Abstract
Legionella pneumophila replicates within alveolar macrophages, and possibly, alveolar epithelial cells and also within protozoa in the aquatic environment. Here we characterize an L. pneumophila mutant defective in the HtrA/DegP stress-induced protease/chaperone homologue and show that HtrA is indispensable for intracellular replication within mammalian macrophages and alveolar epithelial cells and for intrapulmonary replication in A/J mice. Importantly, amino acid substitutions of two conserved residues in the catalytic domain of (H103➛R and S212➛A) and in-frame deletions of either or both of the two conserved PDZ domains of HtrA abolish its function. Interestingly, the htrA mutant exhibits a parental-type phenotype in intracellular replication within the protozoan host Acanthamoeba polyphaga. We used a promoterless lacZ fusion to the htrA promoter to probe the phagosomal microenvironment harboring L. pneumophila within macrophages and within A. polyphaga for the exposure to stress stimuli. The data show that expression through the htrA promoter is induced by 12,000- to 20,000-fold throughout the intracellular infection of macrophages but its induction is by 120- to 500-fold within protozoa compared to in vitro expression. Data derived from confocal laser scanning microscopy reveal that in contrast to the parental strain, phagosomes harboring the htrA mutant within U937 macrophages colocalize with the late endosomal-lysosomal marker LAMP-2, similar to killed L. pneumophila. Coinfection experiments examined by confocal laser scanning microscopy show that in communal phagosomes harboring both the parental strain and the htrA mutant, replication of the mutant is not rescued, while replication of a dotA mutant control, which is normally trafficked into a phagolysosome, is rescued by the parental strain. Our data show, for the first time, that the stress response by L. pneumophila (mediated, at least in part, by HtrA) is indispensable for intracellular replication within mammalian but not protozoan cells.
Legionella pneumophila is a gram-negative, facultative intracellular bacterium that is the causative agent of Legionnaires' disease, a potentially lethal pneumonia. Ubiquitous in the aquatic environment as a parasite of protozoa, the bacteria are transmitted to humans upon inhalation of contaminated aerosols generated by mechanical devices, such as whirlpools, cooling towers, and showerheads (7).
Within the lungs, L. pneumophila replicates intracellularly within macrophages and possibly alveolar epithelial cells (16, 27). Intracellular replication occurs within a phagosome that is blocked from maturation along the “default” endosomal-lysosomal degradation pathway and is surrounded by a ribosome-studded multilayer membrane (1, 34). The ability of L. pneumophila to modulate the biogenesis of its phagosome into this idiosyncratic niche is controlled by the Dot/Icm type IV-like secretion system (40, 48). Interestingly, the life cycle of L. pneumophila within protozoa is highly similar to that within mammalian cells at the morphological and molecular levels (3, 7, 13, 26, 32, 41). In both host cell types, intracellular replication results in lysis of the host cell. Killing of macrophages and alveolar epithelial cells occurs in two phases (21); first, through caspase-3-mediated induction of apoptosis during early stages of the infection (20, 22–24, 28), followed by pore formation-mediated egress of the host cell upon termination of replication (11). In contrast, killing of amoebae does not involve apoptosis, but pore-forming toxin-mediated cytolysis is essential for killing and exiting the protozoan cell (23, 32).
During the course of the infection, intracellular pathogens respond to changes in their microenvironment by a dramatic phenotypic modulation (2, 8, 9, 31). This response is most likely dictated by the nature of the phagosomal microenvironment, which seems to be distinct between intracellular pathogens (8, 31). Although the mechanisms by which intracellular bacterial pathogens modulate the biogenesis of their vacuoles into idiosyncratic replicative niches are now better understood, the biochemical nature of the replicative niches inhabited by intracellular pathogens is not known.
Because the ability to replicate intracellularly within host cells is central to its pathogenesis, L. pneumophila must be able to efficiently adapt to its intracellular microenvironment. In response to its unique intracellular niche within mammalian macrophages, L. pneumophila undergoes a profound phenotypic modulation, increasing expression of at least 30 proteins. At least 13 of these proteins, including GroEL/Hsp60, GroES, and GspA, are also induced by various in vitro stress stimuli (1, 5, 6). This indicates that, although blocked from acidification and endosomal maturation, L. pneumophila encounters a hostile microenvironment within the phagosome (8, 31). The role of the stress response by L. pneumophila in its adaptation to the intracellular microenvironment within macrophages is not known. Whether L. pneumophila manifests a stress response within protozoa and the role of this response in intracellular replication are also not known.
In this study we show that the htrA/degP homologue of L. pneumophila is indispensable for intracellular replication within mammalian cells in vitro and in vivo, yet is dispensable for the intracellular infection of protozoa. Our data provide evidence that, despite the similarity in the intracellular infection of mammalian and protozoan cells by L. pneumophila, significant differences exist in the phagosomal microenvironment of the two evolutionarily distant hosts.
MATERIALS AND METHODS
Bacterial strains, cultures, and vectors.
The virulent strain of L. pneumophila (AA100/130b) is a clinical isolate and has been described previously (4). The GL10 and GT20 mutants have been described previously (25, 26). L. pneumophila strains were grown on buffered charcoal yeast extract (BCYE) agar plates or in buffered yeast extract (BYE) broth, supplemented with kanamycin for the mutants. The plasmid PBC-SK+ was used to subclone L. pneumophila DNA (Stratagene, La Jolla, Calif.). Escherichia coli strain DH5α (BRL, Gaithersburg, Md.) was used for the majority of cloning experiments. The L. pneumophila chromosomal cosmid library has been previously described (44, 45). The plasmid pUC-4K was purchased from Pharmacia (Piscataway, N.J.) and was the source of the kanamycin resistance gene used as a probe for Southern hybridization. Plasmid pLP101 was a pBC-SK+-based plasmid with a 3.9-kb PstI-XbaI insert containing the htrA gene. To generate pLP102, the pLP101 plasmid was digested with NdeI and XbaI to liberate 1.6 kb of DNA downstream of the htrA gene. The noncompatible ends were blunt ended, and the plasmid was religated. Plasmid pLPHtrA was a pBC-SK+-based plasmid containing a 2.1-kb PCR-generated insert with the htrA gene. Plasmid pAM239 containing the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible gfp gene has been described previously (17). Plasmids pH103R, pS212A, pΔPDZ1, pΔPDZ2, and pΔPDZ1+2 were constructed by splice overlap extension PCR and cloned into pBC-SK+ (see below). phtrA-lacZ was constructed by cloning a 4-kb BamHI-EcoRI fragment containing the lacZ gene from pEU730 (19) into the low-copy vector pTLP6 (Cmr) (44), and the resulting plasmid was designated placZ. To generate the promoter fusion construct, the htrA promoter was amplified by PCR using pfu polymerase. Primers used to amplify the htrA promoter were pP1Xb (5′-GGCTGCAGAACCAATCTGG-3′) and pP2BH (5′-GCGGATCCTTATTAATCCTGATTTTACTCATTAATCC-3′). The resulting fragment was ligated into placZ (designated phtrA-lacZ) so that the promoter was inserted immediately upstream of the lacZ gene.
DNA and RNA manipulations.
All DNA manipulations were performed as previously described (44). RNA extraction for Northern analysis was performed using TriReagent (Gibco-BRL) exactly as prescribed by the manufacturer. Transfer of RNA to nitrocellulose was performed according to standard protocols (39). PCR-generated DNA probes for Northern hybridizations were radiolabeled with [32P]-dCTP, and hybridization and detection were performed as described elsewhere (39).
