Abstract
Background
The leopard coral grouper (Plectropomus leopardus) is an important economic species in East Asia-Pacific countries. To meet the market demand, leopard coral grouper is facing overfishing and their population is rapidly declining. With the improvement of the artificial propagation technique, the leopard coral grouper has been successfully cultured by Fisheries Research Institute in Taiwan. However, the skin color of farmed individuals is often lacking bright redness. As such, the market price of farmed individuals is lower than wild-type.
Results
To understand the genetic mechanisms of skin coloration in leopard coral grouper, we compared leopard coral grouper with different skin colors through transcriptome analysis. Six cDNA libraries generated from wild-caught leopard coral grouper with different skin colors were characterized by using the Illumina platform. Reference-guided de novo transcriptome data of leopard coral grouper obtained 24,700 transcripts, and 1,089 differentially expressed genes (DEGs) were found between red and brown skin color individuals. The results showed that nine candidate DEGs (epha2, sema6d, acsl4, slc7a5, hipk1, nol6, timp2, slc25a42, and kdf1) significantly associated with skin color were detected by using comparative transcriptome analysis and quantitative real-time polymerase chain reaction (qRT-PCR).
Conclusions
The findings may provide genetic information for further skin color research, and to boost the market price of farmed leopard coral grouper by selective breeding.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-09091-6.
Keywords: Plectropomus leopardus, Skin color, Transcriptome, DEGs, qRT-PCR
Background
Groupers are one of the most important economic fish species and popular aquaculture species in several Asian countries. About 47 species of grouper are cultivated in East and Southeast Asia [1], such as Epinephelus lanceolatus, E. coioides, E. malabaricus, and Plectropomus leopardus. The leopard coral grouper (P. leopardus) is belongs to the Plectropomus genus of the Serranidae family, which is mainly distributed in the western Pacific Ocean from the south of Japan to Australia, and the east of the Caroline Islands [2]. In nature, leopard coral grouper with different skin colors including red and brown have been found around the sea of Taiwan, Penghu, Okinawa, and the South China Sea [3–5]. In recent years, studies showed that overfishing and the destruction of spawning aggregations significantly declined leopard coral grouper population in the Philippines and Australia [6–8]. Even though the Red Book of the International Union for Conservation of Nature (IUCN) currently lists leopard coral grouper as the least concern [9], it is still essential to establish fishery management and conservation strategies to prevent leopard coral grouper from becoming a species of high-risk level in the future.
Coloration plays an important role in many creatures, which is associated with thermoregulation, social communication, predator avoidance, camouflage, protection from radiation, and selective mating [10, 11]. Previous studies indicated that coloration of teleost fish is determined by six types of pigment cells, including melanophores (black and dark brown), xanthophores (yellow), erythrophores (red and orange), iridophores (reflective), leucophores (white), and cyanophores (blue) [12–14]. Studies also suggested that cAMP (cyclic adenosine monophosphate), MAPK (mitogen-activated protein kinase), PI3k/Akt (phosphatidylinositol 3-kinase-Akt), and Wnt (wingless-type MMTV integration site) signaling pathways are involved in melanin-synthesis related pathways in fish [15–18]. The leopard coral grouper has bright red skin color, which is different from the other species in the same genus. Due to bright red being associated with good luck and happiness in Chinese culture [19], the net price of bright red leopard coral groupers is approximately US$7 per kilogram higher than the brown individuals in the Asian market [5]. As such, for leopard coral grouper, skin color is an important economic trait. Although artificial breeding has been successfully established for leopard coral grouper by Fisheries Research Institute in Taiwan, mostly the skin color of farmed individuals is gray and brown. Several factors, such as habitats, physiological response, food, and genetics, were associated with skin coloration [20–22]. Feed additives and breeding approach have been proposed in order to improve their skin color performance [23, 24]. Previous studies showed that the feed additives of carotenoids contain astaxanthin [4, 25, 26], enhancing the skin color of fish appearing red pigmentation. Besides, gene regulation associated with skin color is also a critical factor. Candidate carotenoid-related genes, including BCO2, LRP11, ANGPTLs, ALAS, PDIL, MED12, SOX, FAX, FATP, SCD, and LDLRA, were involved in carotenoid metabolism and significantly linked to the skin coloration of leopard coral grouper [5, 22, 27, 28]. Several miRNAs including miR-215, miR-2188, miR-194, miR-122, and novel-m0118 may also play a potential role in regulating skin color in the red-colored leopard coral groupers [29].
In 2020, the whole genome sequence of leopard coral grouper has been published [5, 30, 31], and results showed that the length of whole genome sequence is smaller than other grouper species, implying the evolutionary ancient status of leopard coral grouper genome compared to other grouper species [5]. Genetic markers, such as microsatellite loci, were frequently applied to understand the genetic diversity of wild-type leopard coral grouper, which is also useful for breeding program development and fishery management [32–35]. By calculating the heterozygosity ratio of the microsatellite locus of farmed fish, Yang et al. (2020) indicated that the heterozygosity ratio of the leopard coral grouper is 0.42% [5], which is higher than the red-spotted grouper (E. akaara) (0.38%) [36], Murray Cod (Maccullochella peelii) (0.11%) [37], pike-perch (Sander lucioperca) (0.14%) [38], Triplophysa tibetana (0.1%) [39], male grass carp (Ctenopharyngodon idellus) (0.25%) and female grass carp (0.09%) [22], indicating that leopard coral grouper is still a low-level anthropogenic selection species among aquaculture species, and with a high degree of ethnic genetic divergence.
Transcriptome analysis has been widely used to reveal gene expression affecting the skin coloration in non-model organisms, such as common carp (Cyprinus carpio) [17, 40, 41], crucian carp (Carassius auratus) [42], crimson snapper (Lutjanus erythropterus) [43], and catfish-like loach (Triplophysa siluroides) [44]. However, the molecular mechanism of skin coloration is still poorly understood in the Serranidae family. In this study, we characterized the transcriptome sequences of leopard coral grouper with different skin colors (red and brown) collected from Penghu Sea in Taiwan (Fig. 1). We aimed to (i) conduct transcriptome analysis of leopard coral groupers with different skin colors; (ii) validate differentially expressed genes (DEGs) associated with different skin colors of leopard coral groupers from other populations. The purpose of this study was to reveal novel information on candidate DEGs related to skin coloration between red and brown leopard coral groupers. Ultimately, the findings could provide useful genetic information for the artificial breeding of leopard coral grouper with different skin colors.
Fig. 1.
