Abstract
Spinal and bulbar muscular atrophy is caused by polyglutamine (polyQ) expansions in androgen receptor (AR), generating gain-of-function toxicity that may involve phosphorylation. Using cellular and animal models, we investigated what kinases and phosphatases target polyQ-expanded AR, whether polyQ expansions modify AR phosphorylation, and how this contributes to neurodegeneration. Mass spectrometry showed that polyQ expansions preserve native phosphorylation and increase phosphorylation at conserved sites controlling AR stability and transactivation. In small-molecule screening, we identified that CDC25/CDK2 signaling could enhance AR phosphorylation, and the calcium-sensitive phosphatase calcineurin had opposite effects. Pharmacologic and genetic manipulation of these kinases and phosphatases modified polyQ-expanded AR function and toxicity in cells, flies, and mice. Ablation of CDK2 reduced AR phosphorylation in the brainstem and restored expression of Myc and other genes involved in DNA damage, senescence, and apoptosis, indicating that the cell cycle–regulated kinase plays more than a bystander role in SBMA-vulnerable postmitotic cells.
CDC25/CDK2 and calcineurin modify polyQ-expanded AR phosphorylation, transactivation, and toxicity in SBMA.
INTRODUCTION
Spinal and bulbar muscular atrophy (SBMA) is a rare genetically inherited (X-linked) neuromuscular disease with a prevalence of two to five cases per 100,000 men (1). SBMA is caused by ≥38 exonic cytosine, adenine, and guanine (CAG) expansions in the androgen receptor (AR) gene (2), resulting in the expression of an AR with an aberrantly elongated polyglutamine (polyQ) tract. Patients with SBMA show an inverse correlation between CAG repeat length and age at onset (3, 4), sensory and motor dysfunction (5), and disease duration/age at examination (3), as well as between age at onset and muscle strength (5). The absence of neuromuscular symptoms in patients with pure loss-of-function mutations of AR supports the idea that expanded polyQ causes SBMA mainly through gain-of-function mechanisms. SBMA belongs to the family of polyQ diseases that also includes Huntington’s disease, dentatorubral-pallidoluysian atrophy, and six types of spinocerebellar ataxia. Within polyQ diseases, SBMA is the only one that affects an X-linked gene, affects a receptor for steroid hormones, and fully manifests only in males, although AR is widely expressed in both sexes. Females develop very mild or no symptoms even if homozygous for the mutation. The sex bias of SBMA is well-recapitulated in animal models of disease (6–8), which established that SBMA is an androgen-dependent disease. Although the androgen-dependent nature of SBMA suggests therapeutic use of anti-androgens, clinical trials based on androgen-deprivation therapy have not yielded the expected results (9, 10).
Upon activation by its natural ligands, testosterone, and its more potent derivative dihydrotestosterone (DHT), AR acts as a transcription factor to convert sex hormone signaling into cell-specific gene expression. Binding of AR to androgens is a necessary step for neurodegeneration, and this underlies the sex specificity of SBMA (8). Inactive AR resides in the cytosol; in response to androgen binding, AR undergoes a conformational change that leads to nuclear translocation (11). AR integrates hormone signaling with several signal transduction pathways initiated by membrane tyrosine kinase, G protein–coupled receptors, and other types of receptors. These receptors activate cascades of kinases and phosphatases that culminate with phosphorylation of transcription factors to convert extra/intracellular signals into a tightly regulated gene expression program. Changes in phosphorylation status of target transcription factors are usually transient and balanced by timely coordination of kinases and phosphatases. Phosphorylation is the most prominent posttranslational modification of AR and generally causes very rapid responses to environmental cues (12, 13). However, it remains to be established what kinases and phosphatases target polyQ-expanded AR, whether polyQ expansion modifies AR phosphorylation sites and status, and how this contributes to neurodegeneration.
Nonexpanded AR is phosphorylated at multiple residues, mostly serine residues with proline in position +1 (SP site) (14, 15). Three AR SP sites—serine 83 (S83), S96, and S651—are the most conserved and highly phosphorylated sites. S83 and S96 are located near the pathogenic polyQ stretch within the N-terminal domain; S651 is located in the hinge region that acts as a bridge between the DNA binding and ligand-binding domains. S96 phosphorylation occurs constitutively and mainly in the cytosol, is slightly induced by the ligand, and positively contributes to AR stability (15–18). Phosphorylation of S83 and S651 is low and barely detectable in the absence of androgens, is induced by androgen binding, and likely occurs mainly in the nucleus (15, 16, 19, 20). S96 phosphorylation is important for AR biology and toxicity. Phosphorylation at S96 by cyclin-dependent kinase 2 (CDK2) stabilizes AR, and this process is negatively regulated by the adenylyl cyclase (AC)/protein kinase A (PKA) signaling pathway (17). In a mouse model of SBMA, intranasal administration of the neuropeptide pituitary adenylate cyclase–activating polypeptide (PACAP), a potent activator of the AC/PKA pathway, increases cyclic adenosine monophosphate levels in the brainstem and spinal cord and also reduces disease manifestations by inducing the genetic CDK2 inhibitor p21CIP1, decreasing phosphorylation of S96, and enhancing AR degradation. These observations support a fundamental role for CDK2 in regulation of AR function and stability. However, whether CDK2 plays an active role in neurodegeneration caused by polyQ-expanded AR is unknown.
By combining phospho-proteomic analysis of AR with pharmacological drug screening targeting cellular kinases and phosphatases as well as genetic validation in cells, flies, and mice, we provide evidence that polyQ expansion enhances phosphorylation at S83, S96, and S651. These sites are phosphorylated by CDK2, CDK7, and CDK9 and dephosphorylated by a calcium-sensitive phosphatase. Further, pharmacologic and genetic ablation of specific enzymes involved in AR phosphorylation and dephosphorylation modified the toxicity of polyQ-expanded AR in vitro and the SBMA phenotype in vivo, highlighting new disease modifiers and potential therapeutic targets. Last, we showed that CDK2 is overactivated in the brainstem of SBMA mice, resulting in expression of genes involved in cellular senescence and apoptosis. CDK2 ablation reversed these changes, implying an active role for CDK2 in SBMA pathogenesis.
RESULTS
PolyQ expansion enhances AR phosphorylation at S96 and influences other phosphosites
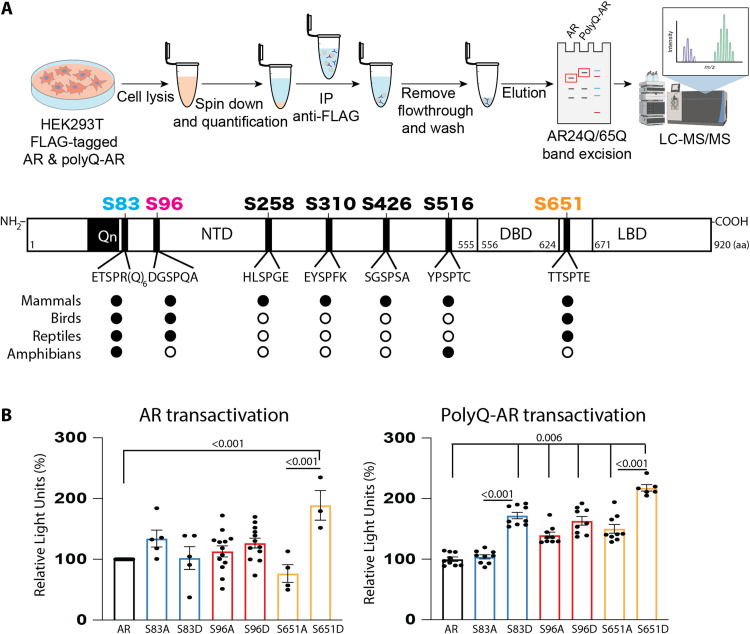
To gain insight into the relationship between pathogenic CAG expansion in AR and phosphorylation, we performed mass spectrometry and Western blot analyses in human embryonic kidney (HEK) 293T cells expressing AR with a nonpathogenic tract (AR12Q or AR24Q) or AR with an expanded polyQ tract (AR55Q or AR65Q) and treated with DHT. Consistent with previous work in COS-1 cells expressing nonexpanded AR (14, 15), nonexpanded AR and polyQ-expanded AR were phosphorylated at SP sites but most notably at S83, S96, and S651 (Fig. 1A and tables S1, A to C), which are the most conserved sites from frog to human (Fig. 1A and fig. S1). PolyQ expansion thus did not preclude native phosphorylation events. To determine whether these phosphosites affect polyQ-expanded AR function, we generated phosphodefective and phosphomimetic mutants by substituting alanine (A) and aspartate (D) for serine at these sites. To measure AR transactivation, we expressed these phosphomutants in cells together with the luciferase reporter gene under the control of an androgen-responsive element, as done previously (17). All phosphomutants were active (Fig. 1B), as previously reported in prostate cancer and SBMA cells (17, 20). For nonexpanded AR, only phosphomimetic substitution of S651 significantly increased activity; for polyQ-expanded AR, phosphomimetic substitution of S83, S96, and S651 significantly increased activity.
Fig. 1. PolyQ expanded AR is highly phosphorylated at conserved phosphosites.
(A) Top: Scheme of experimental workflow. Mass spectrometry analysis in HEK293T cells expressing normal AR (AR24Q) or polyQ-expanded AR (AR65Q) and treated with DHT (10 nM, 24 hours) revealed that both AR and polyQ-expanded AR are phosphorylated at similar serine residues. The bar represents AR (NM_000044) (not to scale). NTD, amino-terminal domain; DBD, DNA-binding domain; LBD, ligand-binding domain. Bottom: The sites highlighted in color are the most conserved phosphosites, as indicated by black circles (white circles indicate lack of conservation) (see fig. S1 for AR sequence alignment). (B) Transactivation luciferase assay in HEK293T cells expressing normal AR (AR12Q, left) or polyQ-expanded AR (AR55Q, right) and the indicated phosphomutants and treated with DHT (0.5 nM, 24 hours) (n = 3 to 13 biological replicates). Graphs show means ± SEM, two-tailed Student’s t test. LC-MS/MS, liquid chromatography–tandem mass spectrometry; m/z, mass/charge ratio; aa, amino acid.
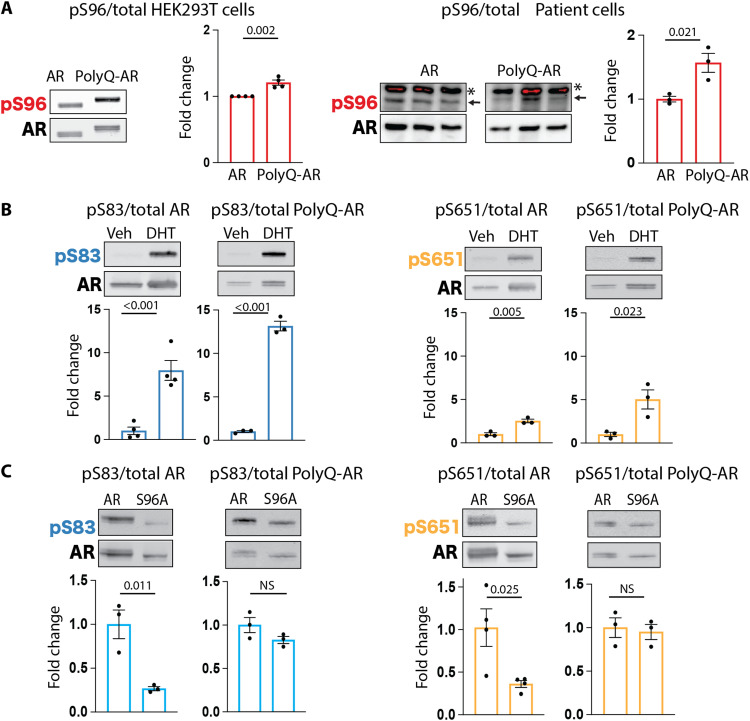
We explored whether polyQ expansion affected phosphorylation at the major phosphosites. In unstimulated cells, phosphorylation at S96 was higher than S83 and S651, both for nonexpanded AR and polyQ-expanded AR (Fig. 2, A and B, and fig. S2, A and B). PolyQ expansion, but not DHT treatment, increased the phosphorylation of AR at S96 in HEK293T cells as well as in neural progenitor cells derived from patients with SBMA (Fig. 2A). DHT treatment increased phosphorylation at S83 and S651, and the effect was larger for polyQ-expanded AR compared to nonexpanded AR (Fig. 2B), suggesting that polyQ expansion facilitates DHT-mediated phosphorylation at S83 and S651.
Fig. 2. AR phosphorylation at serine 96 influences phosphorylation at other major phosphosites and is altered in SBMA cells.
(A to C) Western blot analysis of the phosphorylation status of AR12Q and AR55Q in HEK293T cells treated with vehicle (veh) or DHT (10 nM, 24 hours) (n = 3 to 4 biological replicates). (A) Left: HEK293T cells. Right: isogenic human iPSC-derived neural progenitor cells from an SBMA patient with 54Q and isogenic control with 23Q. Arrow indicates specific signal, and asterisk indicates nonspecific signal. Phosphorylated AR was detected using phospho-specific antibodies, and the total AR was detected with a specific antibody that recognizes AR independently of phosphorylation status. Graphs show means ± SEM, two-tailed Student’s t test. NS, nonsignificant.
To determine whether phosphorylation at one site influences the other two phosphosites, we used phospho-defective mutants. Loss of S83 and S651 did not modify phosphorylation at the other sites (fig. S2C). However, loss of S96 phosphorylation reduced S83 and S651 phosphorylation by ~65% for nonexpanded AR, and this effect was lost for polyQ-expanded AR (Fig. 2C).These results suggest that S96 phosphorylation is required to maintain proper phosphorylation at the other major phosphosites in nonexpanded AR and that this regulation is lost with polyQ expansion.
