Abstract
Transient ischemic attack (TIA) refers to a momentary neurologic deficit caused by focal cerebral, spinal or retinal ischemic insult. TIA is associated with a high risk of impending acute ischemic stroke (AIS), a neurologic dysfunction characterized by focal cerebral, spinal or retinal infarction. Understanding the differences in molecular pathways in AIS and TIA has merit for deciphering the underlying cause for neuronal deficits with long-term effects and high risks of morbidity and mortality. In this study, we performed comprehensive investigations into the circulating microRNA (miRNA) profiles of AIS (n = 191) and TIA (n = 61) patients. We performed RNA-Seq on serum samples collected within 24 hrs of clinical diagnosis and randomly divided the study populations into discovery and validation cohorts. We identified a panel of 11 differentially regulated miRNAs at FDR < 0.05. Hsa-miR-548c-5p, -20a-5p, -18a-5p, -484, -652-3p, -486-3p, -24-3p, -181a-5p and -222-3p were upregulated, while hsa-miR-500a-3p and -206 were downregulated in AIS patients compared to TIA patients. We also probed the previously validated gene targets of our identified miRNA panel to highlight the molecular pathways affected in AIS. Moreover, we developed a multivariate classifier with potential utilization as a discriminative biomarker for AIS and TIA patients. The underlying molecular pathways in AIS compared to TIA may be explored further in functional studies for therapeutic targeting in clinical translation.
Keywords: microRNA, miRNome, ischemic, stroke, acute ischemic stroke, transient ischemic attack
1. Introduction
Stroke is the second leading cause of global mortality and the third leading cause of death and disability combined, responsible for an estimated 6.5 million deaths in 2019 [1]. It is described as a neurological deficit attributed to acute focal injury to the central nervous system (CNS) due to impaired blood perfusion and is broadly classified as ischemic, hemorrhagic or subarachnoid [2]. Ischemic strokes refer to incident neurological dysfunction due to acute cerebral, spinal or retinal infarction by ischemic insult [2] and constitute the majority (62.4%) of all incident stroke cases [1]. Stroke diagnosis and management require clinical investigations and brain imaging to assess the extent of neuronal damage and to determine reperfusion therapies.
Transient ischemic attack (TIA) refers to focal neurological deficits that resolve within minutes to 24 h after onset, and imaging reveals no acute stroke in the majority of cases. Stroke risk is high (~7.5–17.4%) in the initial days following a TIA [3,4,5,6]. TIA occurs in ~20% of ischemic stroke patients in the short time preceding stroke [7]. The risk of myocardial infarction and nonvascular mortality is also high in both TIA and stroke cases [8]. Thus, TIA requires immediate diagnosis and management to mitigate stroke risk. However, a significant proportion of TIA cases remain underdiagnosed due to lack of reporting [9]. The fundamental difference between TIA and AIS is the lack of acute infarction in most patients with TIA. The lack of residual incident neuronal deficits and absence of sustained clinical presentations in TIA hinders the risk stratification for the development of subsequent AIS. Understanding the differences in molecular pathways in AIS and TIA has merit for deciphering the underlying cause of neuronal deficits with long-term risks of morbidity and mortality.
MicroRNAs (miRNAs) are non-coding RNA molecules, which can regulate the expression of specific gene targets by affecting target mRNA stability or repressing translational efficiency. These molecules are detectable in circulation and are recurrently explored as predictive, diagnostic or prognostic biomarkers for various physiological conditions [10]. Investigating the disturbances in miRNA profile (miRNome) can decipher the physiological and molecular disturbances in various diseases, including cardiovascular disease [11]. Several groups have investigated circulating miRNAs in ischemic strokes [12]. However, these studies have predominantly utilized targeted approaches using microarrays to identify distinct circulating miRNAs in AIS patients or excluded other types of strokes including TIAs. Next-generation sequencing (NGS) technologies have emerged as a robust technique in miRNA expression profiling and combination with in silico tools can enhance their utilization as blood-based biomarkers due to high detection sensitivity and accuracy [13]. However, the few studies that have utilized next-generation sequencing technologies for miRNA profiling of stroke patients were limited by the modest patient sample size [14,15,16,17]. Thus, comprehensive miRNA profiling of AIS and TIA patients is warranted for the robust identification of differentially regulated miRNAs, which may be explored as discriminative biomarkers for AIS and TIA.
We have previously reported a panel of 10 differentially regulated circulating miRNAs to distinguish between AIS patients and healthy individuals [18], and a panel of 5 circulating miRNAs between AIS patients with type 2 diabetes mellitus (T2DM), a known risk factor for stroke that is also associated with worse disease outcomes, compared to non-diabetic AIS patients [19]. In the present study, we compared the circulating miRNA profiles of clinically diagnosed AIS and TIA patients using serum samples for identifying differentially regulated circulating miRNAs. We also probed the molecular pathways associated with the experimentally validated gene targets of these miRNAs and investigated the clinical pathology endpoints and diseases associated with disruptions in these pathways. Importantly, we developed a classifier based on our identified panel of dysregulated miRNAs to differentiate between AIS and TIA cases as a potential low-invasive miRNA-based biomarker. The underlying molecular pathways in AIS compared to TIA may be explored further in functional studies for therapeutic targeting and clinical translation.
2. Results
2.1. Identification of Differentially Regulated Circulating miRNAs in AIS Patients Compared to TIA Patients
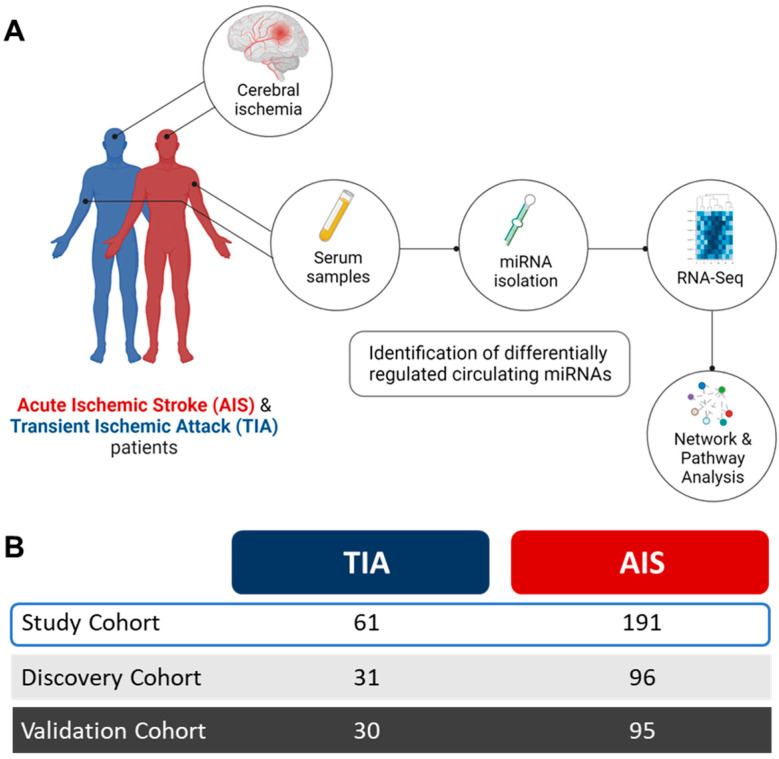
Serum samples from AIS and TIA patients were analyzed for the identification of distinct circulating miRNAs. The study design and analysis workflow are depicted in Figure 1 and the characteristic features of study population are listed in Table 1.
Figure 1.
