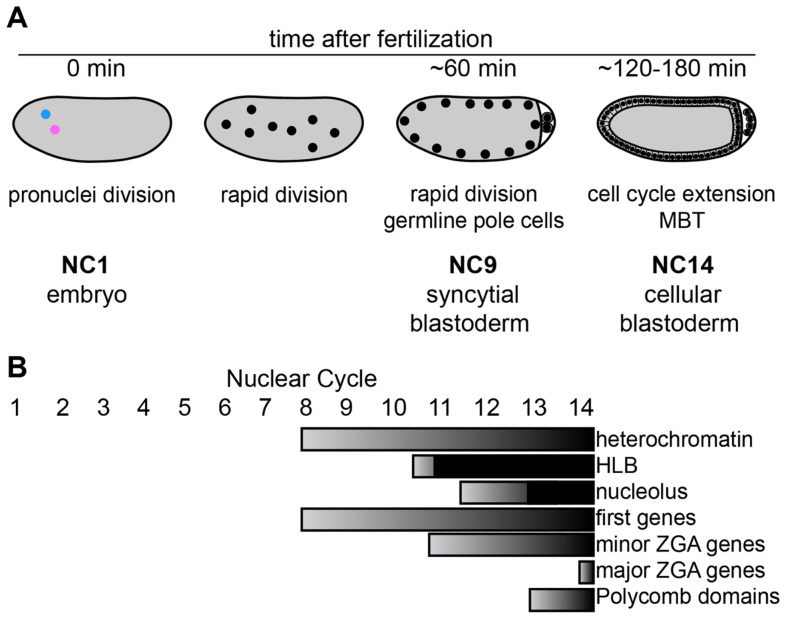
Like most animals, Drosophila females make a large egg that is loaded with proteins and untranslated mRNA. Fertilization by an incoming sperm triggers embryogenesis. A broad outline of chromatin changes in early embryogenesis is shown in Figure 1, where stages are labeled by the number of nuclear cycles (nc) since fertilization. The early stages of embryogenesis rely exclusively on maternally provided material, without any zygotic gene expression. The incoming sperm genome arrives packaged in protamines, which are removed and replaced with the H3.3 histone variant3. In its first hours, the embryo develops as a multinucleate syncytium with totipotent nuclei. Proliferative nuclear cycles occur synchronously and extremely fast, doubling nuclei every 10 minutes, so DNA replication, chromatin assembly, and mitosis occur at what must be close to maximal rates. These first rounds of nuclear replication and division use maternally supplied core histones and a variant histone H1 called bigH14 to duplicate chromatin. The rapidity of these divisions requires nearly simultaneous replication of the entire genome, with replication origins scattered at random roughly every 10 kb5. The speed of early divisions means that syncytial nuclei largely lack any of the localized chromatin features that characterize epigenomes in other cell types, but distinctions appear as nuclear cycles slow down, zygotic transcription begins, and the nuclei are partitioned to form the cellular blastoderm. In this way, the transition from the syncytial to the cellular embryo provides a tabula rasa for observing the onset and order of transcriptional programs and epigenomic specialization.
Figure 1. Chromatin events in early Drosophila embryos.
(A) Outline of the early stages of Drosophila embryogenesis and proliferation. Nuclei are shown in black within the grey cytosol of the embryo. Stages are labeled as the number of nuclear cycles (nc) after fertilization. In nc1, protamines are removed from the male haploid pronucleus (blue) and repackaged with the H3.3 histone variant before DNA replication and mitosis. Fusion of the male and female pronuclei occurs in nc2. The first nuclear cycles up to nc13 occur in a syncytium with rapid progression through the S phase and mitosis without gap phases. (B) The timing of the appearance of chromatin features, including heterochromatin, Polycomb domains, and the histone locus bodies (HLBs), is shown. About 10 genes first activate in nc8, hundreds of genes activate in the minor zygotic genome activation (ZGA) around nc11, and thousands of genes activate in the major ZGA around nc14.