Sequencing was carried out by the University of Kentucky Macromolecular Structure Analysis Facility (Lexington, Ky.). Sequence analysis and comparisons were performed using MacVector (Oxford Molecular Group, Inc., Campbell, Calif.), AssemblyLign, BlastX, GCG SeqWeb, and ProfileScan. Oligonucleotide synthesis for PCR and sequencing was performed by Integrated DNA Technologies Inc. (Coralville, Calif.).
Construction of point mutations and in-frame deletions.
Splice overlap extension PCR was performed using pfu polymerase (Stratagene, La Jolla, Calif.) as we previously described to generate various mutants (6). PCR primers used for the synthesis of pS212A were pCATA-Ps (5′-GGCTGCAGAACCAATCTGG-3′), pS212A-B (5′-CCAAAGCACCTCCTGCATTACCGG-3′), pS212A-C (5′-CCCGGTAATGCAGGAGGTGCTTTGG-3′), and pCATD-Xb (5′-GCTCTAGATGTGAAATGGTGGTATGGG-3′). The underlined bold residues are the point mutations. The PCR product of the pCATA-Ps and pS212A-B primer pair and the pS212A-C and pCATD-Xb primer pair were mixed and amplified to generate S212A-AD. PCR primers used for the synthesis of pH103R were pCATA-Ps (described above), pH103R-B (5′-CGAATAACACGGTCATTCGTTATAATAATGC-3′), pH103R-C (5′-GCATTATTATAACGAATGACCGTGTTATTCG-3′), and pCATD-Xb (described above). The underlined bold residues are the point mutations. The PCR products of the pCATA-Ps and pH103R-B primer pair and the pH103R-C and pCATD-Xb primer pair were mixed and amplified to generate H103R-AD. PCR primers used to generate the deletion construct pΔPDZ1 were pCATA-Ps (described above), pΔPDZ1-B (5′- GATATCTGTGACTTGCTGCGCTACATCTTTTGCCATATTAATTGG-3′), pΔPDZ1-C (5′-GTAGCGCAGCAAGTCACAGATATCAAAAAACATGAA CAAAAATTACAATCC-3′), and pCATD-Xb (described above). The two PCR products were mixed and amplified to generate pΔPDZ1-AD. PCR primers used to generate the deletion construct pΔPDZ2 were pCATA-Ps (described above), pΔPDZ2-B (5′-GGATAAGAAATTTATGTAAGCGGCTTATTATCCCGC-3′), pΔPDZ2-C (5′-GCCGCTTACATAAATTTCTTATCCTAATAACGAAATAACGC-3′), and pCATD-Xb (described above). The two PCR products generated by the pCATA-Ps and pΔPDZ2-B primer pair and the pΔPDZ2-C and pCATD-Xb primer pair were mixed and amplified to generate pΔPDZ2-AD. PCR primers used to generate the deletion construct pΔPDZ1+2 were pCATA-Ps (described above), pΔPDZ-B (5′-GGATAAGAAATTTATTGCTGCGCTACATCTTTTGCCATATTAATTGG-3′), and pΔPDZ-C (5′-GTAGCGCAGCAATAAATTTCTTATCCTAATAACGAAATAACGC-3′). The two PCR products generated by the pCATA-Ps and pΔPDZ-B primer pair and the pΔPDZ-C and pCATD-Xb primer pair were mixed and amplified to generate pΔPDZ-AD. Each mutant construct generated by PCR was confirmed by sequencing.
Infection protocol.
Macrophage-like U937 cells and human type I alveolar epithelial cells (WI-26 VA4; ATCC CCL-95.1) and axenic A. polyphaga were maintained and used for infections as described previously (26, 27). Infections to determine growth kinetics and cytotoxicity were performed as described previously (25). A multiplicity of infection (MOI) of 0.5 was used to determine bacterial growth kinetics and cytopathogenicity in U937 macrophage-like cells, WI-26 alveolar epithelial cells, and A. polyphaga. The number of intracellular bacteria was determined as we previously described (26). At least three independent experiments, in triplicate, were performed for all infections.
An MOI of 1 to 20 was used in laser scanning confocal microscopy experiments for the determination of LAMP-2 colocalization (see below). Coinfection experiments were performed as described previously (17) using AA100 at an MOI of 5, 10, 20, or 40 and GT20(pAM239) or GL10(pAM239) was added simultaneously with AA100 at an MOI of 20, 40, 80, or 160. An MOI of 50 was used for DNA fragmentation assays (see below). An MOI of 10 in six-well plates was used for lacZ promoter fusion studies. At least three independent experiments, in triplicate, were performed.
Antibodies, stains, and laser scanning confocal microscopy.
Confocal laser scanning microscopy and sample analysis using monoclonal anti-LAMP-2 (H4B4) antibody and differential detection of intracellular and extracellular bacteria were performed exactly as we described previously (29). For colocalization experiments of the two strains, all bacteria were labeled with polyclonal anti-L. pneumophila antibody and stained with secondary antibody labeled with Alexa Red or indocarbocyanine (red pseudocolor). Mutant strains harbored the plasmid pAM239 encoding gfp to differentiate them from AA100. Triplicate samples from at least three independent experiments were analyzed.
Promoter expression assays.
AA100 harboring phtrA-lacZ or placZ was used to infect U937 cells or A. polyphaga cells at an MOI of 10 for 1 h, followed by 1 h in gentamicin (50 μg/ml) to kill extracellular bacteria (26). At the end of each time interval, U937 monolayers were lysed hypotonically, and A. polyphaga monolayers were lysed with 0.05% Triton X-100 and treated with 10 μl of 10-mg/ml DNase I for approximately 20 min. The bacteria and cellular debris were pelleted and washed twice with Z-buffer (40 mM NaH2PO4 · H2O, 60 mM Na2HPO4 · 7H2O, 10 mM KCl, 1 mM MgSO4 · H2O, 40 mM β-mercaptoethanol [pH 7]), followed by assay for β-galactosidase activity using 7 mM fluorodiisogalactopyranoside (FDG);(Molecular Probes, Eugene, Oreg.), as described previously (36). The fluorescence was measured using a Perkin Elmer luminescence spectrometer LS50B at an excitation wavelength of 488 nm and an emission wavelength of 520 nm. The β-galactosidase activity of each sample was measured as the increase in fluorescence over time per CFU and calculated as (Δfluorescence per minute per CFU × 5 × dilution factor), as described previously (42). Background fluorescence, determined as the increase in fluorescence generated by the AA100(plac) control under identical conditions, was subtracted from the final calculated value at each time point. The data are presented as the ratio of arbitrary fluorescence units per CFU of intracellular bacteria to arbitrary fluorescence units per CFU of in vitro-grown bacteria.
Infection of A/J mice.
A/J mice were inoculated intratracheally with 106 CFU of L. pneumophila strains, as we previously described (27). The CFU from homogenized lungs were enumerated on BCYE agar containing polymyxin B, cefamandole, and anisomycin (BCYE+PAC; Becton Dickinson, Cockeysville, Md.) after 3 to 4 days of incubation.
DNA fragmentation analysis and hemolysis assays.
DNA fragmentation analysis was performed at 3 h postinfection as we previously described (21). Contact-dependent hemolysis of sheep red blood cells was performed as previously described (11).
