Abstract
Streptococcus pneumoniae remains a major causative agent of serious human diseases. The worldwide increase of antibiotic resistant strains revealed the importance of horizontal gene transfer in this pathogen, a scenario that results in the modulation of the species-specific gene pool. We investigated genomic variation in 20 S. pneumoniae isolates representing major antibiotic-resistant clones and 10 different capsular serotypes. Variation was scored as decreased hybridization signals visualized on a high-density oligonucleotide array representing 1,968 genes of the type 4 reference strain KNR.7/87. Up to 10% of the genes appeared altered between individual isolates and the reference strain; variability within clones was below 2.1%. Ten gene clusters covering 160 kb account for half of the variable genes. Most of them are associated with transposases and are assumed to be part of a flexible gene pool within the bacterial population; other variable loci include mosaic genes encoding antibiotic resistance determinants and gene clusters related to bacteriocin production. Genomic comparison between S. pneumoniae and commensal Streptococcus mitis and Streptococcus oralis strains indicates distinct antigenic profiles and suggests a smooth transition between these species, supporting the validity of the microarray system as an epidemiological and diagnostic tool.
Streptococcus pneumoniae remains a major causative agent of human diseases, which range from otitis media and sinusitis to pneumonia, septicemia, and meningitis. Pneumococci can be divided into more than 90 serotypes according to the immunochemistry of their capsular polysaccharides (18). However, approximately 90% of the invasive diseases worldwide are caused by only 16 different serotypes. Even fewer capsular types are involved in the recent worldwide emergence of penicillin-resistant isolates, where 6B, 9V, 14, 19F, 19A, and 23F are predominant (26, 34). Important clones include the multiple-antibiotic-resistant serotype 23F clone, first described in Spain, which has now spread practically to every continent, and the closely related clonal group of serotype 19A isolates from Hungary and other Eastern European countries (26, 32).
Penicillin binding proteins (PBPs) from penicillin-resistant isolates are encoded by mosaic genes that contain sequence blocks highly divergent from those of sensitive strains. These PBPs were recognized as the product of transformation events. Sequence comparisons of the mosaic genes in S. pneumoniae and the related Streptococcus oralis and Streptococcus mitis revealed horizontal gene transfer events not only among pneumococcal clones but also among pneumococci and commensal streptococci as well (for a review, see reference 13). Other examples have been described since, such as tetracycline and quinolone resistance determinants and, interestingly, genes involved in the regulation of genetic competence (11, 17, 28). These data document that a global gene pool is available to all streptococcal species, but very little is known about the extent of transformation and recombination in the pneumococcal population and their consequences in respect to the genomic variation of individual strains.
The era of bacterial genomes and the methodology developed concomitantly have revolutionized our approaches in molecular biology and epidemiology. The expression level of thousands of genes can now be monitored (8, 40), and allelic variations in the entire yeast genome have been identified (39). In this paper, we used representatives of genetically defined isolates to investigate their genomic variation compared to a type 4 strain. Seventy-five percent of the loci of this particular isolate appeared to be conserved in all 20 isolates tested, thus representing the genetic information common to the pneumococcus. Penicillin-resistant strains could easily be identified on the basis of an altered PBP 2x gene pbp2x, and changes in the dihydrofolate reductase gene correlated with trimethoprim (TMP) resistance, both examples of mosaic genes due to horizontal gene transfer. In contrast, only 144 loci hybridized to all nine S. mitis and S. oralis strains included in the study, many of which were associated with the translational machinery and other cytoplasmic components. Most of the pneumococcal specific virulence gene loci did not hybridize with the oral streptococci. The results demonstrate the power of DNA chip-based hybridization techniques for investigating the overall genetic information of the population of a species.
MATERIALS AND METHODS
Streptococcus spp. strains.
All S. pneumoniae clinical isolates used in this study and their relevant properties are listed in Table 1. Strain KNR.7/87 of serotype 4 represents the reference strain sequenced by The Institute for Genomic Research in collaboration with Human Genome Science. Strains were obtained from Roche (S. pneumoniae serotype 1, KNR.7/87; ATCC 49619, 1711, and 4249); all other strains are from the Kaiserslautern University strain collection. Our sequence information is almost identical to that of the published sequence (http://www.tigr.org) and represents approximately 91% of the total genomic sequence of S. pneumoniae KNR.7/87 (24). Strain R6 is an unencapsulated laboratory strain derived from the type 2 strain D39 (36). Oral streptococci were identified with API 20 STREP (bioMérieux, Marcy l'Etoile, France), and were sent for confirmation to the Statens Serum Institute (Copenhagen, Denmark): S. mitis NCTC10712, strains Hu-o8, B5, and B6; and S. oralis Hu-o2, Hu-o5, Hu-o12 and Hu-o16 (Table 1). Strain M3 from South Africa was initially identified as S. mitis but was suggested to be S. oralis according to more-detailed analysis; the present study again placed it within the S. mitis group.
TABLE 1.
Streptococcus spp. used in this study
| Organism and strain | Place of isolation | Yr of isolation | Serotype | Penicillin MIC | TMP MIC (μg/ml) | Reference or source |
|---|---|---|---|---|---|---|
| S. pneumoniae | ||||||
| Hu-9 | Hungary | 90 | 19A | 8 | >32 | 32 |
| Hu-11 | Hungary | 90 | 19A | 2 | >32 | 32 |
| Hu-15 | Hungary | 91 | 19A | 0.06 | >32 | 32 |
| 456 | Spain | 84 | 23F | 1–2 | >32 | 34 |
| 496 | Spain | 85 | 19F | 1 | >32 | 34 |
| SA17 | South Africa | 87 | 23F | 4 | >32 | 34 |
| 2349 | Spain | 92 | 23F | 2 | >32 | 31 |
| D219 | Germany | 89 | 23F | 0.125 | 0.38 | 32 |
| Fi2303R | Finland | 86 | 23F | 0.5 | 0.25 | 34 |
| F1 | France | 88 | 23F | 0.25 | 1 | 32 |
| 670 | Spain | 88 | 6B | 2 | >32 | 34 |
| B232C2 | PNGc | 69 | 4 | 0.25 | 0.5 | 34 |
| R6 | United States | 50s | 2a | 0.01 | 0.25 | 36 |
| D39 | United States | 16 | 2 | 0.01 | 0.38 | 4 |
| Fi2306 | Finland | 86 | 23F | 0.01 | 3 | 34 |
| ATCC 49619 | United States | 90 | 59 | 0.25 | NDb | 20 |
| 1 | Switzerland | 84 | 22 | ≤0.015 | ND | 24 |
| 1711 | Switzerland | 84 | 3 | ≤0.015 | ND | 24 |
| 4249 | France | 89 | 3 | ≤0.015 | ND | P. Geslin, Créteil, Paris, France |
| KNR.7/87 | United States | 91 | 4 | ND | ND | 1 |
| S. mitis | ||||||
| M3 | South Africa | 86–87 | 0.02 | 1 | 33 | |
| NCTC10712 | Great Britain | late 60s | 0.015 | 4 | 21 | |
| B5 | Germany | 94 | 8 | >32 | 31 | |
| B6 | Germany | 94 | 64 | >32 | 14 | |
| Hu-o8 | Hungary | 90–92 | 6–12 | >32 | 31 | |
| S. oralis | ||||||
| Hu-o2 | Hungary | 90–92 | 12–16 | >32 | 31 | |
| Hu-o5 | Hungary | 90–92 | 32 | >32 | 31 | |
| Hu-o12 | Hungary | 90–92 | 0.25 | >32 | A. Marton, Budapest, Hungary | |
| Hu-o16 | Hungary | 90–92 | 1.5 | >32 | A. Marton |
Serotype of the progenitor, D39.