Study design and analysis workflow. (A). Serum samples were collected from acute ischemic stroke (AIS) and transient ischemic attack (TIA) patients within 24 h of clinical diagnosis, to purify circulating microRNAs (miRNAs) for transcriptomic analysis by RNA-Seq. Stringent analyses criteria enabled identification of differentially regulated circulating miRNAs in AIS patients compared to TIA patients. Downstream analyses for the gene targets of identified miRNA panel were performed to highlight the molecular pathways and networks affected in AIS. (B). The study population comprised TIA (n = 61) and AIS (n = 191) patients who were randomly divided into discovery and validation cohorts for identification of replicated, differentially regulated circulating miRNAs in AIS patients compared to TIA patients.
Table 1.
Characteristic features of the study cohort.
| Characteristic | Discovery Cohort | Validation Cohort | ||
|---|---|---|---|---|
| TIA † | AIS ‡ | TIA | AIS | |
| Number | 31 | 96 | 30 | 95 |
| Age | 48.23 ± 9.68 | 50.38 ± 9.40 | 49.43 ± 10.84 | 50.01 ± 9.50 |
| Gender (Male/Female) | 28/3 | 86/10 | 26/4 | 90/5 |
| Diabetes mellitus (DM) (no/yes) | 19/12 | 49/47 | 17/13 | 50/45 |
| Hypertension (no/yes) | 19/12 | 26/70 * | 13/17 | 25/70 * |
| Smoking (no/yes) | 14/17 | 56/40 | 19/11 | 51/44 |
| DM medication (no/yes) | 23/8 | 73/23 | 22/8 | 66/29 |
| Statin medication (no/yes) | 22/9 | 83/13 * | 21/9 | 78/17 |
| Total cholesterol (mmol/L) | 4.16 ± 0.96 | 4.85 ± 1.22 * | 4.54 ± 0.73 | 5.16 ± 1.20 * |
| LDL-C (mmol/L) | 2.65 ± 1.04 | 3.15 ± 1.08 | 2.84 ± 0.82 | 3.34 ± 1.12 * |
| Triacyl glycerides (mmol/L) | 1.74 ± 0.63 | 1.54 ± 0.81 | 1.38 ± 0.60 | 1.90 ± 1.09 * |
† Transient ischemic attack. ‡ Acute ischemic stroke. * Statistically significant compared to TIA.
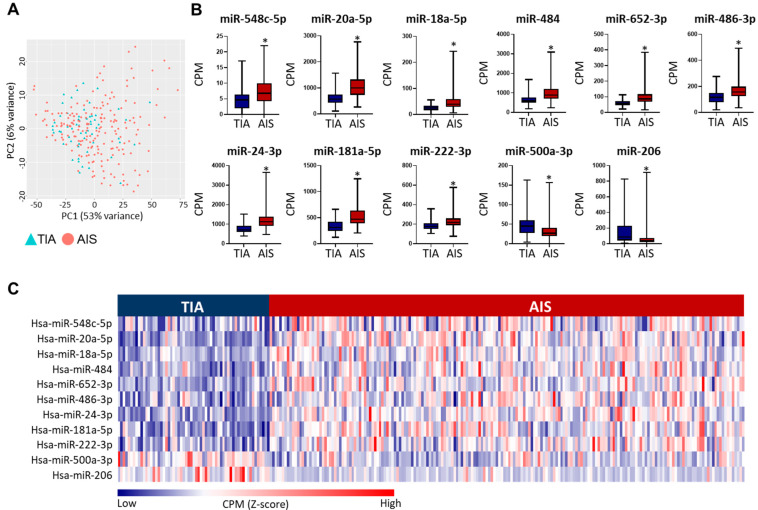
The principal component analysis (PCA) of the miRNA profiles of AIS and TIA patients did not show any distinct clustering of the two cohorts (Figure 2A). The differential expression analyses for miRNA transcripts of AIS and TIA patients were compared in the discovery and validation datasets separately to identify the statistically significant (FDR < 0.05), differentially regulated circulating miRNAs that were replicated in both analyses. In the discovery dataset, 24 miRNAs showed varying degrees of fold change (FC) and fulfilled the analyses criteria (FDR < 0.05, Supplementary Table S1). Out of these, 11 miRNAs were replicated in the validation cohort by meeting the analysis cutoff (Table 2). Moreover, the datasets showed the same direction of effect in the discovery and validation sets for all 11 miRNAs (Table 2). Out of the 11 miRNAs, 9 miRNAs were upregulated and 2 were downregulated in AIS compared to TIA patients (Figure 2B,C). Overall, this panel of 11 miRNAs is presented as differentially regulated miRNAs in AIS versus TIA patients.
Figure 2.
Circulating miRNA profiles of acute ischemic stroke (AIS) and transient ischemic attack (TIA) patients. (A). PCA plot depicts the variability in miRNA expression between TIA and AIS patients. (B). Box and whiskers plots show the differences in counts per million (CPM) in AIS and TIA patients of the 11 differentially regulated, validated miRNAs. Mean with minimum and maximum values, and upper and lower quartiles are depicted for each data set with significant comparisons annotated by an asterisk (*) on top (p < 0.0001). (C). Heatmap shows the dysregulation in identified miRNA panel (Z-scores calculated CPM) between AIS and TIA patients in the overall study population.
Table 2.
Differentially regulated miRNAs in AIS versus TIA patients.
| Discovery | Validation | Combined | ||||
|---|---|---|---|---|---|---|
| miRNA | FC * | FDR ** | FC | FDR | FC | FDR |
| hsa-miR-548c-5p | 1.80 | 2.44 × 10−2 | 1.90 | 1.56 × 10−2 | 1.70 | 1.13 × 10−4 |
| hsa-miR-20a-5p | 1.69 | 1.52 × 10−7 | 1.73 | 1.19 × 10−4 | 1.69 | 1.72 × 10−14 |
| hsa-miR-18a-5p | 1.55 | 2.29 × 10−2 | 1.73 | 1.48 × 10−2 | 1.72 | 5.01 × 10−6 |
| hsa-miR-484 | 1.52 | 1.08 × 10−3 | 1.55 | 8.32 × 10−3 | 1.47 | 2.64 × 10−6 |
| hsa-miR-652-3p | 1.49 | 4.76 × 10−3 | 1.62 | 1.08 × 10−3 | 1.53 | 2.31 × 10−7 |
| hsa-miR-486-3p | 1.46 | 3.93 × 10−3 | 1.49 | 9.51 × 10−3 | 1.46 | 5.73 × 10−7 |
| hsa-miR-24-3p | 1.45 | 3.93 × 10−3 | 1.63 | 6.79 × 10−4 | 1.48 | 1.13 × 10−7 |
| hsa-miR-181a-5p | 1.45 | 4.25 × 10−3 | 1.61 | 1.19 × 10−4 | 1.44 | 2.44 × 10−7 |
| hsa-miR-222-3p | 1.19 | 4.46 × 10−2 | 1.34 | 3.85 × 10−3 | 1.24 | 1.39 × 10−5 |
| hsa-miR-500a-3p | −1.67 | 1.41 × 10−2 | −1.65 | 4.02 × 10−2 | −1.63 | 8.05 × 10−5 |
| hsa-miR-206 | −3.18 | 3.92 × 10−4 | −2.39 | 2.57 × 10−2 | −3.20 | 2.95 × 10−9 |
* Fold change. ** False discovery rate.
