Abstract
Fetal Nuchal fluid collections can manifest with two distinct presentations attributable to the same phenotypic spectrum: increased nuchal translucency (iNT) and cystic hygroma. The prenatal detection of these findings should prompt an accurate assessment through genetic counseling and testing, including karyotype, chromosomal microarray analysis (CMA) and multigene RASopathy panel. We performed a systematic review of the literature and meta-analysis, to calculate diagnostic yields of genetic testing in fetuses with iNT and cystic hygroma. We compared the results with a cohort of 96 fetuses with these isolated findings. Fetuses with isolated NT ≥ 2.5 mm showed karyotype anomalies in 22.76% of cases and CMA presented an incremental detection rate of 2.35%. Fetuses with isolated NT ≥ 3 mm presented aneuploidies in 14.36% of cases and CMA had an incremental detection rate of 3.89%. When the isolated NT measured at least 3.5 mm the diagnostic yield of karyotyping was 34.35%, the incremental CMA detection rate was 4.1%, the incremental diagnostic rate of the RASopathy panel was 1.44% and it was 2.44% for exome sequencing. Interestingly, CMA presents a considerable diagnostic yield in the group of fetuses with NT ≥ 3.5 mm. Similarly, exome sequencing appears to show promising results and could be considered after a negative CMA result.
Keywords: increased nuchal translucency, cystic hygroma, molecular testing, prenatal diagnosis, array, RASopathies
1. Introduction
Fetal body fluid collections occur when the rate of interstitial fluid production by capillary ultrafiltration exceeds the rate of interstitial fluid return to the circulation via lymphatic vessels [1]. The characteristics of prenatal microcirculation and lymphatic system make fetuses more prone to develop interstitial fluid accumulation, due to the higher capillary permeability, the higher interstitial compartment compliance, and the greater influence of venous pressure on lymphatic return. Conditions that alter homeostatic mechanisms can perturb the balance of interstitial fluid movement.
Currently, there is still confusion regarding the etiopathogenesis and the possible associations with chromosomal, genomic, and monogenic conditions following the detection of nuchal fluid collections. These can present as two distinct nosological entities, increased nuchal translucency (iNT) and cystic hygroma. Some authors do not consider the need to distinguish the two ultrasound pictures relevant, as often in pregnancies with these findings the same management is indicated [2]. However, iNT and cystic hygroma appear to present different risks, which must be provided in the genetic counseling, therefore the distinction between the two entities can even play a role in the couple’s decisions regarding the current pregnancy.
The term “nuchal translucency” represents the sonographic measurement of the widest thickness of the physiological collection of fluid under the skin behind the fetal neck that is measured when the crown-rump length ranges between 45 and 84 mm, between 11 and 13 weeks of gestational age (wga) [3,4,5]. This collection has a transient nature and is not considered a structural anomaly. NT is measured in a midsagittal view of the fetal face, defined by the presence of the echogenic tip of the nose and rectangular shape of the palate anteriorly, the translucent diencephalon in the center and the nuchal membrane posteriorly [3,4,5]. Physiological NT and iNT should not present septations in this view [6].
NT ≥ 99th centile (≥3.5 mm), found in around 1% of pregnancies, is associated with chromosomal anomalies, monogenic conditions [7], heart malformations and other malformations in euploid fetuses [8]. NT ≥ 3 mm [9], found in around 5% of screened fetuses [10], is considered a high risk factor for aneuploidies, while the interpretation of the 2.5–2.9 mm NT is controversial.
Cystic hygroma represents a developmental anomaly of the lymphatic system, characterized by a protein-rich fluid [11] located in the nuchal region, behind and around the fetal neck, which can extend the length of the fetus and show septations. It is found in 1:285 fetuses [12] and it arises from the jugular-lymphatic obstruction and can progress determining hydrops. Cystic hygroma is associated with poor prognosis. However, since it is not possible to use the septation to distinguish cystic hygroma and increased nuchal translucency [2], in the literature the criterion used to attribute the ultrasound finding to one or the other entity is not always clear.
These conditions can occur as isolated, or in association with fetal structural anomalies [2,12]. When further anomalies are present, in some cases the association can be ascribed to the co-occurrence of two or more anomalies due to the same underlying cause (e.g., specific genetic disorders) [2,12]. In other cases, the primary anomaly, such as a cardiac malformation or a skeletal dysplasia, might hinder lymphatic development and function, secondarily resulting in a fetal nuchal fluid collection. In these circumstances, the lymphatic anomaly might be regarded as a secondary deformation rather than a primary developmental disorder.
The detection of iNT or cystic hygroma should prompt genetic counseling. Anamnestic reassessment, further sonographic evaluations and cytogenetic and molecular testing play a decisive role in management of fetuses with these findings. It should be noted that cases that are apparently isolated at the time of counseling might be later detected with additional anomalies which were not evident at primary examinations. Standard karyotype and chromosomal microarray analysis (CMA) are usually requested, even if when the NT is in the normal range but above the 95th centile some authors propose non-invasive prenatal screening [13]. In other cases, when cytogenetic testing yields negative results, Next Generation Sequencing (NGS) RASopathy panel or clinical exome sequencing can be proposed to the couple [14].
RASopathies are a group of conditions determined by pathogenic variants in genes involved in the Ras/mitogen-activated protein kinase (RAS-MAPK) signaling pathway, that can be prenatally suspected when sonography identifies iNT, cystic hygroma, pleural/pericardial effusions, polyhydramnios or some subtypes of cardiac malformations, even if these are nonspecific findings [15]. According to the literature, an extensive multigene panel should be offered when an RASopathy is suspected in the prenatal setting [15].
Conflicting results in the literature may be explained by both different NT distributions and parameters used to select the cohort.
We performed a systematic review of the literature and meta-analysis, in order to calculate diagnostic yields of genetic testing in fetuses with iNT, and compared the results with fetuses detected with cystic hygroma. We also collected a dicentric case series of fetuses with these isolated findings that underwent karyotype, CMA and RASopathy panel sequentially.
2. Materials and Methods
2.1. Systematic Review of the Literature and Meta-Analysis
The research was conducted following PRISMA guidelines [16]. We searched the Pubmed database (https://pubmed.ncbi.nlm.nih.gov/), accessed on 21 October 2022, from 2002, for (“fetus” or “fetuses” or “foetus” or “foetuses” or “fetal” or “foetal” or “fetalis” or “foetalis” or “prenatal” or “pre-natal” or “antenatal” or “ante-natal”) and (“hygroma” or “cystic hygroma” or “septated cystic hygroma” or “NT” or “nuchal translucency” or “nuchal thickness”) and (“karyotype” or “karyotyping” or “cytogenetic analysis” or “molecular cytogenetic” or “molecular cytogenetics” or “CMA” or “chromosomal microarray” OR “chromosomal microarrays” or “chromosomal array” or “chromosomal arrays” or “CGH array” or “comparative genomic hybridization” or “SNP array” or “microdeletion” or “microduplication” or “CNV” or “CNVs” or “copy number variant” or “copy number variants” or “copy number variation” or “copy number variations” or “WES” or “CES” or “Exome Sequencing” or “RASopathy” or “RASopathies”). All titles and abstracts were examined. Only papers with full text in English were included. Papers discussing or describing the prenatal diagnostic application of karyotyping, chromosomal microarray, RASopathies molecular testing and/or exome sequencing in iNT and/or cystic hygroma were retained, and full texts were examined. Papers reporting the diagnostic yields of at least one of such techniques in cases of iNT and/or cystic hygroma were deemed eligible for quantitative analysis. A further quantitative analysis was performed for papers reporting the postnatal outcomes of euploid fetuses from these cohorts. iNT cohorts and cystic hygroma cohorts were analyzed separately. Only singleton pregnancies were retained. Cases with post-mortem diagnosis were excluded. Papers in which cystic hygroma cases were included in NT cohorts were excluded if such cases could not be separated from the rest of the series. Papers in which post-mortem cases and twin pregnancies could not be removed from the count were secondarily excluded. The cases were classified as “apparently isolated”, “associated”, or “unspecified”. The classification was retrieved from the reported association status of the primary anomaly at the time of referral. We classified as “apparently isolated” the cases that did not present additional structural or functional anomalies during the first and second trimester and before genetic assessment, regardless of whether other anomalies were later identified or not. Cases in which additional anomalies were identified during the first or second trimester ultrasound scan and before genetic testing were classified as “associated”. These categories were adopted as they represent the two possible initial presentations for cases undergoing genetic counseling and invasive diagnostic procedures in clinical practice. If information on associated anomalies at the time of initial assessment was not provided or could not be retrieved from the cohorts, the cases were classified as “unspecified”. If specified, the values of NT (in centiles or millimeters) of each cohort were reported. For each category, we annotated the diagnostic yield of standard karyotyping, the incremental diagnostic yield of CMA after negative karyotyping, and the subsequent incremental diagnostic yield of RASopathy testing and exome sequencing. For the purpose of these scores, aneuploidies and Copy Number Variations (CNVs) > 10 Mb in extension identified with molecular techniques performed as first-tier testing were considered karyotype-detectable. Only variants classified as Pathogenic or Likely Pathogenic (represented together by the acronym P/LP) according to the American College of Medical Genetics (ACMG) guidelines [17,18] and relevant to the phenotype were considered as diagnostic. When available, we also gathered Variants of Uncertain Significance (VUS) and incidental findings. In addition, pregnancy and postnatal outcomes were recorded for euploid fetuses, specifying whether CNVs or monogenic syndromes were excluded or not. We annotated the rates of miscarriages, intrauterine deaths, live births, perinatal death, major childhood morbidity and apparently isolated intellectual disability.
We then performed a meta-analysis of the diagnostic rates of cytogenetics and molecular prenatal diagnostics in iNT and cystic hygroma. Cases from eligible papers were divided by initial finding (iNT or Cystic Hygroma), association with other anomalies excluding soft markers (Unspecified, Apparently Isolated, Associated), and by the undertaken tests (Standard Karyotyping, CMA, RASopathies testing, Exome Sequencing). Cases with iNT were divided in ranges as 2.5–2.9 mm, 2.5–3.5 mm, 3.0–3.5 mm, ≥2.5 mm, ≥3.0 mm, ≥3.5 mm. The 2.5 mm, 3.0 mm and 3.5 mm cut-offs were chosen as they represent different accession criteria for invasive testing in different countries either in clinical or research settings. Cases belonging to the same category were pooled from reference papers. We then scored the diagnostic yield of standard karyotyping in each class, then incremental diagnostic yield of CMA in karyotype-negative cases, subsequently the further incremental yield of RASopathy testing in karyotype- and CMA-negative cases, and as a final tier the yield of ES. For the purpose of this review, all chromosomal number anomalies and chromosomal imbalances ≥ 10 Mb were considered karyotype-detectable even if originally identified with molecular techniques. Following the same principle, variants in RASopathy genes identified by exome sequencing were classified as RASopathy-panel-detectable. The yield was calculated for each category and tested as “pooled number of cases with a Pathogenic or Likely Pathogenic (P/LP) variant”/“pooled number of recruited cases”. Standard deviations and 95% confidence intervals were scored with the = STDEV.S and = CONFIDENCE functions on Microsoft Excel (Office 365).
2.2. Fetal Cohort
We collected a cohort of 96 fetuses detected with iNT (≥2.5 mm) or cystic hygroma from Policlinico Umberto I and Fatebenefratelli Isola Tiberina-Gemelli Isola Hospitals. After genetic counseling, each couple was offered genetic testing after invasive procedures (chorionic villus sampling or amniocentesis). After informed consent was collected, each sample underwent karyotyping. In cases with normal fetal karyotype, CMA was offered to the couple. In the same way, cases with negative CMA results were offered the NGS RASopathy panel. The fetuses did not show structural anomalies when they underwent invasive procedure and genetic testing.
We excluded from our cohort fetuses diagnosed with malformations at the time the genetic tests were performed (in order to make the data comparable to each other) and families with recurrent iNT or cystic hygroma (with higher suspicion of a monogenic condition). We included fetuses detected with soft markers, noting them.
For karyotyping, the sample was seeded on culture medium with CHANG for 10–15 days and metaphases obtained after treatment with colchicine were G-banded following standard procedures. At least 16 colonies were analyzed.
Genomic screening for CNVs was performed using the Cytoscan HD (Thermo Fisher Scientific, Waltham, MA, USA) or the 180 K oligonucleotide array (Agilent Technologies, Waldbronn, Germany) microarray platform, following the manufacturer’s instructions and using the ChAS (Thermo Fisher Scientific) or Cytogenomics (Agilent Technologies) analysis software, respectively. Both microarray platforms had 75 Kb effective resolution. Rearrangements were confirmed by real-time quantitative PCR on fetal and parental DNA. In accordance with the guidelines of the American College of Medical Genetics (ACMG), the detected CNVs were classified as pathogenic (P), probably pathogenic (LP), variants of uncertain significance (VUS), probably benign (LB) or benign (B) [18].
For the NGS RASopathy panel, the libraries were prepared using a custom HaloPlexHS panel (Agilent, Santa Clara, CA, USA), providing comprehensive coverage of the coding sequence and flanking intronic regions (+/−10 bp) of the BRAF, CBL, HRAS, KRAS, LZTR1, MAP2K1, MAP2K2, NRAS, PPP1CB, PTPN11, RAF1, RIT1, RRAS, SHOC2, SOS1, and SOS2 genes. The enriched libraries were sequenced using a MiSeq sequencing platform (Illumina, San Diego, CA, USA). Sequencing data were processed and analyzed using a custom bioinformatics software pipeline. Reads were aligned to the GRCh37/hg19 reference genome by BWA (v.0.7.17). BAM files were sorted by SAMtools (v.1.7) and purged from duplicates using Mark Duplicates from the Picard suite (v.2.9.0). Mapped reads were locally realigned and base-quality-score recalibrated using GATK 3.8. Reads with mapping quality scores lower than 20 or with more than one-half nucleotides with quality scores less than 30 were filtered out. The GATK’s Haplotype Caller and ANNOVAR were used to identify and annotate single-nucleotide variants and indels. Bidirectional Sanger sequencing was performed using the ABI BigDye Terminator Sequencing Kit v.3.1 (ThermoFisher Scientific) and an ABI 3130 (ThermoFisher Scientific).
3. Results
3.1. Systematic Review of the Literature
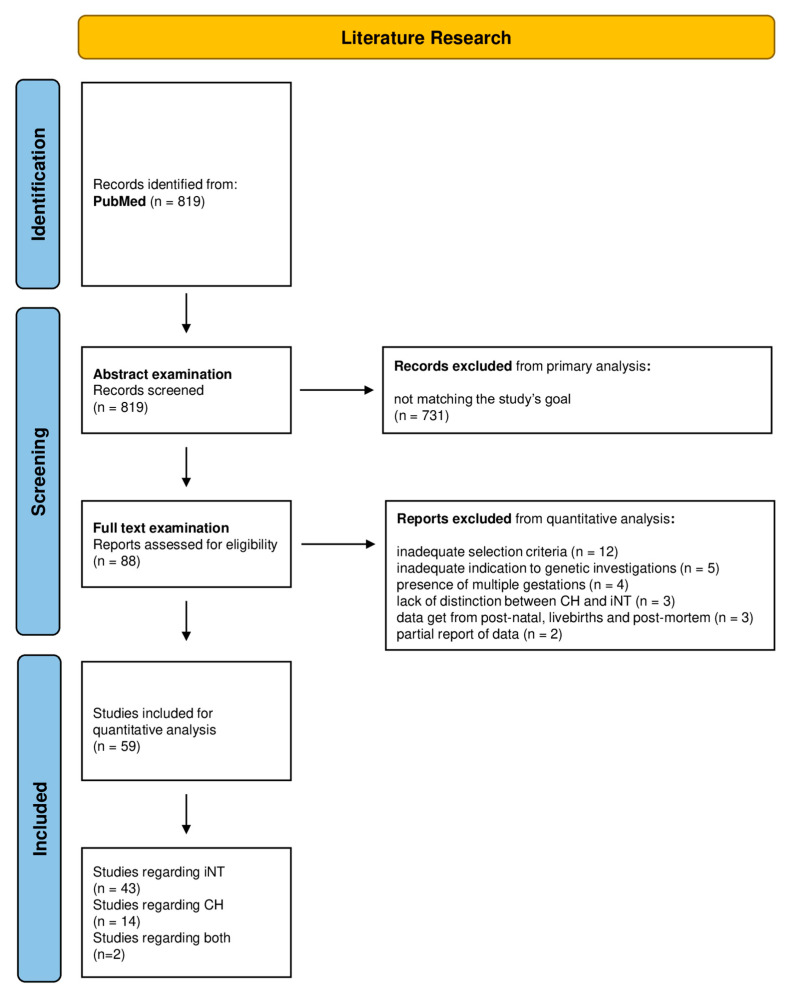
The research results are presented according to the PRISMA guidelines workflow in Figure 1 [16].
