Summary
Reliable, easy-to-handle phenotypic screening platforms are needed for the identification of anti-SARS-CoV-2 compounds. Here, we present caspase 3/7 activity as a readout for monitoring the replication of SARS-CoV-2 isolates from different variants, including a remdesivir-resistant strain, and of other coronaviruses in numerous cell culture models, independently of cytopathogenic effect formation. Compared to other models, the Caco-2 subline Caco-2-F03 displayed superior performance. It possesses a stable SARS-CoV-2 susceptibility phenotype and does not produce false-positive hits due to drug-induced phospholipidosis. A proof-of-concept screen of 1,796 kinase inhibitors identified known and novel antiviral drug candidates including inhibitors of phosphoglycerate dehydrogenase (PHGDH), CDC like kinase 1 (CLK-1), and colony stimulating factor 1 receptor (CSF1R). The activity of the PHGDH inhibitor NCT-503 was further increased in combination with the hexokinase II (HK2) inhibitor 2-deoxy-D-glucose, which is in clinical development for COVID-19. In conclusion, caspase 3/7 activity detection in SARS-CoV-2-infected Caco-2-F03 cells provides a simple phenotypic high-throughput screening platform for SARS-CoV-2 drug candidates that reduces false-positive hits.
Subject areas: Virology, Drugs, Screening in health technology
Graphical abstract
Highlights
-
•
Caco-2-F03 cells are susceptible to SARS-CoV-2 isolates and other coronaviruses
-
•
Coronavirus-infected Caco-2-F03 cells are a screening platform for antiviral drugs
-
•
Caco-2-F03 cells do not produce false-positives due to phosphoplipidosis
-
•
Caspase 3/7 activity is a simple readout assay indicating coronavirus replication
Virology; Drugs; Screening in health technology
Introduction
There is an ongoing search for antiviral drugs against SARS-CoV-2 that can complement the currently available monoclonal antibody preparations and the three approved small-molecule drugs remdesivir, molnupiravir, and nirmatrelvir.1 Effective antiviral drugs and drug combinations will be particularly important for immunocompromized individuals, who cannot effectively be protected by vaccination.2
Previous research has shown that the efficacy of antiviral agents may differ between SARS-CoV-2 variants and cell culture models.3,4,5 Some cell culture models may produce false-positive hits due to unspecific effects on the host cell metabolism such as phospholipidosis that do not translate into in vivo activity.6 Moreover, continued SARS-CoV-2 passaging in cell culture may change virus biology, including virus sensitivity to antiviral drugs.7,8,9 Thus, simple and robust cell culture assays that can cover a broad spectrum of SARS-CoV-2 variants (including primary clinical isolates) are required to accelerate the identification of anti-SARS-CoV-2 drug candidates.
Many assays measure SARS-CoV-2-induced host cell destruction (cytopathogenic effect, CPE) or host cell viability for the identification of antiviral agents.10,11,12,13,14,15,16 However, such assays are not suitable for SARS-CoV-2 culture systems that do not display virus-induced cytotoxicity.17,18,19,20
Antibody-based detection of viral antigens and/or double-stranded RNA is an alternative approach3,21 but requires more manual handling. Assays using genetically modified cells, genetically modified SARS-CoV-2 strains, and SARS-CoV-2 replicons have also been developed15,22,23,24 but cover only the limited number of virus strains that they have been established for.
An ideal assay would enable high-throughput screening of wild-type SARS-CoV-2 including the most current clinical isolates in all available cell culture systems in a very simple format that can be applied by many research groups. Such an assay would also enable the phenotypic resistance testing of virus isolates, which is relevant given that the use of antiviral drugs seems to be inevitably associated with the formation of resistant virus variants.9,25,26
Here, we introduce an effective screening assay for the identification of compounds that inhibit SARS-CoV-2 replication based on measuring caspase 3/7 activity using a one-step readout assay (Caspase-Glo 3/7 Assay System, Promega). This assay works across different coronaviruses including many SARS-CoV-2 strains and clinical isolates as well as across a broad range of cell culture models, including those in which SARS-CoV-2 infection does not result in a recognizable virus CPE. The Caco-2 subline Caco-2-F03 was identified as preferred cell culture model as it is easy-to-handle, displays a stable susceptibility phenotype, and does not produce false positives due to drug-induced phospholipidosis. A validation screen of 1,796 kinase inhibitors confirmed the suitability of our platform and identified 81 compounds that reduced virus-induced caspase activation by more than 90% at 10 μM, including known as well as novel drug candidates such as PHGDH (phosphoglycerate dehydrogenase), CLK-1 (CDC like kinase 1), and CSFR (colony stimulating factor receptor) inhibitors.
