Abstract
Hypertrophic cardiomyopathy (HCM) is a global and relatively common cause of patient morbidity and mortality and is among the first reported monogenic cardiac diseases. For 30 years, the basic etiology of HCM has been attributed largely to variants in individual genes encoding cardiac sarcomere proteins, with the implication that HCM is fundamentally a genetic disease. However, data from clinical and network medicine analyses, as well as contemporary genetic studies show that single gene variants do not fully explain the broad and diverse HCM clinical spectrum. These transformative advances place a new focus on possible novel interactions between acquired disease determinants and genetic context to produce complex HCM phenotypes, also offering a measure of caution against overemphasizing monogenics as the principal cause of this disease. These new perspectives in which HCM is not a uniformly genetic disease but likely explained by multifactorial etiology will also unavoidably impact how HCM is viewed by patients and families in the clinical practicing community going forward, including relevance to genetic counseling and access to healthcare insurance and psychosocial wellness.
“The workings of all our scientific approaches are assumed to be controlled by the same set of rules, [but reductionism] breaks down when confronted with the twin difficulties of scale and complexity.”
–P.W. Anderson in Science, 1972
Introduction
Hypertrophic cardiomyopathy (HCM) is a primary myocardial disorder characterized by substantial biological and clinical variability. Despite 60 years of study in thousands of patients of all ages, an uncertainty as to the cause of HCM persists. Recent scientific progress, including studies of tissue from patients, have caused a shift in the conceptual framework explaining this complex disease. This new direction diverges sharply from the strict traditional dogma tethering HCM to single gene variants (the monogenic hypothesis), now replaced by a more flexible paradigm that allows individual genetic context and acquired determinants of disease to explain phenotypic heterogeneity.
A postmonogenic era of HCM invites a multitude of new opportunities with the potential to advance disease etiology into clinically relevant areas. Achieving this goal, however, requires acknowledging an uncomfortable realization: that overemphasis on the monogenic determinants of HCM over the past 3 decades has diverted time away from efforts to discover alternative disease mechanisms. In this discussion, we present emerging evidence in support of a novel research pathway, distinct from single gene variants toward a contemporary model that integrates genetic context, post-transcriptional events, and acquired determinants of disease to create insights into the true basic cause of HCM.
Background
HCM has an estimated clinical prevalence of 1 : 200 to 1 : 500 in the general population, identified worldwide in at least 125 countries on all continents and is responsible for significant morbidity and mortality in patients of all ages.1 HCM affects patients of both genders and a multitude of cultures, notably with remarkably similar but heterogeneous clinical expression. Despite its clinical and genetic diversity,1 HCM has become a highly treatable condition along personalized care pathways, now resulting in the likelihood of low morbidity and mortality and the possibility of normal life expectancy.1,2 Nevertheless, even in the current era of effective diagnosis and management using sophisticated clinical tools,1,2 understanding the basic mechanisms responsible for HCM at its earliest stages of development remains an important aspiration.
HCM is a complex disease that may be inherited according to an autosomal dominant pattern, albeit with unpredictable expressivity, genotype-phenotype uncoupling, and either variable, low, or no penetrance of clinically overt disease.3 Dominant transmission of HCM is well documented in some families,4-6 sarcomere mutations can precede development of the disease phenotype,1 and left ventricular hypertrophy is an endophenotypic tracking marker of disease inheritance.1,3,4,6
Therefore, it is undeniable that genetic factors play a role in disease evolution for some patients with HCM, presumably as 1 or several potential triggers for susceptibility pathways. Clinically, genetic context has a crucial role in the development of HCM, including commercial genetic testing and cascade screening of families to identify affected relatives without left ventricular hypertrophy who may ultimately develop and/or transmit the disease.4,5
Such observations led to widespread belief for over 30 years that HCM is by definition (always) a genetic disease of autosomal transmission,6 which paradoxically has likely introduced a measure of confusion to patients and relatives, creating uncertainty regarding disease etiology and concerns and guild implicit in harboring and transmitting a genetic “defect” to offspring. Furthermore, this issue poses wider societal implications inherent in the consequences of inaccurate genetic profiling, including discrimination in life and disability insurance as well as other healthcare disparities. Additionally, promoting HCM as a distinctive disease of genetic etiology has led to unnecessary exclusion of patients with HCM from natural history studies of heart failure, potentially stunting research progress.
Contemporary reports leveraging new approaches to analyzing big data have exposed a much more complex pathogenicity for HCM. From these advances, it is increasingly evident that a new global strategy incorporating information on complex genomic context, acquired disease determinants, and post-translational molecular mechanisms will ultimately be required to decipher basic disease causes, advance precision medicine, and develop more personalized early interventions that may prevent progression of HCM.
The Monogenic Hypothesis
The DNA-based hypothesis that is currently used to explain HCM is largely based on genetic studies of circulating lymphocytes from patients with clinically established HCM. This initiative has reported >2,000 variants in 8 core genes encoding contractile myofilament proteins of the cardiac sarcomere and Z-band. Most common are MHY7 and MBPC3, together representing >90% of pathogenic variants.7 More recently, rare protein-altering minor variants (novel sarcomeric and also nonsarcomeric) have been reported with HCM and in some instances inferred as pathogenic, adding to the complex molecular heterogeneity of this disease.8 These genes/variants have been identified by whole genome sequencing studies in single large families (some with linkage analysis) or in individual probands with established HCM and FHOD3 and MLP (CSRP3). These genetic nonsarcomeric observations are additional evidence that HCM may not be exclusively a sarcomeric disease.
Variant assignments for pathogenicity are considered probabalistic probab, that is, with some inherent uncertainty potentially created by the multiple nonuniform criteria and interpretations that can differ among experts and genetic testing laboratories. Ultimately, these approaches introduce the possibility of mutational misclassification and genetic misdiagnosis. At present, much of the mechanistic work related to sarcomeric proteins associated with HCM continues as laboratory experimentation in cell cultures, in murine HCM models, or as part of gene editing initiatives.9,10
Research over the last 3 decades that investigated the determinants of HCM have focused almost exclusively on single gene variants that encode cardiac sarcomere myofilament proteins as being responsible for all disease features. The impetus for this highly specific view of HCM can be traced to genotype-phenotype studies very early in the genomic era identifying familial autosomal dominant inheritance patterns in a few multigenerational kindreds. Such studies marked the beginning of a widely accepted paradigm anchored in the view that a single molecular event (i.e., a gene variant alone) was sufficient evidence to account for HCM, albeit with the basic underlying mechanisms unresolved. The advent of commercial genetic testing for HCM seemed to consolidate the exceedingly broad disease spectrum under a narrow umbrella, defined solely by sarcomere gene mutations.
Consequently, HCM has been described widely and uniformly as a monogenic (Mendelian) disorder. However, novel evidence has emerged in several recent publications (discussed in detail later in the text), providing evidence that deviates from the strict monogenic hypothesis and supports the alternative view that a diverse range of pathobiologic mechanisms beyond a single molecular event may be fundamental to HCM pathogenesis.
