Abstract
A detailed analysis of the complexes of proline-specific peptidases (PSPs) in the midgut transcriptomes of the larvae of agricultural pests Tenebrio molitor and Tribolium castaneum and in the genome of T. castaneum is presented. Analysis of the T. castaneum genome revealed 13 PSP sequences from the clans of serine and metal-dependent peptidases, of which 11 sequences were also found in the gut transcriptomes of both tenebrionid species’ larvae. Studies of the localization of PSPs, evaluation of the expression level of their genes in gut transcriptomes, and prediction of the presence of signal peptides determining secretory pathways made it possible to propose a set of peptidases that can directly participate in the hydrolysis of food proteins in the larvae guts. The discovered digestive PSPs of tenebrionids in combination with the post-glutamine cleaving cysteine cathepsins of these insects effectively hydrolyzed gliadins, which are the natural food substrates of the studied pests. Based on the data obtained, a hypothetical scheme for the complete hydrolysis of immunogenic gliadin peptides by T. molitor and T. castaneum digestive peptidases was proposed. These results show promise regarding the development of a drug based on tenebrionid digestive enzymes for the enzymatic therapy of celiac disease and gluten intolerance.
Keywords: proline-specific peptidases, dipeptidyl peptidase 4, transcriptomic analysis, Tenebrionidae insects, gliadin hydrolysis
1. Introduction
Tribolium castaneum and Tenebrio molitor are genetic and biochemical model insects in the family Tenebrionidae. These insects are stored product pests of grain crops and wheat flour, and their main dietary proteins are therefore proline- and glutamine-rich gliadins—the prolamins of wheat and the main component of gluten. Gliadins contain up to 50% glutamine and up to 30% proline residues [1,2]. Proline is structurally unique and the only imino acid among 20 natural proteinogenic amino acids. Proline bonds cannot be hydrolyzed by most peptidases with broad substrate specificity because their unique conformation provides a rigid structure of proteins and peptides that protects them from degradation. In nature, only a special group of peptidases, proline-specific peptidases (PSPs), almost exclusively hydrolyzes bonds formed by proline. Due to the need to digest gliadin-containing food, tenebrionids should possess enzymes capable of hydrolyzing the peptide bonds formed by proline, which thus makes them a good natural enzymatic system to study the broad spectrum of PSPs.
Proteolysis of proline-rich proteins and peptides is also important for providing insects with free proline. This imino acid is an important component of insect metabolism as oxidation of free proline in the mitochondria may provide an important energy source for overwintering larvae or flying adults [3,4]. It has been shown that increased levels of proline correlate with specific environmental conditions in insects, which can be observed in overwintering insects and those with cold tolerance [3,5,6]. Thus, the study of peptidases that participate in the release of free proline from dietary proteins are of great interest. Such enzymes are especially important in the metabolism of insects that feed on proline-rich proteins.
Information about known eukaryotic PSPs has been summarized in several reviews [7,8,9]. However, the complexes of PSPs in distinct species, including insects, have not been described. This group of peptidases includes enzymes that specifically hydrolyze peptide bonds formed (i) by the carboxyl group of proline (prolyl oligopeptidase (POP) (which includes the so-called prolyl endopeptidase (PEP), which actually is POP), dipeptidyl peptidases (DPP) 2, 4, 8, and 9, fibroblast activation protein α (FAP), and prolyl carboxypeptidase (PRCP)) and (ii) by the imino group of proline (prolidase (XPD, Xaa-Pro dipeptidase) and aminopeptidases P (APP) 1, 2, and 3). Most of the representatives of PSPs that have been described up to now are exopeptidases; POP is an endopeptidase, and FAP can show both exo- and endopeptidase activities. The rest of the PSPs include a rather diverse group of different exopeptidases that cleave amino acids from both ends of the polypeptide chain.
Besides the above, there also exist substrates with N-terminal Pro residue, which are not hydrolyzed by any PSP from the described complex. These substrates can be hydrolyzed by peptidases with broad substrate specificity with no preference to Pro residue (post-proline cleaving peptidases with broad substrate specificity, PPCPbs). Thus, N-terminal Pro from a polypeptide chain of any length can be cleaved by leucyl aminopeptidase (LAP). The dipeptide Pro-X can be cleaved by cytosolic non-specific dipeptidase (CND), which hydrolyzes dipeptides of any composition, except Pro-Pro.
The list of PSPs described in the comprehensive collection of exhaustively characterized peptidases [8] includes 19 types of PSPs from all kingdoms of living organisms. Previously, we reviewed 13 representatives of human PSPs [9] from two clans according to the MEROPS classification [10]: serine peptidases and metallopeptidases. POP, DPP, and FAP belonged to the S9 family of serine peptidases: S9A (POP), S9B (DPP 4, 8, 9, FAP), S9X (DPP 6, 10). PRCP and DPP 2 were included in the S28 family of serine peptidases. APP1, APP2, APP3, and XPD belonged to the M24 family of metallopeptidases. For effective release of Pro residues from dietary proteins, each animal species used a specific complex of digestive PSPs, which is a shorter list than the whole list of animal PSPs. We could not find publications devoted to a description of digestive PSP complexes in animals including insects, while inhibition of its members being an effective way to control insect pests. Among the human intestinal PSPs that could participate in digestion, only DPP 4 [11] and XPD [12,13,14,15] activities have been reported in the literature.
In this paper, we present a detailed analysis of our own data about PSPs complexes in the midgut transcriptomes of stored product pests (T. molitor and T. castaneum larvae), T. castaneum genome [16], in comparison with the PSP complex in the human genome [9].
2. Results
2.1. Search for PSP Sets in Tribolium Castaneum and Tenebrio Molitor
Analysis of the T. castaneum genome using all eukaryotic PSPs as a query revealed 13 PSP sequences (Table 1): single sequences for POP, DPP 8, APP3, and XPD; two sequences for DPP 4, PRCP, and APP1; and three sequences for DPP 10 (inactive DPP 4 homolog). The T. castaneum genome also contained three sequences homologous to human LAP and one sequence homologous to CND. Most of the found sequences were annotated in the UniProt and NCBI databases, but for the DPP4.2_Tc (hereinafter the name before the dot means the type of peptidase, the number after the dot can occur if there are several variants of sequences of one type of peptidase, and the abbreviation after the underscore refers to the organism: Tc—T. castaneum, Tm—T. molitor, and Hs—H. sapiens), DPP10.2_Tc, and DPP10.3_Tc sequences annotation was refined by us during phylogenetic analysis and multiple alignment with human homologs. Comparison of PSP sets found in the T. castaneum genome and T. castaneum larval gut transcriptome showed that some of the found PSPs were not transcribed in the gut, namely POP_Tc, DPP10.3_Tc, LAP.2_Tc, and LAP.3_Tc; thus, the T. castaneum larval gut transcriptome contained 11 PSP sequences and two PPCPbs sequences.
Table 1.
Classification of PSPs found in the T. castaneum genome and gut transcriptome and the T. molitor gut transcriptome compared with human sequences.
| MEROPS Classification |
T. castaneum | T. molitor | H. sapiens | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PSP Name | UniProt ID | NCBI ID | PSP Name | UniProt ID | NCBI ID | PSP Name | UniProt ID | NCBI ID | |
| S9A | POP_Tc * | D6W775 * | XP_015833515 * | POP_Tm | A0A4D5SUD3 | QAY29071 | POP_Hs | P48147 | NP_002717 |
| S9B | DPP4.1_Tc | D6WJ57 | XP_975053 | DPP4.1_Tm | A0A4D5SUH2 | QAY29072 | DPP4_Hs | P27487 | NP_001926 |
| DPP4.2_Tc | D6WL31 | XP_972016 | DPP4.2_Tm | A0A4D5SUB4 | QAY29073 | ||||
| DPP8_Tc | D6WD50 | XP_971949 | DPP8_Tm | A0A4D5SUP4 | QAY29074 | DPP8_Hs | Q6V1X1 | NP_932064 | |
| DPP9_Hs | Q86TI2 | NP_631898 | |||||||
| FAP_Hs | Q12884 | NP_004451 | |||||||
| S9X | DPP6_Hs | P42658 | NP_570629 | ||||||
| DPP10.1_Tc | D6WQR4 | XP_015837625 | DPP10.1_Tm | A0A4D5SUP7 | QAY29075 | DPP10_Hs | Q8N608 | NP_065919 | |
| DPP10.2_Tc | D6WJ59 | XP_008194406 | DPP10.2_Tm | A0A4D5SUC4 | QAY29076 | ||||
| DPP10.3_Tc * | D6WS52 * | XP_008196030 * | |||||||
| S28 | PRCP.1_Tc | D6W8E7 | XP_971305 | PRCP_Tm | A0A4D5SUH1 | QAY29077 | PRCP_Hs | P42785 | NP_005031 |
| PRCP.2_Tc | D6WVZ7 | XP_972807 | |||||||
| DPP2_Hs | Q9UHL4 | NP_037511 | |||||||
| M24 | APP1.1_Tc | D6WLY7 | XP_974698 | APP1_Tm | A0A4D5SVH2 | QAY29078 | APP1_Hs | Q9NQW7 | NP_001161076 |
| APP1.2_Tc | D6X4C7 | EEZ97287 | |||||||
| APP2_Tm | A0A4D5SUC7 | QAY29079 | APP2_Hs | O43895 | NP_003390 | ||||
| APP3_Tc | D6X4Q9 | XP_008199099 | APP3_Tm | A0A4D5SUE5 | QAY29080 | APP3_Hs | Q9NQH7 | NP_071381 | |
| XPD_Tc | A0A139WNR5 | XP_971576 | XPD_Tm | A0A4D5SUD6 | QAY29081 | XPD_Hs | P12955 | NP_000276 | |
| M17 | LAP.1_Tc | D6WAL0 | EEZ97983 | LAP_Tm | A0A4D5SUH6 | QAY29082 | LAP_Hs | P28838 | NP_056991 |
| LAP.2_Tc * | D6WAL5 * | XP_974656 * | |||||||
| LAP.3_Tc * | D6WIH3 * | XP_972664 * | |||||||
| M20 | CND_Tc | D6WGK3 | XP_971451 | CND_Tm | A0A4D5SUC3 | QAY29083 | CND_Hs | Q96KP4 | NP_060705.2 |
* These PSP sequences were found only in the T. castaneum genome and were absent in the gut transcriptome. PSP name: the name before the dot means the type of peptidase, the number after the dot can occur if there are several variants of sequences of one type of peptidase, and the abbreviation after the underscore refers to the organism (Tc—T. castaneum, Tm—T. molitor, Hs—H. sapiens).
The set of T. molitor PSPs was analyzed using the larval gut transcriptome. It contained 11 predicted protein sequences of PSPs similar to those presented in the T. castaneum and human genomes and the T. castaneum larval gut transcriptome (Table 1): single sequences for POP, DPP 8, PRCP, APP1, APP2, APP3, and XPD and two sequences for DPP 4 and DPP 10. We also found sequences homologous to LAP and CND.
The sets of human and insect PSPs are presented in Table 1. Overall, the sets of PSPs for all studied organisms were similar, but insects did not possess some of the PSPs found in humans: DPP 9 (which is very close to DPP 8 and DPP 4), FAP, DPP 6 (an inactive homolog of DPP 4), and DPP 2. Interestingly, insects, unlike humans, had some PSPs duplicated (DPP 4, DPP 10, PRCP, APP1, and LAP), and some of the duplicates were found only in the T. castaneum genome.