Results
Caco-2 cells as SARS-CoV-2 infection model
The human colon carcinoma Caco-2 cell line was established by Jorgen Fogh (Memorial Sloan Kettering Cancer Center, New York) in 197427 and has been used for the cultivation of human pathogenic viruses including influenza viruses and coronaviruses since 1985.28,29,30,31 We already used Caco-2 cells for the cultivation of the close SARS-CoV-2 relative SARS-CoV starting in 2003,32,33 and they also enabled us and others to quickly cultivate SARS-CoV-2 isolates when this novel virus emerged.10,14,34,35,36,37,38,39,40,41
In our hands, Caco-2 cells (obtained from DSMZ, Braunschweig, Germany at the time) have been highly permissive to SARS-CoV and SARS-CoV-2 and developed a pronounced CPE in response to infection with both viruses.32,33,35,39 In other studies, however, Caco-2 cells displayed low SARS-CoV-2 susceptibility and no CPE formation.42,43,44
To further investigate these discrepancies, we ordered fresh Caco-2 cells from the following sources: DSMZ (Braunschweig, Germany, designated as Caco-2A), Sigma (Taufkirchen, Germany, Caco-2B), and CLS (Eppelheim, Germany, Caco-2C). To discriminate our original Caco-2 cell line from these other ones, we will refer to it as Caco-2-F03 from now on.
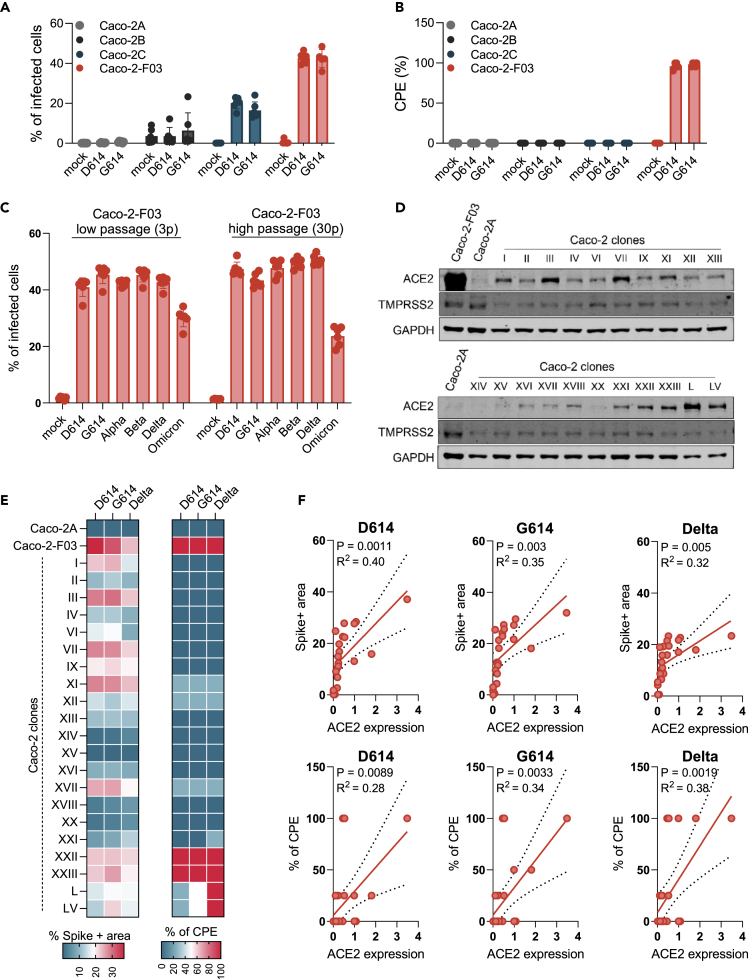
An initial short tandem repeat (STR) analysis confirmed that all Caco-2 cell lines share the reference profile (Table S1). However, Caco-2A, Caco-2B, and Caco-2C cells displayed low SARS-CoV-2 permissiveness as indicated by low viral spike (S) protein levels and a lack of CPE formation compared to Caco-2-F03 (Figure 1A, Figure 1B).