Monogenics in HCM: the Pendulum Swings
Clinical evidence against monogenics
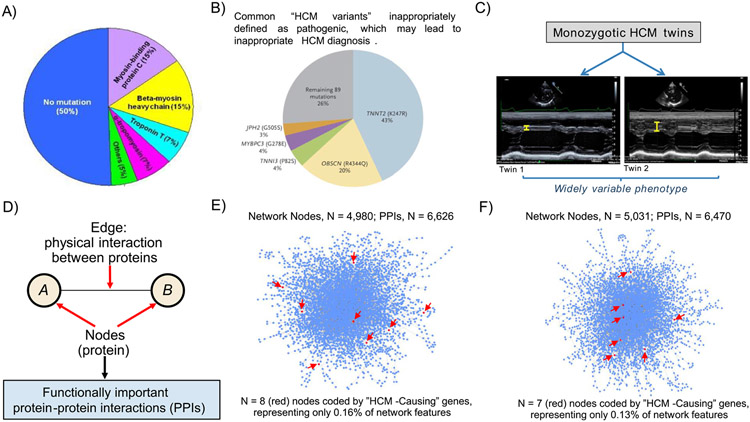
A number of assumptions that are part of the monogenic hypothesis are inconsistent with the clinical expression of HCM. First, and the most compelling, only about 30% of consecutive patients with an established HCM diagnosis demonstrate gene variants that are judged pathogenic (or likely pathogenic) and potentially responsible for the disease process, albeit not increased significantly by factoring in variants of unknown significance (Figure 1).11 Consequently, the vast majority of patients with HCM evaluated in clinical settings (about 70%) do not demonstrate evidence of a (mono)genetic cause.
Figure 1.
Determinants of HCM. (A) Distribution of variants in genes coding for sarcomere proteins in patients with HCM assessed in clinical practice. It is notable that 50% of HCM patients have no sarcomere gene mutation. However, only 30% of consecutive HCM patients have mutations that can be considered pathogenic (disease-causing). (B) Prevalence of putative HCM-causing gene variants is greater than expected in large genetic databases comprising non-affected populations: TNNT2 (K247R), OBSCN (R4344Q), TNNI3 (P82S), MYBPC3 (G278E), and JPH2 (G505S) are top 5 most common gene variants inappropriately defined as pathogenic, and account for most (74%) of HCM diagnoses judged misclassified. Reproduced with permission from Manrai et al.15 (C) Recent data from the Seidman laboratory shows widely divergent HCM phenotypes in monozygotic twins, supporting the hypothesis that factors other than genotype are critical to understanding the totality of HCM. Ventricular septal thickness on echocardiographic imaging differs substantially in the twins. Reproduced from Repetli GG, et al28 with permission. (D) Network features, emphasizing that network connections represent functionally important protein-protein interactions (PPIs). (E, F) Using myectomy specimens from patients with obstructive HCM, PPI networks are dense, complex, and vary widely between patients,31 suggesting that sarcomere genes alone are unlikely to explain the pathogenesis of this disease. Network topology was not informed by putative HCM-causing sarcomere genes, which in fact represented only <0.1% of all network features. Two representative patient networks are shown, which underscore: (i) HCM pathobiology is complex and (ii) a vast majority of functionally important molecular signaling pathways do not involve any of 17 genes commonly designated as HCM disease-causing (red arrows). Images derived from methods and results reported originally in Maron et al.31Heterogeneity in PPI networks is akin to differences observed in wiring maps of other complex systems.
Second, a substantial proportion of HCM family members who carry the same pathogenic sarcomere mutation identified in a relative having a having clinical diagnosis of HCM never develop phenotypic evidence of HCM or the diseaes.1 Third, on the basis of clinical, autopsy, and/or contemporary imaging with echocardiography and CMR in thousands of patients with clinically diagnosed HCM, certain morphologic features of the disease phenotype are fundamentally inconsistent with the presumption that HCM is caused solely by genes encoding proteins of the cardiac sarcomere.12
Such findings inconsistent with strict inference of the monogenic sarcomere hypothesis include: abnormally elongated mitral valve leaflets, causing LV outflow obstruction, phenotypic expression often differing measurably among related patients with identical genetic substrate, vasculopathy involving small intramural coronary arterioles responsible for microvascular ischemia, and the expanded interstitial (collagen) matrix and extracellular space, contributing significantly to increased LV wall thickness. Alternatively, in support of the monogenic hypothesis,13,14 some investigators argue that sarcomere mutations convey more significant disease burden than is evident in the absence of these variants.
Genetic evidence against monogenics
In 2016, Manrai et al15 analyzed sequencing data from 3 large publicly available repositories to estimate the frequency of HCM-causing variants in white and black Americans. Using such accessible exome data, the authors found that variants that were previously considered causal for HCM were over-represented in general (control) populations, whereas patients with HCM of African or unspecified ancestry more frequently had benign variants that were misclassified as pathogenic for HCM. Additionally, a paucity of black Americans in the control populations of previous genetic studies have contributed to the likelihood of misclassified genetic diagnoses (Figure 1).
These data showing the potential for genetic overdiagnosis of HCM in black Americans substantiate that misclassification of gene pathogenicity is not uncommon in clinical HCM practice, with benign variants often misrepresented as HCM-causing. Furthermore, multicenter collaborative data from Australia report that 2/3 of curated genes, many of which are included in commercially available diagnostic tests, offer little or no evidence of an association with HCM.16
The totality of these observations suggests that mutations in genes of the cardiac sarcomere may not be causative of HCM, but possibly facilitators of key susceptibility pathways that could increase the likelihood of developing the HCM phenotype. On the other hand, this does not exclude the possibility that HCM can develop by triggering nonsarcomere mutation mechanisms. This could include patients with a nonsarcomeric genetic context in the setting of other biological or environmental triggers or most likely a multifactorial etiology.17 Indeed, a variety of nongenetic, environmental, or lifestyle variables have been suggested as contributing to development of the HCM phenotype and pathophysiology, including gender, race, obesity, and blood pressure.13,18-24
Furthermore, some investigators have recently pursued a polygenic causative hypothesis for HCM;25-27 however, generalizability of this to patients across the HCM spectrum is unlikely. Additionally, this approach again shifts focus away from modifiable causes of disease. Tadros et al25 underscored the case for the causal polygenic component postulated for the HCM phenotype.26
Additionally, Harper et al27 leveraged 2 independent genome-wide association studies, including 2,780 unselected HCM cases versus 47,486 controls, which identified 13 independent genome-wide common variants in 12 susceptibility loci that are associated significantly with HCM. Results of a 2-sample Mendelian randomization strategy inferred a causal relation of sarcomere mutation-negative HCM with systemic hypertension, such that an increase in diastolic blood pressure by at least 11 mm Hg was associated with a >4-fold increase in the risk for HCM. This report importantly diversifies the framework underlying HCM to include nongenetic factors and doing so is consistent with the recent observation from the Seidman laboratory28 that widely divergent LV hypertrophy patterns can occur in monozygotic twins with HCM (Figure 1).
