Abstract
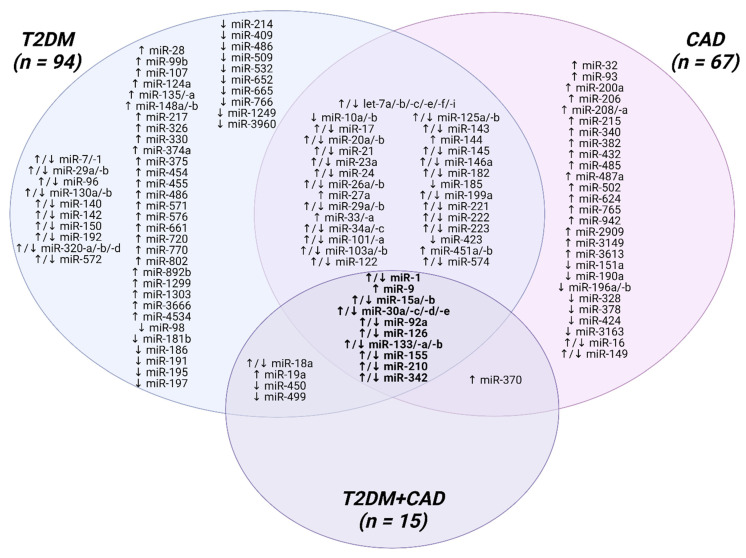
Type 2 diabetes mellitus (T2DM) is a chronic metabolic disease with still growing incidence among adults and young people worldwide. Patients with T2DM are more susceptible to developing coronary artery disease (CAD) than non-diabetic individuals. The currently used diagnostic methods do not ensure the detection of CAD at an early stage. Thus, extensive research on non-invasive, blood-based biomarkers is necessary to avoid life-threatening events. MicroRNAs (miRNAs) are small, endogenous, non-coding RNAs that are stable in human body fluids and easily detectable. A number of reports have highlighted that the aberrant expression of miRNAs may impair the diversity of signaling pathways underlying the pathophysiology of atherosclerosis, which is a key player linking T2DM with CAD. The preclinical evidence suggests the atheroprotective and atherogenic influence of miRNAs on every step of T2DM-induced atherogenesis, including endothelial dysfunction, endothelial to mesenchymal transition, macrophage activation, vascular smooth muscle cells proliferation/migration, platelet hyperactivity, and calcification. Among the 122 analyzed miRNAs, 14 top miRNAs appear to be the most consistently dysregulated in T2DM and CAD, whereas 10 miRNAs are altered in T2DM, CAD, and T2DM-CAD patients. This up-to-date overview aims to discuss the role of miRNAs in the development of diabetic CAD, emphasizing their potential clinical usefulness as novel, non-invasive biomarkers and therapeutic targets for T2DM individuals with a predisposition to undergo CAD.
Keywords: microRNA, type 2 diabetes mellitus, coronary artery disease, atherosclerosis, biomarker, chronic inflammation
1. Introduction
Type 2 diabetes mellitus (T2DM) is one of the most common metabolic disorders worldwide, and its prevalence has been growing among adolescents and young adults at an alarming rate over the past three to four decades [1,2,3]. According to the International Diabetes Federation, 537 million people suffered from diabetes mellitus in 2021, and this number is expected to rise up to 783 million by 2045 [4]. It is estimated that T2DM accounts for approximately 90–95% of all cases of diabetes mellitus, depending on different ethnicity [3,5]. However, it is worth underlining that at least one-third to one-half of people living with T2DM are undiagnosed and untreated [6,7].
T2DM is a chronic, low-grade inflammatory disease characterized by hyperglycemia that results from a progressive beta-cell impairment, insulin secretion deficiency, and concomitant insulin resistance [8]. Long-term exposure to uncontrolled hyperglycemia leads to the development of various micro- and macrovascular complications, including diabetic retinopathy, nephropathy, peripheral neuropathy, and diabetic foot syndrome, as well as cardiovascular and cerebrovascular diseases [9,10]. Initially, the clinical symptoms of the disease may be very subtle, which most likely results in a prolonged diagnostic delay of an average of 5–6 years after the onset of the first signs, therefore the presence of at least one vascular complication can be observed even in newly diagnosed T2DM cases [7,10]. A large cross-sectional study in Denmark revealed that 35% of previously undiagnosed T2DM individuals had complications at the time of diagnosis, of whom 17% had macrovascular ones, with the greater risk of ischemic heart disease occurring in 15% of patients, 12% had microvascular, whereas 6% of participants presented both micro- and macrovascular complications [10].
It should be emphasized that patients with T2DM are two to four times more susceptible to developing coronary artery disease (CAD) than non-diabetic individuals, but on the other hand, the prevalence of diabetes mellitus in CAD patients is up to 40%, and another 6% of them will develop ‘silent’ diabetes in the next few years [11,12,13,14,15]. Anyway, more than half of T2DM individuals die of cardiovascular causes [11,16,17]. Both T2DM and CAD are complex, multifactorial diseases with genetic, epigenetic, and environmental backgrounds [8,18]. Chronic hyperglycemia and insulin resistance stimulate oxidative stress and inflammation, and thus they initiate endothelial dysfunction and contribute to accelerated atherosclerosis, which is the main driver of CAD development [19]. Moreover, persistently high blood glucose levels lead to abnormal activation of platelet, vascular calcification, and deterioration of atherogenic dyslipidemia, that further promote the atherosclerotic process and increase the risk of thrombus formation [19,20]. Therefore, the identification of patients at an early stage of the disease is critical for prevention and adequate treatment.
In recent years, microRNAs (miRNAs, miRs), belonging to an ever-growing family of highly conserved, naturally occurring non-coding RNA (ncRNA) molecules, have emerged as key players in different physiological and pathological processes, such as cell proliferation, differentiation, and apoptosis [21]. Dysregulation of miRNA expression signatures is closely associated with numerous diseases, including diabetes mellitus and cardiovascular disorders. Thus they are considered novel promising biomarkers [22,23,24,25].
The present review is aimed to discuss the role of miRNAs in the pathogenesis of diabetic CAD, moving from preclinical to clinical studies, and to highlight their potential usefulness as early, noninvasive biomarkers and therapeutic targets for CAD in patients with T2DM.
2. Biology of MicroRNAs
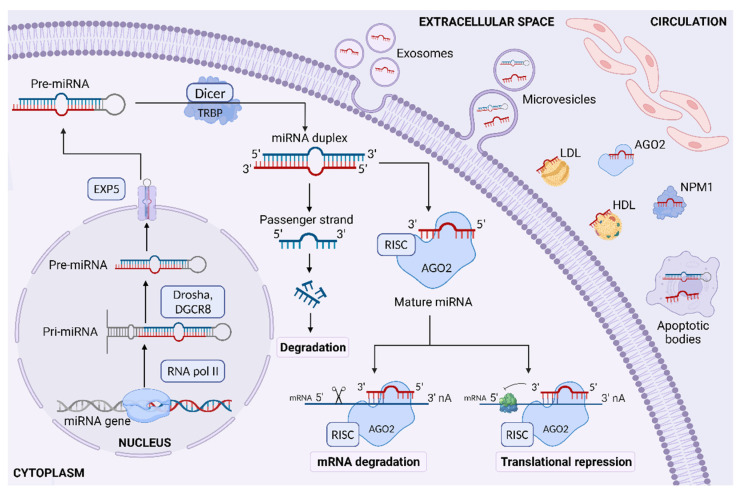
Regulatory ncRNAs could be classified into several types based on their average size involving small ncRNAs (sncRNAs) with less than 200 nucleotides (miRNA, small interfering RNA; siRNA, PIWI-interacting RNA; piRNA), long ncRNAs (lncRNAs) with more than 200 nucleotides, and circular RNAs (circRNAs) with a special closed-loop structure [26]. miRNAs are a dominating class of small endogenous, single-stranded ncRNAs, ranging from 18 to 25 nucleotides in length, that posttranscriptionally regulate gene expression through different pathways [27,28]. The biogenesis of miRNA involves multi-step processes occurring both in the nucleus and in the cytoplasm [28,29]. In the first step of this molecular pathway, miRNA-encoding sequences located in intergenic (40%) or intragenic (60%) regions of the genome are transcribed mostly by RNA polymerase II, which leads to the synthesis of primary miRNA (pri-miRNA) transcript with a 5′-cap and a 3′ poly-A tail in the nucleus [28,29,30]. Pri-miRNAs are long, double-stranded products consisting of approximately 1000 nucleotides having a hairpin-like stem-loop-like structure flanked by single-stranded RNA ends [29]. The nuclear microprocessor protein complex comprised of Drosha, an RNase type III enzyme, and its co-factor known as the double-stranded RNA-binding protein DiGeorge syndrome Critical Region 8 (DGCR8), cleaves pri-miRNA into 70–90 nucleotide precursor miRNA (pre-miRNA), which is then transported through the nuclear membrane to the cytoplasm by the Exportin 5 (EXP5, encoded by XPO5)—RanGTP system [28,29]. In the cytoplasm, pre-miRNAs are further processed by RNase III enzyme Dicer/trans-activation response RNA binding protein (TRBP, encoded by TARBP2) complex to generate shorter, about 22-nucleotide base pair miRNA duplex (miRNA-3p/miRNA-5p) [28,29]. In the last step of this process, miRNA duplex is loaded into the pre-RNA-induced silencing complex (pre-RISC) containing Argonaute proteins (AGO1-4, in humans), of which only AGO2 has the target cleavage activity [28,29,31]. Subsequently, one strand of miRNA duplex, called guide strand or miRNA, remains bound to AGO2 as mature miRNA, whereas another one, passenger strand or miRNA*, is degraded, and thus mature RISC is generated [28,29]. It is worth underlining that the formation of miRNA/pre-RISC complex is an active process that requires an ATP-dependent mechanism involving the heat shock cognate 70 (HSC70)—heat shock protein 90 (HSP90) chaperone complex to mediate conformational changes of AGO proteins and allow them to bind stiff double-stranded miRNA, while the release of the passenger strand is ATP-independent [28,29]. Mature miRNA targets specific messenger RNA (mRNA) by base pairing in the 3′ untranslated regions (3′ UTR) of these transcripts, and in this way, it induces posttranscriptional gene silencing through translational repression and/or mRNA degradation [28,32]. In humans, inhibition of protein synthesis is mediated through perfect or imperfect complementarity in the ‘seed’ region of miRNA composed of 2–8 nucleotides and target mRNA that results in mRNA cleavage or translational repression, respectively [28,32]. Additionally, the degradation of miRNA targets can be accelerated by mRNA deadenylation and decapping via glycine-tryptophan protein of 182 kDa (GW182) interaction with AGO2 [28,33]. Particular steps involved in the biogenesis and functions of miRNAs in humans are shown in Figure 1.
Figure 1.
Canonical pathway of miRNA biogenesis in humans. miRNA genes are transcribed by RNA pol II into pri-miRNA transcripts in the nucleus. Pri-miRNA is processed to pre-miRNA with the Drosha enzyme, and then it is transported to the cytoplasm by EXP5. Dicer endonuclease cleaves it to a short miRNA duplex. Mature miRNA, as a part of the RISC complex, binds with the 3′-untranslated region of target mRNA on different complementarity leading to mRNA degradation or translational repression. Both pre-miRNA and mature miRNA can be released passively or actively into the extracellular space in apoptotic bodies, exosomes, microvesicles, or in the complexes with AGO2, NPM1, HDL, and LDL. miRNA—microRNA; RNA pol II—RNA polymerase II; Pri-miRNA—primary miRNA; Pre-miRNA—precursor miRNA; DGCR8—DiGeorge syndrome Critical Region 8; EXP5—exportin 5; TRBP—trans-activation response RNA binding protein; AGO2—Argonaute 2; RISC—RNA-induced silencing complex; mRNA—messenger RNA; NPM1—nucleophosmin 1; HDL—high-density lipoprotein; LDL—low-density lipoprotein. Created with BioRender.com.
Apart from the above-mentioned canonical pathway of miRNA synthesis, there are some alternative ways of its maturation, including inter alia Drosha/DGCR8-independent and Dicer-independent pathways, which generate about 1% of conserved miRNAs [28,29]. The well-recognized ‘mirtron’ route is responsible for transforming an unconventional class of intragenic miRNAs, so-called mirtrons, escaping from Drosha/DGCR8-mediated process into pre-miRNAs by splicing enzymes (spliceosome) [28,29,34]. In the next step, mirtron-derived pre-miRNAs are exported to the cytoplasm by EXP5 and cleaved by Dicer, similar to the canonical pathway [28,29,34]. In turn, Dicer-mediated processing is only bypassed in the case of pre-miR-451 hairpins, resulting from Drosha cleavage, that are too short (30 nucleotides in length) to be processed by Dicer, and their maturation relies on the catalytic activity of AGO2 [28,29,34,35]. These short substrates are then sliced by AGO2 in the middle of their 3p strand and follow 3′-5′ trimming of the 5p strand to complete their maturation [28,29,34,35].
Although the majority of synthesized miRNAs are found intracellularly, a significant amount appears in the extracellular space [36]. miRNAs are released into the circulation not only passively from damaged cells due to apoptosis or necrosis, but they can also be actively secreted in several extracellular vesicles (EVs), including exosomes (40–100 nm), microvesicles (100–1000 nm) and apoptotic bodies (50–5000 nm) [36,37]. In addition, about 90% of circulating miRNAs form complexes with RNA-binding proteins, including AGO2, nucleophosmin 1 (NPM1) proteins, and lipoproteins, which protect them from RNase-dependent degradation [36].
The first miRNA, lineage-4 (lin-4), was discovered in 1993 by Victor Ambros and colleagues, Rosalind Lee and Rhonda Feinbaum in Caenorhabditis elegans, followed by the first human miRNA lethal-7 (let-7), which was identified 7 years later [38,39]. Since then, more than 2600 human miRNAs have been described according to the online database miRBase (www.mirbase.org, version 22), and the list is still expanding [40]. Although miRNAs have been considered unimportant additions to the transcriptome over decades, currently, a growing body of evidence indicates that they are critical modulators of gene expression [41]. What is interesting, one miRNA may silence from 100 to even 200 genes, and at the same time, multiple miRNAs are able to converge on a single protein-coding gene target [42,43]. According to the data collected in the latest version of miRTarBase (http://miRTarBase.cuhk.edu.cn/, accessed on 24 November 2022), approximately 4,475,477 validated human miRNA-target interactions are known [44]. It is worth emphasizing that miRNAs regulate at least one-third of all genes within the human genome, although the biological functions of only 3.6% of them were confirmed in the experiments [45,46]. miRNAs are involved in a wide range of processes and signaling pathways in different cells and tissues, as well as their disturbed expression profile is observed in many diseases [21,45]. Recent studies underscore their role in the pathogenesis of atherosclerosis under a condition of long-term hyperglycemia that, in consequence, leads to the development of CAD in T2DM [19,47].
3. Type 2 Diabetes Mellitus (T2DM), Atherosclerosis and Coronary Artery Disease (CAD)—The Vicious Circle Paradigm
Atherosclerosis is a chronic inflammatory disease of the inner wall of large- or medium-sized arteries caused by endothelial injury and subendothelial accumulation of lipid, extracellular matrix proteins, and calcium deposits, forming fibroinflammatory lipid plaques, particularly at sites of disturbed blood flow [19]. The formation of atheroma in one or more coronary arteries can narrow the lumen of the blood vessel, which leads to ischemia and metabolic changes in the alimented tissues [19,48]. Furthermore, the subsequent rupture of the coronary artery atherosclerotic plaque may result in coronary thrombus and its inevitable consequences, including unstable angina and acute myocardial infarction [48].
Clinical and experimental data have strongly indicated that the presence of T2DM is associated with a diffuse, more severe, and rapidly progressive form of atherosclerosis. Thereby atherogenesis is a key player linking T2DM with CAD [49,50]. Chronic hyperglycemia and insulin resistance mediate endothelial injury via several pathological pathways involved in the regulation of metabolism-immune system interplay [19,50]. Oxidative stress and chronic low-grade inflammation have been shown to be the main triggers of endothelial dysfunction in prolonged hyperglycemic conditions [47]. Diabetic atherogenesis is a complex, multi-step process characterized by the infiltration of inflammatory cells, monocyte/macrophage activation, vascular smooth muscle cell (VSMC) differentiation, and platelet hyperactivity, along with dysfunctional endothelial cells (ECs) and endothelial-to-mesenchymal transition (EndMT) [8,19,50,51]. Additionally, these events are enhanced by atherogenic dyslipidemia, which commonly accompanies T2DM [19].
The close relationship between T2DM and atherosclerosis is well established, although the epigenetic mechanisms underlying the development of diabetic atherosclerosis are still not completely understood. In the last years, numerous studies have revealed that specific miRNAs are able to modulate almost every step of the atherogenic process from its initiation through progression and, ultimately, thrombotic complications [47,50].
3.1. The Role of MicroRNAs in the Initiation of T2DM-Associated Atherosclerosis
3.1.1. MicroRNAs in Chronic Hyperglycemia-Induced Endothelial Dysfunction
Endothelial dysfunction is considered a major contributor to the initiation of diabetic atherosclerosis, and it requires the orchestration of a series of events, including oxidative stress and inflammatory processes [19]. The enhanced levels of reactive oxygen species (ROS) under hyperglycemic conditions are attributable to both overproduction of oxygen-derived radicals and impaired antioxidant defense resulting from a decline in mitochondrial function, increased expression of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, cyclooxygenase (COX), myeloperoxidase (MPO) as well as decreased expression of superoxide dismutase which then lead to a generation of superoxide anion (O2-), a precursor of the majority of other ROS [52]. In addition, persistent high glucose levels induce the low-grade inflammation reflected by elevated levels of proinflammatory cytokines and chemokines, including interleukin 1β (IL-1β), interleukin 6 (IL-6), interleukin 18 (IL-18), and tumor necrosis factor α (TNF-α) [19,53]. Simultaneously, increased oxidative stress and inflammation in the hyperglycemic milieu accelerate the nonenzymatic glycoxidation of proteins and lipids that results in advanced glycation end-products (AGEs) formation [19,20]. What is more, a predominant precursor of AGEs is also methylglyoxal (MGO), a side-product of glycolysis which accumulates in the cells under diabetic conditions and contributes to insulin resistance [53,54,55]. Upon AGE binding with its surface receptor (RAGE), multiple intracellular signal transduction pathways are activated, while soluble RAGE (sRAGE), a secretory isoform lacking the transmembrane domain, contributes to the neutralization or removal of circulating ligands by competing with membranous RAGE [20,53,56]. In addition to AGEs, RAGE is able to bind several other proinflammatory particles, including high mobility group box-1 (HMGB1), S100 calcium-binding proteins, and amyloid-β-protein, which provokes EC activation and increased expression of surface adhesion molecules, such as vascular cell adhesion molecule-1 (VCAM-1), intercellular adhesion molecule-1 (ICAM-1), and E-selectin via targeting nuclear factor-kappa B (NF-κB) pathway [19,53,57]. Hence, these actions promote the adhesion and entrance of monocytes/macrophages into the subendothelial space, exaggerate inflammatory response and initiate atherosclerotic plaque formation [19]. Long-lasting AGE/RAGE axis activation inhibits biosynthesis of nitric oxide (NO), a major vasodilator, by endothelial NO synthase (eNOS) downregulation and synchronously enhances the production of endothelium-derived COX-dependent contracting factors (EDCFs) and endothelin-1 (ET-1) [58]. The imbalance between vasodilators and vasoconstrictors in favor of the latter leads to disturbances in vascular tone and endothelial dysfunction [58]. Furthermore, AGE-RAGE interaction increases oxidative stress, endoplasmic reticulum stress, and inflammation in ECs [47,56]. It is worth noting that heightened glucose uptake by ECs intensifies the synthesis of diacylglycerol, which along with increased AGEs, ROS, and inflammatory factors, activate protein kinase C (PKC) signaling. This, in consequence, initiates the c-Jun N-terminal kinase (JNK)/extracellular signal-regulated kinases 1/2 (ERK 1/2)/p38 mitogen-activated protein kinase (MAPK) pathway and NADPH oxidase, whereas it inhibits downstream expression of phosphatidylinositol 3-kinase (PI3K)/Akt [19,47,56]. In this way, PKC is able to modulate the migration and proliferation of ECs, increase oxidative stress and inflammation, as well as inhibit angiogenesis and NO production [19,47].
Numerous miRNAs have been implicated in the regulation of endothelial homeostasis, vascular repair, and angiogenesis. Among them, miR-126 was first identified as an endothelial-specific miRNA that is critical for preserving EC integrity, and its level has been found to be decreased in the diabetic microenvironment [59,60,61,62]. The restored expression of miR-126 may suppress inflammation and ROS production by diminishing the expression of downstream components of HMGB1, including TNF-α, NADPH oxidase activity, and triggering AKT-eNOS pathway in ECs treated by high glucose [59]. In addition, miR-126 overexpression in endothelial progenitor cells (EPCs), which are released from the bone marrow in response to endothelium damage or tissue ischemia, may protect against EPC dysfunction induced by hyperglycemia-associated AGEs and promote their proliferation, migration, and inhibit apoptosis that enables repairing the injured endothelium, and reendothelialization [60,61,62]. It was observed that miR-126 is also able to decrease proinflammatory and oxidative stress markers via its target sprouty-related, EVH1 domain-containing protein 1 (Spred-1) as well as through inducing Ras/ERK/vascular endothelial growth factor (VEGF) and PI3K/Akt/eNOS signaling pathways [60,61,62]. Moreover, the recovering of miR-130a level has been found to exert a protective effect on EPCs by targeting Runx3 and JNK, ERK/VEGF, PI3K/Akt pathways, thus increasing proliferation, migration, and differentiation of EPCs, and reducing their apoptosis [63,64,65].
Another miRNA that has been widely studied in the context of high glucose-induced endothelial dysfunction is miR-21 [66,67,68]. Its expression is markedly enhanced in circulating ECs from diabetic patients and in high glucose-treated ECs [66,67,68]. One of the research has suggested an atheroprotective role of miR-21 on ECs exposed to high glucose conditions, probably by inhibiting the expression of death-domain associated protein (DAXX), a factor related to oxidative stress-induced cell death, although miR-21 by itself did not affect high glucose-induced ROS production in ECs [66]. By contrast, other authors have shown the adverse effects of miR-21 on the endothelium [67,68]. The elevated expression of miR-21 in diabetic conditions leads to increased oxidative stress-mediated endothelial dysfunction by downregulation of antioxidant response genes, specifically, SOD2, which induces the release of large amounts of O2− [67]. Anyway, miR-21-3p has been revealed to aggravate the atherosclerotic lesions through the activation of the AGE/RAGE axis and its downstream signaling pathways, including the PKC, MAPK, and NF-κB, that, in consequence, increase ROS generation and intensify inflammatory state reflected by high levels of IL-1β, IL-6, TNF-α, and monocyte chemoattractant protein-1 (MCP-1), also known as chemokine CC-motif ligand 2 (CCL-2) [68]. Paradoxically, miR-21-3p may also be responsible for the degradation of a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10), an enzyme involved in the ectodomain shedding of RAGE, thus leading to reduced sRAGE levels, while no difference was observed in RAGE expression upon miR-21-3p mimicking or inhibition [68].
Notably, the downregulation of miR-214 and miR-190a exert a significant role in MGO-induced endothelial insulin resistance, at least in part, by increasing their specific targets, PH domain leucine-rich repeat protein phosphatase 2 (PHLPP2) levels, a negative regulator of the insulin signaling and GTPase Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS), respectively [54,55]. Meanwhile, the overexpression of these miRNAs rescues the insulin effect on Akt or insulin receptor substrate 1 (IRS1)/Akt/eNOS pathway, improves NO release in response to insulin, and prevents the hyperactivity of ERK1/2 in MGO-treated ECs [54,55].
Intriguingly, upregulation of miR-185 and miR-320, miRNA related to high glucose-induced metabolic memory, protects against vascular endothelial dysfunction via decreasing RAGE, PKC, and/or VEGFA protein levels, which is connected with suppressed proliferation and angiogenesis capacity of ECs, whereas surprisingly they may inhibit (miR-185) or promote (miR-320) EC apoptosis [69,70].
So far, only miR-200a and miR-200c, out of the miR-200 family, have been studied within the context of ROS-induced diabetic endothelial dysfunction [71,72,73]. miR-200a, which is downregulated in hyperglycemia, is engaged in endothelial antioxidant and anti-inflammatory defense by reducing ROS, lipid peroxide marker, VCAM-1, and MCP-1 levels and driving endothelial NO production [71]. However, miR-200c impairs endothelial function by increasing COX-2 and vasoconstrictors, such as prostaglandin E2, thus limiting endothelium-dependent relaxation, and it may also decrease EC proliferation under hyperglycemia-induced oxidative stress [72,73]. Likewise, miR-34a, miR-92a, miR-181c, and miR-210 are involved in regulating vascular endothelial function in diabetes mainly by inducing or reducing levels of oxidative stress markers [74,75,76,77,78].
Several studies have indicated that aberrant expression of the let-7 miRNA family (let-7b and let-7d), miR-149-5p, and miR-190a-5p regulate inflammatory pathways in diabetes-associated endothelial injury, and restoring expression of these miRNAs, similar to downregulation of miR-34a and miR-197 may contribute to decreased TNF-α, IL-6, IL-1β, MCP-1, VCAM-1, and ICAM-1 level [75,79,80,81,82]. It is worth noting that overexpression of miR-1 and miR-181c in hyperglycemic models led to reduced ET-1 and increased eNOS levels that improve endothelium-dependent vasodilatation [83,84].
Moreover, miR-26b-5p, miR-29b-3p, miR-31, miR-34a-5p, miR-93-5p, miR-181a-5p, miR-192-5p, miR-375-3p, and miR-425-5p have been demonstrated to induce EPC or EC apoptosis during sustained exposure to hyperglycemia, whereas miR-26a-5p and miR-29a exerted an opposite effect on EC viability [81,85,86,87,88,89]. The decreased expression of miR-9 and increased level of miR-134-5p are also considered to be possible links between diabetes mellitus and endothelial dysfunction [81,90]. Finally, miR-126, miR-132, miR-139-5p, miR-140-3p, miR-181a/b-5p, miR-221, and miR-342-3p have already been studied to be engaged in EC proliferation, migration, invasion, and tube formation, that presented them as the potential surrogate markers for angiogenesis upon hyperglycemic condition [91,92,93,94,95,96].
3.1.2. MicroRNAs in Diabetes-Associated Endothelial to Mesenchymal Transition
Persistent activation of ECs under hyperglycemic conditions induces EndMT, which contributes to the initiation and progression of diabetes-accelerated atherosclerosis as a consequence [51]. EndMT is the process during which ECs lose their typical phenotypes, including cell-cell contact and cell polarity that results in a spindle-shaped morphology and the acquisition of mesenchymal-like or myofibroblastic phenotypes with gaining migratory and invasive properties [51,97,98]. Throughout EndMT, the expression of endothelial markers such as vascular endothelial cadherin (VE-cadherin), platelet endothelial cell adhesion molecule-1 (PECAM-1, also known as CD31), and von Willebrand Factor (vWF) decreases, whereas the expression of mesenchymal cell markers such as alpha-smooth muscle actin (α-SMA), smooth muscle protein 22 alpha (SM22α), vimentin, fibronectin, N-cadherin, calponin, and fibroblast specific protein-1 (FSP-1) increases [97,98]. EndMT may also lead to the delamination and migration of EC-derived mesenchymal cells into the underlying tissues and thus enhances the extracellular matrix (ECM) production [98]. Remarkably, the involvement of miRNAs in molecular mechanisms driving diabetic atherosclerosis-related EndMT still remains unclear.
Recent studies on culture cell models have revealed that upregulation of miR-142-3p and miR-448-3p alleviated diabetic vascular dysfunction by inhibiting EndMT [99,100]. The expression of the above-mentioned miRNAs is downregulated in diabetic atherosclerosis that promotes EndMT via activation of transforming growth factor β (TGF-β)/Smad signaling pathway, the most well-known inducer of EndMT [99,100]. High glucose-treated endothelial cells manifested a significant decrease in CD31, VE-cadherin, and Smad7, while α-SMA, vimentin, TGF-β1, phospho-Smad2, and phospho-Smad3 were increased [99,100]. In contrast, miR-328 is significantly upregulated in high glucose-induced EndMT, and its effects are related to triggering the mitogen-activated protein kinase kinases 1/2 (MEK1/2) and ERK1/2 phosphorylation [101]. Moreover, EndMT has been found to be abrogated by the transduction of antagomiR-328 [101]. These studies demonstrate the need for further research on the miRNA-EndMT regulatory network in diabetic atherosclerosis.
3.1.3. MicroRNAs in Monocyte Differentiation/Macrophage Activation under Diabetic Condition
It is well documented that macrophage accumulation is a common feature of T2DM, and it is proposed as one of the principal mechanistic cores of diabetes-related atherosclerosis [19]. The recruitment of monocytes from the peripheral blood to the intima of the vessel wall and monocytes subsequent differentiation into macrophages, generating proinflammatory factors and activating other immune cells, promote atherosclerosis in the diabetic microenvironment [102]. In addition, the increased activation and migration of macrophages into different tissues, especially adipose tissue, are involved in the pathogenesis of chronic subclinical inflammation under hyperglycemic conditions [103]. According to the activation state and functions, macrophages can be divided into two heterogenous subsets: M1-type (classically activated macrophages) and M2-type (alternatively activated macrophages) [104,105]. M1 macrophages reveal proinflammatory phenotype and produce proinflammatory cytokines, such as TNF-α, IL-1β, IL-6, interleukin 12 (IL-12), and NO, whereas M2 macrophages are highly expressed in specific anti-inflammatory markers, such as interleukin 10 (IL-10), and argininase (Agr-1) [104,105,106,107].
Until now, only a few studies have explored the role of miRNAs in the context of monocyte differentiation and macrophage activation under a diabetic milieu. The overexpression of miR-99a and miR-448 has been found to prevent M1 phenotype activation and decrease the levels of inflammatory markers in T2DM [105,108]. In contrast, remarkably upregulated miR-130b, miR-330-5p, and miR-495 expression have been reported to enhance macrophage M1 polarization along with attenuating macrophage M2 polarization in the diabetic animal model [104,106,107]. Moreover, miR-483-3p has been observed to be overexpressed in M2-type macrophages in the aortic wall of patients with T2DM, that increased endothelial and macrophage apoptosis and impaired endothelial repair capacity [109]. Intriguingly, increased expression of miR-17-5p and miR-221 in diabetic monocyte-derived macrophages exert pro-proliferative effects on VSMCs by downregulation of cyclin-dependent kinase inhibitors, such as p21Cip1 and p27Kip1, resulting in the development of vascular dysfunction in T2DM [102].
