SUMMARY
Long-term impacts of diet have been well studied; however, the immediate response of the intestinal epithelium to a change in nutrients remains poorly understood. We use physiological metrics and single-cell transcriptomics to interrogate the intestinal epithelial cell response to a high-fat diet (HFD). Within 1 day of HFD exposure, mice exhibit altered whole-body physiology and increased intestinal epithelial proliferation. Single-cell transcriptional analysis on day 1 reveals a cell-stress response in intestinal crypts and a shift toward fatty acid metabolism. By 3 days of HFD, computational trajectory analysis suggests an emergence of progenitors, with a transcriptional profile shifting from secretory populations toward enterocytes. Furthermore, enterocytes upregulate lipid absorption genes and show increased lipid absorption in vivo over 7 days of HFD. These findings demonstrate the rapid intestinal epithelial response to a dietary change and help illustrate the essential ability of animals to adapt to shifting nutritional environments.
Graphical abstract
In brief
Enriquez et al. demonstrate that animals immediately adapt within days to a change in diet, both in whole-body physiology and at the cellular level in the small intestine. Single-cell transcriptional analysis following a shift to HFD shows that cells display unique adaptive responses that suggest striking adaptive ability during nutrient availability.
INTRODUCTION
Profound changes in diet are known to elicit metabolic and physiologic changes over time. For example, caloric imbalance is thought to be one variable contributing to the increased prevalence of obesity within the United States.1 From 1999 through 2018, obesity has increased from 30.5% to 42.4%,2 and metabolic disease, including diabetes and dyslipidemia, has increased from 25.3% to 34.2%.3 While long-term changes in diet are known drivers of obesity and metabolic disease, how rapidly a change in diet can trigger systemic changes is poorly understood.
The gastrointestinal tract is the primary site for dietary sensing and absorption of carbohydrates, proteins, and fats that are then utilized by cells of the body to maintain energy homeostasis. There have been numerous studies using animal models investigating the long-term impact of a high-fat diet (HFD) on metabolic health.4–7 For example, a single-cell RNA sequencing analysis of mice following 12 weeks on a high-fat/high-sugar diet revealed intestinal maladaptation including altered intestinal lineage allocation.8 However, after 12 weeks of HFD, animals are obese and have marked metabolic disease, and the intestine has undergone a significant adaptive response. Previous studies suggest that metabolic changes such as insulin resistance, hepatic steatosis, and ketogenesis may begin within days of HFD exposure.9–13 Therefore, the intestine might also rapidly adapt to immediate changes in diet; however, this has not been previously investigated.
The intestinal epithelium arises from proliferative intestinal stem cells (ISCs) that differentiate into specialized lineages along the crypt-villus axis.14 ISCs first form secretory and absorptive progenitors that migrate through the transit-amplifying (TA) zone and gradually mature to become absorptive enterocytes or various secretory cells. Enterocytes comprise the bulk of the intestine and absorb sugars, small peptides, and lipids. In parallel, secretory cells, such as goblet, Paneth, or enteroendocrine (EEC), secrete mucins, defensins, or hormone peptides, respectively, in response to luminal stimuli. Recently, it has been shown that fasting, caloric restriction, and long-term HFD modulate ISC function and proliferation,8,15–17 suggesting that the intestinal epithelium is exquisitely sensitive to changes in diet. However, the immediate impacts of HFD on ISC differentiation into the various epithelial cells remain unknown.
Here, we sought to investigate the immediate responses of animals to a change in diet by shifting mice from a normal balanced diet to an HFD. We performed a combination of whole-body metabolic analysis, single-cell transcriptomics, and functional analyses to identify systemic and cellular changes that occur immediately following a dietary change. We observed immediate whole-body metabolic changes and intestinal crypt-niche stress after just 1 day of HFD, transcriptional changes indicating altered cell trajectory of progenitors after 3 days of HFD, and functional adaptation to more efficient fatty acid uptake within 7 days of beginning an HFD. These data demonstrate that the intestine rapidly alters its transcriptional, cellular, and functional profile in response to changes in nutrient inputs.
RESULTS
One day of HFD induces changes in mouse metabolism and intestinal proliferation
To investigate the impact of an immediate change in diet, we used indirect calorimetry to monitor adult wild-type mice maintained on normal chow (kcal%: fat, 13%; carbohydrate, 58%; protein, 29%) or switched to an HFD (kcal%: fat, 60%; carbohydrate, 20%; protein, 20%) for 7 days (Figure 1A). We measured oxygen consumption (VO2) and carbon-dioxide expiration (VCO2) (Figures 1B and 1C) to calculate the respiratory exchange ratio (RER), which reflects the primary fuel source metabolized by the body. A ratio closer to 1 indicates predominantly glucose metabolism, whereas a ratio closer to 0.7 suggests reliance on fatty acid metabolism.18 Within the first day of HFD feeding, RER levels decreased from around 0.9 to 0.8, and this difference was maintained over time (Figure 1D). Moreover, we measured the energy expenditure (EE), which measures total energy required for homeostasis.19 Within the first day, EE was increased from around 0.2–0.5 to around 0.4–0.6 kcal/min (Figure 1E). We saw no significant changes in water intake or ambulatory movement over the 7 days (Figures S1A–S1C). We separately measured mouse weight, daily energy intake, and food intake from mice in normal housing. From the first day, mice given an HFD gained weight and had an increased energy intake (Figures 1F and 1G). HFD food intake displayed a significantly decreased slope over time (Figure 1H), suggesting that due to the higher caloric value of an HFD, mice needed to consume less chow to achieve satiety. These data suggested that mice experienced whole-body metabolic changes within 1 day of eating an HFD and that there may be related changes in the intestine.
Figure 1. One day of HFD induces changes in mouse metabolism and intestinal proliferation.
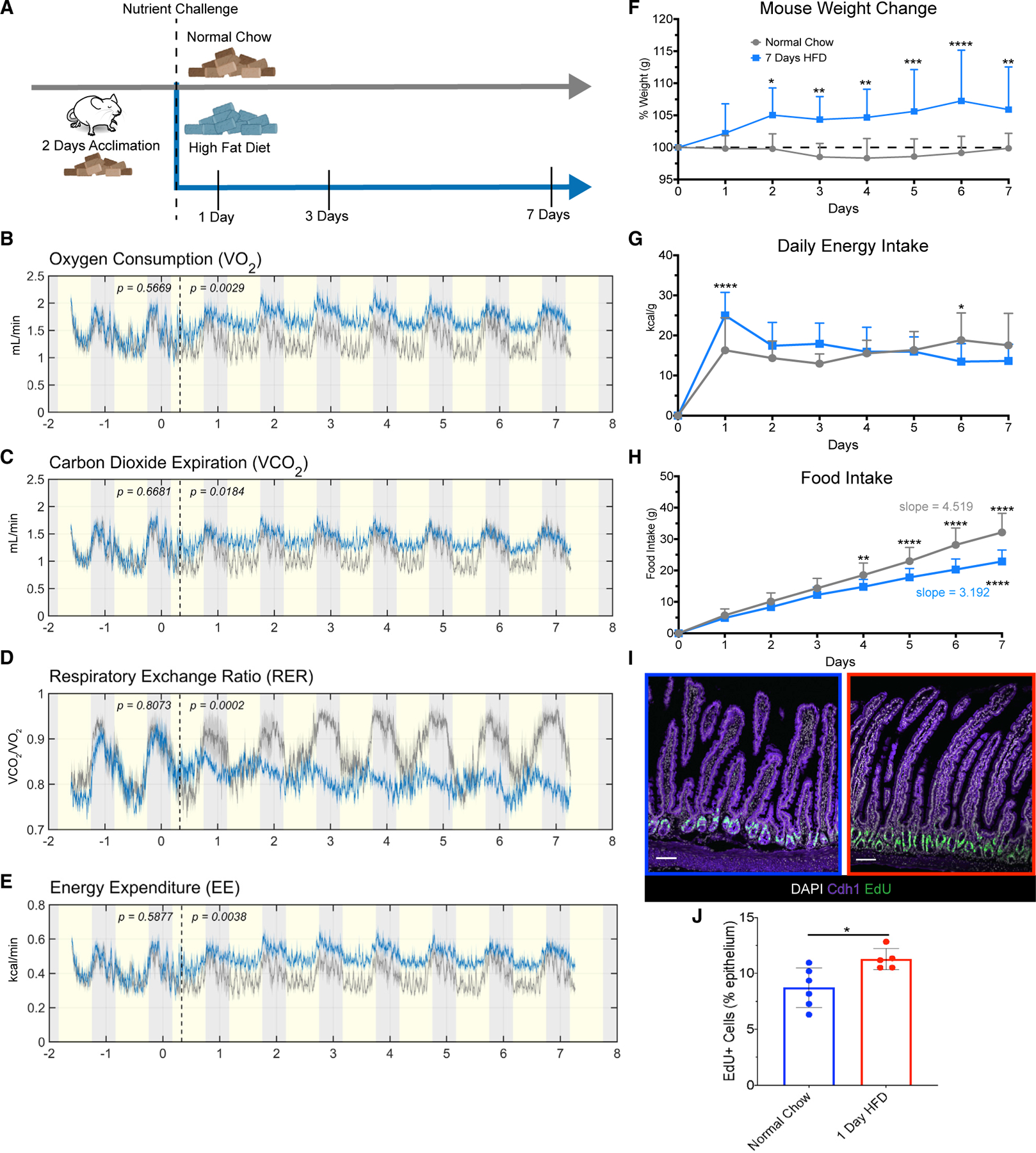
(A) Diagram of feeding schedule in metabolic cages. Mice underwent 2 days of acclimation to the cages, and all consumed normal chow (brown-color pellets) during the acclimation period. The dashed line denotes when mice were divided into two groups, one maintained on normal chow (n = 5), and the other group switched to high-fat diet (HFD; n = 6) for 7 days.
(B) Average oxygen consumption.
(C) Average carbon dioxide expiration rate.
(D) Average respiratory exchange rate ratio calculated by VCO2/VO2.
(E) Average energy expenditure measured in kcal/min captured from animals fed normal chow (gray line) and HFD (blue line). Yellow shading indicates light cycle, whereas gray shading indicates night cycle. The dashed line indicates the end of the acclimation period and beginning of dietary challenge.
(F) Percentage of daily weight change measured in grams (g), n = 11 normal chow, n = 20 HFD.
(G) Average daily energy intake measured in kcal/g, n = 11 normal chow, n = 20 HFD.
(H) Average accumulated food intake over time with calculated average slopes displayed, n = 11 normal chow, n = 20 HFD.
(I) Immunofluorescence images of the proximal jejunum of mice fed normal chow (blue border) and 1 day of HFD (red border) after a 2 h pulse with EdU to label actively proliferating cells (green). Nuclei are stained in white, and Cdh1 marks the epithelium in purple. Scale bars: 100 μm.
(J) Quantification of (I), n = 6 normal chow, n = 5 HFD.
(B–E and J) One-way ANOVA. Error bars are SD. (F–H) Two-way ANOVA; *p < 0.05, **p < 0.0005, ***p < 0.0003, ****p < 0.0001.
