Abstract
Adaptive laboratory evolution (ALE) has long been used as the tool of choice for microbial engineering applications, ranging from the production of commodity chemicals to the innovation of complex phenotypes. With the advent of systems and synthetic biology, the ALE experimental design has become increasingly sophisticated. For instance, implementation of in silico metabolic model reconstruction and advanced synthetic biology tools have facilitated the effective coupling of desired traits to adaptive phenotypes. Furthermore, various multi-omic tools now enable in-depth analysis of cellular states, providing a comprehensive understanding of the biology of even the most genomically perturbed systems. Emerging machine learning approaches would assist in streamlining the interpretation of massive and multiplexed datasets and promoting our understanding of complexity in biology. This review covers some of the representative case studies among the 700 independent ALE studies reported to date, outlining key ideas, principles, and important mechanisms underlying ALE designs in bioproduction and synthetic cell engineering, with evidence from literatures to aid comprehension.
Keywords: Adaptive laboratory evolution, Systems biology, Synthetic biology, Biochemicals, Minimal genome
1. Introduction
Spontaneous errors in DNA replication machineries render countless mutations in genetic codes, enabling biological systems to adapt and evolve in diverse environments. This defining biological feature has long been exploited in bacteria to understand the principles of adaptation and evolutionary dynamics [1]. Currently, this trait is being increasingly harnessed for various biotechnological applications, including, but not limited to, industrial commodity production [2] and optimization of synthetic cells [3]. By leveraging the process of artificial selection, target microbes are subjected to serial propagation in an artificial culture setting that is designed to select for mutant strains with the desired phenotypes. This process, popularly known as adaptive laboratory evolution (ALE), is particularly useful when a limited understanding of complexity in biology precludes conventional engineering efforts [4]. The bespoke nature of ALE allows the integration of various synthetic toolkits, such as biosensor circuitries, as part of the selection machinery to streamline the reconfiguration of microbes towards the desired objectives [5]. In addition, the tempo and mode of adaptive evolution can also be configured by engineering biological mutation rates, or abiotically through random mutagenesis (i.e. chemical mutagens) [6], [7], allowing the expansion of sequence and phenotypic diversities at a faster rate. Therefore, ALE is an important part of the strain engineering pipeline in the field of biotechnology [4], [8].
With the emergence of systems biology techniques such as next-generation sequencing (NGS) and in silico modeling, adaptive changes can be characterized, and the mechanism of adaptation can be defined by identifying causal mutations in unprecedented detail. For instance, independent genome sequencing datasets generated from numerous ALE experiments have now been compiled into a coherent database [9], allowing systematic analyses of the mechanistic effects of mutations on adaptive evolution. Recently, the analysis of large-scale mutational data unveiled the non-random, epigenomic-dependent nature of the mutability of genome, challenging the long-standing notion that genomic mutability is stochastic in nature [10], [11]. A host of multi-omics techniques can help elucidate adaptive remodeling of cellular phenotypes at the genomic, transcriptomic, and translational levels [12], ultimately providing an information-dense knowledge base for building and testing new biological hypotheses. The computational framework of cellular metabolism, known as the genome-scale metabolic model (GEM), provides for a highly accurate description and interpretation of metabolic phenotypes of adaptively evolved strains [13], [14]. As a result, various GEM-derived systems biology frameworks have been used together with the ALE techniques to improve target compound production [15], [16], [17], [18], [19], [20] and acquire complex phenotypes [21], [22]. Thus, ALE demonstrates extended utility in strain engineering and serves a tool that informs users of the best possible optimization scenarios.
In this minireview, we discuss the importance of ALE in biotechnological applications, with particular emphasis on the popular avenues of ALE research: industrial biotechnology and applications in synthetic biology. Further, we outline the variations in ALE methodologies and provide a summary of the key ideas, principles, and mechanisms underlying the proposed approach. We also provide a brief summary of case studies, where appropriate, to aid understanding. Finally, we present future perspectives on ALE-guided strain engineering in association to the emerging technologies.
2. Principles of ALE and its implications in biological engineering
The basis of ALE lies in the continuous propagation of cell populations under specific selection pressures until de novo mutations occur, which help overcome the fitness burden [23], [24] (Fig. 1A). The fast generation time and striking adaptability of microbes make ALE a well-suited, straightforward approach for reconfiguring microbes in a timely manner. A routine ALE workflow involves real-time monitoring of adaptive changes in serially propagating populations, identification of “endpoint” stages of ALE, mutant selection via phenotypic assays, and multi-omics analysis to characterize the molecular basis of adaptation (Fig. 1B, C).
Fig. 1.
Schematic illustration of a general ALE workflow. (A) Serial propagation of wild-type bacteria under sub-optimal culture condition. Spontaneous mutations that alter strain metabolism and regulation in a way that is beneficial under the selective culture conditions are selected during evolution. Intermittent monitoring of strain fitness helps infer the “end-point” of ALE. (B) Phenotypic characterization, typically entailing strain growth rate, is performed to assess adaptive changes in the end-point populations or single clonal isolates (to account for population heterogeneity). (C) Multi-omics analysis, normally with whole-genome resequencing and transcriptome sequencing, is used to account for the genetic basis of adaptation.
From an engineering perspective, ALE primarily serves as a complementary tool for rational genetic engineering. Rational engineering entails knowledge-based design and elaborate genetic toolkits to alter microbial phenotypes. However, inherent complexities and a limited understanding of biological systems, compounded by the lack of efficient tools, often leave microbes at perturbed metabolic and molecular states that prevent them from performing optimally as designed [4], [8]. ALE addresses such impediments and stands out as a powerful interventional approach to complement the shortcomings of biological systems in several aspects.
First, the accumulation of beneficial mutations over time translates to the rewiring of metabolic and regulatory networks of target microbes in ways that are difficult to comprehend using rational engineering design approaches. Taking E. coli ALE in M9 glycerol as an example, mutations in glycerol kinase (glpK) and RNA polymerase subunits (e.g., rpoS, rpoB, and rpoC) are recurrently enriched in glycerol-adapted endpoint strains [6], [24], [25]. The GlpK mutant experiences a growth advantage by evading the inhibitory action induced by fructose-1,6-bisphosphate and IIAGLC, resulting in the hyperactivation of glycerol uptake and a concomitant increase in carbon overflow [26], [27]. In contrast, mutations in RNA polymerase lead to reprogramming of cellular transcriptomic regulation to maximize carbon efficiency, as characterized by enhanced biomass generation, while minimizing carbon overflow [27], [28]. Interestingly, the glpK and rpoC double mutants displayed positive epistatic interactions, imparting a synergistic increase in strain fitness on M9 glycerol [24], [27], ultimately providing the optimal solution for maximizing strain fitness in glycerol-limited conditions. ALE-driven engineering encompasses a broad range of topics, including growth rate improvement in natural and synthetic strains [6], [12], [29], activation of the latent and non-native pathways [21], [30], [31], tolerance against diverse stressors [16], [32], [33], [34], and miscellaneous features such as the reconfiguration of cellular trophic states [22], [35].
ALE allows the investigation of non-intuitively beneficial mutations that are otherwise unpredictable using traditional methods. Thus, with an adequately defined experimental setting, researchers can generate microbes with desired traits, even in the absence of detailed knowledge of the genetic features of target phenotypes [4]. This also translates to the fact that the “knowledge-independent” nature of the ALE technique enables microbial species with limited genetic tractability and genomic information to be “evolutionarily engineered” in a user-defined manner. For example, Eubacterium limosum is an emerging strain with industrial relevance owing to its ability to convert syngas waste feedstock (CO2, CO, H2) into value-added commodities [36]. However, the paucity of efficient genetic tools and the limited understanding of functional genetics have been major bottlenecks in strain engineering of this microorganism [37]. ALE proved to be an effective alternative for constructing chassis strains to produce value-added chemicals from syngas feedstocks, with minimal genetic engineering efforts in E. limosum [32], [34]. ALE-driven engineering is applicable to a broad spectrum of microorganisms, allowing non-conventional microbes with distinct properties such as methane consumption (Methylomonas sp.) [38], bioremediation (Geobacter sulfurreducens) [39], partial oxidation of sugars (Gluconobacter oxydans) [40], antibiotic production (Streptomyces strains) [41], and resistance to extreme temperatures (Metallosphaera sedula) [42], to be investigated for practical applications in biotechnology.