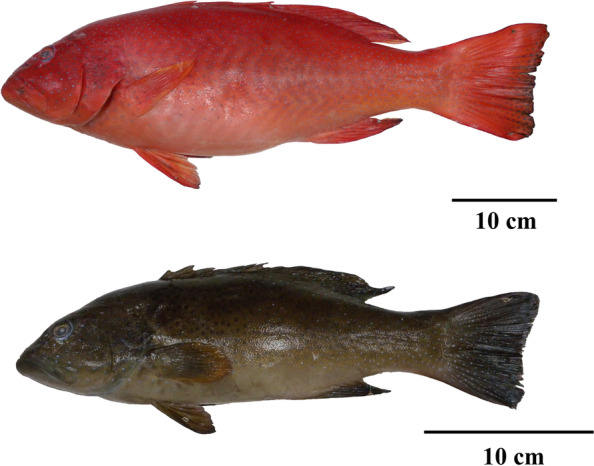
Leopard coral grouper (Plectropumus leopardus). Different skin colors, including red and brown individuals, were involved in this study. The fish were collected from Penghu Sea in Taiwan. Scale bars: 10 cm
Results
Illumina sequencing and genome-guided de novo transcriptome assembly
The transcriptome of six leopard coral groupers, including three red individuals and three brown individuals, were sequenced, respectively (Table 1). The general statistics of the transcriptome sequencing results were presented in Table 1. A total of 47.5 Gb Illumina raw reads were obtained. After the quality control step, 42.6 Gb cleans reads were yielded. The total number of clean reads for six samples were 36,423,012, 40,269,914, 67,288,760, 74,867,838, 39,202,166, and 40,900,758, respectively. The total number of bases for six samples were 5,243,362,538, 5,884,011,219, 9,473,785,507, 10,512,398,825, 5,643,111,214, and 5,888,656,708, respectively. Clean reads with Q20 and Q30 were more than 92%, and the average GC content was 47.5%. The results showed that high-quality reads were used for transcriptome assembly in this study (Table 1).
Table 1.
Summary statistics of sequencing data
| Sample | skin color | Raw reads | Clean reads | Bases (bp) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| R1 | Red | 39,045,732 | 36,423,012 | 5,243,362,538 | 97.12 | 92.33 | 47.62 |
| R2 | Red | 43,187,736 | 40,269,914 | 5,884,011,219 | 96.96 | 92.00 | 46.94 |
| R3 | Red | 68,533,404 | 67,288,760 | 9,473,785,507 | 98.63 | 95.06 | 48.36 |
| B1 | Brown | 76,223,858 | 74,867,838 | 10,512,398,825 | 98.58 | 94.88 | 46.97 |
| B2 | Brown | 42,298,544 | 39,202,166 | 5,643,111,214 | 97.08 | 92.22 | 48.11 |
| B3 | Brown | 43,232,198 | 40,900,758 | 5,888,656,708 | 97.25 | 92.56 | 47.17 |
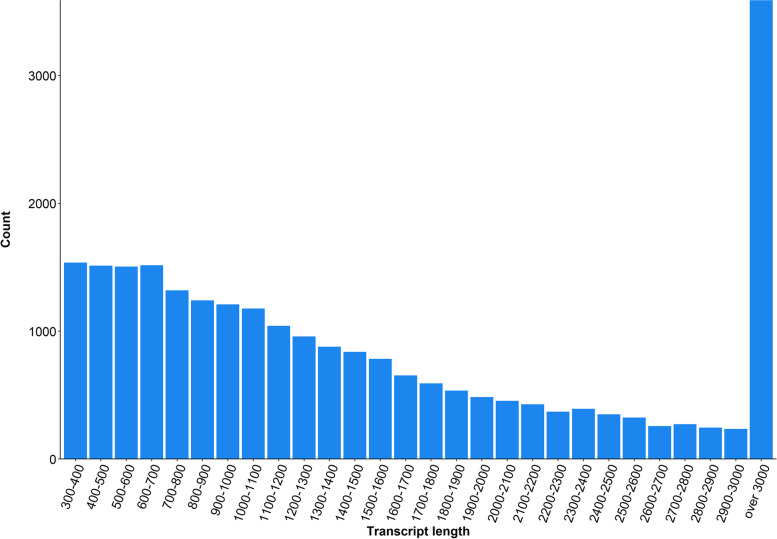
The final contig assembly produced 24,700 transcripts with an N50 length of 2,496 bp, an average contig of 1,776 bp, a medium length of 1,230 bp, and a GC content of 46.7% using CD-HIT-EST. The length distribution of all the transcripts was shown in Fig. 2. The longest and shortest contigs were 65,925 bp and 306 bp, respectively. There were 14,860 transcripts longer than 1,000 bp, accounting for 60% of complete data (Fig. 2).
Fig. 2.
Length distribution of all transcripts
Functional annotation
To annotate these assembled transcripts, which were based on UniProt Knowledgebase (UniProtKB), Cluster of Orthologous Groups database (COG), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genome Orthology (KO), including 14,171 genes, 5,492 genes, 6,411 genes, 8,480 genes accounting for a total of 24,700 genes, were obtained, respectively. 3,049 common genes were annotated by UniProtKB, COG, GO, and KO (Fig. 3). In the COG, the predominant categories were general function prediction only (20%), followed by cell cycle control, cell division, chromosome partitioning (17%), and function unknown (12%) (Additional file 1: Fig. S1). GO was divided into three major function categories: molecular functions (52%), cellular components (8%), and biological processes (40%) (Additional file 1: Fig. S2). KO classification obtained seven categories including 47 pathways. Lipid metabolism (108 genes) and transport and catabolism (128 genes) were the most abundant term in the categories of metabolism and cellular processes (Additional file 1: Fig. S3).
Fig. 3.
