In the current World Health Organization (WHO) Classification of Tumors of the Central Nervous System (CNS), pediatric high-grade gliomas (HGGs), IDH- and histone H3-wildtype (WT) are divided into three molecular subtypes: RTK1, RTK2 and MYCN [2]. HGG-MYCN present recurrent histopathological characteristics (nodular pattern with embryonal cells), genetic features (frequent MYCN and/or ID2 amplification, and somatic TP53 mutations) [1, 5, 6], and a specific DNA-methylation profile. Li-Fraumeni syndrome (LFS) encompasses a wide variety of primary brain tumors. They include HGGs, IDH-WT with MYCN amplification, but only one of them was reported in the literature as a HGG-MYCN by DNA-methylation profiling [3, 4]. The proportion of specimens from the epigenetic subgroup HGG-MYCN associated with LFS remains an unanswered question. The aim of this study was to analyze the somatic and germline status of TP53 and the DNA-methylation profile (using the v12.5 of the Heidelberg Brain Tumor classifier) from a series of HGG-MYCN. From a series of 151 pediatric HGGs, we selected 11 cases suspected of belonging to a MYCN subtype based on histopathology and MYCN amplification (identified by FISH analysis, cf. Additional file 1: Fig. S1).
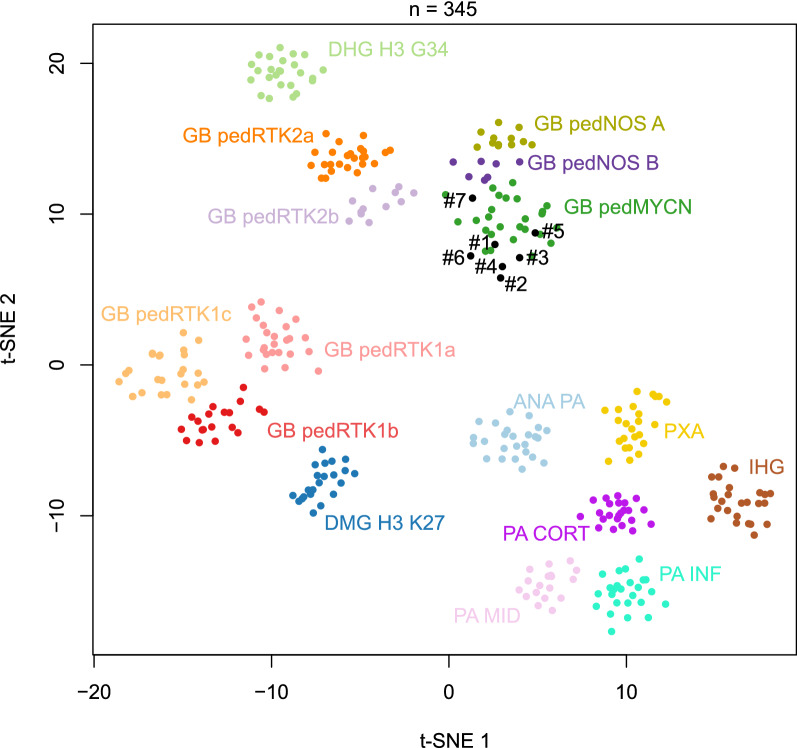
The clinical, genetic and epigenetic characteristics of the cohort are summarized in Table 1. Of the 11 HGGs initially diagnosed as HGG-MYCN, each one presented a p53 overexpression using immunohistochemistry and harbored a somatic TP53 pathogenic variant (PV) (cf. Additional file 2: Methods). The tumor classification based on DNA-methylation confirmed the diagnosis of the pediatric HGG-MYCN subtype in 5/7 cases with high calibrated scores (> 0.9) having sufficient DNA available for the analysis. The two remaining cases were classified as pediatric HGG, not otherwise specified, subtype A (despite the presence of a MYCN amplification detected by FISH and copy number variation of whole exome sequencing data) and HGG-MYCN with a low calibrated score (0.20). However, using t-distributed stochastic neighbor embedding (t-SNE) analysis, all cases clustered within or in close vicinity to HGG-MYCN (Fig. 1).
