Abstract
Sequence analysis of the genome of Neisseria meningititdis serogroup B revealed the presence of an ∼35-kb region inserted within a putative gene coding for an ABC-type transporter. The region contains 46 open reading frames, 29 of which are colinear and homologous to the genes of Escherichia coli Mu phage. Two prophages with similar organizations were also found in serogroup A meningococcus, and one was found in Haemophilus influenzae. Early and late phage functions are well preserved in this family of Mu-like prophages. Several regions of atypical nucleotide content were identified. These likely represent genes acquired by horizontal transfer. Three of the acquired genes are shown to code for surface-associated antigens, and the encoded proteins are able to induce bactericidal antibodies.
Many mobile DNA elements transpose from one chromosomal location to another by a fundamentally similar mechanism. They include IS elements (25), transposons (20), phages (4), and more recently the so-called pathogenicity islands (8). These elements contribute substantially to genetic diversity and genome plasticity. Particularly, in pathogenic bacteria some of these elements may contribute to the exchange of genetic material coding for virulence traits. This mechanism may increase the fitness of bacterial strains through acquisition of virulence factors. Among the mechanisms for transfer of DNA, lysogenic conversion by bacteriophages appears to be advantageous; in fact, bacteriophages can carry large blocks of DNA and can survive harsh conditions. Bacteriophages may also code for virulence factors that allow the host bacterium to enlarge its host range and provide mechanisms to evade immune response. Examples of bacterial virulence factors carried on bacteriophages include the well-studied diphtheria toxin of Corynebacterium diphtheriae (1), cholera toxin (CTX) of Vibrio cholerae (11), the pore-forming toxin CTX of Pseudomonas aeruginosa (9), the erythrogenic toxins of Streptococcus pyogenes (24), the Clostridium botulinum neurotoxin (1), and the Shiga-like toxins and enterohemolysin produced by Escherichia coli (2, 15).
Neisseria meningitidis, a gram-negative capsulated bacterium, is a major cause of septicemia and meningitis that can kill children and young adults within hours. There are five pathogenic N. meningitidis serogroups (A, B, C, Y, and W135) as determined by capsular polysaccharide typing (26). Very recently, the genomic sequences of N. meningitidis serogroup B strain MC58 (22) and serogroup A strain Z2491 (17) have been determined, showing, among other features, a number of open reading frames (ORFs) with homology to phage functions. We analyzed the chromosomal region of serogroup B strain MC58 coding for these genes and compared it to the genomes of N. meningitidis serogroup A strain Z2491 (17) and the closely related bacterium Haemophilus influenzae strain Rd (5). Our analysis indicates that these genomes contain chromosomal regions with similarities to Mu-like phages. These phage DNA regions are clearly mosaic with obvious sequence similarity to phage Mu interspersed with segments that are apparently unrelated. We show that some genes mapping within the phage regions code for surface-exposed proteins capable of eliciting serum bactericidal response. A possible role of these proteins in bacterial virulence and vaccine development is discussed.
MATERIALS AND METHODS
Computer analysis.
The region spanning positions 1,099,626 to 1,134,164 of the serogroup B N. meningitidis genome strain MC58 (22) was analyzed for coding capacity by using databases and computer programs included in the Wisconsin Package (version 10.0; Genetics Computer Group [GCG], Madison, Wis.). We revisited each single ORF in order to assign the correct start codon on the basis of ribosomal binding sequence and promoter regions. Subsequently, the programs Psi-BLAST, FASTA, MOTIFS, FINDPATTERNS, and PSORT (http://psort.nibb.ac.jp), as well as the databases ProDom, Pfam, and Blocks were used to predict protein features and to assign putative functions. The selected region containing a hypothetical Mu-like prophage was screened for conservation against the complete genomes of N. meningitidis serogroup A available at the Sanger Center (17, http://www.genome.ou.edu/gono.html) and H. influenzae Rd available at The Institute for Genomic Research (TIGR) (5) (http://www.tigr.org/tdb/CMR/ghi/htmls/SplashPage.html) and against the partial Neisseria gonorrhoeae genomic sequences available at the Advanced Center for Genome Technology, University of Oklahoma (http://www.genome.ou.edu/gono.html). Identified prophage regions map within positions 1,768,530 to 1,807,766 (PNM1) and 1,207,176 to 1,236,496 (PNM2) of the serogroup A strain Z2491 genome and within positions 1,559,960 to 1,594,298 of the H. influenzae complete genome. The same analysis on coding capacity, ORF reassignments and functional predictions described for MuMenB has been carried out for DNA segments defining PNM1, PNM2, and MuHi.
Nucleotide composition study has been performed using the programs WINDOW and STATPLOT available in the GCG Package. For this analysis we have used a window size of 500 nucleotides with a shift increment of 3 nucleotides.
Cloning, expression, and protein purification.
ORFs were amplified by PCR on chromosomal DNA from strain 2996 (23), with synthetic oligonucleotides used as primers. The amplified DNA fragments were cloned into pGEX-KG vector (7) to express the proteins as NH2-terminal glutathione-S-transferase fusions. Expression of recombinant proteins was evaluated according to the appearance of protein bands in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Recombinant fusion proteins were purified by affinity chromatography on glutathione-Sepharose 4B resin (Pharmacia). Twenty micrograms of each purified protein was mixed with Freund's adjuvant and used to immunize mice at days 1, 21, and 35. Blood samples were taken at days 34 and 49.
Serum analysis. (i) FACScan bacterium binding assay.
