Graphical abstract

Abbreviations: tPOD, Transcriptomics-based point of departure; BMD, Benchmark dose; RNAseq, RNA sequencing; DEGs, Differentially expressed genes; ToxCast, US EPA Toxicity Forecaster; eco-HTTr, Ecological high throughput transcriptomics; HTTr, High throughput transcriptomics; SSRI, Selective serotonin reuptake inhibitor; RIN, RNA integrity number; cDNA, Complementary DNA
Keywords: Computational toxicology, Benchmark dose, ECOTOX knowledgebase, Fish, RNA sequencing, Transcriptomics-based point of departure
Highlights
-
•
Next generation tiered testing framework includes high throughput transcriptomics.
-
•
Developed a high throughput transcriptomics assay with larval fathead minnows.
-
•
Approach was piloted with 10 chemicals; points of departure (PODs) calculated.
-
•
Transcriptomic PODs were <traditional PODs -therefore protective.
-
•
In silico analyses were employed to estimate variability and optimize assay design.
Abstract
Concentrations at which global gene expression profiles in cells or animals exposed to a test substance start to differ significantly from those of controls have been proposed as an alternative point of departure for use in screening level hazard assessment. The present study describes pilot testing of a high throughput compatible transcriptomics assay with larval fathead minnows. One day post hatch fathead minnows were exposed to eleven different concentrations of three metals, three selective serotonin reuptake inhibitors, and four neonicotinoid-like compounds for 24 h and concentration response modeling was applied to whole body gene expression data. Transcriptomics-based points of departure (tPODs) were consistently lower than effect concentrations reported in apical endpoint studies in fish. However, larval fathead minnow-based tPODs were not always lower than concentrations reported to elicit apical toxicity in other aquatic organisms like crustaceans or insects. Random in silico subsampling of data from the pilot assays was used to evaluate various assay design and acceptance considerations such as transcriptome coverage, number of replicate individuals to sequence per treatment, and minimum number of differentially expressed genes to produce a reliable tPOD estimate. Results showed a strong association between the total number of genes for which a concentration response relationship could be derived and the overall variability in the resulting tPOD estimates. We conclude that, for our current assay design and analysis pipeline, tPODs based on fewer than 15 differentially expressed genes are likely to be unreliable for screening and that interindividual variability in gene expression profiles appears to be a more significant driver of tPOD variability than sample size alone. Results represent initial steps toward developing high throughput transcriptomics assays for use in ecological hazard screening.
1. Introduction
In the effort to protect human health and the environment from hazardous effects of chemicals, regulatory agencies and industry alike have heavily relied on animal toxicity testing to identify exposure concentrations at which chemicals can cause adverse effects. Traditionally, these were identified through direct observation of the adverse effect(s) of concern (e.g., impact on survival, growth, reproduction, or organ pathology) in an intact animal model. However, while effective, such testing is costly and time-consuming, making it challenging for apically based hazard evaluations to keep pace with the production and release of new chemicals into the environment. Recognition of these challenges led to the crystallization of a vision for toxicity testing in the 21st century that would employ greater use of mechanistically oriented high throughput assays as a basis for chemical hazard evaluation (Krewski et al., 2010). Use of human cell lines was intended to address human relevance of such testing. However, application to evaluation of ecological hazards of chemicals was not substantively considered in the original human health-focused vision (Villeneuve and Garcia-Reyero, 2011).
A significant response to the proposed vision for high throughput toxicity testing was the US EPA Toxicity Forecaster (ToxCast) program (Kavlock et al., 2012) and the Tox21 consortium (Tice et al., 2013). Despite incorporation of over 1000 assay endpoints representing greater than 300 different gene targets, an acknowledged concern was that the assay battery was still insufficient to cover the diversity of molecular targets and pathways needed to provide a comprehensive evaluation of potential hazards (Thomas et al., 2019). Consequently, as part of an updated blueprint for computational toxicology at the United States Environmental Protection Agency (US EPA), additional broad coverage, high content assays, such as high throughput transcriptomics (HTTr) and high throughput phenotyping utilizing human cell lines, were incorporated as part of a tiered testing framework for hazard characterization (Thomas et al., 2019, Harrill et al., 2021, Nyffeler et al., 2020). High throughput transcriptomics approaches allow near whole human transcriptome coverage, thereby dramatically increasing effective pathway coverage in the screening program. The approach was inspired in part by promising research in mammals suggesting that short-term transcriptomics responses can be effective in predicting concentrations at which much longer term, chronic toxicity can be detected (National Toxicology Program, 2018). In concept, the approach assumes that any stressor that perturbs a biological system sufficiently to yield some kind of adverse outcome (acute or chronic) will cause a “concerted molecular change” detectable as a change in gene expression profile (Johnson et al., 2022). Concentration-response modeling can be applied to determine the benchmark doses at which individual gene expression responses are impacted. In turn the distribution of benchmark doses for individual genes can be used to determine a concentration at which the transcriptome shows some concerted, concentration-dependent, response to the stressor, i.e., a transcriptomic point of departure (tPOD; Thomas et al., 2012, Farmahin et al., 2017, National Toxicology Program, 2018, Johnson et al., 2022). It is postulated that concentrations below the tPOD do not elicit a concerted molecular response in an exposed organism and are thus unlikely to cause adverse effects (Johnson et al., 2022). With this in mind, high throughput transcriptomics approaches were proposed to provide protective point of departure estimates for use in risk-based screening and prioritization as well as pathway-oriented data that can be used to infer putative mode(s) of action that could help guide additional tiers of hazard characterization (Thomas et al., 2019).
A recognized gap in both the ToxCast/Tox21 program and the updated blueprint for computational toxicology has been explicit consideration of ecological hazards (Villeneuve et al., 2019). Conservation of human gene targets, proteins, and pathways across organisms allow for some level of ecological coverage, even with the human oriented testing strategy (LaLone et al., 2018). However, there remain many examples of physiological pathways and processes in other organisms such as plants, invertebrates, “lower” vertebrates, etc. for which no analogous pathways exist in humans (e.g., photosynthesis in plants, molting behaviors in invertebrates, vitellogenesis in fish). These are not only potential incidental targets for chemicals to act on, but in many cases are specifically targeted by designed chemicals such as pest management agents. Consequently, to meet the chemical safety objectives of protecting both human health and the environment, there is a need to develop an appropriate battery of ecologically-focused high throughput assays to incorporate into the overall blueprint for computational toxicology (Thomas et al., 2019, Villeneuve et al., 2019).
With this in mind, US EPA has initiated development and evaluation of a series of ecologically-focused high throughput transcriptomics (eco-HTTr) assays for potential inclusion into its tiered hazard characterization framework. As an initial step in this direction, the present study focused on the development and pilot evaluation of an eco-HTTr assay employing larval fathead minnows. Fathead minnows (Pimephales promelas) are one of the most widely used test species in ecological hazard characterization (Ankley and Villeneuve, 2006). Fish are one of the three main taxa used in the evaluation of hazards to aquatic environments (United Nations, 2015). Additionally, the fathead minnow reference genome has been sequenced, assembled, and annotated (Martinson et al., 2022). Thus, fathead minnow was used as a model species to pilot the approach.
The present study focused on three primary objectives. The first was to devise an experimental design that would facilitate relatively high throughput exposures of larval fathead minnows in a small test volume and provide enough whole-body mRNA to support analysis by RNA sequencing (RNAseq). The second objective was to employ that design to test a preliminary set of 10 chemicals with a range of chemical properties and modes of action. Our final objective was to utilize the pilot results to suggest refinements to the overall experimental design that could improve the approach in terms of efficiency and the ability to judge the quality, acceptability, and fit-for-purpose, of the assay results. In silico subsampling approaches were used to probe the influence of several experimental design variables on the number of genes for which benchmark doses (BMDs) could be defined, and associated levels of variability in the tPOD estimates.
Data from these assays were used to evaluate two hypotheses. First, based on the idea that changes in the transcriptome would indicate that the exposed organism was responding to the presence of the chemical, but that such responses would not necessarily be adverse, we hypothesized that points of departure based on concentration–response modeling of gene expression (i.e., transcriptomics-based points of departure; tPODs) would be lower than adverse effect concentrations observed in acute or chronic toxicity tests. Ideally, tPODs would be protective, but not overly conservative. Second, to test the assumption that eco-HTTr assays with multiple representative taxa may be needed for an effective ecological hazard characterization strategy, we evaluated the hypothesis that tPODs derived from larval fathead minnow exposures would be lower than adverse effect concentrations observed in acute or chronic toxicity tests with invertebrates or algae/plants.
2. Materials and methods
2.1. Assay design and rationale
Our pilot assay design involved exposing 1 day post hatch (equivalent to 5 days post fertilization if incubated at 25 °C) larval fathead minnows in 96-well deep bottom (1 ml well) plates (catalog #502162, NEST Scientific USA, NJ, USA) for 24 h. Exposures were static and conducted in a total of approximately 700 µl of ultraviolet light-treated, 0.4 µM filtered, Lake Superior water, held at 25 °C under a 16 h light, 8 h dark photoperiod. There was one fish per well with eight replicate wells (fish) per concentration, and a total of 11 chemical concentrations and a control (12 treatments) were tested on each plate (Supplementary Fig. S.1). The maximum concentration tested was based on reported LC50s for each of the pilot chemicals, where available (Supplementary Table S.1), and subsequent concentrations represented ½ log serial dilutions from the maximum concentration such that each concentration–response spanned six orders of magnitude. For the pilot investigations, all organism loading and dosing was done manually, so treatments were arrayed systematically on each plate, with concentration increasing across columns on the plate, and each row containing a different replicate fish for each dose (Supplementary Fig. S.1). Consequently, it is recognized that the pilot design confounds position on the plate with treatment, such that any spatially associated variability in temperatures, light intensity, etc. across the plate (e.g., edge effects) may confound the treatment-related transcriptomic responses. That said, we had no prior evidence either supporting or rejecting the potential for spatial location within this system to influence one or more elements of the whole-body transcriptome. The opportunity for errors associated with manual dosing and subsequent processing of a more randomized sample layout was viewed as a greater potential concern for data quality than any speculative impacts of position within the plates on the transcriptomic responses. Thus, the more systematic dosing scheme (Supplementary Fig. S.1) was employed.
Overall attributes of the pilot design were motivated by multiple considerations. First, to achieve throughput needed to eventually test 10 s of chemicals per week in concentration–response, we desired an assay that could be conducted in a 96-well plate which would allow future development of automated dosing and sample processing systems (even though manual processing was employed for the current exposures). Based on some previous larval or adult time-course studies (unpublished results; Garcia-Reyero et al., 2014, Schroeder et al., 2017), 24 h of exposure was identified as generally being sufficient for a robust gene expression response, but still short enough in duration to make the test rapid. It is acknowledged that 24 h may not be the optimal time-point for capturing the transcriptomic response to all chemicals. However, given that the optimal time for measuring a transcriptomic response likely varies based on chemical specific properties associated with adsorption, distribution, metabolism, elimination, mode(s) of action, potency, etc. there likely is no time-point that is universally optimal for all chemicals. Thus, standardization of a practical time-point was viewed as a pragmatic approach. Additionally, it was desirable to limit or eliminate the need for solution renewal during the test to minimize chemical use, labor, and additional opportunities for error and organism stress associated with handling/manipulation. These considerations had to be balanced against the need to maintain water quality and organism health for the duration of the exposure period. We were able to demonstrate water quality could be maintained under static conditions over the 24 h exposure period and control survival was within acceptable limits for aquatic toxicity testing (Flynn et al., 2022, OECD, 2019).
