Abstract
The relative pathogenicities of three Candida albicans strains differing in the function of ADE2 (the gene encoding phosphoribosylaminoimidazole carboxylase) were evaluated in a murine candidiasis model. C. albicans strain CAI7 (ade2/ade2), previously constructed by site-specific recombination, was avirulent in immunosuppressed mice compared to the parent strain, CAF2-1, and a heterozygous ADE2/ade2 strain obtained by transforming CAI7 with a wild-type allele. The reduced virulence of CAI7 was correlated with the inability to proliferate in either synthetic medium or serum without the exogenous addition of >10 μg of adenine/ml. The loss of virulence upon site-specific disruption of the ade2 locus, and the restoration of wild-type virulence with the repair of just one ade2 allele, confirmed that the ADE2 gene and de novo purine biosynthesis were required for Candida pathogenicity. The potential of the phosphoribosylaminoimidazole carboxylase enzyme as a novel target for antifungal drug discovery is discussed.
Candida albicans is an opportunistic human pathogen causing mucosal and cutaneous infections as well as life-threatening systemic infections in immunosuppressed patients. The incidence of candidiasis has increased markedly over the past 20 years, coinciding with an increase in immunosuppressed individuals (6). This increase is primarily due to the expanded use of chemotherapy and organ transplantation and to the rapid increase in the numbers of AIDS patients. Candidiasis is currently treated with amphotericin B or with a number of drugs collectively referred to as azoles. Amphotericin B exerts its antifungal activity by binding to the fungal steroid ergosterol and disrupting membrane integrity (1), whereas the azoles directly inhibit the biosynthesis of ergosterol (9). The potential for widespread resistance to azole drugs is an increasing concern (16). The development of safe, efficacious antifungal drugs that exhibit a novel mechanism of action is an important challenge for medical research. A greater understanding of virulence mechanisms and genes required for C. albicans pathogenesis is needed to meet this challenge.
Recent attention has focused on the identification of virulence factors that dictate the ability of C. albicans to invade tissues and circumvent host defense responses. Tissue adherence (17), cell wall mannan, serine proteases, cellular hydrolases (2), and phospholipases (8) are examples of virulence factors utilized by C. albicans for infection. Also, a number of C. albicans housekeeping and morphogenesis genes are essential for C. albicans pathogenesis. Although not classical virulence factors, these genes are critical for C. albicans survival in a host. For example, purine, pyrimidine, and heme biosynthesis, which are all necessary for cell growth and division in vitro, are required for C. albicans pathogenesis in experimental murine candidiasis (10). Similarly, myristoyl-coenzyme A:protein N-myristoyl transferase, which is involved in protein myristoylation (22), and the PHR1 gene, which functions as a pH-sensitive morphological determinant (7), are essential for pathogenesis. Additional studies of both virulence genes and genes essential for C. albicans survival will provide a basis for future antifungal drug development.
The de novo purine biosynthetic pathway is central to cellular metabolism, providing adenine and guanine nucleotide precursors to DNA synthesis and repair, to RNA transcription, and to histidine biosynthesis. The ADE2 gene encodes phosphoribosylaminoimidazole carboxylase, which catalyzes the sixth step of the de novo purine biosynthesis pathway. The ADE2 gene is essential to fungi and, interestingly, ADE2 mutants appear as red colonies when grown on agar medium (15). The red pigment is thought to result from the accumulation and subsequent polymerization of the substrate, phosphoribosylaminoimidazole (21). The de novo purine biosynthetic pathway has been associated with pathogenesis in C. albicans. Purine auxotrophic mutants obtained after random chemical and UV mutagenesis exhibit reduced virulence in a murine model of systemic candidiasis (12, 14, 19). These results indicate that purine auxotrophy may limit C. albicans virulence, with the caveat that random mutagenesis may have introduced additional unidentified mutations. Recently, the ADE2 gene was more directly linked to reduced virulence in C. albicans (10) and Cryptococcus neoformans (13). In these studies, the ade2 mutant strains of C. albicans and C. neoformans were less virulent than wild-type strains in an immunosuppressed murine model of systemic candidiasis and in an immunosuppressed rabbit model of cryptococcal meningitis, respectively. Although the ade2 mutant strains were again generated by random mutagenesis, the reduced pathogenesis resulted directly from the ade2 mutation since the mutant strains recovered wild-type virulence after transformation with a plasmid containing an active ADE2 gene. These experiments more clearly defined the ADE2 gene as essential for fungal pathogenesis.