Furthermore, there is emerging evidence for a common nonfamilial form of HCM, of genetic etiology.29 In some cohorts, up to 40% of patients with HCM have neither a family history of the disease or a pathogenic sarcomere mutation. In this sporadic non-Mendelian form of HCM, systemic hypertension has also emerged as a contributor to the disease, again reporting a common HCM phenotype that is independent of sarcomere gene variants.1,12,13,16,29-31 Taken together, the evidence for the causative polygenic HCM hypothesis appears to undercut the classical monogenic proposal. Notably, these principles can be particularly relevant to patients and families. The historical mantra that HCM is a genetic (autosomal dominant) disease can misrepresent the true context of this disease to some families and unnecessarily encumber parents with perceived responsibility and guilt for potentially transmitting an inherited disease to offsprings, which may instead ultimately prove to be nongenetic and nonfamilial.
The Case for Network Medicine
It is tempting to simplify a complex biological processes by implicating a single molecular event (e.g., a genetic mutation). However, this view is reductionist and thus, susceptible to major flaws in a complex human disease, such as HCM.32 First, reductionism obligates that a myriad of HCM morphologic and structural features, observed to varying degrees within families and across patients and involving a diverse range of cell and tissue types, is due to a single sarcomere gene mutation. Second, most HCM-causing genetic variants are identified from association studies, which do not yield insight into the biofunctionality of gene variants or their downstream pathways. Third, the role of post-transcriptional events that contribute to genotype-phenotype relations is not evident in data from these analyses. Fourth, information on the effect of acquired environmental determinants of HCM clinical expression are sparse.
Network medicine obviates these restraints of reductionism by considering multiple (functional signaling) pathways simultaneously.33,34 In this approach, a wiring diagram is assembled to map important molecular interactions that underlie disease pathogenesis (Figure 1). The networks use the consolidated human interactome,30,33,34 which represents a compendium of known protein-protein interactions (connections) occurring in human biology, describing functionally important molecular signaling pathways.
Networks can be analyzed further by focusing on pathways that regulate endophenotypes (i.e., disease features) acknowledged as important to the morphology of HCM, such as myocardial fibrosis. Of the multiple signaling pathways that have emerged in HCM networks, those related to fibrosis potentially identify an underappreciated role for collagen formation as a fundamental disease element. Consequently, network medicine is well positioned to uncover unexpected relevant endophenotypes that are important in HCM and thereby generate powerful hypotheses for pursuing novel disease etiologies. Analyzing network topology itself is potentially informative because connectivity patterns can relate to pathobiology.
Finally, it should be underscored that network medicine is not exclusive of genetics. Instead, genetic context may be integrated with post-transcriptional events to localize determinants of the phenotype or understand the true relevance of genes that are absent from the network.30,31,33,34,35
Network Evidence against Monogenics
Our group has innovated a strategy in which next-generation RNA sequencing technology was used to explore the basic mechanisms of HCM by analyzing heart tissue resected from symptomatic patients at surgical myectomy.31 Compared with control tissue from a cohort of rejected heart donors, we identified >2,200 mRNA transcripts in significantly different quantities in patients with HCM, underscoring the molecular complexity of this disease. However, to avoid overstating the importance of any particular mRNA transcript simply on the basis of - fold-change in HCM versus controls or a priori knowledge of gene function, we focused on functional protein-protein interactions (PPIs) and constructed PPI networks unique to individual patients with HCM.
The personalized PPI networks were dense, inclusive of 3,700 functional PPIs on average, and demonstrated basic biological differences among the 18 individual patients with HCM. This approach unmasked 28 different PPI functional categories (i.e., endophenotypes), many of which are related to environmental drivers of cell function, such as inflammation, oxidant stress, and hypoxia signaling (Table 1). Notably, among the myriad of connected proteins, sarcomeric proteins, (preported to be HCM-causing) proved to be exceedingly rare, involving <0.1% of all PPIs in the HCM networks, and representing clear evidence against the causal role of sarcomere protein mutations. Overall, the (1) molecular diversity across patient-specific networks; (2) broad functional diversity of PPIs within individual patients; and (3) insignificant contribution to network topology from classical “disease-causing (pathogenic) genes” of the sarcomere proteins provides further tangible evidence against the monogenic hypothesis as solely and primarily responsible for the development of HCM (Figure 1).
Table 1.
Diverse landscape of HCM endophenotypes
| HCM Endophenotype30 | Relevance to HCM | Selected Supporting References |
|---|---|---|
| Autoimmunity | Association studies linking HCM with Anti-HLA-17 and β1-receptors and Muscarin-2-receptors; Prevalence studies linking HCM with autoimmune connective tissue disease including systemic sclerosis, systemic lupus erythematosus, others. | 39-45 |
| Cell transformation | Endothelial-to-mesenchymal transition and cardiac fibrosis; TGF-β-associated fibrosis in noncardio-myocyte cell types | 46,47 |
| DNA damage | Cardiomyocyte-specific telomere shortening and heart failure in hypertrophic hearts | 48 |
| DNA repair deficiency | G:C base excision repair editing ameliorates HCM phenotype | 49 |
| Fibrosis | Left ventricular thickening due to interstitial collagen deposition, and replacement fibrosis from microvascular ischemia | 50-53 |
| Hyperplasia | Junctophilin-2-mediated cardiomyocyte hyperplasia ex vivo, and hyperplastic remodeling in zebra fish model of HCM in vivo | 54-57 |
| Hypertension | Polygenic risk for HCM associates positively with diastolic blood pressure | 27 |
| Immunological derangement | Tacrolimus use associated with HCM in a pediatric cohort and in other case series in adults | 37, 58, 59 |
| Metabolic derangement | Dysregulated mitochondrial Ca2+ handling linked to abnormal LV contractility; ↑ plasma branched chain amino acids, triglycerides and ether phospholipids in HCM; ↓ phosphocreatine:ATP in HCM vs. control | 60-65 |
| Inflammation | ↑circulating IL-6, IL-1β, IL-1RA, IL-10 | 66, 67 |
| Metaplasia | Juxtaglomerular metaplasia in feline HCM, linked to renin-angiotensin axis overactivation | 68 |
| Mitochondrial dysfunction | ↓cardioplipin à lipotoxicity in HCM | 69,70 |
| Neoplastic signaling | Dysregulated RAS-RAF-MEK-ERK MAPK pathway | 71-73 |
| Neovascularization | Abnormal diastolic coronary vascular reserve related to vasculopathy of microcirculation | 74 |
| Thromboembolism | Impaired von Willebrand factor ↑thrombin generation ↑platelet reactivity |
75-77 |
| Senescence | Leukocyte telomere length correlates with HCM phenotypic parameters (e.g., inversely with LVOT gradient) | 78 |
| Vasculitis | HCM incidence in associations with mixed cryoglobulinemia vasculitis, hepatitis C virus, polyarteritis nodosa | 79-82 |
| Hypoxia | HIF1-α-PPARgamma axis regulates glycolytic and lipid metabolism pathways in hypertrophic cardiomyopathies | 83 |
| Oxidant stress | Disruption to redox potential of cardiomyocytes slinked to fibrosis and LV thickening in HCM | 31, 84-86 |
ATP = adenosine triphosphate; ERK = extracellular signal-regulated kinase; HCM = hypertrophic cardiomyopathy; HIF = hypoxia inducible factor; HLA = human leukocyte antigen; IL = interleukin; LV = left ventricle; LVOT = left ventricular outflow tract; MAPK = mitogen-activated protein kinase; PPAR = peroxisome proliferator-activated receptor; TGF = transforming growth factor.