The proximal small intestine is the primary location for nutrient sensing and absorption. For example, initial intestinal sensing of nutrients and fat emulsification occurs in the duodenum, whereas the bulk of fatty acid absorption occurs in the jejunum.20 Therefore, we investigated whether there was an adaptative response within the proximal intestine to acute HFD mediating these whole-body metabolic changes. Because the stem cell niche is sensitive to changes in dietary nutrients,15–17 we examined intestinal proliferation, crypt depth, and villus height after 1, 3, or 7 days of exposure to an HFD. We only observed increased EdU incorporation after 1 day of HFD and did not observe any changes in crypt depth or villus height (Figures 1I, 1J, and S1D–S1G). We also examined changes related to intestinal cell death using cleaved caspase-3 staining and TUNEL staining and found no difference at any of the time points (Figures S1H–S1K). These data suggested that HFD triggered a proliferative response within 1 day but that this did not result in changes in intestinal surface area over the 1 week time course.
HFD initiates a rapid increase in expression of genes regulating fatty acid metabolism
To investigate how individual cell populations respond to HFD, we performed single-cell RNA sequencing (scRNA-seq) on epithelial cells dissociated from the duodenum and jejunum of adult mice across all time points (Figures S2A–S2E; GEO: GSE199776). Given its essential role in nutrient absorption, we largely focused subsequent analysis on the jejunum. After quality control and filtering of cells, we integrated the datasets of cells from mice eating normal chow and those fed 1, 3, and 7 days of an HFD and utilized the normal chow cells as the reference dataset following previously described computational tools.21 This technique allowed us to not only determine the transcriptional heterogeneity present within each cluster, but also to compare transcriptional changes between conditions through a total number of 10,593 cells (normal chow: 2,214; 1 day HFD: 2,309; 3 day HFD: 2,353; 7 day HFD: 3,717). We performed differential gene expression analysis of each cluster between each dietary condition to identify how each cell type responded to HFD over time (Tables S1 and S2). We identified all expected cell populations including enterocytes, EEC, goblet, tuft, and Paneth cells, as well as enterocyte progenitors (EPs), secretory progenitors (SPs), and stem/early-transit amplifying (TA) zone cells (Figure 2A). The top 5 genes between each cluster verified known genes distinguishing between cell types (Figure 2B).
Figure 2. HFD initiates a rapid increase in expression of genes regulating fatty acid metabolism.
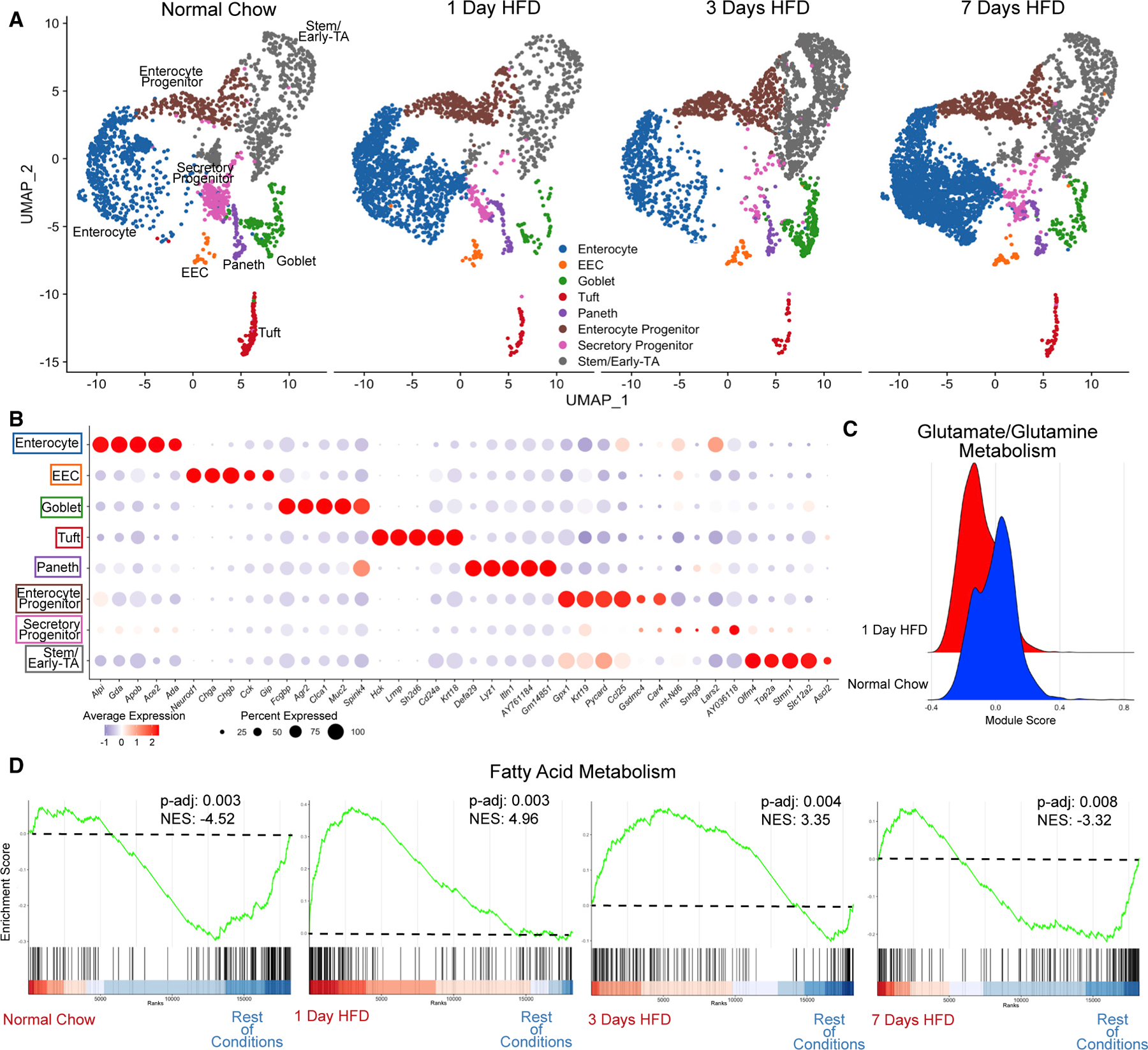
(A) Integrated analysis of jejunal epithelial sorted dataset: normal chow (reference dataset for integration, 2,214 cells), 1 day of HFD (2,309 cells), 3 days of HFD (2,353 cells), and 7 days of HFD (3,717 cells).
(B) Average dot plot expression of top 5 genes per cluster.
(C) Ridge plot between normal chow and 1 day of HFD depicting module gene score calculated for glutamate/glutamine metabolism.
(D) Gene set enrichment analysis (GSEA) using all clusters focusing on Hallmark fatty acid metabolism genes compared between each dietary condition. Green line denotes enrichment score for each cell per condition. Dashed line marks score at 0.0. Black lines above color gradient denote localization of Hallmark fatty acid metabolism genes. Blue (low) to red (high) color gradient depicts enrichment score expression. Adjusted p value (p-adj) and normalized enrichment score (NES) are reported in panel.
Analysis of the transcriptional changes in response to HFD using Biological Process Gene Ontology Terms (GO-Terms) showed increased expression of genes for fatty acid metabolic pathways in several cell types at 1 day of HFD (Figures S2F–S2I), suggesting a shift away from glutamine/glutamate metabolism that is normally utilized by the intestinal epithelium.22 To further investigate this, we used genes collated by the Molecular Signatures Database (MSigDB) to evaluate the transcriptional signature of glutamate/glutamine metabolism and saw an immediate downregulation after 1 day of HFD (Figure 2C). Conversely, using gene set enrichment analysis (GSEA), we observed that most epithelial cells have upregulated genes involved in fatty acid metabolism as indicated by normalized enrichment scores (NESs) (Figure 2D). Specifically, NESs for fatty acid metabolism showed an increase at both 1 and 3 days of HFD, suggesting an immediate metabolic response to the influx of luminal fat. Interestingly, by 7 days, NESs for fatty acid metabolism decreased, suggesting that enterocytes have adapted to the shift to HFD.
HFD triggers an immediate stress response
Aside from immediate metabolic changes, we also observed an upregulation in genes associated with cellular stress (heat-shock protein genes such as Hspa1a, Hsp90aa1, and Hsf1; unfolded protein response genes such as Xbp1, Atf4, and Eif2ak3) for all epithelial cell populations after 1 day of HFD (Figure 3A). To validate this, we repeated the HFD time course and isolated whole epithelial RNA for analysis of heat-shock protein genes via qPCR, which showed a significant upregulation after 1 day of HFD (Figure 3B). From our scRNA-seq, stem/early-TA and Paneth cells trended among the highest gene-expression changes for cellular stress responses (Figure 3B). Therefore, we further examined these specific subsets and observed that the stem/early-TA subset upregulated heat-shock protein genes and, via GSEA, upregulated unfolded protein response (UPR) genes (Figures 3D and 3E). This suggests that stem/early-TA cells have an immediate stress response following exposure to HFD.
Figure 3. HFD triggers an immediate stress response.
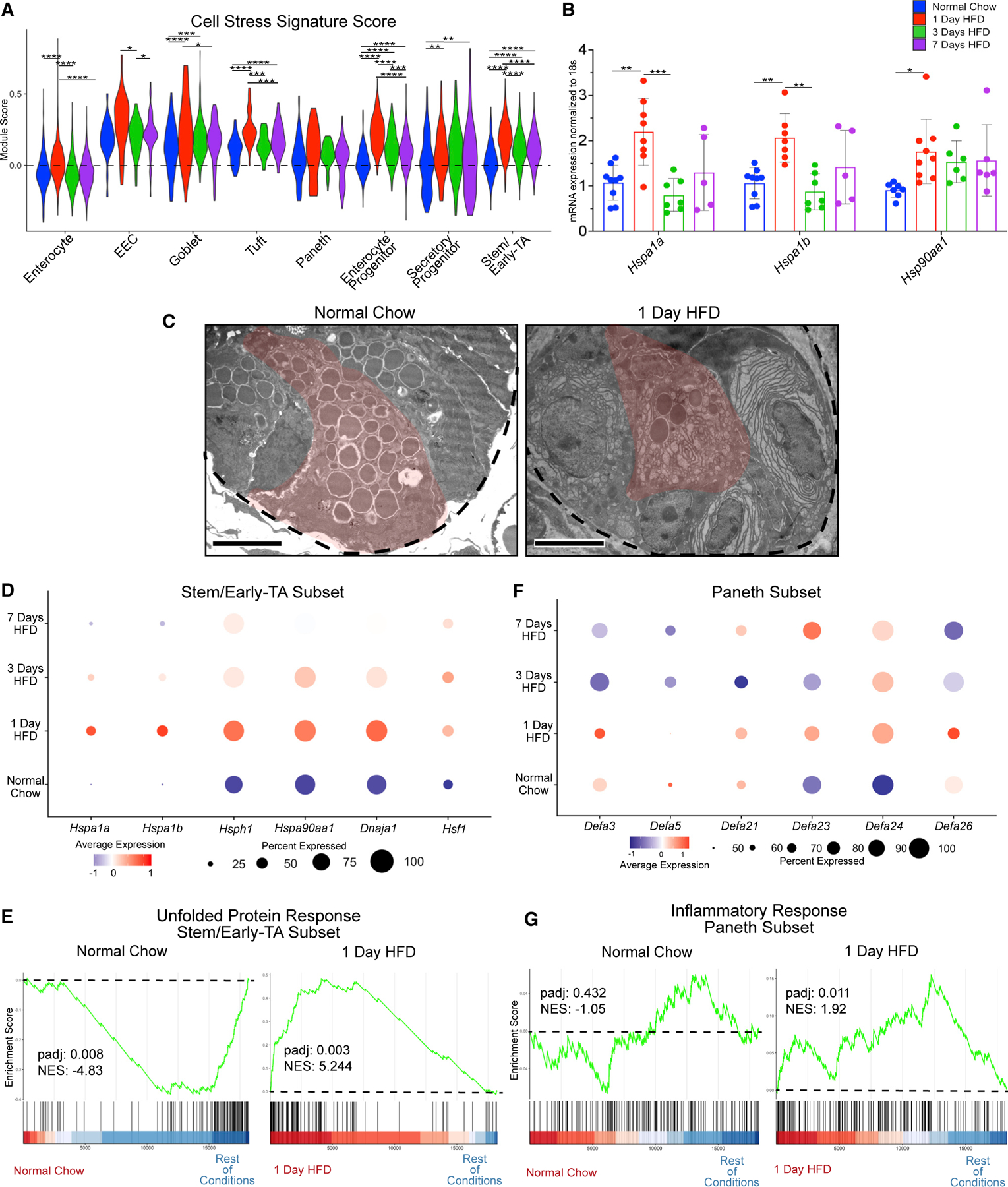
(A) Cell stress module gene scores between dietary conditions in enterocyte, enteroendocrine (EEC), goblet, tuft, Paneth, enterocyte progenitor, secretory progenitor, and stem/early-TA clusters.
