An imbalance between activating and repressive ETS transcription factors transforms genomic GGAA tandem repeats into leukemia-specific enhancers that drive the unique gene expression signature of ETV6-RUNX1+ B-ALL.
Abstract
Distal enhancers play critical roles in sustaining oncogenic gene-expression programs. We identify aberrant enhancer-like activation of GGAA tandem repeats as a characteristic feature of B-cell acute lymphoblastic leukemia (B-ALL) with genetic defects of the ETV6 transcriptional repressor, including ETV6–RUNX1+ and ETV6-null B-ALL. We show that GGAA repeat enhancers are direct activators of previously identified ETV6–RUNX1+/− like B-ALL “signature” genes, including the likely leukemogenic driver EPOR. When restored to ETV6-deficient B-ALL cells, ETV6 directly binds to GGAA repeat enhancers, represses their acetylation, downregulates adjacent genes, and inhibits B-ALL growth. In ETV6-deficient B-ALL cells, we find that the ETS transcription factor ERG directly binds to GGAA microsatellite enhancers and is required for sustained activation of repeat enhancer-activated genes. Together, our findings reveal an epigenetic gatekeeper function of the ETV6 tumor suppressor gene and establish microsatellite enhancers as a key mechanism underlying the unique gene-expression program of ETV6–RUNX1+/− like B-ALL.
Significance:
We find a unifying mechanism underlying a leukemia subtype-defining gene-expression signature that relies on repetitive elements with poor conservation between humans and rodents. The ability of ETV6 to antagonize promiscuous, nonphysiologic ERG activity may shed light on other roles of these key regulators in hematolymphoid development and human disease.
See related commentary by Mercher, p. 2.
This article is highlighted in the In This Issue feature, p. 1
INTRODUCTION
Integrative profiling of B-lymphoblastic leukemia (B-ALL) has identified clinically and biologically distinct subtypes associated with specific genomic alterations and gene-expression signatures (1–3). The ETV6–RUNX1 (E-R) fusion oncogene defines a common B-ALL subtype, representing about 25% of pediatric B-ALL (4). The encoded oncoprotein incorporates a protein–protein interaction domain of ETV6, a transcriptional repressor in the ETS family (5), and nearly the full length of the DNA sequence–specific transcription factor (TF) RUNX1 (6, 7). Somatic mutations or genomic deletions that inactivate ETV6 are frequently seen as secondary events in E-R+ B-ALL (8–10), in B-ALL cases that lack the E-R fusion gene but show an “ETV6–RUNX1-like” signature of aberrant gene-expression (2, 11), and as second-hit events in leukemias that arise in patients with germline loss-of-function ETV6 variants (12, 13). However, the mechanism by which ETV6 dysfunction contributes to leukemia is poorly understood. Here, we identify enhancer-like chromatin state and function of GGAA microsatellite repeats as a characteristic feature of E-R+ and ETV6-null B-ALL. Leukemia-specific GGAA repeat enhancers are bound by the ETS family activator ERG, sustain the expression of known E-R+/E-R–like B-ALL signature genes, including potential leukemia drivers, and are repressed upon restoration of ETV6 expression. Our findings reveal an unexpected mechanism driving the gene-expression signature of a common subtype of childhood leukemia and identify an unanticipated function of ETV6 as an epigenetic chaperone that blocks promiscuous ERG activity at nonphysiologic target sites.
RESULTS
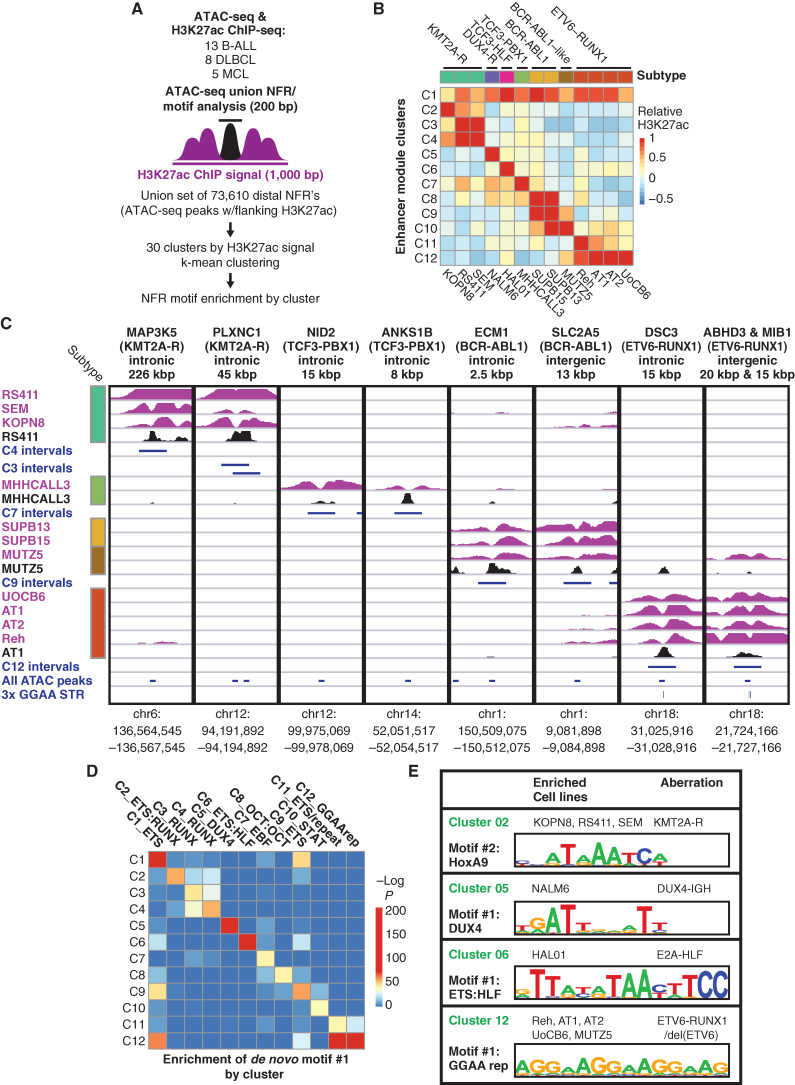
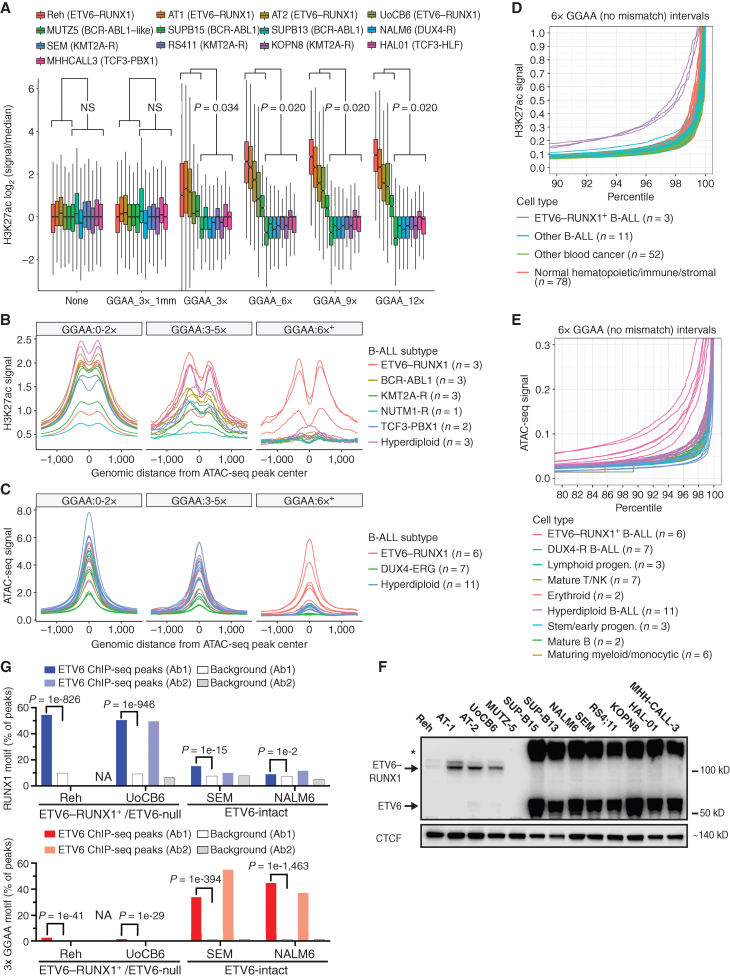
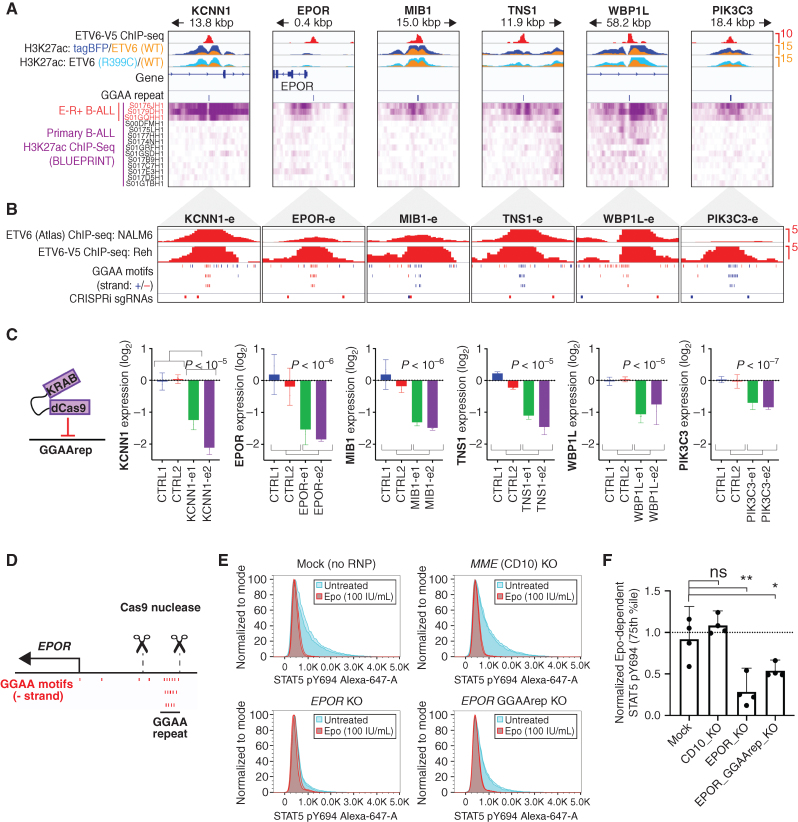
As an exploratory approach to discover genomic sequence motifs associated with enhancer dysregulation in B-cell malignancies, we performed genome-wide k-means clustering of histone H3 lysine 27 acetylation (H3K27ac) chromatin immunoprecipitation sequencing (ChIP-seq) signal centered on using Assay for Transposase-Accessible Chromatin sequencing (ATAC-seq) peaks from 26 B-cell cancer cell lines, including 13 B-ALL cell lines (Fig. 1A and B; Supplementary Fig. S1A; Supplementary Table S1). Hierarchical clustering based on enhancer acetylation readily separated B-cell cancers by both primary type and B-ALL subtype (Supplementary Fig. S1B and S1C). Enhancers active in B-ALL subtype-specific clusters were observed near previously identified subtype-specific signature genes (ref. 1; Fig. 1C; Supplementary Fig. S2A), suggesting that differential enhancer activity could contribute to subtype-specific gene-expression programs. De novo motif enrichment analysis of enhancer clusters that were hyperacetylated in specific B-ALL cell lines revealed strong enrichment for motifs of TFs known to have increased activity in the corresponding subtypes, including DUX4 (B-ALL with DUX4 rearrangement), HLF (TCF3-HLF+ B-ALL), HoxA9 (B-ALL with KMT2A rearrangement), and STAT5 (BCR-ABL1+ and BCR-ABL1-like B-ALL; Fig. 1D and E; Supplementary Fig. S2B and S2C; Supplementary Table S2). Surprisingly, cluster 12 enhancers that were hyperacetylated in E-R+ B-ALL cell lines (n = 4) and in the BCR-ABL1–like B-ALL cell line MUTZ-5 were highly enriched for a motif consisting of tandem repeats of the sequence “GGAA.” Comparing acetylation levels across GGAA repeats of varying lengths in the hg38 reference genome showed increased acetylation in these five cell lines for intervals containing at least three repeats, with stronger effects at intervals with greater than six tandem repeats (Fig. 2A; Supplementary Fig. S3A). Similarly, analysis of ATAC-seq data showed increased chromatin accessibility in the same five cell lines for intervals containing 6× GGAA repeats compared with other B-ALL cell lines (Supplementary Fig. S3B). A similar analysis performed on ATAC-seq (14) and H3K27ac ChIP-seq (15, 16) data from primary B-ALL samples confirmed a strong association between GGAA repeats and an active enhancer-like chromatin state in E-R+ B-ALL (Fig. 2B and C).
Figure 1.
Identification of DNA motifs enriched in enhancers with subtype-specific activity. A, Strategy for identification of enhancer module acetylation clusters across 26 B-cell cancer cell lines. B, Median acetylation signal in each B-ALL cell line for each of the 12 B-ALL–specific enhancer acetylation clusters (C1–C12), relative to all 26 cell lines (mature B-cell lines not shown). Genetic subtypes are listed at the top and cell lines at the bottom. C, H3K27ac ChIP-seq and ATAC-seq data for representative enhancers from subtype-specific clusters. Distance and position with respect to known B-ALL signature gene (Ross et al.; ref. 1) are listed at the top, with the associated B-ALL subtype in parentheses. H3K27ac ChIP-seq tracks are shown in purple (scale, 15 fragments per million, fpm) and ATAC-seq tracks in black (scale, 7.5 fpm). Intervals shown in blue correspond to the cluster-specific enhancer modules defined in (B; 1 kbp), union of all ATAC-seq peaks (200 bp), and position of 3× GGAA tandem repeats. D, Significance of enrichment for the top de novo motif identified by HOMER in each of the clusters from B), reevaluated in all 12 clusters. E, Selected top de novo motifs identified in enhancer acetylation clusters.
