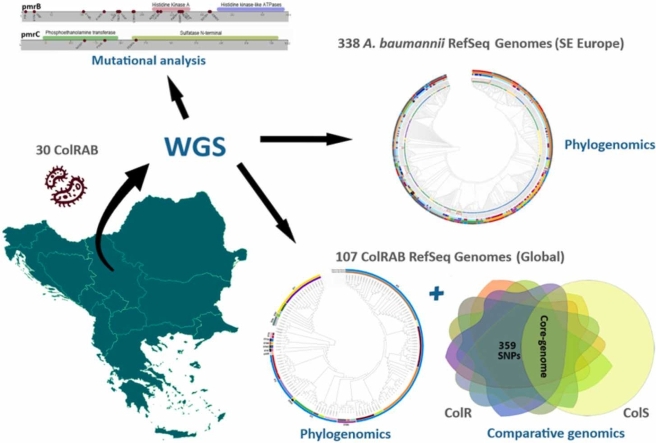
Abstract
This study aimed to investigate the prevalence and resistance mechanisms of colistin-resistant Acinetobacter baumannii (ColRAB) isolates in Serbia, assess their genetic relatedness to other circulating A. baumannii isolates in the neighbouring European countries, and analyse the global genomic epidemiology of ColRAB isolates. A total of 784 isolates of A. baumannii were recovered from hospitalised patients in Serbia between 2018 and 2021. The antimicrobial susceptibility testing was performed using disk diffusion and broth microdilution. All ColRAB isolates were subjected to DNA isolation and whole-genome sequencing (WGS). Overall, 3.94 % (n = 30) isolates were confirmed as ColRAB. Results of mutational and transcriptional analysis of genes associated with colistin resistance indicate the central role of the two-component regulating system, PmrAB, and increased expression of the pmrC gene in ColRAB. Most of the isolates (n = 29, 96.6 %) belonged to international clone II, with the most common sequence type being STPas2 (n = 23, 76.6 %). Based on the WGS analysis, ColRAB isolates belonging to the same ST isolated in various countries were grouped into the same clusters, indicating the global dissemination of several high-risk clonal lineages. Phylogenomic analysis of ColRAB isolates, together with all previously published A. baumannii genomes from South-Eastern European countries, showed that colistin resistance arose independently in several clonal lineages. Comparative genomic analysis revealed multiple genes with various roles (transcriptional regulation, transmembrane transport, outer membrane assembly, etc.), which might be associated with colistin resistance in A. baumannii. The obtained findings serve as the basis for further studies, contributing to a better understanding of colistin resistance mechanisms in A. baumannii.
Keywords: Colistin resistance, Acinetobacter baumannii, WGS, Phylogenomics, Comparative genomics
Graphical Abstract
1. Introduction
Acinetobacter baumannii is an emerging nosocomial pathogen responsible for a wide array of healthcare-associated infections. Due to its intrinsic resistance to numerous antimicrobial agents and its ability to efficiently acquire various mechanisms of resistance to different antibacterial agents, A. baumannii isolates are frequently multidrug-resistant (MDR) or extensively drug-resistant (XDR). Thus, carbapenem-resistant A. baumannii (CRAB) has been recognised as a critical-priority pathogen on the World Health Organization's priority list of antibiotic-resistant bacteria for effective drug development [1]. Currently, CRAB isolates are widespread, with rates reaching or exceeding 90 % in some clinical settings in Southern and Eastern European countries [2]. Reports on CRAB isolates derived from Serbian hospitals approached 93.7 % in 2018 [3].
Therapeutic options for infections caused by CRAB are exceedingly limited, relying in most cases on polymyxins. With the restriction of new drug development, colistin, also referred to as polymyxin E, has been reintroduced into the clinical practice and has become crucial for treating life-threatening CRAB-associated infections. However, the prevalence of colistin-resistant A. baumannii (ColRAB) has been increasing worldwide, with resistance rates reaching 12 % in China in 2015/2016 and 17 % in the Lebanon in 2016 [4]. In addition, a recent study from Serbia reported that 4.3 % of clinical isolates of A. baumannii collected in 2018 were ColRAB [3].
According to current knowledge, two main mechanisms underly colistin resistance in A. baumannii. The first is a phosphoethanolamine modification of lipid A, the primary target of colistin. The lowering of the net negative charge of the lipopolysaccharide (LPS) reduces the colistin's binding affinity and prevents cell membrane leakage [5]. This mechanism has been linked with mutations in the two-component transcriptional regulator genes pmrAB and consequent overexpression of the phosphoethanolamine phosphotransferase PmrC, leading to the lipid A modification. The second mechanism is the complete loss or reduction of LPS due to mutations or insertions of the genes involved in the lipid A biosynthesis pathway (lpxA, lpxC, and lpxD) [6]. In addition, colistin resistance may also result from the overexpression of etpA, a pmrC homolog, by integrating insertion sequence elements upstream of the eptA [7] or due to the insertional inactivation of a gene encoding an H-NS family transcriptional regulator [8].
Furthermore, plasmid-borne mobilised colistin resistance (mcr) genes, encoding phosphoethanolamine transferases, have been described in various Gram-negative bacteria [9]; however, these genes have rarely been described in A. baumannii [10], [11]. A recent study has reported the first occurrence of mcr-1-carrying Escherichia coli from animal farms in Serbia [12]. Still, it has not yet been detected in any isolates from the clinical environment [3], [13]. A recent study has also linked the lpsB gene, which encodes a glycosyltransferase involved in LPS synthesis, with colistin resistance in A. baumannii [14]. Mutations in the lpsB have been detected in ColRAB isolates where mutations in PmrABC and LpxACD operon were missing [15]. Other mechanisms reported include the reduction of osmoprotectant biosynthesis or expression of the efflux pumps [16], [17].
Comparative genomics enables an in-depth understanding of the pathogen's epidemiology and microbial evolution, identifying genomic characteristics responsible for a specific phenotype. A recent comparative genomics study has revealed several genes (pheS, cobS, garK, mutY, tmk and others) that may be associated with colistin resistance [18].
Given that the mechanisms of colistin resistance in A. baumannii have not yet been completely elucidated, this study aimed i) to explore the prevalence of already known resistance mechanisms of ColRAB, ii) to decipher potential novel genetic determinants of colistin resistance, and iii) to analyse genomic epidemiology of circulating ColRAB isolates through the whole-genome sequencing (WGS) and comparative genomics approach.
2. Materials and methods
2.1. Bacterial isolates and identification
This study included a total of 784 A. baumannii isolates obtained from the 15 regional microbiological laboratories of hospitals or clinical centres in nine cities throughout Serbia. The A. baumannii isolates were collected from clinical specimens of patients admitted to the hospitals between August 2018 and August 2021. The isolation and initial identification of Acinetobacter calcoaceticus–baumannii (Acb) complex was performed in the regional microbiological laboratories, using the VITEK®2 system (bioMérieux, Marcy-l’Étoile, France).
All isolates belonging to the Acb complex and the clinical and demographic data were sent to the coordinating laboratory at the Institute of Microbiology and Immunology, Faculty of Medicine, University of Belgrade, for further analysis. Before shipping, all patients were de-identified and re-coded in the regional laboratories. The study was approved by the ethical committee of the Faculty of Medicine, University of Belgrade (1550/II-4; 1550/IX-16).
Genomic DNA from an overnight culture was extracted using a QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. All isolates were screened for the presence of the intrinsic blaOXA-51 gene of A. baumannii [19]. The species identification was then confirmed by sequencing the rpoB, as previously described [20] (Table S1). Minimum inhibitory concentrations (MICs) for colistin were evaluated by ComASP Colistin (Liofilchem, Italy) according to the manufacturer's instruction, and susceptibility categories were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) recommendation (S≤2 μg/mL; R≥4 μg/mL) [21]. Escherichia coli NCTC 13846 was used as a control for colistin broth microdilution. In this study, only A. baumannii isolates resistant to colistin were further analysed.