(B) qRT-PCR between dietary conditions for heat-shock protein 1A (Hspa1a), Hspa1b, and heat-shock protein 90, alpha (cytosolic), class A member 1 (Hsp90aa1). n = 5–9 mice.
(C) Transmission electron microscopy (TEM) images of crypts from animals fed normal chow and 1 day of HFD. Dashed line outlines the bottom of the crypt, while the red-pseudocolor denotes a representative Paneth cell. Scale bars: 4 μm.
(D) Dot plot depicting average gene expression of heat-shock-related genes in stem/early-TA zone cells.
(E) GSEA of Hallmark unfolded protein response genes comparing stem/early-TA zone cells between normal chow and 1 day of HFD.
(F) Dot plot depicting average gene expression of genes involved in defensin regulation in Paneth cells.
(G) GSEA of Hallmark inflammatory response genes comparing Paneth cells between normal chow and 1 day of HFD. Green line denotes enrichment score for each cell per condition. Dashed line marks score at 0.0. Black lines above color gradient denotes localization of Hallmark genes. Blue (low) to red (high) color gradient depicts enrichment score expression.
In addition to a transcriptional stress response, Paneth cells initially upregulate, and then downregulate, defensin-related genes over the course of the HFD, suggesting that a cellular stress response can influence Paneth cell activity (Figure 3F). Previous studies have shown that environmental stresses such as smoking or Crohn’s disease (CD) result in abnormal Paneth cell ultrastructure.23,24 We therefore examined if a HFD alters the structure of Paneth cells via transmission electron microscopy (TEM). After 1 day of HFD, we observed abnormal subcellular structure such as loss of granules and abnormal ER in Paneth cells (Figure 3C). Coincident with stress and degranulation, GSEA suggested an inflammatory response of Paneth cells after 1 day of HFD (Figure 3G). These data are consistent with reports showing that heat-shock proteins help initiate adaptation in response to inflammation.25 Collectively, these data suggest that cell stress is an early response of crypt cells to HFD.
Transcriptional responses of enterocytes correlate with improved lipid absorption and transport
Given that enterocytes mediate absorption of fat, we predicted that they would transcriptionally and functionally adapt to HFD by increasing their ability to handle luminal lipids. We calculated a lipid absorption score with known genes involved in fatty acid absorption. Compared with the other cell types, enterocytes showed a pronounced increase in the lipid absorption score over time (Figures 4A and S3A). Interestingly, the other cell clusters (EEC, Paneth, tuft, EP, SP, goblet, and stem/early TA) also experienced a small increase in score at 1 day of HFD, possibly due to the shift to fatty acid metabolism as a primary energy source. To validate these observations, we repeated the HFD time course, isolated intestinal epithelial RNA, and performed qPCR for Fabp1, Apoa4, and Cpt1a, key genes involved in fatty acid regulation.26 Expression of all three genes was increased in response to HFD feeding.
Figure 4. Transcriptional responses of enterocytes correlate with improved lipid absorption and transport.
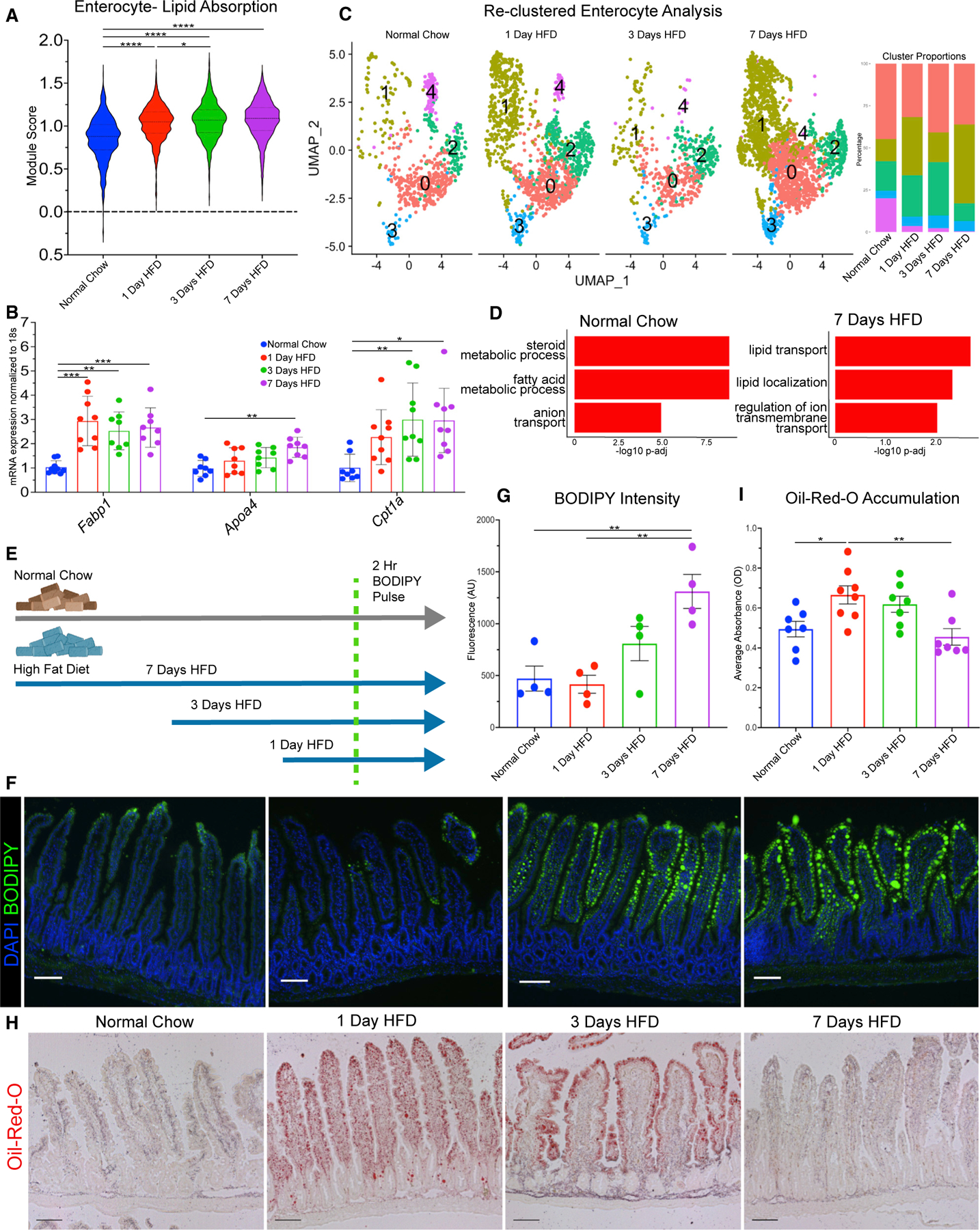
(A) Lipid absorption score compared between enterocytes in each dietary condition.
(B) qRT-PCR between dietary conditions for fatty acid binding protein 1 (Fabp1), apolipoprotein A-IV (Apoa4), and carnitine palmitoyltransferase 1a (Cpt1a).
(C) Uniform manifold approximation and projection (UMAP) of enterocytes reclustered into 5 subsets: 0 (1,735 cells), 1 (1,671 cells), 2 (847 cells), 3 (281 cells), and 4 (200 cells). Barplot visualizes cluster proportion colored by cluster number. Breakdown per condition: normal chow (675 cells), 1 day of HFD (1,308 cells), 3 days of HFD (549 cells), and 7 days of HFD (2,202 cells).
(D) Top 3 upregulated Gene Ontology terms (Biological Process) for normal chow and 7 days of HFD in enterocyte subclusters −0 and −1.
(E) Diagram of BODIPY pulse-chase experiment. Normal chow, gray arrow; HFD, blue arrow; green dashed line denotes time when all mice were gavaged with BODIPY and analyzed 2 h later.
(F) Fluorescence images of BODIPY (green) lipid absorbance in proximal jejunum 2 h post-gavage. DAPI counterstains nuclei in blue. Scale bars: 100 μm.
(G) Quantification of BODIPY fluorescence intensity. n = 4 per condition.
(H) Oil red O staining of proximal jejunum for each condition. Scale bars: 100 μm.
(I) Quantification of (H) reported as optical density (OD) for oil red O accumulation. n = 7 for each condition. (A, F, and H) Error bars are SEM. One-way ANOVA; ns, not significant, *p < 0.05, **p < 0.005, ****p < 0.0001.
We then performed reclustering analysis of enterocytes to determine if specific subpopulations of enterocytes might be specialized for handling the influx of luminal lipids. Our analysis revealed 5 enterocyte subclusters that also transcribe previously identified genes that are regionally located along the crypt-villus axis27 (Figures S3B and S3C). Also, analysis of our 5 subclusters showed distinct functional identity analyzed via differential expression analysis using GO-Term Biological Processes (Figure S3D). We determined that subclusters −0 and −1 seem to specialize in fatty acid or transport-related genes (Figure S3D) and wondered whether these clusters may regulate lipid transport genes during HFD. Using these two specific clusters, we tested for differential gene expression over the HFD time course and observed that during normal chow, these clusters were initially responsible for fatty acid metabolism; however, by 7 days of HFD, these clusters had upregulated genes related toward fatty acid transport and localization (Figure 4D). These data suggest that newly adapted populations of enterocytes transcriptionally improve their lipid absorptive capability.
