Abstract
Yersinia pseudotuberculosis localizes to the distal ileum, cecum, and proximal colon of the gastrointestinal tract after oral infection. Using signature-tagged mutagenesis, we isolated 13 Y. pseudotuberculosis mutants that failed to survive in the cecum of mice after orogastric inoculation. Twelve of these mutants were also attenuated for replication in the spleen after intraperitoneal infection, whereas one strain, mutated the gene encoding invasin, replicated as well as wild-type bacteria in the spleen. Several mutations were in operons encoding components of the type III secretion system, including components involved in translocating Yop proteins into host cells. This indicates that one or more Yops may be necessary for survival in the gastrointestinal tract. Three mutants were defective in O-antigen biosynthesis; these mutants were also unable to invade epithelial cells as efficiently as wild-type Y. pseudotuberculosis. Several other mutations were in genes that had not previously been associated with growth in a host, including cls, ksgA, and sufl. In addition, using Y. pseudotuberculosis strains marked with signature tags, we counted the number of different bacterial clones that were present in the cecum, mesenteric lymph nodes, and spleen 5 days postinfection. We find barriers in the host animal that limit the number of bacteria that succeed in reaching and/or replicating in the mesenteric lymph nodes and spleen after breaching the gut mucosa.
The genus Yersinia includes three species, Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis, that are pathogenic in humans and rodents. All three share a tropism for lymph tissues and carry a 70-kb virulence plasmid, pYV, which is essential for infection in these tissues (8, 11). Y. pestis is the causative agent of plague, while the enteropathogenic Yersinia spp., Y. enterocolitica and Y. pseudotuberculosis, usually cause a self-limiting gastroenteritis and mesenteric lymphadenitis (8). In rodents, and occasionally in humans, the enteropathogenic Yersinia spp. spread to the spleen and liver and cause systemic disease (8, 9). Humans carrying the major histocompatibility complex allele HLA-B27 are susceptible to developing reactive arthritis after infection with Yersinia spp.
After oral ingestion, the enteropathogenic Yersinia spp. localize to the distal ileum and proximal colon (9, 23). The bacteria then invade the M cells of the Peyer's patches (PP) and colonize the underlying lymph tissues (15, 16). The chromosomally encoded protein invasin promotes bacterial entry into the M cells (10, 24, 33), while YadA is important for survival and replication of Y. enterocolitica within the PP (33). The invading bacteria are thought to spread to the mesenteric lymph nodes (MLN) and eventually to the spleen and liver. This model of spread is based on the progressive rise in viable bacteria (defined as CFU) in these tissues after infection and by the fact that certain mutants survive for an abbreviated period of time in the lymph tissues but do not reach the spleen (18).
In the PP, Yersinia encounters immune cells including macrophages and B and T lymphocytes. Many of the pYV genes encode components of a type III secretion system including effector molecules, called Yersinia outer proteins (Yops), which are involved in neutralizing the effects of the immune system (11). The type III secretion system translocates the Yops into host cells when Yersinia binds to host cells via invasin or YadA (6, 31). Yops disrupt normal cellular processes, and several Yops have been implicated in the ability of Yersinia to multiply within PP, spread to deeper tissues, and cause death of mice (11, 18, 30). For instance, YopJ induces apoptosis in macrophages in vitro (29, 31) and in Mac-1-positive cells in the MLN and spleen in vivo (30).
Genetic screens designed to identify the factor(s) involved in interactions of the bacteria with cultured cells, in serum, or Yop secretion have identified genes required for Yersinia infection (4, 19, 28, 35, 37). Alternatively, sequence analysis, particularly of the virulence plasmid, has led to the identification of open reading frames that are important for infection (2, 3, 18). However, additional genes were postulated to be essential for infection in the more complex environment of the animal host. Several experimental strategies have been designed for screening pools of bacterial strains in animal model systems to identify genes whose expression is either induced (in vivo expression technology [IVET] [22] and differential fluorescence induction [40]) or required for survival (signature-tagged mutagenesis [STM] [17]). The IVET strategy has been used to identify 45 chromosomal genes in Y. enterocolitica that are expressed preferentially in the small intestine and/or PP after oral inoculation (43). While some of these genes have no obvious function, many share homologous sequences with genes involved in stress response, iron starvation, and cell envelope maintenance. Moreover, several of these genes are important during Y. enterocolitica oral infection.
The STM strategy allows one to identify avirulent mutants after generating mutant libraries of bacterial strains by transposon mutagenesis (17). Each transposon contains a unique 40-bp sequence. The unique tag can be amplified using PCR, and thus the fate of any one particular mutant can be monitored by the frequency of its tag in the pool of bacteria. Recently, STM has been used to identify Y. enterocolitica genes required for colonization of mouse spleens after intraperitoneal (i.p.) infection (12).
In addition to identifying attenuated strains, infection with signature-tagged (ST) strains allows one to follow the course of infection in the infected animal. Because the bacteria are uniquely marked, one can essentially take snapshots of the infection and determine how many different bacteria seed and are replicating in a tissue at any time during the course of infection. In fact, the success of STM depends on many bacteria seeding and replicating in the tissue of interest. The hypothesis of independent action of pathogens, formulated by Meynell (26) and Meynell and Stocker (27) over 40 years ago, predicts that every bacterium has an equal chance of initiating a lethal infection regardless of the inoculum size and that each bacterium acts independently in a host. One prediction of this model is that many bacteria will replicate and cause disease when the inoculating dose is above the 50% lethal dose (LD50), while only one or a few will cause disease if the dose is below the LD50. Work in the 1950s (26, 27) and more recent studies using pools of ST strains show that many bacteria seed and replicate in the spleen and liver after i.p. inoculation (12, 17). However, little has been done to demonstrate the number of strains seeding various tissues after oral inoculation (39). Infection with a pool of ST strains allows one to use large, marked input populations and thus determine the number of bacterial clones seeding and replicating in various tissues at different times after oral infection.
We have used ST Y. pseudotuberculosis strains to characterize an orogastric infection of Y. pseudotuberculosis at day 5 of infection and to identify genes that are essential in establishing infection in the cecum after oral inoculation. We found several host barriers or constraints that limit the number of bacteria progressing to deeper tissues. We also identified 13 attenuated Y. pseudotuberculosis strains including several with mutations in the type III secretion system and the O-antigen biosynthetic pathway, one mutated in invasin, and several with mutations in genes that had not previously been associated with virulence.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strains SM10λpir and DH10B (Stratagene) were grown in L broth or on L agar plates at 37°C. A mouse passage strain of Y. pseudotuberculosis, YPIIIpIBI, was used as the wild-type (WT) Y. pseudotuberculosis in all experiments. YPIIIpIB71, a kanamycin-resistant mutant that does not secrete the Yops in cell culture or in vitro assays (14), was used in mixed infection experiments designed to assess the feasibility of using STM in oral infections by Yersinia. Y. pseudotuberculosis strains were grown in 2xYT at 26°C; the medium was occasionally supplemented with 5 mM CaCl2 (high Ca2+) or 2 mM MgCl2 and 20 mM sodium oxalate (low Ca2+), as noted. Plasmid pUTminiTn5Kn2+tags (17) (gift from D. Holden) was used as the source of the tagged transposons.