Sensitivity to in vitro stress.
To examine sensitivity to NaCl, bacteria were grown in BYE broth in a 37°C shaker incubator to post-exponential phase (optical density at 550 nm [OD550] = 2.1). Cultures were diluted to an OD550 of 1 and plated on BCYE plates in the presence or absence of 0.6% NaCl for enumeration of bacteria as we described before (26).
To examine sensitivity to heat stress, bacteria grown in BYE broth to post-exponential phase were diluted to an OD550 of 1.0, and aliquots were incubated in triplicate at 37 or 53°C for 1 h, as described previously (6). Following incubation, aliquots were plated on BCYE plates for enumeration of viable bacteria.
To examine sensitivity to oxidative stress, post-exponential-phase broth-grown bacteria were resuspended at an OD550 of 1.0 in BYE broth, pelleted, and resuspended in various concentrations of H2O2 for 30 min at 37°C (6). Following incubation, aliquots were plated on BCYE for enumeration of viable bacteria. Alternatively, bacteria were spread on BCYE plates to generate a lawn of bacteria, and a filter disk saturated with 1 μl of 30% H2O2 was placed in the center of the plate. After 72 h of incubation, the zone of inhibition around the filter disk was measured. At least three independent experiments, in triplicate, were performed.
RESULTS
Intracellular growth of GT20 within mammalian and protozoan cells.
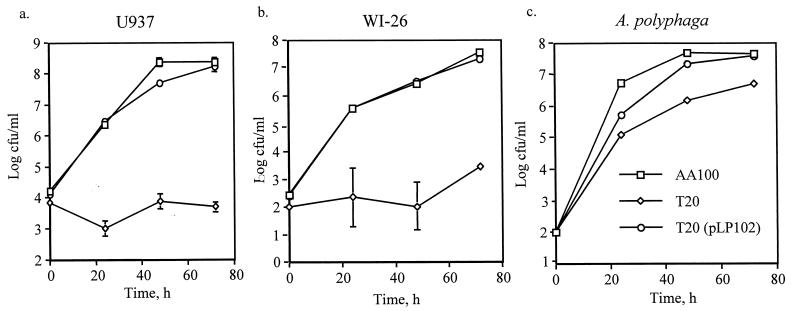
Initially, characterization of the 121 transposon insertion mutants of L. pneumophila (25, 26) was performed using L. pneumophila grown in broth to an OD550 of ∼1 (mid-exponential phase) and showed that GT20 was largely killed by the macrophages by 48 h postinfection (25). Recently, it has been shown that post-exponential-phase L. pneumophila exhibit enhanced virulence compared to mid-log-phase bacteria (14). Therefore, we reexamined the phenotype of GT20 using 3-day plate-grown bacteria or post-exponential-phase broth-grown bacteria (OD550 of 2.1 to 2.2). Under these growth conditions, the mutant GT20 remained severely defective for intracellular replication within U937 macrophages (Fig. 1a) and WI-26 epithelial cells (Fig. 1b). In contrast, post-exponential-phase-grown GT20 was able to replicate within the protozoan host A. polyphaga. There were approximately 10-fold-fewer GT20 compared to AA100 by 72 h postinfection (Fig. 1c), but this difference was not due to an increased sensitivity of GT20 to the A. polyphaga assay buffer used throughout the infection process (data not shown). In vitro growth of GT20 in BYE broth and on solid medium was similar to that of the parental strain (data not shown).
FIG. 1.
Intracellular growth kinetics of GT20 and the parental strain AA100 in (a) U937 macrophage-like cells, (b) WI-26 alveolar epithelial cells, and (c) A. polyphaga. Strain GT20(pLP102) is a plasmid-complemented clone of GT20. These data are representative of at least three independent experiments performed in triplicate. The absence of error bars indicates very small standard deviations that could not be displayed.
Cytopathogenicity to U937 macrophage-like cells.
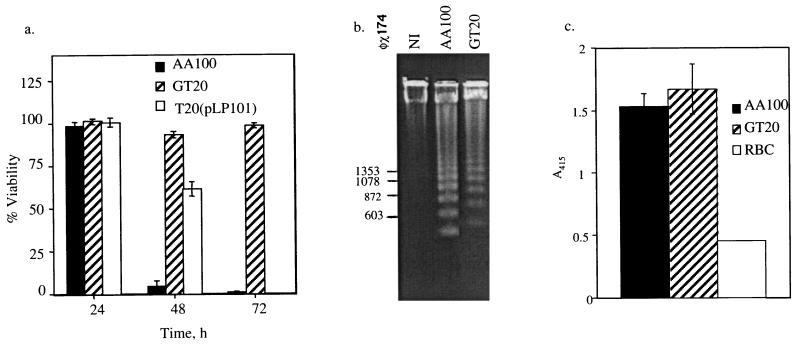
Intracellular replication of L. pneumophila within the host cell is associated with killing of the host cell. The data showed that while AA100 was ∼100% cytopathogenic by 72 h, GT20 was completely noncytopathogenic (Fig. 2a). Since both the induction of apoptosis and the pore-forming activity of L. pneumophila contribute to killing of mammalian cells (11, 21), we examined both modes of killing by GT20. Our results showed that GT20 was as effective at inducing DNA fragmentation, consistent with apoptosis, as the wild-type strain AA100 (Fig. 2b). In addition, GT20 and AA100 exhibited similar levels of contact-dependent pore formation when examined by hemolysis of sheep red blood cells (Fig. 2c). Taken together, the data indicated that the noncytopathogenicity of GT20 was due to the severe defect in intracellular replication.
FIG. 2.
Cytopathogenic defect of GT20 is due to the defect in intracellular replication. (a) Cytopathogenicity of GT20 and the parental strain AA100 to U937 macrophage-like cells as determined by Alamar Blue dye reduction at various time points following infection at an MOI of 0.5. Strain GT20(pLP102) is a plasmid-complemented clone of GT20. Percent killing was normalized to uninfected cells, which were considered 100% viable. These data are representative of at least three independent experiments performed in triplicate. (b) Induction of apoptosis as evidenced by DNA fragmentation in U937 macrophages following 3 h of incubation after 1 h of infection at an MOI of 50. The φχ size standard is shown on the left. (c) Contact-dependent hemolysis of sheep red blood cells following 1 h of incubation at a bacterium-blood cell ratio of 25:1. Hemolysis was measured spectrophotometrically at A415. RBC represents red blood cells incubated in the absence of bacteria. These data are representative of at least three independent experiments performed in triplicate. The absence of error bars indicates very small standard deviations that could not be displayed.
Intrapulmonary replication in A/J mice.
To examine the virulence of GT20 in vivo, A/J mice were intratracheally inoculated with 106 bacteria of either the wild-type AA100 or the mutant GT20 strain. As shown in Table 1, by 48 h postinfection, while there was at least a 100-fold increase in the number of bacteria recovered from the lungs of the mice infected by AA100, there was a 10-fold decrease in the number of GT20 recovered. In addition, while AA100 persisted through 7 days postinfection, GT20 was effectively cleared by 72 h postinfection. The data showed that the severely defective phenotype of GT20 within macrophages and alveolar epithelial cells in vitro correlated with its defect in intrapulmonary replication.
TABLE 1.
