Abstract
Multilocus sequence typing (MLST) is a tool that can be used to study the molecular epidemiology and population genetic structure of microorganisms. A MLST scheme was developed for Streptococcus pyogenes and the nucleotide sequences of internal fragments of seven selected housekeeping loci were obtained for 212 isolates. A total of 100 unique combinations of housekeeping alleles (allelic profiles) were identified. The MLST scheme was highly concordant with several other typing methods. The emm type, corresponding to a locus that is subject to host immune selection, was determined for each isolate; of the >150 distinct emm types identified to date, 78 are represented in this report. For a given emm type, the majority of isolates shared five or more of the seven housekeeping alleles. Stable associations between emm type and MLST were documented by comparing isolates obtained decades apart and/or from different continents. For the 33 emm types for which more than one isolate was examined, only five emm types were present on widely divergent backgrounds, differing at four or more of the housekeeping loci. The findings indicate that the majority of emm types examined define clones or clonal complexes. In addition, an MLST database is made accessible to investigators who seek to characterize other isolates of this species via the internet (http://www.mlst.net).
Group A streptococci (GAS; Streptococcus pyogenes) are highly prevalent bacterial pathogens, having a worldwide distribution, whereby humans serve as their primary biological host. Most often, GAS infect superficial tissue sites, involving the mucosal epithelium of the upper respiratory tract (URT) or the epidermal layer of the skin, leading to pharyngitis or impetigo, respectively. On rare occasions, a GAS infection can lead to invasive disease that includes cellulitis, bacteremia, necrotizing fasciitis, and toxic shock syndrome, which can be life-threatening conditions. In addition, GAS contribute to morbidity through delayed nonsuppurative sequelae, such as rheumatic fever and acute glomerulonephritis.
The M and M-like proteins of GAS form surface fibrils that provide the basis for a widely used serological typing scheme. For many molecules studied in detail, the M serotype (M type) is usually defined by antigenic target sites contained within the distal, amino-terminal ends of these fibrillar proteins, and >80 distinct M types have been identified. M and M-like proteins are also key virulence factors, and protective immunity against GAS infection is type specific (8, 23). More recently, a genotypic typing scheme based on the emm genes that encode M and M-like proteins has become widely used and >150 different emm types have been characterized (15; http://www.cdc.gov/ncidod/biotech/strep/strains.html). The antigenic heterogeneity exhibited by this family of proteins reflects the strong impact of host immunity on the generation of diversity within this bacterial species.
Numerous other genotypic methods have been developed for the typing of GAS isolates. Vir-typing measures restriction fragment length polymorphisms within the emm chromosomal region (18). Pulsed-field gel electrophoresis and arbitrary-primed PCR can provide high levels of resolution between strains by measuring multiple loci for differences that are not necessarily under selection (10, 17, 18, 33). Another important tool for discrimination among strains of GAS is multilocus enzyme electrophoresis (MLEE), which indexes differences in the net charge of housekeeping enzymes resulting from certain mutations (29, 32).
Multilocus sequence typing (MLST) is a nucleotide sequence-based method that is well suited towards characterizing the genetic relationships between the organisms of a bacterial species (12–14, 26). Because it is based on nucleotide sequence, it provides unambiguous results and is easily portable from lab to lab. Housekeeping loci are chosen for analysis because they are present in every organism (i.e., their products serve a vital function), and mutations within them are largely assumed to be selectively neutral (32). Clones, defined as isolates that are descendants of a recent common ancestor, can be identified as having shared alleles at each of the housekeeping loci. In this report, an MLST scheme using seven housekeeping loci was used to evaluate >200 GAS isolates that were derived from several continents, spanning a time period of >50 years and representing 78 distinct emm types.
MATERIALS AND METHODS
Bacterial strains.
The GAS isolates of the MGAS series were kindly provided by Susan Hollingshead (University of Alabama at Birmingham), who had received them from James Musser, and the isolates have been previously described in detail (29, 30). The GAS isolates of the CT98 series were kindly provided by James Hadler and Nancy Barrett (State of Connecticut Department of Public Health, Hartford). Strain 700294 was purchased from the American Tissue Culture Collection (Manassas, Va.). All other GAS isolates have been previously described (6, 17).
Multilocus sequence typing.
Chromosomal DNA was prepared from freshly grown GAS by previously described methods (6). Internal fragments of the glucose kinase (gki), glutamine transporter protein (gtr), glutamate racemase (murI), DNA mismatch repair protein (mutS), transketolase (recP), xanthine phosphoribosyl transferase (xpt), and acetyl coenzyme A (acetyl-CoA) acetyltransferase (yqiL) genes were amplified by PCR using the following primer pairs: gki-up, 5′-GGC ATT GGA ATG GGA TCA CC-3′, and gki-dn, 5′-TCT CCT GCT GCT GAC AC-3′; gtr-up, 5′-GAG GTT GTG GTG ATT ATT GG-3′, and gtr-dn, 5′-GCA AAG CCC ATT TCA TGA GTC-3′; murI-up, 5′-TGC TGA CTC AAA ATG TTA AAA TGA TTG-3′, and murI-dn, 5′-GAT GAT AAT TCA CCG TTA ATG TCA AAA TAG-3′; mutS-up, 5′-GAA GAG TCA TCT AGT TTA GAA TAC GAT-3′, and mutS-dn, 5′-AGA GAG TTG TCA CTT GCG CGT TTG ATT GCT-3′; recP-up, 5′-GCA AAT TCT GGA CAC CCA GG-3′, and recP-dn, 5′-CTT TCA CAA GGA TAT GTT GCC-3′; xpt-up, 5′-TTA CTT GAA GAA CGC ATC TTA-3′, and xpt-dn, 5′-ATG AGG TCA CTT CAA TGC CC -3′; yqiL-up, 5′-TGC AAC AGT ATG GAC TGA CCA GAG AAC AAG ATG C-3′, and yqiL-dn, 5′-CAA GGT CTC GTG AAA CCG CTA AAG CCT GAG-3′. The PCRs were performed in volumes of 50 μl, with an initial denaturation at 95°C for 4 to 5 min, followed by 28 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The amplified DNA fragments were purified either by precipitation with polyethylene glycol or using a PCR purification kit (Qiagen, Valencia, Calif.). The sequence of each fragment was obtained on both strands by using the same primers as those in the initial PCR amplifications and an AB1377 or AB13700 DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.).
For each locus, every different sequence was assigned a distinct allele number, and each isolate was defined by a series of seven integers (the allelic profile) corresponding to the alleles at the seven loci, in the order (alphabetical) of gki, gtr, murI, mutS, recP, xpt, and yqiL. Isolates with an identical allelic profile were assigned to the same sequence type (ST).
emm sequence typing.
emm sequence typing is based on the 5′ end of the central emm gene within the emm chromosomal region (for map, see references 5 and 6). A unique emm type is defined as having <95% sequence identity to any other known emm type over 160 bp near the 5′ end, as specified (http://www.cdc.gov/ncidod/biotech/strep/strains.html). There is a very strong correspondence between M type, as determined by serology, and the emm type that meets the stated definition (3, 15). In addition to a sequence identity of ≥95%, indels of four or fewer codons and/or frameshift mutations relative to the reference emm typing strain are allowed for classification as an established emm type. Until validation is complete, new emm types are assigned the nomenclature “emmst,” which stands for emm sequence type (15) and is not to be confused with “ST,” which refers to the MLST allelic profile.
Computations.
A matrix of pair-wise differences in allelic profiles was constructed, and the similarities between the allelic profiles of the isolates were assessed by cluster analysis using the unweighted pair-group method with arithmetic averages (UPGMA) and the percent disagreement distance measure (Statistica version 5.5; StatSoft, Tulsa, Okla.). The maximum percent nucleotide divergence and average percent nucleotide divergence between pairs of alleles at a given locus were calculated using Mega version 2.0 (http://www.megasoftware.net). The Index of Association (27) was used to test for linkage disequilibrium between alleles at the seven housekeeping loci. The observed variance in the distribution of allelic mismatches in all pair-wise comparisons of the allelic profiles was compared to that expected in a freely recombining population (linkage equilibrium). The significance of the difference in the observed and expected variance was evaluated by computing the maximum variance in the distribution of allelic mismatches obtained using 100 randomizations of the data set. Significant linkage disequilibrium was established if the observed variance obtained with the actual data was greater than that found with any of the 100 randomized data sets; otherwise, there was no evidence of a departure from linkage equilibrium.
RESULTS
Housekeeping loci used for MLST.
Seven housekeeping loci were chosen for the characterization of GAS isolates by MLST and for determining their population genetic structure (Table 1). The nucleotide sequence was determined for an internal portion of about 400 to 500 bp of each gene. The loci that were chosen had been used successfully for pneumococci (14) or were selected with guidance by data from the University of Oklahoma GAS genome sequencing project that is available on the World Wide Web. Large contigs from the database (www.genome.ou.edu) were used in BLASTX searches against the GenBank database. Housekeeping loci were identified based on their putative function. Loci selected for this study were devoid of flanking regions containing genes that are likely to be under selection for variation (e.g., genes encoding cell surface proteins that may be under diversifying selection from the host immune response). The only possible exception was recP, positioned ∼9 kb from a putative penicillin-binding protein gene (pbp2x homologue). However, analysis of a set of 14 isolates showed nucleotide sequence divergence of <1.0% for an internal portion of pbp2x and a lack of evidence for interspecies recombinational events, as has been observed for pneumococcal and meningococcal pbp genes (11) (data not shown). Furthermore, GAS isolates that are resistant to penicillin have not been described as occurring in nature. Ten housekeeping loci were initially examined in a small subset of strains and the least and most polymorphic ones were discarded. The chromosomal distance between any two loci, calculated on the basis of the tentative genome map of strain 700294, ranges from 20 to 600 kb (www.genome.ou.edu); it is possible that for other strains, the genomic location of the loci under study may differ.