Loss-of-function drug screening identifies CDKs that modify polyQ-expanded AR nuclear accumulation and phosphorylation
Protein phosphorylation results from fine balance between the coordinated action of kinases and phosphatases. To investigate the kinases and phosphatases targeting polyQ-expanded AR, we performed an unbiased loss-of-function kinase and phosphatase inhibitor drug screening (tables S2 and S3). We generated HeLa cell subclones that stably expressed AR65Q fused to enhanced green fluorescent protein (EGFP) to identify compounds that modify polyQ-expanded AR phosphorylation and function. As a control, we used EGFP-tagged AR24Q. We first verified that EGFP-tagged AR65Q undergoes androgen-dependent protein stabilization, nuclear translocation, and transactivation similar to EGFP-tagged AR24Q in HeLa cells (fig. S3, A to C). We then set up drug screening conditions using the Opera High-Content Screening System (fig. S3, D to G). Because phosphorylation at S96 enhances AR stabilization and nuclear translocation in response to androgen treatment (17, 18), we used AR nuclear enrichment as a readout. Twenty-seven of 273 kinase inhibitors modified (either enhanced or reduced) AR nuclear accumulation, 40% of which (4% of the entire library) inhibited cellular kinases that target SP sites such as CDKs (Fig. 3A and figs. S4, A to F, and S5). Drug screening revealed that polyQ-expanded AR is the substrate of multiple cellular kinases activated in response to distinct signaling pathways.
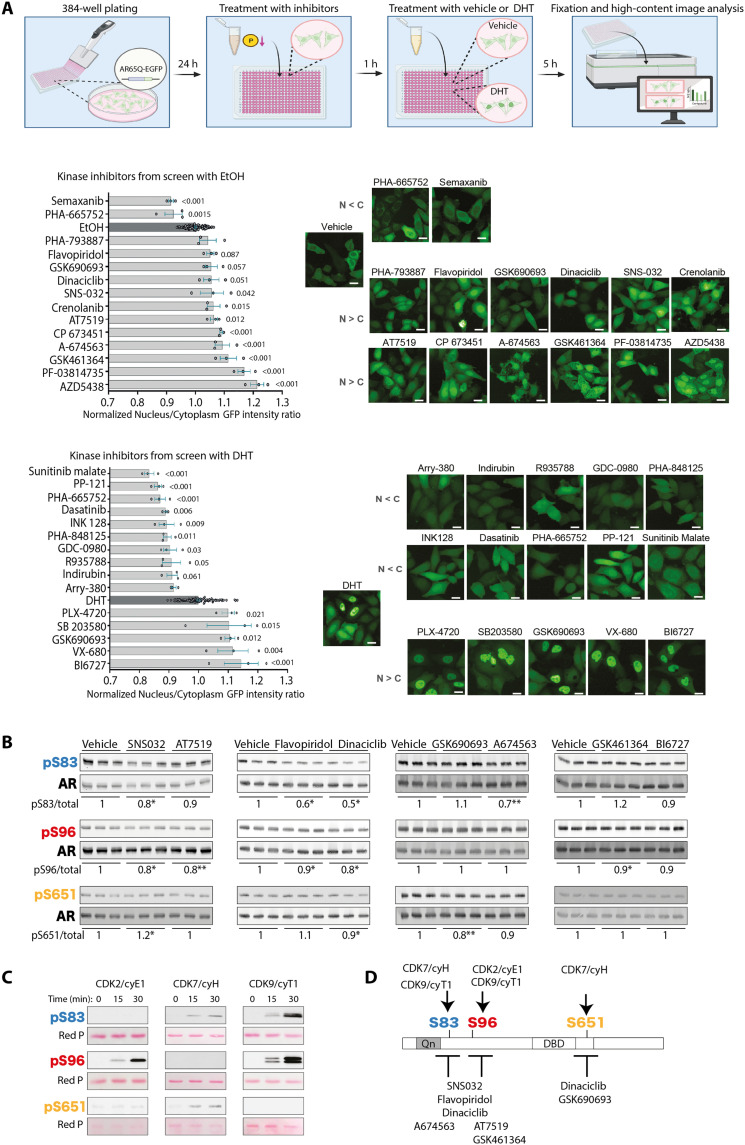
Fig. 3. Loss-of-function drug screening identifies AR as the target of multiple CDKs.
(A) Top: Scheme of experimental workflow. Left: High-throughput loss-of-function screening of small chemical inhibitors targeting kinases carried out in HeLa cells expressing polyQ-expanded AR tagged with EGFP (AR65Q-EGFP) and treated with vehicle [ethanol (EtOH)] or DHT (10 nM) and the indicated drugs (10 μM) (n = 96 wells EtOH/DHT-treated cells; n = 3 wells inhibitor-treated cells). Right: Representative fluorescence images. Scale bars, 25 μm. (B) Western blot analysis of AR phosphorylation after treatment with the indicated inhibitors (200 nM, 5 hours). Quantification of phosphorylated versus total AR is shown at the bottom of each panel (n = 3 biological replicates). (C) Western blot analysis of AR phosphorylation in time course in vitro phosphorylation assays with the AR NTD fragment (amino acids 1 to 153 for S83 and S96) and AR DBD-hinge region (amino acids 556 to 670 for S651) incubated with CDK2/cyE1, CDK7/cyH, or CDK9/cyT1 (n = 2 biological replicates). Total protein is shown by red ponceau (Red P) staining. (D) Schematic representation of AR phosphorylation sites and the respective kinases and small chemical inhibitors identified through drug screening and in vitro phosphorylation assays. Graphs show means ± SEM, one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparisons test, *P < 0.05 and **P < 0.01. Phosphorylated AR was detected using phospho-specific antibodies, and the total AR was detected with a specific antibody that recognizes AR independently of phosphorylation status.
As almost half of the screened compounds are active on CDKs, we validated specific hits targeting CDKs: SNS032, AT7519, flavopiridol hydrochloride, and dinaciclib (Fig. 3B). In addition, we also tested AKT inhibitors (GSK690693 and A674563) and polo-like kinase (PLK) inhibitors (GSK461364 and BI6727). All CDK inhibitors reduced S96 phosphorylation. SNS032 and dinaciclib also reduced phosphorylation of S83 and S651. The PLK inhibitor GSK461364 reduced S96 phosphorylation. These results validate the hits selected from the drug screening, proving their ability to modulate the AR phosphorylation status at the sites of interest.
The validated CDK inhibitors mainly target CDK1 and CDK2, which are involved in cell cycle regulation, and CDK7 and CDK9, which are involved in gene transcription regulation (21). Because SBMA primarily affects postmitotic cells, namely, neurons and myofibers, CDK1 was not further analyzed here, given its involvement in mitosis. We performed in vitro phosphorylation assays using the recombinant AR caspase 3 fragment (amino acids 1 to 153), which contains both S83 and S96 phosphosites, and a fragment that spans the DNA binding domain and hinge region (amino acids 556 to 670) containing S651. By incubating these AR fragments with the selected CDKs in complex with their cognate cyclin (cy), we found that CDK7/cyH and CDK9/cyT1 phosphorylate S83, CDK2/cyE1 and CDK9/cyT1 phosphorylate S96, and CDK7/cyH phosphorylates S651 (Fig. 3, C and D). These results indicate that CDK2/cyE1, CDK7/cyH, and CDK9/cyT1 phosphorylate AR at different SP sites.
CDC25/CDK2 axis enhances polyQ-expanded AR phosphorylation and transactivation
We then tested cellular phosphatases targeting polyQ-expanded AR. From the loss-of-function phosphatase inhibitor screening (Fig. 4A; fig. S6, A to C; and table S3), we identified five compounds that reduced the nuclear enrichment of polyQ-expanded AR by 20 to 30%: three of them, namely, 6-chloro-7-(2-morpholin-4-ylethylamino)quinoline-5,8-dione (also known as NSC-663284) (22), 2,3-bis-(2-hydroxyethylsulfanyl)-1,4-naphthoquinone (also known as NSC-95397) (23), and shikonin (24), which inhibit the phosphatase cell division cycle 25 (CDC25); sanguinarine chloride, which inhibits not only phosphatases, such as protein posphatase 2C (PP2C), but also CDK2 through up-regulation of p21CIP1 (25, 26); and 9,10-phenanthrenequinone, which inhibits CD45 tyrosine phosphatase. The latter compound does not target serine phosphorylation, so it was not investigated further in this work.
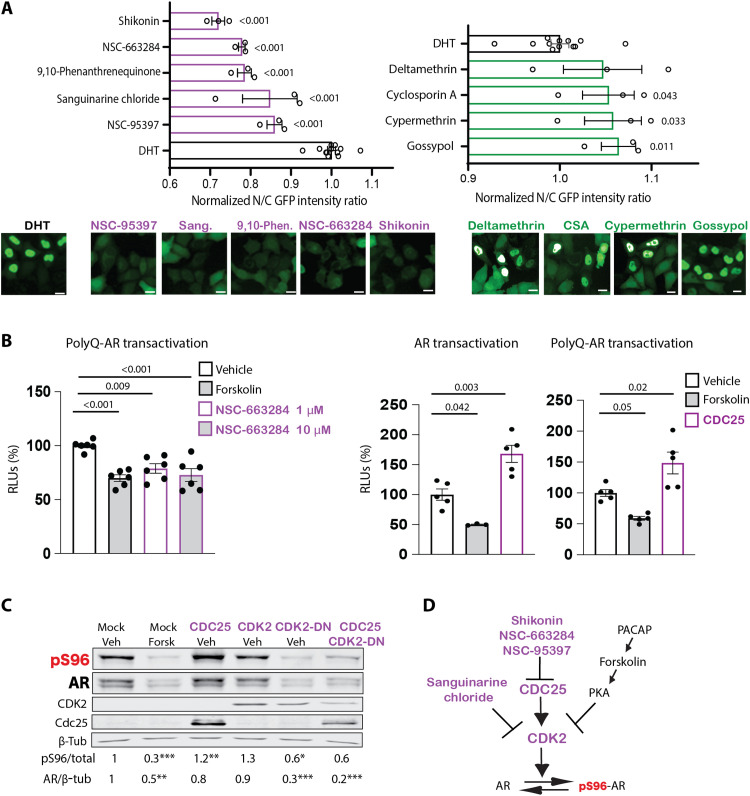
Fig. 4. CDC25/CDK2 axis modifies polyQ-expanded AR transactivation and phosphorylation.
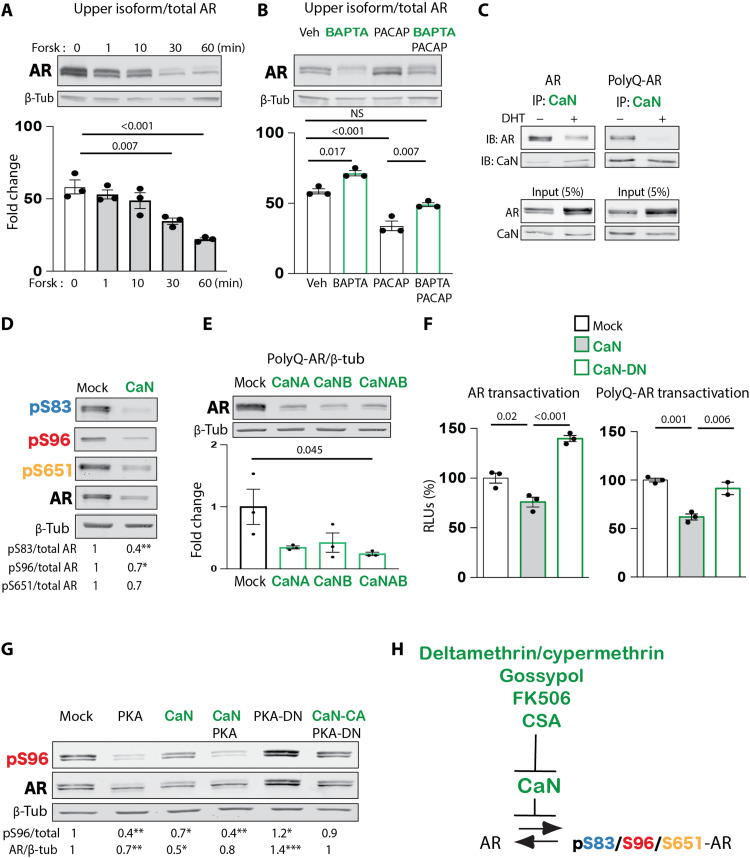
(A) High-throughput loss-of-function screening of small chemical inhibitors targeting phosphatases, as in Fig. 3A. Bottom: Representative fluorescence images. Scale bars, 25 μm. (B) Left: Transactivation luciferase assay in HEK293T cells expressing AR65Q and treated with DHT (0.5 nM, 24 hours) together with vehicle, forskolin (10 μM), and NSC-663284 for 16 hours (n = 6 biological replicates). Right: Transactivation luciferase assay in HEK293T cells expressing AR24Q and AR65Q alone (mock) or in combination with CDC25 and treated with DHT (0.5 nM, 24 hours) together with vehicle or forskolin (n = 3 to 5 biological replicates). (C) Western blot analysis of S96 phosphorylation in HEK293T cells expressing AR24Q alone (mock) or together with CDC25, CDK2, CDK2 dominant-negative (CDK2-DN), or CDC25 + CDK2-DN and treated with vehicle or forskolin (Forsk; 10 μM, 5 hours) (n = 3 to 6 biological replicates). (D) Scheme of drug screening results. Purple is used for results of drug screening; black reports data from previous findings (17). Graphs show means ± SEM, two-tailed Student’s t test (A and C), and one-way ANOVA followed by Tukey’s honest significant difference (HSD) test (B). Phosphorylated AR was detected using phospho-specific antibodies, and the total AR was detected with a specific antibody that recognizes AR independently of phosphorylation status.
CDC25 is a phosphatase that removes inhibitory phosphorylation on specific CDKs, including CDK2, resulting in activation (27). Therefore, we hypothesized that if CDC25 acts upstream of CDK2, then it transactivates polyQ-expanded AR by indirectly enhancing phosphorylation at S96 (17). Thus, inhibition of CDC25 was expected to reduce AR transactivation and S96 phosphorylation. As a control, we used forskolin, which inhibits polyQ-expanded AR transactivation through PKA-mediated up-regulation of p21CIP1 and inhibition of CDK2 (17). Transcriptional assays showed that both forskolin and NSC-663284 reduced polyQ-expanded AR transactivation by ~30% (Fig. 4B, left).