Water quality and organism health was an additional consideration with respect to the life-stage selected for testing. At 1–2 days post hatch, fathead minnows are still dependent on their yolk sac and not independently feeding (US EPA, 1987). Consequently, the fish could be exposed at this stage without adding food to the system which could both compromise water quality and introduce additional organic carbon that could bind exposure chemical(s). At the same time, 1–2 days post hatch fathead minnows are of sufficient size to provide adequate RNA to support individual-level RNAseq analyses (i.e., on average yielding 0.5–1.5 µg RNA). Furthermore, while the fish are not fully mature at this stage, most major organogenesis has taken place and major transcriptional programs associated with early development are complete (US EPA, 1987, US EPA, 1996). Consequently, transcriptomic variability associated with minor differences in developmental stage, earlier in development, were expected to be reduced at the 1–2 days post hatch life-stage.
Standard 96-well microtiter plates have a well volume of 300 µl. Although the 300 µl volume was shown to be sufficient for maintaining water quality over the 24 h, static exposure (Flynn et al., 2022), it was desirable to provide extra volume to help protect against degradation of water quality (e.g., reduced dissolved oxygen, increased ammonia, altered pH, etc.) over the course of chemical exposure. The larger volume also provides a greater reservoir of available exposure chemical to help reduce the impact of chemical uptake by the fish and binding to plate wells. Finally, the additional volume associated with deep-well plates allows for a more robust range of swimming behavior and activity within each well. Consequently, deep well (1 ml) 96 well plates were selected.
2.2. Chemicals
A total of ten chemicals organized into three groups based on mode of action and/or properties were tested in the present study (Supplementary Table S.1). Chemicals were procured from Sigma-Aldrich (St. Louis, MO), and all were >98 % pure. Stock solutions for the metals were prepared in Lake Superior water. Stock solutions of the organic compounds (selective serotonin reuptake inhibitors [SSRIs] and neonicotinoids) were prepared in dimethylsulfoxide (DMSO). Stock solutions were diluted in Lake Superior water to achieve the desired nominal concentrations in the test wells, and in the case of the organic chemical exposures, DMSO concentrations in each well were no greater than 0.5 % DMSO and were equivalent across all wells of the plate for each assay.
2.3. Assay procedure
All larval fathead minnows used for testing were provided by an on-site aquatic culture facility at the US EPA Great Lakes Toxicology and Ecology Division, Duluth, MN. Five days in advance of each exposure, newly fertilized fathead minnow embryos, from several breeding pairs, were gently rolled off polyvinyl chloride breeding tiles and transferred to a bath of aerated Lake Superior water and held at 25 °C. On day five post-fertilization, fish that had hatched and were free swimming were pooled for use in the assay. Larval fish were transferred to a glass petri dish in Lake Superior water. Fish were then individually loaded into each well of 3–4 replicate 96-well deep bottom plates using a small glass capillary tube equipped with a short length of rubber tubing to act as a suction device. Fish were transferred in a minimal volume (generally less than 20 µl) to wells containing 150 µl of Lake Superior water. Plates were visually inspected to assure that all wells were loaded. Subsequently, wells were dosed by pipetting an additional 550 µl volume test chemical dissolved in Lake Superior water to each well. Due to slight variations in the volumes transferred with each fish, an approximate 3 % error in nominal concentration in each well was expected but considered negligible relative to the ½ log concentration series employed. Following dosing, plates were sealed with an air-permeable silicone sealing mat (catalog #506065, NEST Scientific USA, NJ, USA), then placed in a temperature-controlled incubator set to 25 °C for 24 h (±30 min) at a 16:8 light:dark cycle.
Following each exposure period, wells were individually inspected under a dissecting microscope. Mortalities and phenotypic abnormalities such as deformities, cardiac or yolk sac edema, etc. were recorded. Exposure solutions were then removed and either replaced immediately with homogenization buffer (Buffer RLT – Qiagen 79216) or flash frozen at −80 °C. All laboratory procedures involving larval fathead minnows were reviewed and approved by an animal care and use committee in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011).
2.4. RNA extraction and cDNA library preparation
Total RNA was isolated in 96 well format (1 fish per well) using MagMAX-96 Total RNA Isolation kits (ThermoFisher Scientific AM1830) following the manufacturer’s instructions. RNA was quantified on a Synergy HTX plate reader (BioTek, USA) using a Take3 microvolume plate (Biotek). A random set of 16 total RNA samples from each plate was analyzed using an Agilent Tapestation (Agilent, Santa Clara, CA) to confirm the quality of the RNA isolation procedure.
Sequencing libraries were prepared from 250 ng of total RNA using the SENSE mRNA-Seq Library Prep kit (Lexogen) following the manufacturer’s protocol, with 15 cycles of end point PCR amplification. The quality and size of libraries were checked on the D5000 ScreenTape Assay for the TapeStation (Agilent), and concentrations were measured using the Quant-iT PicoGreen dsDNA Assay (ThermoFisher Scientific). Pooled libraries (19–24 samples per pool) were quality checked using either Qubit double stranded DNA HS assay kits (ThermoFischer, Q32851), Agilent 4200 TapeStation HS DNA 1000 assays, or Kapa Illumina Library quantification kits (Roche).
2.5. RNA sequencing
On a per experiment basis, samples, each tagged with a unique adapter sequence, were divided into four pools for sequencing (i.e., 19–24 samples per pool; see Supplementary Table S.2 for total samples sequenced per experiment). Each pool contained samples from across all treatment groups (e.g., n = 2 samples per treatment) wherever possible so that pool and associated sequencing lane was not confounded with treatment. Sequencing was performed at the Research Technology Support Facility Genomics Core at Michigan State University. Each pool was loaded onto one lane of an Illumina 4000 Single Read flow cell (llumina, Foster City, CA). Sequencing was performed in a 1x50 bp single end read format using HiSeq 4000 SBS reagents (Illumina). Base calling was performed with Illumina Real Time Analysis v. 2.7.7 and output was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v.2.20.0.
The fathead minnow genome (Martinson et al., 2022) was indexed with STAR version 2.7.1 and concurrently had in-house gene annotations loaded (Supplementary Information SM.1). Raw reads for all samples were then mapped and counted with STAR to the indexed genome with default parameters, also invoking “--quantMode GeneCounts”. All samples, except two, had at least 50 % of their reads map to a unique genome location (Supplementary Table S.2; Supplementary Information SM.1). Counts assigned to each gene were parsed, and count data for each treatment written to a single tab-delimited matrix file where each column represented a sample and each row represented a gene. Additional quality control screening and filter steps were then applied on an experiment-by-experiment basis (see Supplementary Information SM.1). Based on the QC screening criteria, seven samples were removed across all ten chemicals and twelve doses. There was only one experiment (ZnSO4) where more than a single sample was removed.
Raw FASTQ files, raw count files, and normalized expression matrices have been submitted to the Gene Expression Omnibus (Accession GSE207231).
2.6. Transcriptomic concentration–response modeling and points of departure
Concentration-response modeling was applied to the gene expression datasets to calculate a benchmark dose (BMD) for each gene for which a concentration–response curve could be fit. A tPOD was then calculated based on the distribution of gene-specific BMDs. All concentration–response modeling analyses were conducted using BMDExpress2 (Phillips et al., 2019, Yang et al., 2007). The protocol applied and associated settings in BMDExpress 2 (Supplementary Table S.3) were based on the general approach outlined by the National Toxicology Program (National Toxicology Program, 2018). First, differentially expressed genes (DEGs) were identified by employing a one-way ANOVA test with a Benjamini and Hochberg false discovery rate multiple testing correction (p less than 0.05) and chemical concentration as the single categorical treatment variable (Supplementary Table S.3; Step 1). If at least one feature significantly differed across chemical concentrations as indicated by the ANOVA test (at least one DEG), data were then subjected to a statistical trend test (Williams’ Trend Test; p < 0.05; 100 permutations; Supplementary Table S.3, Step 2). Features that did not show a concentration-dependent trend in expression and at least a twofold change in response were excluded from the more computationally intensive and time-consuming curve fitting analysis. Relationships between remaining chemical-gene pairs were then fit using eight different regression models (Hill, Linear, Power, Polynomial 2°, Exponential 2°, Exponential 3°, Exponential 4°, Exponential 5°; Supplementary Table S.3, Step 3). The model with the lowest Akaike information criterion was selected as the final model and used to calculate the BMD and upper and lower 95 % confidence bounds. The BMD was defined as the modeled dose corresponding to a benchmark response of 1.0-standard deviation over the modeled control response. To retain only transcripts with high quality BMD estimates, additional filters were then applied (Supplementary Table S.3, Step 4): global goodness of fit P-value > 0.1; upper confidence bound of the BMD/ lower confidence bound of the BMD < 40; BMD < highest dose; and BMD > ten times lower than the lowest dose (National Toxicology Program, 2018, Pagé-Larivière et al., 2019). Only BMDs for genes that passed all the filters (filtered DEGs) were included in the determination of a tPOD. While there are multiple methods for calculating a tPOD (Pagé-Larivière et al., 2019, Farmahin et al., 2017) for the present pilot study we used the 10th percentile BMD of filtered DEGs as the tPOD (10th percentile) for the purposes of hypothesis testing the other analyses reported here.
2.7. Effect concentrations from ECOTOX
To support our second objective of comparing tPODs with adverse effect concentrations observed in acute or chronic toxicity tests with fish, invertebrates, or algae/plants, available effects data for the ten pilot chemicals were extracted from US EPA’s Ecotoxicology Knowledgebase (ECOTOX; https://cfpub.epa.gov/ecotox/; Olker et al., 2022). For each chemical, the database was queried by CAS (Chemical Abstracts Service) Registry number. The Aquatic table of results for each query was exported as an Excel file. Using the R statistical environment (R Core Team, 2020), the exported results were filtered to four species groups of interest (Fish, Crustaceans, Algae, Insects) and information fields coded in the ECOTOX outputs (See Supplementary information SM.2; Tables S.4, S.5). Any records for which effect concentrations were reported in units other than mg/L active ingredient were removed from further analyses. Effect types were sorted into two “Tiers”. “Tier 1” included effects coded as mortality, reproduction, behavior, population, growth, development, intoxication, and would generally be viewed as adverse effects in an ecological context. “Tier 2” included remaining types of biological effects (e.g., enzyme(s), genetics, histology, biochemistry, morphology, physiology, feeding behavior, cell(s), immunological, hormone(s), avoidance, injury), which may or may not be directly linked with adversity. It should be noted that effect concentrations reported in ECOTOX are not points of departure based on a benchmark response of one standard deviation from control baseline (as for BMDs), they are a mix of reporting values including lethal LCx, ECx, LOECs, etc. Precise concordance between tPODs and effect concentrations from ECOTOX cannot be expected.