ND, not determined.
PNG, Papua New Guinea.
DNA techniques.
Chromosomal DNA was prepared as previously described (14). Nucleotide sequencing was performed with the ABI Prism dRhodamine Terminator Cycle Sequencing Ready Reaction kit and with AmpliTaq DNA polymerase (Perkin-Elmer–ABI).
Oligonucleotide microarray design.
A sense oligonucleotide array (ROEZ06s) with a feature size of 24 μm, covering both genomes of S. pneumoniae (lower part) and Haemophilus influenzae (upper part), custom designed by Affymetrix (Santa Clara, Calif.), was used. In this paper, we focus on the pneumococcus sequence, for which over 130,000 oligonucleotide probes complementary to S. pneumoniae strain KNR.7/87 were selected. In addition, genes encoding distinct capsular types (3, 14, and 19F) and genes encoding common gram-positive resistance determinants (erythromycin, chloramphenicol, kanamycin, spectinomycin, and tetracycline) were included. Sense refers to the target nucleic acid, i.e., the oligonucleotide probes on microarray have the sequence complementary to the coding strand. In this microarray, feature size was reduced to 24 μm. A total of 1,968 S. pneumoniae gene sequences, as predicted by GeneMark software, and 323 intergenic regions larger than 200 bp were selected. The oligonucleotide probe selection (25-mers) and the array fabrication were performed by Affymetrix according to published procedures (25, 40). Each gene represented on the ROEZ06s microarray has, in general, 25 probe pairs and at least 20 probe pairs for very short genes. A probe pair consists of a perfect match (PM) probe and a mismatch (MM) probe that is identical except for a single base change in the central position (25). The position of the oligonucleotide on each gene is determined by sequence uniqueness criteria and is based on empirical rules for the selection of oligonucleotides likely to hybridize with high specificity and sensitivity (25). Considering an average gene length of 1 kb, 25 probe pairs (25-mers) per gene and an average redundancy of 1.5 in the selection of oligonucleotide probes, an estimated 40% of the genome is covered by oligonucleotide probes.
Only the lower half of the array contains probes for S. pneumoniae genes, whereas the upper half represents H. influenzae genes. Upon analysis by GeneChip software, all pneumococcal genes were unambiguously detected with the exception of two genes for which the original sequence information was of low quality. Moreover, specificity was demonstrated as only 28 H. influenzae genes were scored present due to cross-hybridization, but with a 20-fold-lower intensity value than the mean value for all pneumococcus genes. A high level of cross-hybridization occurred between H. influenzae rRNA and tRNA genes, which was not considered in further analysis. Considering only pneumococcus genes, the mean hybridization signal value was 1,112 arbitrary fluorescence units, and the intensities were found to cluster with 96% of the values within a factor 3 of the mean. Each experiment was performed in duplicate, and measurements of intensity were averaged. The analysis produced intensity data for each feature (2 × 20 × 130,000 features) representing more than 5 million hybridization data points. All intensities were first normalized and filtered on the basis of data quality (standard deviation), and fold change greater than fourfold compared to that of strain KNR.7/87 was used in all analyses.
DNA fragmentation and labeling.
Genomic DNA was first diluted in water to 150 μg in 400 μl and sonicated five times for 1 min each time to produce fragments ranging in size from 300 to 500 bp in length. Subsequently, 89 μl (about 35 μg) of sonicated DNA was partially digested for 8 min at 37°C with DNase I (0.1 U) in RQ1 buffer (Promega) in a final volume of 100 μl. The reaction was then stopped by ethanol precipitation, and the average size of the resulting DNA fragments was between 50 and 100 bp long. We recovered about 25 μg of fragmented DNA. DNA labeling was performed with biotin-labeled dideoxy ATP incorporated at the 3′ end of the fragmented DNA with Terminal-Deoxy-Transferase (Roche Molecular Biochemicals). Labeled fragmented DNA (15 to 20 μg) was mixed with 65 μl of water, 30 μl 5× buffer (1×), 15 μl of CoCl2 (2.5 mM), 10 nmol of biotin-N6-ddATP (NEN), and 10 μl of Terminal-Deoxy-Transferase (250 U). The mixture was incubated for 2 h at 37°C. Then, the labeled DNA fragments were precipitated with ethanol, resuspended in 20 μl of water, and quantified before being used for hybridization experiments.
Hybridization and staining procedures.
Hybridization solutions contained 100 mM N-morpholinoethanesulfonic acid (MES), 1 M Na+, 20 mM EDTA, and 0.01% Tween 20. In addition, the solutions contained 3 mg of unlabeled fragmented yeast RNA (Ambion)/ml, and 1.5 mg of acetylated bovine serum albumin (Sigma)/ml. Prior to hybridization, microarrays were prewarmed at room temperature, rinsed twice with hybridization buffer, and prehybridized for 10 min at 40°C. The hybridization mixture was denatured at 95°C for 5 min, cooled down to hybridization temperature, and centrifuged quickly to pellet all the nonsolubilized material. Finally, 230 μl of this hybridization mixture was loaded onto a chip for an overnight hybridization at 40°C with mixing on a rotisserie at 60 rpm. The hybridization mixture was then removed from the array and stored frozen. The arrays were rinsed twice with 6× SSPE (SSPE is 180 mM NaCl and 10 mM NaH2PO4 [pH 8.3])– 0.01% Tween 20 and washed in the same buffer at 40°C for 20 min. A stringent wash was then performed with 0.5× SSPE–0.01% Tween 20 for 15 min at 45°C. Subsequently, the hybridized DNA was labeled with 3 μg of streptavidin-phycoerythrin conjugate (Molecular Probes)/ml and 2 mg of acetylated bovine serum albumin/ml in 6× SSPE–0.01% Tween 20 for 10 min at 40°C. An additional washing step was performed with 6×SSPE–0.01% Tween 20 for 10 min at 40°C prior to scanning.
