Abstract
Simple Summary
Capybaras are known hosts for various tick species, but there are limited data regarding the tick-borne pathogens they can carry. We assessed the presence of piroplasmids and Ehrlichia spp. in capybaras and their associated ticks in Goiás state, central-western Brazil. Neither capybaras nor ticks were positive for Ehrlichia spp. However, we detected an undescribed species of protozoan in both the capybaras and ticks. Further research is required for a formal delineation of this protozoan species, as well as to investigate the role of these ticks as vectors and the possible pathogenicity of this parasite to other animals, including horses.
Abstract
Capybaras (Hydrochoerus hydrochaeris) are the largest rodents on Earth. While capybaras are hosts for various tick species, there is limited information regarding the tick-borne pathogens they can carry. We investigated the presence of piroplasmids and Ehrlichia spp. in capybaras and their associated ticks in two peri-urban areas in Goiás state, central-western Brazil. Blood samples collected from 23 capybaras were used to investigate the presence of piroplasmids and Ehrlichia spp. in stained-blood smears and by PCR. Ticks collected from the capybaras were identified morphologically and also tested using PCR for the same pathogens. A total of 955 ticks were collected, including 822 (86.1%) Amblyomma sculptum, 132 (13.8%) Amblyomma dubitatum, and one (0.1%) unidentified larva of Amblyomma sp. Neither the capybaras nor ticks were positive for Ehrlichia spp. However, a stained-blood smear examination revealed the presence of ring-stage and pyriform-shaped merozoites in the erythrocytes of one (4.4%) capybara. In the same way, 47.8% (11/23) and 19.9% (36/181) of blood samples and ticks, respectively, were positive for piroplasmids in the PCR. We successfully sequenced a partial 18S rRNA gene fragment of four samples (two capybaras, one A. sculptum, and one A. dubitatum), and the phylogenetic reconstruction disclosed that the organism reported in the present study clusters within the genus Babesia. Further research is required for a formal delineation of this species (designated as Babesia sp. strain Capybara) and to investigate the hypothesis of A. dubitatum and A. sculptum ticks being vectors.
Keywords: Babesia sp., Amblyomma sculptum, Amblyomma dubitatum, rodents
1. Introduction
Capybaras (Hydrochoerus hydrochaeris (Linnaeus, 1766)) are the biggest rodents on Earth and are primarily associated with forests, seasonally flooded savannas, and wetlands. However, the habitat loss due to the extensive urbanization process in the major Brazilian regions has forced capybaras and many other animals to adapt to living in proximity to humans [1,2], posing risks to themselves, domestic animals, and humans. For instance, they can cause crop damage [3]. If they feel threatened, capybaras can attack other animals and humans, and the consequences of bites can be severe [4,5]. Moreover, they can also carry ticks and tick-borne pathogens that may infect animals and humans [6].
In this regard, capybaras are primary hosts for Amblyomma sculptum (Berlese, 1888), the principal vector of Rickettsia rickettsii (Wolbach, 1919), the etiological agent of Rocky Mountain spotted fever or Brazilian spotted fever [7,8,9]. Beyond acting as hosts for A. sculptum, capybaras serve as a source of R. rickettsii infection for ticks, playing an epidemiological role in the transmission of this pathogen in Brazil [7,8,9]. Excluding Rickettsia spp., there is scant information about other tick-borne pathogens (e.g., Anaplasma spp., Babesia spp., Ehrlichia spp., and Theileria spp.) that could possibly infect capybaras. For instance, Criado-Fornelio et al. [10] detected a piroplasmid (order Piroplasmida) in a capybara from southern Brazil that shared a 90% identity with Theileria equi (Laveran, 1901). More recently, Gonçalves et al. [11] detected a piroplasmid in a nymph of Amblyomma dubitatum Neumann, 1899, collected from a black rat (Rattus rattus Linnaeus, 1758), and in a female of the same tick species collected from a capybara. The obtained 18S rRNA sequences shared, respectively, 99.4% and 97.2% identity, respectively, with the sequence previously reported by Criado-Fornelio et al. [10].