In this report, we further investigated the role of de novo purine biosynthesis and specifically the ADE2 gene in C. albicans pathogenesis. The virulence of C. albicans strain CAI7 (ade2/ade2) that was previously constructed by gene disruption (4) was evaluated in an immunosuppressed model of systemic candidiasis. The use of an ade2/ade2 mutant strain generated by targeted gene disruption avoided the potential for secondary mutations inherent with random mutagenesis techniques. CAI7 was unable to proliferate in the kidney and was nonpathogenic compared to its parent strain, CAF2-1, and a heterozygous complemented strain, CAI7-R. The reduced virulence of CAI7 resulted directly from the block in de novo purine biosynthesis and the inability to scavenge purines from the host. Furthermore, only a single copy of the ADE2 gene was required to restore wild-type virulence.
MATERIALS AND METHODS
Organisms and media.
The yeast strains utilized for this study are listed in Table 1. The strains were routinely grown at 30°C on yeast extract-peptone-dextrose (YEPD) medium or synthetic complete medium solidified with 2% agar as needed (20). A heterozygous ADE2/ade2 strain was constructed by repairing the ade2::hisG allele of strain CAI7 by homologous recombination. A XhoI-NotI restriction enzyme fragment from PBSIIZap 25 CA2 containing the C. albicans ADE2 gene (18) was transfected into strain CAI7 spheroplasts essentially as described previously (11). The heterozygous strain, designated CAI7-R, was selected on synthetic complete medium without adenine and uridine. The Ura+ Ade+ phenotype indicated that the transfected ADE2 DNA had recombined with the ade2::hisG allele and not with the ade2::hisG-URA3-hisG allele. The heterozygous genotype was confirmed by PCR (25 cycles; annealing temperature, 50°C) utilizing an antisense primer to the ADE2 coding region, GGTCGATACGAATTCTTATTTTTTCAATTTATCAG, and a primer to the 5′ promoter region, GGTCGGATCCATGGATAGCAAAACTGTTGG, which produced a 1.7-kb DNA fragment.
TABLE 1.
Genotypes of C. albicans strains
Growth rates.
Strains were grown overnight in synthetic complete medium supplemented with 100 μg of adenine (Sigma)/ml. Each strain was diluted to 104 cells/ml into synthetic complete medium lacking adenine. Duplicate dilutions were also made into synthetic complete medium supplemented with 100 μg of adenine/ml. Growth rates were monitored by determining both the optical density at 600 nm and the number of CFU.
Serum growth.
C. albicans strains were grown in YEPD medium, inoculated into fresh mouse or human serum at a concentration of 103 CFU/ml, and incubated at 35°C for up to 24 h.
Experimental murine model.
Female Swiss Webster mice weighing 20 to 25 g were injected with 100 mg of cyclophosphamide (catalog no. C7397; Sigma)/kg of body weight on days 4 and 1 prior to Candida infection and every 3 days during the course of the experiment. Immunosuppression with this dosing regimen was confirmed by determining the differential leukocyte counts with a Coulter counter. Candida strains were grown to saturation (16 h) in YEPD at 30°C. Prior to injection, yeast cells were washed three times in sterile phosphate-buffered saline, diluted, and counted on a hemacytometer. The cell concentrations were adjusted to 106 to 109 cells/ml for CAI7 and 104 to 107 cells/ml for CAF2-1 and CAI7-R. Murine candidiasis was induced by injection of 0.1 ml of yeast cell suspension into the lateral tail vein. Survival was monitored for 21 days.
Histological techniques.