Exploring the genotypic solution space for finding optimal phenotypic traits facilitates the discovery of novel solutions that are difficult to design a priori. This process leverages the inherent errors in genome replication machineries, and the error rates can be adjusted in favor of expanding the genotypic diversity [43]. In an engineering perspective, mutators with a markedly increased genomic mutation rates can be exploited to expedite adaptive evolution, where increased chance acquisition of mutations enables fixation of beneficial mutations in a shorter evolutionary timescale [6], [44]. The expanded genotypic diversity in mutator populations may also lead to ‘fitness readiness’ in non-adaptive environments, where the presence of neutral or deleterious mutations can, in turn, confer benefits on non-adaptive conditions [6], [21], [43]. Similarly, abiotic mutagens such as the alkylating agents help promote sequence diversity, with a downstream enrichment and screening process to select for mutants with desired phenotypes [7]. Most important aspects of ALE, however, lies in the design of appropriate selection pressure to obtain mutants with desired phenotypes. For instance, it took nearly 31,500 generations (∼20 years) of evolution for one of the twelve long-term evolution experiment lineages to develop the ability to digest citrate present in the culture medium [45], while the remaining eleven lineages were yet to acquire the same trait even after three decades of ALE [46]. The principles of experimental design and the criteria for successful ALE have been reviewed extensively elsewhere [2], [4], [8], [47].
2.1. ALE for improving target chemical production
Ease of culture, metabolic plasticity, and the resilience of microbes against various stressors have long been exploited in biotechnological applications. In particular, microbial production of value-added products, including commodity chemicals, heterologous proteins, and biofuels, represent some of the industrially important applications that can be achieved via microbial engineering [48], [49], [50]. Currently, with the growing push towards biosustainability and a carbon-neutral economy, numerous studies are being devoted to the valorization of renewable wastes into commodity chemicals. Industrial by-products and various low-value carbon wastes are emerging candidates for “alternative” feedstocks [51], [52], [53]. However, one of the biggest hurdles in the use of alternative feedstocks lies in the efficient metabolism and innate toxicity of host microbes. For instance, E. coli grows sub-optimally on glycerol, which is a waste stream in biodiesel production [13]. Agro-industrial waste carbons such as sugarcane molasses contain inhibitors that inflict strain fitness and bioconversion efficiency [54]. In addition to feedstock utilization, the functional operation of product biosynthetic machineries also requires substantial optimization [55]. In this regard, ALE has been demonstrated to be an effective means for reconfiguring strain metabolism, tolerance, and target product biosynthetic processes to improve commodity chemical production. The examples covered in this review are listed in Table 1.
Table 1.
A brief summary on ALE studies focusing on bioproduction of commodity chemicals covered in this minireview.
| Host | ALE selective condition | Growth-coupling strategy | Adaptive phenotypes | Applications | ALE type | Ref. |
|---|---|---|---|---|---|---|
| E. coli W | M9 glycerol (0.2% v/v). Serial transfer in batch culture. |
- | ∼40, 47% improvement in specific growth rate and carbon consumption rate | Gamma-aminobutyric acid titer and productivity increased by 3.9 and 4.3-folds | Classic | [57] |
|
E. limosum ECO1 (ALE derivative of ATCC8486) |
DSMZ 135 with 66% CO gas at 200 kPa Serial transfer in batch culture |
- | 2-fold increase in final cell density, 6.2-fold increase in specific growth rate | 6.5-fold improvement in 2,3-butanediol production | [34] | |
| L. delbrueckii NL31 | First phase: Modified Man Rogosa Sharpe broth containing 80–160 g/L of sugarcane molasses in place of D-glucose. Second phase: MRS containing 160 g/L sugarcane molasses and 54.2 g/L soybean meal hydrolysate Serial transfer in batch culture |
- | Nearly 3-fold increase in final cell density on soybean meal | 112.3 g/L D-lactic acid with fed-batch fermentation | [54] | |
| E. coli JCL16 (BW25513 derivative) | M9 glucose (0.4% w/v), anaerobic + 0.1 mM IPTG and a selection marker. Serial transfer in batch culture |
Chromosomal deletion of ΔadhE, ΔldhA, ΔfrdBC Expression of butanol biosynthetic pathway Episomal overexpression of mutD5. |
Restoration of strain growth on minimal medium | 2 g/L of butanol production in minimal medium. | Metabolic | [55] |
| E. coli BW25113 | Glycerol minimal medium (MOPS) | Chromosomal deletion of ΔfrdC | 5-fold increase in specific growth rate 4-fold increase in glycerol dehydrogenase activity |
20- and 5-fold increase in hydrogen and ethanol titer. | [30] | |
| E. coli MG1655 | M9 glycine with increasing concentration of L-serine | Chromosomal deletion of ΔsdaAΔsdaBΔtdcGΔglyA. | Increase tolerance against L-serine (up to 100 g/L) | 37 g/L L-serine titer, 24% mass yield from D-glucose (one of the highest titer reported to date). | [60] | |
| E. coli MG1655 | M9 glucose or glycerol | Integration of ethylene biosynthetic pathway. Chromosomal deletion of ΔproB predicted based on iJO1366. |
Growth-coupled production of ethylene | Optimized ethylene bioproduction for further engineering | Systems metabolic | [17] |
| E. coli W3110 | M9 glucose with increasing concentrations of NiCl2 |
L-lysine-sensitive biosensor with tetA actuator (conferring resistance against Ni2+) Random promoter libraries targeting ppc expression |
Growth-coupled production of L-lysine | Increased L-lysine production (0.6 g/L) | Biosensor-assisted | [5] |
| E. coli W | A modified glycerol minimal medium with increasing concentration of tetracycline | 3-hydroxypropionic acid (3-HB) biosensor with tetA actuator (resistance against tetracycline). | Growth-coupled production of 3-hydroxypropionic acid | 3-HB production yield closest to the theoretical max (0.91 g/g glycerol) | [85] | |
| P. putida KT2400 | M9 glucose | Muconate biosensor with fluorescent reporter actuator | Restoration of fitness defect (but failed to isolate high-muconate producing strain) | 3-fold increase in muconate production FACS-mediated isolation of growth-restored, high muconate producer mutant |
[58] |
* IPTG - isopropyl-β-D-thiogalactopyranoside.