Zygotic transcription of a few genes begins around nc8, reaching a notable level of transcription from about 100 sex determination, cellularization, and early patterning genes around nc11, a time referred to as the minor zygotic genome activation (minor ZGA)6,7. Widespread transcription of thousands of genes begins at nc14, referred to as the major ZGA, during the switch from maternal to zygotic control of morphogenesis. But how does zygotic transcription begin in the early embryo, and how does this alter the chromatin landscape?
Active promoters and enhancers in eukaryotes are characterized by a nucleosome-depleted region (NDR) where transcription factors bind. A small number of transcription-independent NDRs are detected in the nuclei of early syncytial embryos, probably due to intrinsic anti-nucleosomal sequence features8. At this time, developmentally regulated genes lack any nucleosome phasing around promoters or regulatory elements, but NDRs appear when transcription begins. At least three transcription factors have been implicated in driving zygotic transcriptional activation. The zinc-finger protein Zelda (Zld) is first translated beginning around nc8 from maternal mRNA and binds at thousands of sites9–11. These early sites include the promoters and enhancers of genes activated in the minor ZGA but also many regulatory elements of genes that will not be activated until the major ZGA. A second transcription factor implicated in early embryo transcription is the transcription factor GAGA factor (GAF), whose binding first appears in nc912. A second GAGA motif-binding protein called CLAMP is involved in the activation of some major ZGA genes13. These key transcription factors also cooperate with spatially localized transcription factors to establish the body plan of the embryo (for examples, see 14–18).
Pioneer factors… how do they work?
How do Zld and GAF bind chromatin in early nuclei? In many developmental systems, pioneer transcription factors are thought to convert inaccessible chromatin at binding sites into exposed DNA and thereby trigger new programs of gene expression19. For example, the mammalian pluripotency factors Sox2 and Oct4 bind DNA wrapped on the surface of a nucleosome and either shift its position or displace histones, thereby creating an NDR; exposure of this DNA then allows binding of secondary transcription factors. Thus, creating an NDR at binding sites is a key step for activating developmentally regulated enhancers and promoters, underlying the ability of these factors to reprogram cells from diverse types. These properties were first identified for the archetypal pioneer factor FoxA1, which initiates endoderm development19. Recent structural studies of pioneer factors docked onto nucleosomes in vitro have fleshed out factors that may bind specific DNA motifs even when wrapped around a histone octamer and how the binding of one factor may enhance the binding of a second20. However, the mechanism by which pioneer factors first bind sites in vivo has been more difficult to pin down. For example, a large fraction of binding sites for the FoxA1 factor depends on the cell type where it is expressed, implying that other bound transcription factors influence where FoxA1 can bind21,22. Close examination of FoxA1/2-binding sites in differentiating endoderm cells showed that these sites are partially accessible even before these factors are produced23,24. This makes it difficult to determine the relationship between site accessibility and pioneer factor activity, especially as the relevant cell stages in the early embryo cannot be easily observed.
Since the Drosophila embryo starts with a largely featureless epigenome, it provides insight into the chromatin sequence of pioneering. The early factors Zld, GAF, and CLAMP are pioneer factors in that each can bind nucleosomal DNA in vitro13,25,26. The creative use of mutations, engineered constructs, or microinjection into the syncytial embryo is deciphering the molecular activities of these pioneer factors (Box 1). Depletion of Zld, CLAMP, or GAF from early embryos has identified sets of binding sites where new accessibility depends exclusively on only one factor9,12,13,18,27,28 and other sets that require a combination of Zld and CLAMP or of Zld and GAF. Thus, at some sites, a single factor suffices to drive accessibility; at others, multiple transcription factors cooperate. This resembles the interdependency of pioneer factors in mammalian systems that allow for context-specific binding.
Box 1. Systems to manipulate chromatin components in the early Drosophila embryo.