Histological analysis of the liver, lungs, spleen, and kidneys were done for lethally moribund mice or for separate groups of mice infected specifically to provide histological samples for studying time-course infection. Tissue was fixed in 10% phosphate-buffered formalin and processed with standard histological techniques. The sections were stained with hematoxylin and eosin stain (H&E), periodic acid-Schiff stain (PAS), or Gimori's methenamine silver stain (GMS) to visualize the extent of yeast cell invasion and proliferation.
RESULTS
Properties of C. albicans strains.
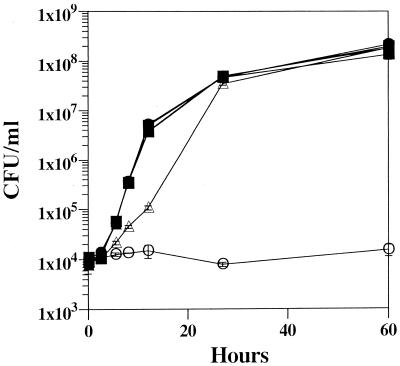
Three strains of C. albicans were used in this study (Table 1). The parent strain, CAF2-1, was phenotypically wild type for in vitro growth and pathogenesis (5). Strain CAI7 was phenotypically an adenine auxotroph and produced red colonies on medium depleted of adenine. Strain CAI7-R was constructed by replacing the ade2::hisG allele of strain CAI7 with a wild-type copy of the C. albicans ADE2 gene. Strain CAI7 was unable to grow in adenine-free medium as expected for a homozygous ade2 mutant (Fig. 1). Interestingly, the heterozygous strain, CAI7-R, exhibited slower growth than CAF2-1 in adenine-free medium, which was reversed with exogenous adenine. The slower growth of the heterozygote indicated that purine biosynthesis was partially rate limiting for growth when only one copy of the ade2 gene was present. All three strains had similar growth rates when cultured in YEPD medium (data not shown) or on synthetic complete medium supplemented with 50 μg of adenine/ml.
FIG. 1.
In vitro growth rates of CAF2-1, CAI7, and CAI7-R. Wild-type CAF2-1 (squares), ade2/ade2 adenine auxotrophic CAI7 (circles), and heterozygous ADE2/ade2 CAI7-R (triangles) were grown in synthetic minimal medium in the presence (closed symbols) or the absence (open symbols) of 50 μg of adenine/ml. At the designated times, growth (CFU) was determined from replicated samples diluted in YEPD broth and grown on YEPD agar for 2 days at 37°C.
Systemic candidiasis in immunosuppressed mice.
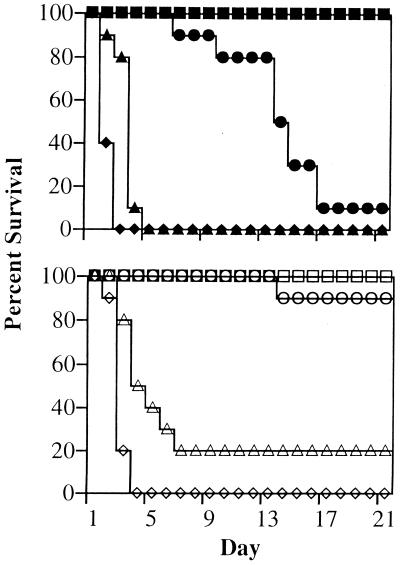
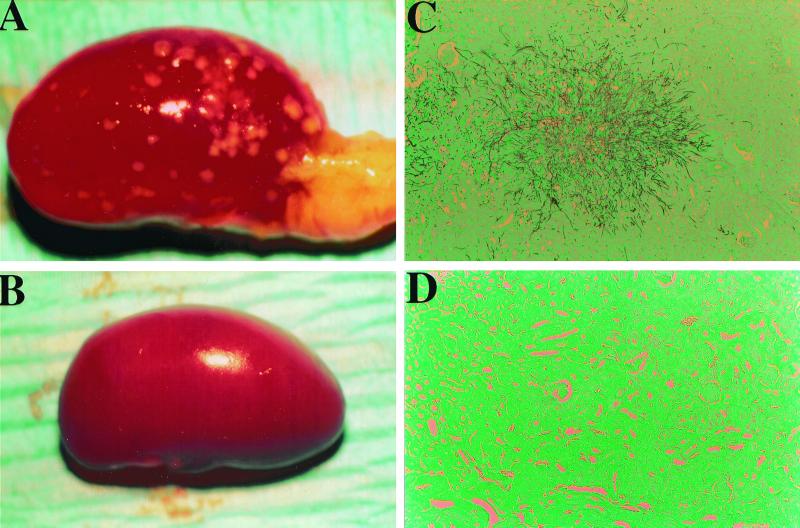
Invasive fungal infections typically occur in immunosuppressed hosts. We compared the virulence of strain CAI7 with that of CAF2-1, its parent strain, in an immunosuppressed murine model of candidiasis. Chronic immune suppression was achieved by administration of cyclophosphamide (100 mg/kg) intraperitoneally every 3 days, starting 4 days prior to Candida injection and continuing for the duration of the experiment. Lymphocyte