Figure 1.
Susceptibility of Caco-2 cells to SARS-CoV-2 infection
(A) Percentage of SARS-CoV-2-infected cells detected in Caco-2 cell lines from different sources infected with different SARS-CoV-2 isolates at a MOI 0.01 as determined by immunostaining for the viral spike (S protein) 48 h postinfection.
(B) Cytopathogenic effect (CPE) formation in SARS-CoV-2 (MOI 0.01)-infected Caco-2 cell lines from different sources as determined 48 h postinfection.
(C) Susceptibility of Caco-2-F03 cells to a broad range of SARS-CoV-2 isolates after different times of cultivation. Cells had been frozen at passage 14 and were resuscitated and cultivated for a further 30 passages. SARS-CoV-2 susceptibility was determined by immunostaining for S 48h after SARS-CoV-2 (MOI 0.01) infection 3 and 30 passages post-resuscitation.
(D) ACE2 and TMPRSS2 levels in Caco-2-F03, Caco-2A, and single-cell derived clones from Caco-2A.
(E) Susceptibility of Caco-2A clones to selected SARS-CoV-2 isolates as indicated by immunostaining for S and CPE formation in SARS-CoV-2 (MOI 0.01)-infected cells 48 h postinfection.
(F) Correlation of S staining and CPE formation with cellular ACE2 levels. Correlations were determined using the Pearson correlation coefficient.
Results are expressed as the mean ± standard deviation.
Caco-2-F03 cells remained permissive to SARS-CoV-2 for 30 passages after the resuscitation of cells that had been frozen at passage 14 (Figure 1C), suggesting that their SARS-CoV-2 permissiveness phenotype is stable during prolonged culturing. In agreement, we have used Caco-2-F03 cells since 2003 for the cultivation of initially SARS-CoV and later SARS-CoV-2.32,33,35,39
Further investigations revealed that Caco-2-F03 cells display higher levels of the cellular SARS-CoV and SARS-CoV-2 receptor ACE2 (angiotensin converting enzyme 2) and the protease TMPRSS2 (transmembrane serine protease 2), which cleaves and activates S for ACE2 binding,45 than Caco-2A, Caco-2B, and Caco-2C (Figure 1D, Figure S1).
The Caco-2A cell line contains SARS-CoV-2-susceptible subpopulations
One explanation for these differences between Caco-2-F03 and Caco-2A, Caco-2B, and Caco-2C is that we may have inadvertently enriched a SARS-CoV-2-permissive subpopulation during Caco-2 cultivation. To test this hypothesis, we established 21 single cell-derived clones from Caco-2A by limited dilution. Four of these clonal sublines were highly susceptible to SARS-CoV-2 infection as demonstrated by high S protein levels and CPE formation (Figure 1E), supporting the hypothesis that Caco-2-F03 has been derived from a SARS-CoV- and SARS-CoV-2-permissive subpopulation of our original Caco-2 cell line. There was some level of correlation between the SARS-CoV-2 susceptibility of Caco-2A clones and the cellular ACE2 levels (Figure 1F) but not between SARS-CoV-2 susceptibility and the cellular TMPRSS2 levels (Figure S1). This suggests that ACE2 levels are more important for the SARS-CoV-2 susceptibility of Caco-2 cells than TMPRSS2 levels and that additional mechanisms are also likely to be involved.
Caspase 3/7 activity for the quantification of the replication of SARS-CoV-2 and other coronaviruses.