TABLE 1.
Housekeeping loci under studya
| Locus | Putative function of gene product | Size of sequenced fragment (bp) | No. of alleles identified | No. of polymorphic nucleotide sites | No. of polymorphic amino acid sites | % Nucleotide divergence between pairs of alleles
|
Chromosomal location (no. of kb from ori)b | GenBank no. | |
|---|---|---|---|---|---|---|---|---|---|
| Maximum | Average | ||||||||
| gki | Glucose kinase | 498 | 34 | 36 | 12 | 2.6 | 1.3 | 1259 | AJ302470–AJ302503 |
| gtr | Glutamine transport ATP-binding protein | 450 | 32 | 30 | 9 | 2.6 | 1 | 1237 | AJ302504–AJ302535 |
| murl | Glutamate racemase | 438 | 24 | 21 | 6 | 1.4 | 0.8 | 313 | AJ406970–AJ406993 |
| mutS | DNA mismatch repair (HexA) | 405 | 21 | 23 | 9 | 2.2 | 0.9 | 1788 | AJ406994–AJ407014 |
| recP | Transketolase | 459 | 35 | 59 | 11 | 6.1 | 1.9 | 1392 | AJ407015–AJ407049 |
| xpt | Xanthine phosphoribosyltransferase | 450 | 29 | 31 | 10 | 2.9 | 0.8 | 931 | AJ291638–AJ291666 |
| yqiL | Acetoacetyl-CoA thiolase (AtoB) | 434 | 22 | 22 | 12 | 1.4 | 0.7 | 129 | AJ291616–AJ291637 |
Alleles of the seven housekeeping loci can be obtained (http://www.mlst.net).
The emm gene is located at kb 1685, relative to the origin, based on the genome of GAS strain 700294 (www.genome.ou.edu; 10 February 2001). The size of the strain 700294 genome is 1,852 kb.
The number of unique alleles identified for each of the seven housekeeping loci ranged from 21 (for mutS) to 35 (for recP) (Table 1). The maximum percent nucleotide sequence divergence between the alleles of a given locus ranged from 1.4% (for yqiL and murI) to 6.1% (for recP). For one housekeeping locus, recP, there were four widely divergent alleles (recP7, recP15, recP21, recP29) which may have arisen by importation of homologous regions from closely related species. As noted above, the recP gene is ∼9 kb from a pbp2x gene; however, pbp2x alleles display low levels of polymorphism, and there were no obvious differences between the pbp2x alleles of isolates recovered in the pre-antibiotic era (early 1940s) and those obtained in recent decades (data not shown). The sequence was determined for part of the pbp2x gene of an isolate containing one of the diverged recP alleles (recP7); this strain (C135) possessed the most prevalent pbp2x allele, and there is no evidence that the increased divergence of some recP alleles is due to hitchhiking driven by selection for interspecies recombination at the pbp2x locus. A more complete analysis of the housekeeping alleles is presented elsewhere (16; A. Kalia, M. C. Enright, B. G. Spratt, and D. E. Bessen, submitted for publication).
MLST of the GAS population.
The collection of 212 GAS isolates (Table 2) was assembled with several goals in mind. First, a genetically diverse group of GAS strains was desired. As will be shown in this report, emm type is a sensitive measure of genetic diversity. Of the >150 emm types characterized to date (http://www.cdc.gov/ncidod/biotech/strep/strains.html), isolates representing 78 emm types were included in the MLST analysis. Secondly, it was of interest to evaluate GAS with large temporal and/or spatial distances between their isolation from human tissue, in order to assess the stability of clones. In addition, the selected GAS isolates were recovered in association with a variety of host tissues and diseases, including deep soft tissue infections. Finally, several GAS that had been previously analyzed using different molecular typing schemes were chosen for comparison to MLST, in order to provide validation of the new method.
TABLE 2.
MLST of 212 GAS isolatesa
| Strain | emm type | codeb | ST | Assigned no. for allele
|
Source | Year | Origin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gki | gtr | murI | mutS | recP | xpt | yqiL | |||||||
| 86-779 | 1 | m1-1 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | URTa | 1986 | Ohio |
| 87-214 | 1 | m1-2 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | URT | 1987 | Chile |
| CT95-111 | 1 | m1-3 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | 1995 | Connecticut |
| CT95-120 | 1 | m1-4 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | 1995 | Connecticut |
| CT95-131 | 1 | m1-5 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | 1995 | Connecticut |
| CT95-132 | 1 | m1-6 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | 1995 | Connecticut |
| CT95-180 | 1 | m1-7 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | 1995 | Connecticut |
| CT98360 | 1 | m1-8 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | 1998 | Connecticut |
| CT98419 | 1 | m1-9 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | 1998 | Connecticut |
| CT98520 | 1 | m1-10 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | 1998 | Connecticut |
| 700294-ATCC | 1 | m1-11 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | NDd | ND |
| MGAS166 | 1 | m1-12 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | ND | Minnesota |
| MGAS2127 | 1 | m1-13 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | ND | ND | India |
| MGAS283 | 1 | m1-14 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | ND | Colorado |
| MGAS302 | 1 | m1-15 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | ND | Washington |
| MGAS307 | 1 | m1-16 | 28 | 4 | 3 | 4 | 4 | 4 | 2 | 4 | Invasive | ND | Texas |
| MGAS2110 | 1 | m1-17 | 91 | 26 | 2 | 18 | 5 | 31 | 3 | 1 | ND | ND | New Zealand |
| MGAS2120 | 1 | m1-18 | 92 | 27 | 25 | 4 | 4 | 32 | 2 | 1 | Impetigo | ND | Australia |
| MGAS2125 | 1 | m1-19 | 92 | 27 | 25 | 4 | 4 | 32 | 2 | 1 | Impetigo | ND | Australia |
| MGAS2109 | 1 | m1-20 | 93 | 28 | 25 | 4 | 4 | 4 | 2 | 1 | URT | ND | New Zealand |
| MGAS2144 | 1 | m1-21 | 93 | 28 | 25 | 4 | 4 | 4 | 2 | 1 | URT | ND | New Zealand |
| MGAS2226 | 1 | m1-22 | 93 | 28 | 25 | 4 | 4 | 4 | 2 | 1 | URT | ND | New Zealand |
| MGAS2123 | 1 | m1-23 | 94 | 28 | 25 | 4 | 4 | 4 | 20 | 1 | Impetigo | ND | Australia |
| B344 | 2 | m2-1 | 55 | 11 | 9 | 1 | 9 | 2 | 3 | 4 | URT | 1950 | Massachusetts |
| D725 | 2 | m2-2 | 55 | 11 | 9 | 1 | 9 | 2 | 3 | 4 | URT | 1973 | New York |