2.2. Antimicrobial susceptibility of colistin-resistant A. baumannii isolates
Antimicrobial susceptibility of ColRAB isolates to ampicillin-sulbactam, amoxicillin-clavulanic acid, piperacillin-tazobactam, ceftazidime, cefepime, cefoxitin, imipenem, meropenem, amikacin, gentamicin, tobramycin, levofloxacin, ciprofloxacin, trimethoprim-sulfamethoxazole and tetracycline was determined by disk diffusion assay (Bio-Rad, UK), following the Clinical and Laboratory Standards Institute (CLSI) guidelines (v.2022) [21]. MICs for tigecycline were determined using the broth microdilution method with Mueller-Hinton broth (Bio-Rad, UK), following the CLSI guidelines [21]. Since there are no established MIC breakpoints for tigecycline in Acinetobacter spp., European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints for Enterobacterales were applied (S≤0.5 μg/mL; R>0.5 μg/mL)(v.2022) [22]. E. coli ATCC 25922 was used as a control strain for antimicrobial susceptibility testing. The isolates of A. baumannii were classified as follows: MDR (resistant to agents from ≥3 different antimicrobial classes), XDR (resistant to ≥1 agent in all but ≤2 antimicrobial classes), and pandrug-resistant (PDR) (resistant to all agents in all antimicrobial classes tested) [23].
2.3. Gene expression analysis by reverse transcription-quantitative PCR
Reverse transcription-quantitative PCR (RT-qPCR) was performed to determine the expression levels of genes associated with colistin resistance (lpxA, lpxC, lpxD, pmrA, pmrB, and pmrC) (Table S1). The overnight cultures of the ColRAB isolates (grown for 12 h at 37 °C) were diluted in fresh Mueller–Hinton broth (Bio-Rad, UK) and grown to the mid-logarithmic growth phase. After that, the RNA isolation, DNase I treatment, and reverse transcription was performed as previously described [24]. RT-qPCR was performed with an IC Green qPCR Universal Kit (Nippon Genetics Europe GmbH, Düren, Germany) in a 7500 Real-Time PCR System thermocycler (Applied Biosystems, Thermo Fischer Scientific, MA). The relative gene expression levels were calculated using the ΔΔCT method with the rpoB gene as an internal control. The obtained values were then normalised against results for colistin-susceptible A. baumannii (ColSAB) isolate 6077/12 [25]. All RT-qPCR experiments were done in triplicate, representing the results as mean values/standard deviations. A two-tailed Student's t-test was performed to compare the two groups. P values of ≤ 0.05 were considered statistically significant.
2.4. Whole genome sequencing and genome assembly
The quality of previously extracted bacterial DNA from all ColRAB isolates was assessed; the DNA concentrations were measured by Qubit (Thermo Fisher, USA), and the DNA integrity was evaluated by gel electrophoresis. The DNA samples were then sent to the Novogene Corporation, Inc. (Amsterdam, Netherlands) for short-read WGS on the MiSeq Illumina platform.
The quality of raw reads was first assessed using FastQC [26], and poor-quality reads were removed (Phread score<20). Afterwards, de novo genome assembly was performed using SPAdes v3.12 [27], and the assemblies' quality was evaluated using Quast v5.0.2 [28]. The genome annotation was then performed using Prokka v1.14.6 [29] and RAST [30].
2.5. Bioinformatic analysis
The species identification was reconfirmed using SpeciesFinder v2.0 from the Center for Genomic Epidemiology (CGE) (https://cge.cbs.dtu.dk/services/SpeciesFinder/) [31]. Sequence types (STs) for all ColRAB isolates were inferred from assembled genomes using mlst v2.19.0 (https://github.com/tseemann/mlst) [32]. The identification of AMR genes was performed using ABRicate v1.0.1 (https://github.com/tseemann/abricate) set against the comprehensive antibiotic resistance database (CARD) (https://card.mcmaster.ca/) [33]. The capsular polysaccharide loci (KL) and lipooligosaccharide outer core loci (OCL) were predicted using KAPTIVE against the A. baumannii databases [34].
In addition, the mutations in the pmrA, pmrB, pmrC, lpxA, lpxC, and lpxD genes were detected using Geneious Prime software with corresponding genes of A. baumannii ATCC 17978, A. baumannii ATCC 19606, A. baumannii ACICU, and A. baumannii AYE used as a negative control. Because mutations can also represent genetic polymorphisms associated with different clonal lineages, additional 280 genomes of ColSAB isolates were included in the analysis (Table S2). Selection criteria for ColSAB genomes included in the analysis were the availability of data on colistin resistance (MICs≤2 μg/mL) and the inclusion of the genomes in the NCBI Reference Sequence project. Furthermore, all ColRAB genomes were manually screened for other colistin resistance mechanisms: the presence of mcr, insertion of ISAba1 upstream of eptA, and insertional inactivation of lpxA, lpxC, and lpxD. In addition, mutations in other genes associated with colistin resistance (CHO1, cobS, cobV, cysH, dcm, DNMT1, DTYMK, eno, ENO, garK, glxK, lpsB, mdh, miaA, MPST, mutY, pheS, pldA, pssA, sseA, tmk, TST, udg, ureC and vacJ) were also analysed.
2.6. Pan-genome and phylogenomic analysis
The pan-genome analysis was performed using Roary v3.13.0 [35], with a 99 % core definition threshold and a minimum 95 % blastp identity. Next, SNP-sites v2.4.1 [36] was used to identify core genome single-nucleotide polymorphisms (SNP). A core-SNP-based phylogenetic tree was constructed using raxmlHPC-PTHREADS v8.2.12 with the neighbour-joining method using 1000 bootstraps [37] and was visualised using iTOL software [38]. Finally, a presence and absence matrix against a phylogenetic tree was generated using the roary_plots script (https://github.com/sanger-pathogens/Roary/tree/master/contrib/roary_plots) and visualised using the Phandango website (https://jameshadfield.github.io/phandango/).
To analyse the global epidemiology of ColRAB isolates, the phylogenetic tree based on core-gene SNPs of 137 ColRAB isolates was built, as described above. Besides 30 ColRAB from the current study, 107 previously published genomes of ColRAB isolates were downloaded from the NCBI genome database (Table S2), and their STs were inferred. Therefore the inclusion criteria for 107 ColRAB genomes were the following: colistin-resistant Acinetobacter baumannii (MICs≥4 μg/mL), data obtained either from the NCBI BioSample database or from the published articles [15], [39], [40], [41], [42], [43], [44], [45], [46]; NCBI quality assurance and validation of assembled genomes and inclusion in the NCBI’s RefSeq database.
In addition, to evaluate genomic relatedness between the Serbian ColRAB isolates and A. baumannii from the region, a total of 368 genomes were included in the phylogenomic analysis. Beside 30 ColRAB genomes from this study, all genomes of A. baumannii from eight neighbouring South-Eastern European countries (Albania, Bosnia and Herzegovina, Croatia, Greece, Hungary, Kosovo, Montenegro, and Romania) available in the NCBI’s Genome database, irrespectively of the susceptibility to colistin (S, R, or unknown) were analysed (n = 338). As previously described, the phylogenetic tree was constructed based on the core-gene SNPs.
2.7. Comparative genome analysis
As A. baumannii is highly susceptible to allelic variations, a comparative genome analysis of isolates with the same genetic background, belonging to the most common ST2 type in Serbia and globally, was performed. A total of 100 ColRAB ST2 (77 available in the NCBI RefSeq Genome Database and 23 from the present study), 107 ColSAB ST2 genomes available in the NCBI RefSeq Genome Database, and the reference strain A. baumannii ACICU (ST2) were analysed. The pan-genome and core-genome analyses were performed as described above for all ColRAB and ColSAB isolates. The pairwise SNP analysis was performed on core genome data of ColRAB isolates aligned with the ColSAB isolates using Geneious Prime MAFFT aligner and SNP calling by Geneious Prime SNP finder.
