Abstract
Simple Summary
Feline calicivirus (FCV) is a common viral pathogen affecting domestic cats, which is responsible for diverse clinical presentations, commonly including upper respiratory tract signs, oral ulcerations, and a fever. In addition, FCV infection can be associated with severe pneumonia, lameness, and virulent systemic disease. In this study, we clinically and pathologically describe a rare case of FCV-associated paw and mouth disease in a febrile household cat. The FCV strains detected in the animal and in an overtly healthy cohabiting cat were analyzed to assess the phenotype and antigenic properties.
Abstract
Feline calicivirus (FCV) infection in cats can led to several diverse clinical presentations, ranging from mild upper respiratory signs to virulent systemic disease. Herein, we report a paw and mouth disease case in a 7-year-old household cat due to an FCV infection. An asymptomatic cat living in the same household was also infected with FCV. Clinical and pathological investigations were combined with the molecular and phenotypical characterization of the FCV strains. The RNA of the FCV was detected using qualitative and quantitative reverse transcription (RT)-PCR assays, and FCV antigen was confirmed by immunohistochemistry. After the whole genome analysis, the strains detected in the two cats appeared to be genetically diverse from FCVs previously detected in association with paw and mouth disease and with virulent systemic disease. Interestingly, the isolates obtained in this study were resistant to low pH conditions and slightly susceptible to bile salts, but they were susceptible to a trypsin treatment, revealing a phenotype pattern that is different from that which has been observed for respiratory FCVs.
Keywords: FCV, paw and mouth disease, immunohistochemistry, WGS, phenotypic and antigenic characterization
1. Introduction
Feline calicivirus (FCV), a member of the genus Vesivirus in the family Caliciviridae, is a small (~35–40 nm), icosahedral, non-enveloped virus that shares a characteristic morphology with approximately 32 cup-like depressions in an icosahedral symmetry. The viral genome is a positive-sense, single-stranded RNA molecule of about 7.7 kilobases (kb) in length, and it is predicted to encode three open reading frames (ORFs) [1]. ORF1 encodes a 200 kDa polyprotein that is processed by a viral proteinase to produce several nonstructural proteins [2,3], ORF2 encodes a 73-kDa capsid precursor protein (pre-VP1) that is post-translationally processed to release the 60 kDa mature capsid protein VP1 [4], and ORF3 encodes a small basic protein (VP2) with a predicted molecular weight of 12 kDa, which has been associated with the stability of the capsid [5,6].
FCV is a highly contagious pathogen with a widespread distribution in the general cat population. The virus was originally isolated in New Zealand in 1957 [7] from the intestinal content of cats showing upper respiratory symptoms, oral ulcerations, and a fever. Since then, FCV infection has been associated with a variety of clinical signs, including acute febrile lameness syndrome, abortion, severe pneumonia, and acute enteritis [8,9,10,11]. Furthermore, in the last two decades, there has been an increasing number of reports of virulent FCV strains, causing outbreaks of severe virulent systemic disease (VSD) with a high mortality [12,13,14,15,16,17,18,19,20,21,22,23]. The clinicopathological features of VSD-FCV differ substantially from those of “classical” FCV disease. The characteristic signs include a high persistent fever, marked subcutaneous oedema, mainly located on the limbs and face, ulcerative dermatitis, icterus, pulmonary oedema, and coagulation abnormalities [12,14,15]. Before the description of VSD-FCV [12], a paw and mouth disease (PMD) characterized by oedema and ulcerative lesions, mainly located on the limbs, head, and mouth had been described in Australia [24,25], and since then, it has been sporadically reported in the USA and Europe [26,27]. Although, the initial clinical presentation of the disease is similar to that reported for VSD, inner organ involvement and high mortality have not been observed in PMD. Additionally, in contrast to VSD-FCV, PMD has been reported in either single cases or in very small outbreaks, without an epizootic course [28].
Herein, we report the clinical and pathological characterization of a case of PMD associated with FCV infection in a household cat. The genome of the FCV strain was sequenced, and its phenotype and antigenic properties were assessed.
2. Materials and Methods
2.1. Case Description
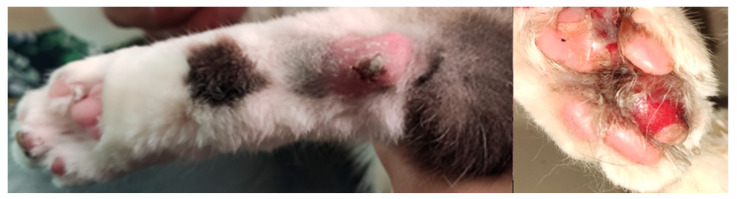
A 7-year-old male neutered domestic shorthair cat (#9284-5) was presented to the Veterinary Teaching Hospital at the Faculty of Veterinary Medicine, University of Teramo (Teramo, Italy) in July 2021 after 2 days of inappetence, lethargy, and lameness. The cat lived exclusively indoors along with a 9-year-old female cat (#9341-1) that did not present any clinical signs. Both of the cats received the last vaccination for feline panleukopenia, rhinotracheitis, and calicivirus in August 2018. Upon physical examination, the cat was febrile (40.7 °C) and showed painful swelling of all of its footpads, which over the next 2 days, progressed to erosion and ulceration (Figure 1), associated with the appearance of limb and tongue ulcerations.
Figure 1.
Severe ulcerative and necrotizing pododermatitis with serocellular crusts.