| 89-465 | 2 | m2-3 | 55 | 11 | 9 | 1 | 9 | 2 | 3 | 4 | URT | 1989 | Nebraska |
| CT95-197 | 2 | m2-4 | 55 | 11 | 9 | 1 | 9 | 2 | 3 | 4 | Invasive | 1995 | Connecticut |
| CT98549 | 2 | m2-5 | 55 | 11 | 9 | 1 | 9 | 2 | 3 | 4 | Invasive | 1998 | Connecticut |
| MGAS286 | 2 | m2-6 | 55 | 11 | 9 | 1 | 9 | 2 | 3 | 4 | Invasive | ND | Colorado |
| MGAS327 | 2 | m2-7 | 55 | 11 | 9 | 1 | 9 | 2 | 3 | 4 | Invasive | ND | Arizona |
| C199 | 3 | m3-1 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | URT | 1942 | New York |
| 1GL90 | 3 | m3-2 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | URT | 1946 | Illinois |
| 2GL215 | 3 | m3-3 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | URT | 1946 | Illinois |
| 1RP228 | 3 | m3-4 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | URT | 1959 | New York |
| D425 | 3 | m3-5 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | 1971 | New York |
| CT95-117 | 3 | m3-6 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | 1995 | Connecticut |
| CT95-119 | 3 | m3-7 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | 1995 | Connecticut |
| CT95-133 | 3 | m3-8 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | 1995 | Connecticut |
| CT95-145 | 3 | m3-9 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | 1995 | Connecticut |
| CT95-186 | 3 | m3-10 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | 1995 | Connecticut |
| CT95-204 | 3 | m3-11 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | 1995 | Connecticut |
| CT98429 | 3 | m3-12 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | 1998 | Connecticut |
| MGAS157 | 3 | m3-13 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | ND | Minnesota |
| MGAS159 | 3 | m3-14 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | ND | Utah |
| MGAS268 | 3 | m3-15 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | ND | Minnesota |
| MGAS277 | 3 | m3-16 | 15 | 2 | 6 | 8 | 5 | 2 | 3 | 2 | Invasive | ND | Colorado |
| 88-019 | 3 | m3-17 | 16 | 2 | 6 | 8 | 16 | 2 | 3 | 2 | URT | 1988 | North Carolina |
| CT95-200 | 3 | m3-18 | 18 | 2 | 27 | 8 | 5 | 2 | 3 | 2 | Invasive | 1995 | Connecticut |
| 2RP113 | 4 | m4-1 | 38 | 5 | 7 | 8 | 5 | 15 | 2 | 1 | Invasive | 1952 | New York |
| 1RP156 | 4 | m4-2 | 38 | 5 | 7 | 8 | 5 | 15 | 2 | 1 | URT | 1954 | New York |
| A837 | 4 | m4-3 | 38 | 5 | 7 | 8 | 5 | 15 | 2 | 1 | Invasive | 1965 | Unknown |
| 87-231 | 4 | m4-4 | 39 | 5 | 11 | 8 | 5 | 15 | 2 | 1 | URT | 1987 | Ohio |
| CT95-195 | 4 | m4-5 | 39 | 5 | 11 | 8 | 5 | 15 | 2 | 1 | Invasive | 1995 | Connecticut |
| YL05 | 4 | m4-6 | 39 | 5 | 11 | 8 | 5 | 15 | 2 | 1 | Invasive | 1997 | Connecticut |
| CT98693 | 4 | m4-7 | 39 | 5 | 11 | 8 | 5 | 15 | 2 | 1 | Invasive | 1998 | Connecticut |
| YL08 | 4 | m4-8 | 39 | 5 | 11 | 8 | 5 | 15 | 2 | 1 | Invasive | 1999 | Connecticut |
| 09C02 | 4 | m4-9 | 39 | 5 | 11 | 8 | 5 | 15 | 2 | 1 | Invasive | ND | ND |
| MGAS320 | 4 | m4-10 | 39 | 5 | 11 | 8 | 5 | 29 | 2 | 1 | Invasive | ND | Texas |
| CT98529 | 4 | m4-11 | 40 | 5 | 11 | 8 | 5 | 29 | 2 | 4 | Invasive | 1998 | Connecticut |
| 1RP144 | 5 | m5-1 | 99 | 33 | 30 | 7 | 5 | 5 | 26 | 3 | URT | 1953 | New York |
| 87-292 | 5 | m5-2 | 99 | 33 | 30 | 7 | 5 | 5 | 26 | 3 | URT | 1987 | Ohio |
| CT95-118 | 5 | m5-3 | 99 | 33 | 30 | 7 | 5 | 5 | 26 | 3 | Invasive | 1995 | Connecticut |
| CT98302 | 5 | m5-4 | 99 | 33 | 30 | 7 | 5 | 5 | 26 | 3 | Invasive | 1998 | Connecticut |
| Manfredo | 5 | m5-5 | 99 | 33 | 30 | 7 | 5 | 5 | 26 | 3 | URT | 1958 | Illinois |
| MGAS254 | 5 | m5-6 | 99 | 33 | 30 | 7 | 5 | 5 | 26 | 3 | Invasive | ND | California |
| MGAS258 | 5 | m5-7 | 99 | 33 | 30 | 7 | 5 | 5 | 26 | 3 | Invasive | ND | California |
| 1RP112 | 6 | m6-1 | 37 | 5 | 2 | 5 | 5 | 5 | 4 | 3 | URT | 1952 | New York |
| D471 | 6 | m6-2 | 37 | 5 | 2 | 5 | 5 | 5 | 4 | 3 | ND | 1971 | Egypt |
| 87-285 | 6 | m6-3 | 37 | 5 | 2 | 5 | 5 | 5 | 4 | 3 | URT | 1987 | Utah |
| 87-376 | 6 | m6-4 | 37 | 5 | 2 | 5 | 5 | 5 | 4 | 3 | URT | 1987 | Colorado |
| CT95-115 | 6 | m6-5 | 37 | 5 | 2 | 5 | 5 | 5 | 4 | 3 | Invasive | 1995 | Connecticut |
| MGAS291 | 6 | m6-6 | 37 | 5 | 2 | 5 | 5 | 5 | 4 | 3 | Invasive | ND | Colorado |
| 29740 | 8 | m8 | 59 | 13 | 2 | 8 | 19 | 1 | 3 | 4 | Impetigo | 1988 | Czech Republic |
| D733 | 9 | m9-1 | 73 | 15 | 14 | 7 | 7 | 19 | 3 | 1 | ND | 1973 | Unknown |
| 1RP278 | 9 | m9-2 | 74 | 15 | 14 | 7 | 16 | 19 | 3 | 1 | URT | 1964 | New York |
| MGAS280 | 9 | m9-3 | 75 | 15 | 14 | 7 | 18 | 19 | 3 | 1 | Invasive | ND | Colorado |
| CT95-126 | 11 | m11-1 | 20 | 3 | 4 | 6 | 5 | 1 | 5 | 1 | Invasive | 1995 | Connecticut |
| CT95-155 | 11 | m11-2 | 20 | 3 | 4 | 6 | 5 | 1 | 5 | 1 | Invasive | 1995 | Connecticut |
| CT98303 | 11 | m11-3 | 20 | 3 | 4 | 6 | 5 | 1 | 5 | 1 | Invasive | 1998 | Connecticut |
| CT98414 | 11 | m11-4 | 20 | 3 | 4 | 6 | 5 | 1 | 5 | 1 | Invasive | 1998 | Connecticut |
| CT98538 | 11 | m11-5 | 21 | 3 | 4 | 6 | 5 | 1 | 5 | 2 | Invasive | 1998 | Connecticut |
| ALAB79 | 11 | m11-6 | 22 | 3 | 4 | 6 | 7 | 1 | 5 | 4 | Impetigo | 1986 | Alabama |
| D691 | 11 | m11-7 | 23 | 3 | 4 | 6 | 7 | 11 | 5 | 4 | URT | 1972 | Trinidad |
| 2RP196 | 12 | m12-1 | 36 | 5 | 2 | 2 | 6 | 6 | 2 | 2 | URT | 1956 | New York |
| A374 | 12 | m12-2 | 36 | 5 | 2 | 2 | 6 | 6 | 2 | 2 | URT | 1960 | Trinidad |
| CT95-127 | 12 | m12-3 | 36 | 5 | 2 | 2 | 6 | 6 | 2 | 2 | Invasive | 1995 | Connecticut |
| CT98386 | 12 | m12-4 | 36 | 5 | 2 | 2 | 6 | 6 | 2 | 2 | Invasive | 1998 | Connecticut |
| CT98647 | 12 | m12-5 | 36 | 5 | 2 | 2 | 6 | 6 | 2 | 2 | Invasive | 1998 | Connecticut |
| MGAS278 | 12 | m12-6 | 36 | 5 | 2 | 2 | 6 | 6 | 2 | 2 | Invasive | ND | Colorado |
| CT95-122 | 13 | m13-1 | 89 | 24 | 2 | 3 | 5 | 1 | 3 | 1 | Invasive | 1995 | Connecticut |
| MGAS248 | 13 | m13-2 | 89 | 24 | 2 | 3 | 5 | 1 | 3 | 1 | Invasive | ND | California |
| 25RS84 | 14 | m14 | 85 | 20 | 22 | 2 | 3 | 27 | 18 | 14 | URT | 1942 | New York |
| 1GL217 | 17 | m17 | 83 | 19 | 21 | 1 | 1 | 5 | 3 | 3 | URT | 1946 | Illinois |
| 88-092 | 18 | m18-1 | 41 | 6 | 5 | 7 | 1 | 5 | 3 | 3 | URT | 1988 | Pennsylvania |
| 1RP62 | 18 | m18-2 | 42 | 6 | 5 | 7 | 1 | 5 | 6 | 3 | URT | 1949 | New York |