Figure 2.
GGAA microsatellites show enhancer-like chromatin state in B-ALL with ETV6–RUNX1 and loss of wild-type ETV6. A, Box plots showing relative H3K27ac levels flanking merged GGAA repeats and ATAC-seq peak-containing intervals identified across 13 B-ALL cell lines. Intervals were grouped according to the longest GGAA tandem repeat present. The “GGAA_3x_1 mm” group contained a motif with a one-base mismatch to a 3× GGAA repeat. “None” indicates the set of ATAC-seq peaks that do not contain a GGAA repeat by any criteria within 300 bp of the peak center. Results shown for Mann–Whitney U test of difference in means in ETV6-altered vs. ETV6-intact cell lines for each repeat class. NS, not significant (P > 0.05). B, Histogram of H3K27ac signal (reads per 10 m total reads/bp/peak) from primary B-ALL samples of the indicated subtypes, centered on the union set of B-ALL cell line ATAC-seq peaks (n = 13). Peaks were grouped as in B. Peaks were grouped according to overlap with 3× or 6× GGAA tandem repeats. C, Histogram of ATAC-seq signal (reads per 10 m total reads/bp/peak) from primary B-ALL samples of the indicated subtypes, centered on the union set of ATAC-seq peaks from those same samples. D, Normalized H3K27ac signal at 6× GGAA repeat–containing intervals for primary leukemias and normal cell types (Blueprint consortium). Signal from genome-wide GGAA repeat–containing intervals and representative non-repeat sites (housekeeping gene promoters and random intervals) was quantile-normalized across all populations and intervals. Signal within each interval set was then ranked by normalized H3K27ac signal within each population (top 10% shown). E, Normalized ATAC-seq signal at 6× GGAA repeat-containing intervals for primary B-ALL (Diedrich et al.; ref. 14) and normal human cell bone marrow cell populations (pseudo-bulk scATAC-seq; Granja et al.; ref. 71). Normalization and data presentation were as described in E. Top 20% of intervals by ranked ATAC-seq signal are shown. See Supplementary Fig. S3B for control regions and additional details. F, Immunoblot of nuclear extracts from 13 B-ALL cell lines with an antibody recognizing the N-terminal portion of ETV6 (Atlas Antibodies, HPA000264). Arrows indicate bands at the expected molecular weight of ETV6 and E-R, respectively. Asterisk indicates an apparent high-molecular-weight form of ETV6. G, Percentage of peaks containing RUNX1 and 3× GGAA repeat motifs (HOMER known motif analysis) in ChIP-seq performed in B-All cell lines with two different ETV6 n-terminal antibodies (Ab1 = Atlas; Ab2 = Santa Cruz). ChIP-seq performed in Reh cells with Ab2 yielded too few peaks for analysis (“NA”). “Background” bars show motif occurrence in randomly selected genomic regions with similar GC content to the corresponding peaks. HOMER motif enrichment P values versus background (binomial test) are shown for Ab1 peaks.
To understand if enhancer-like activation of GGAA repeats occurs in normal development, we performed an analysis of the H3K27ac signal at GGAA repeats and control regions from 78 normal mesenchymal and hematolymphoid cell populations and 66 diverse primary hematologic cancer samples generated through uniform methods by the Blueprint project. Only the three E-R+ B-ALL samples showed substantially increased GGAA repeat acetylation (Fig. 2D; Supplementary Fig. S4A). We performed a similar analysis to compare primary B-ALL bulk ATAC-seq data (n = 24) with pseudobulk ATAC-seq tracks derived from single-cell ATAC-seq analysis of normal human bone marrow hematopoietic and lymphoid progenitors (n = 23 cell populations) and diverse adult and fetal tissues (n = 222 cell populations). This analysis revealed distinctively higher chromatin accessibility of longer GGAA repeats (6×+) in all six E-R+ B-ALL samples, but not in any normal cell type (Fig. 2E; Supplementary Fig. S4B and S4C). A comparable analysis of bulk ATAC-seq data from 90 normal murine hematolymphoid and stromal cell types showed no evidence of a population with substantially increased chromatin accessibility of GGAA repeats (Supplementary Fig. S4D). We concluded that enhancer-like activation of GGAA repeats in E-R+ B-ALL is a cancer-specific aberrant epigenetic state.
The E-R fusion TF is thought to bind primarily to enhancers and promoters enriched for the RUNX1 motif (TGTGG; refs. 17–19), which does not resemble the enriched repeat sequence. Because the sequence GGA(A/T) forms the core of the binding motif for ETS family TFs (20, 21), we hypothesized that aberrant acetylation of GGAA repeats in this subset of B-ALL might be related to deficiency of normal ETV6 repressor function. Western blot confirmed the absence of wild-type ETV6 protein expression in nuclear extracts from MUTZ5 and all four E-R+ cell lines (Fig. 2F). ETV6 gene copy-number analysis combined with ETV6–RUNX1 single-fusion FISH studies indicated deletion of the nonrearranged copy of ETV6 in all four E-R+ cell lines and biallelic ETV6 deletion in MUTZ5 (Supplementary Fig. S5A and Supplementary Table S1). ChIP-seq performed with two different N-terminal ETV6 antibodies yielded peaks that were significantly enriched for the RUNX1 motif, but not GGAA repeats, in B-ALL cell lines that express ETV6–RUNX1, but not wild-type ETV6. In contrast, ChIP-seq performed with the same two antibodies in ETV6-intact cell lines showed enrichment for GGAA repeats, but not RUNX1 motifs (Fig. 2G; Supplementary Table S3), supporting GGAA repeats as a direct binding target for wild-type ETV6 but not for the E-R fusion protein.
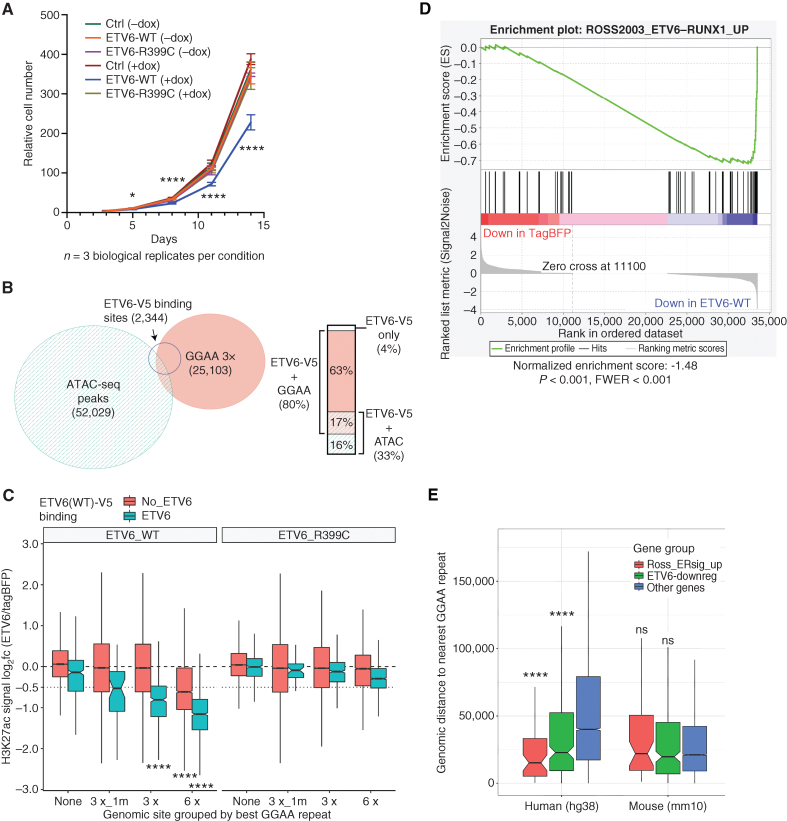
To further investigate ETV6-binding targets and functions, we used a doxycycline-inducible construct to express V5-tagged ETV6WT or ETV6R399C, a variant associated with loss of DNA-binding function and hematopoietic abnormalities (12), in the E-R+ cell line Reh (Supplementary Fig. S5B). Expression of ETV6WT-V5, but not ETV6R399C-V5, significantly reduced the growth of Reh cells compared with tagBFP transgene-expressing controls (Fig. 3A). ChIP-seq with a V5 antibody identified 2,343 significant binding peaks in ETV6WT-V5 expressing Reh cells, of which 80% overlapped genomic sites with at least 3 perfect GGAA tandem repeats (Fig. 3B). To understand the effects of ETV6 on chromatin state, we performed replicate H3K27ac ChIP-seq in Reh cells after the doxycycline induction of ETV6WT, ETV6R399C, or tagBFP control. Expression of ETV6WT was associated with the deacetylation of histones flanking GGAA repeats (Fig. 3C), with a stronger effect seen at sites with longer repeats and at sites where ETV6-WT-V5 binding was detected by ChIP-seq. In contrast, expression of ETV6R399C resulted in minimal acetylation changes at these same sites. These findings support the surprising conclusion that ETV6 chromatin repression activity is primarily directed at GGAA repeats upon restoration in ETV6-deficient B-ALL.
Figure 3.
ETV6 inhibits leukemia cell growth, binds and deacetylates GGAA repeat enhancers, and activates E-R signature genes. A, Relative cell numbers for Reh cells stably transduced with DoxOn-ETV6-V5 constructs or DoxON-tagBFP control and grown with or without 500 ng/mL doxycycline. Error bars show 95% CI of triplicate wells counted for each condition/time point. For each time point, log-transformed cell counts for ETV6-WT + dox were compared with each other sample by one-way ANOVA with Dunnett multiple comparison test. Comparisons at day 3 were nonsignificant, and all comparisons were significant at later time points. B, Area-proportional Euler diagram showing overlap of genome-wide ATAC-seq peaks in parental Reh cells, GGAA tandem repeats (at least 3× GGAA), and sites bound by ETV6-WT-V5 (V5 ChIP-seq peaks) expressed in Reh cells. Bar plot at right details overlaps within the set of ETV6-binding target sites. C, Box plots showing change in acetylation at ATAC-seq peaks and/or GGAA repeats following the induction of ETV6-WT or ETV6-R399C in Reh cells. Peaks were grouped by most stringent repeat class (3× _1m contains 1 mismatch to a 3× GGAA repeat), and further divided by overlap with ETV6-WT-V5 ChIP-seq peaks. Groups with log2 fold change significantly less than −0.5 are indicated (Wilcoxon signed rank test with Holm–Bonferroni correction). D, Gene set enrichment analysis for upregulated E-R signature genes (2) among genes ranked by differential expression after induction of ETV6-WT-V5 or tagBFP (control) expression. E, Comparison of distance to the closest GGAA repeat (3×) for evolutionarily conserved human genes compared with their orthologs in mouse, separated into mutually exclusive sets of previously defined E-R–upregulated signature genes (ref. 1, red), non-ER signature genes that were downregulated by ETV6 expression in REH cells (green), and other genes (blue). Significance of gene sets vs. “other genes” in each species calculated by the Mann–Whitney U test with Holm–Bonferroni correction. P values for all panels: ns, not significant (P > 0.05); *, P < 0.05; ****, P < 10−4.
We next investigated whether restoration of ETV6 would reduce the expression of genes associated with GGAA repeat enhancers. Gene set enrichment analysis of RNA-seq data from the ETV6 restoration model showed strong enrichment of known E-R+ B-ALL signature genes (1) among ETV6-repressed genes (Fig. 3D; Supplementary Fig. S5C). E-R signature gene and other ETV6-repressed gene promotors were located significantly closer to the nearest GGAA repeat compared with nonregulated genes, but this relationship was lost for orthologs of those same genes in mice (Fig. 3E). This finding is consistent with the poor overall conservation of GGAA repeat locations between humans and rodents (Supplementary Fig. S6), suggesting that many gene regulatory consequences of GGAA repeat enhancer formation in human cells are unlikely to be reproducible in mouse models.