N. meningitidis strain M7 (acapsulated) was grown on chocolate agar plates overnight at 37°C with 5% CO2. Bacterial colonies were collected with a sterile Dacron swab and used to inoculate four tubes (8 ml each) of Mueller-Hinton broth (Difco) containing 0.25% glucose. Cells were harvested at an optical density at 620 nm (OD620) of 0.35 to 0.5, washed, and resuspended in blocking buffer (1% bovine serum albumin in phosphate-buffered saline, 0.4% NaN3) at an OD620 of 0.05. One hundred microliters of diluted sera (1:100, 1:200, 1:400) was added to 100 μl of bacterial cells in a 96-well plate (Costar), and incubated for 2 h at 4°C, washed with blocking buffer (200 μl/well), and 100 μl of 1:100 dilution of R-phycoerythrin-conjugated F(ab′)2 goat anti-mouse was added to each well and incubated for 1 h at 4°C. Cells were collected, washed, resuspended in PBS (200 μl/well)–phosphate-buffered saline 0.25% formaldehyde and transferred to FACScan tubes.
(ii) Bactericidal assay.
N. meningitidis strain 2996 was cultivated overnight at 37°C on chocolate agar plates with 5% CO2. Colonies were collected and used to inoculate 7 ml of Mueller-Hinton broth, containing 0.25% glucose, grown at 37°C with shaking to an OD620 of 0.23 to 0.24, and diluted to 105 CFU/ml in assay buffer (50 mM phosphate buffer [pH 7.2] containing 10 mM MgCl2, 10 mM CaCl2, and 0.5% [wt/vol] bovine serum albumin). Serum bactericidal activity determination (18) was carried out in a final volume of 50 μl with 25 μl of serial twofold dilutions of test serum, 12.5 μl of bacteria at the working dilution, and 12.5 μl of baby rabbit complement (final concentration, 25%). Controls included bacteria incubated with complement serum and immune sera incubated with bacteria and with complement inactivated by heating at 56°C for 30 min. Immediately after the addition of the baby rabbit complement, 10 μl of the controls was plated on Mueller-Hinton agar plates using the tilt method (time zero). The 96-well plate was incubated for 1 h at 37°C with rotation. Seven microliters of each sample was plated on Mueller-Hinton agar plates as spots, whereas 10 μl of the controls was plated on Mueller-Hinton agar plates using the tilt method (time one). Agar plates were incubated for 18 h at 37°C, and the colonies corresponding to time zero and time one were counted.
RESULTS AND DISCUSSION
Identification of a Mu-like prophage in the genome of N. meningitidis serogroup B strain MC58.
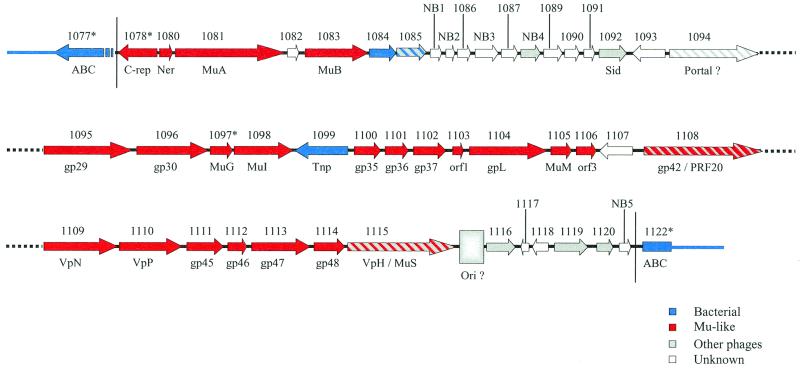
The annotation of the complete genome sequence of N. meningitidis serogroup B strain MC58 revealed the existence of 10 ORFs with striking amino acid similarities (identities ranging from 28.1 to 70.3%) to phage functions (22). These ORFs, interspersed within a genomic region of 20,995 bp spanning coordinates 1,101,155 to 1,131,150, include genes coding for regulatory functions of phage Mu (Ner, MuA, MuB) as well as genes coding for baseplate and tail functions of this phage (MuG, MuI, GpL, VpN, VpP, gp45, VpH). With the exception of a transposase of the IS30 family (NMB1099 in reference 22), these phage functions are surrounded by ORFs of unknown functions (Fig. 1). Moreover, two partial ORFs, NMB1077 and NMB1122, with homologies to ABC transporters map upstream and downstream of the region under study, respectively. Reunion of these truncated ORFs gives rise to a complete ORF which shows 55% amino acid identity to a hypothetical ABC transporter ATP-binding protein of H. influenzae (5). This indicates that the ABC transporter-encoding gene was split upon integration of a DNA segment. Nucleotide sequence analysis of the region flanking the split transporter gene revealed two imperfect septamer direct repeats, 5′-CTCA(A/G)CA-3′. This repeated sequence might arise from a duplication event following integration of a large DNA segment of 34,539 bp spanning positions 1,099,626 to 1,134,164 of the N. meningitidis genome strain MC58 (22). A schematic representation of this DNA region is shown in Fig. 1. This ∼35-kb DNA region includes 46 ORFs, most of which have been recently annotated (22), whereas 5 additional ORFs, named NB1 to NB5 (for new in serogroup B) (Fig. 1), have been identified, including previously unseen duplicated genes (see below).
FIG. 1.
Schematic representation of gene organization in prophage MuMenB of N. meningitidis strain MC58. For each gene, arrows indicate the direction of transcription and are scaled according to gene length. Numbers above arrows correspond to TIGR annotation (the suffix NMB has been omitted for simplicity in all cases). Newly annotated ORFs are marked NB1 to NB5 (for new in serogroup B). Putative functional assignments and correspondences to Mu homologues are reported below the arrows. Hypothetical sources of genes are color coded as indicated below the map. Hatched colored arrows represent genes for which source assignment may fall into two categories.