Figure 1.
PRISMA flow chart of the systematic review and quantitative analysis.
The research conducted on PubMed provided 819 initial results. Of these, a total of 640 papers were excluded at abstract examination as not relevant to the research topic. In total, 179 papers remained and were later examined. Of these, 32 papers did not have complete available text, 22 were not in English and were secondary excluded. In total, 125 were ultimately retained for full-text examination. Of the remaining papers, 37 did not provide quantitative data, while 88 were examined for quantitative analysis. Twenty-nine papers were secondarily excluded from data analysis: 12 papers for inadequate selection criteria to invasive genetic testing [19,20,21,22,23,24,25,26,27,28,29,30]; in 5 papers, the indication to genetic testing was inadequate for incremental yield [31,32,33,34,35]; in 4 papers, it was not possible to distinguish multiple gestations [36,37,38,39]; 3 papers did not provide appropriate differentiation between hygroma and iNT [2,40,41]; 3 papers included data from postnatal, live births and postmortem examination [42,43,44]; 2 papers provided a partial report of data about fetuses’ malformations [45,46]. A total of 59 papers were adequate for quantitative analysis: 43 papers described the diagnostic application of karyotype, CMA, Rasopathy testing and/or ES in iNT cases (Table 1) [14,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88]; 14 papers described their application in cystic hygroma (Table 2) [11,12,89,90,91,92,93,94,95,96,97,98,99,100], and 2 papers their application in both iNT and cystic hygroma (Table 1 and Table 2) [101,102]. Data concerning postnatal outcomes were retrieved from 17 papers for iNT (Table 3) [51,63,64,65,66,68,73,80,81,82,83,84,85,86,87,88,102,103] and from 10 papers for cystic hygroma (Table 4) [12,33,90,91,93,96,97,98,99,102].
Table 1.
Increased Nuchal Translucency and Genetic Testing-systematic review.
| Nuchal Translucency | ||||||
|---|---|---|---|---|---|---|
| Reference | Dimension (mm) |
Association | Yield (Incremental) | |||
| Karyotype | CMA | RASopathy Panel | ES | |||
| [47] | 2.5–2.9 | unspecified | 8/134 (5.97%) |
3/126 (2.38%) |
. | . |
| 3.0–3.4 | unspecified | 8/146 (5.48%) |
2/138 (1.45%) |
. | . | |
| 3.5–4.4 | unspecified | 23/140 (16.43%) |
7/117 (5.98%) |
. | . | |
| 4.5–5.4 | unspecified | 7/32 (21.88%) |
0/25 (0.00%) |
. | . | |
| 5.5–6.4 | unspecified | 7/13 (53.85%) |
1/6 (16.67%) |
. | . | |
| ≥6.5 | unspecified | 18/34 (52.94%) |
2/16 (12.50%) |
. | . | |
| [48] | 3.0–3.4 | unspecified | 15/110 (13.64%) |
3/60 (5.00%) |
. | . |
| 3.5–4.4 | unspecified | 28/83 (33.74%) |
3/37 (8.11%) |
. | . | |
| 4.5–5.4 | unspecified | 18/40 (45.00%) |
1/40 (2.50%) |
. | . | |
| 5.5–6.4 | unspecified | 12/30 (40.00%) |
. | . | . | |
| 6.5–7.4 | unspecified | 9/21 (42.86%) |
. | . | . | |
| ≥7.5 | unspecified | 21/35 (60.00%) |
. | . | . | |
| ≥4.5 | unspecified | 60/126 (47.62%) |
1/40 (2.50%) |
. | . | |
| [49] | ≥3.5 | apparently isolated | . | . | 2/73 (2.74%) |
2/71 (2.82%) |
| [50] | ≥2.5 | unspecified | 18/192 (9.38%) |
. | . | . |
| 2.5–3.4 | apparently isolated | . | 2/119 (1.68%) |
. | . | |
| 2.5–3.4 | associated | . | 0/15 (0.00%) |
. | . | |
| ≥3.5 | apparently isolated | . | 1/43 (2.33%) |
. | . | |
| ≥3.5 | associated | . | 2/12 (16.67%) |
. | . | |
| [51] | ≥3.5 | apparently isolated | . | 1/39 (2.56%) |
. | . |
| [52] | ≥3.5 | apparently isolated | 134/362 (37.02%) |
1/229 (0.44%) |
. | . |
| [14] | ≥3.5 | unspecified | 123/226 (54.42%) |
2/103 (1.94%) |
3/103 (2.74%) |
. |
| [53] | 95ct–3.4 | apparently isolated | 18/114 (15.79%) |
2/96 (2.01%) |
. | . |
| 3.5–4.4 | apparently isolated | 30/150 (20.00%) |
2/120 (1.67%) |
. | . | |
| 4.5–5.4 | apparently isolated | 16/55 (30.09%) |
2/39 (5.13%) |
. | . | |
| ≥5.5 | apparently isolated | 29/55 (52.73%) |
1/26 (3.85%) |
1/22 (4.55%) |
2/21 (9.52%) |
|
| [54] | 3.0–3.4 | apparently isolated | 20/619 (32.31%) |
9/599 (1.50%) |
. | . |
| [55] | ≥3.5 | unspecified | 57/175 (32.57%) |
3/118 (2.54%) |
. | . |
| [56] | 2.5–3.4 | unspecified | 245/1372 (17.86%) |
. | . | . |
| 3.5–4.4 | unspecified | 182/866 (21.02%) |
. | . | . | |
| 4.5–5.4 | unspecified | 77/282 (27.30%) |
. | . | . | |
| 5.5–6.4 | unspecified | 29/109 (26.61%) |
. | . | . | |
| ≥6.5 | unspecified | 27/91 (29.67%) |
. | . | . | |
| [57] | ≥3.5 | apparently isolated | . | . | 0/111 (0.00%) |
2/111 (1.80%) |
| ≥3.5 | associated | . | . | 7/91 (7.69%) |
17/84 (20.24%) |
|
| [58] | 3.0–3.4 | apparently isolated | 8/170 (4.70%) |
3/162 (1.85%) |
. | . |
| ≥3.5 | apparently isolated | 16/138 (42.11%) |
3/122 (2.46%) |
. | . | |
| [50] | 3.5–4.4 | apparently isolated | 16/76 (21.05%) |
3/60 (5.00%) |
. | . |
| ≥4.5 | apparently isolated | 27/56 (48.21%) |
5/29 (17.24%) |
. | . | |
| [60] | 2.5–3.4 | unspecified | 7/57 (12.28%) |
1/49 (2.94%) |
. | . |
| 3.5–4.4 | unspecified | 10/39 (25.64%) |
1/29 (3.45%) |
. | . | |
| 4.5–5.4 | unspecified | 7/19 (36.84%) |
0/12 (0.00%) |
. | . | |
| ≥5.5 | unspecified | 13/24 (54.17%) |
0/11 (0.00%) |
. | . | |
| [61] | 2.5–2.9 | apparently isolated | 10/86 (11.63%) |
1/76 (1.32%) |
. | . |
| 3.0–3.4 | apparently isolated | 11/73 (15.06%) |
2/62 (3.23%) |
. | . | |
| 3.5–4.4 | apparently isolated | 9/50 (18.00%) |
2/41 (4.88%) |
. | . | |
| 4.5–5.4 | apparently isolated | 10/21 (47.62%) |
0/11 (0.00%) |
. | . | |
| ≥5.5 | apparently isolated | 7/11 (63.64%) |
0/4 (0.00%) |
. | . | |
| [62] | ≥95ct | unspecified | 66/287 (23.00%) |
. | . | . |
| [63] | ≥3.5 | unspecified | 179/242 (73.96%) |
. | . | . |
| [64] | ≥95ct | unspecified | 119/541 (22.00%) |
. | . | . |
| [65] | ≥3.5 | unspecified | 16/71 (22.54%) |
. | . | . |
| [66] | ≥3.0 | unspecified | 6/105 (5.71%) |
. | . | . |
| [67] | ≥2.5 | unspecified | 18/122 (14.75%) |
. | . | . |
| [68] | ≥3 | unspecified | 224/1058 (21.17%) |
. | . | . |
| 3.0–3.4 | unspecified | 65/676 (9.62%) |
. | . | . | |
| 3.5–4.4 | unspecified | 51/208 (24.52%) |
. | . | . | |
| 4.5–5.4 | unspecified | 29/67 (43.28%) |
. | . | . | |
| 5.5–6.4 | unspecified | 26/37 (70.27%) |
. | . | . | |
| ≥6.5 | unspecified | 53/70 (75.71%) |
. | . | . | |
| [69] | 95ct–3.4 | unspecified | 124/894 (13.87%) |
8/770 (1.04%) |
. | . |
| ≥3.5 | unspecified | 436/1007 (43.30%) |
30/571 (5.25%) |
. | . | |
| 3.5–4.9 | unspecified | 138/492 (28.05%) |
16/354 (4.52%) |
. | . | |
| 5.0–6.4 | unspecified | 113/199 (56.78%) |
7/86 (8.14%) |
. | . | |
| 6.5–7.9 | unspecified | 93/155 (60.00%) |
5/62 (8.06%) |
. | . | |
| ≥8.0 | unspecified | 92/162 (56.79%) |
2/70 (2.86%) |
. | . | |
| [101] | ≥3 | unspecified | 10/120 (8.33%) |
. | . | . |
| [70] | 95ct–3.4 | unspecified | 40/263 (15.21%) |
. | . | . |
| 3.5–4.4 | unspecified | 32/169 (18.93%) |
. | . | . | |
| 4.5–5.4 | unspecified | 37/79 (46.84%) |
. | . | . | |
| 5.5–6.4 | unspecified | 33/52 (63.46%) |
. | . | . | |
| ≥6.5 | unspecified | 55/85 (64.71%) |
. | . | . | |
| [71] | ≥3.5 | apparently isolated | . | 5/34 (14.70%) |
. | . |
| ≥3.5 | associated | . | 3/16 (18.75%) |
1/13 (7.69%) |
5/12 (41.67%) |
|
| [102] | ≥3.0 | unspecified | 8/46 (17.39%) |
. | . | . |
| 3.0–3.9 | unspecified | 4/35 (11.43%) |
. | . | . | |
| 4.0–4.9 | unspecified | 2/7 (28.57%) |
. | . | . | |
| 5.0–5.9 | unspecified | 1/3 (33.33%) |
. | . | . | |
| ≥6.0 | unspecified | 1/1 (100.00%) |
. | . | . | |
| [72] | 3.5–4.4 | unspecified | . | 4/343 (1.17%) |
. | . |
| 4.5–5.4 | unspecified | . | 3/124 (2.42%) |
. | . | |
| 5.5–6.4 | unspecified | . | 4/73 (5.48%) |
. | . | |
| ≥6.5 | unspecified | . | 0/59 (0.00%) |
. | . | |
| [73] | ≥99ct | unspecified | 94/221 (42.53%) |
1/106 (0.94%) |
. | . |
| [74] | ≥3.0 | apparently isolated | 21/108 (19.44%) |
9/87 (10.34%) |
. | . |
| 3.0–3.9 | apparently isolated | 12/81 (14.81%) |
6/69 (8.70%) |
. | . | |
| ≥4.0 | apparently isolated | 9/27 (33.33%) |
3/18 (16.67%) |
. | . | |
| [75] | ≥3.0 | unspecified | 172/775 (22.19%) |
4/256 (1.56%) |
. | . |
| [76] | ≥3.5 | apparently isolated | . | 1/172 (0.58%) |
. | . |
| [77] | 95ct–3.4 | unspecified | 507/7109 (7.13%) |
. | . | . |
| 3.5–4.4 | unspecified | 423/2101 (20.13%) |
. | . | . | |
| 4.5–5.4 | unspecified | 321/707 (45.40%) |
. | . | . | |
| 5.5–6.4 | unspecified | 219/437 (50.11%) |
. | . | . | |
| 6.5–7.4 | unspecified | 218/309 (70.55%) |
. | . | . | |
| 7.5–8.4 | unspecified | 148/209 (70.81%) |
. | . | . | |
| 8.5–9.4 | unspecified | 126/168 (75.00%) |
. | . | . | |
| 9.5–10.4 | unspecified | 74/88 (84.09%) |
. | . | . | |
| 10.5–11.4 | unspecified | 45/64 (70.31%) |
. | . | . | |
| ≥11.5 | unspecified | 87/123 (70.73%) |
. | . | . | |
| [78] | ≥3.5 | apparently isolated | . | 3/269 (1.12%) |
. | . |
| ≥3.5 | associated | . | 3/31 (9.68%) |
. | . | |
| [79] | ≥6.5 | unspecified | 60/84 (71.43%) |
. | . | . |
| 5.5–6.4 | unspecified | 21/37 (56.76%) |
. | . | . | |
| [80] | ≥3.5 | unspecified | 164/303 (54.13%) |
. | . | . |
| [81] | ≥3.5 | unspecified | 123/222 (55.41%) |
. | . | . |
| ≥3.5 | apparently isolated | 32/107 (29.91%) |
. | . | . | |
| ≥3.5 | associated | 91/115 (79.13%) |
. | . | . | |
| [82] | ≥95ct | unspecified | 154/393 (39.19%) |
. | . | . |
| 95ct–3.4 | unspecified | 31/170 (18.24%) |
. | . | . | |
| 3.5–4.4 | unspecified | 29/81 (35.80%) |
. | . | . | |
| 4.5–5.4 | unspecified | 23/42 (54.76%) |
. | . | . | |
| 5.5–6.4 | unspecified | 15/23 (65.22%) |
. | . | . | |
| ≥6.5 | unspecified | 56/77 (72.73%) |
. | . | . | |
| [83] | ≥95ct | unspecified | 37/186 (19.89%) |
. | . | . |
| 95ct–3.4 | unspecified | 10/92 (10.87%) |
. | . | . | |
| 3.5–4.4 | unspecified | 6/50 (12.00%) |
. | . | . | |
| 4.5–5.4 | unspecified | 4/12 (33.33%) |
. | . | . | |
| 5.5–6.4 | unspecified | 7/15 (46.67%) |
. | . | . | |
| ≥6.5 | unspecified | 10/17 (58.82%) |
. | . | . | |
| [84] | 2.5–4.4 | unspecified | 29/33 (87.88%) |
. | . | . |
| 4.5–6.4 | unspecified | 8/8 (100.00%) |
. | . | . | |
| ≥6.5 | unspecified | 11/13 (84.62%) |
. | . | . | |
| [85] | ≥99ct | unspecified | 64/248 (25.81%) |
. | . | . |
| [86] | ≥4.0 | unspecified | 71/160 (44.38%) |
. | . | . |
| [87] | ≥6.5 | unspecified | 89/120 (74.17%) |
. | . | . |
| [88] | ≥95ct | unspecified | 44/147 (29.93%) |
. | . | . |
Table 2.
