Abstract
Curli fibers are adhesive surface fibers expressed by Escherichia coli and Salmonella enterica that bind several host extracellular matrix and contact phase proteins and were assumed to have a role in pathogenesis. The results presented here suggest that one such role is internalization into host cells. An E. coli K-12 strain transformed with a low-copy vector containing the gene cluster encoding curli fibers (csg operon) was internalized by several lines of eukaryotic cells. The internalization could be correlated with a high level of curli fiber expression and was abolished by disruption of the csg operon. The ability to be internalized by eukaryotic cells could be conferred even by the curli fiber gene cluster of a noninvasive K-12 strain, but the homologous csg cluster from a virulent septicemic E. coli isolate mediated a higher level of internalization. The finding that curli fibers promote bacterial internalization indicates a new role for curli fibers in pathogenesis.
Curli fibers are thin aggregative surface fibers, connected with adhesion, which bind laminin (23), fibronectin (25), plasminogen (31), human contact phase proteins (4), and major histocompatibility complex (MHC) class I molecules (26). Curli fibers are coded for by the csg gene cluster, which is comprised of two divergently transcribed operons. One operon encodes the csgB, csgA, and csgC genes, while the other encodes csgD, csgE, csgF, and csgG. The assembly of the fibers is unique and involves extracellular self-assembly of the curlin subunit (CsgA), dependent on a specific nucleator protein (CsgB) (14). CsgD is a transcriptional activator essential for expression of the two curli fiber operons, and CsgG is an outer membrane lipoprotein involved in extracellular stabilization of CsgA and CsgB (20). The role of the other csg genes has yet to be elucidated.
Curli fibers are expressed by many pathogenic isolates of Escherichia coli, as well as laboratory strains (25). Similar surface proteins were identified in both Salmonella enterica serovar Enteritidis (9) and S. enterica serovar Typhimurium (28). Curli fibers are also present in E. coli strains involved in avian colisepticemia (27)—a serious invasive disease of chickens and turkeys that is characterized by entry of the bacteria into the air sacs, bloodstream, and vital organs (36).
Using PCR, we amplified the curli fiber-encoding (csg) gene cluster from a curli fiber-positive E. coli K-12 strain and cloned it in a low-copy-number vector. The resulting plasmid, when transformed to a noninvasive E. coli strain, conferred the ability to become internalized by eukaryotic cells. We have also cloned the homologous curli fiber-encoding cluster from a virulent isolate of avian E. coli O78 which could mediate a higher level of internalization. The results presented in this communication indicate that high levels of curli fiber expression can mediate entry of bacteria into eukaryotic cells and suggest that these fibers play a role in pathogenesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| E. coli K-12 | ||
| MC4100 | F−araD139 Δ(argF-lac) U169 rpsL150 relA1 flbB5301 deoC1 pts F25 rbsR | 6 |
| C600 | supE hsdR thi thr leu lacY tonA | Laboratory collection |
| VCS257 | tyr258 supE44 glnV44 lacY1 dapD8 tonA53 Δ(gal-UVRB)47 supF58, gyrA29 hsdS3 Δ(thrA57)1 | Stratagene |
| MC1022 | F−araD139 Δ(ara-leu) lacM15 galU galK strA | 7 |
| E. coli O781 | Isolated from bone marrow of chicken with septicemia by E. Z. Ron | |
| Plasmids | ||
| pMMB33 | Kmr | 11 |
| pCL 1920 | Spcr pSC101 ori containing pUC19 polylinker | 19 |
| pMMB33Inv | pMMB33 containing 20-kb fragment; Kmr | This study |
| pMRInv | pMMB33 containing 8.5-kb BamHI-SalI fragment of pMMB33Inv; Kmr | This study |
| pMHSa | Subclone of pMRInv containing 6-kb insert; Kmr | This study |
| pMRBg | pMRInv containing spectinomycin cassette disrupting csgG gene; Kmr Spcr | This study |
| pCLInv | pCL1920 containing BamHI-BamHI fragment of pMRInv; Spcr | This study |
| pCKcsg | pCL1920 containing 9-kb PCR fragment harboring csg genes; Spcr | This study |
| pBCgfp | Cmrgfp+ | 8 |
Construction of genomic libraries of E. coli O78.
