Abstract
Various metals have been associated with the pathogenesis of Alzheimer’s disease (AD), principally heavy metals that are environmental pollutants (such as As, Cd, Hg, and Pb) and essential metals whose homeostasis is disturbed in AD (such as Cu, Fe, and Zn). Although there is evidence of the involvement of these metals in AD, further research is needed on their mechanisms of toxicity. To further assess the involvement of heavy and essential metals in AD pathogenesis, we compared cerebrospinal fluid (CSF) AD biomarkers to macro- and microelements measured in CSF and plasma. We tested if macro- and microelements’ concentrations (heavy metals (As, Cd, Hg, Ni, Pb, and Tl), essential metals (Na, Mg, K, Ca, Fe, Co, Mn, Cu, Zn, and Mo), essential non-metals (B, P, S, and Se), and other non-essential metals (Al, Ba, Li, and Sr)) are associated with CSF AD biomarkers that reflect pathological changes in the AD brain (amyloid β1–42, total tau, phosphorylated tau isoforms, NFL, S100B, VILIP-1, YKL-40, PAPP-A, and albumin). We used inductively coupled plasma mass spectroscopy (ICP-MS) to determine macro- and microelements in CSF and plasma, and enzyme-linked immunosorbent assays (ELISA) to determine protein biomarkers of AD in CSF. This study included 193 participants (124 with AD, 50 with mild cognitive impairment, and 19 healthy controls). Simple correlation, as well as machine learning algorithms (redescription mining and principal component analysis (PCA)), demonstrated that levels of heavy metals (As, Cd, Hg, Ni, Pb, and Tl), essential metals (Ca, Co, Cu, Fe, Mg, Mn, Mo, Na, K, and Zn), and essential non-metals (P, S, and Se) are positively associated with CSF phosphorylated tau isoforms, VILIP-1, S100B, NFL, and YKL-40 in AD.
Keywords: Alzheimer’s disease, heavy metals, essential metals, cerebrospinal fluid, biomarker, arsenic, mercury, cadmium, iron, zinc, calcium
1. Introduction
Macroelements are those elements that the body needs more than any other mineral and include sodium (Na), potassium (K), calcium (Ca), magnesium (Mg), chlorine (Cl), phosphorus (P), and sulfur (S). Microelements are those elements that are required for good health in very small amounts, such as chromium (Cr), copper (Cu), fluorine (F), iodine (I), iron (Fe), manganese (Mn), molybdenum (Mo), selenium (Se), zinc (Zn), and cobalt (Co). Some of these microelements are also metals and are regarded as essential for human health in trace amounts, for example, Fe, Zn, Cu, Mn, Cr, Mo, Se, and Co (Co because it is necessary for the formation of vitamin B12–cobalamin). However, non-essential metals are considered harmful to human health and the environment. These include four heavy metals: arsenic (As), cadmium (Cd), lead (Pb), and mercury (Hg) that can cause neurodegenerative changes [1] and have been also associated with the development of Alzheimer’s disease (AD) [2]. People are exposed to heavy metals through water, soil, and air. Heavy metals can cross the blood–brain barrier (BBB), accumulate in the brain, and bypass BBB, entering the brain directly through the olfactory pathway [3].
There is evidence that essential metal homeostasis is disturbed in the brain of AD patients [4,5,6]. Here, in the first place, we mean metals that are present in the body under normal circumstances and are essential for the functioning of numerous enzymes. Calcium, Na, and Mg are the most abundant metals in the human body, while metals such as Cu, Fe, Zn, Cr, Mo, Mn, and Co are found only in traces. Metal ions stabilize proteins and nucleic acids are crucial for the function of metalloenzymes located in the active site of enzymes and act as secondary messengers.
An increase in the concentration of heavy metals and altered homeostasis of essential metals is observed in AD brains, which contributes to tau protein hyperphosphorylation [7,8] and Aβ aggregation [9,10]. Additionally, it was shown that essential metals such as Fe, Zn, and Cu accumulate within senile plaques and promote Aβ and tau pathology. Increased levels of metals can also contribute to the impairment of the BBB [11], oxidative stress [12], altered calcium homeostasis [13], neuroinflammation [14], apoptosis, and necrosis of neurons [15,16]. Various studies measured both heavy metals and essential metals in plasma and cerebrospinal fluid (CSF) of patients with dementia (reviewed in [17]), but due to high variability among studies, metals are still not used as biomarkers in the diagnostics of AD. However, various metal chelators were tested as potential therapeutic agents in AD (reviewed in [18]), clioquinol in particular, whose action is based on the removal of excess metal ions in the brain. This compound showed good results in the second phase of clinical trials [19], but due to manufacturing difficulties, it did not progress further [20]. PBT2, a homolog of clioquinol, showed better therapeutic effects, but its ability to reduce the pathological changes associated with the accumulation of Aβ in the brain of AD patients could not be demonstrated (http://www.alzforum.org/news/research-news/pbt2-takes-dive-phase-2-alzheimers-trial (accessed on 20 November 2022)). Clioquinol and PBT2 remove excess copper and zinc by acting as ionophores, and clioquinol probably also removes iron by chelation [4,21]. Other iron chelators such as epigallocatechin-3-gallate and M-30 have also shown beneficial effects on pathological changes characteristic of AD in vivo and in vitro, and deferoxamine has also been tested on patients (reviewed in [20]).
The main goal of this study was to test the association of various CSF biomarkers of AD with macro- and microelements measured in CSF and the plasma of AD patients, patients with mild cognitive impairment (MCI), and healthy controls (HC). We assessed such associations with AD-related pathological changes reflected by the levels of eleven AD biomarkers in CSF, whose association with AD is established.
2. Results
2.1. Correlation
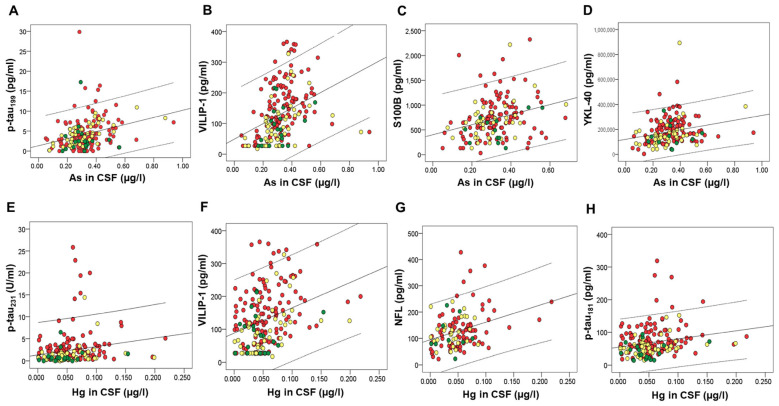
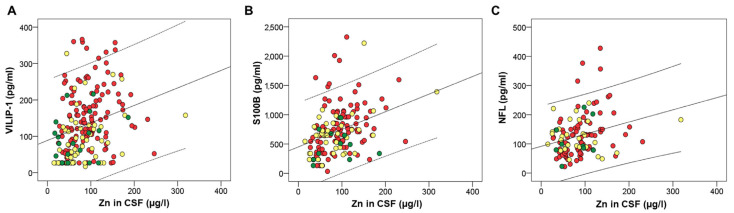
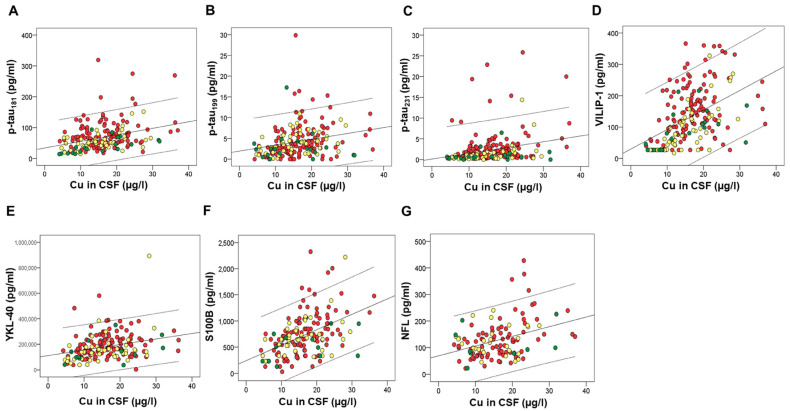
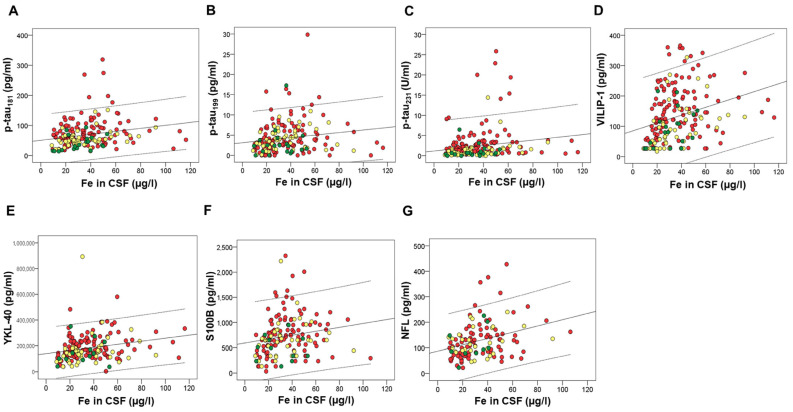
Several macro- and microelements correlated with CSF AD biomarkers. However, after applying Bonferroni correction for multiple comparisons, we considered statistically significant only those correlations with p-values ≤ 0.001. These correlations are presented in Table 1. Macro- and microelements that correlated with the high number of CSF AD biomarkers were all measured in CSF; As and Hg (Figure 1), Zn (Figure 2), Cu (Figure 3), Fe (Figure 4), S, K, Se, Co, Mn, Ni, Na, Mg, Tl, and Li (Table 1).
Table 1.
Correlations of CSF AD biomarkers with macro- and microelements.
| In CSF | p-tau181 | p-tau199 | p-tau231 | VILIP-1 | YKL-40 | S100B | NFL | PAPP-A | Aβ1–42, Aβ1–42/p-tau181 or Albumin |
|---|---|---|---|---|---|---|---|---|---|
| Fe | rS = 0.26, p < 0.001(193) *B |
rs = 0.258, p < 0.001 (193) *F |
rs = 0.236, p = 0.001 (193) *C |
rs = 0.407, p < 0.001 (193)* |
rs = 0.32, p < 0.001 (178)* |
rs = 0.286, p < 0.001 (158) *E |
rs = 0.344, p < 0.001 (118)* |
rs = 0.243, p = 0.001 (178)* |
rs = 0.314, p < 0.001 (142) (albumin)* |
| Cu | rs = 0.321, p < 0.001 (193)* |
rs = 0.334, p < 0.001 (193)* |
rs = 0.303, p < 0.001 (193)* |
rs = 0.537, p < 0.001 (193)* |
rs = 0.309, p < 0.001 (178)* |
rs = 0.483, p < 0.001 (158)* |
rs = 0.323, p < 0.001 (118)* |
rs = 0.435, p < 0.001 (178)* |
rs = 0.439, p < 0.001 (142) (albumin)* |
| S | rs = 0.246, p = 0.001 (193) *A |
rs = 0.322, p < 0.001 (193) *A |
rs = 0.549, p < 0.001 (193)* |
rs = 0.394, p < 0.001 (178)* |
rs = 0.611, p < 0.001 (159)* |
rs = 0.381, p < 0.001 (119)* |
rs = 0.523, p < 0.001 (178)* |
rs = 0.49, p < 0.001 (142) (albumin)* r = −0.25, p < 0.001 (192) (Aβ1–42/p-tau181) *B |
|
| K | rS = 0.239, p = 0.001 (193) *G |
rs = 0.27, p < 0.001 (193)* |
rs = 0.541, p < 0.001 (193)* |
rs = 0.26, p < 0.001 (178) *C |
rs = 0.311, p < 0.001 (159)* |
rs = 0.302, p = 0.001 (119)* |
rs = 0.288, p < 0.001 (178)* |
||
| Se | rs = 0.466, p < 0.001 (193)* |
rs = 0.399, p < 0.001 (193)* |
rs = 0.344, p < 0.001 (193)* |
rs = 0.614, p < 0.001 (193)* |
rs = 0.277, p < 0.001 (178) *H |
rs = 0.398, p < 0.001 (159)* |
rs = 0.322, p < 0.001 (178)* |
r = 0.294, p < 0.001 (142) (albumin) *B |
|
| Co | rS = 0.223, p = 0.002 (193) *B |
rs = 0.359, p < 0.001 (192)* |
rs = 0.446, p < 0.001 (192)* |
rs = 0.284, p < 0.001 (179) *A |
rs = 0.42, p < 0.001 (159)* |
rs = 0.315, p < 0.001 (119)* |
rs = 0.476, p < 0.001 (177)* |
r = 0.349, p < 0.001 (142) (albumin) *A |
|
| Mn | rs = 0.23, p = 0.001 (193) *B |
rs = 0.261, p < 0.001 (193)* |
rs = 0.33, p < 0.001 (193)* |
rs = 0.274, p < 0.001 (159) *B |
rs = 0.318, p < 0.001 (178)* |
||||
| Ni (CSF) | rs = 0.253, p < 0.001 (188)* |
r = 0.307, p < 0.001 (188)* |
rs = 0.309, p < 0.001 (188)* |
rs = 0.252, p = 0.001 (178) *C |
rs = 0.306, p < 0.001 (173)* |
rs = 0.29, p < 0.001 (139) (albumin) *C |
|||
| Ni (plasma) | r = 0.353, p < 0.001 (123)* |
r = 0.376 p < 0.001 (135)* |
rs = 0.288, p = 0.001 (134) (albumin)* |
||||||
| Na (CSF) | rs = 0.3, p < 0.001 (193) *A |
rs = 0.590, p < 0.001 (193) *A |
rs = 0.356, p < 0.001 (178)* |
rs = 0.422, p < 0.001 (159)* |
rs = 0.325, p < 0.001 (119)* |
rs = 0.338, p < 0.001 (178)* |
|||
| Na (plasma) | r = 0.321, p < 0.001 (123) |
||||||||
| Mg | rs = 0.255, p < 0.001 (193) *A |
rs = 0.550, p < 0.001 (193) *A |
rs = 0.29, p < 0.001 (178)* |
rs = 0.37, p < 0.001 (159)* |
rs = 0.366, p < 0.001 (119)* |
rs = 0.345, p < 0.001 (178)* |
|||
| As | rs = 0.384, p < 0.001 (192)* |
rs = 0.521, p < 0.001 (192)* |
rs = 0.32, p < 0.001 (177)* |
rs = 0.344, p < 0.001 (157)* |
|||||
| Hg | rs = 0.228, p = 0.001 (192)* |
rs = 0.262, p < 0.001 (192)* |
rs = 0.347, p < 0.001 (192)* |
rs = 0.331, p < 0.001 (117)* |
|||||
| Tl | r = 0.233, p = 0.001 (193)* |
rs = 0.253, p < 0.001 (193)* |
rs = 0.299, p < 0.001 (159)* |
r = 0.334, p < 0.001 (119)* |
rs = 0.37, p < 0.001 (178)* |
||||
| P | rs = 0.444, p < 0.001 (76)* |
rs = 0.506, p < 0.001 (72)* |
rs = 0.825, p < 0.001 (37)* |
rs = 0.382, p < 0.001 (82)* |
rS = 0.414, p < 0.001 (89) (Aβ)* |
||||
| Li (CSF) | rs = 0.227, p = 0.001 (193) *A |
rs = 0.281, p < 0.001 (178)* |
rs = 0.368, p < 0.001 (159) *B |
rs = 0.290, p = 0.001 (119)* |
rs = 0.318, p < 0.001 (178) *A |
||||
| Li (plasma) | r = −0.413, p < 0.001 (123) |
r = −0.47 p < 0.001 (135) |
rs = −0.287, p = 0.001 (134) (albumin) A |
||||||
| Zn | r = 0.327, p < 0.001 (193)* |
r = 0.376, p < 0.001 (158)* |
r = 0.295, p = 0.001 (118)* |
r = 0.322, p < 0.001 (178)* |
r = 0.375, p < 0.001 (142) (albumin) |
||||
| Mo | r = 0.408, p < 0.001 (193)* |
r = 0.269, p < 0.001 (178)* |
r = 0.29, p < 0.001 (159)* |
r = 0.336, p < 0.001 (178)* |
|||||
| Ba | r = 0.25, p < 0.001 (191) *B |
r = 0.374, p < 0.001 (157) |
r = 0.294, p = 0.001 (117) B |
||||||
| Ca | rs = 0.404, p < 0.001 (193) |
rs = 0.295, p < 0.001 (159)* |
rs = 0.342, p < 0.001 (119) * |
r = 0.25, p = 0.001 (178) |
|||||
| B | rs = 0.258, p < 0.001 (193) *B |
rs = 0.299, p = 0.001 (119) *B |
|||||||
| Pb | r = −0.285, p < 0.001 (178)* |
||||||||
| Sr | r = 0.405, p < 0.001 (119)* |
Data are presented as either Spearman’s or Pearson’s correlation coefficients and p-values, with the number of participants given in the brackets; (* p ≤ 0.001). A Significance was lost after correction for the effect of age, gender, diagnosis, and duration of the disease. Statistically significant after correction for the confounding effect of: B age, gender, and diagnosis; C gender, diagnosis, and duration of the disease; D age and gender; E diagnosis and duration of the disease; F diagnosis; G gender; H gender and diagnosis. Aβ1–42, amyloid β1–42; CSF, cerebrospinal fluid; NFL, neurofilament light chain; PAPP-A, pregnancy-associated plasma protein A; p-tau181, tau protein phosphorylated at threonine 181; p-tau231, tau protein phosphorylated at threonine 231; p-tau199, tau protein phosphorylated at serine 199; S100B, S100 calcium-binding protein B; VILIP-1, Visinin-like protein 1; YKL-40, chitinase-3-like protein 1.