As suggested by Gerson et al. [47], to ensure that all genetic polymorphisms not associated with colistin resistance were excluded from the final results, an additional pairwise SNP analysis was performed against 173 genomes of ColSAB/non-ST2 isolates and three reference strains (A. baumannii ATCC 17978, A. baumannii ATCC 19606 and A. baumannii AYE) (Table S2). Amino acid substitutions found in both ColR and ColS isolates were considered to be genetic polymorphisms not associated with colistin resistance and were excluded. All amino acid substitutions found only in ColRAB isolates and not observed in any other ColSAB (ST2 and non-ST2) isolate were considered to be potentially associated with colistin resistance.
3. Results
3.1. Bacterial isolates
Out of the initial 784 A. baumannii isolates recovered from hospitalised patients, 752 isolates (95.91 %) were molecularly confirmed to be A. baumannii. The prevalence of colistin resistance in clinical isolates of A. baumannii was 3.98 % (n = 30). Detailed information concerning patient characteristics, hospital units, and specimen types are listed in Table 1.
Table 1.
Demographic and clinical characteristics of colistin-resistant Acinetobacter baumannii-infected/colonised study patients.
| Characteristic | No. (%) |
|---|---|
| Gender | |
| Male gender | 17 (56.6) |
| Female gender | 13 (43.4) |
| Region | |
| Belgrade | 8 (26.6) |
| Vojvodina | 14 (46.6) |
| The rest of Serbia | 8 (26.6) |
| Admission ward | |
| Intensive care unit | 12 (40) |
| Birth centre | 1 (3.3) |
| Cardiovascular surgery | 4 (12.2) |
| Neurosurgery | 1 (3.3) |
| Corona care centre | 12 (40) |
| Type of specimen | |
| Tracheal aspirate | 16 (53.3) |
| Bronchoalveolar lavage | 3 (10) |
| Sputum | 2 (6.6) |
| Blood | 9 (30) |
| Comorbidity | |
| Heart insufficiency | 2 (6.6) |
| Chronic kidney disease | 1 (3.3) |
| COVID-19 pneumonia | 12 (40) |
| Invasive procedures | |
| Any surgical procedure | 3 (10) |
| Mechanical ventilation | 12 (40) |
| Central venous catheter | 2 (6.6) |
3.2. Antimicrobial susceptibility of colistin-resistant A. baumannii isolates
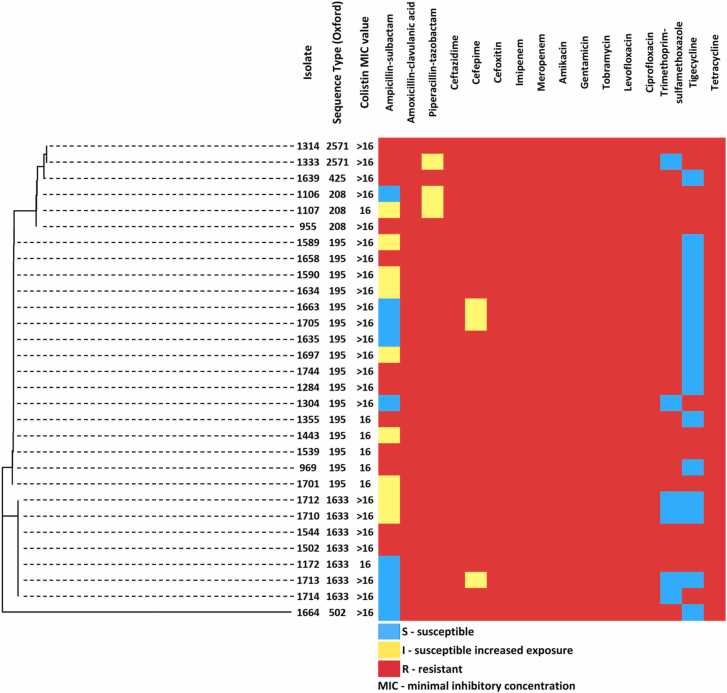
All 30 ColRAB isolates were CRAB and were resistant or had reduced susceptibility to amoxicillin-clavulanic acid, piperacillin-tazobactam, ceftazidime, cefepime, cefoxitin, meropenem, imipenem, amikacin, gentamicin, tobramycin, levofloxacin, ciprofloxacin, and tetracycline (Fig. 1). Antimicrobial susceptibility testing against ampicillin-sulbactam revealed that nine isolates (30 %) were category I and nine other isolates (30 %) were category S. The drug with the highest prevalence of susceptibility was tigecycline (56.7 %), followed by ampicillin-sulbactam (30 %) and trimethoprim-sulfamethoxazole (20 %). Except for seven isolates with MIC values of 16 mg/L (23 %), the majority of isolates exhibited colistin MIC> 16 mg/L (n = 23, 76.7 %). All 30 ColRAB isolates were classified as XDR (100 %), while five matched the PDR criteria (16.7 %). The antibiotic resistance profiles of the 30 ColRAB isolates are shown in Fig. 1.
Fig. 1.
Dendrogram based on core-SNP distances, STs (Oxford scheme), and antimicrobial susceptibility profiles of 30 colistin-resistant Acinetobacter baumannii isolated from 2018 to 2021 in Serbia.
3.3. Expression analysis of the colistin-resistance-associated genes
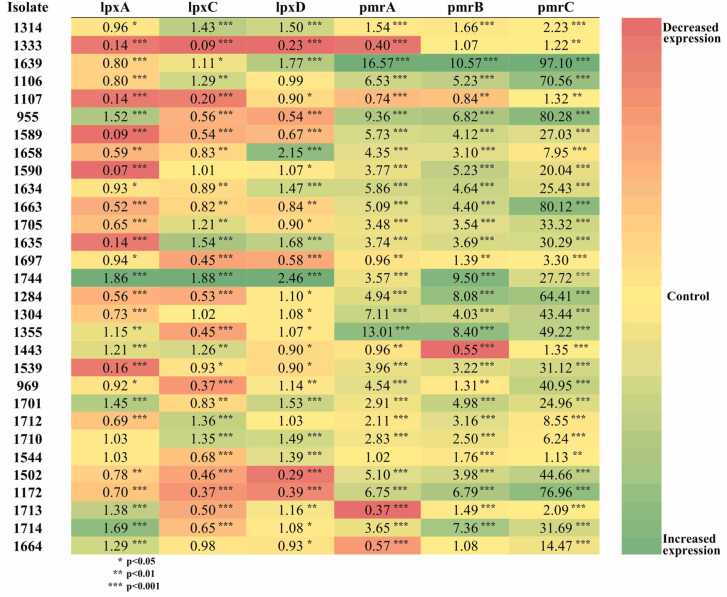
Transcription analysis of the lpxA, lpxC, lpxD, pmrA, pmrB, and pmrC was performed to determine the underlying colistin resistance mechanism in tested ColRAB isolates. The lpxA was statistically significantly downregulated in 20 (66.6 %), lpxC in 19 (63.3 %), and lpxD in 12 (40 %) isolates. Meanwhile, pmrA was significantly upregulated in 23 (76.7 %), pmrB in 27 (90 %), and pmrC in 30 (100 %) isolates (Fig. 2).
Fig. 2.
Heatmap analysis of the relative expression level of colistin-resistance associated genes (lpxA, lpxC, lpxD, pmrA, pmrB, and pmrC) from tested colistin-resistant Acinetobacter baumannii isolates. All expression results were normalised relative to the rpoB gene by the ΔΔCT method. Values are the means from results obtained in triplicate. Green colour - positive fold-change indicates higher expression in the isolate compared with the control (yellow); red colour - negative fold-change indicates lower expression in the isolate compared with the control. A student's t-test was used to compare the results obtained for colistin-resistant isolates with those for colistin-susceptible isolate A. baumannii 6077/12.
Variations in decreased/increased expression of the lpxA (0.09- to 1.86-fold compared with control), lpxD (0.23- to 2.46-fold) and lpxC (0.09- to 1.88-) mRNAs within different isolates were observed (Fig. 2). Expression analysis of the pmrA and pmrB revealed upregulation in 24 isolates (80 %). In contrast, in six isolates (20 %), it was downregulated (Fig. 2). Expression levels of the pmrC were significantly increased in all isolates (100 %) from 1.12- to 97.09-fold (Fig. 2).