At the time of admission, a moderate lymphocytopenia (0.59 K/μL; reference interval (RI) 0.92–6.88 K/μL) and eosinopenia (0.00 K/μL; RI 0.17–1.57 K/μL) were noted on the complete blood count, while the serum chemistry revealed increased activities of creatine kinase (624 U/L; RI 91–326 U/L), alanine aminotransferase (47 U/L; RI 22–45 U/L), and aspartate aminotransferase (44 U/L; RI 14–41 U/L). The cat tested negative for FIV and FeLV on a point-of-care (POC) immunochromatographic test (SNAP FIV/FeLV Combo Test, IDEXX) (IDEXX Laboratories, Inc., Westbrook, ME, USA). Plain radiography of the thorax and abdominal ultrasound were within normal limits. A biopsy specimen was collected at the edge of a footpad ulcerative lesion by using an 8 mm biopsy punch, which was processed for paraffin sectioning and stained with haematoxylin and eosin. The histological examination revealed a severe, diffuse, ulcerative pododermatitis, which was characterized by full-thickness necrosis of the epidermis and dermal granulocytic and lympho-histiocytic inflammation. No fungal elements or bacteria were evident. Additional samples of the affected tissues were collected using cytobrushes, along with blood and serum samples and oropharyngeal and rectal swabs for virological investigation. The sample collection also included an oropharyngeal specimen and a serum sample obtained from the asymptomatic cat living with the diseased animal. Informed consent was obtained from owner of the cats. The presence of common feline viral pathogens including FCV, feline herpesvirus type 1 (FHV-1), feline panleukopenia parvovirus (FPV), and feline coronavirus (FCoV) was investigated either by quantitative (real-time) PCR (qPCR) or qualitative PCR after reverse transcription (RT) [29,30,31,32]. The serum and blood samples were also tested for FeLV and FIV pro-viral DNAs [33,34].
The cat, #9284-5, was hospitalized, and enteral nutrition was guaranteed via a rhino-esophageal feeding tube; the patient was treated with prednisolone 1 mg/kg q24h, subcutaneously, buprenorphine 20 μg/kg q8h EV, and doxycycline oral paste, which were given through feeding tube at the dosage of 10 mg/kg once a day. In addition, a chlorhexidine-based odontostomatological gel was administered once a day. The cat became normothermic after 3 days and restarted to eat voluntarily. After one week, the patient was discharged from the hospital, and the prednisolone treatment was gradually tapered off. Two weeks later, the cat was presented for a control visit, and at the physical examination, the complete healing of the ulcerative lesions was observed. Additionally, the oropharyngeal swabs of both of the cats were negative for FCV on the qualitative and quantitative RT-PCR.
2.2. Real Time RT-PCR (RT-qPCR) for FCV
The FCV RNA quantification was performed using the SuperScript III platinum OneStep Quantitative RT-PCR system (Invitrogen Ltd., Milan, Italy) in a 25 μL volume comprising 5 μL of extracted RNA and 20 μL of master mix. The primers (FCV For: 5′-GTTGGATGAACTACCCGCCAATC-3′ and FCV Rev: 5′- CATATGCGGCTCTGATGGCTTGAAACTG-3′) and a TaqMan probe (FCV Probe: 5′-TCGGTGTTTGATTTGGCCTG-3′) [35,36] were used at concentrations of 200 and 100 nM, respectively. A standard curve was generated using tenfold serial dilutions (from 109 to 100 copies per reaction) of FCV plasmid DNA, which were constructed by cloning 83 bp ORF1 fragment of the strain 160/2015/ITA (GenBank accession no. MT00824650) [11] into the pCR 2.1 vector of the TA Cloning Kit (Invitrogen, Ltd., Milan, Italy). To prevent any contamination occurring during the quantitative RT-PCR assay, three negative controls containing all of the necessary components of the RT-PCR mixtures, except for the template RNA, were included in each run.
2.3. Immunohistochemistry (IHC) Assay
Sections of 5 µm were cut from the formalin-fixed and paraffin-embedded (FFPE) skin biopsies of the footpad ulcerative lesion and mounted on positive-charged slides. Immunohistochemistry was performed using a rabbit polyclonal antibody raised against the FCV-F9 vaccine strain [37]. Briefly, the FFPE tissue sections were deparaffinized, rehydrated, and washed in distilled water. Endogenous peroxidase was blocked with 3% H2O2 for 8 min at room temperature. The slides were then washed in Tris Buffered Saline (TBS) (Invitrogen Ltd., Milan, Italy) and incubated with the primary antibody for 1 h in a humidified chamber at room temperature. The slides were incubated with secondary biotinylated universal secondary antibody, which was followed by sequential incubation with peroxidase-labelled streptavidin (LSAB+/System-HRP, Dako, Glostrup, Denmark). The negative control was treated in the same manner, omitting the primary antibody, and incubating the tissue sections with TBS.
2.4. Whole Genome Sequencing (WGS) and Phylogenetic Analysis
The oropharyngeal swab, and the foot skin and blood samples from cat #9284-5, and the oropharyngeal specimen collected from the cohabiting cat (#9341-1) were subjected to a sequence-independent single-primer amplification (SISPA) enrichment, as previously described [38,39,40], and to deep sequencing to generate the complete genome of the FCV strain. The PCR-enriched samples were cleaned up with AMPure XP beads (Beckman Coulter, Brea, CA, USA), quantified by a Qubit dsDNA HS assay (Thermo Fisher Scientific, Waltham, MA, USA), and used for the libraries preparation and for adapter ligation by the Ligation kit SQK-LSK110 (Oxford Nanopore Technologies, ONT, Oxford, UK) following manufacturer’s guidelines. Each test was run for nine hours on an Oxford Nanopore MinION Mk1C device using FLO-MIN106 R9.4.1 flow cell after loading 75 μL of the sequencing mix (12 μL library, 25.5 μL loading beads II, and 37.5 μL sequencing buffer II). The ONT MinKNOW software (v3.1.5) (Oxford Nanopore Technologies, ONT, Oxford, UK) was used to collect the raw sequencing data, and ONT cloud-based basecaller based on GUPPY (v3.2.8) (Oxford Nanopore Technologies, ONT, Oxford, UK) was used to perform the on-site and real-time basecalling during the sequencing run. Sequence trimming, the assembly of reads, and genome annotation were performed using the Geneious Prime version 2021.1.1 (Biomatters Ltd., Auckland, New Zealand). The FCV-F9 strain (GenBank accession no. M86379) was used to create a reference-based assembly by Minimap2. Open reading frame (ORF) prediction and annotations were performed in Geneious Prime software v. 2021.2.2 (Biomatters Ltd., Auckland, New Zealand), and the alignment of the sequences was conducted using Multiple Alignment based on Fast Fourier Transform (MAFFT) [41]. The phylogenetic analysis was performed in MEGA X software [42].