Generation of transposon library of Y. pseudotuberculosis.
To generate transposon (ST) Y. pseudotuberculosis strains, pUTminiTn5Kn2+tags was electroporated into the E. coli SM10λpir, and the bacteria were grown overnight in 50 ml of L broth supplemented with ampicillin (200 μg/ml). The following morning, the E. coli cultures were washed three times in L broth and mixed with 25 ml of WT Y. pseudotuberculosis grown overnight. The mixed cultures were allowed to stand overnight at room temperature; the following day, 200 μl of culture was spread onto L agar plates containing kanamycin (50 μg/ml) and irgasan (2 μg/ml). Y. pseudotuberculosis is naturally resistant to irgasan, but E. coli is sensitive. To ensure that the kanamycin resistance was due to a transposition event and not to illegitimate integration of the plasmid, kanamycin-resistant Y. pseudotuberculosis was screened for ampicillin resistance, and the ampicillin-resistant colonies were discarded. Two assays were performed to assess the randomness of the transposon insertions. First, we examined the size of the fragments containing the transposon by Southern blotting DNA from 20 different colonies was isolated, digested with EcoRI, and probed with the kanamycin gene. Only one band was detected from each strain, and all sizes were different, indicating that transposition was occurring at different sites in the genome. Second, we assessed the number of mutant bacteria that were incapable of secreting the Yops. Yersinia isolates that are unable to secrete Yops are white on L agar plates containing the dye Congo red when grown at 37°C, while bacteria that secrete Yops bind to the red dye (36). Four of 500 colonies were white, indicating that approximately 0.8% of the transposons disrupted genes involved in secretion.
Infections of mice.
Eight to 10-week-old BALB/c female mice (Charles River) were used for all infections. All mice were denied food for 18 h prior to orogastric infection. Bacteria were grown at 26°C in 96-well plates overnight. ST strains were pooled in groups of 48 and resuspended in phosphate-buffered saline to approximately 5 × 1010 bacteria/ml. Aliguots of 100 μl of the bacterial suspension were delivered to the stomachs of three mice via a feeding tube. Water and food were provided immediately after infection. Mice were killed 5 days after infection, and the cecum, PP, MLN, and spleens were harvested. The tissues were homogenized in stomacher bags containing 1 ml of phosphate-buffered saline. Various dilutions of the mixture were plated on L agar plates containing kanamycin to determine the number of bacteria in each tissue. The remaining suspension was plated on 150-mm-diameter L agar plates containing kanamycin, and bacteria were harvested after 2 days and frozen until needed.
For secondary screening of mutants, strains that had failed to replicate in either the cecum or in the PP and MLN were grouped together into pools of 12 or 16 and tested again in oral infections of two mice. Five days after infection, the mice were killed, tissues were harvested, and the bacteria were recovered. Strains that again failed to colonize the cecum and/or PP and MLN were tested individually in three to six mice for the ability to cause death at a dose of 2 × 109 bacteria. The oral LD50 for the WT Y. pseudotuberculosis used in these studies is 2 × 107 bacteria (30).
In some experiments, competition assays were performed with mixtures of WT and mutant bacteria. The bacteria were grown overnight separately and then mixed together and used to infect mice. In most experiments, equal amounts of WT and mutant bacteria were used in coinfection assays. In the complementation experiments with YPIIIpIB71, the amount of WT was intentionally increased to threefold greater than the amount of mutant. Five days after orogastric infection and 3 days after i.p. infection, mice were killed and the desired tissues were isolated. WT bacteria were distinguished from mutant bacteria based on their sensitivity to kanamycin; all transposon-tagged strains and YPIIIpIB71 are kanamycin resistant, whereas YPIIIpIB1 is kanamycin sensitive.
Colony hybridization assays.
Each member of each pool of 48 ST strains was grown in a well of a 96-well plate, and the pool was replica plated onto Hybond N+ membranes that were then placed on 150-mm L-diameter agar plates containing kanamycin. Colonies formed within 20 h after incubation at room temperature. The colonies were lysed using standard procedures (17), and the membranes were stored at room temperature until needed. Bacterial DNA from the input pool and recovered from each tissue was isolated by the cetyltrimethylammonium bromide method (17) and used as a template in PCRs to generate probes. Probes were prepared as described elsewhere (17) except that only 2 μl of [α-P32]dCTP was used in each reaction. The amplified DNA was digested with HindIII and then added to 3 ml of hybridization buffer containing formamide. The filters were hybridized overnight at 42°C and washed for 1 h in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate.) and 0.2% sodium dodecyl sulfate (SDS) at 65°C. Filters were autoradiographed with Kodak XAR-5 film at −80°C for 3 to 24 h.
To test the sensitivity of the colony hybridization assay, the amount of one of the input colonies in a pool of 48 was intentionally lowered 5-, 10-, 20-, 40-, and 80-fold. Probes were generated from the pools of DNA and hybridized to colony blots. The artificially lowered ST strain was not detected when its concentration was lowered 20-fold or more.
Cloning and sequencing transposon insertion sites.
Genomic DNA was digested with EcoRI, SacI, SaII, PstI, or XbaI, and ligated into pGEM-7zf or pUC19 (New England Biolabs) digested with the same enzyme; the resulting ligation mix was used to transform E. coli DH10B, and kanamycin-resistant colonies were selected. The DNA adjacent to the kanamycin insert was sequenced (Beckman Center PAN Facility, Stanford University). The clones were verified by Southern blotting (data not shown). Sequence homologies were performed using GenBank databases and BLAST programs found at www.molbio.info.nih.gov.
Cell culture assays.
Invasion and survival assays were done as described previously (25) except that bacterial strains were grown in 2xYT containing 5mM CaCl2. Cytotoxicity assays were done using a Cytotox96 kit (Promega) as described elsewhere (31) with the exceptions that the multiplicity of infection was 5 to 10 bacteria/cell and the bacteria were grown in 2xYT with 5 mM CaCl2.
Analysis of Yop secretion and Y. pseudotuberculosis mobility.
To induce the Yops in broth, bacteria were grown in 2xYT at 26°C overnight, diluted 1:40 into 2xYT containing 20 mM sodium oxalate and 2 mM MgCl2, grown at 26°C for 2 hours, and then shifted to 37°C for 2 h. Then 109 bacteria were pelleted, and the supernatant was mixed with 100% trichloroacetic acid to a final concentration of 10% and placed on ice for 30 min. The tubes were spun at 14,000 rpm for 15 min; the pellets were washed in acetone and then resuspended in 50 μl of SDS sample buffer. A 10-μl volume of each sample was run on an SDS–12.5% polyacrylamide gel, and the proteins were visualized with Coomassie blue.