In addition, we divided AIS patients into two groups based on the National Institutes of Health Stroke Scale (NIHSS) scores; minor to moderate stroke (NIHSS score: 0–5, n = 152) and severe stroke (NIHSS score: ≥6, n = 39), and compared the expression levels of our identified miRNA panel. We found that only hsa-miR-652-3p showed dysregulation in AIS patients with severe stroke, but this was with borderline significance (fold change = −1.23, FDR = 0.05). Moreover, we also found that 2 miRNAs out of our identified miRNA panel also showed significant dysregulation in AIS patients compared to healthy controls (Supplementary Table S2), based on our previously reported datasets comparing AIS patients with healthy controls [18].
2.2. Predictive Capacity of the Identified miRNA Panel in AIS Patients
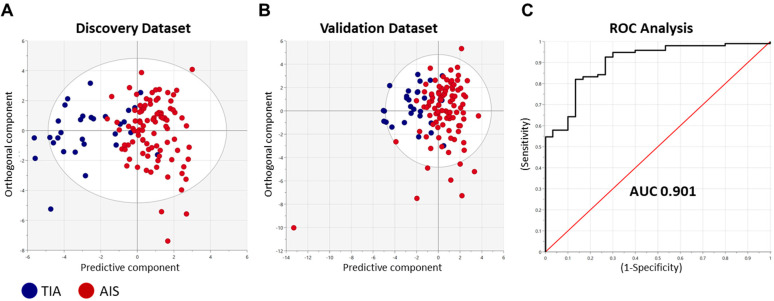
To determine the predictive performance of our identified miRNA panel to distinguish between AIS and TIA patients, we performed multivariate discriminant analyses (Figure 3). The orthogonal projections to latent structures discriminant analysis (OPLS-DA) method was adopted using the top FDR-significant differentially regulated miRNAs (n = 24, Supplementary Table S1) identified from the discovery cohort data (Figure 3A) and testing the classifier model on the validation cohort (Figure 3B) to determine its performance. The classifier showed high predictive performance, evidenced by an AUC value of 0.901 generated on the ROC curve analysis (Figure 3C).
Figure 3.
Discriminative performance of the identified miRNA panel to identify AIS patients. Orthogonal projections to latent structures-discriminant analysis (OPLS-DA) was performed using the top differentially regulated miRNAs (n = 24) in the discovery cohort data. The classifier was trained on data from all participants in (A). discovery cohort (n = 122) and tested on the (B). validation cohort (n = 119). Scatter plots show the predictive component to discriminate AIS cases from TIA cases (blue dots—x-axis) versus the orthogonal component representing a multivariate confounding effect that is independent of stroke (maroon dots—y-axis). (C). ROC curve analysis generated an area under the curve (AUC) of 0.901.
2.3. Identification of Molecular Pathways Modulated by Circulating miRNAs in AIS Patients
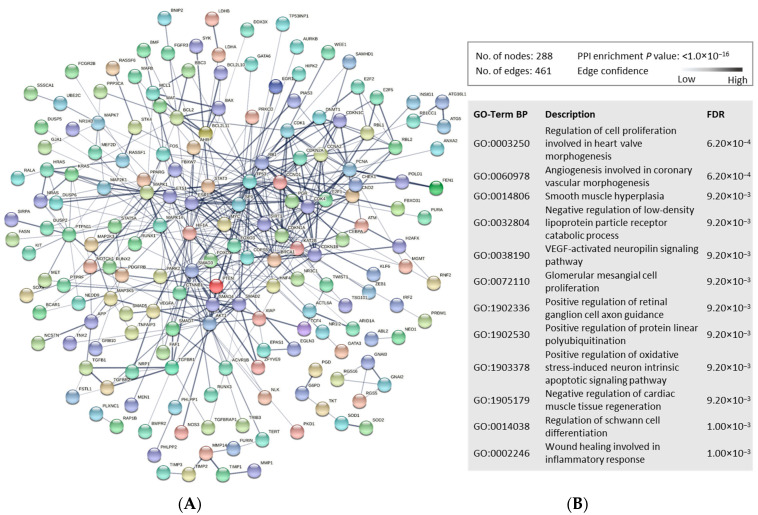
miRNAs can regulate the gene expression of multiple genes, thereby affecting the molecular function of their transcribed proteins. To decipher the potential molecular pathways affected in AIS compared to TIA, we first identified the known gene targets (experimentally validated with strong evidence) of our miRNA panel (Total n = 345 genes, n = 252 unique genes; Supplementary Table S3). We then generated protein–protein interaction (PPI) networks using STRING tool to explore and annotate pathways associated with proteins encoded by the gene targets of our identified miRNA panel (Figure 4). The PPI network showed high enrichment and associations of multiple proteins in mediating several pathways. Gene Ontology (Biological Processes; GO-Term BP) annotations predominantly corresponded to cardiac vascularity and neuronal cell functionality annotations, alongside protein processing and other cellular processes (Figure 4).
Figure 4.
Functional enrichment analysis of the proteins encoded by the gene targets of dysregulated miRNAs in AIS versus TIA patients. (A). The protein–protein interaction (PPI) network generated for the 345 gene targets (unique genes; n = 292, protein coding genes; n = 288) of the identified miRNA panel is shown. Network nodes represent proteins, while edges depict protein–protein associations. They key network statistics are also presented. (B). The top functional enrichment annotations from Gene Ontology (GO) Biological Process are listed.
2.4. Cellular Processes and Clinical Pathology Endpoints Associated with Gene Targets of the Dysregulated miRNAs in AIS Patients
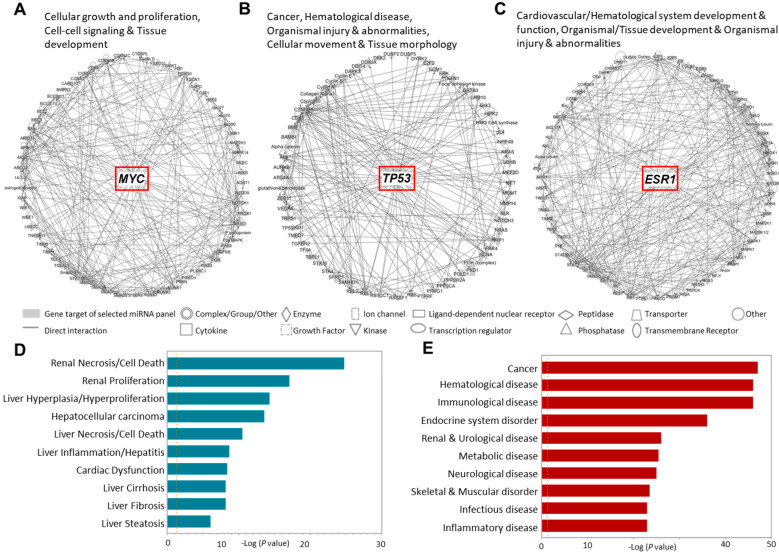
Identifying the molecular and cellular pathways that contribute to the underlying pathologies leading to the high mortality associated with AIS is crucial for therapeutic intervention. We investigated the molecular networks that involved the gene targets of our identified dysregulated miRNA panel in AIS patients (Figure 5A–C). We found that our genes of interest are intensely involved in cellular growth and interactions in tissue development, mediated by MYC activation (Figure 5A). In addition, we found the involvement of these genes in the aberrant molecular networks mediated by p53 and ESR1 in cancer; hematological development and pathologies; organismal injuries that include edema, hemorrhage and lesions; and cellular chemotaxis associated with tissue morphology and development (Figure 5B,C).
Figure 5.