Uptake of fatty acids is mediated via passive diffusion or through active transporters, such as Cd36 or Fatp4 (Slc27a4).26,28 We examined the expression of these transporters in all enterocyte clusters and then focused on subclusters 0 and 1. We observed a significant increase in Slc27a4 but a significant decrease in Cd36 (Figures S3E and S3F). This may indicate differential regulation between active transporters of fatty acids and could suggest that the intestinal epithelium preferentially regulates some fatty acid transporters more than others during acute HFD. Next, to determine if enterocytes functionally increase the efficiency of lipid absorption in response to HFD, we analyzed enterocytes in the jejunum in two ways: BODIPY and Oil-Red-O. First, we took animals that had been on an HFD for 1, 3, and 7 days and synchronously gavaged them with an olive-oil-infused bolus of fluorescent-labeled fatty acid BODIPY dye (Figure 4E). We then isolated the jejunum after 2 h and quantified lipid uptake facilitated by enterocytes. When compared with normal chow, enterocytes adapting to HFD displayed markedly increased capability of absorbing fatty acids (Figures 4F and 4G). The animals that had been on an HFD for 3 and 7 days displayed a significantly higher rate of lipid uptake, and this correlates with the increase in expression of lipid transport genes. Using a separate approach, we visualized lipid droplet accumulation using Oil-Red-O, which measures the steady-state presence of lipids in cells (Figure 4I). In enterocytes from animals maintained on normal chow, lipid accumulation was observed at the villus tips with some diffuse staining in the lamina propria. In response to a shift to an HFD, staining at day 1 was the highest, with lipids accumulating along the whole villus length (Figure 4H). After 3 days of HFD, lipid accumulation in enterocytes began to diminish but increased in the lamina propria. At day 7, the jejunum seems to have adapted to HFD, due to lipid accumulation being similar to normal chow. Together, these data show that the transcriptional increase in genes that control both lipid transport and absorption correlate with a rapid functional increase in lipid transport following a switch to an HFD.
scRNA-seq analysis of rare epithelial cell types using lineage tracing and cell sorting
Absorptive enterocytes are the most abundant cell population in the intestine. Conversely, some secretory lineages, particularly EECs, range from 1% to 3% within the epithelium.14 For example, within our proximal jejunal dataset (Figure 1), out of 10,593 total cells, we only captured 158 total EECs (1.5% of total epithelial cells). However, we were interested in determining the impact of acute HFD on these relatively rare populations, particularly EECs, because EECs are known to secrete hormones in response to luminal stimuli, including fatty acids.29 To increase our power analysis of rare secretory cells, we performed genetic cell labeling and sorting using Neurog3 Cre recombinase and a tdTomato reporter. Previous studies demonstrated that lineage tracing with Neurog3-Cre not only allows for enrichment of EECs but also of progenitors and other secretory cell types.30,31 Therefore, we employed this strategy as a means to enrich for these rare populations. We sorted live epithelial tdTomato+ cells from the duodenum and jejunum of Neurog3Cre-tdTom mice consuming normal chow or mice after 1, 3, or 7 days of HFD and performed scRNA-seq (Figures S4A and S4B). As expected, we enriched for EECs (4,915 total cells, 23% of total secretory enriched cells), as well as other non-endocrine cell types such as Paneth, tuft, and goblet cells (Figures S4C–S4E). As previously reported, we also observed some labeling of intestinal stem and progenitor cell populations (Figures S4C–S4E) as evidenced by rare lineage-traced ribbons emanating from the crypts. Labeled cell lineages were confirmed using immunofluorescence staining with canonical cell-type markers and were colocalized with tdTomato (Figures S4F and S4G). Additionally, we compared our tdTomato+-enriched dataset, our unenriched epithelial-only dataset, and a previously published intestinal single-cell dataset32 and determined that the tdTomato+-sorted populations were substantially enriched for secretory lineages while still containing all other cell types (Figures S4H–S4K).
With this highly powered dataset, we first investigated the impact of short-term HFD on one of the rarest cell types, EECs. There were 9 different clusters segregating into known EEC-hormone subtypes (Figures S5A and S5B). We focused on EECs identified that transcribe glucagon (Gcg), serotonin (Tph1), gastric inhibitory polypeptide (Gip), and cholecystokinin (Cck), respectively. These are known hormones released in response to changes in luminal nutrients.33,34 Given their role in nutrient sensing, we predicted that EEC transcription would be highly impacted by an HFD. Surprisingly, there were modest changes in EEC numbers and expression of hormones in the proximal intestine. For example, K-cell transcription of Gip increased over 7 days of HFD, whereas Gcg, Tph1, and Cck expression decreased slightly after 3 days of HFD before increasing back up by 7 days of HFD (Figure S5C). We validated these gene-expression changes by performing GIP and CCK ELISAs on mouse serum for all time points and saw a similar trend in protein levels (Figure S5D). Interestingly, expression of known EEC fatty acid receptors, such as Ffar1, Ffar2, Ffar3, Ffar4, and Gpr119,35 remained relatively unchanged over the course of the HFD (Figure S5E), suggesting that EEC lipid-sensing mechanisms are not transcriptionally sensitive in response to an immediate change to an HFD.
HFD causes a transcriptional shift in cell lineage trajectory from secretory to absorptive
While our data indicate that individual cell types respond to a change in diet, it is also possible that the allocation of intestinal lineages might be impacted by HFD. To investigate this possibility, we quantified the scRNA-seq data from unenriched epithelial cells to look for any transcriptional changes that indicate a change in the proportions of intestinal lineages. At the transcriptional level, we observed a decrease in all secretory lineages and an increase in enterocytes over the 7 days of HFD in the proximal intestine (Figure S2B). When visualized via cell density plots, Paneth, EEC, goblet, and SP clusters all had fewer cells at 3 days of HFD, whereas EP and enterocyte clusters were all increased over the same time frame (Figures 5A and S2B). Since secretory lineages, like EEC and Paneth cells, are rare, this initial conclusion was based on a relatively small number of cells. We therefore examined the secretory-cell-enriched dataset for shifts in lineage allocation (Figure 5D). Again, via cell density plot, we observed an overall reduction in all secretory cells, with the biggest reduction at 3 days of HFD, whereby EECs, goblet, and Paneth cell clusters were reduced but enterocyte cluster size was increased (Figure 5E). Given that scRNA-seq measures transcriptional changes but not necessarily stable cell-fate changes, we re-examined the HFD time course by staining for canonical markers of secretory lineages including Chga (EEC), lysozyme (Paneth), Dclk1 (tuft), and Alcian blue (goblet) (Figures S5F–S5M). By 3 days of HFD, we observed a similar trend in reduced secretory lineages, aside from tuft cells; however, these did not reach statistical significance. These data suggested that, in response to HFD, stem and progenitor populations shift their transcriptional program from secretory toward absorptive; however, at the protein level, cell lineages are more stable.
Figure 5. HFD causes a transcriptional shift in cell lineage trajectory from secretory to absorptive.
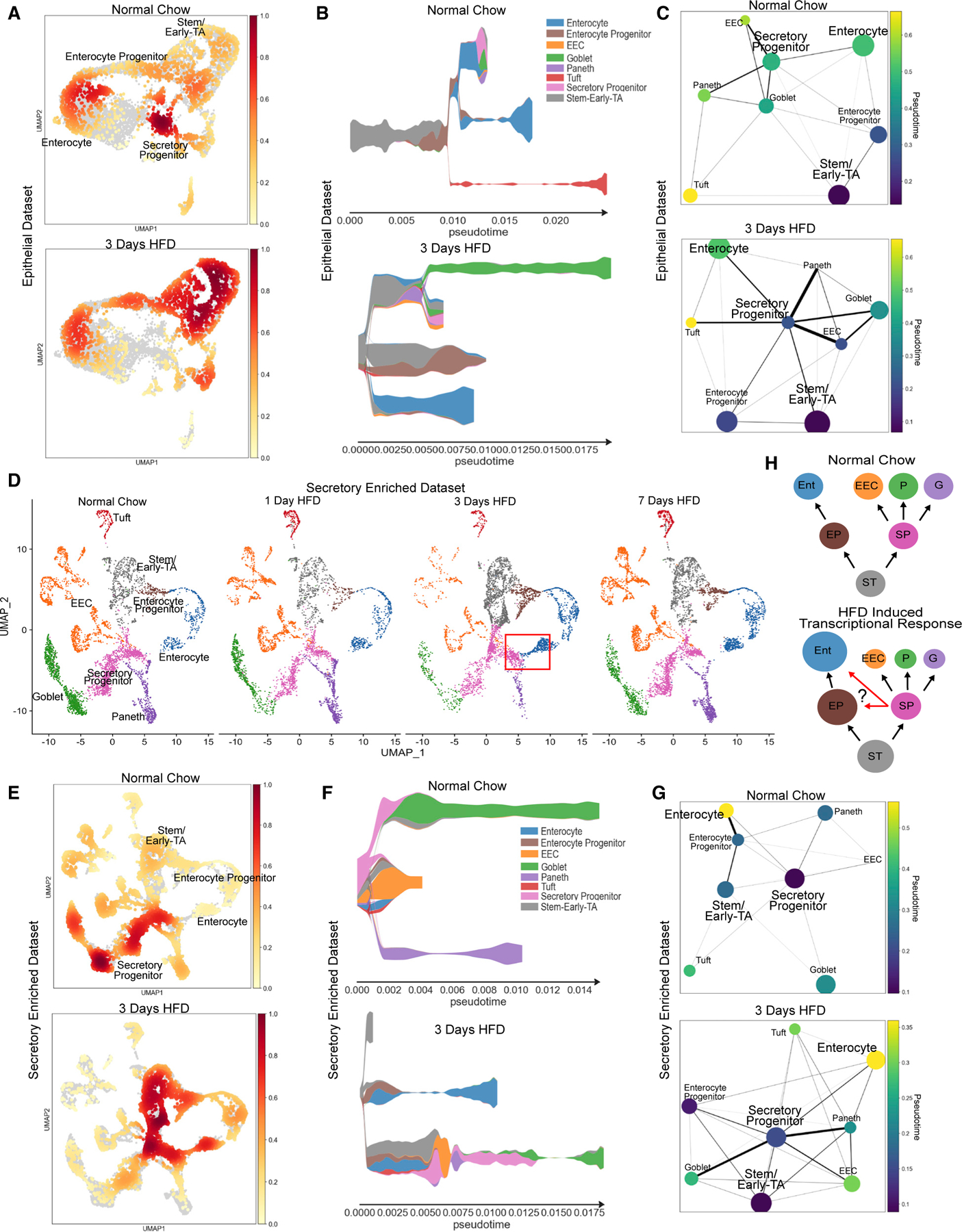
(A) Distribution of epithelial cells depicted as density plot split between mice eating normal chow (top panel) and 3 days of HFD (bottom panel). Yellow (low) to red (high) color gradient depicts density value.
(B) STREAM pseudotime analysis for epithelial cells from mice eating normal chow and 3 days of HFD. Stem cluster was used as the origination node and then unbiased analysis of clustering with 5 branches.
(C) PAGA pseudotime analysis for epithelial cells from mice eating normal chow and 3 days of HFD. Dark purple to yellow color gradient depicts computed trajectory from early- to late-stage differentiation.
(D) Integrated reference analysis for secretory enriched (sorted TdTom+) cells for each condition colored by each cluster. Red box at 3 days of HFD focuses on emerging cluster of new cells bridging between secretory progenitor and enterocyte clusters.
(E) Distribution of secretory-enriched cells depicted as density plot split between mice eating normal chow and 3 days of HFD.
(F) STREAM pseudotime analysis for secretory enriched cells from mice eating normal chow and 3 days of HFD. Stem cluster was used as the origination node and then unbiased analysis of clustering with 3 branches.
(G) PAGA pseudotime analysis for secretory-enriched cells from mice eating normal chow and 3 days of HFD. Dark purple to yellow color gradient depicts computed trajectory from early- to late-stage differentiation.
(H) Diagram delineating intestinal epithelial cell transcriptional differentiation priority during normal chow and the adaptive response to HFD. ST, stem/early-TA zone; EP, enterocyte progenitor; SP, secretory progenitor; Ent, enterocyte; EEC, enteroendocrine cell; P, Paneth; G, goblet. Circle size denotes relative transcriptional cell abundance. Red arrows during HFD indicate potential shift of cells toward enterocyte progenitors and enterocytes.