Figure 1.
Correlation of CSF biomarkers of AD with As (A–D) and Hg (E–H) measured in CSF. Red circles represent AD patients, yellow circles represent MCI patients, while green circles represent healthy controls.
Figure 2.
Correlation of CSF biomarkers VILIP-1 (A), S100B (B), and NFL (C) of AD with Zn measured in CSF. Red circles represent AD patients, yellow circles represent MCI patients, while green circles represent healthy controls.
Figure 3.
Correlation of CSF biomarkers of AD – p-tau181 (A), ptau199 (B), ptau231 (C), VILIP-1 (D), YKL-40 (E), S100B (F), and NFL (G) with Cu measured in CSF. Red circles represent AD patients, yellow circles represent MCI patients, while green circles represent healthy controls.
Figure 4.
Correlation of CSF biomarkers of AD – p-tau181 (A), ptau199 (B), ptau231 (C), VILIP-1 (D), YKL-40 (E), S100B (F), and NFL (G) with Fe measured in CSF. Red circles represent AD patients, yellow circles represent MCI patients, while green circles represent healthy controls.
2.2. Principal Component Analysis
Principal component analysis (PCA) showed differences in all macro- and microelements measured in CSF (except for P, which was measured in the relatively small number of subjects) and plasma. Factor analysis of CSF macro- and microelements included 23 parameters that were classified into 6 groups (factors) after analysis (Table 2). Factors explained 73.2% of the total variance in analyzed CSF macro- and microelements (with Bartlett’s test of sphericity of p < 0.001 and Kaiser–Meyer–Olkin measure of sampling adequacy of 0.846). Factor analysis of plasma macro- and microelements included 21 parameters that were classified into six groups (factors) after analysis (Table 3). Factors explained 66.1% of the total variance in analyzed plasma macro- and microelements (with Bartlett’s test of sphericity of p < 0.001 and Kaiser–Meyer–Olkin measure of sampling adequacy of 0.781).
Table 2.
CSF macro- and microelements and their factor loadings for 6 groups (factors) (given by the PCA).
| Factors | CSF Macro- and Microelements (Factor Loadings) |
|---|---|
| Factor 1 | As (0.470), Ba (0.611), Ca (0.845), Co (0.646), Cu (0.800), Fe (0.712), K (0.897), Mg (0.919), Mn (0.702), Na (0.870), Ni (0.562), S (0.748), Se (0.825), Sr (0.590), Tl (0.584), Zn (0.641) |
| Factor 2 | Al (0.565), Cd (0.664), Pb (0.871) |
| Factor 3 | B (0.595), Li (0.584) |
| Factor 4 | None loaded |
| Factor 5 | Hg (−0.505), Mo (0.601) |
| Factor 6 | None loaded |
Table 3.
Plasma macro- and microelements and their factor loadings for 6 groups (factors) (given by the PCA).
| Factors | Plasma Macro- and Microelements (Factor Loadings) |
|---|---|
| Factor 1 | Ca (0.848), Co (0.491), Cu (0.59), Mg (0.659), Mn (0.577), Na (0.831), P (0.735), S (0.844), Se (0.728), Tl (0.35), Zn (0.569) |
| Factor 2 | B (0.571), Cd (0.604), Li (0.649), Mo (0.580), Pb (0.38) |
| Factor 3 | As (0.634), Hg (0.651) |
| Factor 4 | Ni (0.617) |
| Factor 5 | Sr (−0.595) |
| Factor 6 | Fe (0.478) |
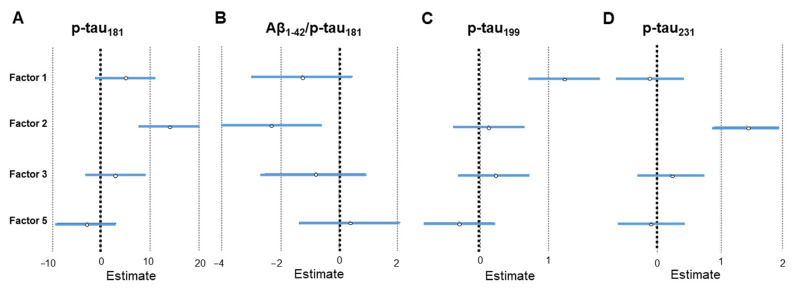
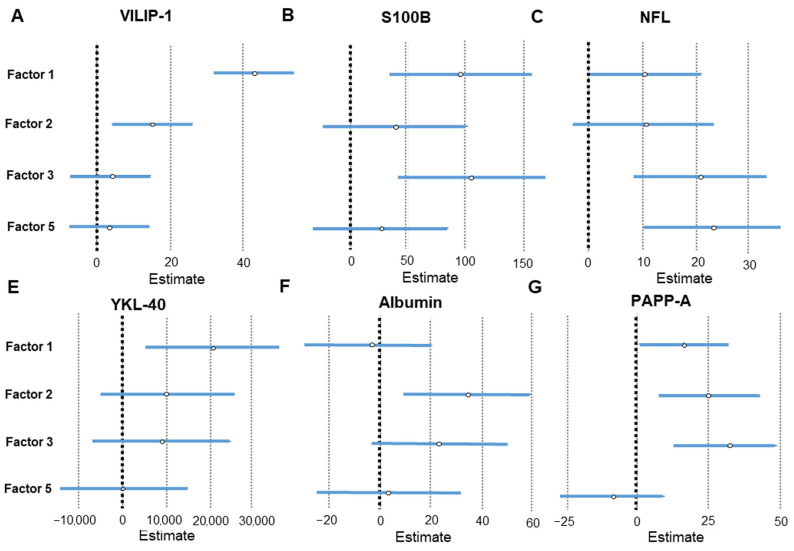
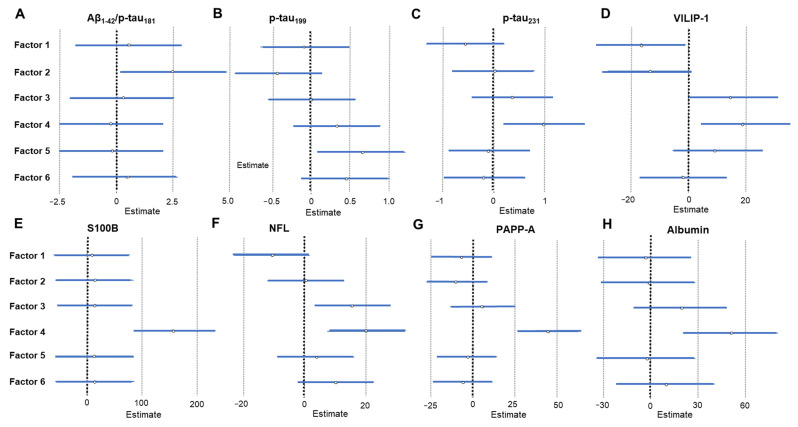
The results of linear regression analysis that compared the factors obtained by PCA analysis of CSF macro- and microelements with CSF AD biomarkers are given in Table 4 and Figure 5 and Figure 6. The results of linear regression analysis that compared the factors obtained by PCA of plasma macro- and microelements with CSF AD biomarkers are given in Table 5 and Figure 7. The most interesting finding is that following PCA heavy metals are distinctly grouped (Cd, Pb, and Al in CSF; As and Hg in plasma). The metals measured in CSF (Cd, Pb, and Al) associated positively with tau phosphorylated at Thr 181 (p-tau181), Thr 231 (p-tau231), visinin-like protein 1 (VILIP-1), pregnancy-associated plasma protein A, pappalysin-1 (PAPP-A) and albumin, and negatively with the Aβ1–42/p-tau181 ratio. The metals measured in plasma (As and Hg) showed a positive association with VILIP-1 and neurofilament light chain (NFL) levels. PCA also showed that Ni measured in plasma was positively associated with various CSF AD biomarkers. In CSF, B and Li grouped together (Factor 3) and showed a positive association with NFL, S100 calcium-binding protein B (S100B), and PAPP-A.
Table 4.
Results of linear regression analysis; comparison of factors obtained by PCA of CSF macro- and microelements with CSF AD biomarkers.
| Factor | CSF AD Biomarker | Linear Regression Analysis |
|---|---|---|
| Factor 2 (Al, Cd, and Pb) |
p-tau181 | β = 0.321, SE = 3.064, p < 0.001 |
| p-tau231 | β = 0.375, SE = 0.263, p < 0.001 | |
| VILIP-1 | β = 0.173, SE = 5.531, p = 0.007 | |
| PAPP-A | β = 0.21, SE = 8.659, p = 0.005 | |
| albumin | β = 0.231, SE = 12.494, p = 0.007 | |
| Aβ1–42/p-tau181 | β = −0.192, SE = 0.873, p = 0.009 | |
| Factor 1 (As, Ba, Ca, Co, Cu, Fe, K, Mg, Mn, Na, Ni, S, Se, Sr, Tl, and Zn) |
p-tau199 | β = 0.331, SE = 0.262, p < 0.001 |
| VILIP-1 | β = 0.495, SE = 5.531, p < 0.001 | |
| YKL-40 | β = 0.205, SE = 7663.241, p = 0.007 | |
| S100B | β = 0.24, SE = 30.022, p = 0.002 | |
| PAPP-A | β = 0.148, SE = 7.893, p = 0.041 | |
| Factor 3 (B and Li) |
S100B | β = 0.259, SE = 31.29, p = 0.001 |
| NFL | β = 0.285, SE = 6.257, p = 0.001 | |
| PAPP-A | β = 0.275, SE = 8.551, p < 0.001 | |
| Factor 5 (Hg and Mo) | NFL | β = 0.312, SE = 6.388, p < 0.001 |
Aβ1–42, amyloid β; AD, Alzheimer’s disease; CSF, cerebrospinal fluid; NFL, neurofilament light chain; PAPP-A, pregnancy-associated plasma protein A; p-tau181, tau protein phosphorylated at threonine 181; p-tau231, tau protein phosphorylated at threonine 231; p-tau199, tau protein phosphorylated at serine 199; S100B, S100 calcium-binding protein B; VILIP-1, Visinin-like protein 1; YKL-40, chitinase-3-like protein 1.
Figure 5.