To assess the motility of Y. pseudotuberculosis strains, the bacteria were inoculated into agar (0.3%) plates and incubated at 26 or 37°C overnight. The following morning, motile strains had grown outward from the inoculating site and covered and area greater than 0.5 cm in diameter, whereas nonmotile strains were confined to a much smaller area.
RESULTS
STM is based on the premises that each member of a population of bacteria has an equal opportunity to establish an infection and that each member acts independently (17). After ST transposon mutagenesis, the vast majority of ST strains will have a transposons genes that are not needed for growth in a tissue and thus should be detected in that tissue. However, if the transposon lies within a gene essential for growth in a tissue, the strain should not be detected. Ideally, 48 to 96 ST bacteria can establish an infection, and mutant strains are not complemented in trans by WT strains. We investigated whether these criteria are met by Y. pseudotuberculosis after oral inoculation.
Numbers of distinct colonies seeding the cecum, MLN, and spleen.
To assess the feasibility of using STM to identify attenuated Y. pseudotuberculosis strains after oral infection of mice, the numbers of different ST strains in the cecum, MLN, and spleen were determined. A pool of 48 ST Y. pseudotuberculosis strains was used to infect BALB/c mice orogastrically at different doses (LD50 = 2 × 107), and the CFU and ST strains recovered from each tissue were enumerated 5 days postinfection (Table 1). Day 5 was chosen because CFU counts in the MLN and spleen are generally high, but the mice are not yet moribund. The presence or absence of each ST strain was ascertained by hybridization of the tags from the bacteria recovered from each tissue to a colony blot of the input ST strains (see Materials and Methods). The number of different ST strains detected in each tissue decreased as the bacteria migrated from the cecum to the MLN and then to the spleen (Table 1). However, the spectrum of ST strains found from mouse to mouse appeared random (data not shown), since the same ST strains were not found in the cecum and MLN of all mice tested. Of the 48 input clones 46 were detected in the cecum of at least one mouse infected at the highest dose. One of the two ST strains was also not detected in the blot probed with tags generated from the input DNA; the second strain had an insertion in yscH and failed to secrete the Yops. Occasionally ST strains were absent in the MLN of a mouse that were present in the spleen, indicating that either these strains reached the spleen without colonizing the MLN or their levels in the MLN were too low to be detected by hybridization.
TABLE 1.
Numbers of CFU and ST strains in tissues 5 days after oral infection
| Dose | Mouse | Cecum
|
MLN
|
Spleen
|
|||
|---|---|---|---|---|---|---|---|
| CFU | STa | CFU | ST | CFU | ST | ||
| 8 × 107 | 1 | 1.0 × 103 | NDb | 4.8 × 104 | 5 | 150 | ND |
| 2 | 200 | ND | 6 | ND | 0 | ||
| 3 | 6.5 × 104 | ND | 1.6 × 103 | 24 | 0 | ||
| 8 × 108 | 1 | 4.2 × 104 | 27 | 4.0 × 103 | 28 | 0 | |
| 2 | 8.9 × 104 | 35 | 1.0 × 104 | 26 | 6.8 × 103 | ND | |
| 3 | 3.0 × 104 | 35 | 1.2 × 104 | 19 | 0 | ||
| 3 × 109 | 1 | 1.3 × 105 | 30 | 9.4 × 103 | 13 | 2.9 × 103 | ND |
| 2 | 2.0 × 106 | 32 | 4.0 × 104 | 25 | 100 | ND | |
| 3 | 5.0 × 104 | 36 | 2.5 × 104 | 22 | 2.5 × 104 | 1 | |
| 8 × 109 | 1 | 6.0 × 105 | 42 | 1.7 × 104 | 31 | 500 | ND |
| 2 | 3.0 × 105 | 36 | 6.0 × 104 | 28 | 3.6 × 103 | 6 | |
| 3 | 6.0 × 105 | 39 | 2.8 × 104 | 27 | 5.5 × 103 | ND | |
Number of different ST strains in each tissue, calculated by probing colony blots as described in Materials and Methods.
ND, not done.
To determine the limit of detection of ST strains, we performed several in vitro reconstruction experiments in which the abundance of one strain in the pool was reduced relative to the rest of the population. The signature tags from these pools were amplified, labeled, and used to probe colony blots. When a strain was present at 0.1% of the total population, its tag was no longer detected by hybridization on colony blots (data not shown). Since our pool contained 48 ST strains, each strain is initially present at approximately 2%; therefore, any strain that decreases 20-fold relative to the whole population will be scored as absent and potentially attenuated.
In summary, there was some selective force at play in the host that reduced the migration of the bacteria from the cecum into the MLN and, especially, into the spleen. However, this selection on the bacteria was independent of where the transposon was inserted into the chromosome since different ST strains reached the MLN and spleen in different mice. We concluded that if pools of 48 ST strains were used to infect three mice, the seeding efficiency would be sufficient to identify mutants unable to survive in the cecum and perhaps the lymph tissues; however, the host constraints on the bacteria reaching the spleen are too severe to identify bona fide attenuated mutants in the spleen.
A Yop-deficient strain is not complemented by WT Yersinia and is defective for growth in the cecum.
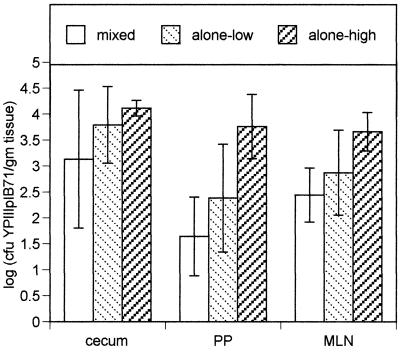
Since the STM technique is performed with a mixture of WT and mutant bacteria, there is the possibility that WT bacteria will complement mutant bacteria and confound our ability to identify mutants. To assess whether the Yops secreted by WT Y. pseudotuberculosis complemented the survival of a strain unable to secrete the Yops, we infected mice with a 3:1 mixture of WT and YPIIIpIB71 bacteria (a strain unable to secrete the Yops) (14). Comparison of the number of bacteria found in mice that were infected with a mixture of WT and YPIIIpIB71 and the number of YPIIIpIB71 bacteria found in mice infected with the mutant strain alone (Fig. 1) demonstrates that WT Y. pseudotuberculosis did not complement YPIIIpIB71.
FIG. 1.