2.2. Classical ALE approach to enhance target compound production
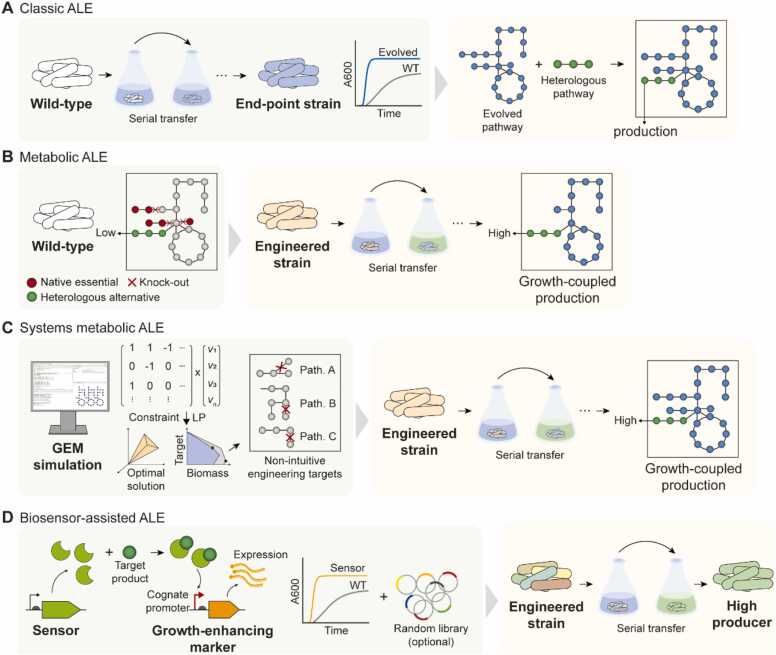
The classical ALE approach that aim to optimize metabolic efficiency has been demonstrated to be effective at improving the titer and productivity of target compounds [56] (Fig. 2A). For instance, the end-point strain of glycerol-adapted E. coli engineered to produce gamma-aminobutyric acid led to a 3.9-fold increase in the final product titer [57]. The fastest-growing clonal isolate of CO (carbon monoxide)-adapted E. limosum showed a 6.5-fold increase in 2,3-butanediol (2,3-BDO) production, with CO as the feedstock [34]. Lactobacillus delbrueckii adaptively evolved under high concentrations of sugarcane molasses and exhibited enhanced D-lactic acid production [54]. The increase in the fitness of alternative carbon sources and the concurrent improvement in target compound production are largely attributed to the optimized carbon metabolism, resilience against toxic compounds, and regulatory changes induced by the fixation of beneficial mutations. However, one caveat of this method is that the mutants that effectively re-direct the metabolic flux into biomass precursors are selected during ALE, rendering the primary objective function as maximization of growth rate, instead of target compound production [58] (Table 2); this is especially prevalent when the target products are metabolically costly, favoring mutations that reduce the metabolic burden by disrupting the biosynthetic capacity of the desired products [59] (Table 2).
Fig. 2.
Summary of representative ALE methods. (A) Classic ALE with the primary objective of optimizing strain fitness in sub-optimal environments (containing alternative carbons or inhibitory compounds). Target biochemical compound production can be enhanced but is not the primary objective of ALE optimization. (B) Metabolic ALE involves a metabolic engineering effort that couples target metabolite production with adaptive phenotypes, rendering target compound production as the primary objective. (C) Systems metabolic ALE entails the use of GEM-based simulations to determine growth-coupling strategies for an enhanced target compound production. The investigation of a feasible solution space within the in silico metabolic network often yields engineering targets that are unpredictable a priori. (D) Biosensor-assisted ALE leverages the ability of sensor domains to sense (target molecule) and actuate response (designed to confer growth advantage). Ideally, the schematic couples target metabolite production with strain fitness in a dose-dependent manner.
Table 2.
Characteristics and limitations of ALE methods outlined in this study.
| Method | Characteristics | Limitations |
|---|---|---|
| Classical ALE | - ALE with the primary objective function designed to select for end-point strains with improved adaptive fitness under a selective environment. | - Maximization of strain fitness (i.e. specific growth rate) as the primary objective function does not necessarily correlate with maximization of target metabolite production. Target compound production can be selected against when target products are metaboically costsly. |
| Metabolic ALE | - Implementation of metabolic engineering schemes to couple target compound production with adaptive phenotypes. May help circumvent negative selection against the production of metabolically costly chemicals. Effectively selects for the end-point adaptive strains with metabolism rewired for the production of target chemicals. | - Metabolic perturbation may impose severe fitness defects in the engineered strain. - Requires a priori knowledge on the metabolic network for the effective coupling of adaptation and target compound production. May limit the application to a relatively few well-known targets |
| Systems metabolic ALE | - Use of genome-scale metabolic models (GEMs) to probe for non-intuitive metabolic engineering targets that may be more effective (in metabolite-growth coupling) compared to traditional targets. - Simulation of growth rate following gene knockouts can help inform the user the feasibility of metabolic engineering designs (i.e. growth defects). - Integration of multi-omic data along with GEM can help infer metabolic bottlenecks (for target compound production) for consideration in ALE design. |
- May not be applicable for non-model strains lacking a high-quality GEM. - The limitations in linear programming may preclude accurate representation of strain metabolism and knockout simulations, resulting in disparities between in silico simulations and empirical results. |
| Biosensor-assisted ALE | - Use of a genetic circuit that sense the target compound (using an orthogonal sensor protein) and respond by controlling the expression of growth-associated genes. | - Non-target 'escapee' mutations can render the biosensor-imposed selection pressure obsolete. |
Metabolic engineering strategies have been established to couple target chemical production with adaptive phenotypes to implement a driving force for the selection of evolved strains with enhanced bioconversion capacities (Fig. 2B). In one instance, synthetic dependency on the heterologous 1-butanol biosynthetic pathway was established in an anaerobically cultured E. coli by inactivating the native NAD+-replenishing pathways (ΔadhEΔldhAΔfrdBC). Notably, 1-butanol biosynthesis was inactive in minimal medium, suggesting an imbalance in cellular resource distribution for the intracellular redox balance. Engineering evolution was conducted in a conducive environment that fine-tuned resource utilization and redox metabolism, demonstrating an effective approach for synthesizing non-native 1-butanol in E. coli [55]. In another instance, E. coli lacking fumarate reductase (frdC), which represses hydrogen synthesis during glycerol fermentation, adaptively evolved in glycerol minimal medium to bioconvert glycerol into biofuels (hydrogen and ethanol). Under alleviated repression of the target compound, ALE led to an evolved strain producing 20- and 5-fold more hydrogen and ethanol, respectively, than those produced by the non-ALE control strain [30]. Innate tolerance towards a target product can be configured with genetic engineering in conjunction with ALE to improve product yield. For example, an E. coli strain lacking the L-serine degradation pathway was adaptively evolved with increasing concentrations of L-serine, which was toxic even at low concentrations. The L-serine tolerant strain achieved an L-serine titer (37 g/L) comparable to the highest L-serine production recorded to date [60]. This workflow of strain engineering – metabolic engineering in tandem with ALE was dubbed “Metabolic engineering to guide evolution (MGE)” [61] and continues to be an effective means to engineer microbes for industrially relevant applications [62].
2.3. Systems biology-driven ALE
The MGE approach is largely reliant on rational engineering design, which requires a priori knowledge of the genetic features of targeted phenotypes, thereby limiting the scope of its application to relatively few well-known targets (Table 2). In this context, GEM was employed in designing growth-coupled ALE for bioproduction (Fig. 2C). The GEM constitutes a matrix of stoichiometry-balanced cellular biochemical reactions represented in a computational framework [63]. With constraint-based approaches, the framework defines a range of metabolic flux solutions that satisfy the reaction stoichiometry. Specific predictions, such as the metabolic flux through biomass generation, can be achieved by implementing an objective function, referred to as flux-balance analysis (FBA) [64]. FBA is used to describe a metabolic system that is akin to the exponential phase of growth, with linear programming to balance the total inputs and outputs of metabolite stoichiometry that satisfy the assumption of maximal cellular growth [65]. Such modeling framework has been proven to be well fitted to describe (laboratory) adopted cellular states, such as growth rate and metabolite secretion rates, across various carbon substrates [13], [14].
The earliest example of GEM-guided engineering involved the use of a bi-level optimization approach, Optknock [15], which simulates metabolic fluxes that satisfy two objective functions: optimization of target metabolite secretion and growth rate [66]. Similar algorithms are available, allowing target metabolite secretion to be effectively coupled with strain fitness resulting from gene knockout(s) [67], [68], [69], [70], [71], [72] (Fig. 2C). Initially applied in E. coli for the overproduction of native and heterologous compounds [15], [16], [17], [18], [19], [20], [70], [73], [74], [75], [76], this approach now extends to a diverse array of “GEM-accessible” microorganisms [77]. Evolutionary engineering is increasingly implemented in tandem with GEMs to enhance the target product titer, largely by optimizing cellular metabolism or compensating for fitness defects that would result from metabolic engineering. For instance, in an attempt to achieve growth-coupled ethylene production in E. coli, a constraint-based simulation using the model, iJO1366, was employed to identify novel growth-coupling targets [17]. Assuming that growth rate was maintained, deletion of proB (glutamate-5-semialdehyde dehydrogenase) was predicted to render the ethylene pathway as the sole passage for L-proline production, thereby coupling ethylene production with growth via L-proline biosynthesis. Subsequently, the deletion strain was subjected to ALE with random mutagenesis in the absence of L-proline to select for a high ethylene-producing strain, resulting in a final strain with enhanced solubility of the ethylene-producing enzyme and a 49-fold increase in ethylene production [17].