Total genomic DNA of strain 781 serotype O78 was prepared and partially digested with Sau3A, and fragments of DNA corresponding to about 20 kb were isolated on a 5 to 40% sucrose gradient. These were ligated into the BamHI site of pMMB33 (11). After in vitro packaging (using the kit GIGAPACK GOLD [Stratagene Cloning Systems]), 5,000 recombinant E. coli K-12 clones were selected.
DNA sequencing.
DNA sequencing was performed as previously described (30).
Construction of plasmids.
A 9-kb fragment harboring the two csg operons from E. coli K-12 strain MC4100 was amplified by using primers C4231 (5′-GTGGATCCGCCCATTCTGAG-3′ [BamHI site underlined]) and C1186 (5′-GCGAGTGGTTGATGGGG-3′) and ExTaq (TaKaRa) DNA polymerase. The resulting 9-kb PCR fragment was purified by ethanolic precipitation (29) and cloned into the SmaI site of pCL1920 previously dephosphorylated by shrimp alkaline phosphatase (Boehringer Mannheim) according to the manufacturer's instructions.
Cell culture.
The human bladder epithelial cell line T24 was maintained in McCoy's 5A medium supplemented with 2 mM glutamine. The human alveolar epithelial cell line A549 was maintained in RPMI medium containing 10% fetal calf serum. The human cervical epithelial cell line HeLa was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 5 mM glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in an atmosphere containing 5% CO2. For invasion and adherence assays, cells were resuspended at a concentration of approximately 5 × 105/ml in DMEM, seeded into a six-well tissue culture plate (Corning) containing the same medium, and then incubated overnight. For confocal laser scanning microscopy, the same concentration of cells was seeded onto 16-well glass cell chambers (Nunc) for the actin labeling experiment, and glass coverslips were added to six-well culture plates before seeding of cells for the tubulin labeling experiment.
In vitro invasion assays.
Bacteria were quantified by a standard antibiotic protection assay (15). Briefly, bacteria were inoculated in Lennox LB broth (Difco) and grown at 37°C for 18 h, diluted 100-fold, and grown to mid-log phase (about 3 × 108 bacteria/ml). Bacteria were then collected by centrifugation (10,000 × g, 5 min) and resuspended in DMEM. Cells were washed three times with phosphate-buffered saline (PBS; pH 7.4), and approximately 108 bacteria were added per well (infection rate, 1:200) unless otherwise stated. Plates were incubated for 2 h at 37°C. The cells were washed three times with PBS, and extracellular bacteria were killed by adding fresh medium containing polymyxin (100 μg/ml). After further incubation for 1.5 h, the cells were washed three times in PBS, scraped off with a disposable cell scraper (Greiner), and lysed by brief sonication (30 in a Transistor/ultrasonic T7 [L&R Manufacturing Company]). Appropriate bacterial dilutions were plated to determine the number of viable internalized bacteria. Results are expressed as the average number of bacteria recovered per well in two independent determinations.
Quantitative Congo red binding assay.
Bacteria were grown on LB agar plates (5 g of NaCl/liter) for 72 h at 37°C. Colonies were scraped off and suspended in saline. Double dilutions were performed, and the bacterial concentration was quantified by measuring optical density at 600 nm against a saline background. Bacteria were then pelleted by centrifugation for 10 min at 14,000 rpm in an Eppendorf centrifuge. A 0.002% solution of Congo red (in saline) was prepared and optical density at 500 nm was measured against a saline background. One milliliter of the Congo red solution was added to each bacterial pellet, and the bacteria were resuspended in the dye solution and left for 10 min of binding at room temperature, followed by a second centrifugation (under the same conditions). The dye solution was recovered, and its optical density at 500 nm was measured to determine the reduction in optical density.
Immunostaining and confocal laser scanning microscopy.
Immunostaining was performed as previously described (33). For actin staining, cells were seeded and grown overnight in cell chambers (Nunc) as described for invasion assays. Following a 2-h incubation with green fluorescent protein (GFP) expressing bacteria, cells were washed with PBS, fixed for 20 min in PBS containing 4% paraformaldehyde and 0.1% Triton X-100, and washed as before. Cells were blocked (1% normal donkey serum and 0.1% bovine serum albumin in PBS) for 1 h, washed, and incubated with 0.3 μg of mouse antiactin antibodies (Boehringer Mannheim) in 40 ml of PBS for 2 h. Cells were then washed and incubated with 0.075 μg of rhodamine-labeled anti-mouse antibodies (Jackson) in 40 ml of PBS.