Comparison of number of YPIIIpIB71 CFU recovered from the cecum, PP, and MLN of mice infected with a 3:1 ratio of WT Y. pseudotuberculosis to the Yop-deficient strain YPIIIpIB71 (mixed) with the number of CFU from mice infected with only YPIIIpIB71 at doses of 3 × 108 (alone-low) and 3 × 109 (alone-high). In the mixed infection experiment, a total of 3 × 109 bacteria were used, of which 7.5 × 108 were YPIIIpIB71. Four mice were infected with the mixed inoculum, and five mice were infected with the low and high doses of YPIIIpIB71. Mice were sacrificed 5 days postinfection.
In the mixed infection, WT Y. pseudotuberculosis grew significantly better than the mutant in all tissues (Table 2); YPIIIpIB71 was not detected in the spleen (limit of detection = 10 CFU). In the lymph tissues, however, the fold enrichment of WT to YPIIIpIB71 was occasionally less than 20 (Table 2). In fact, we occasionally detected a Yop-deficient strain in lymph tissues. These data suggested that the cecum may be the best selective environment to identify attenuated mutants after orogastric infection and furthermore that the type III secretion system and at least one effector molecule is important for survival in the cecum.
TABLE 2.
A strain unable to secrete the Yops (YPIIIpIB71) is outcompeted by WT Y. pseudotuberculosis after orogastric inoculation
| Tissue | Mean log differencea | Pb | Competitive index/mousec |
|---|---|---|---|
| Cecum | −2.86 ± 0.74 | 0.0012 | 0.001, 0.007, 0.003, 0.005 |
| PP | <−2.55 ± 1.11 | 0.0068 | 0.0002, <0.006, 0.026, 0.0002 |
| MLN | −1.74 ± 0.59 | 0.0096 | 0.12, 0.017, 0.013, 0.004 |
| Spleen | <−2.89 ± 0.50 | 0.0014 | <0.002, <0.001, <0.01, <0.007 |
Average of log(CFU of YPIIIpIB71) − log(CFU of WT) recovered from the indicated tissue (n = 4).
From a two-tailed, paired Student's t test.
(CFU of YPIIIpIB71/CFU of WT)output/(CFU of YPIIIpIB71/CFU of WT)input. < indicates no YPIIIpIB71 CFU recovered from that tissue. Limit of detection, 10 CFU/tissue.
Identification of attenuated Y. pseudotuberculosis strains.
We generated a library containing 960 ST Y. pseudotuberculosis strains and screened the strains for reduced virulence in oral infections in BALB/c mice (see Materials and Methods). One hundred and four ST strains were not detected in the cecum and/or the MLN and PP. To further test these strains, the 104 strains were divided into five pools of 16 and two pools of 12, and each pool was mixed with an equal amount of WT Y. pseudotuberculosis and used to reinfect two mice. Nineteen ST strains were not detected in the cecum and/or the PP and MLN after the second round of screening.
The 19 ST strains were tested individually for virulence by infecting mice orogastrically with 2 × 109 bacteria. Table 3 shows the number of mice infected, the number that died during the course of infection, and the day of death after infection. Eight ST strains (425, 426, 427, 429, 431, 497, 498, and 542) were avirulent. Three others (496, 499, and 500) appeared attenuated; one or two mice died after all mice infected with WT had died. Forty days after infection, the mice infected with ST strains 496, 499, and 500 were tested to determine if they were still shedding bacteria. Yersinia was detected in the feces from the two remaining mice infected with the ST strain 500, while no bacteria were detected in the feces of the remaining mice infected with ST strains 496 and 499. Five strains (428, 430, 495, 544, and 555) appeared as virulent as WT Yersinia. Given the high dose used to infect mice, these strains may, in fact, be slightly attenuated; however, they were not further characterized in oral infections or sequenced, and in i.p. infections they appeared as virulent as WT (see below and Table 3). The remaining three strains (493, 494, and 543) killed some but not all mice within the same time frame as WT Y. pseudotuberculosis.
TABLE 3.
Phenotypes of putative ST mutant strains after orogastric and i.p. infection of BALB/c mice
| Strain/genotyped (phenotype) | Orogastric infection with 2 × 109 bacteriaa
|
Mean log difference (log CFU of ST strain − log CFU of WT) after coinfection with equal mixture of WT and mutant Y. pseudotuberculosis (Pe)
|
||||
|---|---|---|---|---|---|---|
| Orogastricb(n = 5)
|
i.p.c(n = 5)
|
|||||
| No. dead/no. infected | Days until death | Cecum | PP | MLN | Spleen | |
| YPIIIpYV (wild type) | 6/6 | 4, 4, 5, 6, 7, 11 | ||||
| 425/yscH (type III secretion) | 0/3 | −2.20 ± 0.27 (<0.0009) | ||||
| 426/yscU (type III secretion) | 0/3 | |||||
| 431/yscB (type III secretion) | 0/3 | |||||
| 542/yscL (type III secretion) | 0/3 | |||||
| 429/lcrR (regulator of type III secretion) | 0/3 | −2.01 ± 1.62 (0.05) | −2.071 ± 1.67 (0.05) | −1.40 ± 0.64 (0.008) | −0.75 ± 0.18 (<0.0009) | |
| 498/lcrR (regulator of type III secretion) | 0/3 | −2.68 ± 0.59 (<0.0009) | ||||
| 427/ddhC (O-Ag biosynthesis) | 0/3 | −1.51 ± 0.12 (<0.0009) | ||||
| 496/wzx (O-Ag biosynthesis) | 2/3 | 12, 16 | −2.71 ± 1.07 (0.004) | −2.42 ± 0.03 (0.032) | −2.67 ± 0.64 <0.0009 | −2.15 ± 0.83 (0.0044) |
| 543/gmd (O-Ag biosynthesis) | 4/6 | 7, 9, 21, 24 | −2.97 ± 1.5 (0.027) | −2.02 ± 0.25 (0.005) | −2.14 ± 0.63 (0.027) | −1.94 ± 0.50 (0.0020) |
| 494/inv (invasin) | 2/3 | 7, 7 | −1.63 ± 0.26 (0.001) | −1.41 ± 1.36 (0.080) | −2.15 ± 0.7 (0.003) | 0.03 ± 0.34 (0.855) |
| 497/sufl (suppressor of ftsI) | 0/3 | −1.70 ± 0.78 (0.008) | ||||
| 499/cls (cardiolipin synthesis) | 2/3 | 12, 15 | −3.42 ± 1.57 (0.004) | −3.31 ± 0.67 (0.032) | −2.16 ± 0.72 (0.001) | −1.21 ± 0.29 (0.0007) |
| 500/ksgA (kasugamycin resistant) | 1/3 | 24 | −0.59 ± 0.39 (0.027) | −0.76 ± 0.68 (0.065) | −2.14 ± 0.63 (0.027) | −0.74 ± 0.36 (0.011) |
| 428 | 3/3 | 6, 6, 10 | ||||
| 430 | 3/3 | 5, 9, 10 | ||||
| 493 | 4/6 | 8, 8, 10, 12 | −1.00 ± 1.24 (0.204) | −0.41 ± 1.01 (0.47) | −0.37 ± 1.50 (0.65) | |
| 495 | 3/3 | 6, 7, 8 | 0.40 ± 1.14 (0.47) | −0.08 ± 0.9 (0.85) | 0.33 ± 1.3 (0.71) | 0.07 ± 0.49 (0.76) |
| 544 | 3/3 | 7, 8, 9 | ||||
| 545 | 3/3 | 6, 8, 8 | ||||
Eight-week-old mice were infected with 2 × 109 bacteria and observed until death or for 40 days.