Mammalian cells express three isoforms of CDC25: CDC25A, CDC25B, and CDC25C. Because CDC25C (hereafter referred to as CDC25) is a target of PKA (28), we focused on this isoform for further characterization. CDC25 overexpression significantly enhanced nonexpanded and polyQ-expanded AR transactivation (Fig. 4B, right). We then verified whether the effect of CDC25 on AR function was correlated with changes in phosphorylation at S96 (Fig. 4C). Treatment of cells with forskolin and overexpression of dominant-negative CDK2 (CDK2-DN) decreased S96 phosphorylation and enhanced AR degradation, as previously reported (17). Overexpression of CDC25 enhanced polyQ-expanded AR phosphorylation at S96 by 30%, and this effect was blocked by CDK2-DN, consistent with CDC25 acting upstream of CDK2 (27). Thus, our kinase inhibitor screening identified that the CDC25/CDK2 axis modifies polyQ-expanded AR function and phosphorylation (Fig. 4D).
Calcium-dependent phosphatase reduces the phosphorylation and transactivation of polyQ-expanded AR
In addition to the inhibitors that prevented AR nuclear translocation and polyQ-expanded AR toxicity, we identified four phosphatase inhibitors that increased nuclear accumulation of AR: deltamethrin, cypermethrin, cyclosporine A (CSA), and gossypol (Fig. 4A and table S3). Deltamethrin and cypermethrin are type II pyrethroids. Gossypol [(2,20-binaphthalene)-8,80-dicarboxaldehyde,1,10,6,60,7,70-hexahydroxy-5,50-diisopropyl-3,30-dimethyl] is a cotton plant derivative. CSA together with FK506 are immunosuppressants that bind cyclophilins and FK binding proteins, which form a complex with AR in the unliganded state. These compounds are unrelated yet share two key features: They alter activity of voltage-gated ion (sodium and calcium) channels (29, 30), and they are potent inhibitors of calcium/calmodulin-sensitive Ser/Thr protein phosphatase calcineurin (CaN, also known as PP2B) (31–33).
We previously showed that prolonged treatment of cells (>5 to 6 hours) and chronic administration in mice (several months) with either the neuropeptide PACAP or forskolin result in S96 dephosphorylation through transcriptional induction of the CDK2 inhibitor p21CIP1 (17). Short incubation (30 min) of HEK293T cells expressing polyQ-expanded AR with forskolin was sufficient to significantly decrease AR phosphorylation by 40% (Fig. 5A). This rapid effect is compatible with the involvement of a phosphatase that directly acts on AR. Our screening suggested that AR dephosphorylation occurs in response to calcium signaling. The calcium chelator 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) increased accumulation of the upper AR isoform by 20%, and it partially inhibited the effect of PACAP (Fig. 5B), consistent with the idea that PACAP reduces AR phosphorylation not only via induction of p21CIP1 (17) but also through activation of a calcium-sensitive phosphatase.
Fig. 5. Calcineurin binds to and reduces polyQ-expanded AR phosphorylation and transactivation.
(A) Western blot in HEK293T cells expressing AR55Q and treated with forskolin (10 μM) for the indicated time points. Quantification of the upper AR isoform over total AR is shown in the graphs (n = 3 biological replicates). (B) Western blot in HEK293T cells expressing AR55Q and treated with BAPTA (10 μM) or PACAP (100 ng/ml). Quantification of the upper AR isoform over total AR is shown in the graphs (n = 3 biological replicates). (C) Immunoprecipitation (IP) assay and corresponding immunoblot (IB) in HEK293T cells expressing AR24Q and AR65Q alone or together with CaN and treated with vehicle or DHT (10 nM, 16 hours) (n = 4 biological replicates). (D) Western blot in HEK293T cells expressing AR55Q alone (mock) and together with CaN showing phosphorylated and total AR (n = 3 biological replicates). (E) Western blot in HEK293T cells expressing AR55Q alone (mock) and together with a vector expressing the catalytic subunit of CaN (CaNA), the regulatory subunit (CaNB), or the holoenzyme (CaNAB) (n = 3 biological replicates). (F) Transactivation luciferase assay in HEK293T cells expressing AR12Q (left) or AR55Q (right) alone (mock) or together with CaN and CaN dominant-negative (CaN-DN) and treated with DHT (0.5 nM, 24 hours) (n = 3 biological replicates). (G) Western blot of S96 phosphorylation in HEK293T cells expressing AR55Q in combination with PKA, CaN, PKA + CaN, PKA-DN, or PKA-DN + CaN constitutively active (CaN-CA) (n = 2 to 7 biological replicates). Quantification of phosphorylated (pS96)/total AR is shown below the blots. (H) Diagram showing effective inhibitors and target phosphatase. Graphs show means ± SEM, one-way ANOVA followed by Tukey HSD tests (A, B, E, and F) and two-tailed Student’s t test (D and G).
On the basis of our screening results, we hypothesized that this phosphatase was CaN. Immunoprecipitation assays showed that both nonexpanded AR and polyQ-expanded AR form a complex with endogenous CaN (Fig. 5C). Androgen treatment reduced the interaction of AR with CaN, indicating that CaN binds before androgen binding or after androgen dissociation. Furthermore, CaN overexpression reduced phosphorylation of polyQ-expanded AR at S83, S96, and S651 (Fig. 5D). Consistent with the effect of AR phosphorylation at S96 on protein stabilization (17), overexpression of CaN reduced accumulation of polyQ-expanded AR (Fig. 5E), and in a transcriptional assay, overexpression of CaN but not CaN dominant-negative (CaN-DN) attenuated normal and polyQ-expanded AR transactivation (Fig. 5F).
Mechanistically, overexpression of PKA reduced S96 accumulation to a similar extent as CaN, whereas overexpression of dominant-negative PKA (PKA-DN) reverted this effect (Fig. 5G) (17). Notably, PKA and CaN overexpression together did not result in an additive effect, whereas PKA-DN–dependent accumulation of S96 phosphorylation was abolished by constitutively active CaN (CaN-CA), suggesting that PKA may dephosphorylate AR through CaN. Together, these observations support a model whereby CaN reduces AR phosphorylation, acting on its accumulation and transactivation in SBMA cells (Fig. 5H).
CDC25/CDK2 enhances and CaN suppresses polyQ-expanded AR toxicity in SBMA motor neuron–derived cells
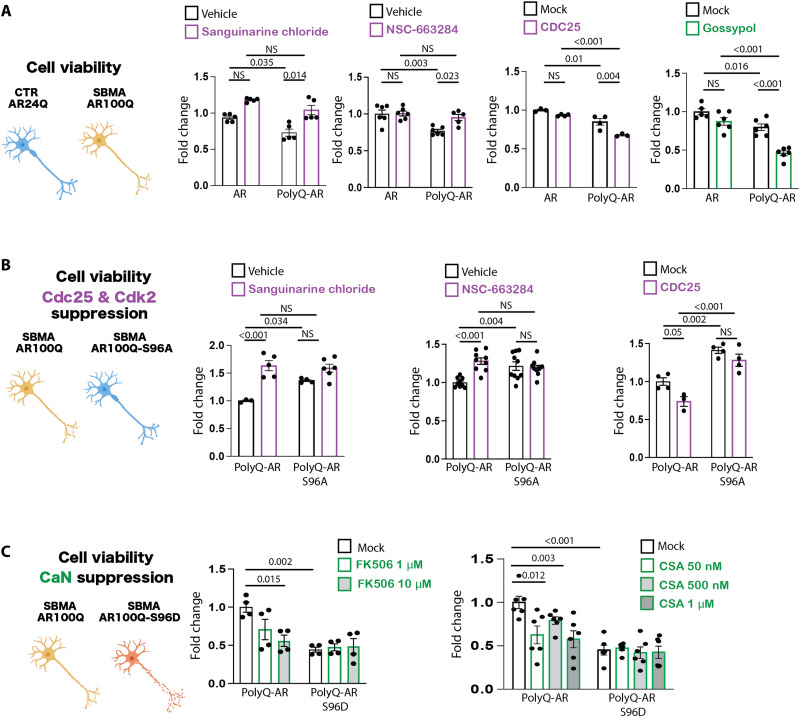
On the basis of the results described above, we asked whether CDC25, CDK2, and CaN modify the toxicity of polyQ-expanded AR. We first addressed this question in cell models of SBMA. Overexpression of ligand-activated polyQ-expanded AR significantly decreased the viability of HEK293T cells compared to cells expressing nonexpanded AR (fig. S7), as previously described (34). Pharmacological inhibition of endogenous CDK2 and CDC25 by sanguinarine chloride and NSC-663284, respectively, restored the viability of HEK293T cells expressing polyQ-expanded AR. SBMA primarily affects brainstem and spinal cord motor neurons. We thus examined the effects of sanguinarine chloride and NSC-663284 on viability in the motor neuron–derived cell line MN1 (35). Overexpression of polyQ-expanded AR decreased MN1 cell viability (Fig. 6A), as previously reported (17). NSC-663284 and sanguinarine chloride reduced polyQ-expanded AR toxicity, indicating that inhibition of CDC25/CDK2 was protective in these cells. However, overexpression of CDC25 had the opposite effect, further supporting the idea that CDC25 enhances polyQ-expanded AR toxicity.
Fig. 6. CDC25/CDK2 and calcineurin modify toxicity in motor neuron–derived cells modeling SBMA.
(A) MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] cell viability assays in MN1 cells stably expressing AR24Q or AR100Q and treated with DHT (10 μM, 48 hours), together with vehicle or sanguinarine chloride (0.1 μM, 16 hours), NSC-663284 (1 μM, 16 hours), or gossypol (10 μM, 16 hours) (n = 3 to 6 biological replicates). (B) MTT cell viability assay in MN1 cells stably expressing AR100Q or AR100Q-S96A treated with sanguinarine chloride or NSC-663284 or transfected with empty vector (mock) or vector expressing CDC25 and treated with DHT (10 μM, 48 hours) (n = 3 to 9 biological replicates). (C) MTT cell viability assay in MN1 cells stably expressing AR100Q or AR100Q-S96D treated with DHT (10 μM, 48 hours), together with FK506 (16 hours) or cyclosporin A (CSA, 16 hours) at the indicated dosages (n = 4 to 6 biological replicates). Graphs show means ± SEM, two-way ANOVA followed by Tukey HSD tests.
In line with evidence that increased nuclear accumulation of polyQ-expanded AR increases toxicity (36), gossypol treatment of MN1 cells expressing AR100Q exacerbated polyQ-expanded AR toxicity. Neither CDC25/CDK2 loss of function (inhibition by sanguinarine chloride and NSC-663284) nor CDC25 gain of function (overexpression) modified the viability of MN1 cells expressing AR100Q-S96A (Fig. 6B), consistent with the idea that the effect of CDC25/CDK2 on polyQ-expanded AR toxicity occurs through regulation of phosphorylation at this site. On the other hand, CaN loss of function (inhibition by CSA and FK506) exacerbated polyQ-expanded AR toxicity in a dose-dependent fashion but had no effect on MN1 cells expressing AR100Q-S96D (Fig. 6C), suggesting that these inhibitors decrease cell viability by phosphorylating polyQ-expanded AR at this site. These results indicate that pharmacologic suppression of CDC25 and CDK2 reduces neurotoxicity, whereas suppression of CaN has the opposite effect, through a mechanism that is affected by the phosphorylation status of polyQ-expanded AR.
CDC25/CDK2 and CaN modify neurotoxicity in a fly model of SBMA
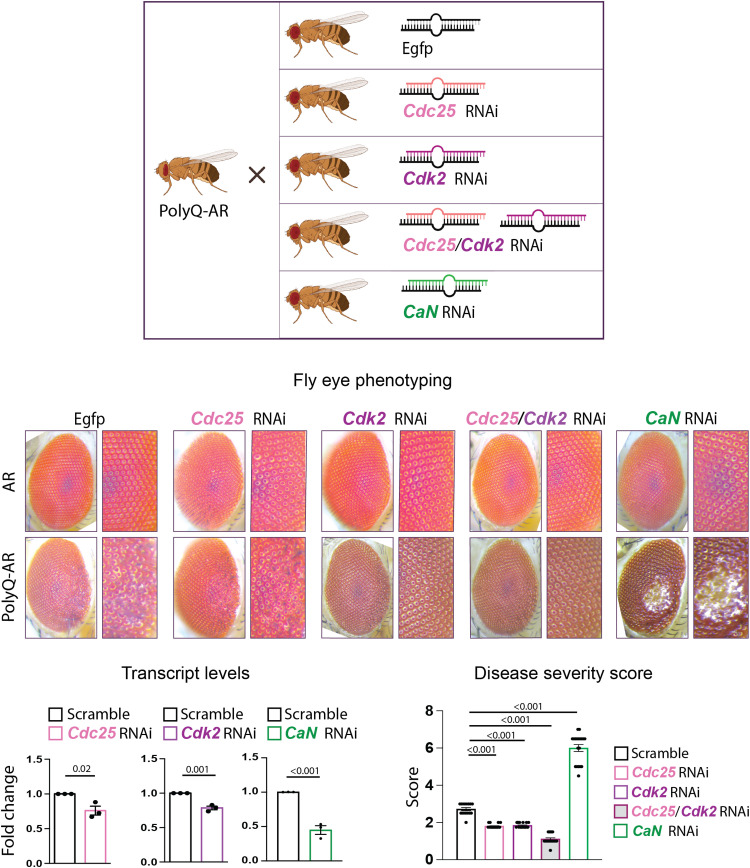
The findings in MN1 cells prompted us to evaluate whether CDC25, CDK2, and CaN modify the SBMA phenotype in vivo. We leveraged a fly model of SBMA that we previously generated and characterized (Fig. 7) (6, 34). We expressed AR0Q and AR52Q in the fly eye while feeding flies androgens and evaluated the effect of genetic manipulation of Cdc25, Cdk2, and CaN on disease severity, which we defined as the degree of ommatidial fusion, bristle disorganization, and depigmentation (6, 34). Consistent with previous findings (34), AR0Q expression did not induce neurodegeneration (Fig. 7, top row), but AR52Q expression significantly altered the retinal structure as evidenced by ommatidia disorganization and degeneration (Fig. 7, bottom row). We crossed AR0Q and AR52Q flies with flies that expressed RNA interference (RNAi) targeting Drosophila orthologs of Cdc25, Cdk2, and CaN. An ~25% decrease in Cdc25 and Cdk2 mRNA transcript levels alone or together did not cause any phenotype in flies expressing AR0Q (Fig. 7, top row). Knockdown of either or both strongly suppressed the toxicity of AR52Q, whereas silencing of the fly CaN ortholog by 50% greatly exacerbated AR52Q toxicity (Fig. 7, bottom row). Together, these observations indicate that genetic suppression of Cdc25 and Cdk2 attenuates the toxicity of ligand-activated polyQ-expanded AR, whereas silencing of CaN has the opposite effect in vivo.