Association of CSF AD biomarkers – p-tau181 (A), Aβ1–42/p-tau181 ratio (B), p-tau199 (C), and p-tau231 (D) with CSF macro- and microelement groups revealed by PCA. Effect sizes and 95% confidence intervals are given by linear regression analysis (statistically significant factors are considered those represented with horizontal lines that do not intersect with the vertical line at 0). Factor 1; CSF As, Ba, Ca, Co, Cu, Fe, K, Mg, Mn, Na, Ni, S, Se, Sr, Tl, and Zn; Factor 2; CSF Al, Cd, and Pb; Factor 3; CSF B and Li; Factor 5; CSF Hg and Mo.
Figure 6.
Association of CSF AD biomarkers – VILIP-1 (A), S100B (B), NFL (C), YKL-40 (E), albumin (F), and PAPP-A (G) with CSF macro- and microelement groups revealed by PCA. Effect sizes and 95% confidence intervals are given by linear regression analysis (statistically significant factors are considered those represented with horizontal lines that do not intersect with the vertical line at 0). Factor 1; CSF As, Ba, Ca, Co, Cu, Fe, K, Mg, Mn, Na, Ni, S, Se, Sr, Tl, and Zn; Factor 2; CSF Al, Cd, and Pb; Factor 3; CSF B and Li; Factor 5; CSF Hg and Mo.
Table 5.
Results of linear regression analysis; comparison of factors obtained by PCA of plasma macro- and microelements with CSF AD biomarkers.
| Factor | CSF AD Biomarker | Linear Regression Analysis |
|---|---|---|
| Factor 1 (Ca, Co, Cu, Mg, Mn, Na, P, S, Se, Tl, and Zn) |
VILIP-1 | β = −0.191, SE = 7.341, p = 0.028 |
| Factor 2 (B, Cd, Li, Mo and Pb) |
Aβ1–42/p-tau181 | β = 0.192, SE = 0.873, p = 0.009 |
| Factor 3 (As and Hg) |
VILIP-1 | β = 0.17, SE = 7.341, p = 0.05 |
| NFL | β = 0.237, SE = 6.109, p = 0.013 | |
| Factor 4 (Ni) | p-tau231 | β = 0.224, SE = 0.39, p = 0.014 |
| VILIP-1 | β = 0.222, SE = 7.341, p = 0.011 | |
| S100B | β = 0.405, SE = 36.065, p < 0.001 | |
| NFL | β = 0.295, SE = 6.450, p = 0.002 | |
| PAPP-A | β = 0.428, SE = 8.945, p < 0.001 | |
| albumin | β = 0.306, SE = 15.086, p = 0.001 | |
| Factor 5 (Sr) | p-tau199 | β = 0.204, SE = 0.284, p = 0.023 |
Aβ1–42, amyloid β; AD, Alzheimer’s disease; CSF, cerebrospinal fluid; NFL, neurofilament light chain; PAPP-A, pregnancy-associated plasma protein A; p-tau181, tau protein phosphorylated at threonine 181; p-tau231, tau protein phosphorylated at threonine 231; p-tau199, tau protein phosphorylated at serine 199; S100B, S100 calcium-binding protein B; VILIP-1, Visinin-like protein 1; YKL-40, chitinase-3-like protein 1.
Figure 7.
Association of CSF AD biomarkers – Aβ1–42/p-tau181 ratio (A), p-tau199 (B), p-tau231 (C), VILIP-1 (D), S100B (E), NFL (F), PAPP-A (G), and albumin (H) with plasma macro- and microelement groups revealed by the PCA. Effect sizes and 95% confidence intervals are given by linear regression analysis (statistically significant factors are considered those represented with horizontal lines that do not intersect with the vertical line at 0). Factor 1; plasma Ca, Co, Cu, Mg, Mn, Na, P, S, Se, Tl, and Zn; Factor 2; plasma B, Cd, Li, Mo, and Pb; Factor 3; plasma As and Hg; Factor 4; plasma Ni; Factor 5; plasma Sr; Factor 6; plasma Fe.
2.3. Redescription Mining
Redescription mining showed an association of CSF AD biomarkers with macro- and microelements measured in CSF (Table 6), and plasma (Table 7), as well as an association of CSF AD biomarkers with macro- and microelements measured both in CSF and in plasma (Table 7). Information on age, Mini-Mental State Examination (MMSE), and APOE (apolipoprotein E) genotype were included in the analysis. Redescription mining gave us in total of 2648 redescriptions. Redescriptions that depict more closely: (1) AD patients, (2) both AD and MCI patients, (3) MCI patients, (4) MCI patients and HC, and (5) HC were extracted (Table 6, Table 7). When analyzing the association of CSF AD biomarkers with macro- and microelements measured in CSF, VILIP-1 occurred together with Se in 81 redescriptions, and Ba and Cu in 69 redescriptions. P-tau181 occurred together with Se in 67 redescriptions and Cu in 59 redescriptions, while chitinase-3-like protein 1 (YKL-40) was associated with Se and Cu in 52 and 41 redescriptions, respectively. When analyzing the association of CSF AD biomarkers with macro- and microelements measured in plasma, PAPP-A occurred together with the following elements (the number of redescriptions is shown in parentheses): Li (281), Ca (279), Na (192), P (191), Ni (177), Hg (172), Se (164), Sr (130), and Mo (125) (only the most significant associations are presented). Total tau (t-tau) occurred together with Ca (181), Li (166), P (126), and Na (125); YKL-40 with Ca (173), Li (171), P (129), and Na (118); p-tau231 with Li (168), Ca (159), P (122), and Se (116); and p-tau181 with Li (122), and Ca (125).
Table 6.
Most significant redescriptions given by the combination of CSF AD biomarkers (first view—W1) and macro- and microelements measured in CSF (second view—W2).
| Redescription Specific for: | HC (%) | MCI (%) | AD (%) | W1R | W2R | JS | p-Value |
|---|---|---|---|---|---|---|---|
| Macro- and Microelements in CSF | |||||||
| AD | 5.3% | 6% | 36% | VILIP-1 (121.28–366.14) MMSE (13–25) Aβ1–42 (275–1246) |
As in CSF (0.34–2.62) S in CSF (14.3–40.2) |
0.56977 | 1.548 × 10−7 * |
| AD | 5.3% | 6% | 31.2% | VILIP-1 (121.28–366.14) MMSE (13–25) Aβ1–42 (275–1246) |
As in CSF (0.34–2.62) K in CSF (120–243) S in CSF (14.3–40.2) Cd in CSF (0.01–0.033) |
0.5584 | 5.711 × 10−8 * |
| AD | 21% | 44% | 70.4% | YKL-40 (102,044.391955–893,335.0) VILIP-1 (62.52–366.14) |
K in CSF (94.0–243) | 0.7125 | 0.003 * |
| AD | 15.7% | 38% | 63.2% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Zn in CSF (33.0–318.1) K in CSF (94–243) |
0.65584 | 0.002 * |
| AD | 15.8% | 38% | 63.2% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Mg in CSF (22.45–52.21) Zn in CSF (33–318.1) |
0.66013 | 0.002 * |
| AD&MCI | 5.3% | 46% | 59.2% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Na in CSF (2545–5335) Li in CSF (0.21–49.93) Cu in CSF (9.9–36.98) |
0.64901 | 0.004 * |
| AD&MCI | 5.3% | 38% | 50.4% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Se in CSF (1.1–3.35) Cu in CSF (10.7–36.27) Co in CSF (0.046–0.219) Ba in CSF (0.98–110.4) Cd in CSF (0.004–0.033) |
0.60145 | 3.545 × 10−4 * |
| AD&MCI | 5.3% | 34% | 52% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Co in CSF (0.046–0.219) Se in CSF (1.1–3.35) Zn in CSF (27.9–246.1) |
0.58865 | 0.003 * |
| AD&MCI | 5.3% | 34% | 51.2% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Li in CSF (0.21–49.93) Zn in CSF (33–318.1) Cu in CSF (10.2–36.98) Na in CSF (2545.0–5335) |
0.59854 | 0.001 * |
| AD&MCI | 5.3% | 42% | 46.4% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Se in CSF (1.1–3.35) Zn in CSF (27.9–111) Ba in CSF (0.62–105.83) |
0.55944 | 0.003 * |
| AD&MCI | 5.3% | 44% | 60.8% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Co in CSF (0.046–0.219) Se in CSF (1.1–3.35) Zn in CSF (27.9–246.1) |
0.66 | 0.002 * |
| AD&MCI | 5.3% | 38% | 56% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Cu in CSF (10.3–36.27) Co in CSF (0.046–0.219) Fe in CSF (19.1–111.3) S in CSF (12.7–32) Ba in CSF (1.61–110.4) |
0.61644 | 0.002 * |
| AD&MCI | 5.3% | 38% | 56% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Cu in CSF (10.3–36.27) Co in CSF (0.046–0.219) Fe in CSF (19.1–111.3) S in CSF (12.7–32) Ba in CSF (1.61–110.4) |
0.616434 | 0.002 * |
| AD&MCI | 5.3% | 36% | 45.6% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Se in CSF (1.1–3.69) Li in CSF (0.39–49.93) Ba in CSF (3.2–110.4) |
0.55882 | 4.295 × 10−4 * |
| AD&MCI | 5.3% | 38% | 52.8% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Se in CSF (1.1–3.35) Zn in CSF (27.9–246.1) Co in CSF (0.046–0.219) Cd in CSF (0.004–0.033) |
0.6014 | 0.001 * |
| AD&MCI | 5.3% | 34% | 50.4% | VILIP-1 (62.52–366.14) MMSE (10–29) |
S in CSF (9.6–32) Se in CSF (1.1–3.35) Co in CSF (0.046–0.219) Ba in CSF (0.98–110.4) Cd in CSF (0.004–0.033) |
0.59124 | 3.177 × 10−4 * |
| AD&MCI | 5.3% | 34% | 56.8% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Zn in CSF (28–246.1) Cu in CSF (9.92–36.27) Na in CSF (2063–4873) Co in CSF (0.046–0.219) |
0.58940 | 0.01 * |
| AD&MCI | 5.3% | 38% | 56.8% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Cu in CSF (10.3–36.27) Co in CSF (0.046–0.219) S in CSF (12.7–32) Ba in CSF (1.61–110.4) |
0.61486 | 0.004 * |
| AD&MCI | 0% | 36% | 48.8% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Hg in CSF (0.025–1.063) Cu in CSF (10.3–36.16) Co in CSF (0.046–0.219) Fe in CSF (19.1–111.3) S in CSF (12.7–32) Ba in CSF (1.61–110.4) |
0.5411 | 0.009 * |
| AD&MCI | 5.3% | 30% | 44% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Se in CSF (1.1–3.35) Cu in CSF (10.7–36.27) Co in CSF (0.046–0.219) Ba in CSF (0.98–110.4) Cd in CSF (0.004–0.033) |
0.568 | 2.255 × 10−4 * |
| AD&MCI | 5.3% | 36% | 48% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Li in CSF (0.25–49.93) Zn in CSF (33–318.1) Cu in CSF (10.2–36.98) Na in CSF (2196–5335) |
0.57246 | 0.002 * |
| AD&MCI | 5.3% | 30% | 44.8% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Se in CSF (1.1–3.35) Zn in CSF (27.9–246.1) Co in CSF (0.046–0.219) Cd in CSF (0.004–0.033) |
0.54545 | 0.002 * |
| AD&MCI | 5.3% | 26% | 50.4% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Zn in CSF (33–246.1) Cu in CSF (10.3–36.27) Co in CSF (0.046–0.219) S in CSF (12.7–32) Ba in CSF (1.61–110.4) |
0.56618 | 0.002 * |
| AD&MCI | 15.8% | 52% | 76% | VILIP-1 (62.52–366.14) MMSE (10–29) |
K in CSF (94–243) | 0.77019 | 0.002 * |
| AD&MCI | 15.8% | 54% | 76.8% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Na in CSF (2527–5492) | 0.76829 | 0.003 * |
| AD&MCI | 15.8% | 46% | 68% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Se in CSF (12.7–40.8) | 0.70253 | 0.005 * |
| AD&MCI | 15.8% | 52% | 71.2% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Se in CSF (1.1–3.69) Zn in CSF (27.9–318.1) |
0.78146 | 1.676 × 10−4 * |
| AD&MCI | 15.8% | 44% | 66.4% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Cd in CSF (0.004–0.033) Mg in CSF (22.45–52.21) Fe in CSF (15.5–116.3) |
0.71053 | 0.001 * |
| AD&MCI | 15.8% | 56% | 78.4% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Cu in CSF (9.35–36.98) S in CSF (7.8–40.8) |
0.7771 | 0.004 * |
| AD&MCI | 15.8% | 52% | 75.2% | VILIP-1 (62.52–366.14) MMSE (10–29) |
Mg in CSF (22.45–52.21) S in CSF (7.9–40.8) |
0.76398 | 0.002 * |
| AD&MCI | 15.8% | 40% | 60% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Zn in CSF (33–318.1) Cu in CSF (9.92–36.98) Fe in CSF (15.5–116.3) Al in CSF (0.9–6.75) Na in CSF (2063–5335) |
0.65772 | 0.001 * |
| AD&MCI | 15.8% | 42% | 64.8% | S100B (234.16–2325.45) VILIP-1 (62.52–366.14) MMSE (10–29) |
Na in CSF (2063–5492) Zn in CSF (28–318.1) Cu in CSF (9.92–36.98) |
0.67308 | 0.002 * |
| AD&MCI | 10.5% | 34% | 50.4% | VILIP-1 (62.52–366.14) YKL-40 (102,044.391955–893,335.0) Aβ1–42 (272.94–1568.0) |
Li in CSF (0.21–49.93) Se in CSF (0.89–3.69) Cu in CSF (10.7–36.98) K in CSF (94–243) Ba in CSF (1.61–110.4) |
0.63077 | 1.050 × 10−4 * |
| HC&MCI | 47.4% | 42% | 28.8% |
APOE genotype = ɛ3ɛ3 Aβ1–42 (275–1672.36) |
Pb in CSF (0.56–14.83) Ba in CSF (0.98–110.4) |
0.51163 | 0.001 * |
| HC&MCI | 42.1% | 50% | 18.4% | VILIP-1 (17.2–89.68) | K in CSF (37–134) S in CSF (4.2–17.6) |
0.51852 | 4.439 × 10−5 * |
| HC&MCI | 42.1% | 50% | 15.2% | VILIP-1 (17.2–89.68) p-tau231 (0–1.978) t-tau (44–598.0) |
Se in CSF (0.36–1.97) K in CSF (37–134) S in CSF (4.2–17.6) |
0.5 | 3.855 × 10−5 * |
| HC&MCI | 47.4% | 30% | 14.4% | VILIP-1 (17.2–89.68) | Se in CSF (0.36–1.09) Mn in CSF (0.4–3.58) |
0.53846 | 5.472 × 10−8 * |
| HC&MCI | 52.6% | 56% | 33.6% | VILIP-1 (17.2–121.08) p-tau181 (16–145.9) |
