Abstract
Simple Summary
Poly (ADP-ribose) polymerase inhibitors (PARPis) have been recently approved by international medicine agencies for the treatment of ovarian cancer patients with either BRCA pathogenic variants or homologous recombination deficiency (HRD), changing the ovarian cancer treatment landscape in both first-line and relapsed disease settings. Thus, assessing HRD is now crucial for patient management, making it a pressing need to offer an alternative to expensive and time-consuming outsourced analysis. The aim of the present study was to demonstrate the feasibility of locally performed HRD testing, using an easy and affordable CE-IVD NGS panel that enabled the analysis of HRD status within 5 days, with perfect concordance with Myriad results. Importantly, this strategy could also be applied in the near future to stratify patients with different tumor types, including breast cancer, pancreatic cancer, and prostatic cancer, thereby expanding the number of patients who could benefit from PARPi treatment.
Abstract
Assessment of HRD status is now essential for ovarian cancer patient management. A relevant percentage of high-grade serous carcinoma (HGSC) is characterized by HRD, which is caused by genetic alterations in the homologous recombination repair (HRR) pathway. Recent trials have shown that not only patients with pathogenic/likely pathogenic BRCA variants, but also BRCAwt/HRD patients, are sensitive to PARPis and platinum therapy. The most common HRD test is Myriad MyChoice CDx, but there is a pressing need to offer an alternative to outsourcing analysis, which typically requires high costs and lengthy turnaround times. In order to set up a complete in-house workflow for HRD testing, we analyzed a small cohort of HGSC patients using the CE-IVD AmoyDx HRD Focus Panel and compared our results with Myriad’s. In addition, to further deepen the mechanisms behind HRD, we analyzed the study cohort by using both a custom NGS panel that analyzed 21 HRR-related genes and FISH analysis to determine the copy numbers of PTEN and EMSY. We found complete concordance in HRD status detected by the Amoy and the Myriad assays, supporting the feasibility of internal HRD testing.
Keywords: BRCA, HRD, HRR, NGS, molecular testing, ovarian cancer, PARPi
1. Introduction
Ovarian cancer is the second most prevalent type of cancer in women over the age of 40 in developed countries, and it is the fifth leading cause of cancer-related death in women [1,2]. Importantly, it is the most lethal gynecologic cancer, having a 5-year survival rate of only 49.7% [3], and because it lacks distinct symptoms and specific biomarkers for early detection, it is frequently diagnosed at an advanced stage, after spreading beyond the pelvis [4,5]. High-grade serous carcinoma (HGSC) represents the most common type of ovarian cancer. HGSC is a genetically unstable tumor with a high mitotic rate that is characterized by ubiquitous pathogenic variants in TP53 and alterations in BRCA1 and BRCA2. In particular, 13–16% of these tumors present germline pathogenic variants of BRCA1/2, and 6% harbor somatic pathogenic variants [6,7,8]. Moreover, up to 51% of HGSCs have shown defects in the homologous recombination repair (HRR) pathway [8].
Deficiency in the HRR pathway leads cells to rely on more error-prone DNA repair systems [9], and therefore, over time, unrepaired double strand breaks (DSBs) induce the accumulation of genomic alterations such as insertions and deletions, copy number variations, and structural chromosomal rearrangements, leaving an irreversible “genomic scar” [10].
In HGSC, homologous recombination deficiency (HRD) is caused by pathogenic germline/somatic variants and epigenetic modifications in either BRCA1/2 or in genes encoding for key actors in the HRR pathway. In fact, alterations in RAD51B/C/D, PALB2, ATM, H2AX, CHK1/2, CDK12, NBN, MRE11, RPA, BRIP1, BARD1, RAD51, Fanconi Anemia genes, PTEN, and EMSY have been shown to potentially confer an HRD or BRCAness phenotype, which is characterized by deficiencies in the DSB repair pathway [11,12].
HRD cells are particularly sensitive to poly (ADP-ribose) polymerase inhibitors (PARPis). In fact, inhibiting PARP1, which is crucial for single-strand DNA break repair, causes cell death in cells with impaired DSB repair capacity; this phenomenon is referred to as synthetic lethality [13].
The addition of PARPis to first-line chemotherapy regimens for women with platinum-sensitive ovarian cancer has improved clinical outcomes in terms of both progression-free and overall survival. Patients with pathogenic/likely pathogenic BRCA variants (BRCAmut) benefit the most from PARPis plus platinum treatment, but it is important to note that BRCAwt/HRD patients are also susceptible to such a therapy. In particular, the PRIMA trial showed median PFS for niraparib of 22.9, 19.6, and 8.1 months, respectively, in BRCAmut, BRCAwt/HRD and HR-proficient (HRp) patients; the PAOLA1 trial reported median PFS for olaparib of 37.2, 28.1, and 16.9 months, respectively, in BRCAmut, BRCAwt/HRD and HRp patients; and the VELIA trial showed median PFS for veliparib of 34.7, 22.9 and 15.0 months, respectively, in BRCAmut, BRCAwt/HRD, and HRp patients [14,15,16,17,18,19,20].