Fig. 7. Silencing Drosophila orthologs of CDC25/CDK2 and CaN modifies the eye phenotype of flies modeling SBMA.
Top: Representative images of the ommatidia of flies expressing AR0Q (top row) or AR52Q (bottom row) with and without the indicated RNAi under the GMR promoter to drive expression in the eye (n = 15 to 20 flies per genotype). Bottom: Cdc25, Cdk2, and CaN mRNA transcript levels in flies overexpressing AR52Q with scramble or target RNAi (n = 3 flies per genotype). Disease severity scores (a function of ommatidial fusion, bristle disorganization, and depigmentation) in flies overexpressing AR52Q with and without the indicated RNAi (n = 15 to 20 flies per genotype). Graphs show means ± SEM, two-tailed Student’s t test (left), and one-way ANOVA followed by Tukey’s HSD test (right).
Ablation of CDK2 ameliorates the phenotype of a severe mouse model of SBMA
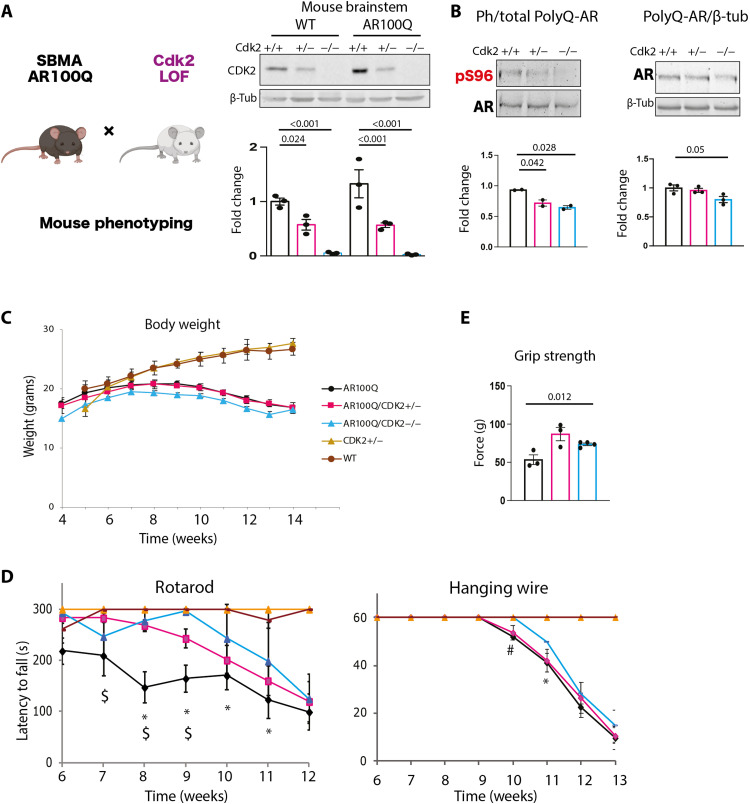
Next, we moved to a mammalian model of SBMA with transgenic mice expressing AR100Q that develop androgen- and age-dependent progressive motor dysfunction by 2 months of age (7). We used specifically male mice, because SBMA fully manifests in males, a disease feature well reproduced in mouse models of the disease (7, 8). Cdk2 deletion is not lethal and does not cause a neurodegenerative phenotype in mice (37), thus offering the opportunity to test the effect of CDK2 partial and complete loss of function in SBMA mice. We crossed transgenic mice expressing AR100Q (7) with Cdk2 hypomorphic or null mice (37) and verified CDK2 reduction at the protein level (Fig. 8A). Western blotting showed that Cdk2 haploinsufficiency and depletion significantly reduced phosphorylation of polyQ-expanded AR at S96 in the brainstem of AR100Q mice (Fig. 8B). We previously showed that S96 phosphorylation stabilizes AR (17). Consistent with these previous observations and with the effect on phosphorylation in the brainstem, Cdk2 deletion reduced accumulation of polyQ-expanded AR in the brainstem and spinal cord but not skeletal muscle (Fig. 8B and fig. S8).
Fig. 8. Haploinsufficiency or deletion of Cdk2 mitigates the phenotype of SBMA mice.
(A) Schematic of mouse cross and Western blot analysis of CDK2 levels in the brainstem of 8-week-old male wild-type (WT) and AR100Q mice (Cdk2 alleles are +/+, +/−, −/−) (n = 3 mice per genotype). (B) Western blot of phosphorylated AR in the brainstem of 8-week-old AR100Q/CDK2+/+, AR100Q/CDK2+/−, and AR100Q/CDK2−/− male mice (n = 2 to 3 mice per genotype). Graphs show means ± SEM. (C) Body weight analysis of mice with the indicated genotypes at 14 weeks [n = 14 (AR100Q/CDK2+/+), n = 10 (AR100Q/CDK2+/−), and n = 7 (AR100Q/CDK2−/−) mice per genotype]. (D) Changes in motor coordination (rotarod) and muscle strength (hanging wire) as mice age [n = 14 (AR100Q/CDK2+/+), n = 10 (AR100Q/CDK2+/−), and n = 7 (AR100Q/CDK2−/−) mice per genotype]. *P (AR100Q/CDK2+/+ versus AR100Q/CDK2−/−) < 0.05; $P (AR100Q/CDK2+/+ versus AR100Q/CDK2+/−) < 0.05; #P (AR100Q/CDK2+/+ versus AR100Q/CDK2−/−) = 0.08. (E) Muscle force measured using grip strength meter in 6-week-old AR100Q/CDK2+/+, AR100Q/CDK2+/−, and AR100Q/CDK2−/− mice (n = 3 mice per genotype). Graphs show means ± SEM, two-way (A) and one-way ANOVA followed by Tukey’s HSD test (B and E) or Fisher’s Least Significant Difference (LSD) (C and D).
We then analyzed the phenotype of SBMA mice. Transgenic mice expressing AR100Q showed a severe phenotype characterized by progressive loss of body weight at 8 weeks of age (Fig. 8C) and signs of motor dysfunction around 6 to 7 weeks of age (Fig. 8D) (7). Although loss of Cdk2 by itself did not improve body weight (Fig. 8C), it significantly improved motor dysfunction assessed by hanging wire, grip strength, and rotarod tasks (Fig. 8, D and E). Notably, the most important effect of Cdk2 loss on mouse phenotype was observed on motor coordination by rotarod, suggesting a central rather than peripheral effect of CDK2 deletion.
Hyperactive CDK2 elicits a stress response in the brainstem of SBMA mice
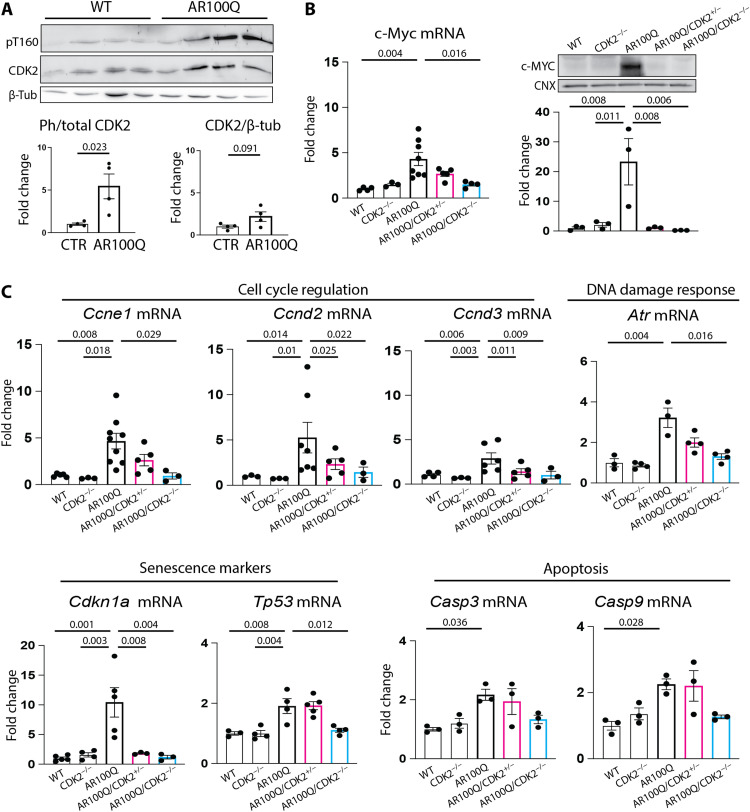
Last, we investigated how CDK2 contributes to the mouse SBMA phenotype. Fully active CDK2 is phosphorylated at threonine 160 (T160) (38). Western blot showed that T160 phosphorylation was significantly increased in the brainstem of male AR100Q mice compared to wild-type (WT) mice, while total CDK2 levels were slightly and not significantly increased in brainstem (Fig. 9A). These findings suggest aberrant CDK2 activity in SBMA brainstem.
Fig. 9. CDK2 triggers a stress response in SBMA brainstem.
(A) Western blot analysis of CDK2 phosphorylation at T160 and total CDK2 in the brainstem of 12-week-old male WT and AR100Q mice (n = 4 mice per genotype). (B) RT-PCR and Western blot analysis of MYC expression in the brainstem of 8-week-old male WT and AR100Q mice (Cdk2 alleles are +/+, +/−, and −/−) (n = 3 to 8 mice per genotype). (C) RT-PCR analysis of expression of the indicated genes in the brainstem of 8-week-old male WT and AR100Q mice (Cdk2 alleles are +/+, +/−, and −/−) (n = 3 to 8 mice per genotype). T160-phosphorylated CDK2 was detected with a specific antibody that recognizes T160 when phosphorylated, while the total CDK2 was detected with an antibody recognizing CDK2 independently of phosphorylation status. Graphs show means ± SEM, two-tailed Student’s t test (A) and one-way ANOVA followed by Tukey’s HSD test (B and C).
To determine whether there was CDK2 gain of function in SBMA brainstem, we analyzed expression of the c-Myc oncogene (39). c-MYC was significantly up-regulated at the transcript and protein levels in the brainstem of male AR100Q mice compared to WT mice (Fig. 9B). CDK2/cyE1 and c-MYC gain of function can synergistically trigger a transcriptional response that activates cell cycle, DNA damage, senescence, and apoptotic pathways (39). Notably, transcript levels of Ccne1 coding cyE1 were significantly up-regulated in the brainstem of AR100Q mice compared to age-matched WT mice, which may in turn be responsible for CDK2 overactivation (40). Moreover, transcript levels of genes involved in cell cycle regulation (Ccnd2 and Ccne1), DNA damage response (Atr), senescence (Tp53 and Cdkn1a, coding for p21CIP1), and apoptosis (Casp3 and Casp9) were also up-regulated in AR100Q mice compared to WT mice (Fig. 9C). Notably, all these brainstem gene expression changes were restored upon Cdk2 ablation, indicating that CDK2 is responsible for inducing these pathological processes in SBMA brainstem.
DISCUSSION
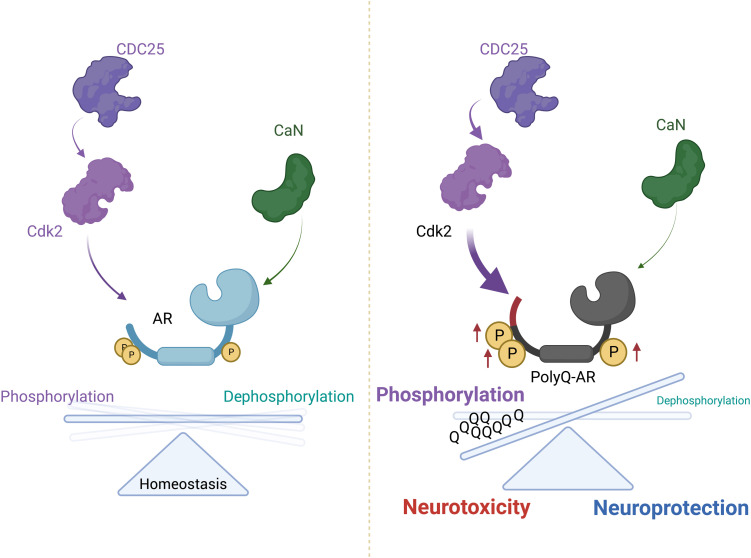
We identified AR as a substrate for CDC25/CDK2 and CaN (Fig. 10). Our results show how AR integrates multiple cellular signaling pathways with changes in phosphorylation, which, in turn, regulate AR biology. PolyQ expansion perturbs the fine-tuned equilibrium of phosphorylation/dephosphorylation, thus enhancing neurotoxicity.
Fig. 10. Opposing effects of CDC25/CDK2 and CaN on polyQ-expanded AR neurotoxicity.
CDC25/CDK2 phosphorylate AR and contribute to toxicity in vitro and in vivo, whereas CaN removes phospho-marks in response to calcium signaling, thus modifying subcellular localization, function, and toxicity. CDK2 triggers a stress response in the brainstem, and its loss of function reduces polyQ-expanded AR phosphorylation and attenuates neurodegeneration.
By comparing the phosphorylation status of normal and polyQ-expanded AR, we showed that polyQ expansion does not alter the pattern of native AR phosphorylation. Rather, polyQ expansion enhances AR phosphorylation at S83, S96, and S651. Most information on the role of AR phosphorylation at specific sites comes from prostate cancer cells. In these cells, phosphorylation of S83 and S651 enhances AR stabilization, transactivation, and cell growth. Analysis of S83 phosphorylation dynamics in prostate cancer cells revealed an effect on AR nuclear translocation and chromatin binding (20). We showed that loss of S96 phosphorylation affected phosphorylation of S83 and S651, suggesting that changes in ligand-independent phosphorylation at S96 have consequences on ligand-dependent phosphorylation at S83 and S651. While loss of S96 phosphorylation reduced phosphorylation at S83 and S651, it did not modify phosphorylation of polyQ-expanded AR, and this may explain why polyQ expansion increases native AR phosphorylation.