Endophenotypes that are implicated as a cause of HCM or a response to HCM.
Interaction between Genetics and Environment to Potentially Explain HCM
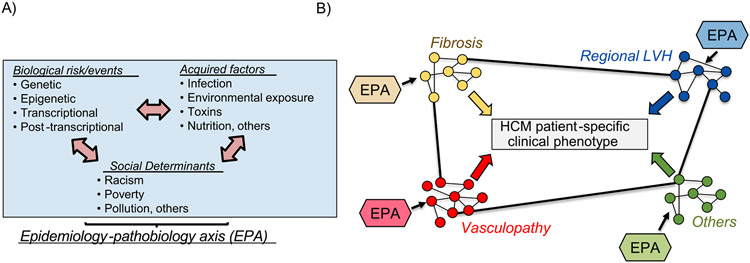
Identifying and integrating the determinants underlying HCM will require an innovative and expansive approach to data collection and analysis. To date, epidemiologic information on HCM has been limited to small case series that linked disease onset to puberty, specific toxic exposures, and systemic hypertension.36-38 By contrast, systematically collected data on environmental, dietary, behavioral, psychosocial, economic, racial, and geographic risk factors that already frame the etiology of other cardiovascular diseases (e.g., atherosclerotic coronary artery disease, myocardial infarction, heart failure, and pulmonary hypertension) have not yet been studied in HCM.30
Therefore, a major knowledge gap exists for identifying and connecting potential environmental triggers with patient-level genetic information to predict HCM onset and disease course (Figure 2). To address this unmet need, research is needed to focus on interactions between genetic context and perinatal events, drug exposure, dietary profile, co-morbidities, and socioeconomic status, all of which are known mediators of LV hypertrophy, myocardial fibrosis, and other HCM endophenotypes.
Figure 2.
HCM is a disease of converging endophenotypes, regulated by complex interaction between genetic, post-transcriptional, and environmental determinants. (A) Genetic, biological, acquired, and social profiles likely influence individual functional protein-protein interaction patterns in individual patients with HCM. (B) This paradigm sets the framework for a model that integrates genetic context with environmental determinants of disease to explain HCM. Cross-talk among PPIs between endophenotypes, which may vary between patients, explains heterogeneity in clinical phenotypes evident in HCM populations. EPA = epidemiologic-pathological axis.
Conclusions
After 60 years of clinical recognition, definitive understanding of the basic mechanisms responsible for the development and expression of the expansive HCM disease spectrum remains largely elusive. Over the last 30 years, much of what we have learned about this complex and heterogeneous disease has been viewed through the restrictive prism of the monogenic sarcomere hypothesis. From our perspective, we do not de-emphasize the significant role of the genetic context in HCM, that is, the genetic profile of patients, which includes variants but also undoubtedly involves nongenetic factors. Recent application of network medicine to HCM demonstrated a previously unappreciated multitude of protein-PPIs underlying disease pathobiology, largely independent of the contractile myofilaments of the sarcomere. Therefore, it is notable and likely that determinants of the HCM phenotype will ultimately prove to be multifactorial, not confined to the sarcomere, and involve environmental and/or other nongenetic factors.
It is now evident that to regard HCM as a disease solely of genetic etiology can be misleading to patients and families. Consequently, a more complex understanding of HCM etiology requires a change in investigative direction beyond strict adherence to the traditional monogenic and (now) polygenic theories. Success in this approach will likely depend on the expansive use of network medicine, functional genetics, and other contemporary methods.
Acknowledgments
Bradley A. Maron has received funding from NIH (Bethesda, MD) 1R01HL139613-01, R01HL153502, R01HL155096-01, U54HL119145, 2021A007243 BWH/Mit-Broad Institute; Cardiovascular Medical Research Foundation. Joseph Loscalzo has received funding from NIH (Bethesda, MD) R01HL155107, U01HG007690, U54HL119145, and AHA 957729 (Dallas, TX) D700382 and CV-19.
Footnotes
Disclosures
Bradley Maron reports funding provided by Harvard Medical School, a relationship with Deerfield that includes: funding grants, relationship with Actelion Pharmaceuticals US Inc that includes: consulting or advisory, a relationship with Regeneron Pharmaceuticals Inc that includes: consulting or advisory, and a relationship with TENAX that includes: consulting or advisory. Joseph Loscalzo reports a relationship with Scipher Medicine Corp that includes: board membership and equity or stocks, a relationship with Applied Biomath LLC that includes: consulting or advisory, a relationship with Naring Health that includes: consulting or advisory, a relationship with Ionis Pharmaceuticals Inc that includes: board membership, a relationship with Leap that includes: board membership and equity or stocks, and a relationship with Mineralys that includes: consulting or advisory. Martin Maron reports a relationship with Cytokinetics Inc that includes: consulting or advisory and a relationship with Imbrium Therapeutics L.P. that includes: consulting or advisory.