Disease annotation and Ingenuity pathway analysis for the gene targets of dysregulated miRNAs in AIS versus TIA patients. The gene targets of the 11 dysregulated miRNAs in AIS compared to TIA patients were analyzed for disease/function annotation and network analysis. (A–C). Ingenuity pathway network analysis of the gene targets of dysregulated miRNAs in AIS patients are shown. (D–E). Bar plots show the clinical pathology endpoints and diseases annotations associated with these genes.
Identifying the clinical pathology endpoints associated with deviations in these molecular networks showed associations with hepatorenal and cardiac pathologies (Figure 5D). In addition, a broad spectrum of diseases covering cancer, hematological, neurological, immunological, endocrinological and metabolic disorders were associated with deviancies in these networks (Figure 5E).
3. Discussion
We investigated the differences in circulating miRNA regulatory network of AIS and TIA patients to highlight the potential molecular pathways affected in AIS patients. We identified a panel of 11 differentially regulated miRNAs in AIS compared to TIA patients using stringent criteria. Many of the identified miRNAs are broadly involved in cerebrovascular integrity or function via neuroprotection or in promoting neuroinflammation. Previous studies have reported dysregulation of many of these miRNAs in various cerebrovascular disorders and suggested them as disease biomarkers [20,21,22] or associated variants of their gene targets with stroke [23] or cardiac diseases [24].
The dysregulation of hsa-miR-548c-5p has been previously associated with intracranial aneurysm [25] and hypertension [26]. Moreover, its downregulation in plasma was associated with venous thrombosis [27]. These findings indicate the roles of miR-548c-5p in the modulation of circulation and vascularity, but it has not been directly linked with stroke. Moreover, numerous putative gene targets of miR-548c-5p have been predicted but none have been experimentally validated till present. Our novel findings of upregulation of miR-548c-5p in AIS patients as one of the most significantly dysregulated miRNAs highlights its relevance to the pathogenesis of stroke, which warrants further investigations.
In agreement with our findings, the upregulation of circulating miR-20a-5p, miR-18a-5p and miR-181a-5p has been previously reported in AIS patients compared to healthy controls [28]. MiR-20a-5p has been presented as a discriminatory biomarker between embolic and thrombotic strokes [29]. Notably, one of the common validated gene targets of miR-20a-5p and miR-206, VEGFA has been previously associated with stroke severity. Circulating VEGF levels were higher in ischemic stroke patients and showed prognostic significance [30]. Moreover, serum VEGF levels were associated with severe disability in AIS patients [31]. In addition, genome-wide association studies (GWAS) have associated variants in VEGFA with coronary heart disease [32] and myocardial infarction [24], and variants in other gene targets of miR-20a-5p, CDKN1A (also a common gene target of miR-181a-5p) [23] and PRKG1 [33] with stroke.
Hsa-miR-18a-5p promotes the differentiation of vascular smooth muscle cells [34] and has been reported as a biomarker for venous malformation [35]. Serum hsa-miR-18a-5p levels have also been associated with cardiovascular anomalies and proposed as a circulating biomarker for atherosclerosis [22]. The protein encoded by one of its gene targets, CTGF (also known as CCN2) is over-expressed in atherosclerotic plaques [36]. CTGF levels were higher in stroke than TIA and are associated with plaque stabilization following stroke [37]. Moreover, GWAS have associated CTGF with coronary artery calcification in type 2 diabetes patients [38]. Upregulation of miR-18a-5p in AIS patients indicates its potential roles in thrombotic or embolic ischemic strokes but needs further investigation.
The upregulation of miR-484 was reported in patients with hyperacute cerebral infarction [39] and its dysregulation has been reported in patients with ruptured intracranial aneurysms compared to those with intact intracranial aneurysm, along with miR-486-3p, miR-181a-5p, miR-222-3p and miR-500a-3p [25]. MiR-652-3p was significantly upregulated in freshly resected brain tissues of stroke patients with severe infarction and life-threatening cerebral swelling, compared to control temporal lobe samples [40]. Notably, the shared gene target of miR-484 and miR-652-3p, ZEB1 was previously explored in relation to ischemic neuronal injury. ZEB1 was induced in response to cerebral ischemia and is involved in neuronal cell survival [41], while its upregulation in microglia associated with alleviating post-AIS cerebral injury by reducing neuroinflammation [42].
The dysregulation of miR-486-3p has been associated with aneurysmal subarachnoid hemorrhage [43], while miR-24-3p has been associated with delayed cerebral vasospasm, which occurs in around one-third of aneurysmal subarachnoid hemorrhage patients [44]. Among the gene targets of miR-486-3p, FASN has been reported as a critical mediator in de novo lipogenesis in astrocytes following cerebral ischemic injury [45], while SYK is involved in cerebral inflammatory activation after ischemic stroke [46]. The upregulation of miR-24-3p in AIS patients concurs with previous reports of its upregulation as exosomal miRNA in AIS patients [47]. Additionally, the neuroprotective role of its gene target ACVR1B in promoting remyelination has been reported previously in a mouse model of ischemic stroke [48]. Importantly, GWAS have associated variants of several of the gene targets of miR-24-3p;FAF1 [49], GATA3 [50], MMP14 [51], NOS3 and PTPRF [23] with stroke.
Hsa-miR-181a-5p is associated with inflammation, upregulated in response to TGFβ signaling [52], and has been identified as a critical regulatory miRNA in ischemic strokes [53]. Serum levels of the protein encoded by its gene targets ATG5 is presented as a biomarker for AIS patients [54], while another gene target, ATM is involved in neuroprotection in the initiation of protective mechanisms in ischemic preconditioning but promotes neuronal cell death following the ischemic insult [55]. Additionally, upregulation of miR-222-3p was previously observed in acute myocardial infarction patients [56]. Moreover, its gene target AR1D1A associated with cerebral lesions of white matter hyperintensities in stroke patients in GWAS [57].
Several gene targets have been predicted for miR-500a-3p, but none experimentally validated. Our findings of its significant downregulation in AIS patients compared to TIA patients suggest its regulation by post-stroke neuroprotective mechanisms but further investigations are warranted to explore its role in the pathophysiology of stroke. In contrast, the levels of miR-206 were higher in cardioembolic stroke patients with hemorrhagic transformation [58]. Moreover, in line with our findings, the downregulation of miR-206 in circulation of AIS patients has been reported previously and was also associated with diagnostic significance [20].
The proteins encoded by the gene targets of our identified miRNA panel showed strong interactions indicating the pathophysiological processes, which may be disrupted in AIS patients compared to TIA patients. These networks showed strong annotations with processes involved in cardiac vascularity and muscle regeneration, neuronal differentiation and apoptosis, angiogenic pathways with possible involvement in wound healing, and catabolic protein biochemical processing. Collectively, these processes highlight the underlying protein pathways responsible for long-term effects and clinical complications of AIS compared to TIA patients. In addition, deciphering the aberrant molecular pathways in AIS patients showed homeostasis-associated processes in tissue maintenance and functionality. In relation to AIS, we found that disruptions in these pathways are associated with hepatorenal and cardiac anomalies, and vast diseases covering metabolic, inflammatory, endocrine and immunological disorders, which may be involved in morbidity and mortality associated with AIS.
Uncovering the pathophysiological and molecular imbalances in stroke has potentials of improving the understanding of the etiopathogenesis of the cerebrovascular neurological events with high risk of permanent neurologic disability and mortality. Tissue infarction due to cerebral ischemia triggers necrotic and apoptotic neuronal cell death. The ischemic core in the brain experiences irreversible insult and necrotic cell death, while the cells in ischemic penumbra undergo apoptosis via mitochondria-mediated intrinsic mechanisms, and cell death receptor-mediated extrinsic mechanisms of apoptosis [59,60]. Additional mechanisms of post-ischemic neuronal damage include necroptosis, autophagy and excitotoxicity [61]. We report that our identified dysregulated miRNAs target several critical mediators of cell death, most notably P53, Bcl-2, tumor necrosis factor family proteins, protein kinases such as Akt, and autophagy-related (ATG) proteins, among others. These findings indicate the involvement of the processes involved in neuronal cell death and may be investigated further in relation to AIS.