Intrapulmonary replication in micea
| Postinfection time | AA100 (CFU/lung) | GT20 (CFU/lung) |
|---|---|---|
| 0 h | 1 × 106 | 9 × 105 |
| 1.5 × 106 | 1 × 106 | |
| 9 × 105 | 9 × 105 | |
| 1 day | 6 × 107 | 5 × 104 |
| 6 × 107 | 8 × 104 | |
| 3 × 107 | 8 × 104 | |
| 2 days | 4 × 108 | 1.4 × 104 |
| 6 × 108 | 1 × 104 | |
| 5 × 108 | 1 × 104 | |
| 3 days | 9.6 × 104 | 2 × 102 |
| 1 × 105 | 5 × 102 | |
| 8 × 104 | 8 × 101 | |
| 7 days | 2 × 103 | 0 |
| 2 × 103 | 0 | |
| 2 × 103 | 0 |
A/J mice were intratracheally inoculated with 106 bacteria per strain, and at the indicated time points, three mice were sacrificed and CFU in the lung were determined.
Genetic characterization and complementation of the defective locus in GT20.
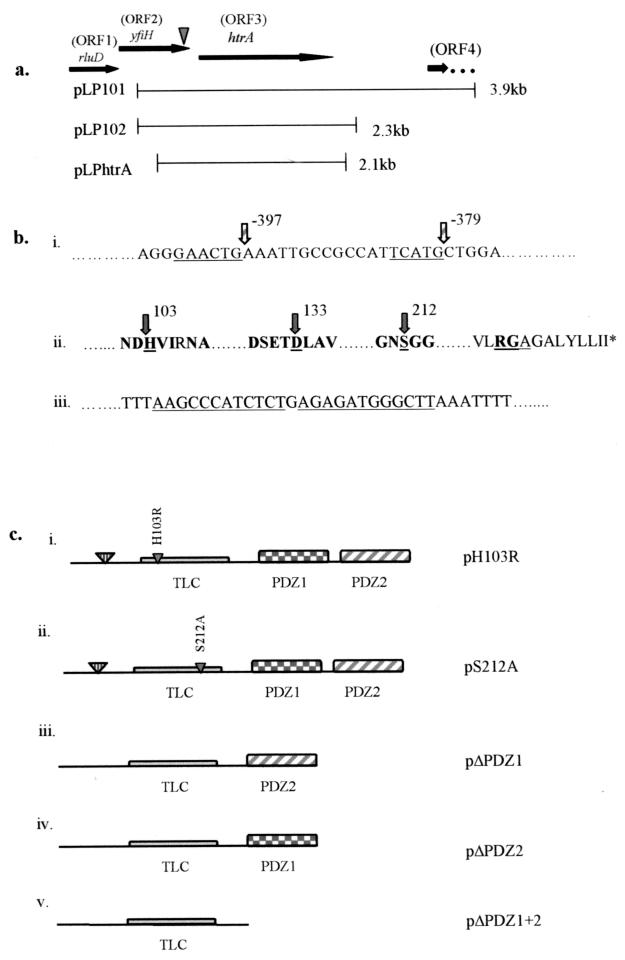
A chromosomal EcoRI fragment containing the mini-Tn10::kan cassette and flanking Legionella DNA was subcloned from GT20 genomic DNA, and the regions flanking the kan cassette were sequenced. The kan cassette was inserted within the C terminus of a 726-nucleotide open reading frame (ORF), ORF 2, 22 nucleotides upstream of the termination codon (Fig. 3a). ORF 2 was preceded immediately upstream by a 474-nucleotide ORF (ORF 1) and followed, 17 nucleotides downstream, by a 1,377-nucleotide ORF (ORF 3). A perfect inverted repeat was located 42 nucleotides downstream of the termination codon of ORF 3, indicating rho-independent termination (Fig. 3b). The next downstream ORF (ORF 4) was located 1,043 nucleotides from the termination codon of ORF 3 (Fig. 3a).
FIG. 3.
Genetic analysis of the disrupted locus in GT20. (a) ORFs and the fragments which were subcloned into pBC to generate plasmids pLP101, pLP102, and pLPHtrA. The inverted arrowhead indicates the location of the kan insert. (b) Conserved sequence elements within promoter regions and predicted HtrA amino acid sequence. (i) Underlined nucleotides indicate the putative ςE −35 and −10 sequences located upstream of the htrA start codon. (ii) Conserved amino acids within the putative catalytic triad as well as conserved surrounding amino acids are indicated in bold. The catalytic triad residues (H, D, and S) are underlined, with arrows and amino acids numbered. The RGA motif is underlined, and the stop codon is indicated by an asterisk. (iii) The perfect direct inverted repeat sequences signifying rho-independent termination downstream of the stop codon are underlined. (c) Conserved domains of HtrA are indicated. The signal sequence cleavage site is indicated by the large inverted triangle, followed by the trypsin-like catalytic domain (TLC) and tandem PDZ domains, designated PDZ1 and PDZ2. Mutant constructs generated are described as follows: (i and ii) Point mutations H103R and S212A, generated by overlap extension PCR to produce the mutant constructs pH103R and pS212A, are indicated by small inverted triangles within the trypsin-like catalytic domain. (iii) Deletion mutant pΔPDZ1, which lacks PDZ domain 1. (iv) Deletion mutant pΔPDZ2, which lacks PDZ domain 2. (v) pΔPDZ1+2, which lacks both PDZ domains.
Comparison of the predicted amino acid sequences of these ORFs with other proteins using the BlastX program revealed that the protein encoded by ORF 1 exhibited significant similarity to Haemophilus influenzae and E. coli RluD, the ribosomal large-subunit pseudouridine synthase D (45% identity and 58% similarity). The protein encoded by ORF 2 exhibited 42% identity and 61% similarity to an E. coli protein of unknown function designated YfiH, which is also present in Mycobacterium tuberculosis (30% identity, 48% similarity). The protein encoded by ORF 3 exhibited similarity to the stress-induced protein HtrA/DegP found in other gram-negative bacteria, including E. coli (42% identity, 60% similarity) and Brucella abortus (40% identity, 59% similarity).
Three cosmid clones harboring the disrupted locus in GT20 were isolated and confirmed by Southern hybridization to contain the corresponding DNA fragment (data not shown). Each of the cosmid clones fully complemented GT20 for intracellular growth and cytopathogenicity to macrophages (data not shown).
The kan insertion of GT20 was located within the 3′ end of yfiH (ORF 2), 42 nucleotides upstream of the htrA (ORF 3) initiation codon (Fig. 3a). Therefore, a 3.9-kb XbaI-PstI restriction fragment containing htrA and 561 nucleotides of DNA upstream of the initiation codon but lacking the complete yfiH gene was subcloned and designated pLP101 (Fig. 3a). The results showed that pLP101 was sufficient to completely complement the intracellular growth and cytopathogenicity defects of GT20 (data not shown). Since a noncoding intervening 1,043 nucleotides were present between htrA and the next downstream ORF, a DNA fragment harboring htrA, 561 nucleotides upstream of the predicted htrA start codon, and 397 nucleotides of noncoding sequence downstream from the termination codon of htrA was subcloned and designated pLP102 (Fig. 3a). Similar to pLP101, this plasmid completely complemented intracellular growth and cytopathogenicity to U937 macrophages (Fig. 1a and 2a). In addition, Northern analysis of mRNA produced by the wild-type strain AA100, GT20, and GT20 complemented with pLP102 demonstrated the absence of htrA mRNA in GT20 (data not shown). These data showed that the defective phenotypes of GT20 were due to the disruption of htrA expression.