All these findings established the key role of both BRCA testing and HRD assessment in the treatment of HGSC patients. The American Society of Clinical Oncology (ASCO) recommends offering germline genetic testing for BRCA1/2 to all women diagnosed with epithelial ovarian cancer, irrespective of their clinical features or family cancer history. Somatic tumor testing for BRCA1/2 should be performed in women who do not carry a germline pathogenic or likely pathogenic BRCA1/2 variant. Women with germline or somatic pathogenic or likely pathogenic BRCA1/2 variants should be offered Food and Drug Administration (FDA)-approved treatments, such as PARPis [21].
Moreover, the FDA has recently approved HRD assays able to detect the related “genomic scar”, in order to stratify BRCAwt patients and consequently predict responses to platinum-based chemotherapy and synthetic lethal agents such as PARPis, and prognosis [10]. The most diffused assay to determine HRD status is MyChoice CDx (Myriad Genetics, Salt Lake City, UT, USA), which calculates a Genomic Instability Score (GIS) based on the evaluation of loss of heterozygosity (LOH), telomeric allelic imbalance (TAI), and large-scale state transitions (LST); furthermore, BRCA1/2 variants are analyzed. Tumors with GIS < 42 are HRp, and tumors with GIS ≥ 42 and/or pathogenic BRCA1/2 variants are defined as HRD.
Given the importance of HRD testing for patient management and treatment decision making, we wanted to assess the feasibility of a locally performed, commercially available HRD assay and to compare our findings to those obtained using Myriad MyChoice CDx. In addition, to further deepen the mechanisms behind HRD, we studied the HRR pathway by using both a custom NGS panel that analyzed 21 HRR-related genes and FISH analysis to determine the copy numbers of PTEN and EMSY.
2. Materials and Methods
2.1. Study Cohort
We analyzed a cohort of 16 patients diagnosed with HGSC at San Raffaele Hospital (Milan, Italy), in 2021. Patients’ clinicopathologic features are summarized in Table 1. The median age at diagnosis was 57.5 years old; the range was between 36 and 69 years old; median progression free survival (PFS) was 6.5 months. For 13 patients, the Myriad MyChoice CDx report was available.
Table 1.
Patients’ clinicopathological features.
| Pt | Age | Sample | pT | G | pN | Therapy | Maintenance | PFS | Progr | Recur | Currently |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID_01 | 47 | SS | 3c | IV | 1a | C + T + B | B + Ola | 13 | NED-maintenance ongoing | ||
| ID_02 | 50 | SS | 3c | IV | x | C + T + B | B | FU lost ° | / | ||
| ID_03 | 68 | SS | 3b | IV | x | C + T | NO | 5 | yes | under evaluation | |
| ID_04 | 59 | SS | 3c | IV | 0 | C + T | Ola | 10 | NED-maintenance ongoing | ||
| ID_05 | 36 | Bx | III | C + T + IDS | Nira | 4 | yes | T + B ongoing | |||
| ID_06 | 43 | Bx | IV | C + T + IDS | Ola | 4 | yes | T + B ongoing | |||
| ID_07 | 61 | Bx | III | C + T + IDS + B | B | 10 | NED-B ongoing | ||||
| ID_08 | 39 | Bx | III | C + T + IDS | Nira * | 7 | NED | ||||
| ID_09 | 51 | SS | 3c | III/IV | 1a | C + T | Ola | 8 | NED-Ola ongoing | ||
| ID_10 | 56 | Bx | III | C + T + IDS | Ola | 6 | NED-Ola ongoing | ||||
| ID_11 | 66 | Bx | - | C + T | Nira | 5 | yes | ||||
| ID_12 | 69 | Bx | IV | C + T + IDS | starting Ola | ||||||
| ID_13 | 67 | SS | 3c | IV | 0 | C + T | Nira | 3 | NED-Nira ongoing | ||
| ID_14 | 53 | Bx | III | C + T | NO ^ | 13 | yes | C + T; Nira ongoing | |||
| ID_15 | 68 | SS | 2b | III | x | C + T | NO ^ | 5 | NED | ||
| ID_16 | 62 | SS | 2b | III | 0 | C + T | NO ^ | 10 | NED |
SS = surgical specimen; Bx = biopsy; G = grade; pT = tumor stage; pN = lymph node stage (according to WHO/TNM); C = carboplatin; T = taxol; B = Bevacizumab; IDS = interval debulking surgery; Ola = Olaparib; Nira = niraparib; PFS = progression-free survival, in months; FU = follow up; Progr = progression; Recur = recurrence; NED = no evidence of disease; ° patient moved to another hospital; * interrupted because of neutropenia; ^ no maintenance therapy because of tumor stage.
2.2. DNA Extraction
DNA from FFPE HGSC was extracted from both surgical specimens (8 patients) and tissue biopsies (8 patients), depending on material availability at the time of analysis. An expert pathologist reviewed each case and selected the most representative areas of the tumor with a percentage of tumor cells above 50%. DNA extraction was performed using thee Maxwell® RSC DNA FFPE kit and Maxwell RSC Instrument (Promega, Milan, Italy) and quantified using the Qubit DNA HS Assay Kit on Qubit 3.0 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA), as previously described [22].