As in CSF (0.05–0.33) Fe in CSF (8.8–74.1) |
0.625 | 7.274 × 10−5 * |
| HC&MCI | 47.4% | 30% | 12.8% | VILIP-1 (17.2–89.68) p-tau231 (0–1.978) t-tau (44–598) |
Mn in CSF (0.4–3.58) Se in CSF (0.36–1.09) |
0.53333 | 6.249 × 10−8 * |
| HC&MCI | 42.1% | 50% | 16% | VILIP-1 (17.2–89.68) p-tau231 (0–1.978) t-tau (44–598) |
Se in CSF (0.36–1.97) K in CSF (37–140) S in CSF (4.2–17.7) |
0.50476 | 3.487 × 10−5 * |
| HC&MCI | 47.4% | 30% | 13.6% | t-tau (44–598) VILIP-1 (17.2–89.68) |
Mn in CSF (0.4–3.58) Se in CSF (0.36–1.09) |
0.53947 | 4.804 × 10−8 * |
| HC&MCI | 42.1% | 50% | 19.2% | VILIP-1 (17.2–89.68) | K in CSF (37–140) S in CSF (4.2–17.7) Se in CSF (0.36–1.97) |
0.53774 | 1.584 × 10−5 * |
| HC&MCI | 42.1% | 50% | 16% | VILIP-1 (17.2–89.68) t-tau (44.0–598) |
Se in CSF (0.36–1.97) K in CSF (37–134) S in CSF (4.2–17.6) Na in CSF (958–3658.0) |
0.50962 | 3.108 × 10−5 * |
| HC&MCI | 42.1% | 50% | 17.6% | VILIP-1 (17.2–89.68) t-tau (44–598) |
Se in CSF (0.36–1.97) K in CSF (37–140) S in CSF (4.2–17.7) |
0.52381 | 1.910 × 10−5 * |
| HC&MCI | 42% | 50% | 18.4% | VILIP-1 (17.2–89.68) | Se in CSF (0.36–1.97) K in CSF (37–140) S in CSF (4.2–17.7) |
0.53333 | 1.682 × 10−5 * |
| HC | 42.1% | 22% | 12.8% | PAPP-A (23.64–345.15) VILIP-1 (17.2–89.12) p-tau231 (0–1.978) p-tau199 (0–5.781) |
Se in CSF (0.36–1.09) Mn in CSF (0.4–3.58) |
0.5303 | 3.716 × 10−8 * |
| HC | 42.1% | 24% | 12.8% | VILIP-1 (17.2–89.12) p-tau231 (0–1.978) PAPP-A (23.64–345.15) |
Mn in CSF (0.4–3.58) Se in CSF (0.36–1.09) |
0.51429 | 1.133 × 10−7 * |
| HC | 42.1% | 22% | 12.8% | VILIP-1 (17.2–89.12) p-tau231 (0–1.978) p-tau199 (0–5.781) PAPP-A (23.64–345.15) |
Mn in CSF (0.4–2.51) Se in CSF (0.36–1.09) As in CSF (0.06–0.34) |
0.55556 | 7.250 × 10−9 * |
Data are presented as percentages of diagnoses and ranges of CSF biomarkers and elements. W1R refers to the first redescription query (constructed using attributes from the first view—W1), W2R refers to the second redescription query (constructed using attributes from the second view—W2), JS refers to the Jaccard similarity coefficient (Jaccard index) and p-value refers to the statistical significance of a redescription [22]. Aβ1–42, amyloid β; AD, Alzheimer’s disease; APOE, apolipoprotein E; CSF, cerebrospinal fluid; HC, healthy control; JS, Jaccard similarity index; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; NFL, neurofilament light chain; PAPP-A, pregnancy-associated plasma protein A; p-tau181, tau protein phosphorylated at threonine 181; p-tau231, tau protein phosphorylated at threonine 231; p-tau199, tau protein phosphorylated at serine 199; S100B, S100 calcium-binding protein B; VILIP-1, Visinin-like protein 1; YKL-40, chitinase-3-like protein 1.* p ≤ 0.05.
Table 7.
Most significant redescriptions given by the combination of CSF AD biomarkers (first view—W1) with macro- and microelements measured in plasma (second view—W2), and macro- and microelements measured both in CSF and in plasma (second view—W2 and third view W3).
| Redescription Specific for: | HC (%) | MCI (%) | AD (%) | W1R | W2R | JS | p-Value | |
|---|---|---|---|---|---|---|---|---|
| Macro- and Microelements in Plasma | ||||||||
| AD | 0% | 8.6% | 30.1% | Albumin (58.44–411) PAPP-A (122.39–511.26) MMSE (5–28) Age (49–82) p-tau181 (23.19–126.71) T-tau (313.77–1992) |
Ca in plasma (67.9–87.5) Na in plasma (3713–4033) Li in plasma (15–48) Cd in plasma (0.02–0.07) Ni in plasma (1.33–2.24) |
0.64583 | 2.475 × 10−8 * | |
| AD | 0% | 8.6% | 32.3% | Albumin (58.44–411) S100B (293.15–1596.64) PAPP-A (141–529.35) MMSE (5–28) Age (53–82) p-tau181 (23.19–126.71) t-tau (207.85–1992) |
Li in plasma (14–29) Sr in plasma (16.1–44.7) Cu in plasma (779–1456) Ca in plasma (67.4–85.3) Pb in plasma (102.8–150) Hg in plasma (0.04–1.51) |
0.6346 | 2.9642 × 10−8 * | |
| AD | 6.6% | 22.9% | 50.5% | t-tau (207.85–1992) YKL-40 (4000–298,634) |
Ca in plasma (73.6–90.4) Mg in plasma (22.3–43.6) Na in plasma (3131–4033) B in plasma (12.3–70.6) Mn in plasma (0.78–2.35) |
0.57143 | 0.002 * | |
| AD | 0% | 5.7% | 32.3% | Albumin (58.44–413.96) S100B (293.15–1925.01) PAPP-A (118.65–529.35) MMSE (5–28) p-tau181 (23.19–126.71) T-tau (211–1992) Aβ1–42 (136.3–1230.43) |
Na in plasma (3716–4023) Li in plasma (15–32) Ca in plasma (67.3–87.5) |
0.51613 | 4.924 × 10−6 * | |
| AD | 0% | 14.3% | 41.9% | PAPP-A (122.39–529.35) YKL-40 (4000–364,460) MMSE (15–28) p-tau231 (0.432–25.83) T-tau (77.46–1992) |
Li in plasma (14–32) Cd in plasma (0.01–0.07) Se in plasma (41.5–95.4) Mn in plasma (0.85–2.35) |
0.53659 | 2.812 × 10−4 * | |
| AD | 6.6% | 8.6% | 40.9% | PAPP-A (122.39–529.35) YKL-40 (67,043.38–364,460) MMSE (5–28) Age (49–82) p-tau181 (23.19–127) T-tau (207.85–1992) |
Ca in plasma (67.3–87.5) Mg in plasma (22.2–32.2) Na in plasma (3671–4039) Mo in plasma (0.72–12.02) As in plasma (0.52–24.8) Zn in plasma (522–942) Ni in plasma (1.33–4.09) Mn in plasma (0.83–2.35) |
0.61111 | 6.127 × 10−6 * | |
| AD | 0% | 22.9% | 41% | T-tau (207.85–1992) MMSE (5–28) Age (49–82) p-tau231 (0.357–9.09) PAPP-A (122.39–529.35) |
Cd in plasma (0–0.07) Ni in plasma (1.33–4.02) Mn in plasma (0.75–2.35) Ca in plasma (67.4–87.5) P in plasma (102.8–160.5) Na in plasma (3139–4033) |
0.64474 | 5.877 × 10−6 * | |
| AD | 6.6% | 17.1% | 41.9% | PAPP-A (122.39–529.35) YKL-40 (4000–384,387.22) p-tau231 (0.432–25.83) p-tau199 (0–15.781) p-tau181 (21.6–319.16) |
Li in plasma (14–32) Cd in plasma (0.01–0.07) Se in plasma (41.5–95.4) Mn in plasma (0.85–2.35) |
0.51111 | 0.001 * | |
| AD | 6.6% | 22.9% | 45.2% | YKL-40 (4000–280,873) T-tau (313.77–2259) NFL (30.607–315.19) |
Li in plasma (15–134) Mo in plasma (0.58–12.02) Mn in plasma (0.78–2.35) Ca in plasma (73.6–90.4) Mg in plasma (22.3–43.6) Na in plasma (3131–4033) B in plasma (12.3–70.6) |
0.54839 | 0.001 * | |
| AD | 6.6% | 20% | 45% | PAPP-A (122.39–529.35) T-tau (313.77–1992) |
Ca in plasma (67.4–87.5) Mg in plasma (22.3–32.2) Na in plasma (3215–4033) B in plasma (12.3–70.6) Hg in plasma (0.04–3.25) Mo in plasma (0.72–12.02) Zn in plasma (522–942) Ni in plasma (1.11–4.09) Mn in plasma (0.8–2.35) |
0.54945 | 8.267 × 10−4 * | |
| AD&MCI | 6.6% | 45.7% | 45.2% | S100B (130.89–1267.47) Age (49–83) NFL (42.473–266.028) |
Mo in plasma (0.83–12.02) Pb in plasma (103.9–223.7) As in plasma (0.48–3.2) |
0.56731 | 0.006 * | |
| AD&MCI | 0% | 22.8% | 41.9% | t-tau (207.85–1992) MMSE (5–28) Age (49–82) p-tau181 (23.19–126.71) PAPP-A (122.39–529.35) |
Cd in plasma (0.01–0.07) Ni in plasma (1.33–4.02) Mn in plasma (0.83–2.35) Ca in plasma (67.4–85.1) Pb in plasma (102.8–160.5) |
0.63514 | 4.880 × 10−6 * | |
| AD&MCI | 6.6% | 48% | 43% | S100B (130.89–1267.47) Age (49–83) NFL (42.473–266.028) |
Se in plasma (70.2–169.9) Co in plasma (0.26–1.07) Pb in plasma (103.9–223.7) |
0.55769 | 0.007 * | |
| AD&MCI | 6.6% | 31.4% | 53.8% | T-tau (235–2259) YKL-40 (4000–298634) MMSE (5–28) |
Pb in plasma (0.12–1.21) Mn in plasma (0.75–2.35) Fe in plasma (742–3939) B in plasma (12.3–56.6) |
0.59048 | 0.008 * | |
| AD&MCI | 6.6% | 28.6% | 47.3% | PAPP-A (132.89–529.35) p-tau181 (21.6–319.16) p-tau199 (0.44–17.266) Aβ1–42 (136.3–696.0) |
Mg in plasma (18.6–30.3) B in plasma (12.3–64.4) Pb in plasma (0.18–3.36) Cd in plasma (0.01–0.07) Zn in plasma (522–942) Ni in plasma (0.95–4.09) Mn in plasma (0.8–3.24) Ca in plasma (66.4–87.5) |
0.56122 | 0.002 * | |
| AD&MCI | 6.6% | 40% | 44.1% | NFL (42.473–266.028) S100B (130.89–1267.47) PAPP-A (23.64–312.85) Age (49–83) |
Li in plasma (16–174) P in plasma (103.9–249.4) As in plasma (0.48–24.8) Mg in plasma (21.8–43.6) Hg in plasma (0.04–2.23) |
0.58333 | 0.001 * | |
| AD&MCI | 6.6% | 31.4% | 46.2% | PAPP-A (132.89–529.35) p-tau181 (21.6–319.16) Aβ1–42 (136.3–696) |
Ca in plasma (66.4–85.3) Mg in plasma (20.8–30.3) B in plasma (12.3–70.6) Cd in plasma (0.01–0.07) Mo in plasma (0.72–12.02) Zn in plasma (522–942) Cu in plasma (500–1120) Ni in plasma (1.11–4.09) Co in plasma (0.26–1.07) |
0.55556 | 0.003 * | |
| Macro- and Microelements in CSF and Plasma | ||||||||
| Redescription specific for: | HC (%) | MCI (%) | AD (%) | W1R | W2R | W3R | JS | p-Value |
| AD&MCI | 6.6% | 45.7% | 66% | Age (60–91) t-tau (148.21–1992) |
Zn in plasma (507–942) S in plasma (654–896) Na in plasma (3248–4043) B in plasma (15.5–213.1) |
Se in CSF (0.69–3.69) | 0.59848 | 0.003 * |
| AD&MCI | 6.6% | 45.7% | 60.2% | NFL (22–220.86) YKL-40 (81,665–483,135.59) p-tau231 (0.357–22.87) p-tau181 (23.19–319.16) |
B in plasma (12.3–70.6) Co in plasma (0.27–1.07) P in plasma (78.1–249.4) Pb in plasma (0.1–3.36) |
Se in CSF (0.74–3.69) Li in CSF (0.03–8.36) |
0.54074 | 1.933 × 10−4 * |
| AD&MCI | 6.6% | 40% | 61.3% | Albumin (73.84–1683.65) Age (64–91) p-tau181 (37.62–319.16) |
S in plasma (452–945) Zn in plasma (492–1115) |
Co in CSF (0.081–0.872) | 0.5 | 0.009 * |
| AD&MCI | 6.6% | 54.3% | 72% | Age (63–91) p-tau181 (23.19–319.16) |
Pb in plasma (0.09–3.36) Mn in plasma (0.82–3.22) Li in plasma (16–174) |
Se in CSF (0.82–3.69) | 0.64444 | 8.697 × 10−4 * |
| AD&MCI | 6.6% | 45.7% | 64.6% | Age (60–91) t-tau (148.21 <= 1992) |
Ca in plasma (67.4–85.1) Mg in plasma (22.2–32.2) Na in plasma (3149–4039) |
Se in CSF (0.69–3.69) | 0.50327 | 0.009 * |
| AD&MCI | 6.6% | 45.7% | 72% | Age (60–91) p-tau231 (0.278–25.83) |
Na in plasma (2578–4012) B in plasma (15.5–213.1) Pb in plasma (0.1–3.36) |
Li in CSF (0.05–21.32) Cu in CSF (10.6–36.98) |
0.5283 | 0.002 * |
| AD&MCI | 26% | 65.7% | 79.6% | p-tau231 (0.278–25.83) t-tau (88–2259) |
Na in plasma (3149–4769) Mo in plasma (0.81–12.02) |
Mg in CSF (13.92–42.7) Se in CSF (0.69–3.15) |
0.66887 | 0.004 * |
| AD&MCI | 20% | 65.7% | 75.4% | p-tau231 (0.278–25.83) t-tau (88–2259) |
Na in plasma (3149–4023) Pb in plasma (0.09–3.36) Mo in plasma (0.81–12.02) |
Cu in CSF (8.77–36.98) | 0.63576 | 0.009 * |
| AD&MCI | 20% | 65.7% | 75.3% | p-tau231 (0–7.941) p-tau181 (21.27–194) Aβ1–42 (136.3–1347) |
B in plasma (7.9–70.6) Hg in plasma (0.04–3.25) |
Ni in CSF (0.26–2.03) Fe in CSF (11.9–111.3) |
0.70073 | 0.006 * |
| AD&MCI | 20% | 60% | 75.3% | p-tau231 (0.288–25.83) t-tau (88–2259) |
Ca in plasma (63.4–85.1) Mo in plasma (0.81–12.02) Zn in plasma (507–1115) |
Mg in CSF (13.92–42.7) Se in CSF (0.69–3.15) |
0.62667 | 0.009 * |
| AD&MCI | 20% | 51.4% | 74.2% | Age (63–91) p-tau181 (23.19–319.16) |
Mg in plasma (15.4–30.4) Mn in plasma (0.51–3.22) |
Se in CSF (0.82–3.69) | 0.65217 | 0.004 * |
| HC | 93.3% | 71.4% | 63.4% | VILIP-1 (17.2–224.29) | Sr in plasma (14.6–44.7) Ca in plasma (71.4–108.2) |
Cd in CSF (0.005–0.045) Na in CSF (958.0–4481.0) |
0.64474 | 0.006 * |
Data are presented as percentages of diagnoses and ranges of CSF biomarkers and elements. W1R refers to the first redescription query (constructed using attributes from the first view—W1), W2R refers to the second redescription query (constructed using attributes from the second view—W2), W3R refers to the third redescription query (constructed using attributes from the third view—W3), JS refers to the Jaccard similarity coefficient (Jaccard index) and p-value refers to the statistical significance of a redescription [22]. Aβ1–42, amyloid β; AD, Alzheimer’s disease; CSF, cerebrospinal fluid; HC, healthy control; JS, Jaccard similarity index; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; NFL, neurofilament light chain; PAPP-A, pregnancy-associated plasma protein A; p-tau181, tau protein phosphorylated at threonine 181; p-tau231, tau protein phosphorylated at threonine 231; p-tau199, tau protein phosphorylated at serine 199; S100B, S100 calcium-binding protein B; VILIP-1, Visinin-like protein 1; YKL-40, chitinase-3-like protein 1.* p ≤ 0.05.
