Abstract
A phoA fusion library of Actinobacillus actinomycetemcomitans genomic DNA has been screened to identify genes encoding exported and secreted proteins. A total of 8,000 colonies were screened, and 80 positive colonies were detected. From these, 48 genes were identified with (i) more than half having homology to known or hypothetical Haemophilus influenzae genes, (ii) 14 having no ascribed function, and (iii) 4 having very limited or no homology to known genes. The proteins encoded by these genes may, by virtue of their presence on the cell surface, be novel virulence determinants.
Actinobacillus actinomycetemcomitans is implicated as a pathogen in one of the most prevalent diseases of humans—periodontitis (9). However, surprisingly little is known about the virulence factors of this organism (16). It is well recognized that bacterial exported proteins play key roles in many bacterial functions and are particularly important in the processes of infection (3). In order to gain some idea of the nature of the genes encoding exported proteins in A. actinomycetemcomitans use has been made of a plasmid-based phoA gene fusion system initially developed for studying protein secretion in Escherichia coli (5). This methodology has been used to identify and characterize exported proteins in a number of gram-negative and gram-positive bacteria (1, 4, 7, 10, 12), including a recent report of exported proteins of A. actinomycetemcomitans (8).
Bacterial strains and media.
A. actinomycetemcomitans NCTC 9710 was grown on brain heart infusion agar (Oxoid) supplemented with 5% (vol/vol) horse blood in a carbon dioxide-rich atmosphere for 48 h. E. coli JM107 was grown on nutrient agar (Oxoid). E. coli CC118 was grown on nutrient agar containing erythromycin (ERY) (150 μg/ml).
Isolation of cell-associated proteins.
A fraction containing the cell surface-associated proteins of A. actinomycetemcomitans was prepared by gentle saline extraction as described in reference (6).
Production of rabbit antisera to A. actinomycetemcomitans cell surface-associated proteins.
Three rabbits were immunized with this saline wash of A. actinomycetemcomitans in a nonulcerogenic adjuvant and boosted with material in Freund's incomplete adjuvant. Animals were bled, and titers of antisera were assessed by enzyme-linked immunosorbent assay at various intervals, until titers peaked. Rabbits were then exsanguinated and sera were prepared using conventional means. The rabbit antisera were pooled and extensively immunoadsorbed with E. coli until binding to this bacterium was extinguished. Animal experimentation was done under United Kingdom Home Office regulations.
Construction of phoA fusion library.
Chromosomal DNA was prepared from A. actinomycetemcomitans using standard methods (described in reference 13). Plasmid libraries containing DNA from A. actinomycetemcomitans were prepared in the plasmid vector pHRM104 (23). Chromosomal DNA was partially digested with Sau3AI for 3 h at 37°C. The vector DNA was extracted from 200 ml of overnight cultures of E. coli JM107 using a Midi plasmid preparation kit (Qiagen Ltd, Crawley, United Kingdom). The plasmid was linearized by digestion with BamH1 for 2 h at 37°C. The partially digested chromosomal DNA was ligated with the linearized pHRM104 overnight in a ligation mixture consisting of 40 μl (5 μg) of Sau3AI fragments, 20 μl (1 μg) of linearized plasmid, 6 μl of ligase buffer, and 2 μl of T4 DNA ligase. To confirm that digestion and ligation had taken place, aliquots were removed from the various reactions and analyzed by agarose gel electrophoresis.
Transformation of E. coli CC118.
Competent E. coli CC118 cells were prepared and transformed with the ligated DNA. The transformed cells were transferred into 5 ml of nutrient broth and incubated at 37°C with shaking for only 1.5 h. Cells were plated onto nutrient agar containing ERY (150 μg/ml) and 5-bromo-chloro-3-indoyl phosphate (XP) (40 μg/ml) a substrate for alkaline phosphatase, and incubated overnight at 37°C. Alkaline phosphatase-positive colonies were picked and subcultured onto fresh ERY-XP plates, and the presence of alkaline phosphatase activity confirmed. Plasmid DNA was extracted from individual alkaline phosphatase positive colonies and prepared using the Qiaprep spin miniprep kit. The sizes of the inserts in the recombinant plasmids were determined by digesting the DNA with KpnI and running the digests on agarose gels.
Sequencing of alkaline phosphatase-positive clones.