For tubulin staining, cells were grown and fixed as described for actin except that glass coverslips were used. Cells were then incubated overnight with 0.5 μg of rat antitubulin antibodies (Serotec) in 50 ml of PBS, washed in PBS, and incubated with 0.5 μg of rhodamine-labeled anti-rat antibodies (Serotec) in 50 ml of, PBS. The bacteria were visualized by green fluorescence conferred by the pBC-GFP plasmid. Stained cells were visualized and photographed using a Zeiss (Oberkochen, Germany) LSM 410 inverted confocal laser scanning microscope equipped with a 25-mW krypton-argon laser (488 and 568 maximum lines). A 40× NA/1.2 C-apochromat water immersion lens (Axiovert 135 M; Zeiss) was used for all imaging.
Transmission electron microscopy.
Microscopy of cells was performed as previously described (21), with the following modifications. Cells were grown on coverslips within culture plates. After the invasion period, cell monolayers were washed three times in PBS. Cells were then fixed for 1 h in 2.5% glutaraldehyde–0.2 M cacodylate buffer. Cells were postfixed for 1.5 h in 2% osmium tetroxide.
RESULTS
Internalization of bacteria carrying cloned curli fiber genes.
Curli fibers are coded for by the csg gene clusters. Preliminary results suggested that curli fibers may be involved in the internalization of curli fiber-expressing bacteria by eukaryotic cells. In order to determine the ability of the csg gene cluster to confer internalization, we performed the antibiotic protection assay (15) that determines in vitro internalization of bacteria. This assay is based on the fact that intracellular bacteria are not killed by antibiotic drugs that do not cross the cellular membrane, such as gentamicin or polymyxin.
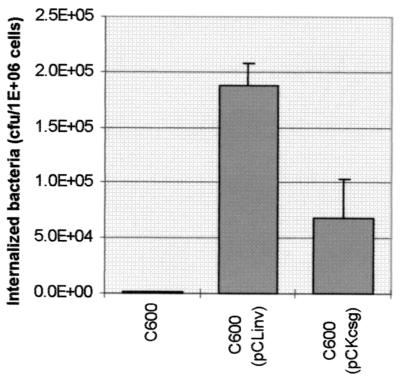
The csg region of curli fiber-positive E. coli K-12 strain MC4100 was amplified using PCR and ligated into very low-copy plasmid pCL. Since curli fiber expression is usually reflected by the ability of colonies to bind the dye Congo red (34), the ligation products were transformed into a curli fiber-negative strain which does not bind Congo red (E. coli K-12 strain MC1022) and Congo red binding colonies were selected. Plasmid DNA was prepared and transformed into E. coli K-12 strain C600. The transformants were examined in the in vitro internalization test using HeLa cells. The results are summarized in Fig. 1 and indicate that all of the transformants carrying the csg cluster were internalized better than the untransformed control.
FIG. 1.
Internalization of clones carrying curli fiber genes by HeLa cells. The experiment was performed as described in Materials and Methods. HeLa cells (5 × 105) were incubated for 120 min with 108 bacteria. The bacteria were (left to right) E. coli K-12 strain C600, E. coli K-12 strain C600 carrying the csg cluster of O78, and E. coli K-12 strain C600 carrying the PCR-amplified csg cluster of E. coli K-12. Each column is the average of two independent determinations. The results for the K-12-derived clones represent the average internalization obtained with four independent transformants carrying a PCR-derived csg cluster. The cells were washed three times, and polymyxin at 100 μg/ml was added. The number of intracellular bacteria was determined by viable count after 90 min of further incubation. Error bars represent standard deviations. The P value was 0.015 as determined by an independent-sample t test.
Isolation of a cosmid harboring the csg cluster from an avian septicemic E. coli O78 strain.
The ability of curli fibers to mediate internalization when expressed from a plasmid suggested that they may play a role in septicemic processes. Therefore, we examined the curli fibers produced by pathogenic septicemic strain O781, an E. coli serotype O78 strain isolated from a chicken with avian colisepticemia. A library of E. coli O781 DNA was constructed in low-copy cosmid pMMB33. The library was used to infect E. coli VCS257, and a clone was isolated that possessed very high Congo red binding and was also internalized by HeLa cells. This cosmid, presumably carrying the genes coding for curli fibers—pMMB33Inv—was also transferred to E. coli C600. Both clones, C600(pMMB33Inv) and VCS257(pMMB33Inv), were internalized by HeLa cells to about the same extent, which was greater than that mediated by the csg operon derived from the K-12 strain (Table 2).