A further analysis of the ORFs contained in this region highlighted an additional 19 ORFs displaying significant amino acid identities to phage proteins (Table 1). A total of 29 ORFs out of 46 (63%) show homologies to phage functions, and 23 of these ORFs (50%) show homologies to functions of phage Mu. We conclude that this region was likely acquired by N. meningitidis strain MC58 upon infection with a Mu-like phage, subsequently referred to as MuMenB phage.
TABLE 1.
ORFs encoded within the prophage region of MuMenB and their genome positionsa
| MuMenB ORFs | Genome position | Length (aab) | ORF in:
|
|||
|---|---|---|---|---|---|---|
| Mu | MuHi | PNM1 | PNM2 | |||
| NMB1077∗ | 1099075–1099616 (−)c | Not annotated | ||||
| NMB1078∗ | 1100312–1099875 (−) | 235 | C repressor | HI1476 | NMA1884 | |
| NMB1080∗ | 1100822–1101061 | 80 | Ner | HI1477 | NMA1883 | |
| NMB1081 | 1101126–1103108 | 661 | MuA | HI1478 | NMA1882 | NMA1284 |
| NMB1082 | 1103120–1103317 | 67 | NMA1285∗ | |||
| NMB1083 | 1103481–1104650 | 390 | MuB | HI1481 | NMA1881∗ | NMA1286 |
| NMB1084° | 1104700–1105173 | 158 | ||||
| NMB1085 | 1105319–1105861 | 181 | NMA1864 | NMA1303 | ||
| NB1 | 1105861–1106062 | 68 | ||||
| NB2 | 1106068–1106230 | 55 | NMA1863 | NMA1304 | ||
| NMB1086 | 1106234–1106467 | 79 | NMA1862 | NMA1305 | ||
| NB3 | 1106442–1106859 | 139 | NMA1861 | NMA1306 | ||
| NMB1087 | 1106758–1107060 | 101 | NMA1860∗ | NMA1307 | ||
| NB4 | 1107160–1107506 | 115 | NMA1858 | |||
| NMB1089 | 1107506–1107841 | 112 | NMA1857 | NMA1308/NMA1309∗ | ||
| NMB1090 | 1107856–1108119 | 88 | NMA1856 | NMA1310 | ||
| NMB1091 | 1108119–1108313 | 65 | NMA1855 | NMA1311 | ||
| NMB1092 | 1108319–1108822 | 168 | NMA1854 | NMA1312 | ||
| NMB1093 | 1109412–1108825 (−) | 196 | ||||
| NMB1094∗ | 1109425–1111045 | 540 | NMA1852∗ | NMA1313 | ||
| NMB1095 | 1111048–1112612 | 522 | gp29 | HI1501 | NMA1851 | NMA1314 |
| NMB1096 | 1112602–1113894 | 431 | gp30 | HI1502 | NMA1850 | NMA1315 |
| NMB1097∗ | 1114006–114417 | 137 | MuG | HI1503 | NMA1849 | NMA1316 |
| NMB1098 | 1114653–1115711 | 353 | Mul | HI1504 | NMA1848 | Not annotated∗ |
| NMB1099 | 1116767–1115805 (−) | 321 | ||||
| NMB1100° | 1116795–1117274 | 160 | gp35 | HI1506 | NMA1845 | |
| NMB1101 | 1117277–1117696 | 140 | gp36 | HI1508 | NMA1844 | |
| NMB1102 | 1117746–1118336 | 197 | gp37 | HI1509 | NMA1843 | |
| NMB1103 | 1118336–1118530 | 65 | gp38 | HI1510 | NMA1842 | |
| NMB1104 | 1118536–1119942 | 469 | gpL | HI1511 | NMA1841 | |
| NMB1105 | 1120010–1120384 | 125 | MuM | HI1512 | NMA1840 | |
| NMB1106 | 1120391–1120753 | 121 | ORF3 | HI1513 | ||
| NMB1107 | 1121610–1121011 (−) | 200 | ||||
| NMB1108 | 1121780–1123933 | 723 | gp42 | HI1514 | NMA1833 (N terminal) | |
| NMB1109 | 1123936–1125264 | 443 | VpN | HI1515 | NMA1831 | NMA1319 (C terminal) |
| NMB1110 | 1125257–1126399 | 381 | VpP | HI1516/fs | NMA1830 | NMA1320 |
| NMB1111 | 1126399–1127064 | 222 | gp45 | HI1518 | NMA1829 | NMA1321 |
| NMB1112 | 1127168–1127512 | 115 | gp46 | HI1519 | NMA1828 | NMA1322 |
| NMB1113∗ | 1127528–1128580 | 350 | gp47 | HI1520 | NMA1827/NMA1826∗ | NMA1323 |
| NMB1114 | 1128580–1129137 | 186 | gp48 | HI1521 | NMA1825 | NMA1324 |
| NMB1115 | 1129151–1131121 | 657 | MuS (N terminal) | HI1522 | NMA1824 (N terminal) | NMA1325 |
| NMB1116 | 1131560–1132084 | 175 | ||||
| NMB1117 | 1132350–1132204 (−) | 49 | NMA1326 | |||
| NMB1118 | 1132762–1132478 (−) | 95 | NMA1327 | |||
| NMB1119 | 1132842–1133444 | 201 | NMA1823 | NMA1328 | ||
| NMB1120 | 1133426–1133719 | 98 | NMA1329 | |||
| NB5 | 1133719–1133926 | 69 | HI1523 (C terminal) | NMA1821 (C terminal) | NMA1330 | |
| NMB1122∗ | 1134173–1135151 (−) | 326 | Not annotated | |||
| Homology(ies) | % Amino acid identity | Function or remarks |
|---|---|---|