In the present study, we investigated the presence of piroplasmids and Ehrlichia spp. in capybaras and their associated ticks in two peri-urban areas in central-western Brazil. We also molecularly characterized a piroplasmid detected in capybaras and their ticks by conducting a comprehensive phylogenetic assessment based on piroplasmid 18S rRNA gene sequences available in GenBank.
2. Materials and Methods
2.1. Study Area
From July 2020 to April 2022, ticks and blood samples were collected from capybaras (n = 23) captured on the campus of the Federal University of Goiás (Site 1) (n = 17) (16°35′42″ S, 49°16′50″ W, 718 m altitude) and in a residential park (Site 2) (n = 6) (16°36′26″ S, 49°10′27″ W, 844 m altitude), Goiânia City, Goiás, central-western Brazil; five capybaras were recaptured (three recaptured once and two twice).
Other animals—i.e., cattle, coatis [Nasua nasua (Linnaeus, 1766)], black-striped capuchin [Sapajus libidinosus (Spix, 1823)]—were present in these areas, but we did not collect ticks from them.
2.2. Ticks and Blood Sample Collection
Capybaras were captured by using 90 m2 corrals baited with corn, corn silage, banana leaves, and sugar cane. The frequency of captures varied according to the year. In particular, captures were carried out weekly from July 2020 to December 2021 and monthly from January to April 2022. Capybaras were physically restrained with a net and anesthetized with an intramuscular injection of ketamine (10 mg/kg) plus xylazine (0.5 mg/kg). Under anesthesia, a 3 mL blood sample was withdrawn from the saphenous vein of each capybara, and a thorough physical was conducted to detect any attached ticks. Capybaras were identified with a subcutaneous microchip (Allflex), clinically monitored during the procedure until recovery from anesthesia, and released at the same capture site. Collected ticks were placed in plastic tubes and sent to the laboratory for species identification, according to the morphological keys for nymphs and adults [12,13]. Because there is no taxonomic key for the larvae of Brazilian Amblyomma spp., larvae were identified on the genus level only [14].
2.3. Blood Smear, DNA Extraction, and PCR Testing
Blood smears were prepared immediately after blood collection and stained with a rapid stain kit (Panótico Rápido, Laborclin, Brazil) for cytological examination under a standard light microscope.
DNA from whole blood samples (200 µL) was extracted using DNeasy® Blood and Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. Tick DNA was extracted individually using the guanidine isothiocyanate protocol for adults [15] and the boiling protocol for nymphs [16].
Extracted DNA from ticks and blood samples were tested using conventional PCR protocols targeting a 378 bp fragment of the Ehrlichia spp. The dsb gene (forward primer: TTGCAAAATGATGTCTGAAGATATGAAACA; reverse primer: GCTGCTCCACCAATAAATGTATCYCCTA) [17] and a 551 bp fragment of the 18S rRNA gene of piroplasmids (forward primer: CCGTGCTAATTGTAGGGCTAATACA; reverse primer: GCTTGAAACACTCTARTTTTCTCAAAG) [18]. Negative DNA samples were further tested using PCR protocols targeting the 16S rDNA gene of ticks or the cytochrome b gene of mammals [19,20] in order to validate the DNA extraction protocol.
2.4. DNA Sequencing and Bioinformatic Analyses
PCR amplicons were purified using the SV Gel and PCR Clean-Up System kit (Wizard, Madison, WI, USA) according to the manufacturer’s instructions. Then, amplicons were prepared using the BigDye terminator v3.1 matrix standard kit (Applied Biosystems, Foster City, CA, USA), and forward and reverse sequencing reactions were carried out in a 3500× L genetic analyzer (Applied Biosystems, Foster City, CA, USA) using the same primers as for the PCR. The yielded reads were analyzed using the Staden package [21], and contig assembly was performed using sense and antisense reads based on a Phred quality score ≥ 30. Sequence similarity searching was performed using the Basic Local Alignment Search Tool (BLASTn) (http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 2 November 2022)) and Piroplasmida (taxid:5863) database.