Cystic Hygroma and Genetic Testing-systematic review.
| Cystic Hygroma | |||||
|---|---|---|---|---|---|
| Reference | Association | Yield (Incremental) | |||
| Karyotype | CMA | RASopathy Panel | ES | ||
| [89] | unspecified | 15/28 (53.50%) |
. | . | . |
| [90] | apparently isolated | 25/50 (50.00%) |
. | . | . |
| associated | 13/22 (59.09%) |
. | . | . | |
| [91] | unspecified | 67/132 (50.76%) |
. | . | . |
| [92] | unspecified | 122/185 (65.95%) |
1/40 (2.50%) |
6/15 (40.00%) |
. |
| [93] | apparently isolated | 3/10 (30.00%) |
. | . | . |
| associated | 13/20 (65.00%) |
. | . | . | |
| [94] | unspecified | 55/85 (64.70%) |
. | . | . |
| [95] | unspecified | 20/37 (54.05%) |
. | . | . |
| [11] | unspecified | 18/50 (36.00%) |
. | . | . |
| [101] | unspecified | 13/27 (48.15%) |
. | . | . |
| [102] | unspecified | 13/30 (43.33%) |
. | . | . |
| unspecified | 0/1 (0.00%) |
. | . | . | |
| unspecified | 1/3 (33.33%) |
. | . | . | |
| unspecified | 4/4 (100.00%) |
. | . | . | |
| unspecified | 11/22 (50.00%) |
. | . | . | |
| [96] | apparently isolated | 12/21 (57.14%) |
. | . | . |
| associated | 13/21 (61.90%) |
. | . | . | |
| [97] | apparently isolated | 5/12 (41.67%) |
. | . | . |
| associated | 7/14 (50.00%) |
. | . | . | |
| [12] | unspecified | 400/729 (54.87%) |
. | . | . |
| [98] | unspecified | 128/194 (65.98%) |
. | . | . |
| [99] | unspecified | 28/69 (40.58%) |
. | . | . |
| [100] | unspecified | 45/100 (45.00%) |
. | . | . |
Table 3.
Increased Nuchal Translucency and Prenatal/Postnatal Outcome-systematic review.
| Nuchal Translucency | ||||||||
|---|---|---|---|---|---|---|---|---|
| Reference | Dimension (mm) |
Association | Miscarriages | Intrauterine Deaths | Live Births | Perinatal Death | Intellectual Disability | Major Childhood Morbidity |
| [52] | ≥3.5 | apparently isolated-euploid | 0/20 (0.00%) |
1/20 (5.00%) |
19/20 (95.00%) |
1/19 (5.26%) |
3/19 (15.79%) |
3/19 (15.79%) |
| [63] | ≥3.5 | unspecified-euploid | . | 5/33 (15.15%) |
28/33 (84.85%) |
. | . | . |
| [64] | ≥3.5 | unspecified-euploid | 4/420 (0.95%) |
. | . | . | 10/270 (3.70%) |
10/370 (2.70%) |
| [65] | ≥3.5 | unspecified-euploid | 6/52 (11.54%) |
. | 46/52 (88.46%) |
. | . | 6/46 (13.04%) |
| [66] | ≤3.0 | unspecified | 45/1395 (3.23%) |
19/1395 (1.36%) |
1305/1395 (93.55%) |
. | . | . |
| ≥3.0 | unspecified | 22/105 (20.95%) |
15/105 (14.29%) |
33/105 (31.43%) |
. | . | . | |
| 3.0–4.0 | unspecified | 10/52 (19.23%) |
5/52 (9.62%) |
28/52 (53.85%) |
. | . | . | |
| 4.0–5.0 | unspecified | 10/33 (30.30%) |
7/33 (21.21%) |
5/33 (15.15%) |
. | . | . | |
| 5.0–6.0 | unspecified | 2/15 (13.33%) |
3/15 (20.00%) |
0/15 (0.00%) |
. | . | . | |
| ≥6.0 | unspecified | 0/5 (0.00%) |
0/5 (0.00%) |
0/5 (0.00%) |
. | . | . | |
| [68] | ≥3.0 | associated-euploid | . | . | 741/834 (88.85%) |
. | . | 43/741 (5.80%) |
| 3–3.4 | associated-euploid | . | . | 562/611 (91.98%) |
. | . | 28/562 (4.98%) |
|
| 3.5–4.4 | associated-euploid | . | . | 141/157 (89.81%) |
. | . | 7/141 (4.96%) |
|
| 4.5–5.4 | associated-euploid | . | . | 30/38 (78.95%) |
. | . | 2/30 (6.67%) |
|
| 5.5–6.4 | associated-euploid | . | . | 5/11 (45.45%) |
. | . | 4/5 (80.00%) |
|
| ≥6.5 | associated-euploid | . | . | 3/17 (17.65%) |
. | . | 2/3 (66.76%) |
|
| [102] | ≥3.0 | unspecified | . | 0/46 (0.00%) |
31/46 (67.39%) |
3/31 (9.68%) |
. | 3/31 (9.68%) |
| [73] | ≥99ct | associated-euploid | . | 4/36 (11.11%) |
13/36 (36.11%) |
1/6 (16.67%) |
. | 6/13 (46.15%) |
| ≥99ct | apparently isolated-euploid | 2/70 (2.86%) |
1/70 (1.73%) |
63/70 (90.00%) |
. | . | 3/63 (4.76%) |
|
| [103] | ≥95ct | unspecified-euploid | 23/625 (3.68%) |
. | . | . | . | . |
| [80] | ≥3.5 | unspecified-euploid | 5/139 (3.60%) |
1/139 (0.72%) |
110/139 (79.14%) |
. | . | 7/110 (6.36%) |
| 3.5–4.4 | unspecified-euploid | 2/86 (2.33%) |
1/86 (1.16%) |
77/86 (89.53%) |
. | . | 3/77 (3.90%) |
|
| 4.5–5.4 | unspecified-euploid | 0/28 (0.00%) |
0/28 (0.00%) |
20/28 (71.43%) |
. | . | 2/20 (10.00%) |
|
| 5.5–6.4 | unspecified-euploid | 1/12 (8.33%) |
0/12 (0.00%) |
7/12 (58.33%) |
. | . | 1/7 (14.29%) |
|
| ≥6.5 | unspecified-euploid | 2/13 (15.38%) |
0/13 (0.00%) |
6/13 (46.15%) |
. | . | 1/6 (16.67%) |
|
| [82] | ≥95ct | unspecified | . | 9/239 (3.77%) |
210/239 (87.87%) |
. | . | 10/210 (4.76%) |
| 95ct–3.4 | unspecified | . | 2/139 (1.44%) |
135/139 (97.12%) |
. | . | 2/135 (1.48%) |
|
| 3.5–4.4 | unspecified | . | 2/52 (3.85%) |
46/52 (88.46%) |
. | . | 2/46 (4.35%) |
|
| 4.5–5.4 | unspecified | . | 2/19 (10.53%) |
16/19 (84.21%) |
. | . | 1/16 (6.25%) |
|
| 5.5–6.4 | unspecified | . | 0/8 (0.00%) |
4/8 (50.00%) |
. | . | 0/4 (0.00%) |
|
| ≥6.5 | unspecified | . | 3/21 (14.29%) |
9/21 (42.86%) |
. | . | 5/9 (55.56%) |
|
| [83] | ≥95ct | unspecified | 7/149 (4.70%) |
. | 110/149 (73.83%) |
. | . | . |
| 95ct–3.4 | unspecified | 1/82 (1.22%) |
. | 71/82 (86.59%) |
. | . | . | |
| 3.5–4.4 | unspecified | 2/43 (4.65%) |
. | 35/43 (81.40%) |
. | . | . | |
| 4.5–5.4 | unspecified | 0/7 (0.00%) |
. | 2/7 (28.57%) |
. | . | . | |
| 5.5–6.4 | unspecified | 3/8 (37.50%) |
. | 1/8 (12.50%) |
. | . | . | |
| ≥6.5 | unspecified | 1/19 (5.26%) |
. | 1/19 (5.26%) |
. | . | . | |
| [84] | 2.5–4.4 | unspecified | 4/33 (12.12%) |
0/33 (0.00%) |
20/33 (60.61%) |
. | . | . |
| 4.5–6.4 | unspecified | 1/8 (12.5%) |
0/8 (0.00%) |
1/8 (12.50%) |
. | . | . | |
| ≥6.5 | unspecified | 2/13 (15.38%) |
4/13 (30.77%) |
1/13 (7.69%) |
. | . | . | |
| [85] | ≥99ct | unspecified-euploid | 1/179 (0.56%) |
5/179 (2.79%) |
162/179 (90.50%) |
. | . | 18/162 (11.11%) |
| [86] | ≥4.0 | unspecified | . | 2/89 (2.25%) |
68/89 (76.40%) |
. | 4/64 (6.25%) |
4/68 (5.88%) |
| [87] | ≥6.5 | unspecified-euploid | 6/27 (22.22%) |
. | 8/27 (29.63%) |
. | . | . |
| [88] | ≥95ct | unspecified-euploid | . | 10/103 (9.71%) |
87/103 (84.47%) |
. | . | 10/87 (11.49%) |
Table 4.
Cystic Hygroma and Prenatal/Postnatal Outcome-systematic review.
| Cystic Hygroma | |||||||
|---|---|---|---|---|---|---|---|
| Reference | Association | Miscarriages | Intrauterine Deaths | Live Births | Perinatal Death | Intellectual Disability | Major Childhood Morbidity |
| [90] | unspecified-euploid | 2/34 (5.88%) |
2/34 (5.88%) |
18/34 (52.94%) |
0/18 (0.00%) |
. | 2/18 (11.11%) |
| [91] | unspecified-euploid | . | 5/36 (13.89%) |
31/36 (86.11%) |
1/31 (3.23%) |
1/31 (3.23%) |
8/31 (25.81%) |
| [93] | associated-euploid | . | 1/3 (33.33%) |
2/3 (66.66%) |
. | . | . |
| apparently isolated-euploid | . | . | 6/6 (100.00%) |
. | . | . | |
| [102] | unspecified | . | 5/30 (16.67%) |
7/30 (23.33%) |
4/7 (57.14%) |
. | 2/7 (28.57%) |
| [96] | unspecified-euploid | . | . | 7/17 (41.18%) |
. | . | . |
| [97] | apparently isolated-euploid | 1/5 (20.00%) |
. | 3/5 (60.00%) |
. | . | 1/3 (33.33%) |
| associated-euploid | 3/7 (42.86%) |
1/7 (14.29%) |
1/7 (14.29%) |
. | . | 1/1 (100.00%) |
|
| [12] | unspecified | 106/295 (14.29%) |
. | 180/295 (24.26%) |
9/180 (5.00%) |
. | . |
| [98] | unspecified-euploid | 7/66 (10.61%) |
. | 27/66 (40.91%) |
. | . | . |
| [99] | unspecified | 5/41 (12.20%) |
. | 12/41 (29.27%) |
. | . | . |
| [100] | unspecified | . | 54/85 (63.53%) |
31/85 (36.47%) |
. | . | 6/31 (19.35%) |
The table illustrates data retrieved from the reference papers on the diagnostic yield of karyotype and the progressively incremental diagnostic yield of CMA in karyotype-negative cases, of RASopathy testing in karyotype- and CMA-negative cases, and ultimately of ES with iNT. For each reference, the cases were divided in categories based on the dimension of the NT and on whether the association with major fetal anomalies was reported or not. For the purpose of this review, chromosomal imbalances ≥ 10 Mb in size were considered karyotype-detectable, even if originally identified by CMA performed as a first-tier test. Following the same principle, variants identified in RASopathy genes by ES were classified under the “RASopathy panel” column.
The table illustrates data retrieved from the reference papers on the diagnostic yield of karyotype and the progressively incremental diagnostic yield of CMA (chromosomal microarray analysis) in karyotype-negative cases, of RASopathy testing in karyotype and CMA-negative cases, and ultimately of ES (exome sequencing) with cystic hygroma. For each reference, the cases were divided in categories based on whether the association with major fetal anomalies was reported or not. As stated in the previous section, the classification is based on the presence or putative absence of further anomalies at US examinations of the first and second trimester and before invasive genetic testing. The term “apparently isolated” identifies cases in which no other anomalies were documented at the time of genetic testing, so it does not indicate cases in which the anomaly appears as isolated throughout the whole pregnancy. For the purpose of this review, chromosomal imbalances ≥ 10 Mb in size were considered karyotype-detectable, even if originally identified by CMA performed as first-tier test. Following the same principle, variants identified in RASopathy genes by ES were classified under the “RASopathy panel” column.
The table illustrates data retrieved from the reference papers on the pregnancy and post-natal outcomes of fetuses with iNT. For each reference, the cases were divided in categories based on NT dimensions, on whether aneuploidies were excluded and whether the association with major fetal anomalies was reported or not.
The table illustrates data retrieved from the reference papers on the pregnancy and post-natal outcomes of fetuses with cystic hygroma. For each reference, the cases were divided into categories based on NT dimensions, on whether aneuploidies were excluded and whether the association with major fetal anomalies was reported or not.
3.2. Meta-Analysis
The results on the diagnostic yield of karyotype, and progressive incremental yield of CMA, RASopathy testing and exome sequencing (ES) iNT (2.5–2.9 mm, 2.5–3.4 mm, 3.0–3.4 mm, ≥2.5 mm, ≥3.0 mm, ≥3.5 mm) and cystic hygroma, either apparently isolated, associated with major fetal anomalies, or unspecified, are presented in Table 5.
Table 5.
Increased Nuchal Translucency and Cystic Hygroma and Genetic testing-meta-analysis.
| Nuchal Translucency | ||||||
|---|---|---|---|---|---|---|
| Dimensions | Association | Yield (Incremental) | REF[Karyo][CMA][RAS][ES] | |||
| Karyotype | CMA | RASopathy Panel | ES | |||
| 2.5–2.9 | unspecified | 8/134 5.97% |
11/896 1.23% (1.17–1.29) |
. | . | [61]; [61]; [.]; [.] |
| apparently isolated | 10/86 11.73% |
1/76 (1.32%) |
. | . | [47,69]; [47]; [.]; [.] | |
| associated | . | . | . | . | [.]; [.]; [.]; [.] | |
| 2.5–3.4 | unspecified | 964/9957 9.68% (9.60–9.76) |
9/819 1.10% (1.05–1.15) |
. | . | [56,60,69,77,82,83]; [60,69]; [.]; [.] |
| apparently isolated | 39/273 14.29% (14.07–14.51) |
4/215 1.86% (1.82–1.90) |
. | . | [53,61]; [50,53]; [.]; [.] | |
| associated | . | 0/15 0.00% |
. | . | [.]; [50]; [.]; [.] | |
| 3.0–3.4 | unspecified | 88/932 9.44% (9.18–9.70) |
5/198 2.53% (2.18–2.88) |
. | . | [47,48,68]; [47,48]; [.]; [.] |
| apparently isolated | 39/862 4.52% (4.09–4.95) |
14/823 1.70% (1.64–1.76) |
. | . | [54,58,61]; [54,58,61]; [.]; [.] | |
| associated | . | . | . | . | [.]; [.]; [.]; [.] | |
| ≥2.5 | unspecified | 4393/19177 22.91% (22.60–23.22) |
61/2098 2.91% (2.88–2.94) |
. | . | [47,50,56,60,62,64,67,69,70,77,82,83,84,88]; [47,50,60,69]; [.]; [.] |
| apparently isolated | 140/615 22.76% (22.46–23.06) |
15/637 2.35% (2.32–2.38) |
. | . | [53,61]; [50,53,61]; [.]; [.] | |
| associated | 2/134 1.49% |
2/27 (7.41%) |
. | . | [50]; [50]; [.]; [.] | |
| ≥3.0 | unspecified | 616/2788 22.09% (21.74–22.44) |
23/695 3.31% (3.18–3.44) |
. | . | [47,48,66,68,75,101,102]; [47,48,75]; [.]; [.] |
| apparently isolated | 82/571 14.36% (13.68–15.04) |
19/489 3.89% (3.50–4.28) |
. | . | [58,61,74]; [58,61,74]; [.]; [.] | |
| associated | . | . | . | . | [.]; [.]; [.]; [.] | |
| ≥3.5 | unspecified | 3680/9204 39.98% (39.64–40.32) |
59/1581 3.73% (3.64–3.82) |
3/103 (2.91%) |
. | [47,48,55,56,60,63,65,68,69,70,77,80,81,82,83]; [47,48,55,60,69,72]; [14]; [.] |
| apparently isolated | 449/1307 34.35% (33.66–35.04) |
56/1367 4.1% (3.50–4.70) |
3/108 1.44% (1.15–1.73) |
5/205 2.44% (2.26–2.62) |
[14,52,53,58,59,61,81]; [14,50,51,52,53,58,59,61,71,76,78]; [49,53,57]; [49,53,57] | |
| associated | 91/115 79.13% |
8/59 13.56% (12.32–14.80) |
8/104 (7.69%) |
22/96 22.92% (19.85–25.99) |
[81]; [50,71,78]; [57,71]; [57,71] | |
| Cystic Hygroma | ||||||
| Association | Yield (incremental) | REF[Karyo][CMA][RAS][ES] | ||||
| Karyotype | CMA | RASopathy panel | ES | |||
| unspecified | 927/1666 55.64% (55.18–56.10) |
1/40 2.50% |
6/15 (40.00%) |
. | [11,12,89,91,92,94,95,99,100,101,102]; [92]; [92]; [.] | |
| apparently isolated |
45/93 48.39% (46.02–50.76) |
. | . | . | [90,93,96,97]; [.]; [.]; [.] | |
| associated | 46/80 57.50% (55.16–59.84) |
. | . | . | [90,93,96,97]; [.]; [.]; [.] | |
CMA: chromosomal microarray analysis; RAS: RASopathy panel; ES: exome sequencing.