2.2. General Characteristics of the PSP Sequences
2.2.1. Serine PSPs
For all peptidases, residues that determine primary specificity were identified. Active sites of active serine PSPs from S9A, S9B, and S28 consisted of a highly conserved catalytic triad in the order Ser, Asp, His. Surrounding residues of all peptidases were also very conservative, with few synonymic substitutions.
Peptidases from the S9A subfamily hydrolyze substrates of the general formula (Xaa)1-15-Xbb-Pro↓Xbb-(Xaa)1-15, where Xaa represents any amino acid and Xbb represents any amino acid except Pro. The active site of the S9A serine peptidase subfamily including POP consists of Ser554, Asp641, and His680 (hereinafter numbering is given by human POP, UniProt P48147) with conserved sequence motifs NGGSNGG, ADHDDRV, and RAGHGAG [17,18,19]. In the sequences found in Tenebrionidae insects (POP_Tc and POP_Tm), composition of the catalytic triad Ser, Asp, His was similar, with surrounding residues and residues forming binding subsites being highly conserved.
Peptidases from the S9B subfamily hydrolyze substrates of the general formula Xbb-Pro↓Xbb-(Xaa)n, where n represents any number. The active site of the S9B serine peptidase subfamily consists of Ser630, Asp708, and His740 (hereinafter numbering is given by human DPP 4, UniProt P27487). Homologous sequences of the S9B serine peptidase subfamily found in Tenebrionidae insects had the same order in the catalytic triad. In the structure of human DPP 4, the Pro (P1) residue of the substrate is located in the hydrophobic S1 subsite surrounded by Tyr631, Val656, Trp659, Tyr662, Tyr666, and Val711. The S2 subsite includes Asn710, Arg125, Glu205, and Glu206 [18,20,21]. Active insect peptidases contained conserved substrate-binding subsite residues, with the only exception being the S1 residue Trp659, which was replaced in DPP4.1_Tc Trp659Leu and DPP4.1_Tm Trp659Phe. All DPP 4-like peptidases found in Tenebrionidae insects, together with human DPP 4, possessed only exopeptidase activity, having Asp or Asn residues in an important Asp663 position similar to human DPP 4. This fact distinguished DPP 4 from FAP, containing in human smaller Ala657 residue [22] in a corresponding position, which explains the presence of endopeptidase activity in addition to exopeptidase activity in FAP [20,22,23,24]. An inactive homolog of DPP 4 (human DPP 10) had a Gly residue instead of Ser630 in the active site [25,26]. In the T. castaneum genome, three sequences of DPP 10 were found. Two of them also had a Gly residue instead of Ser630 and formed homologous pairs with T. molitor sequences. The third—DPP10.3_Tc—had Ser residue in the active site similar to active members of the S9B subfamily, and presumably can thus have enzymatic activity similar to DPP 4, although the sequence structure is characteristic of DPP 10.
PRCP from the S28 subfamily hydrolyzes substrates of the general formula (Xaa)n-Xbb-Pro↓Xbb. DPP 2 hydrolyzes substrates of the same general formula as the S9B subfamily, cleaving Xbb-Pro dipeptide from the N-terminus, as described above; however, no such activity was found in tenebrionids. The catalytic triad of the S28 serine peptidase family contains Ser179, Asp430, and His455 (hereinafter the numbering is given by human PRCP, UniProt P42785) residues. PRCP sequences from Tenebrionidae insects also had conserved residues surrounding the catalytic triad with minor replacements, as well as similar Pro-binding S1 subsites (Table 2).
Table 2.
General characteristics of the serine PSP sequences from the S9 and S28 families.
| PSP | Sequence Name | Active Site Residues |
Residues of S1 Binding Subsite |
Residues of S2 Binding Subsite | Residues of S3 Binding Subsite | ||
|---|---|---|---|---|---|---|---|
| Ser | Asp | His | |||||
| POP | POP_Hs | S554 | D641 | H680 | W595, F476, V644, V580, Y599, Y473 | R643 | F173, M235, C255, I591, A594 |
| POP_Tc | S | D | H | W, F, V, V, Y, Y | R | F, R, C, I, A | |
| POP_Tm | S | D | H | W, F, V, V, Y, Y | R | F, R, C, I, A | |
| DPP 4 | DPP4_Hs | S630 | D708 | H740 | Y631, V656, W659, Y662, Y666, V711 | N710, R125, E205, E206 | - |
| DPP4.1_Tc | S | D | H | Y, V, L, Y, Y, V | N, R, E, E | - | |
| DPP4.2_Tc | S | D | H | Y, V, W, Y, Y, V | N, R, E, E | - | |
| DPP4.1_Tm | S | D | H | Y, V, F, Y, Y, V | N, R, E, E | - | |
| DPP4.2_Tm | S | D | H | Y, V, W, Y, Y, V | N, R, E, E | - | |
| DPP 8 | DPP8_Hs | S | D | H | Y, V, W, Y, Y, V | N, -, E, E | - |
| DPP8_Tc | S | D | H | Y, V, W, Y, Y, V | N, -, E, E | - | |
| DPP8_Tm | S | D | H | Y, V, W, Y, Y, V | N, -, E, E | - | |
| DPP 9 | DPP9_Hs | S | D | H | Y, V, W, Y, Y, V | N, -, E, E | - |
| FAP | FAP_Hs | S | D | H | Y, V, W, Y, Y, V | N, R, E, E | - |
| DPP 6 | DPP6_Hs | D | D | H | Y, I, F, Y, F, I, | K, Q, E, E | - |
| DPP 10 | DPP10_Hs | G | D | H | Y, I, L, Y, F, V | K, H, E, E | - |
| DPP10.1_Tc | G | D | H | Y, I, W, H, F, V | S, K, N, E | - | |
| DPP10.2_Tc | G | D | H | Y, I, W, Y, F, A | T, R, E, D | - | |
| DPP10.3_Tc | S | D | H | Y, V, W, Y, Y, V | N, R, E, E | - | |
| DPP10.1_Tm | G | D | H | Y, I, W, Y, F, V | S, K, N, E | - | |
| DPP10.2_Tm | G | D | H | Y, I, W, Y, F, A | T, R, E, D | - | |
| PRCP | PRCP_Hs | S179 | D430 | H455 | M183, W359, M369, W432 | - | - |
| PRCP.1_Tc | S | D | H | M, W, M, W | - | - | |
| PRCP.2_Tc | S | D | H | M, W, M, W | - | - | |
| PRCP_Tm | S | D | H | M, W, M, W | - | - | |
| DPP 2 | DPP2_Hs | S | D | H | M, W, L, W | ||
2.2.2. Metallodependent PSPs
All metallodependent PSPs (APP1, APP2, APP3, and XPD) belong to the M24B subfamily and are double Mn2+-dependent enzymes, requiring both ions for activity [27]. APP hydrolyzes substrates of the general formula Xbb↓Pro(Xaa)1-9. XPD hydrolyzes dipeptides of the general formula Xbb↓Pro. Insect peptidases contained two conserved metal-binding sites, as well as residues essential for maintaining activity similar to human enzymes (Table 3).
Table 3.
General characteristics of the metallodependent PSP sequences from the M24B subfamily.
| PSP | Sequence Name | Residues of Mn2+-Binding Sites | ||||
|---|---|---|---|---|---|---|
| I | I/II | II | II | I/II | ||
| APP1 | APP1_Hs | D415 | D426 | H489 | E523 | E537 |
| APP1.1_Tc | D | D | H | E | E | |
| APP1.2_Tc | D | D | H | E | E | |
| APP1_Tm | D | D | H | E | E | |
| APP2 | APP2_Hs | D | D | H | E | E |
| APP2_Tm | D | D | H | E | E | |
| APP3 | APP3_Hs | D | D | H | E | E |
| APP3_Tc | D | D | H | E | E | |
| APP3_Tm | D | D | H | E | E | |
| XPD | XPD_Hs | D | D | H | E | E |
| XPD_Tc | D | D | H | E | E | |
| XPD_Tm | D | D | H | E | E | |
2.2.3. Post-Proline Cleaving Peptidases with Broad Substrate Specificity (PPCPbs)
Additional PPCPbs (LAP and CND) are also metallopeptidases. LAP hydrolyzes substrates of the general formula Xaa↓Xbb-(Xaa)n. It is a Zn2+-dependent peptidase from the M17 family and contains two Zn2+ ions. CND hydrolyzes dipeptides of the general formula Xaa↓Xbb. It is from the M20 family and contains two Mn2+ ions. In general, all sequences found in Tenebrionidae insects were highly conserved in metal-binding sites (Table 4, numbering is given by human peptidases).
Table 4.
General characteristics of the LAP and CND sequences from the M17 and M20 families.
| PPCPbs | Sequence Name | Residues of Zn2+-Binding Sites | |||||||
|---|---|---|---|---|---|---|---|---|---|
| II | I/II | II | I | I/II | |||||
| LAP | LAP_Hs | K282 | D287 | K294 | D305 | D364 | E366 | R368 | |
| LAP.1_Tc | K | D | K | D | D | E | R | ||
| LAP.2_Tc | K | D | K | D | D | E | R | ||
| LAP.3_Tc | K | N | K | D | D | A | R | ||
| LAP_Tm | K | D | K | D | D | E | R | ||
| Residues of Mn2+-Binding Sites | |||||||||
| II | I/II | I | I/II | I | |||||
| CND | CND_Hs | H99 | D101 | D132 | E166 | E167 | D195 | H228 | H445 |
| CND_Tc | H | D | D | E | E | D | H | H | |
| CND_Tm | H | D | D | E | E | D | H | H | |
This analysis showed that all found PSP sequences from T. molitor had a conserved structure compared to T. castaneum and human enzymes sequences, which made sequence annotation easier.
2.3. Comparison and Phylogeny of PSPs in T. molitor and T. castaneum
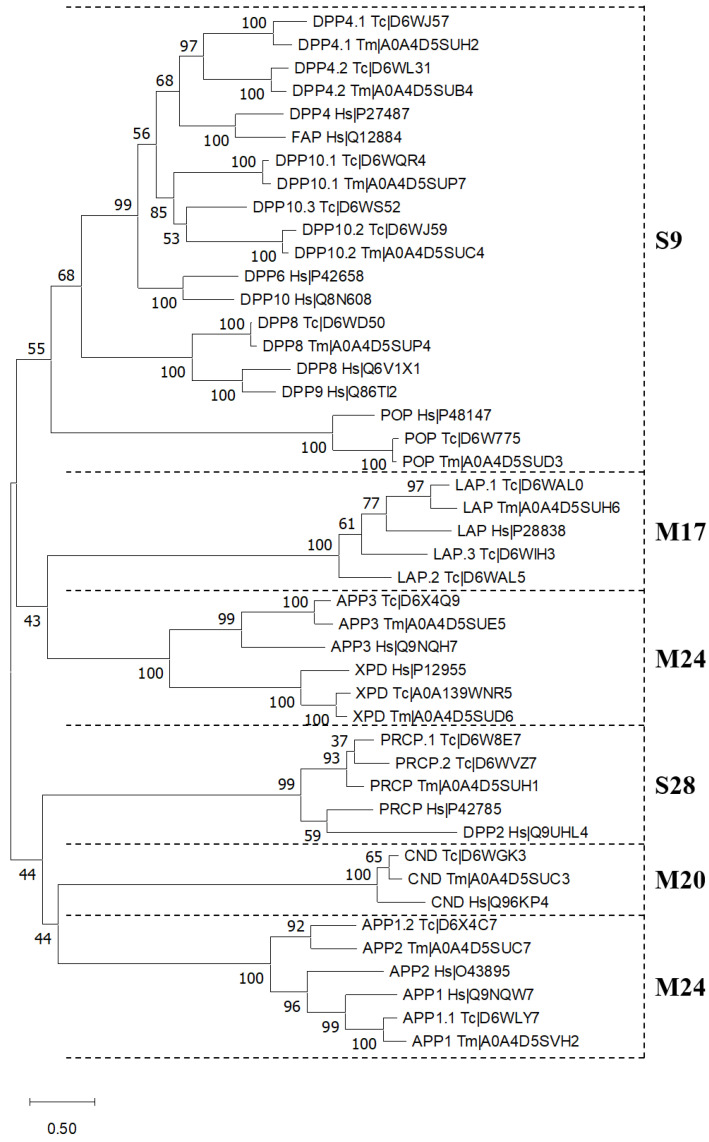
The phylogenetic tree represented in Figure 1 includes H. sapiens, T. castaneum, and T. molitor PSPs. The tree was divided into separate branches, each corresponding to one of the three families of PSPs (S9, S28, M24) or one of the two PPCPbs (M20 and M17). Each branch in turn was subdivided into types of peptidases, which formed orthologous pairs. A clade of DPP 4 proteins was divided into two distinct orthologous pairs: DPP4.1_Tc–DPP4.1_Tm and DPP4.2_Tc–DPP4.2_Tm. Insect DPP 10 formed a separate clade. Among them, DPP10.1_Tc–DPP10.1_Tm and DPP10.2_Tc–DPP10.2_Tm formed two distinct orthologous pairs. DPP10.3_Tc had a Ser residue in the catalytic triad that was located near the second pair. Separate clades were also formed by other PSPs. Distinct groups were formed by the DPP 8, POP, PRCP, APP1, APP2, APP3, XPD, CND, and LAP proteins. In each clade, homologous insect sequences formed orthologous pairs. Human proteins were located near them due to the high structural and functional similarities.