Noteworthy, miRNAs are also involved in the regulation of lipid and lipoprotein metabolism, atherosclerotic plaque formation, as well as its size and stability [103,108,110,111,112]. T2DM is strictly related to atherogenic dyslipidemia, which is characterized by increased levels of triglycerides (TG), decreased high-density lipoprotein (HDL) cholesterol (C), and the existence of small dense low-density lipoprotein (LDL)-C [110]. miR-99a mimics have been shown to improve cholesterol metabolism, which was reflected by a significant decrease in the total cholesterol (TC), HDL-C, and LDL-C levels in the hyperglycemic environment [108]. Kimura et al. revealed that hyperinsulinemia might activate the sterol regulatory element-binding protein (SREBP)-1c/miR-33b pathway that consequently increased TG synthesis [110]. In addition, the elevated expression of miR-33b repressed the level of HDL-C and apolipoprotein A-1 (ApoA-1) by downregulation of the ATP-binding cassette, sub-family A, member 1 (ABCA1) protein which is engaged in the cholesterol efflux and is responsible for the generation of nascent HDL-C [110]. What is more, anti-miR-33 treatment has been observed to promote remodeling of diabetic plaque towards a more stable-appearing phenotype by decreasing macrophage content in the plaque [111]. Lipoprotein uptake by macrophages is believed to be one of the earliest pathogenic events in the nascent plaque and results in the formation of foam cells, a special form of lipid-laden macrophages [112]. miR-27b-3p, whose level is decreased in the diabetic model, may be involved in delaying the progression of atherosclerosis in hyperglycemic conditions as its overexpression reduces the uptake of oxidized low-density lipoprotein (ox-LDL) by macrophages [112]. Additionally, miR-145 overexpression increases apoptosis of monocytes, reduces their proliferation and macrophage infiltration, which attenuate inflammation and markedly reduce plaque size, partially by inhibition of NF-κB activation [103].
3.2. The Role of MicroRNAs in the Progression of T2DM-Associated Atherosclerosis
3.2.1. MicroRNAs in Vascular Smooth Muscle Cell Proliferation and Migration under Diabetic Condition
Under physiological conditions, VSMCs are specialized to maintain a differentiated contractile phenotype which allows them to regulate vascular tone [113]. In response to arterial injury through hyperglycemic or inflammatory stimuli, VSMCs dedifferentiate and adopt a synthetic, proliferative, and migratory phenotype that is the most common pathological change in diabetic atherosclerosis [113]. Principal regulators in the maintenance of the mature VSMC phenotype include the transcription factor serum-response factor (SRF), SRF-associated coactivators such as myocardin, and TGF-β, whereas soluble factors such as platelet-derived growth factor (PDGF) promote dedifferentiation of VSMCs [113]. Moreover, the Krüppel-like factor (KLF) family of transcription factors, and in particular, KLF4, constitutes a key pluripotent transcription factor in VSMC differentiation and dedifferentiation via modulation of KLF4/myocardin/SRF axis [113]. Accumulating evidence has indicated that miRNAs play a pivotal role in the mechanisms determining VSMC phenotype in T2DM-related atherosclerosis.
The miRNA cluster containing miR-143 and miR-145, the most abundant VSMC miRNAs, influence VSMC plasticity in both human culture cells and those from T2DM patients [114,115,116,117]. Deficiency of miR-145 results in the increased migratory and proliferative capacity of VSMCs under high glucose conditions through targeting KLF4/myocardin pathway and Rho-associated coiled-coil forming protein kinase 1 (ROCK1), an important factor involved in atherosclerosis by increasing the permeability of ECs, enhancing the chemotaxis of macrophages, their transformation into foam cells, and VSMC phenotypic switching [114,115]. Interestingly, the divergent saphenous vein VSMC proliferation was observed between T2DM patients and non-diabetic ones undergoing coronary artery bypass grafting [116]. Overexpression of miR-143/-145 decreased VSMC proliferation in healthy individuals, whereas transfection of T2DM-VSMC with anti-miR-143/-145 increased cell proliferation, thus confirming the atheroprotective properties of these miRNAs [116]. Similarly, miR-217 and miR-132 exert their anti-proliferative and anti-migratory effects on VSMCs under high glucose conditions mimicking diabetes via suppression of ROCK1 and E2F transcription factor 5 (E2F5), respectively [118,119]. It should be noted that E2F5, a key member of the E2F family, controls the transcription of proliferation-related genes and the G1/S transition [119].
Another miRNA, which is one of the crucial regulators of high glucose-induced VSMC differentiation, is miR-24 [120,121,122,123]. It has been shown that high glucose-stimulated animal culture cells and/or carotid arteries in diabetic rats transfected with adenovirus-miR-24 precursor exhibited reduced VSMC proliferation, migration, and proinflammatory cytokine secretion, an effect that might be mediated by the inactivation of several pathways, including the HMGB1/NF-κB, PDGF-BB, and PI3K/Akt/mammalian target of rapamycin (mTOR) signaling pathways [120,121,122]. Moreover, miR-24 transfected into the same carotid artery in diabetic rats alleviated VSMC proliferation, and it was related to the regulation of the expression of Cyclin D1 and p21 through the inhibition of Wnt4/Dvl-1/β-catenin signaling pathway [123]. It has also been observed that miR-24 upregulation attenuated diabetic vascular remodeling [121,122,123,124,125].
As it was previously mentioned, the let-7 miRNA family has exerted not only anti-inflammatory effects in ECs but is also proposed as a master regulator of VSMC proliferation and differentiation [79]. In diabetes-associated atherosclerosis, the PDGF and TNF-α induced activation, increased proliferation, and migration of VSMCs are associated with decreased levels of let-7b and let-7d via Lin28b, a negative regulator of let-7 biogenesis [79]. Nevertheless, the restoration of let-7 to normal levels ensures an atheroprotective response [79].
In addition, several studies have proved that platelet-derived miR-223 suppresses the proliferation and dedifferentiation of VSMCs by decreasing platelet-derived growth factor receptor beta (PDGFRβ) and directly targeting the insulin-like growth factor-1 receptor (IGF-1R), which then activates the adenosine monophosphate-activated protein kinase (AMPK) phosphorylation [126,127]. However, miR-223 expression is pathologically reduced under diabetic conditions [126,127]. miR-9, miR-125a, and miR-322-5p have been identified as other miRNAs that contribute to the acquisition of a contractile phenotype of VSMCs [128,129,130]. It has been observed that decreased miR-9 expression in diabetic human VSMCs resulted in increased activity of KLF5, a positive regulator of VSMC dedifferentiation and transformation into synthetic phenotype, and a subsequent decrease in myocardin expression [128].
What is more, the reduced transfer of antiproliferative miR-126-3p from ECs to VSMCs has been observed under the hyperglycemic condition that leads to accelerated proliferation and migration of VSMCs [131]. Most recently, miR-126-3p has occurred to play a crucial role in T2DM VSMC metabolic memory through the activation of MAPK/ERK pathway, enhancing the efficiency of blockers of potassium channels Kv1.3 in VSMCs, thus preventing their proliferation, migration, and vessels remodeling [132].
The data on the relevance of miR-29c in the context of its atherogenic and antiatherogenic properties are inconsistent [133,134]. Torella et al. have revealed that overexpression of miR-29c with contemporaneous miR-204 inhibition upon hyperglycemia prevented exaggerated VSMC proliferation by regulation of epithelial membrane protein 2 (EMP2) and caveolin 1 (CAV1) as direct targets [133]. In contrast, other authors have suggested that hyperglycemia-induced upregulation of miR-29c via inhibition of KLF4 activity, thus stimulating VSMC proliferation [134].
Likewise, miR-19a, belonging to the miR-17-92 cluster, miR-138, and miR-504 promote a switch to the synthetic phenotype, and their expressions have been found to be increased in high glucose-induced culture conditions and diabetic animal models [135,136,137]. Moreover, another miRNA with a proven proatherogenic effect is miR-376b-3p, whose upregulation is observed in a diabetic state and it leads to the increased VSMC proliferation by suppression of KLF15, a negative regulator of proliferative processes [138]. In the given hyperglycemic condition, miR-221/-222 and miR-17-5p, similarly to miR-21-5p, have also been proposed to act upon VSMC proliferation and migration via downregulation of p27Kip1, a cyclin-dependent kinase inhibitor, and tropomyosin 1 (TPM1), a regulator of cytoskeletal actin filaments, respectively [102,139,140,141].
Importantly, T2DM is associated with not only hyperglycemia but also compensatory hyperinsulinemia in the setting of insulin resistance. miR-99a has been shown to be decreased in high-dose insulin environments that stimulated proliferation, migration, and dedifferentiation of VSMCs by attenuation of the inhibitory effects of miR-99a on IGF1-R and mTOR signaling pathways [142].
3.2.2. MicroRNAs in Platelet Hyperactivity under Diabetic Condition
Platelets, although anucleate, are thought to be important cellular components involved in both the initiation and progression of atherosclerosis especially in the ensuing atherothrombotic sequelae [143]. T2DM modulates the function of platelets leading to their increased activation, aggregation, adhesiveness to ECs, and thrombus formation [144,145,146]. Various mechanisms have been proposed to be responsible for the diabetes-induced platelet hyperactivity, including altered calcium homeostasis with the overactivation of calpain, the calcium-activated cysteine protease, abnormal expression of surface glycoprotein receptors and adhesion molecules, and increased binding of fibrinogen [143]. It is worth mentioning that platelets are a rich source of miRNAs and functioning proteins (Dicer, TRBP2, and AGO2), which allow them to convert pre-miRNAs into mature miRNAs [147]. In addition, platelets release abundant miRNAs in the form of microvesicles [145,146]. Accordingly, platelet-specific miRNAs may facilitate the communication between platelets and other immune and vascular cells.
One of the most thoroughly studied miRNAs in relation to platelet hyperactivity under hyperglycemic condition are miR-223, together with miR-126, miR-140, and miR-26b, whose expressions are decreased in both platelets and megakaryocytes from T2DM patients [147]. Moreover, it occurred that altered platelet miR-223, and miR-26b, miR-140 expressions lead to upregulation of mRNA and protein levels of P2Y12 receptor and P-selectin (CD62), known as platelet hyperactivity marker [147]. Investigation of the role of miR-223 in platelet function under diabetic conditions revealed decreased Dicer activity, generating a lower amount of mature miR-223 [147]. Supporting these findings, other researchers observed that inhibition of calpain-dependent cleavage of Dicer normalizes miR-223 processing and restores platelet function [148]. On the other hand, AGO2 levels were comparable in healthy and diabetic platelets [148]. Furthermore, the expression of a subset of platelet-derived miRNAs, such as miR-142/-143 and miR-155, was reduced in human individuals and mice with T2DM [148]. Similar to miR-223, miR-126 may also play a role in the modification of the expression of the P2Y12 receptor in T2DM. Thus miR-126 mimics are able to reverse metallothionein 1 pseudogene 3-mediated upregulation of the P2Y12 receptor [149]. Of note, plasma levels of miR-126 were increased in T2DM and directly correlated with soluble P-selectin [150]. However, it occurred that the administration of aspirin in this group led to the subsequent decline in platelet reactivity and the decrease in miR-126 [150]. Therefore, de Boer et al. hypothesized that other platelet-enriched miRNAs, such as miR-16, miR-223, and miR-423, can be released from activated platelet into circulation in a similar fashion [150]. Recent studies have revealed that patients with T2DM also present significantly diminished expression of miR-30c originating from platelets, and its restoration to the desired level leads to downregulation of plasminogen activator inhibitor-1 (PAI-1) expression as a target, thereby prolonging the time to arterial thrombus formation [151].
Contrary to previous studies, Stratz et al. have demonstrated no major divergences in platelet miRNA profiles between the studied groups, neither T2DM and non-T2DM nor CAD and non-CAD [152]. Nevertheless, the authors have indicated that miR-377-5p, miR-628-3p, and miR-3137 may serve as relatively stable predictors of group membership [152].
3.2.3. MicroRNAs in Diabetes-Associated Calcification
Vascular calcification is an active process strongly associated with atherosclerotic plaque evolution and results in an increased incidence of cardiovascular events and mortality [153,154]. It refers to the pathological deposition of calcium and phosphate minerals in the inner or middle layer of the vascular wall [154].
So far, data concerning the role of miRNAs in the pathogenesis of vascular calcification in T2DM are limited. It has been observed that miR-204 is downregulated in asymptomatic diabetic patients with higher coronary artery calcification (CAC) scores assigned in coronary computed tomography [154]. What is interesting, the expression of miR-204 is negatively correlated with glycated hemoglobin A1c (HbA1c), and it has occurred to be a significant and independent risk factor for the presence of CAC [154]. Similarly, miR-95-5p, miR-6776-5p, miR-3620-5p, and miR-4747-5p are suppressed in high glucose-induced calcification of VSMCs [155]. Nonetheless, the exact mechanisms of the aforementioned miRNAs remain largely uncharacterized [154,155]. The overexpression of miR-34c-5p, but not miR-34c-3p, may alleviate VSMC calcification under hyperglycemic conditions [153]. In turn, miR-128-3p accelerates cardiovascular calcification and insulin resistance in the T2DM animal experimental model through the activation of the Wnt signaling pathway, which has been proved to be involved in the formation of calcium phosphate deposits in the vessels [156].
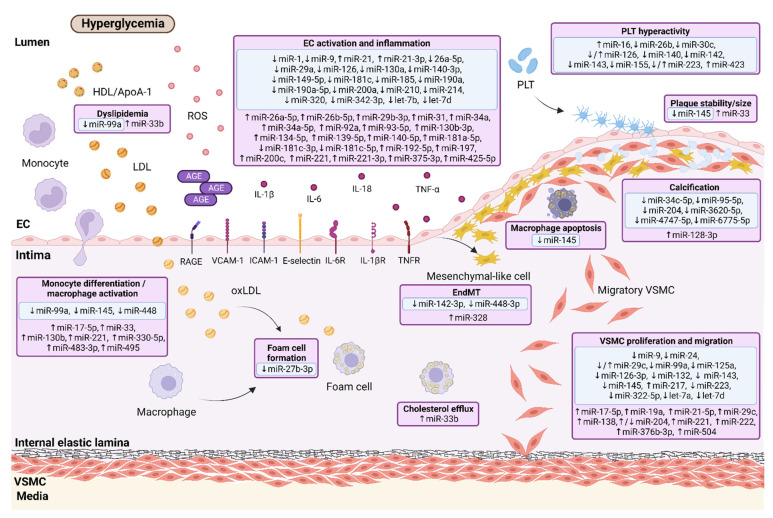
The involvement of various miRNAs in particular stages of the initiation and progression of T2DM-induced atherosclerosis is presented in Figure 2.
Figure 2.
MicroRNAs implicated in T2DM-related atherosclerosis. Positive/atheroprotective (in green frame) and negative/atherogenic (in violet frame) effects of miRNAs on every step of the atherosclerotic process under diabetic conditions are shown. Chronic hyperglycemia induces inflammation, and oxidative stress leading to the cascade of processes, including EC activation, migration of monocytes into the intima and their maturation into macrophages, foam cell formation by oxLDL uptake, PLT hyperactivity, VSMC proliferation and migration into the intima. Single miRNAs are also found to be engaged in EndMT, calcification, and regulation of plaque size/stability. miR—microRNA; EC—endothelial cell; EndMT—endothelial-to-mesenchymal transition; VSMC—vascular smooth muscle cell; PLT—platelet; ROS—reactive oxygen species; IL—interleukin; TNF-α—tumor necrosis factor α; IL-6R—interleukin 6 receptor; IL-1βR—interleukin 1β receptor; TNFR—tumor necrosis factor receptor; AGE—advanced glycation end-product; RAGE—receptor for advanced glycation end-products; VCAM-1—vascular cell adhesion molecule-1; ICAM-1—intercellular adhesion molecule-1; HDL—high-density lipoprotein; LDL—low density lipoprotein; ApoA-1—apolipoprotein A-1; ox-LDL—oxidized low-density lipoprotein. ↑ upregulation; ↓ downregulation. Created with BioRender.com.
4. Clinical Research on Circulating MicroRNAs in T2DM and CAD
Circulating miRNAs were first detected in the samples of human serum and plasma in 2008, and subsequently, they have been found in a variety of body fluids, such as saliva, urine, breast milk, colostrum, bronchial lavage, cerebrospinal fluid, peritoneal fluid, pleural fluid, amniotic fluid, seminal fluid, ovarian follicular fluid, and even tears [28,157,158,159]. In the whole blood, miRNAs are also discovered in peripheral blood mononuclear cells (PBMCs) such as lymphocytes, monocytes, and macrophages, which are suspended in plasma and constitute an additional source of miRNAs [160]. Contrary to proteins and different RNA classes, extracellular miRNAs are highly stable in circulation and resistant not only to enzymatic degradation by RNase but also to deleterious conditions, including boiling, repeated freeze-thaw cycles, and pH fluctuation [28]. Consequently, serum or plasma specimens can be stored at room temperature for up to four days and at −20 °C or −80 °C for several months without remarkable degradation of miRNAs [161]. Moreover, miRNA sequences are conserved among species, and their detection is based on modern, minimally invasive technologies, which ensure high testing sensitivity and specificity [162,163]. miRNAs could be easily and repetitively detectable using reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR), different types of microarrays, and alternative techniques, including northern blotting, next-generation sequencing (NGS) or NanoString nCounter [163,164]. Finally, miRNAs are tissue-specific, which allows the identification of their origin and establishing a disease-specific expression pattern of miRNAs [165]. Interestingly, miRNA signatures seem to be more robust biomarkers than single miRNAs and are more likely to adequately reflect the complexity of disease pathophysiology [165].
The listed features of miRNAs suggest that they are promising as ideal candidates for disease biomarkers. Findings from preclinical and basic research studies highlight that miRNAs may serve as a fingerprint for T2DM-induced atherosclerosis. Therefore, the clinical usefulness of circulating miRNAs as early diagnostic, prognostic, and predictive biomarkers for T2DM, CAD, and CAD related to T2DM have been discussed in detail.
4.1. MicroRNAs as Potential Biomarkers for T2DM
The pathogenesis of T2DM arises from the interplay of genetic, epigenetic, and environmental factors, which may impair insulin sensitivity in target tissues, including mainly the liver, adipose tissue, skeletal muscles, and insulin secretion from pancreatic beta cells [8]. It is commonly known that the compensatory hyperinsulinemia to maintain euglycemia in the setting of peripheral insulin resistance ultimately leads to the deterioration of beta cells, their exhaustion, and eventual death [8]. The state of relative insulin deficiency in chronically elevated blood glucose levels consequently generates the onset of T2DM [8]. Despite the progress made in the exploration of mechanisms underlying the pathogenesis of T2DM, understanding this complex metabolic disorder at the molecular level and searching for miRNA-based biomarkers used for the early detection and identification of highly susceptible individuals seems to be relevant to avoid vascular complications and improve patient quality of life by providing appropriate management.
A growing number of reports have revealed T2DM-related expression changes of circulating miRNAs. However, only seventeen studies have attempted to perform miRNA expression profiling to find a unique miRNA pattern [166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182], while the remaining researchers have focused on miRNA selection based on the review of available scientific literature or analysis of miRNA databases. Therefore, it seems that the concept of initial miRNA screening with their further validation, especially in previously unexplored ethnic groups, holds much potential for the discovery of novel miRNAs associated with T2DM and to select of those molecules that have the greatest discriminatory power.
The pioneering study that established a plasma signature of 13 miRNAs in T2DM individuals based on microarray profiling was the prospective population-based study conducted by Zampetaki et al. [174]. The researchers revealed lower expression of miR-15a, miR-20b, miR-21, miR-24, miR-29b, miR-126, miR-150, miR-191, miR-197, miR-223, miR-320, miR-486, and a modest increase in miR-28-3p expression in diabetic patients compared to controls [174]. Interestingly, it has occurred that 70% of T2DM cases and 92% of the control group were correctly classified using the five most significant miRNAs, including miR-15a, miR-28-3p, miR-126, miR-223, and miR-320 [174]. Moreover, this miRNA panel allowed the prediction of T2DM in normoglycemic patients because their expression was disturbed about 10 years prior to disease diagnosis, confirming the value of miRNAs as potential predictive and diagnostic biomarkers [174]. Similarly, Karolina et al. have identified a set of circulating miRNAs that displayed altered expression in diabetes, but they have observed contrasting expressions of miR-150 and miR-320 in T2DM cohorts, probably due to a distinct sample type than in previous report (plasma vs. whole blood) [172,180]. What is more, among seven diabetes-related serum miRNAs, apparently upregulated miR-34a showed the highest standardized canonical discriminant function coefficients allowing correct judgment of 70.6% of T2DM subjects [183]. Evidence for the clinical usefulness of these miRNA panels was provided by the discovery of miRNA-regulated pathways, including insulin production (miR-30d), insulin secretion (miR-9, miR-124a, miR-375), insulin signaling and action in target tissues (miR-27a, miR-29a, miR-144, miR-146a, miR-150, miR-182, miR-192, miR-320), especially translocation of glucose transporter-4 (GLUT4) receptor from the intracellular storage to the plasma membrane of the insulin-responsive cells to allow uptake of glucose (miR-150) [172,180,183]. Additionally, other miRNAs considered as T2DM biomarkers have been found to be implicated in insulin signaling pathways (let-7f-5p, miR-24-3p, miR-145-5p, miR-214-3p), beta cell function, and insulin secretion (let-7b-5p, miR-375, miR-720, miR-770-5p), insulin resistance (miR-30d, miR-145-5p, miR-199a, miR-330), and glucose or lipid metabolism networks (miR-29, miR-33a, miR-122, miR-155) [175,184,185,186,187,188,189,190,191,192,193].
Recent clinical data have indicated a close link between specific miRNAs and inflammation in T2DM [194]. As observed, miR-126 and miR-146a are among the most commonly reported anti-inflammatory miRNAs with the potential to be used as epigenetic biomarkers for T2DM [195,196,197,198,199,200,201]. It has occurred that normoglycemic individuals with decreased expression of plasma miR-126 with a cut-off point of less than 35 are more likely to develop T2DM in the next two years [197]. Interestingly, the introduction of miR-126 to the established conventional model, including age, gender, body mass index (BMI), blood glucose, and HbA1c, allowed to obtain higher discriminatory power for T2DM patients with an area under the curve (AUC) of 0.893 than for miR-126 or the model alone (0.792 vs. 0.826) [196]. In line with receiver operating characteristic (ROC) analysis results, downregulated expression of endothelial-derived miR-126 in whole blood is suitable for differentiating T2DM subjects from healthy controls with an excellent AUC of 0.932 [198]. On the other hand, Weale et al. have pointed out that increased expression of miR-126, also measured in peripheral blood, achieved a satisfactory overall predictive ability in diagnosing T2DM, although this value was remarkably lower than that calculated for HbA1c commonly recognized as one of the diagnostic indices for diabetes (AUC of 0.646 vs. 0.861) [199]. It is worth mentioning that upregulation of another inflammation-related miRNA, miR-146a, demonstrated a good discriminatory power for T2DM screening with an AUC of 0.725 and significantly increased the risk for new T2DM (crude odds ratio; OR, 4.333; 95% CI, 1.935–9.705; p < 0.001) in patients belonging to the group of the highest miR-146a tertile levels (≥1.339) [195]. Similar to miR-126, adding miR-146a as an auxiliary component of the established conventional model comprising age, sex, BMI, smoking, alcohol drinking, hypertension, family history of diabetes, TG, HDL-C, LDL-C, TC, is able to enhance AUC value from 0.753 to 0.844 [195]. In addition to their potential diagnostic values, the profile of circulating miRNAs could also provide precious information about the pathophysiology of the disease. It merits special mention that both miR-126 and miR-146a are closely linked to generally approved inflammatory and endothelial dysfunction markers, including IL-6, IL-8, TNF-α, NF-κB, VCAM-1 or interleukin 1 receptor-associated kinase 1 (IRAK1), tumor necrosis factor receptor-associated factor 6 (TRAF6) as well as they can lead to the accumulation of dysfunctional endothelial senescent cells and the shift of circulating angiogenic cells from a proangiogenic to a proinflammatory profile [194,202,203,204,205,206,207,208]. Furthermore, these miRNAs have been found to be associated with increased platelet activation, oxidative stress, endoplasmic reticulum stress or apoptosis (caspase-3) indices, and decreased plasma antioxidant capacity [147,204,205]. Aside from the above, miRNAs such as miR-18a, miR-34c, miR-21-5p, miR-103b, miR-122, miR-155, miR-181b, miR-574-3p, and miR-576-3p may act as mediators of inflammatory response in patients with T2DM, although some authors have suggested that miR-34c-5p, miR-146a, miR-155, miR-574-3p, and miR-576-3p did not correlate with the level of proinflammatory cytokines, except for the poor association of miR-574-3p with MCP-1 [181,194,203,206,207,209,210,211,212,213].
Considering that obesity is an established risk factor for T2DM, and adipose tissue is a highly active metabolic and endocrine organ releasing an array of hormones and cytokines (adipocytokines), identifying obesity-related miRNAs as candidate biomarkers for T2DM seems to be essential [8]. So far, circulating miR-130a, miR-326, and miR-3666 have been recognized to modulate the adiponectin pathway [170,182,214]. Notably, upregulated miR-326 is an independent predictor of plasma adiponectin levels, irrespective of age, sex, and BMI [170]. Moreover, significantly altered expression of miR-130a and miR-3666 inhibits adiponectin gene expression, indirectly affecting adipocyte differentiation and reducing insulin sensitivity, promoting apoptosis of pancreatic beta cells, and diminishing their differentiation [182,214]. The observed positive correlations of miR-326 and miR-3666 with HbA1c and/or blood glucose levels are clinically relevant as they can also be used to monitor glycemic control [182,214]. Likewise, other dysregulated miRNAs in T2DM subjects, including pancreatic-islet specific miR-7, miR-21 and miR-148a, miR-217, miR-221, miR-222 displayed positive association with markers of glycemic control, whereas miR-146a, miR-185, miR-222-3p, and miR-342-3p showed the negative one [204,206,208,213,215,216,217,218,219,220].
As presented in Table 1, miR-15a, miR-23a, miR-30c, miR-103a/103b, miR-126, miR-210, miR-320b, miR-499, miR-572, and miR-766-3p have obtained the highest capability to distinguish T2DM from normal glucose tolerant patients with an AUC value above 0.8 and simultaneously sensitivity, specificity reaching even 100% [171,179,197,198,200,221,222,223,224,225,226], while the other miRNAs regarded as T2DM biomarkers have shown the satisfactory or good discriminatory power [171,179,192,195,196,199,218,227,228,229,230]. Therefore, it is proposed that rather than focusing on single miRNAs that are involved in T2DM development, an integrated view should also take into account the combination of several miRNAs whose diagnostic ability, sensitivity, and specificity may be even stronger [166,169,170,173,212,216,231].
Table 1.
Circulating microRNAs as potential biomarkers for type 2 diabetes mellitus.