2.5. Virus Isolation and Phenotypic Characterization
The Crandell-Reese feline kidney (CRFK) [43] cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated foetal bovine serum. Attempts to conduct the virus isolation were performed as previously described [44]. Each isolate was purified for three times by a plaque assay, which was followed by the preparation of virus stocks obtained by the inoculation of the CRFK cells at a multiplicity of infection of 0.1. The suspensions were harvested after 24 h by three cycles of freezing and thawing and clarified by centrifugation at 1800 rpm for 10 min. The resulting titre was calculated by the Reed and Muench endpoint method, which identified the final infectivity at 50% tissue culture infectious doses (TCID50). All of the isolates were assessed to investigate the in vitro stability when they are exposed to a low pH (3.0), trypsin (final concentration of 0.5%), and to a bile salts (final concentration of 0.5%) treatment using protocols that have been previously described [11].
2.6. Seroneutralization Assays
The serum samples collected from both of the cats (#9284-5 and #9341-1) were tested for the presence of neutralizing antibodies against the FCV-F9 vaccine strain by performing seroneutralization (SN) assays as previously described [45]. Briefly, 50 μL of each serial twofold diluted serum and 50 μL of infectious culture medium containing 100 TCID50 of the FCV-F9 strain were mixed and incubated for 1 h at 37 °C. One hundred microliters (1 × 105 cell/mL) of CRFK cell suspension were added to each well. The assay was performed in triplicate for each serum. After 3 days of incubation at 37 °C with 5% CO2, the end point titers were determined as the highest serum dilution that was able to neutralize the cytopathic effect of the virus. To investigate the antigenic correlation between the field isolates and the vaccine strain, the rabbit polyclonal antibody raised against the FCV-F9 [37] was tested for its neutralizing activity against the FCV strains isolated either from the affected cat or from the asymptomatic co-habiting animal following the procedure described above. An internal FCV-F9 homologous control was included in each experiment.
3. Results
3.1. Molecular Identification
By using a nested RT-PCR protocol [32] that was able to amplify a short diagnostic genome fragment (477-nt) of the ORF2 region of FCV, viral RNA was detected in the skin, blood, oropharyngeal, and enteric samples collected from the cat #9284-05, whilst all of the specimens tested negative for FHV-1, FPV, FCoV, FeLV, and FIV, thereby ruling out mixed infections. The subsequent RT-qPCR [35,36] revealed viral loads ranging from 1.1 × 101 to 4.1 × 105 RNA copies/5 μL of the RNA template (Table 1), with the highest titers being found, respectively, in the oropharyngeal swab (4.1 × 105) and in the skin sample (2.6 × 105). The FCV RNA at the viral load of 3.8 × 104 RNA copies/5 μL was also detected in the oropharyngeal specimen collected from the healthy cohabiting cat (#9341-1). All of the negative controls did not produce any detectable fluorescence signal.
Table 1.
Quantification of FCV RNA by qRT-PCR in samples collected from cats #9284-5 and #9341-1.
| Cat ID | Sample | qRT-PCR (Ct) | RNA Copies/5 μL |
|---|---|---|---|
| #9284-5 | Oropharyngeal swab | 13.7 | 4.1 × 105 |
| Footpad skin sample | 15.4 | 2.6 × 105 | |
| Rectal swab | 30.4 | 1.2 × 102 | |
| Blood | 33.6 | 1.1 × 101 | |
| #9341-1 | Oropharyngeal swab | 20.7 | 3.8 × 104 |
3.2. IHC Identification
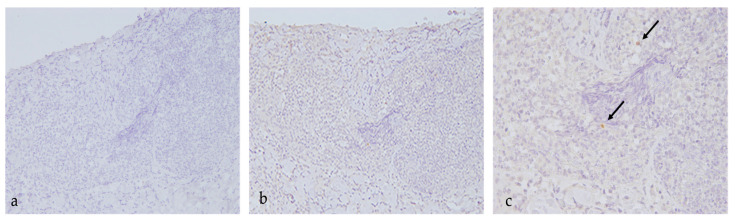
By performing IHC staining on the footpad skin lesion sample, scattered positive cells were evident in the deep lympho-histiocyticinflammatory infiltrate, confirming the presence of the FCV antigen (Figure 2).
Figure 2.
IHC on skin biopsy of the footpad ulcerative lesion using a rabbit polyclonal antibody raised against the FCV-F9 vaccine strain [37]. (a) (20X): negative control; (b) (20X), (c) (40X): positive cells. Arrows indicate positive histiocytes.
3.3. Molecular Characterization
The combination of an SISPA approach with an ONT sequencing platform allowed us to reconstruct the full-length genome sequences, encompassing the entire ORF1, ORF2, and ORF3 genes of strains 9284-5/2020/ITA detected in the oropharyngeal swab (OS) and skin (S) samples. Additionally, the ONT platform was successfully used to generate the complete genome of the strain 9341-1/2020/ITA identified in the oropharyngeal swab from the asymptomatic cohabiting cat. The complete genomes of FCV-S 9284-5/2020/ITA generated from the skin and of FCV-OS 9341-1/2020/ITA identified in the oropharyngeal swab were deposited in GenBank under the accession numbers OP750454 and OP750455, respectively. On pairwise homology and distance analyses, strains FCV-OS 9284-5/2020/ITA and FCV-OS 9341-1/2020/ITA displayed a nucleotide (nt) identity of 99.9% to each other with a deduced amino acid (aa) identity of 100% either in the polyprotein and in the structural proteins VP1 and VP2.A single aa substitution in position 432 (Ala-432-Val) was observed in the capsid protein of the skin virus isolate FCV-S 9284-5/2020/ITA.