In the Drosophila embryo, the first 13 nuclear divisions occur synchronously in a large syncytium with a shared cytoplasm and are accessible for both observing and manipulating chromatin. A wide range of methods and techniques have been used in the studies discussed here. Embryos can be precisely staged by staining fixed material with DNA dyes, and detailed pictures of gene activation and the formation of nuclear bodies have been obtained using antibody staining and DNA in situ hybridization combined with high-resolution microscopy. Other well-developed methods use maternal genotypes that load the egg with fluorescently-labeled histones, chromatin proteins, or transcription factors for live imaging during embryogenesis. Fluorescent protein fusions with transcription factors and RNA polymerase II (RNAPII) subunits have been tracked in this way, and nascent transcripts have been detected with fluorescent MS2 coat protein (MCP) fusions that bind engineered hairpins in mRNAs29. The embryonic syncytium can be manually injected without adversely affecting development, and studies have injected mRNA for fluorescent protein fusions with DNA-binding proteins to image specific DNA sequences or fluorescently-labeled antibody fragments to image specific histone modifications.
Genetics and injection provide strategies to test function in the early embryo. Compounds that interfere with transcription, DNA replication, or chromatin remodeling can be injected. Since the early embryo relies entirely on maternally provided products until the beginning of the cellular blastoderm stage, many components can be altered by modifying the genotype of the mother. A range of genetic strategies are also available. For factors that are required only in the embryo, mutant mothers will produce eggs that lack a specific factor. In other cases, an interesting factor may also be important in other stages of development. Here, mitotic recombination schemes have been designed that produce homozygous mutant germline clones from heterozygous mothers, thereby giving eggs that lack the factor. Finally, for factors that are essential in the female germline, double-stranded RNA targeting a specific factor can be loaded into the egg, and this can accomplish knockdown in the embryo30. More sophisticated strategies are now applied in the embryo using inducible degradation components maternally loaded into the egg31, or optogenetic inactivation25,32 to trigger the elimination of factors during early embryogenesis. One particular method inspired by ‘anchors away’ technology in budding yeast uses the Drosophila Jabba protein to rapidly deplete GFP-labeled factors in the early embryo. Jabba normally localizes to lipid droplets in the early embryo, but a Jabba-anti-GFP nanobody fusion can sequester any target proteins outside of nuclei. Maternally loaded or injected mRNA encoding Jabba-anti-GFP nanobody has been used to acutely deplete histone methyltransferase proteins33, Zld, and Mxc34.
Live imaging has been used to follow the binding of Zld and transcription in the nuclei of early embryos. While earlier studies had shown that DNA replication initiates immediately after mitosis, and transcription slightly later, a recent preprint has detailed the link between replication and transcriptional initiation34. Zld accumulates in small foci in nuclei, and clusters of RNA polymerase II (RNAPII) and nascent gene transcripts appear adjacent to Zld foci17,35. These authors used an innovative sequestration method to rapidly deplete Zld from the embryo. This system uses an anti-GFP nanobody fused to the Jabba protein, which normally binds and sequesters target proteins in cytosolic lipid droplets in the embryo. Expression of this “JabbaTrap” fusion in embryos with a target GFP fusion protein thus sequesters the target away from chromatin33. Injection of JabbaTrap mRNA into embryos with a Zld-GFP fusion showed that Zld is required for the formation of RNAPII clusters. Intriguingly, blocking DNA replication with multiple different inhibitors greatly reduces the formation of Zld clusters and of RNAPII clusters. Clusters that do form are delayed and smaller. Thus, DNA replication appears to enhance the activity of Zld in early embryos.
New nucleosomes behind DNA replication forks are assembled with acetylated H3 and H4 histones, and so while these modifications are enriched in early embryonic nuclei, most other histone modifications associated with transcriptional regulation are absent36. However, when Zld binds, nucleosomes around these sites acquire histone H3K27 acetylation8. CBP is the main H3K27 acetyltransferase, and eliminating the Drosophila homolog Nejire (Nej) in early embryos blocks RNAPII clustering and transcription34. This argues for a sequence of events as follows: first, DNA replication enhances the binding of Zld and clustering of RNAPII on chromatin; second, Zld stimulates histone acetylation by Nej; and, third, acetylation promotes RNAPII elongation34. But why would DNA replication enhance the activity of a nucleosome-binding pioneer factor? Perhaps the binding of Zld is enhanced by cell cycle-coupled protein modifications of the factor. Alternatively, early models for chromatin regulation in development proposed that the chromatin dynamics during DNA replication may provide a “window of opportunity” to bind transcription factors and start new transcriptional programs37. Nucleosome density and positioning are transiently reduced behind replication forks38. For some transcription factors, disorganized nucleosome positioning may delay binding, but others (like GAF) rapidly bind newly replicated DNA and position nucleosomes. Evidence that most Zld and RNAPII signals in early Drosophila embryos depend on DNA replication supports the idea of fast binding behind the replication fork. Importantly, a small fraction of Zld shows delayed binding in blocked embryos, and this might be due to the nucleosomal-binding activity of Zld. While this division between replication-dependent and nucleosomal-binding modes of Zld binding may be more extreme in the fast cycles of the early embryo, DNA replication may promote new transcriptional programming more generally.