The GEM framework can be used in parallel with multi-omics datasets to overcome the shortcomings of the FBA. Although FBA can seamlessly capture biological phenotypes, the inherent limitation of linear programming precludes an accurate representation of the underlying flux distribution in the in silico metabolic network [78], [79]. As a result, efforts have been made to refine GEM by integrating experimental data such as transcriptomic and metabolomic data with the GEM [78], [80], [81]. The central idea is to impose additional constraints, such as mRNA abundance, to minimize the uncertainty of the metabolic flux assigned to each reaction. The resulting model framework, known as a context-specific model, enables condition-specific systems-level analyses of metabolic flux distributions with improved accuracy [82]. In one study, researchers constructed the GEM iCLAU786 developed for Clostridium autoethanogenum, to infer the metabolic basis of ethanol and 2,3-BDO production in the strain [83]. Initially, the FBA calculations failed to predict 2,3-BDO production, which might be due to the fact that FBA selects for the most efficient pathway for optimizing biomass generation [79]. Integration of the RNA-Seq data that reflect the transcriptome state of 2,3-BDO-producing conditions into the GEM enabled in silico 2,3-BDO production, which closely aligned with the experimental results [83]. These findings demonstrate the robustness of the omics-integrated GEM framework in reflecting condition-specific metabolic states and the extended utility of systems metabolic engineering approaches (encompassing GEM, Omics, and ALE) in harnessing highly efficient production strains [20].
2.4. Synthetic biology approach
With the advent of synthetic biology, more innovative strategies have been developed to streamline strain evolution for bioproduction. Genetic biosensors are increasingly being used to couple biochemical production with adaptive phenotypes [5], [58], [84], [85]. Importantly, biosensor-assisted evolution exploits the chemical-sensing and signal-transducing nature of biosensors, where chemical signals are relayed via the sensor domain to regulate the expression of downstream genes [5], [84]. The biosensor output is coupled with fitness-enhancing traits, such as resistance to antibiotics, making cell fitness to be a proxy for assessing target compound production (Fig. 2D).
An L-lysine riboswitch controlling the expression of the tetA gene was used to select L-lysine overproducers using a plasmid-based promoter library designed to alter metabolic flux through phosphoenolpyruvate carboxylase (ppc). The propagation of the library in the presence of NiCl2, which was designed to select clones with high tetA expression, led to a meaningful increase in the final lysine titer (from 0 to 0.6 g/L) [5]. Similarly, ribozyme-guided ALE, which couples 3-hydroxypropionic acid production to cell growth, has led to optimal metabolic flux rewiring in E. coli, resulting in the highest product yield (closest to the theoretical maximum yield) reported to date (0.91 g/g glycerol) [85]. Accordingly, serial propagation of biosensor-embedded strains facilitates the enrichment of “directed evolution” of the desired phenotypes by coupling adaptive fitness to metabolite production. However, non-target mutations that escape selective pressure (and compromise product-growth coupling) can render the biosensor-guided evolution approach infeasible (Table 2), highlighting the need for an additional quality-control measure to maintain low false-positive rates [84]. The target product concentrations can be transduced into a reporter readout using product-specific biosensors, allowing selective enrichment of the desired mutants in a facile manner. For example, active sorting of evolving populations using fluorescence-activated cell sorting (FACS) enables a product concentration-dependent isolation of viable mutant strains in a high-throughput manner [58], [86]. Such a selection system proves to be particularly useful when the selective pressure for the target compound production is insufficient, and the incidence of viable mutants are rare. In an attempt to isolate a hyperproducer of muconate, a muconate-producing Pseudomonas putida suffering a substantial growth defect due to metabolic engineering was subjected to ALE in minimal glucose medium. However, the authors failed to screen for isolates that satisfied both objectives (of high muconate titer and growth) until a fluorescence reporter protein under the control of a muconate biosensor was implemented for high-throughput screening, where they successfully isolated a growth-restored mutant with a 3-fold increase in muconic acid production [58]. The inherent properties of biosensors, including dynamic range, promiscuity, and cross-strain genetic compatibility, are some of the confounding variables that complicate their practical applicability.
The ad hoc nature of ALE can fully utilize enabling technologies in synthetic biology for the design of novel growth-coupling strategies. Notable examples include the substitution of rare codons (that are orthogonal to specific amino acids) in antibiotic markers for the overproduction of target amino acids [87], and the use of phage-assisted continuous evolution (PACE) with a target-sensitive biosensor to optimize target biosynthetic pathways [86]. The aforementioned studies represent only a subset of synthetic biology approaches for designing and guiding ALE for the growth-coupled production of desired products. Further details on the application of synthetic biology to expedite ALE experiments, that is, automated multiplex culture, have been reviewed elsewhere [8].
In summary, engineering evolution guided by synthetic biology has many advantages. First, biosensor-guided selection allows multiplexed phenotype screening [58], which streamlines the screening process for high-performing mutants in an evolving population that can be as large as 1010 CFU/mL [88]. Second, a broad catalog of biosensors orthogonal to diverse ligands is available [89], and novel biosensors can be genome-mined [90] or synthetically engineered [5], enabling biosensor-assisted evolution to serve as a generalizable approach for strain engineering. The use of synthetic biology toolkits, such as synthetic bioparts libraries, as part of the selection machinery also enables the investigation of an even broader spectrum of mutational landscapes, facilitating the fine-tuning of metabolic flux for optimized target compound production.
3. Systems biology approach to understand the adaptive changes in synthetic microbes
The ultimate goal of synthetic biology is to construct a “plug-and-play” chassis strain that is user-tailored to execute specific functions using modularized bioparts. Accordingly, synthetic cells with minimal genetic redundancy [3], [12], recoded genomes [29], and reconfigured trophic states [22], [35], have been highlighted in the past decade, with potential applications in industrially relevant traits such as heterologous protein synthesis [91] and biochemical production [92].
Despite the advances in state-of-the-art manipulation and analytic techniques, avenues that existing techniques struggle to address still remain. Rationally designed genome-wide perturbations in synthetic cells result in severe fitness defects [3], [12], [29]. A prominent example is the de novo synthesized Mycoplasma mycoides genome (JCVI-syn3.0), which was designed to retain minimal genetic components essential for sustaining life [3]. Nonetheless, unexpected phenotypic defects, such as severe growth retardation, called for serial propagation as a contingent protocol to construct a fully viable synthetic cell [3]. This reflected our limited understanding of the complex biology that governs cell fitness, such as synthetic lethality and interlaced regulatory networks. A continued effort to elucidate the remaining biological uncertainties have culminated in the development of a nearly complete whole-cell computer model tailored to simulate the minimal synthetic cell (JCVI-syn3A) which harbors the minimal set of life-supporting 493 genes. Owing to the reduced complexities in the genomic, genetic and metabolic networks, a fully kinetic model incorporating various experimental parameters (i.e. enzyme kinetics, gene expression, spatiotemporal dynamics) was made possible. The model allows simulation of dynamic changes in particular chemicals in terms of its concentrations, spatial locations, rate of diffusion, as well as the energy expenditures simultaneously in a time-course manner. The model also houses 3D spatial reconstructions of macromolecules such as ribosomal coordinates and cognate DNA binding sites, expanding the scope of in silico simulation to ‘real-time’ monitoring and assessment of cellular processes [93]. However, implementing such whole-cell model for lab-grown microbes is challenging (due to biological complexity and uncertainties) and computationally expensive [94]. More generalized systems biology approach is available to aid interpretation of biological processes.