2.8. In silico subsampling to inform assay optimization
Based on the experimental design used for the pilot assays, the full gene expression matrix for each experiment consisted of 79–96 columns and 30,000 + rows. Columns of the matrix represented unique sample identifiers with their corresponding chemical concentration (i.e., generally-eight replicate individuals [samples] per treatment across 10–12 treatments [depending on mortality at the high concentrations tested]). Rows of the matrix represented unique transcript identifiers, and cells within the matrix represented expression levels across the 79–96 samples. To evaluate the effect of transcriptome coverage (number of gene/rows for which counts are included) or sample size (number of individuals analyzed per treatment) on the total DEGs, filtered DEGs, BMDs, and tPOD (10th percentile) for each of the 10 chemicals, in silico subsampling was performed followed by analysis of the subsampled expression matrices via BMDExpress2 using the same approaches detailed in section 2.6.
Expression data for each chemical were subsampled in silico using a custom R script (Supporting Information). Two distinct subsampling events occurred (Table 1; Supplementary Fig. S.2); one focused on evaluating the effect of varying transcriptome coverage (i.e., whole transcriptome versus partial transcriptome coverage), the other focused on the effect of number of replicate samples per treatment. Subsampling was random and performed with replacement between sampling events, such that each time a subsampling event occurred it could contain any replicate/transcript from the original dataset regardless of whether that replicate/transcript had been included in another subsampling event. To generate the subsampled transcript datasets, rows from the original dataset were randomly selected without replacement to contain m transcripts, where m ranged from 100 to 30,000 at various intervals (Table 1). For example, the m = 10,000 dataset consisted of 10,000 randomly selected, distinct, transcripts. All exposure concentrations and the full set of replicates per dose were retained from the original dataset for each sub-sampled transcript dataset.
Table 1.
List of in silico subsampling events performed on the original, full expression datasets. k = 1000×.
| Variable of Interest | Data Sub-sampled | Total Datasets per Chemical |
|---|---|---|
| Transcriptome coverageb | 30 k, 25 k, 20 k, 15 k, 10 k, 5 k – 1 k, 500, 200, 100 genes Genes (rows) included in each set selected randomly All available exposure concentrations (n = 10–12) 8 replicates per concentration |
13 |
| 12x biological replicate sets | 3–7 replicates per dose, 12 iterations of each replicate set Full mapped transcriptome (30,000 + transcripts; all rows) in each set All available exposure concentrations (n = 10–12) Equal number of replicates at each concentration |
60 |
| Background (random) detection of filtered DEGs | Controls (n = 80 samples total) from each of the 10 experiments were randomly assigned to ten mock treatment groups; n = 8 control samples per group. | 30a |
In this case refers to total number of datasets tested, not datasets per chemical.
Refers to the total number of unique transcripts for which count data were used in estimating a tPOD.
To consider the effect of replication (n individuals per treatment) and inter-individual variation, based on inclusion or exclusion of data for specific individuals, sub-sampled replicate datasets were generated by randomly selecting columns from the original matrix, without replacement, such that each dataset contained n distinct replicate samples per treatment group, where n = 3–7. For example, the n = 3 dataset consisted of three randomly selected control samples, three samples for the 0.02 mg/L concentration, three samples for 0.2 mg/L concentration, and so forth for all 10–12 treatment groups. Each subsampling event was performed in 12 iterations per chemical. This generated a total of 60 data matrix files per chemical, with twelve n = 3, n = 4, n = 5, n = 6, and n = 7 datasets. For each replicate set, the full set of genes and all exposure concentrations were retained from the original dataset (Table 1).
Finally, to examine background random detection of filtered DEGs that would yield a BMD estimate an expression matrix consisting of just the control (i.e., Dose = 0 mg/L) samples from all 10 experiments was created. This matrix contained 80 samples total, with 8 replicate controls per chemical exposure, and 34,228 unique features. Original expression values for each control sample were retained; in cases where a feature was not present for a given sample, the expression value was coded as 0 for that sample. Quality assurance metrics (boxplot of raw read counts [Supplementary Fig. S.3], principal component analysis of samples coded by chemical treatment) showed that the data did not require additional downstream processing (e.g., normalization) prior to analysis. Control samples were randomly and arbitrarily assigned to ten “mock treatment groups” ranging from 0 to 10 mg/L (e.g., 0.001 mg/L, 0.03 mg/L, 3.16 mg/L), with eight samples per group (Table 1). This random assignment of control samples to mock treatment groups was repeated 30 times, to create 30 iterations of the control sample matrix. Each of the 30 iterations were imported into the software BMDExpress2 for dose response analysis using the same approaches detailed in Section 2.6.
3. Results
3.1. Assay throughput and performance
The pilot assay design, which was implemented manually, could be viewed as medium throughput. Assay set up, including preparation of chemical dilutions, loading of organisms, and dosing of the plate, took approximately 2.5 h to set up 3–4 plates. Following the 24 h exposure, evaluation of individual survival and phenotypic effects took an average of 10–15 min per plate for manual inspection under a dissecting microscope, followed by an additional 20–30 min per plate for manual removal of test solutions and subsequent homogenization using a bead mixer mill. Extraction of the RNA and cDNA library preparation represented a substantial investment of additional time, taking approximately 10–12 additional hours per plate when performed manually.
Control survival over the course of the 24 h assays was high (Table 2; Supplementary Fig. S.4). For seven of the 10 experiments there were no control mortalities. In two other experiments, a single control fish died. The maximum control mortality observed was 10 %, and in this experiment two fish were verified mortalities while one fish was absent from the well (likely a loading error rather than mortality).
Table 2.
Acute mortality following 24 h static exposure in a 96 well plate format.
| Chemical | Control survival | LC50 (mg/L) | LOEC (mg/L) | NOEC (mg/L) |
|---|---|---|---|---|
| CuSO4 | 32/32 (100 %) | 0.57 | 0.63 | 0.2 |
| NiSO4 | 32/32 (100 %) | 29.29 | 15.81 | 5 |
| ZnSO4 | 24/24 (100 %) | 3.85 | 4 | 1.26 |
| Fluoxetine | 28/31 (90 %)a | 1.42 | 1.5 | 0.5 |
| Paroxetine | 32/32 (100 %) | 4.77 | 5 | 1.58 |
| Sertraline | 31/32 (97 %) | 2.21 | 3 | 0.95 |
| Clothianidin | 23/24 (98 %) | n/a | n/a | n/a |
| Flupyradifurone | 24/24 (100 %) | n/a | n/a | n/a |
| Imidacloprid | 24/24 (100 %) | n/a | n/a | n/a |
| Thiacloprid | 24/24 (100 %) | 194.44 | 250 | 79 |
n/a = not applicable.
Fish missing from one well.
RNA yields were adequate to support RNA sequencing at the level of individual fish. In nearly all cases, at least 1 µg total RNA was obtained with the average sample yielding around 1.2 µg/L.
3.2. – 24 h acute mortality evaluated in 96-well plate format
Seven of the 10 chemicals tested caused acute mortality at the highest concentration tested (Supplementary Fig. S.4; Table 2). Paroxetine, sertraline, thiacloprid, zinc sulfate, and copper sulfate all caused 100 % mortality at the highest concentration tested, while nickel sulfate and fluoxetine caused partial mortality. Concentration response curves were steep, with only copper sulfate causing significant mortality at subsequent diluted concentrations (1/2 log steps). Three test chemicals, clothianidin, flupyradifurone, and imidacloprid, were non-toxic at the nominal concentrations tested (Supplementary Fig. S.4; Table 2). These were all chemicals for which there were limited or no fathead minnow LC50 data upon which to base concentration selection.
3.3. – Transcriptomics-based points of departure
The number of annotated transcripts for each experiment ranged from 30,861–32,996 (Table 3). Among these, the number of genes exhibiting differential expression across treatments (evaluated by ANOVA) and a concentration-dependent trend (based on Willams Trend test), and thus subjected to curve-fitting, ranged from a minimum of 13 for clothianidin to a maximum of 937 for fluoxetine (Table 3). Among the transcripts for which curve-fitting was applied, on average the additional quality filters removed around 70 % DEGs. However, the percentage of DEGs passing the filters, and thus used for final determination of the median BMD and tPOD (10th percentile), varied considerably among experiments, with just 6.3 % of the initial DEGs passing the filters for fluoxetine, while nearly 63 % of those identified for the paroxetine experiment passed the filters (Table 3). Based on median BMD, the rank order of potency among the chemicals was flupyradifurone > ZnSO4 ≈ clothianidin ≈ fluoxetine > CuSO4 > imidacloprid ≈ NiSO4 > sertraline > paroxetine ≫ thiacloprid. By definition, the tPOD (10th percentile BMD) was lower than the median BMD. tPOD (10th percentile) ranged from a factor of 1.5 to 2600-fold lower than the median BMD (Table 3). The notable exception was for flupyradifurone whose median BMD was equivalent to the tPOD (10th percentile), largely due to a highly skewed distribution of best BMDs (Supplementary Fig. S.5). When ranked by 10th percentile-based tPOD, the rank order of potency was quite different with fluoxetine and ZnSO4 ranking as most potent, followed by flupyradifurone ≈ imidacloprid > clothianidin > sertraline ≈ CuSO4 > thiacloprid, with NiSO4 and paroxetine identified as least potent. Median BMD and tPOD (10th percentile) were consistent in ranking ZnSO4 as the most potent of the three metals and fluoxetine as at least 375 times more potent than the other two SSRIs. Similarly, among the neonicotinoid-like compounds flupyradifurone was consistently identified as most potent and thiacloprid least potent, but the relative ranking of imidacloprid and clothianidin varied.
Table 3.