3.4. Sequence types and antimicrobial resistance (AMR) determinants
In the 30 ColRAB isolates, three STs were identified based on the Pasteur MLST scheme. Among them, ST2 was the most prevalent (n = 23; 76.66 %), followed by ST492 (n = 6, 20 %), and ST636 (n = 1; 3.33 %). The Oxford MLST scheme revealed the following six STs: ST195 (n = 16; 53.33 %), ST1633 (n = 7, 23.3 %), ST208 (n = 3, 10 %), ST425 (n = 1, 3.3 %), and ST502 (n = 1, 3 %). The novel type ST2571 was assigned to two isolates (6.6 %) (Table S4).
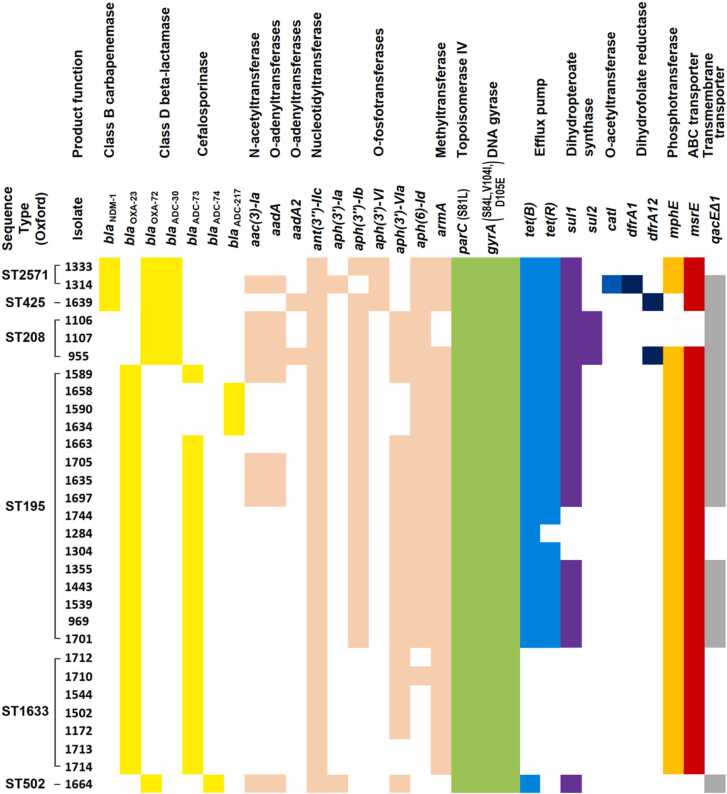
Various AMR determinants associated with resistance to several antimicrobial classes were detected, as illustrated in Fig. 3. Notably, among the ColRAB, several beta-lactamase-encoding genes were detected. Overall, 23 (76.66 %) isolates carried the blaOXA-23 gene, while seven (23.33 %) and three (10 %) isolates harboured the blaOXA-72 and blaNDM-1 as single-copy genes, respectively. The blaADC-73 (n = 20; 66.66 %), blaADC-30 (n = 6; 20 %), blaADC-217 (n = 3; 10 %), and blaADC-74 (n = 1; 3.33 %) genes were also detected. The mcr-1 and other variants conferring colistin resistance, blaKPC and blaGES encoding class A beta-lactamases, and extended-spectrum beta-lactamases, were not detected among tested A. baumannii isolates.
Fig. 3.
Antimicrobial resistance genes detected in the study of 30 colistin-resistant clinical Acinetobacter baumannii isolates. Each colour represents the detected antimicrobial determinant that could deter resistance to different antimicrobial agents: yellow - β-lactams; peach - aminoglycoside; green - fluoroquinolone; sky blue – tetracycline; purple - sulfonamide; blue - chloramphenicol; dark blue - trimethoprim; orange - macrolide; red - macrolide, fluoroquinolone and lincosamide; and grey - disinfecting agents.
Regarding the distribution of AMR genes among detected STs of the ColRAB isolates, all ST2 isolates had the blaOXA-66/blaOXA-23 genotype. In addition, the blaADC-73 and blaADC-217 were detected only among ST2 isolates. The blaNDM-1, blaADC-30, catI, drfA1, dfrA12, aadA2, aph(3')-VI and sul2 genes were only present in ST492 isolates, while blaOXA-72, aac(3)-Ia, aadA and aph(3')-Ia were present in both ST492 and ST636 isolates. The blaADC-74 was detected only in ST636 isolate.
3.5. Capsular polysaccharide locus (KL) and outer core locus (OCL) diversity in colistin-resistant A. baumannii
Six different KL variants were detected among ColRAB isolates. The KL9 (n = 16, 53.3 %) was the most prevalent, followed by KL130 (n = 7, 23.3 %), KL2 (n = 3, 10 %), KL104 (n = 2, 6.6 %), KL40 (n = 1, 3.3 %), and KL4 (n = 1, 3.3 %). The association between STs based on the Oxford MLST scheme and KLs was found, revealing that all isolates belonging to the same ST had the same KL variant (Table S5). The OC locus had less variation among isolates, with only two OC loci being detected, OC1 (n = 29, 96.6 %) and OC2 (n = 1, 3.3 %). Only the ST502 isolate had OC2 loci, while all other detected STs had OC1.
3.6. Mutational analysis of colistin-resistance-associated proteins
Sequence analysis of the genes encoding LpxA, LpxC, LpxD, PmrA, PmrB, and PmrC proteins detected no amino acid substitutions in any of the proteins, with the exception of PmrB and PmrC, when compared 30 ColRAB from the present study to the reference strains and 280 genomes of ColSAB isolates.
PmrB protein substitution T269P was the most prevalent substitution, occurring in 17 isolates (56.6 %), followed by A226V in seven isolates (23.3 %), E185K, K6I and L160F in four isolates (13.3 %), and S17G, I19F, F103L, I163N, I164F, S183F, T232A, R263H, Q265P, H266Y, D313N, and G315V in one isolate (3.3 %) (Table 2.). Substitutions R230Q, R125P and L128I in the PmrC protein were found in eight (26.6 %), seven (23.3 %), and three isolates (10 %), respectivly.
Table 2.
Overview of amino acid substitutions detected in colistin-resistance-associated proteins found in 30 colistin-resistant Acinetobacter baumannii from Serbia.
| Isolate | Sequence Type (Oxford) | Amino acid substitutions |
|||
|---|---|---|---|---|---|
| PmrB | PmrC | PheS | MutY | ||
| 1333 | 2571 | R230Q | |||
| 1314 | 2571 | R230Q | |||
| 1639 | 425 | Q265P | |||
| 1106 | 208 | I163N, I164F, E185K, T269P | |||
| 1107 | 208 | S17G | |||
| 955 | 208 | R263H | |||
| 1589 | 195 | L160F, T269P | R230Q | ||
| 1658 | 195 | K6I, T269P | L128I | ||
| 1590 | 195 | K6I, T269P | L128I | ||
| 1634 | 195 | K6I, T269P | L128I | ||
| 1663 | 195 | L160F, T269P | |||
| 1705 | 195 | T269P | R230Q | ||
| 1635 | 195 | S183F, T269P | |||
| 1697 | 195 | T269P | |||
| 1744 | 195 | K6I, T269P | |||
| 1284 | 195 | L160F, T269P | R230Q | ||
| 1304 | 195 | L160F, T269P | |||
| 1355 | 195 | T269P, G315V | |||
| 1443 | 195 | T269P | R230Q | ||
| 1539 | 195 | E185K, T269P | |||
| 969 | 195 | E185K, T269P | R230Q | ||
| 1701 | 195 | E185K, T269P | |||
| 1712 | 1633 | A226V | R125P | P2S, G94R | |
| 1710 | 1633 | A226V | R125P, R230Q | P2S, G94R | |
| 1544 | 1633 | F103L, A226V | R125P | P2S, G94R | |
| 1502 | 1633 | A226V, D313N | R125P | P2S, G94R | |
| 1172 | 1633 | A226V, H266Y | R125P | P2S, G94R | |
| 1713 | 1633 | A226V | R125P | P2S, G94R | |
| 1714 | 1633 | A226V, T232A | R125P | P2S, G94R | |
| 1664 | 502 | I19F | R101C | G94R | |
In addition, amino acid changes in genes encoding PheS and MutY proteins were detected, while no mutations were found in other genes associated with colistin resistance. Furthermore, insertional inactivation of lpxA, lpxC, and lpxD genes, and insertion of ISAba1 upstream of EptA, have not been detected.