2.3. HRD Assay
HRD status was evaluated using the CE-IVD AmoyDx HRD Focus Panel (Amoy Diagnostics, Xiamen, China), according to the manufacturer’s instructions, using 80–100 ng DNA. DNA libraries were quantified using the Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), and DNA fragment quality control was performed using the 2100 Bioanalyzer System and DNA 1000 kit (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed using the NextSeq500 platform and Mid v2 flow cell (Illumina, San Diego, CA, USA). Raw data were analyzed using the AmoyDx NGS Data Analysis System-ANDAS Software to detect BRCA1/2 variants and HRD status. Samples with GIS ≥ 50 were considered HRD. BRCA1/2 variants were classified according to the ACMG/ENIGMA 5-class system [23,24], thereby giving an accurate description of variants’ clinical significance.
2.4. Evaluation of HRR Pathway Genes Using a Next-Generation Sequencing (NGS) Custom Panel
Somatic alterations involving key players in the HRR pathway were analyzed utilizing a custom NGS panel developed with Thermo Fisher Scientific, the BRCA-Expanded Panel, on the Ion Torrent S5 (ThermoFisher Scientific). The assay analyzes the full-length coding sequences of 21 genes (ATM, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK1, CHEK2, FANCD2, MRE11, NBN, PALB2, PARP1, RAD50, RAD51, RAD51B, RAD51C, RAD51D, RAD52, RAD54L, TP53), using an input of 10 ng of DNA. Targeted libraries were prepared using Ion ChefTM Instrument (ThermoFisher Scientific) following the manufacturer’s instructions and were sequenced on the 530TM Chip (ThermoFisher Scientific). Raw data analysis was performed using Torrent Suite v5.12, as previously described [25]. Variants were classified in accordance with the ACMG/ENIGMA criteria and ClinVar/OncoKB databases.
2.5. Germline BRCA Testing
The presence of germline variants was assessed on genomic DNA extracted from patients’ peripheral whole blood samples (Maxwell® RSC Whole Blood, Promega, Milan, Italy) by Sanger direct sequencing. After amplification, PCR products of the genetic variants and surrounding regions were purified using Clean PCR (CleanNA-PH Waddinxveen, Netherlands) and sequenced in both directions using a Big Dye Terminator v.1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Sequencing products were purified using a Big Dye X-Terminator Kit (Applied Biosystems) and ran on an ABI 3730 Genetic Analyzer (Applied Biosystems). Called sequences were aligned to the reference using the Sequencer V.5.0 Software (Gene Codes Corporation, Ann Arbor, MI, USA) and classified in accordance with the ACMG/ENIGMA criteria [23,24].
2.6. FISH Analysis
PTEN homozygous deletion and EMSY copy number were assessed in all the study cases on 4 μm sections of FFPE tissue by fluorescence in situ hybridization (FISH) as previously described [24], using ZytoLight SPEC PTEN/CEN10 Dual Color Probe (ZytoVysion GmbH, Bremerhaven, Germany) and EMSY FISH Probe (Empire Genomics, Buffalo, NY, USA). FISH was performed according to the probe manufacturers’ suggested protocols, slides were analyzed using a Nikon 90i fluorescence microscope (Nikon Instruments SpA, Italy), and images were captured by Genikon software (Nikon, Tokyo, Japan).
PTEN and CEN10 signals were counted in a minimum of 60 nuclei, and a sample was considered to have deleted PTEN when both copies of the gene were lost in more than 20% of tumor cells. EMSY copies were counted in at least 60 tumor cells, and a sample was considered to have an increased copy number when the EMSY copy number was >4 copies/cell.
2.7. Statistical Analyses
Positive Percent Agreement (PPA), Negative Percent Agreement (NPA), and Overall Percent Agreement (OPA) were calculated to evaluate the performance of the AmoyDx HRD Focus Panel compared with that of the Myriad MyChoice CDx.
3. Results
3.1. Comparison of AmoyDx HRD Focus Panel and Myriad MyChoiceCDx for Assessing HRD Status
A cohort of 16 HGSC patients (Table 1), who were diagnosed at San Raffaele Hospital in 2021, was selected to assess the HRD status utilizing the CE-IVD AmoyDx HRD Focus Panel. This panel evaluates BRCA1/2 status and can detect the HRD genomic scar by analyzing LOH, TAI, and LST. The DNA library concentration ranged between 34.8 and 50.4 ng/μL. The median concentration was 42.2 ng/μL. Thus, all samples were above the manufacturer’s recommended concentration of 20 ng/μL. All the libraries were of outstanding quality. The main peak of the DNA fragment size was between 258 and 268 bp, and the median value was 261 bp (Figure S1). Moreover, we obtained excellent sequencing metrics for all samples (Table S1).
As shown in Table 2 (left panel), thirteen samples were classified as HRD (GIS ≥ 50), whereas three samples (#3, #7, and #13) were identified as HR-proficient (HRD-). Five (#4, #6, #9, #10, and #12) of the thirteen HRD tumors had pathogenic or likely pathogenic BRCA1/2 variants, and one sample (#16) had a variant of uncertain significance (VUS).
Table 2.