Data processing.
Microarrays were scanned at 570 nm with a 3-μm resolution with a gene chip scanner (Affymetrix) and analyzed as previously described (25). The signal intensity for each gene is calculated as the average intensity difference, represented by Σ[(PM−MM)/(number of probe pairs)]. Before comparison, each file was normalized, based on the sum of all signals for each experiment. The average intensity difference value was then averaged for all experiments performed in duplicate. To reduce extreme intensity ratios for genes not detected due to sequence variation, we also arbitrarily set the minimum average intensity difference for each gene to a value of 20, which corresponds to the noise. The intensity ratio corresponds in the case of a signal decrease in strain B to [−(average intensity difference obtained with DNA of strain A)/(average intensity difference obtained with DNA of strain B)], and in the case of a signal increase in strain B to [+(average intensity difference of strain B)/(average intensity difference of strain A)]. Specific details of the individual experiments are discussed in Results.
RESULTS
Reproducibility and validation of genomic hybridization on oligonucleotide chip.
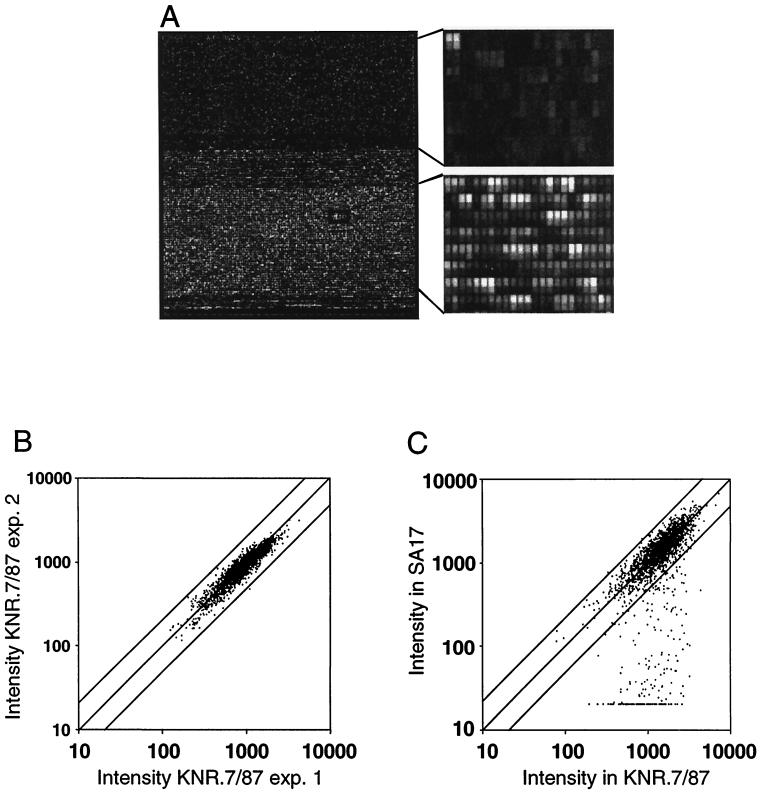
The DNA sequence of S. pneumoniae strain KNR.7/87 was the basis for the design of the oligonucleotide array. To establish the suitability for the hybridization approach on the DNA chip and to estimate the quality of the probes selected on the microarray, we first fragmented, labeled, and hybridized total S. pneumoniae genomic DNA from strain KNR.7/87 in triplicate against the microarray (see Materials and Methods). A typical image obtained after scanning is shown in Fig. 1A. The signal intensity for each gene is given as the sum of all PM intensities subtracted from total number of MM intensities, divided by the number of probes. Therefore, we further analyzed the signals averaged from three parallel genomic DNA hybridization experiments for all pneumococcus genes at the level of each feature instead of looking at the average value across the 25 probe pairs. The distribution of signal intensity for each feature subtracted from the background value shows only 75% of the probes with a signal intensity within a factor 3 of the mean. We considered that a substantial gain in sensitivity could be obtained by not taking into consideration the probe pairs reproducibly producing a very low signal. Therefore, a mask was created, removing 3,310 probe pairs with a signal smaller than 1/6 of the mean value, and all further analysis was performed with this mask. The differences in hybridization intensity across probe sets were then found to be reproducible by repeated independent hybridizations of genomic DNA samples on different chips (Fig. 1B). Only a single gene reproducibly varied by more than twofold, but we know that all probe selection rules could not be used for this gene due to its high A/T content and the small size of the gene. This gene was not considered in further analysis and only changes in intensity by a factor greater than 2.5 were considered relevant. A differential signal intensity of 4 will be used in further analysis to discriminate allelic variations. A total of 1,968 genes were covered in the final analyses. Hybridization of different concentrations of genomic DNA (8 and 24 μg) were also performed, and the results indicate that the probe sets respond quantitatively to a change in DNA concentration (data not shown).
FIG. 1.
Genomic DNA hybridizations on ROEZ06s microarray. (A) Fluorescence image of ROEZ06s 24-μm array containing more than 250,000 oligonucleotide probes complementary to all H. influenzae (top) and S. pneumoniae open reading frames and a selection of intergenic sequences (bottom). The array was hybridized against 15-μg total genomic DNA from S. pneumoniae KNR.7/87. An enlargement of each area shows the low cross-hybridization in the upper part of the chip and the specific signals in the lower part with intense rows of PM probes alternating with MM rows. (B) Scatter plot showing the correlation for the intensities of all genes obtained in two independent labeling reactions and hybridizations of genomic DNA from S. pneumoniae KNR.7/87 (experiment 1 versus 2). The two solid lines flanking the diagonal indicate an intensity ratio of 2. (C) Correlation of intensities for strain KNR.7/87 and a member of the Spanish clone SA17. All genes varying in strain SA17 appear on the lower right part of the graph.
The selection of pneumococcal clinical isolates used for further genomic comparison was based on their clonal relatedness and their serotypes (Table 1). The aim was to compare strains derived from the same ancestor but transferred independently in the laboratory decades ago, members of the same clone, members of distinct clones with the same or a different serotype, and penicillin-resistant strains with sensitive isolates. Nine oral streptococcal strains were also included to get some insight into genes conserved in related species and to estimate their overall relatedness.
Global allelic variations of 20 clinical isolates in comparison to KNR.7/87.