Figure 1.
Phylogenetic tree of T. castaneum, T. molitor, and H. sapiens PSPs and PPCPbs constructed using the maximum likelihood method.
2.4. Localization of PSPs
The predicted molecular masses of serine PSPs from the S9 and S28 families ranged from 54 to 103 kDa, and for metallodependent PSPs from the M24B subfamily these predictions ranged from 54 to 88 kDa. In most cases, calculated molecular masses were similar for enzymes of the same type (Table 5).
Table 5.
Analysis of the localization domains of PSPs.
| PSP | Sequence Name |
Length | Protein Mm (Da) |
SignalP-6.0 * | TargetP-2.0 * | DeepTMHMM Server | Experimental Data (For Mammalian PSPs Only) |
|
|---|---|---|---|---|---|---|---|---|
| Signal Peptide | Signal Peptide | Mitochondrial Transit Peptide |
TMhelix | |||||
| POP | POP_Hs | 710 | 80,700 | Cytoplasm [28] | ||||
| POP_Tc | 704 | 80,212 | ||||||
| POP_Tm | 705 | 80,116 | 1–21 (0.90/0.79) | |||||
| DPP 4 | DPP4_Hs | 766 | 88,279 | 1–22 (0.78/0.40) | 7–27 | Membrane [30,31], soluble [47] | ||
| DPP4.1_Tc | 767 | 87,333 | 1–19 (0.99/0.98) | 1–19 (0.99/0.73) | ||||
| DPP4.2_Tc | 821 | 92,585 | 34–54 | |||||
| DPP4.1_Tm | 768 | 86,576 | 1–19 (0.99/0.98) | 1–19 (0.99/0.78) | ||||
| DPP4.2_Tm | 803 | 90,278 | 34–54 | |||||
| DPP 8 | DPP8_Hs | 898 | 103,358 | Cytoplasm [26,33] | ||||
| DPP8_Tc | 825 | 95,048 | ||||||
| DPP8_Tm | 825 | 94,870 | ||||||
| DPP 10 | DPP10_Hs | 796 | 90,888 | 35–55 | Extracellular membrane [26,35] | |||
| DPP10.1_Tc | 835 | 96,053 | 37–58 | |||||
| DPP10.2_Tc | 845 | 95,598 | 46–66 | |||||
| DPP10.3_Tc | 858 | 97,129 | 25–45 | |||||
| DPP10.1_Tm | 843 | 96,808 | 45–66 | |||||
| DPP10.2_Tm | 882 | 99,854 | 79–100 | |||||
| PRCP | PRCP_Hs | 496 | 55,800 | 1–21 (0.99/0.98) | 1–21 (0.99/0.48) | Lysosome [36,37,38] | ||
| PRCP.1_Tc | 488 | 55,131 | 1–16 (0.99/0.98) | 1–16 (0.92/0.88) | ||||
| PRCP.2_Tc | 478 | 53,843 | 1–17 (0.99/0.98) | 1–17 (0.99/0.96) | ||||
| PRCP_Tm | 488 | 55,105 | 1–15 (0.99/0.98) | 1–15 (0.99/0.87) | ||||
| APP 1 | APP1_Hs | 623 | 69,918 | Cytoplasm [43] | ||||
| APP1.1_Tc | 615 | 69,050 | ||||||
| APP1.2_Tc | 690 | 79,273 | 1–13 (0.99/0.98) | 1–14 (0.89/0.37) | ||||
| APP1_Tm | partial | partial | ||||||
| APP 2 | APP2_Hs | 674 | 75,625 | 1–21 (0.99/0.98) | 1–23 (0.99/0.54) | Plasma membrane [42] | ||
| APP2_Tm | 709 | 81,041 | 1–25 (0.83/0.81) | 1–25 (0.99/0.97) | ||||
| APP 3 | APP3_Hs | 507 | 57,034 | 1–31 (0.89/0.72) | Mitochondrion [44] | |||
| APP3_Tc | 503 | 57,581 | 1–22 (0.58/0.43) | |||||
| APP3_Tm | 504 | 57,356 | –(0.32) | |||||
| XPD | XPD_Hs | 493 | 54,548 | - | ||||
| XPD_Tc | 487 | 54,319 | ||||||
| XPD_Tm | 489 | 54,168 | ||||||
| LAP | LAP_Hs | 519 | 56,166 | 1–27 (0.95/0.60) | Microsomes, cytoplasm [45] | |||
| LAP.1_Tc | 447 | 48,087 | ||||||
| LAP.2_Tc | 530 | 58,105 | 1–26 (0.94/0.71) | |||||
| LAP.3_Tc | 534 | 58,852 | 1–21 (0.93/0.81) | |||||
| LAP_Tm | 513 | 55,124 | 1–24 (0.91/0.31) | |||||
| CND | CND_Hs | 475 | 52,878 | Cytoplasm [46] | ||||
| CND_Tc | 478 | 53,583 | ||||||
| CND_Tm | 478 | 53,794 | ||||||
* After the cleavage site position of the signal peptide or mitochondrial transit peptide in brackets, the number before the slash is the likelihood of the specified localization domain appearing and the number after the slash is the probability of cleavage at the indicated position.
To get closer to understanding the possible functions of PSPs, we analyzed the presence of specific localization domains connected with secretory pathways, which in this case was the signal peptide (with transportation to the gut or lysosomes). We also searched for the mitochondrial transit peptide and transmembrane domains. The results of this study are summarized in Table 5.
Human and insect POP was predicted to lack signal peptide according to SignalP-6.0. However, POP_Tm was predicted to have it by the TargetP-2.0 server. Based on previous studies, human [28] and T. molitor [29] POP were shown to be cytoplasmic enzymes.
Among DPP 4 sequences, DPP4_Hs had a small transmembrane domain (20 amino acid residues). The TargetP-2.0 server predicted signal peptides for this sequence, but the probability of its appearance was not very high. According to experimental data, this protein exists in two forms: membrane-bound with a large extracellular domain on the cell surface [30] or soluble protein resulting from the cleavage of the extracellular domain from the short transmembrane part [31]. Tenebrionidae insects had two pairs of sequences coding DPP 4 proteins. The first homologous pair of sequences (DPP4.1_Tc–DPP4.1_Tm) had signal peptides predicted with high probability and proteins were presumed to be secreted. The other pair of DPP 4 sequences (DPP4.2_Tc–DPP4.2_Tm) had transmembrane domains and no signal peptide, similar to the human protein. The rest of the DPP 4-like proteins (DPP 8 and DPP 10 for both humans and insects) had no signal peptides. Human DPP 8 was shown to be a cytoplasmic peptidase [32,33,34]. All DPP 10 sequences had transmembrane domains, which is supported by previous data for this human protein [25,26,35].
Both human and insect PRCP sequences from the S28 family had signal peptides and were predicted to be secreted in agreement with previous studies [29,36,37,38,39,40].
Almost all metallopeptidase sequences had no specific domains connected to secretion, except for human and T. molitor APP2 and T. castaneum APP1.2_Tc, which had N-terminal signal peptides predicted. The APP1.2_Tc sequence is currently annotated in UniProt and NCBI databases as APP 1-like protein, but due to the predicted localization domain and conducted phylogenetic analysis (Figure 1) it probably belongs to APP2 proteins. Although there are no reliable data on the localization of such peptidases in the studied insect, there is some information about the localization of mammalian peptidases. It is known that signal peptide can serve as a signal-anchor sequence for transmembrane proteins of type III [41] and this is supported by data for human APP2, which was shown to be plasma membrane peptidase [42]. APP1 was localized predominantly in the cytoplasm, and this is supported by data for rat APP1 [43]. Human APP3 had mitochondrial localization [44], which is consistent with the predicted presence of the mitochondrial transit peptide in APP3_Hs and APP3_Tc (high probability), as well as its predicted presence in APP3_Tm (low probability). All LAP sequences, except for LAP.1_Tc, had the predicted mitochondrial transit peptide. Human LAP was found predominantly in microsomal fractions and cytoplasm [45], and CND was found also in cytoplasm with ubiquitous tissue distribution [46].
Overall, there was good agreement between the localization of predicted bioinformatics and experimental data on mammalian PSPs found in the literature (Table 5). Based on the predicted signal peptides for insect peptidases, only DPP4.1 and PRCP were presumed to be secreted proteins.
These bioinformatics predictions correlate with our previous and new biochemical data on the localization of several PSPs in the midgut of T. molitor larvae. We found that DPP 4 [48] and PRCP [29] were located mainly in the AM (anterior mesenteron, anterior part of the midgut), POP [29] was distributed evenly between the AM and PM (posterior mesenteron, posterior part of the midgut), and XPD [49] was localized predominantly in the PM. DPP 4 and PRCP were located in soluble gut contents, while POP and XPD were mainly located in tissue fractions.
We also show that the remaining PSP (APP) is distributed evenly between AM and PM parts of the gut, with more than half of its total activity belonging to soluble tissue fractions (Table 6).
Table 6.
Localization of T. molitor larval midgut peptidases *.
| PSPs | |||||
|---|---|---|---|---|---|
| DPP 4 | PRCP | POP | APP | XPD | |
| Part of the midgut | Peptidase activity, % from the total activity (AM + PM) | ||||
| AM | 60 | 73 | 49 | 52 | 37 |
| PM | 40 | 27 | 51 | 48 | 63 |
| Tissue distribution, % | |||||
| Soluble contents | 81 | 100 | 4 | 18 | 29 |
| Insoluble contents | 3 | - | - | 5 | 4 |
| Soluble tissue fraction | 7 | 0 | 94 | 58 | 57 |
| Insoluble tissue fraction | 9 | 2 | 19 | 10 | |
* The relative content of each peptidase in different fractions is presented.