Results of AmoyDx HRD Focus Panel (left), Myriad MyChioce CDx (central), and BRCA somatic and germline tests (right).
| AmoyDx HRD Focus Panel | Myriad MyChoice CDx | BRCA1/2 Testing | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pt | HRD | GIS | BRCA | Significance | HRD | GIS | BRCA | Significance | Somatic $ | Germline |
| ID_01 | + | 99.2 | - | All benign | + | 72 | - | All benign | WT | |
| ID_02 | + | 100 | - | All benign | + | 51 | - | All benign | WT | |
| ID_03 | - | 45.4 | - | All benign | - | 28 | - | All benign | WT | |
| ID_04 | + | 98.3 | + | Pathogenic | + | 60 | + | Deleterious | * BRCA2 p.Tyr1739Ter (c.5217_5220del) 89% | yes |
| ID_05 | + | 98.3 | - | All benign | + | 66 | - | All benign | WT | |
| ID_06 | + | 97.3 | + | Pathogenic | + | 54 | + | Deleterious | * BRCA1 p.Tyr777Ter (c.2331T > G) 76% | yes |
| ID_07 | - | 36.1 | - | Likely-benign | - | 36 | - | All benign | WT | |
| ID_08 | + | 97.1 | - | All benign | + | 48 | - | All benign | WT | |
| ID_09 | + | 97.6 | + | Likely-pathogenic | + | 60 | + | Deleterious | * BRCA2 p.Ala1327ProfsTer8 (c.3979delG) 76% | no |
| ID_10 | + | 96.8 | + | Pathogenic | + | 57 | + | Deleterious | * BRCA2 p.Val1283LysfsTer2 (c.3847_3848delGT) 79% | yes |
| ID_11 | + | 100 | - | All benign | + | 67 | - | All benign | WT | |
| ID_12 | + | 59.1 | + | Likely-pathogenic | + | 36 | + | Deleterious | * BRCA2 p.Asn615ThrfsTer29 (c.1842delT) 24% | no |
| ID_13 | - | 14.1 | - | All benign | - | 22 | - | All benign | WT | |
| ID_14 | + | 98.9 | - | All benign | na | na | WT | |||
| ID_15 | + | 72 | - | All benign | na | na | WT | |||
| ID_16 | + | 84 | + | Uncertain | na | na | ^ BRCA2 p.Arg2991Cys (c.8971C > T) 31% | |||
HRD+ = HRD; HRD- = HRp; na = not available; BRCA1 (NM_007294.4), BRCA2 (NM_000059); $ evaluated using BRCA-Expanded Panel; * BRCA variants identified by AmoyDx HRD Focus Panel, Myriad MyChoice CDx, and BRCA-Expanded Panel; ^ BRCA variant identified by AmoyDx HRD Focus Panel and BRCA-Expanded Panel.
In addition, 13 out of 16 patients had access to the MyChoice CDx assay, which allowed us to compare the results of the two tests (Table 2, central panel) and obtain complete HRD status concordance, with Positive Percent Agreement (PPA), Negative Percent Agreement (NPA), and Overall Percent Agreement (OPA) values of 100%. The only discordant finding was patient #12: as a result of the existence of a likely pathogenic BRCA2 variant, she was classified as HRD by both assays; nevertheless, the GIS was over the positivity threshold by the Amoy panel and below the positivity threshold by the Myriad panel.
The BRCA1/2 variants identified by the two assays were identical in all cases. The five patients with pathogenic/likely pathogenic BRCA1/2 variants identified on tumor samples were addressed for genetic counseling and germline genetic testing. Peripheral blood analysis revealed that BRCA variants in patients #4, #6, and #10 were germline (Table 2, right panel).
3.2. Investigation of HRR Pathway Gene Alterations
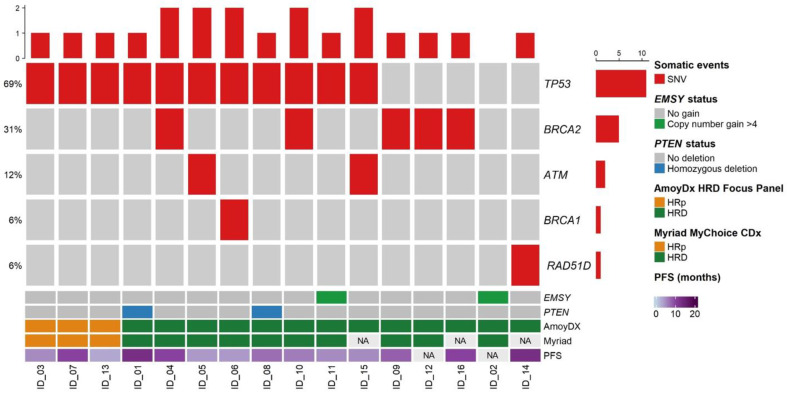
In order to better characterize the HRD phenotype, we analyzed genes encoding for key players in HRR by an NGS custom panel (BRCA-Expanded Panel). Pathogenic/likely pathogenic/uncertain variants are listed in Table 3; no benign/likely benign variants are reported. HR-proficient samples (patients #3, #7, and #13) were wild type for all the analyzed genes, except for TP53 (Figure 1). Among the thirteen HRD patientst, 9 (patients #4, #5, #6, #9, #10, #12, #14, #15, and #16) had mutations in key HRR genes (TP53, BRCA1, BRCA2, ATM, and RAD51D), 3 had only TP53 pathogenic variants (patients #1, #8 and #11), and 1 patient (#2) did not have any variant (Figure 1).
Table 3.