levels were suppressed to <100/μl for the entire course of the experiment compared to >8,000/μl in untreated mice. The observed 50% lethal dose (LD50) was 5 × 103 wild-type C. albicans blastoconidia per mouse for injections into the tail veins of immunosuppressed mice (Fig. 2). This was the lowest LD50 reported for a murine candidiasis model and likely resulted from the extended immunosuppressive treatment. Kidneys from lethally moribund or dead mice displayed numerous severe microabscesses on the surface (Fig. 3A) and exhibited prolific Candida invasion of the cortex, medulla, and papilla regions (Fig. 3B). In this chronic immunosuppressed murine model, microabscess formation on the kidneys enabled reliable and consistent visual scoring for Candida infection.
FIG. 2.
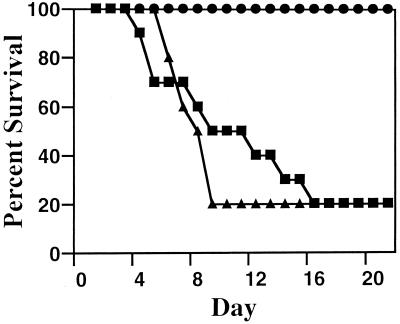
Virulence of C. albicans CAF2-1 and CAI7 in immunosuppressed mice. Mice were injected with 2 × 103 (▪), 2 × 104 (●), 2 × 105 (▴), or 2 × 106 (♦) of wild-type CAF2-1 blastoconidia (top) or 5 × 105 (□), 5 × 106 (○), 5 × 107 (▵), or 5 × 108 (◊) of ade2/ade2 CAI7 blastoconidia (bottom). Ten mice were inoculated through the tail veins with the indicated number of blastoconidia, and mortality was determined over the course of a 21-day experiment. Data were recorded as percent survival on each day.
FIG. 3.
Infection of C. albicans in the kidneys of immunosuppressed mice. Kidneys were dissected, and transverse sections were stained with H&E and GMS to aid in histological examination. Representative kidney (A) and histological (B) sections from mice 4 days after injection with 104 CAF2-1 (wild-type) blastoconidia are shown, as are representative kidney (C) and histological (D) sections from mice 6 days after injection with 105 CAI7 (ade2/ade2) blastoconidia.
Strain CAI7 generally appeared to be nonpathogenic at doses less than 106 blastoconidia per mouse in this model. The LD50 was determined to be 2.5 × 106 compared to the LD50 of 5 × 103 for strain CAF2- (Fig. 2). Histological examination of the kidneys provided an equally striking difference between strains. Kidney sections were examined for every mouse infected with strain CAI7 at the end of the 21-day experiment. No evidence of infection was histologically detectable in the kidney cortex or the medulla, which is the major site of infection by wild-type C. albicans (Fig. 3C and D). Furthermore, no secondary sites of infection were observed upon examination of heart, lung, liver, spleen, and brain tissues. Clearly, the ADE2 gene was required for normal infection in this chronic immunosuppressed murine model.