The molecular and mechanistic basis of fitness restoration during ALE is hidden within the “biological black box,” which can be deciphered, at least partially, by systems analysis of the evolved genome. System analysis largely entails the use of NGS technologies, which allow genome-wide characterization of mutation profiles [23], global changes in transcription [57] and translation levels [95], and the characterization of regulatory interactions in transcription factors [96]. In this section, we briefly describe the systems biology-guided analysis of ALE strains using the smallest minimal E. coli genome (eMS57), which represents some of the most extensively perturbed cell systems [12]. eMS57 adaptively evolved from a minimal E. coli genome MS56, which is an E. coli MG1655-derivative free of all insertion elements (plus other non-essential genes, with ∼25% reduction in the genomic content) constructed to enhance recombinant protein production through genome stabilization and reduction of genetic redundancies [97]. However, MS56 had a pronounced fitness deficit on minimal media and became viable only after an 800-generation ALE experiment in glucose minimal medium supplemented with Luria Bertani broth [12].
3.1. NGS-based approach to identify causal mutations
Whole-genome resequencing allows high-throughput profiling of genome-wide mutations, followed by the determination of the causality of mutations (Fig. 3A). Mutations, which are beneficial drivers or neutral hitchhikers, can be inferred via several approaches. First, recurrent mutations across independent replicate lineages strongly imply causality [23]. Second, a public ALE database depositing mutant data from hundreds of ALE experiments provides users with insights on the causality of mutations [9]. Lastly, manual inspection of gene annotation and pathway information helps to infer the functional attributes of a mutation [6]. In the MS56 ALE experiment, the authors identified a large deletion in the genomic region containing mutS and rpoS. MutS is a part of a post-replicative mismatch repair system, and its inactivation has been demonstrated to confer a hypermutator phenotype that expedites adaptive evolution [6], with proven utility in ALE of genome-recoded synthetic E. coli [29]. In contrast, RpoS is a general stress response regulator that controls over 50 regulons in response to different stressors, including nutrient deprivation [98]. Reverse engineering of the rpoS deletion in MS56 restored its growth rate to 80% of that in eMS57, highlighting the dominant role of rpoS in restoring the fitness of the minimal genome. As such, the cycle of whole-genome resequencing, mutation profiling, and reverse-engineering workflow presents a straightforward yet robust approach for identifying the genetic basis of fitness restoration (Fig. 3A).
Fig. 3.
Systems biology approach to elucidate the molecular basis of adaptation. (A) A workflow for genotype-phenotype characterization of causative mutations. (B) The use of transcriptome-sequencing and ChIP-Seq in tandem help identify changes in TF-mediated transcriptome regulation. (C) Ribosome profiling to indirectly infer regulation at the translation level. Low TE refers to low ribosomal occupancy on mRNA transcripts, where translation presents a bottleneck for protein synthesis in a (highly expressed) gene.
3.2. Elucidation of the altered regulatory landscape using NGS-based omics tools
Notably, mutations in the subunits of RNA polymerase, such as rpoS, rpoD, and rpoC, have been implicated in the global rewiring of transcription regulation during evolution [12], [28]. Understanding multi-layered regulations involving tens, if not hundreds, of TFs and regulons requires specialized sequencing techniques to probe the reprogrammed regulatory landscape. For example, chromatin immunoprecipitation coupled with sequencing (ChIP-Seq) enables the identification of TF binding sites on the genome by sequencing segments of DNA bound to the TF of interest [96] (Fig. 3B). Using RNA-Seq, eMS57 was discovered to harbor a set of unique (mutant) rpoD-binding sites that led to alterations in the expression profiles of several metabolic pathways, including dNTP salvage and redox balance, revealing an adaptive response to the reduced genetic burden in the minimal genome. In addition, to probe for the changes in post-transcriptional regulations that impacted translatome profiles [99], genome-wide translation efficiency (TE) was quantified by selectively sequencing the abundance of ribosome-protected mRNA fragments (Fig. 3C) and comparing them with the corresponding mRNA transcript abundance [95]. Typically, ribosomal occupancy in an actively transcribed mRNA exhibits markedly less variation than mRNA abundance, resulting in highly variable TE levels, a phenomenon known as translational buffering [12], [95], [99]. Unexpectedly, the TE levels in eMS57 remained constant regardless of the gene expression levels, demonstrating reduced translational buffering that led to improved protein production as measured by fluorescence reporter proteins [12].
Overall, the above examples highlight the utility of high-throughput analytic techniques of systems biology for revealing the molecular basis of adaptation, from routine genotype-phenotype profiling to global-scale remodeling of cellular metabolism, transcription, and translation regulations; and to build a novel hypothesis based on data-driven discovery.
4. Summary and outlook
Initially employed in an attempt to understand the mechanism of evolution, ALE now constitutes an important part of the engineering principles in biotechnology, with as many as 18,000 ALE experiments conducted to date [100]. Leveraging the natural process selection, the evolving strains acquire mutations from within the breadth of the genotype solution space, which is often deemed inaccessible (or unpredictable) with our current understanding of biology. Being independent of the need for a priori knowledge of the genetic basis of a target phenotype, ALE has markedly expanded our ability to harness non-model microbes, including those that are highly recalcitrant for engineering. Although growth optimization of ALE aids in improving target product titers, additional metabolic engineering efforts can effectively promote target metabolite production, leading to metabolite production as the main objective of ALE. In silico metabolic models can further refine the metabolic ALE design to achieve stringent coupling of the desired compound with adaptive fitness. On the other hand, the ad hoc nature of ALE experimental design, aligned with the expanding availability of genetic and in silico toolkits, has led the development of various ALE techniques with increasing sophistication. The non-intuitive aspects of the evolved phenotypes, particularly the molecular basis of adaptation that has hitherto remained elusive, are becoming increasingly evident due to advances in NGS analytic tools. Recently, molecular characterization of the adaptive changes in E. coli lacking the electron transport system led to the elucidation of an expanded catalogue of genes associated with oxic respiration, termed ‘Aero-Type System’ [101]. In addition, systems analysis of extensively perturbed cellular systems (such as the minimal genome) provides for an in-depth comprehension of biological mechanisms that have been previously unaccounted for, opening untapped grounds to learn, build, and test novel hypotheses.
Automated, multiplexed ALE experiments are becoming increasingly popular owing to the ease of manipulation, precise control of experimental parameters, and higher throughput (of biological replicates) than in manual transfer [102], [103]. Increasing throughput translates to even larger volumes of downstream data, where strain screening and data analysis present daunting tasks. A high-throughput, rapid screening for high producers among millions of variant libraries can be achieved via the use of biosensors specifically sense and respond to target metabolites [104]. Tethering the target metabolite production with a reporter output (i.e. fluorescence proteins) by means of a biosensor-based genetic circuit followed by a massively parallel screening (i.e. FACS) allows for a rapid and efficient selection of high-producers [104]. On a side note, biosensor-based screening alone stands out as a robust mean to optimize metabolite production, where it was demonstrated that fluorescence-based, real-time screening of product formation facilitated the identification of the process parameter that led to a 3-hydroxypropionate production that was 23-fold higher than previously reported [105]. The past years have seen the development of various analytic tools tailored for high-throughput datasets. For example, the latest updates on the E. coli GEM iML1515 have focused on the extraction and analysis of publicly available high-throughput data, including transcriptomics, metabolomics, and proteomics, ultimately providing an in silico platform to characterize the E. coli metabolic network using constraint-based modeling, protein structure, or genetic variations [106]. Recently, machine learning workflows tailored for the analysis of relevant data have proven to be useful in providing inference on the hidden layers of regulation, such as condition-specifically modulated gene sets and their TFs [101] and evolutionary constraints with different degrees of contribution to adaptive phenotypes [107]. Meta-analysis of large-scale mutational data [9] can also be extrapolated to predict novel genome engineering targets unforeseen in contemporary ALE designs [108]. The AlphaFold deep learning network designed for protein structural prediction [109] enables facile analysis of the implication of structural variants of mutant enzymes on strain phenotype [34]. Similarly, breakthroughs in deep-learning based protein structural prediction has allowed de novo protein design with high accuracy [110]. It is envisaged that ALE will serve as a testbed to assess the properties of artificial enzymes with regards to strain fitness, implications on adaptive phenotypes, and presence of epistatic interactions that can shape evolutionary trajectory of evolving populations. In conclusion, increasing throughput of biological data, in conjunction with data-compatible simulation platforms and effective machine learning workflows, is expected to streamline the deciphering of complex biological phenomena.