References
- 1.Maron BJ. Clinical course and management of hypertrophic cardiomyopathy. N Engl J Med 2018;379:655–668. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Rowin EJ, Casey SA, Maron. How hypertrophic cardiomyopathy became a contemporary and treatable genetic disease with low mortality: shaped by 50 years of clinical research and practice. JAMA Cardiol 2016;1:98–105. [DOI] [PubMed] [Google Scholar]
- 3.De Marvao A, McGurk KA, Zheng SL, Thanaj M, Bai W, Duan J, Biffi C, Mazzarotto F, Statton B, Dawes TJW, Savioli N, Halliday BP, Xu X, Buchan RJ, Baksi AJ, Quinlan M, Tokarczuk P, Tayal U, Francis C, Whiffin N, Theotokis PI, Zhang X, Jang M, Berry A, Pantazis A, Barton PJR, Rueckert D, Prasad SK, Walsh R, Ho CY, Cook SA, Ware JS, O’Regan DP. Phenotypic expression and outcomes in individuals with rare genetic variants of hypertrophic cardiomyopathy. J Am Coll Cardiol 2021;78:1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ommen SR, Mital S, Burke MA, Day SM, Deswal A, Eliott P, Evanovich LL, Hung J, Joglar JA, Kantor P, Kimmelstiel C, Kittleson M, Link MS, Maron MS, Martinez MW, Miyake CY, Schaff HVHV, Semsarian C, Sorajja P, Members ACC/AHA Joint Committee, O'Gara PT, Beckman JA, Levine GN, Al-Khatib SM, Armbruster A, Birtcher KK, Ciggaroa J, Dixon DL, de Las Fuentes L, Deswal A, Fleisher LA, Gentile F, Goldberger ZD, Gorenek B, Haynes N, Hernandez AF, Hlatky MA, Joglar JA, Jones WS, Marine JE, Mark D, Palaniappan L, Piano MR, Tamis-Holland J, Wijeysundera DN, Woo YJ. 2020 AHA/ACC Guidelines for the diagnosis and treatment of patients with hypertrophic cardiomyopathy. J Am Coll Cardiol 2020;76:3022–3055. [DOI] [PubMed] [Google Scholar]
- 5.Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, Hagege AA, Lafont A, Limongelli G, Mahrholdt H, McKenna WJ, Mogensen J, Nihoyannopoulos P, Nistri S, Pieper PG, Pieske B, Rapezzi C, Rutten FH, Tillmanns C, Watkins H. 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the diagnosis and management of hypertrophic cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J 2014;35:2733–2779. [DOI] [PubMed] [Google Scholar]
- 6.Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, Levi T, Donis-Keller H, Seidman JG, Seidman CE. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med 1989;321:1372–1378. [DOI] [PubMed] [Google Scholar]
- 7.Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med 2015;17:880–888. [DOI] [PubMed] [Google Scholar]
- 8.Walsh R, Offerhaus JA, Tadros R, Bezzina CR. Minor hypertrophic cardiomyopathy genes, major insights into the genetics of cardiomyopathies. Nat Rev Cardiol 2022;19:151–167. [DOI] [PubMed] [Google Scholar]
- 9.Toepfer CN, Garfinkel AC, Venturini G, Wakimoto H, Repetti G, Alamo L, Sharma A, Agarwal R, Ewoldt JF, Cloonan P, Letendre J, Lun M, Olivotto I, Colan S, Ashley E, Jacoby D, Michels M, Redwood CS, Watkins HC, Day SM, Staples JF, Padrón R, Chopra A, Ho CY, Chen CS, Pereira AC, Seidman JG, Seidman CE. Myosin sequestration regulates sarcomere function, cardiomyocyte energetics, and metabolism, informing the pathogenesis of hypertrophic cardiomyopathy. Circulation 2020;141:828–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toepfer CN, Wakimoto H, Garfinkel AC, McDonough B, Liao D, Jiang J, Tai AC, Gorham JM, Lunde IG, Lun M, Lynch TL 4th, McNamara JW, Sadayappan S, Redwood CS, Watkins HC, Seidman JG, Seidman CE. Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin. Sci Transl Med 2019;11:eaat119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruner C, Ivanov J, Care M, Williams L, Moravsky G, Yang H, Laczay B, Siminovitch K, Woo A, Rakowski H. Toronto hypertrophic cardiomyopathy genotype score for prediction of a positive genotype in hypertrophic cardiomyopathy. Circ Cardiovasc Genet 2013;6:19–26. [DOI] [PubMed] [Google Scholar]
- 12.Rowin EJ, Maron BJ, Maron MS. The hypertrophic cardiomyopathy phenotype viewed through the prism of multimodality imaging: clinical and etiologic implications. JACC Cardiovasc Imaging 2020;13:2002–2016. [DOI] [PubMed] [Google Scholar]
- 13.Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, Ommen SR, Theis JL, Vaubel RA, Re F, Armentano C, Poggesi C, Torricelli F, Cecchi F. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc 2008;83:630–638. [DOI] [PubMed] [Google Scholar]
- 14.Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Lakdawala NK, Ware JS, Helms AS, Colan SD, Seidman CE, Olivotto I, Investigators SHaRe. Genotype and lifetime burden of disease with hypertrophic cardiomyopathy: insights from the sarcomeric human cardiomyopathy registry (SHaRE). Circulation 2018;138:1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, Kohane IS. Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016;375:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingles J, Goldstein J, Thaxton C, Caleshu C, Corty EW, Crowley SB, Dougherty K, Harrison SM, McGlaughon J, Milko LV, Morales A, Seifert BA, Strande N, Thomson K, Peter van Tintelen J, Wallace K, Walsh R, Wells Q, Whiffin N, Witkowski L, Semsarian C, Ware JS, Hershberger RE, Funke B. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med 2019;12:e002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh R, Buchan R, Wilk A, John S, Felkin LE, Thomson KL, Chiaw TH, Loong CCW, Pua CJ, Raphael C, Prasad S, Barton PJ, Funke B, Watkins H, Ware JS, Cook SA. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur Heart J 2017;38:3461–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olivotto I, Maron BJ, Tomberli B, Appelbaum E, Salton C, Haas TS, Gibson CM, Nistri S, Servettini E, Chan RH, Udelson JE, Lesser JR, Cecchi F, Manning WJ, Maron MS. Obesity and its association to phenotype and clinical course in hypertrophic cardiomyopathy. J Am Coll Cardiol 2013;62:449–457. [DOI] [PubMed] [Google Scholar]
- 19.Lakdawala NK, Olivotto I, Day SM, Han L, Ashley EA, Michels M, Ingles J, Semsarian C, Jacoby D, Jefferies JL, Colan SD, Pereira AC, Rossano JW, Wittekind S, Ware JS, Saberi S, Helms AS, Cirino AL, Leinwand LA, Seidman CE, Ho CY. Associations between female sex, sarcomere variants, and clinical outcomes in hypertrophic cardiomyopathy. Circ Genom Precis Med 2021;14:e003062. [DOI] [PubMed] [Google Scholar]