3. Discussion
In this study, we used three different statistical methods to test the association of macro- and microelements with CSF biomarkers of AD. All three methods (simple correlation, redescription mining, and PCA) demonstrated some association of macro- and microelements with CSF AD biomarkers. Macro- and microelements that positively correlated with a high number of CSF AD biomarkers were As, Hg, Zn, Cu, Fe, S, K, Se, Co, Mn, Ni, Na, Mg, Tl, and Li (all elements measured in CSF). PCA further confirmed the association of these macro- and microelements with CSF AD biomarkers. Following PCA, heavy metals are distinctly grouped (Cd, Pb, and Al in CSF; As and Hg in plasma). The metals measured in CSF (Cd, Pb, and Al) associated positively with p-tau181, p-tau231, VILIP-1, PAPP-A, and albumin, and negatively with the Aβ1–42/p-tau181 ratio. The metals measured in plasma (As and Hg) showed a positive association with VILIP-1 and NFL levels. PCA also showed that Ni measured in plasma was positively associated with various CSF AD biomarkers. The redescription mining algorithm successfully clustered individuals by the combination of CSF AD biomarkers with macro- and microelements measured in CSF, CSF AD biomarkers with macro- and microelements measured in plasma, and CSF AD biomarkers with macro- and microelements measured both in CSF and in plasma. Using this algorithm, we extracted those redescriptions that depict more closely: AD patients, both AD and MCI patients, MCI patients, MCI patients and HC, and HC. Additionally, redescription mining showed the association of Ca, Li, P, and Na measured in plasma with various CSF AD biomarkers.
Several studies investigated the association of heavy metals, essential and non-essential metals, and essential non-metals with CSF AD biomarkers. Hock et al. showed a positive correlation between Hg blood levels and CSF amyloid β1–42 (Aβ1–42) levels [23]. A study on 28 AD patients and 25 HC found a negative correlation between serum Cu levels with CSF Aβ1–42, and a positive correlation with CSF t-tau levels [24], while oral intake of Cu showed no effect on CSF Aβ1–42, t-tau and p-tau levels in 68 AD patients [25]. Strozyk et al. observed a negative correlation between CSF Cu, Zn, Fe, Mn, and Cr levels and CSF Aβ1–42 levels [26]. A recent study that included 20 AD patients, 10 HC, and 10 patients with cerebral amyloid angiopathy (CAA) observed a negative correlation between CSF Fe levels and CSF Aβ1–42 levels (with Ni, Cr, Zn, Mn, Co, Cu, Aβ1–40, t-tau, p-tau181, and NFL being also measured in CSF) [27]. Kushnir et al. did not observe an association between CSF Ca levels and CSF Aβ1–42, t-tau, and p-tau [28], while Ma et al. observed a negative correlation between serum Ca levels and CSF Aβ1–42 levels and no association with CSF t-tau and p-tau181 (811 MCI patients and 413 HC; [29]). Blood Se levels were not associated with plasma Aβ1–42 and t-tau levels [30], while CSF Se levels were negatively associated with CSF Aβ1–42, and showed no association with CSF t-tau and p-tau levels [31]. CSF Mn levels positively correlated with CSF t-tau and p-tau181, while CSF Cs levels correlated negatively with t-tau and p-tau181 levels and positively with CSF Aβ1–42 levels [32]. Blood Mn levels positively correlated with plasma Aβ1–42 and Aβ1–40 levels [33], while serum Mn negatively correlated with serum t-tau levels [34]. Mielke et al. reported that low serum K levels in mid-life are associated with low CSF Aβ1–42 levels later in life [35]. Shams et al. observed a positive association between CSF Fe and Cu levels with CSF Aβ1–42, t-tau, p-tau181, and CSF/serum albumin ratio, and a positive association between CSF Zn levels and CSF/serum albumin ratio [36].
Additionally, it was shown that VILIP-1 is a neuronal calcium sensor protein that contains an EF-hand structural domain. This domain can bind metal ions [37], such as Ca, Mg [38,39], Cd [40], and Zn [41]. S100B is a calcium-binding protein that also contains EF-hand and can bind Ca, Zn [42], Cd [43], Mg, and K [44]. A study in Atlantic sharpnose sharks (Rhizoprionodon terraenovae) showed that brain Hg levels positively correlated with CSF S100B levels [45], while rats prenatally exposed to Hg had a significant increase in S100B expression (the effect was reversed with Zn treatment; [46]). Studies in humans showed that children with acute Hg intoxication had significantly increased serum S100B levels [47], while individuals chronically exposed to Hg had an increase in mRNA S100B expression [48]. Levels of As, Pb, and Cd measured in the blood of the patients with multiple sclerosis positively correlated with serum S100B levels [49]. A study in mice showed that arsenic exposure causes an increase in serum S100B levels [50], while manganese exposure increases the expression of S100B in the brain [51]. Additionally, treatment with magnesium sulfate in patients with aneurysmal subarachnoid hemorrhage did not affect serum S100B levels [52] while in patients with eclampsia [53] and neonatal hypoxic-ischemic encephalopathy [54], this treatment lead to the decrease in CSF and serum S100B levels, respectively. Regarding the association of YKL-40 with metals, we found only one study in patients with bipolar disorder that showed no association between serum Zn and serum YKL-40 levels [55]. Regarding NFL, studies in experimental animals showed that As treatment leads to NFL disappearance [56], while Al treatment reduced NFL mRNA levels [57].
All phosphorylated tau isoforms showed a strong positive correlation with Se, while p-tau181 strongly correlated with Cu, p-tau199 with As, and p-tau231 with Co measured in CSF. Vinceti et al. also reported a positive association of CSF Se with CSF p-tau181 levels [58]. However, the majority of the studies associated Se deficiency with increased risk of AD [59,60,61,62,63], with proposed Se supplementation as valuable in AD treatment [64]. Previous studies showed that both Cu [65,66] and As [7,67,68] induce tau phosphorylation, while exposure to Co induces age-dependent neurodegeneration in mice [69]. The neurodegeneration marker VILIP-1 showed the strongest correlation with CSF Se and Na. Se exerts its biological effects mainly through selenoproteins [70]. Similarly to VILIP-1 [71], selenoproteins are involved in the regulation of calcium homeostasis [72], and as such the strong correlation between VILIP-1 and Se observed in this study is not surprising. Previous studies associated increased Na levels with AD [5,73,74,75]. Another marker of neurodegeneration (NFL) and markers of glial activation (S100B and YKL-40) showed the strongest correlation with S and P CSF levels. Additionally, both CSF S and P levels were significantly increased in AD patients compared to MCI patients and HC, respectively. Higher plasma P levels were observed in AD patients compared to HC [62] and associated with an increased risk of dementia [76]. However, Park et al. showed that serum P levels negatively correlate with cerebral Aβ deposition [77]. To our knowledge, no other study analyzed S levels in AD patients, although the intake of sublimed sulfur was suggested to be protective in AD [78]. Most studies analyzed sulfur-containing compounds in AD patients (reviewed in [79]). Thus, plasma sulfate levels were significantly decreased in 10 AD patients compared to HC [80]. A recent study suggested that the intake of hydrogen sulfide (H2S) is beneficial in AD [81], whereas Disbrow et al. observed an increase in H2S levels in AD patients [82]. Additionally, H2S can be produced by some bacteria [83] that have been associated with a higher risk of AD (such as Porphyromonas gingivalis and Helicobacter pylori; reviewed in [84]). Damage of the BBB during AD pathogenesis can facilitate pathogen entry into the brain; through this route, such pathogens may contribute to neuroinflammation, a key feature of AD [85]. Whether the association between CSF S levels and of S100B and YKL-40 represent an indicator of microbial infections that contribute to AD pathogenesis needs further investigation.
The strength of our study is in the analysis of 24 macro- and microelements measured in CSF and 21 measured in plasma, in addition to 11 CSF AD biomarkers determined in nearly 200 participants. Only two studies [29,32] that investigated the association of macro- and microelements with CSF AD biomarkers included more participants than our current study. We used different statistical methods to test the association of macro- and microelements with CSF AD biomarkers, including redescription mining. Only one recent study used machine learning to classify AD, MCI patients, and HC using CSF Fe and CSF Aβ1–42, p-tau, and t-tau (overall 69 participants) [86]. A limitation of our study is the lack of information on possible confounding variables, such as smoking habits (especially in regard to Cd levels) and intake of over-the-counter dietary supplements (especially regarding essential metals and non-metals).
In conclusion, our study showed that essential metals (Ca, Co, Cu, Fe, Mg, Mn, Mo, Na, K, and Zn), heavy metals (As, Cd, Hg, Ni, Pb, and Tl), and essential non-metals (P, S, and Se) are positively associated with CSF AD biomarkers, mainly phosphorylated tau isoforms, VILIP-1, S100B, NFL, and YKL-40, suggesting new diagnostic opportunities and therapeutic targets in future studies on AD.
4. Materials and Methods
4.1. Participants and Sample Collection
We included 193 patients who were admitted to the University Hospital Center Zagreb and General Hospital Varaždin. Patients underwent thorough neurological testing, including MMSE, complete blood tests (albumin levels, thyroid function, levels of vitamin B12 and electrolytes), and VDRL testing for syphilis, as described previously [87]. NINCDS-ADRDA criteria for AD were fulfilled by 124 patients [88], while 50 patients fulfilled the criteria for MCI [88,89], and 19 were HC. CSF samples were collected from all participants by lumbar puncture (performed at intervertebral spaces L3/L4 or L4/L5). After centrifugation at 2000× g for 10 min, CSF samples were aliquoted in polypropylene tubes and stored at −80 °C. Venous blood samples were collected from 143 participants in the morning on an empty stomach. Samples were collected using plastic syringes (with 1 mL of acid citrate dextrose as an anticoagulant). Thrombocyte-free plasma was collected by centrifugation, first at 1100× g for 3 min and then at 5087 g for 15 min. Plasma samples were stored at −20 °C. Supplementary Table S1 summarizes demographic data. All procedures were approved by the Ethical Committee of the Clinical Hospital Center Zagreb (protocol no. 02/21 AG, class 8.1–18/82-2 from 24 April 2018) and the Central Ethical Committee of the University of Zagreb Medical School (protocol no. 380-59-10106-18-111/126, class 641-01/18-02/01 from 20 June 2018).