| YE67 (H. influenzae) (C terminus) | 58 (on 179 aa) | ABC transporter, putative |
| Repressor protein, Mu-like phage D3112 | 34 | Putative repressor |
| Ner protein phage Mu | 76 | Negative regulator of transcription |
| Transposase A, phage D3112 Transposase A, phage Mu | 30 22 | Transposase |
| None | ?d | |
| DNA transposition protein B, phage Mu | 29 | Bacteriophage integration and replication |
| LCND_LACLA (Lactococcus lactis) | 29 (on 84 aa) | Coded on a plasmid, lipoprotein, secretion of lactococcin A |
| AMIB_ECOLI | 28 (on 88 aa) | Cell wall hydrolase, lytic enzyme |
| None | Outer membrane, periplasmic | |
| None | ? | |
| None | ? | |
| None | Outer membrane, periplasmic | |
| None | Lipoprotein | |
| 10-kDa protein plasmid prfl (Plectonema sp.) | 31 (on 77 aa) | ? |
| None | ? | |
| None | ? | |
| None | ? | |
| Sid protein phage phi-R73 | 34 (on 70 aa) | Head size determination? |
| Outer surface protein OspC (Borrelia spp.) | 30 (on 95 aa) | Antigenic protein |
| None | Portal protein? (deduced by location, size, and aa composition) | |
| gp29 phage Mu | 38 | Head assembly |
| gp30 phage Mu ORF240 (Dichelobacter nodosus) | 35 (on 250 aa) 30 (on 177 aa) | Head assembly |
| G protein phage Mu | 30 | Virion morphogenesis |
| I protein phage Mu | 35 | Virion morphogenesis |
| Transposase for IS1655 | 99 | Transposase |
| gp35 phage Mu | 35 (on 60 aa) | ? |
| gp36 phage Mu Hypothetical protein (Pasteurella multocida) | 35 (on 140 aa) 35 (on 112 aa) | Head-tail junction? |
| gp37 phage Mu Ban protein phage HP1 | 24 (on 95 aa) 28 (on 94 aa) | Helicase? |
| gp38 phage Mu (orf1) | 30 | ? |
| gpL protein phage Mu | 29 | Sheath protein, major tail subunit |
| M protein phage Mu | 24 (on 78 aa) | Tube gene |
| ORF3 phage Mu | 27 | ? |
| None | Lipoprotein | |
| PRF20 (P. aeruginosa) ORF25 (phi-CTX) gp42 phage Mu | 25 (on 139 aa) 21 (on 580 aa) 21 (on 722 aa) | Tail length determination putative |
| VpN phage Mu | 26 (on 157 aa) | Tail, DNA circulation, virion protein |
| VpP phage Mu | 26 | Tail protein |
| gp45 phage Mu | 27 | Baseplate assembly |
| gp46 phage Mu | 41 | Tail |
| gp47 phage Mu PBSX prophage ORF xkdT | 30 24 | ? |
| gp48 phage Mu | 25 | ? |
| Vph bacteriophage HP1 Protein S phage Mu (N terminus) ORF20 (phi-CTX) | 50 (on 528 aa) 23 (on 293 aa) 20 (on 412 aa) | Tail fiber |
| ORF35 (phi-CTX) | 25 (on 103 aa) | ? |
| None | ? | |
| None | ? | |
| ORF21 (phi-CTX) (N terminus) | 46 (on 54 aa) | Tail assembly |
| ORF17 (phi-CTX) | 32 (on 67 aa) | Baseplate? |
| None | ? | |
| YE67 (H. influenzae) (N terminus) | 30 (on 64 aa) | ABC transporter, putative |
Corresponding annotated (5, 17, 22) and not annotated ORFs on serogroup A meningococcus (prophages PNM1 and PNM2) and H. influenzae (prophage MuHi) are also reported. Symbols: ∗, frame-shifted ORF; °, different start codon as compared to TIGR annotation; NB, new ORF in serogroup B meningococcus.
aa, amino acid.
(−), negative sense.
?, undetermined or uncertain.
Similarities of the deduced MuMenB gene products to known sequences and functional assignments.
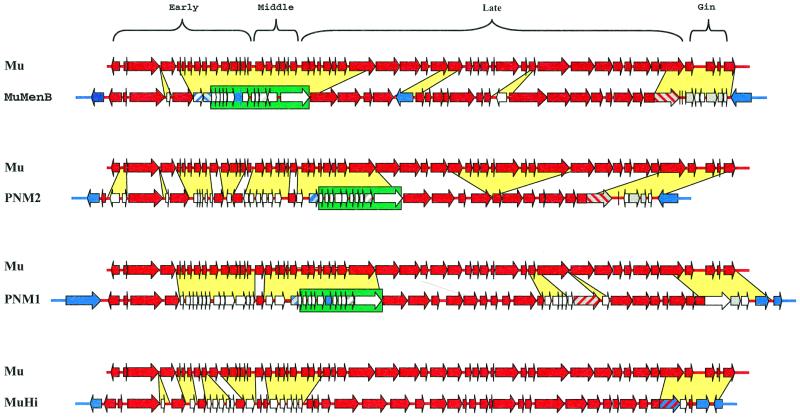
By comparing the genetic map of phage Mu and the genetic map of the newly identified phage MuMenB (Fig. 2), we observed a certain degree of resemblance in the number of ORFs, their amino acid length, and map positions between the two phages. Therefore, the amino acid sequences of the products deduced from the MuMenB ORFs were screened for similarities with sequences from the available databases and between each pair of the corresponding proteins from the two phages. The basic characteristics of the predicted gene products and the significant homologies found that allowed hypothetical functional assignments are described below and summarized in Table 1.