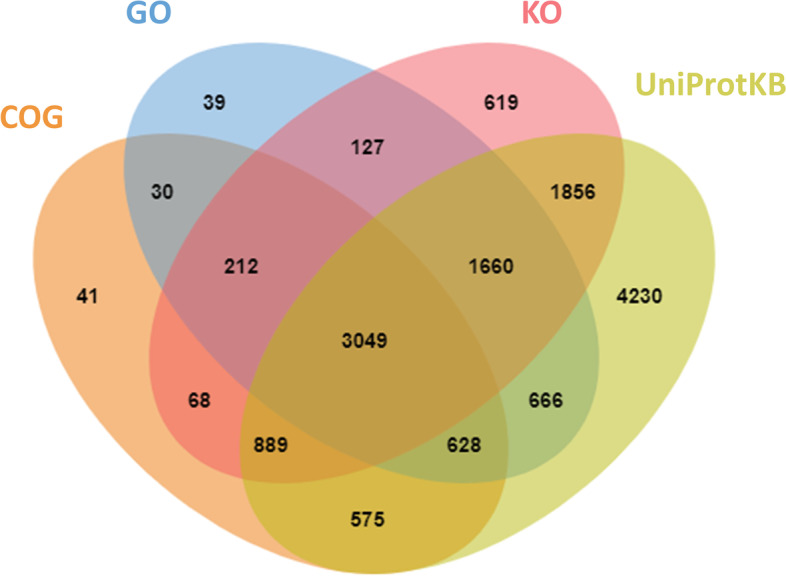
Venn diagram of annotation results based on UniProtKB, GO, COG, and KO databases, respectively. Venn diagram showed common and specific genes from the UniProtKB, GO, COG, and KO databases
Genetic relationship between different skin color fishes
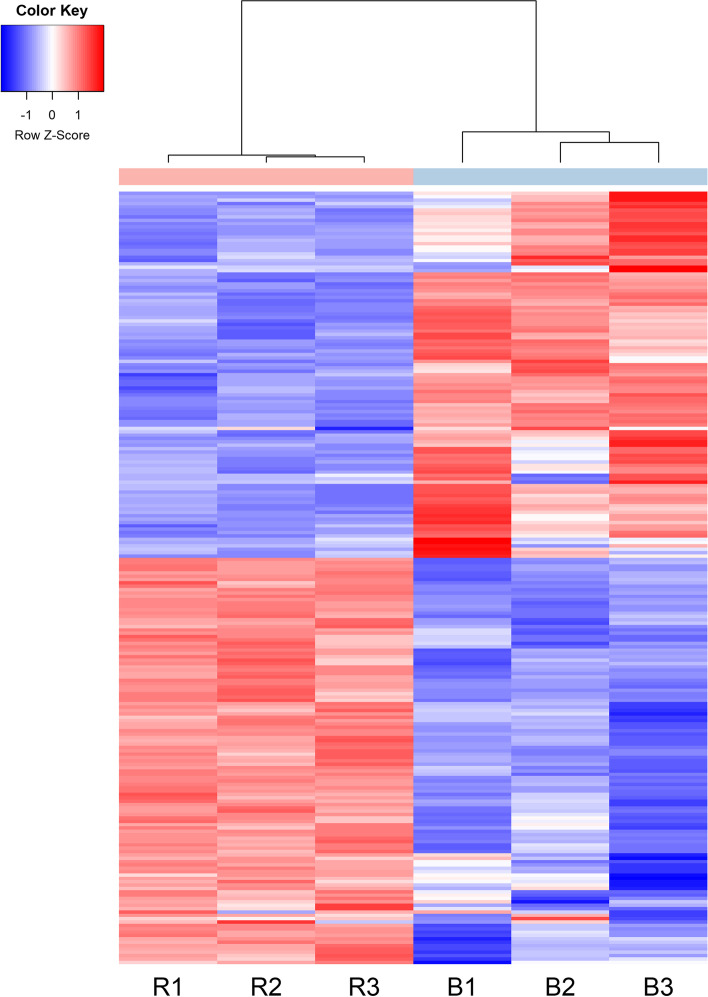
The principal component analysis (PCA) plot showed that the red (R1, R2, and R3) and brown individuals (B1, B2, and B3) were clearly separated with main principal component (PC) scores as follows: PC1 = 34.01% and PC2 = 19.55% (Fig. 4). Heatmaps based on the trimmed mean of M-values (TMM) expression of top 230 DEGs (115 up regulation and 115 down regulation) were generated from six transcriptome sequencing samples. Results showed that the transcripts count of red individuals (R1, R2, and R3) was different from that of the brown individuals (B1, B2, and B3) in Fig. 5 and Additional file 2: Table S1.
Fig. 4.
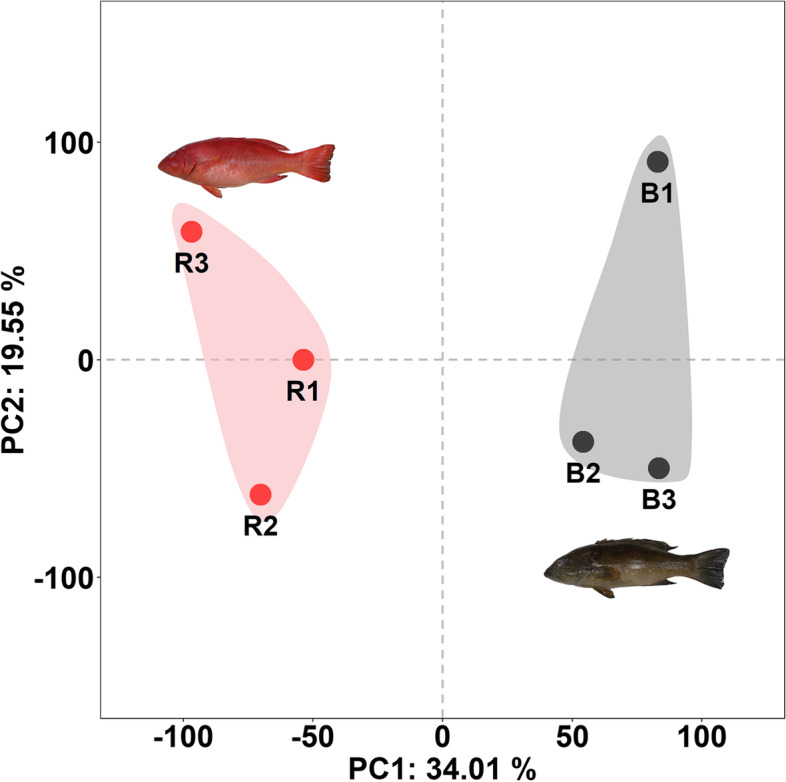
Principal component analysis of TMM expression in fish with different skin colors. Red skin fish: R1, R2, and R3. Brown skin fish: B1, B2, and B3
Fig. 5.
Heatmap analysis of DEGs. Transcriptome analysis showed top 230 DEGs (115 up regulation and 115 down regulation) related to skin color in red (R1, R2, and R3) and brown (B1, B2, and B3) individuals, respectively. The completed name of DEGs were given in Additional file 2: Table S1
Differential expression genes
To analyze the DEGs between the red group (R1, R2, and R3) and the brown group (B1, B2, and B3). There were 1,089 DEGs (613 up-regulated and 476 down-regulated DEGs) obtained based on an absolute value of log2 fold change (FC) ≥ 2 and false discovery rate (FDR) < 0.05 (Additional file 1: Fig. S4). For the GO analysis, up-regulated and down-regulated DEGs were highly enriched in GO terms of protein binding, plasma membrane, nucleus, cytoplasm, and cytosol (Additional file 1: Fig. S5). For the KEGG pathways enrichment analysis, there were significantly enriched signal pathways, including Jak-STAT, cAMP, Rap1, TGF-beta, and PI3K-Akt signaling pathway in up-regulated DEGs. The most significantly enriched KEGG pathways were hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), and cardiac muscle contraction in down-regulated DEGs (Additional file 1: Fig. S6).
Validation of differentially expressed genes by qRT-PCR
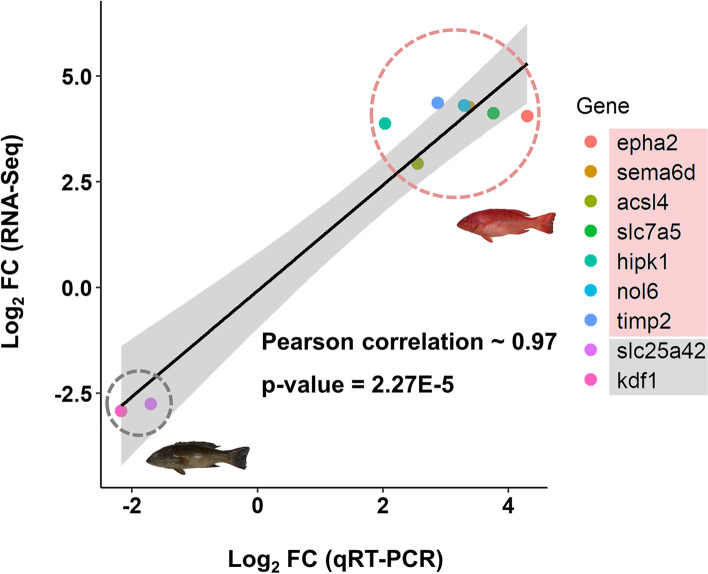
To investigate the DEGs associated with the red and brown skin coloration of leopard coral grouper, nine DEGs (FDR < 0.05 and |log2FC| ≥ 2) may associate with the skin coloration were chosen for qRT-PCR analysis (Table 2). These nine DEGs included eph receptor A2 (epha2), semaphorin 6D (sema6d), acyl-CoA synthetase long-chain family member 4 (acsl4), solute carrier family 7 member 5 (slc7a5), homeodomain interacting protein kinase 1 (hipk1), nucleolar protein 6 (nol6), TIMP metallopeptidase inhibitor 2 (timp2), solute carrier family 25 member 42 (slc25a42), and keratinocyte differentiation factor 1 (kdf1). The primers for qRT-PCR analysis were given in Table 3. The gene expression of epha2, sema6d, acsl4, slc7a5, hipk1, nol6, and timp2 were increased, and the gene expression of slc25a42 and kdf1 were decreased. The log2 FC value of epha2 in red individuals was significantly higher than brown individuals by 17 folds (Fig. 6). There was a high correlation between RNA-Seq and qRT-PCR with Pearson’s correlation of 0.97 (Fig. 7).