CRediT authorship contribution statement
Kangsan Kim: Writing – original draft, Writing – review & editing, Visualization. Minjeong Kang: Writing – review & editing. Sang-Hyeok Cho: Writing – review & editing. Eojin Yoo: Writing – review & editing. Ui-Gi Kim: Writing – review & editing. Suhyung Cho: Writing – review & editing. Bernard Palsson: Writing – review & editing. Byung-Kwan Cho: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the Korea Bio Grand Challenge (2018M3A9H3024759 to B.-K.C.) and the Bio and Medical Technology Development Program (2021M3A9I4024308 to B.-K.C.) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT. This work was also supported by a grant from the Novo Nordisk Foundation (NNF10CC1016517 to B.P.). The authors thank Marc Abrams for editing the manuscript.
References
- 1.Good B.H., McDonald M.J., Barrick J.E., et al. The dynamics of molecular evolution over 60,000 generations. Nature. 2017;551(7678):45–50. doi: 10.1038/nature24287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dragosits M., Mattanovich D. Adaptive laboratory evolution -- principles and applications for biotechnology. Microb Cell Fact. 2013;12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutchison C.A., 3rd, Chuang R.Y., Noskov V.N., et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351(6280):aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 4.Sandberg T.E., Salazar M.J., Weng L.L., et al. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng. 2019;56:1–16. doi: 10.1016/j.ymben.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J., Seo S.W., Jang S., et al. Synthetic RNA devices to expedite the evolution of metabolite-producing microbes. Nat Commun. 2013;4:1413. doi: 10.1038/ncomms2404. [DOI] [PubMed] [Google Scholar]
- 6.Kang M., Kim K., Choe D., et al. Inactivation of a mismatch-repair system diversifies genotypic landscape of Escherichia coli during adaptive laboratory evolution. Front Microbiol. 2019;10:1845. doi: 10.3389/fmicb.2019.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arora N., Lo E., Philippidis G.P. A two-prong mutagenesis and adaptive evolution strategy to enhance the temperature tolerance and productivity of Nannochloropsis oculata. Bioresour Technol. 2022;364 doi: 10.1016/j.biortech.2022.128101. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y., Jameel A., Xing X.H., et al. Advanced strategies and tools to facilitate and streamline microbial adaptive laboratory evolution. Trends Biotechnol. 2022;40(1):38–59. doi: 10.1016/j.tibtech.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Phaneuf P.V., Gosting D., Palsson B.O., et al. ALEdb 1.0: a database of mutations from adaptive laboratory evolution experimentation. Nucleic Acids Res. 2019;47(D1):D1164–D1171. doi: 10.1093/nar/gky983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd M.J., Horton J.S., Taylor T.B. A near-deterministic mutational hotspot in Pseudomonas fluorescens is constructed by multiple interacting genomic features. Mol Biol Evol. 2022;39(6) doi: 10.1093/molbev/msac132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monroe J.G., Srikant T., Carbonell-Bejerano P., et al. Mutation bias reflects natural selection in Arabidopsis thaliana. Nature. 2022;602(7895):101–105. doi: 10.1038/s41586-021-04269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe D., Lee J.H., Yoo M., et al. Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat Commun. 2019;10(1):935. doi: 10.1038/s41467-019-08888-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibarra R.U., Edwards J.S., Palsson B.O. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420(6912):186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 14.Fong S.S., Marciniak J.Y., Palsson B.O. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J Bacteriol. 2003;185(21):6400–6408. doi: 10.1128/JB.185.21.6400-6408.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong S.S., Burgard A.P., Herring C.D., et al. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng. 2005;91(5):643–648. doi: 10.1002/bit.20542. [DOI] [PubMed] [Google Scholar]
- 16.Kim J., Tremaine M., Grass J.A., et al. Systems metabolic engineering of Escherichia coli Improves coconversion of lignocellulose-derived sugars. Biotechnol J. 2019;14(9) doi: 10.1002/biot.201800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaud S., Pearcy N., Hanzevacki M., et al. Engineering improved ethylene production: Leveraging systems biology and adaptive laboratory evolution. Metab Eng. 2021;67:308–320. doi: 10.1016/j.ymben.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen L.T., Lee E.Y. Biological conversion of methane to putrescine using genome-scale model-guided metabolic engineering of a methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. Biotechnol Biofuels. 2019;12:147. doi: 10.1186/s13068-019-1490-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huerta-Beristain G., Cabrera-Ruiz R., Hernandez-Chavez G., et al. Metabolic engineering and adaptive evolution of Escherichia coli KO11 for ethanol production through the Entner-Doudoroff and the pentose phosphate pathways. J Chem Technol Biotechnol. 2017;92(5):990–996. doi: 10.1002/jctb.5138. [DOI] [Google Scholar]
- 20.Tafur Rangel A.E., Rios W., Mejia D., et al. In silico design for systems-based metabolic engineering for the bioconversion of valuable compounds from industrial by-products. Front Genet. 2021;12 doi: 10.3389/fgene.2021.633073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szappanos B., Fritzemeier J., Csorgo B., et al. Adaptive evolution of complex innovations through stepwise metabolic niche expansion. Nat Commun. 2016;7:11607. doi: 10.1038/ncomms11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer F., Keller P., Hartl J., et al. Methanol-essential growth of Escherichia coli. Nat Commun. 2018;9(1):1508. doi: 10.1038/s41467-018-03937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad T.M., Joyce A.R., Applebee M.K., et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10(10):R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herring C.D., Raghunathan A., Honisch C., et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38(12):1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 25.Peabody V.G., Li H., Kao K.C. Sexual recombination and increased mutation rate expedite evolution of Escherichia coli in varied fitness landscapes. Nat Commun. 2017;8(1):2112. doi: 10.1038/s41467-017-02323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Applebee M.K., Joyce A.R., Conrad T.M., et al. Functional and metabolic effects of adaptive glycerol kinase (GLPK) mutants in Escherichia coli. J Biol Chem. 2011;286(26):23150–23159. doi: 10.1074/jbc.M110.195305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng K.K., Lee B.S., Masuda T., et al. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat Commun. 2014;5:3233. doi: 10.1038/ncomms4233. [DOI] [PubMed] [Google Scholar]
- 28.Conrad T.M., Frazier M., Joyce A.R., et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A. 2010;107(47):20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wannier T.M., Kunjapur A.M., Rice D.P., et al. Adaptive evolution of genomically recoded Escherichia coli. Proc Natl Acad Sci U S A. 2018;115(12):3090–3095. doi: 10.1073/pnas.1715530115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu H., Wood T.K. An evolved Escherichia coli strain for producing hydrogen and ethanol from glycerol. Biochem Biophys Res Commun. 2010;391(1):1033–1038. doi: 10.1016/j.bbrc.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Balderas-Hernandez V.E., Hernandez-Montalvo V., Bolivar F., et al. Adaptive evolution of Escherichia coli inactivated in the phosphotransferase system operon improves co-utilization of xylose and glucose under anaerobic conditions. Appl Biochem Biotechnol. 2011;163(4):485–496. doi: 10.1007/s12010-010-9056-3. [DOI] [PubMed] [Google Scholar]
- 32.Kang S., Song Y., Jin S., et al. Adaptive laboratory evolution of Eubacterium limosum ATCC 8486 on carbon monoxide. Front Microbiol. 2020;11:402. doi: 10.3389/fmicb.2020.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deatherage D.E., Kepner J.L., Bennett A.F., et al. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc Natl Acad Sci U S A. 2017;114(10):E1904–E1912. doi: 10.1073/pnas.1616132114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin S., Kang S., Bae J., et al. Development of CO gas conversion system using high CO tolerance biocatalyst. Chem Eng J. 2022;449 doi: 10.1016/j.cej.2022.137678. [DOI] [Google Scholar]