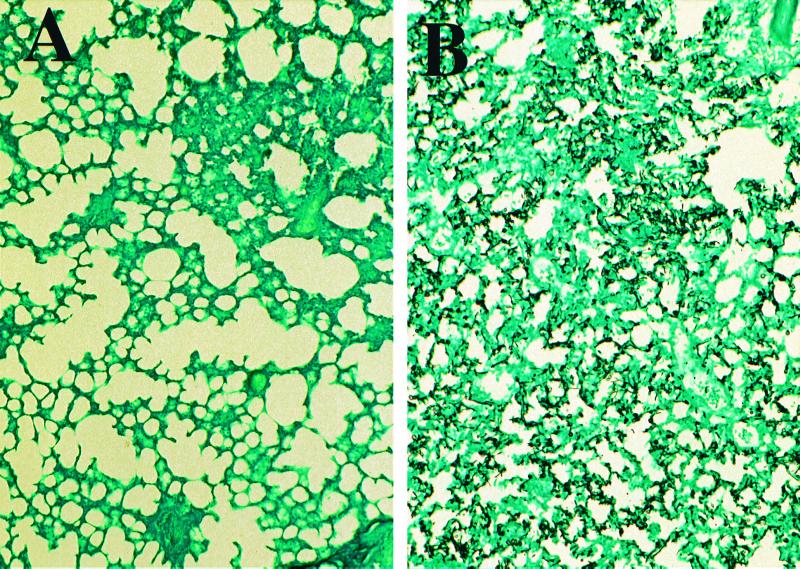
Infection with the highest titer of strain CAI7 (5 × 108 blastoconidia/mouse) resulted in a 100% incidence of mortality in the cyclophosphamide-treated mice (Fig. 2). This mortality at the high doses appeared to be an acute response rather than the result of a progressive fungal infection, and the potential causes were investigated histologically. Mice were injected with 108 yeast cells of strain CAI7/mouse, and tissues were taken 1, 3, and 7 days after injection. Kidney, spleen, liver, and lung sections were stained with PAS and GMS to aid in histological examination. At 108 blastoconidia/mouse, CAI7 was observed in all tissues evaluated. Yeast cells were observed in especially high numbers in the lung at 24 h after injection (Fig. 4). The microcapillary beds in the lung were highly occluded and likely resulted in the acute mortality seen at the high titer. The acute mortality and accumulation of blastoconidia in lung microcapillary beds associated with high-titer infections were also observed for strain CAI4, a nonpathogenic homozygous C. albicans ura3 mutant, and was not specific to strain CAI7 (data not shown).
FIG. 4.
Localization of C. albicans CAI7 in the murine lung. Immunosuppressed mice were injected with 108 C. albicans CAI7 blastoconidia through the tail veins. Lung tissues were dissected 4 days after injection. Sections were stained with PAS and GMS to aid in histological examination. Representative sections are shown for uninfected (A) and CAI7-infected (B) mice.
The necessity of a functional ade2 allele for infection was further established by repairing one ade2 allele in strain CAI7 by using site-specific homologous recombination and evaluating the virulence of the heterozygous complemented strain, CAI7-R. Strains CAF2-1 and CAI7-R were injected at 104 blastoconidia/mouse, and both resulted in similar rates of mortality over the course of the 21-day experiment (Fig. 5). Histological examination of kidneys from infected mice revealed that at the time of death both strains had similarly infected the kidneys, resulting in numerous microabscesses and prolific invasion of the cortex and medulla. Consistent with previous results, strain CAI7 was nonpathogenic in this experiment, even at 105 yeast cells/mouse. These observations demonstrated that the reduced pathogenesis of strain CAI7 resulted directly from the ade2 mutation since full virulence was recovered by specifically repairing the ade2::hisG mutant allele. Furthermore, only a single functioning ADE2 allele was required for apparent wild-type virulence.
FIG. 5.
Restoration of virulence for strain CAI7 in immunosuppressed mice. Mice (10 per treatment) received a tail vein injection of 104 blastoconidia of C. albicans wild-type CAF2-1 (▪), heterozygous ADE2/ade2 CAI7-R (▴), or 105 blastoconidia of ade2/ade2 auxotroph CAI7 (●). Mortality was determined over the course of a 21-day experiment. Data were recorded as percent survival on each day.
Strain CAI7 was unable to proliferate in serum.