Table 2.
Candidate genes involved in skin color between red and brown individuals
| Gene_ID | FPKM (red) |
FPKM (brown) |
FDR | Fold change (log2) |
Annotation | Gene symbol |
|---|---|---|---|---|---|---|
| Up-regulated candidate DEGs associated with skin color in red fish | ||||||
|
maker_0147_ augustus-0.133 |
337.99 | 30.72 | 1.15E-15 | 4.06 | eph receptor A2 | epha2 |
|
maker_0315_ augustus-0.115 |
166.45 | 15.29 | 4.60E-10 | 4.12 | solute carrier family 7 member 5 | slc7a5 |
|
maker_0681_ snap-0.96 |
296.75 | 60.74 | 3.20E-07 | 2.94 | acyl-CoA synthetase long chain family member 4 | acsl4 |
|
maker_0101_ snap-0.156 |
2608.51 | 196.59 | 9.38E-12 | 4.31 | nucleolar protein 6 | nol6 |
|
augustus_0141_ processed-1.34 |
303.58 | 31.55 | 9.05E-12 | 3.88 | homeodomain interacting protein kinase 1 | hipk1 |
|
maker_0248_ augustus-0.220 |
3725.68 | 281.98 | 5.67E-10 | 4.37 | TIMP metallopeptidase inhibitor 2 | timp2 |
|
maker_0156_ augustus-2.210 |
521.19 | 41.71 | 5.80E-13 | 4.27 | semaphorin 6D | sema6d |
| Up-regulated candidate DEGs associated with skin color in brown fish | ||||||
|
maker_0129_ snap-3.241 |
11.56 | 118.54 | 1.85E-08 | -2.76 | solute carrier family 25 member 42 | slc25a42 |
|
augustus_0124_ processed-0.83 |
4.94 | 55.99 | 6.88E-07 | -2.92 | keratinocyte differentiation factor 1 | kdf1 |
Table 3.
List of primers used for qRT-PCR.
| Gene | Primer name | Sequence (5’-3’) | Amplicon size (bp) |
|---|---|---|---|
| β-actin |
β-actin-F β-actin-R |
TACGAGCTGCCTGACGGACA GGCTGTGATCTCCTTCTGCA |
240 bp |
| hipk1 |
hipk1-F hipk1-R |
GGCCAGATTGAGGTGGGTATC AATACCAGGCATGTGTGACCTT |
113 bp |
| epha2 |
epha2-F epha2-R |
GATGGACACGCGTGGATCAAA CTCCAAACGACCTCGCTTCA |
106 bp |
| slc7a5 |
slc7a5-F slc7a5-R |
AAGCCGATCGCACACTCCTT AGCGACCGATCAAGGTGAATA |
109 bp |
| acsl4 |
acsl4-F acsl4-R |
AAAAAGTGCACACAGAAGGCTAC CAATGGGCCGTTCCATATTCTC |
102 bp |
| nol6 |
nol6-F nol6-R |
GCCATTGACTAGGGTGAGATGT CGGCATGTACTATGATGCCCT |
118 bp |
| timp2 |
timp2-F timp2-R |
GTGCAGCGGATGATCTTGCAATC ATCACACTGTGTCACCTCATTCA |
110 bp |
| sema6d |
sema6d-F sema6d-R |
GTCTGGTACTCCTTTACAGGGC CGAGGCATGTCCACAAGGAT |
109 bp |
| slc25a42 |
slc25a42-F slc25a42-R |
ATAAGGGTCATGCCCTACGC GAGGCAGAGTTTTTCCCTGGTA |
100 bp |
| kdf1 |
kdf1-F kdf1-R |
TCATGAGCAGTGACGTTCCA CTCATCTTCCTCGCCAGTTCA |
73 bp |
Fig. 6.
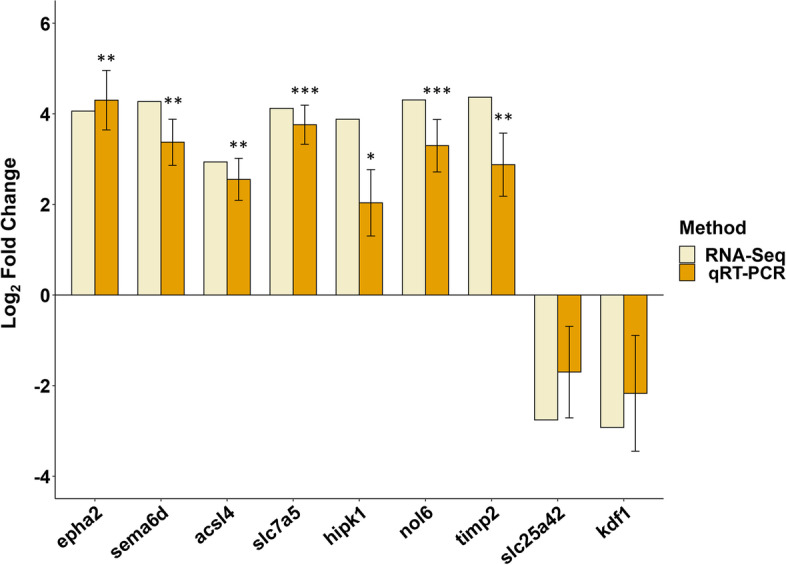
Comparison of gene expression patterns obtained using comparative transcriptome analysis and qRT-PCR. Nine genes were identified as DEGs between brown (control group) and red individuals (experimental group) (n = 4 for each group). Expression of target genes was normalized to β-actin as a reference gene. The Y-axis shows the relative mRNA expression levels (means ± SD). Statistically significant differences compared to control group are presented, with *: P < 0.05; **: P < 0.005; ***: P < 0.001
Fig. 7.