In other bacteria, the htrA gene has been shown to be regulated by the alternative sigma factor ςE (37). Within the sequence upstream of the putative L. pneumophila htrA start codon, a putative ςE promoter region was identified (Fig. 3b) (37). Interestingly, the plasmid pLPHtrA, which lacked the putative −35 region of the predicted ςE promoter, did not complement the defective phenotype of GT20 (data not shown), suggesting a role for this putative promoter in the expression of htrA (Fig. 3a). Further promoter studies, however, must be performed to confirm the role of this putative promoter in the expression of htrA.
Mutational analyses of the conserved regions of HtrA and their effects on function.
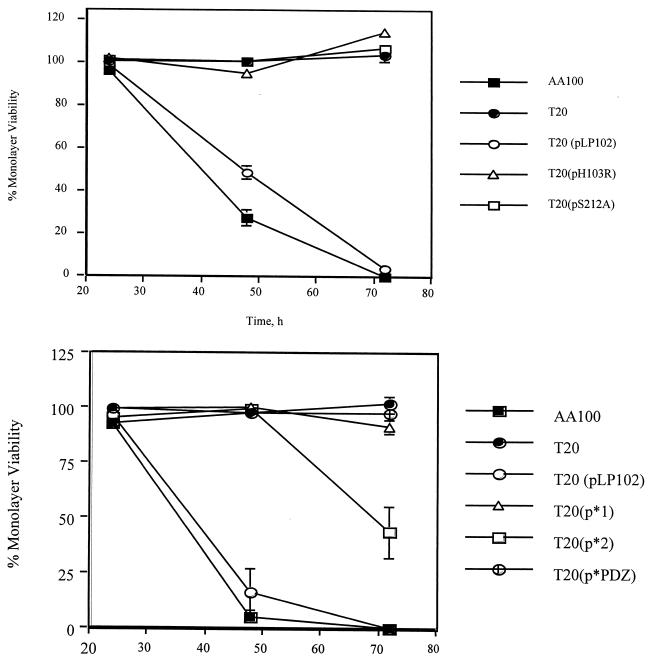
Analysis of the amino acid sequence of the htrA gene revealed the presence of certain features conserved in HtrA that were also conserved in the L. pneumophila HtrA homologue (Fig. 3b and c). These include a hydrophobic leader peptide predicted to be cleaved between residues 23 and 24, an RGD domain, two tandem PDZ domains, and a trypsin-like catalytic domain (Fig. 3c) containing three conserved residues, His, Asp, and Ser, which constitute the catalytic triad (38). We generated amino acid substitutions of the His103 and Ser212 conserved catalytic residues to Arg and Ala, respectively, resulting in the pLP102-based constructs pH103R and pS212A, respectively (Fig. 3c). We also generated in-frame deletions of PDZ1, PDZ2, or both, which were designated pΔPDZ1, pΔPDZ2, and pΔPDZ1+2, respectively (Fig. 3c). The fidelity of all of these constructs was confirmed by DNA sequencing. The single amino acid substitutions as well as the deletion of both PDZ domains (pΔPDZ1+2) or PDZ1 alone (pΔPDZ1) abolished the ability of the htrA gene to complement the intracellular replication defect of GT20 (Fig. 4a and b). However, pΔPDZ2 partially complemented GT20 (Fig. 4b). These data confirmed that these conserved residues are essential for the function of L. pneumophila HtrA during the intracellular infection of macrophages. Although deletion of the PDZ domains in E. coli htrA does not result in unstable proteins (43), we cannot exclude that similar deletions in L. pneumophila htrA may result in unstable proteins.
FIG. 4.
Complementation of GT20 cytopathogenicity defect to macrophages by htrA mutant constructs, with monolayer viability measured by Alamar Blue dye reduction following infection at an MOI of 0.5. (a) Complementation of GT20 (T20) by mutant constructs harboring point mutations within the catalytic domain. (b) Complementation of GT20 (T20) by PDZ deletion mutant constructs. These data are representative of at least three independent experiments.
Increased sensitivity of GT20 to certain in vitro stress stimuli.
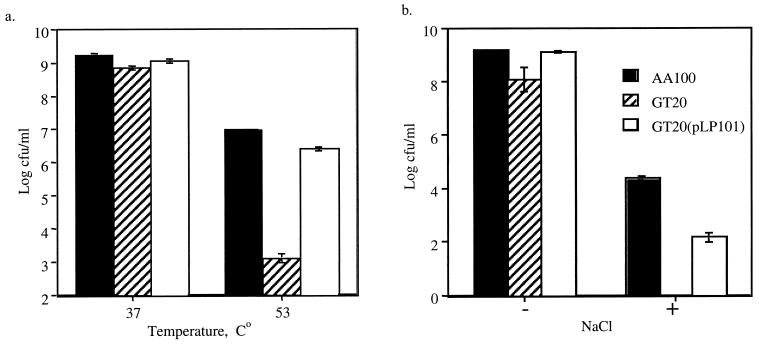
In E. coli, HtrA has been shown to be a periplasmic chaperone/serine protease involved in refolding or degrading misfolded proteins in the periplasm (38) and is indispensable for survival at elevated temperatures (>42°C) (38). Our data showed that GT20 exhibited no difference in viability compared to the wild-type strain AA100 when incubated at 42°C (data not shown). At 53°C, the viability of GT20 was reduced by ∼10,000-fold compared to the wild-type strain (Fig. 5a). In addition, GT20 exhibited significantly higher growth inhibition (>10,000-fold) in the presence of 0.6% NaCl than the wild-type AA100 (Fig. 5b). These temperature and salt sensitivities were complemented by the htrA gene in trans. In contrast, AA100 and GT20 exhibited similar sensitivity to oxidative stress (zones of inhibition by H2O2 were 37.375 ± 1.411, 36.89 ± 0.536, and 37.75 ± 1.494 mm, respectively). In addition, GT20 did not display any increased sensitivity when incubated for 16 h at pH 3, 5.2, or 6.2 compared to the wild-type strain (data not shown).
FIG. 5.
Resistance to in vitro stress stimuli. (a) Viability following 1 h of incubation at 37 or 53°C. (b) Growth inhibition on BCYE agar with (+) or without (−) 0.6% NaCl. Strain GT20(pLP101) is the plasmid-complemented mutant of GT20. These data are representative of at least three independent experiments performed in triplicate.
Intracellular trafficking of GT20.
Laser scanning confocal microscopy was used to examine colocalization of the GT20 phagosomes with the late endosomal-lysosomal marker LAMP-2 at 2 h postinfection of U937 macrophages. The visual assessment of colocalization was corroborated by measurement of fluorescence intensity across the phagosome (29). Colocalization was determined by the presence of two completely overlapping fluorescence peaks (one green for LAMP-2 and one blue for intracellular bacteria) (see, for example, Fig. 4 in reference 29). Of 59 phagosomes examined, 86.5% of the GT20 phagosomes colocalized with LAMP-2. In contrast, of 75 phagosomes examined, only 17.3% of the AA100 phagosomes colocalized with LAMP-2. As expected, of 67 phagosomes examined, 90% of the paraformaldehyde-killed bacterial phagosomes colocalized with LAMP-2. Therefore, the inability of GT20 to replicate intracellularly was associated with a defect in intracellular trafficking.
htrA mutant is unable to replicate within a niche created by parental strain.