| miRNA | Expression Change | Sample Type | Assay Method | Number of Samples |
Ethnicity | Age [Years] | Gender (Male/Female, n) |
BMI [kg/m2] | Duration of T2DM [Years] |
HbA1c [%] | Value of Biomarker (AUC; 95% CI; SV [%]; SP [%]) |
Author, Year (Reference) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| let-7b-5p | Up | Serum | RT-qPCR | T2DM (29) HC (25) |
Emirati | 55.6 ± 9.0 42.8 ± 12.7 |
13/16 9/16 |
31.5 ± 6.0 28.3 ± 6.5 |
Newly diagnosed | 7.6 ± 1.6 5.2 ± 0.4 |
N/A | Aljaibeji et al., 2022 [185] |
| miR-766-3p | Down | Serum | qPCR | T2DM (108) HC (68) | Chinese | 46.80 ± 18.43 46.56 ± 16.85 | 62/46 40/28 |
25.61 ± 6.59 24.14 ± 4.54 |
Newly diagnosed | 9.60 ± 2.48 5.57 ± 0.81 |
0.880 88.9; 75.0 |
Cao et al., 2022 [221] |
| miR-33a, miR-122 | Up | Whole blood | RT-qPCR | T2DM (50) HC (50) |
Iranian | 55.9 ± 8.9 47.4 ± 9.2 |
Only male | 27.0 ± 3.4 25.0 ± 3.3 |
Diagnosed | N/A | N/A | Masoudi et al., 2022 [193] |
| miR-499 | Down | Serum | RT-qPCR | T2DM (60) HC (60) |
Egyptian | Age-matched | Sex-matched | BMI-matched | N/A | N/D | 0.970 90.0; 96.6 |
Oraby et al., 2022 [226] |
| miR-145-5p | Down | Plasma | RT-qPCR | T2DM (20) HC (20) |
Iranian | 57.05 ± 1.99 51.07 ± 2.29 |
N/A | 28.25 ± 0.95 26.94 ± 0.08 |
Diagnosed | 8.15 ± 0.4 5.29 ± 0.06 |
0.77 (0.60–0.93) | Shahrokhi et al., 2022 [192] |
| miR-107 | Up | Serum | RT-qPCR | T2DM (53) HC (54) |
Lithuanian | 65 (44–83) 62 (48–80) |
24/29 25/29 |
34.14 ± 5.92 28.07 ± 5.25 |
15 (5–30) – |
8.23 ± 2.14 5.46 ± 0.49 |
N/A | Šimonienė et al., 2022 [242] |
| miR-21 | Up | Plasma | RT-qPCR | T2DM (24) HC (29) |
Iranian | 54.42 ± 7.76 50.42 ± 6.14 |
15/9 19/10 |
N/A | Newly diagnosed | 7.16 ± 0.16 5.15 ± 0.54 |
0.78 (0.64–0.92) 79.17; 81.48 |
Yazdanpanah et al., 2022 [227] |
| miR-720 | Up | Whole blood | RT-qPCR | T2DM (50) HC (50) |
Chinese | 57 ± 8.2 55 ± 7.8 |
24/26 27/23 |
26.2 ± 4.1 23.1 ± 3.8 |
Newly diagnosed | 9.89 ± 2.74 3.21 ± 1.27 |
N/A | Lu et al., 2021 [191] |
| miR-135a | Up | Saliva | RT-qPCR | T2DM (40) HC (40) | Iranian | 47 ± 1.6 46 ± 1.4 |
26/54 1 | 27.6 ± 1.3 26.4 ± 1.9 |
Diagnosed | 7.6 ± 0.3 4.0 ± 0.2 |
0.007 95.0; 95.0 |
Monfared et al., 2021 [200] |
| miR-126 | Down | 1 100.0; 100.0 |
||||||||||
| miR-33a-5p | Up | Plasma | RT-qPCR | T2DM (20) HC (20) |
Iranian | 57.05 ± 1.99 51.07 ± 2.29 |
10/10 10/10 |
28.25 ± 0.95 26.94 ± 0.08 |
Diagnosed | 8.15 ± 0.4 5.29 ± 0.06 |
0.71 (0.542–0.889) | Saeidi et al., 2021 [230] |
| miR-7-1-5p | Down | NS | ||||||||||
| miR-770-5p | Up | Serum | RT-qPCR | T2DM (20) HC (20) |
Chinese | 32–61 29–64 |
14/8 14/8 |
N/A | Newly diagnosed | N/A | N/A | Wang et al., 2021 [190] |
| miR-30a-5p, miR-126-3p, miR-182-5p, miR-1299 | Up | Whole blood | RT-qPCR | T2DM (92) HC (974) |
South African |
58.15 ± 10.62 45.22 ± 15.3 |
19/73 286/688 |
31.5 ± 8.0 27.4 ± 7.8 |
Newly diagnosed | 7.3 ± 1.9 5.6 ± 0.5 |
N/A | Weale et al., 2021 [232] |
| miR-30a-5p, miR-30e-3p, miR-126-3p, miR-182-5p, miR-1299 | T2DM (188) HC (974) |
57.88 ± 11.97 45.22 ± 15.3 |
37/151 286/688 |
30.7 ± 6.4 27.4 ± 7.8 |
Diagnosed | 8.9 ± 2.4 5.6 ± 0.5 |
||||||
| miR-126-3p | Up | Whole blood | RT-qPCR | T2DM (94) HC (972) |
South African |
58.4 ± 10.6 45.2 ± 15.3 |
19/75 284/688 |
31.3 ± 8.0 27.4 ± 7.9 |
Newly diagnosed | 7.4 5.6 |
0.646 (0.576–0.717) 55.6; 70.8 |
Weale et al., 2021 [199] |
| miR-122 | Up | Whole blood | RT-qPCR | T2DM (30) HC (30) |
Iranian | 53.03 ± 9.66 55.37 ± 8.47 |
15/15 15/15 |
30.27 ± 3.11 29.80 ± 2.89 |
Diagnosed | 7.29 ± 1.22 4.54 ± 0.20 |
N/A | Zeinali et al., 2021 [194] |
| miR-126-3p, miR-146a | Down | |||||||||||
| miR-29, miR-155 | Up | Serum | qPCR | T2DM (59) HC (72) |
Xinjiang Uygurian | 48.45 ± 7.36 44.56 ± 3.58 |
27/32 36/36 |
28.50 ± 4.69 21.94 ± 1.33 |
Diagnosed | N/A | N/A | Zhu et al., 2021 [189] |
| miR-330 | Up | Serum | RT-qPCR | T2DM (100) HC (100) |
Indian | > 50 (40.0%) > 50 (50.0%) |
57/43 55/45 |
> 25 (34.0%) > 25 (15.0%) |
Newly diagnosed | N/A | N/A | Ali Beg et al., 2020 [188] |
| let-7f-5p, miR-24-3p, miR-214-3p | Down | Whole blood | miSript miRNA PCR array, RT-qPCR |
T2DM (40) HC (16) |
Greek | 59 (35–75) 45 (19–52) |
19/21 7/9 |
29.3 (21.5–46.5) 24 (21.3–24.0) |
5 (0–26) – |
6.7 (5.2–12.1) – |
N/A | Avgeris et al., 2020 [175] |
| miR-34a | Up | Plasma | RT-qPCR | T2DM (30) HC (30) |
Indian | 38.9 ± 5.8 40.6 ± 5.95 |
19/11 17/13 |
27.8 ± 6.29 23.33 ± 3.57 |
4.55 ± 4.3 – |
7.51 ± 1.22 4.89 ± 0.29 |
N/A | Banerjee et al., 2020 [235] |
| miR-126-5p, miR-181b | Down | Whole blood | RT-qPCR | T2DM (30) HC (30) |
Iranian | 55.4 ± 5.3 53.5 ± 7.2 |
14/16 16/14 |
N/A | Newly diagnosed | 8.62 ± 1.74 5.1 ± 0.24 |
N/A | Dehghani et al., 2020 [207] |
| miR-103a | Up | Plasma | RT-qPCR | T2DM (48) HC (50) |
Han Chinese | 52.6 ± 9.13 45.62 ± 8.58 |
26/22 26/24 |
25.52 ± 2.89 24.56 ± 3.40 |
Newly diagnosed | 9.16 ± 1.05 5.15 ± 0.32 |
0.998 (0.993–1.0) 97.9; 98.0 |
Luo et al., 2020 [225] |
| miR-103b | Down | 0.964 (0.920–1.0) 98.0; 91.7 |
||||||||||
| miR-135 | Up | Plasma | RT-qPCR | T2DM (40) HC (40) |
Iranian | 53.69 ± 5.69 33.59 ± 7.58 |
N/A | 30.11 ± 1.01 25.23 ± 2.43 |
Newly diagnosed | 7.63 ± 0.41 4.70 ± 2.30 |
N/A 50.5; 91.2 |
Monfared et al., 2020 [229] |
| miR-222 | Up | Plasma | RT-qPCR | T2DM (30) HC (30) |
Iranian | 52.42 ± 8.77 51.44 ± 6.04 |
20/10 21/9 |
28.17 ± 5.46 27.60 ± 3.9 |
Newly diagnosed | 7.34 ± 1.08 5.76 ± 0.41 |
N/A | Sadeghzadeh et al., 2020 [239] |
| miR-15a | Down | |||||||||||
| miR-19a, miR-130a, miR-148b, miR-223 | Up | Serum | RT-qPCR | T2DM (102) HC (68) |
Mongolian (Chinese) | N/A | N/A | N/A | Newly diagnosed | N/A | N/A | Yan et al., 2020 [238] |
| let-7b-5p, miR-1-3p, miR-24-3p, miR-34a-5p, miR-98-5p, 133a-3p | Down | Whole blood | qPCR | T2DM (40) HC (37) |
Greek | 59 (35–75) 49 (19–69) |
19/21 19/18 |
29.3 (21.5–46.5) 26.9 (21.3–36.3) |
5 (0–26) – |
6.7 (5.2–12.1) 5.6 (5.0–6.1) |
N/A | Kokkinopoulou et al., 2019 [234] |
| miR-21 2 | Up | Plasma | RT-qPCR | T2DM (27) HC (44) |
Italian | 61.69 ± 7.59 59.3 ± 9.82 |
10/17 15/29 |
29.26 ± 5.83 25.11 ± 3.32 |
Newly diagnosed | 6.64 ± 0.6 5.80 ± 0.38 |
0.699 93.0; 35.0 |
La Sala et al., 2019 [228] |
| miR-30c | Down | Plasma | qPCR | T2DM (47) HC (32) |
Han Chinese | 60.5 ± 11.1 58.6 ± 8.1 |
23/24 17/15 |
24.76 ± 3.29 24.49 ± 2.30 |
Newly diagnosed | 9.15 ± 1.02 5.36 ± 0.35 |
0.916 (0.853–0.980) 87.9; 87.2 |
Luo et al., 2019 [224] |
| miR-486-3p | Up | Plasma | RT-qPCR | T2DM (29) HC (30) |
Israeli Arab/Jewish | 64 ± 10 31 ± 11 |
18/11 15/15 |
30 ± 5 25 ± 4 |
Newly diagnosed | 7.7 ± 1.9 5.1 ± 0.3 |
N/A | Meerson et al., 2019 [240] |
| miR-423 | Down | |||||||||||
| miR-342 | Up | Serum | RT-qPCR | T2DM (50) HC (50) |
Egyptian | 62.06 ± 1.26 62.22 ± 0.69 |
Only female | 27.58 ± 0.28 23.82 ± 0.14 |
12.06 ± 0.30 – |
10.75 ± 0.17 4.10 ± 0.68 |
N/A | Seleem et al., 2019 [233] |
| miR-450 | Down | |||||||||||
| miR-3666 | Up | Serum | qPCR | T2DM (60) HC (30) |
Chinese | 45.81 ± 5.92 N/A |
36/24 N/A |
25.12 ± 0.31 N/A |
Diagnosed | N/A | N/A | Tan et al., 2019 [214] |
| miR-146a | Down | Plasma, PBMC | RT-qPCR | T2DM (30) HC (30) |
Iranian | 57 (48–61) 50.5 (45.75–61) |
11/19 9/21 |
27.13 ± 4.15 27.22 ± 3.26 |
8.53 ± 1.29 – |
7.4 (6.7–8) 5.1 (5–5.4) |
N/A | Alipoor et al., 2018 [206] |
| miR-9, miR-375 | Up | Whole blood | RT-qPCR | T2DM (30) HC (30) | Bahrainis | 60 ± 12 56 ± 5.1 |
12/18 14/16 |
25.7 ± 5.2 24.2 ± 4.6 |
15 ± 4.4 – |
8.68 ± 2.6 5.03 ± 0.7 |
0.783 (0.665–0.902) 3 | Al-Muhtaresh et al., 2018 [231] |
| miR-210 | Up | Plasma | RT-qPCR | T2DM (54) HC (20) |
Egyptian | 56.5 ± 7.7 58.1 ± 1.1 |
29/25 11/9 |
30.7 ± 5.3 23.2 ± 0.2 |
10.8 ± 7.8 – |
8.3 ± 1.1 4.8 ± 0.4 |
0.950 87.0; 100.0 |
Amr et al., 2018 [223] |
| miR-126 | Down | 0.960 96.3; 95.0 |
||||||||||
| let-7b 3, miR-29a, miR-144 3 | Up | Plasma | Microarray, RT-qPCR | T2DM (112) HC (94) |
Han Chinese | 54.75 ± 7.53 52.84 ± 8.85 |
69/43 53/41 |
27.11 ± 3.17 23.86 ± 3.27 |
Newly diagnosed | 7.58 ± 1.54 5.16 ± 0.39 |
0.871 (0.822–0.919) 3
79.5; 81.9 |
Liang et al., 2018 [173] |
| miR-142 3 | Down | |||||||||||
| let-7e-5p, let-7f-5p, miR-15b-5p, miR-99b-5p, miR-103a-3p | Up | Whole blood | sRNA-Seq, RT-qPCR | T2DM (12) HC (12) |
South African | 54.8 ± 7.5 52.1 ± 7.8 |
Only female | 33.5 ± 8.9 27.3 ± 5.8 |
Newly diagnosed | N/A | N/A | Matsha et al., 2018 [177] |
| miR-30d | Up | Plasma | RT-qPCR | T2DM (30) HC (30) |
Asian Indian | 50.5 ± 6.3 42.1 ± 7.8 |
21/9 21/9 |
27.3 ± 4.6 27.3 ± 4.7 |
3.10 ± 0.99 – |
8.4 ± 2.0 5.5 ± 0.4 |
N/A | Sucharita et al., 2018 [187] |
| miR-126 | Down | Whole blood | RT-qPCR | T2DM (45) HC (45) |
Bahrainis | 61 ± 12 53 ± 8.6 |
23/22 21/24 |
25.4 ± 4.8 24 ± 4.5 |
16 ± 6 – |
7.4 ± 8.3 3.64 ± 1.1 |
0.932 (0.858–1.000) | Al-Kafaji et al., 2017 [198] |
| miR-148a-3p | Up | Plasma | RT-qPCR | T2DM (9) HC (9) |
Italian | 60.2 ± 8.0 57.9 ± 8.9 |
2/7 4/5 |
29.6 ± 7.8 23.7 ± 3.3 |
Newly diagnosed | 6.4 ± 2.7 5.5 ± 2.4 |
N/A | de Candia et al., 2017 [220] |
| miR-222-3p, miR-342-3p | Down | |||||||||||
| miR-26b, miR-126, miR-140, miR-223 | Down | Plasma, Platelet | RT-qPCR | T2DM (28) HC (23) |
Hungarian | 53 (50–59) 53 (34–60) |
15/13 12/11 |
32.9 (30.3–40.2) 24 (22.1–25.9) |
10 (8.0–14.5) – |
7.5 (7.0–8.8) – |
N/A | Fejes et al., 2017 [147] |
| miR-126-3p | Down | Plasma (MPs) | RT-qPCR | T2DM (68) HC (53) |
Italian | 60 ± 1 57 ± 1 |
42/26 30/23 |
30 ± 1.6 25 ± 0.4 |
>5 – |
N/A | N/A | Giannella et al., 2017 [205] |
| miR-223-3p | Down | PBMC | RT-qPCR | T2DM (16) HC (18) |
Han Chinese | 57 ± 9 53 ± 11 |
8/8 12/6 |
N/A | Newly diagnosed | N/A | N/A | Long et al., 2017 [237] |
| miR-217 | Up | Serum | qPCR | T2DM (186) HC (195) |
Chinese | 54.87 ± 11.65 54.12 ± 9.45 |
95/91 99/96 |
25.30 ± 3.11 25.10 ± 3.27 |
6.39 ± 6.31 – |
8.10 ± 2.09 5.36 ± 0.32 |
N/A | Shao et al., 2017 [219] |
| miR-34a, miR-125b | Up | PBMC | RT-qPCR | T2DM (73) HC (52) |
Chinese | 56.81 ± 11.85 Age-matched |
38/35 Sex-matched |
N/A | 4.54 ± 5.41 – |
8.50 ± 2.09 5.82 ± 1.07 |
N/A | Shen et al., 2017 [236] |
| miR-7 | Up | Serum | RT-qPCR | T2DM (76) HC (74) |
Chinese | 48.5 ± 14.5 48.8 ± 15.2 |
50/26 41/33 |
25.2 ± 3.7 23.0 ± 2.7 |
1.8 ± 2.6 – |
9.9 ± 2.9 5.3 ± 0.4 |
0.76 (0.68–0.83) | Wan et al., 2017 [218] |
| Serum (exosome-free) |
0.75 (0.67–0.83) | |||||||||||
| miR-18a | Up | PBMC | RT-qPCR | T2DM (117) HC (105) |
Chinese | 51.68 ± 8.77 49.26 ± 9.09 |
68/49 58/47 |
27.44 ± 3.08 24.18 ± 2.86 |
Newly diagnosed | 7.51 ± 1.42 5.19 ± 0.42 |
0.851 (0.800–0.902) 3 78.6; 80.0 |
Wang et al., 2017 [212] |
| miR-34c | Down | |||||||||||
| miR-96-5p, miR-144-3p, miR-454-3p, miR-455-5p | Up | Serum | miRNA qPCR array, RT-qPCR | T2DM (10) HC (5) |
Chinese | 58.2 ± 7.7 56.4 ± 3.7 |
4/6 2/3 |
N/A | Newly diagnosed | N/A | N/A | Yang et al., 2017 [176] |
| miR-409-3p, miR-665, miR-766-3p | Down | |||||||||||
| miR-574-3p | Down | Serum | RT-qPCR | T2DM (64) HC (44) |
Ecuadorian | 61 (37–85) 53 (32–87) |
24/40 13/31 |
29.5 (22–49) 28.7 (23–42) |
Diagnosed | 7.0 (3.2–12.5) 5.6 (3.9–6.9) |
N/A | Baldeón et al., 2016 [211] |
| miR-451a, miR-4534 | Up | Serum | Microarray, RT-qPCR | T2DM (154) HC (69) |
Chinese | 61.1 ± 12.4 54.2 ± 10.7 |
70/84 30/39 |
N/A | Newly diagnosed | N/A | N/A | Ding et al., 2016 [178] |
| miR-320d, miR-572, miR-3960 | Down | |||||||||||
| miR-221, miR-222 | Up | Serum | RT-qPCR | T2DM (30) HC (20) |
Chinese | 60.79 ± 11.11 59.78 ± 11.23 |
Only female | 28.88 ± 1.18 20.12 ± 1.69 |
Newly diagnosed | 7.60 ± 0.33 4.56 ± 0.45 |
N/A | Li et al., 2016 [217] |
| miR-30c | Down | Plasma, Platelet | RT-qPCR | T2DM (40) HC (50) |
Han Chinese | 58.6 ± 6.2 52.2 ± 5.5 |
21/29 31/19 |
27.3 ± 4.2 23.6 ± 2.8 |
Diagnosed | 7.3 ± 0.5 5.3 ± 0.2 |
N/A | Luo et al., 2016 [151] |
| miR-21, miR-30d 3, miR-34a 3, miR-148a | Up | Plasma | RT-qPCR | T2DM (31) HC (27) |
American | 52.9 ± 2.0 25.3 ± 2.2 |
15/16 15/12 |
34.1 ± 1.3 24.1 ± 0.9 |
Diagnosed | 6.56 ± 0.11 5.24 ± 0.06 |
0.928 3 90.32; 88.89 |
Seyhan et al., 2016 [216] |
| miR-571, miR-661, miR-770-5p, miR-892b, miR-1303 4 | Up | Serum | TLDA, RT-qPCR | T2DM (92) HC (92) |
Chinese | 47.7 ± 13.9 50.2 ± 14.2 |
58/34 56/36 |
25.6 ± 4.5 23.6 ± 2.0 |
2.1 ± 2.7 – |
9.8 ± 2.9 5.3 ± 0.4 |
0.71 (0.64–0.79) 3 | Wang et al., 2016 [166] |
| miR-125b, miR-126, miR-221 5 | N/A | |||||||||||
| miR-572 | Up | Plasma | Microarray, RT-qPCR | T2DM (50) HC (50) |
Han Chinese | 46.22 ± 6.90 45.52 ± 6.22 |
27/23 22/28 |
25.41 ± 0.32 25.36 ± 0.38 |
Newly diagnosed | 8.69 ± 0.36 5.41 ± 0.29 |
0.843 (0.766–0.920) 87.8; 71.4 |
Yan et al., 2016 [179] |
| miR-320b | Down | 0.946 (0.906–0.985) 92.0; 85.7 |
||||||||||
| miR-1249 | 0.784 (0.685–0.883) 86.0; 77.55 |
|||||||||||
| miR-15a | Down | Whole blood | RT-qPCR | T2DM (24) HC (24) |
Bahrainis | 52 ± 6.0 49 ± 9.1 |
10/14 13/11 |
25.3 ± 1.8 24.2 ± 1.0 |
Diagnosed | 7.5 ± 0.8 4.8 ± 0.6 |
0.864 (0.751–0.977) | Al Kafaji et al., 2015 [222] |
| miR-34c-5p, miR-576-3p | Up | PBMC | Microarray, RT-qPCR | T2DM (64) HC (44) |
Ecuadorian | 61 (37–85) 53 (32–87) |
24/40 13/31 |
29.5 (22–49) 28.7 (23–42) |
Diagnosed | 7.0 (3.2–12.5) 5.6 (3.9–6.9) |
N/A | Baldeón et al., 2015 [181] |
| miR-185 | Down | Plasma | qPCR | T2DM (34) HC (30) |
Mongolian (Chinese) |
N/A | N/A | N/A | Diagnosed | N/A | N/A | Bao et al., 2015, [215] |
| miR-142, miR-143, miR-155, miR-223 | Down | Platelet | RT-qPCR | T2DM (22) HC (22) |
German | 45.7 ± 3.1 41.6 ± 7.5 |
10/12 10/12 |
N/A | Diagnosed | 9.01 ± 0.37 4.98 ± 0.58 |
N/A | Elgheznawy et al., 2015 [148] |
| miR-101, miR-375, miR-802 | Up | Serum | sRNA-Seq (mice), RT-qPCR |
T2DM (155) HC (49) | Japanese | 62.3 ± 13.2 46.0 ± 9.67 |
96/59 25/24 |
25.9 ± 4.97 23.6 ± 4.05 |
Diagnosed | 7.31 ± 1.08 6.03 ± 0.39 |
N/A | Higuchi et al., 2015 [167] |
| miR-10b, miR-130a, miR-143 | Down | Whole blood | Microarray, RT-qPCR | T2DM (12) HC (24) |
Xinjiang Uygurian | 56 ± 10 49 ± 13 |
N/A | 30.9 ± 5.8 26.3 ± 3.6 |
Diagnosed | N/A | N/A | Jiao et al., 2015 [182] |
| miR-146a 6 | Down | PBMC | qPCR | T2DM (35) HC (35) |
Indian | 47.3 ± 7 44.7 ± 6 |
19/16 17/18 |
24.6 ± 2 23.9 ± 2 |
Diagnosed | 7.8 ± 1.5 5.5 ± 0.4 |
N/A | Lenin et al., 2015 [204] |
| miR-103b | Down | Platelet | RT-qPCR | T2DM (43) HC (46) |
Han Chinese | 59 ± 9.3 51.4 ± 9.4 |
19/24 17/29 |
23.3 ± 5.4 21.9 ± 2.9 |
Newly diagnosed | 7.0 ± 1.3 5.1 ± 0.5 |
N/A | Luo et al., 2015 [210] |
| miR-155 | Down | PBMC | RT-qPCR | T2DM (20) HC (20) |
Iranian | 46.5 ± 5.8 47.5 ± 4.4 |
10/10 10/10 |
28.7 ± 4.9 26.2 ± 4.0 |
Diagnosed | 7.02 ± 0.5 5.7 ± 0.7 |
N/A | Mazloom et al., 2015 [209] |
| miR-21-5p, miR-126-3p | Down | Plasma | RT-qPCR | T2DM (76) HC (107) |
Italian | 65.56 ± 6.96 64.25 ± 7.56 |
36/40 49/58 |
28.47 ± 4.34 26.67 ± 5.4 |
Diagnosed | 7.34 ± 1.28 5.96 ± 0.41 |
N/A | Olivieri et al., 2015 [203] |
| miR-130b-3p, miR-374a-5p | Up | Serum | miRNA PCR assay, RT-qPCR | T2DM (49) HC (49) |
Asian Indian | 44.4 ± 8.1 44.3 ± 6.9 |
25/24 26/23 |
25.7 ± 3.5 24.5 ± 2.6 |
Newly diagnosed | 7.8 ± 1.6 5.6 ± 0.4 |
N/A | Prabu et al., 2015 [168] |
| miR-126 | Down | Plasma | qPCR | T2DM (20) HC (20) |
Han Chinese | 61.20 ± 10.62 57.25 ± 9.64 |
13/7 9/11 |
24.53 ± 2.87 23.90 ± 2.34 |
Newly diagnosed | N/A | 0.806 77.78; 66.67 |
Zhang et al., 2015 [197] |
| miR-146a | Down | Serum | RT-qPCR | T2DM (56) HC (40) |
Ecuadorian | 62 (38–85) 54 (32–87) |
22/34 12/28 |
29.2 (22–39) 29.3 (23–42) |
Diagnosed | 7.1 (4.8–12.5) 5.7 (3.9–6.7) |
N/A | Baldeón et al., 2014 [202] |
| miR-126 | Down | Serum | RT-qPCR | T2DM (160) HC (138) |
Chinese | 50.2 ± 6.7 46.7 ± 7.2 |
78/82 67/71 |
23.32 ± 0.31 22.87 ± 0.32 |
Newly diagnosed | 9.16 ± 1.64 4.69 ± 0.57 |
0.792 (0.707–0.877) | Liu et al., 2014 [196] |
| miR-140-5p, miR-142-3p 3, miR-222 | Up | Plasma | Microarray, RT-qPCR | T2DM (48) HC (45) |
Spanish | 54 ± 10 7a 57.7 ± 8 7b 48.1 ± 10.1 8a 50.6 ± 14.4 8b |
Only male | 26.4 ± 2.4 7a 33.4 ± 3.3 7b 25.2 ± 1.8 8a 32.2 ± 2.4 8b |
Diagnosed | 7.67 ± 1.46 7a 7.06 ± 2.14 7b 4.73 ± 0.35 8a 4.81 ± 0.33 8b |
0.975 3 | Ortega et al., 2014 [169] |
| miR-125b, miR-126 3, miR-130b, miR-192, miR-195 3, miR-423-5p 3, miR-532-5p | Down | |||||||||||
| miR-326 | Up | Plasma (exosomes) |
Microarray, RT-qPCR | T2DM (18) HC (12) |
Italian | 57.2 ± 9.6 49.5 ± 12.4 |
12/6 6/6 |
31.6 ± 5.1 32.9 ± 5.4 |
Newly diagnosed | 9.6 ± 1.5 5.7 ± 0.5 |
0.912 (0.799–1.000) 3 | Santovito et al., 2014 [170] |
| let-7a, let-7f | Down | |||||||||||
| miR-375 | Up | Plasma | RT-qPCR | T2DM (100) HC (100) |
Chinese Kazak | 51.33 ± 11.75 48.55 ± 12.41 |
54/46 44/56 |
26.30 ± 4.08 24.44 ± 4.63 |
Diagnosed | N/A | N/A | Sun et al., 2014 [184] |
| miR-199a | Up | Plasma | RT-qPCR | T2DM (64) HC (64) |
Han Chinese | 46–62 | N/A | N/A | Newly diagnosed | N/A | N/A | Yan et al., 2014 [186] |
| miR-23a | Down | Serum | Solexa sequencing, RT-qPCR | T2DM (24) HC (20) |
Han Chinese | 51.13 ± 9.21 46.65 ± 16.18 |
16/8 8/12 |
25.27 ± 2.90 25.55 ± 5.27 |
Newly diagnosed | 9.49 ± 2.45 5.98 ± 0.80 |
0.835 (0.717–0.954) 79.2; 75.0 |
Yang et al., 2014 [171] |
| miR-486 | 0.698 (0.540–0.856) 79.2; 60.0 |
|||||||||||
| let-7i | 0.771 (0.629–0.913) 75.0; 70.0 |
|||||||||||
| miR-96, miR-146a, miR-186, miR-191, miR-192 | N/A | |||||||||||
| miR-146a, miR-155 | Down | PBMC | RT-qPCR | T2DM (20) HC (20) |
Méxican | 40–60 18–28 |
11/9 11/9 |
31.9 ± 7.4 23.1 ± 2.5 |
0–20 | 7.9 ± 1.7 4.8 ± 0.7 |
N/A | Corral-Fernández et al., 2013 [213] |
| miR-146a | Up | Plasma | RT-qPCR | T2DM (90) HC (90) |
Han Chinese | 48.5 (42.0–56.0) 48.00 (41.8–55.0) |
47/43 47/43 |
24.58 ± 3.66 23.38 ± 2.95 |
Newly diagnosed | N/A | 0.725 (0.651–0.799) | Rong et al., 2013 [195] |
| miR-126 | Down | Plasma | qPCR | T2DM (30) HC (30) |
Han Chinese | 63 ± 8.6 61 ± 9 |
16/14 16/14 |
N/A | Newly diagnosed | N/A | N/A | Zhang et al., 2013 [201] |
| miR-27a, miR-150, miR-192, miR-320a | Up | Whole blood | Microarray, RT-qPCR | T2DM (29) HC (29) |
Singaporean | 44.2 ± 8.4 45.7 ± 11.3 |
N/A | 26.5 ± 5.9 23.7 ± 3.2 |
Newly diagnosed | N/A | N/A | Karolina et al., 2012 [172] |
| miR-17, miR-92a, miR-130a, miR-195, miR-197, miR-509-5p, miR-652 | Down | |||||||||||
| miR-146a | Down | PBMC | qPCR | T2DM (20) HC (20) |
Indian | 43.7 ± 5.1 42.0 ± 4.7 |
N/A | 26.4 ± 3.7 25.8 ± 4.0 |
Diagnosed | 7.9 ± 1.8 5.5 ± 0.2 |
N/A | Balasubramanyam et al., 2011 [208] |
| miR-29a, miR-144, miR-150, miR-192, miR-320 | Up | Whole blood | Microarray, RT-qPCR | T2DM (8) HC (7) |
Singaporean | 46.7 ± 3.4 46.3 ± 7.5 |
Only male | 24.5 ± 1.1 22.4 ± 2.3 |
Diagnosed | N/A | N/A | Karolina et al., 2011 [180] |
| miR-30d, miR-146a, miR-182 | Down | |||||||||||
| miR-29a, miR-144, miR-150, miR-192, miR-320 | Up | T2DM (13) HC (8) |
41.0 ± 12.1 43.3 ± 5.7 |
28.0 ± 4.9 24.4 ± 3.1 |
Newly diagnosed | |||||||
| miR-30d, miR-146a, miR-182 | Down | |||||||||||
| miR-9, miR-29a, miR-30d, miR-34a, miR-124a, miR-146a, miR-375 | Up | Serum | RT-qPCR | T2DM (18) HC (19) |
Han Chinese | 47.33 ± 2.62 41.00 ± 2.62 |
9/9 12/7 |
26.26 ± 0.79 26.63 ± 0.80 |
Newly diagnosed | N/A | N/A | Kong et al., 2011 [183] |
| miR-28-3p | Up | Plasma | Microarray, RT-qPCR | T2DM (80) HC (80) |
Italian (Bruneck cohort) |
66.3 ± 8.9 66.3 ± 8.9 |
30/50 30/50 |
28.0 ± 4.4 25.0 ± 4.0 |
Diagnosed | 6.5 ± 1.4 5.4 ± 0.3 |
N/A | Zampetaki et al., 2010 [174] |
| miR-15a, miR-20b, miR-21, miR-24, miR-29b, miR-126, miR-150, miR-191, miR-197, miR-223, miR-320, miR-486 | Down |
1 gender distribution assessed for both studied and control groups; 2 miRNA assessed for 39 patients from control group; 3 values for miRNA panel; 4 miRNA assessed for 68 patients from both T2DM and HC groups; 5 miRNA assessed for 31 patients from both T2DM and HC groups; 6 miRNA assessed for 15 patients from both T2DM and HC groups; 7a non-obese T2DM patients; 7b obese T2DM patients; 8a non-obese controls; 8b obese controls. miRNA—microRNA; T2DM—type 2 diabetes mellitus; HC—healthy controls; PBMC—peripheral blood mononuclear cell; MPs—microparticles; qPCR—quantitative real-time polymerase chain reaction; RT-qPCR—reverse transcription quantitative real-time polymerase chain reaction; sRNA-Seq—small RNA sequencing; TLDA—TaqMan Low Density Array; BMI—body mass index; HbA1c—glycated hemoglobin A1c; AUC—area under the curve; 95% CI—95% confidence interval; SV—sensitivity; SP—specificity; NS—non-significant; N/A—not assessed; N/D—no data.
The results obtained in the determination of miRNA from various biological fluids remain controversial, which is why special attention should be paid to the sample type used in the study when miRNAs are considered as T2DM biomarkers. It has been demonstrated that expression of miR-126 and miR-342 in different blood compartments such as serum, plasma, or whole blood can be both upregulated or downregulated [166,169,194,196,199,232,233]. Similarly, miR-34a has presented an opposite pattern of expression between samples, being increased in plasma or PBMCs, and decreased in whole blood [234,235,236]. However, miR-15a and miR-223 have revealed the same expression profile regardless of the biological fluid used (whole blood, plasma vs. PBMC, plasma, platelets), except for the inverse miR-223 expression in serum [147,148,174,222,237,238,239]. Interestingly, Monfared et al. have investigated the expression of miR-126 and miR-135a in saliva for the first time, suggesting that it is an equally promising non-invasive research material [200]. It should also be noted that miRNA profile may be affected by sex- and ethnicity-associated differences, although data in this field are scarce and require further elucidation [240,241].
Additionally, it is emphasized that the expression of miRNAs may be disparate between the same treatment-naïve T2DM individuals and those during anti-diabetic therapy, but the group of anti-diabetic drugs had no effect on the expression profile of the assessed miRNAs [232,242]. Several studies evaluating miRNAs as candidate biomarkers for T2DM have also noticed the possible usefulness of selected miRNAs for monitoring therapeutic responses in patients with diabetes [169,170,196].
The studies showing the potential clinical utility of miRNAs as biomarkers for T2DM are summarized in Table 1.
4.2. MicroRNAs as Potential Biomarkers for CAD
CAD is one of the most common cardiovascular diseases, caused by atherosclerotic plaque accumulation in the epicardial coronary arteries that lead to a different degree of stenosis or obstruction, which consequently restrict blood flow to the myocardium [243,244]. The disease can be stable and asymptomatic for a long time. Nevertheless, its progressive nature makes it serious due to the risk of an acute atherothrombotic event associated with plaque rapture or erosion that may occur even in clinically apparently silent periods [244,245]. Clinically, CAD can be classified into two subsets as acute coronary syndromes (ACS) and chronic coronary syndromes (CCS), previously known as stable CAD [244].
The diagnosis of stable CAD is often delayed because non-invasive diagnostic methods, including resting electrocardiogram and functional imaging, may be without any significant abnormalities [244]. Currently, in spite of the rising significance of cardiac computed tomography angiography, coronary angiography still remains the gold diagnostic standard to anatomically confirm CAD, but it is an invasive, expensive medical procedure associated with the risk of severe complications [244,246]. Therefore, extensive research on non-invasive, blood-based biomarkers is highly necessary for the early detection of CAD and the prevention of its further phases such as unstable angina, myocardial infarction, and sudden cardiac death [244].