To further interrogate the transcriptional shift toward more enterocytes, visual inspection of the secretory enriched dataset identified the emergence of a new cluster of cells at 3 days of HFD that appears to connect clusters between stem/secretory progenitors toward enterocytes (Figure 5D, red box). It is possible that some SPs have transcriptionally activated an enterocyte-like program in response to this significant change in diet. To computationally examine transcriptional trajectory changes, we performed pseudotime analysis of both epithelial-enriched and secretory-enriched datasets for cells at 3 days of HFD and compared them with normal chow cells by using STREAM36 and PAGA.37 First, for the epithelial-enriched dataset, by setting the stem cell/early-TA cluster as the origination node, the analysis revealed distinct changes between lineage trajectories of normal chow and 3 days of HFD (Figures 5B and 5C). In response to HFD, both pseudotime techniques suggest a shift in priority of transcriptional differentiation whereby more cells divert toward a more enterocyte-like trajectory as seen with the prominent enterocyte lineage branch in the STREAM plot (Figure 5B). Furthermore, it seems that at 3 days of HFD, the SP population not only seeks to still differentiate secretory cells but also diverts to a more progenitor-like state to be more closely related toward enterocytes as seen with the trajectory visualization with PAGA (Figure 5C). To explore this further, we performed similar pseudotime analysis on the secretory enriched dataset comparing between normal chow and 3 days of HFD. This highly powered dataset also indicates a transcriptional priority shift in lineage trajectories in response to HFD (Figures 5F and 5G). Specifically, at the 3 days of HFD time point, not only was there was a newly expanded enterocyte branch but also SPs were more closely associated to stem and enterocyte progenitors.
Therefore, using two different single-cell datasets and two different computational approaches, our data suggest that within 1 week of HFD exposure, the intestine transcriptionally restructures itself to better absorb dietary fats by expanding the population of absorptive enterocytes and improving their ability to handle the increased demand for lipid transport (Figure 5H).
DISCUSSION
Animals need to adapt to changes in nutrient availability for organismal survival. For example, these changes might be behavioral by migrating toward food sources, by changing food priorities, or by increasing ingestion rate. The long-term impact of changes in nutrient consumption are well studied; however, how the body immediately responds to changes in diet is still unknown. This study revealed that immediately upon dietary switch from a normal/balanced diet to an HFD, intestinal cell types have unique and robust transcriptional responses. Within 24 h, cells in the crypt exhibited cell stress at the transcriptional and ultrastructural levels. Over the next few days, there was an increase in cellular populations, which we computationally projected to be biased toward an enterocyte fate. By 1 week of HFD, the intestinal epithelium had transcriptionally upregulated molecular pathways involved in lipid metabolism and transport. We mainly focused observations on the jejunum but also observed similar trends in the duodenum (data not shown). Furthermore, we confirmed that the enterocytes of the jejunum had functionally adapted to be highly efficient at lipid absorption. It seems that specific subclusters of enterocytes appear to rapidly adapt to high fat, whereas others do not. This is consistent with the previous observation that there are transcriptionally different enterocyte populations along the crypt villus axis27 that may be differentially involved in lipid transport.
Previous studies using HFD to elicit metabolic phenotypes in animals overlook this initial window of adaptation. Most protocols expose mice to an HFD for periods ranging from 6 weeks to 6 months.4–7 Within the intestine over this prolonged HFD, the epithelial lining thins, secretory lineage abundances alter, and consequential morbidities of obesity and diabetes develop. Our data suggest that the intestine may acutely respond each time there is a major shift in nutritional input and that changes are immediately observed. For this study, we used the experimental paradigm of shifting mice from a normal balanced diet to one that is high in fat to investigate how the intestinal epithelium responds to a major change in diet. We acknowledge that our study is not ideally suited to identify the specific impact of increased lipids because the protein and fiber contents are not identical between normal chow and HFD. In that case, a diet that controls for protein and carbohydrate levels would be needed. Studies that compare the normal balanced diet to the diet that calorically controls for protein content see modest differences in response to an HFD.38,39 Moreover, the experimental paradigm of changing from a balanced diet to one that is high in fat might better model abrupt changes in diet observed in real-world conditions. For example, animals often have profound changes in diet based on availability of nutrients. In particular, omnivores can dramatically change diet from foraging meals comprising grains, fruits, and vegetables to a diet that is high in fat and protein. Humans that intentionally shift from a balanced diet to one that is high in fat (Keto and Atkins diets) exhibit profound metabolic changes.40 Our study suggests that the intestine is one of the first organs to immediately respond to changes in diet.
Coupled with dietary changes, most likely there are other variables accounting for adaptation within the luminal environmental, such as the microbiome. The gut microbiome increases in prevalence distally culminating at the colon and is known to aid in digestion of short chain fatty acids.41 Moreover, it has been previously shown that an HFD not only changes the microbiome but also can immediately affect microbial diversity within days of HFD exposure.42–44 It is therefore likely that changes in the microbiome contribute to our observed changes in the intestinal epithelium.
Our study also suggests that stem and progenitor cells in the crypt experience cellular stress responses during a change to HFD. Previously, it has been shown that induction of endoplasmic reticulum (ER) stress elicits a UPR and is associated with a loss of stem cell activity and an expansion of TA cells.45 Moreover, ER stress and UPR are highly associated with intestinal diseases including inflammatory bowel disease (IBD) and colorectal cancer (CRC).46 Since we observed that HFD triggers a transcriptional signature of stress and inflammation, it is possible that a repeated stress of being exposed to periods of HFD, followed by return to a more balanced diet, could mirror the effects of periodic inflammation and exacerbate gastrointestinal diseases. Alternatively, the intestine may be fully capable of rapidly adapting to dietary changes with no long-term effect. In the future, it will be interesting to map the stability of diet-induced changes in the intestine. For example, following a return from an HFD to a balanced diet, there may be some metabolic or transcriptional parameters that rapidly return to normal, whereas other HFD-induced changes may be more stable.
Previous studies have described changes in ISC function in response to long-term dietary changes.8,15–17 Here, we investigated the immediate response of the intestinal epithelium to a change in diet at the single-cell level. We identified a rapid change in transcriptional signature of intestinal lineages. Not only did an HFD stimulate intestinal proliferation within 24 h, but also we observed a transcriptional shift suggesting that SP cells were acquiring an enterocyte-like signature. Using quantitative immunofluorescent (IF) staining, we observed a similar trend with reduced secretory cells. However, this shift was not statistically significant, suggesting that while transcriptional plasticity is at play, lineage allocation remains more stable. It is possible that scRNA-seq is more sensitive than IF in detecting changes in lineages allocation since our analysis did not detect co-expression of secretory markers in enterocytes. There are now many reports describing an increase in transcriptional plasticity of epithelial cell lineages in response to environmental stressors or injury.47–52 Furthermore, another report using single-cell transcriptomic analysis of mice maintained on a high-sugar/high-fat diet for 12 weeks also observed dietary-induced changes in ISCs and progenitor hyperproliferation.8 Our data suggest that this adaptation may happen within the first week.
To computationally examine how an HFD impacted cell fate plasticity, we performed pseudotime analysis using two approaches: STREAM36 and PAGA.37 The trajectory analysis using the whole epithelial dataset suggested that in response to HFD, there was an emergence of more stem and progenitors coupled with new trajectories for both secretory and enterocyte lineages. One observation was that a population of SPs became more transcriptionally similar to enterocytes (Figure 5C). Given that secretory cells are relatively rare, we enriched for secretory cells using lineage tracing and cell sorting. Analysis of secretory-enriched cell populations showed a new population of progenitors, one that connected SPs to the enterocyte lineage (boxed cell population in Figure 5D). Those data, along with the trajectory analyses, form the basis for the model in Figure 5H suggesting that SPs may begin to express enterocyte genes in response to an HFD. However, as discussed above, identification of cell populations based on transcriptional changes was not reflected in an equivalent change in lineage allocation as measured by IF. Therefore, identification of better markers for secretory and enterocyte progenitors as well as cell lineage-tracing experiments would be needed to show that diet can cause functional changes in cell lineage allocation.
In summary, we took a multi-pronged approach to identify how animals respond to a substantial change in diet, including whole-body metabolism and tissue functional and morphologic changes, as well as single-cell transcriptomics. These transcriptional and functional analyses revealed immediate changes in all intestinal epithelial cell types occurring within 24 h and resulted in a modified intestinal epithelium adapted to maximize absorption of fat within 1 week. This type of intestinal plasticity may be an evolutionary adaptation to periods of nutrient scarcity but, in current times of nutrient excess, could be contributing to rising rates of obesity, metabolic disease, and inflammation.
Limitations of the study
Although we identified many transcriptional changes in the intestine in response to HFD and how they accompany cell morphologic and functional changes, these remain correlative. Moreover, the normal chow diet utilized was not fully controlled for other micronutrients. Lastly, future studies could investigate how stable these adaptations are over time and if repeated HFD challenges cause more permanent changes in cell behaviors.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests may be directed to the lead contact, James M. Wells (james.wells@cchmc.org).
Materials availability
This study did not generate any new unique reagents.
Data and code availability
The single cell RNA sequencing data in this publication have been deposited in the Gene Expression Omnibus (GEO): GSE199776.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal procedures were approved by the Cincinnati Children’s Hospital Research Foundation Institutional Animal Care and Use Committee (IACUC2022–0002) and performed using standard procedures. C57BL6/J and Neurog3Cre x Rosa26tdTom mice under BL6/C57 background were maintained on a 14L: 10D light cycle and had ad libitum access to chow and water. Normal Chow diet (NCD) administered (5010, LabDiet). For the acute high fat diet conditions, mice were given similar food weight of high fat diet (HFD, D12492, Research Diets, Inc.) in the early mornings 1 day, 3 days, and 7 days before time of experimental collection. Mouse weight and diet weight were weighed every morning. All mice were aged 9–14 weeks and both males and females were used in the study.
METHOD DETAILS
Indirect calorimetry
9–14-week-old male C57BL6/J mice were acclimated in metabolic chambers (Promethion, Sable Systems International) for 2 days prior to the start of the study. Mice were continuously recorded for a total of 9 days at ambient room temperature (22°C) with the following measurements taken every 5 minutes: gas exchange (VO2 and VCO2), food intake, water intake, and spontaneous locomotor activity (cm s−1) in the XY plane. All animals were initially fed standard chow until day 2 of the study, when one group was switched to a 60 kcal% high-fat diet (HFD, D12492, Research Diets, Inc.), and one group remained on a standard normal-chow diet (NCD). Otherwise, food and water were available ad libitum. All mice were subjected to a 14L:10D light cycle for the duration of the acclimatization and study period. VO2, VCO2, and energy expenditure (EE) were calculated according to the manufacturer’s guidelines (Metascreen software v.2.3.15.11, Sable Systems International), with energy expenditure estimated via the abbreviated Weir formula. Respiratory exchange ratio (RER) was calculated by the ratio VCO2/VO2. Mass dependent variables (VO2, VCO2, energy expenditure) were not normalized to body weight. Food and water intake were measured by top-fixed load cell sensors, from which food and water containers were suspended into the sealed cage environment. For food consumption, mice demonstrating excessive food grinding behavior were excluded from statistical analyses. Raw data were exported using Sable Systems International ExpeData software v.1.9.27. Published data represent a combination of two independent experiments conducted separately. p values are from one-way repeated measures ANOVA comparing HFD and NCD groups before and after the introduction of the high-fat diet on study day 2.