3.3. Present Fetal Cohort
3.3.1. Cystic Hygroma
Forty-seven out of the 96 fetuses were detected with isolated cystic hygroma (27 males and 20 females) (Table 6). Soft markers and other sonographic findings were noted: 2 ductus venosus agenesis, 1 hypoplastic nasal bone, 1 hyperechoic bowel, 2 absent nasal bone, 1 echoic cardiac focus and 1 fetus with single umbilical artery, ductus venosus agenesis and absent nasal bone.
Table 6.
Present fetal cohort–cystic hygroma.
| ID | Sex | Growth | Soft Marker (s) | Karyotype | CMA | RASopathy Panel |
|---|---|---|---|---|---|---|
| 1 | F | - | - | 46,XX | N | N |
| 2 | F | - | - | 46,XX | N | LZTR1 c.2173_2174insA; p.(Arg725Glnfs*57) mat |
| 3 | F | - | inverted DV flow | mos 47,XX,+21[27]/46,XX [3] | . | . |
| 4 | F | - | - | 47,XX,+21 | . | . |
| 5 | F | - | - | 46,XX | N | SOS1 c.755T>C; p.(Ile252Thr) VUS pat |
| 6 | F | - | - | 46,XX | N | N |
| 7 | F | biometry <5°c | absent nasal bone | 46,XX | N | RAF1: c.770C>T; p.(Ser257Leu) dn |
| 8 | F | - | - | 45,X | . | . |
| 9 | F | - | - | 46,XX | N | N |
| 10 | F | - | - | 46,XX | N | N |
| 11 | F | - | - | 46,XX | N | N |
| 12 | F | - | - | 46,XX | N | N |
| 13 | F | - | - | 46,XX | N | PTPN11 c.179_181delTGG |
| 14 | F | - | absent nasal bone | 47,XX,+21 | . | . |
| 15 | F | - | - | 47,XX,+21,t(2;8)(q33;q22) | . | . |
| 16 | F | - | - | 47,XY,+21 | . | . |
| 17 | F | - | echoic cardiac focus | 47,XX,+21 | . | . |
| 18 | F | - | - | 45,X | . | . |
| 19 | F | - | - | 47,XX,+21 | . | . |
| 20 | F | - | - | 45,X | . | . |
| 21 | M | - | DVA | 46,XY | N | SHOC2 c.807_808delinsTT; p.(Gln269_His270delinsHisTyr) |
| 22 | M | - | hypoplastic nasal bone | 46,XY | N | SOS2 c.1709C>G; p.(Pro570Arg) VUSmat |
| 23 | M | - | - | 47,XY,+21 | n.p. | n.p. |
| 24 | M | - | - | 46,XY | N | N |
| 25 | M | - | - | 46,XY | N | N |
| 26 | M | - | - | 46,XY | 22q11.21(18894835_21464119)x1 | |
| 27 | M | - | - | 46,XY | N | N |
| 28 | M | - | DVA | 46,XY der(4)t(4;7)(4p16.3;7p22.3) | 4p16.3(71552_3872380)x1 7p22.3p22.1(42976_6870943)x3 |
|
| 29 | M | - | hyperechoic bowel | 46,XY | N | N |
| 30 | M | - | - | 46,XY | N | N |
| 31 | M | - | - | 46,XY | N | SOS1 c.643T>C; p.(Tyr215His) mat |
| 32 | M | - | - | 46,XY | N | N |
| 33 | M | - | - | 46,XY | N | N |
| 34 | M | - | - | 47,XXY | . | . |
| 35 | M | - | - | 47,XY,+21 | . | . |
| 36 | M | - | - | 46,XY,del(4)(p15.2)dn | . | . |
| 37 | M | - | - | 47,XY,+21 | . | . |
| 38 | M | - | - | 47,XY,+13 | . | . |
| 39 | M | - | - | 47,XY,+21 | . | . |
| 40 | M | - | - | 47,XY,+21 | . | . |
| 41 | M | - | - | 46,XY | N | N |
| 42 | M | - | - | 46,XY | N | N |
| 43 | M | - | - | 46,XY | N | N |
| 44 | M | - | - | 46,XY | 17q23.2(60033664_60251568)x3 mat | N |
| 45 | M | - | SUA, DVA, absent nasal bone | 47,XY,+21 | . | . |
| 46 | M | - | - | 46,XY | N | N |
| 47 | M | - | absent nasal bone | 47,XY,+21 | . | . |
CMA: chromosomal microarray analysis; N: negative; F: female; M: male; DV: ductus venosus; DVA: ductus venosus agenesis; SUA: single umbilical artery.
Twenty-one of the fetuses (10 males, 11 females) were diagnosed with karyotype anomalies (44.68%): 12 constitutional trisomy of chromosome 21, 1 trisomy 21 mosaicism, 1 trisomy of the chromosome 13, 3 monosomy of X chromosome, 1 Klinefelter syndrome, 1 trisomy 21 with balanced autosomal translocation, 2 4p- (Wolf–Hirschhorn Syndrome).
The 26 cases showing normal standard karyotype underwent chromosomal microarray analysis and in 1 male fetus the 22q11.2 deletion syndrome was detected. In another case, a pathogenic copy number variation was identified.
In the remaining 25 fetuses, the multigene RASopathy panel was performed: in 3 cases (1 male, 2 females), a pathogenic variant was identified: c.179_181delTGG in PTPN11 (NM_002834.5, MIM*176876), c.770C>T, p.(Ser257Leu) in RAF1 (NM_002880.4, MIM*164760) and c.807_808delinsTT, p.(Gln269_His270delinsHisTyr) in SHOC2 (NM_007373.4, MIM*602775) [104]. Four inherited VUSs were noted.
In this cohort, standard karyotype showed a detection rate of 44.68%. CMA presented an incremental diagnostic yield (over karyotype) of 3.8% and the incremental detection rate of the NGS RASopathy panel over CMA was 12%.
3.3.2. Increased Nuchal Translucency
In 19 out of the 49 fetuses (Table 7), standard karyotype identified chromosomal anomalies (12/19 females, 63.12%), with a diagnostic yield of 38.78%, detailed below. No fetal pathogenic or likely pathogenic rearrangement was detected by CMA (0/30, 0%). The multigene RASopathy panel showed a detection rate of 0% (0–1/30).
Table 7.
Present fetal cohort–increased nuchal translucency.
| ID | Sex | NT (mm) | Soft Marker (s) | Karyotype | CMA | RASopathy Panel |
|---|---|---|---|---|---|---|
| NT 2.5–2.9 mm | ||||||
| 1 | F | 2.6 | - | 47,XX,+21 | . | . |
| 2 | M | 2.6 | - | 46,XY | N | SOS1 c.643T>C (het, mat) |
| 3 | F | 2.7 | - | 47,XX,+21 | . | . |
| 4 | F | 2.8 | - | 47,XX,+21 | . | . |
| 5 | F | 2.8 | - | 46,XX | N | N |
| 6 | M | 2.9 | - | 47,XYY | . | . |
| NT 3.0–3.4 mm | ||||||
| 7 | F | 3.0 | - | 47,XX,+21 | . | . |
| 9 | F | 3.0 | - | 46,XX | N | N |
| 10 | F | 3.0 | - | 47,XX,+21 | . | . |
| 11 | M | 3.0 | - | 46,XY | 10p13p12.33(16786323_17726950)x3 mat | . |
| 12 | M | 3.0 | 47,XXY | 10q21.3(68735256_68938523)x3 pat 22q12.2(29911595_30141397)x3 mat |
. | |
| 13 | M | 3.0 | hypoplastic nasal bone | 46,XY | N | N |
| 14 | M | 3.1 | - | 47,XY,+21 | . | . |
| 15 | F | 3.1 | - | 47,XX,+21 | . | . |
| 16 | F | 3.2 | - | mos 47,XX,+18[58]/46,XX [20] | . | . |
| 17 | M | 3.2 | - | 46,XY | N | N |
| 18 | M | 3.2 | - | N | N | N |
| 19 | M | 3.3 | - | 46,XY | N | N |
| 20 | M | 3.3 | - | 46,XY | N | N |
| 21 | F | 3.3 | - | 47,XX,+21 | . | . |
| 22 | M | 3.3 | - | 46,XY | N | N |
| 23 | M | 3.3 | Choroid plexus cyst, hyperechoic cardiac focus | 46,XY | N | N |
| 24 | M | 3.3 | - | 47,XY,+21 | . | . |
| 25 | F | 3.4 | - | 46,XX | 3p26.3(270649_920375)x3mat | N |
| NT 3.5–3.9 mm | ||||||
| 26 | F | 3.5 | - | 46,XX | N | N |
| 27 | M | 3.5 | - | 47,XY,+21 | . | . |
| 28 | M | 3.6 | - | 46,XY | N | N |
| 29 | F | 3.6 | - | 46,XX | N | N |
| 30 | M | 3.6 | - | N | N | N |
| 31 | M | 3.8 | - | N | N | N |
| 32 | F | 3.9 | - | 47,XX,+21 | . | . |
| 33 | F | 3.9 | - | 46,XX | N | LZTR1 c.842del (het, n/a) |
| NT ≥ 4.0 mm | ||||||
| 34 | M | 4.0 | - | 46,XY | N | N |
| 35 | M | 4.0 | - | 46,XY | N | N |
| 36 | M | 4.0 | - | 46,XY,inv(19)(p13.3q13.2)mat | N | N |
| 37 | M | 4–0 | - | 46,XY | N | N |
| 38 | F | 4.1 | - | 46,XX | N | N |
| 39 | M | 4.2 | - | 47,XY,+21 | . | . |
| 40 | M | 4.2 | - | 46,XY | N | N |
| 41 | F | 4.3 | - | N | N | N |
| 42 | F | 4.4 | - | 47,XX,+21 | . | . |
| 43 | F | 4.7 | - | 46,XX,del(3)(p25) | . | . |
| 44 | F | 4.7 | - | 46,XX | N | BRAF c.26G>C (het, mat) |
| 45 | M | 5.3 | - | 46,XY | N | N |
| 46 | M | 5.7 | - | 47,XY,+18 | . | . |
| 47 | F | 5.8 | - | 47,XX,+18 | . | . |
| 48 | F | 7.0 | - | 46,XX | N | N |
| 49 | M | 7.7 | - | 46,XY | 15q11.2(22784523_23179948)x1 | N |
NT: nuchal translucency; CMA: chromosomal microarray analysis; N: negative; F: female; M: male.
2.5–2.9 mm Nuchal Translucency
In 6 out of the 49 fetuses, the NT ranged from 2.5 to 3 mm (two males, four females). Four fetuses were diagnosed with karyotype anomalies (three females, one male): 3 constitutional trisomy 21 and 1 Jacobs syndrome. CMA detected no copy number variations. The NGS RASopathy panel identifies in one fetus the paternally inherited c.643T>C, p.(Tyr215His) variant in SOS1 (NM_005633.4, MIM*182530), classified as VUS.
In this cohort, standard karyotype showed a detection rate of 66.67%. CMA presented an incremental diagnostic yield (over karyotype) of 0% and the incremental detection rate of the NGS RASopathy panel over CMA was 0%.
3.0–3.4 mm Nuchal Translucency
In 18 out of the 49 fetuses, the NT measured 3.0–3.4 mm (11 males, 7 females). In one case, the fetus presented with hyperechoic cardiac focus and choroid plexus cysts; in a second case, hypoplastic nasal bone was noted. Eight fetuses were diagnosed with karyotype anomalies (three males, five females): 6 constitutional trisomy 21, 1 trisomy 18 mosaicism, 1 Klinefelter syndrome. CMA detected copy number variations in three cases, all classified as VUS. The NGS RASopathy panel did not reveal VUS/likely pathogenic/pathogenic variants.
In this cohort, standard karyotype showed a detection rate of 33.33%. CMA presented an incremental diagnostic yield (over karyotype) of 0% and the incremental detection rate of the NGS RASopathy panel over CMA was 0%.
3.5–3.9 mm Nuchal Translucency
In 8 out of the 49 fetuses, the NT measured 3.5–3.9 mm (four males, four females). Two fetuses, a male and a female, presented Trisomy 21. No other karyotype anomaly was identified. CMA did not detect rearrangements. The NGS RASopathy panel did not identify pathogenic variants related to the phenotype. The c.842del, p.(Pro281fs*) variant in LZTR1 (NM_006767.4, MIM*600574) was identified in heterozygosity in a female fetus. The variant was not identified in the mother, but the father was not available for analysis.
In this cohort, standard karyotype showed a detection rate of 25.0%. CMA presented an incremental diagnostic yield (over karyotype) of 0% and the incremental detection rate of the NGS RASopathy panel over CMA was 0%.
≥4.0 mm Nuchal Translucency
In 16 out of the 49 fetuses, the NT measured equal to or more than 4.0 mm (nine males, seven females). Five fetuses (two males, three females) were diagnosed with karyotype anomalies: 2 constitutional trisomy of chromosome 21, 2 trisomy 18 and one with a large deletion of chromosome 3p. CMA detected one copy number variation, classified as VUS. The multigene RASopathy panel identified the maternally inherited heterozygous variant c.26G>C, p.(Gly9Ala) in BRAF (NM_004333.6, MIM*164757), classified as VUS.
In this cohort, standard karyotype showed a detection rate of 31.25%. CMA presented an incremental diagnostic yield (over karyotype) of 0% and the incremental detection rate of the NGS RASopathy panel over CMA was 0%.
4. Discussion
4.1. Physiopathology of Nuchal Fluid Collections
In physiological pregnancies, the NT increases in size with gestational age until about 13 gestational weeks, disappearing after the 14th week [105,106]. Similarly, iNT tends to reabsorb after this period [107], but in some cases it can persist or progress into other fetal fluid collections [108,109,110]. The regression of iNT before the 14th week is not an uncommon event and it has been observed in around 18% of iNT cases [88]. Despite the regression, those fetuses maintain higher risks than the normal population [88]. The presence of higher risk for genetic conditions and adverse outcome despite an apparently reassuring NT regression should be discussed in genetic counseling.
Some authors agree that it can be not easy to differentiate non-septated cystic hygroma and iNT. The first-trimester iNT may also include the initial findings of cystic hygroma, especially when a notch is seen in this area [40]. However, in both sonographic entities genetic testing plays a fundamental role in offering the couple an informed choice in decision-making of the ongoing pregnancy, to formulate the recurrence risks and to define the most appropriate management [111].
Fetal nuchal fluid collection can present as apparently isolated findings, be associated with concurrent malformations or occur as secondary lymphatic drainage anomalies due to cardiovascular malformations or skeletal dysplasia. In fetuses with aneuploidies, an alteration in the drainage of physiologically accumulated nuchal fluid can determine iNT or cystic hygroma. Underlying causes in aneuploid fetuses include cardiac malformations, primary abnormalities of lymphatic vessels and an altered composition of subcutaneous collagen [112,113]. In trisomic fetuses with iNT, increased levels of atrial and brain natriuretic peptide mRNA were identified, suggesting the presence of heart strain [113]. Interestingly, some studies reported that aneuploid fetuses with abnormal blood flow in the ductus venosus showed an umbilical cord diameter above the 95th centile for CRL, if compared to euploid fetuses at 11–14 weeks of gestation [114]. It can be secondary to abnormal ductus venosus flow during presystole, leading to venous congestion, transudation into the Wharton’s jelly and umbilical cord dilatation [115].