3.7. Pan-genome and phylogenomic analysis
The pan-genome analysis of the 30 ColRAB isolates showed that 3118 (59.80 %), 134 (2.57 %), 957 (18.35 %), and 1005 (19.27 %) of 5214 pan-genes were identified as core, soft-core, shell, and cloud genes, respectively. Genes encoding hypothetical proteins represented 41.3 % and 69.4 % of the core and accessory genomes, respectively.
Based on the phylogenomic analysis, isolates have been grouped into six clades according to the detected STs (Fig. S1). Among these clades, isolates have further clustered based on the year of the isolation and the geographical location. The similarity of the tested ColRAB isolates based on the core-genome SNP data ranged from 20 % to 99 %, with isolates in each cluster sharing 99 % pairwise identity (3–79 core-SNPs), showing high relatedness among them (Fig. S2). In addition, the gene presence and absence matrix of the pan-genome profiles of ColRAB isolates indicates higher genetic diversity than the phylogenetic tree based on the core-genome data (Fig. S3).
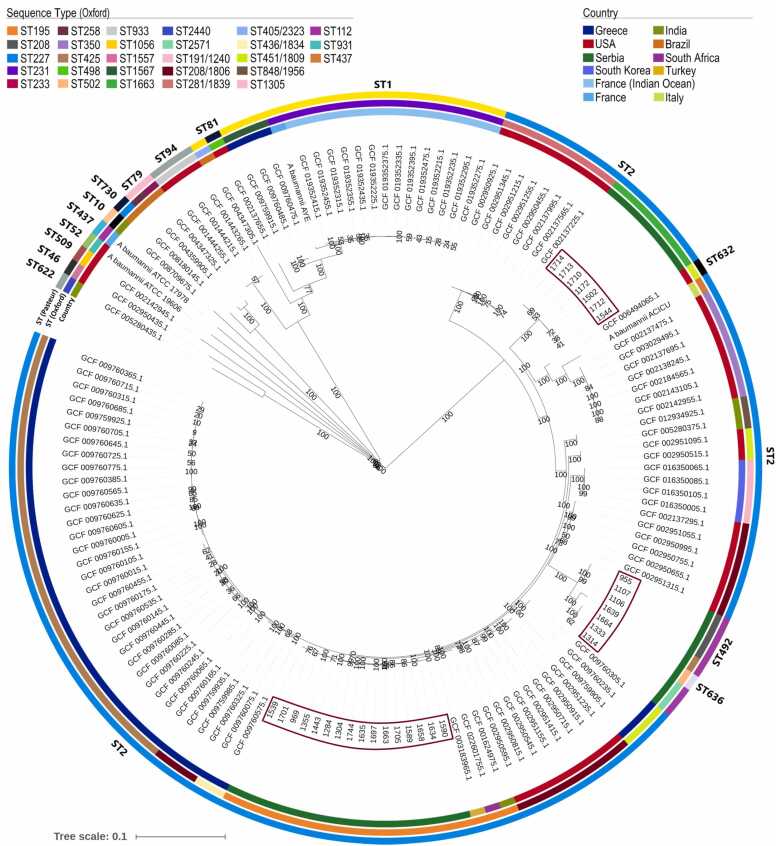
The phylogenetic tree of 137 ColRAB isolates and four ColSAB reference isolates, A. baumannii ATCC 19606, A. baumannii ATCC 17978, A. baumannii ACICU and A. baumannii AYE indicated high diversity of globally detected ColRAB isolates, with the majority (78.26 %) belonging to IC II (Fig. 4). Isolates from different countries mostly clustered together based on the detected STs and geographical location. The ColRAB clonal lineages from Serbia have clustered with isolates from the United States, South Africa and Turkey, indicating the global dissemination of high-risk clones.
Fig. 4.
Global SNP-based phylogenetic tree constructed on 110,717 core gene SNP alignment of the 137 colistin-resistant Acinetobacter baumannii clinical isolates using the neighbour-joining method with 1000 bootstraps. The different countries can be distinguished by colour codes displayed in the figure legend; the fragments with varying colours in the outer rings represent corresponding sequence types (STs), the outer ring depicts detected STs based on the Pasteur scheme, the middle ring shows STs based on the Oxford scheme, while the innermost ring represents the country of origin. Bootstrap values (expressed as percentages of 1000 replications) are shown at the branch points, and branch lengths are proportional to the estimated numer of substitutions per site.
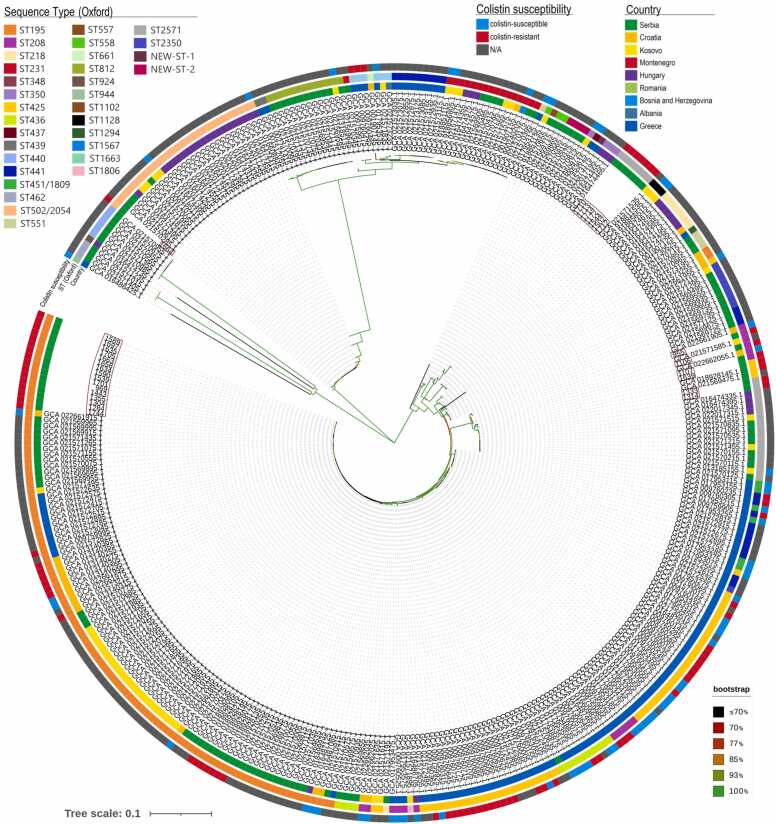
The results of the phylogenomic analysis of the 30 ColRAB genomes from the current study and 338 previously published genomes from the nine neighbouring South-Eastern European countries are shown in Fig. 5. Overall, the phylogenetic tree of the 368 A. baumannii isolates could be divided into many clades based on their detected STs and geographical origin. Furthermore, the ColRAB isolates from this study clustered with other isolates from Serbia, Croatia, Romania, Hungary and Montenegro, indicating their close genetic relatedness.
Fig. 5.
SNP-based phylogenetic tree constructed on 90,445 core gene SNP alignment of the 368 Acinetobacter baumannii isolated in the South-Eastern European region using the neighbour-joining method with 1000 bootstraps. The different countries in the innermost ring can be distinguished by colour codes displayed in the figure legend; the fragments with varying colours in the outer ring represent colistin susceptibility profiles, and the middle ring shows STs based on the Oxford scheme. The branch colour indicates bootstrap value (see right bottom corner), and branch lengths are proportional to the estimated number of substitutions per site.