TABLE 2.
Internalization of bacterial clones by HeLa cells
| Strain | No. of intracellular bacteria/5 × 105 cellsa |
|---|---|
| E. coli VCS257(pMMB33) | 100 |
| E. coli VCS257(pMMB33Inv) | 2 × 105 |
| E. coli C600 (pMMB33) | 12 |
| E. coli C600(pMMB33Inv) | 3.5 × 105 |
The experiment was performed as described in Materials and Methods.
The pMMB33Inv cosmid, which contained a 20-kb insert, was digested with the restriction endonuclease SalI, followed by self-ligation, resulting in a smaller cosmid, pMRInv, containing an 8.5-kb fragment which maintained the high Congo red binding and internalization. (Table 3). More than 2 kb of the 8.5-kb insert was sequenced, confirming that the insert contains the genes corresponding to csgD, csgE, csgF, and csgG of E. coli K-12, which constitute one of the curli fiber operons. A BLASTX search (1) showed complete predicted amino acid identity to the K-12 genes, except for the csgD gene. The protein encoded by this gene was different from the protein encoded by the K-12 homologue in two amino acids: there is a proline in position 19 in O78, as opposed to serine in K-12, as well as an alanine instead of a serine in position 110. It is interesting that in the CsgD homologue of S. enterica serovar Typhimurium, the same substitutions are present (along with nine other substitutions). A comparison of the nonidentical csgD locus of E. coli O78 to the E. coli K-12 and S. enterica serovar Typhimurium homologues can be seen in Fig. 2.
TABLE 3.
Internalization of subclones of pMMB33Inv by HeLa cells
| Strain | Insert length (kb) | Insert genes | No. of intracellular bacteria/5 × 105 cellsa |
|---|---|---|---|
| E. coli C600 | <1 | ||
| E. coli C600(pMMB33Inv) | 20 | Entire csg cluster | 1.58 × 105 |
| E. coli C600(pMRInv) | 8.5 | Entire csg cluster | 3.43 × 105 |
| E. coli C600(pMSa) | 6 | csgBAC csg DEF | <1 |
| E. coli C600(PMRBg) | 10.5 | csgBAC csgDEF (G disrupted) | <1 |
The experiment was performed as described in Materials and Methods.
FIG. 2.
Comparison of the sequences of csgD of E. coli O78, E. coli K-12, and S. typhimurium. A BLASTX 2.0.4 search (1) was conducted using the sequence obtained from E. coli O78 clone pMRInv, which contains the csg cluster of E. coli O78.
We have compared the internalization results obtained with bacteria carrying the K-12 cluster to those obtained with bacteria carrying the homologous cluster from the avian septicemia strain of E. coli serotype O78 cloned into the same vector. The results (Fig. 1) indicate that the csg cluster originating from the virulent strain mediated a level of internalization higher than that conferred by the gene cluster from the nonpathogenic K-12 strain. The difference between the two strains was statistically significant in an independent-sample t test (P = 0.015).
In addition to HeLa cells, the internalization of cells expressing a high level of curli fibers could be shown in T24 cells, HEp-2 cells, and chicken embryo retina cells. Since high expression of curli fibers may cause autoaggregation of bacteria, a control experiment without eukaryotic cells was conducted with mid-logarithmic bacteria of the same concentration used in the antibiotic protection assay. None of the bacteria carrying the plasmid harboring the csg cluster survived the incubation with polymyxin.
The curli fiber genes are essential for internalization.
A subclone of pMRInv (pMSa) which contained a 6-kb fragment harboring a csg cluster lacking the csgG gene was not internalized to a measurable extent (Table 3). Furthermore, when the csg operon was disrupted, the internalization phenotype was also lost. This experiment was performed using the pMRInv cosmid by the insertion of a gene cassette carrying resistance to spectinomycin into the csgG gene, which is involved in secretion and stabilization of the curli fiber subunit; the resulting cosmid—pMRBg—did not promote internalization (Table 3). Since csgG is the last gene in the csgDEFG operon, which is divergently transcribed with respect to the csgBAC operon, it is possible to rule out the possibility of a polar mutation.