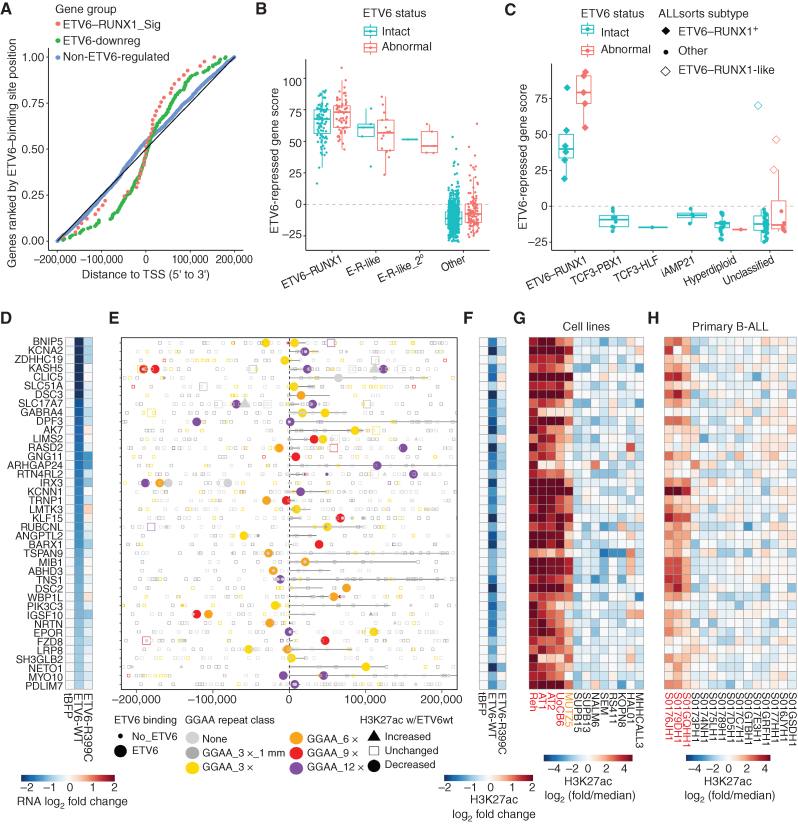
To further evaluate whether ETV6-repressed target genes identified in Reh cells were relevant in primary B-ALL, we focused on significantly repressed protein-coding genes (log2 fold change < 0.5, Padj < 0.001) associated with ETV6-binding sites within 200 kb from the promoter (Supplementary Table S4). ETV6-binding sites showed a tendency for clustering near the promoters of ETV6-repressed genes (±50 kb), whereas no such skew was observed for control genes (Fig. 4A). For 71 ETV6-repressed genes, at least one associated ETV6 binding site showed decreased H3K27ac levels (P < 0.05) upon ETV6-WT-V5 expression (Supplementary Table S4). Of these, 40 genes were significantly overexpressed (FDR Padj < 0.05) in B-ALL classified as E-R+ or E-R–like in a cohort of 1,988 primary B-ALL samples analyzed at St. Jude Children's Research Hospital (ref. 2; Fig. 4B; Supplementary Fig. S7A and S7B). A composite signature of these 40 ETV6-repressed genes robustly separated E-R+ and E-R–like samples (as a primary or secondary subtype) from other B-ALL samples and was significantly higher in E-R+ B-ALL with secondary ETV6 deletion events (P = 0.0018, t test). This ETV6-regulated gene signature was also significantly increased in E-R+ B-ALL from a separate B-ALL cohort [NCI TARGET phase II (ref. 22); Fig. 4C; Supplementary Fig. S7C]. Because the E-R–like subtype had not been defined for the TARGET cohort, we applied ALLSorts (23), a pretrained machine-learning B-ALL classifier, to these data sets and found that all three non–E-R+ cases with high expression of the ETV6-repressed gene signature were classified as E-R–like.
Figure 4.
ETV6-repressed genes and enhancers are hyperactive in ETV6-altered B-ALL A, Plot of distance from TSS to the nearest ETV6-WT-V5–binding site for genes repressed by ETV6 in Reh cells from the set of E-R signature genes, other ETV6-repressed genes (“ETV6-downreg”), and a control set of expressed, non–ETV6-regulated genes. Only genes with an ETV6 binding site ± 200 kbp from the promoter are included. B, Cumulative Z-score for expression of 40 ETV6-repressed genes with associated ETV6-repressed enhancers (“ETV6-repressed gene score”) in 1,141 primary B-ALL samples, categorized by E-R fusion status, E-R-like gene-expression signature (primary or secondary subtype as published; ref. 2), and ETV6 aberration status. “Abnormal” ETV6 status refers to complete ETV6 copy loss in E-R+ B-ALL or any ETV6 fusion or partial/complete copy loss in non–E-R+ B-ALL. C, ETV6-repressed gene score as in B for 102 primary B-ALL samples from the NCI TARGET cohort, categorized by genomic subtype and ETV6 aberration status. Samples classified as E-R+ or E-R–like on the basis of gene expression (ALLSorts) are indicated. For E-R+ B-ALL, “abnormal” ETV6 status refers to partial or complete ETV6 copy loss other than single-copy loss downstream of the E-R fusion breakpoint (see Supplementary Fig. S7C). D–H, Details of the 40 direct ETV6-repressed E-R+/like B-ALL signature gene enhancer–gene pairs. D, Differential gene expression (RNA-seq) for the indicated gene, normalized to tagBFP control. E, Position of analyzed intervals (union of ATAC-seq, GGAA repeats, and ETV6-WT-V5 binding sites) within 200 kbp of the indicated gene TSS, coded by ETV6-WT-V5 binding status, best GGAA repeat class, and differential acetylation. Genes are oriented 5′ to 3′, with annotated gene bodies indicated by a black line. F, Differential acetylation of one element (associated with the gene listed in D) that shows ETV6 binding and significantly decreased acetylation in ETV6-WT-V5–expressing cells (H3K27ac log2 fold change < −0.25, P < 0.05), prioritized by repeat class (longest) and then distance to TSS (shortest, within 200 kbp). G, Relative acetylation of the element from F across 13 B-ALL cell lines (red = E-R+/ETV6-null cell lines, orange = ETV6-null cell line). H, Relative acetylation of the element from F in 15 primary B-ALL samples (Blueprint project; red = E-R+ samples).
We examined the loci of ETV6-repressed genes validated in the primary B-ALL cohorts and found that 39 of 40 were associated with an ETV6-bound GGAA repeat (at least 3× GGAA; Fig. 4D–F; Supplementary Fig. S7D; Supplementary Table S4). The sole exception, CLIC5, was associated with an ETV6-bound low-complexity element containing (AGGGGA)n tandem repeats and 23 nontandem GGAA sequences in 140 bp (Supplementary Fig. S7E). A more inclusive genome-wide list of active enhancer-like elements that were bound and repressed by ETV6 (H3K27ac log2 fold change −0.25, P < 0.05) similarly showed a strong association with ≥ 3× GGAA tandem repeats (1,030 of 1,133 elements, 91%; Supplementary Table S5). Most of the associated ETV6-repressed repeat elements showed increased acetylation across the five ETV6-null B-ALL cell lines (Fig. 4G), and many showed increased acetylation in primary E-R+ B-ALL compared with other subtypes (Fig. 4H). Together, these findings indicate that overexpression of genes associated with ETV6-repressed GGAA microsatellite enhancers is a unifying feature of E-R+ and E-R–like B-ALL.
Next, we directly tested the functional relationship between specific ETV6-regulated GGAA repeats and their putative target genes. We designed sgRNAs to target six ETV6-regulated GGAA repeat enhancers via CRISPR-interference (CRISPRi) in Reh cells (Fig. 5A and B). In each case, doxycycline-induced expression of a dCas9-KRAB repressor led to significant downregulation of the expected target gene, validating these genes as bona fide regulatory targets of GGAA microsatellite enhancers (Fig. 5C). Functionally validated microsatellite enhancer activation targets include PIK3C3 (VPS34), a regulator of vesicle trafficking that has been reported to mediate dependence on autophagy in E-R+ B-ALL (24), and EPOR, which encodes the erythropoietin receptor, confers enhanced STAT5 signaling in primary E-R+ B-ALL cells (25, 26), and whose genetic overexpression cooperates with E-R to generate B-ALL in mice (27). To prove that EPOR-associated GGAA repeat enhancers promote aberrant Epo-dependent signaling in E-R+ B-ALL cells, we performed electroporation of E-R+ AT-2 cells with Cas9 ribonucleoproteins designed to excise this repeat (Fig. 5D; Supplementary Fig. S8A–S8D). GGAA-deleted AT-2 cells showed significantly attenuated STAT5 phosphorylation in response to Epo treatment (Fig. 5E and F), demonstrating that this repeat enhancer is required for maximal EPOR-dependent signaling. EPOR signaling may be important in earlier stages of leukemogenesis, as many B-ALL are known to switch from a STAT5 signaling–dependent pro–B-cell-like state to a MAP kinase signaling-dependent (and STAT5-antagonized) pre–B-cell-like state during their initial evolution or upon recurrence (28). Accordingly, we did not see a consistent growth effect of Epo treatment in E-R+ B-ALL cell lines (Supplementary Fig. S8E), which have been selected for growth in Epo-deficient conditions, nor a growth deficit upon EPOR repeat enhancer repression in Reh cells (Supplementary Fig. S8F).
Figure 5.
GGAA microsatellite enhancers are direct regulators of ETV6–RUNX1 signature gene expression. A, ChIP-seq data from transgene-expressing Reh cells (top) and primary B-ALL samples (bottom) for selected E-R signature gene-linked, ETV6-regulated enhancers. ETV6-WT-V5 ChIP-seq is from doxycycline-induced, transgene-expressing Reh cells. H3K27ac ChIP-seq data were generated from tagBFP, ETV6-WT-V5, and ETV6-R399C-V5 cells, and overlays are color-coded as indicated. Also shown are positions of GGAA repeats called genome-wide in hg38 (3× GGAA). Primary B-ALL H3K27ac ChIP-seq data (Blueprint) is shown at the bottom, with E-R+ B-ALL samples indicated in red. B, Detail (1,000 bp window) of GGAA repeat enhancers shown in A, showing ETV6 ChIP-seq peaks, position of individual GGAA motifs (blue = positive strand, red = negative strand), and position of sgRNA target sequences used in C. C, Relative transcript levels for genes associated with enhancers shown in A–B 72 hours after doxycycline induction of Reh cells expressing doxycycline-inducible dCas9-KRAB and transduced with control sgRNAs or sgRNAs targeting the indicated GGAA microsatellite enhancer. Gene expression is normalized to the average of the two control sgRNAs (error bars = 95% CI of PCR replicates). Significance was calculated as t test of combined replicates for both control sgRNAs versus both enhancer-targeting sgRNAs. D, Genomic position of EPOR TSS, GGAA repeat, and sgRNAs used for GGAA repeat deletion in E–F. E, Representative phospho-STAT5 signal in AT-2 cells electroporated with Cas9-sgRNA complexes targeting the genes MME (CD10-KO) or EPOR (EPOR-KO), flanking the EPOR-adjacent GGAA repeat (EPOR GGAA del), or mock-electroporated (no sgRNA or Cas9). Samples were divided and treated with 100 U/mL Epo (red) or untreated (blue). All samples from one of two independent experiments are shown (two replicates per condition). F, Difference in phospho-STAT5 signal (75th percentile) for Cas9-RNP modified AT-2 cells. Results from two separately conducted experiments with two biological replicates each were normalized to within-experiment control samples and pooled for analysis (two-tailed t test). *, P < 0.05; **, P < 0.01; ns, not significant; P > 0.05.
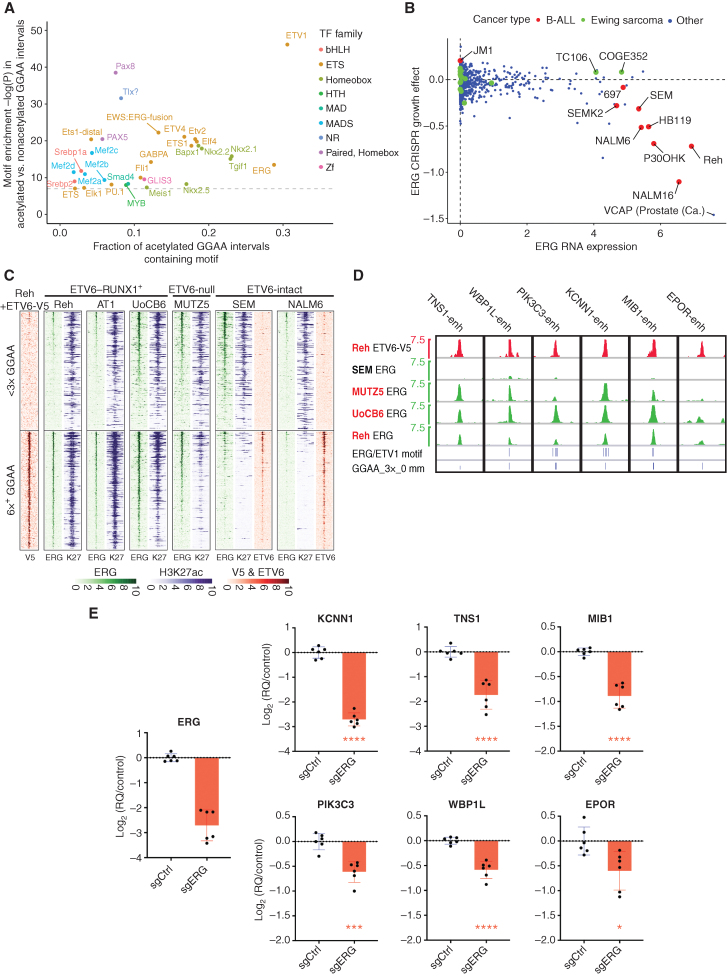
Active enhancers typically require the binding of activating TFs. We therefore sought to identify TFs that contribute positively to GGAA repeat enhancer activation in ETV6-altered B-ALL. Only a minority of GGAA repeats in the hg38 reference genome are accessible and associated with substantial H3K27ac levels in E-R+ B-ALL cell lines (Supplementary Fig. S9A and S9B). Repeat-containing intervals with acetylation that was strong and specific to E-R+ B-ALL cell lines were significantly enriched in several classes of TF motifs (Fig. 6A) compared with nonacetylated repeats, suggesting that binding of specific TFs in the vicinity of repeats might contribute to repeat enhancer activation. TFs that contribute to GGAA repeat enhancer activation likely overlap with those that regulate other B-cell enhancers, as these same motifs were also common in intervals that are strongly acetylated in all types of B-ALL (Supplementary Fig. S9B and S9C). We were interested to note that the motif corresponding to the ETS activator ERG was particularly abundant near acetylated GGAA repeats. Both ERG and its homolog FLI1 are highly expressed in B-ALL, and the fusion oncoproteins EWSR1–ERG and EWSR1–FLI1 are known activators of GGAA microsatellite enhancers in the pediatric bone tumor Ewing sarcoma. However, DepMap CRISPR knockout screens show a substantial growth dependency on ERG, but not FLI1, for the E-R+ cell line Reh and most other B-ALL cell lines screened to date (Fig. 6B; Supplementary Fig. S9D).