FIG. 2.
Pairwise comparison between the structure of Mu (36,717 bp) and the indicated related Mu-like bacteriophages MuMenB (34,538 bp), PNM1 (39,237 bp), PNM2 (29,321 bp), and MuHi (34,339 bp). Green boxes highlight a group of ORFs specifically acquired by neisserial prophages, whereas yellow spaces indicate genes, which either have been inserted or differ from the corresponding region of Mu. Color codes are as described in the legend to Fig. 1 and on Fig. 1 itself.
(i) Early region.
Only 4 (NMB1078, NMB1080, NMB1081, and NMB1083) out of 19 early functions are conserved between MuMenB and Mu bacteriophage genomes (Fig. 1; Table 1). Probably, the missing early functions have been lost during or after phage integration. By contrast, the map positions and orientation of transcription of the conserved genes and of the downstream genes are identical between the two phages.
(ii) Middle region.
A region of about 6 kb that includes 14 ORFs (NMB1084 to NMB1094 and NB1 to NB4) separates hypothetical early and late Mu-related functions. Of these 14 ORFs, NMB1092 shows homology (34% identity on 70 amino acids) to the Sid protein from phage phi-R73 (Fig. 1; Table 1). This protein has been suggested to function as determining head size with a DNA binding activity (21). While ORF NB4 and NMB1084 show similarity to proteins encoded by plasmid-borne genes, the other ORFs detected in this DNA region show no amino acid similarity to known protein. Moreover, it is worth noting that, whereas ORFs NB1 and NB2 are identical to ORFs NMB0988 and NMB0989, respectively, NB3 has a point mutation compared to NMB0990. Therefore, these three ORFs represent duplicated genes with one copy on the bacterial genome and one copy on the phage genome.
(iii) Late region.
The major number of conserved functions between the MuMenB and Mu phages are mainly related to late functions such as to head assembly and to virion morphogenesis and tail proteins (Fig. 1; Table 1). This region spans ORFs NMB1095 to NMB1115. With the exception of ORFs NMB1099, which codes for the IS1655 Tnp transposase, and NMB1107 (of unknown function), the map gene order and direction of transcription parallel those of phage Mu.
The rightmost region of the MuMenB genome contains six ORFs (NMB1116 to -1120 and NB5) either with unknown function or with functions homologous to those of phages different from Mu (Fig. 1; Table 1).
An apparent missing phage function is the lytic enzyme Lys, essential for host cell lysis to release mature phage particles from the cell wall, by breaking down the peptidoglycan. Nevertheless, NMB1085 displays homologies to a number of bacterial hydrolases; thus, it might be involved in bacterial lysis. Completely missing from the MuMenB genome are those functions related to immunity, proteins Mor and C, which function as positive regulators of middle and late transcription, respectively, and the Gin region (Fig. 2).
We conclude that most of the deduced functions mapping within this 35-kb DNA region of the N. meningitidis strain MC58 genome are similar to functions encoded by the bacteriophage Mu genome and, therefore, this region may represent the remnant DNA region evolved with the bacterial genome upon the Mu-like phage infection MuMenB. Evolution of the MuMenB prophage may account for the loss of some Mu functions and for the acquisition of functions related to other infecting phages. Likely, one of these acquired regions lies in the rightmost part of the MuMenB prophage and includes ORFs NMB1116 to NB5. Similarly, a wide region with ORFs of unknown functions (NMB1084 to NMB1094), including chromosomal duplicated genes, has replaced the missing Mu-related early-middle functions. Therefore, the structure of this phage genome is clearly mosaic, with regions of obvious sequence similarity interspersed with segments that are apparently unrelated. This argues not only for the existence of extensive horizontal genetic exchange among members of the Mu phages but also for extensive genetic exchange among phages from different families and with the bacterial genome (10).
Comparison of MuMenB with prophages in group A meningococcus and H. influenzae.
We compared the genetic structure of the Mu functions with those of the two major regions PNM1 and PNM2 of the N. meningitidis serogroup A strain Z2491, which encode putative phage functions (17) as well as with the region coding for phage functions in H. influenzae strain Rd (5), which we call MuHi. Surprisingly, search for a similar region on the partial genomic sequence of N. gonorrhoeae strain FA1090 (http://www.genome.ou.edu/gono.html) revealed no corresponding clusters of Mu-related functions. Accordingly, it has been recently reported (12), that a region corresponding to phage PNM1 of N. meningitidis serogroup A represents a specific genetic island missing in N. gonorrhoeae.
As schematized in Fig. 2, the Mu-like prophages MuMenB, PNM1, PNM2, and MuHi share a similar overall gene organization with most of the phage Mu functions. Interestingly, colinearity of gene order is preferentially and extensively interrupted within the early-middle region of these phages. This region of phage Mu, starting downstream from muB, includes 21 ORFs, which have been described as coding for nonessential or growth-enhancing functions (16). The functions that are not Mu related that were detected in this region include 14 ORFs for MuMenB, 22 ORFs for PNM1, 28 ORFs for PNM2, and 12 ORFs for MuHi.