Correlation of gene expression analysis between comparative transcriptome analysis and qRT-PCR. Gene expression of epha2, sema6d, acsl4, slc7a5, hipk1, nol6, and timp2 increased in red individuals, and gene expression of slc25a42 and kdf1 decreased in red individuals. qRT-PCR and RNA-Seq fold change values were highly correlated (Pearson correlation ~ 0.97 and p-value = 2.27E-5)
Discussion
Leopard coral grouper is a valuable marine species in coral reef ecosystems, and an excellent research model for exploring its special skin color variations [5]. Fish skin coloration is a complex trait that is determined by genetic, cellular, physiological, and environmental factors [16]. The skin color in the fish, such as red leopard coral grouper and red tilapia, is the major factor of commercial value determination in Asian countries [45, 46]. Therefore, it is essential to understand the genetic mechanism of color variations in these aquaculture species. To date, several investigations have reported many candidate genes associated with color changes through RNA-Seq in different non-model teleost fishes, such as common carp, red tilapia, and Taiwanese loach [41, 42, 47, 48], as well as in model organisms such as zebrafish (Danio rerio) and medaka (Oryzias latipes) [49, 50]. Fang et al. (2021) and Zhang et al. (2017) indicated that the candidate skin color genes were related to tyrosine and pteridine metabolism, melanogenesis, ion change, apoptosis, and autophagy [42, 45]. Previous studies reported erythrophore/xanthophore synthesis (slc7a11 and slc24a5), try, tryp1, dct, mitfa, and sox10 are well-recognized melanophore-markers in red crucian carp and red tilapia [42, 51]. Many SLC genes regulating the transport of substances on the cell membrane have been involved in skin pigmentation in humans and fish, such as slc45a2, slc24a5, and slc7a11. slc7a11 encodes the cystine/glutamate exchanger xCT that increases pheomelanin synthesis in red skin formation [42, 51, 52]. slc24a5 plays an essential role in regulating melanophore development in several vertebrates, such as zebrafish and common carp, so knockdown of the slc24a5 could block melanin synthesis to generate albino or golden phenotype [53, 54]. slc45a2 participates in the membrane-associated transporter protein (MATP), which is likely involved in intracellular processing and trafficking of melanosomal proteins [55].
In the present study, we performed a comparative transcriptomic analysis between the red and brown skin of leopard coral grouper, and 1,089 significant DEGs were identified. Through comparative transcriptome analysis and qRT-PCR, we found that mRNA expression levels of epha2, sema6d, acsl4, slc7a5, hipk1, nol6, and timp2 were greater in the red individuals compared to brown individuals, and that mRNA expression levels of slc25a42 and kdf1 were lower in the red individuals compared to brown individuals. Two candidate SLC genes (slc7a5 and slc25a42) were identified as significant DEGs for skin color formation in leopard coral groupers. slc7a5 was an up-regulated DEG in red individuals, and slc25a42 was an up-regulated DEG in brown individuals. slc7a5 is an amino acid transporter that regulates the mTOR pathway, enhancing pheomelanin synthesis through activating xCT expression in mice [56]. The SLC25 family member is the largest solute transporter family in humans and transports solutes across the inner membrane of mitochondria [57]. slc25a38 contributed to red color development in spider mites [58], which may imply that slc25a42 was the potential pigmentation-related gene in leopard coral grouper.
epha2 is a tyrosine kinase, which belongs to the family of Eph receptors. It was reported that epha2 inhibits MAPK and AKT pathways in human lens epithelial (HLE) cells [59], so the gene may prevent the MAPK pathway from activating the downstream gene of melanocyte inducing transcription factor (MITF), which is well known to participate in the enzymatic conversion of tyrosine to melanin [42, 59]. acsl4 encodes a protein associated with lipid biosynthesis and fatty acid degradation [60]. Recent studies have indicated that acsl4 plays a critical role in activating ferroptosis that leads to cell death by iron-dependent lipid peroxidation, and also inhibiting melanin synthesis [61]. epha2 and acsl4 were up-regulated DEGs, which may inhibit melanin synthesis in red skin color individuals. In addition, melanin plays an essential role to protect animals from UV radiation and environmental challenges. Keratinocyte-derived factors are involved in regulating the proliferation and melanogenesis in mammals [62, 63]. In our results, kdf1, a kind of keratinocyte-derived factor, had a higher expression in brown skin color individuals.
Taken together, most skin color-associated genes found in the study are involved in melanin synthesis which has been reported in previous studies. The genetic mechanisms of pigment cells are similar between fish and humans, and some skin color-associated genes in fish models, such as slc25a42 and MATP, have contributed to understanding the genetic mechanism in human skin [64].
Conclusions
This study conducted comparative transcriptome analysis in different skin colors of leopard coral groupers, and results showed that red individuals were different from brown individuals. The results of qRT-PCR showed that nine candidate genes were associated with the formation of skin coloration. To conclude, our results provided useful genetic resources for future studies regarding the genetic mechanisms of skin coloration in leopard coral grouper, and also to assist breeders to conduct molecular assisted selection in leopard coral grouper farming.
Methods
Sample collection
Fish were wild-caught from the exclusive economic zone of Penghu Sea in Taiwan. Skin samples were immediately immersed in the RNA keeper reagent (Protech, Taiwan), and further stored at -80℃. Six fish with different skin colors, including three red individuals and three brown individuals, were selected for RNA extraction. The average body weight of fish was 0.7 ± 0.6 kg, and average body length was 38.1 ± 9.3 cm.
RNA isolation
Total RNA was extracted from the fish skin tissue. The skin sample was homogenized in 1 mL of TRIzol reagent (Invitrogen, USA) containing stainless steel beads, following the manufacturer’s protocol. The quality and quantity of total RNA were determined by a DS-11 spectrophotometer (DeNovix, USA), and run on 1% agarose gel electrophoresis. The RNA samples were stored at -80℃ for further experiments.
Library construction and RNA sequencing
RNA extracted from fish skin tissue was prepared for library construction. The RNA integrity was assessed by a Qsep400 (BiOptic, Taiwan) with quality number (RQN) values ≥ 7.0. A paired-end (PE) sequencing library was constructed using Illumina TruSeq RNA Library Prep Kit v2 (Illumina, USA), following the manufacturer’s protocol. The obtained library was sequenced using the Illumina NovaSeq 6000 platform (Illumina, USA) with 2 × 150 bp PE reads.
Transcriptome assembly
To eliminate errors, adapters and low-quality reads from the raw data were removed by fastp [65]. The Phred score (Q20 and Q30) and GC content of the clean data were calculated. Sequence quality metrics were assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) software. Subsequently, all clean reads were aligned to the reference P. leopardus genome [31] using STAR [66]. The coordinate sorted bam file was assembled using Trinity program [67] for reference-guided de novo assembly. While the assembled data contains abundant duplicate transcripts, CD-HIT-EST clustering application [68] was used to remove redundant transcripts with 95% identity, and establish the final assembled transcripts. Potential protein-coding genes were identified from assembled transcripts using Transdecoder (https://github.com/TransDecoder/TransDecoder).
Gene annotation
Transcriptome assembly annotation was performed with Trinotate pipeline (http://trinotate.github.io). The assemble transcripts were annotated using blastp and blastx program (http://www.ncbi.nlm.nih.gov/BLAST/) with E-value cutoff of 1e-10 against UniprotKB. Functional and pathway annotations were carried out via GO, KO (http://www.genome.jp/kegg) [69], and COG (http://www.ncbi.nlm.nih.gov/COG) databases.