4.2. Analysis of Macro- and Microelements by Inductively Coupled Plasma Mass Spectroscopy
Inductively coupled plasma mass spectroscopy (ICP-MS) was used for the measurement of CSF and plasma levels of As, B, Ca, Cd, Co, Cu, Fe, Hg, Li, Mg, Mn, Mo, Na, Ni, P, Pb, S, Se, Sr, Tl, and Zn, and CSF levels of Al, Ba, and K (Supplementary Table S2). Cr levels were also measured in CSF and plasma, but due to possible contamination of the samples with Cr from the needles used for sample collection, it was excluded from statistical analysis. ICP-MS was performed on Agilent 7500cx (Agilent Technologies, Tokyo, Japan). Before the analysis, CSF samples were diluted at 1:10, while plasma samples were diluted at 1:20 with a solution containing 0.01 mM EDTA, 0.07% (v/v) Triton X-100, 0.7 mM ammonia, and 2 µg/L of internal standards (Ge, Rh, Tb, Lu, and Ir) in ultrapure water. We used a MicroMist nebulizer combined with a Peltier standard quartz spray chamber (Scott–type, cooled at 2 °C) and a quartz torch with a 2.5-mm diameter injector with a Shield Plate system and Ni sampler and skimmer cones. Daily optimization of ICP-MS working conditions was achieved using a tuning solution of 1 µg/L 7Li, 59Co, 89Y, 140Ce, and 205Tl. HVAC systems (Heating, Ventilating, and Air Conditioning) combined with HEPA filters were used for sample preparation and analysis. Quantification of elements concentration in samples was done by the standard addition method (matrix-matched calibration). Commercially available reference materials were used to confirm the accuracy of the measurements: ClinChek Serum Controls (Level I and II) and ClinChek Plasma Controls (Level I and II) from RECIPE (Munich, Germany); Seronorm Trace Elements Serum (Level I and II) (Sero AS, Billingstad, Norway).
4.3. Analysis of AD Biomarkers in CSF
Eleven AD biomarkers were measured in CSF by the enzyme-linked immunosorbent assays (ELISA); Aβ1–42, VILIP-1, t-tau, p-tau181, p-tau231, tau phosphorylated at Ser 199 (p-tau199), NFL, S100B, YKL-40, PAPP-A, and albumin. Following ELISA kits were used for the measurement of CSF AD biomarkers; Aβ1–42 (Innotest β-amyloid1–42, Fujirebio, Gent, Belgium), p-tau181 (Innotest Phospho-Tau (181P), Fujirebio), p-tau231 (Tau (pT231) Phospho-ELISA Kit, Human, Thermo Fisher Scientific, Waltham, MA, USA), p-tau199 (TAU [pS199] Phospho-ELISA Kit, Human, Thermo Fisher Scientific), t-tau (Innotest hTau Ag, Fujirebio), VILIP-1 (VILIP-1 Human ELISA, BioVendor, Brno, Czech Republic), NFL (Human NF-L/NEFL ELISA Kit, LifeSpan BioSciences, Seattle, WA, USA), S100B (Human S100B, R&D Systems, Minneapolis, MN, USA), YKL-40 (Chitinase 3-like 1 Quantikine ELISA Kit R&D Systems), PAPP-A (Human Pappalysin-1/PAPP-A Quantikine ELISA Kit, R&D Systems), and albumin (Human Albumin ELISA Kit, Abcam, Cambridge, UK).
4.4. Statistical Analysis
4.4.1. Correlation and Principal Component Analysis
Correlation (Spearman’s and Pearson’s correlation coefficient) and linear regression were used for testing the association between CSF AD biomarkers and macro- and microelements measured in CSF and plasma. Additionally, principal component analysis (PCA, unsupervised machine learning method) was used to reduce the dimensionality of macro- and microelements measured in CSF and plasma. PCA analyses were performed for the macro- and microelements measured in CSF, and for the macro- and microelements measured in plasma. Orthogonal (varimax) rotation was used in PCA, while the adequacy of test items and sample size for factor analysis was assessed by Bartlett’s χ2 test of sphericity and the Kaiser–Meyer–Olkin index. The list of 23 macro- and microelements measured in CSF (we excluded P from the PCA because it was measured in a relatively small number of participants) and 21 macro- and microelements measured in plasma were reduced to key factors (groups). If a factor loading for an element was ≥0.4, it was considered that the element loads in the given group (factor). Factor loading measures the correlation between the element and a factor. Additionally, factor-specific scores were calculated for each individual. Multiple linear regression analyses were used for testing the association between PCA-obtained macro- and microelements’ groups (factors) and CSF AD biomarkers (with CSF AD biomarkers as dependent variables and macro- and microelements’ groups as independent variables). Statistical analyses were performed using SPSS 19.0.1 (SPSS Inc., Chicago, IL, USA) and R (R Foundation for Statistical Computing, Vienna, Austria), with the α value set at 0.05 for statistical significance.
4.4.2. Redescription Mining
Three redescription sets were created using the redescription mining algorithm CLUS-RM [90,91]. The first redescription set describes patients that share properties of various CSF AD biomarkers and macro- and microelements measured in CSF. The second redescription set describes patients that share properties of various CSF AD biomarkers and macro- and microelements measured in plasma. The third redescription set describes patients that share properties of various CSF AD biomarkers with macro- and microelements measured in CSF and macro- and microelements measured in plasma. All three discovered redescription sets enable association analyses between indicators from the three groups of attributes: (a) CSF AD biomarkers, (b) macro- and microelements measured in CSF, and (c) macro- and microelements measured in plasma.
In all experiments, we performed 10 runs with different random initialization and 30 iterations of the CLUS-RM algorithm for each run. In each run, the algorithm creates one starting initial clustering of patients that is used to create initial pair of Predictive Clustering trees (PCTs). Initial pairs of PCTs are used as a starting point for a sequence of iterations (called alternations) that create two pairs of matching PCTs per iteration, used to construct redescriptions. A supplement random forest, consisting of twenty trees and a conjunctive refinement procedure was used to obtain more diverse and accurate redescriptions (for more details see [90,91]). The final result of the methodology is a set of redescriptions that are tuples of logical rules. Each redescription describes a set of patients (the support set of a redescription). A redescription describes a patient if every rule from the corresponding tuple describes this patient, and a rule describes a patient if this patient has measurements and concentrations that satisfy logical conditions specified in the rule. For example, a rule PAPP-A (122.39–511.26) describes all patients whose measured PAPP-A level is in the interval [122.39, 511.26]. A rule PAPP-A (122.39–511.26) AND MMSE (5–28) AND AGE (49–82) describes all patients that additionally have MMSE measured concentration in the interval (5, 28), are at least 49 years old and maximally 82 years old.
In this work, we use a minimal support set size of 30 and a maximal support set size of 155 for the first set, a minimal ort set size of 30 and a maximal support set size of 115 for the second set, and a minimal support set size of 20 and a maximal support set size of 110 for the third set. Redescription accuracy measures what fraction of patients, described by either of the rules forming some redescription, is described by all these rules. The corresponding measure that captures this property is called the Jaccard index [92]. In this work, we use the minimal accuracy threshold of 0.5. The statistical significance of a redescription (reported through a corresponding p-value) measures how probable it would be to obtain a redescription at random (by a random choice of rules that form it), so that each rule in a randomly created redescription describes the same number of patients as the original, and that the resulting redescription has a support set size equal or larger to the support set size of the original redescription [93]. In this work, we use a maximal p-value of 0.01. To maximize interpretability and to allow analyses of strong associations between different indicators (measurements), we construct redescriptions with rules containing only logical AND operator. As it can be seen from the example above, when only AND operator is used, each patient described by a rule must have all measurements for all indicators in the exactly specified interval. On the other hand, the rule NOT PAPP-A (122.39–511.26) describes all patients such that either PAPP-A < 122.39 OR PAPP-A >511.26, and understanding such rules would be much more difficult.
Abbreviations
Aβ1–42, amyloid β1–42; Al, aluminum; ApoE, apolipoprotein E; APP, amyloid precursor protein; As, arsenic; B, boron; Ba, barium; BBB, blood–brain barrier; Ca, calcium; Co, cobalt; Cu, copper; CAA, cerebral amyloid angiopathy; Cd, cadmium; Cs, cesium; CSF, cerebrospinal fluid; DOHaD, developmental origins of health and disease; ELISA, enzyme-linked immunosorbent assay; Fe, iron; HC, healthy controls; Hg, mercury; ICP-MS, inductively coupled plasma mass spectroscopy; JS, Jaccard similarity coefficient; K, potassium; Li, lithium; MCI, mild cognitive impairment; Mg, magnesium; MMSE, Mini-Mental State Examination; Mn, manganese; Mo, molybdenum; Na, sodium; NFL, neurofilament light chain; NFT, neurofibrillary tangles; Ni, nickel; P, phosphorus; PAPP-A, pregnancy-associated plasma protein A, pappalysin-1; Pb, lead; PCA, principal component analysis; PTCs, Predictive Clustering trees; p-tau181, tau phosphorylated at Thr 181; p-tau231, tau phosphorylated at Thr 231; p-tau199, tau phosphorylated at Ser 199; S, sulfur; SCI, subjective cognitive impairment; Se, selenium; SP, senile plaques; Sr, strontium; S100B, S100 calcium-binding protein B; Tl, thallium; t-tau, total tau; VaD, vascular dementia; VILIP-1, visinin-like protein 1; W1R, first redescription query; W2R, second redescription query; W3R, third redescription query; YKL-40, chitinase-3-like protein 1; Zn, zinc.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24010467/s1.
Author Contributions
Conceptualization, G.Š., M.B.L. and M.M.; methodology, M.B.L., J.J., M.N.P., E.Š., A.S., T.O., K.Z., L.L.H., S.K.-P. and Ž.V.; software, M.M.; validation, G.Š., N.P. (Nela Pivac), P.R.H. and A.D.; formal analysis, M.B.L., M.M. and N.P. (Nikolina Pleić); investigation, M.B.L., M.M., J.J., M.N.P., E.Š., A.S., T.O., K.Z., L.L.H., S.K.-P., F.B. and Ž.V.; resources, G.Š.; data curation, M.B.L. and M.M.; writing—original draft preparation, M.B.L. and M.M.; writing—review and editing, G.Š., A.D. and P.R.H.; visualization, M.B.L., M.M. and G.Š.; supervision, G.Š.; project administration, G.Š.; funding acquisition, G.Š. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Central Ethical Committee of the University of Zagreb Medical School (case no. 380-59-10106-18-111/126, class 641-01/18-02/01 from 20 June 2018) and Ethical Committee of the Clinical Hospital Center Zagreb (case no. 02/21 AG, class 8.1-18/82-2 from 24 April 2018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patients to publish this paper.
Data Availability Statement
All data are presented in this article. Original data are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. Goran Šimić is a guest co-editor of the IJMS Special Issue “Neuropathological Advances in Brain Disorders from MNS2022” and co-author of this article. They were excluded from all editorial decision-making related to the acceptance of this article for publication.