The genomic comparison was performed with 20 clinical isolates to identify regions of the sequence which vary most frequently in the pneumococcus genome (Table 1). Most isolates were of known clonal relationship and from the Kaiserslautern University strain collection. They included penicillin-resistant as well as -sensitive isolates covering 14 clonal groups and 10 different serotypes. Two major multiple-antibiotic-resistant and highly penicillin-resistant clones were represented by four and three members, respectively: the intercontinental 23F clone first recognized in Spain and the 19A clone predominant in Hungary and Eastern European countries. Strain 670 represented the multiple-antibiotic-resistant 6B clone, and other penicillin-resistant strains included the early resistant isolate B232C2 from Papua (type 4) and members of another three distinct 23F clones from different geographic areas (D219, F1, and Fi2303R). Penicillin-sensitive isolates covered a variety of diverse serotypes including 2, 3, 22, 23F, and 59. According to the previous analysis, data were filtered with the change factor value to limit and mask nonsignificant variations. Genes with a signal reduction of more than fourfold in comparison with KNR.7/87 were selected. These changes are visualized on the scatter graph for the comparison KNR.7/87 against SA17 (Fig. 1C).
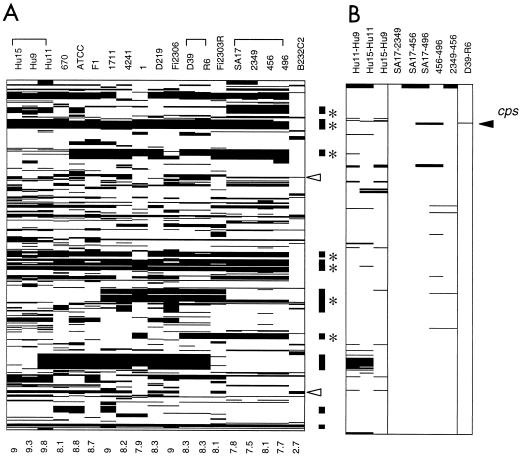
Figure 2 lists the 470 genes (24% of those represented on the microarray) where variation was detected in at least one of the strains, listed according to the unfinished genome of KNR.7/87 (http://www.tigr.org). About 50% are genes of unknown function, 35-+% encode genes involved in different metabolic pathways (especially in sugar metabolism), and about 15% code for surface-located proteins, transferases, phosphotransferase systems, ABC transporters, or efflux pumps for cell detoxification. Other highly variable regions relate to biologically active peptides: one is the bacteriocin cluster recently described (9, 30), and another one is homologous to the Enterococcus faecalis cytolysin locus cyl (35, 41) (Fig. 2A). In summary, all but one of the strains differed from strain KNR.7/87 by 8 to 11% of their genes (Fig. 2A). The exception was the type 4 strain B232C2, isolated in Papua, where only 54, or 2.7%, of the genes produced a reduced signal.
FIG. 2.
Cluster image showing genes with different intensity ratios in pairwise comparisons between S. pneumoniae strains. The genes are vertically sorted by position according to the unfinished genome of KNR.7/87 (http://www.tigr.org). Each column represents a pairwise comparison of the strains indicated on top of each lane; the number at the bottom indicates the pecentage of genes varying. (A) Comparison of 19 S. pneumoniae strains to KNR.7/87. All genes varying by an intensity ratio smaller than −4 in at least one of the comparisons are listed and marked by black lines (470 genes in total). Bars on the right side indicate clusters of variable genes covering more than 9 kb; *, the presence of transposases. Open arrowheads indicate the position of the bacteriocin cluster (blp or pcn) (9, 30) (top) and a Enterococcus cytolysin-related gene cluster (bottom) (41). (B) Comparison of D39 and its derivative R6 and members of the Spanish and Hungarian clone groups. All genes varying by an intensity ratio of less than −4 or greater than 4 in at least one of the comparisons are marked.
Ten major gene clusters where the hybridization signals are uniformly low in all affected strains cover more than 9 kb and up to 37 kb each; they encompass 130 loci and cover approximately 160 kb in total (Fig. 2A). Seven of them contain one or more transposases, and one resembles a prophage, i.e., they mark large regions of mobile elements. Two of the variable gene clusters include homologues of the virulence gene for immunoglobulin A1 (IgA1) protease and neuraminidase nanC (42), respectively.
With the exception of IgA1 protease genes, none of the genes implicated in virulence—those for hyaluronidase, neuraminidase A and B, autolysin, and pneumolysin (29)—showed any degree of variation. In addition to the IgA1 protease gene mentioned above, another two homologues were found in strain KNR.7/87, both of which showed variable and low hybridization signals, indicating sequence variation of the gene previously reported for one of them (16). The S. pneumoniae choline binding proteins (CBPs) have been implicated in adhesion; 12 CBPs have been identified (27). They are surface-exposed proteins associated with the pneumococcal choline-containing wall teichoic acid via C-terminal repeats. Four of the 10 CBP genes included on the microarray indicated sequence variation (encoded by pspA, cpbJ, pspC and cbpI), in agreement with reported variability in pspC (7).
Variation within clones. (i) Comparison of strain D39 and R6.
The serotype 2 strain D39 originally isolated by Avery in 1916 represents the parental strain of the unencapsulated R6 derivative commonly used in pneumococcal research worldwide (4, 36). Only one gene, cpsB, the second gene in the capsule biosynthesis cluster, presented a high 12-fold reduction in the hybridization signal for strain R6. cpsA is detected in both strains, while the genes cpsCDE that are not detected at all are probably different in D39 and KNR.7/87 (type 4). The R6 strain contains a 7.5-kb deletion starting from cpsB to cpsG (19). The microarray analysis confirms this without ambiguity (Fig. 2B). Other loci where the signal appeared different in the two strains were within the fourfold range that was not considered significant.
(ii) Variation within multiple resistant S. pneumoniae.
Two groups of strains were analyzed in more detail for intraclonal divergence, since they represent two major multiple-antibiotic-resistant clones: the 23F clone, first recognized in Spain and now isolated all over Europe, South Africa, the United States, and Asia (referred to as the Spanish clone); and the 19A clone predominant in Hungary and isolated so far only in other Eastern European countries. The four members of the Spanish clone differed by isolation date (1984 to 1992), geographic area (Spain or South Africa), and serotype (the isolate 496 represented a type 19F variant that was indistinguishable in all other parameters from other members of the Spanish clone by multilocus enzyme electrophoresis analysis and PBP properties) (31, 34). The three members of the multiple-antibiotic-resistant serotype 19A clonal group included a penicillin-sensitive isolate Hu-15 and the two high-level penicillin-resistant strains Hu-9 and Hu-11.
The comparison of the four members of the 23F clone with KNR.7/87 produced a list of 174 genes that differed in at least one of the strains, but only 19 of these varied in intensity within the Spanish clone (Fig. 2B). Strain 496 has sustained a serotype shift which was identified from this analysis as cps19Fb cps19Fg cps19Fi, also present on the array as control genes, and differed in another three genes related to capsular biosynthesis; this strain showed the highest degree of variation with 13 different genes (including the 6 capsule-related ones) compared to strain 456 or 2349. The type 19A clonal group was more variable, with 19 to 41 genes (1 to 2.1%) in the pairwise comparisons or a total of 55 genes (2.8%), but one has to consider that 18 loci were contained in one cluster that differentiated Hu-11 from the other two 19A strains.