Sequence alignment was performed using the MAFFT algorithm [22], approaching the integrative refinement method FFT-NS-i. Phylogenetic analysis was performed as described elsewhere [23]. In brief, phylogenetic reconstruction was performed using maximum likelihood inference using IQ-TREE version 1.6.12 [24] approaching an ultrafast bootstrap (1000 replicates) method. The best-fit evolutive model was calculated using ModelFinder [25], implemented in IQ-TREE, and selected according to the Bayesian Information Criterion (BIC). The ML tree was visualized and edited using the iTOL v.4 tool [26]. Genetic distances were calculated using MEGA7 [27] between intra- and interspecific groups (i.e., between the Babesia sensu stricto clade and the clade encompassing parasites detected in rodents from Brazil and the organism reported in the present study).
3. Results
A total of 955 ticks were collected, including 822 (86.1%) A. sculptum (460 nymphs, 236 males, and 126 females), 132 (13.8%) A. dubitatum (45 nymphs, 42 males, and 45 females), and one (0.1%) unidentified larva of Amblyomma sp. (Table 1). Amblyomma sculptum predominated in both sites, representing 83.7% and 97.6% of the ticks collected on the university campus (Site 1) and in the residential park (Site 2), respectively.
Table 1.
Number of tick-infested and piroplasmid-positive capybaras according to each collection site. The number and species of ticks identified, as well as the number of positive ticks, is also shown.
| Site | Number of Capybaras Examined (n) | Number of Piroplasmid-Positive Capybaras (n, %) | Number of Tick-Infested Capybaras (n, %) | Number of Ticks Collected (n) | Number of Ticks Tested (n)/Number of Piroplasmid-Positive Ticks (n, %) | Total Number of Ticks Tested/Total Number Piroplasmid-Positive Ticks (n, %) | |||
|---|---|---|---|---|---|---|---|---|---|
| Amblyomma sculptum | Amblyomma dubitatum | Amblyomma sp. | Amblyomma sculptum | Amblyomma dubitatum | |||||
| Site 1 | 17 | 11, 64.7% | 17, 100% | 656 | 128 | 1 | 99/19, 19.2% | 26/12, 46.1% | 125/31, 24.8% |
| Site 2 | 6 | 0, 0% | 6, 100% | 166 | 4 | 0 | 56/5, 8.9% | 0/0, 0% | 56/5, 8.9% |
| Total | 23 | 11, 47.8% | 23, 100% | 822 | 132 | 1 | 155/24, 15.4% | 26/12, 46.1% | 181/36, 19.9% |
Overall, 181 ticks were used for DNA extraction, including 149 adults (66 females and 83 males) and six nymphs of A. sculptum and 26 adults (11 females and 15 males) of A. dubitatum. None of the ticks contained Ehrlichia spp. DNA. On the other hand, a Piroplasmida 18S rRNA gene fragment was successfully amplified from 19.9% (36/181) of the ticks, 66.7% (24/36) of which were A. sculptum (13 females and 11 males), and 33.3% (12/36) were A. dubitatum (three females and nine males).
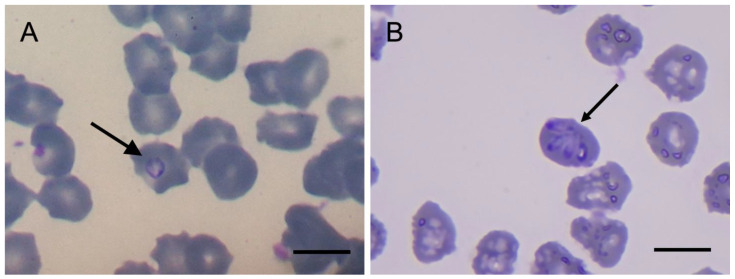
Stained-blood smear examinations revealed intra-erythrocytic inclusions similar to the trophozoite (ring stage) and merozoites (pyriform shape) of Babesia sp. in one (4.4%) capybara (Figure 1). All capybaras were negative for Ehrlichia spp. However, 11 samples (39.3; n = 28; two samples from animals recaptured twice were not tested by PCR) were positive for the Piroplasmida 18S rRNA gene fragment, all of which were captured at the university campus (Site 1). All DNA samples from ticks and blood samples that had negative results for piroplasmids and Ehrlichia spp. were positive for the 16S rDNA gene of ticks and the cytochrome b gene of vertebrates, respectively.