Our loss-of-function kinase screening identified several inhibitors that target CDKs involved in transcription, namely, CDK7 and CDK9, which is an intriguing observation considering that AR is a transcription factor. Our in vitro phosphorylation assays showed that CDK9/cyT1 phosphorylated AR at S83, as previously shown (41, 42), and at S96. CDK7/cyH phosphorylates AR at S516 (43). Here, we showed in vitro that CDK7 can also phosphorylate AR at S83 and S651. CDK9/cyT1 is a subunit of the positive transcription elongation factor b (pTEFb), and CDK7/cyH is a subunit of the basal transcription factor II H (TFIIH). CDK9 and CDK7 phosphorylate the C-terminal domain of RNA polymerase II, thus regulating transcription initiation/promoter clearance and elongation. In addition to targeting components of the basal transcription machinery, TFIIH and pTEFb also bind to and phosphorylate specific transcription factors including AR (44, 45). Phosphorylation of AR may then facilitate transcription of androgen-target genes. CDK7 can promote AR activity by direct phosphorylation or through phosphorylation of effectors involved in transcription. CDK7 is a CDK-activating kinase (CAK) and phosphorylates CDK2. Further, CDK7 phosphorylates CDK9, as well as the Mediator complex subunit 1 (MED1), component of the mediator complex, enhancing the interaction with AR (46). AR phosphorylation by CDK7/cyH and CDK9/cyT1 may represent a molecular link between activity of basal transcription factors/RNA polymerase II and the nuclear hormone receptor (43, 45).
Our drug screening also identified inhibitors of CDKs involved in cell cycle regulation, such as CDK2, consistent with our previous evidence that CDK2 phosphorylates AR (17). In our assays, CDK2 was selective for S96. CDK2 has previously been shown to target S83 (47). Although CDK2 did not phosphorylate S83 in our in vitro phosphorylation assay, we cannot rule out the possibility that CDK2 targets this site as well as other sites. CDK2 activity is tightly regulated in cells by phosphorylation at threonine 14 (T14) and tyrosine 15 (Y15), which is inhibitory and removed by CDC25 (27), and at T160, which is activatory and added by CAKs (38). Phosphorylation at T160 can be enhanced by CDK7, suggesting a link and cross-talk between these two CDKs. CDC25 activates CDK2, but not CDK7 and CDK9, which have amino acid positions that differ from T14 and Y15. CDC25 inhibitors identified by our screening may act through CDK2 to modify the AR response to androgens.
Furthermore, CDC25 is regulated through phosphorylation, with phosphorylation at specific sites leading to enzyme inhibition and at other sites resulting in enzyme activation. CDC25 is phosphorylated at multiple sites by kinases that, in our loss-of-function screening, modified the AR response to the ligand, notably PLK (48), PKA (49), and aurora-A kinase (50). We previously showed that AR phosphorylation is modified by the AC/PKA pathway (17). The phosphatase inhibitor okadaic acid also reduced AR phosphorylation. PKA activates an okadaic acid–sensitive serine/threonine phosphatase that leads to dephosphorylation and inactivation of CDC25 (49). Thus, it is possible that PKA modifies AR phosphorylation through CDC25, which, in turn, acts upstream of CDK2, resulting in decreased phosphorylation of AR at specific sites. On the other hand, PKA has been reported to directly phosphorylate and inhibit CDC25 (28). Thus, in addition to inhibiting CDK2 by up-regulating p21CIP1 expression (17), PKA can also directly act on CDC25 to ultimately inhibit CDK2.
Another key finding is that CaN reduced phosphorylation of AR at multiple sites. CaN is activated by calcium signaling and is widely expressed, at particularly high levels in the brain, to target solvent-exposed serine and threonine residues, with specificity from transients in calcium signaling and specific protein-protein interactions. CaN interacts with PKA through an A kinase–anchoring protein (AKAP), which recruits both the phosphatase and kinase in close proximity for posttranslational modification of the target protein (51). Coordinated dynamic action of PKA and CaN mediated by AKAP regulates function and calcium signaling at the synapse (52). In neurons, CaN is activated by calcium transients in response to membrane depolarization. It follows that altered calcium signaling results in excitotoxicity and involves several proteins sensitive to calcium, such as CaN. Aberrant CaN toxic gain-of-function mutations due to calcium overload have been reported in Alzheimer’s disease (53), Parkinson’s disease (54, 55), and Huntington’s disease (56). Notably, CaN directly modifies the phosphorylation status of tau (57), huntingtin (56), TAR DNA-binding protein 43 (TDP-43) (58), and AR as shown here. Regulation of AR phosphorylation by CaN implies that AR is subject to calcium signaling.
CDK2-activating phosphorylation was increased in the brainstem of SBMA mice. Aberrant activation of CDKs and CDC25 has previously been implicated in the pathogenesis of several neurodegenerative diseases. Expression and function of CDK5 and CDK2 are altered in Alzheimer’s disease (59) and Parkinson’s disease cells (60), while an inhibitor of the cell division cycle kinase 7 and CDK9 mitigates disease manifestations in animal models of amyotrophic lateral sclerosis (61) and spinocerebellar ataxia type 17 (62). Moreover, CDC25 gain-of-function mutations have been reported in Alzheimer’s disease (63), and CDC25 loss-of-function mutations are neuroprotective in a fly model of ataxia telangiectasia (64). Therefore, dysfunction of cell cycle–regulated kinases in postmitotic cells, such as neurons and myofibers, may induce cell cycle reentry, leading to cell death by apoptosis. In SBMA brainstem, CDK2 is responsible for induction of genes like c-MYC and genes involved in cell cycle regulation, senescence, and apoptosis. Thus, CDK2 overactivation in SBMA brainstem causes a stress response that may lead to neuronal damage, supporting an active role for this and maybe other CDKs in the neurodegenerative process induced by polyQ-expanded AR.
Expansions of tandem repeats often result in repeat-associated non-AUG (RAN) translation. The pathogenic (CAG)n of AR is located in the N-terminal domain, making it possible that the CUG repeat is expressed in two other reading frames. Recent studies show a very low level of its expression potentially being responsible for the X-linked neurodegenerative disease outcomes, such as fragile X–associated tremor ataxia syndrome or X-linked dystonia-parkinsonism (65, 66). Investigation of RAN translation in SBMA vulnerable tissues might provide insights on how CAG expansions cause disease.
Study limitations
Because AR is targeted by several CDKs and because other CDKs (e.g., CDK4) can compensate for loss of CDK2 function, suppressing several kinases would likely have a stronger effect on the SBMA phenotype. Nevertheless, these data establish a causal link between elevated AR phosphorylation and pathogenesis and identify new therapeutic targets for SBMA. Moreover, targeted inhibition of CDK2 in the brainstem and spinal cord will inform cell-autonomous versus nonautonomous effects of CDK2 toxicity in SBMA. AR is involved in several disease conditions, spanning from androgen-insensitivity syndrome to prostate cancer and other types of cancers with a sex bias (bladder and renal) and to neurodegeneration—our findings are likely to be relevant for these conditions as well. Pharmacological CDK inhibitors are in clinical trials for different types of cancer, but their applicability to chronic conditions such as neurodegenerative diseases requires deep investigation of safety, tolerability, and efficacy.
MATERIALS AND METHODS
Experimental animal models
Mouse strains
Animal care and experimental procedures were conducted by the Ethics Committees of the University of Trento and the University of Padova and were approved by the Italian Ministry of Health (1289/2015-PR). Mice were housed in filtered cages in pathogen-free facilities, in a temperature-controlled room with a 12-hour light/12-hour dark cycle and ad libitum access to water and a standard diet. All mice were kept in congenic C57BL/6J background. Because SBMA is a sex-specific disease, only male mice were used for the study. Mouse ages are reported for each experiment in the figure legend. Sample size was assessed through power analysis. Mouse genotyping was performed as previously described (7). Genomic DNA was extracted from tails or ears and amplified using a RED Extract-N-Amp Tissue Polymerase Chain Reaction (PCR) kit (Sigma-Aldrich). AR100Q mouse primers were 5′-CTTCTGGCGTGTGACCGGCG-3′ (forward) and 5′-TGAGCTTGGCTGAATCTTCC-3′ (reverse); PCR conditions were 94°C for 30 s for denaturation, 60°C for 1 min, 30 s for annealing, and 72°C for 1 min 30 s for extension; and all steps were repeated for 30 cycles. PCR products were visualized on a 1% agarose gel. CDK2+/+, +/−, and −/− mice were distinguished by PCR product amplification with the following primers: 5′-CAAGTTGACGGGAGAAGTTG-3′ (CDK2a), 5′-ACGAACAGCCCTGGACCCCTC-3′ (CDK2b), and 5′-GCGATAAGCTTCGAGGGACC-3′ (CDK2c). CDK2a in combination with CDK2b was used to detect the WT CDK2 allele. CDK2a in combination with CDK2c was used to detect the knockout allele. PCR conditions were set at 94°C for 30 s of denaturation, at 60°C for 30 s of annealing, and at 72°C for 1 min and 30 s for extension, and all steps were repeated for 35 cycles. PCR products were visualized on a 1 or 2% agarose gel for knockout mice or WT, respectively.
Motor coordination was measured by rotarod analysis (Ugo Basile Instruments). AR100Q, AR100Q/CDK2+/−, and AR100Q/CDK2−/− mice underwent weekly sessions on the rotarod apparatus, which included three test trials at 15 to 30 rpm progressively increasing in speed for a maximum period of 300 s and a recovery time of 300 s between each test. The first week was considered as training, and recording was started in the second week. The best performance was used to assess motor coordination.
A grip strength meter (Ugo Basile Instruments) was used to measure the grip strength of the forelimb. The grip strength meter was horizontally placed, and the mice were held by the tail and lowered toward the apparatus. Mice were allowed to grasp the smooth metal triangular pull bar with their forelimbs and then pulled backward. The force applied to the bar at the moment the grasp was released was recorded as the maximum tension. Mice received a weekly session that included three test trials, and the best performance was used to assess muscle force production.
A cage grid was used for the assessment of the muscle force of the hanging wire, in which the mice were allowed to grasp and immediately gently reversed. The test lasted a maximum of 60 s, and the number of seconds the mice lasted hanging on the grid was recorded. Three test trials were performed every week, and the average of the recordings for each mouse was used to assess muscle strength.
For tissue collection, mice were euthanized by CO2 inhalation. After euthanasia, the tissues were snap-frozen in precooled isopentane in liquid nitrogen and then stored at −80°C until further processing.
Fly strains
All Drosophila stocks were maintained on a standard cornmeal medium and fed 2 mM DHT at 28°C in light/dark-controlled incubators. The Cdc25 (34831), Cdk2 (28952), gmr-Gal4, and EGFP lines were obtained from the Bloomington Drosophila Stock Center. The AR52Q line was previously described (34). Eye images were taken with a Leica M205C dissection microscope equipped with a Leica DFC450 camera. Eye degeneration was quantified as previously described (6).
RNA was extracted in triplicate from Cdc25, Cdk2, CaN (Pp2B-14D), and EGFP expressing fly heads using TRIzol (Ambion, #15596026) in 1-bromo-3-chloropropane (BCP; Sigma-Aldrich, #MKCB0830V). Brains were dissected from larvae, snap-frozen on dry ice, ground in TRIzol, and centrifuged to pellet. BCP was added, and samples were centrifuged again. The upper aqueous layer was used to precipitate RNA with isopropanol, followed by pelleting RNA through centrifugation. RNA pellets were washed with 70% ethanol and allowed to air-dry before resuspending in ribonuclease-free water. RNA samples were quantified on a NanoDrop ND-1000 spectrophotometer, and purity was assessed using 260/280 and 260/230 ratios. Samples were run on a 1% agarose gel with ethidium bromide to rule out RNA degradation. Complementary DNA (cDNA) was then generated from RNA samples with the Bio-Rad iScript Select cDNA Synthesis Kit (#170-8897) in a Thermo Hybaid Omn-E PCR machine. All cDNA samples were run on a 96-well plate (Applied Biosystems, #4306737) on an Applied Biosystems 7300 Real-Time PCR system with Bio-Rad iQ Supermix (#170-8862). cDNA-specific PrimeTime quantitative PCR (qPCR) Assay primers (Integrated DNA Technologies) were used for qPCR reactions. Tubulin was used as a housekeeping control. The comparative C(T) method was used to analyze results. GraphPad Prism six software was used for statistical analyses. Primers (5′-to-3′) used are as follows: Pp2b-14D: AGGCGGTACTATTGGCATCTG (forward), TAACCAGAGCAGCAGCGTT (reverse), and AACAAGTGCCCGCACCGTCA (probe); Cdc25: GTGGGAAACTATTGTGGAGGAA (forward), CACGACGAGATCCACTCATTT (reverse), and ACACCAGCAGTTCGAGTAGCATCA (probe); Cdk2: CGAGTACCTGAACATGGATCTAAAG (forward), GGATACGATTCGTGTGGCAA (reverse), TGCATCAGATATTAGATGCCGTCGGC (probe).
Cell lines
HEK293T [American Type Culture Collection (ATCC), CRL-3216) and HeLa (ATCC, CCL-2) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS), penicillin/streptomycin (100 U/ml), and l-glutamine (2 mM) at 37°C in a humidified atmosphere containing 5% CO2. MN1 cells that stably expressed AR24Q and AR100Q with and without the S96A mutation were cultured in DMEM with 10% FBS, penicillin/streptomycin (100 U/ml), and l-glutamine (2 mM) at 37°C in a humidified atmosphere containing 5% CO2 (17). HEK293T cells were transfected with a polyethyleneimine (PEI) linear molecular weight of 25,000Da (Sigma-Aldrich) according to the dimensions of the well. DNA:PEI (0.5%, v/v) ratio was 1:1. MN1 cells were transfected with Lipofectamine 2000 according to the manufacturer’s instructions (Thermo Fisher Scientific).