Gene variants identified by BRCA-Expanded Panel.
| Pt | HRD | BRCA-Expanded Panel | ||
|---|---|---|---|---|
| Variant | VAF | Significance | ||
| ID_01 | + | TP53 p.Pro190Thr (c.568C > A) | 82% | VUS |
| ID_02 | + | WT | ||
| ID_03 | - | TP53 p.Arg175His (c.524G > A) | 73% | Pathogenic |
| ID_04 | + | BRCA2 p.Tyr1739Ter (c.5217_5220del) | 89% | Pathogenic |
| ID_05 | + | ATM p.His2552Asn (c.7654C > A) | 44% | VUS |
| ID_06 | + | BRCA1 p.Tyr777Ter (c.2331T > G) | 76% | Pathogenic |
| ID_07 | - | TP53 p.Arg175His (c.524G > A) | 64% | Pathogenic |
| ID_08 | + | TP53 p.Cys141Tyr (c.422G > A) | 90% | Pathogenic |
| ID_09 | + | BRCA2 p.Ala1327ProfsTer8 (c.3979delG) | 76% | Likely-pathogenic |
| ID_10 | + | BRCA2 p.Val1283LysfsTer2 (c.3847_3848delGT) | 79% | Pathogenic |
| ID_11 | + | TP53 p.Tyr163Cys (c.488A > G) | 48% | Pathogenic |
| ID_12 | + | BRCA2 p.Asn615ThrfsTer29 (c.1842delT) | 24% | Likely-pathogenic |
| ID_13 | - | TP53 p.Cys182AlafsTer65 (c.544delT) | 72% | Likely-pathogenic |
| ID_14 | + | RAD51D p.Cys9Ser (c.26G > C) | 71% | VUS |
| ID_15 | + | ATM p.Tyr454His (c.1360T > C) | 40% | VUS |
| ID_16 | + | BRCA2 p.Arg2991Cys (c.8971C > T) | 31% | VUS |
HRD+ = HRD; HRD- = HRp VAF = variant allele frequency; VUS = variant of uncertain significance; TP53 (NM_000546.5); BRCA1 (NM_007294.4); BRCA2 (NM_000059); ATM (NM_000051.3); RAD51D (NM_133629.2).
Figure 1.
Oncoprint of mutated HRR genes identified in the study cohort. Cases sorted by HRD status. SNV were evaluated by BRCA-Expanded Panel, PTEN homozygous deletion, and EMSY copy number gain were evaluated by FISH analysis. NA = not available.
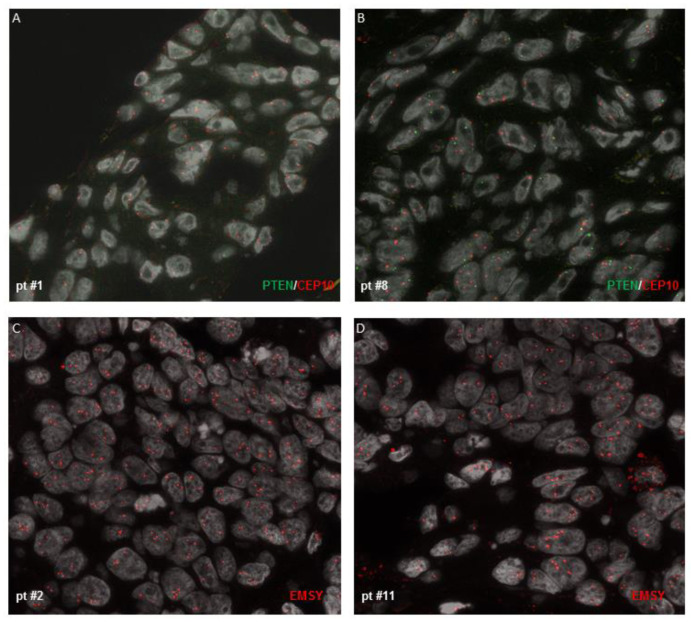
To further deepen the understanding of the mechanisms behind HRD, we investigated PTEN deletion and EMSY copy number by FISH analysis (Figure 1). We found homozygous deletion of PTEN in patients #1 and #8 (Figure 2A,B) and elevated EMSY copy numbers in patients #2 and #11 (Figure 2C,D). In all the other samples, we observed neither PTEN homozygous deletion nor EMSY copy number gain.
Figure 2.
FISH analyses for PTEN and EMSY. PTEN homozygous deletion was observed in patient #1 (A) and in patient #8 (B). An elevated EMSY copy number was observed in patient #2 (C) and in patient #11 (D).
4. Discussion
Assessment of HRD status is now essential for ovarian cancer patient management. In fact, up to 51% of HGSCs have shown defects in the HRR pathway [8], and recent trials have demonstrated that not only patients with pathogenic/likely pathogenic BRCA variants, but also BRCAwt/HRD patients, are sensitive to PARPis and platinum therapy. PARPis have been recently approved by international medicine agencies for the treatment of ovarian cancer patients with either BRCA pathogenic variants or HRD, changing the ovarian cancer treatment landscape in both the first-line and relapsed disease settings [14,15,16,17,18,19,20]. The most common HRD test is Myriad MyChoice CDx, but there is a pressing need to offer an alternative to outsourcing analysis [26], which typically requires high costs and lengthy turnaround times.