Funding Statement
This work was funded by The Croatian Science Foundation grants IP-2019-04-3584 (“Role of the blood–brain barrier, innate immunity, and tau protein oligomerization in the pathogenesis of Alzheimer’s disease“) and IP-2014-09-9730 (“Tau protein hyperphosphorylation, aggregation, and trans-synaptic transfer in Alzheimer’s disease: cerebrospinal fluid analysis and assessment of potential neuroprotective compounds”) to G.Š., and by the Scientific Centre of Excellence for Basic, Clinical, and Translational Neuroscience CoRE-NEURO (“Experimental and clinical research of hypoxic-ischemic damage in perinatal and adult brain”; GA KK01.1.1.01.0007 funded by the European Union through the European Regional Development Fund), and in part by the NIH grant P30 AG066514 to PRH. The work was additionally funded by the Research Cooperability Program of the Croatian Science Foundation, Croatia, funded by the European Union from the European Social Fund under the Operational Programme Efficient Human Resources 2014–2020, through Grant 8525: “Augmented Intelligence Workflows for Prediction, Discovery, and Understanding in Genomics and Pharmacogenomics”.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ashok A., Rai N.K., Tripathi S., Bandyopadhyay S. Exposure to As-, Cd-, and Pb-mixture induces Aβ, amyloidogenic APP processing and cognitive impairments via oxidative stress-dependent neuroinflammation in young rats. Toxicol. Sci. 2015;143:64–80. doi: 10.1093/toxsci/kfu208. [DOI] [PubMed] [Google Scholar]
- 2.Elonheimo H.M., Andersen H.R., Katsonouri A., Tolonen H. Environmental substances associated with Alzheimer’s disease —A scoping review. Int. J. Environ. Res. Public Health. 2021;18:11839. doi: 10.3390/ijerph182211839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunderman F., Jr. Nasal toxicity, carcinogenicity, and olfactory uptake of metals. Ann. Clin. Lab. Sci. 2001;31:3–24. [PubMed] [Google Scholar]
- 4.Bush A.I. The metal theory of Alzheimer’s disease. J. Alzheimers Dis. 2013;33((Suppl. S1)):S277–S281. doi: 10.3233/JAD-2012-129011. [DOI] [PubMed] [Google Scholar]
- 5.Babić Leko M., Jurasović J., Nikolac Perković M., Španić E., Sekovanić A., Orct T., Lukinović Škudar V., Bačić Baronica K., Kiđemet-Piskač S., Vogrinc Ž., et al. The association of essential metals with APOE genotype in Alzheimer’s disease. J. Alzheimers Dis. 2021;82:661–672. doi: 10.3233/JAD-210158. [DOI] [PubMed] [Google Scholar]
- 6.Zubčić K., Hof P.R., Šimić G., Jazvinšćak Jembrek M. The role of copper in tau-related pathology in Alzheimer’s disease. Front. Mol. Neurosci. 2020;13:572308. doi: 10.3389/fnmol.2020.572308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wisessaowapak C., Visitnonthachai D., Watcharasit P., Satayavivad J. Prolonged arsenic exposure increases tau phosphorylation in differentiated SH-SY5Y cells: The contribution of GSK3 and ERK1/2. Environ. Toxicol. Pharmacol. 2021;84:103626. doi: 10.1016/j.etap.2021.103626. [DOI] [PubMed] [Google Scholar]
- 8.Shati A.A., Alfaifi M.Y. Trans-resveratrol inhibits tau phosphorylation in the brains of control and cadmium chloride-treated rats by activating PP2A and PI3K/Akt induced-inhibition of GSK3β. Neurochem. Res. 2019;44:357–373. doi: 10.1007/s11064-018-2683-8. [DOI] [PubMed] [Google Scholar]
- 9.Yano K., Hirosawa N., Sakamoto Y., Katayama H., Moriguchi T. Aggregations of amyloid β-proteins in the presence of metal ions. Toxicol. Lett. 2003;144:134. doi: 10.1016/S0378-4274(03)90499-1. [DOI] [Google Scholar]
- 10.Wallin C., Sholts S.B., Österlund N., Luo J., Jarvet J., Roos P.M., Ilag L., Gräslund A., Wärmländer S.K.T.S. Alzheimer’s disease and cigarette smoke components: Effects of nicotine, PAHs, and Cd(II), Cr(III), Pb(II), Pb(IV) ions on amyloid-β peptide aggregation. Sci. Rep. 2017;7:14423. doi: 10.1038/s41598-017-13759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu H., Territo P.R., Persohn S.A., Bedwell A.A., Eldridge K., Speedy R., Chen Z., Zheng W., Du Y. Evaluation of chronic lead effects in the blood brain barrier system by DCE-CT. J. Trace Elem. Med. Biol. 2020;62:126648. doi: 10.1016/j.jtemb.2020.126648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Augusti P.R., Conterato G.M.M., Somacal S., Sobieski R., Spohr P.R., Torres J.V., Charão M.F., Moro A.M., Rocha M.P., Garcia S.C., et al. Effect of astaxanthin on kidney function impairment and oxidative stress induced by mercuric chloride in rats. Food Chem. Toxicol. 2008;46:212–219. doi: 10.1016/j.fct.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Yin X., Sun J., Mei Y., Guo X., Chen S.-I., Wang Z.-I., Yang L. Effect of Hg2+ on voltage-dependent calcium channels and intracellular free calcium in trigeminal ganglion neurons of rats. Chin. J. Ind. Hyg. Occup. Dis. 2008;26:542–545. [PubMed] [Google Scholar]
- 14.Mao J., Yang J., Zhang Y., Li T., Wang C., Xu L., Hu Q., Wang X., Jiang S., Nie X., et al. Arsenic trioxide mediates HAPI microglia inflammatory response and subsequent neuron apoptosis through p38/JNK MAPK/STAT3 pathway. Toxicol. Appl. Pharmacol. 2016;303:79–89. doi: 10.1016/j.taap.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Chattopadhyay S., Bhaumik S., Purkayastha M., Basu S., Nag Chaudhuri A., Das Gupta S. Apoptosis and necrosis in developing brain cells due to arsenic toxicity and protection with antioxidants. Toxicol. Lett. 2002;136:65–76. doi: 10.1016/S0378-4274(02)00282-5. [DOI] [PubMed] [Google Scholar]
- 16.Bashir S., Sharma Y., Irshad M., Gupta S.D., Dogra T.D. Arsenic-induced cell death in liver and brain of experimental rats. Basic Clin. Pharmacol. Toxicol. 2006;98:38–43. doi: 10.1111/j.1742-7843.2006.pto_170.x. [DOI] [PubMed] [Google Scholar]
- 17.Babić Leko M. Ph.D. Thesis. School of Medicine, University of Zagreb; Zagreb, Croatia: 2017. Predictive Value of Biomarkers in Early Detection and Differential Diagnosis of Alzheimer’s Disease. [Google Scholar]
- 18.Fasae K.D., Abolaji A.O., Faloye T.R., Odunsi A.Y., Oyetayo B.O., Enya J.I., Rotimi J.A., Akinyemi R.O., Whitworth A.J., Aschner M. Metallobiology and therapeutic chelation of biometals (copper, zinc and iron) in Alzheimer’s disease: Limitations, and current and future perspectives. J. Trace Elem. Med. Biol. 2021;67:126779. doi: 10.1016/j.jtemb.2021.126779. [DOI] [PubMed] [Google Scholar]
- 19.Ritchie C.W., Bush A.I., Mackinnon A., Macfarlane S., Mastwyk M., MacGregor L., Kiers L., Cherny R., Li Q.-X., Tammer A., et al. Metal-protein attenuation with iodochlorhydroxyquin (clioquinol) targeting Aβ amyloid deposition and toxicity in Alzheimer disease: A pilot phase 2 clinical trial. Arch. Neurol. 2003;60:1685–1691. doi: 10.1001/archneur.60.12.1685. [DOI] [PubMed] [Google Scholar]
- 20.Ayton S., Lei P., Bush A.I. Metallostasis in Alzheimer’s disease. Free Radic. Biol. Med. 2013;62:76–89. doi: 10.1016/j.freeradbiomed.2012.10.558. [DOI] [PubMed] [Google Scholar]
- 21.Li C., Wang J., Zhou B. The metal chelating and chaperoning effects of clioquinol: Insights from yeast studies. J. Alzheimers Dis. 2010;21:1249–1262. doi: 10.3233/JAD-2010-100024. [DOI] [PubMed] [Google Scholar]
- 22.Galbrun E. Ph.D. Thesis. University of Helsinki; Helsinki, Finland: 2013. Methods for Redescription Mining. [Google Scholar]
- 23.Hock C., Drasch G., Golombowski S., Müller-Spahn F., Willershausen-Zönnchen B., Schwarz P., Hock U., Growdon J.H., Nitsch R.M. Increased blood mercury levels in patients with Alzheimer’s disease. J. Neural Transm. 1998;105:59–68. doi: 10.1007/s007020050038. [DOI] [PubMed] [Google Scholar]
- 24.Squitti R., Barbati G., Rossi L., Ventriglia M., Dal Forno G., Cesaretti S., Moffa F., Caridi I., Cassetta E., Pasqualetti P., et al. Excess of nonceruloplasmin serum copper in AD correlates with MMSE, CSF β-amyloid, and h-tau. Neurology. 2006;67:76–82. doi: 10.1212/01.wnl.0000223343.82809.cf. [DOI] [PubMed] [Google Scholar]
- 25.Kessler H., Pajonk F.G., Bach D., Schneider-Axmann T., Falkai P., Herrmann W., Multhaup G., Wiltfang J., Schäfer S., Wirths O., et al. Effect of copper intake on CSF parameters in patients with mild Alzheimer’s disease: A pilot phase 2 clinical trial. J. Neural Transm. 2008;115:1651–1659. doi: 10.1007/s00702-008-0136-2. [DOI] [PubMed] [Google Scholar]
- 26.Strozyk D., Launer L.J., Adlard P.A., Cherny R.A., Tsatsanis A., Volitakis I., Blennow K., Petrovitch H., White L.R., Bush A.I. Zinc and copper modulate Alzheimer Aβ levels in human cerebrospinal fluid. Neurobiol. Aging. 2009;30:1069–1077. doi: 10.1016/j.neurobiolaging.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee G., Forsgard N., Ambler G., Keshavan A., Paterson R.W., Foiani M.S., Toombs J., Heslegrave A., Thompson E.J., Lunn M.P., et al. Cerebrospinal fluid metallomics in cerebral amyloid angiopathy: An exploratory analysis. J. Neurol. 2022;269:1470–1475. doi: 10.1007/s00415-021-10711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kushnir M.M., Michno W., Rockwood A.L., Blennow K., Strathmann F.G., Hanrieder J. Association of PTHrP levels in CSF with Alzheimer’s disease biomarkers. Clin. Mass Spectrom. 2018;14:124–129. doi: 10.1016/j.clinms.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma L.Z., Wang Z.X., Wang Z.T., Hou X.H., Shen X.N., Ou Y.N., Dong Q., Tan L., Yu J.T. Serum calcium predicts cognitive decline and clinical progression of Alzheimer’s disease. Neurotoxicol. Res. 2021;39:609–617. doi: 10.1007/s12640-020-00312-y. [DOI] [PubMed] [Google Scholar]
- 30.Krishnan S., Rani P. Evaluation of selenium, redox status and their association with plasma amyloid/tau in Alzheimer’s disease. Biol. Trace Elem. Res. 2014;158:158–165. doi: 10.1007/s12011-014-9930-x. [DOI] [PubMed] [Google Scholar]
- 31.Vinceti M., Chiari A., Eichmüller M., Rothman K.J., Filippini T., Malagoli C., Weuve J., Tondelli M., Zamboni G., Nichelli P.F., et al. A selenium species in cerebrospinal fluid predicts conversion to Alzheimer’s dementia in persons with mild cognitive impairment. Alzheimers Res. Ther. 2017;9:100. doi: 10.1186/s13195-017-0323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerhardsson L., Blennow K., Lundh T., Londos E., Minthon L. Concentrations of metals, β-amyloid and tau-markers in cerebrospinal fluid in patients with Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 2009;28:88–94. doi: 10.1159/000233353. [DOI] [PubMed] [Google Scholar]
- 33.Tong Y., Yang H., Tian X., Wang H., Zhou T., Zhang S., Yu J., Zhang T., Fan D., Guo X., et al. High manganese, a risk for Alzheimer’s disease: High manganese induces amyloid-β related cognitive impairment. J. Alzheimers Dis. 2014;42:865–878. doi: 10.3233/JAD-140534. [DOI] [PubMed] [Google Scholar]
- 34.Mohammed R.S., Ibrahim W., Sabry D., El-Jaafary S.I. Occupational metals exposure and cognitive performance among foundry workers using tau protein as a biomarker. Neurotoxicology. 2020;76:10–16. doi: 10.1016/j.neuro.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Mielke M.M., Zandi P.P., Blennow K., Gustafson D., Sjögren M., Rosengren L., Skoog I. Low serum potassium in mid life associated with decreased cerebrospinal fluid Aβ42 in late life. Alzheimer Dis. Assoc. Disord. 2006;20:30–36. doi: 10.1097/01.wad.0000201848.67954.7d. [DOI] [PubMed] [Google Scholar]
- 36.Shams M., Martola J., Charidimou A., Granberg T., Ferreira D., Westman E., Wintermark M., Iv M., Larvie M., Kristoffersen Wiberg M., et al. Cerebrospinal fluid metals and the association with cerebral small vessel disease. J. Alzheimer’s Dis. 2020;78:1229–1236. doi: 10.3233/JAD-200656. [DOI] [PubMed] [Google Scholar]
- 37.Biekofsky R.R., Feeney J. Cooperative cyclic interactions involved in metal binding to pairs of sites in EF-hand proteins. FEBS Lett. 1998;439:101–106. doi: 10.1016/S0014-5793(98)01349-0. [DOI] [PubMed] [Google Scholar]
- 38.Gifford J.L., Walsh M.P., Vogel H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix–loop–helix EF-hand motifs. Biochem. J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 39.Li C., Pan W., Braunewell K.H., Ames J.B. Structural analysis of Mg2+ and Ca2+ binding, myristoylation, and dimerization of the neuronal calcium sensor and visinin-like protein 1 (VILIP-1) J. Biol. Chem. 2011;286:6354–6366. doi: 10.1074/jbc.M110.173724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akke M., Forsén S., Chazin W.J. 15N NMR assignments of (Cd2+,)2-calbindin D9k and comparison with (Ca2+)2-calbindin D9k. Cadmium as a substitute for calcium in calcium-binding proteins. Magn. Reson. Chem. 1993;31:S128–S132. doi: 10.1002/mrc.1260311324. [DOI] [Google Scholar]
- 41.Baksheeva V.E., Tsvetkov P.O., Zalevsky A.O., Vladimirov V.I., Gorokhovets N.V., Zinchenko D.V., Permyakov S.E., Devred F., Zernii E.Y. Zinc modulation of neuronal calcium sensor proteins: Three modes of interaction with different structural outcomes. Biomolecules. 2022;12:956. doi: 10.3390/biom12070956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baudier J., Glasser N., Gerard D. Ions binding to S100 proteins. I. Calcium- and zinc-binding properties of bovine brain S100αα, S100a (αβ), and S100b (ββ) protein: Zn2+ regulates Ca2+ binding on S100b protein. J. Biol. Chem. 1986;261:8192–8203. doi: 10.1016/S0021-9258(19)83895-4. [DOI] [PubMed] [Google Scholar]
- 43.Donato H., Mani R.S., Kay C.M. Spectral studies on the cadmium-ion-binding properties of bovine brain S-100b protein. Biochem. J. 1991;276:13–18. doi: 10.1042/bj2760013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogoma Y., Kobayashi H., Fujii T., Kondo Y., Hachimori A., Shimizu T., Hatano M. Binding study of metal ions to S100 protein: 43Ca, 25Mg, 67Zn and 39K n.m.r. Int. J. Biol. Macromol. 1992;14:279–286. doi: 10.1016/S0141-8130(05)80041-8. [DOI] [PubMed] [Google Scholar]