Figure 6.
ERG contributes to GGAA repeat enhancer activity in ETV6-altered B-ALL. A, Enrichment of known TF motifs (HOMER motif library) in 200 bp intervals centered on GGAA repeats with selective strong acetylation in E-R+ B-ALL cell lines, compared with repeat intervals not associated with acetylation (see Supplementary Fig. S9B for thresholds used). All motifs with enrichment −log(P) > 7 are shown. Note that motif analysis may have a limited ability to discriminate between factors within a given family. B, DepMap data showing ERG expression and CRISPR knockout growth effect for B-ALL cell lines vs. other cancer types. The TMPRSS2–ERG+ prostate cancer cell line VCAP is also labeled. C, Heat map of ERG, ETV6, and H3K27ac ChIP-seq signal at repeat-containing (at least 6× GGAA) and non–repeat-containing (<3× GGAA) ATAC-seq peaks in B-ALL cell lines. Peaks shown had H3K27ac fragment counts > 5 per million in at least one of 13 cell lines. The <3× GGAA group was randomly downsampled to the same number of peaks as the 6× GGAA group. ATAC-seq peaks were sorted according to the maximum ERG signal across the four ChIP-seq data sets. V5 ChIP-seq was performed in Reh cells induced to express ETV6-WT-V5; all other ChIP-seq studies were performed in parental cell lines. D, ERG and ETV6-V5 ChIP-seq signal at CRISPRi-validated GGAA repeat enhancers shown in Fig. 5. Positions of 3× GGAA repeats and predicted high-affinity motifs for ERG and ETV1 within ± 250 bp of the central GGAA repeat (HOMER motifs and thresholds) are shown at the bottom. Cell lines in red text are E-R+/ETV6-null. E, Gene-expression differences (qRT-PCR) 72 hours after doxycycline induction in Reh cells expressing doxycycline-inducible dCas9-KRAB and promoter-targeting sgRNA against ERG versus control (nontargeting) sgRNA. Values are pooled from two biological replicate experiments with three PCR replicates each (two-tailed t test). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
We performed ERG ChIP-seq on B-ALL cell lines, which showed frequent ERG binding to acetylated GGAA repeats in the E-R+ B-ALL cell lines Reh, UoCB6, and AT-1, as well as the ETV6-null cell line MUTZ5. Far less ERG binding to GGAA repeat enhancers was observed in two ETV6-intact cell lines, which instead showed frequent ETV6 binding at these same sites (Fig. 6C; Supplementary Table S5). We observed ERG binding to all six functionally validated GGAA repeat enhancers (Fig. 6D), with four of the six enhancers showing predicted high-affinity ETS factor binding sites in addition to GGAA repeats. CRISPRi-mediated ERG knockdown resulted in significantly decreased transcript levels for 36 of 40 genes in the ETV6-repression signature by RNA-seq (Supplementary Table S4), confirmed by qRT-PCR for the 6 functionally validated targets (Fig. 6E), supporting a model of mutually antagonistic regulation of GGAA repeat enhancers by the ERG activator and ETV6 repressor. In contrast, we saw no consistent additional effect on the expression of GGAA repeat enhancer-regulated genes when we knocked down FLI1, either alone or in combination with ERG knockdown (Supplementary Fig. S9E).
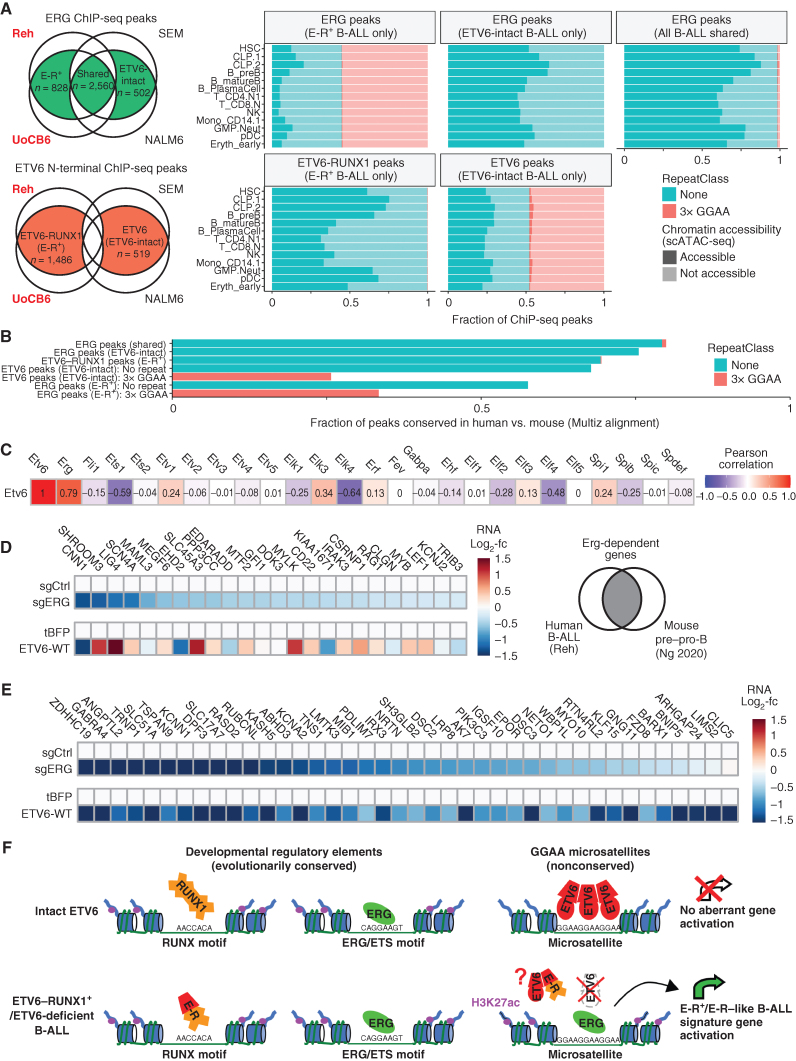
In addition to being strongly implicated in B-ALL pathogenesis (29, 30), ERG is a key regulator of normal hematopoietic stem cell (31, 32) and B-cell progenitor (33) biology. Although our global analysis of GGAA repeat chromatin state showed little evidence of GGAA repeat enhancer activation in normal development, we wondered if this remained true of the specific GGAA repeats that are bound by ETV6 or ERG in our ETV6-intact and ETV6-null B-ALL cell lines, respectively. To explore this question, we identified consensus ERG and ETV6 ChIP-seq peaks specific to ETV6–RUNX1+/ETV6-deficient and ETV6-intact B-ALL, and examined their chromatin accessibility across human hematolymphoid progenitor and mature cell populations identified in scATAC-seq data (Fig. 7A; Supplementary Fig. S10A). ERG-binding sites in ETV6-intact B-ALL cell lines and ETV6–RUNX1–binding sites in E-R+ B-ALL were mostly accessible in common lymphoid progenitors and pre-B cells, with less accessibility seen in most other bone marrow populations, consistent with many of these sites representing physiologic developmental enhancers. In contrast, ERG peaks unique to E-R+ B-ALL and ETV6 peaks unique to ETV6-intact B-ALL were highly enriched in GGAA repeats, nearly all of which were nonaccessible in any available adult bone marrow population. Due to purifying selection, developmental enhancers typically show greater evolutionary conservation than nonfunctional regions. Accordingly, GGAA repeats bound by ERG in E-R+ B-ALL and by ETV6 in ETV6-intact B-ALL showed much less frequent evolutionary conservation than ETV6–RUNX1-binding sites and ERG-binding sites in ETV6-intact B-ALL (Fig. 7B; Supplementary Fig. S10B–S10C). Thus, both epigenetic data from normal human hematopoietic subpopulations and sequence conservation analysis argue against a conserved function for ERG-activated B-ALL GGAA repeat enhancers in normal cells.
Figure 7.
B-ALL microsatellite enhancers regulated by ERG and ETV6 lack epigenetic and genetic features of normal developmental enhancers. A, Left, schematic Euler diagrams showing strategy for defining ETV6, ETV6–RUNX1, and ERG consensus binding sites specific to ETV6–RUNX1+ B-ALL (cell line names in red) or ETV6-intact B-ALL (cell line names in black), using overlaps of ETV6 (Atlas N-terminal antibody) and ERG ChIP-seq peaks identified in each individual cell line. Right, Chromatin accessibility in selected normal bone marrow populations (pseudo-bulk scATAC-seq; Granja et al.; ref. 71) for ERG or ETV6-binding sites identified in multiple E-R+ and/or ETV6-intact cell lines as defined at left. GGAA repeat status is indicated by color and accessibility is indicated by shading. See Supplementary Fig. S10A for chromatin accessibility thresholds. B, Fraction of ETV6 and ERG consensus binding sites, defined as in A, that are conserved between hg38 and mm10 (at least 10% base mapping ratio, Multiz). Peaks that overlap at least a 3× GGAA repeat are shown as separate subgroups for ER+ B-ALL ERG peaks and ETV6-intact B-ALL ETV6 peaks. C, Correlation between expression of Etv6 and all ETS transcription factors in mouse hematopoietic stem/progenitor and lymphoid populations (n = 72, Immgen RNA-seq). D, List of candidate conserved ERG target genes in mouse B-lymphopoiesis and in a human B-ALL cell line, defined as decreased both in Erg conditional knockout mouse pre–pro-B cells (Ng et al.; ref. 33) and by ERG knockdown in Reh cells (this study). Heat maps show differential expression upon ERG knockdown or ETV6-WT reexpression. E, Developmental expression in mouse (Immgen RNA-seq data) for conserved B-ALL/pro-B cell ERG target genes (defined in D) versus the 36 direct ETV6-repressed E-R+/like signature genes that are significantly ERG-dependent in Reh cells. F, Summary model for binding of ETV6, ETV6–RUNX1 (E-R), and ERG at developmental enhancers and GGAA microsatellites in B-ALL. ETV6 gene inactivation (due to ETV6–RUNX1 gene fusion formation and secondary ETV6 deletion) eliminates ETV6-mediated repression of GGAA repeats, allowing for ERG binding and neoenhancer activation in E-R+ B-ALL. Cancer-specific GGAA repeat enhancers activate many genes with minimal expression in normal B-cell progenitors, likely resulting in more distinctive changes to the transcriptome than the direct effects of the E-R fusion itself, which binds mainly at physiologic developmental enhancers. GGAA repeat enhancer formation might also be promoted by biallelic ETV6 inactivation in the absence of an E-R fusion (e.g., the MUTZ5 cell line and some E-R–like B-ALL). Although the E-R fusion does not bind directly to most GGAA repeat enhancers, it might contribute indirectly to their activation via dominant-negative interactions with ETV6 (43) in the subset of E-R+ B-ALL with one intact ETV6 allele.
Genetic studies in mice have shown that Etv6 (34) and Erg (31, 32) are both essential for the maintenance of normal adult hematopoietic stem cells, and Erg is specifically required for B-lymphopoiesis (33), raising the question of whether ETV6 might also antagonize ERG activity at ERG-dependent developmental enhancers, similar to its repressive activity at GGAA repeats in B-ALL. Etv6 and Erg expression is highly correlated in murine hematopoietic populations, being high in stem cells and early lymphoid progenitors, but low in differentiated populations (Fig. 7C; Supplementary Fig. S11A), suggestive of a complementary rather than antagonistic function. Only six genes identified as Erg-dependent in murine hematopoietic stem cells (32) were repressed by Erg knockdown in Reh human B-ALL cells (Supplementary Fig. S11B-S11C), but 26 Erg-dependent homologous gene pairs were identified in murine pre–pro-B cells (33) and Reh cells (Fig. 7D; Supplementary Fig. S11C). We focused on the two most strongly expressed of these conserved Reh/pre–pro-B-cell ERG target genes, MYB and LEF1, which encode TFs with important roles at the pro-B to pre-B transition (35–37). The genomic loci surrounding these genes showed strong ATAC-seq peaks in human lymphoid progenitors that were bound by ERG in both ETV6-deficient and ETV6-intact B-ALL cell lines but showed absent or minimal ETV6 binding (Supplementary Fig. S11D–S11F). These candidate enhancers were conserved (alignable) between the human and mouse genome and showed both ERG binding and accessibility by ATAC-seq in mouse B-cell progenitors, further supporting these as evolutionarily conserved physiologic ERG targets. However, MYB, LEF1, and many of the other conserved physiologic ERG target genes were not repressed by ETV6 in Reh cells (Fig. 7D; Supplementary Fig. S11B), in contrast to the repeat enhancer-linked genes (Fig. 7E). Although these findings require further exploration in developmental models, they suggest that ETV6 is not an equal antagonist of ERG at all of its physiologic regulatory sites but preferentially inhibits aberrant ERG activity at sites with repetitive low-affinity ETS target sequences, such as GGAA tandem repeats (Fig. 7F).
DISCUSSION
Distal regulatory elements are key drivers of oncogenic gene-expression programs. Although many well-characterized oncogene enhancers derive from evolutionarily conserved enhancers with normal functions in the cancer's tissue of origin (38–40), large-scale efforts to map cancer chromatin landscapes have revealed only partial overlap with enhancers known to be active in normal developing tissues (41). The frequency with which human cancers utilize true de novo enhancers arising from nonconserved elements remains an open question. Here, we identified a class of cancer-specific enhancers that become active in E-R+ B-ALL, are distinct from the binding targets of the E-R fusion protein itself, and show minimal evidence for developmental function or evolutionary conservation.