| 4RP104 | 18 | m18-3 | 42 | 6 | 5 | 7 | 1 | 5 | 6 | 3 | URT | 1951 | New York |
| 1RP268 | 18 | m18-4 | 42 | 6 | 5 | 7 | 1 | 5 | 6 | 3 | URT | 1963 | New York |
| A941 | 18 | m18-5 | 42 | 6 | 5 | 7 | 1 | 5 | 6 | 3 | URT | 1967 | New York |
| 87-282 | 18 | m18-6 | 42 | 6 | 5 | 7 | 1 | 5 | 6 | 3 | URT | 1987 | Ohio |
| 87-373 | 18 | m18-7 | 42 | 6 | 5 | 7 | 1 | 5 | 6 | 3 | URT | 1987 | Utah |
| MGAS300 | 18 | m18-8 | 42 | 6 | 5 | 7 | 1 | 5 | 6 | 3 | Invasive | ND | Washington |
| 1RP118 | 19 | m19-1 | 65 | 13 | 7 | 8 | 1 | 13 | 9 | 8 | URT | 1952 | New York |
| 1RP232 | 19 | m19-2 | 65 | 13 | 7 | 8 | 1 | 13 | 9 | 8 | URT | 1959 | New York |
| 1GL205 | 19 | m19-3 | 69 | 13 | 7 | 8 | 10 | 13 | 9 | 8 | URT | 1946 | Illinois |
| D709 | 19 | m19-4 | 69 | 13 | 7 | 8 | 10 | 13 | 9 | 8 | URT | 1973 | New York |
| B243 | 22 | m22-1 | 45 | 9 | 8 | 1 | 1 | 1 | 3 | 3 | ND | 1947 | United Kingdom |
| MGAS275 | 22 | m22-2 | 46 | 9 | 8 | 1 | 1 | 1 | 3 | 4 | Invasive | ND | Colorado |
| MGAS330 | 22 | m22-3 | 46 | 9 | 8 | 1 | 1 | 1 | 3 | 4 | Invasive | ND | ND |
| MGAS339 | 22 | m22-4 | 46 | 9 | 8 | 1 | 1 | 1 | 3 | 4 | Invasive | ND | Texas |
| 22RS72 | 24 | m24-1 | 66 | 13 | 7 | 8 | 1 | 13 | 12 | 8 | URT | 1941 | New York |
| 1RP284 | 24 | m24-2 | 70 | 13 | 18 | 8 | 1 | 13 | 9 | 8 | URT | 1964 | New York |
| D316 | 25 | m25 | 54 | 11 | 7 | 15 | 7 | 17 | 3 | 4 | Invasive | 1969 | New York |
| 11RS100 | 26 | m26 | 67 | 13 | 7 | 8 | 1 | 26 | 3 | 13 | URT | 1942 | New York |
| CT95-189 | 28 | m28-1 | 52 | 11 | 6 | 14 | 5 | 9 | 17 | 19 | Invasive | 1995 | Connecticut |
| CT98220 | 28 | m28-2 | 52 | 11 | 6 | 14 | 5 | 9 | 17 | 19 | Invasive | 1998 | Connecticut |
| CT98601 | 28 | m28-3 | 52 | 11 | 6 | 14 | 5 | 9 | 17 | 19 | Invasive | 1998 | Connecticut |
| MGAS255 | 28 | m28-4 | 52 | 11 | 6 | 14 | 5 | 9 | 17 | 19 | Invasive | ND | California |
| MGAS325 | 28 | m28-5 | 52 | 11 | 6 | 14 | 5 | 9 | 17 | 19 | Invasive | ND | ND |
| 3RP70 | 29 | m29-1 | 65 | 13 | 7 | 8 | 1 | 13 | 9 | 8 | URT | 1949 | New York |
| D470 | 29 | m29-2 | 65 | 13 | 7 | 8 | 1 | 13 | 9 | 8 | ND | 1971 | Egypt |
| 10RS101 | 32 | m32 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | URT | 1942 | New York |
| 13RS60 | 33 | m33-1 | 3 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | URT | 1941 | New York |
| A982 | 33 | m33-2 | 3 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | Impetigo | 1968 | New York |
| 6010-5 | 33 | m33-3 | 3 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | Impetigo | 1971 | Alabama |
| 29487 | 33 | m33-4 | 3 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | Impetigo | 1987 | Czech Republic |
| MGAS306 | 33 | m33-5 | 3 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | Invasive | ND | ND |
| C142 | 34 | m34 | 64 | 13 | 7 | 8 | 1 | 9 | 22 | 3 | ND | 1942 | Unknown |
| A457 | 36 | m36 | 47 | 9 | 16 | 1 | 20 | 25 | 3 | 16 | ND | 1961 | Yugoslavia |
| 19RS14 | 39 | m39 | 32 | 4 | 17 | 2 | 5 | 1 | 25 | 1 | Invasive | 1941 | New York |
| 1RS79 | 42 | m42 | 80 | 16 | 15 | 19 | 7 | 1 | 2 | 17 | Invasive | 1941 | New York |
| D407 | 43 | m43 | 4 | 2 | 2 | 1 | 2 | 2 | 3 | 2 | Invasive | 1971 | New York |
| C135 | 49 | m49-1 | 29 | 4 | 6 | 2 | 7 | 7 | 7 | 5 | URT | 1942 | New York |
| A945 | 49 | m49-2 | 29 | 4 | 6 | 2 | 7 | 7 | 7 | 5 | Impetigo | 1966 | Minnesota |
| B737 | 49 | m49-3 | 30 | 4 | 6 | 2 | 7 | 21 | 7 | 1 | Impetigo | 1957 | Minnesota |
| MGAS163 | 49 | m49-4 | 34 | 4 | 29 | 2 | 7 | 7 | 7 | 4 | Invasive | ND | Louisiana |
| A203 | 50/62 | m50 | 2 | 1 | 2 | 20 | 5 | 35 | 3 | 4 | Mouse | 1959 | New York |
| MGAS323 | 50/62 | m62-1 | 2 | 1 | 2 | 20 | 5 | 35 | 3 | 4 | Invasive | ND | Idaho |
| MGAS324 | 50/62 | m62-2 | 2 | 1 | 2 | 20 | 5 | 35 | 3 | 4 | Invasive | ND | Idaho |
| A291 | 51 | m51 | 84 | 20 | 22 | 2 | 3 | 27 | 18 | 12 | ND | 1960 | Delaware |
| A946 | 52 | m52 | 43 | 7 | 2 | 8 | 3 | 8 | 2 | 2 | Impetigo | 1971 | Trinidad |
| D948 | 53 | m53-1 | 11 | 2 | 6 | 1 | 2 | 2 | 2 | 2 | Impetigo | 1976 | Trinidad |
| ALAB49 | 53 | m53-2 | 11 | 2 | 6 | 1 | 2 | 2 | 2 | 2 | Impetigo | 1986 | Alabama |
| AP53 | 53 | m53-3 | 11 | 2 | 6 | 1 | 2 | 2 | 2 | 2 | Impetigo | ND | Minnesota |
| D488 | 55 | m55 | 100 | 34 | 2 | 2 | 21 | 1 | 29 | 16 | Impetigo | 1971 | Trinidad |
| D306 | 57 | m57 | 33 | 4 | 26 | 23 | 7 | 1 | 28 | 3 | ND | 1968 | Trinidad |
| 29454 | 58 | m58 | 19 | 3 | 2 | 3 | 3 | 3 | 3 | 3 | Impetigo | 1987 | Czech Republic |
| 4500-S | 60 | m60 | 53 | 11 | 6 | 22 | 7 | 9 | 2 | 17 | Impetigo | 1960 | Alabama |
| A956 | 63 | m63 | 88 | 23 | 24 | 2 | 3 | 23 | 19 | 18 | ND | 1967 | Unknown |
| ALAB53 | 66 | m66-1 | 44 | 8 | 7 | 8 | 8 | 9 | 3 | 1 | Impetigo | 1986 | Alabama |
| MGAS167 | 66 | m66-2 | 44 | 8 | 7 | 8 | 8 | 9 | 3 | 1 | Invasive | ND | Texas |
| D795 | 67 | m76 | 61 | 13 | 2 | 16 | 1 | 5 | 3 | 1 | ND | 1975 | Georgia |
| 5552-S | 68 | m68 | 86 | 21 | 31 | 1 | 3 | 28 | 3 | 4 | Impetigo | 1971 | Alabama |
| D998 | 70 | m70 | 24 | 3 | 4 | 24 | 5 | 4 | 23 | 6 | ND | 1976 | Japan |
| CT95-121 | 73 | m73 | 7 | 2 | 2 | 9 | 3 | 18 | 3 | 4 | Invasive | 1995 | Connecticut |
| 86-809 | 75 | m75-1 | 49 | 11 | 2 | 1 | 3 | 12 | 3 | 7 | URT | 1986 | Utah |
| CT95-154 | 75 | m75-2 | 49 | 11 | 2 | 1 | 3 | 12 | 3 | 7 | Invasive | 1995 | Connecticut |
| CT95-171 | 75 | m75-3 | 49 | 11 | 2 | 1 | 3 | 12 | 3 | 7 | Invasive | 1995 | Connecticut |
| MGAS264 | 75 | m75-4 | 49 | 11 | 2 | 1 | 3 | 12 | 3 | 7 | Invasive | ND | California |
| CT98200 | 76 | m76-1 | 50 | 11 | 6 | 3 | 6 | 6 | 27 | 4 | Invasive | 1998 | Connecticut |
| CS110 | 76 | m76-2 | 51 | 11 | 6 | 12 | 6 | 6 | 2 | 4 | ND | ND | Unknown |
| MGAS261 | 77 | m77-1 | 35 | 4 | 31 | 2 | 13 | 34 | 3 | 21 | Invasive | ND | California |
| CT95-159 | 77 | m77-2 | 63 | 13 | 6 | 2 | 3 | 23 | 3 | 11 | Invasive | 1995 | Connecticut |
| CT98594 | 77 | m77-3 | 63 | 13 | 6 | 2 | 3 | 23 | 3 | 11 | Invasive | 1998 | Connecticut |
| MGAS331 | 77 | m77-4 | 63 | 13 | 6 | 2 | 3 | 23 | 3 | 11 | Invasive | ND | New York |
| D812 | 78 | m78-1 | 76 | 16 | 2 | 8 | 3 | 1 | 3 | 1 | Impetigo | 1975 | Trinidad |