An oligonucleotide (5′-CGGTTTTCCAGAACAGG-3′) specific to the 5′ end of the truncated phoA gene in pHRM104 was used to sequence over the fusion junction and into the 3′ end of the A. actinomycetemcomitans insert DNA. Double-stranded plasmid DNA was sequenced using dye terminator chemistry and cycle sequencing using the BigDye terminator kit according to the manufacturer's instructions (ABI Perkin Elmer). The reactions were run on an ABI 377 sequencer.
Bioinformatics.
The DNA sequences were analyzed using BLAST searches of the A. actinomycetemcomitans database at the University of Oklahoma (http://www.genome.ou.edu/act.html) and also using the PEDANT database, which contains complete and partial genome sequences of bacteria, including A. actinomycetemcomitans and Haemophilus influenzae (http://pedant.mips.biochem.mpg.de/). PEDANT was also the source of the numbering for the A. actinomycetemcomitans open reading frames (ORFs). The database of all derived protein sequences was also searched at the NCBI database. Segments containing the first 70 amino acids were searched for signal sequences using the SignalP program (http://www.expasy.ch/tools/). For those proteins that were negative on the SignalP programe, the transmembrane protein sequence analysis program DAS, TMpred, TMHMM, and HMMTOP on the Expasy Tools site (http://www.expasy.ch/tools/) were used.
Immunoscreening of phoA clones.
A volume of 5 μl of each phoA clone was spotted onto nutrient agar containing ERY (150 μg/ml) and grown for 18 h at 37°C. Colony lifts were made onto 0.45-μm-pore-size nitrocellulose membranes (Nitrocellulose Extra; Sartorius), and these were blocked by immersion for 30 min in phosphate-buffered saline (PBS) containing 0.5% Tween 20 (PBS-T) and 5% skim milk powder. Membranes were washed three times in PBS-T for 10 min per wash. They were then incubated for 1 h at room temperature with immunoadsorbed rabbit antiserum (1:200 dilution) to the saline wash of A. actinomycetemcomitans. Membranes were then washed three times for 10 min each in PBS-T before being incubated for 1 h at room temperature in a 1:5,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (Sigma A2074). Following a further three washes in PBS-T, membranes were incubated with a commercial ECL Western blotting detection reagent (Amersham Pharmacia Biotech), and the enzymic reaction was determined by exposing the treated membranes to X-ray film.
Positive clones and analysis of insert sizes.
A total of 80 clones, out of 8,000 screened, producing alkaline phosphatase fusion proteins were identified, with insert sizes ranging from 300 bp to 6 kb. Of these, 77 positive clones were sequenced.
Sequencing of clones.
The protein sequences derived from the DNA sequences across the fusion junction were used in BLAST searches of the A. actinomycetemcomitans database and the full-length ORFs were obtained. The derived full-length protein sequence was then used in a BLAST search of all protein sequences at the NCBI database. Data analysis was confirmed by using the PEDANT database, which in most cases provided an annotation of the primary data from the University of Oklahoma A. actinomycetemcomitans genome database. The amount of protein fused to PhoA in each of the recombinants and the size of the full-length ORF, provided from genome databases, is shown in Tables 1 to 3. A number of the clones could not be sequenced, or the sequences showed that two different pieces of A. actinomycetemcomitans genomic DNA had become fused to phoA, and were not interpretable. A number of clones revealed the presence of identical DNA sequences. For example four clones contained the signal sequence of peptide methionine sulphoxide reductase. Tables 1 to 3 and Fig. 1 show the 48 nonredundant clones identified in this study.
TABLE 1.