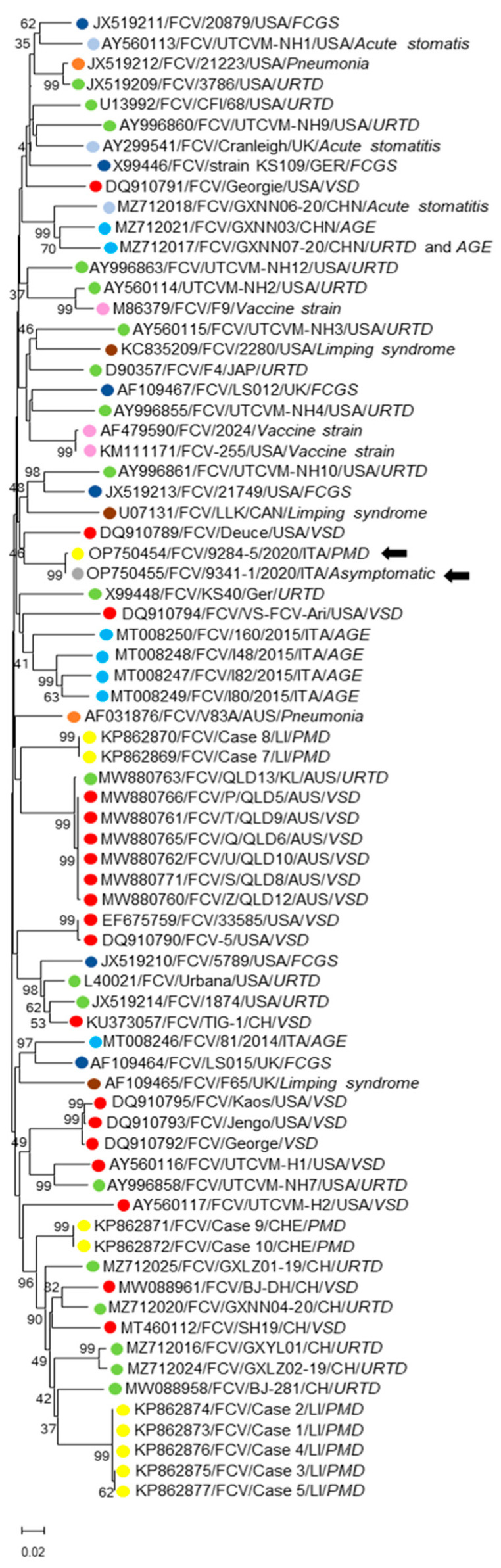
Upon the preliminary analyses with BLAST (www.ncbi.nlm.nih.gov/blast, accessed on 10 September 2022) and FASTA (www.ebi.ac.uk/fasta33, accessed on 10 September 2022), the overall nt identity similarities of the FCV-OS strains 9284-5/2020/ITA and 9341-1/2020/ITA to the FCV complete sequences currently available in the databases ranged from 76.2% to 81.2%. The distance and maximum likelihood-based phylogenetic analyses were performed using a selection of VP1 capsid sequences consisting of three FCV vaccine strains (FCV F9, FCV 255, and FCV 2024) and 68 field strains retrieved from GenBank and isolated from distinct disease manifestations. In the VP1 aa sequence, the similarities of the identities of strains 9284-5/2020/ITA and 9341-1/2020/ITA to other FCVs were 83.1–91.3%, with the highest identity being attributed (91.1–91.3%) to a VSD isolate (FCV/Deuce/USA/VSD DQ910789) [18]. The overall similarities of the aa identities of all of the FCV capsid sequences available in the databases ranged from 81.6% to 92.3%. The similarities of the identities of the two Italian strains to FCV sequences associated with PMD, VSD, and upper respiratory tract disease (URTD) greatly overlapped with each other, with values, respectively, of 85.2–90.4%, 86.3–91.3%, and 85.5–90.5%. Additionally, upon visual inspection of the alignments, the aa changes unique to the PMD or VSD isolates were not found. In the VP1-based tree (Figure 3), the FCVs were grouped into several polyphyletic clades, without any evidence of clustering patterns correlated to a clinical disease. The strains 9284-5/2020/ITA and 9341-1/2020/ITA were grouped alone with FCV/Deuce/USA (DQ910789), although not tightly. A similar phylogenetic topography was also observed in the tree performed only on the region E (aa 426-523) (data not shown) of the VP1 protein.
Figure 3.
Phylogenetic analysis based on the aa sequence of the full-length VP1 capsid protein was generated using the maximum likelihood. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of aa substitutions per site, supplying statistical support with bootstrapping of 1000 replicates. Arrows indicate the FCV strains detected in this study. Abbreviations: VSD = virulent systemic disease; PMD = paw and mouth disease; URTD = upper respiratory tract disease; FCGS = feline chronic gingivostomatitis; AGE = acute gastroenteritis. Disease phenotype is colour coded.
To further investigate the possible genetic relationship of our strains with VSD-FCVs, the inferred aa sequences of the hypervariable region E of the Italian viruses were mapped to identify seven aa residue positions (438, 440, 448, 452, and 455, 465, and 492), whose physical and chemical properties were previously shown to be statistically significant for differentiation between the URTD and VSD FCV pathotypes [46]. The alignments were performed, including the corresponding sequences extracted from nine FCV strains previously detected in cats showing paw and mouth signs (KP862869–KP862877) [27] (Table 2). In our analysis, the predicted properties for the virulent pathotypes were found in five of the seven residues (positions 438, 440, 452, 455, and 492) of the hypervariable region E of both of the strains 9284-5/2020/ITA and 9341-1/2020/ITA and in only three positions of the paw and mouth disease FCVs.
Table 2.