| CT95-157 | 78 | m78-2 | 79 | 16 | 2 | 11 | 3 | 1 | 11 | 1 | Invasive | 1995 | Connecticut |
| CT95-104 | 80 | m80 | 8 | 2 | 2 | 9 | 3 | 18 | 11 | 2 | Invasive | 1995 | Connecticut |
| 29665 | 81 | m81 | 57 | 11 | 23 | 2 | 7 | 24 | 3 | 2 | Impetigo | 1988 | Czech Republic |
| 2RSC3 | 38/40 | m38/40 | 81 | 17 | 19 | 1 | 1 | 1 | 15 | 3 | URT | 1942 | New York |
| 3RP141 | 44/61 | m44/61-1 | 25 | 4 | 2 | 3 | 11 | 17 | 3 | 1 | URT | 1953 | New York |
| D938 | 44/61 | m44/61-2 | 25 | 4 | 2 | 3 | 11 | 17 | 3 | 1 | Impetigo | 1976 | Trinidad |
| 1RP18RN | 44/61 | m44/61-3 | 31 | 4 | 10 | 3 | 6 | 14 | 10 | 4 | URT | 1946 | New York |
| 5569-S | 44/61 | m44/61-4 | 31 | 4 | 10 | 3 | 6 | 14 | 10 | 4 | Impetigo | 1971 | Alabama |
| MGAS296 | 82 (pt180) | m82 | 26 | 4 | 2 | 21 | 16 | 17 | 3 | 1 | Invasive | ND | Washington |
| ALAB55 | 83 (pt2110) | m83-1 | 5 | 2 | 2 | 2 | 3 | 2 | 3 | 2 | Impetigo | 1986 | Alabama |
| 29689 | 83 (pt2110) | m83-2 | 5 | 2 | 2 | 2 | 3 | 2 | 3 | 2 | Impetigo | 1988 | Czech Republic |
| D943 | 84 (pt2233) | m84-1 | 58 | 12 | 2 | 1 | 3 | 12 | 8 | 7 | Impetigo | 1976 | Trinidad |
| MGAS1914A | 84 (pt2233) | m84-2 | 58 | 12 | 2 | 1 | 3 | 12 | 8 | 7 | ND | ND | ND |
| D964 | 86 (pt2631) | m86 | 9 | 2 | 2 | 9 | 3 | 18 | 14 | 2 | Impetigo | 1976 | Trinidad |
| 10RS57 | 87 (pt2841) | m87 | 62 | 13 | 3 | 10 | 12 | 20 | 2 | 3 | URT | 1941 | New York |
| D976 | 88 (pt3875) | m88 | 90 | 25 | 2 | 13 | 6 | 30 | 25 | 15 | ND | 1976 | Trinidad |
| MGAS2017 | 89 (pt4245) | m89-1 | 77 | 16 | 2 | 8 | 3 | 1 | 11 | 1 | URT | 1991 | United States |
| MGAS2018 | 89 (pt4245) | m89-2 | 77 | 16 | 2 | 8 | 3 | 1 | 11 | 1 | URT | 1992 | United States |
| CT95-176 | 89 (pt4245) | m89-3 | 77 | 16 | 2 | 8 | 3 | 1 | 11 | 1 | Invasive | 1995 | Connecticut |
| CT98677 | 89 (pt4245) | m89-4 | 77 | 16 | 2 | 8 | 3 | 1 | 11 | 1 | Invasive | 1998 | Connecticut |
| D424 | 89 (pt4245) | m89-5 | 78 | 16 | 2 | 8 | 7 | 1 | 13 | 1 | Invasive | 1971 | New York |
| 89-456 | 90 (pt4931) | m90 | 56 | 11 | 9 | 2 | 5 | 2 | 5 | 4 | URT | 1989 | Kuwait |
| C506 | 91 (pt5757) | m91-1 | 12 | 2 | 6 | 1 | 5 | 2 | 2 | 2 | URT | 1943 | Unknown |
| D821 | 91 (pt5757) | m91-2 | 13 | 2 | 6 | 2 | 5 | 22 | 3 | 2 | Impetigo | 1975 | Trinidad |
| 4426-S | 92 (st2974) | m92-1 | 82 | 18 | 20 | 7 | 9 | 2 | 2 | 15 | Impetigo | 1970 | Alabama |
| MGAS270 | 92 (st2974) | m92-2 | 82 | 18 | 20 | 7 | 9 | 2 | 2 | 15 | Invasive | ND | Minnesota |
| D466 | 93 (potter41) | m93-1 | 10 | 2 | 2 | 9 | 13 | 2 | 14 | 2 | ND | 1971 | Egypt |
| D502 | 93 (potter41) | m93-2 | 17 | 2 | 13 | 8 | 5 | 2 | 3 | 10 | Impetigo | 1971 | Trinidad |
| 2GL32 | st1RP31 | strp31-1 | 65 | 13 | 7 | 8 | 1 | 13 | 9 | 8 | URT | 1946 | Illinois |
| 1RP31 | st1RP31 | strp31-2 | 68 | 13 | 7 | 8 | 1 | 26 | 9 | 8 | URT | 1947 | New York |
| CT95-169 | st2346 | st2346-1 | 60 | 13 | 2 | 14 | 1 | 9 | 3 | 1 | Invasive | 1995 | Connecticut |
| MGAS319 | st2346 | st2346-2 | 60 | 13 | 2 | 14 | 1 | 9 | 3 | 1 | Invasive | ND | Idaho |
| 29486 | st2370.1 | st2370.1 | 6 | 2 | 2 | 8 | 3 | 2 | 16 | 12 | Impetigo | 1987 | Czech Republic |
| MGAS341 | st3365 | st3365 | 97 | 31 | 2 | 2 | 5 | 35 | 24 | 12 | Invasive | ND | Missouri |
| MGAS308 | st4529 | st4529 | 72 | 14 | 28 | 2 | 17 | 4 | 5 | 1 | Invasive | ND | Washington, D.C. |
| 6250-S | st4935 | st4935 | 27 | 4 | 3 | 2 | 5 | 2 | 26 | 1 | Impetigo | 1972 | Alabama |
| D997 | st4973 | st4973 | 98 | 32 | 6 | 8 | 3 | 2 | 8 | 22 | ND | 1976 | Japan |
| MGAS273 | st64/14 | m64/14-1 | 14 | 2 | 6 | 8 | 3 | 9 | 3 | 1 | Invasive | ND | California |
| MGAS2111 | st64/14 | m64/14-2 | 14 | 2 | 6 | 8 | 3 | 9 | 3 | 1 | ND | ND | ND |
| MGAS2140 | st833 | st833 | 95 | 29 | 32 | 2 | 5 | 33 | 21 | 4 | ND | ND | ND |
| A756 | stA207 | stA207-1 | 87 | 22 | 10 | 17 | 14 | 14 | 3 | 20 | Invasive | 1964 | New York |
| MGAS288 | stA207 | stA207-2 | 87 | 22 | 10 | 17 | 14 | 14 | 3 | 20 | Invasive | ND | Colorado |
| D432 | stD432 | stD432 | 71 | 14 | 12 | 2 | 7 | 16 | 5 | 9 | ND | 1971 | Egypt |
| D626e | stD626 | stD626 | 9 | 2 | 2 | 9 | 3 | 18 | 14 | 2 | Impetigo | 1972 | Trinidad |
| D631 | stD631 | stD631 | 96 | 30 | 2 | 9 | 13 | 18 | 14 | 2 | Impetigo | 1972 | Trinidad |
| D633 | stD633 | stD633 | 48 | 10 | 6 | 2 | 6 | 10 | 3 | 6 | Impetigo | 1972 | Trinidad |
| D641 | stNS5 | stNS5 | 11 | 2 | 6 | 1 | 2 | 2 | 2 | 2 | Impetigo | 1972 | Trinidad |
Note: emm13 is equivalent to emm13W, emm77 is equivalent to emm27L, and emm50 is equivalent to emm62 (http://www.cdc.gov/ncidod/biotech/strep/strains.html).
Codes for strains were used in dendrogram (Fig. 1).
URT, upper respiratory tract.
ND, not determined.
The sequences of the seven loci were determined for each of the 212 GAS isolates, and their allelic profiles were assigned. One hundred different allelic profiles were found, corresponding to ST1 through ST100. Sixty-six of the 100 STs were represented by only a single isolate; the number of isolates assigned to the other STs ranged from 2 to 16.
The average number of alleles per locus was 28.1, and therefore, the GAS MLST scheme is able to distinguish >13 billion different allelic profiles. An isolate with the most common allele at each of the seven loci is expected to occur, by chance, at a frequency of 7.5 × 10−5 (no isolates with this allelic profile were found among the 212 strains); most allelic profiles will occur by chance at much lower frequencies. Thus, it is extremely unlikely that two unrelated GAS isolates will have the same allelic profile.
Relationships between emm type and MLST.
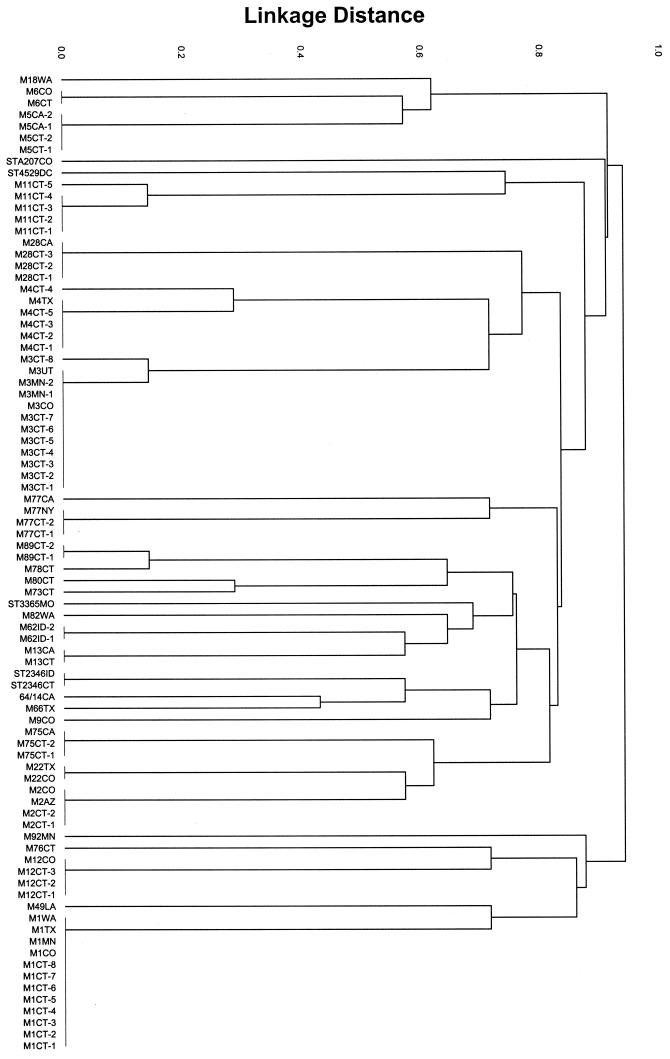
A matrix of pair-wise differences in allelic profiles was determined, and a dendrogram displaying the genetic linkage distance between the 212 isolates was constructed by cluster analysis using UPGMA (Fig. 1). In the dendrogram presented in Fig. 1, the 15 STs that are represented by four or more isolates are depicted. In 13 of these STs, all isolates are of a singular emm type. Is was of interest to further ascertain the strength of the associations between emm types and ST among GAS. Or, in other words, how well does emm type equate to clone?