3.8. Comparative genomic analysis
The comparative genomic analysis of 1864 core-genes present in all isolates (100 ColRAB/ST2 and 284 ColSAB isolates) revealed 246,360 SNPs, of which 359 non-synonymous SNPs were detected only in ColRAB isolates (Table S6). These SNPs were located in 262 different genes, involved in outer membrane assembly, peptidoglycan synthesis, transmembrane transport, transcription, DNA repair, stress response, protein biosynthesis, lipid metabolism, tRNA processing, rRNA processing, as well as encoding for various oxidoreductases and hydrolases (Table S6). The highest number of different SNPs was detected in the PmrB protein (31 SNP). In addition, amino acid substitution A226V in PmrB protein was detected in 44.6 % of the ColRAB isolates, followed by amino acid substitutions in proteins involved in the regulation of transcription, transmembrane transport and signal transduction (Table 3).
Table 3.
Highest frequency polymorphisms detected in core genes of colistin-resistant Acinetobacter baumannii isolates compared to the colistin-sensitive Acinetobacter baumannii isolates.
| Protein name | Function | Gene locus tag in A. baumannii ACICU | Frequency | Amino Acid Change |
|---|---|---|---|---|
| Acetyl-coenzyme A synthetase Acs | Acetyl-CoA biosynthesis | ACICU_RS17860 | 37.40 % | A565V |
| Aromatic ring-hydroxylating dioxygenase subunit alpha | Oxidoreductase | ACICU_RS10105 | 17.20 % | R349H |
| NADPH-dependent 2,4-dienoyl-CoA reductase | Flavoprotein oxidoreductase | ACICU_RS10315 | 16.20 % | A155V |
| Hypothetical protein | Unknown function | ACICU_RS11690 | 16.20 % | R77H |
| AarF/UbiB family protein | Protein phosphorylation | ACICU_RS11940 | 37.40 % | P82S |
| LemA family protein | Integral component of membrane | ACICU_RS17225 | 23.20 % | A167E |
| ATP-binding protein | Unknown function | ACICU_RS13425 | 23.20 % | D640A |
| Penicillin-binding protein 1B MrcB | Regulation of cell shape, peptidoglycan synthesis | ACICU_RS12655 | 16.20 % | P545L |
| GntR family transcriptional regulator | Transcription regulation | ACICU_RS09030 | 16.20 % | S39T |
| Alkene reductase NemA | Xenobiotic reductase | ACICU_RS17885 | 16.20 % | D692G |
| PepP | Proteolysis | ACICU_RS05710 | 16.20 % | Y800F |
| Peptide chain release factor 3 | Protein biosynthesis | ACICU_RS02980 | 17.20 % | K231N |
| PmrB | Signal transduction, LPS modification |
ACICU_RS15230 | 44.40 % | A226V |
| 16.20 % | T269P | |||
| RpoA | Transcription regulation | ACICU_RS16580 | 37.40 % | V59L |
| MotA/TolQ/ExbB proton channel family protein | Integral component of membrane, transmembrane transport | ACICU_RS02510 | 37.40 % | A127T |
| PDR/VanB family oxidoreductase | Oxidoreductase | ACICU_RS10090 | 16.20 % | I181M |
4. Discussion
The emergence of infections caused by ColRAB poses a significant problem in hospital settings worldwide and has reached critical levels in some countries, with the highest rates reported in the Lebanon (17.5 % in 2016) and China (11.8 % in 2015/16) [4]. Since the vast majority of ColRAB isolates are CRAB or even pan-resistant, managing these infections is challenging. Thus, it is of utmost importance to explore and understand the mechanisms of colistin resistance in A. baumannii isolates and evaluate the dissemination of ColRAB strains. To address the genetic relatedness of the isolates and the molecular basis of resistance, WGS was applied to explore and study ColRAB isolates collected from hospitals throughout Serbia.
Among collected A. baumannii isolates, the prevalence of ColRAB (3.98 %) was similar to that reported in Serbia in 2018 (4.3 %) [3]. In the present study, all ColRAB isolates were XDR, while five isolates were classified as PDR. Furthermore, three out of five PDR strains were isolated in the same hospital within a three-week time span, two being closely genetically related, indicating the possibility of intra-hospital dissemination.
In the current study, the mutations in genes conferring resistance to colistin were detected only in pmrB and pmrC, with 17 different mutations detected in pmrB. Amino acid substitutions in the PmrB histidine kinase have been described as the primary mechanism for the development of colistin resistance in clinical isolates of A. baumannii [8], [47], [48]. Multiple mutations detected in the histidine kinase domain of PmrB protein could play a role in colistin resistance (A226V, T232A, R263S, Q265, H266Y and T269P), as the same or different substitutions in these positions were previously described in ColRAB isolates [25], [49]. Furthermore, the substitution of the 315 amino acid position (G to V) in PmrB protein could also contribute to colistin resistance, as glycine at that position has been shown to be required for autophosphorylation of PmrC protein [50]. The R125P substitution in the PmrC protein was previously correlated with increased expression of pmrC [47]. Other substitutions detected in PmrC have not been previously reported, so their effect on colistin resistance requires further investigation.
All tested ColRAB isolates had upregulated expression of the pmrC, indicating its central role in colistin resistance through lipid A modification. Thus, transcriptional analysis of genes encoding the two-component regulating system, PmrAB, revealed increased and decreased expression compared with the ColSAB isolates. In addition, the downregulation of genes involved in LPS biosynthesis could contribute to colistin resistance in analysed isolates through decreased LPS production.
The globally disseminated class D carbapenemase-encoding gene, blaOXA-23 [51], [52], was detected in most ColRAB isolates, whereas the blaOXA-72 was less prevalent. In addition, some blaOXA-72-positive isolates also co-harboured a metallo-β-lactamase, the blaNDM-1 gene. These findings are in concordance with previous reports on A. baumannii isolates from Serbia [3], [13], [51], [53], [54]. Additionally, ISAba125 was located upstream of the blaNDM-1 gene, providing a promoter sequence for the expression of blaNDM-1 [55].
The pan-genome analysis revealed that ColRAB isolates from this study shared 59.80 % core genes and contained 19.27 % cloud genes, indicating high genetic relatedness of A. baumannii clinical isolates from Serbia. Furthermore, a high level of hypothetical proteins was detected in the core and accessory genome. These genes could have a significant role in pathogen survival and adaptation mechanisms. Furthermore, the presence and absence matrix of the pan-genome revealed a higher genomic diversity among isolates than the phylogenetic tree based only on core-genome SNP data. As such, the inclusion of an accessory genome provided additional discrimination and illuminated possible epidemiological links.
This study presented the characterised collection of all MDR A. baumannii genomes from the South-Eastern European region deposited in the NCBI database up to September 2022. Phylogenomic analysis of ColRAB genomes from this study and previously published genomes from eight neighbouring European countries revealed high diversity among circulating A. baumannii strains, with 32 different STs detected (Fig. 5). Isolates belonging to ST425 and ST208 were genetically diverse and formed different clusters. These results showed that the cgSNP analysis is instrumental as a typing marker tool with very high discriminatory power, revealing a diversity that the conventional MLST method overlooked. Isolates from this study clustered independently from previously reported ColRAB isolates from Serbia and were more closely related to isolates from neighbouring countries. Based on the data available, colistin resistance arose independently in South-Eastern Europe in several lineages and was, in some cases, clonally disseminated locally.
Most of the reported ColRAB isolates in South-Eastern Europe and globally reported ColRAB isolates belonged to the IC II, a globally disseminated high-risk clone frequently implicated in nosocomial infections. Several ColRAB clonal lineages from Serbia were more closely related to the US ColRAB isolates than those collected from Greece, including those carrying the blaNDM-1 gene. These results indicate that human migration and travel facilitate the global spread of high-risk clones and antimicrobial resistance and the ease of cross-border clonal dissemination.
The result of comparative genomics revealed polymorphisms in 262 core genes of ColRAB isolates compared to the ColSAB isolates. Among them, 73 genes with SNPs detected are genes engaged in the transmembrane transport, cell membrane and outer membrane biosynthesis or are an integral part of the membrane. Given the complexity of the pathways by which Gram-negative bacteria acquire colistin resistance, some of these genes might be associated with colistin resistance. On the other hand, some of the detected polymorphisms might be a result of the natural evolution of A. baumannii and might not be involved in the resistance phenotype. Nonetheless, further study is needed to define these genes and their association with colistin resistance.