Table 1.
Clinical, genetic and epigenetic details of the cohort
| Reference case | Age at diagnosis (y), sex | Tumor location | Somatic TP53 pathogenic variation (NM_000546.5) | Methylation-based classification (calibrated score) v12.5 | Germline TP53 status | Status at the end of follow-up, OS (y) |
|---|---|---|---|---|---|---|
| 1 | 4.3, F | Pons | c.469G>T; p.(Val157Phe), Exon 5 | Diffuse paediatric-type HGG, MYCN subtype (0.99) | Mutated: c.469G>T; p.(Val157Phe), Exon 5 | Dead (0.4) |
| 2 | 5.2, M | Multifocal (cerebellum. mesencephalic. bulbar and thalamic) | c.742C>G; p.(Arg248Gly), Exon 07 | Diffuse paediatric-type HGG, MYCN subtype (0.90) | Mutated: c.742C>G; p.(Arg248Gly), Exon 07 | Dead (1) |
| 3 | 3.4, M | Left fronto-parietal lobe | c.701A>G; p.(Tyr234Cys), Exon 07 | Diffuse paediatric-type HGG, MYCN subtype (0.99) | Mutated: c.701A>G; p.(Tyr234Cys), Exon 07 | Dead (1.7) |
| 4 | 3.3, M | Left frontal lobe | c.731G>A; p.(Gly244Asp), Exon 07 | Diffuse paediatric-type HGG, MYCN subtype (0.99) | Mutated; c.731G>A, p.(Gly244Asp) | Dead (1.5) |
| 5 | 4.5, M | Left thalamus | c. 743G>A; p.(Arg248Gln), Exon 07 | Diffuse paediatric-type HGG, MYCN subtype (0.99) | WT | Dead (0.3) |
| 6 | 7.6, F | Pons | c.817C>T; p.(Arg273Cys), Exon 8 | Diffuse paediatric-type HGG, MYCN subtype (0.20) | WT | Dead (0) |
| 7 | 3.2, F | Right thalamus | c.853G>A; p.(Glu285Lys), Exon 08 | Diffuse paediatric-type HGG, H3 wildtype and IDH WT, Subtype A (0.99) | WT | Dead (0.7) |
| 8 | 3.1, M | Pons | c.916C>T; (p.Arg306Ter), Exon 8 c.632C>T; p.(Thr211Ile), Exon 6 | NA | WT | Dead (0.5) |
| 9 | 1.3, F | Pons | c.524G>A; p.(Arg175His), Exon 08 | NA | WT | Dead (0.2) |
| 10 | 2.8, F | Pons | c.742C>T; p.(Arg248Trp), Exon 7 | NA | WT | Dead (NA) |
| 11 | 4.4, M | Pons | c.844C>A; p.(Arg282Trp), Exon 8 | NA | WT | Dead (0.7) |
F female, HGG high-grade glioma, M male, NA not available, OS overall survival, WT wildtype, y years-old
Fig. 1.