Differential expression analysis
Each PE read from the RNA-Seq was mapped with Bowtie2 [70] to the final assembled transcripts, and the quantification of the read count was generated using RSEM [71]. The read counts were normalized by the TMM method implemented in the R package “edgeR” [72], and generated PCA and heatmap. Differential expression analysis was performed using “edgeR”, and p-values were adjusted using the Benjamini-Hochberg (BH) method to control the FDR. Genes with FDR < 0.05 and an absolute value of log2 FC ≥ 2 were considered to be DEGs, and were used in the subsequent analysis. The GO and KEGG enrichment analysis of DEGs were performed using KOBAS [73].
Quantitative real-time polymerase chain reaction (qRT-PCR)
cDNA synthesis was generated from total RNA using SuperScript® IV First-Strand cDNA Synthesis Reaction (Invitrogen, USA), and random hexamer primers (Invitrogen, USA). Diluted cDNA (10 ng/µL) was used as a template for qRT-PCR. β-actin was used as the housekeeping gene for the normalization of gene expression levels. The primers used for target and reference genes were designed based on transcriptome sequence using Primer-BLAST [74] with an amplicon size of approximately 70–240 bp. qRT-PCR analysis was performed in the QuantStudio™ 3 real-time PCR systems (Thermo Fisher Scientific, USA) and the reactions were carried out using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, USA). The conditions for reactions were 50℃ for 2 min, 95℃ for 2 min, followed by 40 cycles of 95℃ for 15 s, 60℃ for 15 min, 72℃ for 1 min. To ensure a single amplicon reaction, a melting curve analysis was performed at the end of each run. All the experiments were conducted with three biological replicates, and each group was conducted with four samples. The relative gene expression was calculated using the comparative threshold cycle (2−△△CT) method [75]. Student t-test was performed to determine the significant differences between red and brown groups. The Pearson correlation coefficient between RNA-Seq and qRT-PCR was calculated using “cor.test” function in R.
Supplementary Information
Additional file 1: Fig. S1. COG classification analysis. Fig. S2. GO categorization of the annotation genes. Fig. S3. KO classification. Fig. S4. Differential expression analysis. Fig. S5. GO enrichment of DEGs between red and brown groups. Fig. S6. KEGG pathways enrichment analysis of DEGs between red and brown groups.
Additional file 2: Table S1. Heatmap analysis of top 230 DEGs (115 up regulation and 115 down regulation).
Acknowledgements
Transcriptome sequencing was generated by Genomics Bioscience Technology Co., Ltd. (Taipei, Taiwan).
Ethical guidelines
The study is reported in accordance with ARRIVE guidelines. All methods were carried out in accordance with relevant guidelines and regulations.
Author’s contributions
HYW and HYT analyzed the data and prepared the manuscript. KSC, YSH, HYH, and HYT conceived the investigation. All authors have read and agreed to the manuscript.
Funding
This study was supported by Council of Agriculture and Marine National Park Headquarters in Taiwan.
Availability of data and materials
The Sequence Read Archive (SRA) has been deposited at GenBank in NCBI. The datasets generated during the current study are available in the NCBI GenBank repository, with accession number PRJNA826254. Please see link below for details (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA826254).
Declarations
Ethics approval and consent to participate
The animal trial was carried out at the Department of Marine Biotechnology and Resources in National Sun Yat-Sen University (Taiwan) and approved by the ethical review committee for animal project in National Sun Yat-Sen University (Taiwan).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hung-Yi Wu and Kao-Sung Chen contributed equally to this work.
References
- 1.Rimmer MA, Glamuzina B. A review of grouper (Family Serranidae: Subfamily Epinephelinae) aquaculture from a sustainability science perspective. Rev Aquac. 2019;11:58–87. doi: 10.1111/raq.12226. [DOI] [Google Scholar]
- 2.Zeller DC. Home range and activity patterns of the coral trout Plectropomus leopardus (Serranidae) Mar Ecol Prog Ser. 1997;154:65–77. doi: 10.3354/meps154065. [DOI] [Google Scholar]
- 3.Cai X, Qu M, Ding S, Wang H, Wang H, Hu L, et al. Differentiation of coral trout (Plectropomus leopardus) based on an analysis of morphology and complete mitochondrial DNA: are cryptic species present? Acta Oceanol Sin. 2013;32:40–6. doi: 10.1007/s13131-013-0304-6. [DOI] [Google Scholar]
- 4.Maoka T, Sato W, Nagai H, Takahashi T. Carotenoids of red, brown, and black specimens of plectropomus leopardus, the coral trout (Suziara in Japanese) J Oleo Sci. 2017;66:579–84. doi: 10.5650/jos.ess16179. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Wu LN, Chen JF, Wu X, Xia JH, Meng ZN, et al. Whole-genome sequencing of leopard coral grouper (Plectropomus leopardus) and exploration of regulation mechanism of skin color and adaptive evolution. Zool Res. 2020;41:328–40. doi: 10.24272/j.issn.2095-8137.2020.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padilla JE, Mamauag S, Braganza G, Brucal N, Yu D, Morales A. Sustainability Assessment of the live reef-fish for Food Industry in Palawan, Philippines. Quezon City: WWF-Philippines; 2003.
- 7.Mclean DL, Harvey ES, Meeuwig JJ. Declines in the abundance of coral trout (Plectropomus leopardus) in areas closed to fishing at the Houtman Abrolhos Islands, Western Australia. J Exp Mar Bio Ecol. 2011;406:71–8. doi: 10.1016/j.jembe.2011.06.009. [DOI] [Google Scholar]
- 8.Yin X. Sustainability of Coral Trout (Plectropomus leopardus) Fisheries in the Philippines and Indonesia (MSc thesis). Hong Kong: University of Hong Kong; 2014.
- 9.Choat JH, Samoilys M.A. “Plectropomus leopardus,” The IUCN Red List of Threatened Species. 2018. Available online: http://www.iucnredlist.org/details/44684/0. Accessed 25 June 2019.