Figure 1.
Intra-erythrocytic inclusions (arrowed) found in one of the infected capybaras. (A), a ring stage. (B), pyriform shape merozoites (note: the whitish structure over one of the parasites is a staining artifact). Scalebars = 10 µm.
We successfully sequenced the Piroplasmida 18S rRNA gene fragment from two capybaras and two female ticks (one A. sculptum and one A. dubitatum); other positive samples contained a very low amount of DNA, and sequencing was not possible. The four sequences produced were identical to each other, and a consensus sequence was deposited in GenBank under the accession number OP714347. The BLAST search revealed that the organism detected herein presented a high identity (99.3–100%) with two sequences referred to as Babesia sp., available in GenBank (accession numbers: EF222255 and MW290045).
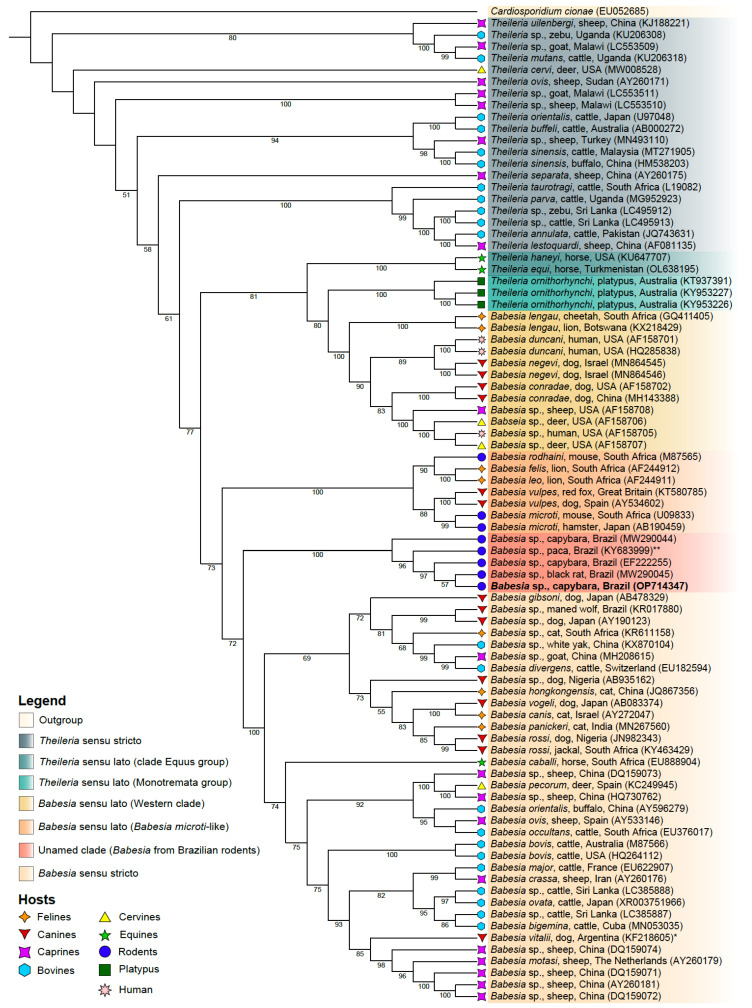
Our phylogenetic reconstruction, using sequences available for numerous Babesia and Theileria spp., disclosed that the organism reported in the present study can be placed within a clade (branch support = 100%) that includes other parasites detected in rodents from Brazil (i.e., capybara, paca, and rat) (Figure 2). This clade shares a common ancestor (branch support = 72%) with the Babesia sensu stricto clade. As such, the organism detected in capybaras reported in the present study was designated Babesia sp. strain Capybara. In addition, our analyses revealed a high inter-clade distance (11.5%) between these two clades in contrast to the intra-clade distance observed within the Babesia sensu stricto clade (5.4%) and within the clade comprising parasites detected in rodents in Brazil (~1.0%).