Fichtlscherer et al. were the first to investigate the potential role of circulating miRNAs as biomarkers in patients with stable CAD [247]. The study has revealed that mainly endothelial-related miRNAs exerted significant differences in CAD individuals compared with controls [247]. Eight differentially expressed miRNAs, identified in microarray-based miRNA profiling, were validated in larger cohorts, and these data demonstrated that endothelial-enriched miRNAs miR-126, miR-17, and miR-92a, VSMC-enriched miR-145, inflammatory cell-enriched miR-155 were significantly reduced in both serum and plasma of patients with CAD, whereas cardiomyocyte-enriched miR-133 was elevated only in plasma [247]. These findings suggest that miRNA expression patterns may be distinctive between particular peripheral blood compartments, including serum, plasma, and PBMCs [247,248,249]. Additionally, this research indicates the need to perform pilot miRNA profiling to obtain a set of miRNAs exhibiting the most significant population-specific changes, which is why other ten studies have assessed the differences in hundreds of miRNAs, using mostly microarray profiling and NGS in a single case, in stable CAD as compared to non-CAD subjects [250,251,252,253,254,255,256,257,258,259].
Although many miRNAs show a satisfactory power to discriminate CAD patients from healthy ones as a single parameter [260,261,262,263,264,265,266], it is pertinent to note that the combination of several miRNAs can improve their diagnostic utility [252,253,257,259,267,268,269,270]. Interestingly, a 3-miRNA signature, consisting of miR-29a-3p, miR-574-3p, and miR-574-5p, has been proposed as a reliable marker for the diagnosis of CAD with AUC estimated at 0.916, which turned out to be higher than those calculated for each single miRNAs (0.830, 0.792, and 0.789, respectively) [267]. Similarly, a panel of four plasma-derived miRNAs, including let-7i-5p, miR-26a-5p, miR-32-3p, and miR-3149, has appeared quite promising in distinguishing between CAD and non-CAD patients [252]. The combination of these four miRNAs exhibited better diagnostic performance compared with any individual miRNA, with an AUC of 0.837 [252]. In the study conducted by Dong et al., a set of four lipometabolism-related miRNAs (miR-24, miR-33a, miR-103a, miR-122) isolated from PBMCs revealed the high discriminatory performance for CAD with 84.5% sensitivity and 81.9% specificity (AUC = 0.911) [269]. What is more, all of these parameters, when analyzed together, had better diagnostic accuracy with respect to sensitivity (69.6%, 72.0%, 64.6%, 68.3%) and specificity (65.1%, 67.1%, 60.4%, 65.1%) assessed for every single miRNA [269].
Considering that advanced age is a relevant risk factor for CAD development, the discovery of age-related miRNAs as stable CAD biomarkers seems to be promising due to the atypical symptomatology of the disease and the limitation of invasive diagnostics in elderly patients [271]. It has occurred that upregulated miR-765 had a significant association with the aging of the heart and may be a useful tool in the detection of CAD in the geriatric population (AUC = 0.959) [272]. In turn, the downregulation of serum miR-145-3p, miR-190a-5p, miR-196b-5p, miR-3163-3p, and upregulation of platelet-derived miR-340, miR-624, and endothelial-related miR-451b can help to discriminate patients with atypical, early-onset CAD diagnosed at a young age (at or before 55 years in men or 65 years in women) thus improving the prevention strategies [254,259,273]. Furthermore, Ali Sheikh et al. have revealed that alterations in miR-149, miR-424, and miR-765 expressions might be novel, sensitive (71.8%, 68.7%, 81.5%), and specific (95.3%, 92.3%, 93.7%) predictors for the diagnosis of middle-aged CAD patients [274]. In addition, miR-126 and miR-143 may serve as independent risk factors of CAD [275].
The growing role of miRNAs in the regulation of genes involved in the initiation and progression of coronary occlusion indicates that miRNAs may serve not only as diagnostic biomarkers but also to determine the severity and complexity of CAD. Currently, the extension of coronary atherosclerotic lesions is estimated according to the Gensini score (GS), and the Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery (SYNTAX) score during invasive coronary angiography [268,276,277]. It has been proved that miR-17-5p (r = 0.489), miR-34a (r = 0.327), miR-133a (r = 0.303), miR-155 (r = 0.612), miR-208a (r = 0.853), miR-223 (r = 0.729), and miR-2909 (r = 0.943) expressions gradually increase with the severity of coronary occlusion reflected by GS, whereas downregulated expressions of miR-16 (r = −0.514), miR-126 (r = −0.351 or r = −0.416 according to different authors), miR-210 (r = −0.367), and miR-378 (r = −0.235 or r = −0.422 based on two independent studies), were negatively correlated with GS in patients with CAD (all p < 0.05) [249,264,268,277,278,279,280,281,282,283,284]. Moreover, miR-101a and miR-126-5p were significantly reduced in progressing stages of CAD in accordance with the SYNTAX score. Thus they may be considered biomarkers for evaluating the presence and severity of CAD [276,285]. It has also been reported that miR-145 negatively correlated with higher SYNTAX scores, indicating decreased plasma miR-145 expression with an increase in the severity of CAD [286]. Surprisingly, miR-33 showed notable differences only between the mild form of CAD (SYNTAX score ≤ 22) compared to controls, however, the authors have suggested that it may be the result of the low number of subjects analyzed in case of moderate and severe CAD [287]. That is why the possibility for the role of miR-33 at later stages of CAD cannot be ruled out [287]. Additionally, it has appeared that changes in the expressions of miR-23a, miR-27a, miR-126-5p, and miR-206 were associated with the number of vessels with angiographically documented atherosclerosis [285,288,289,290].
Interestingly, it has occurred that additional use of miR-223 may increase the diagnostic value of established cardiovascular risk factors, including LDL-C, HDL-C, and TG [264]. Similarly, Li et al. displayed a positive correlation between miR-34a and TC, LDL-C, and TG in patients suffering from CAD, highlighting that higher miR-34a expression and LDL-C levels with lower HDL-C levels were independently linked with increased CAD risk [282]. Anyway, a panel of these three parameters has been found to predict CAD risk with AUC greater than those calculated for miRNA alone (AUC = 0.912 vs. AUC = 0.899) [282]. It is worth noting that miR-2909 increases lipid peroxidation and ox-LDL uptake, thereby contributing significantly to the initiation and progression of the atherogenic process in individuals with CAD [277]. Among other miRNAs considered potential biomarkers for stable CAD, miR-20a, miR-30e, miR-92a, miR-101a, miR-122, miR-133a-5p, miR-144-3p, miR-222-5p, and miR-223 are involved in the regulation of cholesterol metabolism [276,291,292,293,294]. Apart from being closely associated with blood lipids, several miRNAs, including miR-16, miR-34a, miR-126, miR-342-5p, and miR-2909, also disclose a correlation with inflammatory- (C-reactive protein, TNF-α, interferon-γ (IFN-γ), IL-1β, IL-6, IL-8, IL-10, IL-17, VCAM-1, ICAM-1) and oxidative stress-related indexes (ROS) [277,279,281,282,295]. The studies showing the potential relevance of miRNAs as biomarkers for stable CAD are summarized in Table 2.
Table 2.
Circulating microRNAs as potential biomarkers for stable coronary artery disease.
| miRNA | Expression Change | Sample Type | Assay Method | Number of Samples |
Ethnicity | Age [Years] | Gender (Male/Female, n) |
BMI [kg/m2] | Value of Biomarker AUC (95% CI); SV [%]; SP [%] |
Author, Year (Reference) |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-9-5p | Up | Serum | RT-qPCR | CAD (40) HC (20) |
Iranian | 42.36 ± 5.2 42.9 ± 4.6 |
19/21 4/16 |
N/A | 0.693 (0.530–0.857) 74.07; 53.33 |
Gholipour et al., 2022 [260] |
| miR-182-5p | 0.752 (0.608–0.897) 74.19; 66.67 |
|||||||||
| miR-27a | Up | Plasma | qPCR | CAD (30) HC (30) |
Iranian | 57.6 ± 20.32 55.30 ± 8.40 |
Only male | 24.8–30.0 24.66–27.0 |
0.67 (0.54–0.81) 86.7; 46.7 |
Hosseinpor et al., 2022 [288] |
| miR-146a | Down | N/A | ||||||||
| miR-34a | Up | Plasma | RT-qPCR | CAD (203) HC (100) |
Chinese | 61.5 ± 9.4 62.0 ± 6.7 |
158/45 75/25 |
24.2 ± 3.0 23.8 ± 3.1 |
0.899 (0.865–0.934) 76.4; 90.0 |
Li et al., 2022 [282] |
| miR-122 | Up | Serum | RT-qPCR | CAD (100) HC (100) |
Indian | 52.15 ± 1.13 50.90 ± 2.08 |
75/25 74/26 |
>26 (44.0%) >26 (33.0%) |
0.806 64.0; 84.0 |
Ali et al., 2021 [261] |
| miR-126 | Down | 0.806 61.54; 80.0 |
||||||||
| miR-200a-3p, miR-382-3p, miR-432-5p, miR-3613-3p | Up | Plasma (exosomes) |
NGS | CAD (52) HC (52) |
Han Chinese | 65.04 ± 10.68 60.65 ± 11.26 |
43/9 34/18 |
26.87 ± 3.80 26.42 ± 3.57 |
N/A | Chang et al., 2021 [250] |
| miR-125a-5p, miR-151a-3p, miR-185-5p, miR-328-3p | Down | |||||||||
| miR-122 | Down | Serum | RT-qPCR | CAD (78) HC (60) |
Indian | 52.07 ± 9.94 50.13 ± 8.12 |
N/A | 25.9 ± 4.8 25.2 ± 4.7 |
N/A | Mishra et al., 2021 [291] |
| miR-101a | Down | Serum | RT-qPCR | CAD (200) HC (100) |
Chinese | 62 (31–87) 1a 59 (37–80) 1c 60 (40–86) |
74/26 1a 78/22 1c 73/27 |
N/A | N/A | Yu et al., 2021 [276] |
| miR-23a, miR-27a | Up | PBMC | qPCR | CAD (82) HC (80) |
Iranian | 60.24 ± 0.91 57.40 ± 0.94 |
35/47 41/39 |
27.60 ± 0.48 26.23 ± 0.50 |
N/A | Babaee et al., 2020 [289] |
| miR-21 | Up | Plasma | RT-qPCR | CAD (24) HC (54) |
Indian | N/A | N/A | N/A | 0.780 (0.670–0.890) | Kumar et al., 2020 [262] |
| miR-133b | Down | CAD (28) HC (54) |
0.746 (0.620–0.870) | |||||||
| miR-21 | Up | PBMC | RT-qPCR | CAD (56) HC (29) |
Turkish | 58.96 ± 8.95 56.93 ± 6.35 |
41/15 9/20 |
N/A | N/A | Sanlialp et al., 2020 [248] |
| miR-155, miR-221 | Down | |||||||||
| miR-10a-5p | Down | Serum | sRNA-Seq, RT-qPCR |
CAD (39) HC (39) |
Chinese | 63.1 ± 7.4 59.3 ± 6.8 |
21/18 21/18 |
24.2 ± 2.3 23.2 ± 2.2 |
0.817 (0.715–0.918) | Wang et al., 2020 [251] |
| miR-423-3p | 0.656 (0.532–0.779) | |||||||||
| miR-423-3p | CAD (30) HC (21) |
63.9 ± 7.95 57.24 ± 5.35 |
15/15 9/12 |
24.57 ± 2.37 25.01 ± 2.02 |
0.808 (0.684–0.932) | |||||
| miR-16 | Down | Plasma, PBMC | RT-qPCR | CAD (40) HC (40) |
Chinese | 63.33 ± 5.63 61.20 ± 5.82 |
Only male | 26.69 ± 2.85 25.85 ± 3.12 |
N/A | Wang et al., 2020 [281] |
| miR-29a-3p, miR-574-3p, miR-574-5p | Up | Plasma | qPCR | CAD (88) HC (67) |
Chinese | 61.66 ± 1.32 63.72 ± 0.99 |
55/33 40/27 |
26.18 ± 0.50 24.89 ± 0.68 |
0.916 (0.856–0.957) 2 | Zhang et al., 2020 [267] |
| let-7i-5p, miR-26a-5p, miR-32-3p, miR-3149 | Up | Plasma | Microarray, RT-qPCR |
CAD (40) HC (69) |
Chinese | 56.2 ± 7.6 55.0 ± 6.5 |
30/10 43/26 |
N/A | 0.837 (0.763–0.911) 2 | Zhang et al., 2020 [252] |
| miR-32-5p | Up | Serum (exosomes) |
qPCR | CAD (20) HC (20) |
Chinese | 64 (52–68) 57 (52–62) |
14/6 12/8 |
24.7 ± 2.9 24.0 ± 3.0 |
0.691 (0.525–0.858) 85.0; 55.0 |
Zhang et al., 2020 [263] |
| miR-149-5p | 0.702 (0.536–0.869) 70.0; 75.0 |
|||||||||
| miR-942-5p | 0.693 (0.527–0.858) 80.0; 60.0 |
|||||||||
| miR-133a-5p, miR-144-3p, miR-222-5p | Up | Plasma | RT-qPCR | CAD (46) HC (43) |
Turkish | 60.02 ± 10.01 55.26 ± 13.85 |
34/12 28/15 |
27.87 (25.04–30.43) 27.10 (24.38–29.42) |
N/A | Gorur et al., 2019 [292] |
| miR-378 | Down | Plasma | RT-qPCR | CAD (215) HC (52) |
Chinese | 61 ± 10 61 ± 12 |
153/62 30/22 |
23 ± 10 22 ± 10 |
0.789 (0.728–0.851) | Li et al., 2019 [283] |
| miR-451b | Up | Serum | RT-qPCR | CAD (30) HC (30) |
Chinese | 46–59 45–58 |
15/15 15/15 |
N/A | N/A | Lin et al., 2019 [273] |
| miR-30c | Down | Plasma | qPCR | CAD (34) HC (32) |
Han Chinese | 60.9 ± 5.3 58.6 ± 8.1 |
18/16 17/15 |
24.57 ± 3.01 24.49 ± 2.30 |
0.895 (0.811–0.978) | Luo et al., 2019 [224] |
| miR-33 | Up | Plasma | RT-qPCR | CAD (30) HC (30) |
Indian | 54.7 ± 8.7 56.17 ± 9.18 |
15/15 15/15 |
26.67 ± 3.7 27.47 ± 4.35 |
N/A | Reddy et al., 2019 [287] |
| miR-30e, miR-92a | Up | Plasma (exosomes) |
RT-qPCR | CAD (42) HC (42) |
Chinese | 63 63 |
22/20 22/20 |
N/A | N/A | Wang et al., 2019 [293] |
| miR-206 | Up | Plasma | qPCR | CAD (100) HC (30) |
Iranian | 57 ± 9 55 ± 8 |
87/13 16/14 |
27.78 ± 3.45 27.45 ± 2.09 |
N/A | Zehtabian et al., 2019 [290] |
| miR-342-5p | Up | PBMC | qPCR | CAD (82) HC (80) |
Iranian | 60.10 ± 0.89 57.86 ± 0.97 |
35/47 41/39 |
27.56 ± 0.47 26.14 ± 0.49 |
0.702 (0.620–0.783) | Ahmadi et al., 2018 [295] |
| miR-20a, miR-92a, miR-223 | Up | Plasma | RT-qPCR | CAD (19) HC (6) |
Australian | 65.2 ± 10.7 59.0 ± 5.1 |
19/0 5/1 |
N/A | N/A | Choteau et al., 2018 [294] |
| miR-223 | Up | Plasma | RT-qPCR | CAD (300) HC (100) |
Chinese | 56.2 | N/A | N/A | 0.933 (0.905–0.961)86.0; 91.3 | Guo et al., 2018 [264] |
| miR-155 | Up | Serum | RT-qPCR | CAD (300) HC (100) |
Chinese | N/A | N/A | N/A | N/A | Qiu et al., 2018 [249] |
| miR-126 | Down | PBMC | qPCR | CAD (119) HC (96) |
Chinese | 59 ± 11 57 ± 10 |
36/83 27/69 |
24.6 ± 3.9 23.8 ± 3.4 |
0.801 (0.740–0.861) 70.6; 85.4 |
Wu at al., 2018 [279] |
| miR-221-3p | Down | Serum | qPCR | CAD (89) HC (93) |
Turkish | 58.97 ± 13.79 57.07 ± 9.80 |
N/A | NS | 0.623 (0.539–0.702) 76.27; 49.43 |
Yilmaz et al., 2018 [265] |
| miR-222-3p | 0.654 (0.571–0.731) 69.49; 54.02 |
|||||||||
| miR-17-5p, miR-92a, miR-126, miR-210, miR-378 | Down | Plasma | RT-qPCR | CAD (102) HC (92) |
Chinese | 60.2 ± 11.4 57.9 ± 14.8 |
21/81 26/66 |
24.2 ± 3.7 23.6 ± 3.5 |
0.756 (0.687–0.725) 2 84.3; 60.9 |
Zhang et al., 2018 [268] |
| miR-24, miR-33a, miR-103a, miR-122 | Up | PBMC | RT-qPCR | CAD (161) HC (149) |
Chinese | 61.35 ± 7.10 61.08 ± 7.51 |
86/75 72/77 |
25.77 ± 3.06 24.81 ± 3.29 |
0.911 (0.880–0.942) 2 84.5; 81.9 |
Dong et al., 2017 [269] |
| let-7c, miR-145, miR-155 | Down | Plasma | OpenArray RT-qPCR, RT-qPCR |
CAD (69) HC (32) |
French (South-western) | 58.4 ± 9.0 57.3 ± 11.6 |
Only male | 27.3 ± 4.4 26.6 ± 3.1 |
0.708 (0.600–0.811) 2 75.76; 63.33 |
Faccini et al., 2017 [253] |
| miR-126, miR-143, miR-145 3 |
Up | PBMC | RT-qPCR | CAD (450) HC (450) |
Chinese | < 60 (26.8%/25.6%) 60–80 (68.0%/67.8%) ≥ 80 (5.2%/6.7%) |
234/216 215/235 |
18.5–24.9 (89.9%/94.3%) 25.0–29.9 (10.1%/5.7%) |
N/A | Lin et al., 2017 [275] |
| miR-208a | Up | Plasma | RT-qPCR | CAD (290) HC (110) |
Chinese | N/A | N/A | N/A | 0.919 (0.893–0.945) 75.5; 93.6 |
Zhang et al., 2017 [278] |
| miR-133a | Up | Plasma | RT-qPCR | CAD (79) HC (63) |
Chinese | 58 ± 12 55 ± 11 |
57/22 39/24 |
24.8 ± 4.12 24.0 ± 3.7 |
0.597 (0.504–0.691) 29.1; 92.5 |
Zhu, 2017 [284] |
| miR-145-3p | Down | Serum | miSript miRNA PCR Array, RT-qPCR |
CAD (40) HC (40) |
Han Chinese | 34.20 ± 5.93 36.58 ± 3.96 |
37/3 36/4 |
28.01 ± 4.90 27.33 ± 2.75 |
0.753 (0.643–0.863) 67.50; 82.10 |
Du et al., 2016 [254] |
| miR-190a-5p | 0.782 (0.680–0.884) 70.00; 75.00 |
|||||||||
| miR-196b-5p | 0.824 (0.731–0.917) 85.00; 72.50 |
|||||||||
| miR-3163-3p | 0.758 (0.651–0.864) 57.50; 84.60 |
|||||||||
| miR-126-5p | Down | Plasma | qPCR | CAD (110) HC (40) |
Chinese | 66.5 ± 11.7 1a 67.4 ± 9.7 1b 68.9 ± 11.3 1c 64.0 ± 10.4 |
67/43 28/12 |
24.9 ± 2.7 1a 24.4 ± 3.0 1b 25.2 ± 3.2 1c 23.9 ± 3.5 |
N/A | Li et al., 2016 [285] |
| miR-208a, miR-370 | Up | Plasma | RT-qPCR | CAD (95) HC (50) |
Chinese | 65 (44–78) 65 (46–75) |
65/30 34/16 |
23 (20–26) 22 (20–24) |
0.856 (0.796–0.917) 2 73.7; 86.0 |
Liu et al., 2016 [270] |
| miR-15a-5p | Up | Plasma | RT-qPCR | CAD (50) HC (50) |
Irish | 65 ± 9 60 ± 13 |
43/7 42/8 |
27.69 ± 3.31 26.96 ± 2.96 |
0.67 | O’Sullivan et al., 2016 [266] |
| miR-16-5p | 0.68 | |||||||||
| miR-93-5p | 0.75 | |||||||||
| miR-146a-5p | Down | 0.65 | ||||||||
| miR-206 | Up | Plasma | Microarray, RT-qPCR |
CAD (67) HC (67) |
Chinese | 64.70 ± 6.79 63.69 ± 5.96 |
43/24 32/35 |
N/A | 0.607 (0.508–0.706) | Zhou et al., 2016 [255] |
| miR-574-5p | 0.696 (0.609–0.787) | |||||||||
| miR-765 | Up | Plasma | RT-qPCR | CAD (37) HC (20) |
Chinese | 72.97 ± 4.28 71.7 ± 5.2 |
25/12 10/10 |
23.08 ± 3.03 22.29 ± 1.49 |
0.959 | Ali Sheikh et al., 2015 [272] |
| miR-149 | Down | 0.938 | ||||||||
| miR-765 | Up | Plasma | RT-qPCR | CAD (65) HC (32) |
Chinese | 53 (49–57) 53 (49–57) |
38/27 16/16 |
22 (19–25) 22 (20–23) |
0.968 (0.939–0.996) 81.5; 93.7 |
Ali Sheikh et al., 2015 [274] |
| miR-149 | Down | 0.938 (0.894–0.983) 71.8; 95.3 |
||||||||
| miR-424 | 0.919 (0.863–0.975) 68.7; 92.3 |
|||||||||
| miR-17-5p | Up | Plasma | qPCR | CAD (59) NS-CAD (33) HC (20) |
Chinese | 65.07 ± 10.55 65.23 ± 7.46 55.90 ± 4.72 |
40/19 18/15 7/13 |
N/A | 0.894 (0.780–0.968) | Chen et al., 2015 [280] |
| miR-145 | Down | Plasma | RT-qPCR | CAD (26) HC (28) |
Chinese | N/A | N/A | N/A | N/A | Gao et al., 2015 [286] |
| miR-21, miR-34a | Up | Plasma | Microarray (mice), RT-qPCR |
CAD (32) HC (20) |
Chinese | 67 ± 11 62 ± 8 |
Only male | 24.1 ± 3.7 23.6 ± 4.0 |
N/A | Han et al., 2015 [256] |
| miR-23a | Down | |||||||||
| miR-2909 | Up | PBMC | RT-qPCR | CAD (80) HC (20) |
Iranian | 50 ± 4 49 ± 8 |
Only male | N/A | N/A | Arora et al., 2014 [277] |
| miR-208, miR-215, miR-487a, miR-502 | Up | Serum | TLDA, RT-qPCR |
CAD (92) HC (34) |
Chinese | 65.2 ± 10.5 59.4 ± 13.1 |
53/39 15/19 |
25.83 ± 1.48 24.82 ± 2.72 |
0.909 (0.858–0.960) 2 83.7; 82.4 |
Wang et al., 2014 [257] |
| miR-29b | Down | |||||||||
| miR-1 | Up | Plasma | Microarray, RT-qPCR |
CAD (34) HC (20) |
Italian | 60.0 ± 10.6 62.5 ± 2.1 |
30/4 19/1 |
N/A | 0.918 | D’Alessandra et al., 2013 [258] |
| miR-126 | 0.929 | |||||||||
| miR-485-3p | 0.851 | |||||||||
| miR-340, miR-624 | Up | Platelet | Microarray, RT-qPCR |
CAD (40) HC (40) |
Dutch | 51.4 ± 4.7 51.0 ± 4.6 |
Only male | N/A | 0.71 (0.59–0.83) 2 (combined with miR-451, miR-454) |
Sondermeijer et al., 2011 [259] |
| miR-133 | Up | Plasma | RT-qPCR | CAD (36) HC (17) |
German | 67.69 ± 11.07 32.18 ± 8.78 |
25/11 6/11 |
> 25 (38.7%) > 25 (23.5%) |
N/A | Fichtlscherer et al., 2010 [247] |
| miR-17, miR-92a, miR-126, miR-145, miR-155, miR-199a | Down | |||||||||
| miR-17, miR-92a, miR-126, miR-145, miR-155 | Down | Serum | CAD (31) HC (14) |
68.06 ± 9.66 39.28 ± 17.52 |
21/10 5/9 |
N/A |
1a Syntax Score ≤ 22; 1b Syntax Score > 22 and ≤ 32; 1c Syntax Score > 32; 2 values for miRNA panel; 3 miRNA assessed for 70 patients from CAD and HC groups. miRNA—microRNA; CAD—coronary artery disease; HC—healthy controls; PBMC—peripheral blood mononuclear cell; qPCR—quantitative real-time polymerase chain reaction; RT-qPCR—reverse transcription quantitative real-time polymerase chain reaction; NGS—next-generation sequencing; sRNA-Seq—small RNA sequencing; TLDA—TaqMan Low Density Array; BMI—body mass index; AUC—area under the curve; 95% CI—95% confidence interval; SV—sensitivity; SP—specificity; NS—non-significant; N/A—not assessed.
4.3. MicroRNAs as Potential Biomarkers for CAD Related to T2DM
Accumulating evidence has demonstrated that patients with T2DM are more prone to develop subsequent CAD than individuals without diabetes [296,297]. It is worth noting that T2DM shares a number of well-established risk factors with CAD, involving mainly dyslipidemia, hypertension, and obesity [297,298]. These cardiometabolic features, along with coexisting chronic low-grade inflammation, hypercoagulability, and increased oxidative stress under a hyperglycemic milieu, may contribute to the approximate doubling of CAD risk in patients with T2DM [298]. It has led to the ‘common soil’ hypothesis, postulating that both conditions have a common pathogenetic background [296]. Thus, revealing the underlying molecular mechanism of T2DM promoting the pathological progression of cardiovascular disease will not only help to alleviate the cardiovascular damage caused by diabetes-accelerated atherosclerosis but also to identify the candidate molecules for stratifying the risk of CAD in T2DM.
To date, more than 120 studies (Table 1 and Table 2) have attempted to explore the potential of miRNAs as a diagnostic tool for T2DM or CAD in clinical practice. However, there are only a few reports on miRNA-based biomarkers for the early detection of CAD in asymptomatic patients with T2DM (Table 3). As shown in Figure 3, only ten of the 39 miRNAs overlapping in T2DM, and CAD were evaluated for their relevance in the detection of CAD in T2DM. Nonetheless, distinct sets of unique circulating miRNAs were also discovered for each disease (94 miRNAs for T2DM and 67 miRNAs for CAD).
Table 3.
Circulating microRNAs as potential biomarkers for stable coronary artery disease related to type 2 diabetes mellitus.
| miRNA | Expression Change | Sample Type | Assay Method | Number of Samples |
Ethnicity | Age [Years] | Gender (Male/Female, n) |
BMI [kg/m2] | Duration of T2DM [Years] |
HbA1c [%] | Value of Biomarker AUC (95% CI); SV [%]; SP [%] |
Author, Year (Reference) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-499 | Down | Serum | RT-qPCR | T2DM+CAD (60) T2DM (60) HC (60) |
Egyptian | Age-matched | Sex-matched | BMI-matched | N/A | N/D | 0.720 73.0; 70.0 |
Oraby et al., 2022 [226] |
| miR-19a-3p | Up | Plasma (EVs) |
sRNA-Seq, RT-qPCR |
T2DM+CAD (32) HC (20) |
Chinese | Age-matched | Sex-matched | N/A | N/A | N/D | 0.698 (0.530–0.866) 2 53.9; 85.7 |
Zhang et al., 2022 [303] |
| miR-15a-3p | Down | 0.874 (0.765–0.982) 2 88.5; 71.4 |
||||||||||
| miR-18a-5p | 0.871 (0.760–0.982) 2 80.8; 78.6 |
|||||||||||
| miR-133a-3p | 0.745 (0.567–0.922) 2 88.5; 57.1 |
|||||||||||
| miR-155-5p | 0.901 (0.800–1.000) 2 92.3; 78.6 |
|||||||||||
| miR-210-3p | 0.786 (0.647–0.925) 2 61.5; 92.9 |
|||||||||||
| miR-1, miR-133 | Up | Whole blood | RT-qPCR | T2DM+CAD (30) T2DM (30) HC (30) |
Bahrainis | 60 ± 12 58 ± 11.5 56 ± 5.1 |
15/15 12/18 14/16 |
25.35 ± 4.4 25.7 ± 5.2 24.2 ± 4.6 |
15 ± 4.4 14 ± 9.3 – |
8.68 ± 2.6 7.09 ± 1.06 5.03 ± 0.7 |
0.752 (0.626–0.879) 1,3 | Al-Muhtaresh et al., 2019 [301] |
| miR-1 | 0.912 (0.828–0.995) 2 | |||||||||||
| miR-133 | 0.920 (0.842–0.998) 2 | |||||||||||
| miR-30c | Down | Plasma | qPCR | T2DM+CAD (27) T2DM (47) CAD (34) HC (32) |
Han Chinese |
64.5 ± 6.5 60.5 ± 11.1 60.9 ± 5.3 58.6 ± 8.1 |
17/10 23/24 18/16 17/15 |
25.02 ± 3.12 24.76 ± 3.29 24.57 ± 3.01 24.49 ± 2.30 |
Newly diagnosed |
9.13 ± 1.01 9.15 ± 1.02 6.28 ± 0.69 5.36 ± 0.35 |
0.474 (0.355–0.593) 1 70.2; 52.0 0.972 (0.940–1.000) 2 90.9; 85.2 |
Luo et al., 2019 [224] |
| miR-342 | Up | Serum | RT-qPCR | T2DM+CAD (50) T2DM (50) CAD (50) HC (50) |
Egyptian | 62.30 ± 0.61 62.06 ± 1.26 62.32 ± 0.56 62.22 ± 0.69 |
Only female | 28.87 ± 0.33 27.58 ± 0.28 27.88 ± 0.23 23.82 ± 0.14 |
12.06 ± 0.30 12.06 ± 0.30 – – |
11.92 ± 0.18 10.75 ± 0.17 9.73 ± 0.17 4.10 ± 0.68 |
0.781 1 80.0; 72.0 |
Seleem et al., 2019 [233] |
| miR-450 | Down | 0.824 1 72.0; 78.0 |
||||||||||
| miR-92a | Up | Serum | RT-qPCR | T2DM+CAD (117) T2DM (69) HC (68) |
Chinese | 64.73 ± 8.22 64.29 ± 3.77 62.98 ± 7.42 |
79/38 48/21 45/23 |
26.44 ± 3.31 25.61 ± 5.76 24.08 ± 2.42 |
N/A | 8.22 ± 2.64 6.80 ± 2.41 5.29 ± 0.33 |
0.866 1 76.9; 88.4 0.958 2 78.6; 98.5 |
Wang et al., 2019 [300] |
| miR-210 | Up | Plasma | RT-qPCR | T2DM+CAD (46) T2DM (54) HC (20) |
Egyptian | 57.0 ± 6.2 56.5 ± 7.7 58.1 ± 1.1 |
23/23 29/25 11/9 |
29.7 ± 3.5 30.7 ± 5.3 23.2 ± 0.2 |
11.2 ± 5.2 10.8 ± 7.8 – |
9.4 ± 1.0 8.3 ± 1.1 4.8 ± 0.4 |
0.980 1 93.5; 100.0 0.980 2 97.8; 100.0 |
Amr et al., 2018 [223] |
| miR-126 | Down | 0.970 1 91.3; 100.0 0.980 2 97.8; 95.0 |
||||||||||
| miR-126 | Down | Whole blood | RT-qPCR | T2DM+CAD (45) T2DM (45) HC (45) |
Bahrainis | 64 ± 11.7 61 ± 12 53 ± 8.6 |
24/21 23/22 21/24 |
26.1 ± 4.3 25.4 ± 4.8 24 ± 4.5 |
18 ± 9.3 16 ± 6 – |
9.6 ± 3.2 7.4 ± 8.3 3.64 ± 1.1 |
0.807 (0.714–0.900) 1 0.948 (0.894–1.000) 2 |
Al-Kafaji et al., 2017 [198] |
| miR-9 4, miR-370 | Up | Serum | RT-qPCR | T2DM+CAD (50) T2DM (50) CAD (50) HC (50) |
Egyptian | 62.30 ± 0.45 62.06 ± 1.26 62.32 ± 0.56 62.22 ± 0.69 |
35/15 32/18 38/12 36/14 |
28.87 ± 0.32 27.58 ± 0.27 27.88 ± 0.23 23.82 ± 0.13 |
12.06 ± 0.30 12.22 ± 0.30 – – |
11.0–12.5 5 | N/A 84.0; 84.0 3 |
Motawae et al., 2015 [302] |
1 value of miRNA to distinguish T2DM+CAD patients from T2DM ones; 2 value of miRNA to distinguish T2DM+CAD patients from HC; 3 values for miRNA panel; 4 significant differences were observed between T2DM+CAD patients and those from both CAD and HC groups; 5 HbA1c assessed for all T2DM patients. miRNA—microRNA; T2DM—type 2 diabetes mellitus; CAD—coronary artery disease; T2DM+CAD—type 2 diabetes mellitus with coronary artery disease; HC—healthy controls; EVs—extracellular vesicles; qPCR—quantitative real-time polymerase chain reaction; RT-qPCR—reverse transcription quantitative real-time polymerase chain reaction; sRNA-Seq—small RNA sequencing; BMI—body mass index; AUC—area under the curve; 95% CI—95% confidence interval; SV—sensitivity; SP—specificity; N/A—not assessed; N/D—no data.