A. actinomycetemcomitans clones showing homology to known Haemophilus influenzae proteinsa
| Clonee | No. of fused aa | ORF (full length) | Protein identity | % Similarity | % Identity | SP or TMb |
|---|---|---|---|---|---|---|
| 1 | 43 | 1242 (356) | Peptide methionine sulfoxide reductase | 85 | 73 | SP |
| 4 | 132 | 892 (476) | Murein peptide ligase | 87 | 78 | TM |
| 11 | 191 | 2224 (461) | Anaerobic dimethylsulfoxide reductase (DmsA) | 96 | 92 | TM |
| 14 | 388 | 306 (769) | Thiol:disulfide interchange protein (DsbD) | 66 | 54 | TM |
| 15 | 373 | (404 in HI) | Aminotransferasec | 95 | 88 | TM3 |
| 19 | 29 | 1171 (259) | Amino acid ABC transporter periplasmic component | 66 | 49 | SP |
| 22 | 122 | 788 (356) | Type B outer membrane protein | 69 | 58 | SP |
| 27 | 47 | (162) | Hypothetical lipoprotein2 | 83 | 70 | SP |
| 28 | 47 | 604 (626) | Signal peptidase | 75 | 60 | SP |
| 30 | 110 | 1365 (311) | Malate dehydrogenase | 93 | 89 | SP |
| 31 | 152 | (738) | Soluble lytic transglycosylase2 | 69 | 59 | SP |
| 35 | 85 | 1082 (514) | Oligopeptide transporter periplasmic binding protein (putative) | 65 | 50 | SP |
| 37 | 58 | 2003 (423) | UDP-N-acetylglucosamine-1-carboxy vinyl transferase | 92 | 87 | TM |
| 38 | 23 | 498 (809) | Recombination protein rec 2 | 67 | 52 | TM |
| 41 | 120 | 1715 (332) | d-XyloseABC transporter/periplasmic binding protein | 92 | 85 | SP |
| 46 | 32 | 346 (127) | Thiol:disulfide interchange protein (DsbE) | 83 | 72 | SP |
| 47 | 66 | 802 (810) | Probable organic solvent tolerance protein precursor | 81 | 67 | TM |
| 56 | 37 | 1619 (470) | Aspartate aminotransferase | 91 | 84 | No |
| 59 | 59 | 853 (396) | Bicyclomycin resistance protein homologue | 85 | 72 | SP |
| 69 | 71 | 403 (591) | Penicillin binding protein 3 | 84 | 72 | SP |
| 72 | 51 | 778 (172) | Cytochrome c biogenesis protein | 80 | 72 | SP |
| 73 | 77 | 1290 (313) | Cell division protein FTSH homologue 1 | 92 | 85 | SP |
| 79 | 170 | 1442 (484) | Cytochrome c552 formate-dependent nitrite reductase | 89 | 80 | SP |
| 80 | 234 | (487 in HI) | Colicin tolerance protein2 | 86 | 72 | SP |
The table provides information on the number of amino acids in the protein fused to the PhoA, the ORF number (derived from the PEDANT database), the number of amino acids encoded by the full-length ORF, the identity of the protein, the identity and similarity to the same protein in the H. influenzae genome database, and whether the cloned protein has a signal peptide (SP) or transmembrane spanning (TM) segment.
SP denotes the presence of a signal sequence within the N-terminal 50 to 70 amino acids using the program SignalP V1.1 (http://www.expasy.ch/tools/). TM denotes the presence of a transmembrane spanning segment using the programs DAS, HMMTOP, TMHMM, Tmpred, and TopPred 2 at http://www.expasy.ch/tools/.
These DNA sequences are not yet in the PEDANT annotated A. actinomycetemcomitans database and thus do not have an ORF number.
A weak transmembrane spanning segment is seen via the DAS programme in the segment fused to phoA.
Numbers in boldface type represent clones that were recognized by the rabbit antiserum to a saline wash of A. actinomycetemcomitans.
TABLE 3.
A. actinomycetemcomitans clones showing homology to other bacterial proteinsa
| Cloneb | No. of fused aa | ORF (full length) | Protein identity | % Similarity | % Identity | SP or TM |
|---|---|---|---|---|---|---|
| 8 | 119 | 1058 (333) | Methanosarcina barkeri hypothetical protein | 46 | 26 | SP |
| 12 | 66 | 1752 (478) | E. coli 46-kDa membrane protein | 69 | 52 | TM |
| 34 | 98 | 793 (300) | H. somnus 31-kDa antigen gene | 76 | 61 | SP |
| 40 | 132 | 1701 (690) | E. coli α-amylase precursor | 72 | 60 | SP |
| 44 | 102 | 2059 (639) | Y. pestis conserved hypothetical protein TP0792 | 74 | 61 | SP |
| 55 | 80 | 1613 (4000) | P. aeruginosa oprN gene | 45 | 27 | SP |
| 63 | 25 | 362 (143) | Cytolethal distending toxin protein C | 100 | 100 | SP |
| 65 | 138 | 1787 (252) | E. coli hydrogenase isoenzyme formation protein HypB | 81 | 70 | No |
| 74 | 45 | 770 (76) | E. coli YnbE protein precursor | 73 | 52 | SP |
| 78 | 38 | 1745 (198) | P. multocida Skp 26 protein | 73 | 64 | SP |
The table provides information on the: number of amino acids in the protein fused to the PhoA, the ORF number (derived from the PEDANT database) the number of amino acids encoded by the full-length ORF, the identity of the protein, the identity and similarity to the same protein in the H. influenzae genome database, and whether the cloned protein has a signal peptide (SP) or transmembrane spanning (TM) segment.