Some authors speculate that a slight iNT in male fetuses can represent a physiological gender-related characteristic in early development of the cardiovascular system and in the permeability of fetal skin. Testosterone production may reduce the epidermal thickness, lamellar body production and stratum corneum integrity [116] and the possibility of a favorable outcome was considered 1.8-fold higher in males than in females [117].
The definition of cystic hygroma is often reported ambiguously in the analyzed articles, which is why it is not possible to exclude selection bias of the fetuses included in the cohorts. Several authors defined that incomplete obstruction of lymphatic drainage, which can resolve spontaneously, may determine the formation of a non-septated cystic hygroma, while the complete obstruction may account for a septated cystic hygroma [118,119]. In some studies, the cohort was selected based on the presence of the septation, trying to make a further distinction within the category of cystic hygroma.
It is accepted that cystic hygroma is associated with high rates of aneuploidies, major structural anomalies, fetal demise, and poor outcome. Additionally, some studies reported that as the width increases, the odds of anomalies increase [12]. Interestingly, a paper demonstrated lower rates of chromosomal anomalies, and a higher number of good birth outcomes, in fetuses in which the cystic hygroma was detected with CRL measuring <45 mm, if compared to those detected later, without different prevalence of major malformations and intrauterine death among fetuses with normal karyotype [98]. However, these are data to be considered cautiously, as it would be necessary to know the causes for which the pregnant women performed this ultrasound before the scheduled time. Several authors observed that some cases with early disappearance of the cystic hygroma progressed to have normal outcomes [98,100]. It has been discussed that usually there is not a clear distinction between the “cystic hygroma” and the “iNT” phenotypes and that NT thickness may be the best independent predictor of aneuploidies [2].
iNT is detected in around 4.4% of euploid fetuses [10], and NT ≥ 3.0 mm, both in the unselected cohort and in high-risk populations, increased the risk of malformation almost 10-fold and the risk of miscarriage about 5-fold, in proportion to the NT width and without an association between iNT and perinatal death [103]. In the literature, the NT cut-off levels are extremely varied; therefore, the criteria for enrollment of cases are heterogeneous and not all papers could be included in the meta-analysis. Some studies recommended using MoM or delta-NT [120]; however, in most cases, for practicality, the absolute value of the measurement continued to be used. In fetuses with karyotype anomalies, further measurements of NT confirm the same alteration or reveal an increase in this thickness, while in fetuses with normal karyotyping the size generally decreases [62].
Many disorders are associated with iNT, suggesting that there may not be a single underlying mechanism for this condition: cardiovascular anomalies (also showing reduced diastolic function [121]), venous congestion in the head and neck from constriction of the fetal body, altered composition of the extracellular matrix, failure of lymphatic drainage, fetal anemia or hypoproteinemia, and infections (Parvovirus B19) [122]. Many proteins of the extracellular matrix are encoded on chromosomes 21, 18, or 13. In the nuchal skin of fetuses diagnosed with trisomy of chromosome 21, there is a substantial increase in hyaluronic acid and collagen type IV, which present hydrophilic properties leading to fluid accumulation [123].
iNT has been associated with fetal structural anomalies, in particular with cardiac malformations, and it has even been suggested that NT measurement can be used as a screening tool for fetal cardiac defects [124]. It is to be noted that, given the high prenatal prevalence of cardiac anomalies, some cases of apparently isolated iNT or cystic hygroma might actually underlie a cardiovascular defect [124]. For this reason, early echocardiographic assessment should be suggested in each fetus presenting with iNT or cystic hygroma. Obviously, some fetal anomalies may manifest at later stages of development and not in the gestational age in which NT screening is performed, which is why a careful sonographic morphological evaluation and an echocardiographic follow-up (even if no cardiovascular malformation has been detected) are required.
An unbiased evaluation of diagnostic yield of chromosomal, genomic or monogenic conditions in twin pregnancies is difficult to obtain, due to the increased risk of structural anomalies that could not be seen at the gestational age in which the invasive procedure is performed. iNT can also represent an early manifestation of the complications of placental vascular anastomoses in monochorionic twins [125,126], which would explain the higher incidence of chromosomal anomalies in dichorionic twins with one fetus detected with NT above the 95th centile than monochorionic twins [127].
Some authors showed that fetuses with isolated iNT and normal CMA present an increased risk for developing placental-related disorders in the following stages of the pregnancy [128].
4.2. Meta-Analysis on Genetic Testing and Present Fetal Cohort Considerations
The results of the meta-analysis highlight that, although increased nuchal translucency and cystic hygroma are both attributable to nuchal fluid collection, they appear to confer different risks for chromosomal, genomic and monogenic disorders.
In order to provide reliable information for prenatal counseling, we will discuss and compare meta-analysis data concerning cohorts with apparently isolated or explicitly associated iNT or cystic hygroma, as the unspecified/not reported cohorts might not reflect the actual presentation of cases in clinical practice.
In the reported cohorts, the indication to genetic testing is based on prenatal ultrasonographic findings. Ultrasound examination has inherent limitations, and a small but not negligible amount of anomalies might not be identified by initial ultrasound scans. For this reason, apparently isolated cases might have structural anomalies that become apparent only later in pregnancy or at birth. However, information from apparently isolated cases retrieved from the literature can be confidently proposed when counseling for fetuses with apparently isolated iNT or cystic hygroma in clinical practice, as both instances share the same limitations.
Standard karyotype revealed chromosomal anomalies in 48.39% (46.02–50.7) of fetuses with apparently isolated cystic hygroma included in the meta-analysis and in 44.68% of the fetuses included in the multicentric cohort. In the cohort we report, the incremental yield of CMA over karyotyping was 3.8%, while the NGS RASopathy panel presented a detection rate over CMA of 12%.
Regarding iNT, the parameters used are heterogeneous in the various cohorts; therefore, it was possible to carry out the meta-analysis by grouping the cases in the most frequently reported ranges, although these were partly overlapping with each other. NT ≥ 3.5 mm was the most common cut-off, followed by NT ≥ 3.0 mm. We scored the diagnostic yield of karyotyping, CMA, RASopathy testing and ES in the gathered cases by adopting these three cut-offs (all fetuses with NT ≥ 2.5, all fetuses with NT ≥ 3.0, all fetuses with NT ≥ 3.5), and also analyzed groups of fetuses with NT values in intervals within these cut-offs (2.5–2.9, 2.5–3.4, 3.0–3.4).
Fetuses with apparently isolated iNT that measured 2.5–2.9 mm showed chromosomal anomalies in 11.73% of cases involved in the meta-analysis and in 66.67% of fetuses enrolled in the cohort. To date, there are no univocal guidelines regarding the genetic investigations to be performed in the case of mild iNT; therefore, it can be complicated to define the selection criterion. It is possible that a proportion of pregnant women with iNT between 2.5 and 2.9 mm have performed screening testing (e.g., Non-invasive prenatal screening, combined screening of first trimester) resulting in high risk and are not included in these series as they refer to invasive investigation with a different indication. The data emerging from the meta-analysis regarding fetuses with this range of nuchal translucency can also be considered less realistic than the other ranges as they involve a smaller number of cases. CMA revealed a pathogenic rearrangement in 1.32% of cases (from only one article) and in no case of the present cohort.
Among the fetuses in which the apparently isolated iNT measured 3.0–3.4 mm included in the meta-analysis, 4.52% showed chromosomal anomalies, while in the retrospective cohort the detection rate of karyotype was 33.33% and the different diagnostic yield can easily be explained by the different sample size. CMA presented an incremental diagnostic yield over karyotype of 1.7% in the meta-analysis and of 0% in the present cohort.
It was also possible to group the data present in the literature on the basis of the minimum cut-off indicated for the iNT. In this way, overlaps have been created between the three groups.
In the meta-analysis, fetuses with apparently isolated NT ≥ 2.5 mm showed karyotype anomalies in 22.76% of cases and CMA presented an incremental detection rate of 2.35%. Fetuses with isolated NT ≥ 3 mm presented chromosomal anomalies in 14.36% of cases and CMA had an incremental detection rate of 3.89%. When the isolated NT measured at least 3.5 mm, the diagnostic yield of karyotyping was 34.35%, the incremental CMA detection rate was 4.1%, the incremental diagnostic rate of the RASopathy panel over CMA was 1.44%, while the incremental diagnostic rate of Clinical Exome Sequencing over the multigene RASopathy panel was 2.44%. Information for genetic counseling in apparently isolated iNT and cystic hygroma cases is provided in Table 8.
Table 8.
Apparently isolated iNT and cystic hygroma: summary of the meta-analysis findings.
| Apparently Isolated iNT | Apparently Isolated Cystic Hygroma | |||
|---|---|---|---|---|
| 2.5–2.9 mm | 3–3.4 mm | ≥3.5 mm | ||
| Karyotype | 11.73% | 4.52% | 34.35% | 48.39% |
| CMA | 1.32% | 1.7% | 4.1% | |
| RASopathy panel | 1.44% | |||
| ES | 2.44% | |||
A limit of this methodology can be found in the different number of cases undergoing different tests due to a sequential and non-parallel approach that progressively reduces the sample size.
When considering associated cases, information is lacking for NT values between 2.5 and 3.5 mm. For cases with NT ≥ 3.5 mm and associated anomalies, chromosomal disorders are extremely probable, with a rate of aneuploidies as high as 79.13% [20], and and incremental yield of 13.56% for submicroscopic chromosomal imbalances identified by CMA. The frequency of RASopathies, 7.69%, also appears to be higher than in apparently isolated cases, being only 1.44%. The incremental yield of ES in cases with NT ≥ 3.5 mm and associated anomalies (22.92%) is also noteworthy, especially if compared to the 2.44% yield of ES in fetuses with apparently isolated iNT in the same ranges.
From the analysis of these data, it appears that the rate of chromosomal anomalies in fetuses with NT below 3.5 mm, which is instead a widely spread cut-off to define iNT, is far from negligible, and that even values between 2.5 and 2.9 present such risk. Interestingly, from these data it emerges that CMA presents a considerable diagnostic rate in the group of fetuses with NT ≥ 3.5 mm, while for lower values it has a marginal incremental yield over karyotype. Similarly, in the same group ES appears to show promising results and could be considered after a negative CMA result. For associated cases, the extremely high rates of chromosomal and genetic anomalies should be provided to consultants, and ES should be recommended in karyotype- and CMA- negative cases.
4.3. Long-Term Follow-Up after Pregnancies with Nuchal Fluid Collections
The evaluation of the pregnancy and, especially, post-natal outcomes of fetuses detected with iNT and cystic hygroma is heterogeneous in the literature, in methods and results, with a lack of proper cohorts. This is due to the marked difficulties of a years-long follow-up, and to the differences in the timing of the presentation of the possibly associated conditions. Terminations of pregnancy also negatively affect this field of research, as the recourse to termination varies greatly depending not only on ultrasound findings and genetic data, but also on cultural and law differences across countries. Data retrieved from the literature concerning pregnancy and post-natal outcomes are provided in Table 3 and Table 4. These evaluations are limited to euploid fetuses. In most cases, submicroscopic CNVs and RASopathies had not been excluded. Terminations of pregnancy were excluded from the count. It is possible that voluntary termination of pregnancy with severe presentations, even in euploid fetuses, skewed the results of pregnancy and post-natal outcomes towards an overall better prognosis [70,129]. There is an increased risk in iNT for miscarriage and intrauterine death (the conditions are often merged in the sources), the live-birth rate being 88.85% in the largest cohort (853 fetuses) of iNT ≥ 3.0 mm [68]. Some authors suggested the rate of miscarriage/intrauterine deaths to be proportional to the NT width, increasing from 1.6% when NT measured between the 95th and 99th centiles to about 20% for NT ≥ 6.5 mm [70,122]. The rate of fetal loss appears to be more consistent in cystic hygroma, with 7 out of 11 papers reporting a live birth rate below 50%. Perinatal deaths are only occasionally assessed and reported. One of the main concerns in ongoing pregnancies for prospective parents in these cases is the risk for neurodevelopmental anomalies. The evaluations have been performed in heterogeneous ways, usually without comparison with a control group, yielding different results [85]. A large study has documented a statistically significant association with intellectual disability and autism spectrum disorder with OR of 6.16 (95% CI, 1.51–25.0) and 2.48 (95% CI,1.02–5.99), respectively, in euploid fetuses with NT > 99th centile but not in lower increases [31]. Another interesting paper documented that the developmental quotient was significantly lower than controls, but still remained in the normal range at two years of age [130]. The association has not been extensively examined for cystic hygroma.
Both iNT and cystic hygroma can represent an early manifestation of different diseases, so each case must be scrupulously evaluated in order to define the indication to search for mutations in a panel of genes or perform whole exome sequencing. Major childhood morbidities, including malformations and genetic syndromes, are often reported in the literature, with a rate of 4.58% (28/611) and a 2.7% (10/370) in the largest cohorts [64,68]. It is to be noted that RASopathies can account for some of them [90]. Clearly, one of the limitations is that an extensive examination of all genes allows the identification of variants of unknown significance and can create difficulties in interpreting the results obtained [131,132]. Moreover, the fear of undetectable anomalies can induce anxiety even after negative genetic testing [79].
4.4. Future Perspectives
Interesting insights emerged from this analysis that could serve as a starting point for new studies. In particular, the path of genetic investigations which is usually performed sequentially was analyzed, defining the incremental diagnostic yield of the individual tests in fetuses with cystic hygroma and iNT.
Data concerning fetuses with NT measuring between 2.5 and 2.9 mm are less representative than those of the other ranges, due to a smaller sample size. It is possible that in this group there is a bias due to pregnant women who have performed screening tests resulting in high risk and have opted for voluntary termination of pregnancy before referring to genetic counseling or it is possible that they were not included in this study for a different indication to the invasive procedure.
The risk for chromosomal anomalies appears to be relevant even from NT values of 2.5 mm. The diagnostic yield of the CMA increases and becomes consistent above 3.5 mm.
Recently, promising studies have been performed in the literature regarding the diagnostic yield of the exome sequencing in the event of an ultrasound finding of fetal fluid collections, especially in the case of fetal hydrops, which can appear in the first trimester with an ultrasound picture attributable to iNT or cystic hygroma [133,134,135,136,137,138]. In up to 29% of cases, pathogenic variants had been identified, including RASopathies, inborn errors of metabolism, musculoskeletal disorders, lymphatic, cardiovascular, neurodevelopmental and hematologic diseases [133]. These insights may lead to consider the possibility of performing CMA and the RASopathy panel in parallel, in order to reach an earlier diagnosis. However, data concerning RASopathy panels or ES sequencing large cohorts of fetuses with isolated iNT, especially below 3.5 mm, are still lacking.
Future research should better explore the diagnostic rate of CMA at all iNT levels, and focus on the evaluation of monogenic conditions in fetuses presenting NT measurement between 2.5 and 2.9 mm or 2.5 and 3.4 mm. In this range, the rearrangements identified with CMA appear to be not frequent and the main chromosomal abnormalities responsible can also be identified by NIPT. Even if NIPS is recommended as the first choice for fetal diagnosis in pregnant women with smaller iNT who do not opt for invasive procedures [13], conventional cfDNA testing can miss the diagnosis in a high percentage of fetuses with iNT [38].
If the rate for submicroscopic chromosomal anomalies and for monogenic disorders is confirmed to be low, in that case, in countries where NIPS is offered as a public service it could be an approach to be offered to pregnant women with only slightly iNT, reducing unnecessary invasive procedures and parental anxiety.
Further studies may be decisive in determining the advantages of performing exome sequencing for iNT or cystic hygroma, also considering that these findings may represent characteristics of a fetal phenotype not yet evident in such an early gestational age.
Moreover, based on our experience, frequently in those cases in which the nuchal fluid collection remains an isolated finding and the genetic investigations are negative, an adequate follow-up is not performed after birth, which instead can prove to be important for the evaluation of the individual case.
5. Conclusions
Cystic hygroma and iNT, albeit belonging to the same phenotypic spectrum attributable to the fetal nuchal fluid collections, appear to present with a different prevalence of chromosomal, genomic and monogenic conditions both in the literature and in the present cohort. Karyotype abnormalities are the most frequent finding in these fetuses, even when the cut-off for iNT is a value below the common threshold of 3.5 mm or even below 3 mm, while rearrangements identified with CMA are more consistent when the NT is above 3.5 mm. The RASopathy panel detected pathogenic variants especially in fetuses diagnosed with cystic hygroma, with a diagnostic yield that is sometimes higher than in CMA.