Summary of BMDExpress2 results for larval FHM high throughput exposure assays involving the full transcriptome and 10–12 doses per chemical with 8 replicates per dose. Transcriptomic point of departure (tPOD) calculated as the 10th percentile of the benchmark dose (BMD). Differentially expressed genes (DEGs), lower bound of the 95% confidence interval of the BMD (BMDL), upper bound of the 95% confidence interval of the BMD (BMDU), transcriptomics-based point of departure (tPOD).
| Chemical name | Input transcripts | DEGS pre-filter1 | DEGs post-filter2 | Median BMD (mg/L) | Median BMDL (mg/L) | Median BMDU (mg/L) | 10th Percentile tPOD (mg/L) |
|---|---|---|---|---|---|---|---|
| CuSO4 | 32,996 | 209 | 104 | 2.6e−2 | 1.8e−2 | 4.9e−2 | 5.1e−3 |
| NiSO4 | 31,449 | 158 | 33 | 2.2e−1 | 1.6e−1 | 4.6e−2 | 1.5e−1 |
| ZNso4 | 32,297 | 206 | 25 | 3.2e−4 | 1.1e−4 | 1.4e−3 | 6.3e−5 |
| Fluoxetine | 31,865 | 937 | 24 | 4.6e−4 | 1.1e−4 | 1.4e−3 | 3.2e−6 |
| Paroxetine | 31,884 | 498 | 313 | 7.2e−1 | 5.5e−1 | 1.0 | 4.4e−1 |
| Sertraline | 31,711 | 309 | 148 | 4.4e−1 | 3.1e−1 | 6.4e−1 | 1.2e−3 |
| Clothianidin | 31,158 | 13 | 3 | 3.8e−4 | 1.1e−4 | 2.6e−3 | 2.3e−4 |
| Flupyradifurone | 30,861 | 54 | 10 | 2.3e−5 | 2.3e−5 | 2.3e−5 | 2.3e−5 |
| Imidacloprid | 31,182 | 72 | 10 | 9.2e−2 | 2.9e−2 | 3.0e−1 | 3.5e−5 |
| Thiacloprid | 32,776 | 209 | 97 | 37 | 28 | 56 | 3.6e−2 |
Refers to number of genes significant by ANOVA and showing a trend via William’s trend test.
DEGs remaining after removing those with a global goodness of fit P-value ≤ 0.1; upper 95 % confidence bound of BMD/ lower 95 % confidence bound of BMD ≥ 40; BMD > highest concentration; and BMD < ten times lower than the lowest concentration tested.
3.4. – Comparison with effect concentrations from ECOTOX
One or more ECOTOX records (accessed January 2021) meeting our filtering criteria were available for all ten chemicals tested in the current study. However, effects in fish were not available for flupyradifurone, thiacloprid, or paroxetine. Fish data were also limited for clothianidin and sertraline with just six records, all Tier 1, available for sertraline and just two records, both for Tier 2 endpoints, available for clothianidin (Fig. 1, Supplementary Fig. S.6). Regarding our hypothesis that tPODs will be protective of apical adverse effects, tPODs derived using the pilot assay design with fathead minnow were uniformly lower than Tier 1 effect concentrations reported in ECOTOX for all chemicals for which fish data were available (Fig. 1). The tPODs (10th percentile) were generally well below the distribution of Tier 1 effect concentrations. The only exception was the tPOD for CuSO4 which corresponded to the approximate 5th percentile of the nearly 2500 records for effects in fish in ECOTOX. The tPODs were anywhere from 0.7 to 5 orders of magnitude lower than the than the 25th percentile of Tier 1 effect concentrations for fish available in ECOTOX (Fig. 1). Comparing with effect concentrations reported for fish Tier 2 endpoints from ECOTOX, tPODs were universally protective, with only the tPODs for CuSO4 (4.5th percentile of 1,374 Tier 2 records) and fluoxetine (2nd percentile of 43 Tier 2 records; Supplementary Fig. S.6) overlapping the distribution of Tier 2 effect concentrations. The tPODs ranged from 0.9 to 3.7 orders of magnitude lower than 25th percentile of the ECOTOX Tier 2 effect concentrations.
Fig. 1.
Comparison of FHM tPOD (10th percentile) estimates with traditional fish Tier 1 ECOTOX points of departure (i.e., for effects on survival, growth, reproduction, behavior). Numbers in parentheses indicate the number of relevant ECOTOX records. Center line of box and whisker plot = median; box = interquartile range; whiskers = 1.5x interquartile range; points values outside 1.5x interquartile range.
Given the different order of potency when ranked based on median BMD versus 10th percentile tPOD, it was important to compare the transcriptomics-based potency estimates to those based on more traditional toxicity testing. Tier 1 (Fig. 1) or Tier 2 (Supplementary Fig. S.6) data for fish were only available for six out of the 10 chemicals, with clothianidin, flupyradifuone, paroxetine, and thiacloprid lacking Tier 1 data, while Tier 2 data were available for clothianidin, but not sertraline. Based on median ECOTOX effect concentrations for Tier 1, CuSO4 was always the most potent of the three metals tested, followed by ZnSO4 and NiSO4. However, based on both median BMD and 10th percentile tPOD, ZnSO4 was ranked as the most potent metal, although the distribution of effect concentrations in ECOTOX overlap considerably between ZnSO4 and CuSO4 (Fig. 1).
With regard to our second hypothesis, that fish-based tPODs would be lower than effect concentrations reported for other aquatic taxa as well, fish-based tPODs were not always protective of other aquatic organisms (Fig. 2). Specifically, the tPODs for paroxetine and thiacloprid overlapped the 50th and 38th percentile of Tier 1 effect concentrations for crustaceans while those for NiSO4 and CuSO4 overlapped the 10th and 7th percentile, respectively (Fig. 2A). The tPOD for thiacloprid corresponded with the 77th percentile of Tier 1 effect concentrations for insects (Fig. 2B). With the exceptions noted previously, the rest of the fish-based 10th percentile tPODs were anywhere from one to approximately-four orders of magnitude lower than the 25th percentile of ECOTOX Tier 1 effect concentrations for crustaceans and insects (Fig. 2).
Fig. 2.
Comparison of FHM tPOD (10th percentile) estimates with traditional Tier 1 ECOTOX points of departure for crustaceans (A) and insects (B). Numbers in parentheses indicate the number of relevant ECOTOX records. Center line of box and whisker plot = median; box = interquartile range; whiskers = 1.5x interquartile range; points values outside 1.5x interquartile range.
3.5. In silico subsampling results
The effect of transcriptome coverage on the ability to derive a tPOD and the estimated tPOD concentration was evaluated via concentration–response modeling of randomly selected subsets of transcripts from each experiment using BMDExpress2 (Supplementary Fig. S.2). As one might expect, the number of filtered DEGs was relatively proportional with the total number of transcripts analyzed (Supplementary Fig. S.7). For random transcript set sizes of at least 10,000 unique transcripts, it was always possible to derive a tPOD (10th percentile) estimate, even in cases where just a handful of filtered DEGs were detected (e.g., clothianidin, flupyradifurone, imidacloprid; Supplementary Figs. S.7 and S.8). Below a random coverage of 10,000 unique transcripts, no tPOD estimates could be derived for imidacloprid or flupyradifurone. Using 1,000 or less randomly selected transcripts, tPODs could only be estimated for approximately 50 % of the chemicals tested (Supplementary Fig. S.8). With the exception of fluoxetine, for chemicals that yielded over 10 filtered DEGs the tPOD estimated based on whole transcriptome was qualitatively similar to that based on a random subset of the transcriptome until 5000 or less unique transcripts were included (Supplementary Fig. S.8). There was generally a trend toward the tPOD increasing in concentration (becoming less sensitive) as the size of the transcript set was reduced, although there were some exceptions.
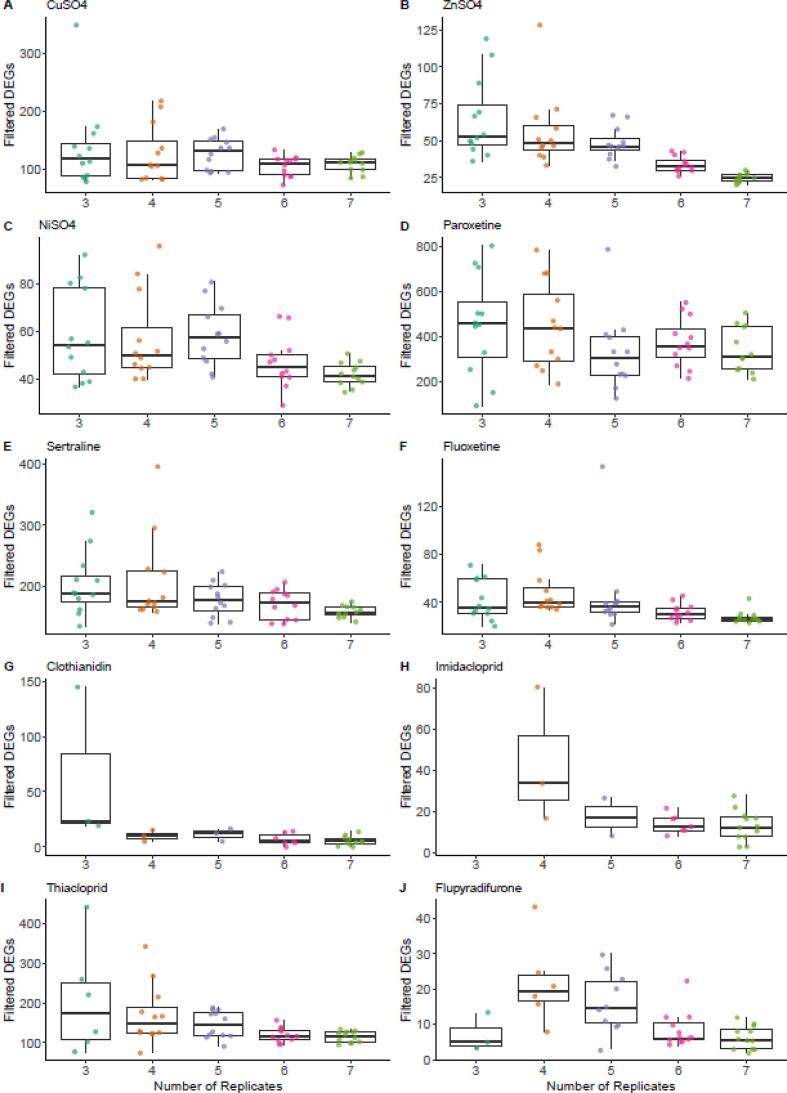
A second application of in silico subsampling was to evaluate the potential effect of a smaller sample size per treatment group on the tPOD estimates and their relative variability. Twelve random selections of n = 3, 4, 5, 6, or 7 individuals from each treatment group were generated for each chemical, then the resulting sixty files for each chemical were evaluated in BMDExpress2. This analysis provided the means to both evaluate the effect of different sample sizes on the overall variability in the tPOD estimate (Fig. 3; Supplementary Fig. S.9) and explore how the inclusion or exclusion of certain individual samples from the data set can skew the tPOD determinations due to impacts on total filtered DEGs (Fig. 4) and overall shape of the best BMD distribution (e.g., Supplementary Fig. S.5). Ultimately, there was not a clear trend in terms of the effect of numbers of replicates tested per treatment versus overall variability in the derived tPOD across 12 random iterations (Supplementary Fig. S.9). Variability in the number of filtered DEGs identified tended to increase with smaller sample sizes per treatment (Fig. 4). Correspondingly, in general, chemicals with larger numbers of filtered DEGs had less variability among the tPOD estimates for the 12 random iterations (Supplementary Fig. S.10). For paroxetine, sertraline, and CuSO4, which were three of the top four chemicals in terms of median number of filtered DEGs, the derived tPODs varied by less than a factor of three (Fig. 2). In contrast, for the three chemicals with the least filtered DEGs, flupyaradifurone, imidacloprid, and clothianidin, the tPOD estimates varied by 5–6 orders of magnitude (Fig. 2). There were some deviations from the general trend toward more variable tPOD estimation when based on fewer filtered DEGs. For example, thiacloprid and fluoxetine had greater numbers of filtered DEGs than NiSO4 or ZnSO4, respectively, but showed 10–500 fold variability in their tPOD estimates versus approximately 3-fold for the metals (Fig. 2). The lack of obvious trend between sample size and tPOD variability suggests that interindividual variability in gene expression, and associated variations in the numbers and composition of filtered DEGs identified (Fig. 4) and used in calculating the tPOD, lead to the somewhat unpredictable variability observed in the current in silico sub-sampled data sets (Supplementary Fig. S.9).