We found that the combination of ETV6 repressor insufficiency and ERG activator expression facilitates aberrant activation of GGAA microsatellite enhancers that represent a key mechanism underlying the unique gene activation program of E-R+ B-ALL. Our findings suggest a mechanistic explanation for phenocopying of the E-R+ B-ALL gene-expression program in E-R–like B-ALL, which is highly enriched for ETV6-inactivating mutations and deletions (2, 11) and has been defined as a distinct B-ALL subtype in the 5th World Health Organization Classification of Haematolymphoid Tumors (42). We saw some degree of GGAA repeat chromatin activation and repeat enhancer target gene overexpression in all primary E-R+ B-ALL data sets we examined, although not all E-R+ B-ALL cases show biallelic ETV6 inactivation. ETV6 haploinsufficiency, along with the known dominant-negative effects of E-R protein (43) and other altered forms of ETV6 that lack the ETS domain (43–45) on wild-type ETV6 repressor function, may compromise the silencing of repeats in cases without biallelic ETV6 alteration. Further work is indicated to investigate a potential role for microsatellite enhancers in B-ALL occurring in the setting of germline ETV6 mutations, which are reported to be heterogeneous in their biology and gene-expression signatures (13), and in diverse (albeit rare) leukemias and solid tumors that bear rearrangements between ETV6 and gene partners other than RUNX1 (46, 47), at least some of which involve biallelic ETV6 inactivation (48–50).
The in vitro DNA sequence affinity of the ETV6 DNA-binding domain is similar to that of other endogenous human ETS factors (20, 21), all of which recognize a core motif of GGA(A/T). However, ETV6 (and its homolog ETV7) are distinct in their ability to oligomerize via their N-terminal PNT domain (51–53), as the PNT domains present in ERG and many other ETS factors do not self-associate (21, 54). The self-associating property of the ETV6 PNT domain has been shown to confer cooperative binding of ETV6 to DNA sequences containing two ETS-binding sites in vitro (53), and models of ETV6 oligomers binding to greater numbers of low-affinity core binding sites have previously been proposed (52). These properties may explain our unexpected observation that ETV6 shows a strong in vivo preference for binding to GGAA repeat elements rather than canonical ETS motif–containing enhancers in B-ALL and is able to selectively antagonize ERG activity at repeat sites. Comparison of diverse ETS factor ChIP-seq data sets from the ENCODE project and various other sources further support an association of ETV6, and to a lesser extent ERG and FLI1, with GGAA repeat elements in cell types of endothelial or hematolymphoid origin (Supplementary Fig. S12A and S12B; Supplementary Table S6).
Our findings imply that a significant function of ETV6 is to sustain or restore the epigenetic silencing of GGAA repeats that can serve as low-affinity ETS-binding sites and could otherwise be prone to aberrant enhancer-like activity. Although GGAA repeat enhancer activation is a well-documented function of ERG and FLI1 fusion oncoproteins in Ewing sarcoma (55, 56), our findings indicate that the nonfusion form of ERG contributes to GGAA repeat enhancer activation in the absence of ETV6 and that repeats with adjacent high-affinity binding sites for ERG or other factors may be particularly prone to aberrant activation. A role for ERG in driving GGAA repeat-regulated genes is also supported by frequent overexpression of the ETV6-repressed gene signature in ETV6-altered cases of iAMP21 B-ALL, in which the ERG gene is amplified and overexpressed (Supplementary Fig. S7A and S7B). The genome-wide binding and distribution of individual ETS factors are known to be affected by many variables, including lineage and developmental stage-specific chromatin accessibility as well as competition or cooperation with other ETS factors. The fraction of ChIP-seq peaks for ETV6, ERG, and FLI1 that overlap GGAA repeats is variable in published data sets from hematopoietic and endothelial cells but seems consistently low in epithelial cells, or for other ETS factors regardless of cell type (Supplementary Fig. S12). Further work is needed to identify the developmental states and cooperating chromatin factors that facilitate or inhibit ETS factor binding to GGAA repeats.
Identification of microsatellite enhancers as key drivers of the E-R+/like B-ALL gene-expression program has several other important implications. Genetic mouse models of E-R expression alone yield either absent or very rare B-ALL (57–60), whereas B-ALL generated by the combination of E-R and transposon-mediated random gene disruption failed to recapitulate the transcriptional program of human E-R+ B-ALL and showed no selection for Etv6-inactivating second hits (27). Our findings suggest limitations of engineered mouse models and widely used mouse pro–B-cell lines such as Ba/F3 for studying the biology of ETV6-deficient B-ALL, as most GGAA microsatellites present in the human genome lack similarly positioned orthologs in rodents. Notably, attempts to model Ewing sarcoma in mice have also yielded very limited results (55). However, direct genetic activation of human GGAA repeat–activated genes in mice may represent an alternate modeling strategy, and it is notable that leukemias arising in the aforementioned E-R/transposase model showed selection for recurrent transposase insertions adjacent to Epor that lead to its overexpression (27).
In summary, we find strong evidence for GGAA repeat enhancer activation and target gene overexpression in ETV6-altered B-ALL cell lines and primary samples, identifying a unifying mechanism for the activation of many genes in the E-R+/−like gene-expression signature, and an unanticipated function of the ETV6 repressor that may have implications for other cancers with recurrent ETV6 alterations. Future investigations in appropriate models will be required to identify specific repeat enhancers that promote the development and/or maintenance of B-ALL, as repression of the repeat enhancers that we functionally validated did not affect the fitness of the Reh cell line. Repeat enhancer-dependent EPOR expression is a strong candidate for such a role in leukemogenesis, as it promotes activation of STAT5 signaling, an essential pathway in pro-B cells and in a subset of fully evolved B-ALL (27), and because artificial Epor overexpression appears to cooperate with E-R to generate leukemia in mice (27). Furthermore, both EPOR and STAT5 target genes are upregulated in an in vitro directed-differentiation human cell model of E-R+ leukemic progenitors (61). Our findings lay a mechanistic groundwork for future studies to determine whether this cancer-specific mechanism might represent a feasible target for selective intervention in either B-ALL therapy or prevention.
METHODS
Cell Lines
See Supplementary Table S1 for details of cell line source and validation. The identity of all B-ALL cell lines was verified by short tandem repeat profiling. All cell lines matched the expected profile from public databases (when available), and all cell line profiles were unique, with the exception of pairs of cell lines derived from the same donor [AT-1 and AT-2 (62, 63), and SUP-B13 and SUP-B15 (64)], which showed mutually identical profiles as expected. All cell lines were cultured in RPMI-1640 + glutamax supplemented with 10% fetal calf serum, penicillin/streptomycin, nonessential amino acids, 1 mmol/L sodium pyruvate, and 55 μmol/L 2-mercaptoethanol.
Cell line cytospins were evaluated using FISH probes for ETV6 (Chr12p13; SpectrumOrange; Abbott) and RUNX1 (Chr21q22; SpectrumGreen; Abbott) by standard techniques. Cell lines with one or more fusion signals per cell were considered positive for an E-R rearrangement, with 1F1R2G representing the expected pattern for a single balanced E-R rearrangement. Note that the ETV6 probe covers the 5′ portion of the ETV6 gene and a large region of chromosome 12 (486 kb). The absence of a red signal therefore indicates the absence of a nonrearranged copy of ETV6, but a focal ETV6 deletion is not excluded by the presence of a red signal.
Genomic copy-number analyses of cell lines were carried out as previously described. Briefly, depth coverages for the input lowpass WGS (ChIP-seq chromatin input) libraries were quantified at a 50-kb resolution (bins), the resulting data were segmented, and a probabilistic model (65, 66) was fit to assign absolute copy-number states to the observed coverages. The models were constrained assuming 100% neoplastic cellularity.
ChIP-seq
Protocols used for new ChIP-seq data sets were similar to those previously described (67, 68). Briefly, five million cells were cross-linked in PBS + 1% formaldehyde for 10 minutes at room temperature (histone marks) or 37°C (TFs), quenched with 1/20th volume of 2.5M glycine, washed twice in cold PBS with protease inhibitors, and lysed in cold cytoplasmic lysis buffer (20 mmol/L Tris-HCl ph8.0, 85 mmol/L KCl, 0.5% NP 40 + PI). Nuclei were pelleted at 3,000 × g and resuspended in cold SDS lysis buffer (0.3% SDS for H3K27ac and 1% SDS for TFs, 10 mmol/L EDTA, 50 mmol/L Tris-HCl, pH 8.1 + PI) for 10 minutes. Nuclei were fragmented on a Q800R2 Sonifier (QSonica) as follows: three cycles of amplitude = 50, pulse times: 30 s on/30 s off, total on time = 3:20 m, temperature = 8°C (histone marks) or two cycles of amplitude = 70, pulse times: 45 s on/15 s off, total on time = 8:50 m, temperature = 4°C (TFs). Samples were diluted 1:3 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mmol/L EDTA, 16.7 mmol/L Tris-HCl, pH 8.1, 167 mmol/L NaCl + PI), and rotated at 4°C overnight with 2–5 μg of antibody (H3K27ac, Active Motif; cat. #39133; V5 tag, Thermo Fisher; #R960-25; ETV6 n-terminal (two antibodies), Atlas Antibodies; #HPA000264, Santa Cruz Biotechnology, #sc-166835x; ERG, Cell Signaling Technologies; #97249). Subsequent chromatin capture, washing, DNA elution, purification, and Illumina library preparation steps were performed as previously described. Libraries were sequenced on NextSeq High-output flow cell 75 cycles (2 × 38 bp paired-end) or NovaSeq (2 × 150 bp paired-end).
See Supplementary Table S1 for the list of new and previously published H3K27ac ChIP-seq data sets used in this study, as well as references to corresponding protocols used (67, 68). Data sets generated on a subset of cell lines with both new and old methods showed qualitatively equivalent results.
ATAC-seq
Nuclei were isolated from 50,000 cells for each sample using Nuclei EZ prep-Nuclei Isolation Kit (Sigma-Aldrich). The transposition reaction mix (25 μL of 2× TD buffer, 2.5 μL of Tn5 transposase (Illumina), 15 μL of PBS and 7.05 μL of nuclease-free water) was added to nuclei and incubated at 37°C for 1 hour in an orbital shaker at 300 RPM. 50 μL Qiagen buffer PB was added to each sample to stop the reaction and DNA was isolated with AMPure XP beads (Beckman Coulter). Fifteen cycles of PCR were performed with transposed DNA using the dual index primers and NEBNext PCR Master Mix, followed by AMPure XP purification. After quantification and fragment size analysis, libraries were sequenced on Illumina NextSeq with 2 × 38 bp paired-end sequencing.
ETV6 Transgene Experiments
The lentiviral vector DoxON-ETV6-V5-GFP was a kind gift from the lab of Dr. Arul Chinniayan. The ETV6 coding sequence was cloned into pCW57.1 (Addgene; cat. #41393) and modified as previously described to incorporate a GFP reporter (69). DoxON-tagBFP was generated from that vector by restriction cloning tagBFP in place of ETV6 after digestion with BstBI and BmtI. DoxON-ETV6(R399C)-V5-GFP was generated with the Q5 site–directed mutagenesis kit (NEB) per the manufacturer's protocol.
Lentivirus was produced in 293T cells by standard protocols. To generate uniform doxycycline-inducible populations, Reh cells were transduced with DoxON constructs via spinfection at 2,250 rpm for 90 minutes at 37°C in the presence of 6 μg/mL polybrene and sorted for GFP+ cells on a BD MoFlo Astrios EQ. Uniformly sorted Reh cell populations were induced with 500 ng/mL doxycycline (dox) for 48 hours prior to harvest for western blot or V5 ChIP-seq. Induction was performed in duplicate for H3K27 ChIP-seq or triplicate for RNA-seq. To determine the effect of transgene expression on cell growth, each population was plated at equal density in triplicate wells with and without doxycycline. Cells were counted every 4 days on a DeNovix CellDrop BF counter with trypan blue staining and replated at equal densities.
RNA-seq
RNA was isolated via RNeasy columns with on-column DNAase digestion. RNA-seq libraries were generated with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina per the manufacturer's instructions and sequenced on an Illumina NextSeq with 2 × 38 bp paired-end sequencing.
Western Blotting
Western blotting of nuclear extracts or whole-cell extracts was performed by standard methods using antibodies specific for the N-terminal portion of ETV6 (Sigma, # HPA000264; Santa Cruz, #sc-166835x), CTCF (Cell Signaling Technology, #3418), and actin (Santa Cruz, #sc-8432).
Chromatin Data Analysis
Paired-end ChIP-seq and ATAC-seq reads were aligned to hg38 using BWA-ALN (v 0.7.17) and filtered to remove PCR duplicates and read-pairs mapping to >2 sites genome-wide. Display files were generated with deepTools bamCoverage and visualized with IGV. Scaling for all ChIP-seq and ATAC-seq tracks in figures is equal to local paired-end fragment coverage × (1,000,000/totalCount). ERG, endogenous ETV6, and ETV6-V5 ChIP-seq peak calling was performed with HOMER findPeaks using the “factor” style and FDR < 0.001 for ERG, FDR < 0.01 for ETV6-V5, and FDR <0.05 for endogenous ETV6. ATAC-seq peak summits were identified with MACS2 using default parameters. All peak sets were post-filtered against hg38 blacklist regions (available at https://github.com/Boyle-Lab/Blacklist/blob/master/lists/hg38-blacklist.v2.bed.gz).