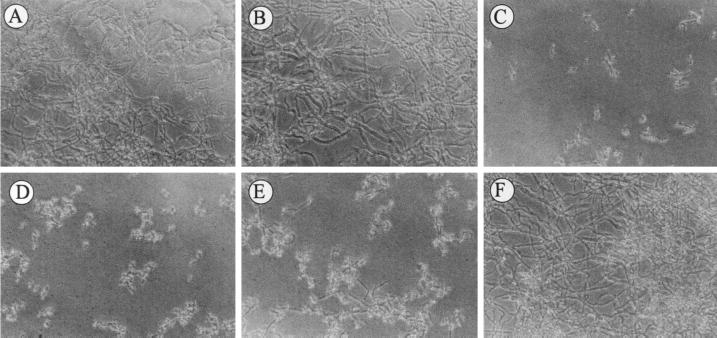
The virulence of the mutant was attenuated presumably because serum purine levels were inadequate to support growth. To test this hypothesis, each strain was evaluated for growth in human serum. Strains CAF2-1 and CAI7-R rapidly underwent a dimorphic switch and proliferated in normal human serum in both the presence and the absence of exogenous adenine (Fig. 6). Strain CAI7 initiated a dimorphic switch but was unable to proliferate in normal serum. The ability of strain CAI7 to proliferate in serum was restored when serum was supplemented with 50 μg of exogenous adenine/ml. Interestingly, the addition of <10 μg of adenine/ml had little effect on CAI7 proliferation in serum, suggesting that a threshold concentration of adenine was required for growth. These experiments were repeated using murine serum with similar results (data not shown).
FIG. 6.
Growth of C. albicans in whole human serum. Strains CAF2-1 (A) and CAI7-R (B) were inoculated in whole human serum without the addition of exogenous adenine. Similar results were obtained for both strains in the presence of up to 50 μg of adenine/ml (data not shown). Strain CAI7 was inoculated into human serum with 0 (C), 0.5 (D), 5 (E), and 50 (F) μg of exogenous adenine/ml. Observations were made at 24 h after inoculation.
DISCUSSION
The de novo purine biosynthetic pathway is an essential pathway in fungi. The ADE2 gene encodes phosphoribosylaminoimidazole carboxylase, a bifunctional enzyme that catalyzes the sixth step of de novo purine biosynthesis. Homozygous C. albicans ade2 mutants are adenine auxotrophs in vitro and appear as red colonies due to the accumulation and spontaneous polymerization of the substrate aminoimidazole ribosyl phosphate. The requirement of the ADE2 gene for infection was previously shown for both C. albicans and C. neoformans (10, 13) with the use of ade2 mutants obtained by random mutagenesis procedures. Recently, a homozygous ade2 C. albicans strain, CAI7, was generated by targeted gene disruption, which is a direct and selective means of creating isogenic strains to study gene function. The availability of strain CAI7 provided a new opportunity to study the role of the ADE2 gene and de novo purine biosynthesis in pathogenesis. The virulence of strain CAI7 was compared to the phenotypically wild-type parent strain, CAF2-1, from which it was derived. This was a critical experiment to determine whether strain CAI7 could scavenge purines from the host, circumventing the block in de novo biosynthesis. The titer of C. albicans needed to kill 50% of the mice was >100-fold higher for strain CAI7 than for strain CAF2-1 in an immunosuppressed murine candidiasis model. Correlated with the reduced virulence, strain CAI7 was unable to proliferate in the cortex or medulla of the kidney, which is the primary site of infection for wild-type C. albicans in the murine experimental model. Furthermore, strain CAI7 was unable to infect the heart, spleen, lungs, or nervous system under the same conditions, which result in lethal systemic and disseminated candidiasis by a wild-type strain. The inability of CAI7 to proliferate in human serum was directly correlated to the observed reduced virulence. These results provided a more-detailed confirmation of a previous report that C. albicans ade2 mutants are avirulent (10). Clearly, de novo purine biosynthesis is required for C. albicans infection and ade2 mutant strains are unable to scavenge sufficient purines from host tissues to overcome the block in de novo biosynthesis.