In order to set up a complete in-house workflow for HRD testing, we analyzed a cohort of 16 HGSC patients using the CE-IVD AmoyDx HRD Focus Panel. This panel perfectly matches the requirements of a molecular diagnostic laboratory because it is simple to use, allows a 50% cost reduction when compared to the Myriad MyChoice CDx, and enables results with a turnaround time of 5 working days. It is important to emphasize that proper tumor sampling and fixation [27] allowed us (i) to recover DNA with sufficient quality and quantity for the assay from both surgical samples and biopsies, (ii) to obtain excellent sequencing metrics, and (iii) to successfully analyze all samples. Thirteen patients showed HRD, and three samples were HR-proficient. We found complete concordance in HRD status detected by the Myriad assay, which was available for 13 out of 16 patients. We are aware of the limited number of patients included in the study. This investigation was intended to be a proof of concept to determine both the feasibility and the performance of local HRD testing. Despite the small cohort, our results are extremely encouraging and in line with previous studies [28,29], highlighting the utility of such an approach and allowing us to include this test in the clinical molecular diagnostics routine.
The two assays identified the same variants in all patients with BRCA1/2 alterations, but while the Myriad assay does not classify BRCA variants, grouping both pathogenic and likely pathogenic variants together as deleterious, the AmoyDx HRD assay adheres to the ACMG/ENIGMA 5-class system, giving a more accurate description of variants’ clinical significance [23].
It is important to underline that one of the limitations of HRD tests is the absence of information on the HRR-related genes beyond BRCA1/2. Thus, we analyzed 21 genes involved in the HRR pathway by using the BRCA-Expanded Panel. We detected the same pathogenic/likely pathogenic BRCA1/2 variants as in the HRD tests, but we also found SNV variants in ATM (patient #5 and patient #15), and in RAD51D (patient #14). Further, we investigated the homozygous deletion of PTEN and copy number gain of EMSY by FISH analysis. In fact, PTEN deficiency leads to impaired RAD51-mediated DSB repair and genomic instability, thereby causing HR deficiency and leading to sensitivity to PARPis, both in vitro and in vivo [30,31]. The copy number gain of EMSY, which binds to BRCA2, exon 3, has been described in 17% of high-grade ovarian cancer, and it has been reported as an alternative mechanism for HRD, even if its role is controversial and it does not seem to confer sensitivity to PARPis [32,33,34]. We found that patients #1 and #8 had homozygous deletion of PTEN, and patients #2 and #11 had elevated EMSY copy numbers, suggesting that these alterations may have been the causes of HRD in the four patients. Additionally, the presence of either PTEN homozygous deletion or increased EMSY copy number was not concurrent with alterations in other HRR key players. However, due to the small size of the study group, a larger cohort should be used to further corroborate this observation.
Among the nine HRD patients treated with PARPis, three (33%) experienced tumor progression, and six (67%) do not show evidence of disease. PARPi resistance has already been reported in the literature; therefore, for patient #6, carrying a BRCA1 pathogenic variant, we could hypothesize several causing events, such as the occurring of alterations in PARP1, the loss of PARG, and reversion mutations in BRCA1 or other HRR genes; moreover, loss of TP53BP1, RIF1, REV7, or DYNLL1 has been associated with PARPi resistance in BRCA1-deficient cells [35,36]. For patients #5 and #11, as the ATM variant is of uncertain significance (VUS) (patient #5) and the role of EMSY gene copy number increase (patient #11) is still under investigation, further studies are required to establish their significance in drug responses.
The patients with no evidence of disease after at least 6 months were characterized by PTEN homozygous deletion (#1 and #8), BRCA2 pathogenic variants (#4, #9, and #10), and RAD51D VUS (#14). This last patient experienced recurrence after chemotherapy and is now showing sensitivity to niraparib. Interestingly, such a finding could support the in silico prediction of a deleterious effect of this variant on protein structure/function, in line with the association between RAD51D loss-of-function variants and sensitivity to PARPis [37,38].
In conclusion, this is one of the first studies to compare the Myriad assay with the results of a locally performed test, showing excellent concordance and short turnaround times. Despite the restriction of a small cohort of samples, our findings support the feasibility of internal HRD testing, which has now become essential for the management of ovarian cancer. Importantly, this strategy could be also applied in the near future to stratify patients with different tumor types [39], including breast cancer, pancreatic cancer, and prostate cancer, thereby expanding the number of patients who could benefit from PARPi treatment.
5. Conclusions
Updated ovarian cancer patient therapy selection requires assessment of the HRD status; therefore, it is advantageous that such analysis is locally available, avoiding the higher costs and lengthy turnaround times of outsourcing. Our data demonstrated the feasibility of internal HRD testing for HGSC and designed an HRD testing strategy that may be extended to other tumor types that could benefit from PARPi treatment.
Acknowledgments
We thank Cecilia Lencioni and Gianmarco Azara, LCM-Genect “Enabling Precision Medicine” Sesto San Giovanni (MI) Italy, for the technical support in this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15010043/s1. Figure S1: Bioanalyzer densitometry plots of DNA libraries. Gel-like images show the expected main peak of 258–268 bp for all samples. Table S1: Sample sequencing metrics of AmoyDx HRD Focus Panel.