Numbers in boldface type represent clones that were recognized by the rabbit antiserum to a saline wash of A. actinomycetemcomitons.
FIG. 1.
Protein sequences of the four proteins with no homologies to other proteins in current sequence databases. A vertical arrow indicates the site of signal sequence cleavage using the SignalP V1.1 program. A horizontal arrow indicates that the C terminus of the protein is not known and the amino acid sequence shown is fused to the truncated PhoA at this point. Clone 26 ORF 1440 has no signal sequence cleavage site but has a weak membrane-spanning segment just prior to the point of fusion to PhoA. The clones which bound to the rabbit antisera raised to saline washes of A. actinomycetemcomitans have been identified.
Identification of proteins fused to phoA.
Proteins fused to phoA could be divided into four groups. The first were those proteins that had homology to known H. influenzae proteins (Table 1). The second were those proteins that had homology to hypothetical proteins in the H. influenzae database (Table 2). The third were those proteins which had no homology to H. influenzae proteins but could be recognized by homology to other bacterial proteins (Table 3). The final group consisted of those proteins that had no homology to any proteins in the current sequence databases (Fig 1).
TABLE 2.
A. actinomycetemcomitans clones showing homology to predicted H. influenzae proteinsa
| Clonec | No. of fused aa | ORF (full length) | Protein identity | % Similarity | % Identity | SP or TM |
|---|---|---|---|---|---|---|
| 2 | 74 | 2007 (169) | HI1085 | 89 | 79 | TM |
| 10 | 82 | 437 (227) | HI1603 | 89 | 80 | No |
| 15 | 378 | 1091 (404) | HI1701 | 66 | 49 | TM |
| 21 | 31 | 2011 (191) | HI1150 | 74 | 62 | SP |
| 42 | 145 | 543 (269) | HI0693 | 89 | 78 | SP |
| 51 | 65 | 802 (204) | HI0370 | 74 | 51 | TM |
| 53b | 43 | (367) | HI1236 | 24 | 44 | SP |
| 57b | 246 | (371) | HI1126.1 | 87 | 80 | TM |
| 66 | 25 | 1301 (136) | HI1628 | 58 | 46 | TM |
| 67 | 101 | 748 (185) | HI0389 | 65 | 50 | SP |
The table provides information on the number of amino acids in the protein fused to the PhoA, the ORF number (derived from the PEDANT database), the number of amino acids encoded by the full-length ORF, the identity of the protein, the identity to the same protein in the H. influenzae genome database, and whether the cloned protein has a signal peptide (SP) or transmembrane spanning (TM) segment.
DNA sequence not yet in the PEDANT annotated A. actinomycetemcomitans database so no ascribed ORF number is available.
Numbers in boldface type represent clones that were recognized by the rabbit antiserum to a saline wash of A. actinomycetemcomitans.
Immunoscreening of phoA clones.
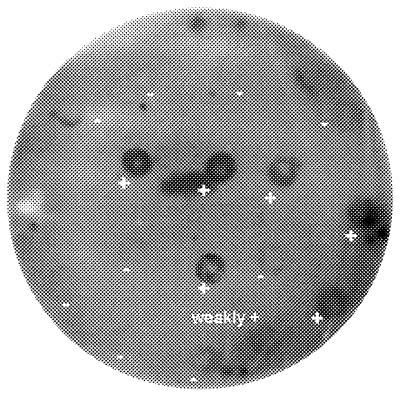
A high-titer polyclonal antiserum to a saline wash of A. actinomycetemcomitans was used to immunoscreen the phoA clones. Negative controls of E. coli CC118 containing unligated vector, as expected, failed to bind the antiserum. This antiserum bound to 22 out of the 48 clones identified in this study (Fig 2). These antibody-binding clones are identified in the tables by the emboldened clone numbers. All four of the clones showing no homology to proteins in sequence databases were recognised by this antiserum (Fig 1).
FIG. 2.
Typical results of screening phoA clones with a polyclonal antiserum to a saline wash of A. actinomycetemcomitans. Some clones bind to the antibody (+) while others show no reaction (−). Clones which react with this antiserum have been denoted in the tables by emboldened clone numbers.