- 20.Eberly LA, Day SM, Ashley EA, Jacoby DL, Jefferies JL, Colan SD, Rossano JW, Semsarian C, Pereira AC, Olivotto I, Ingles J, Seidman CE, Channaoui N, Cirino AL, Han L, Ho CY, Lakdawala NK. Association of race with disease expression and clinical outcomes among patients with hypertrophic cardiomyopathy. JAMA Cardiol 2020;5:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nijenkamp LLAM, Bollen IAE, Niessen HWM, Dos Remedios CG, Michels M, Poggesi C, Ho CY, Kuster DWD, van der Velden J. Sex-specific cardiac remodeling in early and advanced stages of hypertrophic cardiomyopathy. PLoS One 2020;15:e0232427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sridharan A, Maron MS, Carrick RT, Madias CA, Huang D, Cooper C, Drummond J, Maron BJ, Rowin EJ. Impact of comorbidities on atrial fibrillation and sudden cardiac death in hypertrophic cardiomyopathy. J Cardiovasc Electrophysiol 2022;33:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fumagalli C, Maurizi N, Day SM, Ashley EA, Michels M, Colan SD, Jacoby D, Marchionni N, Vincent-Tompkins J, Ho CY, Olivotto I, Investigators SHARE. Association of obesity with adverse long-term outcomes in hypertrophic cardiomyopathy. JAMA Cardiol 2020;5:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells S, Rowin EJ, Bhatt V, Maron MS, Maron BJ. Association between race and clinical profile of patients referred for hypertrophic cardiomyopathy. Circulation 2018;137:1973–1975. [DOI] [PubMed] [Google Scholar]
- 25.Tadros R, Francis C, Xu X, Vermeer AMC, Harper AR, Huurman R, Kelu Bisabu K, Walsh R, Hoorntje ET, Te Rijdt WP, Buchan RJ, van Velzen HG, van Slegtenhorst MA. Vermeulen JM, Offerhaus JA, Bai W, de Marvao A, Lahrouchi N, Beekman L, Karper JC, Veldink JH, Kayvanpour E, Pantazis A, Baksi AJ, Whiffin N, Mazzarotto F, Sloane G, Suzuki H, Schneider-Luftman D, Elliott P, Richard P, Ader F, Villard E, Lichtner P, Meitinger T, Tanck MWT, van Tintelen JP, Thain A, McCarty D, Hegele RA, Roberts JD, Amyot J, Dubé MP, Cadrin-Tourigny J, Giraldeau G, L’Allier PL, Garceau P, Tardif JC, Boekholdt SM, Lumbers RT, Asselbergs FW, Barton PJR, Cook SA, Prasad SK, O'Regan DP, van der Velden J, Verweij KJH, Talajic M, Lettre G, Pinto YM, Meder B, Charron P, de Boer RA, Christiaans I, Michels M, Wilde AAM, Watkins H, Matthews PM, Ware JS, Bezzina CR. Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat Genet 2021;53:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watkins H, Ashrafian H, Redwood C. Inherited cardiomyopathies N Engl J Med 2011;364:1643–1656. [DOI] [PubMed] [Google Scholar]
- 27.Harper AR, Goel A, Grace C, Thomson KL, Petersen SE, Xu X, Waring A, Ormondroyd E, Kramer CM, Ho CY, Neubauer S, Investigators HCMR, Tadros R, Ware JS, Bezzina CR, Farrall M, Watkins H. Common genetic variants and modifiable risk factors underpin hypertrophic cardiomyopathy susceptibility and expressivity. Nat Genet 2021;53:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Repetti GG, Kim Y, Pereira AC, Ingles J, Russell MW, Lakdawala NK, Ho CY, Day S, Semsarian C, McDonough B, DePalma SR, Quiat D, Green EM, Seidman CE, Seidman JG. Discordant clinical features of identical hypertrophic cardiomyopathy twins. Proc Natl Acad Sci U S A 2021;118:e2021717118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingles J, Burns C, Bagnall RD, Lam L, Yeates L, Sarina T, Puranik R, Briffa T, Atherton JJ, Driscoll T, Semsarian C. Nonfamilial hypertrophic cardiomyopathy: prevalence, natural history, and clinical implications. Circ Cardiovasc Genet 2017;10:e001620. [DOI] [PubMed] [Google Scholar]
- 30.Maron BJ, Maron MS, Maron BA, Loscalzo J. Moving beyond the sarcomere to explain heterogeneity in hypertrophic cardiomyopathy: JACC review topic of the week. J Am Coll Cardiol 2019;73:1978–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maron BA, Wang RS, Shevtsov S, Drakos SG, Arons E, Wever-Pinzon O, Huggins GS, Samokhin AO, Oldham WM, Aguib Y, Yacoub MH, Rowin EJ, Maron BJ, Maron MS, Loscalzo J. Individualized interactomes for network-based precision medicine in hypertrophic cardiomyopathy with implications for other clinical pathophenotypes. Nat Commun 2021;12:873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leopold JA, Maron BA, Loscalzo J. The application of big data to cardiovascular disease: paths to precision medicine. J Clin Invest 2020;130:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maron BA, Altucci L, Balligand JL, Baumbach J, Ferdinandy P, Filetti S, Parini P, Petrillo E, Silverman EK, Barabási AL, Loscalzo J. International Network Medicine Consortium. A global network for network medicine. NPJ Syst Biol Appl 2020;6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samokhin AO, Stephens T, Wertheim BM, Wang RS, Vargas SO, Yung LM, Cao M, Brown M, Arons E, Dieffenbach PB, Fewell JG, Matar M, Bowman FP, Haley KJ, Alba GA, Marino SM, Kumar R, Rosas IO, Waxman AB, Oldham WM, Khanna D, Graham BB, Seo S, Gladyshev VN, Yu PB, Fredenburgh LE, Loscalzo J, Leopold JA, Maron BA. NEDD9 targets COL3A1 to promote endothelial fibrosis and pulmonary arterial hypertension. Sci Transl Med 2018;10:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng F, Zhao J, Wang Y, Lu W, Liu Z, Zhou Y, Martin WR, Wang R, Huang J, Hao T, Yue H, Ma J, Hou Y, Castrillon JA, Fang J, Lathia JD, Keri RA, Lightstone FC, Antman EM, Rabadan R, Hill DE, Eng C, Vidal M, Loscalzo J. Comprehensive characterization of protein-protein interactions perturbed by disease mutations. Nat Genet 2021;53:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maron BJ, Desai MY, Nishimura RA, Spirito P, Rakowski H, Towbin JA, Dearani JA, Rowin EJ, Maron MS, Sherrid MV. Management of hypertrophic cardiomyopathy: JACC state-of-the-art review. J Am Coll Cardiol 2022;79:390–414. [DOI] [PubMed] [Google Scholar]
- 37.Atkison P, Joubert G, Barron A, Grant D, Paradis K, Seidman E, Wall W, Rosenberg H, Howard J, Williams S. Hypertrophic cardiomyopathy associated with tacrolimus in paediatric transplant patients. Lancet 1995;345:894–896. [DOI] [PubMed] [Google Scholar]
- 38.Watkins H Time to think differently about sarcomere-negative hypertrophic cardiomyopathy. Circulation 2021;143:2415–2417. [DOI] [PubMed] [Google Scholar]