Detection of penicillin and TMP resistance and mosaic gene structures. (i) Penicillin resistance.
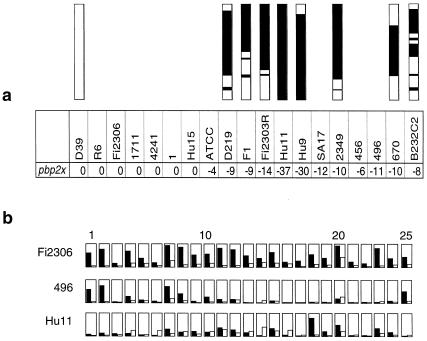
PBP2x represents a primary resistance determinant, and all penicillin-resistant isolates contain a low-affinity PBP2x encoded by a mosaic pbp2x gene, due to localized interspecies recombinational events (13, 23). Figure 3a shows the intensity ratios between KNR.7/87 and all 20 strains for PBP2x. pbp2x presents a very low intensity for all penicillin-resistant isolates, which was predicted from the high degree of mosaic structure within this gene; no difference in intensity was apparent for the sensitive strains. In fact, it was through this analysis that the ATCC 46619 strain was recognized as being penicillin resistant, which was subsequently confirmed by MIC determination (Table 1). A schematic representation of the intensity for all features against PBP2x is shown for strains Fi2306 (penicillin sensitive), 496, and Hu-11 (penicillin resistant) (Fig. 3b). It clearly highlights that most probe pairs did not hybridize against the resistant genes and that the hybridization pattern for the different oligonucleotides is specific for each of the pbp2x variants. Thus, pbp2x proves a significant marker for penicillin resistance. In this context it is also important to note that in all cases the pbp2x of the S. mitis and S. oralis strains did not hybridize with the reference S. pneumoniae strain (see Results).
FIG. 3.
Detailed analysis of pbp2x genes. (a) Intensity ratios for pbp2x genes in pairwise comparisons of 19 S. pneumoniae strains to KNR.7/87. Intensity ratios are 0 in penicillin-sensitive strains. The values for pbp2x range from −4 to −37 for pbp2x in all penicillin-resistant strains. Mosaic structures of the pbp2x genes are shown above the strain designation, with white bars representing conserved sequences in sensitive S. pneumoniae, and black bars representing mosaic blocks. Sequence differences between the mosaic blocks are not distinguished. (b) Probe performance graph for pbp2x in three strains. The results obtained for each of the 25 oligonucleotides representing the pbp2x gene are shown for the penicillin-sensitive strain Fi2306 and two resistant-strains, 496 and Hu-11. Signal intensity for the PM probe is shown in black, while MM probe signals are shown in white. Probe pairs are sorted from 5′ (left) to 3′.
For high-level resistance, alterations of at least pbp2b and pbp1a are also required. However, we discovered significant changes in these two genes, but not in all resistant isolates. It is possible that the oligonucleotides do not cover the sequences representing the mosaic block which, at least in the case of pbp2b, is often fairly small (data not shown). The pbp3 gene does not present significant intensity differences when compared to KNR.7/87 (less than 2.5-fold intensity variation), similar to pbp2a and pbp1b (data not shown). These genes play only a minor role, if any, in resistance development in clinical isolates, and mosaic structures for these genes have yet not been described.
TMP resistance.
Alterations in the dihydrofolate reductase gene are responsible for TMP resistance, which has increased in S. pneumoniae worldwide in the last decades and is frequently associated with penicillin resistance (22). A low intensity for the dhfr region was observed in all members of the Spanish 23F and the Hungarian 19A clones and for the multiple-antibiotic-resistant 6B strain 670 also. DNA sequence analysis confirmed highly altered dhfr genes (Fig. 4), and all these strains were indeed TMP resistant. All TMP-resistant strains had an altered dhfr codon 100 resulting in the substitution Leu100 to Ile, confirming the importance of this change for TMP resistance suggested recently (2). Four allelic variants were distinguishable, two of them found in the Hungarian 19A strains. It has been postulated that altered dhfr sequences are the result of horizontal gene transfer (2). We therefore determined the DNA sequence of the dhfr gene from five S. mitis strains, three of which were TMP and penicillin resistant (Table 1). The dhfr gene of the Hungarian S. mitis strain Hu-08 was almost identical to that of S. pneumoniae Hu-11, once more documenting that both species have access to a common gene pool.
FIG. 4.
DNA sequences of dihydrofolate reductase genes in S. pneumoniae, S. mitis, and S. oralis strains. Only sites where at least one sequence differed from that of S. pneumoniae R6 dhfr are shown. The codons are indicated vertically in the first three rows and numbered according to the published sequence (2). The numbers 1, 2, and 3 refer to the position in the respective codon.
Comparison between S. pneumoniae and oral Streptococcus spp.
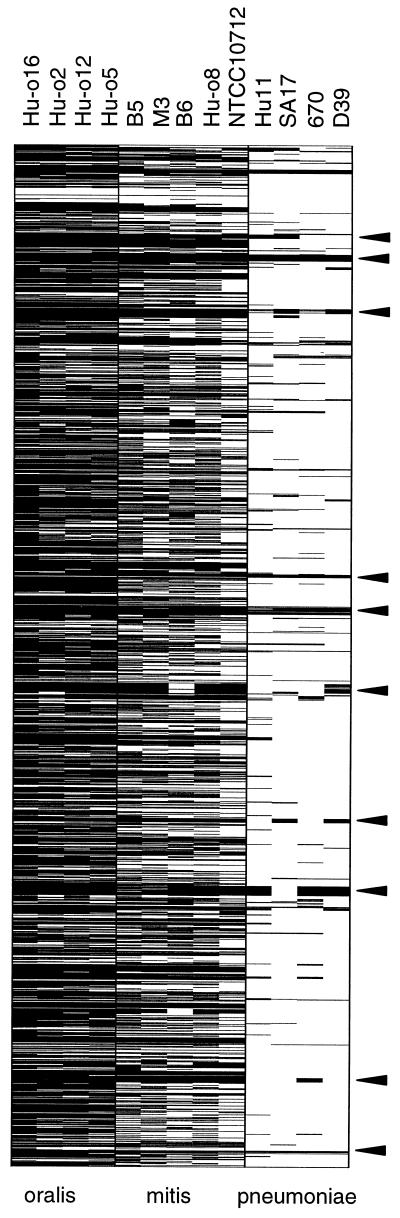
Nine strains of oral streptococci isolated in Spain, Hungary, and Germany were used for the present analysis. The global analysis showed that genes in all nine strains were highly divergent from the KNR.7/87 strain, ranging from 39% (759 loci) in S. mitis NCTC10712 up to 85% (1,671 loci) in S. oralis Hu-016. Strains previously identified as S. mitis all clustered in the group which differed by 39 to 56% of their genes when compared to S. pneumoniae KNR.7/87, whereas the four S. oralis strains showed reduced a hybridization signal in over 79% and up to 85% of their genes compared to KNR.7/87. The strain M3, previously identified as S. oralis, clearly clustered within the S. mitis group of strains (Fig. 5).