To analyze H3K27ac ChIP-seq signal associated with individual enhancer modules, we resized ATAC-seq peaks to 200 bp around MACS2 peak summits, discarded peaks with low signalValue (<5), and then used GenomicRanges “reduce” and “resize” to generate consensus union ATAC-seq peak sets of 200 bp intervals for the samples of interest (26 B-cell cancer cell lines or 13 B-ALL cell lines depending on the analysis). HOMER annotatePeaks was used to annotate union ATAC-seq peaks with normalized H3K27ac ChIP-seq signal from the relevant cell lines in a 1,000 bp window around each interval center.
To identify clusters of enhancers with correlated acetylation levels across the 26 B-cell cancer cell lines, we filtered out ATAC-seq union peak intervals located < 2 kb upstream or < 1 kb downstream of an annotated TSS, as well as intervals associated with low H3K27ac signal in all cell lines. The acetylation signal was square root-transformed, centered, and scaled by genomic region across all cell lines. K-means clustering (k = 30) was used to identify enhancer clusters. HOMER findMotifsGenome was used to identify both known and de novo enriched TF motifs in each cluster, with the set of all enhancers used as a background (option -b). The top two de novo motifs identified in each B-ALL–specific cluster were then used as custom-known motifs to calculate enrichment in all B-ALL–specific clusters. For known motif enrichment analysis of endogenous (N-terminal) ETV6 ChIP-seq peaks, the custom “GGAA_3x_0mm” motif described below was appended to the Homer known motif library.
Identification of Chromatin Features and Genes Associated with GGAA Repeat Intervals
We used HOMER seq2profile.pl to generate HOMER custom motif files corresponding to a specified number of GGAA tandem repeats and a permitted number of mismatches, e.g., motif “GGAA_3x_1mm” corresponds to a genomic sequence with no more than one mismatch to the sequence “GGAAGGAAGGAA,” whereas “GGAA_3x” corresponds to exact matches to that same genomic sequence. We then used HOMER scanMotifsGenomeWide.pl to identify all occurrences in the hg38 reference genome for the motifs GGAA_3x_1mm, GGAA_3x, GGAA_6x, GGAA_9x, and GGAA_12x. We used HOMER mergePeaks to merge identified motifs into uniform 200 bp genomic intervals (−d 200), each of which was centered on one or more motif instances and was annotated with the most stringent contained motif. This set of annotated repeat-containing intervals was then overlapped with the union set of ATAC-seq peaks in 13 B-ALL cell lines. HOMER annotatePeaks was then used to annotate each interval in the union ATAC-seq/GGAA repeat set with normalized H3K27ac ChIP-seq signal in an 800-bp window.
To generate histograms of ATAC-seq or H3K27ac ChIP-seq signal associated with nonrepeat and repeat-containing nucleosome-free regions, we assigned each peak in the union set of distal ATAC-seq peaks from 13 B-ALL cell lines or 24 primary B-ALL samples (ref. 14; reprocessed as described above) to one of three groups based on whether it contained a 6× GGAA motif, 3× (but not 6×) GGAA motif, or neither. HOMER annotatePeaks (-hist 25 -size 3,000) was used to generate normalized histograms for the appropriate ATAC-seq or H3K27ac ChIP-seq data sets for each of the three repeat groups.
To analyze chromatin effects of ETV6 restoration in Reh cells, Reh ATAC-seq peaks were merged into a union interval set with ETV6-WT-V5 peaks and GGAA repeat–containing intervals (defined as above, a total of 147,399 intervals). Normalized H3K27ac ChIP-seq signal from Reh cells expressing tagBFP, ETV6-WT-V5, and ETV6-R399C-V5 (two replicates each) was calculated in an 800-bp window around each peak. DESeq2 was used to calculate log2 fold-change values for acetylation associated with each peak, using default parameters with apeglm shrinkage. ggplot2 was used to display boxplots for differential acetylation data according to the repeat and ETV6 binding status of each interval.
RNA-seq transcript levels for DoxON-tagBFP, ETV6-WT-V5, and ETV6-R399C-V5 expressing cells were quantified with Salmon and collapsed to gene level (Ensembl, Feb2014) with tximport. Differential gene-expression analysis was performed with DESeq2. The same approach was used to analyze RNA-seq data from doxycycline-inducible dCas9-KRAB–expressing Reh cells transduced with an ERG promoter-targeting sgRNA or nontargeting control (3 replicates each, 72 hours after doxycycline treatment).
The ROSS2003_ETV6-RUNX1_UP gene set was derived from Ross and colleagues (1), Supplementary Information, section II “Top 100 chi-square probe sets selected for TEL-AML1, decision tree format,” including all genes with HD>50 above mean that could be successfully converted to Ensembl 2014 gene symbols. For gene set enrichment analysis, normalized gene-level RNA-seq counts for tagBFP and ETV6-WT triplicates were exported by DESeq2 and converted to .gct format. GSEA_4.1 software was then used to calculate enrichment for the ROSS2003_ETV6-RUNX1_UP gene set.
To link differentially expressed genes to candidate regulatory elements, runSeq2gene (Bioconductor package: seq2pathway) was used to link each interval in the Reh ATAC-seq/ETV6-V5 ChIP-seq/GGAA repeat union interval set to each hg38 Ensembl 2014 gene TSS within 200 kbp. Intervals were annotated (HOMER annotatePeaks) with normalized acetylation signal for ETV6 transgene or tagBFP control transgene-expressing cells, 13 B-ALL cell lines, and Blueprint primary B-ALL samples.
For genome-wide comparison of GGAA repeat element acetylation and motif associations in ETV6–RUNX1+ B-ALL, ETV6-intact B-ALL, and Ewing sarcoma, HOMER annotatePeaks was used to annotate hg38 genome-wide repeat-containing intervals (as defined above) with normalized H3K27ac ChIP-seq signal (800 bp window) from B-ALL cell lines plus the Ewing sarcoma cell lines A673 and SKNMC (data from Riggi and colleagues; ref. 56). B-ALL cell lines were grouped as ETV6-null (Reh, AT-1, UoCB6, and MUTZ-5; AT-2 was omitted due to shared origin with AT-1), or ETV6-intact (SUP-B15, NALM-6, SEM, RS4;11, KOPN-8, HAL-01, and MHH-CALL-3; SUP-B13 was omitted due to shared origin with SUP-B15), and median acetylation values determined for each interval in each group. Intervals with median acetylation value log2(tags per million + 1) < 2 for both the ETV6-null and ETV6-intact groups were defined as being “non-acetylated,” intervals with median acetylation value log2(tags per million + 1) > 4 for both the ETV6-null and ETV6-intact groups were defined as having “shared acetylation” and intervals with log2(tags per million + 1) < 4 in ETV6-intact, >4 in ETV6-null, and log2(ETV6-null tags per million + 1) − log2(ETV6-intact tags per million + 1) > 2 were defined as “ETV6-null-specific acetylation.” Homer findMotifsGenome was then used to determine enrichment of known TF motifs in the ETV6-null–specific acetylation intervals or shared acetylation intervals (200-bp window), versus the nonacetylated intervals used as a custom background (option -b).
To generate heat maps of ETV6, ERG, and H3K27ac ChIP-seq signal, the union set of ATAC-seq peaks from 13 B-ALL cell lines was annotated with H3K27ac signal for 13 B-ALL cell lines (800-bp window) and filtered to retain peaks with normalized H3K27ac signal >5 fragments per million in at least one cell line. Peaks were then annotated with signal profiles from each ChIP-seq data set using HOMER annotatePeaks with options -size 8,000 -hist 20 -ghist. Groups of peaks containing 6× GGAA repeats or no repeats (<3× GGAA) were retained, with the latter group randomly downsampled such that each group had equal numbers of peaks. Peaks were then sorted according to maximum ERG ChIP-seq signal (400-bp window) across all six cell lines.
Comparison of GGAA Repeat Acetylation and Accessibility Across Diverse Cell Types
To compare GGAA repeat acetylation and accessibility across diverse cell types, we generated a set of 400-bp genomic intervals containing all GGAA repeats for hg38 (6× GGAA_0mismatch, n = 5,426; 3× GGAA_0mismatch, n = 16,897; 3× GGAA_1mismatch, n = 72,678), as well as control intervals consisting of 100,000 random genomic intervals (bedtools random) and a published set of universally chromatin-accessible housekeeping gene promoters (n = 6,440; ref. 70). An identical approach was used to generate corresponding intervals for mm10. We used deepTools multiBigWigSummary to generate signal matrices for these intervals from ATAC-seq or H3k27ac ChIP-seq bigwig files (intervals were expanded to 1 kb for H3K27ac).
Bigwig inputs for Fig. 2D/Supplementary Fig. S4A consisted of uniformly processed primary human H3K27ac ChIP-seq bigwig files from the Blueprint consortium (ETV6–RUNX1+ B-ALL n = 3; other B-ALL n = 11; other blood cancer n = 52; sorted normal hematopoietic, immune, and stromal cell populations n = 78). The signal matrix was quantile normalized across all samples and intervals. For each interval subset (GGAA repeat sets, random control regions, and housekeeping promoters), signal values were then ranked within each population and plotted.
Bigwig inputs for Fig. 2E/Supplementary Fig. S4B consisted of pseudobulk ATAC-seq tracks generated from normal human bone marrow single-cell ATAC-seq data sets as described in the original publication (71) by filtering, normalization, and clustering of single-cell data, followed by assignment of clusters to 23 known populations. Because preprocessed data sets from this work were only available for hg19, we used UCSC liftOver to convert the midpoint of all intervals to hg19 coordinates, and then reexpanded to 400 bp. We generated a single signal matrix for these intervals from normal bone marrow scATAC-seq data sets and hg19-aligned bulk ATAC-seq data sets from primary ETV6–RUNX1+, DUX4-rearranged, and hyperdiploid B-ALL samples (14), and then quantile normalized, ranked, and plotted data as described above.
Bigwig inputs for Supplementary Fig. S4C consisted of pseudobulk ATAC-seq tracks (hg38 bigwigs, processed as originally described) generated by scATAC-seq from diverse normal human adult and fetal tissues (72), with clusters assigned to 222 known cell types. The same primary B-ALL bulk ATAC-seq data sets used above (ref. 14; but aligned to hg38) were included in the signal matrix for hg38 intervals, which was then quantile normalized and plotted as described above.
Bigwig inputs for Supplementary Fig. S4D consisted of mm10-aligned bulk ATAC-seq data sets from sorted mouse hematopoietic and immune system-related cell populations (n = 90) generated via uniform methods by the Immgen Consortium. A signal matrix was generated for all samples for 400-bp GGAA repeat and control region intervals. Quantile normalization and plotting were performed as described above.
Chromatin Accessibility and Evolutionary Conservation of Consensus ETV6–RUNX1, ETV6, and ERG Motifs
Peaks from ChIP-seq with ETV6 n-terminal (Atlas #HPA000264) and ERG antibodies in E-R+/ETV6WT-null cell lines (Reh, UoCB6) and ETV6-intact cell lines (SEM, NALM6) were resized to a uniform 300 bp. Note that ETV6 n-terminal ChIP-seq peaks in E-R+/ETV6WT-null cell lines were interpreted as binding sites for ETV6–RUNX1 and peaks obtained with the same antibody in ETV6-intact cell lines were interpreted as binding sites for ETV6WT. GenomicRanges findOverlaps was used to identify consensus peaks shared in the E-R+/ETV6WT-null cell lines but not the ETV6-intact cell lines and vice versa, whereas peaks shared by all four cell lines were identified for ERG data sets. We then used deepTools multiBigWigSummary to generate signal matrices for these intervals from normal human bone marrow cell populations derived from TSS-normalized single-cell ATAC-seq data sets (71) as described above. Intervals were further subdivided for some figures by overlaps with 3× GGAA and 6× GGAA repeat-containing intervals.
To generate sequence conservation profiles for consensus ETV6 and ERG peak sets (divided into GGAArep+ and GGAArep− for peak sets with >2% GGAA overlaps), we used deepTools computeMatrix reference-point (-referencePoint center -a 1,000 -b 1,000) to summarize phyloP base-wise conservation signal derived from Cactus 241-placental mammal multialignment (bigwig accessed at). To look at factor binding interval conservation between humans and specific species, hg38 consensus TF binding intervals were resized to 200 bp, and we then used the UCSC liftOver command line tool to map all intervals in multiz chain files for hg38 and 5 other species (chimpanzee PanTro5, rhesus macaque RheMac10, dog CanFam5, mouse Mm10, and opossum MonDom5) with a minimum base remapping ratio of 0.1 (intervals returning corresponding intervals or the error “Duplicated in new” were considered alignable, whereas errors “Deleted in new,” “Partially deleted in new,” and “Split in new” were considered nonalignable.
Analysis of GGAA Repeat Binding across Diverse ETS Factor Data Sets and Cell Types
ChIP-seq binding peaks for ETV6, ERG, FLI1, ETS fusion proteins, and other wild-type ETS factors were obtained from several sources (73–78). See Supplementary Table S6 for details of data source and processing. Peaks were resized to a uniform width of 300 bp and were overlapped with 300-bp intervals containing all hg38 3× GGAA repeats using genomicRanges findOverlaps.