The fast nuclear division cycle of early embryos has also revealed factor-specific distinctions for mitotic inheritance. While many transcription factors dissociate as mitotic chromatin condenses, a long-standing question in mammalian systems has been whether some factors persist in maintaining stable patterns of gene expression in cell lineages39. Indeed, both properties appear in the early Drosophila embryo. Live imaging of Zld protein shows that the factor releases from chromatin as mitosis initiates40, and the chromatin accessibility of Zld-bound sites is reduced as nuclei divide28. In contrast, GAF binds both interphase and mitotic chromatin, and mitotic GAF-bound sites remain accessible41. This difference appears to result from the longer residence time of GAF on chromatin that allows persistent binding to mitotic chromosomes. Thus, GAF fits the definition of a mitotic bookmarking transcription factor and appears to be functionally important, as imaging nascent transcription of a target promoter has shown GAF-dependent mitotically heritable expression41.
There are additional transcription factors that activate genes in the major ZGA42–44. These can be understood in the framework that pioneering activity is a matter of degree, as transcription factors vary in affinity from site to site and cooperatively interact with other chromatin proteins45. However, one distinct class comprises about 2,000 promoters for housekeeping genes that are activated in the major ZGA but lack binding by any known pioneer factors. Instead, these promoters are enriched for the H2AV histone variant (the Drosophila ortholog of H2A.Z and H2A.X variants)46. H2AV is deposited at these promoters well before these genes are zygotically expressed by a conserved SWR1-type chromatin remodeler called Domino. While the elimination of Domino from the early embryo prevents H2AV accumulation and gene activation, chromatin accessibility is not altered. It remains to be determined whether these sites are targeted by pioneering core promoter factors or whether intrinsic nucleosome-positioning features of their DNA sequences make them accessible.
Accretion of nuclear organization
The de novo establishment of nuclear organization in the early embryo includes the appearance of nuclear bodies and distinctive chromatin regions, including the histone locus body (HLB), the nucleolus, and heterochromatin. Observation of how these features appear reveals the steps in establishing phase-separated compartments within the nucleus. The HLB first appears in nc11 when the histone genes are transcribed from a gene array on chromosome 2, but DNA-binding factors — including the CLAMP pioneer factor and the NPAT homolog Mxc — first arrive at the histone genes one cycle earlier in nc1047. This nuclear body appears to be anchored by a DNA-bound seed, while processing factors may localize into the HLB through interactions with histone mRNA48. A similar sequence drives the formation of the nucleolus, where transcription of the rDNA genes from arrays on the X and Y chromosomes begins in nc11, and processing factors appear in nc1349 (Figure 1). These studies have led to a model where the early nucleus is supersaturated for nuclear body components, and factor binding followed by transcription triggers reliable formation of structured bodies.
Observations in the early embryo have also detailed the temporal relationship between histone modifications and gene regulation. Chromatin in the early embryo is globally hyperacetylated because of replication-coupled nucleosome assembly, but active modifications, such as H3K4 and H3K36 methylations, first appear after gene transcription is initiated in the major ZGA36. Markers of Polycomb gene silencing, such as H3K27 methylation and Polycomb complexes, first accumulate in nc148,50 (although a conflicting report described localized H3K27 methylation at all cycles of the early embryo51). In Drosophila, regulatory elements called Polycomb Response Elements (PREs) are required for gene silencing, and PREs first become accessible in nc1152. Many PREs are binding sites for GAF, so the general mechanism of pioneering activity initiating zygotic gene expression also applies to the establishment of developmental gene silencing.