Fig. 3.
Distribution of transcriptomic points of departure (tPOD; 10th percentile) determined from twelve random selections of n = 7 individuals from each treatment group. Center line of box and whisker plot = median; box = interquartile range; whiskers = 1.5x interquartile range; points indicate the values determined for each individual trial.
Fig. 4.
Variation in the number of differentially expressed genes that passed the filters in BMDExpress2 (filtered DEGs) as determined from twelve random selections of n = 3, 4, 5, 6, or 7 individuals from each treatment group. Center line of box and whisker plot = median; box = interquartile range; whiskers = 1.5x interquartile range; points indicate the values determined for each individual trial. Missing points indicate cases where no filtered DEGs were obtained for the trial.
3.6. Determination of background rate of random filtered DEG detection
Thirty iterations of parsing control samples into randomly assigned to mock treatments were analyzed to evaluate the background rate of random detection of filtered DEGs that would contribute to a BMD and thus influence tPOD calculation. Among the 30 iterations, there were only two cases (6.6 %) for which the randomly grouped sample set passed the initial ANOVA test, and thus would have proceeded to BMD estimation (Supplementary Table S.5). In these two cases, the background number of filtered DEGs detected was 3 and 34. Among the 30 iterations, when the initial ANOVA results were ignored and the randomly grouped control datasets were subjected to analysis in BMDExpress2, random detection of filtered DEGs ranged from 0 to 34 with a mean of 5, median of 2.5, mode of 1, and 95th percentile of 14.5 (Supplementary Table S.5). Thus, for 95 % of assays conducted using the present design, less than 15 BMDs would be randomly detected and estimated, and the majority of samples would have less than three random BMDs included in the dataset.
4. Discussion
The aim of the present study was pilot testing of a high throughput compatible transcriptomics assay with larval fathead minnows and use of the resulting data to inform further refinement and optimization of a method suitable for incorporation into a tiered hazard screening framework (e.g., Thomas et al., 2019). The promise of transcriptomics in toxicology and ecotoxicology has been heralded for nearly-two decades (Snape et al., 2004, Ankley et al., 2006, Boverhof and Zacharewski, 2006, National Research Council (US) Committee on Applications of Toxicogenomic Technologies to Predictive Toxicology. Applications of Toxicogenomic Technologies to Predictive Toxicology and Risk Assessment. Washington (DC): National Academies Press (US), 2007). However, optimism about the potential advances that toxicogenomic approaches could facilitate has been tempered by the challenges of interpreting and applying such data in a regulatory context (e.g., Boverhof and Zacharewski, 2006, Kroeger, 2006, Goetz et al., 2011). Some of the notable, and often cited, challenges include the lack of standardized, repeatable, approaches for processing and analyzing data, difficult data interpretation particularly with regard to linking effects to adversity, and costs limiting experimental design in relation to dose–response characterization, time-course, and/or statistical power. However, a broader role for toxicogenomics in a regulatory hazard screening context is starting to emerge.
Thomas et al. (2007) proposed integration of the rich biological information of transcriptomics data sets with statistically rigorous calculation of BMDs to facilitate integration of transcriptomics data into chemical risk assessment. The approach leverages the capacity to concurrently examine effects on all biological pathways represented in the expressed / functional portion of the transcriptome of a given cell type or model organism, thereby addressing many of the concerns regarding pathway coverage in current high throughput testing batteries. The complexity of the output is greatly reduced from a long list of DEGs, dense heat map, or indecipherable “hairball” network, to a concentration estimate, which risk assessors and regulators are accustomed to working with. The initial demonstration of concept (Thomas et al., 2007) was followed by a series of case studies (e.g., Thomas et al., 2012, Thomas et al., 2013, Moffat et al., 2015, Auerbach et al., 2015, Pagé-Larivière et al., 2019) and evaluations of various technologies and approaches for BMD modeling (Webster et al., 2015, Farmahin et al., 2017, Pagé-Larivière et al., 2019). These efforts have led to a National Toxicology Program-authored approach and guidance for genomic dose response modeling (National Toxicology Program, 2018), along with proposed integration of this approach into high throughput chemical screening approaches (Harrill et al., 2019, Harrill et al., 2021). Notably, because HTTr is proposed as an initial tier in a screening battery, challenges associated with linking gene expression changes to adversity are alleviated in that it is assumed that if exposure concentrations are likely to exceed the tPOD, additional targeted testing will be conducted to evaluate potential hazard in more detail (Thomas et al., 2019). Integration of HTTr into a screening framework is facilitated by standardized approaches for BMD estimation, standardized software to conduct the analyses, and emerging reporting frameworks (Gant et al., 2017, Harrill et al., 2021) that support increased transparency and reproducibility in transcriptomics data analysis. Along with advances in RNAseq technology that have lowered the costs of transcriptomic profiling to a point where generation of extensive concentration–response data are feasible, the opportunity for integration of toxicogenomics into regulatory hazard assessment practices has never been better. However, given that the large majority of effort in this area to date has focused on human health, it is important to make sure that consideration of ecological hazards does not get left behind.
4.1. Adaptation to a high throughput compatible assay format
Results of pilot testing with an initial set of 10 chemicals suggest that the life stage of fathead minnows selected for testing and the overall exposure format are reasonable for a high throughput capable experimental design. Control survival was greater than 90 % (Table 2). Water quality (i.e., dissolved oxygen, pH, ammonia concentrations) was maintained at acceptable levels for testing, even at less than half the volume used in the current design (Flynn et al., 2022). Additionally, observed treatment related toxicity was consistent with previous literature values, where available. The pilot testing in the present study would be characterized as medium throughput rather than high throughput, as loading, dosing, and processing of the plates was performed manually. However, given the 96-well plate format, all steps, with the possible exception of organism loading, can be automated using commercial liquid handlers and automated dispensers. Automated dosing would not only make assay set up more rapid, but would also allow for randomization (e.g., Harrill et al., 2021), thereby eliminating spatial position on the plate as a confounding factor with concentration. The greatest current barrier to higher throughput is not the time required for the assay itself, but rather the time and labor required for RNA extraction and cDNA library preparation. Current HTTr with human cell lines eliminate these steps by employing methods that can be applied directly to crude cell lysates (Harrill et al., 2021). Similar, significant gains in throughput could be achieved if the downstream analysis methods were compatible with the use of whole-body homogenates, rather than purified RNA. Thus, while adaptation to a 96-well plate compatible format represents an important first step to increase throughput, there is ample opportunity to improve the assay further.
4.2. Suitability as an alternative to traditional testing
To evaluate the overall suitability and role for a fathead minnow HTTr assay, tPODs were compared with biological effect concentrations compiled in the ECOTOX knowledgebase (Olker et al., 2022). No specific attempts were made to match the species, exposure duration, testing conditions (e.g., static versus flow through) or other design factors. Only filtering and sorting of the data to remove studies deemed wholly irrelevant to the current comparison, such as bioaccumulation or field studies, and broad taxonomic grouping of the data was applied. Based on the seven chemicals for which comparison of tPODs to either Tier 1 or Tier 2 fish effect concentrations from ECOTOX were possible, tPODs were less than the vast majority of effect concentrations from the published literature compiled in the knowledgebase. In those few cases where tPODs overlapped the distribution of effect concentrations in fish, they generally fell at the 5th percentile or lower (Fig. 1, Supplementary Fig. S.7). Consequently, the tPODs do appear quite protective, and thus could serve as reasonable lower bound toxicity estimates for initial risk-based screening and prioritization (e.g., similar to Paul Friedman et al., 2020).
The current question is whether the current tPODs (10th percentile) are too protective. In relation to Tier 1 effect concentrations for fish, there were several cases where the tPODs were over 1000 times lower (e.g., ZnSO4, imidacloprid; Fig. 1). Even in relation to Tier 2 fish effect concentrationss, the tPODs were in some cases two or more orders of magnitude lower than the 25th percentile of effect concentrations from ECOTOX (e.g., clothianidin; fluoxetine; imidacloprid; ZnSO4; Supplementary Fig. S.7). This level of conservativism contrasts with the results compiled by Pagé-Larivière et al. (2019), that found tPODs to be within a factor of 10 for four of the five chemicals they evaluated. However, it is noted that the chemicals considered by Pagé-Larivière et al. (2019) all act through an estrogenic mode of action, and the transcriptomic analyses included in that study were generally derived from single tissues from adults, rather than whole body larval homogenates. Similarly, results compiled in the National Toxicology Program’s approach to genomic dose–response modeling (National Toxicology Program, 2018) generally yielded transcriptomics-based points of departure (termed BEPOD in that document) that were within one order of magnitude of the most sensitive apical potency values. But once again, these data were from in vivo studies that applied transcriptomics to a single tissue. Ultimately, the testing of a much larger set of chemicals will be needed to robustly evaluate how conservative the larval fathead minnow tPODs may be relative to benchmarks from traditional aquatic toxicity tests.
A second question that we evaluated with our pilot data was whether fish-based tPODs would be protective for other aquatic taxa, or whether eco-HTTr assays with multiple representative taxa may be needed in a testing framework. Upon comparing the fish-based tPODs with Tier 1 ECOTOX effect concentrations for crustaceans and insects, it was evident that the fish tPODs were not uniformly protective (Fig. 2). The fish tPOD for thiacloprid exceeded the 35th percentile of crustacean Tier 1 effect concentrations and the 75th percentile of Tier 1 effect concentrations for insects. Given that thiacloprid and other neonicotinoids were specifically designed for high toxicity to target insect pests, but low toxicity to vertebrates (Tomizawa and Casida, 2005), it might be expected that fish would not necessarily be as sensitive. Nonetheless, the fish tPODs for clothianidin and imidacloprid were protective. Somewhat surprisingly, the fish tPOD for paroxetine overlapped Tier 1 effect concentrations reported in crustacea (Fig. 2A). Based on the intended therapeutic use of SSRIs as antidepressants, one might assume that fish (which have a more complex central nervous system) would be as, if not more, sensitive to an SSRI than crustaceans. Nonetheless, analysis in US EPA’s Sequence Alignment to Predict Across Species Sensitivity tool (SeqAPASS; LaLone et al., 2016) suggest that orthologs for paroxetine’s target protein, solute carrier family 6, member 4 (slc6a4), in humans is conserved across a range of invertebrate taxa (Supplementary Fig. S.12). The number of ECOTOX records for effects of paroxetine on crustaceans was limited, but overall the effect concentrations reported in ECOTOX were quite consistent (Fig. 2A). Ultimately, pilot testing results suggest that fish tPODs alone may not be protective of the full diversity of aquatic taxa, particularly for compounds that act through modes of action associated with selective sensitivity to non-vertebrate or non-animal physiology.