The genome segment corresponding to the gin invertase region of phage Mu appears to be replaced by functions likely acquired from other phages. Twenty-nine tandem repeats of 13 bp that could represent an origin of DNA replication are detected in this region of the MuMenB genome (Fig. 1) but not in the other phages. By contrast, late functions could represent the target for gene variability by means of gene insertions and/or deletions and/or substitutions (Fig. 2). While most of the acquired ORFs display no homology to known proteins in databases and are not conserved among the phages, a subset of 12 ORFs mapping within the early-middle region are found in the three phages of N. meningitidis (Fig. 2). Their features, as deduced by computer algorithms (Motifs and PSORT) are reported in Table 2. Intriguingly, some of these ORFs are predicted to encode membrane-associated proteins, and some of them are duplicated within the N. meningitidis genomes. From the evolutionary point of view, some of these ORFs may have been acquired simultaneously as a cluster of genes, with others being acquired as a single gene acquisition. Therefore, evolution of these phages could have been achieved by a stepwise mechanism of gene acquisition, thus generating a mosaic genome structure whose products may contribute to N. meningitidis pathogenicity.
TABLE 2.
Hypothetical location for ORFs mapping within the conserved region acquired by the N. meningitidis phages. Symbols are as in Table 1.
| Features | MenB
|
MenA
|
|||
|---|---|---|---|---|---|
| MuMenB | Genome-encoded | PNM1 | PNM2 | Genome-encoded | |
| Outer/periplasmic | NB1 | NMB0988 | NA1 | NA1' | |
| NB2 | NMB0989 | NMA1863 | NMA1304 | ||
| NMB1086 | NMA1862 | NMA1305 | NMA1190 | ||
| Outer/periplasmic | NB3 | NMB0990∗ | NMA1861 | NMA1306 | |
| Lipoprotein | NMB1087 | NMA1860∗ | NMA1307∗ | NMA1192∗ | |
| Outer/periplasmic | NB4 | NMA1858 | |||
| NMB1089 | NMA1857 | NMA1308 | |||
| NMB1090 | NMA1856 | NMA1310 | |||
| NMB1091 | NMA1855 | NMA1311 | NMA1197 | ||
| DNA binding | NMB1092 | NMA1854 | NMA1312 | NMA1198 | |
| NMB1093∗ | |||||
| Portal protein | NMB1094 | NMA1852 | NMA1313 | ||
Horizontally acquired regions.
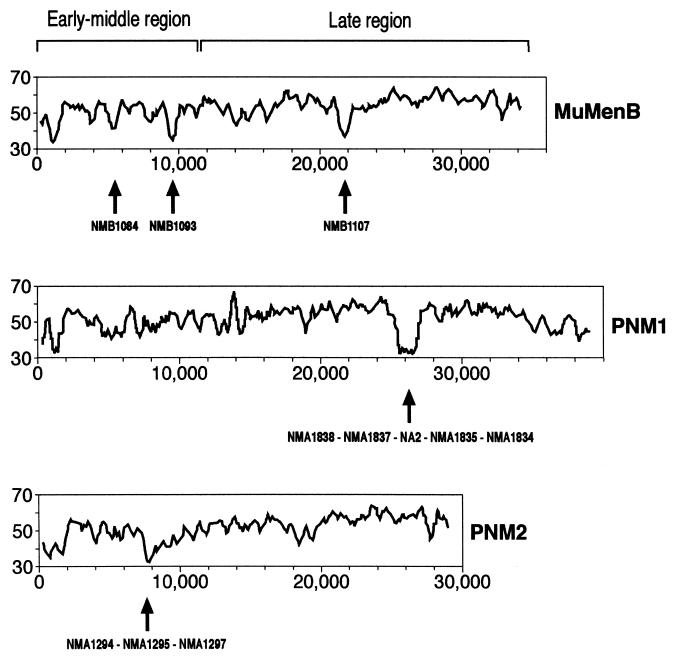
Horizontal transfer of DNA between species is well documented and is often associated with evolution of pathogenicity and drug resistance (6, 13). Bacteriophages may play an important role in acquisition of new genetic information, acting either as carriers of DNA fragments or as specialized systems for virulence-related genes (3, 4). Regions of DNA that have been acquired by horizontal transfer are often characterized by atypical DNA composition relative to the rest of the genome. One example is the cytotoxin-converting phage phi-CTX of P. aeruginosa (9). This phage shows an extensive homology to and a gene arrangement similar to that of coliphage P2 and P2-related Mu-like phages, and it carries the cytotoxin gene coding for the pore-forming toxin inserted within a region of atypical nucleotide content (14). Therefore, G+C composition study was used to identify recently acquired regions within neisserial prophages with results reported in Fig. 3.
FIG. 3.
G+C content plots derived for neisserial prophages. The x and y axes report base positions and percentage of G+C, respectively. Arrows indicate peaks of atypical G+C composition along with the ORFs encoded therein. The graphs were obtained by using the computer programs WINDOWS and STATPLOT of the GCG Package.
In all bacteriophages here reported, as well as in phage Mu, the region located at the leftmost terminus displays a lower G+C content relative to the overall corresponding phage genomes and includes the genes coding for the C-repressor and Ner proteins (Fig. 1 and 2). The intrinsic property of a low G+C content in this region may indicate an evolutionary constraint unrelated to DNA transfer.
The early-middle regions of MuMenB and PNM2 display a low G+C content, with peaks corresponding to a few specific DNA fragments. In MuMenB, these regions include ORFs NMB1084 and NMB1093, with a G+C value of 38 and 40%, respectively. Noteworthy, NMB1084 shows a putative leader peptide characteristic of lipoproteins and shares no amino acid homologies to known proteins, whereas NMB1093, although lacking a predicted signal peptide, shows significant similarity (Table 1) to the variable outer surface protein C (OspC) of Borrelia species. Both ORFs are absent from the corresponding early-middle region of PNM2, thus explaining why no peaks of low GC content are present within this region of PNM2. Nevertheless, another segment of atypical composition (G+C = 36.8%) is evident in the early-middle portion of PNM2, and this includes ORFs NMA1294, NMA1295, and NMA1297, specific for prophage PNM2. Interestingly, the three ORFs share a significant (44 to 54% identity) degree of amino acid similarity to each other, thus suggesting that these genes have evolved from a common ancestor gene. By contrast, the early-middle region of PNM1 shows a nucleotide composition that, on the whole, approximates the average value calculated for the whole genome (G+C = 52%).