Epithelial single cell dissociation and FACS sorting
Approximately 5–7 cms of duodenal intestine was dissected and then subsequently after the duodenal-jejunal flexure another 5–7 cms of proximal jejunal intestine was dissected for all dietary conditions. Each piece was filleted open to expose crypts and villi and rinsed with PBS before digested into single cells with 5 mLs of 1x TrypLE (Gibco) and 5 uls of 10 μM Y-27632 dihydrochloride (Tocris). Single cells were filtered through a 40-um cell strainer and then recorded using LSR Fortessa flow cytometer and analyzed with FACSDiva software. In all experiments, samples were labeled with Anti-EpCam-APC (BD Biosciences) to delineate epithelial cells and stained with SYTOX Blue dead cell stain (Life Technologies). The forward and side scatter plots were used to discriminate doubles and cellular debris. The secretory enriched datasets were additionally sorted for TdTom+ fluorescence.
Single-cell RNA sequencing pipeline and analysis
Single cell RNA-Sequencing (scRNA-seq) library preparation was performed by the CCHMC Gene Expression Core using the Chromium 3′v3 GEM Kit (10x Genomics, CG000183RevC). Approximately 12,800 cells were loaded to achieve 8,000 captured cells per sample to be sequenced. Sequencing was performed by the CCHMC DNA Sequencing core using the NovaSeq 6000 (Illumina) sequencing platform with an S4 flow cell to obtain approximately 320 million reads per sample. Raw scRNA-seq data was converted to FASTQ files and then aligned to the mouse genome [mm10] using default parameters of CellRanger (v3.0.2, 10x Genomics). Reads were aligned to mouse genome [mm10]. Quality control and clustering were performed using Seurat21 [v3.2.3] in R. Basic filtering parameters included cells with unique features of minimum 100 and maximum 7500. Cells expressing less than 50 percent mitochondrial related genes were included. Cell cycle effect was regressed out using previously established methods in Seurat. After initial filtering, each individual dataset was run through DoubletDecon53 using default parameters. Rhop values for each sample are as follows: Epithelial Datasets-Normal Chow: Duo- 1 & Jej- 1.2; 1 Day HFD: Duo- 1.1 & Jej- 1.2; 3 Days HFD: Duo- 1.2 & Jej 1.1; 7 Days HFD: Duo- 1.2 & Jej 1.1; Secretory Enriched Datasets- Normal Chow: Duo- 1.2 & Jej- 1.2; 1 Day HFD: Duo- 1.2 & Jej- 1.2; 3 Days HFD: Duo- 1.2 & Jej- 1.1; 7 Days HFD: Duo- 1.2 & Jej- 1.1. After identification of doublets and removal from each dataset, the newly filtered datasets were subsequently used for all downstream analysis. The Epithelial dataset samples were integrated using standard SCTransform Seurat protocols with Normal Chow Condition as Reference Dataset using 2000 integration features. Likewise, the Secretory Enriched Datasets were also integrated using standard SCTransform Seurat protocols with Normal Chow Condition as Reference Dataset using 2000 integration features. After clustering, cells were visualized using UMAP63 and color customized using ggplot2.59 Marker genes were determined using ‘FindAllMarkers’ function (Wilcoxon rank-sum test) in Seurat. The initial Integrated Epithelial dataset showed some contaminating mesenchymal cells that were not removed during the sort. These cells were distinct from the other clusters and were subsequently removed from the final analysis.
To perform Gene Ontology- Term analysis based on biological processes, we first used differentially expressed genes from Seurat ‘FindAllMarkers’ function and used org.Mm.eg.db58 to retrieve gene names associated with gene identifiers. Next, we used cluster-Profiler,57 enrichplot,61 and ReactomePA62 to analyze and visualize data that was sorted using dplyr60 package.
To perform analysis in comparison to Haber et al.32 related to Figure S4, first we downloaded the Regional_UMIcounts.txt file from GSE92332. We then performed the standard SCTransform Seurat pipeline and subsequently filtered out only the Jejunum. We then performed SCTransform Seurat Integration between Normal Chow jejunal Epithelial, Normal Chow jejunal Secretory Enriched, and the Haber dataset with the Haber dataset as the Reference Dataset using 2000 integration features. Subsequent clustering and marker gene expression was performed as previously described.
To perform GSEA analysis, we used the msigdbr59 package (which pulls from MSigDB Collections) to determine lists of genes from the Hallmark Gene Sets (H: Fatty Acid Metabolism, Unfolded Protein Response, and Inflammatory Response). We then used presto56 to determine differentially expressed genes between conditions and then performed gene set enrichment analysis using fgsea.55
To determine Glutamate/Glutamine Metabolism score, the integrated Epithelial dataset was used with the default setting for AddModuleScore() function of Seurat. We pulled genes from MSigDB Curated Gene Sets (C2: Reactome- Glutamate/Glutamine Metabolism). We then plotted results using RidgePlot() function of Seurat.
To determine Cell Stress Signature Score, the integrated Epithelial dataset was used with the default settings for AddModuleScore() function of Seurat. The following genes were used: Hspa1a, Hspa1b, Hsp90aa1, Dnaja1, Hsf1, Xbp1, Atf4, Atf6, Ern1, Hspa5, Eif2ak3, Bcl2.
To determine Lipid Absorption Signature Score, the integrated Epithelial dataset was used with the default settings for AddModuleScore() function of Seurat. The following genes were used: Mgat4a, Dgat1, Dgat2, Npc1l1, Abcg5, Abcg8, Apoa1, Apoa4, Mttp, Slc27a4, Apob, Apoc3, Fabp6, Fabp2, Plin3, Sar1b, Cd36.
To visualize cell density plots, both the integrated Epithelial dataset and the Secretory Enriched dataset was migrated from Seurat to Scanpy.54 Once data was formatted correctly in Scanpy, default settings for sc.tl.embedding_density were used to calculate Gaussian kernel density for all cells. Plots were generated by using sc.pl.embedding density grouped by condition.
Pseudotime analysis was performed with default parameters of STREAM63 or utilization of PAGA37 from Scanpy. With STREAM, visualization of river plots was performed using st.plot_stream as log normalized gene expression. With PAGA, visualization of plots was performed using sc.tl.paga and sc.pl.paga colored by clusters that were initially identified via Seurat.
RNA isolation and qRT-PCR
Mouse proximal intestines were harvested and enriched for epithelial cells as described above and flash frozen. Total RNA was isolated using the NucleoSpin RNA kit (Macherey-Nagel) and reverse transcribed to cDNA using the SuperScript VILO cDNA synthesis kit (Invitrogen). For qRT-PCR, we used PowerUp SYBR Green Master mix (Applied Biosystems) and ran the reaction on a QuantStudio 6 machine (ThermoFisher Scientific). List of primers are included in qPCR primer list supplemental excel sheet.
Tissue harvest and immunofluorescence imaging
Mouse proximal intestines were harvested and prepared for ‘swiss-roll technique’ to be fixed in 4% paraformaldehyde (PFA) at 4C overnight. The fixed samples were washed with PBS for 10 mins three times. The intestines were then immersed in 30% sucrose/PBS for 48 hrs. Afterwards, tissue sections were prepped by embedding each region in OCT and cryosectioned (8um) onto Superfrost Plus slides. Immunofluorescence staining was performed by treating PBS re-hydrated tissue slides with 0.1% Triton X-100/PBS and then incubated with 5% normal donkey serum. Tissues were incubated overnight at 4C with the following primary antibodies: ECAD (rat; R&D 1:1000), Ace-2 (gt, 1:500), Lysozyme (Dako, rab, 1:200), Chga (ImmunoStar, rab, 1:200), Olfm4 (CellSignaling, rb, 1:200), Ckrt18 (Abcam, biotin, 1:200), Dclk1 (Novus Biologicals, rab,1:200). The next day, slides were washed with PBS for 10 mins three times, and then incubated with fluorescently conjugated secondary antibodies for the corresponding animal. The Nikon A1 confocal with DUG detector on TiE microscope was used for imaging.
Crypt depth/villus height measurements
Processed and imaged mouse proximal intestine was measured using NIS Elements by two independent viewers with blinded samples. Per mouse, between 70 and 150 crypts and villi were counted. Crypt depth was measured from the base of the crypt to the top of the transit-amplifying zone. Villus height was measured from the top of the transit-amplifying zone to the tip of a villus. Average values for each mouse were then plotted for each condition.
EdU- cell proliferation
On the day of collection after nutrient challenge, 9–14-week-old male and female mice were intraperitoneally injected with 100 mg/kg of 5-Ethynyl-2′-deoxyuridine (EdU). After waiting for 2 hours, intestines were collected for ‘swiss-roll technique’ as described in previous section. The standard protocol for the Click-iT™ EdU Cell Proliferation Kit (647) was then followed and subsequently imaged.
Alcian Blue staining
Stock solution was made by mixing 1-gram Alcian Blue, 8GX in 3% Acetic Acid and adjusted to pH 2.5. Then specimens, that were similarly prepared for immunofluorescence imaging on glass slides, were rehydrated in H2O for 5 mins, washed in 3% acetic acid for 3 mins, stained with Alcian Blue solution for 30 mins, and finally rinsed with running H2O. Then a counter stain with Nuclear Fast Red was used and each slide was then cover-slipped with Fluoromount-G. Imaging was performed with the Andor Zyla 4.2 plus camera with Lumencor Lida RGB transmitted source. To perform quantification, area of intestinal epithelium was outlined, and Alcian Blue positive labeled cells were manually counted within each area. Proportion of cells within area epithelium is reported.
Oil-Red-O staining
Oil-Red-O Stock (ScienCell, 0843a) was diluted 3:2 using deionized H2O to make a working solution. The working solution was then filtered with 0.22 um syringe driven filter unit (Millipore). Specimens, similarly prepared for immunofluorescence imaging on glass slides, were rehydrated in H2O for 5 mins, subsequently washed in 60% ethanol, and then exposed to filtered working Oil-Red-O solution for 20 minutes. After another 60% ethanol rinse, slides were rehydrated with H2O and then cover-slipped with Fluoromount-G. Imaging was performed with the Andor Zyla 4.2 plus camera with Lumencor Lida RGB transmitted source. To perform quantification, slides were processed the same but were not rehydrated; rather eluted with 100% ethanol for 1 hour. Then eluted dye from slides were transferred onto a 96-well imaging plate and absorption was measured in triplicate at 510 nm using a plate reader.
Transmission electron microscopy (TEM)
Proximal jejunum was dissected and fixed in 3% glutaraldehyde and 0.175 M sodium cacodylate buffer, pH 7.4, at 4°C for one hour. The samples were then post fixed in 1% osmium tetroxide in 0.2 M sodium cacodylate buffer for 1 hour at 4°C, processed through a graded series of alcohols, infiltrated, and embedded in LX-112 resin. After polymerization at 60°C for three days, ultrathin sections (100 nm) were cut using a Leica EM UC7 microtome and counterstained in 2% aqueous uranyl acetate and Reynolds’s lead citrate. Images were taken with a transmission electron microscope (TEM, Hitachi H-6750) equipped with a digital camera (AMT 2k×2K tem CCD).