2.5. Analysis of the Expression Levels of Genes Coding PSPs in T. castaneum and T. molitor Guts
Data on expression levels of genes coding the identified T. castaneum and T. molitor gut PSPs are presented in Table 7 as reads per kilobase per million mapped reads (RPKM). For better comparison of the possible digestive role of PSPs between two insects, we also calculated the expression levels as a percentage from the sum of expressions of all active PSPs. This calculation involved only active PSPs: POP, DPP 4, DPP 8, PRCP, APP1, APP2, APP3, and XPD. DPP 10 was not included in this calculation because it is an inactive DPP 4 homolog and does not possess peptidase activity, and LAP and CND were not included because they possess broad substrate specificity and the cleavage of post-proline bonds is only additional, not primary.
Table 7.
Gut mRNA expression levels of T. castaneum and T. molitor PSPs and PPCPbs.
| Peptidase | T. castaneum | T. molitor | ||||
|---|---|---|---|---|---|---|
| Sequence Name | Expression (RPKM) | Percentage from the Sum of Expressions of All Active PSPs * | Sequence Name | Expression (RPKM) | Percentage from the Sum of Expressions of All Active PSPs * | |
| POP | POP_Tc | - | 0 | POP_Tm | 25 | 4.2 |
| DPP 4 | DPP4.1_Tc | 1688 | 25.1 | DPP4.1_Tm | 147 | 24.6 |
| DPP4.2_Tc | 533 | 7.9 | DPP4.2_Tm | 102 | 17.1 | |
| DPP 8 | DPP8_Tc | 75 | 1.1 | DPP8_Tm | 20 | 3.3 |
| DPP 10 | DPP10.1_Tc | 8 | - | DPP10.1_Tm | 370 | - |
| DPP10.2_Tc | 5 | - | DPP10.2_Tm | 7 | - | |
| PRCP | PRCP.1_Tc | 27 | 0.4 | PRCP_Tm | 53 | 8.9 |
| PRCP.2_Tc | 675 | 10.0 | ||||
| APP1 | APP1.1_Tc | 2420 | 35.9 | APP1_Tm | 88 | 14.7 |
| APP1.2_Tc | 8 | 0.1 | ||||
| APP2 | APP2_Tm | 3 | 0.5 | |||
| APP3 | APP3_Tc | 58 | 0.9 | APP3_Tm | 12 | 2.0 |
| XPD | XPD_Tc | 1254 | 18.6 | XPD_Tm | 148 | 24.7 |
| LAP | LAP.1_Tc | 86 | - | LAP_Tm | 15 | - |
| CND | CND_Tc | 2462 | - | CND_Tm | 625 | - |
* This calculation involved only active PSPs: POP, DPP 4, DPP 8, PRCP, APP1, APP2, APP3, and XPD. DPP 10 was not included because it is an inactive DPP 4 homolog and does not possess peptidase activity. LAP and CND were not included because cleavage of post-proline bonds is not their main specificity.
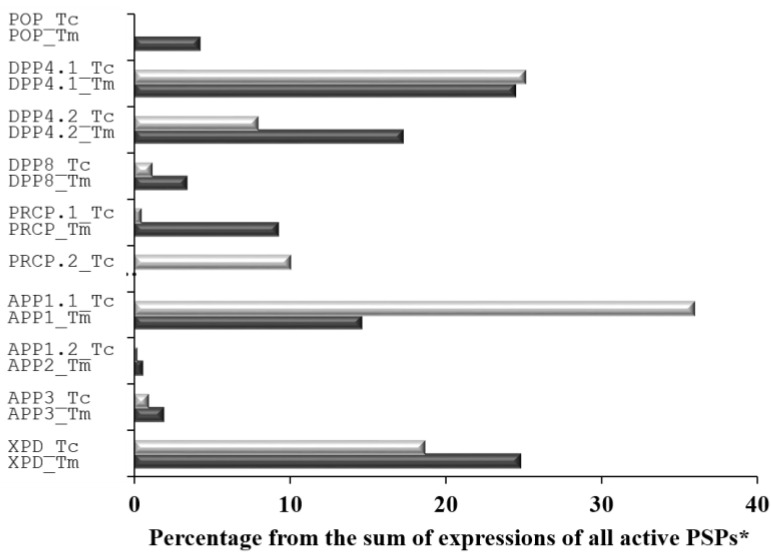
The highest expression level of S9 PSP mRNA was observed in the orthologous pair of secreted DPP4.1_Tc–DPP4.1_Tm (25% each from the sum of expressions of all active PSPs). The membrane pair DPP4.2_Tc–DPP4.2_Tm had lower expression levels of 8% and 17%, respectively.
The expression level of mRNA in T. castaneum PRCP (PRCP.1_Tc) was only 0.4%, whereas orthologous mRNA in T. molitor PRCP (PRCP_Tm) exhibited a 9% expression level. However, T. castaneum had another sequence (PRCP.2_Tc) with an expression level of 10%, for which there was no ortholog in the T. molitor transcriptome.
Among metal-dependent PSP mRNA, the APP1.1_Tc–APP1_Tm pair possessed high expression levels of about 36% and 15%. High expression was also evaluated for the XPD_Tc–XPD_Tm pair, about 19% and 25%, respectively.
All other mRNA of active PSPs from both organisms had expression levels of less than 5% of the sum of expressions of all active PSPs, including the single oligopeptidase POP that was expressed only in the gut of T. molitor (POP_Tm, 4%).
Among the rest of the mRNA sequences, there was a highly expressed inactive DPP 4 homolog in T. molitor (DPP10.1_Tm) whereas both orthologs of T. castaneum were low in terms of expression. PPCPbs mRNA (LAP and CND) had comparable expression levels in both insects, and CND had the highest expression levels among studied sequences in T. molitor and T. castaneum.
In general, there were similar expression profiles of PSP mRNA in both T. castaneum and T. molitor (Figure 2). Prospective orthologs had comparable expression levels.
Figure 2.
Gut mRNA expression levels of T. castaneum (light grey columns) and T. molitor (dark grey columns) active PSPs. * This calculation involved only active PSPs: POP, DPP 4, DPP 8, PRCP, APP1, APP2, APP3, and XPD. DPP 10 was not included because it is an inactive DPP 4 homolog and does not possess peptidase activity. LAP and CND were not included because cleavage of post-proline bonds is not their main specificity.
The highest level of gut expression among active PSPs belonged to the exopeptidases DPP 4, PRCP, APP1, and XPD, which may indicate their important role in tenebrionid physiology, particularly participation in the digestive process. It should be noted that POP had a low or non-existent level of gut expression, which makes its participation in the proteolysis of food proteins unlikely.
Localization studies together with the evaluation of the expression level of genes coding T. molitor PSPs in the gut transcriptome, as well as the presence of signal peptides connected with secretion, allowed for confirmation of the set of peptidases that can directly participate in the hydrolysis of food proteins in insect larval guts. This set consists of DPP4.1_Tm and PRCP_Tm, which are characterized as secreted peptidases localized in soluble gut contents with high expression levels of their mRNA in the gut transcriptome.
2.6. Gliadin Hydrolysis
Gliadin, a major storage protein of wheat, cannot be completely hydrolyzed by human digestive peptidases with the usual broad specificity. Proline- and glutamine-rich peptides, with the high content of proline and glutamine residues in their composition [1,2], remain unhydrolyzed [50] and can cause the development of diseases of the human gastrointestinal tract in predisposed people. In this regard, digestive peptidases of Tenebrionidae insects, in particular PSPs, may present the potential to hydrolyze these remaining fragments by serving as a proposed oral medicine as tenebrionids are stored product pests and gliadins are their major dietary proteins.
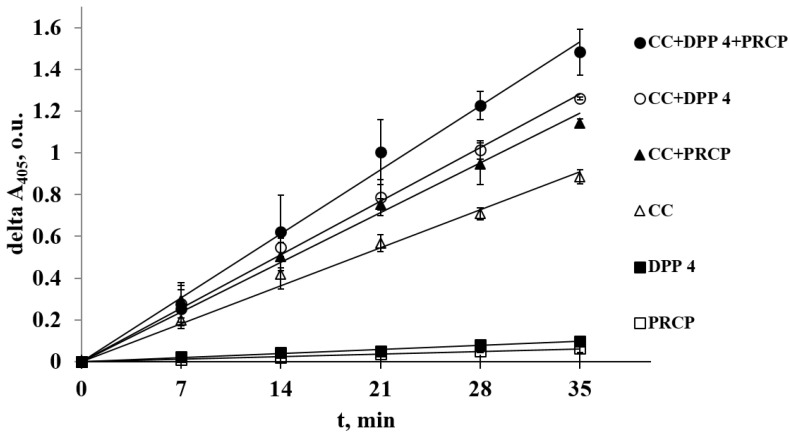
We studied the effect of the major luminal digestive T. molitor peptidases DPP4.1_Tm and PRCP_Tm, which are capable of cleaving bonds formed by proline on the total preparation of wheat gliadins (Figure 3). In addition, the effect of both individual PSPs and their combination with digestive post-glutamine cleaving peptidases of T. molitor (the total preparation of larval digestive cysteine cathepsins (CC)) [51] was studied. Proline-specific luminal enzymes (DPP 4 and PRCP) are exopeptidases, and CC are endopeptidases. Thus, we were able to study the combined action of endo- and exopeptidases on gliadins.
Figure 3.
Proteolysis of gliadins by six samples of T. molitor digestive peptidase preparations: 1. PRCP (white squares), 2. DPP 4 (black squares), 3. CC (white triangles), 4. CC + PRCP mixture (black triangles), 5. CC + DPP 4 mixture (white circles), and 6. CC + DPP 4 + PRCP mixture (black circles) (description of the curves from bottom to top). Proteolysis of gliadins was detected by an increase in free amino groups using 2,4,6-trinitrobenzene sulfonic acid (TNBS). The error bars represent standard deviation.
The enzymes were taken in equal amounts per larvae. Among the individual enzymes, the highest intensity of gliadin hydrolysis was achieved by the action of post-glutamine cleaving endopeptidases: CC preparation (62% of which belongs to the major digestive cathepsin L of T. molitor NCBI ID AJF94885 according to [52]) was eight times more active than studied PSPs. Among the used PSPs, DPP 4 exhibited 1.5-fold higher activity with gliadins than PRCP. Separate addition of DPP 4 and PRCP to CC increased hydrolysis by 1.4 and 1.3 times, respectively. The combination of all three peptidase preparations improved the rate of hydrolysis by 1.7 times compared to single CC preparation (Figure 3). It should be noted that, in all cases, the combined effect of endo- and exopeptidases was greater than the sum of the separate enzyme actions. This indicates that hydrolysis with endopeptidases forms additional substrates for exopeptidases, and this leads to an increase in the hydrolysis rate of the original protein.