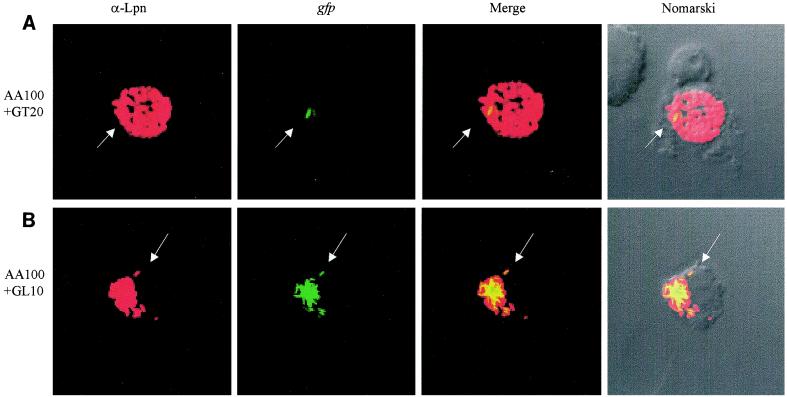
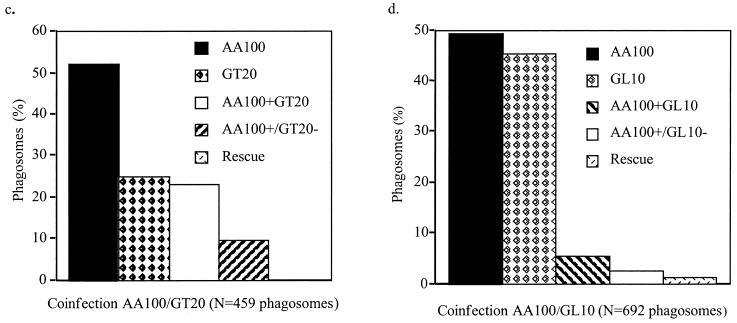
To examine whether the defect in trafficking of the htrA mutant was due to intolerance of the microenvironment within the phagosome, we coinfected U937 macrophages with the wild-type strain AA100 and either the htrA mutant GT20 or the dotA mutant GL10 derivative of AA100 (26) to examine whether colocalization with AA100 could rescue each mutant for intracellular replication. Coinfections were performed using AA100 and gfp-expressing GT20 or GL10. Phagosomes were examined at 2, 10, and 16 h postinfection using laser scanning confocal microscopy. As expected, at 2 h there was no evidence of intracellular replication of any of the strains (data not shown). In single infections, there was no evidence of replication of GT20 or GL10 at any time point tested (data not shown). When GT20 or GL10 was present in phagosomes lacking AA100 in the dual infections, there was no detectable replication of either of the mutants at any time point examined (data not shown). In coinfection experiments with AA100 and GL10, of 692 phagosomes examined at 16 h postinfection, 5.5% colocalization of AA100 and GL10 was observed. In 23.6% of the coinhabited phagosomes, rescue of GL10 was clearly observed (Fig. 6b and d). Results were similar at 10 h postinfection (673 phagosomes examined). In coinfection experiments using AA100 and GT20, of 472 phagosomes examined at 16 h postinfection, 23% colocalization of AA100 and GT20 was observed. In contrast to the dotA mutant, no detectable replication of GT20 was ever observed in communal phagosomes (Fig. 6a and c). Similar results were observed at 10 h postinfection (262 phagosomes examined). In qualitative examination of thousands of coinfected cells at 10 h and 16 h postinfection, we never observed any detectable replication of GT20. Our data showed that rescue of the htrA mutant for the defect in intracellular trafficking was not sufficient to rescue its intracellular replication and that the replication defect of the htrA mutant was most likely due to an inability to withstand the phagosomal microenvironment in mammalian cells.
FIG. 6.
Rescue of intracellular replication of the dotA mutant (GL10) but not the htrA mutant (GT20) by the parental strain AA100. Colocalization of (A) AA100 and GT20(gfp) and (B) AA100 and GL10(gfp) within communal phagosomes, determined by scanning laser confocal microscopy. Red, anti-L. pneumophila antiserum (α-Lpn); green, gfp-GT20 or gfp-GL10 (gfp). Arrows indicate GT20 or GL10 (a dotA mutant) which cannot be rescued within a communal phagosome coinhabited by AA100. (c and d) Infected phagosomes were scored as follows. AA100, AA100 alone within a phagosome; GT20 or GL10, mutants alone within a phagosome; AA100+GT20 or AA100+GL10, AA100 and mutants colocalized within the same phagosome; AA100+/GT20− or AA100+/GL10−, colocalization of AA100 and mutants within the same phagosome, with proficient replication of AA100 but no replication of the mutant; Rescue, intracellular replication of mutants within a communal phagosome with AA100. Phagosomes were scored for all macrophages in randomly selected fields. These data are representative of three independent experiments performed in triplicate.
Expression of htrA within macrophages and within protozoa.
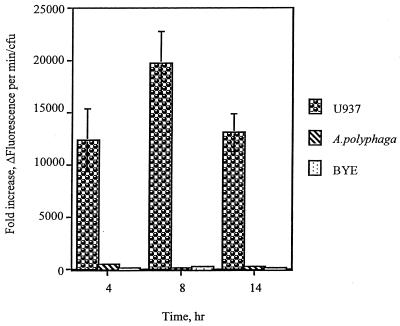
Although it is well documented that L. pneumophila is exposed to stress stimuli within macrophages (8, 31), it has never been examined whether a similar stress response is also manifested within protozoa. This prompted us to use the htrA promoter to probe the phagosomal microenvironment for exposure of L. pneumophila to stress stimuli within protozoa and contrast that to mammalian cells. We constructed a promoter fusion of the L. pneumophila htrA to the promoterless lacZ gene on a low-copy-number vector (designated phtrA-lacZ) to monitor the kinetics of expression of htrA during the intracellular infection of macrophages. The use of FDG fluorescence substrate for β-galactosidase facilitated detection of expression during early stages of the infection, when the number of intracellular bacteria is small. The background of β-galactosidase activity from the vector with the promoterless lacZ gene was subtracted from the β-galactosidase activity of phtrA-lacZ under the same experimental conditions and each time point examined (4, 8, and 14 h). The β-galactosidase activity increased by 12,320- to 20,545-fold throughout the intracellular infection of macrophages compared to bacteria incubated in tissue culture medium or in BYE broth (Fig. 7). There was no significant difference in β-galactosidase expression between in vitro-grown bacteria incubated in BYE and in tissue culture medium or during different phases of growth in BYE (Fig. 7). In addition, the macrophage lysates had no detectable effect on expression of β-galactosidase by in vitro-grown bacteria (data not shown). When AA100 harboring the htrA-lacZ promoter construct was used to infect A. polyphaga, the β-galactosidase activity was induced by 120- to 500-fold compared to bacteria incubated in assay medium or in BYE (Fig. 7). The lysate of amoebae had no detectable effect on expression of β-galactosidase by in vitro-grown bacteria (data not shown). These data showed that although htrA was induced within both macrophages and protozoa, levels were consistently much higher within macrophages (by approximately 100—fold) than in protozoa.
FIG. 7.