Clone harboring the csg cluster from E. coli O78 express a high level of curli fibers at 37°C.
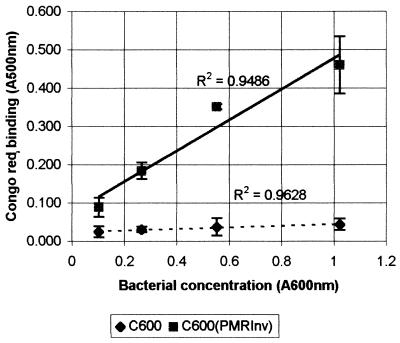
To determine the level of curli fiber expression by E. coli C600(pMRInv), a quantitative Congo red binding assay was performed. Various curli fiber-expressing cells were incubated in a Congo red solution, and the decrease in the Congo red color of the solution was determined. As can be seen from Fig. 3, E. coli K-12 strain C600(pMRInv) Congo red binding was up to 10-fold higher than that of the host strain. Furthermore, the Congo red binding of the clone was directly correlated with the concentration of bacteria, unlike that of the host strain, indicating specificity. The low level of Congo red binding of the host strain and the lack of a substantial increase with the concentration of bacteria seem to indicate nonspecific binding and low, if any, curli fiber expression. These results were substantiated by immunoblot analysis.
FIG. 3.
Curli fiber expression level of a clone harboring the csg cluster from E. coli O78. Curli fiber expression was determined by the ability to bind the dye Congo red as described in Materials and Methods. Briefly, bacterial colonies were scraped off, subjected to twofold dilutions, and incubated with a solution of Congo red in saline. The bacteria were pelleted, and the solution's optical density at 500 nm was compared to that obtained at the same wavelength prior to incubation with bacteria. Congo red absorption in this figure is defined as the difference in optical density before and after incubation with the bacteria. The results are averages of two independent determinations. Error bars represent standard deviations.
Visualization of internalized bacteria.
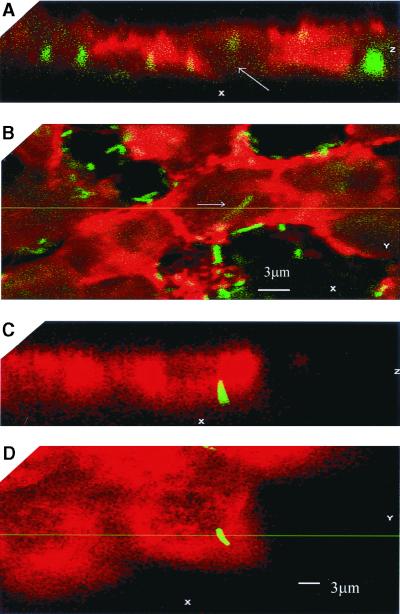
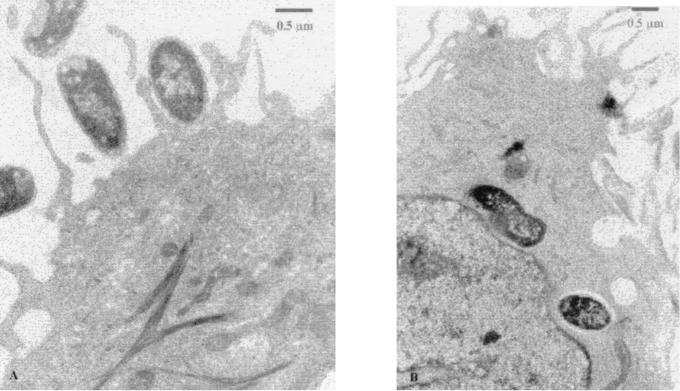
Since curli fibers have been shown to mediate adherence and autoaggregation (13), the possibility arose that the escape from the effect of polymyxin in the antibiotic protection assay may be due to aggregation and intimate adherence rather than internalization of the bacteria. To further establish evidence for microbial entry into the cells, confocal laser scanning microscopy was conducted. Bacteria containing the plasmid pCLInv were cotransformed with the plasmid pBCGFP (kindly provided by Ann Matthysse), carrying a modification of the gene coding for the GFP from the jellyfish Aquoria victoria (8). These bacteria were used to infect HeLa cells that were fixed after infection. The cells were treated with antiactin (Fig. 4A and B) or antitubulin (Fig. 4C and D) antibodies and visualized with rhodamine-labeled secondary antibodies. The results presented in Fig. 4 demonstrate that the bacteria are located within the labeled cells and are in close association with actin. As can be seen in Fig. 4A and B, many adherent extracellular bacteria are also present, which is to be expected when adhesive surface fibers such as curli fibers are expressed at a relatively high level. The internalized bacteria could also be visualized by transmission electron microscopy in thin sections (Fig. 5).