4.3. Assay refinement
In considering the development of an eco-HTTr testing protocol, there were several experimental design considerations that were difficult to optimize in the absence of a set of concentration–response transcriptomic datasets. Consequently, a third major aim of the current pilot testing was to use the generated data to evaluate several questions related to assay design, optimization, and acceptability.
One of the first questions was whether a reduced transcriptome approach would be viable for eco-HTTr. Human health oriented HTTr-based screening to date has employed the TempOSeq® assay (Yeakley et al., 2017, Harrill et al., 2021). TempOSeq® is a targeted approach that is based on hybridization and sequencing of gene-specific detector oligonucleotides. For humans, such detectors have been developed for nearly the entire transcriptome. However, for other species, the approach would not be viable until sizeable enough sets of detector oligos were developed to provide broad pathway coverage, particularly for pathways not well conserved with humans. Thus, a whole transcriptome RNAseq-based approach was employed in the current work.
With the possibility to transition to a targeted approach in the future, we were interested in using the current data sets to estimate the number of genes that would be needed for reliable generation of tPODs. Overall, the current results suggest tPODs could be derived in all cases using a random selection of 10,000 or more transcripts (Supplementary Fig. S.8; Table 4). However, excluding flupyradifurone, imidacloprid, and clothianidin for which 10 or fewer filtered DEGs were detected overall, a tPOD could generally be estimated using even as few as 2,000 transcripts. Likewise, in cases where greater than 10 filtered DEGs were identified for the full data set, estimated tPODs were relatively stable when the evaluated number of unique transcripts exceeded 5000 (Supplementary Fig. S.8). Note, the present results are for completely random selection of transcripts. One would assume that a more strategic selection based on transcripts that have been previously shown to be differentially expressed following exposure to various stressors and/or known to be strongly correlated with the expression of other transcripts, would further enhance the utility of a more targeted sub-set for tPOD derivation. Indeed, that was the conceptual underpinning leading to the development of the Tox21 program’s sentinel gene set (S1500+; Mav et al., 2018). Based on these results, it is expected that once critical mass of whole transcriptome data has been generated, the eco-HTTr approach could be transitioned to a targeted, sentinel gene approach to further reduce costs and potentially increase throughput.
Table 4.
Assay design and acceptance recommendations based on pilot testing results.2
| Design Parameter or Acceptance Criterion | Recommendation | Elaboration |
|---|---|---|
| Fathead minnow life stage | 5–6 days post fertilization | Sufficient RNA; not yet independently feeding; advanced stage of organogenesis and tissue differentiation. |
| Minimum transcriptome coverage | 10,000 unique transcripts | Until sufficient data to support strategic design and evaluation of sentinel gene sets are available. |
| Replicate samples per treatment1 | N = 4 | Minimum of 3. Inclusion of additional sample allows for estimation of uncertainty using in silico subsampling. |
| Pooled individuals per replicate1 | N ≥ 3 | Multiple individuals across at least three replicate plates are needed for LC50 estimates and other phenotypic data. More individuals per pool expected to lead to reduced variability in final number of filtered DEGs. |
| Minimum number of filtered DEGs | N = 15 | Average false discovery of filtered DEGs was 3-fold lower; all tPODs from pilot testing with filtered DEGs ≤ 10 resulted in tPODs with an uncertainty range exceeding four orders of magnitude, based on in silico subsampling. |
Experimental design recommendations based on inference from pilot study results. Design has not yet been tested.
All recommendations subject to change as additional data are generated and alternative designs are evaluated.
In the near term, the largest cost-savings could potentially be achieved through a reduction in the number of samples that are sequenced. Consequently, a second question we aimed to address through analysis of the pilot datasets was the viable minimum sample size that could be employed to estimate a tPOD with reasonable levels of variability. In general, the number of filtered DEGs for which a BMD could be estimated became more variable across the 12 random iterations of sample selection as the number of replicate individuals included (sample size) decreased (Fig. 4). However, greater variability in the number of filtered DEGs identified did not necessarily translate to greater variability in the tPOD estimates as a function of sample size (Supplementary Fig. S.9). This presumably reflects that the tPOD depends not only on the number of filtered DEGs, but the identities of the filtered DEGs in each set and the associated distribution of BMDs. As certain individuals are randomly included or excluded from the data set for various treatments, the overall composition of genes that have concentration-dependent expression and pass the various quality filters in BMD express shifts, resulting in a less predictable relationship between tPOD variability and sample size.
Based on this interpretation, we hypothesize it would be feasible to reduce the number of samples sequenced at each concentration considerably, provided the number of individuals represented in each sample was increased. An example would be a pooling strategy in which a minimum of three pooled samples were sequenced, with each pool representing a minimum of three individual fish, although potentially up to eight or more per pool. Such an approach would reduce cost, potentially without increasing the variability of the tPOD, as estimated via iterations of random sampling. Such a pooling strategy would have an additional advantage of making the assay less dependent on the mass of RNA that can be extracted from each individual, potentially allowing use of RNA extraction methods that may sacrifice some extraction efficiency in the interest of higher throughput.
Tentatively, three pooled replicates per treatment would be the minimum we would recommend (Table 4). However, inclusion of a fourth sample would allow for the kind of in silico subsampling applied here, which would allow variability of the tPODs to be assessed, which is important when optimizing assay design. Ideally, a boot-strapping approach with hundreds of random iterations would be applied. However, even the less computationally costly approach of simulating a dozen random iterations can provide a reasonable sense of variability in the tPOD. Without such a subsampling or boot-strapping approach, only the variability associated with the curve-fitting (represented as the upper and lower confidence bounds of the BMD) can be estimated. Thus, preliminary recommendation would be a sample size of four replicate pooled samples per treatment, with a minimum of three individuals per pool (n ≥ 12 individuals exposed per treatment; Table 4). However, additional testing will be needed to determine whether this design truly improves efficiency while reducing variability as hypothesized.
A third consideration for assay refinement was whether a set of assay acceptance criteria could be proposed. Given that the tPOD estimate is based on the distribution of gene specific BMDs calculated for the filtered DEGs, it is intuitive that greater numbers of filtered DEGs would result in more confident tPOD determination (e.g., Supplementary Fig. S.10). In essence, the more values in the BMD distribution, the more confidently-one can determine the 10th percentile. Empirically, we found that chemicals for which less than 25 filtered DEGs were determined produced highly variable tPOD estimates over 12 iterations of random in silico sample selection (Fig. 3). Likewise, background identification of random filtered DEGs was quite low; on average, just five false discoveries per experiment and less than 15 false filtered DEGs in 95 % of cases were identified (Supplementary Table S.5). Together, these two lines of evidence suggest that a minimum of 15 filtered DEGs (15 BMDs) would be reasonable as an assay acceptance criterion (Table 4), at least for the current assay design and analysis pipeline employed. Transcriptomic points of departure calculated based on less than 15 BMDs would be considered highly unreliable in the present assay. Given this consideration, our current estimates for three of the four neonicotinoid-related compounds should be viewed as unreliable estimates for fish. We would not recommend their use for risk-based screening and prioritization without re-running the assay over a refined concentration range, or with a different design, such as the pooled design suggested above, that would yield more filtered DEGs.
4.4. Conclusions and next steps for Eco-HTTr
Overall, the pilot testing of the larval fathead minnow-based eco-HTTr assay suggests that continued development of this assay for potential inclusion in a tiered hazard characterization framework is warranted. In its present iteration, the assay was not substantially more rapid than a 96 h acute toxicity test. That said, the richness of the data generated in terms of ability to capture not only lethal effects, but a broader suite of biological impacts that could lead to more subtle, chronic impacts on fitness is an advantage. At the same time the data complexity was reduced to a single point estimate that could be used in risk-based screening. Based on initial comparisons with traditional toxicity data, the method appears protective, and in that respect useful as an early screening and prioritization tier, but perhaps more conservative than might be optimal. As additional chemicals with diverse modes of action are tested, it will be possible to better evaluate the overall degree of conservatism of the eco-HTTr approach.
It is noted that the current comparison of tPODs to ECOTOX Tier 1 and Tier 2 effect concentrations are based on nominal concentrations in the eco-HTTr tests. While this is consistent with the general lack of exposure verification in the high throughput screening program (Thomas et al., 2019), it is a recognized potential source of error. Free concentrations in the test well may be substantially lower than nominal, depending on the properties of the chemical being tested (e.g., hydrophobicity, stability, volatility, etc.) as well as the assay conditions (surface area to volume ratios, dissolved organic matter in the test well, etc.; Henneberger et al., 2021, Proença et al., 2021, Fischer et al., 2017). The parameterization and use of models for predicting the freely dissolved chemical concentration in the larval fathead minnow eco-HTTr assay would allow for more reliable comparison to traditional toxicity values. Likewise, sufficient analytical exposure verification to either establish or reject the modeled estimates of free concentrations will also be needed.
Although based on a limited number of chemicals, the present results suggest fish-derived tPODs are not protective for all aquatic organisms. Thus, the development of parallel assays for additional surrogate aquatic taxa, such as plants/algae and invertebrates, appears warranted. This adaptation is already underway (Flynn et al., 2022). Additional testing of a broader chemical set in eco-HTTr assays with multiple species will help to better inform how many different eco-HTTr assays may be needed in a testing battery, and will also yield datasets that can support selection of more targeted sentinel gene sets that could be employed in future testing.
Perhaps the most critical insights gained from pilot testing of the eco-HTTr approach were those into assay design (Table 4). A sample size of n = 8 individuals (allowing for a maximum of n = 7 for subsampling) per treatment did not lead to reliable reductions in tPOD variability/uncertainty, as estimated via in silico subsampling, compared to smaller sample sizes. Instead, variations in the number and composition of the filtered DEGs determined for different iterations of sample composition were a greater determinant. Consequently, we hypothesize that variability in the DEG composition could be minimized by including as many exposed individuals as is feasible in the sample population and pooling individuals into a smaller number of replicates for sequencing. We hypothesize that a relatively small number of pooled samples at each concentration would appear be sufficient for the concentration–response modeling needed for BMD and tPOD estimation, although these hypotheses need to be tested.