Figure 3.
Venn diagram of differentially expressed microRNAs in patients with T2DM, CAD, and CAD related to T2DM. Overlapping fields indicate miRNAs that have been associated with two of these pathologies. In the center of the figure, ten miRNAs are highlighted with different expressions that have been detected in T2DM, CAD and T2DM with CAD. Created with BioRender.com.
One of the most thoroughly studied miRNAs in both T2DM and CAD is the anti-inflammatory miR-126, which is responsible for maintaining endothelial homeostasis and vascular integrity. The expression of miR-126 in whole blood and plasma has been observed to be remarkably decreased by almost 1.9–4.6-fold between T2DM patients with and without CAD and 7.7–13.1-fold between T2DM with CAD and healthy individuals [198,223]. The obtained intergroup differences allowed miR-126 to be proposed as a potential biomarker of CAD occurrence in T2DM patients with a discriminatory ability ranging from AUC 0.807 to 0.970, according to two different studies [198,223]. Moreover, miR-126 showed a negative correlation with the concentration of LDL-C (r = −0.575, p < 0.0001 or r = −0.46, p = 0.001), TC (r = −0.48, p = 0.001), fasting plasma glucose (r = −0.92, p < 0.001), HbA1c (r = −0.81, p < 0.001), whereas a positive one was observed in case of HDL-C (r = 0.41, p = 0.005), which directly proves association of this miRNA with known risk factors of CAD in T2DM [198,223]. It is worth emphasizing that patients with simultaneously reduced expression of miR-126 and HDL-C levels were more likely to develop diabetes complicated by CAD [223]. In addition, it has been reported that individuals with T2DM exhibiting lower levels of miR-126 were at a higher risk of comorbid CAD and previous major cardiovascular events compared to those subjects suffering from other diabetes-related complications [203,299].
According to recent scientific reports, other promising candidates, miR-92a, miR-342, and miR-450, seem to play a beneficial role in the prediction of CAD in patients with T2DM because, such as miR-126, they are strongly involved in inflammatory and oxidative stress pathways [233,300]. Seelem et al. have revealed significant upregulation of miR-342 expression and downregulation of miR-450 expression in the serum of individuals with T2DM, CAD, and T2DM-related CAD compared to controls [233]. Additionally, these miRNAs displayed a proatherogenic nature due to the observed valid correlations with dyslipidemic (TG, TC, LDL-C, HDL-C), anthropometric (BMI, waist-to-hip ratio; WHR) variables, and glucose homeostasis indices (HbA1c, fasting plasma glucose, diabetes duration) [233]. Interestingly, miR-342 and miR-450 were associated with the activity of NADPH oxidase 4 (NOX-4), an enzyme involved in promoting ROS production and CRP concentration, unrevealing the underlying molecular pathomechanisms during the development of CAD in patients with T2DM [233]. Both miR-342 and miR-450 demonstrated good or very good discriminatory power for CAD in T2DM (AUC = 0.781, 80.0% sensitivity, 72.0% specificity and AUC = 0.824, 72.0% sensitivity, 78.0% specificity, respectively) [233]. In turn, Wang et al. have explored miR-92a as a potential biomarker for CAD in diabetes and a contributor to CAD onset through the activation of NF-κB, a critical regulator of inflammation, and its downstream inflammatory pathways [300]. Progressively increasing expression of miR-92a in serum from healthy individuals via T2DM to T2DM with CAD showed positive correlations with NF-κB p65 expression and the level of proinflammatory cytokines, including ET-1, MCP-1, and ICAM-1, in patients with T2DM and CAD [300]. What is more, logistic regression analysis revealed a strong association of miR-92a expression with CAD in T2DM, showing an OR of 15.835 (95% CI, 6.307–39.754; p < 0.001), thus indicating miR-92a as an independent risk factor for T2DM-related CAD [300]. miR-92a may help predict CAD in T2DM with a calculated AUC of 0.866 (76.9% sensitivity and 88.4% specificity) [300].
It is pertinent to note that individuals with T2DM are characterized by abnormal fibrinolytic activity with elevated levels of PAI-1, known as an independent risk factor for hypercoagulable and thrombotic events in this group of patients [224]. Luo et al. have observed decreased expression of plasma miR-30c in T2DM patients with CAD as compared to those without this vascular complication [224]. Furthermore, miR-30c negatively correlated with circulating PAI-1 (r = −0.733, p < 0.0001) and the degree of coronary artery lesions evaluated by the GS criteria (r = −0.782, p < 0.0001) in the T2DM with CAD group [224]. Interestingly, patients with T2DM and CAD had more advanced coronary lesions, even than those diagnosed with CAD, supporting the hypothesis of T2DM-accelerated atherosclerosis [224]. In line with ROC analysis results, miR-30c exerted excellent capability to distinguish T2DM-CAD from healthy controls with an AUC of 0.972 (90.9% sensitivity and 85.2% specificity) [224]. Yet, the diagnostic value of miR-30c in the early detection of CAD among T2DM patients occurred to be poor (AUC of 0.474, 70.2% sensitivity and 52.0% specificity) [224].
Regarding miRNAs as possible diagnostic biomarkers for CAD in highly susceptible individuals with T2DM, it is relevant to consider especially those miRNAs that strictly correspond to their origin and the injury of a given tissue. So far, three cardiomyocyte-enriched miRNAs, miR-1, miR-133, and miR-499, revealed a good or very good discriminatory power to identify CAD in patients with T2DM [226,301]. Although several miRNAs having obtained good performance in detecting CAD in T2DM patients, it should be mentioned that miRNAs may target multiple genes and regulate different signaling pathways. Therefore, a single miRNA might be insufficient for diagnostic purposes, and the miRNA panels can better reflect the complex pathophysiology of T2DM-related CAD. It turned out that the combined signature composed of two upregulated miRNAs, miR-9 and miR-370, demonstrated higher sensitivity and specificity (84.0%, 84.0%) in the identification of CAD in T2DM compared to each miRNA alone (72.0% sensitivity, 82.0% specificity and 26.0% sensitivity, 82.0% specificity, respectively) [302]. Although the two-miRNA panel consisting of miR-1 and miR-133 achieved good discriminatory performance for CAD in T2DM (AUC = 0.752), the diagnostic value for miR-1 (AUC = 0.802) alone was even better than for the miRNA panel, probably due to the only marginally statistically significant AUC for miR-133 [301].
So far, only Zhang et al. have aimed to explore miRNA expression patterns in T2DM-CAD patients and identify novel miRNA molecules as disease biomarkers [303]. Based on miRNA profiling, five downregulated EV-carried miRNAs (miR-15a-3p, miR-18a-5p, miR-133a-3p, miR-155-5p, miR-210-3p) and upregulated miR-19a-3p have been established as promising biomarkers for CAD related to T2DM with satisfactory or even excellent values of AUC [303].
As mentioned previously, a distinct miRNA profile may be observed depending on the body fluids used. It has been demonstrated that miR-210 (downregulated in EVs or upregulated in plasma according to different studies) showed better predictive value when assessed in plasma (AUC = 0.786 vs. AUC = 0.980, respectively), therefore further research is needed to determine the exact blood compartment for future clinical evaluation [223,303].
5. MicroRNAs as Potential Therapeutic Targets in T2DM and CAD
Given the emerging role of miRNAs in the regulation of several steps in the disease pathway, they may not only pave the way for novel diagnostic strategies in T2DM and CAD but also seem to be attractive therapeutic targets. The ability to modulate miRNA expression by repressing pathological miRNAs or overexpressing protective miRNAs has led to the development of therapies based on miRNA inhibitors, such as locked nucleic acid (LNA) antimiRs, antagomiRs, and miRNA sponges, or miRNA mimics, respectively [304]. Currently, miRNA inhibitors are the most common in vivo approach when designing miRNA-based therapies [304].
One of the pioneering studies on the potential application of antimiRs was the study conducted by Trajkovski et al., who administered a recombinant adenovirus expressing 2′O-methylmodified antimiR-103 and -107 (15 mg/kg for two consecutive days) to ob/ob mice [305]. It has been observed that this therapy leads to a decrease in plasma glucose and insulin levels and improves glucose tolerance and sensitivity, especially in the liver and adipose tissue [305]. Of note, a GalNAc-conjugated oligonucleotide targeting miR-103/-107 (RG-125, AZD4076) has entered phase I/IIa randomized, single-blind, placebo-controlled clinical trial (NCT02826525) investigating the safety and tolerability of AZD4076 and assessing its effect on insulin sensitivity and liver fat content in patients with T2DM and non-alcoholic fatty liver disease, although there are still no results in the available literature. Recent studies have demonstrated that intravenous, subcutaneous, or intraperitoneal injections with antimiR-34a, antagomiR-132, LNA-29, and LNA-181a antimiRs reduce blood glucose levels, decrease inflammation, improve insulin secretion and its hepatic sensitivity in a mouse model of obesity [306,307,308,309]. Interestingly, subcutaneous administration of antagomir-22 (APT-110) at a dose of 15 mg/kg on days 0, 2, 4 of week 1 of treatment and then once a week for a total of 8 or 12 weeks in mice with diet-induced obesity provides a sustained increase in energy expenditure, reduction in body mass of 30% and liver steatosis, decreases blood glucose, insulin, cholesterol, leptin levels, and alleviates insulin resistance [310,311]. Based on in vivo experimental studies (db/db mice, rats with streptozocin-induced T2DM), it turned out that long-term injections with LNA-21 or antagomiR-21 may reduce body weight, pericardial fat, adipocyte size, improve glucose homeostasis (decrease HOMA-IR, HbA1c, blood glucose concentration, elevate plasma adiponectin level) and lipid metabolism parameters [312,313]. Moreover, this potential miRNA-based drug has revealed no apparent liver and kidney toxicity nor negative effects on cardiac function [312]. Among the miRNA mimics tested so far in the T2DM animal model, miR-125a, miR-145, and miR-383 agomiRs have been found to be able to improve glucose tolerance and lipid homeostasis indices as well as decrease inflammation in T2DM [103,314,315,316].
In vivo studies have also indicated that the regulation of miRNA expression may influence the development and progression of atherosclerosis and, thus, CAD. It has been observed that subcutaneous or intravenous administration of antagomiR-155 to mice with atherosclerosis leads to a reduction in macrophages in atherosclerotic plaque, suppresses its development, and decreases the level of serum inflammatory markers without significant effect on blood lipids and body weight [317,318]. Moreover, Wei et al. have revealed diminished macrophage and VSMC content in atherosclerotic plaque after injection of LNA-342 antimiR (25 mg/kg), which also inhibited miR-155 expression [319]. Similar effects have been obtained after intravenous administration of antagomiR-133b, resulting in a wider vascular lumen, more stable plaque size, thicker fibrous cap, smaller lipid core, and reduced macrophage immune response [320]. Another tested miRNA-based therapy with a potential vasoprotective effect in both diabetic and atherosclerotic mice is antagomiR-92a, which reduces oxidative stress and the content of macrophages, T lymphocytes in atherosclerotic plaque, but without a positive impact on lipid parameters [76,321,322]. Preclinical studies on the usefulness of antimiR-33 seemed promising, as short-term subcutaneous injections of antimiR-33 at a dose of 5–10 mg/kg led to a reduction in local and systemic inflammation as well as atherosclerotic plaque size with a simultaneous improvement in plaque stability or plasma HDL-C level [111,323,324,325,326]. Nevertheless, long-term antimiR-33 therapy might cause deleterious effects, including moderate steatosis and hypertriglyceridemia [327]. Furthermore, administration of antimiR-122, LNA-148a, or miR-30c mimic has been found to reduce TC, LDL-C, and TG levels, increase HDL-C level, diminish plaque size and hepatic steatosis, and prolong time to occlusion in in vivo mouse experimental models [151,328,329,330]. There are also several in vivo studies assessing the potential anti-inflammatory and atheroprotective ability of therapy based on antagomiR-17-5p, agomiR-188-3p, agomiR-200a, or agomiR-532-3p [331,332,333,334]. Interestingly, the administration of antagomiR-449a has revealed protective effects against atherosclerosis in diabetic mice [335].
Despite many studies that have been conducted in vivo on an animal experimental model of T2DM and CAD, there are still no ongoing registered clinical trials at clinicaltrials.gov which aimed to test miRNA-based therapeutics in humans. The clinical application of these promising next-generation drugs requires solving several critical issues, including the potential of miRNAs to interact with multiple pathways/targets, the risks of off-target toxicity, and the need for efficient and safe vectors or delivery systems to transport miRNA-regulating agents into target cells.
6. Conclusions and Future Directions
Over the past decade, an increasing number of reports have highlighted that aberrant expression of miRNAs may impair diverse signaling pathways underlying the pathophysiology of cardiometabolic diseases such as T2DM and CAD. Findings from preclinical evidence collected in this review strongly indicate both atheroprotective and atherogenic effects of different miRNAs targeting specific steps in the initiation and progression of hyperglycemia-induced atherosclerosis as a key player linking T2DM with CAD. Although a list of circulating miRNAs involved in T2DM and CAD development is still expanding, the existing data concerning their expression patterns are conflicting. Hence, to better evaluate the potential usefulness of miRNAs as diagnostic and prognostic tools in CAD related to T2DM, the group of candidate miRNA-based biomarkers in T2DM and CAD alone has also been provided. Among the 122 differentially expressed miRNAs that were analyzed in this paper, 14 top miRNAs appear to be the most consistently dysregulated miRNAs in T2DM (miR-135/-a, miR-148a/-b, miR-191, miR-195, miR-197, miR-375, miR-486, miR-766, miR-770) and CAD (miR-32, miR-206, miR-208/-a, miR-378, miR-765) posing them as promising biomarker candidates. miR-1, miR-9, miR-15a, miR-30, miR-92a, miR-126, miR-133a, miR-155, miR-210, and miR-342 were found to have the potential to arrive at miRNA-based risk stratification and early, non-invasive diagnosis of CAD among T2DM patients. Of note, these miRNAs are involved in the core processes associated with T2DM-induced atherosclerosis, mainly endothelial dysfunction, inflammation, VSMC proliferation/migration, and platelet hyperactivity. It may be a helpful suggestion for further studies in the search for novel miRNA-based biomarkers and therapeutic strategies in T2DM patients with CAD.
Nevertheless, the translation of these molecules into clinical practice faces many challenges. To avoid the discrepancies between miRNA expression profiles with generally poor reproducibility observed in available studies, establishing standardized miRNA detection technologies, efficient normalization, and validation methods are required. Based on the fact that miRNAs exhibit unique and stable patterns in various body fluids, determining the appropriate biological material (whole blood, plasma, serum, PBMCs, or saliva) for miRNA measurement, is necessary to incorporate miRNAs into clinical management. The ‘one gene, one protein, one disease’ paradigm may not fully reflect the complex pathophysiology of CAD related to T2DM. Thus, the use of a panel of two or more selected circulating miRNAs can be more effective and guarantee higher specificity and sensitivity in the detection of CAD among T2DM individuals. Therefore, the success of the implementation of miRNAs in a personalized approach to the diagnosis of T2DM-related CAD will mostly depend on further simultaneous studies in larger and well-defined populations to evaluate and strengthen their significance as promising biomarkers.
Abbreviations
| ABCA1 | ATP-binding cassette: sub-family A, member 1 |
| ACS | acute coronary syndrome |
| ADAM10 | a disintegrin and metalloproteinase domain-containing protein 10 |
| AGEs | advanced-glycation end-products |
| AGO | Argonaute |
| Agr-1 | argininase-1 |
| AMPK | adenosine monophosphate-activated protein kinase |
| ApoA-1 | apolipoprotein A-1 |
| AUC | area under the curve |
| BMI | body mass index |
| CAC | coronary artery calcification |
| CAD | coronary artery disease |
| CAV-1 | caveolin 1 |
| CCL-2 | chemokine CC-motif ligand 2 |
| CCS | chronic coronary syndrome |
| CI | confidence interval |
| circRNA | circular RNA |
| COX | cyclooxygenase |
| DAXX | death-domain associated protein |
| DGCR8 | DiGeorge syndrome Critical Region 8 |
| EC | endothelial cell |
| ECM | extracellular matrix |
| EDCF | endothelium-derived cyclooxygenase-dependent contracting factor |
| EMP2 | epithelial membrane protein 2 |
| EndMT | endothelial-to-mesenchymal transition |
| eNOS | endothelial nitric oxide synthase |
| EPC | endothelial progenitor cell |
| ERK | extracellular signal-regulated kinase |
| ET-1 | endothelin-1 |
| EVs | extracellular vesicles |
| EXP5 | Exportin-5 |
| FSP-1 | fibroblast specific protein-1 |
| GLUT-4 | glucose transporter-4 |
| GS | Gensini score |
| HbA1c | glycated hemoglobin A1c |
| HDL-C | high-density lipoprotein cholesterol |
| HMGB1 | high mobility group box-1 |
| HSC70 | heat shock cognate 70 |
| HSP90 | heat shock protein 90 |
| ICAM-1 | intercellular adhesion molecule-1 |
| IGF-1R | insulin-like growth factor-1 receptor |
| IFN-γ | interferon-γ |
| IL | interleukin |
| IRAK1 | interleukin 1 receptor associated kinase 1 |
| IRS-1 | insulin receptor substrate 1 |
| JNK | c-Jun N-terminal kinase |
| KLF | Krüppel-like factor |
| KRAS | Kirsten Rat Sarcoma Viral Oncogene Homolog |
| LDL-C | low-density lipoprotein cholesterol |
| LNA | locked nucleic acid |
| lncRNA | long non-coding RNA |
| α-SMA | alpha smooth muscle actin |
| MAPK | mitogen-activated protein kinase |
| MCP-1 | monocyte chemoattractant protein-1 |
| MEK | mitogen-activated protein kinase |
| MGO | methylglyoxal |
| MPO | myeloperoxidase |
| mRNA | messenger RNA |
| miRNA, miR | microRNA |
| mTOR | mammalian target of rapamycin |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| ncRNA | non-coding RNA |
| NF-κB | nuclear factor-kappa B |
| NGS | next-generation sequencing |
| NO | nitric oxide |
| NPM1 | nucleophosmin 1 |
| OR | odds ratio |
| ox-LDL | oxidized low-density lipoprotein |
| PAI-1 | plasminogen activator inhibitor-1 |
| PBMC | peripheral blood mononuclear cell |
| PDGF | platelet-derived growth factor |
| PDGFRβ | platelet-derived growth factor receptor beta |
| PECAM-1 | platelet endothelial cell adhesion molecule-1 |
| PHLPP2 | PH domain leucine-rich repeat protein phosphatase 2 |
| PI3K | phosphatidylinositol 3-kinase |
| piRNA | PIWI-interacting RNA |
| PKC | protein kinase C |
| pre-miRNA | precursor miRNA |
| pri-miRNA | primary miRNA |
| RAGE | receptor for advanced glycation end-products |
| RISC | RNA-induced silencing complex |
| RNA pol II | RNA polymerase II |
| ROC | receiver operating characteristic |
| ROCK1 | Rho-associated coiled-coil forming protein kinase 1 |
| ROS | reactive oxygen species |
| RT-qPCR | real-time polymerase chain reaction |
| siRNA | small interfering RNA |
| SM22α | smooth muscle protein 22 alpha |
| sncRNA | small non-coding RNA |
| Spred-1 | sprouty-related, EVH1 domain-containing protein 1 |
| sRAGE | soluble receptor for advanced glycation end-products |
| SREBP | sterol regulatory element-binding protein |
| SRF | serum-response factor |
| SYNTAX | Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery |
| T2DM | type 2 diabetes mellitus |
| TC | total cholesterol |
| TG | triglycerides |
| TGF-β | transforming growth factor-β |
| TNF-α | tumor necrosis factor α |
| TPM1 | tropomyosin 1 |
| TRAF6 | tumor necrosis factor receptor associated factor 6 |
| TRBP | trans-activation response RNA binding protein |
| 3′ UTR | 3′ untranslated region |
| WHR | waist-to-hip ratio |
| VCAM-1 | vascular cell adhesion molecule-1 |
| VE-cadherin | vascular endothelial cadherin |
| VEGF | vascular endothelial growth factor |
| VSMC | vascular smooth muscle cell |
| vWF | von Willebrand Factor |
Author Contributions
Conceptualization, J.S. and B.M.-M.; resources, J.S.; writing—original draft preparation, J.S.; writing—review and editing, J.S. and B.M.-M.; visualization, J.S.; supervision, B.M.-M.; funding acquisition, J.S. and B.M.-M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the statutory funds (DS 358/2020-2022 and PBsd 120/2021-2022) of the Medical University of Lublin.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Wild S., Roglic G., Green A., Sicree R., King H. Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Chen L., Magliano D.J., Zimmet P.Z. The worldwide epidemiology of type 2 diabetes mellitus—Present and future perspectives. Nat. Rev. Endocrinol. 2011;8:228–236. doi: 10.1038/nrendo.2011.183. [DOI] [PubMed] [Google Scholar]
- 3.Lascar N., Brown J., Pattison H., Barnett A.H., Bailey C.J., Bellary S. Type 2 diabetes in adolescents and young adults. Lancet Diabetes Endocrinol. 2018;6:69–80. doi: 10.1016/S2213-8587(17)30186-9. [DOI] [PubMed] [Google Scholar]
- 4.Sun H., Saeedi P., Karuranga S., Pinkepank M., Ogurtsova K., Duncan B.B., Stein C., Basit A., Chan J.C.N., Mbanya J.C., et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2021;183:109119. doi: 10.1016/j.diabres.2021.109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henning R.J. Type-2 diabetes mellitus and cardiovascular disease. Future Cardiol. 2018;14:491–509. doi: 10.2217/fca-2018-0045. [DOI] [PubMed] [Google Scholar]
- 6.Peer N., Balakrishna Y., Durao S. Screening for type 2 diabetes mellitus. Cochrane Database Syst. Rev. 2020;5:CD005266. doi: 10.1002/14651858.CD005266.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogurtsova K., Guariguata L., Barengo N.C., Ruiz P.L.-D., Sacre J.W., Karuranga S., Sun H., Boyko E.J., Magliano D.J. IDF diabetes Atlas: Global estimates of undiagnosed diabetes in adults for 2021. Diabetes Res. Clin. Pract. 2021;183:109118. doi: 10.1016/j.diabres.2021.109118. [DOI] [PubMed] [Google Scholar]
- 8.Galicia-Garcia U., Benito-Vicente A., Jebari S., Larrea-Sebal A., Siddiqi H., Uribe K.B., Ostolaza H., Martín C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020;21:6275. doi: 10.3390/ijms21176275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chawla A., Chawla R., Jaggi S. Microvasular and macrovascular complications in diabetes mellitus: Distinct or continuum? Indian J. Endocrinol. Metab. 2016;20:546–553. doi: 10.4103/2230-8210.183480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gedebjerg A., Almdal T.P., Berencsi K., Rungby J., Nielsen J.S., Witte D.R., Friborg S., Brandslund I., Vaag A., Beck-Nielsen H., et al. Prevalence of micro- and macrovascular diabetes complications at time of type 2 diabetes diagnosis and associated clinical characteristics: A cross-sectional baseline study of 6958 patients in the Danish DD2 cohort. J. Diabetes Complicat. 2018;32:34–40. doi: 10.1016/j.jdiacomp.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Dal Canto E., Ceriello A., Rydén L., Ferrini M., Hansen T.B., Schnell O., Standl E., Beulens J.W. Diabetes as a cardiovascular risk factor: An overview of global trends of macro and micro vascular complications. Eur. J. Prev. Cardiol. 2019;26:25–32. doi: 10.1177/2047487319878371. [DOI] [PubMed] [Google Scholar]
- 12.Goldfine A.B., Phua E.-J., Abrahamson M.J. Glycemic Management in Patients With Coronary Artery Disease and Prediabetes or Type 2 Diabetes Mellitus. Circulation. 2014;129:2567–2573. doi: 10.1161/CIRCULATIONAHA.113.006634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartnik M., Rydén L., Ferrari R., Malmberg K., Pyörälä K., Simoons M., Standl E., Soler-Soler J., Öhrvik J., The Euro Heart Survey Investigators The prevalence of abnormal glucose regulation in patients with coronary artery disease across Europe. The Euro Heart Survey on diabetes and the heart. Eur. Heart J. 2004;25:1880–1890. doi: 10.1016/j.ehj.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 14.Khot U.N., Khot M.B., Bajzer C.T., Sapp S.K., Ohman E.M., Brener S.J., Ellis S.G., Lincoff A.M., Topol E.J. Prevalence of Conventional Risk Factors in Patients With Coronary Heart Disease. JAMA. 2003;290:898–904. doi: 10.1001/jama.290.7.898. [DOI] [PubMed] [Google Scholar]
- 15.Munkhaugen J., Hjelmesæth J., Otterstad J.E., Helseth R., Sollid S.T., Gjertsen E., Gullestad L., Perk J., Moum T., Husebye E., et al. Managing patients with prediabetes and type 2 diabetes after coronary events: Individual tailoring needed—A cross-sectional study. BMC Cardiovasc. Disord. 2018;18:160. doi: 10.1186/s12872-018-0896-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laakso M. Cardiovascular disease in type 2 diabetes from population to man to mechanisms: The Kelly West Award Lecture 2008. Diabetes Care. 2010;33:442–449. doi: 10.2337/dc09-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Einarson T.R., Acs A., Ludwig C., Panton U.H. Prevalence of cardiovascular disease in type 2 diabetes: A systematic literature review of scientific evidence from across the world in 2007–2017. Cardiovasc. Diabetol. 2018;17:83. doi: 10.1186/s12933-018-0728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan L., Liu C., Hu J., Liu Y., Wang J., Chen G., Li Z., Chen H. Epigenetic mechanisms in coronary artery disease: The current state and prospects. Trends Cardiovasc. Med. 2018;28:311–319. doi: 10.1016/j.tcm.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Poznyak A., Grechko A.V., Poggio P., Myasoedova V.A., Alfieri V., Orekhov A.N. The Diabetes Mellitus–Atherosclerosis Connection: The Role of Lipid and Glucose Metabolism and Chronic Inflammation. Int. J. Mol. Sci. 2020;21:1835. doi: 10.3390/ijms21051835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazzana N., Ranalli P., Cuccurullo C., Davì G. Diabetes mellitus and thrombosis. Thromb. Res. 2012;129:371–377. doi: 10.1016/j.thromres.2011.11.052. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Chen J., Sen S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016;231:25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y., Ding Y., Liang B., Lin J., Kim T.-K., Yu H., Hang H., Wang K. A Systematic Study of Dysregulated MicroRNA in Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2017;18:456. doi: 10.3390/ijms18030456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi R., Chen Y., Liao Y., Li R., Lin C., Xiu L., Yu H., Ding Y. Research Status of Differentially Expressed Noncoding RNAs in Type 2 Diabetes Patients. Biomed Res. Int. 2020;2020:3816056. doi: 10.1155/2020/3816056. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.González-Sánchez L.E., Ortega-Camarillo C., Contreras-Ramos A., Barajas-Nava L.A. miRNAs as biomarkers for diagnosis of type 2 diabetes: A systematic review. J. Diabetes. 2021;13:792–816. doi: 10.1111/1753-0407.13166. [DOI] [PubMed] [Google Scholar]
- 25.Romaine S.P.R., Tomaszewski M., Condorelli G., Samani N.J. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart. 2015;101:921–928. doi: 10.1136/heartjnl-2013-305402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P., Wu W., Chen Q., Chen M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019;16:20190027. doi: 10.1515/jib-2019-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Q., Chen Y., Zhou X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019;51:11–17. doi: 10.1016/j.cbpa.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 30.Meola N., Gennarino V.A., Banfi S. microRNAs and genetic diseases. Pathogenetics. 2009;2:7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niaz S. The AGO proteins: An overview. Biol. Chem. 2018;399:525–547. doi: 10.1515/hsz-2017-0329. [DOI] [PubMed] [Google Scholar]