5. Conclusions
In conclusion, this study provides insight into the diversity and complexity of colistin resistance in A. baumannii, presenting novel candidate genes that may be associated with colistin resistance. Mutations in the PmrB protein and subsequent overexpression of the phosphoethanolamine transferase PmrC were found to be the main mechanism of colistin resistance among the tested isolates. Furthermore, obtained results showed that colistin resistance emerged in Serbia and this part of the Europe as the result of the accumulation of chromosomal mutations independently in several global high-risk clonal lineages rather than due to the dissemination of resistant strains, which were then locally disseminated. Global phylogenomic analysis of ColRAB isolates revealed the extent to which some MDR high-risk clones have disseminated. Therefore, the rising resistance rates and the independent emergence of ColRAB clones point out the need for a global surveillance system for tracking high-risk clones, the continuous surveillance of infections caused by A. baumannii and the development of control and prevention strategies in Serbia and the Balkans.
Ethical approval
The study was approved by the ethical committee of the Medical Faculty, University of Belgrade (permission number No. 1550/IX-16).
Funding
This work was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia, Serbia [Grant No. 451-03-68/2022-14/200110, Grant No. 451-03-68/2022-14/200042 and 451-03-68/2022-14/200178] and the Science Fund of the Republic of Serbia, Serbia [IN-DEPTH, Grant No. 6059147].
CRediT authorship contribution statement
BJ and IG conceived and designed the study. All authors contributed to the study's elaboration and design. JK and KN carried out the experiments; JK conducted the analysis and interpretation of the data. All of the authors drafted the manuscript. BJ and IG revised the manuscript and gave the final approval of the manuscript. All authors have read and agreed with the final version of the manuscript.
Conflict of Interest
The authors declare that they have no conflict of interests.
Acknowledgements
The authors wish to thank the microbiologists from all participating laboratories for supplying the isolates used in this study: Lazar Ranin, Anita Sente Zigmanovic, Sanja Zornic, Snezana Delic, Bojana Lukovic, Lidija Boskovic, Dragana Andjelkovic and Tatjana Boskovic.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2022.12.045.
Contributor Information
Branko Jovcic, Email: bjovcic@bio.bg.ac.rs.
Ina Gajic, Email: ina.gajic@med.bg.ac.rs.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Talebi Bezmin Abadi A., Rizvanov A.A., Haertlé T., Blatt N.L. World Health Organization Report: current crisis of antibiotic resistance. Bionanoscience. 2019;9:778–788. doi: 10.1007/S12668-019-00658-4/TABLES/1. [DOI] [Google Scholar]
- 2.ECDC Antimicrobial resistance surveillance in Antimicrobial resistance surveillance in Europe, Stockholm. 2022 doi: 10.2900/112339. [DOI] [Google Scholar]
- 3.Lukovic B., Gajic I., Dimkic I., Kekic D., Zornic S., Pozder T., et al. The first nationwide multicenter study of Acinetobacter baumannii recovered in Serbia: emergence of OXA-72, OXA-23 and NDM-1-producing isolates. Antimicrob Resist Infect Control. 2020;9 doi: 10.1186/s13756-020-00769-8. Article 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pormohammad A., Mehdinejadiani K., Gholizadeh P., Nasiri M.J., Mohtavinejad N., Dadashi M., et al. Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: a systematic review and meta-analysis. Microb Pathog. 2020;139 doi: 10.1016/j.micpath.2019.103887. Article 103887. [DOI] [PubMed] [Google Scholar]
- 5.Poirel L., Jayol A., Nordmanna P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nurtop E., Baylndlr Bilman F., Menekse S., Kurt Azap O., Gönen M., Ergonul O., et al. Promoters of colistin resistance in Acinetobacter baumannii infections. Microb Drug Resist. 2019;25:997–1002. doi: 10.1089/MDR.2018.0396. [DOI] [PubMed] [Google Scholar]
- 7.Potron A., Vuillemenot J.B., Puja H., Triponney P., Bour M., Valot B., et al. ISAba1-dependent overexpression of eptA in clinical strains of Acinetobacter baumannii resistant to colistin. J Antimicrob Chemother. 2019;74:2544–2550. doi: 10.1093/JAC/DKZ241. [DOI] [PubMed] [Google Scholar]
- 8.Lucas D.D., Crane B., Wright A., Han M.L., Moffatt J., Bulach D., et al. Emergence of high-level colistin resistance in an Acinetobacter baumannii clinical isolate mediated by inactivation of the global regulator H-NS. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.02442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khuntayaporn P., Thirapanmethee K., Chomnawang M.T. An update of mobile colistin resistance in non-fermentative Gram-Negative Bacilli. Front Cell Infect Microbiol. 2022;0 doi: 10.3389/FCIMB.2022.882236. Article 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan R., Li C., Duan R., Qin S., Liang J., Xiao M., et al. Retrospective screening and analysis of mcr-1 and blaNDM in Gram-Negative Bacteria in China, 2010–2019. Front Microbiol. 2020;11 doi: 10.3389/FMICB.2020.00121/BIBTEX. Article 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyman Y., Reuter S., Whitelaw A.C., Stein L., Maloba M.R.B., Newton-Foot M. Characterisation of mcr-4.3 in a colistin-resistant Acinetobacter nosocomialis clinical isolate from Cape Town, South Africa. J Glob Antimicrob Resist. 2021;25:102–106. doi: 10.1016/J.JGAR.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Mišić D., Kiskaroly F., Szostak M.P., Cabal A., Ruppitsch W., Bernreiter-Hofer T., et al. The first report of mcr-1-carrying Escherichia coli originating from animals in Serbia. Antibiotics. 2021;10:1063. doi: 10.3390/ANTIBIOTICS10091063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ušjak D., Novović K., Filipić B., Kojić M., Filipović N., Stevanović M.M., et al. In vitro colistin susceptibility of pandrug-resistant Ac. baumannii is restored in the presence of selenium nanoparticles. J Appl Microbiol. 2022 doi: 10.1111/JAM.15638. [DOI] [PubMed] [Google Scholar]
- 14.Vijayakumar S., S B.A., Kanthan K., Veeraraghavan B. Whole-genome shotgun sequences of seven colistin-resistant Acinetobacter baumannii isolates from bacteraemia. J Glob Antimicrob Resist. 2018;12:155–156. doi: 10.1016/J.JGAR.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Nogbou N.D., Ramashia M., Nkawane G.M., Allam M., Obi C.L., Musyoki A.M. Whole-genome sequencing of a colistin-resistant Acinetobacter baumannii strain isolated at a tertiary health facility in Pretoria, South Africa. Antibiotics. 2022;11:594. doi: 10.3390/ANTIBIOTICS11050594/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin M.F., Lin Y.Y., Lan C.Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J Microbiol. 2017;55:130–136. doi: 10.1007/S12275-017-6408-5. [DOI] [PubMed] [Google Scholar]
- 17.Thi Khanh Nhu N., Riordan D.W., Do Hoang Nhu T., Thanh D.P., Thwaites G., Huong Lan N.P., et al. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci Rep. 2016;6 doi: 10.1038/srep28291. Article 28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahm C., Chung H.S., Lee M. Whole-genome sequencing for the characterization of resistance mechanisms and epidemiology of colistin-resistant Acinetobacter baumannii. PLoS One. 2022;17 doi: 10.1371/JOURNAL.PONE.0264335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turton J.F., Woodford N., Glover J., Yarde S., Kaufmann M.E., Pitt T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to This Species. J Clin Microbiol. 2006;44 doi: 10.1128/JCM.01021-06. Article 2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Scola B., Gundi V.A.K.B., Khamis A., Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter Species. J Clin Microbiol. 2006;44 doi: 10.1128/JCM.44.3.827-832.2006. Article 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute . 32nd ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2022. M100: Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 22.The European Committee on Antimicrobial Susceptibility Testing., Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0., (2022). 〈http://www.eucast.org〉 (accessed February 23, 2022).