Generation of neuronal progenitors
For the derivation of induced pluripotent stem cells (iPSCs), primary fibroblast cells were obtained according to the National Institutes of Health protocol 00-N-0043 with the approval of the Institutional Review Board from forearm skin biopsies of male patients with SBMA and controls. iPSCs from patients with SBMA and controls were grown and expanded on Matrigel-coated 60-mm plates using StemFlexmedium (Thermo Fisher Scientific) and were passaged using ReleSR (StemCell Technologies). After three passages (P3), iPSCs were split into Matrigel-coated 24-well plates at a cell density of ~0.5 × 106 cells/ml to begin neural induction. The differentiation of iPSCs into neuronal precursor cells (NPCs) was performed using PSC neural induction medium (Invitrogen) and the published PSC neural induction medium (NIM) protocol (#MAN0008031). After 7 days, newly differentiated NPCs were passaged with Accutase and replated in six-well plates coated with Matrigel at ~1.0 × 106 cells per well. The NPCs were passed using an NIM (50:50 neurobasal:advanced DMEM/F12 and 1× neural induction supplement). NPCs were passaged in NIM medium three times before cryopreservation at P3. After three passages, the NPC medium was changed to neural maintenance medium: 50:50 neurobasal:advanced DMEM/F12, heparin (5 μg/ml; Sigma-Aldrich), fibroblast growth factor 2 (20 ng/ml; Thermo Fisher Scientific), and epidermal growth factor (20 ng/ml; Thermo Fisher Scientific). The NPCs were maintained until P5 before performing the experiments.
Sequence alignment
Orthologous AR sequences were retrieved from the Orthologous MAtrix (OMA) Browser database (67) and aligned with tCoffee (www.ebi.ac.uk/Tools/msa/tcoffee/) (68). The resulting alignment was visualized with ESPript (69).
Expression vectors
Vectors expressing human AR (NM_000044) tagged with FLAG were generated from the Origene vector (catalog no. RC215316). The full-length AR sequence was subcloned into a pEGFP-N1 vector from Clontech (catalog no. 6085-1) to generate EGFP-tagged AR at the C terminus. Vectors expressing pARE-luciferase, TK-Renilla, CDK2 variants, and PKA variants were previously described (17). The CDC25C coding sequence was cloned in a mammalian expression vector in-frame with an myc tag and under the control of the cytomegalovirus promoter. CaN expression vectors were a gift from F. Saudou (Grenoble Institute of Neuroscience, France) and D. Medina (TIGEM, Italy). Single phospho-defective and phospho-mimetic variants of AR12Q/55Q/100Q were generated as previously described (17). In summary, phospho-defective and phosphomimetic mutants were generated using site-directed mutagenesis using Q5 polymerase (M0491S, New England Biolabs) in combination with the Kinase, Ligase, and Dpnl (KLD) enzyme mix (M0554, New England Biolabs) or a QuickChange II site-directed mutagenesis kit (Agilent Technologies) according to manufacturer’s protocols. PCR primers (either overlapping or not overlapping) are listed here: S83D: GCAGCAGCAGCAAGAGACTGACCCCAGGCAGCAGCAGC(forward) and GCTGCTGCTGCCTGGGGTCAGTCTCTTGCTGCTGCTGC (reverse); S96D: GCAGGGTGAGGATGGTGATCCCCAAGCCCATCGTAGAGG (forward) and CCTCTACGATGGGCTTGGGGATCACCATCCTCACCCTGC (reverse); S651D: GGCTTCCAGCACCACCGACCCCACTGAGGAGACAACCC (forward) and GGGTTGTCTCCTCAGTGGGGTCGGTGGTGCTGGAAGCC (reverse).
Mutagenesis at S83 and S96 was performed using site-directed mutagenesis by PCR overlap extension. PCR was carried out by amplifying the first tract using a forward primer (ATGGAAGTGCAGTTAGGGCTGGGAA) and reverse primer of S83D; in a separate reaction, the second fragment from the mutagenesis site was amplified using a forward primer of S83D and a reverse primer (GACACCGACACTGCCTTAC). The PCR products were then used as templates and amplified using specific primers. Products were purified and digested with restriction enzymes to be inserted into AR-containing vectors. The same approach was used for S96D mutagenesis.
To generate vectors for bacterial expression of the AR1-153 fragment with 13Q, the AR caspase fragment region was amplified by PCR using the Gtw Tobacco Etch Virus protease (TEV) AR forward primers (from 5′-to-3′): GGGGACAAGTTTGTACAAAAAAGCAGGCTGGGAAAACCTGTACTTCCAGGGCATGGAAGTGCAGTTAGGGCTGGG and Gtw XbaI AR reverse (from 5′-to-3′):
GGGGACCACTTTGTACAAGAAAGCTGGGTTCTAGACTACGGAGGTGCTGGCAGCTG. PCR amplified products were subcloned into the pDONR221 vector (Thermo Fisher Scientific) using Gateway technology (Thermo Fisher Scientific). The resulting pDEST vector was used for the bacterial expression of the AR caspase fragment (AR1-153) tagged at the N terminus with a histidine tag and maltose-binding protein (His6–MBP–AR1-153). The DNA sequence corresponding to the AR DBD-hinge region for recombinant protein expression was amplified by PCR from full-length AR using the following primers: DBD-h Nde forward: 5′-GGGGGGCATATGCCCCAGAAGACCTGCCTG and DBD-h end Xho reverse: 5′-GGGGGCTCGAGTCAACATTCATAGCCTTCAATGTGTGAC and cloned into pET28b (+) (Novagen) for Escherichia coli protein expression.
Protein expression and purification
The AR caspase fragment (AR1-153) with 13Q was expressed and purified as previously described (70). Briefly, E. coli Rosetta (DE3) competent cells (Merck, catalog no. 70954-3) were transformed with pDEST vectors containing His6–MBP–AR1-153. An overnight culture grown in lysogeny broth (LB) supplemented with antibiotics was diluted into LB and grown at 37°C until it reached an optical density of 0.6. Protein expression was triggered by the addition of 0.75 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and the culture was transferred at 20°C overnight. The bacteria pellets were frozen and thawed on the day of purification in core buffer [20 mM sodium phosphate (pH 8.0), 500 mM sodium chloride, 5% glycerol, and 1 mM β-mercaptoethanol), supplemented with protease inhibitors (protease inhibitor cocktail and phenylmethylsulfonyl fluoride; Sigma-Aldrich). Lysis was achieved after incubation of the resuspended pellet with lysozyme and deoxyribonuclease (DNase) for 1 hour at 4°C, and then the lysate was sonicated and cleared by centrifugation and filtration before loading on the 2 × 5 ml HisTrap column (GE Healthcare) for affinity purification. After column washing for nonspecifically bound proteins, His6–MBP–AR1-153 was eluted with a gradient of imidazole [from 4 to 100% elution buffer in 10 column volumes, where elution buffer is 20 mM sodium phosphate (pH 8.0), 500 mM NaCl, 5% glycerol, 1 mM dithiothreitol, and 500 mM imidazole], concentrated, and loaded into a preparative gel filtration column HiLoad 26/600 S75 (GE Healthcare) equilibrated with 20 mM sodium phosphate (pH 7.5), 500 mM sodium chloride, 5% glycerol, and 1 mM β-mercaptoethanol. Fractions corresponding to the correct molecular weight were collected for cleavage of TEV (Blirt) to release tags; TEV cleavage was carried out in a dialysis bag in TEV cleavage buffer [50 mM sodium phosphate (pH 8.0), 100 mM NaCl, 0.5 mM EDTA, and 1 mM β-mercaptoethanol] to release AR1-153. After cleavage, urea was added to the protein solution up to a concentration of 8 M, and the sample was loaded into 2 × 5 6m HisTrap columns for reverse affinity chromatography. The flow sample was collected and concentrated, and the purity was checked by SDS–polyacrylamide gel electrophoresis (SDS-PAGE). The sample was dialyzed overnight to remove excess salt, then quantified, and lyophilized.
The AR DBD-hinge was expressed in E. coli Rosetta cells transformed with the His-tagged AR DBD-hinge carrying vector. An overnight starter was diluted in antibiotic-supplied LB medium and grown at 37°C after shaking. Protein expression was activated at optical density (OD) of 0.6 with the addition of 1 mM IPTG and 0.1 mM ZnCl2 in culture medium. Protein expression was carried out overnight at 16°C. The cells were harvested, and the wet pellet was frozen before further processing. Bacterial lysis was achieved by adding lysozyme and DNase to the thawed pellet resuspended in binding buffer [20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 5% glycerol, and 5 mM imidazole] supplemented with a protease inhibitor cocktail (Sigma-Aldrich). After 1 hour of incubation under stirring conditions, the lysate was cleared by centrifugation and loaded onto a 5-ml HisTrap column (GE Healthcare) for affinity purification. A wash step [20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 5% glycerol, and 25 mM imidazole] was done before elution in one step in elution buffer [20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 5% glycerol, and 250 mM imidazole]. Eluted fractions were analyzed by SDS-PAGE followed by Coomassie Blue staining. Fractions containing His-tagged DBD-hinge were collected and dialyzed overnight in kinase reaction buffer [20 mM tris-HCl (pH 7.4), 150 mM NaCl, 2 mM β-mercaptoethanol, 5% glycerol, and 0.1 mM ZnCl2]. Soluble protein after dialysis was quantified by ultraviolet absorbance at 280 nm (molar extinction coefficient at 280 nm 5960 M−1 cm−1, Expasy ProtParam tool), aliquoted, and snap-frozen in liquid nitrogen.
In vitro kinase assay
Five micrograms of purified AR caspase fragment or AR DBD-hinge has been incubated for 0′, 30′, or 60′ with equal amounts of recombinant commercially available CDK2/cyclin E (Sigma-Aldrich, catalog no. C0999), CDK7/cyclin H/MNAT1 (Thermo Fisher Scientific, catalog no. PV3868) CDK9/cyclin T (EMD Millipore, catalog no. 14-685), or extracellular signal–regulated kinase 2 (EMD Millipore, catalog no. 14-550) in buffer containing 50 μM final concentration of adenosine 5′-triphosphate as recommended by manufacturer’s instructions. Reaction has been conducted at 30°C and stopped by addition of Laemmli sample buffer. Time points have been loaded on an SDS-PAGE and transferred onto a nitrocellulose membrane for Western blot analysis. Membranes have been stained with Red Ponceau solutions before antibody incubation for detection.
Biochemical analysis
HEK293T cells were cultured in DMEM supplemented with charcoal-stripped FBS and vehicle (ethanol) or 10 nM DHT and harvested 24 hours after treatment. In the case of treatment with kinase inhibitors (SNS-032 from Cayman Chemicals, catalog no. 17904; AT7519 from Cayman Chemicals, catalog no. 16231; dinaciclib from MCE MedChem Express, catalog no. HY-10492; flavopiridol from Sigma-Aldrich, catalog no. F3055; GSK690693 from TOCRIS, catalog no. 937174-76-0; A674563 from Selleckchem, catalog no. S2670; GSK461364 from MedChemExpress, catalog no. HY-50877; and BI6727 from Selleckchem, catalog no. S2235), compounds or vehicle solution was added to cells at final concentration of 200 nM 5 hours before lysis. 1,2-bis(2-aminophenoxy)ethane-N,N,N',N'-tetraacetic acid-acetoxymethyl ester (BAPTA-AM) (Sigma-Aldrich, catalog no. A1076) was added to cells for 5 hours. Cells are treated with DHT, forskolin, or PACAP, as previously described (17). Cells are treated with phosphatase inhibitors (1 or 10 μM NSC-663284 from Sigma-Aldrich, catalog no. N7537; 0.1 μM sanguinarine chloride from Sigma-Aldrich, catalog no S5890; 10 μM gossypol from Sigma-Aldrich, catalog no. G8761; 1 or 10 μM FK506 from Selleckchem, catalog no. S5003; and 50 nM, 500 nM, or 1 μM cyclosporin from Santa Cruz Biotechnology, catalog no. sc-3503) for 16 hours. Cells were rinsed in ice-cold phosphate-buffered saline (PBS) and lysed in the appropriate volume of radioimmunoprecipitation assay lysis buffer [25 mM tris (pH 7.5), 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% NP-40] supplemented with protease inhibitor cocktail (Halt protease inhibitor cocktail, Thermo Fisher Scientific, or Complete tablets, Roche) and phosphatase inhibitor cocktail III (Sigma-Aldrich). Lysates were cleared by centrifugation, and protein content was assessed using a Pierce BCA kit.
For the biochemical analysis of mouse tissues, the brainstem and spinal cord were pulverized using mortar and pestle. Proteins were extracted from pulverized tissues using a lysis buffer containing 2% SDS, 150 mM NaCl, 2 mM EDTA, and 10 mM Hepes (pH 7.4) supplemented with a protease inhibitor cocktail (Roche) and a phosphatase inhibitor cocktail (Sigma-Aldrich). Brainstem and spinal cord lysates were sonicated and centrifuged at 15,000 rpm for 15 min at room temperature (RT). Equal amounts of protein extract were loaded onto a 7 to 10% SDS-PAGE and electrotransferred to a nitrocellulose membrane (Bio-Rad).
Immunoblotting was performed in 5% nonfat dry milk in tris-buffered saline (TBS) for all antibodies as follows: total AR (1:1000; catalog no. sc-13062 or catalog no. sc-7305, Santa Cruz Biotechnology, RRID AB_633881 and AB_626671, respectively), phospho-S96 AR (1:5000; custom made from Biomatik), phospho-S83 (1:1000; catalog no. 07-1375, Merck-Millipore, RRID AB_11211980), phospho-S651 (1:1000; catalog no. PA5-36617, Thermo Fisher Scientific, RRID AB_2553616), CDC25c (1:1000; catalog no. ab226958), pan CaN A (1:1000; catalog no. 2614, Cell Signaling Technology, RRID AB_2168458), and CDK2 (1:1000; catalog no. ab7954, Abcam, RRID AB_2078401). β-Tubulin (1:5000; catalog no. T7816, Sigma-Aldrich, RRID AB_261770) and calnexin (1:1000; catalog no. ADI-SPA-860, Enzo LifeScience, RRID AB_10616095) were used as loading controls. Immunoreactivity was detected using IRDye secondary antibodies for the Odyssey Imaging system (LI-COR Biosciences), following the manufacturer’s instructions. Alternatively, horseradish peroxidase–conjugated secondary antibodies (Thermo Fisher Scientific) coupled with Enhanced chemiluminescence (ECL) reagents (Pierce) were used for detection by ChemiDoc (Bio-Rad). Quantifications were done using ImageJ 1.51j8 software (National Institutes of Health).