- 32.Witkos T.M., Koscianska E., Krzyzosiak W.J. Practical Aspects of microRNA Target Prediction. Curr. Mol. Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 34.Stavast C.J., Erkeland S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells. 2019;8:1465. doi: 10.3390/cells8111465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrera-Carrillo E., Berkhout B. Dicer-independent processing of small RNA duplexes: Mechanistic insights and applications. Nucleic Acids Res. 2017;45:10369–10379. doi: 10.1093/nar/gkx779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gareev I., Beylerli O., Yang G., Sun J., Pavlov V., Izmailov A., Shi H., Zhao S. The current state of MiRNAs as biomarkers and therapeutic tools. Clin. Exp. Med. 2020;20:349–359. doi: 10.1007/s10238-020-00627-2. [DOI] [PubMed] [Google Scholar]
- 37.Redis R.S., Calin S., Yang Y., You M.J., Calin G.A. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol. Ther. 2012;136:169–174. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 39.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 40.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He L., Hannon G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 42.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 43.Gennarino V.A., Sardiello M., Mutarelli M., Dharmalingam G., Maselli V., Lago G., Banfi S. HOCTAR database: A unique resource for microRNA target prediction. Gene. 2011;480:51–58. doi: 10.1016/j.gene.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H.-Y., Lin Y.-C.-D., Li J., Huang K.-Y., Shrestha S., Hong H.-C., Tang Y., Chen Y.-G., Jin C.-N., Yu Y., et al. miRTarBase 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2020;48:D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 46.Alles J., Fehlmann T., Fischer U., Backes C., Galata V., Minet M., Hart M., Abu-Halima M., Grässer F.A., Lenhof H.-P., et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019;47:3353–3364. doi: 10.1093/nar/gkz097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan T., Yang T., Chen H., Fu D., Hu Y., Wang J., Yuan Q., Yu H., Xu W., Xie X. New insights into oxidative stress and inflammation during diabetes mellitus-accelerated atherosclerosis. Redox Biol. 2019;20:247–260. doi: 10.1016/j.redox.2018.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansson G.K., Libby P., Tabas I. Inflammation and plaque vulnerability. J. Intern. Med. 2015;278:483–493. doi: 10.1111/joim.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aronson D., Edelman E.R. Revascularization for coronary artery disease in diabetes mellitus: Angioplasty, stents and coronary artery bypass grafting. Rev. Endocr. Metab. Disord. 2010;11:75–86. doi: 10.1007/s11154-010-9135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.La Sala L., Prattichizzo F., Ceriello A. The link between diabetes and atherosclerosis. Eur. J. Prev. Cardiol. 2019;26:15–24. doi: 10.1177/2047487319878373. [DOI] [PubMed] [Google Scholar]
- 51.Giordo R., Ahmed Y.M.A., Allam H., Abusnana S., Pappalardo L., Nasrallah G.K., Mangoni A.A., Pintus G. EndMT Regulation by Small RNAs in Diabetes-Associated Fibrotic Conditions: Potential Link With Oxidative Stress. Front. Cell Dev. Biol. 2021;9:683594. doi: 10.3389/fcell.2021.683594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H., Xu Q., Thakur A., Alfred M.O., Chakraborty M., Ghosh A., Yu X. Endothelial dysfunction in diabetes and hypertension: Role of microRNAs and long non-coding RNAs. Life Sci. 2018;213:258–268. doi: 10.1016/j.lfs.2018.10.028. [DOI] [PubMed] [Google Scholar]
- 53.Petrie J.R., Guzik T.J., Touyz R.M. Diabetes, Hypertension, and Cardiovascular Disease: Clinical Insights and Vascular Mechanisms. Can. J. Cardiol. 2018;34:575–584. doi: 10.1016/j.cjca.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nigro C., Mirra P., Prevenzano I., Leone A., Fiory F., Longo M., Cabaro S., Oriente F., Beguinot F., Miele C. miR-214-Dependent Increase of PHLPP2 Levels Mediates the Impairment of Insulin-Stimulated Akt Activation in Mouse Aortic Endothelial Cells Exposed to Methylglyoxal. Int. J. Mol. Sci. 2018;19:522. doi: 10.3390/ijms19020522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirra P., Nigro C., Prevenzano I., Procopio T., Leone A., Raciti G.A., Andreozzi F., Longo M., Fiory F., Beguinot F., et al. The role of miR-190a in methylglyoxal-induced insulin resistance in endothelial cells. Biochim. Biophys. Acta-Mol. Basis Dis. 2017;1863:440–449. doi: 10.1016/j.bbadis.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 56.Piperi C., Goumenos A., Adamopoulos C., Papavassiliou A.G. AGE/RAGE signalling regulation by miRNAs: Associations with diabetic complications and therapeutic potential. Int. J. Biochem. Cell Biol. 2015;60:197–201. doi: 10.1016/j.biocel.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Geroldi D., Falcone C., Emanuele E. Soluble Receptor for Advanced Glycation End Products: From Disease Marker to Potential Therapeutic Target. Curr. Med. Chem. 2006;13:1971–1978. doi: 10.2174/092986706777585013. [DOI] [PubMed] [Google Scholar]
- 58.Shi Y., Vanhoutte P.M. Macro- and microvascular endothelial dysfunction in diabetes. J. Diabetes. 2017;9:434–449. doi: 10.1111/1753-0407.12521. [DOI] [PubMed] [Google Scholar]
- 59.Tang S., Wang F., Shao M., Wang Y., Zhu H. MicroRNA-126 suppresses inflammation in endothelial cells under hyperglycemic condition by targeting HMGB1. Vascul. Pharmacol. 2017;88:48–55. doi: 10.1016/j.vph.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 60.Meng S., Cao J.-T., Zhang B., Zhou Q., Shen C.-X., Wang C.-Q. Downregulation of microRNA-126 in endothelial progenitor cells from diabetes patients, impairs their functional properties, via target gene Spred-1. J. Mol. Cell. Cardiol. 2012;53:64–72. doi: 10.1016/j.yjmcc.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Jansen F., Yang X., Hoelscher M., Cattelan A., Schmitz T., Proebsting S., Wenzel D., Vosen S., Franklin B.S., Fleischmann B.K., et al. Endothelial Microparticle–Mediated Transfer of MicroRNA-126 Promotes Vascular Endothelial Cell Repair via SPRED1 and Is Abrogated in Glucose-Damaged Endothelial Microparticles. Circulation. 2013;128:2026–2038. doi: 10.1161/CIRCULATIONAHA.113.001720. [DOI] [PubMed] [Google Scholar]
- 62.Li Y., Zhou Q., Pei C., Liu B., Li M., Fang L., Sun Y., Li Y., Meng S. Hyperglycemia and Advanced Glycation End Products Regulate miR-126 Expression in Endothelial Progenitor Cells. J. Vasc. Res. 2016;53:94–104. doi: 10.1159/000448713. [DOI] [PubMed] [Google Scholar]
- 63.Meng S., Cao J., Zhang X., Fan Y., Fang L., Wang C., Lv Z., Fu D., Li Y. Downregulation of MicroRNA-130a Contributes to Endothelial Progenitor Cell Dysfunction in Diabetic Patients via Its Target Runx3. PLoS ONE. 2013;8:e68611. doi: 10.1371/journal.pone.0068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Q., Meng S., Liu B., Li M.-Q., Li Y., Fang L., Li Y.-G. MicroRNA-130a regulates autophagy of endothelial progenitor cells through Runx3. Clin. Exp. Pharmacol. Physiol. 2014;41:351–357. doi: 10.1111/1440-1681.12227. [DOI] [PubMed] [Google Scholar]
- 65.Ye M., Li D., Yang J., Xie J., Yu F., Ma Y., Zhu X., Zhao J., Lv Z. MicroRNA-130a Targets MAP3K12 to Modulate Diabetic Endothelial Progenitor Cell Function. Cell. Physiol. Biochem. 2015;36:712–726. doi: 10.1159/000430132. [DOI] [PubMed] [Google Scholar]
- 66.Zeng J., Xiong Y., Li G., Liu M., He T., Tang Y., Chen Y., Cai L., Jiang R., Tao J. MiR-21 is Overexpressed in Response to High Glucose and Protects Endothelial Cells from Apoptosis. Exp. Clin. Endocrinol. Diabetes. 2013;121:425–430. doi: 10.1055/s-0033-1345169. [DOI] [PubMed] [Google Scholar]
- 67.La Sala L., Mrakic-Sposta S., Micheloni S., Prattichizzo F., Ceriello A. Glucose-sensing microRNA-21 disrupts ROS homeostasis and impairs antioxidant responses in cellular glucose variability. Cardiovasc. Diabetol. 2018;17:105. doi: 10.1186/s12933-018-0748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao M., Yu M., Zhao J., Mei J., Pan Y., Zhang J., Wu H., Yu M., Liu F., Chen G. miR-21-3p regulates AGE/RAGE signalling and improves diabetic atherosclerosis. Cell Biochem. Funct. 2020;38:965–975. doi: 10.1002/cbf.3523. [DOI] [PubMed] [Google Scholar]
- 69.Liu X., Wang D., Yang X., Lei L. Huayu Tongmai Granules protects against vascular endothelial dysfunction via up-regulating miR-185 and down-regulating RAGE. Biosci. Rep. 2018;38:BSR20180674. doi: 10.1042/BSR20180674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao J., Ailifeire M., Wang C., Luo L., Zhang J., Yuan L., Zhang L., Li X., Wang M. miR-320/VEGFA axis affects high glucose-induced metabolic memory during human umbilical vein endothelial cell dysfunction in diabetes pathology. Microvasc. Res. 2020;127:103913. doi: 10.1016/j.mvr.2019.103913. [DOI] [PubMed] [Google Scholar]
- 71.Jiang Z., Wu J., Ma F., Jiang J., Xu L., Du L., Huang W., Wang Z., Jia Y., Lu L., et al. MicroRNA-200a improves diabetic endothelial dysfunction by targeting KEAP1/NRF2. J. Endocrinol. 2020;245:129–140. doi: 10.1530/JOE-19-0414. [DOI] [PubMed] [Google Scholar]
- 72.Zhang H., Liu J., Qu D., Wang L., Luo J.-Y., Lau C.W., Liu P., Gao Z., Tipoe G.L., Lee H.K., et al. Inhibition of miR-200c Restores Endothelial Function in Diabetic Mice Through Suppression of COX-2. Diabetes. 2016;65:1196–1207. doi: 10.2337/db15-1067. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y., Guan Q., Jin X. miR-200c serves an important role in H5V endothelial cells in high glucose by targeting Notch1. Mol. Med. Rep. 2017;16:2149–2155. doi: 10.3892/mmr.2017.6792. [DOI] [PubMed] [Google Scholar]
- 74.Li Q., Kim Y.-R., Vikram A., Kumar S., Kassan M., Gabani M., Lee S.K., Jacobs J.S., Irani K. P66Shc-Induced MicroRNA-34a Causes Diabetic Endothelial Dysfunction by Downregulating Sirtuin1. Arterioscler. Thromb. Vasc. Biol. 2016;36:2394–2403. doi: 10.1161/ATVBAHA.116.308321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu J., Liang W., Tian Y., Ma F., Huang W., Jia Y., Jiang Z., Wu H. Inhibition of P53/miR-34a improves diabetic endothelial dysfunction via activation of SIRT1. J. Cell. Mol. Med. 2019;23:3538–3548. doi: 10.1111/jcmm.14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gou L., Zhao L., Song W., Wang L., Liu J., Zhang H., Huang Y., Lau C.W., Yao X., Tian X.Y., et al. Inhibition of miR-92a Suppresses Oxidative Stress and Improves Endothelial Function by Upregulating Heme Oxygenase-1 in db/db Mice. Antioxid. Redox Signal. 2018;28:358–370. doi: 10.1089/ars.2017.7005. [DOI] [PubMed] [Google Scholar]
- 77.Shen X., Li Y., Sun G., Guo D., Bai X. miR-181c-3p and -5p promotes high-glucose-induced dysfunction in human umbilical vein endothelial cells by regulating leukemia inhibitory factor. Int. J. Biol. Macromol. 2018;115:509–517. doi: 10.1016/j.ijbiomac.2018.03.173. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Z., Collado A., Sun C., Tratsiakovich Y., Mahdi A., Winter H., Chernogubova E., Seime T., Narayanan S., Jiao T., et al. Downregulation of Erythrocyte miR-210 Induces Endothelial Dysfunction in Type 2 Diabetes. Diabetes. 2022;71:285–297. doi: 10.2337/db21-0093. [DOI] [PubMed] [Google Scholar]
- 79.Brennan E., Wang B., McClelland A., Mohan M., Marai M., Beuscart O., Derouiche S., Gray S., Pickering R., Tikellis C., et al. Protective Effect of let-7 miRNA Family in Regulating Inflammation in Diabetes-Associated Atherosclerosis. Diabetes. 2017;66:2266–2277. doi: 10.2337/db16-1405. [DOI] [PubMed] [Google Scholar]
- 80.Yuan J., Chen M., Xu Q., Liang J., Chen R., Xiao Y., Fang M., Chen L. Effect of the Diabetic Environment On the Expression of MiRNAs in Endothelial Cells: Mir-149-5p Restoration Ameliorates the High Glucose-Induced Expression of TNF-α and ER Stress Markers. Cell. Physiol. Biochem. 2017;43:120–135. doi: 10.1159/000480330. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Q., Xiao X., Zheng J., Li M., Yu M., Ping F., Wang T., Wang X. Vildagliptin, a dipeptidyl peptidase-4 inhibitor, attenuated endothelial dysfunction through miRNAs in diabetic rats. Arch. Med. Sci. 2019;17:1378–1387. doi: 10.5114/aoms.2019.86609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng J., Liu Q., Hu N., Zheng F., Zhang X., Ni Y., Liu J. Downregulation of hsa_circ_0068087 ameliorates TLR4/NF-κB/NLRP3 inflammasome-mediated inflammation and endothelial cell dysfunction in high glucose conditioned by sponging miR-197. Gene. 2019;709:1–7. doi: 10.1016/j.gene.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 83.Feng B., Cao Y., Chen S., Ruiz M., Chakrabarti S. miRNA-1 regulates endothelin-1 in diabetes. Life Sci. 2014;98:18–23. doi: 10.1016/j.lfs.2013.12.199. [DOI] [PubMed] [Google Scholar]
- 84.Yang G., Wu Y., Ye S. MiR-181c restrains nitration stress of endothelial cells in diabetic db/db mice through inhibiting the expression of FoxO1. Biochem. Biophys. Res. Commun. 2017;486:29–35. doi: 10.1016/j.bbrc.2017.02.083. [DOI] [PubMed] [Google Scholar]
- 85.Silambarasan M., Tan J.R., Karolina D.S., Armugam A., Kaur C., Jeyaseelan K. MicroRNAs in Hyperglycemia Induced Endothelial Cell Dysfunction. Int. J. Mol. Sci. 2016;17:518. doi: 10.3390/ijms17040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lian W., Hu X., Shi R., Han S., Cao C., Wang K., Li M. MiR-31 regulates the function of diabetic endothelial progenitor cells by targeting Satb2. Acta Biochim. Biophys. Sin. 2018;50:336–344. doi: 10.1093/abbs/gmy010. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Q., Xiao X., Zheng J., Li M. A glucagon-like peptide-1 analog, liraglutide, ameliorates endothelial dysfunction through miRNAs to inhibit apoptosis in rats. PeerJ. 2019;7:e6567. doi: 10.7717/peerj.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo E., Wang D., Yan G., Qiao Y., Zhu B., Liu B., Hou J., Tang C. The NF-κB/miR-425-5p/MCT4 axis: A novel insight into diabetes-induced endothelial dysfunction. Mol. Cell. Endocrinol. 2020;500:110641. doi: 10.1016/j.mce.2019.110641. [DOI] [PubMed] [Google Scholar]
- 89.Huang Z., Li N., Shan Y., Liang C. Hsa-miRNA-29a protects against high glucose-induced damage in human umbilical vein endothelial cells. J. Cell. Biochem. 2019;120:5860–5868. doi: 10.1002/jcb.27871. [DOI] [PubMed] [Google Scholar]
- 90.Chen H., Feng Z., Li L., Fan L. MicroRNA-9 rescues hyperglycemia-induced endothelial cell dysfunction and promotes arteriogenesis through downregulating Notch1 signaling. Mol. Cell. Biochem. 2021;476:2777–2789. doi: 10.1007/s11010-021-04075-8. [DOI] [PubMed] [Google Scholar]
- 91.Rawal S., Munasinghe P.E., Shindikar A., Paulin J., Cameron V., Manning P., Williams M.J.A., Jones G.T., Bunton R., Galvin I., et al. Down-regulation of proangiogenic microRNA-126 and microRNA-132 are early modulators of diabetic cardiac microangiopathy. Cardiovasc. Res. 2017;113:90–101. doi: 10.1093/cvr/cvw235. [DOI] [PubMed] [Google Scholar]
- 92.Luo Y.-F., Wan X.-X., Zhao L.-L., Guo Z., Shen R.-T., Zeng P.-Y., Wang L.-H., Yuan J.-J., Yang W.-J., Yue C., et al. MicroRNA-139-5p upregulation is associated with diabetic endothelial cell dysfunction by targeting c-jun. Aging. 2020;13:1186–1211. doi: 10.18632/aging.202257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang D., Wang H., Liu C., Mu X., Cheng S. Hyperglycemia inhibition of endothelial miR-140-3p mediates angiogenic dysfunction in diabetes mellitus. J. Diabetes Complicat. 2019;33:374–382. doi: 10.1016/j.jdiacomp.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 94.Sun T., Yin L., Kuang H. miR-181a/b-5p regulates human umbilical vein endothelial cell angiogenesis by targeting PDGFRA. Cell Biochem. Funct. 2020;38:222–230. doi: 10.1002/cbf.3472. [DOI] [PubMed] [Google Scholar]
- 95.Li Y., Song Y.-H., Li F., Yang T., Lu Y.W., Geng Y.-J. microRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem. Biophys. Res. Commun. 2009;381:81–83. doi: 10.1016/j.bbrc.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng S., Cui Y., Fan L., Mu X., Hua Y. T2DM inhibition of endothelial miR-342-3p facilitates angiogenic dysfunction via repression of FGF11 signaling. Biochem. Biophys. Res. Commun. 2018;503:71–78. doi: 10.1016/j.bbrc.2018.05.179. [DOI] [PubMed] [Google Scholar]
- 97.Kim J. MicroRNAs as critical regulators of the endothelial to mesenchymal transition in vascular biology. BMB Rep. 2018;51:65–72. doi: 10.5483/BMBRep.2018.51.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Souilhol C., Harmsen M.C., Evans P.C., Krenning G. Endothelial–mesenchymal transition in atherosclerosis. Cardiovasc. Res. 2018;114:565–577. doi: 10.1093/cvr/cvx253. [DOI] [PubMed] [Google Scholar]
- 99.Zhu G.-H., Li R., Zeng Y., Zhou T., Xiong F., Zhu M. MicroRNA-142-3p inhibits high-glucose-induced endothelial-to-mesenchymal transition through targeting TGF-β1/Smad pathway in primary human aortic endothelial cells. Int. J. Clin. Exp. Pathol. 2018;11:1208–1217. [PMC free article] [PubMed] [Google Scholar]
- 100.Guan G., Wei N., Song T., Zhao C., Sun Y., Pan R., Zhang L., Xu Y., Dai Y., Han H. miR-448-3p alleviates diabetic vascular dysfunction by inhibiting endothelial–mesenchymal transition through DPP-4 dysregulation. J. Cell. Physiol. 2020;235:10024–10036. doi: 10.1002/jcp.29817. [DOI] [PubMed] [Google Scholar]
- 101.Chen Y., Yang Q., Zhan Y., Ke J., Lv P., Huang J. The role of miR-328 in high glucose-induced endothelial-to-mesenchymal transition in human umbilical vein endothelial cells. Life Sci. 2018;207:110–116. doi: 10.1016/j.lfs.2018.05.055. [DOI] [PubMed] [Google Scholar]
- 102.Chen T.-C., Sung M.-L., Kuo H.-C., Chien S.-J., Yen C.-K., Chen C.-N. Differential Regulation of Human Aortic Smooth Muscle Cell Proliferation by Monocyte-Derived Macrophages from Diabetic Patients. PLoS ONE. 2014;9:e113752. doi: 10.1371/journal.pone.0113752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He M., Wu N., Leong M.C., Zhang W., Ye Z., Li R., Huang J., Zhang Z., Li L., Yao X., et al. miR-145 improves metabolic inflammatory disease through multiple pathways. J. Mol. Cell Biol. 2020;12:152–162. doi: 10.1093/jmcb/mjz015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang M., Zhou Z., Wang J., Li S. MiR-130b promotes obesity associated adipose tissue inflammation and insulin resistance in diabetes mice through alleviating M2 macrophage polarization via repression of PPAR-γ. Immunol. Lett. 2016;180:1–8. doi: 10.1016/j.imlet.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 105.Zhao Q., Wang X., Hu Q., Zhang R., Yin Y. Suppression of TLR4 by miR-448 is involved in Diabetic development via regulating Macrophage polarization. J. Pharm. Pharmacol. 2019;71:806–815. doi: 10.1111/jphp.13048. [DOI] [PubMed] [Google Scholar]
- 106.Sun J., Huang Q., Li S., Meng F., Li X., Gong X. miR-330-5p/Tim-3 axis regulates macrophage M2 polarization and insulin resistance in diabetes mice. Mol. Immunol. 2018;95:107–113. doi: 10.1016/j.molimm.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 107.Hu F., Tong J., Deng B., Zheng J., Lu C. MiR-495 regulates macrophage M1/M2 polarization and insulin resistance in high-fat diet-fed mice via targeting FTO. Pflügers Arch.-Eur. J. Physiol. 2019;471:1529–1537. doi: 10.1007/s00424-019-02316-w. [DOI] [PubMed] [Google Scholar]
- 108.Jaiswal A., Reddy S.S., Maurya M., Maurya P., Barthwal M.K. MicroRNA-99a mimics inhibit M1 macrophage phenotype and adipose tissue inflammation by targeting TNFα. Cell. Mol. Immunol. 2019;16:495–507. doi: 10.1038/s41423-018-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kuschnerus K., Straessler E.T., Müller M.F., Lüscher T.F., Landmesser U., Kränkel N. Increased Expression of miR-483-3p Impairs the Vascular Response to Injury in Type 2 Diabetes. Diabetes. 2019;68:349–360. doi: 10.2337/db18-0084. [DOI] [PubMed] [Google Scholar]
- 110.Kimura Y., Tamasawa N., Matsumura K., Murakami H., Yamashita M., Matsuki K., Tanabe J., Murakami H., Matsui J., Daimon M. Clinical Significance of Determining Plasma MicroRNA33b in Type 2 Diabetic Patients with Dyslipidemia. J. Atheroscler. Thromb. 2016;23:1276–1285. doi: 10.5551/jat.33670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Distel E., Barrett T.J., Chung K., Girgis N.M., Parathath S., Essau C.C., Murphy A.J., Moore K.J., Fisher E.A. miR33 Inhibition Overcomes Deleterious Effects of Diabetes Mellitus on Atherosclerosis Plaque Regression in Mice. Circ. Res. 2014;115:759–769. doi: 10.1161/CIRCRESAHA.115.304164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Su Y., Guan P., Li D., Hang Y., Ye X., Han L., Lu Y., Bai X., Zhang P., Hu W. Intermedin attenuates macrophage phagocytosis via regulation of the long noncoding RNA Dnm3os/miR-27b-3p/SLAMF7 axis in a mouse model of atherosclerosis in diabetes. Biochem. Biophys. Res. Commun. 2021;583:35–42. doi: 10.1016/j.bbrc.2021.10.038. [DOI] [PubMed] [Google Scholar]
- 113.Porter K.E., Riches K. The vascular smooth muscle cell: A therapeutic target in Type 2 diabetes? Clin. Sci. 2013;125:167–182. doi: 10.1042/CS20120413. [DOI] [PubMed] [Google Scholar]
- 114.Shyu K.-G., Cheng W.-P., Wang B.-W. Angiotensin II Downregulates MicroRNA-145 to Regulate Kruppel-like Factor 4 and Myocardin Expression in Human Coronary Arterial Smooth Muscle Cells under High Glucose Conditions. Mol. Med. 2015;21:616–625. doi: 10.2119/molmed.2015.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen M., Zhang Y., Li W., Yang J. MicroRNA-145 alleviates high glucose-induced proliferation and migration of vascular smooth muscle cells through targeting ROCK1. Biomed. Pharmacother. 2018;99:81–86. doi: 10.1016/j.biopha.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 116.Riches K., Alshanwani A.R., Warburton P., O’Regan D.J., Ball S.G., Wood I.C., Turner N.A., Porter K.E. Elevated expression levels of miR-143/5 in saphenous vein smooth muscle cells from patients with Type 2 diabetes drive persistent changes in phenotype and function. J. Mol. Cell. Cardiol. 2014;74:240–250. doi: 10.1016/j.yjmcc.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chettimada S., Ata H., Rawat D.K., Gulati S., Kahn A.G., Edwards J.G., Gupte S.A. Contractile protein expression is upregulated by reactive oxygen species in aorta of Goto-Kakizaki rat. Am. J. Physiol. Heart Circ. Physiol. 2014;306:H214–H224. doi: 10.1152/ajpheart.00310.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou W., Ye S., Wang W. miR-217 alleviates high-glucose-induced vascular smooth muscle cell dysfunction via regulating ROCK1. J. Biochem. Mol. Toxicol. 2021;35:e22668. doi: 10.1002/jbt.22668. [DOI] [PubMed] [Google Scholar]
- 119.Xu Q., Liang Y., Liu X., Zhang C., Liu X., Li H., Liang J., Yang G., Ge Z. miR-132 inhibits high glucose-induced vascular smooth muscle cell proliferation and migration by targeting E2F5. Mol. Med. Rep. 2019;20:2012–2020. doi: 10.3892/mmr.2019.10380. [DOI] [PubMed] [Google Scholar]
- 120.Yang J., Chen L., Ding J., Fan Z., Li S., Wu H., Zhang J., Yang C., Wang H., Zeng P., et al. MicroRNA-24 inhibits high glucose-induced vascular smooth muscle cell proliferation and migration by targeting HMGB1. Gene. 2016;586:268–273. doi: 10.1016/j.gene.2016.04.027. [DOI] [PubMed] [Google Scholar]
- 121.Yang J., Zeng P., Yang J., Liu X., Ding J., Wang H., Chen L. MicroRNA-24 regulates vascular remodeling via inhibiting PDGF-BB pathway in diabetic rat model. Gene. 2018;659:67–76. doi: 10.1016/j.gene.2018.03.056. [DOI] [PubMed] [Google Scholar]
- 122.Cai W., Zhang J., Yang J., Fan Z., Liu X., Gao W., Zeng P., Xiong M., Ma C., Yang J. MicroRNA-24 attenuates vascular remodeling in diabetic rats through PI3K/Akt signaling pathway. Nutr. Metab. Cardiovasc. Dis. 2019;29:621–632. doi: 10.1016/j.numecd.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 123.Yang J., Fan Z., Yang J., Ding J., Yang C., Chen L. MicroRNA-24 Attenuates Neointimal Hyperplasia in the Diabetic Rat Carotid Artery Injury Model by Inhibiting Wnt4 Signaling Pathway. Int. J. Mol. Sci. 2016;17:765. doi: 10.3390/ijms17060765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang J., Cai W., Fan Z., Yang C., Wang W., Xiong M., Ma C., Yang J. MicroRNA-24 inhibits the oxidative stress induced by vascular injury by activating the Nrf2/Ho-1 signaling pathway. Atherosclerosis. 2019;290:9–18. doi: 10.1016/j.atherosclerosis.2019.08.023. [DOI] [PubMed] [Google Scholar]
- 125.Fan Z., Yang J., Yang C., Zhang J., Cai W., Huang C. MicroRNA-24 attenuates diabetic vascular remodeling by suppressing the NLRP3/caspase-1/IL-1β signaling pathway. Int. J. Mol. Med. 2020;45:1534–1542. doi: 10.3892/ijmm.2020.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zeng Z., Xia L., Fan X., Ostriker A.C., Yarovinsky T., Su M., Zhang Y., Peng X., Xie Y., Pi L., et al. Platelet-derived miR-223 promotes a phenotypic switch in arterial injury repair. J. Clin. Investig. 2019;129:1372–1386. doi: 10.1172/JCI124508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Su M., Fan S., Ling Z., Fan X., Xia L., Liu Y., Li S., Zhang Y., Zeng Z., Tang W.H. Restoring the Platelet miR-223 by Calpain Inhibition Alleviates the Neointimal Hyperplasia in Diabetes. Front. Physiol. 2020;11:742. doi: 10.3389/fphys.2020.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lu X., Ma S.-T., Zhou B., Li T. MiR-9 promotes the phenotypic switch of vascular smooth muscle cells by targeting KLF5. Turk. J. Med. Sci. 2019;49:928–938. doi: 10.3906/sag-1710-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ye D., Lou G., Li A., Dong F., Chen G., Xu W., Liu Y., Hu S. MicroRNA-125a-mediated regulation of the mevalonate signaling pathway contributes to high glucose-induced proliferation and migration of vascular smooth muscle cells. Mol. Med. Rep. 2020;21:165–174. doi: 10.3892/mmr.2020.11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang W., Chen S., Zhang Z., Wang C., Liu C. FAM3B mediates high glucose-induced vascular smooth muscle cell proliferation and migration via inhibition of miR-322-5p. Sci. Rep. 2017;7:2298. doi: 10.1038/s41598-017-02683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jansen F., Zietzer A., Stumpf T., Flender A., Schmitz T., Nickenig G., Werner N. Endothelial microparticle-promoted inhibition of vascular remodeling is abrogated under hyperglycaemic conditions. J. Mol. Cell. Cardiol. 2017;112:91–94. doi: 10.1016/j.yjmcc.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 132.Arevalo-Martinez M., Cidad P., Moreno-Estar S., Fernández M., Albinsson S., Cózar-Castellano I., López-López J.R., Pérez-Garcia M.T. miR-126 contributes to the epigenetic signature of diabetic vascular smooth muscle and enhances antirestenosis effects of Kv1.3 blockers. Mol. Metab. 2021;53:101306. doi: 10.1016/j.molmet.2021.101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Torella D., Iaconetti C., Tarallo R., Marino F., Giurato G., Veneziano C., Aquila I., Scalise M., Mancuso T., Cianflone E., et al. miRNA Regulation of the Hyperproliferative Phenotype of Vascular Smooth Muscle Cells in Diabetes. Diabetes. 2018;67:2554–2568. doi: 10.2337/db17-1434. [DOI] [PubMed] [Google Scholar]
- 134.Hien T.T., Garcia-Vaz E., Stenkula K.G., Sjögren J., Nilsson J., Gomez M.F., Albinsson S. MicroRNA-dependent regulation of KLF4 by glucose in vascular smooth muscle. J. Cell. Physiol. 2018;233:7195–7205. doi: 10.1002/jcp.26549. [DOI] [PubMed] [Google Scholar]
- 135.Sun G., Song H., Wu S. miR-19a promotes vascular smooth muscle cell proliferation, migration and invasion through regulation of Ras homolog family member B. Int. J. Mol. Med. 2019;44:1991–2002. doi: 10.3892/ijmm.2019.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu J., Li L., Yun H., Han Y. MiR-138 promotes smooth muscle cells proliferation and migration in db/db mice through down-regulation of SIRT1. Biochem. Biophys. Res. Commun. 2015;463:1159–1164. doi: 10.1016/j.bbrc.2015.06.076. [DOI] [PubMed] [Google Scholar]
- 137.Reddy M.A., Das S., Zhuo C., Jin W., Wang M., Lanting L., Natarajan R. Regulation of Vascular Smooth Muscle Cell Dysfunction Under Diabetic Conditions by miR-504. Arterioscler. Thromb. Vasc. Biol. 2016;36:864–873. doi: 10.1161/ATVBAHA.115.306770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang B., Gao X., Sun Y., Zhao J., Chen J., Gao L., Zhao L., Li Y. Dihydroartemisinin alleviates high glucose-induced vascular smooth muscle cells proliferation and inflammation by depressing the miR-376b-3p/KLF15 pathway. Biochem. Biophys. Res. Commun. 2020;530:574–580. doi: 10.1016/j.bbrc.2020.07.095. [DOI] [PubMed] [Google Scholar]