Finally, the pilot approach was agnostic to gene identity. All filtered DEGs and associated BMDs were included in the estimation of the tPOD. This largely limits application of the results to potency evaluation. However, given the richness of the transcriptomic data set, there is a recognized opportunity to identify pathway-specific points of departure (e.g., Thomas et al., 2012, Pagé-Larivière et al., 2019, Harrill et al., 2021). Derivation of pathway-specific points of departure can facilitate increased mode of action insight from the data, which may support an additional range of risk assessment applications. With updates and annotation of the fathead minnow genome (Martinson et al., 2022), a pathway-based approach should be feasible for the present assay, provided there are adequate numbers of filtered DEGs to support further subdivision of the BMDs into pathway groups. However, an analogous approach may prove more challenging for species with less developed genomes and/or with substantial unannotated portions of their genome (e.g., Colbourne et al., 2011, Lee et al., 2019).
Overall, the current data set represents the most extensive generation of ecologically focused transcriptomics-based point of departure data to date. Significant insights were gained into testing parameters that could improve throughput, reduce cost, and enhance quality evaluation of the resulting data. Nonetheless, current conclusions regarding the overall utility of the approach are tentative. More robust testing of the approach with a larger battery of chemicals representing diverse modes of action are needed to draw firm conclusions about the future role of the assay as part of an overall approach to 21st century toxicity assessment.
CRediT authorship contribution statement
Daniel L. Villeneuve: Conceptualization, Methodology, Validation, Writing – original draft, Supervision, Project administration, Funding acquisition. Michelle Le: Investigation, Methodology, Data curation, Supervision. Monique Hazemi: Methodology, Software, Data curation, Formal analysis, Writing – review & editing, Visualization. Adam Biales: Resources, Writing – review & editing. David C. Bencic: Investigation, Resources, Writing – review & editing. Kendra Bush: Investigation, Methodology. Robert Flick: Investigation, Resources, Writing – review & editing. John Martinson: Methodology, Validation, Formal analysis, Data curation, Writing – review & editing. Mackenzie Morshead: Investigation. Kelvin Santana Rodriguez: Investigation. Kelsey Vitense: Methodology, Software, Formal analysis, Data curation, Writing – review & editing. Kevin Flynn: Conceptualization, Methodology, Investigation, Resources, Writing – review & editing, Resources, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors thank J. Harrill, L. Everett, R. Judson, I. Shah, K. Jensen, J. Cavallin, E. Maloney, R. Hockett, T. Norberg King, J. Olker, J. O’Brien, J. Prindiville, and C. Inglis for technical discussions that helped inform the pilot experiments and analysis. We thank J. Harrill and M. Meier for comments on an earlier version of this manuscript.
Disclaimer: The contents of this manuscript neither constitute nor necessarily reflect US EPA policy. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Data Availability: All data reported in this article are accessible via DOI: 10.23719/1527818, Data.gov, and the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/; GSE207231).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crtox.2022.100099.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
All research data/code have been made publicly accessible via GEO and US EPA Clowder; they will also be available through data.gov
References
- Ankley G.T., Daston G.P., Degitz S.J., Denslow N.D., Hoke R.A., Kennedy S.W., Miracle A.L., Perkins E.J., Snape J., Tillitt D.E., Tyler C.R., Versteeg D. Toxicogenomics in regulatory ecotoxicology. Environ. Sci. Tech. 2006;40(13):4055–4065. doi: 10.1021/es0630184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankley G.T., Villeneuve D.L. The fathead minnow in aquatic toxicology: past, present and future. Aquat. Toxicol. 2006;78(1):91–102. doi: 10.1016/j.aquatox.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Auerbach S.S., Phadke D.P., Mav D., Holmgren S., Gao Y., Xie B., Shin J.H., Shah R.R., Merrick B.A., Tice R.R. RNA-Seq-based toxicogenomic assessment of fresh frozen and formalin-fixed tissues yields similar mechanistic insights. J. Appl. Toxicol. 2015;35(7):766–780. doi: 10.1002/jat.3068. [DOI] [PubMed] [Google Scholar]
- Boverhof D.R., Zacharewski T.R. Toxicogenomics in risk assessment: applications and needs. Toxicol. Sci. 2006;89(2):352–360. doi: 10.1093/toxsci/kfj018. [DOI] [PubMed] [Google Scholar]
- Colbourne J.K., Pfrender M.E., Gilbert D., Thomas W.K., Tucker A., Oakley T.H., Tokishita S., Aerts A., Arnold G.J., Basu M.K., Bauer D.J., Cáceres C.E., Carmel L., Casola C., Choi J.H., Detter J.C., Dong Q., Dusheyko S., Eads B.D., Fröhlich T., Geiler-Samerotte K.A., Gerlach D., Hatcher P., Jogdeo S., Krijgsveld J., Kriventseva E.V., Kültz D., Laforsch C., Lindquist E., Lopez J., Manak J.R., Muller J., Pangilinan J., Patwardhan R.P., Pitluck S., Pritham E.J., Rechtsteiner A., Rho M., Rogozin I.B., Sakarya O., Salamov A., Schaack S., Shapiro H., Shiga Y., Skalitzky C., Smith Z., Souvorov A., Sung W., Tang Z., Tsuchiya D., Tu H., Vos H., Wang M., Wolf Y.I., Yamagata H., Yamada T., Ye Y., Shaw J.R., Andrews J., Crease T.J., Tang H., Lucas S.M., Robertson H.M., Bork P., Koonin E.V., Zdobnov E.M., Grigoriev I.V., Lynch M., Boore J.L. The ecoresponsive genome of Daphnia pulex. Science. 2011;331(6017):555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmahin R., Williams A., Kuo B., Chepelev N.L., Thomas R.S., Barton-Maclaren T.S., Curran I.H., Nong A., Wade M.G., Yauk C.L. Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch. Toxicol. 2017;91(5):2045–2065. doi: 10.1007/s00204-016-1886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F.C., Henneberger L., König M., Bittermann K., Linden L., Goss K.U., Escher B.I. Modeling Exposure in the Tox21 in Vitro Bioassays. Chem. Res. Toxicol. 2017;30(5):1197–1208. doi: 10.1021/acs.chemrestox.7b00023. [DOI] [PubMed] [Google Scholar]
- Flynn K.M., Le M., Hazemi M., Bush K., Morsehead M., Santana Rodriguez K., Hoang J., Villeneuve D.L. In Preparation. 2022. Development of high throughput assays in four species relevant to ecological hazard assessment. [Google Scholar]
- Gant T.W., Sauer U.G., Zhang S.D., Chorley B.N., Hackermüller J., Perdichizzi S., Tollefsen K.E., van Ravenzwaay B., Yauk C., Tong W., Poole A. A generic Transcriptomics Reporting Framework (TRF) for 'omics data processing and analysis. Regul. Toxicol. Pharm. 2017;91(Suppl 1):S36–S45. doi: 10.1016/j.yrtph.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N., Ekman D.R., Habib T., Villeneuve D.L., Collette T.W., Bencic D.C., Ankley G.T., Perkins E.J. Integrated approach to explore the mechanisms of aromatase inhibition and recovery in fathead minnows (Pimephales promelas) Gen. Comp. Endocrinol. 2014;1(203):193–202. doi: 10.1016/j.ygcen.2014.03.022. [DOI] [PubMed] [Google Scholar]
- Goetz A.K., Singh B.P., Battalora M., Breier J.M., Bailey J.P., Chukwudebe A.C., Janus E.R. Current and future use of genomics data in toxicology: opportunities and challenges for regulatory applications. Regul. Toxicol. Pharm. 2011;61(2):141–153. doi: 10.1016/j.yrtph.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Harrill J., Shah I., Setzer R.W., Haggard D., Auerbach S., Judson R., Thomas R.S. Considerations for Strategic Use of High-Throughput Transcriptomics Chemical Screening Data in Regulatory Decisions. Curr Opin Toxicol. 2019;15:64–75. doi: 10.1016/j.cotox.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J.A., Viant M.R., Yauk C.L., Sachana M., Gant T.W., Auerbach S.S., Beger R.D., Bouhifd M., O'Brien J., Burgoon L., Caiment F., Carpi D., Chen T., Chorley B.N., Colbourne J., Corvi R., Debrauwer L., O'Donovan C., Ebbels T.M.D., Ekman D.R., Faulhammer F., Gribaldo L., Hilton G.M., Jones S.P., Kende A., Lawson T.N., Leite S.B., Leonards P.E.G., Luijten M., Martin A., Moussa L., Rudaz S., Schmitz O., Sobanski T., Strauss V., Vaccari M., Vijay V., Weber R.J.M., Williams A.J., Williams A., Thomas R.S., Whelan M. Progress towards an OECD reporting framework for transcriptomics and metabolomics in regulatory toxicology. Regul. Toxicol. Pharm. 2021;125 doi: 10.1016/j.yrtph.2021.105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J.A., Everett L.J., Haggard D.E., Sheffield T., Bundy J.L., Willis C.M., Thomas R.S., Shah I., Judson R.S. High-Throughput Transcriptomics Platform for Screening Environmental Chemicals. Toxicol. Sci. 2021;181(1):68–89. doi: 10.1093/toxsci/kfab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger L., Huchthausen J., Wojtysiak N., Escher B.I. Quantitative In Vitro-to-In Vivo Extrapolation: Nominal versus Freely Dissolved Concentration. Chem. Res. Toxicol. 2021;34(4):1175–1182. doi: 10.1021/acs.chemrestox.1c00037. [DOI] [PubMed] [Google Scholar]
- Johnson KJ, Auerbach SS, Stevens T, Barton-Maclaren TS, Costa E, Currie RA, Dalmas Wilk D, Haq S, Rager JE, Reardon AJF, Wehmas L, Williams A, O'Brien J, Yauk C, LaRocca JL, Pettit S. A transformative vision for an omics-based regulatory chemical testing paradigm. Toxicol Sci. 2022 27:kfac097. doi: 10.1093/toxsci/kfac097. [DOI] [PMC free article] [PubMed]
- Kavlock R., Chandler K., Houck K., Hunter S., Judson R., Kleinstreuer N., Knudsen T., Martin M., Padilla S., Reif D., Richard A., Rotroff D., Sipes N., Dix D. Update on EPA's ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 2012;25(7):1287–1302. doi: 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Krewski D., Acosta D., Jr, Andersen M., Anderson H., Bailar J.C., 3rd, Boekelheide K., Brent R., Charnley G., Cheung V.G., Green S., Jr, Kelsey K.T., Kerkvliet N.I., Li A.A., McCray L., Meyer O., Patterson R.D., Pennie W., Scala R.A., Solomon G.M., Stephens M., Yager J., Zeise L. Toxicity testing in the 21st century: a vision and a strategy. J. Toxicol. Environ. Health B Crit. Rev. 2010;13(2–4):51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger M. How omics technologies can contribute to the '3R' principles by introducing new strategies in animal testing. Trends Biotechnol. 2006;24(8):343–346. doi: 10.1016/j.tibtech.2006.06.003. [DOI] [PubMed] [Google Scholar]