The bacteria causing the periodontal diseases form biofilms between the teeth and the gums, in lesional sites called periodontal pockets, and there is limited contact between these organisms and the cells and tissues in which the disease is manifest (14). It is therefore assumed that the periodontal diseases are driven largely by secreted virulence factors emanating from the causative bacteria. We have attempted to isolate and identify the secreted proteins of A. actinomycetemcomitans, a major periodontopathogen (9, 17) by growing cells on agar and washing them with saline. Separating the extracted material by two-dimensional polyacrylamide gel electrophoresis revealed between 150 and 200 individual proteins (6). Isolating and identifying the virulence proteins in this population has proved to be difficult and, to date, we have only identified chaperonin 60 (6) as a putative virulence factor.
A generic strategy for the identification of at least a proportion of the genes encoding secreted bacterial proteins is based on the generation of translational fusions to a truncated gene for alkaline phosphatase (PhoA) lacking a functional signal sequence (5). Expression of alkaline phosphatase within the reducing environment of the cytoplasm is incompatible with correct folding and results in an inactive enzyme. It is only if the enzyme is translocated across the cytoplasmic membrane, into the oxidizing environment of the periplasm, that enzymic activity is generated. This can only occur if the DNA encoding the PhoA fuses with the coding regions of heterologous signal sequences or a transmembrane spanning sequence which allows the alkaline phosphatase to fold in the periplasm and be anchored to the cytoplasmic membrane. A genomic library of the bacterium of interest is prepared in the truncated phoA-containing vector and colonies are screened for the presence of alkaline phosphatase activity using a colorimetric substrate. Positive colonies can then be expanded, the plasmid can be isolated, and the DNA encoding the heterologous signal sequence can be sequenced.
In the present study, 8,000 colonies of a phoA fusion library of A. actinomycetemcomitans genomic DNA were screened, and 80 positive colonies were identified. Of these, 77 had inserts and were sequenced. The sequences of the A. actinomycetemcomitans DNA inserts (signal sequence plus variable amount of the ORFs of the individual genes) were compared to the genome sequence of A. actinomycetemcomitans to identify the full length ORFs. The sequencing of the genome of A. actinomycetemcomitans is almost completed (http://www.genome.ou.edu/act.html). These ORFs were then subject to further analysis to identify the nature of the genes. Of the 77 clones subject to sequence analysis, 72 gave unambiguous sequences, and when these sequences were analyzed 68 could be identified as full-length ORFs. A number of the clones contained the same or similar DNA sequences such that the total number of genes identified was 48. These proteins divide into those that can be ascribed a function based upon homology to proteins produced by other bacteria (34 proteins) and those that are either hypothetical proteins or proteins not found in any sequence databases (14 proteins). Of the 48 proteins identified, all bar three had either predicted signal peptides or a transmembrane spanning segment.
The assumption made in this and in other papers using the phoA cloning strategy is that the proteins identified are those exported in E. coli. In order to strengthen this assumption we have utilized a high-titer rabbit polyclonal antiserum to a saline wash of A. actinomycetemcomitans to immunoscreen the phoA clones. The saline wash contains protein secreted and loosely associated with the bacterial surface. The antiserum recognized a large proportion of the proteins in this saline wash fraction. The bacteria were not lysed during the immunoscreening and thus any colonies binding antibody must have had the recombinant protein or protein fragment associated with the cell surface. Of the 48 colonies screened, 22 bound the antiserum. This included all four of the proteins listed in Fig. 1, which, as yet, have no homology to proteins in sequence databases. This shows that these genes are encoding immunoreactive proteins. The antiserum also picked up a protein homologous to the aspartate aminotransferase of H. influenzae. This protein had no predicted signal sequence or transmembrane spanning region, which suggests that in spite of the absence of these characteristics this protein can gain access to the bacterial surface. The failure of the antiserum to recognise all clones is no doubt due to the fact that many of the inserts are small and therefore antigenic sites on these proteins are not expressed.
A recent report has also used the PhoA fusion methodology to identify secreted proteins of A. actinomycetemcomitans (8). These workers screened 2,500 colonies and found 28 alkaline phosphatase-positive colonies. Five of these clones proved to have homology to known proteins, and a number appear to be similar to the proteins identified in this study, such as the hypothetical H. influenza protein HI0370 (A. actinomycetemcomitans ORF 802). However, the identities of the other 23 proteins were not presented in this paper.
The proteins identified in this study as being exported/secreted include periplasmic binding proteins involved in nutrient transport, redox proteins such as MrsA (known to be involved in bacterial virulence [2, 18]) and DsbD and resistance proteins (to organic solvents or antibiotics). One protein, CdtC is a toxin and putative virulence factor (11). Fourteen proteins have no ascribed function and would therefore be worth further study to see if they are involved in bacterial virulence.