Interestingly, strain CAI7 was not completely avirulent but instead exhibited 100-fold attenuation of virulence. Injection of 108 CAI7 yeast cells in the immunosuppressed murine model resulted in significant acute mortality within 48 h that was distinct from mortality resulting from the time-dependent appearance of systemic candidiasis. Acute mortality at higher doses may have reflected a limitation of the experimental animal model resulting from several possible explanations. High doses of avirulent bacterial strains often lead to acute mortality in experimental models as the result of endotoxin-induced septic shock. Although not known to produce endotoxins, C. albicans does produce a number of tissue-damaging proteases and hydrolases which may have contributed to the observed acute mortality at high titer (2). Additionally, the number of blastoconidia, which can be introduced into the bloodstream of mice, may have an upper limit before the shear volume of blastoconidia results in occluded capillaries. In fact, our observations confirmed that >107 CAI7 yeast cells per mouse resulted in highly occluded capillary beds in the lungs. The acute mortality resulting from tissue-degrading enzymes and physical occlusion of capillary beds likely has less clinical relevance than the time-dependent systemic infection of organs, such as the kidneys, which is observed at lower titers. When the objective is an experimental assessment of virulence or drug efficacy, we suggest that the C. albicans titer be more appropriately limited to ensure that the host survives the first 48 h.
C. albicans ade2 mutants exhibited some ability to survive in vivo, albeit with attenuated pathogenicity. First, C. albicans ade2 mutants were able to undergo a dimorphic switch and initiated hyphae in serum in vitro and in murine kidney sections in vivo. Prior to the experiments, strain CAI7 was grown in rich medium containing adenine and then introduced as blastoconidia. The blastoconidia appeared to contain enough purine reserves to make a dimorphic switch and initiate hyphae. Second, some red CAI7 colonies were obtained from kidneys of chronically immunosuppressed mice up to 21 days after injection, although a majority of the kidneys were sterile at this time. The recovery of viable CAI7 colonies from minced kidneys without histological evidence of the ability to proliferate in the kidney cortex suggests that strain CAI7 may survive in a static state for several weeks.
A prediction from the murine models was that de novo purine biosynthesis and specifically the ADE2 gene will be required for C. albicans infection in humans. This prediction assumes that free purine levels in human tissues are similar to those in murine tissues; otherwise, free adenine or adenosine might circumvent a block in de novo synthesis. In fact, the C. albicans ade2/ade2 strain was unable to proliferate in human serum unless it was supplemented with exogenous adenine. These results confirm that human serum does not have enough free purines to support growth and, in combination with the murine candidiasis experimental data, support the prediction that the ADE2 gene is required for pathogenesis in humans. The identification of the ADE2 gene as potentially critical for human pathogenesis has increased importance in light of recent publications that suggest a divergent catalytic mechanism between human and bacterial phosphorylribosylaminoimidazole carboxylase enzymes (3). The bacterial enzymes catalyze the ATP- and bicarbonate-dependent carboxylation of phosphoribosylaminoimidazole to carboxyphosporibosylaminoimidazole through an N5-carbamate intermediate, whereas the vertebrate ADE2 enzymes catalyze CO2-dependent, ATP-independent direct carboxylation of the imidazole ring, foregoing the carbamate intermediate. Sequence analysis and complementation testing confirm that the C. albicans ADE2 gene is similar to the bacterial genes (18). The divergent catalytic mechanisms may provide a basis to develop selective mechanism-based antifungal compounds.
ACKNOWLEDGMENTS
We acknowledge the excellent technical assistance provided by Jana Dodge, Jim Nuckolls, and Suzanne Davis at the Pharmacia Animal Research Facility. We also appreciate the critical comments provided by Thomas Sanderson of Pharmacia Product Safety Assessment. Finally, we acknowledge the efforts of Ruth Berger at Histotechniques for the preparation and staining of histological sections.