- 10.Cheney KL, Grutter AS, Marshall NJ. Facultative mimicry: cues for colour change and colour accuracy in a coral reef fish. Proc R Soc B Biol Sci. 2008;275:117–22. doi: 10.1098/rspb.2007.0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J, Bhushan B, Tong J. Structural coloration in nature. RSC Adv. 2013;3:14862–89. doi: 10.1039/c3ra41096j. [DOI] [Google Scholar]
- 12.Fujii R. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 2000;13:300–19. doi: 10.1034/j.1600-0749.2000.130502.x. [DOI] [PubMed] [Google Scholar]
- 13.Braasch I, Schartl M, Volff JN. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol Biol. 2007;7:1–18. doi: 10.1186/1471-2148-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelsh RN, Inoue C, Momoi A, Kondoh H, Furutani-Seiki M, Ozato K, et al. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech Dev. 2004;121:841–59. doi: 10.1016/j.mod.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Henning F, Jones JC, Franchini P, Meyer A. Transcriptomics of morphological color change in polychromatic Midas cichlids. BMC Genomics. 2013;14:1–14. doi: 10.1186/1471-2164-14-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Zhang S, Xu J, Feng J, Mahboob S, Al-Ghanim KA, et al. Comparative transcriptome analysis reveals the genetic basis of skin color variation in common carp. PLoS ONE. 2014;9:e108200. doi: 10.1371/journal.pone.0108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Wachholtz M, Wang J, Liao X, Lu G. Analysis of the skin transcriptome in two Oujiang color varieties of common carp. PLoS ONE. 2014;9:1–8. doi: 10.1371/journal.pone.0090074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oka M, Nagai H, Ando H, Fukunaga M, Matsumura M, Araki K, et al. Regulation of melanogenesis through phosphatidylinositol 3-kinase-akt pathway in human G361 melanoma cells. J Invest Dermatol. 2000;115:699–703. doi: 10.1046/j.1523-1747.2000.00095.x. [DOI] [PubMed] [Google Scholar]
- 19.Fabinyi M. Historical, cultural and social perspectives on luxury seafood consumption in China. Environ Conserv. 2012;39:83–92. doi: 10.1017/S0376892911000609. [DOI] [Google Scholar]
- 20.Kingsford MJ. Spatial and temporal variation in predation on reef fishes by coral trout (Plectropomus leopardus, Serranidae) Coral Reefs. 1992;11:193–8. doi: 10.1007/BF00301993. [DOI] [Google Scholar]
- 21.Lerebours A, Chapman EC, Sweet MJ, Heupel MR, Rotchell JM. Molecular changes in skin pigmented lesions of the coral trout Plectropomus leopardus. Mar Environ Res. 2016;120:130–5. doi: 10.1016/j.marenvres.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Yu C, Guo L, Lin H, Meng Z. In silico comparative transcriptome analysis of two color morphs of the common coral trout (Plectropomus leopardus) PLoS ONE. 2015;10:1–15. doi: 10.1371/journal.pone.0145868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelsh RN. Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 2004;17:326–36. doi: 10.1111/j.1600-0749.2004.00174.x. [DOI] [PubMed] [Google Scholar]
- 24.Melianawati R, Astuti NWW, Suwirya K. The use of copepods to improve juveniles production of coral trout Plectropomus leopardus (Lacepède, 1802) Middle East J Sci Res. 2013;16:237–44. [Google Scholar]
- 25.Liu X, Wang H, Chen Z. Effect of carotenoids on body colour of discus fish (Symphysodon aequifasciatus axelrodi Schultz, 1960) Aquac Res. 2016;47:1309–14. doi: 10.1111/are.12591. [DOI] [Google Scholar]
- 26.Kusumawati D, Setiawati KM. The use of carotene materials as the source of red color pigmentation on leopard grouper larvae (Plectropomus leopardus) Aquac Indones. 2017;17:35. doi: 10.21534/ai.v17i2.50. [DOI] [Google Scholar]
- 27.Zhu X, Hao R, Tian C, Zhang J, Zhu C, Li G. Integrative transcriptomics and metabolomics analysis of body color formation in the Leopard Coral Grouper (Plectropomus leopardus) Front Mar Sci. 2021;8:726102. doi: 10.3389/fmars.2021.726102. [DOI] [Google Scholar]
- 28.Hao R, Zhu X, Tian C, Zhu C, Li G. Analysis of body color formation of leopard coral grouper Plectropomus leopardus. Front Mar Sci. 2022;9:964774. doi: 10.3389/fmars.2022.964774. [DOI] [Google Scholar]
- 29.Hao R, Zhu X, Tian C, Jiang M, Huang Y, Zhu C. Integrated analysis of the role of miRNA-mRNA in determining different body colors of leopard coral grouper (Plectropomus leopardus) Aquaculture. 2022;548:737575. doi: 10.1016/j.aquaculture.2021.737575. [DOI] [Google Scholar]
- 30.Zhou Q, Guo X, Huang Y, Gao H, Xu H, Liu S, et al. De novo sequencing and chromosomal-scale genome assembly of leopard coral grouper, Plectropomus leopardus. Mol Ecol Resour. 2020;20:1403–13. doi: 10.1111/1755-0998.13207. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Wen X, Zhang X, Fu S, Liu J, Tan W, et al. Chromosome Genome Assembly of the Leopard Coral Grouper (Plectropomus leopardus) with Nanopore and Hi-C sequencing data. Front Genet. 2020;11:876. doi: 10.3389/fgene.2020.00876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding SX, Zeng HS, Wang Y, Pan Y, Shi XF. Characterization of eight polymorphic microsatellite loci for the leopard coral grouper (Plectropomus leopardus Lacepède) Mol Ecol Resour. 2009;9:1485–7. doi: 10.1111/j.1755-0998.2009.02690.x. [DOI] [PubMed] [Google Scholar]
- 33.Van Herwerden L, Howard Choat J, Newman SJ, Leray M, Hillersøy G. Complex patterns of population structure and recruitment of Plectropomus leopardus (Pisces: Epinephelidae) in the Indo-West Pacific: implications for fisheries management. Mar Biol. 2009;156:1595–607. doi: 10.1007/s00227-009-1195-0. [DOI] [Google Scholar]
- 34.Zhang J, Liu H, Song Y. Development and characterization of polymorphic microsatellite loci for a threatened reef fish Plectropomus leopardus. Conserv Genet Resour. 2010;2:101–3. doi: 10.1007/s12686-010-9210-9. [DOI] [Google Scholar]
- 35.Ma KY, Van Herwerden L, Newman SJ, Berumen ML, Howard Choat J, Chu KH, et al. Contrasting population genetic structure in three aggregating groupers (Percoidei: Epinephelidae) in the Indo-West Pacific: the importance of reproductive mode. BMC Evol Biol. 2018;18:1–15. doi: 10.1186/s12862-018-1284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge H, Lin K, Shen M, Wu S, Wang Y, Zhang Z, et al. De novo assembly of a chromosome-level reference genome of red-spotted grouper (Epinephelus akaara) using nanopore sequencing and Hi-C. Mol Ecol Resour. 2019;19:1461–9. doi: 10.1111/1755-0998.13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Austin CM, Tan MH, Harrisson KA, Lee YP, Croft LJ, Sunnucks P, et al. De novo genome assembly and annotation of Australia’s largest freshwater fish, the Murray cod (Maccullochella peelii), from Illumina and Nanopore sequencing read. Gigascience. 2017;6:1–6. doi: 10.1093/gigascience/gix063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguinkal JA, Brunner RM, Verleih M, Rebl A, Ríos-Pérez L de los, Schäfer N, et al. The first highly contiguous genome assembly of pikeperch (Sander lucioperca), an emerging aquaculture species in Europe. Genes. 2019;10:708. [DOI] [PMC free article] [PubMed]