- 23.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/J.1469-0691.2011.03570.X. [DOI] [PubMed] [Google Scholar]
- 24.Novović K., Trudić A., Brkić S., Vasiljević Z., Kojić M., Medić D., et al. Molecular epidemiology of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae in Serbia from 2013 to 2016. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02550-16/ASSET/9193E84A-5805-4C2B-A007-3266FE225F9B/ASSETS/GRAPHIC/ZAC0051761330001.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jovcic B., Novovic K., Dekic S., Hrenovic J. Colistin resistance in environmental isolates of Acinetobacter baumannii. Microb Drug Resist. 2021;27:328–336. doi: 10.1089/mdr.2020.0188. [DOI] [PubMed] [Google Scholar]
- 26.S. Andrews, Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data, (2010). 〈https://www.bioinformatics.babraham.ac.uk/projects/fastqc/〉 (accessed June 9, 2022).
- 27.Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinforma. 2020;70 doi: 10.1002/CPBI.102. Article e102. [DOI] [PubMed] [Google Scholar]
- 28.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/BIOINFORMATICS/BTT086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/BIOINFORMATICS/BTU153. [DOI] [PubMed] [Google Scholar]
- 30.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST. Nucleic Acids Res. 2014;42 doi: 10.1093/NAR/GKT1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen M.V., Cosentino S., Lukjancenko O., Saputra D., Rasmussen S., Hasman H., et al. Benchmarking of methods for genomic taxonomy. J Clin Microbiol. 2014;52:1529–1539. doi: 10.1128/JCM.02981-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3 doi: 10.12688/WELLCOMEOPENRES.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/NAR/GKZ935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyres K.L., Cahill S.M., Holt K.E., Hall R.M., Kenyon J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb Genom. 2020;6 doi: 10.1099/mgen.0.000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/BIOINFORMATICS/BTV421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Page A.J., Taylor B., Delaney A.J., Soares J., Seemann T., Keane J.A., et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2 doi: 10.1099/MGEN.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/BIOINFORMATICS/BTU033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camargo C.H., Cunha M.P.V., de Barcellos T.A.F., Bueno M.S., de A.M., Bertani J., et al. Genomic and phenotypic characterisation of antimicrobial resistance in carbapenem-resistant Acinetobacter baumannii hyperendemic clones CC1, CC15, CC79 and CC25. Int J Antimicrob Agents. 2020;56 doi: 10.1016/J.IJANTIMICAG.2020.106195. Article 106195. [DOI] [PubMed] [Google Scholar]
- 40.Palmieri M., D’Andrea M.M., Pelegrin A.C., Perrot N., Mirande C., Blanc B., et al. abundance of colistin-resistant, OXA-23- and ArmA-Producing Acinetobacter baumannii belonging to International Clone 2 in Greece. Front Microbiol. 2020;11 doi: 10.3389/FMICB.2020.00668/BIBTEX. Article 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon E.J., Kim H.S., Woo H., Choi Y.J., Won D., Choi J.R., et al. Trajectory of genetic alterations associated with colistin resistance in Acinetobacter baumannii during an in-hospital outbreak of infection. J Antimicrob Chemother. 2021;77:69–73. doi: 10.1093/JAC/DKAB363. [DOI] [PubMed] [Google Scholar]
- 42.Veeraraghavan B., Anandan S., Ragupathi N.K.D., Vijayakumar S., Sethuvel D.P.M., Biswas I. Draft genome sequence of colistin-resistant Acinetobacter baumannii strain VB22595 isolated from a central line-associated bloodstream infection. Genome Announc. 2016;4:835–851. doi: 10.1128/GENOMEA.00835-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naha A., Vijayakumar S., Lal B., Shankar B.A., Chandran S., Ramaiah S., et al. Genome sequencing and molecular characterisation of XDR Acinetobacter baumannii reveal complexities in resistance: Novel combination of sulbactam–durlobactam holds promise for therapeutic intervention. J Cell Biochem. 2021;122:1946–1957. doi: 10.1002/JCB.30156. [DOI] [PubMed] [Google Scholar]
- 44.Mustapha M.M., Li B., Pacey M.P., Mettus R.T., McElheny C.L., Marshall C.W., et al. Phylogenomics of colistin-susceptible and resistant XDR Acinetobacter baumannii. J Antimicrob Chemother. 2018;73 doi: 10.1093/JAC/DKY290. Article 2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miltgen G., Bour M., Allyn J., Allou N., Vedani T., Vuillemenot J.B., et al. Molecular and epidemiological investigation of a colistin-resistant OXA-23-/NDM-1-producing Acinetobacter baumannii outbreak in the Southwest Indian Ocean Area. Int J Antimicrob Agents. 2021;58 doi: 10.1016/J.IJANTIMICAG.2021.106402. Article 106402. [DOI] [PubMed] [Google Scholar]
- 46.Vijayakumar S., Jacob J.J., Vasudevan K., Mathur P., Ray P., Neeravi A., et al. Genomic characterization of mobile genetic elements associated with carbapenem resistance of Acinetobacter baumannii From India. Front Microbiol. 2022;13 doi: 10.3389/FMICB.2022.869653/BIBTEX. Article 2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerson S., Lucaßen K., Wille J., Nodari C.S., Stefanik D., Nowak J., et al. Diversity of amino acid substitutions in PmrCAB associated with colistin resistance in clinical isolates of Acinetobacter baumannii. Int J Antimicrob Agents. 2020;55 doi: 10.1016/J.IJANTIMICAG.2019.105862. Article 105862. [DOI] [PubMed] [Google Scholar]
- 48.Gerson S., Betts J.W., Lucaßen K., Nodari C.S., Wille J., Josten M., et al. Investigation of novel pmrB and eptA mutations in isogenic Acinetobacter baumannii isolates associated with colistin resistance and increased virulence in vivo. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.01586-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olaitan A.O., Morand S., Rolain J.M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5 doi: 10.3389/FMICB.2014.00643/XML/NLM. Article 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghahraman M.R.K., Hosseini-Nave H., Azizi O., Shakibaie M.R., Mollaie H.R., Shakibaie S. Stereochemical trajectories of a two-component regulatory system PmrA/B in a colistin-resistant Acinetobacter baumannii Clinical Isolate. Iran Biomed J. 2021;25 doi: 10.52547/IBJ.25.3.193. Article 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kostyanev T., Xavier B.B., García-Castillo M., Lammens C., Bravo-Ferrer Acosta J., Rodríguez-Baño J., et al. Phenotypic and molecular characterizations of carbapenem-resistant Acinetobacter baumannii isolates collected within the EURECA study. Int J Antimicrob Agents. 2021;57 doi: 10.1016/J.IJANTIMICAG.2021.106345. Article 106345. [DOI] [PubMed] [Google Scholar]
- 52.Hamidian M., Nigro S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom. 2019;5 doi: 10.1099/MGEN.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novovic K., Mihajlovic S., Vasiljevic Z., Filipic B., Begovic J., Jovcic B. Carbapenem-Resistant Acinetobacter baumannii from Serbia: Revision of CarO Classification. PLoS One. 2015;10 doi: 10.1371/journal.pone.0122793. Article e0122793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gajic I., Jovicevic M., Milic M., Kekic D., Opavski N., Zrnic Z., et al. Clinical and molecular characteristics of OXA-72-producing Acinetobacter baumannii ST636 outbreak at a neonatal intensive care unit in Serbia. J Hosp Infect. 2021;112:54–60. doi: 10.1016/j.jhin.2021.02.023. [DOI] [PubMed] [Google Scholar]
- 55.Chukamnerd A., Pomwised R., Paing Phoo M.T., Terbtothakun P., Hortiwakul T., Charoenmak B., et al. In vitro synergistic activity of fosfomycin in combination with other antimicrobial agents against carbapenem-resistant Klebsiella pneumoniae isolated from patients in a hospital in Thailand. J Infect Chemother. 2021;27:507–514. doi: 10.1016/J.JIAC.2020.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material