Further studies are desirable, in order to define the most suitable diagnostic algorithm, determining the most appropriate path based on the ultrasound and dimensional characteristics, and also considering the possibility of performing exome sequencing.
Author Contributions
Conceptualization, G.M., D.G. and N.K.H.; methodology, G.M., D.G. and N.K.H.; formal analysis, D.G. and N.K.H.; investigation, L.B., G.P., A.D.L. and A.P.; resources, L.B., A.G., G.P., A.D.L. and A.P.; data curation, G.M.; writing—original draft preparation, G.M., D.G. and N.K.H.; writing—review and editing, L.B., G.P., A.D.L. and A.P.; supervision, A.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the articles cited in the References section.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Apkon M. Pathophysiology of hydrops fetalis. Semin. Perinatol. 1995;19:437–446. doi: 10.1016/S0146-0005(05)80051-5. [DOI] [PubMed] [Google Scholar]
- 2.Molina F.S., Avgidou K., Kagan K.O., Poggi S., Nicolaides K.H. Cystic hygromas, nuchal edema, and nuchal translucency at 11–14 weeks of gestation. Obstet. Gynecol. 2006;107:678–683. doi: 10.1097/01.AOG.0000201979.23031.32. [DOI] [PubMed] [Google Scholar]
- 3.Evans M.I., Krantz D.A., Hallahan T.W., Sherwin J. Impact of nuchal translucency credentialing by the FMF, the NTQR or both on screening distributions and performance. Ultrasound Obstet. Gynecol. 2012;39:181–184. doi: 10.1002/uog.9023. [DOI] [PubMed] [Google Scholar]
- 4.Sahota D.S., Chen M., Leung T.Y., Chan L.W., Fung T.Y., Ting Y.H., Lau T.K. Assessment of sonographer nuchal translucency measurement performance central tendency and dispersion. J. Matern. Fetal Neonatal Med. 2011;24:812–816. doi: 10.3109/14767058.2010.531310. [DOI] [PubMed] [Google Scholar]
- 5.Sun Q., Xu J., Hu S.-Q., Chen M., Ma R.-M., Lau T.K., Zhang L., Xiao X., Qian Y., Guo Z. Distribution and normal reference range of fetal nuchal translucency thickness in Kunming pregnant women in the first trimester. Zhonghua Fu Chan Ke Za Zhi. 2012;47:514–517. [PubMed] [Google Scholar]
- 6.Mack L.M., Lee W., Mastrobattista J.M., Belfort M.A., Van den Veyver I.B., Shamshirsaz A.A., Ruano R., Sanz Cortes M., Espinoza A., Thiam Diouf A., et al. Are First Trimester Nuchal Septations Independent Risk Factors for Chromosomal Anomalies? J. Ultrasound Med. 2017;36:155–161. doi: 10.7863/ultra.16.01066. [DOI] [PubMed] [Google Scholar]
- 7.Alamillo C.M., Fiddler M., Pergament E. Increased nuchal translucency in the presence of normal chromosomes: What’s next? Curr. Opin. Obstet. Gynecol. 2012;24:102–108. doi: 10.1097/GCO.0b013e3283505b25. [DOI] [PubMed] [Google Scholar]
- 8.Wright D., Kagan K.O., Molina F.S., Gazzoni A., Nicolaides K.H. A mixture model of nuchal translucency thickness in screening for chromosomal defects. Ultrasound Obstet. Gynecol. 2008;31:376–383. doi: 10.1002/uog.5299. [DOI] [PubMed] [Google Scholar]
- 9.Pandya P.P., Kondylios A., Hilbert L., Snijders R., Nicolaides K. Chromosomal defects and outcome in 1015 fetuses with increased nuchal translucency. Ultrasound Obstet. Gynecol. 1995;5:15–19. doi: 10.1046/j.1469-0705.1995.05010015.x. [DOI] [PubMed] [Google Scholar]
- 10.Snijders R., Noble P., Sebire N., Souka A., Nicolaides K. UK multicentre project on assessment of risk of trisomy 21 by maternal age and fetal nuchal-translucency thickness at 10–14 weeks of gestation. Lancet. 1998;352:343–346. doi: 10.1016/S0140-6736(97)11280-6. [DOI] [PubMed] [Google Scholar]
- 11.Gedikbasi A., Oztarhan K., Aslan G., Demirali O., Akyol A., Sargin M.A., Ceylan Y. Multidisciplinary approach in cystic hygroma: Prenatal diagnosis, outcome and postnatal follow-up. Pediatr. Int. 2009;51:670–677. doi: 10.1111/j.1442-200X.2009.02846.x. [DOI] [PubMed] [Google Scholar]
- 12.Scholl J., Durfee S.M., Russell M.A., Heard A.J., Iyer C., Alammari R., Coletta J., Craigo S.D., Fuchs K.M., D’Alton M., et al. First-trimester cystic hygroma: Relationship of nuchal translucency thickness and outcomes. Obstet. Gynecol. 2012;120:551–559. doi: 10.1097/AOG.0b013e318264f829. [DOI] [PubMed] [Google Scholar]
- 13.Xie X., Zhou H., Zhao Q., Lu Y., Meng Y. Application of expanded noninvasive prenatal test in prenatal diagnosis of fetuses with increased nuchal translucency. J. Matern. Fetal. Neonatal. Med. 2021;35:6213–6218. doi: 10.1080/14767058.2021.1909564. [DOI] [PubMed] [Google Scholar]
- 14.Sinajon P., Chitayat D., Roifman M., Wasim S., Carmona S., Ryan G., Noor A., Kolomietz E., Chong K. Microarray and RASopathy-disorder testing in fetuses with increased nuchal translucency. Ultrasound Obstet. Gynecol. 2020;55:383–390. doi: 10.1002/uog.20352. [DOI] [PubMed] [Google Scholar]
- 15.Scott A., Di Giosaffatte N., Pinna V., Daniele P., Corno S., D’Ambrosio V., Andreucci E., Marozza A., Sirchia F., Tortora G., et al. When to test fetuses for RASopathies? Proposition from a systematic analysis of 352 multicenter cases and a postnatal cohort. Genet. Med. 2021;23:1116–1124. doi: 10.1038/s41436-020-01093-7. [DOI] [PubMed] [Google Scholar]
- 16.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021;88:105906. doi: 10.1016/j.ijsu.2021.105906. [DOI] [PubMed] [Google Scholar]
- 17.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riggs E.R., Andersen E.F., Cherry A.M., Kantarci S., Kearney H., Patel A., Raca G., Ritter D.I., South S.T., Thorland E.C., et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet. Med. 2020;22:245–257. doi: 10.1038/s41436-019-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen O., Smith E., Van Opstal D., Polak M., Knapen M.F.C.M., Diderich K.E.M., Bilardo C.M., Arends L.R., Vogel I., Srebniak M.I. Nuchal translucency of 3.0–3.4 mm an indication for NIPT or microarray? Cohort analysis and literature review. Acta Obstet. Gynecol. Scand. 2020;99:765–774. doi: 10.1111/aogs.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Li R., Fu F., Zhang Y., Li D., Liao C. Submicroscopic chromosomal abnormalities in fetuses with increased nuchal translucency and normal karyotype. J. Matern. Fetal Neonatal Med. 2017;30:194–198. doi: 10.3109/14767058.2016.1168394. [DOI] [PubMed] [Google Scholar]
- 21.Lichtenbelt K.D., Diemel B.D.M., Koster M.P.H., Manten G.T.R., Siljee J., Schuring-Blom G.H., Page-Christiaens G.C.M.L. Detection of fetal chromosomal anomalies: Does nuchal translucency measurement have added value in the era of non-invasive prenatal testing? Prenat. Diagn. 2015;35:663–668. doi: 10.1002/pd.4589. [DOI] [PubMed] [Google Scholar]
- 22.Li L., Fu F., Li R., Liu Z., Liao C. Prenatal diagnosis and pregnancy outcome analysis of thickened nuchal fold in the second trimester. Medicine. 2018;97:e13334. doi: 10.1097/MD.0000000000013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grande M., Solernou R., Ferrer L., Borobio V., Jimenez J.M., Bennasar M., Soler A., Borrell A. Is nuchal translucency a useful aneuploidy marker in fetuses with crown-rump length of 28–44 mm? Ultrasound Obstet. Gynecol. 2014;43:520–524. doi: 10.1002/uog.13203. [DOI] [PubMed] [Google Scholar]
- 24.Arigita M., Grande M., Mula R., Borobio V., Sanchez A., Soler A., Borrell A. Nuchal translucency thickness in the prediction of unbalanced translocations. Prenat. Diagn. 2014;34:982–985. doi: 10.1002/pd.4409. [DOI] [PubMed] [Google Scholar]
- 25.Özcan H Ç., Uğur M.G., Balat Ö., Sucu S., Bayramoğlu Tepe N., Öztürk E., Kömürcü Karuserci Ö., Kazaz T.G. Analysis of cystic hygroma diagnosed in the prenatal period: 5-years’ experience at a tertiary hospital in Southeastern Turkey. J. Matern. Fetal Neonatal Med. 2019;32:1800–1805. doi: 10.1080/14767058.2017.1418315. [DOI] [PubMed] [Google Scholar]
- 26.Cheng P.-J., Chang S.-D., Shaw S.-W., Soong Y.-K. Nuchal translucency thickness in fetuses with chromosomal translocation at 11–12 weeks of gestation. Obstet. Gynecol. 2005;105:1058–1062. doi: 10.1097/01.AOG.0000158862.84467.d7. [DOI] [PubMed] [Google Scholar]
- 27.Chitty L.S., O Kagan K., Molina F.S., Waters J.J., Nicolaides K.H. Fetal nuchal translucency scan and early prenatal diagnosis of chromosomal abnormalities by rapid aneuploidy screening: Observational study. BMJ. 2006;332:452–455. doi: 10.1136/bmj.38730.655197.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christiansen M., Ekelund C.K., Petersen O.B., Hyett J., Eastwood N., Ball S., Tabor A., Vogel I. Nuchal translucency distributions for different chromosomal anomalies in a large unselected population cohort. Prenat. Diagn. 2016;36:49–55. doi: 10.1002/pd.4711. [DOI] [PubMed] [Google Scholar]
- 29.Kievskaya J., Shilova N., Kanivets I., Kudryavtseva E., Pyankov D., Korostelev S. SNP-Based Chromosomal Microarray Analysis for Detecting DNA Copy Number Variations in Fetuses with a Thickened Nuchal Fold. Sovrem. Tekhnologii V Meditsine. 2021;13:72–76. doi: 10.17691/stm2021.13.6.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Has R., Kalelioglu I., Ermis H., Ibrahimoglu L., Yuksel A., Yildirim A., Basaran S. Screening for fetal chromosomal abnormalities with nuchal translucency measurement in the first trimester. Fetal Diagn. Ther. 2006;21:355–359. doi: 10.1159/000092465. [DOI] [PubMed] [Google Scholar]
- 31.Hellmuth S.G., Pedersen L.H., Miltoft C.B., Petersen O., Kjaergaard S., Ekelund C.K., Tabor A. Increased nuchal translucency thickness and risk of neurodevelopmental disorders. Ultrasound Obstet. Gynecol. 2017;49:592–598. doi: 10.1002/uog.15961. [DOI] [PubMed] [Google Scholar]
- 32.Schou K.V., Kirchhoff M., Nygaard U., Jørgensen C., Sundberg K. Increased nuchal translucency with normal karyotype: A follow-up study of 100 cases supplemented with CGH and MLPA analyses. Ultrasound Obstet. Gynecol. 2009;34:618–622. doi: 10.1002/uog.7468. [DOI] [PubMed] [Google Scholar]
- 33.Noia G., Maltese P.E., Zampino G., D’Errico M., Cammalleri V., Convertini P., Marceddu G., Mueller M., Guerri G., Bertelli M. Cystic Hygroma: A Preliminary Genetic Study and a Short Review from the Literature. Lymphat. Res. Biol. 2019;17:30–39. doi: 10.1089/lrb.2017.0084. [DOI] [PubMed] [Google Scholar]
- 34.Ali M.M., Chasen S.T., Norton M.E. Testing for Noonan syndrome after increased nuchal translucency. Prenat. Diagn. 2017;37:750–753. doi: 10.1002/pd.5076. [DOI] [PubMed] [Google Scholar]
- 35.Hui L., Pynaker C., Bonacquisto L., Lindquist A., Poulton A., Kluckow E., Hutchinson B., Norris F., Pertile M.D., Gugasyan L., et al. Reexamining the optimal nuchal translucency cutoff for diagnostic testing in the cell-free DNA and microarray era: Results from the Victorian Perinatal Record Linkage study. Am. J. Obstet. Gynecol. 2021;225:527.e1–527.e12. doi: 10.1016/j.ajog.2021.03.050. [DOI] [PubMed] [Google Scholar]
- 36.Axt-Fliedner R., Hartge D., Chiriac A., Krapp M., Berg C., Geipel A., Germer U., Gembruch U. Long-term outcome for children born after a first-trimester measurement of increased nuchal translucency with a normal karyotype: A retrospective analysis. Ultraschall Medizin. 2009;30:558–563. doi: 10.1055/s-2008-1027948. [DOI] [PubMed] [Google Scholar]
- 37.Goncé A., Borrell A., Meler E., Arigita M., Martínez J.M., Botet F., Sánchez A., Gratacós E. Prevalence and perinatal outcome of dichorionic and monochorionic twins with nuchal translucency above the 99(th) percentile and normal karyotype. Ultrasound Obstet. Gynecol. 2010;35:14–18. doi: 10.1002/uog.7498. [DOI] [PubMed] [Google Scholar]
- 38.Miranda J., Paz YMiño F., Borobio V., Badenas C., Rodriguez-Revenga L., Pauta M., Borrell A. Should cell-free DNA testing be used in pregnancy with increased fetal nuchal translucency? Ultrasound Obstet. Gynecol. 2020;55:645–651. doi: 10.1002/uog.20397. [DOI] [PubMed] [Google Scholar]
- 39.Şahin Uysal N., Gülümser Ç., Çelik Z.Y., Yanık F.B. Increased nuchal translucency and pregnancy outcomes: Experience of Başkent University Ankara Hospital. Turk. J. Obstet. Gynecol. 2019;16:100–106. doi: 10.4274/tjod.galenos.2019.51482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida S., Miura K., Yamasaki K., Miura S., Shimada T., Tanigawa T., Yoshida A., Nakayama D., Masuzaki H. Does increased nuchal translucency indicate a fetal abnormality? A retrospective study to clarify the clinical significance of nuchal translucency in Japan. J. Hum. Genet. 2008;53:688. doi: 10.1007/s10038-008-0299-6. [DOI] [PubMed] [Google Scholar]
- 41.Cicatiello R., Pignataro P., Izzo A., Mollo N., Pezone L., Maruotti G.M., Sarno L., Sglavo G., Conti A., Genesio R., et al. Chromosomal Microarray Analysis versus Karyotyping in Fetuses with Increased Nuchal Translucency. Med. Sci. 2019;7:40. doi: 10.3390/medsci7030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zalel Y., Zemet R., Kivilevitch Z. The added value of detailed early anomaly scan in fetuses with increased nuchal translucency. Prenat. Diagn. 2017;37:235–243. doi: 10.1002/pd.4997. [DOI] [PubMed] [Google Scholar]
- 43.BBehera S., Bawa M., Kanojia R.P., Saha P.K., Singh T., Samujh R. Outcome of antenatally diagnosed cystic hygroma—Lessons learnt. Int. J. Pediatr. Otorhinolaryngol. 2020;138:110227. doi: 10.1016/j.ijporl.2020.110227. [DOI] [PubMed] [Google Scholar]
- 44.Huang J., Poon L.C., Akolekar R., Choy K.W., Leung T.Y., Nicolaides K.H. Is high fetal nuchal translucency associated with submicroscopic chromosomal abnormalities on array CGH? Ultrasound Obstet. Gynecol. 2014;43:620–624. doi: 10.1002/uog.13384. [DOI] [PubMed] [Google Scholar]