- 35.Gleizer S., Ben-Nissan R., Bar-On Y.M., et al. Conversion of Escherichia coli to Generate All Biomass Carbon from CO2. Cell. 2019;179(6):1255–1263. doi: 10.1016/j.cell.2019.11.009. e1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae J., Song Y., Lee H., et al. Valorization of C1 gases to value-added chemicals using acetogenic biocatalysts. Chem Eng J. 2022;428 doi: 10.1016/j.cej.2021.131325. [DOI] [Google Scholar]
- 37.Shin J., Kang S., Song Y., et al. Genome engineering of Eubacterium limosum Using expanded genetic tools and the CRISPR-Cas9 system. ACS Synth Biol. 2019;8(9):2059–2068. doi: 10.1021/acssynbio.9b00150. [DOI] [PubMed] [Google Scholar]
- 38.Lee J.K., Kim S., Kim W., et al. Efficient production of d-lactate from methane in a lactate-tolerant strain of Methylomonas sp. DH-1 generated by adaptive laboratory evolution. Biotechnol Biofuels. 2019;12:234. doi: 10.1186/s13068-019-1574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tremblay P.L., Summers Z.M., Glaven R.H., et al. A c-type cytochrome and a transcriptional regulator responsible for enhanced extracellular electron transfer in Geobacter sulfurreducens revealed by adaptive evolution. Environ Microbiol. 2011;13(1):13–23. doi: 10.1111/j.1462-2920.2010.02302.x. [DOI] [PubMed] [Google Scholar]
- 40.Lu L., Wei L., Zhu K., et al. Combining metabolic engineering and adaptive evolution to enhance the production of dihydroxyacetone from glycerol by Gluconobacter oxydans in a low-cost way. Bioresour Technol. 2012;117:317–324. doi: 10.1016/j.biortech.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Harwani D., Begani J., Barupal S., et al. Adaptive laboratory evolution triggers pathogen-dependent broad-spectrum antimicrobial potency in Streptomyces. J Genet Eng Biotechnol. 2022;20(1):1. doi: 10.1186/s43141-021-00283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ai C., McCarthy S., Eckrich V., et al. Increased acid resistance of the archaeon, Metallosphaera sedula by adaptive laboratory evolution. J Ind Microbiol Biotechnol. 2016;43(10):1455–1465. doi: 10.1007/s10295-016-1812-0. [DOI] [PubMed] [Google Scholar]
- 43.Sprouffske K., Aguilar-Rodriguez J., Sniegowski P., et al. High mutation rates limit evolutionary adaptation in Escherichia coli. PLoS Genet. 2018;14(4) doi: 10.1371/journal.pgen.1007324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overbeck T.J., Welker D.L., Hughes J.E., et al. Transient muts-based hypermutation system for adaptive evolution of Lactobacillus casei to Low pH. Appl Environ Microbiol. 2017;83(20) doi: 10.1128/AEM.01120-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blount Z.D., Borland C.Z., Lenski R.E. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A. 2008;105(23):7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leon D., D'Alton S., Quandt E.M., et al. Innovation in an E. coli evolution experiment is contingent on maintaining adaptive potential until competition subsides. PLoS Genet. 2018;14(4) doi: 10.1371/journal.pgen.1007348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winkler J.D., Kao K.C. Recent advances in the evolutionary engineering of industrial biocatalysts. Genomics. 2014;104(6 Pt A):406–411. doi: 10.1016/j.ygeno.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Lee S.Y., Kim H.U., Chae T.U., et al. A comprehensive metabolic map for production of bio-based chemicals. Nat Catal. 2019;2(1):18–33. doi: 10.1038/s41929-018-0212-4. [DOI] [Google Scholar]
- 49.de Souza F.M., Ingsel T., Gupta R.K. In: Application of Microbes in Environmental and Microbial Biotechnology. Inamuddin, Ahamed MI, Prasad R, editors. Springer Nature; Singapore, Singapore: 2022. Applications of microbes for energy; pp. 153–190. [Google Scholar]
- 50.Tripathi N.K., Shrivastava A. Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front Bioeng Biotechnol. 2019;7:420. doi: 10.3389/fbioe.2019.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarsaiya S., Jain A., Kumar Awasthi S., et al. Microbial dynamics for lignocellulosic waste bioconversion and its importance with modern circular economy, challenges and future perspectives. Bioresour Technol. 2019;291 doi: 10.1016/j.biortech.2019.121905. [DOI] [PubMed] [Google Scholar]
- 52.Kiefer D., Merkel M., Lilge L., et al. From acetate to bio-based products: underexploited potential for industrial biotechnology. Trends Biotechnol. 2021;39(4):397–411. doi: 10.1016/j.tibtech.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 53.da Silva G.P., Mack M., Contiero J. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv. 2009;27(1):30–39. doi: 10.1016/j.biotechadv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Liang S., Jiang W., Song Y., et al. Improvement and metabolomics-based analysis of d-Lactic acid production from agro-industrial wastes by Lactobacillus delbrueckii submitted to adaptive laboratory evolution. J Agric Food Chem. 2020;68(29):7660–7669. doi: 10.1021/acs.jafc.0c00259. [DOI] [PubMed] [Google Scholar]
- 55.Pontrelli S., Fricke R.C.B., Sakurai S.S.M., et al. Directed strain evolution restructures metabolism for 1-butanol production in minimal media. Metab Eng. 2018;49:153–163. doi: 10.1016/j.ymben.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Hua Q., Joyce A.R., Fong S.S., et al. Metabolic analysis of adaptive evolution for in silico-designed lactate-producing strains. Biotechnol Bioeng. 2006;95(5):992–1002. doi: 10.1002/bit.21073. [DOI] [PubMed] [Google Scholar]
- 57.Kim K., Hou C.Y., Choe D., et al. Adaptive laboratory evolution of Escherichia coli W enhances gamma-aminobutyric acid production using glycerol as the carbon source. Metab Eng. 2022;69:59–72. doi: 10.1016/j.ymben.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 58.Bentley G.J., Narayanan N., Jha R.K., et al. Engineering glucose metabolism for enhanced muconic acid production in Pseudomonas putida KT2440. Metab Eng. 2020;59(64–75) doi: 10.1016/j.ymben.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 59.Rugbjerg P., Myling-Petersen N., Porse A., et al. Diverse genetic error modes constrain large-scale bio-based production. Nat Commun. 2018;9(1):787. doi: 10.1038/s41467-018-03232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mundhada H., Seoane J.M., Schneider K., et al. Increased production of L-serine in Escherichia coli through Adaptive Laboratory Evolution. Metab Eng. 2017;39:141–150. doi: 10.1016/j.ymben.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 61.Schwentner A., Feith A., Munch E., et al. Metabolic engineering to guide evolution - Creating a novel mode for L-valine production with Corynebacterium glutamicum. Metab Eng. 2018;47:31–41. doi: 10.1016/j.ymben.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 62.Lee S., Kim P. Current status and applications of adaptive laboratory evolution in industrial microorganisms. J Microbiol Biotechnol. 2020;30(6):793–803. doi: 10.4014/jmb.2003.03072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thiele I., Palsson B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long C.P., Antoniewicz M.R. How adaptive evolution reshapes metabolism to improve fitness: recent advances and future outlook. Curr Opin Chem Eng. 2018;22:209–215. doi: 10.1016/j.coche.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis. Nat Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burgard A.P., Pharkya P., Maranas C.D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84(6):647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 67.von Kamp A., Klamt S. Growth-coupled overproduction is feasible for almost all metabolites in five major production organisms. Nat Commun. 2017;8:15956. doi: 10.1038/ncomms15956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patil K.R., Rocha I., Forster J., et al. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinformatics. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lun D.S., Rockwell G., Guido N.J., et al. Large-scale identification of genetic design strategies using local search. Mol Syst Biol. 2009;5:296. doi: 10.1038/msb.