Figure 2.
Phylogenetic reconstruction of Theileria and Babesia species based on a 610-bp alignment of the 18S rRNA gene. Maximum likelihood tree inferred using TN + F + I + G4 evolutive model. The support of branches above 50 is shown (based on 1000 bootstrap replicates). Symbols at branch tips refer to the hosts from which parasites were detected. The sequence generated in the present study is highlighted in bold. The 18S rRNA gene sequence (EU052685) of Cardiosporidium cionae (Van Gaver and Stephan, 1907) was used as an outgroup. Sequences marked * or ** are referred to as Rangelia vitalii (Pestana, 1910) and Theileria sp. in GenBank, respectively. The nomenclature of the clades is according to Jalovecka et al. [28].
4. Discussion
We reported the presence of a piroplasmid parasite infecting capybaras, A. sculptum, and A. dubitatum in central-western Brazil. Interestingly, 64.7% (11/17) of the capybaras captured at the university campus were positive for this organism, whereas all capybaras from the residential park were negative. Possible explanations for this difference could be the low number of capybaras captured and the comparatively low level of tick infestation in the residential park. Another aspect to consider was the higher number of A. dubitatum collected at the university campus as compared with the residential park. Nonetheless, this does not seem to be a factor in explaining the absence of the piroplasmid in capybaras from the residential park, as ticks from both species were positive for the parasite detected in the capybaras.
Morphologically, the parasite detected herein resembles a large Babesia. BLAST similarity and phylogenetic analyses support its inclusion in the genus Babesia, particularly in a clade containing other undescribed parasites found infecting rodents in other Brazilian states [10]. The phylogenetic relationship of piroplasmids is still an open question, with both Babesia and Theileria genera segregating into several clades each [28]. In fact, the current knowledge suggests that host interaction may be the major driving force of piroplasmid diversification [28]. The fragment of the 18S RNA gene amplified herein offers a glimpse into the phylogeny of this clade of undescribed parasites from rodents in Brazil, which appears to be related to Babesia sensu stricto [29,30]. However, further research is needed to determine the phylogenetic position of this species, which was also discussed by Criado-Fornelio et al. [10].
As such, it is plausible to suppose these organisms represent a putative novel Babesia sp. that parasites capybaras and other rodents in Brazil. An organism designated “Babesia sp. isolate LR5” (GenBank accession number: MW290045) (100% similarity with Babesia sp. strain Capybara) was detected from an infected A. dubitatum tick found on a black rat. Our data reinforce the putative role of A. dubitatum as a vector of Babesia sp. strain Capybara. Amblyomma dubitatum, popularly known as the capybara tick, is well adapted to wet environments and typically feeds on capybaras, although different developmental stages of this tick may infest other animals, including small rodents and birds [31,32,33].
Amblyomma sculptum is another tick commonly found on capybaras in Brazil, being the primary vector of R. rickettsii in Brazil [6]. Altogether, our data suggest that both A. dubitatum and A. sculptum could play a role as vectors of Babesia sp. strain Capybara in the regions where this parasite may be found. This hypothesis deserves further research, especially considering that some positive ticks found in this study were feeding on negative capybaras. Finally, further research is needed to investigate whether Babesia sp. strain Capybara is pathogenic to capybaras or to other animals, such as horses, which are frequently parasitized by these ticks.
5. Conclusions
The present study reports on the presence of an undescribed parasite (Babesia sp. strain Capybara) in capybaras in central-western Brazil. Further research is required for a formal delineation of this species and to investigate the hypothesis of A. dubitatum and A. sculptum ticks as vectors of this parasite.