Coimmunoprecipitation
Coimmunoprecipitation experiments were performed from HEK293T cells transfected with AR and CaN. Cells were lysed in immunoprecipitation buffer [20 mM Hepes (pH 7.4), 150 mM NaCl, and 5 mM CHAPS supplemented with protease inhibitors] using a syringe with a narrow-gauge (29-gauge) hypodermic needle. Lysates have been cleared by centrifugation, and protein content has been quantified by a Pierce BCA Protein quantification kit. Six hundred micrograms of total protein lysate has been incubated with 2 μg of 441 anti-AR antibody (Santa Cruz Biotechnology, catalog no. sc-7305) for 4 hours at 4°C and then incubated overnight with Pierce protein A magnetic beads (catalog no. 88845). Beads have been washed three times with TBS 0.05% Tween-20. Elution was achieved adding Laemmli sample buffer.
Transcriptional assay
Transcriptional assays were performed following the manufacturer’s instructions (Dual-Luciferase assay kit, Promega). Briefly, HEK293T and MN1 cells were transfected with AR-expressing plasmids together with pARE-Luciferase and TK-Renilla plasmids. On the day of the assay, cells were washed in PBS at RT, lysed in passive lysis buffer, and rocked for 30 min at RT. Each sample was transferred to a 96-well plate, and luciferase substrate was added to each well. The plate was analyzed with Infinite 200Pro (Tecan Instruments). After the acquisition, the Renilla substrate was added, and a second reading was carried out.
Mass spectrometry analysis
For immunoprecipitation and mass spectrometry analysis, HEK293T cells were transfected in triplicate with empty vectors or vectors expressing AR24Q and AR65Q tagged with FLAG tag at the C-terminal end of the protein. Three hours after transfection, the medium was replaced with complete DMEM supplemented with 10 nM DHT. Cells were harvested 24 hours after transfection and lysed with FLAG beads lysis buffer [50 mM tris HCl (pH 7.4), with 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100] supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail III (Sigma-Aldrich). Protein content was assessed using a Pierce BCA kit. Then, proteins (2 mg/ml) were added to preconditioned FLAG beads (Sigma-Aldrich) and incubated at 4°C with gentle shaking overnight following the manufacturer’s instructions. The beads were washed three times with TBS before elution with the addition of Laemmli sample buffer. The beads were moved to a new tube before elution to avoid elution of unspecific proteins. No reducing agents were added to prevent the release of immunoglobulins from the beads. The samples were loaded into a precast gel (4 to 12% bis-tris protein gel; Thermo Fisher Scientific). After electrophoresis, the gels were stained with Coomassie blue brilliant, and bands corresponding to AR were excised for mass spectrometry analysis. Excised bands were treated for free cysteine alkylation and digested with trypsin as previously reported (71). The resulting peptide mixtures were subjected to a phosphopeptide enrichment procedure before injection into a nano-LC system (Ultimate 3000, Dionex, Thermo Fisher Scientific) coupled online with an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Phosphopeptide enrichment, liquid chromatography–tandem mass spectrometry, and data analysis were performed as previously reported (72).
High-throughput screening of kinase inhibitors and phosphatase inhibitors
HeLa AR65Q-GFP cells were seeded in 384-well plates. The following day, cells were treated with 10 μM phosphatase and kinase inhibitors, taken from the SCREEN-WELL Phosphatase Inhibitor Library (Enzo Life Sciences) and Kinase Inhibitor Library (Selleck Chemicals), respectively, for 5 hours. After 1 hour, DHT or vehicle was added. The cells were then fixed with 4% paraformaldehyde for 10 min and counterstained with 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific) and CellTracker (Thermo Fisher Scientific) to visualize the nucleus and cytosol, respectively. The images were taken with a high-content imaging system, Opera (PerkinElmer), and analyzed with the Columbus image data storage and analysis system (PerkinElmer).
MTT assay for cell viability
The cells were seeded at a concentration of 5000 to 200,000 cells per well. Cells were incubated with DHT for 48 hours and phosphatase inhibitors for 16 hours. On the day of the test, 1:10 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich, catalog no. M5655) was added to cell culture medium and incubated for 30 min to 1 hour, according to cell types. Dimethyl sulfoxide was added to solubilize formazan crystals. The maximum absorbance peak was recorded at 570 nm and the reference absorbance at 690 nm with Infinite 200Pro (Tecan Instruments).
Quantitative real-time PCR analysis
Total RNA was extracted with TRIzol (Invitrogen). RNA was retrotranscribed using the SuperScript III First-Strand Synthesis System Kit (Invitrogen) following the manufacturer’s instructions. Gene expression analyses were made by reverse transcription qPCR with SYBR Green (ssoAdvanced Universal Sybr Green Supermix, Bio-Rad) with the C1000 Touch Thermal Cycler–CFX96 Real-Time System (Bio-Rad). The level of each transcript was measured with the threshold cycle (Ct) method using as endogenous control 18S ribosomal RNA (18S) or actin. The specific mouse primers (Eurofins Genomics) used are listed here: ATR, TTGCCTGATCATCCAGAATTAGA (forward) and GCTGGAGAGTGGTTTGAAGA (reverse); C-MYC, GCTGTTTGAAGGCTGGATTTC (forward) and GATGAAATAGGGCTGTACGGAG (reverse); CYCLIN E1, GAGGATGAGAGCAGTTCTTCTG (forward) and AAGAAGTCCTGTGCCAAGTAG (reverse); CYCLIN D2, CTGTGCATTTACACCGACAAC (forward) and ACTTCAGCTTACCCAACACTAC (reverse); CYCLIN D3, AGGAAGTCGTGCGCAATC (forward) and GCTTTGCATCTATACGGACCA (reverse); p21CIP1, AGGACCACGTGGCCTTGTC (forward) and TTTTCGGCCCTGAGATGTTC (reverse);
p53, GGGACGGGACAGCTTTGAG (forward) and AGGACTTCCTTTTTGCGGAAA (reverse); CASPASE 3, TCTGACTGGAAAGCCGAAAC (forward) and AGTCCCACTGTCTGTCTCAA (reverse); CASPASE 9, TTTGTGGTGGTCATCCTCTC (forward) and GAGCATCCATCTGTCCCATAG (reverse).
Statistical analysis
To compare continuous measures distributed normally between groups, Student’s two-sample t tests and one-way analysis of variance (ANOVA) tests were used for comparisons of two groups and more than two groups, respectively. Two-way ANOVA was used to test the effects of two independent categorical predictors on a continuous normal dependent variable. ANOVAs were followed by Tukey’s honest significant difference (HSD) post hoc tests. For behavioral data and body weight analysis, we used a two-way factorial ANOVA with time and genotype as predictors, followed by Fisher’s least significant difference post hoc tests. All data are presented as means ± SEM. The number of experimental replicates and tests used is reported in figure legends for each experiment. For all tests, the significance threshold was set at P < 0.05.
Acknowledgments
We thank M. Barbacid and B. Amati (IIT, Milan, Italy) for providing CDK2 null mice. We thank M. Simonato for technical support. Workflow illustrations and Figure 10 are generated with BioRender.
Funding: This work was supported by the following: Muscular Dystrophy Association (479363 to M.P.), Telethon-Italy (GGP19128 to M.P.), Association Française contre les Myopathies (22221 to M.P. and M.B.), PRIN-MIUR Italy (2017F2A2C5 to M.P.), Kennedy’s Disease Association (to L.T., E.Z., M.B., and A.F.), FONDAZIONE AIRC-Italy (24423 to M.P.), Akira Arimura Foundation (to L.T. and E.Z.), CNCCS Scarl Pomezia (to M.P.), National Institutes of Health (1R21NS111768 to M.P. and U.B.P.; R01 NS100023 and R35 NS122140 to A.R.L.S.), Fondazione Caritro (to L.T.), Fondazione Umberto Veronesi Fellowship (to L.T. and E.Z.), European Research Council (CONCERT, contract number 648201 to X.S.).
Author contributions: Conceptualization: D.P., L.T., E.Z., M.B., and M.P. Methodology: D.P., L.T., E.Z., M.P., E.N.A., A.F., M.J.P., C.M., F.L., J.W., E.B., G.M., S.P., G.A., D.L.M., and M.P. Investigation: D.P., L.T., E.Z., F.S., D.L.M., X.S., and M.P. Visualization: D.P., L.T., E.Z., D.L.M., and M.P. Supervision: E.Z., L.T., A.B., A.R.L.S., S.C.E.T., D.L.M., U.B.P., M.B., and M.P. Writing—original draft: M.P. Writing—review and editing: DP., L.T., E.Z., and M.P.
Competing interests: A.B. is cofounder of Casma Therapeutics and advisory board member of Next Generation Diagnostics, Avilar Therapeutics, and Coave Therapeutics. X.S. is a founder and scientific advisor of Nuage Therapeutics. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.Kennedy W. R., Alter M., Sung J. H., Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology 18, 671–680 (1968). [DOI] [PubMed] [Google Scholar]
- 2.La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H., Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352, 77–79 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Ni W., Chen S., Qiao K., Wang N., Wu Z. Y., Genotype-phenotype correlation in Chinese patients with spinal and bulbar muscular atrophy. PLOS ONE 10, e0122279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejager S., Bry-Gauillard H., Bruckert E., Eymard B., Salachas F., LeGuern E., Tardieu S., Chadarevian R., Giral P., Turpin G., A comprehensive endocrine description of Kennedy's disease revealing androgen insensitivity linked to CAG repeat length. J. Clin. Endocrinol. Metab. 87, 3893–3901 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Kim H., Lim Y. M., Lee E. J., Oh Y. J., Kim K. K., Correlation between the CAG repeat size and electrophysiological findings in patients with spinal and bulbar muscular atrophy. Muscle Nerve 57, 683–686 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., Schwartz S. L., DiProspero N. A., Knight M. A., Schuldiner O., Padmanabhan R., Hild M., Berry D. L., Garza D., Hubbert C. C., Yao T. P., Baehrecke E. H., Taylor J. P., HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447, 859–863 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Chivet M., Marchioretti C., Pirazzini M., Piol D., Scaramuzzino C., Polanco M. J., Romanello V., Zuccaro E., Parodi S., D’Antonio M., Rinaldi C., Sambataro F., Pegoraro E., Soraru G., Pandey U. B., Sandri M., Basso M., Pennuto M., Polyglutamine-expanded androgen receptor alteration of skeletal muscle homeostasis and myonuclear aggregation are affected by sex, age and muscle metabolism. Cells 9, 325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsuno M., Adachi H., Kume A., Li M., Nakagomi Y., Niwa H., Sang C., Kobayashi Y., Doyu M., Sobue G., Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron 35, 843–854 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Rhodes L. E., Kokkinis A. D., White M. J., Watts C. A., Auh S., Jeffries N. O., Shrader J. A., Lehky T. J., Li L., Ryder J. E., Levy E. W., Solomon B. I., Harris-Love M. O., Pean A. L., Schindler A. B., Chen C., Di Prospero N. A., Fischbeck K. H., Efficacy and safety of dutasteride in patients with spinal and bulbar muscular atrophy: A randomised placebo-controlled trial. Lancet Neurol. 10, 140–147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hijikata Y., Hashizume A., Yamada S., Ito D., Banno H., Suzuki K., Sobue G., Katsuno M., Long-term effects of androgen deprivation in a patient with spinal and bulbar muscular atrophy - a case report with 14 years of follow-up. Intern. Med. 58, 2231–2234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennuto M., Rinaldi C., From gene to therapy in spinal and bulbar muscular atrophy: Are we there yet? Mol. Cell. Endocrinol. 465, 113–121 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Sambataro F., Pennuto M., Post-translational modifications and protein quality control in motor neuron and polyglutamine diseases. Front. Mol. Neurosci. 10, 82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y., Chen S., Proline-directed androgen receptor phosphorylation. J. Mol. Genet. Med. 7, 75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong H. Y., Burghoorn J. A., van Leeuwen M., de Ruiter P. E., Schippers E., Blok L. J., Li K. W., Dekker H. L., de Jong L., Trapman J., Grootegoed J. A., Brinkmann A. O., Phosphorylation of androgen receptor isoforms. Biochem. J. 383, 267–276 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gioeli D., Ficarro S. B., Kwiek J. J., Aaronson D., Hancock M., Catling A. D., White F. M., Christian R. E., Settlage R. E., Shabanowitz J., Hunt D. F., Weber M. J., Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J. Biol. Chem. 277, 29304–29314 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Kesler C. T., Gioeli D., Conaway M. R., Weber M. J., Paschal B. M., Subcellular localization modulates activation function 1 domain phosphorylation in the androgen receptor. Mol. Endocrinol. 21, 2071–2084 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Polanco M. J., Parodi S., Piol D., Stack C., Chivet M., Contestabile A., Miranda H. C., Lievens P. M.-J., Espinoza S., Jochum T., Rocchi A., Grunseich C., Gainetdinov R. R., Cato A. C. B., Lieberman A. P., La Spada A. R., Sambataro F., Fischbeck K. H., Gozes I., Pennuto M., Adenylyl cyclase activating polypeptide reduces phosphorylation and toxicity of the polyglutamine-expanded androgen receptor in spinobulbar muscular atrophy. Sci. Transl. Med. 8, 370ra181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gioeli D., Black B. E., Gordon V., Spencer A., Kesler C. T., Eblen S. T., Paschal B. M., Weber M. J., Stress kinase signaling regulates androgen receptor phosphorylation, transcription, and localization. Mol. Endocrinol. 20, 503–515 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Chen S., Xu Y., Yuan X., Bubley G. J., Balk S. P., Androgen receptor phosphorylation and stabilization in prostate cancer by cyclin-dependent kinase 1. Proc. Natl. Acad. Sci. U.S.A. 103, 15969–15974 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S., Gulla S., Cai C., Balk S. P., Androgen receptor serine 81 phosphorylation mediates chromatin binding and transcriptional activation. J. Biol. Chem. 287, 8571–8583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim S., Kaldis P., Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 140, 3079–3093 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Lazo J. S., Aslan D. C., Southwick E. C., Cooley K. A., Ducruet A. P., Joo B., Vogt A., Wipf P., Discovery and biological evaluation of a new family of potent inhibitors of the dual specificity protein phosphatase Cdc25. J. Med. Chem. 44, 4042–4049 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Han Y., Shen H., Carr B. I., Wipf P., Lazo J. S., Pan S. S., NAD(P)H:quinone oxidoreductase-1-dependent and -independent cytotoxicity of potent quinone Cdc25 phosphatase inhibitors. J. Pharmacol. Exp. Ther. 309, 64–70 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Pu L., Amoscato A. A., Bier M. E., Lazo J. S., Dual G1 and G2 phase inhibition by a novel, selective Cdc25 inhibitor 6-chloro-7-[corrected](2-morpholin-4-ylethylamino)-quinoline-5,8-dione. J. Biol. Chem. 277, 46877–46885 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Zhong M., Li X., Zhao F., Huang Y., Long Y., Chen K., Tian X., Liu M., Ma X., Natural compound library screening identifies Sanguinarine chloride for the treatment of SCLC by upregulating CDKN1A. Transl. Oncol. 17, 101345 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adhami V. M., Aziz M. H., Reagan-Shaw S. R., Nihal M., Mukhtar H., Ahmad N., Sanguinarine causes cell cycle blockade and apoptosis of human prostate carcinoma cells via modulation of cyclin kinase inhibitor-cyclin-cyclin-dependent kinase machinery. Mol. Cancer Ther. 3, 933–940 (2004). [PubMed] [Google Scholar]