Our panel of dysregulated miRNAs showed high discriminative performance in assessing AIS and TIA patients, which shows promising clinical utilization. Importantly, despite identification of several candidate biomarkers, TIA remains a clinical diagnosis and development of robust diagnostic protocols is necessitated [62]. Moreover, the current individual blood-based biomarkers for AIS diagnosis also lack robust performance for use in clinical settings [63]. Notably, while these biomarkers predominantly discriminate between TIA or AIS and healthy controls, our identified miRNA panel distinguishes between AIS and TIA patients and benefits more from superior discriminatory performance than candidate biomarkers for stroke, such as C-reactive protein levels (AUC 0.73 [64,65]). However, further experimental validations for confirming expression are required. Moreover, we highlighted the crucial pathways potentially disrupted or exploited in AIS, showed potential utilization of differentially regulated circulating miRNAs as discriminatory biomarkers for AIS and TIA patients and identified several gene targets and pathways, which may be investigated further. However, the lack of confirmation of miRNA/gene/protein expression, functional validation, modest sample size of TIA patients and absence of validation in an external dataset necessitate additional investigations. Overall, the remarkably distinct circulating miRNA profiles of AIS patients compared to TIA patients strengthens the rationale for further research into their clinical application as low-invasive disease biomarkers.
4. Materials and Methods
4.1. Study Population
The study population (n = 252) comprised clinically diagnosed acute ischemic stroke (AIS; n = 191) and transient ischemic attack (TIA; n = 61) patients admitted to Hamad General Hospital (Doha, Qatar). All patients provided written informed consent prior to sample donation. This study was approved by the institutional review boards of Qatar Biomedical Research Institute, Doha, Qatar (Protocol no. 2019-013) and Hamad Medical Corporation (Protocol no. 15304/15), Doha, Qatar. Patients’ medical records were also accessed to retrieve relevant information. Characteristic features of the study population are listed in Table 1. AIS patients were also divided into two groups based on the clinical severity of stroke as assessed on the NIHSS scale.
4.2. Study Design
Fresh serum samples (200 µL) were collected from AIS and TIA patients within 24 h of the cerebrovascular event and stored at −80 °C for subsequent miRNA profiling by RNA-Seq. The study population was divided randomly into discovery and validation cohorts with proportionate distribution of covariates (age, gender, prior administration of DM management and cholesterol lowering drugs).
The miRNA profiles of AIS patients (n = 96) were first compared with TIA patients (n = 31) in the discovery cohort. The differential expression analyses were performed while adjusting for age, gender, diabetes and statin treatment to identify significant (false discovery rate (FDR) < 0.05) miRNAs in the discovery cohort. Similar analysis was performed on the validation cohort (AIS; n = 95, TIA; n = 30) to identify the FDR-significant miRNAs, which were replicated from analyses of the discovery cohort. These miRNAs were presented as the panel of differentially regulated miRNAs in AIS versus TIA patients. A metanalysis for the overall datasets was also performed to confirm our findings. The study design and analysis workflow are depicted in Figure 1. Downstream analyses to explore the potential pathways affected by the panel of dysregulated miRNAs in AIS patients were also performed (explained in succeeding sections).
4.3. miRNA Isolation and Sequencing
The miRNA transcripts from serum samples were sequenced as described previously [18]. Briefly, miRNeasy Serum/Plasma Advanced Kit (Qiagen, Hilden, Germany) was used to purify total RNA from serum samples. RNA quantification was carried out using Qubit RNA Broad Range Assay Kit (Invitrogen, Carlsbad, CA, USA). The libraries were prepared using QIAseq miRNA next-generation sequencing (NGS) Library Kit (Qiagen) using QIAseq miRNA NGS 96 Index IL kit (Qiagen) for indexing. The quality-passed libraries, assessed using Qubit dsDNA HS assay kit (Invitrogen) and Agilent 2100 Bioanalyzer DNA1000 chip (Agilent Technologies, Santa Clara, CA, USA), were pooled using TruSeq PE Cluster Kit v3-cBot-HS kit (illumina, San Diego, CA, United States). HiSeq 3000/4000 SBS kit (illumina) was used to perform sequencing on illumina HiSeq 4000 system (10 million reads per sample).
4.4. Data Curation and Analyses
The sequencing data were generated as single reads (at 75 cycles) and alignment was carried out on CLC Genomics Workbench (v.21.0.5, Qiagen) utilizing the human miRbase v22 reference genome. miRNA transcript expression levels were computed as counts per million (CPM) from the total counts of mapped miRNA reads. Calibration for RNA spike-in (RNA transcript of known sequence and quantity) was also performed. The differential expression analyses were carried out on RStudio (version 4.1.1; RStudio, Boston, MA, USA) utilizing the DSEq2 method [66], while adjusting for covariates (age, gender, DM and statin treatment). Statistical analyses and representations were performed using GraphPad Prism 9.1.2 (GraphPad Software, Columbia, ML, USA). A p value of <0.05 was considered statistically significant.
4.5. Discriminant Analyses
Discriminant analysis and classification was performed to determine the predictive performance of our identified panel of miRNAs to discriminate between AIS and TIA patients. The orthogonal projections to latent structures discriminant analysis (OPLS-DA) classifier was first trained on the findings of the discovery datasets (statistically significant differentially regulated miRNAs in AIS versus TIA patients) using SIMCA multivariate data analysis software (version 16; Umetrics, Vasterbottens Lan, Sweden). The model was then tested on the validation dataset and the performance was assessed by determining the area under the curve (AUC) value of the receiver operating characteristic (ROC) curve.
4.6. Downstream Pathway and Network Analyses
The previously identified gene targets with strong evidence of experimental validation of the identified miRNA panel dysregulated in AIS patients compared to TIA patients were retrieved from the miRTargetLink 2.0 database [67]. Target gene enrichment and protein–protein interaction (PPI) network analysis was performed by STRING [68]. The functional pathway network analyses were performed on QIAGEN Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., https://digitalinsights.qiagen.com/IPA (accessed on 12 September 2022)) [69].
Acknowledgments
We would like to thank the Genomics Core Facility at Qatar Biomedical Research Institute, Hamad Bin Khalifa University, Qatar Foundation for performing RNA-Sequencing, and all participants for donation of their samples.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24010108/s1.