Residues of region E of the VP1 capsid protein statistically associated with the VSD-FCV pathotype [46].
| Physicochemical Properties of Amino Acids Associated with VSD Pathotype | |||||||
|---|---|---|---|---|---|---|---|
| Strain | 438 | 440 | 448 | 452 | 455 | 465 | 492 |
| Non Polar, Aliphatic Chain |
Non Small | Polar, Positive Charged |
Non Small | Non Negative |
Polar | Small | |
| VSD FCV | V9T8 | Q6G5E4SK | K7A2E2G2P2R2 | E11D6 | T6D4M2I2NES | S14G3 | V16R |
| URTD FCV | T37V2I | G22S6Q4R2A2ENDT | A30P4G3K3 | D36E3N | D27T5S3V2GRE | G26S14 | V18L8I7R5K2 |
| 9284-5/2020/ITA/PMD (OP750454) |
V | H | L | E | I | G | V |
| 9341-1/2020/ITA/ Asyntomatic (OP750455) |
V | H | L | E | I | G | V |
| Case 1/LI/PMD (KP862873) | T | E | P | D | K | S | I |
| Case 2/LI/PMD (KP862874) | T | E | P | D | K | S | I |
| Case 3/LI/PMD (KP862875) | T | E | P | D | K | S | I |
| Case 4/LI/PMD (KP862876) | T | E | P | D | K | S | I |
| Case 5/LI/PMD (KP862877) | T | E | P | D | K | S | I |
| Case 7/LI/PMD (KP862869) | I | G | R | E | D | G | L |
| Case 8/LI/PMD (KP862870) | I | G | R | E | D | G | L |
| Case 9/LI/PMD (KP862871) | T | G | P | D | A | S | V |
| Case 10/LI/PMD (KP862872) | T | G | P | D | A | S | V |
Residues in bold indicate amino acids matching with the VSD configuration.
3.4. Biochemical and Antigenic Characterization
FCV was successful isolated from the skin and oropharyngeal samples of the affected cat (#9284-5), as well as from the oropharyngeal swab of the asymptomatic cat (#9341-1). The three isolates were titrated and analyzed to investigate their in vitro sensitivity to the pH, bile salts, and trypsin treatments. As reported in Table 3, all of the isolates were completely resistant at pH 3.0, and they were slightly susceptible to the bile salts, with a titres reduction of 1.25 log10, whilst after exposure to trypsin, a decrease in the titres of 3.0 log10 was observed. By assessing the serum samples from both of the cats by the SN assay, specific neutralizing antibodies anti-FCV-F9 were detected, respectively, at titers of 1:64 in the affected cat and 1:128 in the cohabiting healthy animal. When we tested the anti-FCV-F9 serum against the three isolates, all of the strains were neutralized at titres of 1:8, while the neutralizing activity against the homologous virus occurred at 1:256.
Table 3.
Titre of FCV (log10 TCID50) and log10 reduction obtained after application of different treatments.
| Oropharyngeal Swab 9284-5/2020/ITA |
Footpad Skin Sample 9284-5/2020/ITA |
Oropharyngeal Swab 9341-1/2020/ITA |
||||
|---|---|---|---|---|---|---|
| Control | Treated | Control | Treated | Control | Treated | |
| HCl—pH3 | 7.25 | 7.25 | 7.25 | 7.25 | 7.25 | 7.25 |
| Bile salts (0.5%) | 7.25 | 6.0 | 7.25 | 6.0 | 7.25 | 6.0 |
| Trypsin (0.5%) | 7.25 | 4.25 | 7.25 | 4.25 | 7.25 | 4.25 |
4. Discussion
The high degree of genomic plasticity of FCV accounts for the emergence of several variants, some of which are associated with a severe clinical disease. This is exemplified by the increasing number of reports on VSD-FCVs, which are divergent from the “classical mild FCVs” for their marked tissue tropism for epithelial and endothelial cells, multisystemic target, the induction of systemic vascular failure, and for involvement of visceral organs [12,13,14,15,16,17,18,19,20,21,22,23]. In contrast, FCV-associated PMD has been described on only a few occasions [24,25,26,27], and it is still unclear whether these cases represented mild forms of VSD or they were rather the clinical presentation of a distinct syndrome.
In this study, we documented a case of FCV infection in a 7-year-old cat showing ulcerative lesions on the skin of their paws and in and around the mouth, but this occurred without inner organs involvement. The presence of the FCV antigen in the skin lesion was confirmed by the IHC assay. Furthermore, the molecular screening by qualitative and quantitative RT-PCRs detected FCV RNA in all of the tested specimens, with the highest viral RNA loads being in the oropharyngeal and skin samples, and the lowest ones being in blood and rectal specimens. A high viral load was also found in the oropharyngeal specimen collected from a clinically healthy cat living in contact with the animal with PMD. After the complete genome analysis, the viruses of oropharyngeal origin identified in the two cats were identical to the deduced aa sequence (100% aa identities in polyprotein, VP1 and VP2), whilst the isolate detected in the skin sample differed for a single aa substitution (432) which was located in the 5′ hypervariable region E (426–460 aa residues) of the VP1. Attempts were made to investigate the genetic relationship between the FCV strains detected in this study and FCV strains previously found in association with PMD, VSD, URTD, acute and chronic gingivostomatitis, limping syndrome, and acute gastroenteritis. As expected, the VP1-based phylogenetic analysis failed to identify the well-defined pathotype-associated clusters. The intrinsic molecular mechanisms by which some FCV isolates show a highly virulent phenotype are still unclear. The analysis of the FCV sequences from VP1 region E has been used to differentiate VSD from classical respiratory FCVs, and we have identified seven key residue positions (438, 440, 448, 453, 455, 465, and 492) which are statistically significant for pathotype differentiation, and these are mainly located in the N-terminal hypervariable part of region E [46]. This domain displays the highest variability along the VP1 protein, and it includes immunodominant neutralizing epitopes that are able to interact with the feline cell receptor JAM-1 during the viral attachment [47,48,49,50,51,52]. In our analysis, after the residue mapping of the hypervariable region E of the strains 9284-5/2020/ITA and 9341-1/2020/ITA, only five of the seven key residue positions showed the predicted physical and chemical properties. Likewise, none of FCV strains previously detected in the cats with suspected PMD [27] showed the VSD combination pattern.