Intracellular expression of htrA. β-Galactosidase activity, measured by hydrolysis of the fluorescence substrate FDG, resulting from the expression of lacZ driven by the htrA promoter during the intracellular infection of macrophages and A. polyphaga and compared to bacteria grown in BYE. Background fluorescence in arbitrary units per CFU generated by a promoterless lacZ construct was subtracted at each time point. The values shown are the ratios of the fluorescence units of intracellular bacteria to those in in vitro-grown bacteria incubated in tissue culture medium. The absence of error bars indicates error values too small to be shown. At least two independent experiments, in duplicate, were performed.
DISCUSSION
Despite evasion of the major microbicidal mechanisms of macrophages (35) and the profound modifications of the phagosome (1), L. pneumophila manifests the induction of many stress proteins during intracellular replication (1, 4, 18). However, the role of the stress response in the intracellular survival of L. pneumophila is not known. In addition, whether L. pneumophila is exposed to stress stimuli within protozoa has never been demonstrated. Here we show that a stress-induced gene (htrA) of L. pneumophila is indispensable for the intracellular replication of L. pneumophila within mammalian but not protozoan cells. We show that regardless of the growth phase at which the infection is initiated, the htrA mutant is severely impaired in its ability to replicate intracellularly within macrophages and epithelial cells, yet exhibits a parental strain-like phenotype within protozoa. Importantly, the inability of the htrA mutant to replicate within mammalian cells in vitro correlates with a similar defect in intrapulmonary replication in A/J mice. Similarly, htrA mutants of other bacteria also exhibit a defect in intracellular survival and replication within macrophages and are also attenuated in animal models (12, 15). These observations provide strong evidence that although these intracellular bacteria inhabit distinct niches within their host cells, they are all exposed to stressful conditions within their phagosomal microenvironments (8, 31), but the nature of these stress stimuli is likely to be different for different pathogens.
Although alveolar epithelial cells are not professional phagocytic cells, the htrA mutant is similarly defective for intracellular replication within these cells. Although it is well established that numerous stress proteins are induced by L. pneumophila during the intracellular infection of macrophages, the essential role of HtrA during the intracellular infection of alveolar epithelial cells provides the first evidence that the stress response (mediated at least in part by HtrA) is indispensable for the intracellular survival and replication of L. pneumophila within macrophages and epithelial cells.
We have demonstrated that amino acid substitution of either of two of the conserved catalytic residues, His103 and Ser212, renders the L. pneumophila HtrA nonfunctional during the intracellular infection of macrophages. Similar substitutions in the E. coli HtrA also results in loss of function. Our data also show that deletion of either both PDZ domains or PDZ1 alone completely disrupts the function of HtrA during intracellular infection of macrophages, while deletion of PDZ2 alone resulted in partial function of HtrA in the intracellular infection. Similar deletions in E. coli HtrA result in a similar pattern of loss of function (43). Our data confirm an essential role of the L. pneumophila HtrA during the intracellular infection of macrophages and epithelial cells.
We have shown that the htrA mutant is unable to exclude the late endosomal-lysosomal marker LAMP-2 from its phagosome. In addition, we have previously shown, by electron microscopy, that the GT20 phagosome does not associate with the rough endoplasmic reticulum within macrophages, yet does recruit this organelle during replication within protozoa (25). Therefore, the defect in intracellular replication within macrophages is associated with the inability of the htrA mutant to perform essential modifications of its phagosome at early stages of the infection.
The observation that the L. pneumophila htrA mutant is found in a phagosome that colocalizes with LAMP-2 suggests that the mutant may be unable to properly fold the proteins (including the Dot/Icm proteins) necessary for the rapid establishment of its protective replicative niche, resulting in trafficking of the phagosome along the “default” endosomal-lysosomal degradation pathway (38, 43). It has recently been demonstrated that several dot/icm mutants, including a dotA mutant, which are defective in early phagosomal modulation and are trafficked into a phagolysosome, can be rescued by the parental strain when they occupy a communal phagosome (17). Similarly, we have demonstrated that a dotA mutant can also be rescued when it resides within a communal phagosome with the parental strain. In contrast, the htrA mutant cannot be rescued by the parental strain when they inhabit the same phagosome. Thus, the defect in intracellular trafficking of the htrA mutant is most likely due to an intolerance of the phagosomal microenvironment inhabited by the wild-type strain within the macrophage.
Our data suggest that the intracellular niche inhibited by L. pneumophila within protozoa is less stressful than that of mammalian cells. This is supported by several lines of evidence. First, the number of GT20 recovered at 72 h postinfection of A. polyphaga is consistently 10-fold less than that of AA100, suggesting that L. pneumophila may be exposed to a low level of stress stimuli within protozoa, or that HtrA plays a minor role in this host. In contrast, there is no detectable replication of the htrA mutant within mammalian cells. Second, promoter fusion studies revealed that the induction levels of the htrA promoter are approximately 100-fold higher during the intracellular infection of macrophages compared to protozoa. It is possible that different stress stimuli are encountered by L. pneumophila within protozoa, such that additional proteins induced within protozoa, but not within macrophages, may compensate for the absence of HtrA within protozoa. Our data favor the prediction that it is more likely that L. pneumophila is exposed to a significantly lesser degree of stress stimuli within protozoa than within macrophages. We have recently shown that the Rep helicase of L. pneumophila, which is required for DNA repair following damage by stress stimuli, is also indispensable with mammalian cells but not within protozoa (30). These observations provide compelling evidence that although the intracellular infection is similar at the molecular and ultrastructural levels (3, 13, 26, 41), significant differences exist between the phagosomal microenvironments within protozoan and mammalian host cells. It is possible that the different receptors (10, 33, 46, 47) utilized in both host cells to internalize legionellae result in different signaling events that result in exposure to stress stimuli within mammalian but not protozoan cells.
In summary, we have provided evidence that differences exist in the phagosomal microenvironments within protozoan and mammalian cells. Despite its dispensability within protozoa, HtrA is essential for L. pneumophila replication within macrophages and epithelial cells and for intrapulmonary replication, the hallmarks of Legionnaires' disease. The data show a critical role for the stress response mediated, at least in part, by HtrA, in the intracellular replication of L. pneumophila within mammalian cells.
ACKNOWLEDGMENTS
Y.A.K. is supported by Public Health Service grants R29AI38410 and RO1AI43965.
We thank Craig R. Roy for kindly providing the pAM239 plasmid. We thank Lian-Yong Gao for technical help. We also thank Bruce Maley, Omar S. Harb, Richard Watson, and Mary Gail Engle for technical assistance with laser scanning confocal microscopy.