Author Contributions
S.M.T.: Formal analysis, Visualization, Writing—original draft. E.K.A.: Data curation, Formal analysis, Investigation. A.P. and N.A.: Resources, Investigation, Sample preparation. Y.A.-S.: Investigation. E.M.A., A.A., O.E.-A. and P.J.T.: Resources. S.V.P., G.J.P. and R.A.: Investigation, sample preparation. A.S.: Resources, Investigation, Writing—review and editing. N.M.A.: Formal analysis, Writing—review and editing. O.M.E.A.: Conceptualization, Funding acquisition, Project administration, Resources, Investigation, Supervision, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board from Qatar Biomedical Research Institute (2019-013) and Hamad Medical Corporation (15304/15), Doha, Qatar.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files) or are available from the corresponding author by reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Interdisciplinary Research Project grant (IDRP) from Qatar Biomedical Research Institute and Hamad Bin Khalifa University, Qatar Foundation. Patients were recruited through a grant awarded to NA from Hamad Medical Corporation, Qatar (15304/15). The APC was funded by College of Health and Life Sciences, Hamad Bin Khalifa University, Qatar Foundation.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Collaborators G.B.D.S. Global, regional, and national burden of stroke and its risk factors, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet Neurol. 2021;20:795–820. doi: 10.1016/S1474-4422(21)00252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacco R.L., Kasner S.E., Broderick J.P., Caplan L.R., Connors J.J., Culebras A., Elkind M.S., George M.G., Hamdan A.D., Higashida R.T., et al. An updated definition of stroke for the 21st century: A statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2013;44:2064–2089. doi: 10.1161/STR.0b013e318296aeca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shahjouei S., Sadighi A., Chaudhary D., Li J., Abedi V., Holland N., Phipps M., Zand R. A 5-Decade Analysis of Incidence Trends of Ischemic Stroke After Transient Ischemic Attack: A Systematic Review and Meta-analysis. JAMA Neurol. 2021;78:77–87. doi: 10.1001/jamaneurol.2020.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appelros P., Hals Berglund M., Strom J.O. Long-Term Risk of Stroke after Transient Ischemic Attack. Cerebrovasc. Dis. 2017;43:25–30. doi: 10.1159/000451061. [DOI] [PubMed] [Google Scholar]
- 5.Hill M.D., Yiannakoulias N., Jeerakathil T., Tu J.V., Svenson L.W., Schopflocher D.P. The high risk of stroke immediately after transient ischemic attack: A population-based study. Neurology. 2004;62:2015–2020. doi: 10.1212/01.WNL.0000129482.70315.2F. [DOI] [PubMed] [Google Scholar]
- 6.Mendelson S.J., Prabhakaran S. Diagnosis and Management of Transient Ischemic Attack and Acute Ischemic Stroke: A Review. JAMA. 2021;325:1088–1098. doi: 10.1001/jama.2020.26867. [DOI] [PubMed] [Google Scholar]
- 7.Rothwell P.M., Warlow C.P. Timing of TIAs preceding stroke: Time window for prevention is very short. Neurology. 2005;64:817–820. doi: 10.1212/01.WNL.0000152985.32732.EE. [DOI] [PubMed] [Google Scholar]
- 8.Touze E., Varenne O., Chatellier G., Peyrard S., Rothwell P.M., Mas J.L. Risk of myocardial infarction and vascular death after transient ischemic attack and ischemic stroke: A systematic review and meta-analysis. Stroke. 2005;36:2748–2755. doi: 10.1161/01.STR.0000190118.02275.33. [DOI] [PubMed] [Google Scholar]
- 9.Johnston S.C., Fayad P.B., Gorelick P.B., Hanley D.F., Shwayder P., van Husen D., Weiskopf T. Prevalence and knowledge of transient ischemic attack among US adults. Neurology. 2003;60:1429–1434. doi: 10.1212/01.WNL.0000063309.41867.0F. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y., Li Q., Zhang R., Dai X., Chen W., Xing D. Circulating microRNAs: Biomarkers of disease. Clin. Chim. Acta. 2021;516:46–54. doi: 10.1016/j.cca.2021.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S.K., Bang C., Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ. Cardiovasc. Genet. 2010;3:484–488. doi: 10.1161/CIRCGENETICS.110.958363. [DOI] [PubMed] [Google Scholar]
- 12.Eyileten C., Wicik Z., De Rosa S., Mirowska-Guzel D., Soplinska A., Indolfi C., Jastrzebska-Kurkowska I., Czlonkowska A., Postula M. MicroRNAs as Diagnostic and Prognostic Biomarkers in Ischemic Stroke-A Comprehensive Review and Bioinformatic Analysis. Cells. 2018;7:249. doi: 10.3390/cells7120249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coenen-Stass A.M.L., Magen I., Brooks T., Ben-Dov I.Z., Greensmith L., Hornstein E., Fratta P. Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol. 2018;15:1133–1145. doi: 10.1080/15476286.2018.1514236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X.W., Shi Y.H., Liu Y.S., Li G.F., Zhao R., Hu Y., Lin C.C., Zhuang M.T., Su J.J., Liu J.R. Increased plasma levels of miR-124-3p, miR-125b-5p and miR-192-5p are associated with outcomes in acute ischaemic stroke patients receiving thrombolysis. Atherosclerosis. 2019;289:36–43. doi: 10.1016/j.atherosclerosis.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Mick E., Shah R., Tanriverdi K., Murthy V., Gerstein M., Rozowsky J., Kitchen R., Larson M.G., Levy D., Freedman J.E. Stroke and Circulating Extracellular RNAs. Stroke. 2017;48:828–834. doi: 10.1161/STROKEAHA.116.015140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen T.T.M., van der Bent M.L., Wermer M.J.H., van den Wijngaard I.R., van Zwet E.W., de Groot B., Quax P.H.A., Kruyt N.D., Nossent A.Y. Circulating tRNA Fragments as a Novel Biomarker Class to Distinguish Acute Stroke Subtypes. Int. J. Mol. Sci. 2020;22:135. doi: 10.3390/ijms22010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X., Zhuang C., Chen L. Exosomal Long Non-Coding RNA Expression from Serum of Patients with Acute Minor Stroke. Neuropsychiatr. Dis. Treat. 2020;16:153–160. doi: 10.2147/NDT.S230332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aldous E.K., Toor S.M., Parray A., Al-Sarraj Y., Diboun I., Abdelalim E.M., Arredouani A., El-Agnaf O., Thornalley P.J., Akhtar N., et al. Identification of Novel Circulating miRNAs in Patients with Acute Ischemic Stroke. Int. J. Mol. Sci. 2022;23:3387. doi: 10.3390/ijms23063387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toor S.M., Aldous E.K., Parray A., Akhtar N., Al-Sarraj Y., Abdelalim E.M., Arredouani A., El-Agnaf O., Thornalley P.J., Pananchikkal S.V., et al. Identification of Distinct Circulating miRNAs in Acute Ischemic Stroke Patients with Type 2 Diabetes Mellitus. Front. Cardiovasc. Med. 2022;9:1024790. doi: 10.3389/fcvm.2022.1024790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X., Liu X., Liu Y., Chang W., Song Y., Zhu S. Uncovering the Potential Differentially Expressed miRNAs and mRNAs in Ischemic Stroke Based on Integrated Analysis in the Gene Expression Omnibus Database. Eur. Neurol. 