Both of the strains (9284-5/2020/ITA and 9341-1/2020/ITA) detected in the oropharyngeal swabs and in the footpad skin sample (9284-5/2020/ITA) were adapted to grow in the CRFK line, while the viruses identified from the blood and rectal swabs could not be adapted to replicate in vitro. This could be accounted for by the low viral loads found in the faecal swab (1.2 × 102) and the blood sample (1.1 × 101). There is evidence that FCVs associated with atypical clinical signs possess in vitro phenotype properties that are different from FCV strains which are responsible for the classical respiratory disease [11,18,53]. The in vitro characteristics of the three isolates obtained in this study revealed a high resistance to a low pH treatment and only a moderate susceptibility to the bile salts, yet, they were susceptible to the trypsin treatment, differing in their phenotype patterns from FCV strains of respiratory origin. Cats #9284-5 and #9341-1 had been immunized against FCV with a vaccine based on the live attenuated strain F9, although the vaccination dated back to 2018, i.e., 3 years before the PMD case. Neutralizing antibodies versus FCV F9 were detected in the serum collected from both of the animals at titers, respectively, of 1:64 in the PMD cat and of 1:128 in the cohabiting healthy animal. However, when we were testing the neutralizing activity of an F9 antisera, all of the three isolates were neutralized at titers of 1:8, suggesting a low antigenic cross-reactivity between the field strains obtained in this study and the vaccine FCV-F9. Additionally, these findings confirmed the remarkable variability of FCVs in terms of the virus neutralization profile [27,54].
In the present case report, the origin of the infection remains unknown since both of the cats were exclusively kept indoors. However, the contact cat was hospitalized for an orthopedic problem two weeks before the onset of the clinical signs in cat #9284-5. Accordingly, we speculated that cat #9341-1 could have acquired an asymptomatic infection during hospitalization, acting as carrier for FCV.
5. Conclusions
We described a rare case of PMD due to an FCV infection in a vaccinated cat that received its last booster 3 years before. The same virus was detected in a contact animal that was overtly healthy and that likely introduced the virus in the household after a nosocomial inapparent infection. Genetic hallmarks accounting for the PMD clinical form were not found in PMD-associated FCVs, although the FCV strain sequenced from the skin lesions differed in a unique aa mutation (Ala-432-Val) in the hypervariable region of the capsid. The whole genome sequencing of virus pairs or clones, i.e., viral strains obtained from different lesions/tissues of the same animal, could help us to decipher the mechanisms driving phenotype shifts in FCV.
Author Contributions
Conceptualization, F.D.P. and B.D.M.; methodology, B.D.M., F.M., M.C., A.S. and V.M.; validation, V.M., M.C., A.S. and G.L.; investigation, A.P., V.S., M.V.G., P.F., L.B., V.R.; data curation, A.P., V.S., F.D.P.; writing—original draft preparation, B.D.M., F.D.P., A.P., P.E.C.; writing—review and editing, V.M., M.C., A.S., G.L., G.A.; supervision B.D.M., G.A., F.D.P.; funding acquisition, B.D.M. and F.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical review and approval were waived for this study because all of the samples were collected for diagnostic purposes. Furthermore, blood sampling was conducted during necessary clinical investigations with the written consent of the owner during the routinely clinical activity at the Veterinary Teaching Hospital of the University of Teramo.
Informed Consent Statement
Written informed consent was asked for and signed by the owner of animals.
Data Availability Statement
The data presented in this study are available in this manuscript. Sequence data presented in this study are openly available in the GenBank database.
Conflicts of Interest
All of the authors declare that there are no financial or other relationships that might lead to a conflict of interest.
Funding Statement
The present study has been carried out in the framework of the Project “Demetra” (Dipartimenti di Eccellenza 2018–2022, CUP_C46C18000530001), and it was funded by the Italian Ministry for Education, University and Research.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Carter M.J., Milton I.D., Meanger J., Bennett M., Gaskell R.M., Turner P.C. The complete nucleotide sequence of a feline calicivirus. Virology. 1992;190:443–448. doi: 10.1016/0042-6822(92)91231-I. [DOI] [PubMed] [Google Scholar]
- 2.Sosnovtseva S.A., Sosnovtsev S.V., Green K.Y. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 1999;73:6626–6633. doi: 10.1128/JVI.73.8.6626-6633.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sosnovtsev S.V., Garfield M., Green K.Y. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 2002;76:7060–7072. doi: 10.1128/JVI.76.14.7060-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sosnovtsev S.V., Sosnovtseva S.A., Green K.Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 1998;72:3051–3059. doi: 10.1128/JVI.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosnovtsev S.V., Belliot G., Chang K.O., Onwudiwe O., Green K.Y. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Martino B., Marsilio F. Feline calicivirus VP2 is involved in the self-assembly of the capsid protein into virus-like particles. Res. Vet. Sci. 2010;89:279–281. doi: 10.1016/j.rvsc.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Fastier L.B. A new feline virus isolated in tissue culture. Am. J. Vet. Res. 1957;18:382–389. [PubMed] [Google Scholar]
- 8.Ellis T.M. Jaundice in a Siamese cat with in utero feline calicivirus infection. Aust. Vet. J. 1981;57:383–385. doi: 10.1111/j.1751-0813.1981.tb00527.x. [DOI] [PubMed] [Google Scholar]