- 45.Baer R.J., Norton M.E., Shaw G.M., Flessel M.C., Goldman S., Currier R.J., Jelliffe-Pawlowski L.L. Risk of selected structural abnormalities in infants after increased nuchal translucency measurement. Am. J. Obstet. Gynecol. 2014;211:675.e1–675.e19. doi: 10.1016/j.ajog.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Westin M., Saltvedt S., Bergman G., Almström H., Grunewald C., Valentin L. Is measurement of nuchal translucency thickness a useful screening tool for heart defects? A study of 16,383 fetuses. Ultrasound Obstet. Gynecol. 2006;27:632–639. doi: 10.1002/uog.2792. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z., Hu T., Wang J., Li Q., Wang H., Liu S. Prenatal Diagnostic Value of Chromosomal Microarray in Fetuses with Nuchal Translucency Greater than 2.5 mm. BioMed Res. Int. 2019;2019:6504159. doi: 10.1155/2019/6504159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X.-R., Gao L., Wu Y., Wang Y.-L. Application of chromosomal microarray in fetuses with increased nuchal translucency. J. Matern. -Fetal Neonatal Med. 2020;33:1749–1754. doi: 10.1080/14767058.2019.1569622. [DOI] [PubMed] [Google Scholar]
- 49.Yang X., Huang L.Y., Pan M., Xu L.L., Zhen L., Han J., Li D.Z. Exome sequencing improves genetic diagnosis of fetal increased nuchal translucency. Prenat. Diagn. 2020;40:1426–1431. doi: 10.1002/pd.5789. [DOI] [PubMed] [Google Scholar]
- 50.Su L., Huang H., An G., Cai M., Wu X., Li Y., Xie X., Lin Y., Wang M., Xu L. Clinical application of chromosomal microarray analysis in fetuses with increased nuchal translucency and normal karyotype. Mol. Genet. Genom. Med. 2019;7:e811. doi: 10.1002/mgg3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuurman K.E., van der Mespel-Brouwer M.H., Engels M.A.J., Elting M.W., Bhola S.L., Meijers-Heijboer H. Isolated Increased Nuchal Translucency in First Trimester Ultrasound Scan: Diagnostic Yield of Prenatal Microarray and Outcome of Pregnancy. Front. Med. 2021;8:737936. doi: 10.3389/fmed.2021.737936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srebniak M.I., de Wit M.C., Diderich K.E., Govaerts L.C., Joosten M., Knapen M.F., Bos M.J., Looye-Bruinsma G.A., Koningen M., Go A.T., et al. Enlarged NT (≥3.5 mm) in the first trimester—Not all chromosome aberrations can be detected by NIPT. Mol. Cytogenet. 2016;9:69. doi: 10.1186/s13039-016-0279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue S., Yan H., Chen J., Li N., Wang J., Liu Y., Zhang H., Li S., Zhang W., Chen D., et al. Genetic Examination for Fetuses with Increased Fetal Nuchal Translucency by Genomic Technology. Cytogenet. Genome Res. 2020;160:57–62. doi: 10.1159/000506095. [DOI] [PubMed] [Google Scholar]
- 54.Sagi-Dain L., Singer A., Ben Shachar S., Ben Yehoshua S.J., Feingold-Zadok M., Greenbaum L., Maya I. Risk of Clinically Significant Chromosomal Microarray Analysis Findings in Fetuses With Nuchal Translucency From 3.0 mm Through 3.4 mm. Obstet. Gynecol. 2021;137:126–131. doi: 10.1097/AOG.0000000000004195. [DOI] [PubMed] [Google Scholar]
- 55.Pan M., Han J., Zhen L., Yang X., Li R., Liao C., Li D.-Z. Prenatal diagnosis of fetuses with increased nuchal translucency using an approach based on quantitative fluorescent polymerase chain reaction and genomic microarray. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016;197:164–167. doi: 10.1016/j.ejogrb.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 56.Long N.H., Cuong T.D., Anh N.T. Relation Between Increased Fetal Nuchal Translucency Thickness and Chromosomal Defects in Northern Vietnam. Cureus. 2021;13:e18446. doi: 10.7759/cureus.18446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mellis R., Eberhardt R., Hamilton S., McMullan D., Kilby M.D., Maher E., Hurles M., Giordano J., Aggarwal V., Goldstein D., et al. Fetal exome sequencing for isolated increased nuchal translucency: Should we be doing it? BJOG Int. J. Obstet. Gynaecol. 2022;129:52–61. doi: 10.1111/1471-0528.16869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maya I., Yacobson S., Kahana S., Yeshaya J., Tenne T., Agmon-Fishman I., Cohen-Vig L., Shohat M., Basel-Vanagaite L., Sharony R. Cut-off value of nuchal translucency as indication for chromosomal microarray analysis. Ultrasound Obstet. Gynecol. 2017;50:332–335. doi: 10.1002/uog.17421. [DOI] [PubMed] [Google Scholar]
- 59.Lund I.C.B., Christensen R., Petersen O., Vogel I., Vestergaard E.M. Chromosomal microarray in fetuses with increased nuchal translucency. Ultrasound Obstet. Gynecol. 2015;45:95–100. doi: 10.1002/uog.14726. [DOI] [PubMed] [Google Scholar]
- 60.Lan L., Wu H., She L., Zhang B., He Y., Luo D., Wang H., Zheng Z. Analysis of copy number variation by sequencing in fetuses with nuchal translucency thickening. J. Clin. Lab. Anal. 2020;34:e23347. doi: 10.1002/jcla.23347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang C., Tang J., Tong K., Huang D., Tu H., Zhu J. Chromosomal microarray analysis versus noninvasive prenatal testing in fetuses with increased nuchal translucency. J. Hum. Genet. 2022;67:533–539. doi: 10.1038/s10038-022-01041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zoppi M.A., Ibba R.M., Floris M., Manca F., Axiana C., Monni G. Changes in nuchal translucency thickness in normal and abnormal karyotype fetuses. BJOG Int. J. Obstet. Gynaecol. 2003;110:584–588. doi: 10.1046/j.1471-0528.2003.02180.x. [DOI] [PubMed] [Google Scholar]
- 63.Holzer I., Husslein P.W., Bettelheim D., Scheidl J., Kiss H., Farr A. Value of increased nuchal translucency in the era of noninvasive prenatal testing with cell-free DNA. Int. J. Gynaecol. Obstet. 2019;145:319–323. doi: 10.1002/ijgo.12808. [DOI] [PubMed] [Google Scholar]
- 64.Iuculano A., Pagani G., Stagnati V., Floris M., Ibba R.M., Monni G. Pregnancy outcome and long-term follow-up of fetuses with isolated increased NT: A retrospective cohort study. J. Perinat. Med. 2016;44:237–242. doi: 10.1515/jpm-2015-0157. [DOI] [PubMed] [Google Scholar]
- 65.Socolov D., Socolov R., Gorduza V.E., Butureanu T., Stanculescu R., Carauleanu A., Pavaleanu I. Increased nuchal translucency in fetuses with a normal karyotype-diagnosis and management: An observational study. Medicine. 2017;96:e7521. doi: 10.1097/MD.0000000000007521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abotorabi S., Moeini N., Moghbelinejad S. High Frequency of Fetal Loss in Fetuses With Normal Karyotype and Nuchal Translucency ≥ 3 Among the Iranian Pregnant Women. J. Fam. Reprod. Health. 2020;14:81–87. doi: 10.18502/jfrh.v14i2.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alexioy E., Trakakis E., Kassanos D., Farmakidis G., Kondylios A., Laggas D., Salamalekis E., Florentin L., Kanavakis E., Basios G., et al. Predictive value of increased nuchal translucency as a screening test for the detection of fetal chromosomal abnormalities. J. Matern. -Fetal Neonatal Med. 2009;22:857–862. doi: 10.1080/14767050902994572. [DOI] [PubMed] [Google Scholar]
- 68.Ayras O., Tikkanen M., Eronen M., Paavonen J., Stefanovic V. Increased nuchal translucency and pregnancy outcome: A retrospective study of 1063 consecutive singleton pregnancies in a single referral institution. Prenat. Diagn. 2013;33:856–862. doi: 10.1002/pd.4143. [DOI] [PubMed] [Google Scholar]
- 69.Bardi F., Bosschieter P., Verheij J., Go A., Haak M., Bekker M., Sikkel E., Coumans A., Pajkrt E., Bilardo C. Is there still a role for nuchal translucency measurement in the changing paradigm of first trimester screening? Prenat. Diagn. 2020;40:197–205. doi: 10.1002/pd.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bilardo C.M., Müller M.A., Pajkrt E., Clur S.A., van Zalen M.M., Bijlsma E.K. Increased nuchal translucency thickness and normal karyotype: Time for parental reassurance. Ultrasound Obstet. Gynecol. 2007;30:11–18. doi: 10.1002/uog.4044. [DOI] [PubMed] [Google Scholar]
- 71.Choy K.W., Wang H., Shi M., Chen J., Yang Z., Zhang R., Yan H., Wang Y., Chen S., Chau M.H.K., et al. Prenatal Diagnosis of Fetuses With Increased Nuchal Translucency by Genome Sequencing Analysis. Front. Genet. 2019;10:761. doi: 10.3389/fgene.2019.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Egloff M., Hervé B., Quibel T., Jaillard S., Le Bouar G., Uguen K., Saliou A.-H., Valduga M., Perdriolle E., Coutton C., et al. Diagnostic yield of chromosomal microarray analysis in fetuses with isolated increased nuchal translucency: A French multicenter study. Ultrasound Obstet. Gynecol. 2018;52:715–721. doi: 10.1002/uog.18928. [DOI] [PubMed] [Google Scholar]
- 73.Gouas L., Kémény S., Beaufrère A.-M., Eymard-Pierre E., Pebrel-Richard C., Tchirkov A., Lemery D., Laurichesse-Delmas H., Vago P., Goumy C. Prenatal Screening of 21 Microdeletion/Microduplication Syndromes and Subtelomeric Imbalances by MLPA in Fetuses with Increased Nuchal Translucency and Normal Karyotype. Cytogenet. Genome Res. 2015;146:28–32. doi: 10.1159/000435865. [DOI] [PubMed] [Google Scholar]
- 74.Jin H., Wang J., Zhang G., Jiao H., Zhu J., Li Z., Chen C., Zhang X., Huang H., Wang J. A Chinese multicenter retrospective study of isolated increased nuchal translucency associated chromosome anomaly and prenatal diagnostic suggestions. Sci. Rep. 2021;11:5596. doi: 10.1038/s41598-021-85108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang L.-Y., Pan M., Han J., Zhen L., Yang X., Li D.-Z. What would be missed in the first trimester if nuchal translucency measurement is replaced by cell free DNA foetal aneuploidy screening? J. Obstet. Gynaecol. 2018;38:498–501. doi: 10.1080/01443615.2017.1391755. [DOI] [PubMed] [Google Scholar]
- 76.Hui A.S., Chau M.H.K., Chan Y.M., Cao Y., Kwan A.H., Zhu X., Kwok Y.K., Chen Z., Lao T.T., Choy K.W., et al. The role of chromosomal microarray analysis among fetuses with normal karyotype and single system anomaly or nonspecific sonographic findings. Acta Obstet. Gynecol. Scand. 2021;100:235–243. doi: 10.1111/aogs.14003. [DOI] [PubMed] [Google Scholar]
- 77.Kagan K.O., Avgidou K., Molina F.S., Gajewska K., Nicolaides K.H. Relation between increased fetal nuchal translucency thickness and chromosomal defects. Obstet. Gynecol. 2006;107:6–10. doi: 10.1097/01.AOG.0000191301.63871.c6. [DOI] [PubMed] [Google Scholar]
- 78.Leung T.Y., Yeung K.C.A., Leung W.C., Leung K.Y., Lo T.K., To W.W.K., Lau W.L., Chan L.W., Sahota D., Choy R.K.W. Prenatal diagnosis of pathogenic genomic imbalance in fetuses with increased nuchal translucency but normal karyotyping using chromosomal microarray. Hong Kong Med. J. 2019;25:30–32. [PubMed] [Google Scholar]
- 79.Nakamura Y., Fujita S., Yamada K., Song M., Tajima A., Matsumoto J., Kihira C., Kurata Y., Arakawa H., Tamura C. Outcome of the fetuses with severely increased nuchal translucency thickness in the first trimester. J. Obstet. Gynaecol. Res. 2020;46:1977–1981. doi: 10.1111/jog.14411. [DOI] [PubMed] [Google Scholar]
- 80.Lithner C.U., Kublickas M., Ek S. Pregnancy outcome for fetuses with increased nuchal translucency but normal karyotype. J. Med. Screen. 2016;23:1–6. doi: 10.1177/0969141315595826. [DOI] [PubMed] [Google Scholar]
- 81.Tekesin I. The diagnostic value of a detailed first trimester anomaly scan in fetuses with increased nuchal translucency thickness. J. Perinat. Med. 2019;47:241–246. doi: 10.1515/jpm-2018-0145. [DOI] [PubMed] [Google Scholar]
- 82.Tekesin I. Pregnancy outcome in foetuses with increased nuchal translucency—10-years’ experience in a prenatal medical practice. J. Obstet. Gynaecol. 2020;40:455–460. doi: 10.1080/01443615.2019.1621822. [DOI] [PubMed] [Google Scholar]
- 83.Tahmasebpour A., Rafiee N.B., Ghaffari S., Jamal A. Increased nuchal translucency and pregnancy outcome. Iran. J. Public Health. 2012;41:92–97. [PMC free article] [PubMed] [Google Scholar]
- 84.Shakoor S., Dileep D., Tirmizi S., Rashid S., Amin Y., Munim S. Increased nuchal translucency and adverse pregnancy outcomes. J. Matern. -Fetal Neonatal Med. 2017;30:1760–1763. doi: 10.1080/14767058.2016.1224836. [DOI] [PubMed] [Google Scholar]
- 85.Senat M.V., Bussières L., Couderc S., Roume J., Rozenberg P., Bouyer J., Ville Y. Long-term outcome of children born after a first-trimester measurement of nuchal translucency at the 99th percentile or greater with normal karyotype: A prospective study. Am. J. Obstet. Gynecol. 2007;196:53.e1–53.e536. doi: 10.1016/j.ajog.2006.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Senat M.-V., Bussières L., Couderc S., Roume J., Rozenberg P., Bouyer J., Ville Y. Pregnancy outcome in fetuses with increased nuchal translucency and normal karyotype. Prenat. Diagn. 2002;22:345–349. doi: 10.1002/pd.321. [DOI] [PubMed] [Google Scholar]
- 87.Scott F., Evans J., McLennan A. Perinatal outcome in fetuses with extremely large nuchal translucency measurement. Aust. New Zealand J. Obstet. Gynaecol. 2009;49:254–257. doi: 10.1111/j.1479-828X.2009.00997.x. [DOI] [PubMed] [Google Scholar]
- 88.Müller M.A., Pajkrt E., Bleker O.P., Bonsel G.J., Bilardo C.M. Disappearance of enlarged nuchal translucency before 14 weeks’ gestation: Relationship with chromosomal abnormalities and pregnancy outcome. Ultrasound Obstet. Gynecol. 2004;24:169–174. doi: 10.1002/uog.1103. [DOI] [PubMed] [Google Scholar]
- 89.Orgul G., Ozyuncu O., Oktem A., Beksac M.S. Management and outcomes of cystic hygromas: Experience of a tertiary center. J. Ultrasound. 2017;20:127–131. doi: 10.1007/s40477-017-0251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Graesslin O., Derniaux E., Alanio E., Gaillard D., Vitry F., Quéreux C., Ducarme G. Characteristics and outcome of fetal cystic hygroma diagnosed in the first trimester. Acta Obstet. Gynecol. Scand. 2007;86:1442–1446. doi: 10.1080/00016340701644843. [DOI] [PubMed] [Google Scholar]