- 139.Coleman C.B., Lightell D.J., Moss S.C., Bates M., Parrino P.E., Woods T.C. Elevation of miR-221 and -222 in the internal mammary arteries of diabetic subjects and normalization with metformin. Mol. Cell. Endocrinol. 2013;374:125–129. doi: 10.1016/j.mce.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lightell D.J., Moss S.C., Woods T.C. Upregulation of miR-221 and -222 in response to increased extracellular signal-regulated kinases 1/2 activity exacerbates neointimal hyperplasia in diabetes mellitus. Atherosclerosis. 2018;269:71–78. doi: 10.1016/j.atherosclerosis.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jia S., Ma W., Zhang C., Zhang Y., Yao Z., Quan X., Guo X., Wang C. Tanshinone IIA attenuates high glucose induced human VSMC proliferation and migration through miR-21-5p-mediated tropomyosin 1 downregulation. Arch. Biochem. Biophys. 2019;677:108154. doi: 10.1016/j.abb.2019.108154. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Z., Guo R., Lv J., Wang X., Ye J., Lu N., Liang X., Yang L. MicroRNA-99a inhibits insulin-induced proliferation, migration, dedifferentiation, and rapamycin resistance of vascular smooth muscle cells by inhibiting insulin-like growth factor-1 receptor and mammalian target of rapamycin. Biochem. Biophys. Res. Commun. 2017;486:414–422. doi: 10.1016/j.bbrc.2017.03.056. [DOI] [PubMed] [Google Scholar]
- 143.Kaur R., Kaur M., Singh J. Endothelial dysfunction and platelet hyperactivity in type 2 diabetes mellitus: Molecular insights and therapeutic strategies. Cardiovasc. Diabetol. 2018;17:121. doi: 10.1186/s12933-018-0763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Randriamboavonjy V., Fleming I. Platelet Function and Signaling in Diabetes Mellitus. Curr. Vasc. Pharmacol. 2012;10:532–538. doi: 10.2174/157016112801784639. [DOI] [PubMed] [Google Scholar]
- 145.Pordzik J., Pisarz K., De Rosa S., Jones A.D., Eyileten C., Indolfi C., Malek L., Postula M. The Potential Role of Platelet-Related microRNAs in the Development of Cardiovascular Events in High-Risk Populations, Including Diabetic Patients: A Review. Front. Endocrinol. 2018;9:74. doi: 10.3389/fendo.2018.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pordzik J., Jakubik D., Jarosz-Popek J., Wicik Z., Eyileten C., De Rosa S., Indolfi C., Siller-Matula J.M., Czajka P., Postula M. Significance of circulating microRNAs in diabetes mellitus type 2 and platelet reactivity: Bioinformatic analysis and review. Cardiovasc. Diabetol. 2019;18:113. doi: 10.1186/s12933-019-0918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fejes Z., Póliska S., Czimmerer Z., Káplár M., Penyige A., Gál Szabó G., Beke Debreceni I., Kunapuli S.P., Kappelmayer J., Nagy Jr B. Hyperglycaemia suppresses microRNA expression in platelets to increase P2RY12 and SELP levels in type 2 diabetes mellitus. Thromb. Haemost. 2017;117:529–542. doi: 10.1160/TH16-04-0322. [DOI] [PubMed] [Google Scholar]
- 148.Elgheznawy A., Shi L., Hu J., Wittig I., Laban H., Pircher J., Mann A., Provost P., Randriamboavonjy V., Fleming I. Dicer Cleavage by Calpain Determines Platelet microRNA Levels and Function in Diabetes. Circ. Res. 2015;117:157–165. doi: 10.1161/CIRCRESAHA.117.305784. [DOI] [PubMed] [Google Scholar]
- 149.Zhou M., Gao M., Luo Y., Gui R., Ji H. Long non-coding RNA metallothionein 1 pseudogene 3 promotes p2y12 expression by sponging miR-126 to activate platelet in diabetic animal model. Platelets. 2019;30:452–459. doi: 10.1080/09537104.2018.1457781. [DOI] [PubMed] [Google Scholar]
- 150.De Boer H.C., van Solingen C., Prins J., Duijs J.M.G.J., Huisman M.V., Rabelink T.J., van Zonneveld A.J. Aspirin treatment hampers the use of plasma microRNA-126 as a biomarker for the progression of vascular disease. Eur. Heart J. 2013;34:3451–3457. doi: 10.1093/eurheartj/eht007. [DOI] [PubMed] [Google Scholar]
- 151.Luo M., Li R., Ren M., Chen N., Deng X., Tan X., Li Y., Zeng M., Yang Y., Wan Q., et al. Hyperglycaemia-induced reciprocal changes in miR-30c and PAI-1 expression in platelets. Sci. Rep. 2016;6:36687. doi: 10.1038/srep36687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stratz C., Nührenberg T., Fiebich B.L., Amann M., Kumar A., Binder H., Hoffmann I., Valina C., Hochholzer W., Trenk D., et al. Controlled type II diabetes mellitus has no major influence on platelet micro-RNA expression. Results from micro-array profiling in a cohort of 60 patients. Thromb. Haemost. 2014;111:902–911. doi: 10.1160/TH13-06-0476. [DOI] [PubMed] [Google Scholar]
- 153.Lin X., Zhan J.-K., Zhong J.-Y., Wang Y.-J., Wang Y., Li S., He J.-Y., Tan P., Chen Y.-Y., Liu X.-B., et al. lncRNA-ES3/miR-34c-5p/BMF axis is involved in regulating high-glucose-induced calcification/senescence of VSMCs. Aging. 2019;11:523–535. doi: 10.18632/aging.101758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ding Y., Pei Y., Rui-Wang, Yang J., Zhao Y., Liu X., Shen H., Ma Q., Zhang S., Ge H. Association of Plasma MiRNA-204 and the Presence and Severity of Coronary Artery Calcification in Patients With Type 2 Diabetes. Angiology. 2021;72:451–458. doi: 10.1177/0003319720984592. [DOI] [PubMed] [Google Scholar]
- 155.Zhong J.-Y., Cui X.-J., Zhan J.-K., Wang Y.-J., Li S., Lin X., Xiang Q.-Y., Ni Y.-Q., Liu L., Liu Y.-S. LncRNA-ES3 inhibition by Bhlhe40 is involved in high glucose–induced calcification/senescence of vascular smooth muscle cells. Ann. N. Y. Acad. Sci. 2020;1474:61–72. doi: 10.1111/nyas.14381. [DOI] [PubMed] [Google Scholar]
- 156.Wang X.-Y., Zhang X.-Z., Li F., Ji Q.-R. MiR-128-3p accelerates cardiovascular calcification and insulin resistance through ISL1-dependent Wnt pathway in type 2 diabetes mellitus rats. J. Cell. Physiol. 2019;234:4997–5010. doi: 10.1002/jcp.27300. [DOI] [PubMed] [Google Scholar]
- 157.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 158.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., Wang K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bhatia P., Raina S., Chugh J., Sharma S. miRNAs: Early prognostic biomarkers for Type 2 diabetes mellitus? Biomark. Med. 2015;9:1025–1040. doi: 10.2217/bmm.15.69. [DOI] [PubMed] [Google Scholar]
- 161.Mraz M., Malinova K., Mayer J., Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem. Biophys. Res. Commun. 2009;390:1–4. doi: 10.1016/j.bbrc.2009.09.061. [DOI] [PubMed] [Google Scholar]
- 162.Tijsen A.J., Pinto Y.M., Creemers E.E. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am. J. Physiol. Heart Circ. Physiol. 2012;303:H1085–H1095. doi: 10.1152/ajpheart.00191.2012. [DOI] [PubMed] [Google Scholar]
- 163.Ali Sheikh M.S., Alduraywish A., Almaeen A., Alruwali M., Alruwaili R., Alomair B.M., Salma U., Hedeab G.M., Bugti N., Abdulhabeeb I.A.M. Therapeutic Value of miRNAs in Coronary Artery Disease. Oxid. Med. Cell. Longev. 2021;2021:8853748. doi: 10.1155/2021/8853748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pritchard C.C., Cheng H.H., Tewari M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Backes C., Meese E., Keller A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016;20:509–518. doi: 10.1007/s40291-016-0221-4. [DOI] [PubMed] [Google Scholar]
- 166.Wang C., Wan S., Yang T., Niu D., Zhang A., Yang C., Cai J., Wu J., Song J., Zhang C.-Y., et al. Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Sci. Rep. 2016;6:20032. doi: 10.1038/srep20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Higuchi C., Nakatsuka A., Eguchi J., Teshigawara S., Kanzaki M., Katayama A., Yamaguchi S., Takahashi N., Murakami K., Ogawa D., et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism. 2015;64:489–497. doi: 10.1016/j.metabol.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 168.Prabu P., Rome S., Sathishkumar C., Aravind S., Mahalingam B., Shanthirani C.S., Gastebois C., Villard A., Mohan V., Balasubramanyam M. Circulating MiRNAs of ‘Asian Indian Phenotype’ Identified in Subjects with Impaired Glucose Tolerance and Patients with Type 2 Diabetes. PLoS ONE. 2015;10:e0128372. doi: 10.1371/journal.pone.0128372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ortega F.J., Mercader J.M., Moreno-Navarrete J.M., Rovira O., Guerra E., Esteve E., Xifra G., Martínez C., Ricart W., Rieusset J., et al. Profiling of Circulating MicroRNAs Reveals Common MicroRNAs Linked to Type 2 Diabetes That Change With Insulin Sensitization. Diabetes Care. 2014;37:1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 170.Santovito D., De Nardis V., Marcantonio P., Mandolini C., Paganelli C., Vitale E., Buttitta F., Bucci M., Mezzetti A., Consoli A., et al. Plasma Exosome MicroRNA Profiling Unravels a New Potential Modulator of Adiponectin Pathway in Diabetes: Effect of Glycemic Control. J. Clin. Endocrinol. Metab. 2014;99:E1681–E1685. doi: 10.1210/jc.2013-3843. [DOI] [PubMed] [Google Scholar]
- 171.Yang Z., Chen H., Si H., Li X., Ding X., Sheng Q., Chen P., Zhang H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014;51:823–831. doi: 10.1007/s00592-014-0617-8. [DOI] [PubMed] [Google Scholar]
- 172.Karolina D.S., Tavintharan S., Armugam A., Sepramaniam S., Pek S.L.T., Wong M.T.K., Lim S.C., Sum C.F., Jeyaseelan K. Circulating miRNA Profiles in Patients with Metabolic Syndrome. J. Clin. Endocrinol. Metab. 2012;97:E2271–E2276. doi: 10.1210/jc.2012-1996. [DOI] [PubMed] [Google Scholar]
- 173.Liang Y.-Z., Dong J., Zhang J., Wang S., He Y., Yan Y.-X. Identification of Neuroendocrine Stress Response-Related Circulating MicroRNAs as Biomarkers for Type 2 Diabetes Mellitus and Insulin Resistance. Front. Endocrinol. 2018;9:132. doi: 10.3389/fendo.2018.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zampetaki A., Kiechl S., Drozdov I., Willeit P., Mayr U., Prokopi M., Mayr A., Weger S., Oberhollenzer F., Bonora E., et al. Plasma MicroRNA Profiling Reveals Loss of Endothelial MiR-126 and Other MicroRNAs in Type 2 Diabetes. Circ. Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 175.Avgeris M., Kokkinopoulou I., Maratou E., Mitrou P., Boutati E., Scorilas A., Fragoulis E.G., Christodoulou M.-I. Blood-based analysis of 84 microRNAs identifies molecules deregulated in individuals with type-2 diabetes, risk factors for the disease or metabolic syndrome. Diabetes Res. Clin. Pract. 2020;164:108187. doi: 10.1016/j.diabres.2020.108187. [DOI] [PubMed] [Google Scholar]
- 176.Yang Z.-M., Chen L.-H., Hong M., Chen Y.-Y., Yang X.-R., Tang S.-M., Yuan Q.-F., Chen W.-W. Serum microRNA profiling and bioinformatics analysis of patients with type 2 diabetes mellitus in a Chinese population. Mol. Med. Rep. 2017;15:2143–2153. doi: 10.3892/mmr.2017.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Matsha T.E., Kengne A.P., Hector S., Mbu D.L., Yako Y.Y., Erasmus R.T. MicroRNA profiling and their pathways in South African individuals with prediabetes and newly diagnosed type 2 diabetes mellitus. Oncotarget. 2018;9:30485–30498. doi: 10.18632/oncotarget.25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ding L., Ai D., Wu R., Zhang T., Jing L., Lu J., Zhong L. Identification of the differential expression of serum microRNA in type 2 diabetes. Biosci. Biotechnol. Biochem. 2016;80:461–465. doi: 10.1080/09168451.2015.1107460. [DOI] [PubMed] [Google Scholar]
- 179.Yan S., Wang T., Huang S., Di Y., Huang Y., Liu X., Luo Z., Han W., An B. Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes. Acta Diabetol. 2016;53:693–702. doi: 10.1007/s00592-016-0837-1. [DOI] [PubMed] [Google Scholar]
- 180.Karolina D.S., Armugam A., Tavintharan S., Wong M.T.K., Lim S.C., Sum C.F., Jeyaseelan K. MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE. 2011;6:e22839. doi: 10.1371/annotation/698b7123-174f-4a09-95c9-fd6f5017d622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Baldeón R.L., Weigelt K., de Wit H., Ozcan B., van Oudenaren A., Sempértegui F., Sijbrands E., Grosse L., van Zonneveld A.-J., Drexhage H.A., et al. Type 2 Diabetes Monocyte MicroRNA and mRNA Expression: Dyslipidemia Associates with Increased Differentiation-Related Genes but Not Inflammatory Activation. PLoS ONE. 2015;10:e0129421. doi: 10.1371/journal.pone.0129421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jiao Y., Zhu M., Mao X., Long M., Du X., Wu Y., Abudureyimu K., Zhang C., Wang Y., Tao Y., et al. MicroRNA-130a expression is decreased in Xinjiang Uygur patients with type 2 diabetes mellitus. Am. J. Transl. Res. 2015;7:1984–1991. [PMC free article] [PubMed] [Google Scholar]
- 183.Kong L., Zhu J., Han W., Jiang X., Xu M., Zhao Y., Dong Q., Pang Z., Guan Q., Gao L., et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011;48:61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 184.Sun K., Chang X., Yin L., Li J., Zhou T., Zhang C., Chen X. Expression and DNA methylation status of microRNA-375 in patients with type 2 diabetes mellitus. Mol. Med. Rep. 2014;9:967–972. doi: 10.3892/mmr.2013.1872. [DOI] [PubMed] [Google Scholar]
- 185.Aljaibeji H., Elemam N.M., Mohammed A.K., Hasswan H., Thahyabat M.A., Alkhayyal N., Sulaiman N., Taneera J. Let7b-5p is Upregulated in the Serum of Emirati Patients with Type 2 Diabetes and Regulates Insulin Secretion in INS-1 Cells. Exp. Clin. Endocrinol. Diabetes. 2022;130:22–29. doi: 10.1055/a-1261-5282. [DOI] [PubMed] [Google Scholar]
- 186.Yan S.-T., Li C.-L., Tian H., Li J., Pei Y., Liu Y., Gong Y.-P., Fang F.-S., Sun B.-R. MiR-199a is overexpressed in plasma of type 2 diabetes patients which contributes to type 2 diabetes by targeting GLUT4. Mol. Cell. Biochem. 2014;397:45–51. doi: 10.1007/s11010-014-2170-8. [DOI] [PubMed] [Google Scholar]
- 187.Sucharita S., Ashwini V., Prabhu J.S., Avadhany S.T., Ayyar V., Bantwal G. The Role of Circulating MicroRNA in the Regulation of Beta Cell Function and Insulin Resistance among Indians with Type 2 Diabetes. Indian J. Endocrinol. Metab. 2018;22:770–773. doi: 10.4103/ijem.IJEM_162_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ali Beg M.M., Verma A.K., Saleem M., Saud Alreshidi F., Alenazi F., Ahmad H., Joshi P.C. Role and Significance of Circulating Biomarkers: miRNA and E2F1 mRNA Expression and Their Association with Type-2 Diabetic Complications. Int. J. Endocrinol. 2020;2020:6279168. doi: 10.1155/2020/6279168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhu J., Wang C., Zhang X., Qiu T., Ma Y., Li X., Pang H., Xiong J., Yang X., Pan C., et al. Correlation analysis of microribonucleic acid-155 and microribonucleic acid-29 with type 2 diabetes mellitus, and the prediction and verification of target genes. J. Diabetes Investig. 2021;12:165–175. doi: 10.1111/jdi.13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang M., Wei J., Ji T., Zang K. miRNA-770-5p expression is upregulated in patients with type 2 diabetes and miRNA-770-5p knockdown protects pancreatic β-cell function via targeting BAG5 expression. Exp. Ther. Med. 2021;22:664. doi: 10.3892/etm.2021.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lu C., Wang D., Feng Y., Feng L., Li Z. miR-720 Regulates Insulin Secretion by Targeting Rab35. Biomed Res. Int. 2021;2021:6662612. doi: 10.1155/2021/6662612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shahrokhi S.Z., Saeidi L., Sadatamini M., Jafarzadeh M., Rahimipour A., Kazerouni F. Can miR-145-5p be used as a marker in diabetic patients? Arch. Physiol. Biochem. 2022;128:1175–1180. doi: 10.1080/13813455.2020.1762657. [DOI] [PubMed] [Google Scholar]
- 193.Masoudi F., Sharifi M.R., Pourfarzam M. Investigation of the relationship between miR-33a, miR-122, erythrocyte membrane fatty acids profile, and serum lipids with components of metabolic syndrome in type 2 diabetic patients. Res. Pharm. Sci. 2022;17:242–251. doi: 10.4103/1735-5362.343078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zeinali F., Aghaei Zarch S.M., Jahan-Mihan A., Kalantar S.M., Vahidi Mehrjardi M.Y., Fallahzadeh H., Hosseinzadeh M., Rahmanian M., Mozaffari-Khosravi H. Circulating microRNA-122, microRNA-126-3p and microRNA-146a are associated with inflammation in patients with pre-diabetes and type 2 diabetes mellitus: A case control study. PLoS ONE. 2021;16:e0251697. doi: 10.1371/journal.pone.0251697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rong Y., Bao W., Shan Z., Liu J., Yu X., Xia S., Gao H., Wang X., Yao P., Hu F.B., et al. Increased MicroRNA-146a Levels in Plasma of Patients with Newly Diagnosed Type 2 Diabetes Mellitus. PLoS ONE. 2013;8:e73272. doi: 10.1371/journal.pone.0073272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liu Y., Gao G., Yang C., Zhou K., Shen B., Liang H., Jiang X. The Role of Circulating MicroRNA-126 (miR-126): A Novel Biomarker for Screening Prediabetes and Newly Diagnosed Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2014;15:10567–10577. doi: 10.3390/ijms150610567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang T., Li L., Shang Q., Lv C., Wang C., Su B. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem. Biophys. Res. Commun. 2015;463:60–63. doi: 10.1016/j.bbrc.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 198.Al-Kafaji G., Al-Mahroos G., Abdulla Al-Muhtaresh H., Sabry M.A., Abdul Razzak R., Salem A.H. Circulating endothelium-enriched microRNA-126 as a potential biomarker for coronary artery disease in type 2 diabetes mellitus patients. Biomarkers. 2017;22:268–278. doi: 10.1080/1354750X.2016.1204004. [DOI] [PubMed] [Google Scholar]
- 199.Weale C.J., Matshazi D.M., Davids S.F.G., Raghubeer S., Erasmus R.T., Kengne A.P., Davison G.M., Matsha T.E. MicroRNAs-1299, -126-3p and -30e-3p as Potential Diagnostic Biomarkers for Prediabetes. Diagnostics. 2021;11:949. doi: 10.3390/diagnostics11060949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Monfared Y.K., Mirzaii-Dizgah M.-R., Khodabandehloo E., Sarookhani M.R., Hashemipour S., Mirzaii-Dizgah I. Salivary microRNA-126 and 135a: A potentially non-invasive diagnostic biomarkers of type- 2 diabetes. J. Diabetes Metab. Disord. 2021;20:1631–1638. doi: 10.1007/s40200-021-00914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang T., Lv C., Li L., Chen S., Liu S., Wang C., Su B. Plasma miR-126 Is a Potential Biomarker for Early Prediction of Type 2 Diabetes Mellitus in Susceptible Individuals. Biomed Res. Int. 2013;2013:761617. doi: 10.1155/2013/761617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Baldeón R.L., Weigelt K., de Wit H., Ozcan B., van Oudenaren A., Sempértegui F., Sijbrands E., Grosse L., Freire W., Drexhage H.A., et al. Decreased Serum Level of miR-146a as Sign of Chronic Inflammation in Type 2 Diabetic Patients. PLoS ONE. 2014;9:e115209. doi: 10.1371/journal.pone.0115209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Olivieri F., Spazzafumo L., Bonafè M., Recchioni R., Prattichizzo F., Marcheselli F., Micolucci L., Mensà E., Giuliani A., Santini G., et al. MiR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: Relationship with type 2 diabetes complications. Oncotarget. 2015;6:35372–35382. doi: 10.18632/oncotarget.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lenin R., Sankaramoorthy A., Mohan V., Balasubramanyam M. Altered immunometabolism at the interface of increased endoplasmic reticulum (ER) stress in patients with type 2 diabetes. J. Leukoc. Biol. 2015;98:615–622. doi: 10.1189/jlb.3A1214-609R. [DOI] [PubMed] [Google Scholar]
- 205.Giannella A., Radu C.M., Franco L., Campello E., Simioni P., Avogaro A., de Kreutzenberg S.V., Ceolotto G. Circulating levels and characterization of microparticles in patients with different degrees of glucose tolerance. Cardiovasc. Diabetol. 2017;16:118. doi: 10.1186/s12933-017-0600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Alipoor B., Ghaedi H., Meshkani R., Omrani M.D., Sharifi Z., Golmohammadi T. The rs2910164 variant is associated with reduced miR-146a expression but not cytokine levels in patients with type 2 diabetes. J. Endocrinol. Investig. 2018;41:557–566. doi: 10.1007/s40618-017-0766-z. [DOI] [PubMed] [Google Scholar]
- 207.Dehghani M., Aghaei Zarch S.M., Vahidi Mehrjardi M.Y., Nazari M., Babakhanzadeh E., Ghadimi H., Zeinali F., Talebi M. Evaluation of miR-181b and miR-126-5p expression levels in T2DM patients compared to healthy individuals: Relationship with NF-κB gene expression. Endocrinol. Diabetes Nutr. 2020;67:454–460. doi: 10.1016/j.endinu.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 208.Balasubramanyam M., Aravind S., Gokulakrishnan K., Prabu P., Sathishkumar C., Ranjani H., Mohan V. Impaired miR-146a expression links subclinical inflammation and insulin resistance in Type 2 diabetes. Mol. Cell. Biochem. 2011;351:197–205. doi: 10.1007/s11010-011-0727-3. [DOI] [PubMed] [Google Scholar]
- 209.Mazloom H., Alizadeh S., Pasalar P., Esfahani E.N., Meshkani R. Downregulated microRNA-155 expression in peripheral blood mononuclear cells of type 2 diabetic patients is not correlated with increased inflammatory cytokine production. Cytokine. 2015;76:403–408. doi: 10.1016/j.cyto.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 210.Luo M., Li R., Deng X., Ren M., Chen N., Zeng M., Yan K., Xia J., Liu F., Ma W., et al. Platelet-derived miR-103b as a novel biomarker for the early diagnosis of type 2 diabetes. Acta Diabetol. 2015;52:943–949. doi: 10.1007/s00592-015-0733-0. [DOI] [PubMed] [Google Scholar]
- 211.Baldeón Rojas L., Weigelt K., de Wit H., Ozcan B., van Oudenaren A., Sempértegui F., Sijbrands E., Grosse L., van Zonneveld A.-J., Drexhage H.A., et al. Study on inflammation-related genes and microRNAs, with special emphasis on the vascular repair factor HGF and miR-574-3p, in monocytes and serum of patients with T2D. Diabetol. Metab. Syndr. 2016;8:6. doi: 10.1186/s13098-015-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wang S.-S., Li Y.-Q., Liang Y.-Z., Dong J., He Y., Zhang L., Yan Y.-X. Expression of miR-18a and miR-34c in circulating monocytes associated with vulnerability to type 2 diabetes mellitus and insulin resistance. J. Cell. Mol. Med. 2017;21:3372–3380. doi: 10.1111/jcmm.13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Corral-Fernández N.E., Salgado-Bustamante M., Martínez-Leija M.E., Cortez-Espinosa N., García-Hernández M.H., Reynaga-Hernández E., Quezada-Calvillo R., Portales-Pérez D.P. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Exp. Clin. Endocrinol. Diabetes. 2013;121:347–353. doi: 10.1055/s-0033-1341516. [DOI] [PubMed] [Google Scholar]
- 214.Tan J., Tong A., Xu Y. Pancreatic β-cell function is inhibited by miR-3666 in type 2 diabetes mellitus by targeting adiponectin. Brazilian J. Med. Biol. Res. 2019;52:e8344. doi: 10.1590/1414-431x20198344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bao L., Fu X., Si M., Wang Y., Ma R., Ren X., Lv H. MicroRNA-185 Targets SOCS3 to Inhibit Beta-Cell Dysfunction in Diabetes. PLoS ONE. 2015;10:e0116067. doi: 10.1371/journal.pone.0116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Seyhan A.A., Nunez Lopez Y.O., Xie H., Yi F., Mathews C., Pasarica M., Pratley R.E. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci. Rep. 2016;6:31479. doi: 10.1038/srep31479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Li M.Y., Pan S.R., Qiu A.Y. Roles of microRNA-221/222 in type 2 diabetic patients with post-menopausal breast cancer. Genet. Mol. Res. 2016;15:gmr.15027259. doi: 10.4238/gmr.15027259. [DOI] [PubMed] [Google Scholar]
- 218.Wan S., Wang J., Wang J., Wu J., Song J., Zhang C.-Y., Zhang C., Wang C., Wang J.-J. Increased serum miR-7 is a promising biomarker for type 2 diabetes mellitus and its microvascular complications. Diabetes Res. Clin. Pract. 2017;130:171–179. doi: 10.1016/j.diabres.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 219.Shao Y., Ren H., Lv C., Ma X., Wu C., Wang Q. Changes of serum Mir-217 and the correlation with the severity in type 2 diabetes patients with different stages of diabetic kidney disease. Endocrine. 2017;55:130–138. doi: 10.1007/s12020-016-1069-4. [DOI] [PubMed] [Google Scholar]
- 220.De Candia P., Spinetti G., Specchia C., Sangalli E., La Sala L., Uccellatore A., Lupini S., Genovese S., Matarese G., Ceriello A. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS ONE. 2017;12:e0188980. doi: 10.1371/journal.pone.0188980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Cao Z., Yao F., Lang Y., Feng X. Elevated Circulating LINC-P21 Serves as a Diagnostic Biomarker of Type 2 Diabetes Mellitus and Regulates Pancreatic β-cell Function by Sponging miR-766-3p to Upregulate NR3C2. Exp. Clin. Endocrinol. Diabetes. 2022;130:156–164. doi: 10.1055/a-1247-4978. [DOI] [PubMed] [Google Scholar]
- 222.Al-Kafaji G., Al-Mahroos G., Alsayed N.A., Hasan Z.A., Nawaz S., Bakhiet M. Peripheral blood microRNA-15a is a potential biomarker for type 2 diabetes mellitus and pre-diabetes. Mol. Med. Rep. 2015;12:7485–7490. doi: 10.3892/mmr.2015.4416. [DOI] [PubMed] [Google Scholar]
- 223.Amr K.S., Abdelmawgoud H., Ali Z.Y., Shehata S., Raslan H.M. Potential value of circulating microRNA-126 and microRNA-210 as biomarkers for type 2 diabetes with coronary artery disease. Br. J. Biomed. Sci. 2018;75:82–87. doi: 10.1080/09674845.2017.1402404. [DOI] [PubMed] [Google Scholar]
- 224.Luo M., Wang G., Xu C., Zeng M., Lin F., Wu J., Wan Q. Circulating miR-30c as a predictive biomarker of type 2 diabetes mellitus with coronary heart disease by regulating PAI-1/VN interactions. Life Sci. 2019;239:117092. doi: 10.1016/j.lfs.2019.117092. [DOI] [PubMed] [Google Scholar]
- 225.Luo M., Xu C., Luo Y., Wang G., Wu J., Wan Q. Circulating miR-103 family as potential biomarkers for type 2 diabetes through targeting CAV-1 and SFRP4. Acta Diabetol. 2020;57:309–322. doi: 10.1007/s00592-019-01430-6. [DOI] [PubMed] [Google Scholar]
- 226.Oraby H.E., Elshaer S.S., Rashed L.A., Eldesoky N.A. MicroRNA-499 Gene Expression in Egyptian Type 2 Diabetes Mellitus Patients with and without Coronary Heart Disease. Azhar Int. J. Pharm. Med. Sci. 2022;2:73–81. doi: 10.21608/aijpms.2021.210569. [DOI] [Google Scholar]
- 227.Yazdanpanah Z., Kazemipour N., Kalantar S.M., Vahidi Mehrjardi M.Y. Plasma miR-21 as a potential predictor in prediabetic individuals with a positive family history of type 2 diabetes mellitus. Physiol. Rep. 2022;10:e15163. doi: 10.14814/phy2.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.La Sala L., Mrakic-Sposta S., Tagliabue E., Prattichizzo F., Micheloni S., Sangalli E., Specchia C., Uccellatore A.C., Lupini S., Spinetti G., et al. Circulating microRNA-21 is an early predictor of ROS-mediated damage in subjects with high risk of developing diabetes and in drug-naïve T2D. Cardiovasc. Diabetol. 2019;18:18. doi: 10.1186/s12933-019-0824-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Monfared Y.K., Honardoost M., Sarookhani M.R., Farzam S.A. Circulating miR-135 May Serve as a Novel Co-biomarker of HbA1c in Type 2 Diabetes. Appl. Biochem. Biotechnol. 2020;191:623–630. doi: 10.1007/s12010-019-03163-2. [DOI] [PubMed] [Google Scholar]
- 230.Saeidi L., Shahrokhi S.Z., Sadatamini M., Jafarzadeh M., Kazerouni F. Can circulating miR-7-1-5p, and miR-33a-5p be used as markers of T2D patients? Arch. Physiol. Biochem. 2021:1–7. doi: 10.1080/13813455.2021.1871762. [DOI] [PubMed] [Google Scholar]
- 231.Al-Muhtaresh H.A., Al-Kafaji G. Evaluation of Two-Diabetes Related microRNAs Suitability as Earlier Blood Biomarkers for Detecting Prediabetes and type 2 Diabetes Mellitus. J. Clin. Med. 2018;7:12. doi: 10.3390/jcm7020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Weale C.J., Matshazi D.M., Davids S.F.G., Raghubeer S., Erasmus R.T., Kengne A.P., Davison G.M., Matsha T.E. Expression Profiles of Circulating microRNAs in South African Type 2 Diabetic Individuals on Treatment. Front. Genet. 2021;12:702410. doi: 10.3389/fgene.2021.702410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Seleem M., Shabayek M., Ewida H.A. MicroRNAs 342 and 450 together with NOX-4 activity and their association with coronary artery disease in diabetes. Diabetes. Metab. Res. Rev. 2019;35:e3130. doi: 10.1002/dmrr.3130. [DOI] [PubMed] [Google Scholar]
- 234.Kokkinopoulou I., Maratou E., Mitrou P., Boutati E., Sideris D.C., Fragoulis E.G., Christodoulou M.-I. Decreased expression of microRNAs targeting type-2 diabetes susceptibility genes in peripheral blood of patients and predisposed individuals. Endocrine. 2019;66:226–239. doi: 10.1007/s12020-019-02062-0. [DOI] [PubMed] [Google Scholar]