Primary B-ALL Gene-Expression Analysis
Normalized RNA-seq gene-expression values, subtype assignments, fusion genes, and ETV6 copy-number abnormalities for 1,988 B-ALL samples were obtained from published Supplementary Data Tables (2) and the St Jude Cloud website (https://pecan.stjude.cloud/proteinpaint/study/PanALL). The set of upregulated signature genes for the ETV6–RUNX1 and ETV6–RUNX1-like B-ALL were defined as published (log2 fold change > 1 and Padj <0.05 for the whole cohort (2). Samples with corresponding copy-number data (n = 1,141) were used for further analysis. Uniformly processed NCI TARGET B-ALL phase II data were downloaded from cBioPortal, including gene-expression Z-scores, hg19 genomic copy-number segmentation, ETV6–RUNX1 FISH results, and assigned molecular subtype. Analysis was performed on a subset of TARGET samples (102 total) from unique patients with both RNA-seq and CNA segmentation data available, after the removal of patients also represented in the St. Jude data set based on a published key of matched St Jude/PCGP and TARGET patient identifiers (79). Molecular subtypes were used as provided except that the groups “Trisomy of both chromosomes 4 and 10,” “Hyperdiploidy without trisomy of both chromosomes 4 and 10,″ and “Hyperdiploid; status of 4 and 10 unknown” were merged into a single “Hyperdiploid” group. Samples were assigned an “abnormal” ETV6 status if segmentation data showed monoallelic or biallelic deletion of any portion of the ETV6 gene, except for three E-R+ samples for which monoallelic deletion of ETV6 on the 3′ side of the fifth intron could represent loss of the der(12)t(12;21) chromosome without affecting the intact ETV6 gene. As these three ambiguous samples showed repeat enhancer–gene signature scores intermediate between the samples with no ETV6 deletions and those with definitive secondary ETV6 deletions, including them in either the ETV6-intact or ETV6-deleted groups did not affect the statistical significance of our conclusions.
To identify a signature of ETV6-repressed genes, we identified protein-coding genes that met the following criteria in our Reh ETV6-WT-V5–overexpression experiments: RNA log2 fold change(tagBFP/ETV6-WT-V5) < −0.5, Padj < 0.001, and RNA log2 fold change(tagBFP/ETV6-WT-V5) < log2 fold change(tagBFP/ETV6-R399C-V5). We further filtered for genes linked to an ETV6-binding site within 200 kbp of the promoter that showed decreased H3K27ac signal in ETV6-WT-V5 versus tagBFP (1 kb window, H3K27ac log2 fold change < −0.25, P < 0.05). Seventy-one genes met these criteria, of which 40 were significantly overexpressed in ETV6-RUNX1+/− like B-ALL from the St. Jude data set according to the published analysis (2).
Comparative Genomic Analysis
The following approach was used to compare the relationship between GGAA repeats and gene promoters across mammalian species. HOMER scanMotifsGenomeWide was used to independently identify GGAA repeat-containing intervals in the hg38 (human), panTro5 (chimpanzee), Mmul10 (rhesus macaque), and mm10 (house mouse) genomes. UROPA (80) was used to annotate the distance from each repeat to the start sites of all genes within 1 Mbp, using .gtf gene annotation files from ENSEMBL version 102. Gene-repeat linkages were filtered to retain only ENSEMBL genes with annotated homologs across all 4 species in the ENSEMBL 102 Biomart database. For pairwise comparisons between species, further filtering retained only the pair of gene homolog-repeat linkages with the shortest genomic distance in humans, and only one pair of gene homologs per HUGO gene symbol, selected for the least difference in genomic distance from gene homolog to repeat between the two species.
CRISPR Interference
To design sgRNAs targeting GGAA microsatellite enhancers, we used FlashFry (81) to identify and score all candidate sgRNAs in a 2-kb window around repeats of interest. Candidates were kept that met the following scoring criteria: Doench2014OnTarget > 0.1, Hsu2013 > 50, JostCRISPRi_specificityscore > 0.1, dangerous_GC = = “NONE,” dangerous_polyT = = “NONE,” dangerous_in_genome = = “IN_GENOME = 1”, otCount < 500. The final sgRNAs used for experiments were selected on the bases of shortest distance to GGAA repeat and highest Doech2014 on-target score (see Supplementary Table S1). Complementary oligonucleotides encoding sgRNA sequences plus appropriate overhangs were annealed and cloned into BsmBI-digested sgOpti (Addgene #85681).
A CRISPRi-ready Reh cell population with dox-inducible dCas9-KRAB and a GFP reporter (Reh-CiG) was generated as follows. Reh cells were transduced with lentivirus produced from TRE3-KRAB-dCas9-IRES-GFP and pLVX-EF1alpha-Tet3G vectors. Cells were serially sorted for GFP+ cells after doxycycline induction, for GFP-negative cells without doxycycline induction, and again for GFP+ cells after doxycycline induction.
For enhancer-targeting sgRNA experiments, Reh-CiG cells were transduced with control and repeat enhancer-targeting sgRNA lentivirus by spinfection. Cells were treated 48 hours after transduction with 1 μg/mL puromycin and 100 ng/mL doxycycline and were harvested 5 days after transduction for RNA extraction and qRT-PCR.
Optimized promoter-targeting sgRNA sequences for the knockdown of ERG and FLI1 were selected from the “Dolcetto” genome-wide human CRISPRi library (82). For the knockdown of ERG and/or FLI1, variants of the sgOpti vector were generated by cloning tagBFP (sgMW-tagBFP) or tagRFP (sgMW-tagRFP) into BamHI and MluI-digested sgOpti in place of the PuroR gene. An ERG-targeting sgRNA sequence or nontargeting control was cloned into sgMW-tagRFP and an FLI1-targeting sequence was cloned into sgMW-tagBFP. Reh-CiG cells were transduced with appropriate combinations of control, ERG, FLI1, or ERG + FLI1 targeting sgRNA lentivirus and were flow sorted to ensure uniform expression of the appropriate fluorescent reporter(s). Cells were then induced with 500 ng/mL doxycycline for 3 days prior to RNA harvest and qPCR analysis.
Cas9 Ribonucleoprotein-Based Genome Editing and Assessment of EPOR-Dependent Signaling
Cas9 protein, tracerRNA, and custom sgRNAs were purchased from IDT DNA (ALT-R), and Cas9 ribonucleoprotein complexes (RNP) were generated according to the manufacturer's instructions. Mock electroporation (no RNP) or electroporation with RNP targeting MME (CD10), EPOR, or the EPOR-associated GGAA repeat (50/50 mix of two flanking sgRNAs) were electroporated into AT-2 cells with the NEON system (10 μL tips, pulse voltage: 1750 V, pulse width: 20 ms, pulse number: 1). Each biological replicate consisted of two sequential electroporations of 5 × 105 cells done in the same tip and pooled into a single recovery well containing 1 mL of prewarmed media. Two replicates were performed per modification, per experiment, and data from two separately conducted experiments were pooled for the final analysis.
Biallelic genome modification efficiency for a single sgRNA was estimated at >80% of cells based on loss of CD10 expression at > 1 week post-electroporation in samples electroporated with MME-targeting RNP. Efficiency of GGAA repeat excision in dual-sgRNA experiments was calculated by PCR amplification of genomic DNA containing the deletion target with primers P1-F and P1-R, visualized by gel-electrophoresis on a 2% agarose/TAE gel prestained with 1× GelGreen (41005, Biotium). Bands were quantified with the Fiji package (ImageJ) and normalized by base-pair length to calculate the relative concentration of intact versus GGAArep-deleted amplicons.
Erythropoietin-dependent phospho-STAT5 activation was measured via flow cytometry 4 days after electroporation. 300,000 Cas9-modified and mock-electroporated cells were equilibrated overnight in 0.5 mL fresh media. Cells were then treated with 100 IU/mL of recombinant erythropoietin (Amgen NDC55513028301) for 30 minutes. Fixable viability stain (BD 562247) was added during the final 5 minutes of EPO treatment. Cells were fixed using Cytofix Fixation Buffer (BD 554655) and incubated for 10 minutes at 37°C. After washing, cells were permeabilized with Perm Buffer III (BD 558050) on ice for 30 minutes, washed, and stained with AF647-conjugated phospho-Stat5 (pY694; BD 612599) and read out with a Bio-Rad ZE5 Analyzer. Results were consistent across two experiments conducted and assayed on separate occasions, with all replicates from both experiments pooled for the final analysis.
Data Availability
New sequencing data sets produced for this work are available at GEO under accession number GSE186942. Previously published data are available under accession numbers GSE69558 and GSE97541. BLUEPRINT epigenome consortium H3K27ac ChIP-seq data were accessed at http://dcc.blueprint-epigenome.eu/#/data sets. NCI TARGET transcriptomic data were accessed at https://portal.gdc.cancer.gov/projects.
Supplementary Material
Supplementary Table 1 lists oligonucleotide sequences and cell line information
Supplementary Table 2 details known and de novo motif analysis for distal acetylation clusters
Supplementary Table 3 summarizes TF ChIP-Seq results and motif enrichment statistics
Supplementary table 4 lists ETV6-regulated gene-enhancer linkages
Supplementary Table 5 annotates genome-wide candidate ETV6-repressed enhancers
Supplementary Table 6 details GGAA repeat overlap analysis of new and previously published ETS factor datasets.
Contains Supplementary Figures 1-12 and additional information about Supplementary tables
Acknowledgments
We thank M. Le Beau and E. Davis for validating and providing B-ALL cell lines, D. Boyer for assistance with FISH, and A. Chinniayan, R. Khoriaty, X. Wang, J. Kidd, P. Hsu, and E. Lawlor for helpful discussions. R.J.H. Ryan acknowledges support from the NCI (K08-CA208013) and a Hollis Brownstein Research Grant from the Leukemia Research Foundation. J.W. Goldman acknowledges support from NIH training grant T32HL007622. A.C. Monovich acknowledges support from NIH training grant T32CA140044. The results published here are in whole or part based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments (https://ocg.cancer.gov/programs/target) initiative, phs000218.