BODIPY gavage
Mice 9–14 weeks were placed on dietary challenge (Normal Chow or 1, 3, 7 Days HFD). On the final day of diet, mice were then gavaged with BODIPY (50 ug/mg) and swiss-rolled intestines were flash-frozen harvested 2 hours later. Sections were fixed with 4% PFA for 5 minutes and then stained with DAPI for 10 min. Slides were imaged using the Nikon Ti2 with Andor Zyla 4.2 plus camera and Lumencor SpectraX illuminator.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all confocal quantification, such as EdU and BODIPY analysis, each dot represents an individual ‘‘N’’ number of mice shown as reported in legend. For Indirect Calorimetry measurements, p values are from one-way repeated measures ANOVA comparing HFD and NCD groups before and after the introduction of the high-fat diet on study day 2. For mouse weight changes, daily energy intake, and food intake, p values are from two-way ANOVA Sidak’s multiple comparisons test. For comparison between conditions of module gene scores, p values are from one-way repeated measures ANOVA Tukey’s multiple comparisons test. All error bars are reported as Standard Deviation (SD). Statistical analysis was performed in Prism 9 [v9.3.1].
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rat monoclonal Ecadherin (Cdh11) | R&D Systems | Cat# MAB7481; RRID: AB_2076679 |
| Goat polyclonal Ace-2 | R&D Systems | Cat# AF3437 RRID: AB_223140 |
| Rabbit polyclonal Lysozyme | Dako | Cat# A0099 RRID: AB_2341230 |
| Rabbit polyclonal SP-1 Chromogranin A (Bovine) | Immunostar | Cat# 20085 RRID: AB_572227 |
| Rabbit monoclonal Olfm4 | CellSignaling Technology | Cat# 39141 RRID: AB_2650511 |
| Biotin Anti-Cytokeratin 18 | Abcam | Cat# ab27553 RRID: AB_471034 |
| Rabbit polyclonal Dclk1 | Novus Biologicals | Cat# NBP1–77127 RRID: AB_873537 |
| Rabbit Cleaved Caspase-3 (Asp175) | Cell Signaling Technology | Cat# 9611 RRID: AB_234118 |
| APC Rat Anti-Mouse (EpCAM) CD326 | BD Biosciences | Cat# 563478 RRID: AB_2738234 |
| SYTOX Blue Dead Cell Stain | Invitrogen | Cat# S34857 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Donkey anti-Rabbit Alexa Fluor 647 | Invitrogen | Cat# A31573 RRID: AB_2536183 |
| Donkey anti-Rat Alexa Fluor 488 | Invitrogen | Cat# A21208 RRID: AB_141709 |
| Donkey anti-Goat Alexa Fluor 488 | Invitrogen | Cat# A11055 RRID: AB_2534102 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| TrypLE Select Enzyme (1x), no phenol red | Gibco | Cat# 12563011 |
| Y-27632 dihydrochloride | Tocris | Cat# 1254 |
| 5-Ethynyl-2′-deoxyuridine,95% | Sigma-Aldrich | Cat# 900584–50MG |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Cat# A25777 |
| BODIPY | Invitrogen | Cat# D3922 |
| Oil-Red-O | ScienCell | Cat# 0843a |
|
| ||
| Critical commercial assays | ||
|
| ||
| Click-iT™ EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 647 dye | Invitrogen | Cat# C10340 |
| NucleoSpin RNA | Macherey-Nagel | Cat# 740955.250 |
| SuperScript VILO cDNA Synthesis Kit | Invitrogen | Cat# 11754250 |
| Click-iT™ Plus TUNEL Assay Kits for In Situ Apoptosis Detection | Invitrogen | Cat# C10619 |
| Alcian Blue Stain Kit | Vector Laboratories | Cat# H-3501 |
| Rat/Mouse GIP (total) ELISA | Sigma-Aldrich | Cat# EZRMGIP-55K |
| Cholecystokinin (CCK) EIA Kit | Sigma-Aldrich | Cat# RAB0039–1KT |
|
| ||
| Deposited data | ||
|
| ||
| Raw and Analyzed C57/Bl6 Wild Type-Intestine scRNA-seq Normal, 1 Day HFD, 3 Days HFD, 7 Days HFD | This paper | GEO: GSE199776 |
| Raw and Analyzed Neurog3Cre-tdTom-Intestine scRNA-seq Normal, 1 Day HFD, 3 Days HFD, 7 Days HFD | This Paper | GEO: GSE199776 |
| Single Cell Comparison related to Figure S4 | Haber et al.32 | GEO: GSE92332 |
| Enterocyte Marker genes related to Figure S3 | Moor et al.27 | GEO: GSE109413 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | Jackson Laboratories | Strain #000664 |
| Mouse: Tg(Neurog3-cre)C1Able | Jackson Laboratories | Strain #005667 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | Jackson Laboratories | Strain #007909 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primer: Hspa1a Forward: TGG TGC AGT CCG ACA TGA AG Reverse: GCT GAG AGT CGT TGA AGT AGG C |
This paper | N/A |
| Primer: Hspa1b Forward: GAG ATC GAC TCT CTG TTC GAG G Reverse: GCC CGT TGA AGA AGT CCT G |
This paper | N/A |
| Primer: Hspa1b Forward: GAG ATC GAC TCT CTG TTC GAG G Reverse: GCC CGT TGA AGA AGT CCT G |
This paper | N/A |
| Primer: Hsp90aa1 Forward: AAT TGC CCA GTT AAT GTC CTT GA Reverse: CGT CCG ATG AAT TGG AGA TGA |
This paper | N/A |
| Primer: Fabp1 Forward: ATG AAC TTC TCC GGC AAG TAC C Reverse: CTG ACA CCC CCT TGA TGT CC |
This paper | N/A |
| Primer: Apoa4 Forward: ATG CCA AGG AGG CTG TAG AA Reverse: CAG TTT CCT GGG CTA GAT GC |
This paper | N/A |
| Primer: Cpt1a Forward: CTG ATG ACG GCT ATG GTG TTT Reverse: GTG AGG CCA AAC AAG GTG ATA |
This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| NIS Elements | Nikon | N/A |
| R | R Core Team (2020) | v3.6.3, http://www.R-project.org/ |
| R Studio | RStudioTeam (2020) | v1.2.5033, https://www.rstudio.com/products/rstudio/download/ |
| CellRanger | 10x Genomics | v3.0.2, https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| DoubletDecon | DePasquale et al.53 | v1.1.6, https://github.com/EDePasquale/DoubletDecon |
| Seurat | Butler et al.21 | v3.2.3, https://satijalab.org/seurat/ |
| STREAM | Chen et al.36 | v0.4.1, https://github.com/pinellolab/STREAM |
| python | Python Software Foundation | v3.8.6, https://www.python.org/downloads/ |
| Scanpy | Wolf et al.54 | v1.6.1, https://scanpy.readthedocs.io/en/stable/ |
| fgsea | Korotkevich et al.55 | v1.20.0, https://github.com/ctlab/fgsea |
| presto | Korsunsky et al.56 | v1.0.0, https://github.com/immunogenomics/presto |
| clusterProfiler | Yu et al.57 | v4.2.0, https://github.com/YuLab-SMU/clusterProfiler |
| org.Mm.eg.db | Carlson58 | v3.14.0, https://bioconductor.org/packages/release/data/annotation/html/org.Mm.eg.db.html |
| ggplot2 | Wickham59 | v3.3.5, https://ggplot2.tidyverse.org |
| msigdbr | Dolgalev58 | v7.4.1, https://cran.r-project.org/web/packages/msigdbr/vignettes/msigdbr-intro.html |
| dplyr | Wickham et al.60 | v1.0.7, https://dplyr.tidyverse.org |
| enrichplot | Yu et al.61 | v1.14.1, https://www.bioconductor.org/packages/release/bioc/html/enrichplot.html |
| ReactomePA | Yu and He62 | v1.38.0, https://bioconductor.org/packages/release/bioc/html/ReactomePA.html |
| Prism 9 GraphPad | www.graphpad.com | v9.3.1 |
| Metascreen | Sable Systems | v2.3.15.11 |
| ExpeData | Sable Systems | v1.9.27 |
Highlights.
High-fat diet (HFD) changes whole-body metabolism within 24 h
Within the intestinal epithelium, 1 day of HFD causes a stress response in cells of the crypt
By 3 days of HFD, cells computationally shift lineage allocation toward absorptive transcripts
By 7 days of HFD, enterocytes functionally adapt to robust lipid absorption
ACKNOWLEDGMENTS
We thank Dr. Kelli VanDussen, Dan Schnell, Jay Stone, and Praneet Chaturvedi for valuable guidance and discussion. We also thank support provided by the Confocal Imaging Core, Research Flow Cytometry Core, and Gene Expression Core at CCHMC. We thank the members of the Wells and Zorn lab for reagents and feedback. All figures were created with Adobe Illustrator or with BioRender.com. This research was supported by the grants from the NIH, U19 AI116491 (J.M.W.), P01 HD093363 (J.M.W.), and UH3 DK119982 (J.M.W.), and the Allen Foundation (J.M.W.). We also received support from the Digestive Disease Research Center (P30 DK078392).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111641.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Botchlett R, and Wu C (2018). Diet composition for the management of obesity and obesity-related disorders. J. Diabetes Mellit. Metab. Syndr 3, 10–25. 10.28967/JDMMS.2018.01.18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hales CM, Carroll MD, Fryar CD, and Ogden CL (2017). Prevalence of obesity and severe obesity among adults: United States, 2017–2018 key findings data from the national health and nutrition examination survey. NCHS Data Brief, 1–8. https://www.cdc.gov/nchs/products/index.htm. (Accessed 3 December 2021).