2020;83:404–414. doi: 10.1159/000507364. [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Zhang H., Li X., Wang L., Yu H., Huang J., Liu Q., Wang C., Jiang A. Diagnostic and prognostic significance of aberrant miR-652-3p levels in patients with acute decompensated heart failure and acute kidney injury. J. Int. Med. Res. 2020;48:300060520967829. doi: 10.1177/0300060520967829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng P., Liu Y., Zhang M., Ji W. Diagnostic and Prognostic Significance of serum miR-18a-5p in Patients with Atherosclerosis. Clin. Appl. Thromb. Hemost. 2021;27:10760296211050642. doi: 10.1177/10760296211050642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik R., Rannikmae K., Traylor M., Georgakis M.K., Sargurupremraj M., Markus H.S., Hopewell J.C., Debette S., Sudlow C.L.M., Dichgans M., et al. Genome-wide meta-analysis identifies 3 novel loci associated with stroke. Ann. Neurol. 2018;84:934–939. doi: 10.1002/ana.25369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartiala J.A., Han Y., Jia Q., Hilser J.R., Huang P., Gukasyan J., Schwartzman W.S., Cai Z., Biswas S., Tregouet D.A., et al. Genome-wide analysis identifies novel susceptibility loci for myocardial infarction. Eur. Heart J. 2021;42:919–933. doi: 10.1093/eurheartj/ehaa1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao B., Zhou M.X., Zhou F.K., Luo X.M., Zhong S.X., Zhou Y.F., Qin Y.S., Li P.P., Qin C. Exosome-Derived MiRNAs as Biomarkers of the Development and Progression of Intracranial Aneurysms. J. Atheroscler. Thromb. 2020;27:545–610. doi: 10.5551/jat.51102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z., Chyr J., Jia Z., Wang L., Hu X., Wu X., Song C. Identification of Hub Genes Associated with Hypertension and Their Interaction with miRNA Based on Weighted Gene Coexpression Network Analysis (WGCNA) Analysis. Med. Sci. Monit. 2020;26:e923514. doi: 10.12659/MSM.923514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Rius A., Lopez S., Martinez-Perez A., Souto J.C., Soria J.M. Identification of a Plasma MicroRNA Profile Associated with Venous Thrombosis. Arterioscler. Thromb. Vasc. Biol. 2020;40:1392–1399. doi: 10.1161/ATVBAHA.120.314092. [DOI] [PubMed] [Google Scholar]
- 28.Tiedt S., Prestel M., Malik R., Schieferdecker N., Duering M., Kautzky V., Stoycheva I., Bock J., Northoff B.H., Klein M., et al. RNA-Seq Identifies Circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as Potential Biomarkers for Acute Ischemic Stroke. Circ. Res. 2017;121:970–980. doi: 10.1161/CIRCRESAHA.117.311572. [DOI] [PubMed] [Google Scholar]
- 29.Chen L.T., Jiang C.Y. MicroRNA Expression Profiles Identify Biomarker for Differentiating the Embolic Stroke from Thrombotic Stroke. BioMed Res. Int. 2018;2018:4514178. doi: 10.1155/2018/4514178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Escudero C., Acurio J., Lopez E., Rodriguez A., Benavente A., Lara E., Korzeniewski S.J. Vascular endothelial growth factor and poor prognosis after ischaemic stroke. Eur. J. Neurol. 2021;28:1759–1764. doi: 10.1111/ene.14641. [DOI] [PubMed] [Google Scholar]
- 31.Bhasin A., Srivastava M.V.P., Vivekanandhan S., Moganty R., Talwar T., Sharma S., Kuthiala N., Kumaran S., Bhatia R. Vascular Endothelial Growth Factor as Predictive Biomarker for Stroke Severity and Outcome; An Evaluation of a New Clinical Module in Acute Ischemic Stroke. Neurol. India. 2019;67:1280–1285. doi: 10.4103/0028-3886.271241. [DOI] [PubMed] [Google Scholar]
- 32.Davies R.W., Wells G.A., Stewart A.F., Erdmann J., Shah S.H., Ferguson J.F., Hall A.S., Anand S.S., Burnett M.S., Epstein S.E., et al. A genome-wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circ. Cardiovasc. Genet. 2012;5:217–225. doi: 10.1161/CIRCGENETICS.111.961243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keene K.L., Hyacinth H.I., Bis J.C., Kittner S.J., Mitchell B.D., Cheng Y.C., Pare G., Chong M., O’Donnell M., Meschia J.F., et al. Genome-Wide Association Study Meta-Analysis of Stroke in 22 000 Individuals of African Descent Identifies Novel Associations With Stroke. Stroke. 2020;51:2454–2463. doi: 10.1161/STROKEAHA.120.029123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kee H.J., Kim G.R., Cho S.N., Kwon J.S., Ahn Y., Kook H., Jeong M.H. miR-18a-5p MicroRNA Increases Vascular Smooth Muscle Cell Differentiation by Downregulating Syndecan4. Korean Circ. J. 2014;44:255–263. doi: 10.4070/kcj.2014.44.4.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Wang D., Wang Z., Li X., Xia W., Han Y., Su L., Fan X. MiR-18a-5p acts as a novel serum biomarker for venous malformation and promotes angiogenesis by regulating the thrombospondin-1/P53 signaling axis. Am. J. Transl. Res. 2021;13:11271–11286. [PMC free article] [PubMed] [Google Scholar]
- 36.Cicha I., Yilmaz A., Klein M., Raithel D., Brigstock D.R., Daniel W.G., Goppelt-Struebe M., Garlichs C.D. Connective tissue growth factor is overexpressed in complicated atherosclerotic plaques and induces mononuclear cell chemotaxis in vitro. Arterioscler. Thromb. Vasc. Biol. 2005;25:1008–1013. doi: 10.1161/01.ATV.0000162173.27682.7b. [DOI] [PubMed] [Google Scholar]
- 37.Leeuwis J.W., Nguyen T.Q., Theunissen M.G., Peeters W., Goldschmeding R., Pasterkamp G., Vink A. Connective tissue growth factor is associated with a stable atherosclerotic plaque phenotype and is involved in plaque stabilization after stroke. Stroke. 2010;41:2979–2981. doi: 10.1161/STROKEAHA.110.589036. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y., Dimitrov L., Chen S.H., Bielak L.F., Bis J.C., Feitosa M.F., Lu L., Kavousi M., Raffield L.M., Smith A.V., et al. Multiethnic Genome-Wide Association Study of Subclinical Atherosclerosis in Individuals with Type 2 Diabetes. Circ. Genom. Precis Med. 2021;14:e003258. doi: 10.1161/CIRCGEN.120.003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian C., Li Z., Yang Z., Huang Q., Liu J., Hong B. Plasma MicroRNA-16 Is a Biomarker for Diagnosis, Stratification, and Prognosis of Hyperacute Cerebral Infarction. PLoS ONE. 2016;11:e0166688. doi: 10.1371/journal.pone.0166688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlson A.P., McKay W., Edwards J.S., Swaminathan R., SantaCruz K.S., Mims R.L., Yonas H., Roitbak T. MicroRNA Analysis of Human Stroke Brain Tissue Resected during Decompressive Craniectomy/Stroke-Ectomy Surgery. Genes. 2021;12:1860. doi: 10.3390/genes12121860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bui T., Sequeira J., Wen T.C., Sola A., Higashi Y., Kondoh H., Genetta T. ZEB1 links p63 and p73 in a novel neuronal survival pathway rapidly induced in response to cortical ischemia. PLoS ONE. 2009;4:e4373. doi: 10.1371/journal.pone.0004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D., Lang W., Zhou C., Wu C., Zhang F., Liu Q., Yang S., Hao J. Upregulation of Microglial ZEB1 Ameliorates Brain Damage after Acute Ischemic Stroke. Cell Rep. 2018;22:3574–3586. doi: 10.1016/j.celrep.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Powers C.J., Dickerson R., Zhang S.W., Rink C., Roy S., Sen C.K. Human cerebrospinal fluid microRNA: Temporal changes following subarachnoid hemorrhage. Physiol. Genom. 2016;48:361–366. doi: 10.1152/physiolgenomics.00052.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W.X., Springer J.E., Xie K., Fardo D.W., Hatton K.W. A Highly Predictive MicroRNA Panel for Determining Delayed Cerebral Vasospasm Risk Following Aneurysmal Subarachnoid Hemorrhage. Front. Mol. Biosci. 2021;8:657258. doi: 10.3389/fmolb.2021.657258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei H., Zhen L., Wang S., Zhang Y., Wang K., Jia P., Zhang Y., Wu Z., Yang Q., Hou W., et al. De novo Lipogenesis in Astrocytes Promotes the Repair of Blood-Brain Barrier after Transient Cerebral Ischemia Through Interleukin-33. Neuroscience. 2022;481:85–98. doi: 10.1016/j.neuroscience.2021.11.026. [DOI] [PubMed] [Google Scholar]