Author Contributions
Conceptualization, supervision, and project administration, M.G.C.; methodology, G.M. (Gilda Magliacane), E.B., G.M. (Giovanna Marra), G.G., M.R., S.C. and G.B.P.; software, validation, writing—original draft preparation, investigation, and visualization, G.M. (Gilda Magliacane) and E.B.; data curation, G.M. (Gilda Magliacane), E.B. and M.G.C.; formal analysis, G.M. (Gilda Magliacane), E.B., G.T., M.G.C. and L.P.; resources, C.D., M.G.C., P.C., A.B., E.R. and G.M. (Giorgia Mangili); writing—review and editing, M.G.C., L.P., P.C., S.C., A.B., E.R., G.M. (Giorgia Mangili) and C.D.; funding acquisition, C.D., M.G.C. and G.M. (Giorgia Mangili). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of IRCCS San Raffaele Scientific Institute (approved 3 December 2018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Vargas A.N. Natural history of ovarian cancer. J. Cancer Sci. Ther. 2014;6:465. doi: 10.4172/1948-5956.1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Institutes of Health. National Cancer Institute . Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Female Breast Cancer. National Cancer Institute, NIH; Bethesda, MD, USA: 2020. [(accessed on 15 November 2022)]. Available online: https://seer.cancer.gov/statfacts/html/ovary.html. [Google Scholar]
- 4.Stewart C., Ralyea C., Lockwood S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019;35:151–156. doi: 10.1016/j.soncn.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Chiang Y.-C., Lin P.-H., Cheng W.-F. Homologous Recombination Deficiency Assays in Epithelial Ovarian Cancer: Current Status and Future Direction. Front. Oncol. 2021;11:675972. doi: 10.3389/fonc.2021.675972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George S.H.L., Shaw P. BRCA and early events in the development of serous ovarian cancer. Front. Oncol. 2014;4:5. doi: 10.3389/fonc.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed A.A., Etemadmoghadam D., Temple J., Lynch A.G., Riad M., Sharma R., Stewart C., Fereday S., Caldas C., Defazio A., et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 10.Watkins J.A., Irshad S., Grigoriadis A., Tutt A.N.J. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16:211. doi: 10.1186/bcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creeden J.F., Nanavaty N.S., Einloth K.R., Gillman C.E., Stanbery L., Hamouda D.M., Dworkin L., Nemunaitis J. Homologous recombination proficiency in ovarian and breast cancer patients. BMC Cancer. 2021;21:1154. doi: 10.1186/s12885-021-08863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ngoi N.Y.L., Tan D.S.P. The role of homologous recombination deficiency testing in ovarian cancer and its clinical implications: Do we need it? ESMO Open. 2021;6:100144. doi: 10.1016/j.esmoop.2021.100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzese E., Centonze S., Diana A., Carlino F., Guerrera L.P., Di Napoli M., De Vita F., Pignata S., Ciardiello F., Orditura M. PARP inhibitors in ovarian cancer. Cancer Treat. Rev. 2019;73:1–9. doi: 10.1016/j.ctrv.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Del Campo J.M., Matulonis U.A., Malander S., Provencher D., Mahner S., Follana P., Waters J., Berek J.S., Woie K., Oza A.M., et al. Niraparib maintenance therapy in patients with recurrent ovarian cancer after a partial response to the last platinum-based chemotherapy in the ENGOT-OV16/NOVA trial. J. Clin. Oncol. 2019;37:2968–2973. doi: 10.1200/JCO.18.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray-Coquard I., Pautier P., Pignata S., Pérol D., González-Martín A., Berger R., Fujiwara K., Vergote I., Colombo N., Mäenpää J., et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019;381:2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 16.González-Martín A., Pothuri B., Vergote I., DePont Christensen R., Graybill W., Mirza M.R., McCormick C., Lorusso D., Hoskins P., Freyer G., et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 17.Moore K.N., Secord A.A., Geller M.A., Miller D.S., Cloven N., Fleming G.F., Wahner Hendrickson A.E., Azodi M., DiSilvestro P., Oza A.M., et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): A multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019;20:636–648. doi: 10.1016/S1470-2045(19)30029-4. [DOI] [PubMed] [Google Scholar]
- 18.Coleman R.L., Fleming G.F., Brady M.F., Swisher E.M., Steffensen K.D., Friedlander M., Okamoto A., Moore K.N., Efrat Ben-Baruch N., Werner T.L., et al. Veliparib with First-Line Chemotherapy and as Maintenance Therapy in Ovarian Cancer. N. Engl. J. Med. 2019;381:2403–2415. doi: 10.1056/NEJMoa1909707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coleman R.L., Oza A.M., Lorusso D., Aghajanian C., Oaknin A., Dean A., Colombo N., Weberpals J.I., Clamp A., Scambia G., et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): Phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennington K.P., Walsh T., Harrell M.I., Lee M.K., Pennil C.C., Rendi M.H., Thornton A., Norquist B.M., Casadei S., Nord A.S., et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konstantinopoulos P.A., Lacchetti C., Annunziata C.M. Germline and Somatic Tumor Testing in Epithelial Ovarian Cancer: ASCO Guideline Summary. JCO Oncol. Pract. 2020;16:e835–e838. doi: 10.1200/JOP.19.00773. [DOI] [PubMed] [Google Scholar]