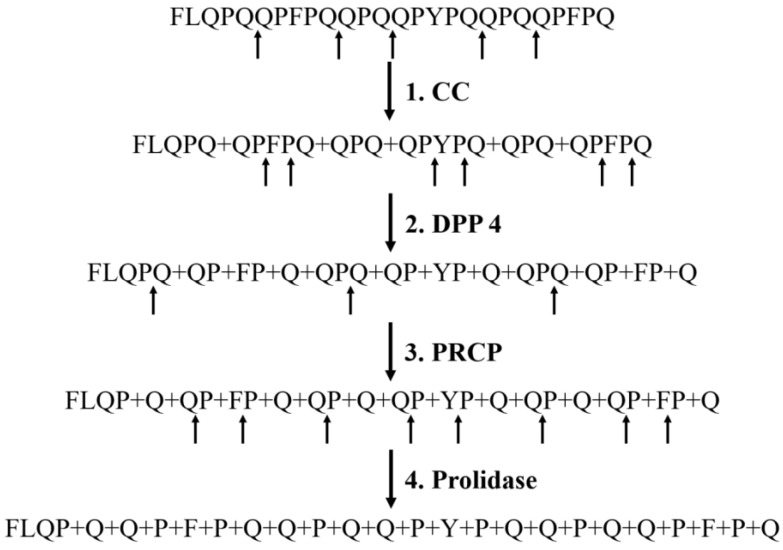
2.7. Hypothetical Scheme for the Complete Hydrolysis of Glutamine- and Proline-Rich 26-Mer Immunogenic Gliadin Fragments by T. molitor and T. castaneum Digestive Peptidases
Based on the obtained data, we suggested a hypothetical scheme for the complete hydrolysis of some gliadin fragments by T. molitor and T. castaneum digestive peptidases (Figure 4). As an example, we have taken the 26-mer FLQPQQPFPQQPQQPYPQQPQQPFPQ of γ-gliadin (UniProt: Q8L6B2), which along with the 33-mer peptide is a strong immunogenic peptide [50]. During the first stage of proteolysis, digestive post-glutamine cleaving endopeptidase CC can most effectively hydrolyze peptide bonds between two glutamine residues. Despite the ability of CC to hydrolyze bonds after the glutamine residue [53], the QP bond should not be cleaved by them, since the bonds formed by proline residue are hydrolyzed by the PSPs almost exclusively. Resulting fragments can be the substrates for DPP 4, which cleaves dipeptides such as QP, FP, and YP from their N-terminus. The remaining tripeptides (QPQ) can be effectively hydrolyzed by PRCP. Prolidase XPD may complete gliadin degradation, cleaving final dipeptides to single amino acids in the cytoplasm of the epithelial cells in the gut [49]. The remaining fragment FLQP is not immunogenic [50].
Figure 4.
Hypothetical scheme for the complete hydrolysis of glutamine- and proline-rich 26-mer immunogenic gliadin fragments by T. molitor and T. castaneum digestive CC and PSPs.
According to the scheme, the cleavage of bonds formed by glutamine residues by CC is an important first step in the hydrolysis process, and each of the found PSPs is essential for complete digestion of glutamine- and proline-rich gliadin fragments in the midgut of tenebrionids.
3. Discussion
A bioinformatic search for PSPs in the T. castaneum genome and both Tenebrionidae larvae gut transcriptomes revealed 14 distinct types of sequences in both insects: one POP, two DPP 4, one DPP 8, two PRCP, two APP1, one APP2, one APP3, one XPD sequence (all active peptidases), and three DPP 10 (inactive homologs of DPP 4). There also were two types of peptidases found with broad substrate specificity, possessing the ability to cleave bonds formed by proline residue (three LAP and one CND). The main difference in PSP sets in Tenebrionidae larvae gut transcriptomes was that T. castaneum, unlike T. molitor, had two PRCP sequences, two APP1 sequences, and lacked APP2 and POP sequences. Generally, Tenebrionidae insect PSP sets were similar to those in humans, lacking only DPP 9, FAP, DPP 2, as well as DPP 6, which is inactive homolog of DPP 4 together with DPP 10 found in all three species. Inactive homologs are represented in all major peptidase families. According to the MEROPS database, it is the serine peptidases that have the highest number of identified inactive enzymes, especially in insects. It has been suggested that a large number of both inactive and active serine peptidases indicates that the evolution of inactive peptidases and their new functions were beneficial for insects [54]. We have also shown a large number of serine peptidases and their inactive homologs in T. molitor [55], which can indicate that the emergence of inactive homologs is not an accident, but rather a natural process.
Phylogenetic analysis and multiple alignment of Tenebrionidae insect PSPs with those of humans allowed us to annotate all T. molitor peptidase sequences and refine annotation for several T. castaneum peptidase sequences based on analysis of cladograms of the tree, active centers, and substrate binding subsites.
The major digestive organ of T. molitor larvae is the midgut, where pH varies from 5.2 to 6.5 in the AM and 7.8 to 8.2 in the PM [56,57,58]. This gradient determines localization of the activity of different digestive enzymes along the midgut in accordance with their pH optima [59]. Gut peptidases may be localized either inside the gut lumen contents, and thus directly participate in digestion, or in the epithelial tissues lining the gut lumen. The combination of gene expression studies in the larval gut transcriptome, bioinformatic prediction of signal peptides, and biochemical localization experiments demonstrate the digestive function of the highly expressed secreted PSPs.
Using our previous and new data on the localization of T. molitor PSPs, we can summarize that DPP 4 [48] and PRCP [29] were located mainly in the AM, POP [29] and newly studied APP were distributed evenly between the AM and PM parts of the gut, and XPD [49] was localized predominantly in the PM. Analysis of the tissue distribution of the PSPs showed that DPP 4 [48] and PRCP [29] were located in soluble gut contents and can participate in T. molitor digestion, and this is in agreement with bioinformatic data (these peptidases were predicted to be secreted and highly expressed). The remaining POP, APP, and XPD were localized in tissues. Among them, XPD definitely seems to participate in digestion during the final stages as the substrates for XPD are dipeptides that can be absorbed by epithelial tissues [49]. There is also the possibility of APP participating in digestion because it can cleave oligopeptides and tripeptides, and tripeptides also can be absorbed by midgut epithelia [60]. POP hydrolyzes only oligopeptides, which cannot be absorbed by tissues. Compared to POP and APP, the participation of XPD in gliadin hydrolysis seems most likely and is supported by its high expression level. Since about 60% of XPD activity was located in the PM, it can act during the final stages of digestion and thus provide complete hydrolysis of gliadins.
Information from various sources about PSPs that can effectively break down gliadins was summarized in a previous review [61]. Attempts to hydrolyze gliadins were mostly limited by the search for peptidases capable of cleaving only peptide bonds formed by proline, such as prolyl oligopeptidases from Myxococcus xanthus, Sphingomonas capsulata, and Flavobacterium meningosepticum [62], prolyl endopeptidase from Aspergillus niger [63], and PSPs from the lactic acid bacteria Lactobacillus plantarum and Pediococcus pentosaceus [64]. However, the use of single PSPs did not achieve the complete hydrolysis of gliadins, and only the discovery of cysteine peptidase from Hordeum vulgare (EP-B2) in 2006 [65,66] allowed for the creation and testing of a dosage form for celiac disease treatment, which was based on a mixture of prolyl oligopeptidase from S. capsulata and cysteine peptidase from H. vulgare (EP-B2) (ALV003 Alvine, USA) and which is now under clinical trial [67,68]. The other cysteine peptidase capable of participating in the hydrolysis of gliadins was later described from Triticum aestivum [69].
Previously, we showed that T. molitor DPP 4 is able to hydrolyze gliadins more efficiently than human DPP 4 [48]. In the present study, we tested a natural complex of digestive enzymes of tenebrionids to discover whether this would increase the efficiency of hydrolysis by providing efficient hydrolysis of their proline-rich natural food substrates (such as gliadins) in natural conditions and ensuring sufficient intake of proline. Gliadin hydrolysis was studied using single digestive T. molitor peptidases and their mixtures capable of cleaving bonds formed by glutamine and proline residues. Among the separate enzymes, the highest intensity of hydrolysis was achieved by the action of post-glutamine cleaving cysteine cathepsins (CC), which corresponds with the hypothesis that they arose during evolution for the efficient hydrolysis of gliadins [51]. Individual digestive PSPs (DPP 4 and PRCP) were less active on intact gliadins, but the combined action of endo- and exopeptidases demonstrated the synergistic effect; this indicates that, after hydrolysis by endopeptidases, additional substrates for exopeptidases were formed, thus leading to an increase in the hydrolysis rate of the original protein.
Based on the obtained data, we suggested a hypothetical scheme for the complete hydrolysis of glutamine- and proline-rich 26-mer immunogenic gliadin fragments by T. molitor and T. castaneum digestive peptidases. The first stage of hydrolysis was carried out with endopeptidase CC, which can hydrolyze bonds formed by glutamine residues. DPP 4 then cleaved dipeptides from the resulting oligopeptides. The remaining tripeptides were hydrolyzed by PRCP. During the final stage, XPD degraded dipeptides to single amino acids in the cytoplasm of midgut epithelial cells.
The suggested hypothetical scheme is original and interesting because it can explain the hydrolysis of glutamine- and proline-rich gliadin fragments from different gliadins, including not only γ-gliadins (UniProt: P21292) containing PQQPFPQ repeats and ω-gliadins (UniProt: Q9FUW7) with PQQPFPQQ (which are the richest in glutamine and proline residues) but also α-gliadins (UniProt: P02863) with PQPQPFP and PQQPY repeats. Importantly, considering the suggested scheme, several proline- and glutamine-rich gliadin peptides may be degraded, such as QLQPFPQPQLPY, PQPQLPYPQPQLP, LGQQQPFPPQQPYPQPQPF, LQLQPF(PQPQLPY)3PQPQPF, and FLQPQQPF(PQQ)2PY(PQQ)2PFPQ that are not hydrolyzed by human digestive peptidases [50,70] and which cause autoimmune celiac disease. This indicates that it may be possible to develop an enzyme therapy drug for celiac disease and gluten intolerance that is based on tenebrionid digestive enzymes.
4. Materials and Methods
4.1. Preparation of the Biological Material and Sequencing of cDNA
Preparation of biological material from T. molitor and T. castaneum larvae guts and sequencing of cDNA was described previously [52]. The preparation of total gut RNA from T. molitor larvae from three independent biological replicates was sent to a sequencing facility (National Center for Genome Resources (NCGR), Santa Fe, NM, USA), where mRNA was isolated by polyA, standard libraries were made, and paired-end sequencing was performed on an Illumina HiSeq 2000 (San Diego, CA, USA) using standard protocols from the manufacturer. We obtained approximately 240 million sequence reads with a 250 bp insert.
Paired-end sequencing on the Illumina HiSeq 2000 of T. castaneum cDNA was performed by the High Throughput Genomics Center, Seattle, USA. We obtained approximately 340 million sequence reads, and insert size was approximately 250 bp.
4.2. Assembly of Contigs
A custom assembly of T. molitor sequences combined from all replicates was made by the NCGR, resulting in 197,800 contigs (minimum length = 100 and maximum length = 51328; Q1 = 123, Q2 = 153, Q3 = 335; N50 = 2232, B1000 = 71.9%, B2000 = 54.1%). For T. molitor sequences, we also combined the replicate data and included previous Sanger sequencing [71] and pyrosequencing [72] databases of mRNA from the larval gut and performed additional de novo assemblies using SeqManNGen (v. 4.0.1.4, DNAStar, Madison, WI USA) and custom assembly programs. For T. castaneum sequences, we used SeqManNGen to map sequences to the T. castaneum genome (Tcas3, NCBI; parameters for alignment were merSize = 19; 309,572,610 bp submitted, 263,305,494 aligned, 17,268,742 unaligned; sequence count score was >90%), as well as the Galaxy platform [73,74]. Potential coding sequences, starting at methionine and covering at least 20% of the mRNA sequence, were found in the T. molitor contigs using custom software.