Another region displaying an atypical nucleotide content maps within the middle portion of the late transcriptional unit and is present in MuMenB and PNM1 prophages (Fig. 3). In MuMenB the inserted DNA fragment (G+C = 39%) contains a single gene (nmb1107) in the reverse orientation, which codes for a predicted lipoprotein with no significant homologies to know proteins. The corresponding region of PNM1 shows a G+C content of 38.6% and codes for five ORFs (NMA1838, NMA1837, NA2 [for new in serogroup A], NMA1835, and NMA1834), including one which was not reported in the previous annotation (17). Of these, NMA1838 shows a homology to a regulatory protein of Streptomyces coelicolor, NMA1837 is characterized by a zinc-metalloendopeptidase motif, and NA2 is predicted to be a membrane protein. These segments of the two MuMenB and PNM1 phages very likely correspond to recent insertion events occurring upstream of the genes coding for the tail length determination proteins.
Some genes acquired by MuMenB encode surface-exposed antigens.
The mosaic genetic architecture of the neisserial phages indicates the existence of acquired conserved genes as well as of genes unique to each phage with hypothetical assigned or unknown functions (Fig. 1, Fig. 2, and Table 1). We selected three ORFs mapping to different positions in the phage genome for further characterization. ORF NB3 is in common with the three phages and maps within the region of conserved ORFs into the hypervariable early-middle region (Fig. 2); it was originated by gene duplication from the genome, and it may code for an outer membrane protein (Table 2). NMB1107 is likely to represent a recently acquired gene (Fig. 3), it is MuMenB specific (Fig. 2), and it may code for a lipoprotein (Table 1). NMB1119 is in common to MuMenB and PNM1 and has a homolog in PNM2 (27% amino acid identity); it maps within the 3′ end of the genomes, with features similar to a phage function different from that of Mu, and it may code for a tail assembly protein. These proteins are likely to be membrane associated on the bacterial envelope.
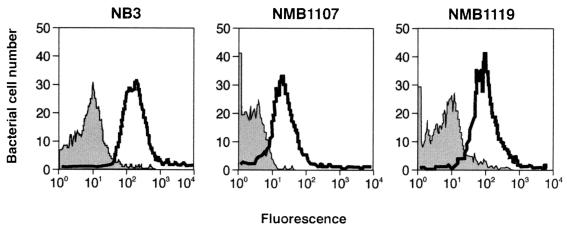
To assess whether these proteins are exposed on the bacterial surface, we raised antibodies against recombinant proteins in mice and used the immune sera in fluorescence-activated cell sorter (FACS) analysis (Materials and Methods). As shown in Fig. 4, the three immune sera recognized the heterologous N. meningitidis strain M7, suggesting that these proteins are exposed on the surface of the cell, therefore confirming the predicted computer search and analyses.
FIG. 4.
FACS analyses showing binding of polyclonal NB3, NMB1107, and NMB1119 antisera to the ethanol-treated homologous strain 2996. Gray profiles show binding of preimmune sera; white profiles show binding of immune sera.
Very recently, we have reported that surface-exposed proteins can be used as vaccine components against group B N. meningitidis strains (19). Therefore, FACS results prompted us to test whether immune sera obtained against proteins NB3, NMB1107, and NMB1119 can exert bactericidal activity, which in turn correlates with protection in humans. Immune sera have been tested for complement-mediated bacteriolysis against strain 2996 as described by Pizza et al. (19) and in Materials and Methods of the present work. Interestingly, antisera against proteins NB3, NMB1107, and NMB1119 showed a bacterial killing activity, reducing to 50% the number of viable bacterial cells, at 1:32, 1:32, and 1:64 dilutions, respectively. We conclude that these recombinant proteins may elicit bactericidal immune response, and therefore, these should be considered for vaccine development studies.
This conclusion is further substantiated by preliminary data obtained on the high degree of amino acid conservation for protein NMB1119 among meningococcus strains. Deduced amino acid sequences of gene NMB1119 from five different serogroup B strains (MC58, 1000, 2996, BZ133, and NGH38) and two serogroup A strains (Z2491 and F6124) (17, 22) revealed an amino acid conservation ranging from 93.7 to 97.0% amino acid identity (data not shown). This, suggests that at least this protein is conserved among serogroup B strains.
Conclusions.
We have reported the identification of chromosomal DNA regions of N. meningitidis strains that represent remnants of a Mu-like phage infection. Likely, this phage originally infected the bacterium and subsequently acquired specific genes to spread itself among a population of different strains. This is supported by the observation that a few of these genes are duplicated genes with one copy still residing within the bacterial chromosome. By contrast, some of the acquired genes seem to be unique to a given strain, thus suggesting a peculiar function in the host strain. Interestingly, computer search and comparison analyses suggest that both specific and common genes might code for membrane-associated proteins. This suggests that these proteins contribute to the variability in envelope structure and composition and may influence virulence and pathogenicity. Also, three of these proteins can be added to the list of vaccine candidates recently discovered among the meningococcal genome by Pizza and coworkers (19).