- 22.Schiavo Lena M., Cangi M.G., Pecciarini L., Francaviglia I., Grassini G., Maire R., Partelli S., Falconi M., Perren A., Doglioni C. Evidence of a common cell origin in a case of pancreatic mixed intraductal papillary mucinous neoplasm–neuroendocrine tumor. Virchows Arch. 2021;478:1215–1219. doi: 10.1007/s00428-020-02942-1. [DOI] [PubMed] [Google Scholar]
- 23.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Anesthesia Analg. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francaviglia I., Magliacane G., Lazzari C., Grassini G., Brunetto E., Dal Cin E., Girlando S., Medicina D., Smart C.E., Bulotta A., et al. Identification and monitoring of somatic mutations in circulating cell-free tumor DNA in lung cancer patients. Lung Cancer. 2019;134:225–232. doi: 10.1016/j.lungcan.2019.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Redegalli M., Grassini G., Magliacane G., Pecciarini L., Lena M.S., Smart C.E., Johnston R.L., Waddell N., Maestro R., Macchini M., et al. Routine molecular profiling in both resectable and unresectable pancreatic adenocarcinoma: Relevance of cytological samples. Clin. Gastroenterol. Hepatol. 2022 doi: 10.1016/j.cgh.2022.10.014. in press . [DOI] [PubMed] [Google Scholar]
- 26.Capoluongo E.D., Pellegrino B., Arenare L., Califano D., Scambia G., Beltrame L., Serra V., Scaglione G.L., Spina A., Cecere S.C., et al. Alternative academic approaches for testing homologous recombination deficiency in ovarian cancer in the MITO16A/MaNGO-OV2 trial. ESMO Open. 2022;7:100585. doi: 10.1016/j.esmoop.2022.100585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magliacane G., Grassini G., Bartocci P., Francaviglia I., Cin E.D., Barbieri G., Arrigoni G., Pecciarini L., Doglioni C., Cangi M.G. Rapid targeted somatic mutation analysis of solid tumors in routine clinical diagnostics. Oncotarget. 2015;6:30592–30603. doi: 10.18632/oncotarget.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weichert W., Lukashchuk N., Yarunin A., Riva L., Easter A., Bannister H., Qiu P., French T. An evaluation of the performance of molecular assays to identify homologous recombination deficiency-positive tumors in ovarian cancer. Int. J Gynecol. Cancer. 2021;31:A366. [Google Scholar]
- 29.Fumagalli C., Betella I., Ranghiero A., Guerini-Rocco E., Bonaldo G., Rappa A., Vacirca D., Colombo N., Barberis M. In-house testing for homologous recombination repair deficiency (HRD) testing in ovarian carcinoma: A feasibility study comparing AmoyDx HRD Focus panel with Myriad myChoiceCDx assay. Pathologica. 2022;114:288–294. doi: 10.32074/1591-951X-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendes-Pereira A.M., Martin S.A., Brough R., McCarthy A., Taylor J.R., Kim J., Waldman T., Lord C.J., Ashworth A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dedes K.J., Wilkerson P.M., Wetterskog D., Weigelt B., Ashworth A., Reis-Filho J.S. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10:1192–1199. doi: 10.4161/cc.10.8.15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkerson P.M., Dedes K.J., Wetterskog D., Mackay A., Lambros M.B., Mansour M., Frankum J., Lord C.J., Natrajan R., Ashworth A., et al. Functional characterization of EMSY gene amplification in human cancers. J. Pathol. 2011;225:29–42. doi: 10.1002/path.2944. [DOI] [PubMed] [Google Scholar]
- 33.Hughes-Davies L., Huntsman D., Ruas M., Fuks F., Bye J., Chin S.-F., Milner J., Brown L.A., Hsu F., Gilks B., et al. EMSY Links the BRCA2 Pathway to Sporadic Breast and Ovarian Cancer. Cell. 2003;115:523–535. doi: 10.1016/S0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 34.Konstantinopoulos P.A., Ceccaldi R., Shapiro G.I., D’Andrea A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noordermeer S.M., van Attikum H. PARP Inhibitor Resistance: A Tug-of-War in BRCA-Mutated Cells. Trends Cell Biol. 2019;29:820–834. doi: 10.1016/j.tcb.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Giudice E., Gentile M., Salutari V., Ricci C., Musacchio L., Carbone M.V., Ghizzoni V., Camarda F., Tronconi F., Nero C., et al. PARP Inhibitors Resistance: Mechanisms and Perspectives. Cancers. 2022;14:1420. doi: 10.3390/cancers14061420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loveday C., Breast Cancer Susceptibility Collaboration (UK) Turnbull C., Ramsay E., Hughes D., Ruark E., Frankum J.R., Bowden G., Kalmyrzaev B., Warren-Perry M., et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat. Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang X., Li X., Li W., Bai H., Zhang Z. PARP inhibitors in ovarian cancer: Sensitivity prediction and resistance mechanisms. J. Cell. Mol. Med. 2019;23:2303–2313. doi: 10.1111/jcmm.14133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pacheco-Barcia V., Muñoz A., Castro E., Ballesteros A.I., Marquina G., González-Díaz I., Colomer R., Romero-Laorden N. The Homologous Recombination Deficiency Scar in Advanced Cancer: Agnostic Targeting of Damaged DNA Repair. Cancers. 2022;14:2950. doi: 10.3390/cancers14122950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.