Acknowledgments
We thank Paulo Sérgio Cardoso Neves and João Lucas Andrade de Faria for their support during the field collections.
Author Contributions
Conceptualization L.C.N., A.C.B., F.d.S.K. and F.D.-T.; methodology, A.C.B., L.C.N., L.C.d.S.-P., S.A.D., B.B.F.d.S., W.V.d.F.P., L.G.F.d.P., B.G.P., G.T.P., E.R.N.C., F.d.S.K. and F.D.-T.; formal analysis, L.C.d.S.-P., F.d.S.K. and F.D.-T.; investigation, L.C.N., A.C.B., F.d.S.K. and F.D.-T.; resources, F.d.S.K. and F.D.-T.; data curation, L.C.d.S.-P., F.d.S.K. and F.D.-T.; writing—original draft preparation, L.C.N., F.d.S.K. and F.D.-T.; writing—review and editing, L.C.N., F.d.S.K. and F.D.-T.; visualization, F.d.S.K. and F.D.-T.; supervision, F.d.S.K. and F.D.-T.; funding acquisition, F.d.S.K. and F.D.-T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was authorized by the Chico Mendes Institute for Biodiversity (ICMBio Permit No. 70679-5) and was approved by the Ethical Committee of Animal Use of the Federal University of Goiás (protocol No. 092/19).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data generated or analyzed during this study are included in this published case study. The consensus sequence generated in this study is available in GenBank (OP714347).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This project was financed by the Fundação de Amparo à Pesquisa do Estado de Goiás (FAPEG) (202110267000287) and by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (317557/2021-1). This study also was financed (scholarships to LCN) in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brazil (CAPES)—Finance Code 001. This work was supported in part by the Division of Intramural Research, NIAID, NIH (Z01 AI001337-01).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Dias T.C., Stabach J.A., Huang Q., Labruna M.B., Leimgruber P., Ferraz K.M.P.M.B., Lopes B., Luz H.R., Costa F.B., Benatti H.R., et al. Habitat selection in natural and human-modified landscapes by capybaras (Hydrochoerus hydrochaeris), an important host for Amblyomma sculptum ticks. PLoS ONE. 2020;15:e0229277. doi: 10.1371/journal.pone.0229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreira J.R., Alvarez M.R., Tarifa T., Pacheco V., Taber A., Tirira D.G., Herrera E.A., Ferraz K.M.P.M.B., Aldana-Domínguez J., Macdonald D.W. Capybara: Biology, Use and Conservation of an Exceptional Neotropical Species. Volume 1. Springer; New York, NY, USA: 2013. Taxonomy, natural history and distribution of the capybara; pp. 3–37. [Google Scholar]
- 3.Ferraz K.M.P.M.B., Lechevalier M.A., Couto H.T.Z., Verdade L.M. Damage caused by capybara on a corn field. Sci. Agrocola. 2003;60:191–194. doi: 10.1590/S0103-90162003000100029. [DOI] [Google Scholar]
- 4.de Oliveira Vieira C., Bernardes Filho F., Azulay-Abulafia L. Capybara bites: Report of human injury caused by a Hydrochoerus hydrochaeris. J. Emerg. Med. 2015;49:e179–e182. doi: 10.1016/j.jemermed.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Rossetto A.L., Amarante L.F., Rossetto A.L., Haddad Junior V. Injuries and infection caused by capybara bites in a human. Rev. Soc. Bras. Med. Trop. 2021;54:e0043–e2021. doi: 10.1590/0037-8682-0043-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luz H.R., Costa F.B., Benatti H.R., Ramos V.N., Serpa M.C.A., Martins T.F., Acosta I.C.L., Ramirez D.G., Muñoz-Leal S., Ramirez-Hernandez A., et al. Epidemiology of capybara-associated Brazilian spotted fever. PLoS Negl. Trop. Dis. 2019;13:e0007734. doi: 10.1371/journal.pntd.0007734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krawczak F.S., Nieri-Bastos F.A., Nunes F.P., Soares J.F., Moraes-Filho J., Labruna M.B. Rickettsial infection in Amblyomma cajennense ticks and capybaras (Hydrochoerus hydrochaeris) in a Brazilian spotted fever-endemic area. Parasites Vectors. 