Mice were infected with an equal mixture of 2 × 109 WT Y. pseudotuberculosis and the indicated ST strain. After 5 days, the numbers of WT and ST bacteria in the cecum, PP, and MLN were determined.
Mice were infected i.p. with an equal mixture of 106 WT Y. pseudotuberculosis and the ST mutant. After 3 days, the numbers of WT Y. pseudotuberculosis and ST bacteria in the spleens was determined.
The insertions within the yscH, yscB, lcrV, ddhC, wzx, gmd, cls, and ksgA genes may be polar on downstream open reading frames.
From a two-tailed, paired Student's t test.
To further assess the virulence of the strains that caused death in some mice (493, 494, 496, 499, 500, and 543), coinfection experiments with WT Y. pseudotuberculosis were performed. Table 3 shows the ratio of WT to ST strain recovered from the cecum, PP, and MLN 5 days postinfection. Strains 496, 499, 500, and 543 could not effectively compete with WT bacteria in all tissues tested, whereas 493 was clearly as effective as WT in colonizing all tissues. Strain 494 grew as well as WT in the PP but could not effectively compete with WT bacteria in the cecum and MLN. In another set of orogastric coinfection experiments (n = 3), 494 did not effectively compete WT in any host tissue (data not shown). In summary, of 960 ST strains tested, 13 were attenuated after oral challenge of BALB/c mice.
Infectivity of the ST mutants when delivered intraperitoneally.
Although the attenuated ST strains were all impaired for survival in the cecum, it remained possible that they retained the ability to replicate and cause disease if they bypassed the intestinal epithelial barrier. To test this hypothesis, each mutant was mixed with an equal amount of WT Y. pseudotuberculosis and then injected i.p. into five BALB/c mice. Three days postinfection, we assessed the ratio of WT to mutant bacteria recovered from the spleens (Table 3). Strain 494 was able to colonize the spleen as effectively as WT. The other attenuated ST strains were defective in surviving and/or replicating in the spleen, while the ST strains, 428, 430, and 495, which did not appear attenuated in oral infections, colonized the spleen as efficiently as WT Y. pseudotuberculosis.
Sequence analysis of the virulence attenuated strains.
To determine the loci disrupted by the ST transposon, the transposon and adjacent DNA were cloned into either pUC19 or pGEM-7Zf, and the adjacent DNA was sequenced. Sequence analysis indicated that six of the ST strains had transposons within genes in operons encoding components of the type III secretion system (Table 3). These mutants fell into two classes, those that were incapable of secreting the Yops (425, 426, 431, and 542) and those that were capable of secreting at least some of the Yops (429 and 498) (11). Three transposons disrupted genes encoding enzymes involved in biosynthesis of O antigen (427, 496, and 543). These strains showed a rough colony morphology on L plates, and broth-grown cultures exhibited marked cell clumping. The remaining four strains had transposons within the invasin gene, inv (494); sufl, a gene which when overexpressed suppresses a ftsl mutation (497); cardiolipin synthase (cls), which makes one of the three primary phospholipids in the bacterial envelope (499); and ksgA, a ribosomal protein which is targeted by the drug kasugamycin (500). Further analysis of the regions surrounding these genes indicated that the transposon insertions in many of these loci (yscB, yscH, IcrV, cls, and ksgA) are likely polar on downstream genes (Table 3).
Phenotypes of the attenuated strains in vitro and in cell culture assays.
Several in vitro and cell culture phenotypes of Y. pseudotuberculosis have been associated with virulence. These phenotypes include the ability of Yersinia to secrete the Yops into broth when the bacteria is grown in low-Ca2+ conditions at 37°C, to invade epithelial cells efficiently when grown at 26°C, and to deliver Yops to mammalian cells when grown at 37°C (11). SDS-polyacrylamide gel electrophoresis indicated that the Yops were secreted by all the attenuated ST strains we isolated except for strains 425, 426, 431 and 542, which had insertions in operons encoding structural components of the type III secretion system (data not shown). Strain 498 (lcrV) secreted all of the Yops except LcrV, YopB, and YopD. Notably, 429 (lcrR) behaved like WT Y. pseudotuberculosis; it secreted Yops under low-Ca2+ conditions but not under high-Ca2+ conditions.
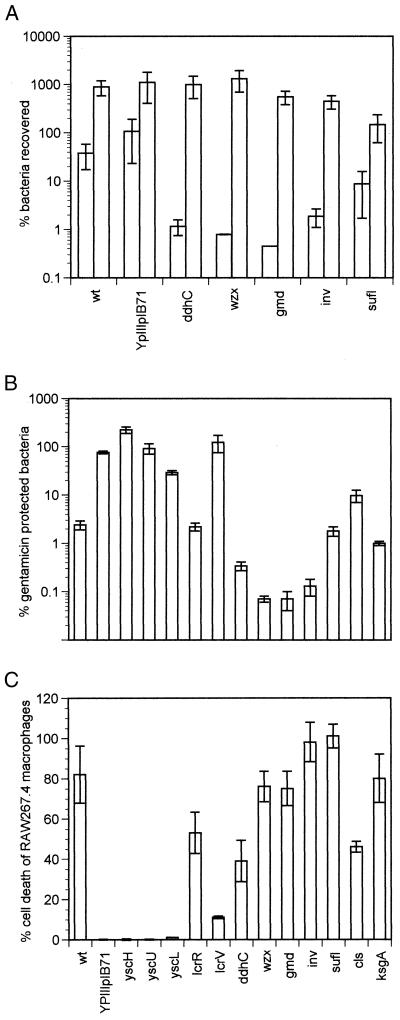
The attenuated ST mutants were tested for the ability to invade epithelial cells when grown at 26°C using a gentamicin protection assay (Fig. 2A and data not shown). The gentamicin protection assay distinguishes intracellular bacteria from extracellular bacteria because intracellular bacteria are protected from the bactericidal effects of the drug. As expected, the invasin mutant did not invade HeLa cells, and this defect was complemented with a plasmid expressing invasin, pRI203 (20) (data not shown). Surprisingly, strains containing mutations in the O-antigen biosynthetic pathway appeared as defective as the invasin-defective strain in invading epithelial cells. Control experiments indicated that when gentamicin was not added to the cultures, O-antigen mutants survived and replicated in the assay conditions as well as WT Y. pseudotuberculosis (Fig. 2A) and that the O-antigen mutants were not hypersensitive to lethal effects of gentamicin (data not shown). One strain, the ksgA mutant strain, displayed a slight growth defect in the control experiments; it did not double during the course of the assay, whereas the other strains all increased two- to fourfold (Fig. 2A). The remaining attenuated ST strains invaded HeLa cells as efficiently and replicated as much as WT Y. pseudotuberculosis (data not shown).