- 3.Moore JX, Chaudhary N, and Akinyemiju T (2017). Metabolic syndrome prevalence by race/ethnicity and sex in the United States, national health and nutrition examination survey, 1988–2012. Prev. Chronic Dis 14, E24. 10.5888/PCD14.160287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mana MD, Hussey AM, Tzouanas CN, Imada S, Barrera Millan Y, Bahceci D, Saiz DR, Webb AT, Lewis CA, Carmeliet P, et al. (2021). High-fat diet-activated fatty acid oxidation mediates intestinal stemness and tumorigenicity. Cell Rep 35, 109212. 10.1016/J.CELREP.2021.109212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrich DE, Ou Y, Melbouci L, Leduc-Gaudet JP, Auclair N, Mercier J, Secco B, Tomaz LM, Gouspillou G, Danialou G, et al. (2018). Altered lipid metabolism impairs skeletal muscle force in young rats submitted to a short-term high-fat diet. Front. Physiol 9, 1327. 10.3389/FPHYS.2018.01327/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Do GM, Oh HY, Kwon EY, Cho Y.y., Shin S.k., Park HJ, Jeon SM, Kim E, Hur CG, Park TS, et al. (2011). Long-term adaptation of global transcription and metabolism in the liver of high-fat diet-fed C57BL/6J mice. Mol. Nutr. Food Res 55, S173–S185. 10.1002/MNFR.201100064. [DOI] [PubMed] [Google Scholar]
- 7.Lundsgaard AM, Holm JB, Sjøberg KA, et al. (2019). Mechanisms preserving insulin action during high dietary fat intake. Cell Metab 29, 50–63.e4. 10.1016/J.CMET.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Aliluev A, Tritschler S, Sterr M, Oppenländer L, Hinterdobler J, Greisle T, Irmler M, Beckers J, Sun N, Walch A, et al. (2021). Diet-induced alteration of intestinal stem cell function underlies obesity and prediabetes in mice. Nat. Metab 3, 1202–1216. 10.1038/s42255-021-00458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuipers EN, Held NM, Panhuis WIH, Modder M, Ruppert PMM, Kersten S, Kooijman S, Guigas B, Houtkooper RH, Rensen PCN, et al. (2019). A single day of high-fat diet feeding induces lipid accumulation and insulin resistance in brown adipose tissue in mice. Am. J. Physiol. Endocrinol. Metab 317, E820–E830. 10.1152/AJPENDO.00123.2019. [DOI] [PubMed] [Google Scholar]
- 10.Clara R, Schumacher M, Ramachandran D, Fedele S, Krieger JP, Langhans W, and Mansouri A (2017). Metabolic adaptation of the small intestine to short- and medium-term high-fat diet exposure. J. Cell. Physiol 232, 167–175. 10.1002/jcp.25402. [DOI] [PubMed] [Google Scholar]
- 11.Edwards LM, Murray AJ, Holloway CJ, Carter EE, Kemp GJ, Codreanu I, Brooker H, Tyler DJ, Robbins PA, and Clarke K (2011). Short-term consumption of a high-fat diet impairs whole-body efficiency and cognitive function in sedentary men. FASEB J 25, 1088–1096. 10.1096/fj.10-171983. [DOI] [PubMed] [Google Scholar]
- 12.Tremblay AJ, Lamarche B, Guay V, Charest A, Lemelin V, and Couture P (2013). Short-term, high-fat diet increases the expression of key intestinal genes involved in lipoprotein metabolism in healthy men. Am. J. Clin. Nutr 98, 32–41. 10.3945/ajcn.113.060251. [DOI] [PubMed] [Google Scholar]
- 13.Wiedemann MSF, Wueest S, Item F, Schoenle EJ, and Konrad D (2013). Adipose tissue inflammation contributes to short-term high-fat diet-induced hepatic insulin resistance. Am. J. Physiol. Endocrinol. Metab 305, 388–395. 10.1152/AJPENDO.00179.2013. [DOI] [PubMed] [Google Scholar]
- 14.Beumer J, and Clevers H (2021). Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol 22, 39–53. 10.1038/s41580-020-0278-0. [DOI] [PubMed] [Google Scholar]
- 15.Yilmaz ÖH, Katajisto P, Lamming DW, Gültekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. (2012). MTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495. 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mihaylova MM, Cheng CW, Cao AQ, Tripathi S, Mana MD, Bauer-Rowe KE, Abu-Remaileh M, Clavain L, Erdemir A, Lewis CA, et al. (2018). Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell 22, 769–778.e4. 10.1016/J.STEM.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, et al. (2016). High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58. 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tschöp MH, Speakman JR, Arch JRS, Auwerx J, Brüning JC, Chan L, Eckel RH, Farese RV Jr., Galgani JE, Hambly C, et al. (2011). A guide to analysis of mouse energy metabolism. Nat. Methods 9, 57–63. 10.1038/NMETH.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speakman JR (2013). Measuring energy metabolism in the mouse – theoretical, practical, and analytical considerations. Front. Physiol 4, 34. 10.3389/FPHYS.2013.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall JE, and Hall ME (2021). Digestion and absorption in the gastrointestinal tract. In Guyton and Hall Textbook of Medical Physiology, 14th ed. (Elsevier; ), pp. 823–832. https://www.clinicalkey.com/#!/content/book/3-s2.0-B9780323597128000667?scrollTo=%23hl0000316. [Google Scholar]
- 21.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeds PJ, and Burrin DG (2001). Glutamine and the bowel. J. Nutr 131, 2505S, discussion.2523S-4S. 10.1093/jn/131.9.2505S. [DOI] [PubMed] [Google Scholar]
- 23.Liu TC, Kern JT, VanDussen KL, Xiong S, Kaiko GE, Wilen CB, Rajala MW, Caruso R, Holtzman MJ, Gao F, et al. (2018). Interaction between smoking and ATG16L1T300A triggers Paneth cell defects in Crohn’s disease. J. Clin. Invest 128, 5110–5122. 10.1172/JCI120453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. (2008). A unique role for autophagy and Atg16L1 in Paneth cells in murine and human intestine. Nature 456, 259–263. 10.1038/NATURE07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoter A, and Naim HY (2019). The functions and therapeutic potential of heat shock proteins in inflammatory bowel disease—an update. Int. J. Mol. Sci 20, E5331. 10.3390/IJMS20215331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko CW, Qu J, Black DD, and Tso P (2020). Regulation of intestinal lipid metabolism: current concepts and relevance to disease. Nat. Rev. Gastroenterol. Hepatol 17, 169–183. 10.1038/s41575-019-0250-7. [DOI] [PubMed] [Google Scholar]
- 27.Moor AE, Harnik Y, Ben-Moshe S, Massasa EE, Rozenberg M, Eilam R, Bahar Halpern K, and Itzkovitz S (2018). Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus Axis. Cell 175, 1156–1167.e15. 10.1016/j.cell.2018.08.063. [DOI] [PubMed] [Google Scholar]
- 28.Wang TY, Liu M, Portincasa P, and Wang DQH (2013). New insights into the molecular mechanism of intestinal fatty acid absorption. Eur. J. Clin. Invest 43, 1203–1223. 10.1111/ECI.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gribble FM, and Reimann F (2016). Enteroendocrine cells: chemosensors in the intestinal epithelium. Annu. Rev. Physiol 78, 277–299. 10.1146/annurev-physiol-021115-105439. [DOI] [PubMed] [Google Scholar]
- 30.Schonhoff SE, Giel-Moloney M, and Leiter AB (2004). Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev. Biol 270, 443–454. 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Gehart H, van Es JH, Hamer K, Beumer J, Kretzschmar K, Dekkers JF, Rios A, and Clevers H (2019). Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell 176, 1158–1173.e16. 10.1016/j.cell.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 32.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCauley HA (2020). Enteroendocrine regulation of nutrient absorption. J. Nutr 150, 10–21. 10.1093/jn/nxz191. [DOI] [PubMed] [Google Scholar]
- 34.Gribble FM, and Reimann F (2019). Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat. Rev. Endocrinol 15, 226–237. 10.1038/s41574-019-0168-8. [DOI] [PubMed] [Google Scholar]
- 35.Meijerink J (2021). The intestinal fatty acid-enteroendocrine interplay, emerging roles for olfactory signaling and Serotonin conjugates. Molecules 26, 1416. 10.3390/MOLECULES26051416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Albergante L, Hsu JY, Lareau CA, Lo Bosco G, Guan J, Zhou S, Gorban AN, Bauer DE, Aryee MJ, et al. (2019). Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat. Commun 10, 1903. 10.1038/s41467-019-09670-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, and Theis FJ (2019). PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 20, 59. 10.1186/S13059-019-1663-X/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beyaz S, Mana MD, and Yilmaz ÖH (2021). High-fat diet activates a PPAR-d program to enhance intestinal stem cell function. Cell Stem Cell 28, 598–599. 10.1016/J.STEM.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almeida-Suhett CP, Scott JM, Graham A, Chen Y, and Deuster PA (2019). Control diet in a high-fat diet study in mice: regular chow and purified low-fat diet have similar effects on phenotypic, metabolic, and behavioral outcomes. Nutr. Neurosci 22, 19–28. 10.1080/1028415X.2017.1349359. [DOI] [PubMed] [Google Scholar]
- 40.Batch JT, Lamsal SP, Adkins M, Sultan S, and Ramirez MN (2020). Advantages and disadvantages of the ketogenic diet: a review article. Cureus 12, e9639. 10.7759/CUREUS.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shreiner AB, Kao JY, and Young VB (2015). The gut microbiome in health and in disease. Curr. Opin. Gastroenterol 31, 69–75. 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, and Gordon JI (2009). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med 1, 6ra14. 10.1126/SCITRANSLMED.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. (2013). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bisanz JE, Upadhyay V, Turnbaugh JA, Ly K, and Turnbaugh PJ (2019). Meta-analysis reveals reproducible gut microbiome alterations in response to a high-fat diet. Cell Host Microbe 26, 265–272.e4. 10.1016/J.CHOM.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heijmans J, Van Lidth de Jeude JF, Koo BK, Rosekrans SL, Wielenga MCB, van de Wetering M, Ferrante M, Lee AS, Onderwater JJM, Paton JC, et al. (2013). ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep 3, 1128–1139. 10.1016/J.CELREP.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 46.Coleman OI, and Haller D (2019). ER stress and the UPR in shaping intestinal tissue homeostasis and immunity. Front. Immunol 10, 2825. 10.3389/FIMMU.2019.02825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A, et al. (2017). Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell 21, 78–90.e6. 10.1016/J.STEM.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt M, Schewe M, Sacchetti A, Feijtel D, van de Geer WS, Teeuwssen M, Sleddens HF, Joosten R, van Royen ME, van de Werken HJG, et al. (2018). Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep 24, 2312–2328.e7. 10.1016/J.CELREP.2018.07.085. [DOI] [PubMed] [Google Scholar]
- 49.Jones JC, Brindley CD, Elder NH, Myers MG Jr., Rajala MW, Dekaney CM, McNamee EN, Frey MR, Shroyer NF, and Dempsey PJ (2019). Cellular plasticity of Defa4Cre-expressing Paneth cells in response to notch activation and intestinal injury. Cell. Mol. Gastroenterol. Hepatol 7, 533–554. 10.1016/J.JCMGH.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murata K, Jadhav U, Madha S, van Es J, Dean J, Cavazza A, Wucherpfennig K, Michor F, Clevers H, and Shivdasani RA (2020). Ascl2-Dependent cell dedifferentiation drives regeneration of ablated intestinal stem cells. Cell Stem Cell 26, 377–390.e6. 10.1016/j.stem.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buczacki SJA, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, and Winton DJ (2013). Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69. 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 52.Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, et al. (2016). Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203–213. 10.1016/J.STEM.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 53.DePasquale EAK, Schnell DJ, Van Camp PJ, Valiente-Alandí Í, Blaxall BC, Grimes HL, Singh H, and Salomonis N (2019). Double-Decon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Rep 29, 1718–1727.e8. 10.1016/J.CELREP.2019.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. 10.1186/S13059-017-1382-0/FIGURES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, and Sergushichev A (2021). Fast gene set enrichment analysis. Preprint at bioRxiv 10.1101/060012. [DOI]
- 56.Korsunsky I, Nathan A, Millard N, and Raychaudhuri S (2019). Presto scales Wilcoxon and auROC analyses to millions of observations. Preprint at bioRxiv 10.1101/653253. [DOI]
- 57.Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS 16, 284–287. 10.1089/OMI.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carlson M org.Mm.eg.db: Genome Wide Annotation for Mouse 2016; R Package Version 3.14.0. https://bioconductor.org/packages/release/data/annotation/html/org.Mm.eg.db.
- 59.Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; New York: )978–3-319–24277-4. [Google Scholar]
- 60.Wickham H, Francois R, Henry L, and Muller K (2021). Dplyr: A Grammar of Data Manipulation R package version 1.0.7. https://dplyr.tidyverse.org.
- 61.Yu G (2021). Enrichplot: Visualization of Functional Enrichment Result R package version 1.14.1. https://yulab-smu.top/biomedical-knowledge-mining-book/.
- 62.Yu G, and He QY (2016). ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst 12, 477–479. 10.1039/C5MB00663E. [DOI] [PubMed] [Google Scholar]
- 63.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single cell RNA sequencing data in this publication have been deposited in the Gene Expression Omnibus (GEO): GSE199776.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