4.3. Search for Insect PSP Sequences
Reference sequences were taken from the human genome and downloaded from the UniProt database [75,76]. The list of reference human PSPs is given in Table 1. Initially, translated T. molitor ORFs homologous to human PSP query sequences were determined using the algorithm tblastx [77,78]. Multiple alignment was constructed by means of pairwise alignments using the Wagner–Fisher algorithm [79]. ORFs that were grouped into blocks with an identity of at least 95% and that overlapped with another block of at least 10 amino acid residues were considered as referring to one unique peptidase. Multiple sequence alignment (MAFFT) [80] was used to refine and build consensus sequences. The predicted start of the T. molitor mature enzyme sequences was identified by sequence homology through alignment with mature human PSPs. Multiple alignments of the analyzed amino acid sequences were performed using the program Clustal Omega [81,82].
T. castaneum peptidase sequences were found in the NCBI database [83,84] via Protein BLAST, using human PSPs as queries. Found sequences that lacked annotation in the database (namely DPP4.2_Tc, DPP10.2_Tc, and DPP10.3_Tc) were annotated during the subsequent construction of a phylogenetic tree and the analysis of multiple alignments.
4.4. Phylogenetic Analysis and Orthology Predictions
A phylogenetic tree of human, T. castaneum, and T. molitor PSPs was constructed in MEGA X [85,86] using the maximum likelihood method and 200 bootstrap repetitions. Based on the cladogram, pairs of sequences from T. castaneum and T. molitor were considered orthologous if they formed a single clade.
4.5. Analysis of the Expression Levels
Analysis of the expression levels was carried out as written in [52]. To analyze the expression of peptidase transcripts in T. castaneum, we used reads mapped to the genome of T. castaneum (Tcas3) by SeqManPro (DNAStar) to obtain expression values in normalized reads per kilobase per million mapped reads (RPKM) [87].
For T. molitor, about 40 million reads were obtained from each of the three RNA-Seq datasets. BLAST was used to obtain all nucleotide sequences in the contigs that potentially encode peptidases. The contigs were used to assemble and refine sequences of complete peptidase mRNA. Refined peptidase mRNA sequences were used to identify contigs in each assembly with at least 97% sequence identity, which were then used for expression calculations. If a contig aligned only partially to the mRNA, its contribution to expression was proportional to the aligned part if it was more than 50% of the contig’s length. RPKM was used to calculate the number of reads mapped to a contig when calculating each multiread as one unit.
4.6. Analysis of the Structure of PSP Sequences
Active centers and substrate binding subsites were analyzed using multiple alignments of studied peptidases (performed in Clustal Omega [81,82] and visualized in GeneDoc [88,89]). The SignalP-6.0 server [90,91] and TargetP-2.0 [92,93] were used to analyze N-terminal parts in the translated mRNA sequences, predict the presence and location of signal peptide cleavage sites and the subsequent secretion of proteins, and predict the presence of mitochondrial transit peptides. Trans-membrane domains were predicted using the DeepTMHMM server [94,95]. Molecular masses were calculated using the online server ExPASy [96]. Molecular masses and sequence lengths for human and T. castaneum PSPs were taken from the UniProt and NCBI databases.
4.7. Enzyme Assays Using Chromogenic p-Nitroanilide Substrates
Peptidase activity with chromogenic p-nitroanilide substrates was determined spectrophotometrically according to [97] at 405 nm with an ELx808 Absorbance Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA) and was calculated using the initial rates of hydrolysis.
Exact protocols for studied peptidases were described earlier: CC with 0.5 mM Glp-Phe-Gln-pNA (was obtained according to [53]) at pH 5.6 with 6 mM Cys [53], DPP 4 with 0.25 mM Ala-Pro-pNA (was synthesized according to standard procedures [98]) at pH 7.9 [48], and PRCP with 0.5 mM Z-Ala-Ala-Pro-pNA (was synthesized according to standard procedures [98]) at pH 5.6 [99].
4.8. Enzyme Assays Using Fluorogenic Substrates
The reaction mixture was placed in a microplate well and included 5-50 µL of APP enzyme, 10 µL of 15 mM MnCl2, and 50 mM Tris-HCl buffer (pH 7.5) to a total volume of 195 µL. After 20 min of preincubation at room temperature, 5 µL of 3 mM Lys(Abz)-Pro-Pro-pNA (Bachem, Bubendorf BL, Switzerland) dissolved in DMF was added. APP activity toward the fluorogenic substrate Lys(Abz)-Pro-Pro-pNA was determined at λex 340 nm and λem 400 nm with a FLx800 Fluorescence Microplate Reader (BioTek Instruments, Inc., USA), and was calculated as described in the previous section.
4.9. Localization Studies
For localization studies, fractionation of AM (anterior mesenteron, anterior part of the midgut) and PM (posterior mesenteron, posterior part of the midgut) parts from T. molitor larvae was performed as described in [48]. AM and PM sections were isolated from fourth-instar T. molitor larvae and were split longitudinally. The contents were extruded and retained, while the tissue was rinsed with 0.9% saline. The lumen contents or the gut tissues were pooled from 50 AM or PM (for each of the four biological replicates) and were homogenized separately at 4 °C in 0.5 mL of double distilled water in a glass Dounce homogenizer and centrifuged for 1 h at 35,000× g. There were two fractions after centrifugation of the lumen contents: supernatant (soluble contents) and precipitate (insoluble contents). After centrifugation of the tissues there were also two fractions: supernatant (soluble tissues fraction) and precipitate (insoluble tissues fraction). All fractions were stored at −70 °C. The activity of the studied peptidase was measured in each fraction using the same larval gut equivalents of material and the corresponding substrate. Measurements were carried out in 3 biological replicates with 3–5 technical repetitions. Standard deviation did not exceed 5–10%. The relative content of each peptidase in the fractions was then calculated.
4.10. Gliadin Proteolysis Assay
To isolate digestive peptidases, extract from 200 pooled AM gut sections (2–2.5 mL) was applied to a Sephadex G-150 column (2.7 × 110 cm). Elution was performed with 0.01 M phosphate buffer (pH 5.6) containing 0.5 M NaCl at 4 °C. Fractions of 9.0 mL in volume were collected and analyzed for protein content at 280 nm and for hydrolytic activity with a specific chromogenic substrate for each peptidase: 0.5 mM Glp-Phe-Gln-pNA at pH 5.6 with 6 mM Cys for CC, 0.25 mM Ala-Pro-pNA at pH 7.9 for DPP 4, and 0.5 mM Z-Ala-Ala-Pro-pNA at pH 5.6 for PRCP. Active fractions corresponding to the major peak of each peptidase’s activity were pooled separately, concentrated, and desalted on Amicon YM3 membranes (Amicon, Winsum, Netherlands) at 4 °C and either used immediately for further analysis or stored at −70 °C.
Proteolysis of gliadins was measured by an increase in free amino groups with 2,4,6-trinitrobenzene sulfonic acid (TNBS) (Reachem, Moscow, Russia) [100]. For a comparative assay of activity, the enzymes were taken in equal amounts per larvae. We used an aliquot of studied peptidases (CC, DPP 4, or PRCP) corresponding to the activity of 1.7 T. molitor larval guts. This was calculated using data regarding the activity of the enzymes in the initial midgut extract in the presence of 2 M urea. The activity of CC was measured with 0.5 mM Glp-Phe-Gln-pNA at pH 5.6 with 6 mM Cys, DPP 4 was measured with 0.25 mM Ala-Pro-pNA at pH 7.9, and PRCP was measured with 0.5 mM Z-Ala-Ala-Pro-pNA at pH 5.6. The gliadin proteolysis reaction mixture included 750 µg of gliadins (MP Biomedicals, Illkirch, France) dissolved in 0.1 M universal acetate–phosphate–borate buffer (UB) [101] at a pH of 6.5, 3 mM DTT, 2 M urea, and T. molitor purified enzymes equal to 1.7 larvae. The final volume was 307.5 µL. In total, 20 μL of the reaction mixture was removed at time zero and at 7 min intervals over a 35 min incubation period at 37 °C. The reaction with TNBS was carried out immediately according to the protocol described in [100]. The optical absorbance of the colored reaction product was measured at 405 nm with an ELx808 Absorbance Microplate Reader (BioTek Instruments, Inc., USA).
Acknowledgments
We are grateful to Brenda Oppert for participating in the discussion of our results and providing valuable comments.
Author Contributions
Conceptualization, E.N.E.; data curation, I.Y.F.; formal analysis, Y.E.D.; investigation, V.F.T. and I.A.G.; methodology, V.F.T.; validation, M.A.B.; writing—original draft, V.F.T.; writing—review and editing, I.Y.F., Y.E.D. and E.N.E. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was supported by the Russian Science Foundation (project № 22-24-00553).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Shewry P.R., Tatham A.S. The prolamin storage proteins of cereal seeds: Structure and evolution. Biochem. J. 1990;267:1–12. doi: 10.1042/bj2670001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shewry P.R., Halford N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002;53:947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- 3.Misener S.R., Chen C., Walker V.K. Cold tolerance and proline metabolic gene expression in Drosophila melanogaster. J. Insect Physiol. 2001;47:393–400. doi: 10.1016/S0022-1910(00)00141-4. [DOI] [PubMed] [Google Scholar]
- 4.Teulier L., Weber J.M., Crevier J., Darveau C.A. Proline as a fuel for insect flight: Enhancing carbohydrate oxidation in hymenopterans. Proc. Biol. Sci. 2016;283:20160333. doi: 10.1098/rspb.2016.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnot G., Peypelut L., Gérard Febvay L., Fleurat-Lessard F., Fields P.G. The effect of cold acclimation and deacclimation on cold tolerance, trehalose and free amino acid levels in Sitophilus granarius and Cryptolestes ferrugineus (Coleoptera) J. Insect Physiol. 1998;44:955–965. doi: 10.1016/s0022-191000055-9. [DOI] [PubMed] [Google Scholar]
- 6.Bonnett T.R., Robert J.A., Pitt C., Fraser J.D., Keeling C.I., Bohlmann J., Huber D.P. Global and comparative proteomic profiling of overwintering and developing mountain pine beetle, Dendroctonus ponderosae (Coleoptera: Curculionidae), larvae. Insect Biochem. Mol. Biol. 2012;42:890–901. doi: 10.1016/j.ibmb.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham D.F., O’Connor B. Proline specific peptidases. Biochim. Biophys. Acta. 1997;1343:160–186. doi: 10.1016/S0167-4838(97)00134-9. [DOI] [PubMed] [Google Scholar]
- 8.Rawlings N.D., Salvesen G., editors. Handbook of Proteolytic Enzymes. 3rd ed. Academic Press; London, UK: 2013. p. 4104. [Google Scholar]