Footnotes
Note Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
J.W. Goldman reports grants from NIH during the conduct of the study. A.C. Monovich reports grants from NIH during the conduct of the study. R.J. Ryan reports grants from NCI and Leukemia Research Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
R. Kodgule: Formal analysis, validation, investigation, writing–original draft, writing–review and editing. J.W. Goldman: Formal analysis, validation, investigation, writing–original draft, writing–review and editing. A.C. Monovich: Formal analysis, validation, visualization, investigation, writing–original draft, writing–review and editing. T. Saari: Software, formal analysis. A.R. Aguilar: Investigation. C.N. Hall: Investigation. N. Rajesh: Investigation. J. Gupta: Investigation. S.-C.A. Chu: Formal analysis. L. Ye: Formal analysis. A. Gurumurthy: Investigation. A. Iyer: Investigation. N.A. Brown: Resources, supervision, investigation. M.Y. Chiang: Resources, writing–review and editing. M.P. Cieslik: Resources, software, formal analysis, visualization, methodology, writing–review and editing. R.J.H. Ryan: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood 2003;102:2951–9. [DOI] [PubMed] [Google Scholar]
- 2. Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet 2019;51:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lilljebjörn H, Fioretos T. New oncogenic subtypes in pediatric B-cell precursor acute lymphoblastic leukemia. Blood 2017;130:1395–401. [DOI] [PubMed] [Google Scholar]
- 4. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al., editors. WHO classification of tumors of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon: World Health Organization; 2017. [Google Scholar]
- 5. Lopez RG, Carron C, Oury C, Gardellin P, Bernard O, Ghysdael J. TEL is a sequence-specific transcriptional repressor. J Biol Chem 1999;274:30132–8. [DOI] [PubMed] [Google Scholar]
- 6. Romana S, Mauchauffe M, Le Coniat M, Chumakov I, Le Paslier D, Berger R, et al. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood 1995;85:3662–70. [PubMed] [Google Scholar]
- 7. Golub TR, Barker GF, Bohlander SK, Hiebert SW, Ward DC, Bray-Ward P, et al. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci 1995;92:4917–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raynaud S, Cave H, Baens M, Bastard C, Cacheux V, Grosgeorge J, et al. The 12;21 translocation involving TEL and deletion of the other TEL allele: two frequently associated alterations found in childhood acute lymphoblastic leukemia. Blood 1996;87:2891–9. [PubMed] [Google Scholar]
- 9. Kim DH, Moldwin RL, Vignon C, Bohlander SK, Suto Y, Giordano L, et al. TEL-AML1 translocations with TEL and CDKN2 inactivation in acute lymphoblastic leukemia cell lines. Blood 1996;88:785–94. [PubMed] [Google Scholar]
- 10. Stegmaier K, Pendse S, Barker GF, Bray-Ward P, Ward DC, Montgomery KT, et al. Frequent loss of heterozygosity at the TEL gene locus in acute lymphoblastic leukemia of childhood. Blood 1995;86:38–44. [PubMed] [Google Scholar]
- 11. Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, et al. Identification of ETV6–RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang MY, Churpek JE, Keel SB, Walsh T, Lee MK, Loeb KR, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet 2015;47:180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishii R, Baskin-Doerfler R, Yang W, Oak N, Zhao X, Yang W, et al. Molecular basis of ETV6-mediated predisposition to childhood acute lymphoblastic leukemia. Blood 2021;137:364–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diedrich JD, Dong Q, Ferguson DC, Bergeron BP, Autry RJ, Qian M, et al. Profiling chromatin accessibility in pediatric acute lymphoblastic leukemia identifies subtype-specific chromatin landscapes and gene regulatory networks. Leukemia 2021;35:3078–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pradel LC, Vanhille L, Spicuglia S. The European Blueprint project: towards a full epigenome characterization of the immune system. Med Sci (Paris) 2015;31:236–8. [DOI] [PubMed] [Google Scholar]
- 16. Vijayakrishnan J, Studd J, Broderick P, Kinnersley B, Holroyd A, Law PJ, et al. Genome-wide association study identifies susceptibility loci for B-cell childhood acute lymphoblastic leukemia. Nat Commun 2018;9:1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Niebuhr B, Kriebitzsch N, Fischer M, Behrens K, Günther T, Alawi M, et al. Runx1 is essential at two stages of early murine B-cell development. Blood 2013;122:413–23. [DOI] [PubMed] [Google Scholar]
- 18. Linka Y, Ginzel S, Krüger M, Novosel A, Gombert M, Kremmer E, et al. The impact of TEL-AML1 (ETV6-RUNX1) expression in precursor B cells and implications for leukaemia using three different genome-wide screening methods. Blood Cancer J 2013;3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teppo S, Laukkanen S, Liuksiala T, Nordlund J, Oittinen M, Teittinen K, et al. Genome-wide repression of eRNA and target gene loci by the ETV6-RUNX1 fusion in acute leukemia. Genome Res 2016;26:1468–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J 2010;29:2147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 2011;80:437–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmidt B, Brown LM, Ryland GL, Lonsdale A, Kosasih HJ, Ludlow LEA, et al. ALLSorts: an RNA-seq subtype classifier for B-cell acute lymphoblastic leukemia. Blood Advances 2022;6:4093–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polak R, Bierings MB, van der Leije CS, Sanders MA, Roovers O, Marchante JRM, et al. Autophagy inhibition as a potential future targeted therapy for ETV6-RUNX1-driven B-cell precursor acute lymphoblastic leukemia. Haematologica 2019;104:738–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inthal A, Krapf G, Beck D, Joas R, Kauer MO, Orel L, et al. Role of the erythropoietin receptor in ETV6/RUNX1-positive acute lymphoblastic leukemia. Clin Cancer Res 2008;14:7196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Torrano V, Procter J, Cardus P, Greaves M, Ford AM. ETV6-RUNX1 promotes survival of early B lineage progenitor cells via a dysregulated erythropoietin receptor. Blood 2011;118:4910–8. [DOI] [PubMed] [Google Scholar]
- 27. van der Weyden L, Giotopoulos G, Rust AG, Matheson LS, van Delft FW, Kong J, et al. Modeling the evolution of ETV6-RUNX1–induced B-cell precursor acute lymphoblastic leukemia in mice. Blood 2011;118:1041–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chan LN, Murakami MA, Robinson ME, Caeser R, Sadras T, Lee J, et al. Signalling input from divergent pathways subverts B cell transformation. Nature 2020;583:845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet 2016;48:1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qian M, Xu H, Perez-Andreu V, Roberts KG, Zhang H, Yang W, et al. Novel susceptibility variants at the ERG locus for childhood acute lymphoblastic leukemia in Hispanics. Blood 2019;133:724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taoudi S, Bee T, Hilton A, Knezevic K, Scott J, Willson TA, et al. ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification. Genes Dev 2011;25:251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Knudsen KJ, Rehn M, Hasemann MS, Rapin N, Bagger FO, Ohlsson E, et al. ERG promotes the maintenance of hematopoietic stem cells by restricting their differentiation. Genes Dev 2015;29:1915–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ng AP, Coughlan HD, Hediyeh-zadeh S, Behrens K, Johanson TM, Low MSY, et al. An Erg-driven transcriptional program controls B cell lymphopoiesis. Nat Commun 2020;11:3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hock H, Meade E, Medeiros S, Schindler JW, Valk PJM, Fujiwara Y, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev 2004;18:2336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reya T, O'Riordan M, Okamura R, Devaney E, Willert K, Nusse R, et al. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity 2000;13:15–24. [DOI] [PubMed] [Google Scholar]
- 36. Jin ZX, Kishi H, Wei XC, Matsuda T, Saito S, Muraguchi A. Lymphoid enhancer-binding factor-1 binds and activates the recombination-activating gene-2 promoter together with c-Myb and Pax-5 in immature B cells. J Immunol 2002;169:3783–92. [DOI] [PubMed] [Google Scholar]
- 37. Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K., Bender TP. C-Myb Is critical for B cell development and maintenance of follicular B cells. Immunity 2005;23:275–86. [DOI] [PubMed] [Google Scholar]
- 38. Shi J, Whyte W, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 2013;27:2648–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Herranz D, Ambesi-Impiombato A, Palomero T, Schnell S, Belver L, Wendorff A, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 2014;20:1130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bunting KL, Soong TD, Singh R, Jiang Y, Béguelin W, Poloway DW, et al. Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center-specific locus control region. Immunity 2016;45:497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, et al. The chromatin accessibility landscape of primary human cancers. Science 2018;362:eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IB de O, Berti E, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia 2022;36:1720–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gunji H, Waga K, Nakamura F, Maki K, Sasaki K, Nakamura Y, et al. TEL/AML1 shows dominant-negative effects over TEL as well as AML1. Biochem Biophys Res Commun 2004;322:623–30. [DOI] [PubMed] [Google Scholar]
- 44. Sasaki K, Nakamura Y, Maki K, Waga K, Nakamura F, Arai H, et al. Functional analysis of a dominant-negative ΔeTS TEL/ETV6 isoform. Biochem Biophys Res Commun 2004;317:1128–37. [DOI] [PubMed] [Google Scholar]
- 45. Van Waalwijk Van Doorn-Khosrovani SB, Spensberger D, De Knegt Y, Tang M, Löwenberg B, Delwel R. Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene 2005;24:4129–37. [DOI] [PubMed] [Google Scholar]
- 46. De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res 2012;36:945–61. [DOI] [PubMed] [Google Scholar]
- 47. Biswas A, Rajesh Y, Mitra P, Mandal M. ETV6 gene aberrations in non-haematological malignancies: a review highlighting ETV6 associated fusion genes in solid tumors. Biochim Biophys Acta Rev Cancer 2020;1874:188389. [DOI] [PubMed] [Google Scholar]
- 48. Suto Y, Sato Y, Smith SD, Rowley JD, Bohlander SK. A t(6;12)(q23;p13) results in the fusion of ETV6 to a novel gene, STL, in a B-cell ALL cell line. Genes Chromosomes and Cancer 1997;18:254–68. [DOI] [PubMed] [Google Scholar]
- 49. Golub TR, Goga A, Barker GF, Afar DE, McLaughlin J, Bohlander SK, et al. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol Cell Biol 1996;16:4107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yagasaki F, Jinnai I, Yoshida S, Yokoyama Y, Matsuda A, Kusumoto S, et al. Fusion of TEL/ETV6 to a novel ACS2 in myelodysplastic syndrome and acute myelogenous leukemia with t(5;12)(q31;p13). Genes Chromosomes and Cancer 1999;26:192–202. [DOI] [PubMed] [Google Scholar]
- 51. Potter MD, Buijs A, Kreider B, Van Rompaey L, Grosveld GC. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood 2000;95:3341–8. [PubMed] [Google Scholar]
- 52. Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, et al. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J 2001;20:4173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Green SM, Coyne HJ, McIntosh LP, Graves BJ. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J Biol Chem 2010;285:18496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mackereth CD, Schärpf M, Gentile LN, MacIntosh SE, Slupsky CM, McIntosh LP. Diversity in structure and function of the Ets family PNT domains. J Mol Biol 2004;342:1249–64. [DOI] [PubMed] [Google Scholar]
- 55. Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci U S A 2008;105:10149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvà ML, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in ewing sarcoma. Cancer Cell 2014;26:668–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsuzuki S, Seto M, Greaves M, Enver T. Modeling first-hit functions of the t(12;21) TEL-AML1 translocation in mice. Proc Natl Acad Sci 2004;101:8443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fischer M, Schwieger M, Horn S, Niebuhr B, Ford A, Roscher S, et al. Defining the oncogenic function of the TEL/AML1 (ETV6/RUNX1) fusion protein in a mouse model. Oncogene 2005;24:7579–91. [DOI] [PubMed] [Google Scholar]
- 59. Schindler JW, Van Buren D, Foudi A, Krejci O, Qin J, Orkin SH, et al. TEL-AML1 corrupts hematopoietic stem cells to persist in the bone marrow and initiate leukemia. Cell Stem Cell 2009;5:43–53. [DOI] [PubMed] [Google Scholar]
- 60. Rodríguez-Hernández G, Casado-García A, Isidro-Hernández M, Picard D, Raboso-Gallego J, Alemán-Arteaga S, et al. The second oncogenic hit determines the cell fate of ETV6-RUNX1 positive leukemia. Front Cell Dev Biol 2021;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Böiers C, Richardson SE, Laycock E, Zriwil A, Turati VA, Brown J, et al. A human IPS model implicates embryonic B-myeloid fate restriction as developmental susceptibility to B acute lymphoblastic leukemia-associated ETV6-RUNX1. Dev Cell 2018;44:362–77.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang LQ, Downie PA, Goodell WR, McCabe NR, LeBeau MM, Morgan R, et al. Establishment of cell lines from B-cell precursor acute lymphoblastic leukemia. Leukemia 1993;7:1865–74. [PubMed] [Google Scholar]
- 63. Fears S, Chakrabarti SR, Nucifora G, Rowley JD. Differential expression of TCL1 during pre-B-cell acute lymphoblastic leukemia progression. Cancer Genet Cytogenet 2002;135:110–9. [DOI] [PubMed] [Google Scholar]
- 64. Naumovski L, Morgan R, Hecht F, Link MP, Glader BE, Smith SD. Philadelphia chromosome-positive acute lymphoblastic leukemia cell lines without classical breakpoint cluster region rearrangement. Cancer Res 1988;48:2876–9. [PubMed] [Google Scholar]
- 65. Riester M, Singh AP, Brannon AR, Yu K, Campbell CD, Chiang DY, et al. PureCN: copy number calling and SNV classification using targeted short read sequencing. Source Code Biol Med 2016;11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang C, Chen L, Savage SR, Eguez RV, Dou Y, Li Y, et al. Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer Cell 2021;39:361–79.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ryan RJH, Drier Y, Whitton H, Cotton MJ, Kaur J, Issner R, et al. Detection of enhancer-associated rearrangements reveals mechanisms of oncogene dysregulation in B-cell lymphoma. Cancer Discov 2015;5:1058–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ryan RJH, Petrovic J, Rausch DM, Zhou Y, Lareau CA, Kluk MJ, et al. A B cell regulome links notch to downstream oncogenic pathways in small B cell lymphomas. Cell Rep 2017;21:784–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Parolia A, Cieslik M, Chu SC, Xiao L, Ouchi T, Zhang Y, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 2019;571:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fan K, Moore JE, Zhang XO, Weng Z. Genetic and epigenetic features of promoters with ubiquitous chromatin accessibility support ubiquitous transcription of cell-essential genes. Nucleic Acids Res 2021;49:5705–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Granja JM, Klemm S, McGinnis LM, Kathiria AS, Mezger A, Corces MR, et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol 2019;37:1458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang K, Hocker JD, Miller M, Hou X, Chiou J, Poirion OB, et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 2021;184:5985–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Neveu B, Caron M, Lagacé K, Richer C, Sinnett D.. Genome wide mapping of ETV6 binding sites in pre-B leukemic cells. Scientific Reports 2018;8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kalna V, Yang Y, Peghaire CR, Frudd K, Hannah R, Shah AV, et al. The transcription factor ERG regulates super-enhancers associated with an endothelial-specific gene expression program. Circ Res 2019;124:1337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nagai N, Ohguchi H, Nakaki R, Matsumura Y, Kanki Y, Sakai J, et al. Downregulation of ERG and FLI1 expression in endothelial cells triggers endothelial-to-mesenchymal transition. PLOS Genetics 2018;14:e1007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xiao L, Parolia A, Qiao Y, Bawa P, Eyunni S, Mannan R, et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature 2022;601:439–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sotoca AM, Prange KHM, Reijnders B, Mandoli A, Nguyen LN, Stunnenberg HG, et al. The oncofusion protein FUS-ERG targets key hematopoietic regulators and modulates the all-trans retinoic acid signaling pathway in t(16;21) acute myeloid leukemia. Oncogene 2016;35:1965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thoms JAI, Truong P, Subramanian S, Knezevic K, Harvey G, Huang Y, et al. Disruption of a GATA2-TAL1-ERG regulatory circuit promotes erythroid transition in healthy and leukemic stem cells. Blood 2021;138:1441–55. [DOI] [PubMed] [Google Scholar]
- 79. Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in ph-like acute lymphoblastic leukemia. N Engl J Med 2014;371:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kondili M, Fust A, Preussner J, Kuenne C, Braun T, Looso M. UROPA: a tool for universal robust peak annotation. Sci Rep 2017;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McKenna A, FlashFry SJ.: A fast and flexible tool for large-scale CRISPR target design. BMC Biol 2018;16:4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 2018;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 lists oligonucleotide sequences and cell line information
Supplementary Table 2 details known and de novo motif analysis for distal acetylation clusters
Supplementary Table 3 summarizes TF ChIP-Seq results and motif enrichment statistics
Supplementary table 4 lists ETV6-regulated gene-enhancer linkages
Supplementary Table 5 annotates genome-wide candidate ETV6-repressed enhancers
Supplementary Table 6 details GGAA repeat overlap analysis of new and previously published ETS factor datasets.
Contains Supplementary Figures 1-12 and additional information about Supplementary tables
Data Availability Statement
New sequencing data sets produced for this work are available at GEO under accession number GSE186942. Previously published data are available under accession numbers GSE69558 and GSE97541. BLUEPRINT epigenome consortium H3K27ac ChIP-seq data were accessed at http://dcc.blueprint-epigenome.eu/#/data sets. NCI TARGET transcriptomic data were accessed at https://portal.gdc.cancer.gov/projects.