- 45.Ehnert-Russo S.L., Gelsleichter J. Mercury accumulation and effects in the brain of the Atlantic sharpnose shark (Rhizoprionodon terraenovae) Arch. Environ. Contam. Toxicol. 2020;78:267–283. doi: 10.1007/s00244-019-00691-0. [DOI] [PubMed] [Google Scholar]
- 46.Weng Z., Liu Z., Zhang S., Tao H., Ji X. Zinc protection in fetal rats for maternal mercury exposure-induced growth retardation is probably associated with S100B expression. J. Obstet. Gynaecol. Res. 2017;43:73–77. doi: 10.1111/jog.13175. [DOI] [PubMed] [Google Scholar]
- 47.Yilmaz F.M., Yilmaz H., Tutkun E., Uysal S., Carman K.B., Ilber D., Ercan M. Serum biochemical markers of central nerve system damage in children with acute elemental mercury intoxicatıon. Clin. Toxicol. 2014;52:32–38. doi: 10.3109/15563650.2013.860986. [DOI] [PubMed] [Google Scholar]
- 48.de Paula Fonseca Arrifano G., Del Carmen Rodriguez Martin-Doimeadios R., Jiménez-Moreno M., Augusto-Oliveira M., Souza-Monteiro J.R., Paraense R., Machado C.R., Farina M., Macchi B., do Nascimento J.L.M., et al. Assessing mercury intoxication in isolated/remote populations: Increased S100B mRNA in blood in exposed riverine inhabitants of the Amazon. Neurotoxicology. 2018;68:151–158. doi: 10.1016/j.neuro.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 49.Paknejad B., Shirkhanloo H., Aliomrani M. Is there any relevance between serum heavy metal concentration and BBB leakage in multiple sclerosis patients? Biol. Trace Elem. Res. 2019;190:289–294. doi: 10.1007/s12011-018-1553-1. [DOI] [PubMed] [Google Scholar]
- 50.Golmohammadi J., Jahanian-Najafabadi A., Aliomrani M. Chronic oral arsenic exposure and its correlation with serum S100B concentration. Biol. Trace Elem. Res. 2019;189:172–179. doi: 10.1007/s12011-018-1463-2. [DOI] [PubMed] [Google Scholar]
- 51.Baek S.Y., Cho J.H., Kim E.S., Kim H.J., Yoon S., Kim B.S., Kim J.B., Lee C.R., Yoo C., Lee J.H., et al. CDNA array analysis of gene expression profiles in brain of mice exposed to manganese. Ind. Health. 2004;42:315–320. doi: 10.2486/indhealth.42.315. [DOI] [PubMed] [Google Scholar]
- 52.Hassan T., Nassar M., Elhadi S.M., Radi W.K. Effect of magnesium sulfate therapy on patients with aneurysmal subarachnoid hemorrhage using serum S100B protein as a prognostic marker. Neurosurg. Rev. 2012;35:421–427. doi: 10.1007/s10143-011-0368-8. [DOI] [PubMed] [Google Scholar]
- 53.Li X., Han X., Yang J., Bao J., Di X., Zhang G., Liu H. Magnesium sulfate provides neuroprotection in eclampsia-like seizure model by ameliorating neuroinflammation and brain edema. Mol. Neurobiol. 2017;54:7938–7948. doi: 10.1007/s12035-016-0278-4. [DOI] [PubMed] [Google Scholar]
- 54.El Farargy M.S., Soliman N.A. A randomized controlled trial on the use of magnesium sulfate and melatonin in neonatal hypoxic ischemic encephalopathy. J. Neonatal. Perinatal. Med. 2019;12:379–384. doi: 10.3233/NPM-181830. [DOI] [PubMed] [Google Scholar]
- 55.Jonsson B.H., Orhan F., Bruno S., Oliveira A.O., Sparding T., Landen M., Sellgren C.M. Serum concentration of zinc is elevated in clinically stable bipolar disorder patients. Brain Behav. 2022;12:e2472. doi: 10.1002/brb3.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vahidnia A., van der Voet G.B., de Wolff F.A. Arsenic neurotoxicity—A review. Hum. Exp. Toxicol. 2007;26:823–832. doi: 10.1177/0960327107084539. [DOI] [PubMed] [Google Scholar]
- 57.Strong M.J., Gaytan-Garcia S. Proximal sciatic axotomy does not inhibit the induction of neurofilamentous inclusions following intracisternal aluminum chloride exposure. J. Neuropathol. Exp. Neurol. 1996;55:419–423. doi: 10.1097/00005072-199604000-00004. [DOI] [PubMed] [Google Scholar]
- 58.Vinceti M., Michalke B., Malagoli C., Eichmüller M., Filippini T., Tondelli M., Bargellini A., Vinceti G., Zamboni G., Chiari A. Selenium and selenium species in the etiology of Alzheimer’s dementia: The potential for bias of the case-control study design. J. Trace Elem. Med. Biol. 2019;53:154–162. doi: 10.1016/j.jtemb.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 59.González-Domínguez R., García-Barrera T., Gómez-Ariza J.L. Characterization of metal profiles in serum during the progression of Alzheimer’s disease. Metallomics. 2014;6:292–300. doi: 10.1039/C3MT00301A. [DOI] [PubMed] [Google Scholar]
- 60.Paglia G., Miedico O., Cristofano A., Vitale M., Angiolillo A., Chiaravalle A.E., Corso G., Di Costanzo A. Distinctive pattern of serum elements during the progression of Alzheimer’s disease. Sci. Rep. 2016;6:22769. doi: 10.1038/srep22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardoso B.R., Hare D.J., Bush A.I., Li Q.X., Fowler C.J., Masters C.L., Martins R.N., Ganio K., Lothian A., Mukherjee S., et al. Selenium levels in serum, red blood cells, and cerebrospinal fluid of Alzheimer’s disease patients: A report from the Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing (AIBL) J. Alzheimers Dis. 2017;57:183–193. doi: 10.3233/JAD-160622. [DOI] [PubMed] [Google Scholar]
- 62.Ashraf A., Stosnach H., Parkes H.G., Hye A., Powell J., Soinine H., Tsolaki M., Vellas B., Lovestone S., Aarsland D., et al. Pattern of altered plasma elemental phosphorus, calcium, zinc, and iron in Alzheimer’s disease. Sci. Rep. 2019;9:3147. doi: 10.1038/s41598-018-37431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Socha K., Klimiuk K., Naliwajko S.K., Soroczyńska J., Puścion-jakubik A., Markiewicz-żukowska R., Kochanowicz J. Dietary habits, selenium, copper, zinc and total antioxidant status in serum in relation to cognitive functions of patients with Alzheimer’s disease. Nutrients. 2021;13:287. doi: 10.3390/nu13020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo X., Lie Q., Liu Y., Jia Z., Gong Y., Yuan X., Liu J. Multifunctional selenium quantum dots for the treatment of Alzheimer’s disease by reducing Aβ-neurotoxicity and oxidative stress and alleviate neuroinflammation. ACS Appl. Mater. Interfaces. 2021;13:30261–30273. doi: 10.1021/acsami.1c00690. [DOI] [PubMed] [Google Scholar]
- 65.Kitazawa M., Cheng D., Laferla F.M. Chronic copper exposure exacerbates both amyloid and tau pathology and selectively dysregulates cdk5 in a mouse model of AD. J. Neurochem. 2009;108:1550–1560. doi: 10.1111/j.1471-4159.2009.05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou L.-X., Du J.-T., Zeng Z.-Y., Wu W.-H., Zhao Y.-F., Kanazawa K., Ishizuka Y., Nemoto T., Nakanishi H., Li Y.-M. Copper (II) modulates in vitro aggregation of a tau peptide. Peptides. 2007;28:2229–2234. doi: 10.1016/j.peptides.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 67.Giasson B.I., Sampathu D.M., Wilson C.A., Vogelsberg-Ragaglia V., Mushynski W.E., Lee V.M.Y. The environmental toxin arsenite induces tau hyperphosphorylation. Biochemistry. 2002;41:15376–15387. doi: 10.1021/bi026813c. [DOI] [PubMed] [Google Scholar]
- 68.Pakzad D., Akbari V., Sepand M.R., Aliomrani M. Risk of neurodegenerative disease due to tau phosphorylation changes and arsenic exposure via drinking water. Toxicol. Res. 2021;10:325–333. doi: 10.1093/toxres/tfab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng F., Li Y., Zhang F., Sun Y., Zheng C., Luo Z., Wang Y.L., Aschner M., Zheng H., Lin L., et al. Cobalt induces neurodegenerative damages through Pin1 inactivation in mice and human neuroglioma cells. J. Hazard. Mater. 2021;419:126378. doi: 10.1016/j.jhazmat.2021.126378. [DOI] [PubMed] [Google Scholar]
- 70.Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigó R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 71.Burgoyne R.D., Haynes L.P. Understanding the physiological roles of the neuronal calcium sensor proteins. Mol. Brain. 2012;5:2. doi: 10.1186/1756-6606-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pitts M.W., Hoffmann P.R. Endoplasmic reticulum-resident selenoproteins as regulators of calcium signaling and homeostasis. Cell Calcium. 2018;70:76–86. doi: 10.1016/j.ceca.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faraco G., Hochrainer K., Segarra S.G., Schaeffer S., Santisteban M.M., Menon A., Jiang H., Holtzman D.M., Anrather J., Iadecola C. Dietary salt promotes cognitive impairment through tau phosphorylation. Nature. 2019;574:686–690. doi: 10.1038/s41586-019-1688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Souza L.A.C., Trebak F., Kumar V., Satou R., Kehoe P.G., Yang W., Wharton W., Earley Y.F. Elevated cerebrospinal fluid sodium in hypertensive human subjects with a family history of Alzheimer’s disease. Physiol. Genomics. 2020;52:133–142. doi: 10.1152/physiolgenomics.00093.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mohamed S.A., Herrmann K., Adlung A., Paschke N., Hausner L., Frölich L., Schad L., Groden C., Ulrich H.K. Evaluation of sodium (23Na) MR-imaging as a biomarker and predictor for neurodegenerative changes in patients with Alzheimer’s disease. In Vivo. 2021;35:429–435. doi: 10.21873/invivo.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li T., Xie Y., Bowe B., Xian H., Al-Aly Z. Serum phosphorus levels and risk of incident dementia. PLoS ONE. 2017;12:e0171377. doi: 10.1371/journal.pone.0171377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park J.C., Han S.H., Byun M.S., Yi D., Lee J.H., Park K., Lee D.Y., Mook-Jung I. Low serum phosphorus correlates with cerebral Aβ deposition in cognitively impaired subjects: Results from the KBASE study. Front. Aging Neurosci. 2017;9:362. doi: 10.3389/fnagi.2017.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiu S.S., Khan A.S., Badmaev V., Terpstra K., Cernovsky Z., Varghese J., Khazaeipool Z., Elias H., Carriere A., Husni M., et al. Exploratory study of sublimed sulfur, in cognitively normal subjects and in Alzheimer’s dementia (AD) subjects: Implications for sulfur targeting hydrogen sulfide (H2S)/homocysteine (Hcy) and beta-galactosidase (GALAC)/autophagy signaling in AD. J. Syst. Integr. Neurosci. 2017;3:1–10. doi: 10.15761/JSIN.1000158. [DOI] [Google Scholar]
- 79.Paul B.D. Neuroprotective roles of the reverse transsulfuration pathway in Alzheimer’s disease. Front. Aging Neurosci. 2021;13:659402. doi: 10.3389/fnagi.2021.659402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heafield M.T., Fearn S., Steventon G.B., Waring R.H., Williams A.C., Sturman S.G. Plasma cysteine and sulphate levels in patients with motor neurone, Parkinson’s and Alzheimer’s disease. Neurosci. Lett. 1990;110:216–220. doi: 10.1016/0304-3940(90)90814-P. [DOI] [PubMed] [Google Scholar]
- 81.Giovinazzo D., Bursac B., Sbodio J.I., Nalluru S., Vignane T., Snowman A.M., Albacarys L.M., Sedlak T.W., Torregrossa R., Whiteman M., et al. Hydrogen sulfide is neuroprotective in Alzheimer’s disease by sulfhydrating GSK3β and inhibiting tau hyperphosphorylation. Proc. Natl. Acad. Sci. USA. 2021;118:e2017225118. doi: 10.1073/pnas.2017225118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Disbrow E., Stokes K.Y., Ledbetter C., Patterson J., Kelley R., Pardue S., Reekes T., Larmeu L., Batra V., Yuan S., et al. Plasma hydrogen sulfide: A biomarker of Alzheimer’s disease and related dementias. Alzheimers Dement. 2021;17:1391–1402. doi: 10.1002/alz.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakamura S., Shioya K., Hiraoka B.Y., Suzuki N., Hoshino T., Fujiwara T., Yoshinari N., Ansai T., Yoshida A. Porphyromonas gingivalis hydrogen sulfide enhances methyl mercaptan-induced pathogenicity in mouse abscess formation. Microbiology. 2018;164:529–539. doi: 10.1099/mic.0.000640. [DOI] [PubMed] [Google Scholar]
- 84.Ekundayo T.C., Olasehinde T.A., Okaiyeto K., Okoh A.I. Microbial pathogenesis and pathophysiology of Alzheimer’s disease: A systematic assessment of microorganisms’ implications in the neurodegenerative disease. Front. Neurosci. 2021;15:648484. doi: 10.3389/fnins.2021.648484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Španić E., Langer Horvat L., Ilić K., Hof P.R., Šimić G. NLRP1 inflammasome activation in the hippocampal formation in Alzheimer’s disease: Correlation with neuropathological changes and unbiasedly estimated neuronal loss. Cells. 2022;11:2223. doi: 10.3390/cells11142223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ficiarà E., Boschi S., Ansari S., D’Agata F., Abollino O., Caroppo P., Di Fede G., Indaco A., Rainero I., Guiot C. Machine learning profiling of Alzheimer’s disease patients based on current cerebrospinal fluid markers and iron content in biofluids. Front. Aging Neurosci. 2021;13:52. doi: 10.3389/fnagi.2021.607858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boban M., Malojčić B., Mimica N., Vuković S., Zrilić I., Hof P.R., Šimić G. The reliability and validity of the mini-mental state examination in the elderly Croatian population. Dement. Geriatr. Cogn. Disord. 2012;33:385–392. doi: 10.1159/000339596. [DOI] [PubMed] [Google Scholar]
- 88.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petersen R.C., Smith G.E., Waring S.C., Ivnik R.J., Tangalos E.G., Kokmen E. Mild cognitive impairment: Clinical characterization and outcome. Arch. Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 90.Mihelčić M., Džeroski S., Lavrač N., Šmuc T. Redescription mining augmented with random forest of multi-target predictive clustering trees. J. Intell. Inf. Syst. 2018;50:63–96. doi: 10.1007/s10844-017-0448-5. [DOI] [Google Scholar]
- 91.Mihelčić M., Džeroski S., Lavrač N., Šmuc T. A framework for redescription set construction. Expert Syst. Appl. 2017;68:196–215. doi: 10.1016/j.eswa.2016.10.012. [DOI] [Google Scholar]
- 92.Jaccard P. The distribution of the flora in the Alpine zone. New Phytol. 1912;11:37–50. doi: 10.1111/j.1469-8137.1912.tb05611.x. [DOI] [Google Scholar]
- 93.Mihelčić M., Šmuc T. Approaches for multi-view redescription mining. IEEE Access. 2021;9:19356–19378. doi: 10.1109/ACCESS.2021.3054245. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented in this article. Original data are available from the corresponding author upon reasonable request.