The PhoA fusion cloning method described in this paper is a relatively rapid methodology for identifying secreted proteins in bacteria of interest. We have recently used it to identify secreted proteins in Staphylococcus aureus (15) and identified 18 novel genes, including a novel group of genes encoding exotoxins (16). Here we show the utility of the system for A. actinomycetemcomitans and describe four exported proteins (Fig. 1) not identified in current databases, of which all appear to be secreted. Further work is under way to determine if these proteins are involved in the virulence characteristics of A. actinomycetemcomitans.
Acknowledgments
We acknowledge the invaluable help of the Actinobacillus Genome Sequencing Project, University of Oklahoma, and B. A. Roe, F. Z. Najar, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer (project is supported by USPHS/NIH grant from the National Institute of Dental Research).
REFERENCES
- 1.Bina J E, Nano F, Hancock R E. Utilisation of alkaline phosphatase fusions to identify secreted proteins, including potential efflux proteins and virulence factors, from Heliocobacter pylori. FEMS Microbiol Lett. 1997;148:63–68. doi: 10.1111/j.1574-6968.1997.tb10268.x. [DOI] [PubMed] [Google Scholar]
- 2.El Hassouni M, Chambost J P, Expert D, Van Gijsegem F, Barras F. The minimal gene set member mrsA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc Natl Acad Sci USA. 1999;96:887–892. doi: 10.1073/pnas.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guitierrez C, Barondess J, Manoil C, Beckwith J. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. J Mol Biol. 1987;195:289–297. doi: 10.1016/0022-2836(87)90650-4. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman C S, Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc Natl Acad Sci USA. 1985;82:5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirby A C, Meghji S, Nair S P, White P A, Reddi K, Nishihara T, Nakashima K, Willis A C, Sim R, Wilson M, Henderson B. The potent bone resorbing mediator of Actinobacillus actinomycetemcomitans is homologous to the molecular chaperone, groEL. J Clin Investig. 1995;96:1185–1194. doi: 10.1172/JCI118150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 8.Mintz K P, Fives-Taylor P M. Identification of genes encoding exported proteins of Actinobacillus actinomycetemcomitans. Infect Immun. 1999;67:6217–6220. doi: 10.1128/iai.67.11.6217-6220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman H N, Wilson M. Dental plaque revisited: oral biofilms in health and disease. Cardiff, Wales: Bioline; 1999. [Google Scholar]
- 10.Pearce B J, Yin Y B, Masure H R. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol. 1993;9:1037–1050. doi: 10.1111/j.1365-2958.1993.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 11.Sugai M, Kawamoto T, Peres S J, Ueno Y, Komatsuzawa H, Fujiwara T, Kurihara H, Suginawa H, Oswald E. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitans is a cytolethal distending toxin. Infect Immun. 1998;66:5008–5019. doi: 10.1128/iai.66.10.5008-5019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor R T, Manoil C, Mekalanos J L. Broad host range mobilization system for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White P A, Nair S P, Kim M-J, Wilson M, Henderson B. Molecular characterization of an outer membrane, putative virulence protein of Actinobacillus actinomycetemcomitans belonging to the OmpA family. Infect Immun. 1998;66:369–372. doi: 10.1128/iai.66.1.369-372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams R C. Periodontal disease. N Engl J Med. 1990;322:373–376. doi: 10.1056/NEJM199002083220606. [DOI] [PubMed] [Google Scholar]
- 15.Williams R J, Ward J M, Henderson B, Wilson M, Nair S P. Rapid screening for putative exported proteins from Staphylococcus aureus using alkaline phosphatase as a reporter molecule. Mol Biotechnol. 2000;15:11–20. doi: 10.1385/MB:15:1:11. [DOI] [PubMed] [Google Scholar]
- 16.Williams R J, Ward J M, Henderson B, Poole S, O'Hara B P, Wilson M, Nair S P. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: characterization of the prototypic gene and it protein product SET1. Infect Immun. 2000;68:4407–4415. doi: 10.1128/iai.68.8.4407-4415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson M, Henderson B. Virulence factors of Actinobacillus actinomycetemcomitans. FEMS Rev Microbiol. 1995;17:365–379. doi: 10.1111/j.1574-6976.1995.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 18.Wizeman T M, Moskowitz J, Pearce B J, Cundell D, Arvidson C G, So M, Weissbach H, Brot N, Masure H R. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]