- 9.Dunaevsky Y.E., Tereshchenkova V.F., Oppert B., Belozersky M.A., Filippova I.Y., Elpidina E.N. Human proline specific peptidases: A comprehensive analysis. Biochim. Biophys. Acta Gen. Subj. 2020;1864:129636. doi: 10.1016/j.bbagen.2020.129636. [DOI] [PubMed] [Google Scholar]
- 10.Rawlings N.D., Waller M., Barrett A.J., Bateman A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darmoul D., Voisin T., Couvineau A., Rouyer-Fessard C., Salomon R., Wang Y., Swallow D.M., Laburthe M. Regional expression of epithelial dipeptidyl peptidase IV in the human intestines. Biochem. Biophys. Res. Commun. 1994;203:1224–1229. doi: 10.1006/bbrc.1994.2313. [DOI] [PubMed] [Google Scholar]
- 12.Rubino A., Vetrella M., Pierro M., Rey J., Jos J., Auricchio S. Intestinal glutaminyl-proline dipeptidase activity in children with treated coeliac disease. Rev. Eur. Etud. Clin. Biol. 1972;17:993–997. [PubMed] [Google Scholar]
- 13.Grävinghoff J., Hütter H.J. The activities of alanine aminopeptidase, leucine aminopeptidase, proline dipeptidase and prolyl dipeptidase in the mucosa of the small intestine. Investigations on normal children and patients with the malabsorption syndrome. Eur. J. Pediatr. 1977;127:57–62. doi: 10.1007/BF00465566. [DOI] [PubMed] [Google Scholar]
- 14.Sjöström H., Norén O., Krasilnikoff P.A., Gudmand-Høyer E. Intestinal peptidases and sucrase in coeliac disease. Clin. Chim. Acta. 1981;109:53–58. doi: 10.1016/0009-8981(81)90136-4. [DOI] [PubMed] [Google Scholar]
- 15.Ganapathy V., Pashley S.J., Roesel R.A., Pashley D.H., Leibach F.H. Inhibition of rat and human prolidases by captopril. Biochem. Pharmacol. 1985;34:1287–1291. doi: 10.1016/0006-2952(85)90507-6. [DOI] [PubMed] [Google Scholar]
- 16.Tribolium Genome Sequencing Consortium The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 17.Polgár L. The prolyl oligopeptidase family. Cell Mol. Life Sci. 2002;59:349–362. doi: 10.1007/s00018-002-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rea D., Fülöp V. Structure-function properties of prolyl oligopeptidase family enzymes. Cell Biochem. Biophys. 2006;44:349–365. doi: 10.1385/CBB:44:3:349. [DOI] [PubMed] [Google Scholar]
- 19.Szeltner Z., Polgár L. Structure, function and biological relevance of prolyl oligopeptidase. Curr. Protein Pept. Sci. 2008;9:96–107. doi: 10.2174/138920308783565723. [DOI] [PubMed] [Google Scholar]
- 20.Aertgeerts K., Ye S., Tennant M.G., Kraus M.L., Rogers J., Sang B.C., Skene R.J., Webb D.R., Prasad G.S. Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Sci. 2004;13:412–421. doi: 10.1110/ps.03460604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiramatsu H., Yamamoto A., Kyono K., Higashiyama Y., Fukushima C., Shima H., Sugiyama S., Inaka K., Shimizu R. The crystal structure of human dipeptidyl peptidase IV (DPPIV) complex with diprotin A. Biol. Chem. 2004;385:561–564. doi: 10.1515/BC.2004.068. [DOI] [PubMed] [Google Scholar]
- 22.Meadows S.A., Edosada C.Y., Mayeda M., Tran T., Quan C., Raab H., Wiesmann C., Wolf B.B. Ala657 and conserved active site residues promote fibroblast activation protein endopeptidase activity via distinct mechanisms of transition state stabilization. Biochemistry. 2007;46:4598–4605. doi: 10.1021/bi062227y. [DOI] [PubMed] [Google Scholar]
- 23.Park J.E., Lenter M.C., Zimmermann R.N., Garin-Chesa P., Old L.J., Rettig W.J. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J. Biol. Chem. 1999;274:36505–36512. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 24.Aertgeerts K., Levin I., Shi L., Snell G.P., Jennings A., Prasad G.S., Zhang Y., Kraus M.L., Salakian S., Sridhar V., et al. Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha. J. Biol. Chem. 2005;280:19441–19444. doi: 10.1074/jbc.C500092200. [DOI] [PubMed] [Google Scholar]
- 25.Yokotani N., Doi K., Wenthold R.J., Wada K. Non-conservation of a catalytic residue in a dipeptidyl aminopeptidase IV-related protein encoded by a gene on human chromosome 7. Hum. Mol. Genet. 1993;2:1037–1039. doi: 10.1093/hmg/2.7.1037. [DOI] [PubMed] [Google Scholar]
- 26.Qi S.Y., Riviere P.J., Trojnar J., Junien J.L., Akinsanya K.O. Cloning and characterization of dipeptidyl peptidase 10, a new member of an emerging subgroup of serine proteases. Biochem. J. 2003;373 (Pt 1):179–189. doi: 10.1042/bj20021914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Lou Z., Li X., Zhou W., Ma M., Cao Y., Geng Y., Bartlam M., Zhang X.C., Rao Z. Structure of human cytosolic X-prolyl aminopeptidase: A double Mn(II)-dependent dimeric enzyme with a novel three-domain subunit. J. Biol. Chem. 2008;283:22858–22866. doi: 10.1074/jbc.M710274200. [DOI] [PubMed] [Google Scholar]
- 28.Walter R. Partial purification and characterization of post-proline cleaving enzyme: Enzymatic inactivation of neurohypophyseal hormones by kidney preparations of various species. Biochim. Biophys. Acta. 1976;422:138–158. doi: 10.1016/0005-2744(76)90015-2. [DOI] [PubMed] [Google Scholar]
- 29.Goptar I.A., Filippova I.Y., Lysogorskaya E.N., Oksenoit E.S., Vinokurov K.S., Zhuzhikov D.P., Bulushova N.V., Zalunin I.A., Dunaevsky Y.E., Belozersky M.A., et al. Localization of post-proline cleaving peptidases in Tenebrio molitor larval midgut. Biochimie. 2008;90:508–514. doi: 10.1016/j.biochi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Alfalah M., Jacob R., Naim H.Y. Intestinal dipeptidyl peptidase IV is efficiently sorted to the apical membrane through the concerted action of N- and O-glycans as well as association with lipid microdomains. J. Biol. Chem. 2002;277:10683–10690. doi: 10.1074/jbc.M109357200. [DOI] [PubMed] [Google Scholar]
- 31.Ghersi G., Zhao Q., Salamone M., Yeh Y., Zucker S., Chen W.T. The protease complex consisting of dipeptidyl peptidase IV and seprase plays a role in the migration and invasion of human endothelial cells in collagenous matrices. Cancer Res. 2006;66:4652–4661. doi: 10.1158/0008-5472.CAN-05-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbott C.A., Yu D.M., Woollatt E., Sutherland G.R., McCaughan G.W., Gorrell M.D. Cloning, expression and chromosomal localization of a novel human dipeptidyl peptidase (DPP) IV homolog, DPP8. Eur. J. Biochem. 2000;267:6140–6150. doi: 10.1046/j.1432-1327.2000.01617.x. [DOI] [PubMed] [Google Scholar]
- 33.Ajami K., Abbott C.A., McCaughan G.W., Gorrell M.D. Dipeptidyl peptidase 9 has two forms, a broad tissue distribution, cytoplasmic localization and DPIV-like peptidase activity. Biochim. Biophys. Acta. 2004;1679:18–28. doi: 10.1016/j.bbaexp.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Yu D.M., Ajami K., Gall M.G., Park J., Lee C.S., Evans K.A., McLaughlin E.A., Pitman M.R., Abbott C.A., McCaughan G.W., et al. The in vivo expression of dipeptidyl peptidases 8 and 9. J. Histochem. Cytochem. 2009;57:1025–1040. doi: 10.1369/jhc.2009.953760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen M., Heinzmann A., Noguchi E., Abecasis G., Broxholme J., Ponting C.P., Bhattacharyya S., Tinsley J., Zhang Y., Holt R., et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat. Genet. 2003;35:258–263. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- 36.Kakimoto T., Oshima G., Yeh H.S., Erdös E.G. Purification of lysosomal prolycarboxypeptidase angiotensinase C. Biochim. Biophys. Acta. 1973;302:178–182. doi: 10.1016/0005-2744(73)90021-1. [DOI] [PubMed] [Google Scholar]
- 37.Odya C.E., Marinkovic D.V., Hammon K.J., Stewart T.A., Erdös E.G. Purification and properties of prolylcarboxypeptidase (angiotensinase C) from human kidney. J. Biol. Chem. 1978;253:5927–5931. doi: 10.1016/S0021-9258(17)34557-X. [DOI] [PubMed] [Google Scholar]
- 38.Tan F., Morris P.W., Skidgel R.A., Erdös E.G. Sequencing and cloning of human prolylcarboxypeptidase (angiotensinase C). Similarity to both serine carboxypeptidase and prolylendopeptidase families. J. Biol. Chem. 1993;268:16631–16638. doi: 10.1016/S0021-9258(19)85465-0. [DOI] [PubMed] [Google Scholar]
- 39.Chiravuri M., Agarraberes F., Mathieu S.L., Lee H., Huber B.T. Vesicular localization and characterization of a novel post-proline-cleaving aminodipeptidase, quiescent cell proline dipeptidase. J. Immunol. 2000;165:5695–5702. doi: 10.4049/jimmunol.165.10.5695. [DOI] [PubMed] [Google Scholar]
- 40.Maes M.B., Lambeir A.M., Gilany K., Senten K., Van der Veken P., Leiting B., Augustyns K., Scharpé S., De Meester I. Kinetic investigation of human dipeptidyl peptidase II (DPPII)-mediated hydrolysis of dipeptide derivatives and its identification as quiescent cell proline dipeptidase (QPP)/dipeptidyl peptidase 7 (DPP7) Biochem. J. 2005;386 (Pt 2):315–324. doi: 10.1042/BJ20041156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyde R.J., Hooper N.M., Turner A.J. Molecular cloning and expression in COS-1 cells of pig kidney aminopeptidase P. Biochem. J. 1996;319 (Pt 1):197–201. doi: 10.1042/bj3190197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venema R.C., Ju H., Zou R., Venema V.J., Ryan J.W. Cloning and tissue distribution of human membrane-bound aminopeptidase P. Biochim. Biophys. Acta. 1997;1354:45–48. doi: 10.1016/S0167-4781(97)00126-7. [DOI] [PubMed] [Google Scholar]
- 43.Czirják G., Burkhart W.A., Moyer M.B., Antal J., Shears S.B., Enyedi P. Cloning and functional expression of the cytoplasmic form of rat aminopeptidase P. Biochim. Biophys. Acta. 1999;1444:326–336. doi: 10.1016/S0167-4781(99)00005-6. [DOI] [PubMed] [Google Scholar]
- 44.O’Toole J.F., Liu Y., Davis E.E., Westlake C.J., Attanasio M., Otto E.A., Seelow D., Nurnberg G., Becker C., Nuutinen M., et al. Individuals with mutations in XPNPEP3, which encodes a mitochondrial protein, develop a nephronophthisis-like nephropathy. J. Clin. Investig. 2010;120:791–802. doi: 10.1172/JCI40076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohno H., Kanno T. Properties and activities of aminopeptidases in normal and mitogen-stimulated human lymphocytes. Biochem. J. 1985;226:59–65. doi: 10.1042/bj2260059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teufel M., Saudek V., Ledig J.P., Bernhardt A., Boularand S., Carreau A., Cairns N.J., Carter C., Cowley D.J., Duverger D., et al. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J. Biol. Chem. 2003;278:6521–6531. doi: 10.1074/jbc.M209764200. [DOI] [PubMed] [Google Scholar]
- 47.Durinx C., Lambeir A.M., Bosmans E., Falmagne J.B., Berghmans R., Haemers A., Scharpé S., De Meester I. Molecular characterization of dipeptidyl peptidase activity in serum: Soluble CD26/dipeptidyl peptidase IV is responsible for the release of X-Pro dipeptides. Eur. J. Biochem. 2000;267:5608–5613. doi: 10.1046/j.1432-1327.2000.01634.x. [DOI] [PubMed] [Google Scholar]