The establishment of heterochromatin in the embryo may operate by a different mechanism. In most cell types, repetitive transposons and short satellite repeats replicate very late in the S phase and become wrapped in nucleosomes marked by histone H3K9 methylation, comprising a repressive heterochromatic state. In the fast cycles of the early embryo, all sequences replicate at the same time, but repetitive sequence blocks begin to show heterochromatic properties at different times in later stages. Some satellite blocks become compacted in nc8 and gain H3K9 methylation in nc12, which then accumulates to high levels in successive cycles as interphases get longer33. More widespread heterochromatic features, including delayed replication and then HP1 binding, are apparent in nc13 and nc1453,54. Intriguingly, the embryonic establishment of heterochromatin on a specific satellite block depends on its maternal presence, suggesting that maternally-loaded RNAs from this satellite are required to establish heterochromatin in the embryo55. Similarly, maternal PIWI-interacting RNAs (piRNAs) may work on other repetitive sequences in the nucleus56. These dependencies may be distinct from the activities that maintain mature heterochromatin in later cell types.
The power of observation in the early embryo is now being applied to deciphering how the 3D organization of the nucleus is established. Chromatin in the nucleus displays a hierarchy of interactions, where loops between regulatory elements organize genes within topologically associated domains (TADs), which are further aggregated into inactive and active compartments57. This organization is thought to result from both cohesin binding that constrains the chromatin fiber and from non-specific associations between decondensed transcribed chromatin and inactive chromatin. As the early Drosophila embryo is transcriptionally silent, this stage provides a natural setting to track the acquisition of nuclear organization. Mapping of 3D organization in Drosophila embryos revealed that only a few boundaries are present in very early embryos, and these coincide with constitutive H2AZ-containing promoters46. Additional TADs and compartments appear in nc8 when Zld binds to chromatin58,59. This order of events demonstrates that factor binding alone can create domains and that transcription is not needed. Factor binding at TAD boundaries and at looping elements creates a framework of invariant cell type-independent domains in embryos, but only a subset of these interactions are productive in specific cell types59–61. Direct imaging of individual nuclei in the Drosophila embryo has further refined our understanding of nuclear organization. While contact maps have led to the impression of extensive 3D organization within nuclei, tracing chromatin paths62–64 or imaging the juxtaposition of transcribing genes35 shows that 3D organization in individual nuclei is very diverse, with structures detected by contact mapping being rare and transient. This places important limits on the functional significance of higher-order structures for gene expression, where 3D interactions appear to be a pre-condition for gene activation but do not determine it.
Conclusions
Whereas the core questions in chromatin biology were posed many years ago, the intricacy of many eukaryotic systems has complexified the answers. The blank slate of chromatin in the early embryo has provided a simple setting for probing the mechanics of how transcription factors first interact with chromatin, how phase-separated bodies within the nucleus are first established, and how 3D nuclear organization comes to be. The Drosophila embryo has revealed that the binding of a small number of key DNA-binding proteins can trigger the organization of much of the nucleus, which successive gene activity elaborates. The sequence by which nuclear features emerge implies that they are consequences of “sticky” weak electrostatic interactions. These issues even apply to universally accepted concepts, such as how enhancers act on promoters to activate transcription. While textbooks describe that promoters and enhancers loop together in 3D space to form an activating structure, such contacts are rare and transient in direct imaging. How can transient contacts promote the slower process of transcriptional activation? One class of models suggests that an enhancer leaves a long-lasting mark on promoters, perhaps as histone modifications or stabilizing bound factors, and the persistence of these effects at promoters stimulates transcription62. More radical models propose that enhancers produce diffusible molecules that move across nuclear distance to activate promoters65. The ability to resolve the sequence of nuclear events in time and space in the early embryo holds promise in understanding these fundamental issues in gene regulation.
Acknowledgments
We thank Nadiya Khyzha (Fred Hutchinson Cancer Center, Seattle WA) for comments on the manuscript.
The peer reviewers who approve this article are:
Alistair Boettiger, Department of Developmental Biology, Stanford University, Stanford, CA, USA
Melissa Harrison, Department of Biomolecular Chemistry, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI, USA
Christine Rushlow, Department of Biology, New York University, New York, NY, USA
Funding Statement
The authors declare that no grants were involved in supporting this work.