- 46.Ye X.C., Hao Q., Ma W.J., Zhao Q.C., Wang W.W., Yin H.H., Zhang T., Wang M., Zan K., Yang X.X., et al. Dectin-1/Syk signaling triggers neuroinflammation after ischemic stroke in mice. J. Neuroinflamm. 2020;17:17. doi: 10.1186/s12974-019-1693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X., Xu C., Chao D., Chen Z., Li S., Shi M., Pei Y., Dai Y., Ji J., Ji Y., et al. Acute Cerebral Ischemia Increases a Set of Brain-Specific miRNAs in Serum Small Extracellular Vesicles. Front. Mol. Neurosci. 2022;15:874903. doi: 10.3389/fnmol.2022.874903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng J., Zhang T., Han S., Liu C., Liu M., Li S., Li J. Activin A improves the neurological outcome after ischemic stroke in mice by promoting oligodendroglial ACVR1B-mediated white matter remyelination. Exp. Neurol. 2021;337:113574. doi: 10.1016/j.expneurol.2020.113574. [DOI] [PubMed] [Google Scholar]
- 49.Cheng Y.C., Stanne T.M., Giese A.K., Ho W.K., Traylor M., Amouyel P., Holliday E.G., Malik R., Xu H., Kittner S.J., et al. Genome-Wide Association Analysis of Young-Onset Stroke Identifies a Locus on Chromosome 10q25 Near HABP2. Stroke. 2016;47:307–316. doi: 10.1161/STROKEAHA.115.011328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carty C.L., Keene K.L., Cheng Y.C., Meschia J.F., Chen W.M., Nalls M., Bis J.C., Kittner S.J., Rich S.S., Tajuddin S., et al. Meta-Analysis of Genome-Wide Association Studies Identifies Genetic Risk Factors for Stroke in African Americans. Stroke. 2015;46:2063–2068. doi: 10.1161/STROKEAHA.115.009044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma S.S., Bergmeijer T.O., Gong L., Reny J.L., Lewis J.P., Mitchell B.D., Alexopoulos D., Aradi D., Altman R.B., Bliden K., et al. Genomewide Association Study of Platelet Reactivity and Cardiovascular Response in Patients Treated With Clopidogrel: A Study by the International Clopidogrel Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 2020;108:1067–1077. doi: 10.1002/cpt.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vishnubalaji R., Alajez N.M. Epigenetic regulation of triple negative breast cancer (TNBC) by TGF-beta signaling. Sci. Rep. 2021;11:15410. doi: 10.1038/s41598-021-94514-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X., Yan S., Wang P., Wang G. Identification of Hub Genes in the Pathogenesis of Ischemic Stroke Based on Bioinformatics Analysis. J. Korean Neurosurg. Soc. 2022;65:697–709. doi: 10.3340/jkns.2021.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ajoolabady A., Shademan B., Avci C.B., Nikanfar M., Nourazarian A., Laghousi D. Diagnostic Potential of Autophagy-5 Protein, Apolipoprotein B-48, and Oxidative Stress Markers in Serum of Patients with Early-Stage Ischemic Stroke. World Neurosurg. 2022;167:e656–e663. doi: 10.1016/j.wneu.2022.08.063. [DOI] [PubMed] [Google Scholar]
- 55.Xie G.H., Dai H.J., Liu F., Zhang Y.P., Zhu L., Nie J.J., Wu J.H. A Dual Role of ATM in Ischemic Preconditioning and Ischemic Injury. Cell Mol. Neurobiol. 2020;40:785–799. doi: 10.1007/s10571-019-00773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Degano I.R., Camps-Vilaro A., Subirana I., Garcia-Mateo N., Cidad P., Munoz-Aguayo D., Puigdecanet E., Nonell L., Vila J., Crepaldi F.M., et al. Association of Circulating microRNAs with Coronary Artery Disease and Usefulness for Reclassification of Healthy Individuals: The REGICOR Study. J. Clin. Med. 2020;9:1402. doi: 10.3390/jcm9051402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Traylor M., Zhang C.R., Adib-Samii P., Devan W.J., Parsons O.E., Lanfranconi S., Gregory S., Cloonan L., Falcone G.J., Radmanesh F., et al. Genome-wide meta-analysis of cerebral white matter hyperintensities in patients with stroke. Neurology. 2016;86:146–153. doi: 10.1212/WNL.0000000000002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng L., Xiong Y., Liu J., Yang X., Wang L., Zhang S., Liu M., Wang D. MMP-9-Related microRNAs as Prognostic Markers for Hemorrhagic Transformation in Cardioembolic Stroke Patients. Front. Neurol. 2019;10:945. doi: 10.3389/fneur.2019.00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radak D., Katsiki N., Resanovic I., Jovanovic A., Sudar-Milovanovic E., Zafirovic S., Mousad S.A., Isenovic E.R. Apoptosis and Acute Brain Ischemia in Ischemic Stroke. Curr. Vasc. Pharmacol. 2017;15:115–122. doi: 10.2174/1570161115666161104095522. [DOI] [PubMed] [Google Scholar]
- 60.Broughton B.R., Reutens D.C., Sobey C.G. Apoptotic mechanisms after cerebral ischemia. Stroke. 2009;40:e331–e339. doi: 10.1161/STROKEAHA.108.531632. [DOI] [PubMed] [Google Scholar]
- 61.Sekerdag E., Solaroglu I., Gursoy-Ozdemir Y. Cell Death Mechanisms in Stroke and Novel Molecular and Cellular Treatment Options. Curr. Neuropharmacol. 2018;16:1396–1415. doi: 10.2174/1570159X16666180302115544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nouri-Vaskeh M., Khalili N., Sadighi A., Yazdani Y., Zand R. Biomarkers for Transient Ischemic Attack: A Brief Perspective of Current Reports and Future Horizons. J. Clin. Med. 2022;11:1046. doi: 10.3390/jcm11041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dagonnier M., Donnan G.A., Davis S.M., Dewey H.M., Howells D.W. Acute Stroke Biomarkers: Are We There Yet? Front. Neurol. 2021;12:619721. doi: 10.3389/fneur.2021.619721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glickman S.W., Phillips S., Anstrom K.J., Laskowitz D.T., Cairns C.B. Discriminative capacity of biomarkers for acute stroke in the emergency department. J. Emerg. Med. 2011;41:333–339. doi: 10.1016/j.jemermed.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 65.Worthmann H., Tryc A.B., Goldbecker A., Ma Y.T., Tountopoulou A., Hahn A., Dengler R., Lichtinghagen R., Weissenborn K. The temporal profile of inflammatory markers and mediators in blood after acute ischemic stroke differs depending on stroke outcome. Cerebrovasc. Dis. 2010;30:85–92. doi: 10.1159/000314624. [DOI] [PubMed] [Google Scholar]
- 66.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kern F., Aparicio-Puerta E., Li Y., Fehlmann T., Kehl T., Wagner V., Ray K., Ludwig N., Lenhof H.P., Meese E., et al. miRTargetLink 2.0-interactive miRNA target gene and target pathway networks. Nucleic Acids Res. 2021;49:W409–W416. doi: 10.1093/nar/gkab297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files) or are available from the corresponding author by reasonable request.