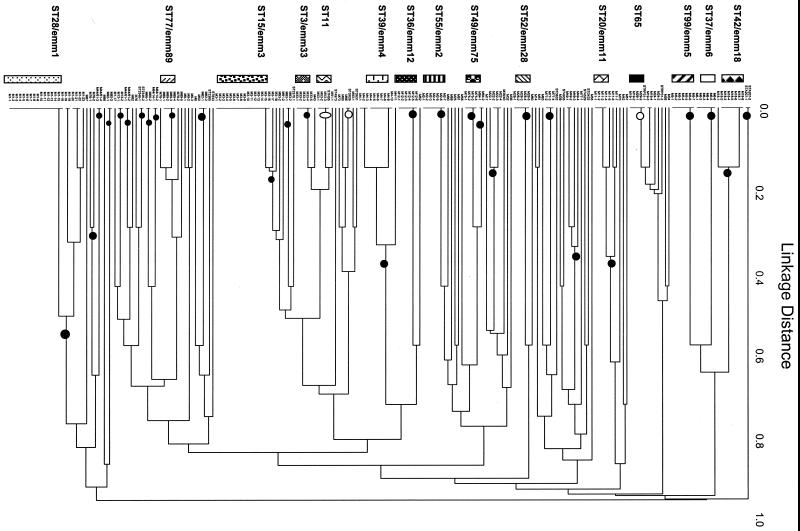
FIG. 1.
Dendrogram showing UPGMA cluster analysis of 212 GAS isolates. Bars to the left show allelic profiles (STs) represented by four or more isolates. Codes for strain designations at branch tips are listed in Table 2. Filled circles (n = 28) mark branches in which multiple descendants are all represented by a single emm type. Open circles (n = 3) mark branches containing isolates with different emm types but sharing identical allelic profiles.
For analysis of the relationships between emm type and MLST, selection criteria for GAS isolates were set to minimize the inclusion of epidemiologically related clones. Therefore, our analysis was specifically intended to provide a conservative estimate of the strength of the association between emm type and allelic profile. Multiple isolates of the same emm type and ST combination were included in the analysis only if they were recovered from subjects located on different continents or isolated >1 year apart within the same continent. Also, at least one representative of all unique emm type-ST combinations were included. emm types represented by four or more isolates satisfying the above-stated epidemiologic criteria (n = 15 emm types and 81 isolates in total) were assessed for the genetic distances between all possible pair-wise comparisons of alleles of the seven housekeeping loci (Table 3). This provides a measure of the genetic diversity at multiple loci within a set of epidemiologically unrelated organisms that share an emm type.
TABLE 3.
Pair-wise comparisons of housekeeping alleles among isolates of the same emm type
| emm typea | No. of isolates | No. of pair-wise comparisons | % of pairs with identical loci:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 7 of 7 | 6 of 7 | 5 of 7 | 4 of 7 | 3 of 7 | 2 of 7 | 1 of 7 | 0 of 7 | |||
| 1 | 9 | 36 | 28 | 3 | 3 | 17 | 28 | 0 | 8 | 4 |
| 2 | 5 | 10 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 9 | 36 | 58 | 39 | 3 | 0 | 0 | 0 | 0 | 0 |
| 4 | 8 | 28 | 32 | 43 | 14 | 11 | 0 | 0 | 0 | 0 |
| 5 | 4 | 6 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 4 | 6 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 5 | 10 | 10 | 30 | 30 | 30 | 0 | 0 | 0 | 0 |
| 12 | 5 | 10 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 7 | 21 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 4 | 6 | 33 | 67 | 0 | 0 | 0 | 0 | 0 | 0 |
| 33 | 5 | 10 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 44/61 | 4 | 6 | 33 | 0 | 0 | 0 | 0 | 67 | 0 | 0 |
| 49 | 4 | 6 | 17 | 0 | 67 | 17 | 0 | 0 | 0 | 0 |
| 77 | 4 | 6 | 50 | 0 | 0 | 0 | 0 | 50 | 0 | 0 |
| 89 | 4 | 6 | 50 | 0 | 50 | 0 | 0 | 0 | 0 | 0 |
| Total | 81 | 203 | ||||||||
| Average | 56.7 | 16.7 | 7.9 | 6.4 | 4.9 | 3.4 | 1.5 | 2.5 | ||
| All STs | ||||||||||
| Total | 100 | 4,950 | ||||||||
| Average | 0 | 0.4 | 0.7 | 1.1 | 3.4 | 13 | 35 | 47 | ||
In order to avoid comparisons between epidemiologically related clones, for each emm type, all isolates were collected ≥1 year apart or from different continents, or isolates differed in allelic profile. All emm types listed in Table 2, of which four or more isolates meet the above-stated criteria, are included in the analysis.
For six of the 15 emm types assessed (emm2, emm5, emm6, emm12, emm18, emm33), representing a total of 30 isolates, all isolates within an emm type displayed identical allelic profiles and can be regarded as clones (Table 3). Identical allelic profiles were observed for some organisms isolated >50 years apart (Table 2), indicating that GAS clones can be stable over this prolonged time period. One emm type (emm19) had isolates differing at one locus only, whereas two emm types had isolates differing at two loci (emm3, emm89). Isolates differing at two or fewer housekeeping loci (out of seven) can be regarded as clones or clonal complexes (16).
For epidemiologically distant organisms, as defined above, that were represented by only two or three isolates of the same emm type (n = 18 emm types), 11 emm types had identical allelic profiles, whereas five emm types differed at only one or two of the seven loci (Table 2). Although in some instances the sample size was small, emm type appears to closely correlate with clone or clonal complex for the majority (25 out of 33, or 76%) of emm types studied.
For several emm types represented by four or more epidemiologically distant isolates, there was a higher degree of genetic diversity. For three emm types—emm4, emm11, and emm49—pair-wise comparisons showed differences among three of the seven housekeeping loci (Table 3). An additional three emm types displayed differences at five or more of the housekeeping loci: emm1, emm44/61, and emm77 (also known as emm27L/77). Perhaps it is of biological relevance that isolates of two of the emm types (emm44/61 and emm77) were recently reported to be found in association with more that one sof allele, which provides the basis for a second major serological typing scheme for GAS (4). For emm1 isolates, pair-wise comparisons indicated that this group is the most genetically diverse (Table 3). However, of the nine epidemiologically distant isolates evaluated, eight differed from one another at three or fewer of the seven loci (Table 2); furthermore, the emm1 isolates cluster together, and there is a single node on the dendrogram from which all but one of the 23 emm1 isolates descend (Fig. 1). One emm1 isolate (MGAS2110; ST91) differs from the other emm1 isolates at six or seven of the seven housekeeping loci. In addition to the emm1, emm44/61, and emm77 isolates, the only other examples found for a single emm type on widely divergent genetic backgrounds are emm91 and emm93, whereby two isolates of each type differ at three and five of the seven housekeeping loci, respectively (Table 2).
The genetic distances within an emm type can be compared to the genetic distance between the 100 different STs identified. By definition, none of the isolates representing each of the 100 unique STs shared alleles at all seven of the housekeeping loci. Whereas the majority of epidemiologically distant isolates within an emm type differed at two or fewer loci, 95% of the distinct allelic profiles (i.e., ST1 through ST100) differed from each other at five or more loci (Table 3). Furthermore, nearly half of the 4,950 possible pair-wise comparisons among the 100 STs differed at all seven housekeeping loci. Thus, comparisons between individual GAS clones most often reveal large genetic distances, contrasting sharply with the similar genotypes typically found within an emm type.
There were several examples of isolates with identical allelic profiles that differed in emm type: emm86 and emmstD626 (ST9), emm53 and emmstNS5 (ST11), and emm19, emm29, and emmstRP31 (ST65) (Fig. 1). It is extremely unlikely that these examples of multiple emm types within a clone are due to a lack of discrimination of the GAS MLST scheme. For example, a single isolate with the allelic profile of ST65 was expected to occur by chance in the data set at a frequency of 2.2 × 10−8, and the likelihood of unrelated emm19, emm29, and emmstRP31 isolates having this allelic profile is essentially zero.
One emm type present on two or more genetically distant backgrounds, or multiple emm types present on a single genetic background, may have arisen as a consequence of the lateral movement of emm genes between different GAS strains. In GAS, generalized transduction by bacteriophage is the most probable mechanism for horizontal gene transfer.
Levels of linkage disequilibrium within the GAS population.
The extent of recombination within the GAS population was assessed by the Index of Association (27). Using one isolate of each of the 100 STs, there was significant linkage disequilibrium between the alleles at each of the seven housekeeping loci. However, in populations in which recombination is sufficient to randomize the alleles at different loci over a longer term, the recent expansion of clones can result in the appearance of multiple isolates with similar genotypes (27). Therefore, the Index of Association was recalculated using one isolate of each of the 72 STs obtained by truncating the dendrogram (Fig. 1) at a genetic distance of 0.3; no significant linkage disequilibrium between alleles was observed. The truncation effectively reduced each clonal complex to a single representative strain and thereby diminished any bias introduced by the oversampling of select emm types.
Comparison of MLST to other typing methods.