ACKNOWLEDGMENTS
We are grateful to M. Pizza, G. Grandi, and all members of the MenB group of the IRIS Research Center for sharing data and for useful discussions. We also thank G. Corsi for artwork and C. Mallia for editing the manuscript.
REFERENCES
- 1.Barksdale L, Arden S B. Persisting bacteriophage infections, lysogeny, and phage conversions. Annu Rev Microbiol. 1974;28:265–299. doi: 10.1146/annurev.mi.28.100174.001405. [DOI] [PubMed] [Google Scholar]
- 2.Beutin L, Stroeher U H, Manning P A. Isolation of enterohemolysin (Ehly2)-associated sequences encoded on temperate phages of Escherichia coli. Gene. 1993;132:95–99. doi: 10.1016/0378-1119(93)90519-9. [DOI] [PubMed] [Google Scholar]
- 3.Bishai W R, Murphy J R. Bacteriophage gene products that cause human disease. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum Press; 1988. pp. 683–724. [Google Scholar]
- 4.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 5.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs T M. Molecular mechanisms of bacterial pathogenicity. Naturwissenschaften. 1998;85:99–108. doi: 10.1007/s001140050463. [DOI] [PubMed] [Google Scholar]
- 7.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 8.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi T, Matsumoto H, Ohnishi M, Terawaki Y. Molecular analysis of a cytotoxin-converting phage, phiCTX, of Pseudomonas aeruginosa: structure of the attP-cos-ctx region and integration into the serine tRNA gene. Mol Microbiol. 1993;7:657–667. doi: 10.1111/j.1365-2958.1993.tb01157.x. [DOI] [PubMed] [Google Scholar]
- 10.Hendrix R W, Smith M C M, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karaolis D K, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 12.Klee S R, Nassif X, Kusecek B, Merker P, Beretti J-L, Achtman M, Tinsley C R. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect Immun. 2000;68:2082–2095. doi: 10.1128/iai.68.4.2082-2095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morschhauser J, Kohler G, Ziebuhr W, Blum-Oehler G, Dobrindt U, Hacker J. Evolution of microbial pathogens. Phil Trans R Soc Lond B Biol Sci. 2000;355:695–704. doi: 10.1098/rstb.2000.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama K, Kanaya S, Ohnishi M, Terawaki Y, Hayashi T. The complete nucleotide sequence of phi CTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol Microbiol. 1999;31:399–419. doi: 10.1046/j.1365-2958.1999.01158.x. [DOI] [PubMed] [Google Scholar]
- 15.Newland J W, Strockbine N A, Miller S F, O'Brien A D, Holmes R K. Cloning of Shiga-like toxin structural genes from a toxin converting phage of Escherichia coli. Science. 1985;230:179–181. doi: 10.1126/science.2994228. [DOI] [PubMed] [Google Scholar]
- 16.Paolozzi L, Symonds N. The SE region. In: Symonds N, Toussaint A, van de Putte P, Howe M M, editors. Phage Mu. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1987. pp. 53–62. [Google Scholar]
- 17.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, Davies R M, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail M A, Rajandream M A, Rutherford K M, Simmonds M, Skelton J, Whitehead S, Spratt B G, Barrel B G. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 18.Peeters C C, Claassen I J, Schuller M, Kersten G F, van der Voort E M, Poolman J T. Immunogenicity of various presentation forms of PorA outer membrane protein of Neisseria meningitidis in mice. Vaccine. 1999;17:2702–2712. doi: 10.1016/s0264-410x(99)00011-0. [DOI] [PubMed] [Google Scholar]
- 19.Pizza M, Scarlato V, Masignani V, Giuliani M M, Aricò B, Comanducci M, Jennings G T, Baldi L, Bartolini E, Capecchi B, Galeotti C L, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood D W, Jeffries A C, Saunders N J, Granoff D M, Venter C J, Moxon E R, Grandi G, Rappuoli R. Whole genome sequencing to identify novel vaccine candidates against serogroup B meningococcus. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 20.Salyers A A, Shoemaker N B, Stevens A M, Li L Y. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J, Inouye M, Inouye S. Association of a retroelement with a P4-like cryptic prophage (retronphage phi R73) integrated into the selenocystyl tRNA gene of Escherichia coli. J Bacteriol. 1991;173:4171–4181. doi: 10.1128/jb.173.13.4171-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, Nelson W C, Gwinn M L, DeBoy R, Peterson J D, Hickey E K, Haft D H, Salzberg S L, White O, Fleischmann R D, Dougherty B A, Mason T, Ciecko A, Parksey D S, Blair E, Cittone H, Clark E B, Cotton M D, Utterback T R, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith H O, Fraser C M, Moxon E R, Rappuoli R, Venter J C. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 23.van der Ley P, Poolman J T. Construction of a multivalent meningococcal vaccine strain based on the class 1 outer membrane protein. Infect Immun. 1992;60:3156–3161. doi: 10.1128/iai.60.8.3156-3161.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weeks C R, Ferretti N N. Nucleotide sequence of the type A streptococcal exotoxin (erythrogenic toxin) gene from Streptococcus pyogenes bacteriophage T12. Infect Immun. 1986;52:144–150. doi: 10.1128/iai.52.1.144-150.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinert T A, Schaus N A, Grindley N D. Insertion sequence duplication in transpositional recombination. Science. 1983;222:755–765. doi: 10.1126/science.6314502. [DOI] [PubMed] [Google Scholar]
- 26.Yakubu D E, Abadi F J, Pennington T H. Molecular typing methods for Neisseria meningitidis. J Med Microbiol. 1999;48:1055–1064. doi: 10.1099/00222615-48-12-1055. [DOI] [PubMed] [Google Scholar]