- 27.Rudolph J., Cdc25 phosphatases: Structure, specificity, and mechanism. Biochemistry 46, 3595–3604 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Duckworth B. C., Weaver J. S., Ruderman J. V., G2 arrest in Xenopus oocytes depends on phosphorylation of cdc25 by protein kinase A. Proc. Natl. Acad. Sci. U.S.A. 99, 16794–16799 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark J. M., Symington S. B., Pyrethroid action on calcium channels: Neurotoxicological implications. Invert. Neurosci. 7, 3–16 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Jan C. R., Lin M. C., Chou K. J., Huang J. K., Novel effects of gossypol, a chemical contraceptive in man: Mobilization of internal Ca(2+) and activation of external Ca(2+) entry in intact cells. Biochim. Biophys. Acta 1496, 270–276 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Enan E., Matsumura F., Specific inhibition of calcineurin by type II synthetic pyrethroid insecticides. Biochem. Pharmacol. 43, 1777–1784 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Baumgrass R., Weiwad M., Erdmann F., Liu J. O., Wunderlich D., Grabley S., Fischer G., Reversible inhibition of calcineurin by the polyphenolic aldehyde gossypol. J. Biol. Chem. 276, 47914–47921 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Liu J., Farmer JD Jr., Lane W. S., Friedman J., Weissman I., Schreiber S. L., Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66, 807–815 (1991). [DOI] [PubMed] [Google Scholar]
- 34.Scaramuzzino C., Casci I., Parodi S., Lievens P. M. J., Polanco M. J., Milioto C., Chivet M., Monaghan J., Mishra A., Badders N., Aggarwal T., Grunseich C., Sambataro F., Basso M., Fackelmayer F. O., Taylor J. P., Pandey U. B., Pennuto M., Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron 85, 88–100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennuto M., Basso M., In vitro and in vivo modeling of spinal and bulbar muscular atrophy. J. Mol. Neurosci. 58, 365–373 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Montie H. L., Cho M. S., Holder L., Liu Y., Tsvetkov A. S., Finkbeiner S., Merry D. E., Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 18, 1937–1950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortega S., Prieto I., Odajima J., Martín A., Dubus P., Sotillo R., Barbero J. L., Malumbres M., Barbacid M., Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35, 25–31 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Desai D., Gu Y., Morgan D. O., Activation of human cyclin-dependent kinases in vitro. Mol. Biol. Cell 3, 571–582 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poon E., Liang T., Jamin Y., Walz S., Kwok C., Hakkert A., Barker K., Urban Z., Thway K., Zeid R., Hallsworth A., Box G., Ebus M. E., Licciardello M. P., Sbirkov Y., Lazaro G., Calton E., Costa B. M., Valenti M., de Haven Brandon A., Webber H., Tardif N., Almeida G. S., Christova R., Boysen G., Richards M. W., Barone G., Ford A., Bayliss R., Clarke P. A., de Bono J., Gray N. S., Blagg J., Robinson S. P., Eccles S. A., Zheleva D., Bradner J. E., Molenaar J., Vivanco I., Eilers M., Workman P., Lin C. Y., Chesler L., Orally bioavailable CDK9/2 inhibitor shows mechanism-based therapeutic potential in MYCN-driven neuroblastoma. J. Clin. Invest. 130, 5875–5892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bretones G., Delgado M. D., Leon J., Myc and cell cycle control. Biochim. Biophys. Acta 1849, 506–516 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Gao X., Liang J., Wang L. Y., Zhang Z., Yuan P., Wang J., Gao Y., Ma F., Calagua C., Ye H., Voznesensky O., Wang S., Wang T., Liu J., Chen S., Liu X., Phosphorylation of the androgen receptor at Ser81 is co-sustained by CDK1 and CDK9 and leads to AR-mediated transactivation in prostate cancer. Mol. Oncol. 15, 1901–1920 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon V., Bhadel S., Wunderlich W., Zhang J., Ficarro S. B., Mollah S. A., Shabanowitz J., Hunt D. F., Xenarios I., Hahn W. C., Conaway M., Carey M. F., Gioeli D., CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol. Endocrinol. 24, 2267–2280 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chymkowitch P., Le May N., Charneau P., Compe E., Egly J. M., The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 30, 468–479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee D. K., Duan H. O., Chang C., From androgen receptor to the general transcription factor TFIIH. Identification of cdk activating kinase (CAK) as an androgen receptor NH(2)-terminal associated coactivator. J. Biol. Chem. 275, 9308–9313 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Lee D. K., Duan H. O., Chang C., Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 276, 9978–9984 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Rasool R. U., Natesan R., Deng Q., Aras S., Lal P., Sander Effron S., Mitchell-Velasquez E., Posimo J. M., Carskadon S., Baca S. C., Pomerantz M. M., Siddiqui J., Schwartz L. E., Lee D. J., Palanisamy N., Narla G., den R. B., Freedman M. L., Brady D. C., Asangani I. A., CDK7 inhibition suppresses castration-resistant prostate cancer through MED1 inactivation. Cancer Discov. 9, 1538–1555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorda R., Buckova Z., Reznickova E., Bouchal J., Krystof V., Selective inhibition reveals cyclin-dependent kinase 2 as another kinase that phosphorylates the androgen receptor at serine 81. Biochim. Biophys. Acta Mol. Cell Res. 1865, 354–363 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Roshak A. K., Capper E. A., Imburgia C., Fornwald J., Scott G., Marshall L. A., The human polo-like kinase, PLK, regulates cdc2/cyclin B through phosphorylation and activation of the cdc25C phosphatase. Cell. Signal. 12, 405–411 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Grieco D., Avvedimento E. V., Gottesman M. E., A role for cAMP-dependent protein kinase in early embryonic divisions. Proc. Natl. Acad. Sci. U.S.A. 91, 9896–9900 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu C., Lyu J., Yang E. J., Liu Y., Zhang B., Shim J. S., Targeting AURKA-CDC25C axis to induce synthetic lethality in ARID1A-deficient colorectal cancer cells. Nat. Commun. 9, 3212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coghlan V. M., Perrino B. A., Howard M., Langeberg L. K., Hicks J. B., Gallatin W. M., Scott J. D., Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science 267, 108–111 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Sanderson J. L., Gorski J. A., Dell'Acqua M. L., NMDA receptor-dependent LTD requires transient synaptic incorporation of Ca2+ -permeable AMPARs mediated by AKAP150-anchored PKA and calcineurin. Neuron 89, 1000–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdul H. M., Sama M. A., Furman J. L., Mathis D. M., Beckett T. L., Weidner A. M., Patel E. S., Baig I., Murphy M. P., LeVine H. III, Kraner S. D., Norris C. M., Cognitive decline in Alzheimer's disease is associated with selective changes in calcineurin/NFAT signaling. J. Neurosci. 29, 12957–12969 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caraveo G., Auluck P. K., Whitesell L., Chung C. Y., Baru V., Mosharov E. V., Yan X., Ben-Johny M., Soste M., Picotti P., Kim H., Caldwell K. A., Caldwell G. A., Sulzer D., Yue D. T., Lindquist S., Calcineurin determines toxic versus beneficial responses to α-synuclein. Proc. Natl. Acad. Sci. U.S.A. 111, E3544–E3552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo J., Sun L., Lin X., Liu G., Yu J., Parisiadou L., Xie C., Ding J., Cai H., A calcineurin- and NFAT-dependent pathway is involved in α-synuclein-induced degeneration of midbrain dopaminergic neurons. Hum. Mol. Genet. 23, 6567–6574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xifro X., Garcia-Martinez J. M., Del Toro D., Alberch J., Perez-Navarro E., Calcineurin is involved in the early activation of NMDA-mediated cell death in mutant huntingtin knock-in striatal cells. J. Neurochem. 105, 1596–1612 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Luo J., Ma J., Yu D.-Y., Bu F., Zhang W., Tu L.-H., Wei Q., Infusion of FK506, a specific inhibitor of calcineurin, induces potent tau hyperphosphorylation in mouse brain. Brain Res. Bull. 76, 464–468 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Liachko N. F., Saxton A. D., McMillan P. J., Strovas T. J., Currey H. N., Taylor L. M., Wheeler J. M., Oblak A. L., Ghetti B., Montine T. J., Keene C. D., Raskind M. A., Bird T. D., Kraemer B. C., The phosphatase calcineurin regulates pathological TDP-43 phosphorylation. Acta Neuropathol. 132, 545–561 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patrick G. N., Zukerberg L., Nikolic M., de la Monte S., Dikkes P., Tsai L. H., Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature 402, 615–622 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Diaz-Corrales F. J., Asanuma M., Miyazaki I., Miyoshi K., Hattori N., Ogawa N., Dopamine induces supernumerary centrosomes and subsequent cell death through Cdk2 up-regulation in dopaminergic neuronal cells. Neurotox. Res. 14, 295–305 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Liachko N. F., McMillan P. J., Guthrie C. R., Bird T. D., Leverenz J. B., Kraemer B. C., CDC7 inhibition blocks pathological TDP-43 phosphorylation and neurodegeneration. Ann. Neurol. 74, 39–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung Y.-H., Lin C.-W., Huang H.-Y., Chen S.-L., Huang H.-J., Sun Y.-C., Lee G.-C., Lee-Chen G.-J., Chang Y.-C., Hsieh-Li H. M., Targeting inflammation, PHA-767491 shows a broad spectrum in protein aggregation diseases. J. Mol. Neurosci. 70, 1140–1152 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Ding X. L., Husseman J., Tomashevski A., Nochlin D., Jin L. W., Vincent I., The cell cycle Cdc25A tyrosine phosphatase is activated in degenerating postmitotic neurons in Alzheimer's disease. Am. J. Pathol. 157, 1983–1990 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rimkus S. A., Katzenberger R. J., Trinh A. T., Dodson G. E., Tibbetts R. S., Wassarman D. A., Mutations in String/CDC25 inhibit cell cycle re-entry and neurodegeneration in a Drosophila model of Ataxia telangiectasia. Genes Dev. 22, 1205–1220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh C. R., Glineburg M. R., Moore C., Tani N., Jaiswal R., Zou Y., Aube E., Gillaspie S., Thornton M., Cecil A., Hilgers M., Takasu A., Asano I., Asano M., Escalante C. R., Nakamura A., Todd P. K., Asano K., Human oncoprotein 5MP suppresses general and repeat-associated non-AUG translation via eIF3 by a common mechanism. Cell Rep. 36, 109376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reyes C. J., Asano K., Todd P. K., Klein C., Rakovic A., Repeat-associated non-AUG translation of AGAGGG repeats that cause X-linked dystonia-parkinsonism. Mov. Disord. 37, 2284–2289 (2022). [DOI] [PubMed] [Google Scholar]
- 67.Altenhoff A. M., Train C.-M., Gilbert K. J., Mediratta I., Mendes de Farias T., Moi D., Nevers Y., Radoykova H.-S., Rossier V., Warwick Vesztrocy A., Glover N. M., Dessimoz C., OMA orthology in 2021: Website overhaul, conserved isoforms, ancestral gene order and more. Nucleic Acids Res. 49, D373–D379 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang J. M., Di Tommaso P., Notredame C., TCS: A new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol. Biol. Evol. 31, 1625–1637 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eftekharzadeh B., Piai A., Chiesa G., Mungianu D., García J., Pierattelli R., Felli I. C., Salvatella X., Sequence context influences the structure and aggregation behavior of a PolyQ Tract. Biophys. J. 110, 2361–2366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Resmini G., Rizzo S., Franchin C., Zanin R., Penzo C., Pegoraro S., Ciani Y., Piazza S., Arrigoni G., Sgarra R., Manfioletti G., HMGA1 regulates the plasminogen activation system in the secretome of breast cancer cells. Sci. Rep. 7, 11768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franchin C., Borgo C., Cesaro L., Zaramella S., Vilardell J., Salvi M., Arrigoni G., Pinna L. A., Re-evaluation of protein kinase CK2 pleiotropy: New insights provided by a phosphoproteomics analysis of CK2 knockout cells. Cell. Mol. Life Sci. 75, 2011–2026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S3