t-Distributed stochastic neighbor embedding (t-SNE) analysis of DNA methylation profiles of the investigated tumors alongside selected reference samples. Reference DNA methylation classes: high-grade astrocytoma with piloid features (ANA PA); diffuse high-grade glioma, H3.3 G34 mutant (DHG H3 G34); diffuse midline glioma H3 K27M mutant (DMG H3 K27); pediatric glioblastoma, IDH wildtype, subclass MYCN (GB pedMYCN); pediatric glioblastoma, IDH wildtype, subclass not otherwise specified sutbype A (GB pedNOS A); pediatric glioblastoma, IDH wildtype, subclass not otherwise specified sutbype B (GB pedNOS B); pediatric glioblastoma, IDH wildtype, subclass RTK1a (GB pedRTK1a); pediatric glioblastoma, IDH wildtype, subclass RTK1b (GB pedRTK1b); pediatric glioblastoma, IDH wildtype, subclass RTK1c (GB pedRTK1c); pediatric glioblastoma, IDH wildtype, subclass RTK2a (GB pedRTK2a); pediatric glioblastoma, IDH wildtype, subclass RTK2b (GB pedRTK2b); infant-type hemispheric glioma (IHG); hemispheric pilocytic astrocytoma (PA CORT); infratentorial pilocytic astrocytoma (PA INF); midline pilocytic astrocytoma (PA MID); pleomorphic xanthoastrocytoma (PXA)
Four out of seven (57%) patients had a TP53 germline PV. All four of these four cases presented a high calibrated score for the HGG-MYCN methylation class. The information concerning a family history, or for a predisposition to cancer was not available for all patients. Among the families explored in genetic counseling, one patient presented a de novo PV (case #4) and genetic analyses are currently in progress for parents of another patient (case #2) having a family history of malignant sarcoma in the grandfather. No TP53 germline mutation was observed in the other cases.
Previously, only one case report of HGG-MYCN in LFS has been reported in the literature [3]. Based on the high prevalence of somatic TP53 PV in epigenetically confirmed HGG-MYCN (100% of cases in our series, 67% in the study which initially described the methylation class [1]), we demonstrate for the first time that this tumor type is frequently associated with LFS and may constitute the mode of revelation for this genetic predisposition syndrome. LFS cases do not seem to form a distinct subcluster in the DNA-methylation based classification of CNS tumors compared to those without a TP53 germline mutation. Whereas MYCN amplification and TP53 PV are enriched in HGG-MYCN, these alterations are not specific to this subgroup and may be encountered within other subtypes of pediatric HGGs (RTK1, RTK2) [1]. In this current study, one case was classified as a pediatric HGG, not otherwise specified, subtype A (with a high calibrated score) but clustered in close vicinity to HGG-MYCN using t-SNE analysis. This result highlights the fact that the epigenetic boundaries between all subtypes (eight different methylation classes defined in the v12.5 of the DKFZ classifier) of pediatric HGGs are still in progress, and potentially argues that the three initial subgroups defined by Korshunov et al. (and particularly cases included in the HGG-MYCN subgroup which do not present MYCN amplification and TP53 mutation) are probably redefined in other methylation classes.
To conclude, a constitutional analysis of TP53 and a genetic counseling should be proposed to all patients with proven HGG-MYCN harboring a TP53 somatic alteration. Additional cases are needed to determine if the HGG-MYCN associated with LFS forms a distinct methylation subclass from those without a germline mutation of TP53, as was described for Primary mismatch repair deficient IDH-mutant astrocytoma in the v12.3 of the DKFZ classifier. Moreover, further studies are needed to determine if clinical (pediatric tumor), histopathological (HGG with embryonal features), and genetic (TP53 PV and MYCN amplification) features may constitute alternative diagnostic criteria by DNA-methylation profiling for a diagnosis of HGG-MYCN.
Supplementary Information
Additional file 1. Fig. S1: Histopathological features. Black scale bars represent 1 mm (A), 100 μm (B) and 50 μm (C to K). (A-C-E-G-H-I-K-M) Diffuse and solid proliferation with several nodules infiltrating the brain parenchyma. Undifferenciated proliferation composed of hyperchromatic cells presenting anisocaryotic nuclei with numerous apoptotic bodies (HPS, x400 magnification). (B-D-F-H-J-L-N) Nuclear accumulation of p53 (x400 magnification). Black scale bars represent 50 μm.
Additional file 2. Methods used in this series.
Acknowledgements
We are thankful for the laboratory technicians at GHU Paris Neuro Sainte-Anne Hospital for their assistance, and the Integragen platform for their technical assistance with DNA-methylation analyses.