FIG. 5.
Global allelic variations in oral streptococci. All;1,968 pneumococcus genes present on the microarray are represented vertically according to their position on the unfinished genome of KNR.7/87 (http://www.tigr.org). Genes varying by an intensity ratio smaller than −4 in at least one of the comparisons were marked. For comparison, genes with low intensity of the major multiple-antibiotic-resistant clones (Hu-11, SA17, 670) as well as the ancestor of the laboratory strain R6 (D39) are also listed, and the major variable gene clusters as indicated in Fig. 2 are marked by arrows on the right side.
One cluster of genes that appeared almost identical in all oral streptococci, compared to S. pneumoniae (Fig. 5), represents a 50S ribosomal gene cluster; other genes that did not differ between the species include 30S ribosomal proteins, housekeeping genes such as pyruvate oxidase, glutamate racemase, glyceraldehyde 3-phosphate dehydrogenase, and the heat shock protein dnaK. In total, 144 genes or 7.3% did not reveal changes within the fourfold range of their hybridization signals. It is striking that none of these conserved loci include surface proteins, indicating a completely different antigenic potential of the commensal species compared to the pathogen.
We investigated three groups of functionally related genes more closely: those associated with pneumococcal pathogenicity, to see whether virulence factors moved into nonpathogenic species; peptidoglycan biosynthesis, since murein biochemistry has served as a species-specific marker; and competence induction that include genes suggested to undergo allelic variation via horizontal gene transfer (Fig. 6).
FIG. 6.
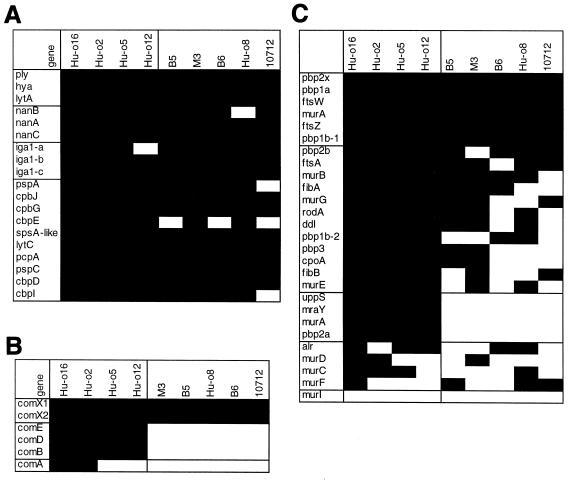
Variation of pneumococcal virulence genes and choline binding proteins (A), early competence genes (B), peptidoglycan biosynthesis-related genes (C) in oral streptococci. The genes are listed on the left side; black boxes indicate intensity ratios smaller than −4. Genes with low intensity in both S. mitis and S. oralis, as well as those with low intensity in S. oralis only, are boxed.
All pneumococcal virulence factors gave very low or no hybridization signals in the S. oralis and S. mitis strains, with three exceptions: one of the three IgA1 proteases was recognizable in Hu-o12 and Hu-o16, nanB appeared to be present in Hu-o8, and the lytA gene reacted low but significantly in strains B5 and Hu-o8 (Fig. 6A). Like the virulence factors, choline binding proteins were almost all discriminated in the streptococcal strains; exceptions were most notable in S. mitis NCTC10712, and cbpE was apparent in another two S. mitis strains (Fig. 6A).
The early competence genes comABDE of S. mitis showed intensity signals very similar to thethose of the S. pneumoniae reference strain (Fig. 6B). DNA sequencing of comDE confirmed differences to the S. pneumoniae genes of approximately 20% in S. oralis Hu-o5 and less than 4% in S. mitis B6, M3, and NCTC10712, except for a mosaic block located in the 5′ region encoding the receptor part of the ComD histidine protein kinase that was apparently not covered by the comD representing oligonucleotides (K. Kaminski, C. Bergmann, and R. Hakenbeck, unpublished results). The similarity between S. mitis and S. pneumoniae comDE sequences indicates that horizontal gene transfer and recombination in this region is easily possible between S. mitis and S. pneumoniae, thus allowing for pheromone switch in these species. The alternative sigma factors comX1and comX2 produced very low intensities in all streptococci.
The peptidoglycan biosynthetic proteins considered here include the genes required for synthesis of muropeptides (mur genes, including the recently described fibAB genes, also known as murMN genes, responsible for branched muropeptides [10, 37], alanine racemase alr, d-Ala-d-Ala, and d-Ala-ligase ddl), all PBPs and components of the division machinery (ftsZ, ftsA, ftsW, and rodA); genes related to the undecaprenyl-dependent translocation (uppS undecaprenyl pyrophosphate synthase, mraY lipid-linked MurNAc-pentapeptide synthesis, and murG); and cpoA, suggested to be involved in cell surface polysaccharide biosynthesis (12). Most of the genes related to peptidoglycan synthesis appeared variable within the species (Fig. 6C). Genes that discriminated between S. pneumoniae and all commensal streptococci included those described as the “divisome,” i.e., pbp2x, ftsA, ftsW, and ftsZ; those that distinguished between the S. oralis and S. mitis strains included one of the two UDP-N-acetylglucosamine enolpyruvyl transferases murA, uppS, mraY, and pbp2a.
DISCUSSION
The methodology provided by the high-density oligonucleotide microarray representing 1,968 genes of the S. pneumoniae KNR.7/87 genome in combination with the availability of a collection of strains used previously in comparative genetic analyses provided an ideal setting for investigating the variability of the genome within this species and for identification of genes potentially relevant for the pneumococcus as a pathogenic organism. It is clear that the present analysis can only be the start of a series of much more detailed investigations, including the annotations and the comparison between the two pneumococcal genomes that are expected in the near future, the KNR.7/87 sequence (http://www.tigr.org) and that of the laboratory strain R6 (5).
The reliability of the data could clearly be demonstrated by the reproducibility of fluorimetric representations in independent hybridization experiments with the reference DNA. Also, the comparison of the type 2 strain D39 isolated by Avery in 1916 with its unencapsulated derivative R6 isolated in the 1950s confirmed that the only detectable difference between these two strains is the deletion within the capsular gene cluster, described recently (19).