REFERENCES
- 1.Brajtburg J, Powderly W G, Kobayashi G S, Medoff G. Amphotericin B: current understanding of mechanisms of action. Antimicrob Agents Chemother. 1990;34:183–188. doi: 10.1128/aac.34.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1992;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 3.Firestine S M, Poon S-W, Mueller E J, Stubbe J, Davisson V J. Reactions catalyzed by 5-aminoimidazole ribonucleotide carboxylases from Escherichia coli and Gallus gallus: a case for divergent catalytic mechanisms. Biochemistry. 1994;33:11927–11934. doi: 10.1021/bi00205a031. [DOI] [PubMed] [Google Scholar]
- 4.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 6.Georgopapadakou N H, Walsh T J. Human mycoses: drugs and targets for emerging pathogens. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 7.Ghannoum M A, Spellberg B, Saporito-Irwin S M, Fonzi W A. Reduced virulence of Candida albicans PHR1 mutants. Infect Immun. 1995;63:4528–4530. doi: 10.1128/iai.63.11.4528-4530.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahim A S, Mirbod F, Filler S G, Banno Y, Cole G T, Kitajima Y, Edwards J E, Jr, Nozawa Y, Ghannoum M A. Evidence implicating phospholipase as a virulence factor in Candida albicans. Infect Immun. 1995;63:1993–1998. doi: 10.1128/iai.63.5.1993-1998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joly V, Bolar J, Yeni P. In vitro models for studying toxicity of antifungal agents. Antimicrob Agents Chemother. 1992;36:1799–1804. doi: 10.1128/aac.36.9.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirsch D R, Whitney R R. Pathogenicity of Candida albicans auxotrophs in experimental infections. Infect Immun. 1991;59:3297–3300. doi: 10.1128/iai.59.9.3297-3300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurtz M B, Cortelyou M W, Miller S M, Lai M, Kirsch D R. Development of autonomously replicating plasmids for Candida albicans. Mol Cell Biol. 1987;7:209–217. doi: 10.1128/mcb.7.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon-Chung K J, Hill W B. Studies on the pink, adenine-deficient strains of Candida albicans. 1. Cultural and morphological characteristics. Sabouraudia. 1970;8:48–59. [PubMed] [Google Scholar]
- 13.Perfect J R, Toffaletti D L, Rude T H. The gene encoding phosphoribosylaminoimidazole carboxylase (ade2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61:4446–4451. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulter R T M, Rikkerink E H A. Genetic analysis of red, adenine-requiring mutants of Candida albicans. J Bacteriol. 1983;156:1066–1077. doi: 10.1128/jb.156.3.1066-1077.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reaume S E, Tatum E L. Spontaneous and nitrogen mustard-induced nutritional deficiencies in Saccharomyces cerevisiae. Arch Biochem. 1948;22:331–338. [PubMed] [Google Scholar]
- 16.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rotrosen D, Calderone R A, Edwards J E., Jr Adherence of Candida species to host tissues and plastic surfaces. Rev Infect Dis. 1986;8:73–85. doi: 10.1093/clinids/8.1.73. [DOI] [PubMed] [Google Scholar]
- 18.Schmuke J J, Davisson V J, Gheesling-Mullis K, Bonar S L, Dotson S B. Sequence analysis of the Candida albicans ADE2 gene and physical separation of the two functionally distinct domains of the phosphoribosylaminoimidazole carboxylase. Yeast. 1997;13:769–776. doi: 10.1002/(SICI)1097-0061(19970630)13:8<769::AID-YEA133>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 19.Shepard M G. Pathogenicity of morphological and auxotrophic mutants of Candida albicans in experimental infections. Infect Immun. 1985;50:541–544. doi: 10.1128/iai.50.2.541-544.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratories; 1986. [Google Scholar]
- 21.Smirnov M N, Smirnov V N, Budowsky E I, Inge-Vechtomov S G, Serebrjakov N G. Red pigment of adenine-deficient yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1967;3:299–304. doi: 10.1016/s0006-291x(67)80096-2. [DOI] [PubMed] [Google Scholar]
- 22.Weinberg R A, McWherter C A, Freeman S K, Wood D C, Gordon J I, Lee S C. Genetic studies reveal that myristoylCoA:protein N-myristoyl transferase is an essential enzyme in Candida albicans. Mol Microbiol. 1995;16:241–250. doi: 10.1111/j.1365-2958.1995.tb02296.x. [DOI] [PubMed] [Google Scholar]