References
- 1. Zeitlinger J, Stark A: Developmental gene regulation in the era of genomics. Dev Biol. 2010; 339(2): 230–9. 10.1016/j.ydbio.2009.12.039 [DOI] [PubMed] [Google Scholar]
- 2. Nüsslein-Volhard C, Wieschaus E: Mutations affecting segment number and polarity in Drosophila. Nature. 1980; 287(5785): 795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- 3. Loppin B, Bonnefoy E, Anselme C, et al. : The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005; 437(7063): 1386–90. 10.1038/nature04059 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 4. Pérez-Montero S, Carbonell A, Morán T, et al. : The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev Cell. 2013; 26(6): 578–90. 10.1016/j.devcel.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 5. Blumenthal AB, Kriegstein HJ, Hogness DS: The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974; 38: 205–23. 10.1101/sqb.1974.038.01.024 [DOI] [PubMed] [Google Scholar]
- 6. Kwasnieski JC, Orr-Weaver TL, Bartel DP: Early genome activation in Drosophila is extensive with an initial tendency for aborted transcripts and retained introns. Genome Res. 2019; 29(7): 1188–97. 10.1101/gr.242164.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lott SE, Villalta JE, Schroth GP, et al. : Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011; 9(2): e1000590. 10.1371/journal.pbio.1000590 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 8. Li XY, Harrison MM, Villalta JE, et al. : Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife. 2014; 3: e03737. 10.7554/eLife.03737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harrison MM, Li XY, Kaplan T, et al. : Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011; 7(10): e1002266. 10.1371/journal.pgen.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 10. Nien CY, Liang HL, Butcher S, et al. : Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011; 7(10): e1002339. 10.1371/journal.pgen.1002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larson ED, Komori H, Gibson TJ, et al. : Cell-type-specific chromatin occupancy by the pioneer factor Zelda drives key developmental transitions in Drosophila. Nat Commun. 2021; 12(1): 7153. 10.1038/s41467-021-27506-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaskill MM, Gibson TJ, Larson ED, et al. : GAF is essential for zygotic genome activation and chromatin accessibility in the early Drosophila embryo. eLife. 2021; 10: e66668. 10.7554/eLife.66668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duan J, Rieder L, Colonnetta MM, et al. : CLAMP and Zelda function together to promote Drosophila zygotic genome activation. eLife. 2021; 10: e69937. 10.7554/eLife.69937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamada S, Whitney PH, Huang SK, et al. : The Drosophila Pioneer Factor Zelda Modulates the Nuclear Microenvironment of a Dorsal Target Enhancer to Potentiate Transcriptional Output. Curr Biol. 2019; 29(8): 1387–1393.e5. 10.1016/j.cub.2019.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yáñez-Cuna JO, Dinh HQ, Kvon EZ, et al. : Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 2012; 22(10): 2018–30. 10.1101/gr.132811.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu Z, Chen H, Ling J, et al. : Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 2014; 28(6): 608–21. 10.1101/gad.234534.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mir M, Stadler MR, Ortiz SA, et al. : Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife. 2018; 7: e40497. 10.7554/eLife.40497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun Y, Nien CY, Chen K, et al. : Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 2015; 25(11): 1703–14. 10.1101/gr.192542.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iwafuchi-Doi M, Zaret KS: Cell fate control by pioneer transcription factors. Development. 2016; 143(11): 1833–7. 10.1242/dev.133900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Balsalobre A, Drouin J: Pioneer factors as master regulators of the epigenome and cell fate. Nat Rev Mol Cell Biol. 2022; 23(7): 449–64. 10.1038/s41580-022-00464-z [DOI] [PubMed] [Google Scholar]