- 91.Malone F.D., Ball R.H., Nyberg D.A., Comstock C.H., Saade G.R., Berkowitz R.L., Gross S.J., Dugoff L., Craigo S.D., Timor-Tritsch I.E., et al. First-trimester septated cystic hygroma: Prevalence, natural history, and pediatric outcome. Obstet. Gynecol. 2005;106:288–294. doi: 10.1097/01.AOG.0000173318.54978.1f. [DOI] [PubMed] [Google Scholar]
- 92.Schreurs L., Lannoo L., De Catte L., Van Schoubroeck D., Devriendt K., Richter J. First trimester cystic hygroma colli: Retrospective analysis in a tertiary center. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018;231:60–64. doi: 10.1016/j.ejogrb.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 93.Chen M., Lee C.P., Lin S.M., Lam Y.H., Tang R.Y.K., Tse H.Y., Tang M.H.Y. Cystic hygroma detected in the first trimester scan in Hong Kong. J. Matern. -Fetal Neonatal Med. 2014;27:342–345. doi: 10.3109/14767058.2013.818122. [DOI] [PubMed] [Google Scholar]
- 94.Yakıştıran B., Altınboğa O., Canpolat E., Çakar E., Çelen Ş., Çağlar A.T., Üstün Y.E. Analysis of cystic hygroma diagnosed in the first trimester: Single-center experience. J. Turk. Ger. Gynecol. Assoc. 2020;21:107–110. doi: 10.4274/jtgga.galenos.2019.2019.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ganapathy R., Guven M., Sethna F., Vivekananda U., Thilaganathan B. Natural history and outcome of prenatally diagnosed cystic hygroma. Prenat. Diagn. 2004;24:965–968. doi: 10.1002/pd.991. [DOI] [PubMed] [Google Scholar]
- 96.Kharrat R., Yamamoto M., Roume J., Couderc S., Vialard F., Hillion Y., Ville Y. Karyotype and outcome of fetuses diagnosed with cystic hygroma in the first trimester in relation to nuchal translucency thickness. Prenat. Diagn. 2006;26:369–372. doi: 10.1002/pd.1423. [DOI] [PubMed] [Google Scholar]
- 97.Tanriverdi H., Ertan A., Hendrik H., Remberger K., Schmidt W. Outcome of cystic hygroma in fetuses with normal karyotypes depends on associated findings. Eur. J. Obstet. Gynecol. Reprod. Biol. 2005;118:40–46. doi: 10.1016/j.ejogrb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 98.Scholl J., Chasen S.T. First trimester cystic hygroma: Does early detection matter? Prenat. Diagn. 2016;36:432–436. doi: 10.1002/pd.4799. [DOI] [PubMed] [Google Scholar]
- 99.Sanhal C., Mendilcioglu I., Özekinci M., Yakut S., Merdun Z., Şimşek M., Lüleci G. Prenatal management, pregnancy and pediatric outcomes in fetuses with septated cystic hygroma. Braz. J. Med. Biol. Res. 2014;47:799–803. doi: 10.1590/1414-431X20143895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Noia G., Pellegrino M., Masini L., Visconti D., Manzoni C., Chiaradia G., Caruso A. Fetal cystic hygroma: The importance of natural history. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013;170:407–413. doi: 10.1016/j.ejogrb.2013.07.043. [DOI] [PubMed] [Google Scholar]
- 101.Beke A., Joó J.G., Csaba A., Lázár L., Bán Z., Papp C., Tóth-Pál E., Papp Z. Incidence of chromosomal abnormalities in the presence of fetal subcutaneous oedema, such as nuchal oedema, cystic hygroma and non-immune hydrops. Fetal Diagn. Ther. 2009;25:83–92. doi: 10.1159/000201946. [DOI] [PubMed] [Google Scholar]
- 102.Esmer A.C., Kalelioglu I., Keyif B., Ozsurmeli M., Yüksel A., Has R. Significance of septa in first trimester increased nuchal translucency thickness. J. Med. Ultrason. 2014;41:51–56. doi: 10.1007/s10396-013-0464-1. [DOI] [PubMed] [Google Scholar]
- 103.Westin M., Saltvedt S., Almström H., Grunewald C., Valentin L. By how much does increased nuchal translucency increase the risk of adverse pregnancy outcome in chromosomally normal fetuses? A study of 16,260 fetuses derived from an unselected pregnant population. Ultrasound Obstet. Gynecol. 2007;29:150–158. doi: 10.1002/uog.3905. [DOI] [PubMed] [Google Scholar]
- 104.Motta M., Giancotti A., Mastromoro G., Chandramouli B., Pinna V., Pantaleoni F., Di Giosaffatte N., Petrini S., Mazza T., D’Ambrosio V., et al. Clinical and functional characterization of a novel RASopathy-causing SHOC2 mutation associated with prenatal-onset hypertrophic cardiomyopathy. Hum. Mutat. 2009;40:1046–1056. doi: 10.1002/humu.23767. [DOI] [PubMed] [Google Scholar]
- 105.Pajkrt E., De Graaf I.M., Mol B.W., Van Lith J.M., Bleker O.P., Bilardo C.M. Weekly nuchal translucency measurements in normal fetuses. Obstet. Gynecol. 1998;91:208–211. doi: 10.1016/S0029-7844(97)00658-3. [DOI] [PubMed] [Google Scholar]
- 106.Pajkrt E., Bleker O.P., Bilardo C.M., Van Lith J.M., Mol B.W. Nuchal translucency measurement in normal fetuses. Obstet. Gynecol. 1995;86:994–997. doi: 10.1016/0029-7844(95)00310-N. [DOI] [PubMed] [Google Scholar]
- 107.Pandya P.P., Snijders R., Johnson S., Nicolaides K. Natural history of trisomy 21 fetuses with increased nuchal translucency thickness. Ultrasound Obstet. Gynecol. 1995;5:381–383. doi: 10.1046/j.1469-0705.1995.05060381.x. [DOI] [PubMed] [Google Scholar]
- 108.van Zalen-Sprock R.M., van Vugt J.M., van Geijn H.P. First-trimester diagnosis of cystic hygroma--course and outcome. Am. J. Obstet. Gynecol. 1992;167:94–98. doi: 10.1016/S0002-9378(11)91634-2. [DOI] [PubMed] [Google Scholar]
- 109.Chervenak F.A., Isaacson G., Blakemore K.J., Breg W.R., Hobbins J.C., Berkowitz R.L., Tortora M., Mayden K., Mahoney M.J. Fetal cystic hygroma. Cause and natural history. N. Engl. J. Med. 1983;309:822–825. doi: 10.1056/NEJM198310063091403. [DOI] [PubMed] [Google Scholar]
- 110.Gustavii B., Edvall H. First-trimester diagnosis of cystic nuchal hygroma. Acta Obstet. Gynecol. Scand. 1984;63:377–378. doi: 10.3109/00016348409155537. [DOI] [PubMed] [Google Scholar]
- 111.Mastromoro G., Hashemian N.K., Guadagnolo D., Giuffrida M.G., Torres B., Bernardini L., Ventriglia F., Piacentini G., Pizzuti A. Chromosomal Microarray Analysis in Fetuses Detected with Isolated Cardiovascular Malformation: A Multicenter Study, Systematic Review of the Literature and Meta-Analysis. Diagnostics. 2022;12:1328. doi: 10.3390/diagnostics12061328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moscoso G. Fetal nuchal translucency: A need to understand the physiological basis. Ultrasound Obstet. Gynecol. 1995;5:6–8. doi: 10.1046/j.1469-0705.1995.05010006.x. [DOI] [PubMed] [Google Scholar]
- 113.Hyett J.A., Brizot M.L., Von Kaisenberg C.S., McKie A.T., Farzaneh F., Nicolaides K.H. Cardiac gene expression of atrial natriuretic peptide and brain natriuretic peptide in trisomic fetuses. Obstet. Gynecol. 1996;87:506–510. doi: 10.1016/0029-7844(95)00486-6. [DOI] [PubMed] [Google Scholar]
- 114.Schwarze A., Kreiselmaier P., Krapp M., Smrcek J., Axt-Fliedner R. Umbilical cord diameter at 11–14 weeks of gestation: Relationship to nuchal translucency, ductus venous blood flow and chromosomal defects. Fetal Diagn. Ther. 2006;21:390–395. doi: 10.1002/uog.2147. [DOI] [PubMed] [Google Scholar]
- 115.Ghezzi F., Raio L., Di Naro E., Franchi M., Buttarelli M., Schneider H. First-trimester umbilical cord diameter: A novel marker of fetal aneuploidy. Ultrasound Obstet. Gynecol. 2002;19:235–239. doi: 10.1046/j.1469-0705.2002.00650.x. [DOI] [PubMed] [Google Scholar]
- 116.El Daouk M., Brustman L., Langer O., Lysikiewicz A. Male-to-female gender ratio in fetuses with increased nuchal translucency. J. Matern. -Fetal Neonatal Med. 2012;25:2613–2615. doi: 10.3109/14767058.2012.704444. [DOI] [PubMed] [Google Scholar]
- 117.Timmerman E., Pajkrt E., Bilardo C.M. Male gender as a favorable prognostic factor in pregnancies with enlarged nuchal translucency. Ultrasound Obstet. Gynecol. 2009;34:373–378. doi: 10.1002/uog.6397. [DOI] [PubMed] [Google Scholar]
- 118.Gbrumfield C., Dwenstrom K., Odavis R., Owen J., Cosper P. Second-trimester cystic hygroma: Prognosis of septated and nonseptated lesions. Obstet. Gynecol. 1996;88:979–982. doi: 10.1016/S0029-7844(96)00358-4. [DOI] [PubMed] [Google Scholar]
- 119.Bernstein H.S., Filly R.A., Goldberg J.D., Golbus M.S. Prognosis of fetuses with a cystic hygroma. Prenat. Diagn. 1991;11:349–355. doi: 10.1002/pd.1970110603. [DOI] [PubMed] [Google Scholar]
- 120.Maymon R., Tercanli S., Dreazen E., Sartorius G., Holzgreve W., Herman A. Comparison of pregnancy outcome of euploid fetuses with increased nuchal translucency (NT) expressed in NT MoM or delta-NT. Ultrasound Obstet. Gynecol. 2004;23:477–481. doi: 10.1002/uog.1060. [DOI] [PubMed] [Google Scholar]
- 121.Rizzo G., Muscatello A., Angelini E., Capponi A. Abnormal cardiac function in fetuses with increased nuchal translucency. Ultrasound Obstet. Gynecol. 2003;21:539–542. doi: 10.1002/uog.137. [DOI] [PubMed] [Google Scholar]
- 122.Souka A.P., von Kaisenberg C.S., Hyett J.A., Sonek J.D., Nicolaides K.H. Increased nuchal translucency with normal karyotype. Am. J. Obstet. Gynecol. 2005;192:1005–1021. doi: 10.1016/j.ajog.2004.12.093. [DOI] [PubMed] [Google Scholar]
- 123.Bohlandt S., von Kaisenberg C., Wewetzer K., Christ B., Nicolaides K., Brand-Saberi B. Hyaluronan in the nuchal skin of chromosomally abnormal fetuses. Hum. Reprod. 2000;15:1155–1158. doi: 10.1093/humrep/15.5.1155. [DOI] [PubMed] [Google Scholar]
- 124.Hyett J., Perdu M., Sharland G., Snijders R., Nicolaides K. Using fetal nuchal translucency to screen for major congenital cardiac defects at 10–14 weeks of gestation: Population based cohort study. BMJ. 1999;318:81–85. doi: 10.1136/bmj.318.7176.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sebire N., Souka A., Skentou H., Geerts L., Nicolaides K. Early prediction of severe twin-to-twin transfusion syndrome. Hum. Reprod. 2000;15:2008–2010. doi: 10.1093/humrep/15.9.2008. [DOI] [PubMed] [Google Scholar]
- 126.Sebire N.J., D’Ercole C., Hughes K., Carvalho M., Nicolaides K. Increased nuchal translucency thickness at 10–14 weeks of gestation as a predictor of severe twin-to-twin transfusion syndrome. Ultrasound Obstet. Gynecol. 1997;10:86–89. doi: 10.1046/j.1469-0705.1997.10020086.x. [DOI] [PubMed] [Google Scholar]
- 127.Shi X., Liu Q., He W., Liu Y., Rao T., Fang L., Wu J. Prenatal and perinatal outcomes of twin pregnancy discordant for one fetus with nuchal translucency above the 95th percentile. J. Matern. -Fetal Neonatal Med. 2022;35:3152–3157. doi: 10.1080/14767058.2020.1814242. [DOI] [PubMed] [Google Scholar]
- 128.Krispin E., Kushnir A., Shemer A., Rienstein S., Berkenstadt M., Yinon Y., Weisz B. Abnormal nuchal translucency followed by normal microarray analysis is associated with placental pathology-related complications. Prenat. Diagn. 2021;41:855–860. doi: 10.1002/pd.5896. [DOI] [PubMed] [Google Scholar]
- 129.Quaresima P., Homfray T., Greco E. Obstetric complications in pregnancies with life-limiting malformations. Curr. Opin. Obstet. Gynecol. 2019;31:375–387. doi: 10.1097/GCO.0000000000000583. [DOI] [PubMed] [Google Scholar]
- 130.Buffin R., Fichez A., Decullier E., Roux A., Bin S., Combourieu D., Diez B.P., Huissoud C., Picaud J., Basson E., et al. Neurodevelopmental outcome at 2 years of corrected age in fetuses with increased nuchal translucency thickness and normal karyotype compared with matched controls. Ultrasound Obstet. Gynecol. 2021;57:790–797. doi: 10.1002/uog.22009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guadagnolo D., Mastromoro G., Di Palma F., Pizzuti A., Marchionni E. Prenatal Exome Sequencing: Background, Current Practice and Future Perspectives-A Systematic Review. Diagnostics. 2021;11:224. doi: 10.3390/diagnostics11020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mastromoro G., Guadagnolo D., Khaleghi Hashemian N., Marchionni E., Traversa A., Pizzuti A. Molecular Approaches in Fetal Malformations, Dynamic Anomalies and Soft Markers: Diagnostic Rates and Challenges-Systematic Review of the Literature and Meta-Analysis. Diagnostics. 2022;12:575. doi: 10.3390/diagnostics12030575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sparks T.N., Lianoglou B.R., Adami R.R., Pluym I.D., Holliman K., Duffy J., Downum S.L., Patel S., Faubel A., Boe N.M., et al. Exome Sequencing for Prenatal Diagnosis in Nonimmune Hydrops Fetalis. N. Engl. J. Med. 2020;383:1746–1756. doi: 10.1056/NEJMoa2023643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Han J., Li D.-Z. Further genetic testing in prenatal cases of nonimmune hydrops fetalis with a normal array: A targeted panel or exome? Am. J. Obstet. Gynecol. 2022;226:276–277. doi: 10.1016/j.ajog.2021.09.040. [DOI] [PubMed] [Google Scholar]
- 135.Mone F., Eberhardt R.Y., Hurles M.E., Mcmullan D.J., Maher E.R., Lord J., Chitty L.S., Dempsey E., Homfray T., Giordano J.L., et al. Fetal hydrops and the Incremental yield of Next-generation sequencing over standard prenatal Diagnostic testing (FIND) study: Prospective cohort study and meta-analysis. Ultrasound Obstet. Gynecol. 2021;58:509–518. doi: 10.1002/uog.23652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Quinn A.M., Valcarcel B.N., Makhamreh M.M., Al-Kouatly H.B., Berger S.I. A systematic review of monogenic etiologies of nonimmune hydrops fetalis. Genet. Med. 2021;23:3–12. doi: 10.1038/s41436-020-00967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Norton M.E., Ziffle J.V., Lianoglou B.R., Hodoglugil U., Devine W.P., Sparks T.N. Exome sequencing vs targeted gene panels for the evaluation of nonimmune hydrops fetalis. Am. J. Obstet. Gynecol. 2022;226:128.e1–128.e11. doi: 10.1016/j.ajog.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Al-Kouatly H.B., Makhamreh M.M., Rice S.M., Smith K., Harman C., Quinn A., Valcarcel B.N., Firman B., Liu R., Hegde M., et al. High diagnosis rate for nonimmune hydrops fetalis with prenatal clinical exome from the Hydrops-Yielding Diagnostic Results of Prenatal Sequencing (HYDROPS) Study. Genet. Med. 2021;23:1325–1333. doi: 10.1038/s41436-021-01121-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in the articles cited in the References section.