The overall degree of genomic variation concerned roughly 25% of the genes, whereas between the reference strain KNR.7/87 and any one of the 19 other strains, 8 to 10% of the genes differed. Members of two predominant multiple-antibiotic-resistant clones—the Spanish clone (<1%) and the Hungarian 19A clone (≤2.1%) were much more uniform. This latter value is close to that between KNR.7/87 and B232C2, both type 4 strains (2.7%), suggesting that they are possibly representatives of a global serotype 4 clone. Using a large number and a wide spectrum of genetically different strains, one could expect a gradual variability among the pneumococcal population, since despite the recognition of clonal spread, population analysis suggested a freely recombining structure characteristic of transformable organisms (15). Considering that the genome of the R6 strain is approximately 2 Mb in size, i.e., roughly 10% less than that of the KNR.7/87 strain (5), the apparent differences between strains may be to a considerable degree the result of missing genes represented by the gene clusters that appeared as low-intensity signals.
The use of the S. pneumoniae KNR.7/87 DNA sequence as a reference for the present studies poses obvious limitations in that it excludes genes missing from this particular strain and unique allelic variants as well. Especially, such genes could be relevant for specific features related to virulence profiles or the successful spread of multiple-antibiotic-resistant clones. Potential candidates for such distinctions could be the IgA1 protease gene and the nanC gene that are both present in variable gene clusters.
In Helicobacter pylori, genomic sequence comparison between two unrelated strains revealed only eight genes with >98% nucleotide identity, and the overall sequence variation appeared much more substantial than in S. pneumoniae (3). In contrast, only deletions were apparent between two Mycobacterium strains (6). In our study, the absence of genes as well as variability within genes could be detected. We could not distinguish between deletion of part of the genes versus variable sequences within the genes. Two genes with very low comparative fluorescence intensity were therefore further investigated by PCR and DNA sequence analysis, confirming the presence of deletions in the particular strains affected. The examples include a hypothetical transporter protein which was absent in five strains (D39, R6, D219, 1, and ATCC 49619) and the Spanish clone and another gene of unknown function where all strains except KNR.7/87, D39-R6, 1, and Fi2303R contained a 690-bp deletion. Nevertheless, it is also clear that single point mutations were not detected in our analysis, and since the oligonucleotides cover only parts of the genes, some variable regions may also pass detection as was obvious with the comD gene, where none of the known allelic variants were found.
An example that could be exploited for diagnosing antibiotic resistance is the pbp2x gene, which must be present in all strains, since it is an essential gene (13). In fact, all penicillin-resistant strains were recognized here on the basis of sequence variation in their pbp2x gene, making this gene a tool suitable for diagnostic chip design (Fig. 3). Similarly, the dihydrofolate reductase gene revealed a reduced signal in all TMP-resistant strains, and the variation was confirmed by DNA sequence analysis (Fig. 4). The presence of resistance determinants mediated by transposons (erythromycin, tetracycline, and chloramphenicol resistance) was also efficiently detected on the microarray in agreement with the resistance profile of the strains (results not shown). The Hungarian group and strain 670 were identified as containing an erythromycin resistance gene, and chloramphenicol resistance was detected in the same strains and in the Spanish clone (SA17, 2349, 456, and 496).
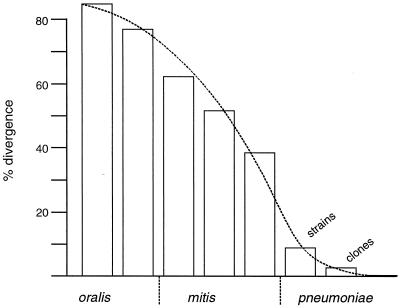
The distinction between the pathogenic S. pneumoniae and the commensal oral streptococci also became evident. Important pneumococcal virulence genes were discriminated in the nine streptococcal strains used in this study. Commensal strains that have acquired pneumococcal virulence factors, as described previously (38), could be identified easily using microarray-based hybridization techniques. The high number of genes differing between S. pneumoniae and the streptococci (40 to 85%) reflect any degree of variation ranging from a few point mutations within a gene up to its complete absence. We propose that a smooth transition across the species border reflects the relationship between commensal and pathogenic species more appropriately than organizing them into three distinct species (Fig. 7). The M3 strain poses a perfect example of this problem: the API system does not allow a distinction between S. mitis and S. oralis; M3 has then been specified as S. oralis because of a surface antigen that reacts with an antiserum raised against an ATCC S. oralis strain (33). According to the result presented here, it would be classified as S. mitis. A larger number of strains should be tested eventually, and it remains to be seen whether individual genes can be used as reliable markers to distinguish between S. mitis and S. oralis.
FIG. 7.
Genomic variation of S. mitis, S. oralis, and S. pneumoniae strains. The percentage of genes with low intensity compared to S. pneumoniae KNR.7/87 is indicated; the last bar represents the variation observed between the two multiple-antibiotic-resistant S. pneumoniae clones.
The possibility for exploiting sequence information via DNA chip-based comparative genomics for investigating the pathogenicity potential of individual species and predominant clones within species has become evident. In a next step, detailed characterization of the function of the genes is needed to fully appreciate such distinctions.
ACKNOWLEDGMENTS
We gratefully acknowledge Detlef Wolf, Clemens Broger, and Martin Neeb for their help with bioinformatics and Kurt Amrein for helpful discussions. We also thank Karin Kuratli, Nathalie Moulin, Katharina Rupp, Brigitte Rosenberg, and Ulrike Klein for excellent technical assistance.
Part of this work was supported by the Deutsche Forschungsemeinschaft, the Stiftung Rheinland Pfalz für Innovation, and the European Community grant No. BI04-CT98–0424.
N.B. and B.W. contributed equally to the work.