- 9.Pedersen N.C., Laliberte L., Ekman S. A transient febrile limping syndrome of kittens caused by two different strains of feline calicivirus. Feline Pract. 1983;13:26–35. [Google Scholar]
- 10.Dawson S., Bennett D., Carter S.D., Bennett M., Meanger J., Turner P.C., Carter M.J., Milton I., Gaskell R.M. Acute arthritis of cats associated with feline calicivirus infection. Res. Vet. Sci. 1994;56:133–143. doi: 10.1016/0034-5288(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 11.Di Martino B., Lanave G., Di Profio F., Melegari I., Marsilio F., Camero M., Catella C., Capozza P., Bányai K., Barrs V.R., et al. Identification of feline calicivirus in cats with enteritis. Transbound. Emerg. Dis. 2020;67:2579–2588. doi: 10.1111/tbed.13605. [DOI] [PubMed] [Google Scholar]
- 12.Pedersen N.C., Elliott J.B., Glasgow A., Poland A., Keel K. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Vet. Microbiol. 2000;73:281–300. doi: 10.1016/S0378-1135(00)00183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schorr-Evans E.M., Poland A., Johnson W.E., Pedersen N.C. An epizootic of highly virulent feline calicivirus disease in a hospital setting in New England. J. Feline Med. Surg. 2003;5:217–226. doi: 10.1016/S1098-612X(03)00008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley K.E., Pesavento P.A., Pedersen N.C., Poland A.M., Wilson E., Foley J.E. An outbreak of virulent systemic feline calicivirus disease. J. Am. Vet. Med. Assoc. 2004;224:241–249. doi: 10.2460/javma.2004.224.241. [DOI] [PubMed] [Google Scholar]
- 15.Pesavento P.A., MacLachlan N.J., Dillard-Telm L., Grant C.K., Hurley K.F. Pathologic, immunohistochemical, and electron microscopic findings in naturally occurring virulent systemic feline calicivirus infection in cats. Vet. Pathol. 2004;41:257–263. doi: 10.1354/vp.41-3-257. [DOI] [PubMed] [Google Scholar]
- 16.Abd-Eldaim M., Potgieter L., Kennedy M. Genetic analysis of feline caliciviruses associated with a hemorrhagic-like disease. J. Vet. Diagn. Invest. 2005;17:420–429. doi: 10.1177/104063870501700503. [DOI] [PubMed] [Google Scholar]
- 17.Coyne K.P., Jones B.R., Kipar A., Chantrey J., Porter C.J., Barber P.J., Dawson S., Gaskell R.M., Radford A.D. Lethal outbreak of disease associated with feline calicivirus infection in cats. Vet. Rec. 2006;158:544–550. doi: 10.1136/vr.158.16.544. [DOI] [PubMed] [Google Scholar]
- 18.Ossiboff R.J., Sheh A., Shotton J., Pesavento P.A., Parker J.S.L. Feline caliciviruses (FCVs) isolated from cats with virulent systemic disease possess in vitro phenotypes distinct from those of other FCV isolates. J. Gen. Virol. 2007;88:506–517. doi: 10.1099/vir.0.82488-0. [DOI] [PubMed] [Google Scholar]
- 19.Reynolds B.S., Poulet H., Pingret J.L., Jas D., Brunet S., Lemeter C., Etievant M., Boucraut-Baralon C. A nosocomial outbreak of feline calicivirus associated virulent systemic disease in France. J. Feline Med. Surg. 2009;11:633–644. doi: 10.1016/j.jfms.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H., Miao Q., Zhu J., Yang Z., Liu G. Isolation and molecular characterization of a virulent systemic feline calicivirus isolated in China. Infect. Genet. Evol. 2018;65:425–429. doi: 10.1016/j.meegid.2018.08.029. [DOI] [PubMed] [Google Scholar]
- 21.Caringella F., Elia G., Decaro N., Martella V., Lanave G., Varello K., Catella C., Diakoudi G., Carelli G., Colaianni M.L., et al. Feline calicivirus infection in cats with virulent systemic disease, Italy. Res. Vet. Sci. 2019;124:46–51. doi: 10.1016/j.rvsc.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z., Xin T., Wei J., Jiang Y., Liu X., Song W., Guo X., Yuan W., Cui Y., Zhu H., et al. Isolation and phylogenetic analysis of strains of feline calicivirus in Beijing, China. Arch. Virol. 2021;166:2521–2527. doi: 10.1007/s00705-021-05163-2. [DOI] [PubMed] [Google Scholar]
- 23.Bordicchia M., Fumian T.M., Van Brussel K., Russo A.G., Carrai M., Le S.J., Pesavento P.A., Holmes E.C., Martella V., White P., et al. Feline Calicivirus Virulent Systemic Disease: Clinical Epidemiology, Analysis of Viral Isolates and In Vitro Efficacy of Novel Antivirals in Australian Outbreaks. Viruses. 2021;13:2040. doi: 10.3390/v13102040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper L.M., Sabine M. Paw and mouth disease in a cat. Aust. Vet. J. 1972;48:644. doi: 10.1111/j.1751-0813.1972.tb05107.x. [DOI] [PubMed] [Google Scholar]
- 25.Love D.N., Zuber R.M. Feline calicivirus associated with pyrexia, profound anorexia and oral and perianal ulceration in a cat. Aust. Vet. Pract. 1987;17:136–137. [Google Scholar]
- 26.Meyer A., Kershaw O., Klopfleisch R. Feline calicivirus-associated virulent systemic disease: Not necessarily a local epizootic problem. Vet. Rec. 2011;168:589. doi: 10.1136/vr.d160. [DOI] [PubMed] [Google Scholar]
- 27.Willi B., Spiri A.M., Meli M.L., Samman A., Hoffmann K., Sydler T., Cattori V., Graf F., Diserens K.A., Padrutt I., et al. Molecular characterization and virus neutralization patterns of severe, non-epizootic forms of feline calicivirus infections resembling virulent systemic disease in cats in Switzerland and in Liechtenstein. Vet. Microbiol. 2016;182:202–212. doi: 10.1016/j.vetmic.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann-Lehmann R., Hosie M.J., Hartmann K., Egberink H., Truyen U., Tasker S., Belák S., Boucraut-Baralon C., Frymus T., Lloret A., et al. Calicivirus infection in cats. Viruses. 2022;14:937. doi: 10.3390/v14050937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gut M., Leutenegger C.M., Huder J.B., Pedersen N.C., Lutz H. One-tube fluorogenic reverse transcription-polymerase chain reaction for the quantitation of feline coronaviruses. J. Virol. Methods. 1999;77:37–46. doi: 10.1016/S0166-0934(98)00129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buonavoglia C., Martella V., Pratelli A., Tempesta M., Cavalli A., Buonavoglia D., Bozzo G., Elia G., Decaro N., Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 2001;82:3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- 31.Marsilio F., Di Martino B., Di Francesco C. Use of a duplex-PCR assay to screen for Feline Herpesvirus-1 and Chlamydophila spp. in mucosal swabs from cats. New Microbiol. 2004;27:287–292. [PubMed] [Google Scholar]