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fowler Z.L., Gikandi W.W., Koffas M.A. Increased malonyl coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavanone production. Appl Environ Microbiol. 2009;75(18):5831–5839. doi: 10.1128/AEM.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J., Reed J.L. OptORF: Optimal metabolic and regulatory perturbations for metabolic engineering of microbial strains. BMC Syst Biol. 2010;4:53. doi: 10.1186/1752-0509-4-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J., Reed J.L. RELATCH: relative optimality in metabolic networks explains robust metabolic and regulatory responses to perturbations. Genome Biol. 2012;13(9):R78. doi: 10.1186/gb-2012-13-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alper H., Miyaoku K., Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005;23(5):612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 74.Alper H., Jin Y.S., Moxley J.F., et al. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng. 2005;7(3):155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 75.Yim H., Haselbeck R., Niu W., et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7(7):445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 76.Pharkya P., Burgard A.P., Maranas C.D. Exploring the overproduction of amino acids using the bilevel optimization framework OptKnock. Biotechnol Bioeng. 2003;84(7):887–899. doi: 10.1002/bit.10857. [DOI] [PubMed] [Google Scholar]
- 77.Gu C., Kim G.B., Kim W.J., et al. Current status and applications of genome-scale metabolic models. Genome Biol. 2019;20(1):121. doi: 10.1186/s13059-019-1730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Åkesson M., Förster J., Nielsen J. Integration of gene expression data into genome-scale metabolic models. Metab Eng. 2004;6(4):285–293. doi: 10.1016/j.ymben.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 79.Brynildsen M.P., Winkler J.A., Spina C.S., et al. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat Biotechnol. 2013;31(2):160–165. doi: 10.1038/nbt.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Becker S.A., Palsson B.O. Context-specific metabolic networks are consistent with experiments. PLoS Comput Biol. 2008;4(5) doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmidt B.J., Ebrahim A., Metz T.O., et al. GIM3E: condition-specific models of cellular metabolism developed from metabolomics and expression data. Bioinformatics. 2013;29(22):2900–2908. doi: 10.1093/bioinformatics/btt493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis N.E., Nagarajan H., Palsson B.O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol. 2012;10(4):291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valgepea K., de Souza Pinto Lemgruber R., Meaghan K., et al. Maintenance of ATP homeostasis triggers metabolic shifts in gas-fermenting acetogens. Cell Syst. 2017;4(5):505–515. doi: 10.1016/j.cels.2017.04.008. e505. [DOI] [PubMed] [Google Scholar]
- 84.Raman S., Rogers J.K., Taylor N.D., et al. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci U S A. 2014;111(50):17803–17808. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seok J.Y., Han Y.H., Yang J.S., et al. Synthetic biosensor accelerates evolution by rewiring carbon metabolism toward a specific metabolite. Cell Rep. 2021;36(8) doi: 10.1016/j.celrep.2021.109589. [DOI] [PubMed] [Google Scholar]
- 86.Johnston C.W., Badran A.H., Collins J.J. Continuous bioactivity-dependent evolution of an antibiotic biosynthetic pathway. Nat Commun. 2020;11(1):4202. doi: 10.1038/s41467-020-18018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng B., Ma X., Wang N., et al. Utilization of rare codon-rich markers for screening amino acid overproducers. Nat Commun. 2018;9(1):3616. doi: 10.1038/s41467-018-05830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gresham D., Dunham M.J. The enduring utility of continuous culturing in experimental evolution. Genomics. 2014;104(6 Pt A):399–405. doi: 10.1016/j.ygeno.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.d'Oelsnitz A.D.E S. GroovDB: A database of ligand-inducible transcription factors. bioRxiv. 2022 doi: 10.1101/2022.07.18.500503. [DOI] [PubMed] [Google Scholar]
- 90.Daeffler K.N., Galley J.D., Sheth R.U., et al. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol Syst Biol. 2017;13(4):923. doi: 10.15252/msb.20167416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim K., Choe D., Lee D.H., et al. Engineering biology to construct microbial chassis for the production of difficult-to-express proteins. Int J Mol Sci. 2020;21(3) doi: 10.3390/ijms21030990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chi H., Wang X., Shao Y., et al. Engineering and modification of microbial chassis for systems and synthetic biology. Synth Syst Biotechnol. 2019;4(1):25–33. doi: 10.1016/j.synbio.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thornburg Z.R., Bianchi D.M., Brier T.A., et al. Fundamental behaviors emerge from simulations of a living minimal cell. Cell. 2022;185(2):345–360. doi: 10.1016/j.cell.2021.12.025. e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Landon S., Chalkley O., Breese G., et al. Understanding metabolic flux behaviour in whole-cell model output. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.732079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ingolia N.T., Brar G.A., Rouskin S., et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8):1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson D.S., Mortazavi A., Myers R.M., et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 97.Park M.K., Lee S.H., Yang K.S., et al. Enhancing recombinant protein production with an Escherichia coli host strain lacking insertion sequences. Appl Microbiol Biotechnol. 2014;98(15):6701–6713. doi: 10.1007/s00253-014-5739-y. [DOI] [PubMed] [Google Scholar]
- 98.Lombardo M.J., Aponyi I., Rosenberg S.M. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166(2):669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jeong Y., Kim J.N., Kim M.W., et al. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2) Nat Commun. 2016;7:11605. doi: 10.1038/ncomms11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van den Bergh B., Swings T., Fauvart M., et al. Experimental design, population dynamics, and diversity in microbial experimental evolution. Microbiol Mol Biol Rev. 2018;82(3) doi: 10.1128/MMBR.00008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sastry A.V., Gao Y., Szubin R., et al. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat Commun. 2019;10(1):5536. doi: 10.1038/s41467-019-13483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Si T., Chao R., Min Y., et al. Automated multiplex genome-scale engineering in yeast. Nat Commun. 2017;8:15187. doi: 10.1038/ncomms15187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.LaCroix R.A., Palsson B.O., Feist A.M. A model for designing adaptive laboratory evolution experiments. Appl Environ Microbiol. 2017;83(8) doi: 10.1128/AEM.03115-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rogers J.K., Taylor N.D., Church G.M. Biosensor-based engineering of biosynthetic pathways. Curr Opin Biotechnol. 2016;42:84–91. doi: 10.1016/j.copbio.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 105.Rogers J.K., Church G.M. Genetically encoded sensors enable real-time observation of metabolite production. Proc Natl Acad Sci U S A. 2016;113(9):2388–2393. doi: 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Monk J.M., Lloyd C.J., Brunk E., et al. iML1515, a knowledgebase that computes Escherichia coli traits. Nat Biotechnol. 2017;35(10):904–908. doi: 10.1038/nbt.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maeda T., Iwasawa J., Kotani H., et al. High-throughput laboratory evolution reveals evolutionary constraints in Escherichia coli. Nat Commun. 2020;11(1):5970. doi: 10.1038/s41467-020-19713-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Phaneuf P.V., Zielinski D.C., Yurkovich J.T., et al. Escherichia coli data-driven strain design using aggregated adaptive laboratory evolution mutational data. ACS Synth Biol. 2021;10(12):3379–3395. doi: 10.1021/acssynbio.1c00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wicky B.I.M., Milles L.F., Courbet A., et al. Hallucinating symmetric protein assemblies. Science. 2022;378(6615):56–61. doi: 10.1126/science.add1964. [DOI] [PMC free article] [PubMed] [Google Scholar]