FIG. 2.
Phenotypes of ST strains with cells in culture. (A) Percentage of ST strains recovered after bacteria were grown at 26°C in 2xYT containing 5 mM CaCl2 and exposed to HeLa cells at a multiplicity of infection of 10:1. After a 30-min infection, gentamicin was added to the medium (first box in each pair) for an additional 90 min, the cells were washed and lysed with 1% Triton X-100, and the bacteria were plated. In a side-by-side control, gentamicin was not added to the medium cells were lysed with a final concentration of 1% Triton X-100 and bacteria were plated (second box in each pair). All experiments were done in triplicate, and each strain was tested at least two times in both assays. (B) Percentage of gentamicin-resistant bacteria of various ST strains after infection of HeLa cells with bacteria pregrown at 37°C for 2 h. HeLa cells were infected as described above and in Materials and Methods. (C) Percentage of RAW264.7 macrophage cell death due to infection with Y. pseudotuberculosis relative to complete lysis of the macrophages. The bacteria were pregrown at 37°C for 2 h, the cells were infected for 2 h, gentamicin was added to the medium, and culture supernatants were assayed 7 h after the addition of gentamicin. All experiments were performed in triplicate and repeated twice. The strain labeled lcrR is YPIIIpIB71, which does not secrete any Yops.
To determine if the Yops can be effectively delivered to the cytoplasm of mammalian cells by the ST mutants, their ability to deliver YopE and YopJ to epithelial cells and macrophages, respectively, was assessed. Delivery of the Yops requires that Y. pseudotuberculosis be grown at 37°C to induce synthesis of the type III secretion system and the Yops and that the bacteria adhere tightly to cells via invasin or YadA (11). When YopE is delivered to epithelial cells, the cells can no longer internalize Y. pseudotuberculosis, and thus the bacteria are susceptible to gentamicin (6, 25, 37). When YopJ is delivered to macrophages, the macrophages undergo programmed cell death (29, 31). Gentamicin protection assays indicated that the HeLa cells rapidly internalized all of the mutants with insertions in type III secretion system except for the lcrR mutant (Fig. 2B). Likewise, the same strains were unable to induce macrophage cell death (Fig. 2C). The lcrR mutant delivered the Yops as efficiently as WT in both assays, consistent with its ability to secrete the Yops into the media. The mutants in O-antigen biosynthesis (427, 496, and 543) and invasin (494) induced cell death in macrophages, suggesting that at least one adherence factor, possibly YadA, was capable of binding to macrophages to deliver YopJ. These mutants displayed an increase in sensitivity to gentamicin compared to WT Y. pseudotuberculosis, which was likely due to a combination of their inability to invade epithelial cells using invasin and to YopE function. The epithelial cells rounded up as occurs when YopE is delivered to epithelial cells (data not shown). The strains with mutations in the cls, sufI, and ksgA genes did not have any notable defect in delivering Yops to mammalian cells.
Additional phenotypes tested included growth at 26°C and motility. All strains tested grew as well as WT in 2xYT broth at 26°C except for the ksgA mutant, which had slightly greater doubling time. All strains were motile at 26°C but not as 37°C (data not shown), as judged by diffusion of bacteria from the inoculation site after 18 h of incubation in soft agar.
DISCUSSION
In this study, we used a library of Y. pseudotuberculosis ST strains to characterize an oral infection of BALB/c mice and identify attenuated mutants at day 5 postinfection. Since 50 to 80% of the input pool was present in the cecum of individual mice, we decided to identify mutants unable to survive in the cecum. We initially anticipated that we might also identify mutants attenuated for survival in the PP and MLN. In practice, however, all of the ST strains that were absent in the MLN and PP but present in the cecum in the preliminary screening were found in all tissues in the secondary screening or proved as virulent as WT when used to infect mice. Thus, only mutants unable to thrive in the cecum were identified in our STM screen.
We found that 12 of 13 mutants had defects that also rendered the bacteria unable to colonize the spleen as efficiently as WT Y. pseudotuberculosis after i.p. infection. Hence, in general, a failure to thrive in the small intestine appears due to defects that leave the bacteria incapable of surviving in other host environments. This does not necessarily imply that the environment in the small intestine or cecum is the same as that in the spleen; more likely, bacteria can more easily survive the small intestine than the spleen, and therefore mutants that cannot thrive in the small intestine are also impaired in other host environments. Notably, we did not identify any auxotrophs in our screen even though we did not intentionally remove them from our pools, suggesting that the contents of the small intestine are rich enough in nutrients so that most auxotrophs can survive.
One gene, inv, was only necessary for survival in the small intestine or cecum but was not required after i.p. infection. While invasin mutants in Y. enterocolitica have an LD50 similar to that of WT Yersinia, they are delayed in colonizing the PP (33). Likewise, invasin mutants in Y. pseudotuberculosis remain in the mucus layer of the gut in ligated-loop experiments rather than homing to the M cells of the PP (24). Presumably by day 5 postinfection, the invasin-defective bacteria have been expelled from or killed within the gut. The identification of inv in this selection demonstrates another subtlety of STM. Depending on the inoculation site, genes that enhance the survival of an organism in a particular environment but which are not essential for disease can be identified. This type of observation can point to the possibility that there are alternative modes of causing disease in the host.
A number of mutants in the type III secretion system were identified, which confirmed our results with the YPIIIpIB71 strain that type III secretion is important for survival in the cecum. Previous work in Y. enterocolitica demonstrated that factors encoded on pYV are important for binding to the intestinal mucus and brush border in rabbits (23, 32). While YadA is one pYV-encoded factor that is important for binding to mucus (24, 32), another factor on the pYV might aid in binding to the brush border membranes or increase the survival of Yersinia in the lumen. Isolation of an insertion in lcrV, which knocks out secretion only of LcrV, YopB, and YopD and eliminates the transfer of a number of Yops into cells, suggests that LcrV, YopB, and/or YopD are required for survival in the small intestine and/or that transfer of another effector molecule into host cells is crucial for survival in the cecum.