Coronavirus replication, including that of SARS-CoV-2, has been shown to result in the activation of caspases including the initiator caspases 8 and 9 and the effector caspase 3.34,35,46,47,48
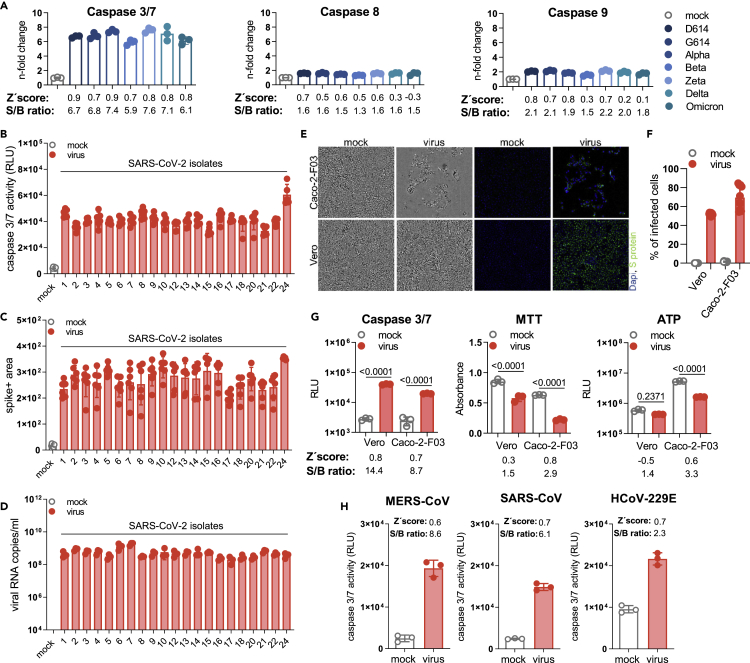
Quantification of caspase activity in Caco-2-F03 cells infected with different SARS-CoV-2 isolates at a MOI of 0.01 48 h postinfection using the Caspase-Glo assay kit (Promega) resulted in substantially higher signal-to-basal (S/B) ratios for caspase-3/7 (5.9- to 7.7-fold) than for caspase-8 (1.3- to 1.6-fold) and caspase-9 (1.5- to 2.2-fold) (Figure 2A). Caspase 3/7 activity also resulted in higher Z′ scores (0.7–0.9) than caspase 8 (0.5–0.7) and caspase 9 (0.3–0.8) activity (Figure 2A), indicating higher assay robustness.49 Hence, caspase 3/7 activity detection was selected for further investigation as potential screening endpoint method.
Figure 2.
Caspase 3/7 activity for the quantification of the replication of SARS-CoV-2 and other coronaviruses
(A) Caspase 3/7, caspase 8, and caspase 9 activity in Caco-2-F03 cells infected with a range of different SARS-CoV-2 isolates (MOI 0.01), as determined by Caspase-Glo assay (Promega) 48 h postinfection. Higher signal-to-basal (S/B) ratios and Z′ scores indicate higher assay robustness.
(B–D) Caspase 3/7 activity as determined by Caspase-Glo assay, C) SARS-CoV-2 Spike (S) protein staining, and D) virus titers as indicated by genomic RNA copy numbers determined by qPCR in Caco-2-F03 cells infected with a wide range of uncharacterized SARS-CoV-2 isolates (MOI 0.01) 48 h postinfection.
(E) Representative images indicating CPE formation in G614 (MOI 0.01)-infected Caco-2-F03 and Vero cells 48 h postinfection as indicated by phase contrast microscopy and immunofluoresce staining for the viral S protein in combination with DAPI-stained nuclei.
(F) Quantification of cellular S protein levels in Caco-2-F03 cells infected with G614 (MOI 0.01) 48 h postinfection by immunostaining.
(G) Only caspase 3/7 activity but not viability assays (MTT, CellTiter-Glo measuring ATP production) reflects G614 (MOI 0.01) replication 48h postinfection in Vero cells, which do not display a virus-induced CPE. G614 (MOI 0.01)-infected Caco-2-F03 cells served as a control that displays a CPE. p values were calculated by one-way ANOVA.
(H) Caspase 3/7 activity in Caco-2-F03 cells infected with MERS-CoV, SARS-CoV, and HCoV-229E (MOI 0.01) as determined 48 h postinfection including S/B ratios and Z′ scores.