FIG. 4.
Internalization of E. coli K-12 strain C600 (pCLINV) by HeLa cells. HeLa cells were infected for 2 h with E. coli K-12 strain C600(pCLINV) expressing GFP, fixed and stained with antiactin antibody (A and B) or antitubulin antibody (C and D). The cells were visualized using secondary antibodies and analyzed by confocal microscopy. The images obtained are presented in panels B and D, and the Z sections perpendicular to the focal planes of the images panels in B and D are presented in panels A and C, respectively.
FIG. 5.
Transmission electron micrographs of cells infected with E. coli K-12 strain C600(pMRInv) harboring the csg cluster. Panels: A, a bacterium engulfed by an A549 cell; B, intracellular bacteria within a T24 cell.
DISCUSSION
Curli fibers and their Salmonella homologues (termed thin aggregative fimbriae) bind laminin (24), fibronectin, and plasminogen (31) and have been shown to be important for adhesion to solid surfaces (34). Although these fimbriae were first characterized in clinical isolates (9, 25), their role in pathogenesis has not yet been established. The ability of curli fibers to bind extracellular matrix molecules, MHC class I molecules (26), and human contact phase proteins (4) led to the suggestion that they have a role in invasion (31). In this paper, we present data demonstrating that high levels of curli fibers mediate internalization of E. coli by eukaryotic cells in tissue cultures.
The finding that an adherence factor mediates internalization by eukaryotic cells is not unique. Several other bacterial proteins have been shown to mediate both adhesion and invasion—the Inv, YadA, and Ail proteins of Yersinia enterocolitica (22) and AfaE of uropathogenic and diarrhea-associated E. coli strains (16). Recently, evidence was presented suggesting an involvement of fimbriae in internalization. This has been shown for Dr fimbriae of uropathogenic E. coli (12) and for fimbriae of Porphyromonas gingivalis (35).
The internalization mediated by curli fibers was moderate (0.19% to 0.35%), in comparison with invasin-mediated internalization of enteroinvasive bacteria such as Y. entercolitica (about 27%) (22) or enteroinvasive E. coli (about 3%) (32). Nevertheless, the observed uptake was substantial and of the same order of magnitude as that conferred by the ail gene of Y. entercolitica for HEp-2 cells—0.37% (22).
Curli fibers are encoded by a gene cluster containing two divergently transcribed operons—csgB csgA csgC and csgD csgE csgF csgG (13). This cluster is present and expressed in many E. coli strains, including nonpathogenic strains such as E. coli K-12 strain C600 (25) that are internalized poorly. However, a higher expression of the genes obtained by a higher copy number in cosmid clones has been shown here to increase uptake of the bacteria, even if the cloned genes are from E. coli K-12 strains. The results presented here indicated that a higher expression level of the csg gene cluster from E. coli K-12 strain MC4100, cloned on a low-copy-number vector, resulted in a higher level of internalization (Fig. 1).
The internalization of bacteria carrying a plasmid with cloned curli fiber genes of E. coli O781 was higher than that of bacteria carrying the same plasmid but with cloned curli fiber genes of E. coli K-12. These results support the possibility that there are differences between the pathogenic O781 and the nonpathogenic K-12 strains in the csg cluster, whether structural or regulatory. Such differences have already been found in two amino acid substitutions in the activator CsgD that in avian septicemic E. coli O781 positions 19 and 110 contains proline and alanine, respectively, instead of serine as in K-12. These differences are probably significant, since the same substitutions are found in the CsgD protein of the pathogen S. enterica serovar Typhimurium (Fig. 2).