Only one mutation in the virulence plasmid, lcrR, did not have a measurable effect on Yop secretion or function in cell culture assays, yet this mutation did alter the course of infection in the mouse. While our result show that the lcrR mutant strain is attenuated both after oral and i.p. infection in BALB/c mice, the degree of attenuation after i.p. infection was not as severe as for lcrV or ysc mutants, suggesting that in vivo the Yops are at least partially effective. lcrR has been studied in both Y. pestis and Y. pseudotuberculosis, and investigators have drawn different conclusions about its role in virulence (4, 7). In Y. pestis, insertions in lcrR increase levels of the proteins encoded by the downstream operon, lcrGVH-yopBD, increase the temperature sensitivity of the bacteria when Y. pestis is grown in defined media, and reduce the virulence of Y. pestis in mice (4). In contrast, the Y. pseudotuberculosis lcrR mutant had no virulence phenotype after oral infection of Swiss albino mice (7). We have not examined whether our strain is temperature sensitive in defined media, although we noted that it behaved like WT Yersinia in 2xYT regardless of Ca2+ concentration. The differences between our lcrR allele and the 019-4 allele (7) in Y. pseudotuberculosis could be due to differences in animal model systems, location of the insertion (our insertion is 360 bases downstream of the ATG, whereas the 019-4 insertion is 390 downstream of the ATG), or expression of the downstream operon due to the location of or sequences in the transposon. Studies with small inframe deletions or point mutations in the lcrR gene would conclusively resolve these discrepancies about the role of LcrR in type III secretion and infection.
Three insertions that we identified were in genes involved in the O-antigen biosynthetic pathway. O-antigen biosynthesis has been shown to be crucial for enteropathogenic Yersinia virulence after both oral and i.p. inoculation (1, 38). Surprisingly, the O-antigen mutants behaved similarly to the invasin mutant in cell culture assays in that they appeared unable to invade epithelial cells efficiently; however, a defect in invasion is not the only deficiency in the O-antigen strains because the inv mutant is fully virulent after i.p. infection, whereas the O-antigen mutants are severely attenuated.
Finally, three insertions were in genes, ksgA, sufI, and cls, which have not previously been associated with virulence. None of these mutants were defective in Yop synthesis or delivery. The insertion in the ksgA locus occurs at the very 3′ end of the gene, so the phenotype observed in vivo may be due to a defect in ksgA or in the downstream genes, apaGH, which are involved in cobalt resistance and Mg2+ transport (5). We observed a growth defect in the ksgA mutant strain under some conditions; growth defects have also been observed in E. coli ksgA strains (42). The virulence defect may be due to the reduced growth rate, which could enable the immune system of the mouse to keep the strain in check. Our observation that mice were still shedding the strain 40 days postinfection suggests that the strain is capable of surviving and most likely replicating in the cecum for long periods of time despite its reduced growth rate. sufI was originally identified as a multicopy suppressor of a temperature-sensitive ftsI (PBP-3) allele in E. coli (21); however, its role in replication and peptidoglycan biosynthesis has not been studied. sufI is the third gene of a tricistronic operon containing parC or plsC, which are essential proteins in E. coli. Thus, the virulence defect is likely due to the lack of sufI and may be a result of faulty peptidoglycan structure, unless the insertion destabilizes the RNA. cls is the first gene in a bicistronic operon, and so in-frame deletions are needed to verify that the virulence defect is due to cls.
In all, 1.3% of the ST strains that we tested were attenuated. This is at the low end of the reported 1 to 4% range found in other STM studies (for examples, see references 12, 13, 17, 34, and 39). Our low yield may be a result of a more permissive selective environment in the cecum or greater stringency of the screen. We scored each potential mutant in five mice and excluded candidates that appeared fully lethal at a dose 100-fold above the LD50. Nonetheless, we were successful in identifying several novel genes required for survival in the gastrointestinal tract.
With doses 40 to 400-fold greater than the LD50, only 27 to 42 of the input 48 ST strains were detected in the cecum of an individual mouse. Even fewer ST strains reached the MLN and spleen. This finding suggests that host barriers—anatomic, biochemical and/or cellular—exist in the bowel or the PP that prevent most of the invading bacteria from colonizing the MLN and spleen. Furthermore, from one spleen we isolated 25,000 bacteria that all appeared to be progeny of one ST strain. This suggests that passage through the gut is not the only limiting barrier to cause systemic disease, but in fact, additional barriers exist in other tissues. Moreover, two ST strains were detected in the spleen but not in the MLN of that mouse, suggesting either that they had passed through the MLN but not established an efficient infectious focus or that they had bypassed the MLN altogether and reached the spleen by an alternative route. Alternative routes for reaching the spleen and liver that do not involve colonizing the PP have been postulated for both Y. pseudotuberculosis (24) and Salmonella enterica serovar Typhimurium (41).
The hypothesis of independent action of pathogens during infection predicts that each bacterium acts independently and each has a certain probability of causing a lethal infection regardless of the inoculating dose (26, 27). When testing this hypothesis using mixtures of several Salmonella strains, Meynell and Stocker found all strains in the postmortem blood in roughly equal proportion to their input ratios at doses above the LD50 after i.p. inoculation (27). In contrast, after orogastric inoculation of doses 250-fold above the LD50, often only one strain was isolated. The two strains used in the oral infections did not have the same growth rate in vitro or in vivo; however, the slower-growing Salmonella strain was sometimes the only strain detected in the postmortem blood of some mice. Hence, the authors predicted that fewer than 250 bacteria breach the barrier of the gut. Our findings that around 40 Y. pseudotuberculosis strains remained in the gut 5 days postinfection and fewer are in the MLN and spleen are consistent with the number predicted for Salmonella.
The observation that restricted numbers of bacteria seed a variety of tissues during infection may extend to other experimental systems designed to detect virulence-specific genes. A variety of screens (IVET and differential fluorescence induction) which allow investigators to assess bacterial gene expression in complex biological environments like animals or in a mixed microbial community in biofilms have been devised (22, 40). Our data indicate that at least with Y. pseudotuberculosis, around 40 bacteria seed the cecum at doses 40 to 400-fold above the LD50. Thus, in some screens, which occur in complex environments, inoculation with large pools of strains need not necessarily guarantee that all of these strains reach and replicate in the biological site of importance during the assay. The use of ST strains may be a useful experimental tool for establishing parameters of seeding and replication efficiency in different biological systems and estimating the number of strains screened in various complex biological systems.
ACKNOWLEDGMENTS
We thank David Holden for the gift of pUTminiTn5Kn2+tags, Barbel Raupach and Denise Monack for help with mice experiments, and Denise Monack and Penelope Barnes for critical reading of the manuscript.
This work was supported by grants from the American Cancer Society (PF-4477) to J.M. and the Defense Advance Research Projects Agency to S.F.