REFERENCES
- 1.Abu Kwaik Y. Fatal attraction of mammalian cells to Legionella pneumophila. Mol Microbiol. 1998;30:689–696. doi: 10.1046/j.1365-2958.1998.01092.x. [DOI] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection. Infect Immun. 1998;66:203–212. doi: 10.1128/iai.66.1.203-212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y. The phagosome containing Legionella pneumophila within the protozoan Hartmanella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, Eisenstein B I, Engleberg N C. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect Immun. 1993;61:1320–1329. doi: 10.1128/iai.61.4.1320-1329.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu Kwaik Y, Engleberg N C. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress stimuli. Mol Microbiol. 1994;13:243–251. doi: 10.1111/j.1365-2958.1994.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 6.Abu Kwaik Y, Gao L-Y, Harb O S, Stone B J. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol Microbiol. 1997;24:629–642. doi: 10.1046/j.1365-2958.1997.3661739.x. [DOI] [PubMed] [Google Scholar]
- 7.Abu Kwaik Y, Gao L-Y, Stone B J, Venkataraman C, Harb O S. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64:3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abu Kwaik Y, Harb O S. Phenotypic modulation by intracellular bacterial pathogens. Electrophoresis. 1999;20:2248–2258. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2248::AID-ELPS2248>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 9.Abu Kwaik Y, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 10.Abu Kwaik Y, Venkataraman C, Gao L-Y, Harb O S. Signal transduction in the protozoan host Hartmannella vermiformis upon attachment and invasion by its bacterial parasite, the Legionnaires' disease agent, Legionella micdadei. Appl Environ Microbiol. 1998;64:3134–3139. doi: 10.1128/aem.64.9.3134-3139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alli O A T, Gao L-Y, Pedersen L L, Zink S, Radulic M, Doric M, Abu Kwaik Y. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect Immun. 2000;68:6431–6440. doi: 10.1128/iai.68.11.6431-6440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bozue J A, Johnson W. Interaction of Legionella pneumophila with Acanthamoeba catellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne B, Swanson M S. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatfield S N, Strahan K, Pickard D, Charles I G, Hormaeche C E, Dougan G. Evaluation of Salmonella typhimurium strains harbouring defined mutations in htrA and aroA in the murine salmonellosis model. Microb Pathog. 1992;12:145–151. doi: 10.1016/0882-4010(92)90117-7. [DOI] [PubMed] [Google Scholar]
- 16.Cianciotto N P, Stamos J K, Kamp D W. Infectivity of Legionella pneumophila mip mutant for alveolar epithelial cells. Curr Microbiol. 1995;30:247–250. doi: 10.1007/BF00293641. [DOI] [PubMed] [Google Scholar]
- 17.Coers J, Monahan C, Roy C R. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat Cell Biol. 1999;1:451–453. doi: 10.1038/15687. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez R C, Logan S, Lee S H S, Hoffman P S. Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlated with virulence. Infect Immun. 1996;64:1968–1976. doi: 10.1128/iai.64.6.1968-1976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Froehlich B, Husmann L, Caron J, Scott J R. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J Bacteriol. 1994;176:5385–5392. doi: 10.1128/jb.176.17.5385-5392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao L-Y, Abu Kwaik Y. Activation of caspase-3 in Legionella pneumophila-induced apoptosis in macrophages. Infect Immun. 1999;67:4886–4894. doi: 10.1128/iai.67.9.4886-4894.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao L-Y, Abu Kwaik Y. Apoptosis in macrophages and alveolar epithelial cells during early stages of infection by Legionella pneumophila and its role in cytopathogenicity. Infect Immun. 1999;67:862–870. doi: 10.1128/iai.67.2.862-870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao L-Y, Abu Kwaik Y. Hijacking the apoptotic pathways of the host cell by bacterial pathogens. Microb Infect. 2000;2:1705–1719. doi: 10.1016/s1286-4579(00)01326-5. [DOI] [PubMed] [Google Scholar]
- 23.Gao L-Y, Abu Kwaik Y. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ Microbiol. 2000;2:79–90. doi: 10.1046/j.1462-2920.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- 24.Gao L-Y, Abu Kwaik Y. The modulation of host cell apoptosis by intracellular bacterial pathogens. Trends Microbiol. 2000;8:306–313. doi: 10.1016/s0966-842x(00)01784-4. [DOI] [PubMed] [Google Scholar]
- 25.Gao L-Y, Harb O S, Abu Kwaik Y. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect Immun. 1998;66:883–892. doi: 10.1128/iai.66.3.883-892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao L-Y, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao L-Y, Stone B J, Brieland J K, Abu Kwaik Y. Different fates of Legionella pneumophila pmi and mil mutants within human-derived macrophages and alveolar epithelial cells. Microb Pathog. 1998;25:291–306. doi: 10.1006/mpat.1998.0237. [DOI] [PubMed] [Google Scholar]
- 28.Hägele S, Hacker J, Brand B C. Legionella pneumophila kills human phagocytes but not protozoan host cells by inducing apoptotic cell death. FEMS Microbiol Lett. 1998;169:51–58. doi: 10.1111/j.1574-6968.1998.tb13298.x. [DOI] [PubMed] [Google Scholar]
- 29.Harb O S, Abu Kwaik Y. Characterization of a macrophage-specific infectivity locus (milA) of Legionella pneumophila. Infect Immun. 2000;68:368–376. doi: 10.1128/iai.68.1.368-376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harb O S, Abu Kwaik Y. An essential role for the rep helicase of Legionella pneumophila in intracellular infection. Infect Immun. 2000;68:6970–6978. doi: 10.1128/iai.68.12.6970-6978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harb O S, Abu Kwaik Y. Probing the microenvironment of intracellular bacterial pathogens. Microb Infect. 1999;1:445–453. doi: 10.1016/s1286-4579(99)80048-3. [DOI] [PubMed] [Google Scholar]
- 32.Harb O S, Gao L-Y, Abu Kwaik Y. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ Microbiol. 2000;2:251–265. doi: 10.1046/j.1462-2920.2000.00112.x. [DOI] [PubMed] [Google Scholar]
- 33.Harb O S, Venkataraman C, Haack B J, Gao L-Y, Abu Kwaik Y. Heterogeneity in the attachment and uptake mechanisms of the Legionnaires' disease bacterium, Legionella pneumophila, by protozoan hosts. Appl Environ Microbiol. 1998;64:126–132. doi: 10.1128/aem.64.1.126-132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horwitz M A. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob T, Escallier J C, Sanguedolce M V, Chicheportiche C, Bongrand P, Capo C, Mege J L. Legionella pneumophila inhibits superoxide generation in human monocytes via the down-modulation of α and β protein kinase C isotypes. J Leukocyte Biol. 1994;55:310–312. doi: 10.1002/jlb.55.3.310. [DOI] [PubMed] [Google Scholar]
- 36.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 37.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 38.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Segal G, Purcell M, Shuman H A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila chromosome. Proc Natl Acad Sci USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segal G, Shuman H A. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slauch J M, Mahan M J, Mekalanos J J. Measurement of transcriptional activity in pathogenic bacteria recovered directly from infected host tissue. Biotechniques. 1994;16:641–644. [PubMed] [Google Scholar]
- 43.Spiess C, Bell A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 44.Stone B J, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone B J, Abu Kwaik Y. Natural competency for DNA uptake by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181:1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venkataraman C, Gao L-Y, Bondada S, Abu Kwaik Y. Identification of putative cytoskeletal protein homologues in the protozoan Hartmannella vermiformis as substrates for induced tyrosine phosphatase activity upon attachment to the Legionnaires' disease bacterium, Legionella pneumophila. J Exp Med. 1998;188:505–514. doi: 10.1084/jem.188.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkataraman C, Haack B J, Bondada S, Abu Kwaik Y. Identification of a Gal/GalNAc lectin in the protozoan Hartmannella vermiformis as a potential receptor for attachment and invasion by the Legionnaires' disease bacterium, Legionella pneumophila. J Exp Med. 1997;186:537–547. doi: 10.1084/jem.186.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel J P, Andrews H L, Wong S K, Isberg R R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]