The expression of the genes coding for curli fibers is complex and involves several control elements, such as H-NS, RpoS, and OmpR (2, 34). As a result, in most known strains, the expression of curli fibers is greatly reduced at temperatures higher than 30°C and at high osmolarity (2). However, mutations leading to higher expression can occur by genetic changes in any one of these elements. One such mutation has already been identified in E. coli, where a point mutation in ompR resulted in significant curli fiber overexpression (34). Recently, it has been shown that several E. coli isolates from humans with sepsis also produce curli fibers at 37°C (5). The results presented here indicate that the curli fiber genes of the pathogenic and nonpathogenic strains of E. coli can promote internalization when present in multiple copies, thus bypassing the tight control of curli fiber expression. The avian pathogenic E. coli O78 strain, from which we cloned the csg operon reported here, appears to differ in the control of curli fiber expression, as it produces high levels of curli fibers constitutively from a chromosomal one-copy gene. This strain is also invasive to tissue cultures, but internalization is lower than that of the recombinant strain that carries multiple copies of the csg operon. In the avian E. coli O78 strain, curli fiber production was observed in all of the media tested (even those of high osmolarity, such as Lennox LB broth) and at temperatures ranging from 25 to 42°C. As already mentioned, these findings indicate that curli fibers of the O78 strain differ from those of K-12 strains in structure or in the regulation of expression. The ability to express curli fibers under host conditions may be of critical importance upon bacterial entry into the host and is probably common in septicemic E. coli strains. These findings suggest that curli fibers constitute an significant virulence factor.
Avian colisepticemia is a systemic disease involving bacterial entrance into the bloodstream and organs. The results showing that curli fiber-encoding genes from avian colisepticemic strains bring about efficient internalization are compatible with the nature of the disease. Additional support is found in a recent publication (17) demonstrating that natural avian O78 isolates defective in curli fiber expression, due to a natural insertional inactivation by an IS1 element, exhibited reduced persistence in poultry, presumably due to less efficient colonization. Moreover, a study conducted with the avian pathogen S. enterica serovar enteritidis demonstrated that mutants with changes in the curli fiber homologue SEF17, showed significantly reduced internalization by epithelial cells and that the invasion of cells by the wild-type bacteria could be inhibited by anti-SEF17 serum (10). On the other hand, insertional inactivation of the csgA gene in an E. coli isolate from avian colisepticemia, which completely abolished curli fiber expression, resulted in only a marginal decrease in internalization (18). This result suggests that curli fibers are not the only virulence factor involved in the internalization of avian E. coli strains. One possibility was that many avian E. coli O78 isolates produce several virulence factors, including a fimbrial adhesin of the S-fimbria family termed AC/I (3). Although the increase in the uptake of bacteria when E. coli K-12 strain 600 was transformed with a cosmid coding for AC/I fimbriae was minor, it is possible that in the wild type a synergy exists between the two adhesins, contributing to invasion and virulence. It is also assumed that the wild-type septicemic strain contains additional internalization factors that, together with curli fibers, as well as AC/I fimbriae, participate in the initial attachment and internalization of the bacteria and could affect their virulence.
Although internalization by itself is clearly insufficient for pathogenesis, the demonstration that curli fibers can mediate bacterial internalization labels them as a significant virulence factor.
ACKNOWLEDGMENTS
This work was supported by the German Israeli Foundation (GIF), by the Israel Science Foundation founded by the Israeli Academy of Sciences & Humanities, and by the Manja and Morris Leigh Chair for Biophysics and Biotechnology.