The high degree of concordance between ST and emm type provides strong evidence that the MLST typing scheme leads to accurate identification of clones or clonal complexes. The MLST scheme can be further validated by comparison to other typing methods. Isolates that had been previously assessed by MLEE, as reported by others (22, 29, 30), were compared for emm type, ST, and electrophoretic type (ET) (Table 4). For organisms represented by one or more isolates of the same emm type-ST combination, 20 were also concordant for ET, whereas 9 were discordant with ET; however, for the discordant ETs, several were genetically close in their relationship. For organisms represented by one or more isolates of the same emm type-ET combination, 20 out of 21 were also concordant for ST.
TABLE 4.
Comparison of MLST to other typing methods
| Straina | emm type | MLST-ST | ET based on MLEE | RAPD profile |
|---|---|---|---|---|
| MGAS166 | 1 | 28 | 1 | |
| MGAS307 | 1 | 28 | 1 | |
| MGAS302 | 1 | 28 | 15 | |
| MGAS283 | 1 | 28 | 25 | |
| MGAS2127 | 1 | 28 | 1D | |
| MGAS2110 | 1 | 91 | 1B | |
| MGAS2120 | 1 | 92 | 1C | |
| MGAS2125 | 1 | 92 | 1C | |
| MGAS2109 | 1 | 93 | 1F | |
| MGAS2144 | 1 | 93 | 1F | |
| MGAS2226 | 1 | 93 | 1F | |
| MGAS2123 | 1 | 94 | 1F | |
| MGAS327 | 2 | 55 | 4 | |
| MGAS286 | 2 | 55 | 6 | |
| MGAS157 | 3 | 15 | 2 | |
| MGAS159 | 3 | 15 | 2 | |
| MGAS268 | 3 | 15 | 2 | |
| MGAS277 | 3 | 15 | 20 | |
| MGAS254 | 5 | 99 | 28 | |
| MGAS258 | 5 | 99 | 28 | |
| MGAS300 | 18 | 42 | 20 | |
| 1RP232 | 19 | 65 | 45 | |
| MGAS330 | 22 | 46 | 23 | |
| MGAS339 | 22 | 46 | 23 | |
| MGAS275 | 22 | 46 | 24 | |
| 1RP284 | 24 | 70 | 45 | |
| D316 | 25 | 54 | 55 | |
| MGAS255 | 28 | 52 | 10 | |
| MGAS325 | 28 | 52 | 10 | |
| D470 | 29 | 65 | 46 | |
| MGAS323 | 50/62 | 2 | 9 | |
| MGAS324 | 50/62 | 2 | 9 | |
| MGAS264 | 75 | 49 | 18 | |
| 86-809 | 75 | 49 | 59 | |
| MGAS261 | 77 | 35 | 4 | |
| MGAS331 | 77 | 63 | 13 | |
| MGAS2017 | 89 | 77 | 67 | |
| MGAS2018 | 89 | 77 | 67 | |
| CT95-120 | 1 | 28 | 1 | |
| CT95-111 | 1 | 28 | 1 | |
| CT95-131 | 1 | 28 | 1 | |
| CT95-132 | 1 | 28 | 1 | |
| CT95-180 | 1 | 28 | 1 | |
| CT95-133 | 3 | 15 | 2 | |
| CT95-145 | 3 | 15 | 3 | |
| CT95-204 | 3 | 15 | 4 | |
| CT95-117 | 3 | 15 | 5 | |
| CT95-119 | 3 | 15 | 5 | |
| CT95-186 | 3 | 15 | 8 | |
| CT95-200 | 3 | 18 | 5 | |
| CT95-155 | 11 | 20 | 19 | |
| CT95-126 | 11 | 20 | 19 | |
| CT95-154 | 75 | 49 | 21 | |
| CT95-171 | 75 | 49 | 22 | |
| CT95-159 | 77 | 63 | 15 | |
| CT95-176 | 89 | 77 | 15 |
Arbitrary-primed PCR, yielding random amplified polymorphic DNA (RAPD) profiles, has been previously conducted on another subset of the GAS isolates reported here (17). For organisms represented by one or more isolates of the same emm type-ST combination, nine also had concordant RAPD profiles, whereas seven displayed distinct RAPD profiles (Table 4). However, for organisms represented by one or more isolates of the same emm type-RAPD profile combination, 9 out of 10 were also concordant for ST.
Although the level of strain resolution differs for emm typing, MLEE, and RAPD analysis, each method displays high levels of concordance with the new MLST scheme.
GAS causing invasive disease.
A total of 84 GAS isolates associated with invasive disease in the United States between 1986 and 1999 were included in this study. Thirty distinct emm types were represented by 34 unique allelic profiles (Fig. 2). Among the subset of invasive disease isolates, there was a high one-to-one correspondence between emm type and ST. However, for the vast majority of pair-wise comparisons between invasive disease isolates of different emm types, there were differences at four or more loci. Therefore, invasive disease caused by GAS can be attributed to a large number of genetically diverse strains or clones, confirming other reports (2, 17, 29, 35). However, two major clusters of isolates with identical or very similar allelic profiles were identified. These two clusters contained isolates of emm1 and emm3, which are the emm types most commonly recovered from invasive disease in the United States during the 1990s (2, 17, 35).
FIG. 2.
Dendrogram of invasive isolates from the United States (1986 to 1999). UPGMA cluster analysis of all 84 isolates derived from normally sterile tissue sites, as listed in Table 2, is shown. The nomenclature at the branch tips indicate emm type, followed by the two-letter abbreviation of the state of origin and a unique isolate number (where necessary).
DISCUSSION
A primary objective of this report is to provide the foundation for a new typing scheme for GAS that can be readily expanded upon by other investigators. In general terms, the value of molecular typing schemes lies in their ability to discriminate between the various strains within a bacterial species. However, high levels of discrimination are often achieved by indexing variation that accumulates very rapidly, making it difficult to demonstrate the relatedness of isolates that have diversified from a common ancestor that existed many decades ago. Variation within the nucleotide sequences of housekeeping genes accumulates relatively slowly, and as demonstrated in this report, isolates with the same allelic profile can be recovered many decades apart. Although the genetic variation indexed by MLST accumulates slowly, the multilocus approach allows for a vast number of distinct genotypes to be distinguished. Furthermore, MLST has high resolving power and, in many instances, it can discriminate among isolates of a single emm type.
The clustering of isolates achieved by MLST was in good agreement with those obtained using other typing procedures, and thus, the GAS MLST scheme provides a validated method for the unambiguous identification of GAS isolates. Since it is based on nucleotide sequence data, MLST allows different laboratories to compare their results via the internet. A website containing an initial database of the allelic profiles and molecular properties of the 212 GAS isolates and associated epidemiological data, together with interrogation and analysis software, is available (http://www.mlst.net).
The organisms initially selected for analysis by MLST represented a total of 78 emm types, and their isolation from human subjects dates back nearly 60 years. A future goal is to apply the MLST scheme to at least one isolate of every known emm type, collected from worldwide sources. A thorough documentation of existing GAS clones will lay the groundwork for gaining a better understanding of the epidemiological trends underlying GAS disease and aid in deciphering the molecular basis for biological diversity within this species.
emm type provides the basis for a serological typing scheme that differentiates between antigenic epitopes contained within the amino-terminal, distal region of M-protein surface fibrils. Serum immunoglobulin G directed to M-type-specific epitopes leads to protective immunity for most strains that have been studied (1, 9, 25). Furthermore, the M proteins are key virulence factors, displaying a wide array of functional activities that act to promote disease (8). Unlike the housekeeping loci, emm genes are highly variable as a consequence of diversifying selection applied by the host immune response. It might therefore be expected that emm type would change more rapidly than alleles at housekeeping loci, resulting in variation within emm type among isolates of a clone or clonal complex. However, emm type is not defined by a unique nucleotide sequence but by ≥95% sequence identity. Consequently, descendants of an ancestral strain may accumulate as many as eight nucleotide changes (and small indels or frameshifts) within the 160-bp sequenced region of the emm gene without altering the emm type, whereas even a single nucleotide change in the ∼450 bp sequenced regions of any of the seven housekeeping loci results in a change in allelic profile. There are a few examples of isolates with identical allelic profiles having different emm types, such as ST65, which includes isolates of emm19, emm29, and emmst1RP31. Presumably, in these isolates, recombinational exchanges have resulted in the replacement of the region of the emm gene that defines emm type with the corresponding region from isolates of different emm types, since their divergence in emm type far exceeds 5%. Another multilocus typing method—MLEE—has also uncovered examples of isolates of the same genotype having different emm types (24, 30, 34).
A striking finding of this report is the degree to which multiple isolates of a given emm type share identical or highly similar allelic profiles (Table 3). Isolates of these emm types are considered to be clones or to form a clonal complex consisting of isolates with closely related allelic profiles. A much more extensive sampling of the GAS population will confirm the validity of this concept. The finding of a high one-to-one correspondence between emm type and clones or clonal complex suggests that GAS clones typically emerge and begin to diversify without changing their ancestral emm type. Recent studies using statistical tests of congruence between different housekeeping loci have indicated that recombination may be relatively common in GAS (16). This view was also supported by the lack of significant linkage disequilibrium between alleles that was observed when multiple isolates with similar genotypes were removed from the MLST data set, as measured by the Index of Association (27). Given this evidence for a major impact of recombination in the evolution of GAS populations, it is surprising that horizontal gene transfer appears to have rarely resulted in the presence of the same emm type in distantly related lineages. There are examples of this phenomenon, but they are uncommon. For example, among emm1 isolates (the most intensively sampled emm type), 22 of the 23 isolates form a cluster of lineages that all descend from the same relatively deep node (genetic distance of 0.5), whereas the other emm1 isolate differed from the former emm1 isolates at six or seven of the seven loci (Fig. 1; Table 2) (30).