- 235.Banerjee J., Roy S., Dhas Y., Mishra N. Senescence-associated miR-34a and miR-126 in middle-aged Indians with type 2 diabetes. Clin. Exp. Med. 2020;20:149–158. doi: 10.1007/s10238-019-00593-4. [DOI] [PubMed] [Google Scholar]
- 236.Shen Y., Xu H., Pan X., Wu W., Wang H., Yan L., Zhang M., Liu X., Xia S., Shao Q. miR-34a and miR-125b are upregulated in peripheral blood mononuclear cells from patients with type 2 diabetes mellitus. Exp. Ther. Med. 2017;14:5589–5596. doi: 10.3892/etm.2017.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Long Y., Zhan Q., Yuan M., Duan X., Zhou J., Lu J., Li Z., Yu F., Zhou X., Yang Q., et al. The Expression of microRNA-223 and FAM5C in Cerebral Infarction Patients with Diabetes Mellitus. Cardiovasc. Toxicol. 2017;17:42–48. doi: 10.1007/s12012-015-9354-7. [DOI] [PubMed] [Google Scholar]
- 238.Yan L.-N., Zhang X., Xu F., Fan Y.-Y., Ge B., Guo H., Li Z.-L. Four-microRNA signature for detection of type 2 diabetes. World J. Clin. Cases. 2020;8:1923–1931. doi: 10.12998/wjcc.v8.i10.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sadeghzadeh S., Dehghani Ashkezari M., Seifati S.M., Vahidi Mehrjardi M.Y., Dehghan Tezerjani M., Sadeghzadeh S., Ladan S.A.B. Circulating miR-15a and miR-222 as Potential Biomarkers of Type 2 Diabetes. Diabetes Metab. Syndr. Obes. Targets Ther. 2020;13:3461–3469. doi: 10.2147/DMSO.S263883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Meerson A., Najjar A., Saad E., Sbeit W., Barhoum M., Assy N. Sex Differences in Plasma MicroRNA Biomarkers of Early and Complicated Diabetes Mellitus in Israeli Arab and Jewish Patients. Non-Coding RNA. 2019;5:32. doi: 10.3390/ncrna5020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wang X., Sundquist J., Zöller B., Memon A.A., Palmér K., Sundquist K., Bennet L. Determination of 14 Circulating microRNAs in Swedes and Iraqis with and without Diabetes Mellitus Type 2. PLoS ONE. 2014;9:e86792. doi: 10.1371/journal.pone.0086792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Šimonienė D., Stukas D., Daukša A., Veličkienė D. Clinical Role of Serum miR107 in Type 2 Diabetes and Related Risk Factors. Biomolecules. 2022;12:558. doi: 10.3390/biom12040558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Montalescot G., Sechtem U., Achenbach S., Andreotti F., Arden C., Budaj A., Bugiardini R., Crea F., Cuisset T., Di Mario C., et al. 2013 ESC guidelines on the management of stable coronary artery disease: The Task Force on the management of stable coronary artery disease of the European Society of Cardiology. Eur. Heart J. 2013;34:2949–3003. doi: 10.1093/eurheartj/eht296. [DOI] [PubMed] [Google Scholar]
- 244.Knuuti J., Wijns W., Saraste A., Capodanno D., Barbato E., Funck-Brentano C., Prescott E., Storey R.F., Deaton C., Cuisset T., et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur. Heart J. 2020;41:407–477. doi: 10.1093/eurheartj/ehz425. [DOI] [PubMed] [Google Scholar]
- 245.Ferrari R., Pavasini R., Censi S., Squeri A., Rosano G. The New ESC Guidelines for the Diagnosis and Management of Chronic Coronary Syndromes: The Good and the Not So Good. Curr. Probl. Cardiol. 2021;46:100554. doi: 10.1016/j.cpcardiol.2020.100554. [DOI] [PubMed] [Google Scholar]
- 246.Bertolone D.T., Gallinoro E., Esposito G., Paolisso P., Bermpeis K., De Colle C., Fabbricatore D., Mileva N., Valeriano C., Munhoz D., et al. Contemporary Management of Stable Coronary Artery Disease. High Blood Press. Cardiovasc. Prev. 2022;29:207–219. doi: 10.1007/s40292-021-00497-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Fichtlscherer S., De Rosa S., Fox H., Schwietz T., Fischer A., Liebetrau C., Weber M., Hamm C.W., Röxe T., Müller-Ardogan M., et al. Circulating MicroRNAs in Patients With Coronary Artery Disease. Circ. Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 248.Sanlialp M., Dodurga Y., Uludag B., Alihanoglu Y.I., Enli Y., Secme M., Bostanci H.E., Cetin Sanlialp S., Tok O.O., Kaftan A., et al. Peripheral blood mononuclear cell microRNAs in coronary artery disease. J. Cell. Biochem. 2020;121:3005–3009. doi: 10.1002/jcb.29557. [DOI] [PubMed] [Google Scholar]
- 249.Qiu X.-K., Ma J. Alteration in microRNA-155 level correspond to severity of coronary heart disease. Scand. J. Clin. Lab. Investig. 2018;78:219–223. doi: 10.1080/00365513.2018.1435904. [DOI] [PubMed] [Google Scholar]
- 250.Chang S.-N., Chen J.-J., Wu J.-H., Chung Y.-T., Chen J.-W., Chiu C.-H., Liu C.-J., Liu M.-T., Chang Y.-C., Li C., et al. Association between Exosomal miRNAs and Coronary Artery Disease by Next-Generation Sequencing. Cells. 2021;11:98. doi: 10.3390/cells11010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang X., Dong Y., Fang T., Wang X., Chen L., Zheng C., Kang Y., Jiang L., You X., Gai S., et al. Circulating MicroRNA-423-3p Improves the Prediction of Coronary Artery Disease in a General Population―Six-Year Follow-up Results from the China-Cardiovascular Disease Study. Circ. J. 2020;84:1155–1162. doi: 10.1253/circj.CJ-19-1181. [DOI] [PubMed] [Google Scholar]
- 252.Zhang X., Cai H., Zhu M., Qian Y., Lin S., Li X. Circulating microRNAs as biomarkers for severe coronary artery disease. Medicine. 2020;99:e19971. doi: 10.1097/MD.0000000000019971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Faccini J., Ruidavets J.-B., Cordelier P., Martins F., Maoret J.-J., Bongard V., Ferrières J., Roncalli J., Elbaz M., Vindis C. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci. Rep. 2017;7:42916. doi: 10.1038/srep42916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Du Y., Yang S.H., Li S., Cui C.J., Zhang Y., Zhu C.G., Guo Y.L., Wu N.Q., Gao Y., Sun J., et al. Circulating MicroRNAs as Novel Diagnostic Biomarkers for Very Early-onset (≤40 years) Coronary Artery Disease. Biomed. Environ. Sci. 2016;29:545–554. doi: 10.3967/bes2016.073. [DOI] [PubMed] [Google Scholar]
- 255.Zhou J., Shao G., Chen X., Yang X., Huang X., Peng P., Ba Y., Zhang L., Jehangir T., Bu S., et al. miRNA 206 and miRNA 574-5p are highly expression in coronary artery disease. Biosci. Rep. 2016;36:e00295. doi: 10.1042/BSR20150206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Han H., Qu G., Han C., Wang Y., Sun T., Li F., Wang J., Luo S. MiR-34a, miR-21 and miR-23a as potential biomarkers for coronary artery disease: A pilot microarray study and confirmation in a 32 patient cohort. Exp. Mol. Med. 2015;47:e138. doi: 10.1038/emm.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wang J., Pei Y., Zhong Y., Jiang S., Shao J., Gong J. Altered Serum MicroRNAs as Novel Diagnostic Biomarkers for Atypical Coronary Artery Disease. PLoS ONE. 2014;9:e107012. doi: 10.1371/journal.pone.0107012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.D’Alessandra Y., Carena M.C., Spazzafumo L., Martinelli F., Bassetti B., Devanna P., Rubino M., Marenzi G., Colombo G.I., Achilli F., et al. Diagnostic Potential of Plasmatic MicroRNA Signatures in Stable and Unstable Angina. PLoS ONE. 2013;8:e80345. doi: 10.1371/journal.pone.0080345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Sondermeijer B.M., Bakker A., Halliani A., de Ronde M.W.J., Marquart A.A., Tijsen A.J., Mulders T.A., Kok M.G.M., Battjes S., Maiwald S., et al. Platelets in Patients with Premature Coronary Artery Disease Exhibit Upregulation of miRNA340* and miRNA624*. PLoS ONE. 2011;6:e25946. doi: 10.1371/journal.pone.0025946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Gholipour A., Shakerian F., Zahedmehr A., Irani S., Mowla S.J., Malakootian M. Downregulation of Talin-1 is associated with the increased expression of miR-182-5p and miR-9-5p in coronary artery disease. J. Clin. Lab. Anal. 2022;36:e24252. doi: 10.1002/jcla.24252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ali W., Mishra S., Rizvi A., Pradhan A., Perrone M.A. Circulating microRNA-126 as an Independent Risk Predictor of Coronary Artery Disease: A Case-Control Study. EJIFCC. 2021;32:347–362. [PMC free article] [PubMed] [Google Scholar]
- 262.Kumar D., Narang R., Sreenivas V., Rastogi V., Bhatia J., Saluja D., Srivastava K. Circulatory miR-133b and miR-21 as Novel Biomarkers in Early Prediction and Diagnosis of Coronary Artery Disease. Genes. 2020;11:164. doi: 10.3390/genes11020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zhang P., Liang T., Chen Y., Wang X., Wu T., Xie Z., Luo J., Yu Y., Yu H. Circulating Exosomal miRNAs as Novel Biomarkers for Stable Coronary Artery Disease. Biomed Res. Int. 2020;2020:3593962. doi: 10.1155/2020/3593962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Guo J.-F., Zhang Y., Zheng Q.-X., Zhang Y., Zhou H.-H., Cui L.-M. Association between elevated plasma microRNA-223 content and severity of coronary heart disease. Scand. J. Clin. Lab. Investig. 2018;78:373–378. doi: 10.1080/00365513.2018.1480059. [DOI] [PubMed] [Google Scholar]
- 265.Yilmaz S.G., Isbir S., Kunt A.T., Isbir T. Circulating microRNAs as Novel Biomarkers for Atherosclerosis. In Vivo. 2018;32:561–565. doi: 10.21873/invivo.11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.O′Sullivan J.F., Neylon A., McGorrian C., Blake G.J. miRNA-93-5p and other miRNAs as predictors of coronary artery disease and STEMI. Int. J. Cardiol. 2016;224:310–316. doi: 10.1016/j.ijcard.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 267.Zhang L., Zhang Y., Xue S., Ding H., Wang Y., Qi H., Wang Y., Zhu W., Li P. Clinical significance of circulating microRNAs as diagnostic biomarkers for coronary artery disease. J. Cell. Mol. Med. 2020;24:1146–1150. doi: 10.1111/jcmm.14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zhang H., Hao J., Sun X., Zhang Y., Wei Q. Circulating pro-angiogenic micro-ribonucleic acid in patients with coronary heart disease. Interact. Cardiovasc. Thorac. Surg. 2018;27:336–342. doi: 10.1093/icvts/ivy058. [DOI] [PubMed] [Google Scholar]
- 269.Dong J., Liang Y.-Z., Zhang J., Wu L.-J., Wang S., Hua Q., Yan Y.-X. Potential Role of Lipometabolism-Related MicroRNAs in Peripheral Blood Mononuclear Cells as Biomarkers for Coronary Artery Disease. J. Atheroscler. Thromb. 2017;24:430–441. doi: 10.5551/jat.35923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Liu H., Yang N., Fei Z., Qiu J., Ma D., Liu X., Cai G., Li S. Analysis of plasma miR-208a and miR-370 expression levels for early diagnosis of coronary artery disease. Biomed. Rep. 2016;5:332–336. doi: 10.3892/br.2016.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Madhavan M.V., Gersh B.J., Alexander K.P., Granger C.B., Stone G.W. Coronary Artery Disease in Patients ≥80 Years of Age. J. Am. Coll. Cardiol. 2018;71:2015–2040. doi: 10.1016/j.jacc.2017.12.068. [DOI] [PubMed] [Google Scholar]
- 272.Ali Sheikh M.S., Xia K., Li F., Deng X., Salma U., Deng H., Wei L., Yang T.-L., Peng J. Circulating miR-765 and miR-149: Potential Noninvasive Diagnostic Biomarkers for Geriatric Coronary Artery Disease Patients. Biomed Res. Int. 2015;2015:740301. doi: 10.1155/2015/740301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lin J., Jiang J., Zhou R., Li X., Ye J. MicroRNA-451b participates in coronary heart disease by targeting VEGFA. Open Med. 2019;15:1105–1111. doi: 10.1515/med-2020-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Ali Sheikh M.S., Xia K., Li F., Deng X., Salma U., Li T., Deng H., Yang D., Haoyang Z., Yang T.-L., et al. The diagnostic value of circulating microRNAs for middle-aged (40–60-year-old) coronary artery disease patients. Clinics. 2015;70:257–263. doi: 10.6061/clinics/2015(04)07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lin D.-C., Lin J.-B., Chen Z., Chen R., Wan C.-Y., Lin S.-W., Ruan Q.-S., Li H.-Y., Wu S.-Y. Independent and combined effects of environmental factors and miR-126, miR-143, and miR-145 on the risk of coronary heart disease. J. Geriatr. Cardiol. 2017;14:688–695. doi: 10.11909/j.issn.1671-5411.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Yu H., Tu Y.-F., Liu H.-M., Xu M.-E. Diagnostic utility of circulating plasma microRNA-101a in severity of coronary heart disease. Ir. J. Med. Sci. 2021;190:1391–1396. doi: 10.1007/s11845-021-02512-7. [DOI] [PubMed] [Google Scholar]
- 277.Arora M., Kaul D., Sharma Y.P. Human coronary heart disease: Importance of blood cellular miR-2909 RNomics. Mol. Cell. Biochem. 2014;392:49–63. doi: 10.1007/s11010-014-2017-3. [DOI] [PubMed] [Google Scholar]
- 278.Zhang Y., Li H.-H., Yang R., Yang B.-J., Gao Z.-Y. Association between circulating microRNA-208a and severity of coronary heart disease. Scand. J. Clin. Lab. Investig. 2017;77:379–384. doi: 10.1080/00365513.2017.1328740. [DOI] [PubMed] [Google Scholar]
- 279.Wu H., Zhang J. miR-126 in Peripheral Blood Mononuclear Cells Negatively Correlates with Risk and Severity and is Associated with Inflammatory Cytokines as well as Intercellular Adhesion Molecule-1 in Patients with Coronary Artery Disease. Cardiology. 2018;139:110–118. doi: 10.1159/000484236. [DOI] [PubMed] [Google Scholar]
- 280.Chen J., Xu L., Hu Q., Yang S., Zhang B., Jiang H. MiR-17-5p as circulating biomarkers for the severity of coronary atherosclerosis in coronary artery disease. Int. J. Cardiol. 2015;197:123–124. doi: 10.1016/j.ijcard.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 281.Wang M., Li J., Cai J., Cheng L., Wang X., Xu P., Li G., Liang X. Overexpression of MicroRNA-16 Alleviates Atherosclerosis by Inhibition of Inflammatory Pathways. Biomed Res. Int. 2020;2020:8504238. doi: 10.1155/2020/8504238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Li H., Chen M., Feng Q., Zhu L., Bai Z., Wang B., Guo Z., Hou A., Li H. MicroRNA-34a in coronary heart disease: Correlation with disease risk, blood lipid, stenosis degree, inflammatory cytokines, and cell adhesion molecules. J. Clin. Lab. Anal. 2022;36:e24138. doi: 10.1002/jcla.24138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Li H., Gao F., Wang X., Wu J., Lu K., Liu M., Li R., Ding L., Wang R. Circulating microRNA-378 levels serve as a novel biomarker for assessing the severity of coronary stenosis in patients with coronary artery disease. Biosci. Rep. 2019;39:BSR20182016. doi: 10.1042/BSR20182016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Zhu L. The correlations of circulating microRNA-133a with the risk and severity of coronary heart disease. Int. J. Clin. Exp. Med. 2017;10:972–978. [Google Scholar]
- 285.Li H.-Y., Zhao X., Liu Y.-Z., Meng Z., Wang D., Yang F., Shi Q.-W. Plasma MicroRNA-126-5p is Associated with the Complexity and Severity of Coronary Artery Disease in Patients with Stable Angina Pectoris. Cell. Physiol. Biochem. 2016;39:837–846. doi: 10.1159/000447794. [DOI] [PubMed] [Google Scholar]
- 286.Gao H., Guddeti R.R., Matsuzawa Y., Liu L.-P., Su L.-X., Guo D., Nie S.-P., Du J., Zhang M. Plasma Levels of microRNA-145 Are Associated with Severity of Coronary Artery Disease. PLoS ONE. 2015;10:e0123477. doi: 10.1371/journal.pone.0123477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Reddy L.L., Shah S.A.V., Ponde C.K., Rajani R.M., Ashavaid T.F. Circulating miRNA-33: A potential biomarker in patients with coronary artery disease. Biomarkers. 2019;24:36–42. doi: 10.1080/1354750X.2018.1501760. [DOI] [PubMed] [Google Scholar]
- 288.Hosseinpor S., Khalvati B., Safari F., Mirzaei A., Hosseini E. The association of plasma levels of miR-146a, miR-27a, miR-34a, and miR-149 with coronary artery disease. Mol. Biol. Rep. 2022;49:3559–3567. doi: 10.1007/s11033-022-07196-5. [DOI] [PubMed] [Google Scholar]
- 289.Babaee M., Chamani E., Ahmadi R., Bahreini E., Balouchnejadmojarad T., Nahrkhalaji A.S., Fallah S. The expression levels of miRNAs- 27a and 23a in the peripheral blood mononuclear cells (PBMCs) and their correlation with FOXO1 and some inflammatory and anti-inflammatory cytokines in the patients with coronary artery disease (CAD) Life Sci. 2020;256:117898. doi: 10.1016/j.lfs.2020.117898. [DOI] [PubMed] [Google Scholar]
- 290.Zehtabian S.H., Alibakhshi R., Seyedena S.Y., Rai A.R. Relationship between microRNA-206 plasma levels with the severity of coronary artery conflicts in patients with coronary artery disease. Bratisl. Med. J. 2019;120:581–585. doi: 10.4149/BLL_2019_095. [DOI] [PubMed] [Google Scholar]
- 291.Mishra S., Rizvi A., Pradhan A., Perrone M.A., Ali W. Circulating microRNA-126 &122 in patients with coronary artery disease: Correlation with small dense LDL. Prostaglandins Other Lipid Mediat. 2021;153:106536. doi: 10.1016/j.prostaglandins.2021.106536. [DOI] [PubMed] [Google Scholar]
- 292.Gorur A., Celik A., Yildirim D.D., Gundes A., Tamer L. Investigation of possible effects of microRNAs involved in regulation of lipid metabolism in the pathogenesis of atherosclerosis. Mol. Biol. Rep. 2019;46:909–920. doi: 10.1007/s11033-018-4547-3. [DOI] [PubMed] [Google Scholar]
- 293.Wang Z., Zhang J., Zhang S., Yan S., Wang Z., Wang C., Zhang X. MiR-30e and miR-92a are related to atherosclerosis by targeting ABCA1. Mol. Med. Rep. 2019;19:3298–3304. doi: 10.3892/mmr.2019.9983. [DOI] [PubMed] [Google Scholar]
- 294.Choteau S.A., Cuesta Torres L.F., Barraclough J.Y., Elder A.M.M., Martínez G.J., Chen Fan W.Y., Shrestha S., Ong K.L., Barter P.J., Celermajer D.S., et al. Transcoronary gradients of HDL-associated MicroRNAs in unstable coronary artery disease. Int. J. Cardiol. 2018;253:138–144. doi: 10.1016/j.ijcard.2017.09.190. [DOI] [PubMed] [Google Scholar]
- 295.Ahmadi R., Heidarian E., Fadaei R., Moradi N., Malek M., Fallah S. miR-342-5p Expression Levels in Coronary Artery Disease Patients and its Association with Inflammatory Cytokines. Clin. Lab. 2018;64:603–609. doi: 10.7754/Clin.Lab.2017.171208. [DOI] [PubMed] [Google Scholar]
- 296.De Rosa S., Arcidiacono B., Chiefari E., Brunetti A., Indolfi C., Foti D.P. Type 2 Diabetes Mellitus and Cardiovascular Disease: Genetic and Epigenetic Links. Front. Endocrinol. 2018;9:2. doi: 10.3389/fendo.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Vesa C.M., Popa L., Popa A.R., Rus M., Zaha A.A., Bungau S., Tit D.M., Corb Aron R.A., Zaha D.C. Current Data Regarding the Relationship between Type 2 Diabetes Mellitus and Cardiovascular Risk Factors. Diagnostics. 2020;10:314. doi: 10.3390/diagnostics10050314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Merino J., Jablonski K.A., Mercader J.M., Kahn S.E., Chen L., Harden M., Delahanty L.M., Araneta M.R.G., Walford G.A., Jacobs S.B.R., et al. Interaction Between Type 2 Diabetes Prevention Strategies and Genetic Determinants of Coronary Artery Disease on Cardiometabolic Risk Factors. Diabetes. 2020;69:112–120. doi: 10.2337/db19-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Jansen F., Wang H., Przybilla D., Franklin B.S., Dolf A., Pfeifer P., Schmitz T., Flender A., Endl E., Nickenig G., et al. Vascular endothelial microparticles-incorporated microRNAs are altered in patients with diabetes mellitus. Cardiovasc. Diabetol. 2016;15:49. doi: 10.1186/s12933-016-0367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wang W.-Y., Zheng Y.-S., Li Z.-G., Cui Y.-M., Jiang J.-C. MiR-92a contributes to the cardiovascular disease development in diabetes mellitus through NF-κB and downstream inflammatory pathways. Eur. Rev. Med. Pharmacol. Sci. 2019;23:3070–3079. doi: 10.26355/eurrev_201904_17589. [DOI] [PubMed] [Google Scholar]
- 301.Al-Muhtaresh H.A., Salem A.H., Al-Kafaji G. Upregulation of Circulating Cardiomyocyte-Enriched miR-1 and miR-133 Associate with the Risk of Coronary Artery Disease in Type 2 Diabetes Patients and Serve as Potential Biomarkers. J. Cardiovasc. Transl. Res. 2019;12:347–357. doi: 10.1007/s12265-018-9857-2. [DOI] [PubMed] [Google Scholar]
- 302.Motawae T.M., Ismail M.F., Shabayek M.I., Seleem M.M. MicroRNAs 9 and 370 Association with Biochemical Markers in T2D and CAD Complication of T2D. PLoS ONE. 2015;10:e0126957. doi: 10.1371/journal.pone.0126957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Zhang L., Zhang J., Qin Z., Liu N., Zhang Z., Lu Y., Xu Y., Zhang J., Tang J. Diagnostic and Predictive Values of Circulating Extracellular Vesicle-Carried microRNAs in Ischemic Heart Disease Patients With Type 2 Diabetes Mellitus. Front. Cardiovasc. Med. 2022;9:813310. doi: 10.3389/fcvm.2022.813310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Zogg H., Singh R., Ro S. Current Advances in RNA Therapeutics for Human Diseases. Int. J. Mol. Sci. 2022;23:2736. doi: 10.3390/ijms23052736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 306.Choi S.-E., Fu T., Seok S., Kim D.-H., Yu E., Lee K.-W., Kang Y., Li X., Kemper B., Kemper J.K. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell. 2013;12:1062–1072. doi: 10.1111/acel.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Bijkerk R., Esguerra J.L.S., Ellenbroek J.H., Au Y.W., Hanegraaf M.A.J., de Koning E.J., Eliasson L., van Zonneveld A.J. In Vivo Silencing of MicroRNA-132 Reduces Blood Glucose and Improves Insulin Secretion. Nucleic Acid Ther. 2019;29:67–72. doi: 10.1089/nat.2018.0763. [DOI] [PubMed] [Google Scholar]
- 308.Hung Y.-H., Kanke M., Kurtz C.L., Cubitt R., Bunaciu R.P., Miao J., Zhou L., Graham J.L., Hussain M.M., Havel P., et al. Acute suppression of insulin resistance-associated hepatic miR-29 in vivo improves glycemic control in adult mice. Physiol. Genomics. 2019;51:379–389. doi: 10.1152/physiolgenomics.00037.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Zhou B., Li C., Qi W., Zhang Y., Zhang F., Wu J.X., Hu Y.N., Wu D.M., Liu Y., Yan T.T., et al. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia. 2012;55:2032–2043. doi: 10.1007/s00125-012-2539-8. [DOI] [PubMed] [Google Scholar]
- 310.Thibonnier M., Esau C., Ghosh S., Wargent E., Stocker C. Metabolic and energetic benefits of microRNA-22 inhibition. BMJ Open Diabetes Res. Care. 2020;8:e001478. doi: 10.1136/bmjdrc-2020-001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Thibonnier M., Esau C. Metabolic Benefits of MicroRNA-22 Inhibition. Nucleic Acid Ther. 2020;30:104–116. doi: 10.1089/nat.2019.0820. [DOI] [PubMed] [Google Scholar]
- 312.Seeger T., Fischer A., Muhly-Reinholz M., Zeiher A.M., Dimmeler S. Long-term Inhibition of miR-21 Leads to Reduction of Obesity in db/db Mice. Obesity. 2014;22:2352–2360. doi: 10.1002/oby.20852. [DOI] [PubMed] [Google Scholar]
- 313.Wang Y., Yang L.-Z., Yang D.-G., Zhang Q.-Y., Deng Z.-N., Wang K., Mao X.-J. MiR-21 antagomir improves insulin resistance and lipid metabolism disorder in streptozotocin-induced type 2 diabetes mellitus rats. Ann. Palliat. Med. 2020;9:394–404. doi: 10.21037/apm.2020.02.28. [DOI] [PubMed] [Google Scholar]
- 314.Xu L., Li Y., Yin L., Qi Y., Sun H., Sun P., Xu M., Tang Z., Peng J. miR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics. 2018;8:5593–5609. doi: 10.7150/thno.27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Liu R., Wang M., Li E., Yang Y., Li J., Chen S., Shen W.-J., Azhar S., Guo Z., Hu Z. Dysregulation of microRNA-125a contributes to obesity-associated insulin resistance and dysregulates lipid metabolism in mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158640. doi: 10.1016/j.bbalip.2020.158640. [DOI] [PubMed] [Google Scholar]
- 316.Cheng X., Huang Y., Yang P., Bu L. miR-383 ameliorates high glucose-induced β-cells apoptosis and hyperglycemia in high-fat induced diabetic mice. Life Sci. 2020;263:118571. doi: 10.1016/j.lfs.2020.118571. [DOI] [PubMed] [Google Scholar]
- 317.Tian F.-J., An L.-N., Wang G.-K., Zhu J.-Q., Li Q., Zhang Y.-Y., Zeng A., Zou J., Zhu R.-F., Han X.-S., et al. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc. Res. 2014;103:100–110. doi: 10.1093/cvr/cvu070. [DOI] [PubMed] [Google Scholar]
- 318.Yang Y., Yang L., Liang X., Zhu G. MicroRNA-155 Promotes Atherosclerosis Inflammation via Targeting SOCS1. Cell. Physiol. Biochem. 2015;36:1371–1381. doi: 10.1159/000430303. [DOI] [PubMed] [Google Scholar]
- 319.Wei Y., Nazari-Jahantigh M., Chan L., Zhu M., Heyll K., Corbalán-Campos J., Hartmann P., Thiemann A., Weber C., Schober A. The microRNA-342-5p Fosters Inflammatory Macrophage Activation Through an Akt1- and microRNA-155–Dependent Pathway During Atherosclerosis. Circulation. 2013;127:1609–1619. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- 320.Zheng C.-G., Chen B.-Y., Sun R.-H., Mou X.-Z., Han F., Li Q., Huang H.-J., Liu J.-Q., Tu Y.-X. miR-133b Downregulation Reduces Vulnerable Plaque Formation in Mice with AS through Inhibiting Macrophage Immune Responses. Mol. Ther. Nucleic Acids. 2019;16:745–757. doi: 10.1016/j.omtn.2019.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Loyer X., Potteaux S., Vion A.-C., Guérin C.L., Boulkroun S., Rautou P.-E., Ramkhelawon B., Esposito B., Dalloz M., Paul J.-L., et al. Inhibition of MicroRNA-92a Prevents Endothelial Dysfunction and Atherosclerosis in Mice. Circ. Res. 2014;114:434–443. doi: 10.1161/CIRCRESAHA.114.302213. [DOI] [PubMed] [Google Scholar]
- 322.Gao F., Chen X., Xu B., Luo Z., Liang Y., Fang S., Li M., Wang X., Lin X. Inhibition of MicroRNA-92 alleviates atherogenesis by regulation of macrophage polarization through targeting KLF4. J. Cardiol. 2022;79:432–438. doi: 10.1016/j.jjcc.2021.10.015. [DOI] [PubMed] [Google Scholar]
- 323.Rayner K.J., Suárez Y., Dávalos A., Parathath S., Fitzgerald M.L., Tamehiro N., Fisher E.A., Moore K.J., Fernández-Hernando C. MiR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Rayner K.J., Esau C.C., Hussain F.N., McDaniel A.L., Marshall S.M., van Gils J.M., Ray T.D., Sheedy F.J., Goedeke L., Liu X., et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and reduces VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Rayner K.J., Sheedy F.J., Esau C.C., Hussain F.N., Temel R.E., Parathath S., van Gils J.M., Rayner A.J., Chang A.N., Suarez Y., et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Investig. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Ouimet M., Ediriweera H.N., Gundra U.M., Sheedy F.J., Ramkhelawon B., Hutchison S.B., Rinehold K., van Solingen C., Fullerton M.D., Cecchini K., et al. MicroRNA-33–dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Investig. 2015;125:4334–4348. doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Goedeke L., Salerno A., Ramírez C.M., Guo L., Allen R.M., Yin X., Langley S.R., Esau C., Wanschel A., Fisher E.A., et al. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol. Med. 2014;6:1133–1141. doi: 10.15252/emmm.201404046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M., Watts L., Booten S.L., Graham M., McKay R., et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 329.Goedeke L., Rotllan N., Canfrán-Duque A., Aranda J.F., Ramírez C.M., Araldi E., Lin C.-S., Anderson N.N., Wagschal A., de Cabo R., et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 2015;21:1280–1289. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Soh J., Iqbal J., Queiroz J., Fernandez-Hernando C., Hussain M.M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Tan L., Liu L., Jiang Z., Hao X. Inhibition of microRNA-17-5p reduces the inflammation and lipid accumulation, and up-regulates ATP-binding cassette transporterA1 in atherosclerosis. J. Pharmacol. Sci. 2019;139:280–288. doi: 10.1016/j.jphs.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 332.Zhang X.-F., Yang Y., Yang X.-Y., Tong Q. MiR-188-3p upregulation results in the inhibition of macrophage proinflammatory activities and atherosclerosis in ApoE-deficient mice. Thromb. Res. 2018;171:55–61. doi: 10.1016/j.thromres.2018.09.043. [DOI] [PubMed] [Google Scholar]
- 333.Wang J., Li P., Xu X., Zhang B., Zhang J. MicroRNA-200a Inhibits Inflammation and Atherosclerotic Lesion Formation by Disrupting EZH2-Mediated Methylation of STAT3. Front. Immunol. 2020;11:907. doi: 10.3389/fimmu.2020.00907. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 334.Huang R., Cao Y., Li H., Hu Z., Zhang H., Zhang L., Su W., Xu Y., Liang L., Melgiri N.D., et al. miR-532-3p-CSF2RA Axis as a Key Regulator of Vulnerable Atherosclerotic Plaque Formation. Can. J. Cardiol. 2020;36:1782–1794. doi: 10.1016/j.cjca.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 335.Jiang L., Hao C., Li Z., Zhang P., Wang S., Yang S., Wei F., Zhang J. miR-449a induces EndMT, promotes the development of atherosclerosis by targeting the interaction between AdipoR2 and E-cadherin in Lipid Rafts. Biomed. Pharmacother. 2019;109:2293–2304. doi: 10.1016/j.biopha.2018.11.114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.