2014;7:7. doi: 10.1186/1756-3305-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramírez-Hernández A., Uchoa F., Serpa M.C.A., Binder L.C., Souza C.E., Labruna M.B. Capybaras (Hydrochoerus hydrochaeris) as amplifying hosts of Rickettsia rickettsii to Amblyomma sculptum ticks: Evaluation during primary and subsequent exposures to R. rickettsii infection. Ticks Tick Borne Dis. 2020;11:101463. doi: 10.1016/j.ttbdis.2020.101463. [DOI] [PubMed] [Google Scholar]
- 9.de Paula L.G.F., do Nascimento R.M., Franco A.O., Szabó M.P.J., Labruna M.B., Monteiro C., Krawczak F.S. Seasonal dynamics of Amblyomma sculptum: A review. Parasites Vectors. 2022;15:193. doi: 10.1186/s13071-022-05311-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Criado-Fornelio A., Buling A., Casado N., Gimenez C., Ruas J., Wendt L., da Rosa-Farias N., Pinheiro M., Rey-Valeiron C., Barba-Carretero J.C. Molecular characterization of arthropod-borne hematozoans in wild mammals from Brazil, Venezuela and Spain. Acta Parasitol. 2009;54:187–193. doi: 10.2478/s11686-009-0031-5. [DOI] [Google Scholar]
- 11.Gonçalves L.R., Paludo G., Bisol T.B., Perles L., de Oliveira L.B., de Oliveira C.M., da Silva T.M.V., Nantes W.A.G., Duarte M.A., Santos F.M., et al. Molecular detection of piroplasmids in synanthropic rodents, marsupials, and associated ticks from Brazil, with phylogenetic inference of a putative novel Babesia sp. from white-eared opossum (Didelphis albiventris) Parasitol. Res. 2021;120:3537–3546. doi: 10.1007/s00436-021-07284-8. [DOI] [PubMed] [Google Scholar]
- 12.Martins T.F., Onofrio V.C., Barros-Battesti D.M., Labruna M.B. Nymphs of the genus Amblyomma (Acari: Ixodidae) of Brazil: Descriptions, redescriptions, and identification key. Ticks Tick Borne Dis. 2010;1:75–99. doi: 10.1016/j.ttbdis.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Dantas-Torres F., Fernandes Martins T., Muñoz-Leal S., Onofrio V.C., Barros-Battesti D.M. Ticks (Ixodida: Argasidae, Ixodidae) of Brazil: Updated species checklist and taxonomic keys. Ticks Tick Borne Dis. 2019;10:101252. doi: 10.1016/j.ttbdis.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Clifford C.M., Anastos G., Elbl A. The larval ixodid ticks of the Eastern United States (Acarina-Ixodidae) Misc. Publ. Entomol. Soc. Am. 1961;1:213–237. [Google Scholar]
- 15.Sangioni L.A., Horta M.C., Vianna M.C., Gennari S.M., Soares R.M., Galvão M.A., Schumaker T.T., Ferreira F., Vidotto O., Labruna M.B. Rickettsial infection in animals and Brazilian spotted fever endemicity. Emerg. Infect. Dis. 2005;11:265–270. doi: 10.3201/eid1102.040656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horta M.C., Labruna M.B., Pinter A., Linardi P.M., Schumaker T.T. Rickettsia infection in five areas of the state of São Paulo, Brazil. Mem. Inst. Oswaldo Cruz. 2007;102:793–801. doi: 10.1590/s0074-02762007000700003. [DOI] [PubMed] [Google Scholar]