- 39.Dawood M, Lateef N, Tauseef A, Patel J. Association of hypertrophic obstructive cardiomyopathy with rheumatoid arthritis. Cureus 2018;10:e2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peukert S, Fu ML, Eftekhari P, Poepping I, Voss A, Thalhammer C, Hempel A, Menz M, Dietz R, Osterziel KJ. The frequency of occurrence of anti-cardiac receptor autoantibodies and their correlation with clinical manifestation in patients with hypertrophic cardiomyopathy. Autoimmunity 1999;29:291–297. [DOI] [PubMed] [Google Scholar]
- 41.Moyssakis I, Papadopoulos DP, Anastasiadis G, Vlachoyannopoulos P. Hypertrophic cardiomyopathy in systemic sclerosis. A report of two cases. Clin Rheumatol 2006;25:404–406. [DOI] [PubMed] [Google Scholar]
- 42.Murata I, Takenaka K, Shinohara S, Suzuki T, Sasaki T, Yamamoto K. Diversity of myocardial involvement in systemic sclerosis: an 8-year study of 95 Japanese patients. Am Heart J 1998;135:960–969. [DOI] [PubMed] [Google Scholar]
- 43.Frayha R, Jubran F, Partamian L, Sawaya J. Hypertrophic nonobstructive crardiomyopathy in scleroderma. Arthritis Rheum 1978;21:609–611. [DOI] [PubMed] [Google Scholar]
- 44.Asherson RA, Ames D, Coltart J, Byrne C, Hughes GR. Hypertrophic cardiomyopathy in systemic lupus erythematosus and “lupus-like” disease. Chance association? A report of 2 cases. J Rheumatol 1992;19:1973–1977. [PubMed] [Google Scholar]
- 45.Anastasiadis GP, Moyssakis I, Boki K, Kyriakidis M. Hypertrophic cardiomyopathy in systemic lupus erythematosus. Mayo Clin Proc 2001;76:111. [DOI] [PubMed] [Google Scholar]
- 46.Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, Neilson EG, Sayegh MH, Izumo S, Kalluri R. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med 2007;13:952–961. [DOI] [PubMed] [Google Scholar]
- 47.Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, Nayor M, Konno T, Gorham JM, Wolf CM, Kim JB, Schmitt JP, Molkentin JD, Norris RA, Tager AM, Hoffman SR, Markwald RR, Seidman CE, Seidman JG. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-β J Clin Invest 2010;120:3520–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharifi-Sanjani M, Oyster NM, Tichy ED, Bedi KC Jr, Harel O, Margulies KB, Mourkioti F. Cardiomyocyte-specific telomere shortening is a distinct signature of heart failure in humans. J Am Heart Assoc 2017;6:e005086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Park JE, Paa P, Rajakumar PD, Prekop H-T, Ting Chew YT, Manivannan SN, Chew WL. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat Commun 2021;12:1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirani J, Pick R, Roberts WC, Maron BJ. Morphology and significance of the left ventricular collagen network in young patients with hypertrophic cardiomyopathy and sudden cardiac death. J Am Coll Cardiol 2000;35:36–44. [DOI] [PubMed] [Google Scholar]
- 51.Chan RH, Maron BJ, Olivotto I, Pencina MJ, Assenza GE, Haas T, Lesser JR, Gruner C, Crean AM, Rakowski H, Udelson JE, Rowin E, Lombardi M, Cecchi F, Tomberli B, Spirito P, Formisano F, Biagini E, Rapezzi C, De Cecco CN, Autore C, Cook EF, Hong SN, Gibson CM, Manning WJ, Appelbaum E, Maron MS. Prognostic value of quantitative contrast-enhanced cardiovascular magnetic resonance for the evaluation of sudden death risk in patients with hypertrophic cardiomyopathy. Circulation 2014;130:484–495. [DOI] [PubMed] [Google Scholar]
- 52.Ho CY, López B, Coelho-Filho OR, Lakdawala NK, Cirino AL, Jarolim P, Kwong R, González A, Colan SD, Seidman JG, Díez J, Seidman CE. Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N Engl J Med 2010;363:552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maron BJ, Maron MS. LGE means better selection of HCM patients for primary prevention implantable defibrillators. JACC Cardiovasc Imaging 2016;9:1403–1406. [DOI] [PubMed] [Google Scholar]
- 54.Landstrom AP, Weisleder N, Batalden KB, Bos JM, Tester DJ, Ommen SR, Wehrens XHT, Claycomb WC, Ko JK, Hwang M, Pan Z, Ma J, Ackerman MJ. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol 2007;42:1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Becker JR, Deo RC, Werdich AA, Panàkovà D, Coy S, MacRae CA. Human cardiomyopathy mutations induce myocyte hyperplasia and activate hypertrophic pathways during cardiogenesis in zebrafish. Dis Model Mech 2011;4:400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferrans VJ, Rodriguez ER. Evidence of myocyte hyperplasia in hypertrophic cardiomyopathy and other disorders with myocardial hypertrophy. In: Kaltenbach M, Hopf R, Kunkel Bed. New Aspects of Hypertrophic Cardiomyopathy. Steinkopff; 1988:33–41. [PubMed] [Google Scholar]
- 57.Fujiwara H, Hoshino T, Yamana K, Fujiwara T, Furuta M, Hamashima Y, Kawai C. Number and size of myocytes and amount of interstitial space in the ventricular septum and in the left ventricular free wall in hypertrophic cardiomyopathy. Am J Cardiol 1983;52:818–823. [DOI] [PubMed] [Google Scholar]
- 58.Roberts CA, Stern DL, Radio SJ. Asymmetric cardiac hypertrophy at autopsy in patients who received FK506 (tacrolimus) or cyclosporine A after liver transplant. Transplantation 2002;74:817–821. [DOI] [PubMed] [Google Scholar]
- 59.Scott JS, Boyle GJ, Daubeney PE, Miller SA, Law Y, Pigula F, Griffith BP, Webber SA. Tacrolimus: a cause of hypertrophic cardiomyopathy in pediatric heart transplant recipients? Transplant Proc 1999;31:82–83. [DOI] [PubMed] [Google Scholar]
- 60.Vakrou S, Liu Y, Zhu L, Greenland GV, Simsek B, Hebl VB, Guan Y, Woldemichael K, Talbot CC, Aon MA, Fukunaga R, Abraham MR. Differences in molecular phenotype in mouse and human hypertrophic cardiomyopathy. Sci Rep 2021;11:13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jørgenrud B, Jalanko M, Heliö T, Jääskeläinen P, Laine M, Hilvo M, Nieminen MS, Laakso M, Hyötyläinen T, Orešič M, Kuusisto J. The metabolome in Finnish carriers of the MYBPC3-Q1061X mutation for hypertrophic cardiomyopathy. PLoS One 2015;10:e0134184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crilley JG, Boehm EA, Blair E, Rajagopalan B, Blamire AM, Styles P, McKenna WJ, Ostman-Smith I, Clarke K, Watkins H. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J Am Coll Cardiol 2003;41:1776–1782. [DOI] [PubMed] [Google Scholar]
- 63.Jung WI, Sieverding L, Breuer J, Hoess T, Widmaier S, Schmidt O, Bunse M, van Erckelens F, Apitz J, Lutz O, Dietze GJ. 31P NMR spectroscopy detects metabolic abnormalities in asymptomatic patients with hypertrophic cardiomyopathy. Circulation 1998;97:2536–2542. [DOI] [PubMed] [Google Scholar]