- 48.Tereshchenkova V.F., Goptar I.A., Kulemzina I.A., Zhuzhikov D.P., Serebryakova M.V., Belozersky M.A., Dunaevsky Y.E., Oppert B., Filippova I.Y., Elpidina E.N. Dipeptidyl peptidase 4—An important digestive peptidase in Tenebrio molitor larvae. Insect Biochem. Mol. Biol. 2016;76:38–48. doi: 10.1016/j.ibmb.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Tereshchenkova V.F., Goptar I.A., Zhuzhikov D.P., Belozersky M.A., Dunaevsky Y.E., Oppert B., Filippova I.Y., Elpidina E.N. Prolidase is a critical enzyme for complete gliadin digestion in Tenebrio molitor larvae. Arch. Insect Biochem. Physiol. 2017;95:e21395. doi: 10.1002/arch.21395. [DOI] [PubMed] [Google Scholar]
- 50.Shan L., Qiao S.W., Arentz-Hansen H., Molberg Ø., Gray G.M., Sollid L.M., Khosla C. Identification and analysis of multivalent proteolytically resistant peptides from gluten: Implications for celiac sprue. J. Proteome Res. 2005;4:1732–1741. doi: 10.1021/pr050173t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goptar I.A., Semashko T.A., Danilenko S.A., Lysogorskaya E.N., Oksenoit E.S., Zhuzhikov D.P., Belozersky M.A., Dunaevsky Y.E., Oppert B., Filippova I.Y., et al. Cysteine digestive peptidases function as post-glutamine cleaving enzymes in tenebrionid stored-product pests. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012;161:148–154. doi: 10.1016/j.cbpb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Martynov A.G., Elpidina E.N., Perkin L., Oppert B. Functional analysis of C1 family cysteine peptidases in the larval gut of Тenebrio molitor and Tribolium castaneum. BMC Genom. 2015;16:75. doi: 10.1186/s12864-015-1306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Filippova I.Y., Dvoryakova E.A., Sokolenko N.I., Simonyan T.R., Tereshchenkova V.F., Zhiganov N.I., Dunaevsky Y.E., Belozersky M.A., Oppert B., Elpidina E.N. New Glutamine-Containing Substrates for the Assay of Cysteine Peptidases From the C1 Papain Family. Front. Mol. Biosci. 2020;7:578758. doi: 10.3389/fmolb.2020.578758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pils B., Schultz J. Inactive enzyme-homologues find new function in regulatory processes. J. Mol. Biol. 2004;340:399–404. doi: 10.1016/j.jmb.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 55.Zhiganov N.I., Tereshchenkova V.F., Oppert B., Filippova I.Y., Belyaeva N.V., Dunaevsky Y.E., Belozersky M.A., Elpidina E.N. The dataset of predicted trypsin serine peptidases and their inactive homologs in Tenebrio molitor transcriptomes. Data Brief. 2021;38:107301. doi: 10.1016/j.dib.2021.107301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terra W.R., Ferreira C., Bastos F. Phylogenetic consideration of insect digestion. Disaccharidases and the spatial organization of digestion in the Tenebrio molitor larvae. Insect Biochem. 1985;15:443–449. doi: 10.1016/0020-1790(85)90056-3. [DOI] [Google Scholar]
- 57.Vinokurov K.S., Elpidina E.N., Oppert B., Prabhakar S., Zhuzhikov D.P., Dunaevsky Y.E., Belozersky M.A. Diversity of digestive proteinases in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006;145:126–137. doi: 10.1016/j.cbpb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 58.Elpidina E.N., Goptar I.A. Digestive peptidases in Tenebrio molitor and possibility of use to treat celiac disease. Entomol. Res. 2007;37:139–147. doi: 10.1111/j.1748-5967.2007.00103.x. [DOI] [Google Scholar]
- 59.Vinokurov K.S., Elpidina E.N., Oppert B., Prabhakar S., Zhuzhikov D.P., Dunaevsky Y.E., Belozersky M.A. Fractionation of digestive proteinases from Tenebrio molitor (Coleoptera: Tenebrionidae) larvae and role in protein digestion. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006;145:138–146. doi: 10.1016/j.cbpb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Silk D.B. Digestion and absorption of dietary protein in man. Proc. Nutr. Soc. 1980;39:61–70. doi: 10.1079/PNS19800009. [DOI] [PubMed] [Google Scholar]
- 61.Dunaevsky Y.E., Tereshchenkova V.F., Belozersky M.A., Filippova I.Y., Oppert B., Elpidina E.N. Effective Degradation of Gluten and Its Fragments by Gluten-Specific Peptidases: A Review on Application for the Treatment of Patients with Gluten Sensitivity. Pharmaceutics. 2021;13:1603. doi: 10.3390/pharmaceutics13101603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shan L., Marti T., Sollid L.M., Gray G.M., Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: Implications for coeliac sprue. Biochem. J. 2004;383 (Pt 2):311–318. doi: 10.1042/BJ20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.ClinicalTrials.gov. [(accessed on 30 November 2022)]; Available online: https://clinicaltrials.gov/ct2/show/NCT00810654.
- 64.Gerez C.L., Font de Valdez G., Rollán G.C. Functionality of lactic acid bacteria peptidase activities in the hydrolysis of gliadin-like fragments. Lett. Appl. Microbiol. 2008;47:427–432. doi: 10.1111/j.1472-765X.2008.02448.x. [DOI] [PubMed] [Google Scholar]
- 65.Gass J., Vora H., Bethune M.T., Gray G.M., Khosla C. Effect of barley endoprotease EP-B2 on gluten digestion in the intact rat. J. Pharmacol. Exp. Ther. 2006;318:1178–1186. doi: 10.1124/jpet.106.104315. [DOI] [PubMed] [Google Scholar]
- 66.Gass J., Bethune M.T., Siegel M., Spencer A., Khosla C. Combination enzyme therapy for gastric digestion of dietary gluten in patients with celiac sprue. Gastroenterology. 2007;133:472–480. doi: 10.1053/j.gastro.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 67.Tye-Din J.A., Anderson R.P., Ffrench R.A., Brown G.J., Hodsman P., Siegel M., Botwick W., Shreeniwas R. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin. Immunol. 2010;134:289–295. doi: 10.1016/j.clim.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 68.Lähdeaho M.L., Kaukinen K., Laurila K., Vuotikka P., Koivurova O.P., Kärjä-Lahdensuu T., Marcantonio A., Adelman D.C., Mäki M. Glutenase ALV003 attenuates gluten-induced mucosal injury in patients with celiac disease. Gastroenterology. 2014;146:1649–1658. doi: 10.1053/j.gastro.2014.02.031. [DOI] [PubMed] [Google Scholar]
- 69.Savvateeva L.V., Gorokhovets N.V., Makarov V.A., Serebryakova M.V., Solovyev A.G., Morozov S.Y., Reddy V.P., Zernii E.Y., Zamyatnin A.A., Jr., Aliev G. Glutenase and collagenase activities of wheat cysteine protease Triticain-α: Feasibility for enzymatic therapy assays. Int. J. Biochem. Cell Biol. 2015;62:115–124. doi: 10.1016/j.biocel.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 70.Shan L., Molberg Ø., Parrot I., Hausch F., Filiz F., Gray G.M., Sollid L.M., Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 71.Prabhakar S., Chen M.S., Elpidina E.N., Vinokurov K.S., Smith C.M., Marshall J., Oppert B. Sequence analysis and molecular characterization of larval midgut cDNA transcripts encoding peptidases from the yellow mealworm, Tenebrio molitor L. Insect Mol. Biol. 2007;16:455–468. doi: 10.1111/j.1365-2583.2007.00740.x. [DOI] [PubMed] [Google Scholar]
- 72.Oppert B., Dowd S.E., Bouffard P., Li L., Conesa A., Lorenzen M.D., Toutges M., Marshall J., Huestis D.L., Fabrick J., et al. Transcriptome profiling of the intoxication response of Tenebrio molitor larvae to Bacillus thuringiensis Cry3Aa protoxin. PLoS ONE. 2012;7:e34624. doi: 10.1371/journal.pone.0034624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goecks J., Nekrutenko A., Taylor J., Galaxy Team Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.UniProt Consortium UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.UniProtKB. [(accessed on 2 November 2022)]. Available online: https://www.uniprot.org.
- 77.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 78.Basic Local Alignment Search Tool. [(accessed on 2 November 2022)]; Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome.
- 79.Wagner R.A., Fischer M.J. The string-to-string correction problem. JACM. 1974;21:168–173. doi: 10.1145/321796.321811. [DOI] [Google Scholar]
- 80.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clustal Omega. [(accessed on 2 November 2022)]. Available online: https://www.ebi.ac.uk/Tools/msa/clustalo/
- 83.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.The National Center for Biotechnology Information (NCBI) [(accessed on 2 November 2022)]; Available online: https://www.ncbi.nlm.nih.gov.
- 85.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Molecular Evolutionary Genetics Analysis (MEGA) [(accessed on 23 November 2022)]. Available online: https://www.megasoftware.net/home.
- 87.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 88.Nicholas K.B., Nicholas H.B., Jr., Deerfield D.W., II GeneDoc: Analysis and Visualization of Genetic Variation. EMBNEW.NEWS. 1997;4:14. [Google Scholar]
- 89.GeneDoc. [(accessed on 2 November 2022)]. Available online: https://genedoc.software.informer.com.
- 90.Teufel F., Almagro Armenteros J.J., Johansen A.R., Gíslason M.H., Pihl S.I., Tsirigos K.D., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022;40:1023–1025. doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.SignalP-6.0. [(accessed on 23 November 2022)]. Available online: https://biolib.com/DTU/SignalP-6/
- 92.Almagro Armenteros J.J., Salvatore M., Emanuelsson O., Winther O., von Heijne G., Elofsson A., Nielsen H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance. 2019;2:e201900429. doi: 10.26508/lsa.201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.TargetP-2.0. [(accessed on 23 November 2022)]. Available online: https://services.healthtech.dtu.dk/service.php?TargetP-2.0.
- 94.Hallgren J., Tsirigos K.D., Pedersen M.D., Almagro Armenteros J.J., Marcatili P., Nielsen H., Krogh A., Winther O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv. 2022. in press . [DOI]
- 95.DeepTMHMM. [(accessed on 23 November 2022)]. Available online: https://dtu.biolib.com/DeepTMHMM.
- 96.Compute pI/Mw Tool. [(accessed on 2 November 2022)]. Available online: http://web.expasy.org/compute_pi/
- 97.Erlanger B.F., Kokowsky N., Cohen W. The preparation and properties of two new chromogenic substrates of trypsin. Arch. Biochem. Biophys. 1961;95:271–278. doi: 10.1016/0003-9861(61)90145-X. [DOI] [PubMed] [Google Scholar]
- 98.Pennington M.W., Dunn B.M., editors. Peptide Synthesis Protocols. Humana Press; Totowa, NJ, USA: 1995. p. 321. [DOI] [Google Scholar]
- 99.Goptar I.A., Shagin D.A., Shagina I.A., Mudrik E.S., Smirnova Y.A., Zhuzhikov D.P., Belozersky M.A., Dunaevsky Y.E., Oppert B., Filippova I.Y., et al. A digestive prolyl carboxypeptidase in Tenebrio molitor larvae. Insect Biochem. Mol. Biol. 2013;43:501–509. doi: 10.1016/j.ibmb.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 100.Fields R. The rapid determination of amino groups with TNBS. Methods Enzymol. 1972;25:464–468. doi: 10.1016/S0076-687925042-X. [DOI] [PubMed] [Google Scholar]
- 101.Frugoni J.A.C. Tampone universale di Britton e Robinson a forza ionica constante. Gazz. Chem. Ital. 1957;87:403–407. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.