Author contributions
LGR and ATE participated in conception, design, collection and assembly of data. LGR, JG, KB, TB and FB provided medical care to patients. ATE and PV conducted the neuropathological examinations. DC, ER, RS, PS, MB and MAD, conducted the molecular somatic and germline analyses. LGR and ATE drafted the manuscript. All authors reviewed the manuscript and approved the final version.
Funding
The authors declare that they have been supported by the Pediatric Campaign of the Fondation Gustave Roussy “Guérir les Cancers des Enfants au XXI siècle”.
Declarations
Ethics approval and consent to participate
This study was approved by the local ethical committees from GHU Paris Psychiatry and Neurosciences, Sainte-Anne Hospital, of Necker Enfants Malades Hospital and of Gustave Roussy. Informed consent was obtained specifically from each patient/family for the constitutional genetic study.
Competing interests
The authors declare that they have no conflicts of interest directly related to the topic of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Léa Guerrini-Rousseau and Arnault Tauziède-Espariat have contributed equally to this work
References
- 1.Korshunov A, Schrimpf D, Ryzhova M, Sturm D, Chavez L, Hovestadt V, Sharma T, Habel A, Burford A, Jones C, Zheludkova O, Kumirova E, Kramm CM, Golanov A, Capper D, von Deimling A, Pfister SM, Jones DTW. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017;134:507–516. doi: 10.1007/s00401-017-1710-1. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-Oncology. 2021;6:66. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoof M, Kordes U, Volk AE, Al-Kershi S, Kresbach C, Schüller U. Malignant gliomas with H3F3A G34R mutation or MYCN amplification in pediatric patients with Li Fraumeni syndrome. Acta Neuropathol. 2021;142:591–593. doi: 10.1007/s00401-021-02346-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sloan EA, Hilz S, Gupta R, Cadwell C, Ramani B, Hofmann J, Kline CN, Banerjee A, Reddy A, Oberheim Bush NA, Chang S, Braunstein S, Chang EF, Raffel C, Gupta N, Sun PP, Kim JYH, Moes G, Alva E, Li R, Bruggers CS, Alashari M, Wetmore C, Garg S, Dishop M, Van Ziffle J, Onodera C, Devine P, Grenert JP, Lee JC, Phillips JJ, Pekmezci M, Tihan T, Bollen AW, Berger MS, Costello JF, Perry A, Solomon DA. Gliomas arising in the setting of Li-Fraumeni syndrome stratify into two molecular subgroups with divergent clinicopathologic features. Acta Neuropathol. 2020;139:953–957. doi: 10.1007/s00401-020-02144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tauziède-Espariat A, Debily M-A, Castel D, Grill J, Puget S, Roux A, Saffroy R, Pagès M, Gareton A, Chrétien F, Lechapt E, Dangouloff-Ros V, Boddaert N, Varlet P. The pediatric supratentorial MYCN-amplified high-grade gliomas methylation class presents the same radiological, histopathological and molecular features as their pontine counterparts. Acta Neuropathol Commun. 2020;8:104. doi: 10.1186/s40478-020-00974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tauziède-Espariat A, Debily M-A, Castel D, Grill J, Puget S, Sabel M, Blomgren K, Gareton A, Dangouloff-Ros V, Lechapt E, Boddaert N, Varlet P. An integrative radiological, histopathological and molecular analysis of pediatric pontine histone-wildtype glioma with MYCN amplification (HGG-MYCN) Acta Neuropathol Commun. 2019;7:87. doi: 10.1186/s40478-019-0738-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Fig. S1: Histopathological features. Black scale bars represent 1 mm (A), 100 μm (B) and 50 μm (C to K). (A-C-E-G-H-I-K-M) Diffuse and solid proliferation with several nodules infiltrating the brain parenchyma. Undifferenciated proliferation composed of hyperchromatic cells presenting anisocaryotic nuclei with numerous apoptotic bodies (HPS, x400 magnification). (B-D-F-H-J-L-N) Nuclear accumulation of p53 (x400 magnification). Black scale bars represent 50 μm.
Additional file 2. Methods used in this series.