MLST studies of Streptococcus pneumoniae have also shown that isolates with identical or closely related allelic profiles almost invariably have the same serotype. However, in contrast to the findings on GAS, there are often multiple examples of distantly related clones or clonal complexes sharing the same pneumococcal serotype. The paucity of distantly related GAS lineages sharing the same emm type may reflect differences in the strength of the immune response against pneumococcal capsular polysaccharides compared to that against M proteins, leading to differences in the strength of competitive exclusion between clones with the same capsular serotype or emm type. However, it might also be explained by the likelihood that changes in GAS serotype (i.e., emm type) occur by both mutation and recombination, whereas recombination involving the capsular biosynthetic genes is the only known mechanism underlying serotype changes in pneumococci (7). In the presence of strong selective immunological pressures, the diversification of emm genes might be further promoted by highly mutable processes, such as frameshift mutation and DNA slipped-strand mispairing (21, 28, 31). Unless recombinational exchanges that result in the presence of the same emm type in different lineages have occurred relatively recently, the diversifying selection applied by the host immune system is likely to result in the divergence of the emm types of the parental and recipient lineages. Thus, descendants of ancient horizontal genetic transfer events that distributed a particular pneumococcal capsular locus into multiple lineages may have retained the same serotype, whereas it is far less likely that the descendants of a similar ancient horizontal distribution of an emm gene will have retained the original emm type. The different extent to which the same capsular or M type is found in different lineages of pneumococci or GAS may rest more on the ease with which serotypes can change in these species, rather than differences in the rates of horizontal gene transfer.
The GAS MLST scheme provides a new and unambiguous method for characterizing GAS isolates for epidemiological purposes by using the internet. The MLST data can be used to address several epidemiological issues concerning GAS disease. Changes in epidemiological trends can be more readily ascribed to the emergence of new clones. Vaccine design strategies can be further refined, and vaccine efficacy can be measured with greater precision. The sequences of fragments of seven housekeeping genes from hundreds of GAS isolates provide data that can be used to address aspects of the population and evolutionary biology of the species. For example, the ancestral relationships and patterns of descent among closely related isolates can be deduced, although relationships between more distantly related isolates are likely to be obscured by a history of recombination (16). The population genetic structure of GAS, based on neutral housekeeping loci, will provide a framework upon which to measure the distribution of adaptive loci. This, in turn, should provide new insights into the molecular basis for biological diversity among GAS, as well as the role of cell surface antigens in structuring the population (19, 20).
ACKNOWLEDGMENTS
We thank Yury Nunez, Eric Peterson, and Michelle Benitez for expert technical assistance, Susan Hollingshead (UAB) for supplying the MGAS strains, and Jim Hadler and Nancy Barrett (CT DOH) for providing the invasive isolates collected in Connecticut during 1998 (CT98 series) and the emm-typing data. We also acknowledge the Streptococcal Genome Sequencing Project funded by USPHS/NIH grant AI-38406 and the work performed by B. A. Roe, S. P. Linn, L. Song, X. Yuan, S. Clifton, R. E. McLaughlin, M. McShan, and J. Ferretti.
This work was supported by grants from the Wellcome Trust (to B.G.S.), the National Institutes of Health (AI-28944 to D.E.B. and GM-60793 to D.E.B. and B.G.S.), the American Heart Association (grant-in-aid to D.E.B.), and a Brown-Coxe Postdoctoral Fellowship (to A.K.). M.C.E. is a Royal Society University Research Fellow. D.E.B. is an Established Investigator of the American Heart Association.
REFERENCES
- 1.Beachey E H, Seyer J M, Dale J B, Simpson W A, Kang A H. Type-specific protective immunity evoked by synthetic peptide of Streptococcus pyogenes M protein. Nature. 1981;292:457–459. doi: 10.1038/292457a0. [DOI] [PubMed] [Google Scholar]
- 2.Beall B, Facklam R, Hoenes T, Schwartz B. Survey of emm sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J Clin Microbiol. 1997;35:1231–1235. doi: 10.1128/jcm.35.5.1231-1235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beall B, Gherardi G, Lovgren M, Forwick B, Facklam R, Tyrrell G. Emm and sof gene sequence variation in relation to serological typing of opacity factor positive group A streptococci. Microbiology. 2000;146:1195–1209. doi: 10.1099/00221287-146-5-1195. [DOI] [PubMed] [Google Scholar]
- 5.Bessen D E, Carapetis J R, Beall B, Katz R, Hibble M, Currie B J, Collingridge T, Izzo M W, Scaramuzzino D A, Sriprakash K S. Contrasting molecular epidemiology of group A streptococci causing tropical and non-tropical infections of the skin and throat. J Infect Dis. 2000;182:1109–1116. doi: 10.1086/315842. [DOI] [PubMed] [Google Scholar]
- 6.Bessen D E, Izzo M W, Fiorentino T R, Caringal R M, Hollingshead S K, Beall B. Genetic linkage of exotoxin alleles and emm gene markers for tissue tropism in group A streptococci. J Infect Dis. 1999;179:627–636. doi: 10.1086/314631. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham M W. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dale J, Simmons M, Chiang E, Chiang E. Recombinant, octavalent group A streptococcal M protein vaccine. Vaccine. 1996;14:944–948. doi: 10.1016/0264-410x(96)00050-3. [DOI] [PubMed] [Google Scholar]
- 10.Desai M, Tanna A, Efstratiou A, George R, Clewley J, Stanley J. Extensive genetic diversity among clinical isolates of Streptococcus pyogenes serotype M5. Microbiology. 1998;144:629–637. doi: 10.1099/00221287-144-3-629. [DOI] [PubMed] [Google Scholar]
- 11.Dowson C, Coffey T, Spratt B. Penicillin-binding protein mediated resistance to beta-lactam antibiotics in naturally-transformable pathogens. Trends Microbiol. 1994;2:361–366. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 12.Enright M, Day N, Davies C, Peacock S, Spratt B. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright M, Spratt B. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 14.Enright M C, Spratt B G. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 15.Facklam R, Beall B, Efstratiou A, Fischetti V, Kaplan E, Kriz P, Lovgren M, Martin D, Schwartz B, Totolian A, Bessen D, Hollingshead S, Rubin F, Scott J, Tyrrell G. Report on an international workshop: demonstration of emm typing and validation of provisional M-types of group A streptococci. Emerg Infect Dis. 1999;5:247–253. doi: 10.3201/eid0502.990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feil E J, Holmes E C, Bessen D E, Chan M-S, Day N P J, Enright M C, Goldstein R, Hood D, Kalia A, Moore C E, Zhou J, Spratt B G. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci USA. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiorentino T R, Beall B, Mshar P, Bessen D E. A genetic-based evaluation of principal tissue reservoir for group A streptococci isolated from normally sterile sites. J Infect Dis. 1997;176:177–182. doi: 10.1086/514020. [DOI] [PubMed] [Google Scholar]
- 18.Gardiner D, Hartas J, Currie B, Mathews J D, Kemp D J, Sriprakash K S. Vir typing: a long-PCR typing methods for group A streptococci. PCR Methods App. 1995;4:288–293. doi: 10.1101/gr.4.5.288. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S, Anderson R. Population structure of pathogens: the role of immune selection. Parasitol Today. 1999;15:497–501. doi: 10.1016/s0169-4758(99)01559-8. [DOI] [PubMed] [Google Scholar]
- 20.Gupta S, Maiden M C J, Feavers I M, Nee S, May R M, Anderson R M. The maintenance of strain structure in populations of recombining infectious agents. Nat Med. 1996;2:437–442. doi: 10.1038/nm0496-437. [DOI] [PubMed] [Google Scholar]
- 21.Harbaugh M P, Podbielski A, Hugl S, Cleary P P. Nucleotide substitutions and small-scale insertion produce size and antigenic variation in group A streptococcal M1 protein. Mol Microbiol. 1993;8:981–991. doi: 10.1111/j.1365-2958.1993.tb01642.x. [DOI] [PubMed] [Google Scholar]
- 22.Kapur V, Topouzis S, Majesky M W, Li L-L, Hamrick M R, Hamill R J, Patti J M, Musser J M. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 23.Kehoe M A. Cell wall-associated proteins in Gram-positive bacteria. New Comphr Biochem. 1995;27:217–261. [Google Scholar]
- 24.Kehoe M A, Kapur V, Whatmore A M, Musser J M. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 1996;4:436–443. doi: 10.1016/0966-842x(96)10058-5. [DOI] [PubMed] [Google Scholar]
- 25.Lancefield R C. Current knowledge of the type specific M antigens of group A streptococci. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 26.Maiden M, Bygraves J, Feil E, Morelli G, Russell J, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D, Feavers I, Achtman M, Spratt B. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maynard Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 29.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D M. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of human disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Relf W A, Martin D R, Sriprakash K S. Antigenic diversity within a family of M proteins from group A streptococci: evidence for the role of frameshift and compensatory mutations. Gene. 1994;144:25–30. doi: 10.1016/0378-1119(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 32.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Upton M, Carter P, Orange G, Pennington T. Genetic heterogeneity of M type 3 group A streptococci causing severe infections in Tayside, Scotland. J Clin Microbiol. 1996;34:196–198. doi: 10.1128/jcm.34.1.196-198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whatmore A M, Kapur V, Sullivan D J, Musser J M, Kehoe M A. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol Microbiol. 1994;14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 35.Zurawski C A, Bardsley M, Beall B, Elliot J A, Facklam R, Schwartz B, Farley M M. Invasive group A streptococcal disease in metropolitan Atlanta: a population-based assessment. Clin Infect Dis. 1998;27:150–157. doi: 10.1086/514632. [DOI] [PubMed] [Google Scholar]