REFERENCES
- 1.Aaberge I S, Eng J, Lermark G, Lovik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–152. doi: 10.1016/s0882-4010(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 2.Adrian P V, Klugman K P. Mutations in the dihydrofolate reductase gene of trimethoprim-resistant isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2406–2413. doi: 10.1128/aac.41.11.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noona B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 4.Avery O T, MacLeod C M, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltz R H, Norris F H, Matsushima P, DeHoff B S, Rockey P, Porter G, Burgett S, Peery R, Hoskins J, Braverman L, Jenkins I, Solenburg P, Young M, McHenney M A, Skatrud P L, Rosteck P R., Jr DNA sequence sampling of the Streptococcus pneumoniae genome to identify novel targets for antibiotic development. Microb Drug Resist. 1998;4:1–9. doi: 10.1089/mdr.1998.4.1. [DOI] [PubMed] [Google Scholar]
- 6.Behr A M, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 7.Brooks-Walter A, Briles D E, Hollingshead S K. The pspC gene of Streptococcus pneumoniae encodes a poolymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect Immun. 1999;67:6533–6542. doi: 10.1128/iai.67.12.6533-6542.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 9.de Saizieu A, Gardès C, Flint N, Wagner C, Kamber M, Mitchell T J, Keck W, Amrein K E, Lange R. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipe S R, Tomasz A. Inhibition of the expression of penicillin-resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc Natl Acad Sci USA. 2000;97:4891–4896. doi: 10.1073/pnas.080067697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González I, Georgiou M, Alcaide F, Balas D, Liñares J, de la Campa A G. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob Agents Chemother. 1998;42:2792–2798. doi: 10.1128/aac.42.11.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grebe T, Paik J, Hakenbeck R. A novel resistance mechanism for β-lactams in Streptococcus pneumoniae involves CpoA, a putative glycosyltransferases. J Bacteriol. 1997;179:3342–3349. doi: 10.1128/jb.179.10.3342-3349.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakenbeck R, Kaminski K, König A, van der Linden M, Paik J, Reichmann P, Zähner D. Penicillin-binding proteins in β-lactam-resistant Streptococcus pneumoniae. Microb Drug Resist. 1999;5:91–99. doi: 10.1089/mdr.1999.5.91. [DOI] [PubMed] [Google Scholar]
- 14.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall L M, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halter R, Pohlner J, Meyer T F. Mosaic-like organization of IgA protease genes in Neisseria gonorrhoeae generated by horizontal genetic exchange in vivo. EMBO J. 1989;8:2737–2744. doi: 10.1002/j.1460-2075.1989.tb08415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Håvarstein L S, Hakenbeck R, Gaustad P. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol. 1997;179:6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iannelli F, Pearce B J, Pozzi G. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol. 1999;181:2652–2654. doi: 10.1128/jb.181.8.2652-2654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen J H, Doern G V, Ferraro M-J, Knapp C C, Swenson J M, Washington J A., II Multicenter evaluation of the use of Haemophilus test medium for broth microdilution antimicrobial susceptibility testing of Streptococcus pneumoniae and development of quality control limits. J Clin Microbiol. 1992;30:961–966. doi: 10.1128/jcm.30.4.961-966.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilian M, Mikkelsen L, Henrichsen J. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946). Streptococcus oralis (Bridge and Sneath 1982) and Streptococcus mitis (Andrewes and Horder 1906) Int J Syst Bacteriol. 1989;39:471–484. [Google Scholar]
- 22.Klugman K P. Pneumococcal resistance to antibiotics. Clin Microbiol Rev. 1990;3:171–196. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laible G, Spratt B G, Hakenbeck R. Inter-species recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 24.Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, Stieger M, Caspers P, Kamber M, Keck W, Amrein K E. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene. 1999;237:223–234. doi: 10.1016/s0378-1119(99)00266-8. [DOI] [PubMed] [Google Scholar]
- 25.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 26.Muñóz R, Musser J M, Crain M, Briles D E, Marton A, Parkinson A J, Sorensen U, Tomasz A. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 27.Novak R, Tuomanen E. Pathogenesis of pneumococcal pneumoniae. Semin Respir Infect. 1999;14:209–217. [PubMed] [Google Scholar]
- 28.Oggioni R M, Dowson C G, Maynard Smith J, Provvedi R, Pozzi G. The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid. 1996;35:156–163. doi: 10.1006/plas.1996.0018. [DOI] [PubMed] [Google Scholar]
- 29.Paton J C, Berry A M, Lock R A. Molecular analysis of putative pneumococcal virulence proteins. Microb Drug Resist. 1997;3:1–10. doi: 10.1089/mdr.1997.3.1. [DOI] [PubMed] [Google Scholar]
- 30.Reichmann P, Hakenbeck R. Allelic variation in a peptide-inducible two component system of Streptococcus pneumoniae. FEMS Microbiol Lett. 2000;190:231–236. doi: 10.1111/j.1574-6968.2000.tb09291.x. [DOI] [PubMed] [Google Scholar]
- 31.Reichmann P, König A, Liñares J, Alcaide F, Tenover F C, McDougal L, Swidsinski S, Hakenbeck R. A global gene pool for high-level cephalosporin resistance in commensal Streptococcus spp. and Streptococcus pneumoniae. J Infect Dis. 1997;176:1001–1012. doi: 10.1086/516532. [DOI] [PubMed] [Google Scholar]
- 32.Reichmann P, Varon E, Günther E, Reinert R R, Lütticken R, Marton A, Geslin P, Wagner J, Hakenbeck R. Penicillin-resistant Streptococcus pneumoniae in Germany: genetic relationship to clones from other European countries. J Med Microbiol. 1995;43:377–385. doi: 10.1099/00222615-43-5-377. [DOI] [PubMed] [Google Scholar]
- 33.Sibold C, Henrichsen J, König A, Martin C, Chalkley L, Hakenbeck R. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol. 1994;12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 34.Sibold C, Wang J, Henrichsen J, Hakenbeck R. Genetic relationship of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect Immun. 1992;60:4119–4126. doi: 10.1128/iai.60.10.4119-4126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siezen R J, Kuipers O P, de Vos W M. Comparison of lantibiotic gene clusters and encoded proteins. Antonie Leeuwenhoek. 1996;69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- 36.Smith M D, Guild W R. A plasmid in Streptococcus pneumoniae. J Bacteriol. 1979;137:735–739. doi: 10.1128/jb.137.2.735-739.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber B, Ehlert K, Diehl A, Reichmann P, Labischinski H, Hakenbeck R. The fib locus in Streptococcus pneumoniae is required for peptidoglycan crosslinking and PBP-mediated beta-lactam resistance. FEMS Microbiol Lett. 2000;188:81–85. doi: 10.1111/j.1574-6968.2000.tb09172.x. [DOI] [PubMed] [Google Scholar]
- 38.Whatmore A M, Efstratiou A, Pickerill A P, Broughton K, Woodward G, Sturgeon D, George R, Dowson C G. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect Immun. 2000;68:1374–1382. doi: 10.1128/iai.68.3.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winzeler E A, Richards D R, Conway A R, Goldstein A L, Kalman S, McCullough M J, McCusker J H, Stevens D A, Wodicka L, Lockhart D J, Davis R W. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 40.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 41.Zähner D. Identifizierung von Zielgenen des signaltransduzierenden Zwei-Komponenten-Systems cia von Streptococcus pneumoniae. Ph.D. thesis. Kaiserslautern, Germany: Universität Kaiserslautern; 1999. [Google Scholar]
- 42.Zysk G, Bongaerts R J M, ten Thoren E, Bethe G, Hakenbeck R, Heinz H-P. Detection of 23 immunogenic pneumococcal proteins using convalescent-phase serum. Infect Immun. 2000;68:3740–3743. doi: 10.1128/iai.68.6.3740-3743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]