- 64.Jung WI, Dietze GJ. 31P nuclear magnetic resonance spectroscopy: a noninvasive tool to monitor metabolic abnormalities in left ventricular hypertrophy in human. Am J Cardiol 1999;83:19H–24H. [DOI] [PubMed] [Google Scholar]
- 65.Sakuma H, Takeda K, Tagami T, Nakagawa T, Okamoto S, Konishi T, Nakano T. 31P MR spectroscopy in hypertrophic cardiomyopathy: comparison with TI-201 myocardial perfusion imaging. Am Heart J 1993;125:1323–1328. [DOI] [PubMed] [Google Scholar]
- 66.Kuusisto J, Kärjä V, Sipola P, Kholová I, Peuhkurinen K, Jääskeläinen P, Naukkarinen A, Ylä-Herttuala S, Punnonen K, Laakso M. Low-grade inflammation and the phenotypic expression of myocardial fibrosis in hypertrophic cardiomyopathy. Heart 2012;98:1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Höogye M, Máandi Y, Csanády M, Sepp R, Buzás K. Comparison of circulating levels of interleukin-6 and tumor necrosis factor-alpha in hypertrophic cardiomyopathy and in idiopathic dilated cardiomyopathy. Am J Cardiol 2004;94:249–251. [DOI] [PubMed] [Google Scholar]
- 68.Taugner FM. Stimulation of the renin-angiotensin system in cats with hypertrophic cardiomyopathy. J Comp Pathol 2001;125:122–129. [DOI] [PubMed] [Google Scholar]
- 69.Cole LK, Mejia EM, Sparagna GC, Vandel M, Xiang B, Han X, Dedousis N, Kaufman BA, Dolinsky VW, Hatch GM. Cardiolipin deficiency elevates susceptibility to a lipotoxic hypertrophic cardiomyopathy. J Mol Cell Cardiol 2020;144:24–34. [DOI] [PubMed] [Google Scholar]
- 70.van der Velden J, Tocchetti CG, Varricchi G, Bianco A, Sequeira V, Hilfiker-Kleiner D, Hamdani N, Leite-Moreira AF, Mayr M, Falcão-Pires I, Thum T, Dawson DK, Balligand JL, Heymans S. Metabolic changes in hypertrophic cardiomyopathies: scientific update from the Working Group of Myocardial Function of the European Society of Cardiology. Cardiovasc Res 2018;114:1273–1280. [review]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sala V, Gallo S, Leo C, Gatti S, Gelb BD, Crepaldi T. Signaling to cardiac hypertrophy: insights from human and mouse RASopathies. Mol Med 2012;18:938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu X, Simpson J, Hong JH, Kim KH, Thavarajah NK, Backx PH, Neel BG, Araki T. MEK-ERK pathway modulation ameliorates disease phenotypes in a mouse model of Noonan syndrome associated with the RAF1(L613V) mutation. J Clin Invest 2011;121:1009–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong JC, Perez-Mancera PA, Huang TQ, Kim J, Grego-Bessa J, Del Pilar Alzamora M, Kogan SC, Sharir A, Keefe SH, Morales CE, Schanze D, Castel P, Hirose K, Huang GN, Zenker M, Sheppard D, Klein OD, Tuveson DA, Braun BS, Shannon K. KrasP34R and KrasT58I mutations induce distinct RASopathy phenotypes in mice. JCI Insight 2020;5:e140495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krams R, Ten Cate FJ, Carlier SG, Van Der Steen AFW, Serruys PW. Diastolic coronary vascular reserve: a new index to detect changes in the coronary microcirculation in hypertrophic cardiomyopathy. J Am Coll Cardiol 2004;43:670–677. [DOI] [PubMed] [Google Scholar]
- 75.Le Tourneau T, Susen S, Caron C, Millaire A, Maréchaux S, Polge AS, Vincentelli A, Mouquet F, Ennezat PV, Lamblin N, de Groote P, Van Belle E, Deklunder G, Goudemand J, Bauters C, Jude B. Functional impairment of von Willebrand factor in hypertrophic cardiomyopathy: relation to rest and exercise obstruction. Circulation 2008;118:1550–1557. [DOI] [PubMed] [Google Scholar]
- 76.Dimitrow PP, Undas A, Bober M, Tracz W, Dubiel JS. Obstructive hypertrophic cardiomyopathy is associated with enhanced thrombin generation and platelet activation. Heart 2008;94:e21. [DOI] [PubMed] [Google Scholar]
- 77.Jandrey KE, Norris JW, MacDonald KA, Kittleson MD, Tablin F. Platelet function in clinically healthy cats and cats with hypertrophic cardiomyopathy: analysis using the Platelet Function Analyzer-100. Vet Clin Pathol 2008;37:385–388. [DOI] [PubMed] [Google Scholar]
- 78.Chatterjee S, de Gonzalo-Calvo D, Derda AA, Schimmel K, Sonnenschein K, Bavendiek U, Bauersachs J, Bär C, Thum T. Leukocyte telomere length correlates with hypertrophic cardiomyopathy severity. Sci Rep 2018;8:11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Terrier B, Karras A, Cluzel P, Collet JP, Sène D, Saadoun D, Cacoub P. Presentation and prognosis of cardiac involvement in hepatitis C virus-related vasculitis. Am J Cardiol 2013;111:265–272. [DOI] [PubMed] [Google Scholar]
- 80.Papadopoulos DP, Moyssakis I, Votteas VE. Polyarteritis nodosa and hypertrophic obstructive cardiomyopathy. A true association? Clin Rheumatol 2004;23:57–58. [DOI] [PubMed] [Google Scholar]
- 81.Satish OS, Ravikumar A, Koshy G, Patnaik AN, Rao DS. Apical hypertrophic cardiomyopathy in association with Takayasu’s arteritis. Indian Heart J 2002;54:208–211. [PubMed] [Google Scholar]
- 82.Menacho K, Ramirez S, Segura P, Nordin S, Abdel-Gadir A, Illatopa V, Bhuva A, Benedetti G, Boubertakh R, Abad P, Rodriguez B, Medina F, Treibel T, Westwood M, Fernandes J, Walker JM, Litt H, Moon JC. INCA (Peru) study: impact of non-invasive cardiac magnetic resonance assessment in the developing world. J Am Heart Assoc 2018;7:e009891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krishnan J, Suter M, Windak R, Krebs T, Felley A, Montessuit C, Tokarska-Schlattner M, Aasum E, Bogdanova A, Perriard E, Perriard JC, Larsen T, Pedrazzini T, Krek W. Activation of a HIF1alpha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab 2009;9:512–524. [DOI] [PubMed] [Google Scholar]
- 84.Marian AJ, Senthil V, Chen SN, Lombardi R. Antifibrotic effects of antioxidant N-acetylcysteine in a mouse model of human hypertrophic cardiomyopathy mutation. J Am Coll Cardiol 2006:47827–47834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Senthil V, Chen SN, Tsybouleva N, Halder T, Nagueh SF, Willerson JT, Roberts R, Marian AJ. Prevention of cardiac hypertrophy by atorvastatin in a transgenic rabbit model of human hypertrophic cardiomyopathy. Circ Res 2005;97:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakamura K, Kusano KF, Matsubara H, Nakamura Y, Miura A, Nishii N, Banba K, Nagase S, Miyaji K, Morita H, Saito H, Emori T, Ohe T. Relationship between oxidative stress and systolic dysfunction in patients with hypertrophic cardiomyopathy. J Card Fail 2005;11:117–123. [DOI] [PubMed] [Google Scholar]