We thank Martin Woodward for helpful comments, Hilde Merkert for invaluable help with the electron microscopy, and Rom Altstock and Ilan Tsarfaty of the Department of Human Microbiology, Sackler Faculty of Medicine, Tel-Aviv University, and Leonid Mittelman of the Interdepartmental Core Facility, Sackler School of Medicine, Tel-Aviv University, for guidance in the use of confocal microscopy. We also thank Orlev Levi for help with tissue cultures.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnqvist A, Olsen A, Normark S. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 3.Babai R, Stern B E, Hacker J, Ron E Z. New fimbrial gene cluster of S-fimbrial adhesin family. Infect Immun. 2000;68:5901–5907. doi: 10.1128/iai.68.10.5901-5907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben Nasr A, Olsen A, Sjobring U, Muller-Esteri W, Bjorck L. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol Microbiol. 1996;20:927–935. doi: 10.1111/j.1365-2958.1996.tb02534.x. [DOI] [PubMed] [Google Scholar]
- 5.Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 8.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 9.Collinson S K, Clouthier S C, Doran J L, Banser P A, Kay W W. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dibb-Fuller M P, Allen-Vercoe E, Thorns C J, Woodward M J. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology. 1999;145:1023–1031. doi: 10.1099/13500872-145-5-1023. [DOI] [PubMed] [Google Scholar]
- 11.Frey J, Bagdasarian M, Feiss D, Franklin F C, Deshusses J. Stable cosmid vectors that enable the introduction of cloned fragments into a wide range of gram-negative bacteria. Gene. 1983;24:299–308. doi: 10.1016/0378-1119(83)90090-2. [DOI] [PubMed] [Google Scholar]
- 12.Goluszko P, Popov V, Selvarangan R, Nowicki S, Pham T, Nowicki B J. Dr fimbriae operon of uropathogenic Escherichia coli mediate microtubule-dependent invasion to the HeLa epithelial cell line. J Infect Dis. 1997;176:158–167. doi: 10.1086/514018. [DOI] [PubMed] [Google Scholar]
- 13.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin-and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 14.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isberg R R, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 16.Jouve M, Garcia M-I, Courcoux P, Labigne A, Gounon P, Le Bouguénec C. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3 gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect Immun. 1997;65:4082–4089. doi: 10.1128/iai.65.10.4082-4089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Ragione R M, Collighan R J, Woodward M J. Non-curliation of Escherichia coli O78:K80 isolates associated with IS1 insertion in csgB and reduced persistence in poultry infection. FEMS Microbiol Lett. 1999;175:247–253. doi: 10.1111/j.1574-6968.1999.tb13627.x. [DOI] [PubMed] [Google Scholar]
- 18.La Ragione R M, Cooley W A, Woodward M J. The role of fimbriae and flagella in the adherence of avian strains of Escherichia coli O78:K80 to tissue culture cells and tracheal and gut explants. J Med Microbiol. 2000;49:327–338. doi: 10.1099/0022-1317-49-4-327. [DOI] [PubMed] [Google Scholar]
- 19.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 21.Meier C, Oelschlaeger T A, Merkert H, Korhonen T K, Hacker J. Ability of Escherichia coli isolates that cause meningitis in newborns to invade epithelial and endothelial cells. Infect Immun. 1996;64:2391–2399. doi: 10.1128/iai.64.7.2391-2399.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller V L, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen A, Arnqvist A, Hammar M, Normark S. Environmental regulation of curli production in Escherichia coli. Infect Agents Dis. 1993;2:272–274. [PubMed] [Google Scholar]
- 24.Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 25.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 26.Olsén A, Wick M J, Mörgelin M, Björck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect Immun. 1998;66:944–994. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Provence D L, Curtiss R., III Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or curli production. Infect Immun. 1992;60:4460–4467. doi: 10.1128/iai.60.11.4460-4467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Römling U, Bian Z, Hammar M, Sierralta W D, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Segal G, Ron E Z. Cloning, sequencing, and transcriptional analysis of the gene coding for the vegetative sigma factor of Agrobacterium tumefaciens. J Bacteriol. 1993;175:3026–3030. doi: 10.1128/jb.175.10.3026-3030.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sjobring U, Pohl G, Olsen A. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA) Mol Microbiol. 1994;14:443–452. doi: 10.1111/j.1365-2958.1994.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 32.Small P L C, Isberg R R, Falkow S. Comparison of the ability of enteroinvasive Escherichia coli, Salmonella typhimurium, Yersinia pseudotuberculosis, and Yersinia enterocolitica to enter and replicate within HEp-2 cells. Infect Immun. 1987;55:1674–1679. doi: 10.1128/iai.55.7.1674-1679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsarfaty I, Altstock R T, Mittelman L, Sandovsky-Losica H, Jadoun J, Fabian I, Segal E, Sela S. Confocal microscopy in the study of the interactions between microorganisms and cells. In: Rosenberg E, editor. Microbial ecology and pathogenesis. Washington, D.C.: ASM Press; 1999. pp. 75–88. [Google Scholar]
- 34.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinberg A, Belton C M, Park Y, Lamont R J. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yerushalmi Z, Smorodinsky N I, Naveh M W, Ron E Z. Adherence pili of avian strains of Escherichia coli O78. Infect Immun. 1990;58:1129–1131. doi: 10.1128/iai.58.4.1129-1131.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]