REFERENCES
- 1.al-Hendy A, Toivanen P, Skurnik M. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect Immun. 1992;60:870–875. doi: 10.1128/iai.60.3.870-875.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaoui A, Schulte R, Cornelis G R. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 3.Allaoui A, Woestyn S, Sluiters C, Cornelis G R. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barve S S, Straley S C. lcrR, a low-Ca2+-response locus with dual Ca2+-dependent functions in Yersinia pestis. J Bacteriol. 1990;172:4661–4671. doi: 10.1128/jb.172.8.4661-4671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchin-Roland S, Blanquet S, Schmitter J M, Fayat G. The gene for Escherichia coli diadenosine tetraphosphatase is located immediately clockwise to folA and forms an operon with ksgA. Mol Gen Genet. 1986;205:515–522. doi: 10.1007/BF00338091. [DOI] [PubMed] [Google Scholar]
- 6.Bliska J B, Copass M C, Falkow S. The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into HEp-2 cells. Infect Immun. 1993;61:3914–3921. doi: 10.1128/iai.61.9.3914-3921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolin I, Portnoy D A, Wolf-Watz H. Expression of the temperature-inducible outer membrane proteins of yersiniae. Infect Immun. 1985;48:234–240. doi: 10.1128/iai.48.1.234-240.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottone E J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997;10:257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter P B. Pathogenecity of Yersinia enterocolitica for mice. Infect Immun. 1975;11:164–170. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark M A, Hirst B H, Jepson M A. M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells. Infect Immun. 1998;66:1237–1243. doi: 10.1128/iai.66.3.1237-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin A J, Miller V L. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol Microbiol. 1999;32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 13.Edelstein P H, Edelstein M A, Higa F, Falkow S. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc Natl Acad Sci USA. 1999;96:8190–8195. doi: 10.1073/pnas.96.14.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsberg A, Wolf-Watz H. The virulence protein Yop5 of Yersinia pseudotuberculosis is regulated at transcriptional level by plasmid-plB1-encoded trans-acting elements controlled by temperature and calcium. Mol Microbiol. 1988;2:121–133. doi: 10.1111/j.1365-2958.1988.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 15.Fujimura Y, Kihara T, Mine H. Membranous cells as portal of Yersinia pseudotuberculosis entry into rabbit ileum. J Clin Electron Microsc. 1992;25:35–45. [Google Scholar]
- 16.Grutzkau A, Hanski C, Hahn H, Riecken E O. Involvement of M cells in the bacterial invasion of Peyer's patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut. 1990;31:1011–1015. doi: 10.1136/gut.31.9.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 18.Holmstrom A, Rosqvist R, Wolf-Watz H, Forsberg A. Virulence plasmid-encoded YopK is essential for Yersinia pseudotuberculosis to cause systemic infection in mice. Infect Immun. 1995;63:2269–2276. doi: 10.1128/iai.63.6.2269-2276.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isberg R R, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 20.Isberg R R, Voorhis D L, Falkow S. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell. 1987;50:769–778. doi: 10.1016/0092-8674(87)90335-7. [DOI] [PubMed] [Google Scholar]
- 21.Kato J, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahan M J, Slauch J M, Hanna P C, Camilli A, Tobias J W, Waldor M K, Mekalanos J J. Selection for bacterial genes that are specifically induced in host tissues: the hunt for virulence factors. Infect Agents Dis. 1993;2:263–268. [PubMed] [Google Scholar]
- 23.Mantle M, Basaraba L, Peacock S C, Gall D G. Binding of Yersinia enterocolitica to rabbit intestinal brush border membranes, mucus, and mucin. Infect Immun. 1989;57:3292–3299. doi: 10.1128/iai.57.11.3292-3299.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marra A, Isberg R R. Invasin-dependent and invasin-independent pathways for translocation of Yersinia pseudotuberculosis across the Peyer's patch intestinal epithelium. Infect Immun. 1997;65:3412–3421. doi: 10.1128/iai.65.8.3412-3421.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mecsas J, Raupach B, Falkow S. The Yersinia Yops inhibit invasion of Listeria, Shigella and Edwardsiella but not Salmonella into epithelial cells. Mol Microbiol. 1998;28:1269–1281. doi: 10.1046/j.1365-2958.1998.00891.x. [DOI] [PubMed] [Google Scholar]
- 26.Meynell G G. The applicability of the hypothesis of independent action to fatal infections in mice given Salmonella typhimurium by mouth. J Gen Microbiol. 1957;16:396–404. doi: 10.1099/00221287-16-2-396. [DOI] [PubMed] [Google Scholar]
- 27.Meynell G G, Stocker B A D. Some hypotheses on the aetiology of fatal infections in partially resistant hosts and their application to mice challenged with Salmonella paratyphi-B or Salmonella typhimurium by intraperitoneal injection. J Gen Microbiol. 1957;16:38–58. doi: 10.1099/00221287-16-1-38. [DOI] [PubMed] [Google Scholar]
- 28.Miller V L, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills S D, Boland A, Sory M P, van der Smissen P, Kerbourch C, Finlay B B, Cornelis G R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monack D M, Mecsas J, Bouley D, Falkow S. Yersinia-induced apoptosis in vivo aids in the establishment of a systemic infection of mice. J Exp Med. 1998;188:2127–2137. doi: 10.1084/jem.188.11.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monack D M, Mecsas J, Ghori N, Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paerregaard A, Espersen F, Jensen O M, Skurnik M. Interactions between Yersinia enterocolitica and rabbit ileal mucus: growth, adhesion, penetration, and subsequent changes in surface hydrophobicity and ability to adhere to ileal brush border membrane vesicles. Infect Immun. 1991;59:253–260. doi: 10.1128/iai.59.1.253-260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pepe J C, Wachtel M R, Wagar E, Miller V L. Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect Immun. 1995;63:4837–4848. doi: 10.1128/iai.63.12.4837-4848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Revell P A, Miller V L. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol Microbiol. 2000;35:677–685. doi: 10.1046/j.1365-2958.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 36.Riley G, Toma S. Detection of pathogenic Yersinia enterocolitica by using Congo red-magnesium oxalate agar medium. J Clin Microbiol. 1989;27:213–214. doi: 10.1128/jcm.27.1.213-214.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosqvist R, Forsberg A, Rimpilainen M, Bergman T, Wolf-Watz H. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol. 1990;4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 38.Skurnik M, Venho R, Bengoechea J A, Moriyon I. The lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3 is required for virulence and plays a role in outer membrane integrity. Mol Microbiol. 1999;31:1443–1462. doi: 10.1046/j.1365-2958.1999.01285.x. [DOI] [PubMed] [Google Scholar]
- 39.Tsolis R M, Townsend S M, Miao E A, Miller S I, Ficht T A, Adams L G, Baumler A J. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun. 1999;67:6385–6393. doi: 10.1128/iai.67.12.6385-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 41.Vazquez-Torres A, Jones-Carson J, Baumler A J, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks W T, Fang F C. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 42.Vila-Sanjurjo A, Squires C L, Dahlberg A E. Isolation of kasugamycin resistant mutants in the 16 S ribosomal RNA of Escherichia coli. J Mol Biol. 1999;293:1–8. doi: 10.1006/jmbi.1999.3160. [DOI] [PubMed] [Google Scholar]
- 43.Young G M, Miller V L. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]