- 32.Marsilio F., Di Martino B., Decaro N., Buonavoglia C. A novel nested PCR for the diagnosis of calicivirus infections in the cat. Vet. Microbiol. 2005;105:1–7. doi: 10.1016/j.vetmic.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 33.Quackenbush S.L., Dean G.A., Mullins J.I., Hoover E.A. Analysis of FeLV-FAIDS provirus burden and productive infection in lymphocyte subsets in vivo. Virology. 1996;223:1–9. doi: 10.1006/viro.1996.0449. [DOI] [PubMed] [Google Scholar]
- 34.Stiles J., Bienzle D., Render J.A., Buyukmihci N.C., Johnson E.C. Use of nested polymerase chain reaction (PCR) for detection of retroviruses from formalin-fixed, paraffin-embedded uveal melanomas in cats. Vet. Ophthalmol. 1999;2:113–116. doi: 10.1046/j.1463-5224.1999.00066.x. [DOI] [PubMed] [Google Scholar]
- 35.Helps C., Lait P., Tasker S., Harbour D. Melting curve analysis of feline calicivirus isolates detected by real-time reverse transcription PCR. J. Virol. Methods. 2002;106:241–244. doi: 10.1016/S0166-0934(02)00167-2. [DOI] [PubMed] [Google Scholar]
- 36.Brunner C., Kanellos T., Meli M.L., Sutton D.J., Gisler R., Gomes-Keller M.A., Hofmann-Lehmann R., Lutz H. Antibody induction after combined application of an adjuvanted recombinant FeLV vaccine and a multivalent modified live virus vaccine with a chlamydial component. Vaccine. 2006;24:1838–1846. doi: 10.1016/j.vaccine.2005.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Martino B., Marsilio F., Roy P. Assembly of feline calicivirus-like particle and its immunogenicity. Vet. Microbiol. 2007;120:173–178. doi: 10.1016/j.vetmic.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 38.Reyes G.R., Kim J.P. Sequence-independent, single-primer amplification (SISPA) of complex DNA populations. Mol. Cell. Probes. 1991;5:473–481. doi: 10.1016/S0890-8508(05)80020-9. [DOI] [PubMed] [Google Scholar]
- 39.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Djikeng A., Halpin R., Kuzmickas R., Depasse J., Feldblyum J., Sengamalay N., Afonso C., Zhang X., Anderson N.G., Ghedin E., et al. Viral genome sequencing by random priming methods. BMC Genom. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crandell R.A., Fabricant C.G., Nelson-Rees W.A. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK) In Vitro. 1973;9:176–185. doi: 10.1007/BF02618435. [DOI] [PubMed] [Google Scholar]
- 44.Di Martino B., Di Rocco C., Ceci C., Marsilio F. Characterization of a strain of feline calicivirus isolated from a dog faecal sample. Vet. Microbiol. 2009;139:52–57. doi: 10.1016/j.vetmic.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Povey C., Ingersoll J. Cross-protection among feline caliciviruses. Infect. Immun. 1975;11:877–885. doi: 10.1128/iai.11.5.877-885.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunet S., Sigoillot-Claude C., Pialot D., Poulet H. Multiple Correspondence Analysis on Amino Acid Properties within the Variable Region of the Capsid Protein Shows Differences between Classical and Virulent Systemic Feline Calicivirus Strains. Viruses. 2019;11:1090. doi: 10.3390/v11121090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tohya Y., Yokoyama N., Maeda K., Kawaguchi Y., Mikami T. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J. Gen. Virol. 1997;78:303–305. doi: 10.1099/0022-1317-78-2-303. [DOI] [PubMed] [Google Scholar]
- 48.Radford A.D., Willoughby K., Dawson S., McCracken C., Gaskell R.M. The capsid gene of feline calicivirus contains linear B-cell epitopes in both variable and conserved regions. J. Virol. 1999;73:8496–8502. doi: 10.1128/JVI.73.10.8496-8502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geissler K., Schneider K., Truyen U. Mapping neutralizing and non-neutralizing epitopes on the capsid protein of feline calicivirus. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2002;49:55–60. doi: 10.1046/j.1439-0450.2002.00529.x. [DOI] [PubMed] [Google Scholar]
- 50.Makino A., Shimojima M., Miyazawa T., Kato K., Tohya Y., Akashi H. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 2006;80:4482–4490. doi: 10.1128/JVI.80.9.4482-4490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhella D., Gatherer D., Chaudhry Y., Pink R., Goodfellow I.G. Structural insights into calicivirus attachment and uncoating. J. Virol. 2008;82:8051–8058. doi: 10.1128/JVI.00550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhella D., Goodfellow I.G. The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1. J. Virol. 2011;85:11381–11390. doi: 10.1128/JVI.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Martino B., Ceci C., Di Profio F., Marsilio F. In vitro inactivation of feline calicivirus (FCV) by chemical disinfectants: Resistance variation among field strains. Arch. Virol. 2010;155:2047–2051. doi: 10.1007/s00705-010-0795-9. [DOI] [PubMed] [Google Scholar]
- 54.Huang C., Hess J., Gill M., Hustead D. A dual-strain feline calicivirus vaccine stimulates broader cross-neutralization antibodies than a single-strain vaccine and lessens clinical signs in vaccinated cats when challenged with a homologous feline calicivirus strain associated with virulent systemic disease. J. Feline Med. Surg. 2010;12:129–137. doi: 10.1016/j.jfms.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in this manuscript. Sequence data presented in this study are openly available in the GenBank database.