- 21. Lupien M, Eeckhoute J, Meyer CA, et al. : FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008; 132(6): 958–70. 10.1016/j.cell.2008.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 22. Larson ED, Marsh AJ, Harrison MM: Pioneering the developmental frontier. Mol Cell. 2021; 81(8): 1640–1650. 10.1016/j.molcel.2021.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meers MP, Janssens DH, Henikoff S: Pioneer Factor-Nucleosome Binding Events during Differentiation Are Motif Encoded. Mol Cell. 2019; 75(3): 562–575.e5. 10.1016/j.molcel.2019.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cernilogar FM, Hasenöder S, Wang Z, et al. : Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 2019; 47(17): 9069–9086. 10.1093/nar/gkz627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDaniel SL, Gibson TJ, Schulz KN, et al. : Continued Activity of the Pioneer Factor Zelda Is Required to Drive Zygotic Genome Activation. Mol Cell. 2019; 74(1): 185–195.e4. 10.1016/j.molcel.2019.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 26. Tsukiyama T, Becker PB, Wu C: ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994; 367(6463): 525–32. 10.1038/367525a0 [DOI] [PubMed] [Google Scholar]
- 27. Schulz KN, Bondra ER, Moshe A, et al. : Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 2015; 25(11): 1715–26. 10.1101/gr.192682.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blythe SA, Wieschaus EF: Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. eLife. 2016; 5: e20148. 10.7554/eLife.20148 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 29. Garcia HG, Tikhonov M, Lin A, et al. : Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol. 2013; 23(21): 2140–5. 10.1016/j.cub.2013.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Staller MV, Yan D, Randklev S, et al. : Depleting gene activities in early Drosophila embryos with the "maternal-Gal4-shRNA" system. Genetics. 2013; 193(1): 51–61. 10.1534/genetics.112.144915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caussinus E, Kanca O, Affolter M: Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol. 2011; 19(1): 117–21. 10.1038/nsmb.2180 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 32. Huang A, Amourda C, Zhang S, et al. : Decoding temporal interpretation of the morphogen Bicoid in the early Drosophila embryo. eLife. 2017; 6: e26258. 10.7554/eLife.26258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seller CA, Cho CY, O'Farrell PH: Rapid embryonic cell cycles defer the establishment of heterochromatin by Eggless/SetDB1 in Drosophila. Genes Dev. 2019; 33(7–8): 403–417. 10.1101/gad.321646.118 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 34. Cho CY, Kemp JP, Jr, Duronio RJ, et al. : Mandatory coupling of zygotic transcription to DNA replication in early Drosophila embryos. bioRxiv. 2022. 10.1101/2022.01.04.474810 [DOI] [Google Scholar]
- 35. Huang SK, Whitney PH, Dutta S, et al. : Spatial organization of transcribing loci during early genome activation in Drosophila. Curr Biol. 2021; 31(22): 5102–5110.e5. 10.1016/j.cub.2021.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 36. Rudolph T, Yonezawa M, Lein S, et al. : Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007; 26(1): 103–15. 10.1016/j.molcel.2007.02.025 [DOI] [PubMed] [Google Scholar]
- 37. Wolffe AP: Implications of DNA replication for eukaryotic gene expression. J Cell Sci. 1991; 99(Pt 2): 201–6. 10.1242/jcs.99.2.201 [DOI] [PubMed] [Google Scholar]
- 38. Ramachandran S, Henikoff S: Transcriptional Regulators Compete with Nucleosomes Post-replication. Cell. 2016; 165(3): 580–92. 10.1016/j.cell.2016.02.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zaidi SK, Young DW, Montecino MA, et al. : Mitotic bookmarking of genes: A novel dimension to epigenetic control. Nat Rev Genet. 2010; 11(8): 583–9. 10.1038/nrg2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dufourt J, Trullo A, Hunter J, et al. : Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat Commun. 2018; 9(1): 5194. 10.1038/s41467-018-07613-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bellec M, Dufourt J, Hunt G, et al. : The control of transcriptional memory by stable mitotic bookmarking. Nat Commun. 2022; 13(1): 1176. 10.1038/s41467-022-28855-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koromila T, Gao F, Iwasaki Y, et al. : Odd-paired is a pioneer-like factor that coordinates with Zelda to control gene expression in embryos. eLife. 2020; 9: e59610. 10.7554/eLife.59610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soluri IV, Zumerling LM, Payan Parra OA, et al. : Zygotic pioneer factor activity of Odd-paired/Zic is necessary for late function of the Drosophila segmentation network. eLife. 2020; 9: e53916. 10.7554/eLife.53916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hannon CE, Blythe SA, Wieschaus EF: Concentration dependent chromatin states induced by the bicoid morphogen gradient. eLife. 