- LaLone C.A., Villeneuve D.L., Lyons D., Helgen H.W., Robinson S.L., Swintek J.A., Saari T.W., Ankley G.T. Editor's Highlight: Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS): A Web-Based Tool for Addressing the Challenges of Cross-Species Extrapolation of Chemical Toxicity. Toxicol. Sci. 2016;153(2):228–245. doi: 10.1093/toxsci/kfw119. [DOI] [PubMed] [Google Scholar]
- LaLone C.A., Villeneuve D.L., Doering J.A., Blackwell B.R., Transue T.R., Simmons C.W., Swintek J., Degitz S.J., Williams A.J., Ankley G.T. Evidence for Cross Species Extrapolation of Mammalian-Based High-Throughput Screening Assay Results. Environ. Sci. Tech. 2018;52(23):13960–13971. doi: 10.1021/acs.est.8b04587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.Y., Choi B.S., Kim M.S., Park J.C., Jeong C.B., Han J., Lee J.S. The genome of the freshwater water flea Daphnia magna: A potential use for freshwater molecular ecotoxicology. Aquat. Toxicol. 2019;210:69–84. doi: 10.1016/j.aquatox.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Martinson J.W., Bencic D.C., Toth G.P., Kostich M.S., Flick R.W., See M.J., Lattier D., Biales A.D., Huang W. De Novo Assembly of the Nearly Complete Fathead Minnow Reference Genome Reveals a Repetitive but Compact Genome. Environ. Toxicol. Chem. 2022;41(2):448–461. doi: 10.1002/etc.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mav D., Shah R.R., Howard B.E., Auerbach S.S., Bushel P.R., Collins J.B., Gerhold D.L., Judson R.S., Karmaus A.L., Maull E.A., Mendrick D.L., Merrick B.A., Sipes N.S., Svoboda D., Paules R.S. A hybrid gene selection approach to create the S1500+ targeted gene sets for use in high-throughput transcriptomics. PLoS One. 2018;13(2):e0191105. doi: 10.1371/journal.pone.0191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat I., Chepelev N., Labib S., Bourdon-Lacombe J., Kuo B., Buick J.K., Lemieux F., Williams A., Halappanavar S., Malik A., Luijten M., Aubrecht J., Hyduke D.R., Fornace A.J., Jr, Swartz C.D., Recio L., Yauk C.L. Comparison of toxicogenomics and traditional approaches to inform mode of action and points of departure in human health risk assessment of benzo[a]pyrene in drinking water. Crit. Rev. Toxicol. 2015;45(1):1–43. doi: 10.3109/10408444.2014.973934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. 8th edition. Washington (DC): National Academies Press (US); 2011. Available from: https://www.ncbi.nlm.nih.gov/books/NBK54050/ doi: 10.17226/12910. [PubMed]
- National Research Council (US) Committee on Applications of Toxicogenomic Technologies to Predictive Toxicology. Applications of Toxicogenomic Technologies to Predictive Toxicology and Risk Assessment. Washington (DC): National Academies Press (US); 2007. [PubMed]
- National Toxicology Program NTP Research Report on National Toxicology Program Approach to Genomic Dose-Response Modeling. NTP RR 5. Research Triangle Park. NC National Toxicology Program. 2018;5:1–44. [PubMed] [Google Scholar]
- Nyffeler J., Willis C., Lougee R., Richard A., Paul-Friedman K., Harrill J.A. Bioactivity screening of environmental chemicals using imaging-based high-throughput phenotypic profiling. Toxicol. Appl. Pharmacol. 2020;15(389) doi: 10.1016/j.taap.2019.114876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (2019), Test No. 203: Fish, Acute Toxicity Test, OECD Guidelines for the Testing of Chemicals, Section 2, OECD Publishing, Paris, https://doi.org/10.1787/9789264069961-en.
- Olker J.H., Elonen C.M., Pilli A., Anderson A., Kinziger B., Erickson S., Skopinski M., Pomplun A., LaLone C.A., Russom C.L., Hoff D. The ECOTOXicology Knowledgebase: A Curated Database of Ecologically Relevant Toxicity Tests to Support Environmental Research and Risk Assessment. Environ. Toxicol. Chem. 2022;41(6):1520–1539. doi: 10.1002/etc.5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagé-Larivière F., Crump D., O'Brien J.M. Transcriptomic points-of-departure from short-term exposure studies are protective of chronic effects for fish exposed to estrogenic chemicals. Toxicol. Appl. Pharmacol. 2019;378 doi: 10.1016/j.taap.2019.114634. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K., Gagne M., Loo L.H., Karamertzanis P., Netzeva T., Sobanski T., Franzosa J.A., Richard A.M., Lougee R.R., Gissi A., Lee J.J., Angrish M., Dorne J.L., Foster S., Raffaele K., Bahadori T., Gwinn M.R., Lambert J., Whelan M., Rasenberg M., Barton-Maclaren T., Thomas R.S. Utility of In Vitro Bioactivity as a Lower Bound Estimate of In Vivo Adverse Effect Levels and in Risk-Based Prioritization. Toxicol. Sci. 2020;173(1):202–225. doi: 10.1093/toxsci/kfz201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.R., Svoboda D.L., Tandon A., Patel S., Sedykh A., Mav D., Gift J.S. BMDExpress 2: enhanced transcriptomic dose-response analysis workflow. Bioinformatics. 2019;35(10):1780–1782. doi: 10.1093/bioinformatics/bty878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proença S., Escher B.I., Fischer F.C., Fisher C., Grégoire S., Hewitt N.J., Nicol B., Paini A., Kramer N.I. Effective exposure of chemicals in in vitro cell systems: A review of chemical distribution models. Toxicol. In Vitro. 2021;73 doi: 10.1016/j.tiv.2021.105133. [DOI] [PubMed] [Google Scholar]
- R Core Team . In. (4.0.3 ed.). R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Schroeder A.L., Ankley G.T., Habib T., Garcia-Reyero N., Escalon B.L., Jensen K.M., Kahl M.D., Durhan E.J., Makynen E.A., Cavallin J.E., Martinovic-Weigelt D., Perkins E.J., Villeneuve D.L. Rapid effects of the aromatase inhibitor fadrozole on steroid production and gene expression in the ovary of female fathead minnows (Pimephales promelas) Gen. Comp. Endocrinol. 2017;1(252):79–87. doi: 10.1016/j.ygcen.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape J.R., Maund S.J., Pickford D.B., Hutchinson T.H. Ecotoxicogenomics: the challenge of integrating genomics into aquatic and terrestrial ecotoxicology. Aquat. Toxicol. 2004;67(2):143–154. doi: 10.1016/j.aquatox.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Thomas R.S., Allen B.C., Nong A., Yang L., Bermudez E., Clewell H.J., 3rd, Andersen M.E. A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicol. Sci. 2007;98(1):240–248. doi: 10.1093/toxsci/kfm092. [DOI] [PubMed] [Google Scholar]
- Thomas R.S., Clewell H.J., 3rd, Allen B.C., Yang L., Healy E., Andersen M.E. Integrating pathway-based transcriptomic data into quantitative chemical risk assessment: a five chemical case study. Mutat. Res. 2012;746(2):135–143. doi: 10.1016/j.mrgentox.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Thomas R.S., Wesselkamper S.C., Wang N.C., Zhao Q.J., Petersen D.D., Lambert J.C., Cote I., Yang L., Healy E., Black M.B., Clewell H.J., 3rd, Allen B.C., Andersen M.E. Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol. Sci. 2013;134(1):180–194. doi: 10.1093/toxsci/kft094. [DOI] [PubMed] [Google Scholar]
- Thomas R.S., Bahadori T., Buckley T.J., Cowden J., Deisenroth C., Dionisio K.L., Frithsen J.B., Grulke C.M., Gwinn M.R., Harrill J.A., Higuchi M., Houck K.A., Hughes M.F., Hunter E.S., Isaacs K.K., Judson R.S., Knudsen T.B., Lambert J.C., Linnenbrink M., Martin T.M., Newton S.R., Padilla S., Patlewicz G., Paul-Friedman K., Phillips K.A., Richard A.M., Sams R., Shafer T.J., Setzer R.W., Shah I., Simmons J.E., Simmons S.O., Singh A., Sobus J.R., Strynar M., Swank A., Tornero-Valez R., Ulrich E.M., Villeneuve D.L., Wambaugh J.F., Wetmore B.A., Williams A.J. The Next Generation Blueprint of Computational Toxicology at the U.S. Environmental Protection Agency. Toxicol. Sci. 2019;169(2):317–332. doi: 10.1093/toxsci/kfz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice R.R., Austin C.P., Kavlock R.J., Bucher J.R. Improving the human hazard characterization of chemicals: a Tox21 update. Environ. Health Perspect. 2013;121(7):756–765. doi: 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa M., Casida J.E. Neonicotinoid insecticide toxicology: mechanisms of selective action. Annu. Rev. Pharmacol. Toxicol. 2005;45:247–268. doi: 10.1146/annurev.pharmtox.45.120403.095930. [DOI] [PubMed] [Google Scholar]
- US EPA . US EPA; Duluth, MN: 1987. Guidelines for the culture of fathead minnows Pimephales promelas for use in toxicity tests. EPA/600/3-87/001. [Google Scholar]
- US EPA . Office of Research and Development; Washington, DC: 1996. Prehatching development of the fathead minnow Pimephales promelas rafinesqe. EPA/600/R-96/079. [Google Scholar]
- United Nations. 2015. Globally harmonized system of classification and labelling of chemicals (GHS). Sixth Revised Edition. ST/SG/AC.10/30/Rev.6. United Nations, New York, NY. Part 4 – Environmental Hazards.
- Villeneuve D.L., Coady K., Escher B.I., Mihaich E., Murphy C.A., Schlekat T., Garcia-Reyero N. High-throughput screening and environmental risk assessment: State of the science and emerging applications. Environ. Toxicol. Chem. 2019;38(1):12–26. doi: 10.1002/etc.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve D.L., Garcia-Reyero N. Vision & strategy: Predictive ecotoxicology in the 21st century. Environ. Toxicol. Chem. 2011;30(1):1–8. doi: 10.1002/etc.396. [DOI] [PubMed] [Google Scholar]
- Webster A.F., Chepelev N., Gagné R., Kuo B., Recio L., Williams A., Yauk C.L. Impact of Genomics Platform and Statistical Filtering on Transcriptional Benchmark Doses (BMD) and Multiple Approaches for Selection of Chemical Point of Departure (PoD) PLoS One. 2015;10(8):e0136764. doi: 10.1371/journal.pone.0136764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Allen, B. C., & Thomas, R. S. (2007). BMDExpress: a software tool for the benchmark dose analyses of genomic data. In (Vol. 8, pp. 1-8): BMC Genomics. [DOI] [PMC free article] [PubMed]
- Yeakley J.M., Shepard P.J., Goyena D.E., VanSteenhouse H.C., McComb J.D., Seligmann B.E. A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLoS One. 2017;12(5):e0178302. doi: 10.1371/journal.pone.0178302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All research data/code have been made publicly accessible via GEO and US EPA Clowder; they will also be available through data.gov