- 39.Yang X, Liu H, Ma Z, Zou Y, Zou M, Mao Y, et al. Chromosome-level genome assembly of Triplophysa tibetana, a fish adapted to the harsh high‐altitude environment of the Tibetan Plateau. Mol Ecol Resour. 2019;19:1027–36. doi: 10.1111/1755-0998.13021. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Song YN, Xiao GB, Zhu BH, Xu GC, Sun MY, et al. Gene expression variations of red—white skin coloration in common carp (Cyprinus carpio) Int J Mol Sci. 2015;16:21310–29. doi: 10.3390/ijms160921310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du J, Chen X, Wang J, Chen H, Yue W, Lu G, et al. Comparative skin transcriptome of two Oujiang color common carp (Cyprinus carpio var. Color) varieties. Fish Physiol Biochem. 2019;45:177–85. doi: 10.1007/s10695-018-0551-8. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Liu J, Peng L, Ren L, Zhang H, Zou L, et al. Comparative transcriptome analysis of molecular mechanism underlying gray-to-red body color formation in red crucian carp (Carassius auratus, red var. Fish Physiol Biochem. 2017;43:1387–98. doi: 10.1007/s10695-017-0379-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhang YP, Wang ZD, Guo YS, Liu L, Yu J, Zhang S, et al. Morphological characters and transcriptome profiles associated with black skin and red skin in crimson snapper (Lutjanus erythropterus) Int J Mol Sci. 2015;16:26991–7004. doi: 10.3390/ijms161126005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Gong Q, Lai J, Song M, Liu Y, Wu Y, et al. Transcriptome analysis identifies candidate genes associated with skin color variation in Triplophysa siluroides. Comp Biochem Physiol - Part D Genomics Proteomics. 2020;35:100682. doi: 10.1016/j.cbd.2020.100682. [DOI] [PubMed] [Google Scholar]
- 45.Fang W, Huang J, Li S, Lu J. Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis. Aquaculture. 2021;547:737429. doi: 10.1016/j.aquaculture.2021.737429. [DOI] [Google Scholar]
- 46.Wang Z, Jia Y, Huang X, Zhu D, Liu H, Wang W. Transcriptome profiling towards understanding of the morphogenesis in the scale development of blunt snout bream (Megalobrama amblycephala) Genomics. 2021;113:983–91. doi: 10.1016/j.ygeno.2020.12.043. [DOI] [PubMed] [Google Scholar]
- 47.Huang D, Lewis VM, Toomey MB, Corbo JC, Parichy DM. Development and genetics of red coloration in the zebrafish relative Danio albolineatus. Elife. 2021;10:e70253. doi: 10.7554/eLife.70253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan X, Wei L, Huang J, Wang J, Yang Z, Gan B, et al. Comparative skin transcriptome between common carp and the variety jinbian carp (Cyprinus carpio v. jinbian) Aquac Res. 2020;51:187–96. doi: 10.1111/are.14363. [DOI] [Google Scholar]
- 49.Tsetskhladze ZR, Canfield VA, Ang KC, Wentzel SM, Reid KP, Berg AS, et al. Functional Assessment of Human coding mutations affecting skin pigmentation using zebrafish. PLoS One. 2012;7:e47398. [DOI] [PMC free article] [PubMed]
- 50.Hirata M, Nakamura K, Kanemaru T, Shibata Y, Kondo S. Pigment cell organization in the hypodermis of zebrafish. Dev Dyn. 2003;227:497–503. doi: 10.1002/dvdy.10334. [DOI] [PubMed] [Google Scholar]
- 51.Zhu W, Wang L, Dong Z, Chen X, Song F, Liu N, et al. Comparative transcriptome analysis identifies candidate genes related to skin color differentiation in red tilapia. Sci Rep. 2016;6:1–12. doi: 10.1038/srep31347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu P, Zhang X, Wang X, Li J, Liu G, Kuang Y, et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet. 2014;46:1212–9. doi: 10.1038/ng.3098. [DOI] [PubMed] [Google Scholar]
- 53.Lamason RL, Mohideen MPK, Mest JR, Wong AC, Norton HL, Aros MC, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–7. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 54.Yu L, Chen H, Hu X, Chen X, Liu Z, Wang J, et al. SLC24A5 plays fundamental roles in regulating melanophore development in Cyprinidae fish. Reprod Breed. 2021;1:167–73. doi: 10.1016/j.repbre.2021.11.001. [DOI] [Google Scholar]
- 55.Graf J, Voisey J, Hughes I, Daal A. Promoter polymorphisms in the MATP (SLC45A2) gene are associated with normal human skin color variation. Hum Mutat. 2006;28:710–7. doi: 10.1002/humu.20504. [DOI] [PubMed] [Google Scholar]
- 56.Li C, Chen H, Lan Z, He S, Chen R, Wang F, et al. mTOR-dependent upregulation of xCT blocks melanin synthesis and promotes tumorigenesis. Cell Death Differ. 2019;26:2015–28. doi: 10.1038/s41418-019-0274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruprecht JJ, Kunji ERS. The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem Sci. 2020;45:244–58. doi: 10.1016/j.tibs.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huo SM, Zhang YY, Song ZR, Xiong XH, Hong XY. The potential pigmentation-related genes in spider mites revealed by comparative transcriptomes of the red form of Tetranychus urticae. Insect Mol Biol. 2021;30:580–93. doi: 10.1111/imb.12727. [DOI] [PubMed] [Google Scholar]
- 59.Ma X, Ma Z, Jiao X, Hejtmancik JF. Functional non-coding polymorphism in an EPHA2 promoter PAX2 binding site modifies expression and alters the MAPK and AKT pathways. Sci Rep. 2017;7:1–15. doi: 10.1038/s41598-017-10117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mashek DG, Li LO, Coleman RA. Future Lipidology Long-Chain acyl-coa synthetases and fatty acid channeling. Future Lipidol. 2017;2:465–76. doi: 10.2217/17460875.2.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu X, Jin S, Yang Y, Lu X, Dai X, Xu Z, et al. Altered expression of ferroptosis markers and iron metabolism reveals a potential role of ferroptosis in vitiligo. Pigment Cell Melanoma Res. 2022;35:328–41. doi: 10.1111/pcmr.13032. [DOI] [PubMed] [Google Scholar]
- 62.Hirobe T. Role of keratinocyte-derived factors involved in regulating the proliferation and differentiation of mammalian epidermal melanocytes. Pigment Cell Res. 2005;18:2–12. doi: 10.1111/j.1600-0749.2004.00198.x. [DOI] [PubMed] [Google Scholar]
- 63.Costin G-E, Hearing VJ. Human skin pigmentation: melanocytes modulate skin color in response to stress. FASEB J. 2007;21:976–94. doi: 10.1096/fj.06-6649rev. [DOI] [PubMed] [Google Scholar]
- 64.Cheng KC. Skin color in Fish and humans: impacts on Science and Society. Zebrafish. 2008;5:242. doi: 10.1089/zeb.2008.0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Philip D, Bowden J, et al. De novo transcript sequence recostruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc. 2013;8:1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2022;27:gkac963. [DOI] [PMC free article] [PubMed]
- 70.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li B, Dewey CN, RSEM Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:1–16. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49:W317–25. doi: 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 – ∆∆CT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. COG classification analysis. Fig. S2. GO categorization of the annotation genes. Fig. S3. KO classification. Fig. S4. Differential expression analysis. Fig. S5. GO enrichment of DEGs between red and brown groups. Fig. S6. KEGG pathways enrichment analysis of DEGs between red and brown groups.
Additional file 2: Table S1. Heatmap analysis of top 230 DEGs (115 up regulation and 115 down regulation).
Data Availability Statement
The Sequence Read Archive (SRA) has been deposited at GenBank in NCBI. The datasets generated during the current study are available in the NCBI GenBank repository, with accession number PRJNA826254. Please see link below for details (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA826254).