- 17.Doyle C.K., Labruna M.B., Breitschwerdt E.B., Tang Y.W., Corstvet R.E., Hegarty B.C., Bloch K.C., Li P., Walker D.H., McBride J.W. Detection of medically important Ehrlichia by quantitative multicolor TaqMan real-time polymerase chain reaction of the dsb gene. J. Mol. Diagn. 2005;7:504–510. doi: 10.1016/S1525-1578(10)60581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almeida A.P., Marcili A., Leite R.C., Nieri-Bastos F.A., Domingues L.N., Martins J.R., Labruna M.B. Coxiella symbiont in the tick Ornithodoros rostratus (Acari: Argasidae) Ticks Tick Borne Dis. 2012;3:203–206. doi: 10.1016/j.ttbdis.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Kocher T.D., Thomas W.K., Meyer A., Edwards S.V., Pääbo S., Villablanca F.X., Wilson A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangold A.J., Bargues M.D., Mas-Coma S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae) Parasitol. Res. 1998;84:478–484. doi: 10.1007/s004360050433. [DOI] [PubMed] [Google Scholar]
- 21.Staden R., Beal K.F., Bonfield J.K. The Staden Package, 1998. Methods Mol. Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 22.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sousa-Paula L.C., da Silva L.G., da Silva Junior W.J., Figueirêdo Júnior C.A.S., Costa C.H.N., Pessoa F.A.C., Dantas-Torres F. Genetic structure of allopatric populations of Lutzomyia longipalpis sensu lato in Brazil. Acta Trop. 2021;222:106031. doi: 10.1016/j.actatropica.2021.106031. [DOI] [PubMed] [Google Scholar]
- 24.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I., Bork P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jalovecka M., Sojka D., Ascencio M., Schnittger L. Babesia life cycle—When phylogeny meets biology. Trends Parasitol. 2019;35:356–368. doi: 10.1016/j.pt.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Soares H.S., Marcili A., Barbieri A.R.M., Minervino A.H.H., Moreira T.R., Gennari S.M., Labruna M.B. Novel piroplasmid and Hepatozoon organisms infecting the wildlife of two regions of the Brazilian Amazon. Int. J. Parasitol. Parasites Wildl. 2017;6:115–121. doi: 10.1016/j.ijppaw.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnittger L., Ganzinelli S., Bhoora R., Omondi D., Nijhof A.M., Florin-Christensen M. The Piroplasmida Babesia, Cytauxzoon, and Theileria in farm and companion animals: Species compilation, molecular phylogeny, and evolutionary insights. Parasitol. Res. 2022;121:1207–1245. doi: 10.1007/s00436-022-07424-8. [DOI] [PubMed] [Google Scholar]
- 31.Nava S., Venzal J.M., Labruna M.B., Mastropaolo M., González E.M., Mangold A.J., Guglielmone A.A. Hosts, distribution and genetic divergence (16S rDNA) of Amblyomma dubitatum (Acari: Ixodidae) Exp. Appl. Acarol. 2010;51:335–351. doi: 10.1007/s10493-009-9331-6. [DOI] [PubMed] [Google Scholar]
- 32.Dantas-Torres F., Melo M.F., Sales K.G.S., Sousa-Paula L.C., da Silva F.J., Figueredo L.A., Labruna M.B. Seasonal dynamics and rickettsial infection in free-living Amblyomma dubitatum in the Atlantic forest biome in north-eastern Brazil. Acta Trop. 2021;217:105854. doi: 10.1016/j.actatropica.2021.105854. [DOI] [PubMed] [Google Scholar]
- 33.de Paula L.G.F., Zeringóta V., Sampaio A.L.N., Bezerra G.P., Barreto A.L.G., dos Santos A.A., Miranda V.C., Paula W.V.F., Neves L.C., Secchis M.V., et al. Seasonal dynamics of Amblyomma sculptum in two areas of the Cerrado biome midwestern Brazil, where human cases of rickettsiosis have been reported. Exp. Appl. Acarol. 2021;84:215–225. doi: 10.1007/s10493-021-00615-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated or analyzed during this study are included in this published case study. The consensus sequence generated in this study is available in GenBank (OP714347).