2017; 6: e28275. 10.7554/eLife.28275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hansen JL, Loell KJ, Cohen BA: A test of the pioneer factor hypothesis using ectopic liver gene activation. eLife. 2022; 11: e73358. 10.7554/eLife.73358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ibarra-Morales D, Rauer M, Quarato P, et al. : Histone variant H2A.Z regulates zygotic genome activation. Nat Commun. 2021; 12(1): 7002. 10.1038/s41467-021-27125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. White AE, Burch BD, Yang XC, et al. : Drosophila histone locus bodies form by hierarchical recruitment of components. J Cell Biol. 2011; 193(4): 677–94. 10.1083/jcb.201012077 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 48. Hur W, Kemp JP, Jr, Tarzia M, et al. : CDK-Regulated Phase Separation Seeded by Histone Genes Ensures Precise Growth and Function of Histone Locus Bodies. Dev Cell. 2020; 54(3): 379–394.e6. 10.1016/j.devcel.2020.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 49. Falahati H, Pelham-Webb B, Blythe S, et al. : Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr Biol. 2016; 26(3): 277–85. 10.1016/j.cub.2015.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 50. Chen K, Johnston J, Shao W, et al. : A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife. 2013; 2: e00861. 10.7554/eLife.00861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zenk F, Loeser E, Schiavo R, et al. : Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science. 2017; 357(6347): 212–216. 10.1126/science.aam5339 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 52. Alhaj Abed J, Ghotbi E, Ye P, et al. : De novo recruitment of Polycomb-group proteins in Drosophila embryos. Development. 2018; 145(23): dev165027. 10.1242/dev.165027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shermoen AW, McCleland ML, O'Farrell PH: Developmental control of late replication and S phase length. Curr Biol. 2010; 20(23): 2067–77. 10.1016/j.cub.2010.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 54. Yuan K, Shermoen AW, O'Farrell PH: Illuminating DNA replication during Drosophila development using TALE-lights. Curr Biol. 2014; 24(4): R144–5. 10.1016/j.cub.2014.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yuan K, O'Farrell PH: TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev. 2016; 30(5): 579–93. 10.1101/gad.272237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wei KHC, Chan C, Bachtrog D: Establishment of H3K9me3-dependent heterochromatin during embryogenesis in Drosophila miranda. eLife. 2021; 10: e55612. 10.7554/eLife.55612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rowley MJ, Corces VG: Organizational principles of 3D genome architecture. Nat Rev Genet. 2018; 19(12): 789–800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 58. Hug CB, Grimaldi AG, Kruse K, et al. : Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell. 2017; 169(2): 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- 59. Espinola SM, Götz M, Bellec M, et al. : Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat Genet. 2021; 53(4): 477–86. 10.1038/s41588-021-00816-z [DOI] [PubMed] [Google Scholar]
- 60. Ing-Simmons E, Vaid R, Bing XY, et al. : Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nat Genet. 2021; 53(4): 487–99. 10.1038/s41588-021-00799-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Batut PJ, Bing XY, Sisco Z, et al. : Genome organization controls transcriptional dynamics during development. Science. 2022; 375(6580): 566–70. 10.1126/science.abi7178 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 62. Xiao JY, Hafner A, Boettiger AN: How subtle changes in 3D structure can create large changes in transcription. eLife. 2021; 10: e64320. 10.7554/eLife.64320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mateo LJ, Murphy SE, Hafner A, et al. : Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature. 2019; 568(7750): 49–54. 10.1038/s41586-019-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 64. Cardozo Gizzi AM, Cattoni DI, Fiche JB, et al. : Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Mol Cell. 2019; 74(1): 212–222.e5. 10.1016/j.molcel.2019.01.011 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 65. Karr JP, Ferrie JJ, Tjian R, et al. : The transcription factor activity gradient (TAG) model: Contemplating a contact-independent mechanism for enhancer-promoter communication. Genes Dev. 2022; 36(1–2): 7–16. 10.1101/gad.349160.121 [DOI] [PMC free article] [PubMed] [Google Scholar]