Results are expressed as the mean ± standard deviation.
Caspase 3/7 activity displayed an MOI-dependent increase in Caco-2-F03 cells 24 h postinfection (Figure S2A), which mirrored CPE formation (Figure S2B). 48 h postinfection, such differences were not detectable anymore (Figures S2A and S2B). Moreover, infection of Caco-2-F03 cells with an additional 21 clinical SARS-CoV-2 isolates (after a maximum of two passages in Caco-2-F03 cells) also resulted in effective caspase 3/7 activation (Figure 2B), which reflected S protein levels and virus titers (as indicated by genomic RNA copy numbers) (Figures 2C and 2D). UV-inactivated virus did not cause caspase 3/7 activation (Figure S2C), further confirming that SARS-CoV-2-induced caspase 3/7 activation depends on virus replication.
However, caspase 3/7 activation does not appear to be critically involved in SARS-CoV-2 replication as the clinically approved pan-caspase inhibitor emricasan50 did not interfere with SARS-CoV-2 replication and CPE formation, despite suppressing caspase 3/7 activity (Figure S3).
Caspase 3/7 activity for the monitoring of SARS-CoV-2 replication in the presence and absence of virus-induced CPEs
The caspase 3/7 assay also enabled the monitoring of SARS-CoV-2 replication in Vero cells, in which SARS-CoV-2 does not induce a CPE (Figure 2E), although cells are infected (Figure 2F). SARS-CoV-2- and mock-infected Vero cells displayed a smaller difference in viability as indicated by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (measures oxidative phosphorylation in the mitochondria) and CellTiterGlo assay (Promega, measures cellular ATP production) than SARS-CoV-2- and mock-infected Caco-2-F03 cells (Figure 2G). In contrast, caspase 3/7 activation was higher in SARS-CoV-2-infected Vero cells relative to mock-infected than in SARS-CoV-2- and mock-infected Caco-2-F03 cells (Figure 2G). This shows that the ability of the caspase 3/7 assay to monitor SARS-CoV-2 infection is (in contrast to the MTT and CellTiter Glo assays) not affected by a lack of CPE formation.
Caspase 3/7-induction by additional coronaviruses
Moreover, we detected caspase 3/7 activation also in Caco-2-F03 cells infected with the additional human-pathogenic coronaviruses Middle East respiratory syndrome–related coronavirus (MERS-CoV), SARS-CoV, and HCoV-229E (Figure 2H). All three additional coronaviruses induced considerable caspase 3/7 activity (Figure 2H). These findings suggest that Caco-2-F03 cells are susceptible to a broader range of coronaviruses and that caspase 3/7 activity is a potential readout measure for monitoring Caco-2-F03 cells infected by different coronaviruses.
Caspase 3/7 activity for the monitoring of SARS-CoV-2 replication in primary human cell cultures
Furthermore, SARS-CoV-2-induced caspase 3/7 activity was detected in primary cultures of normal human cells, including induced pluripotent stem cell-derived cardiomyocytes (CMS), air liquid interface (ALI) cultures of bronchial epithelial (HBE) cells, and primary human hepatocytes (PHH) (Figure S4A). Immunoblots for the viral nucleoprotein (NP) were used to confirm SARS-CoV-2 infection in these primary cell cultures (Figure S4B). ALI HBE did not display disruption of cellular barrier during SARS-CoV-2 infection as measured by transepithelial electrical resistance (TEER) and LDH (lactate dehydrogynase) release (Figures S4C and S4D), whereas CMS and PHH displayed a CPE in response to SARS-CoV-2 infection (Figures S4E and S4F). This agrees with our previous results that caspase 3/7 activity is a suitable readout method for monitoring SARS-CoV-2 infection both in the presence and absence of a virus-induced CPE.
Taken together, detection of caspase 3/7 activity does not only enable the monitoring of the replication of a wide range of SARS-CoV-2 variants and clinical isolates (and of other coronaviruses) but also is a suitable readout for SARS-CoV-2 replication across many different susceptible permanent cell lines and human primary cultures, independently of whether SARS-CoV-2 induces a CPE in these systems.
Caspase 3/7 activity for the identification of antiviral drugs
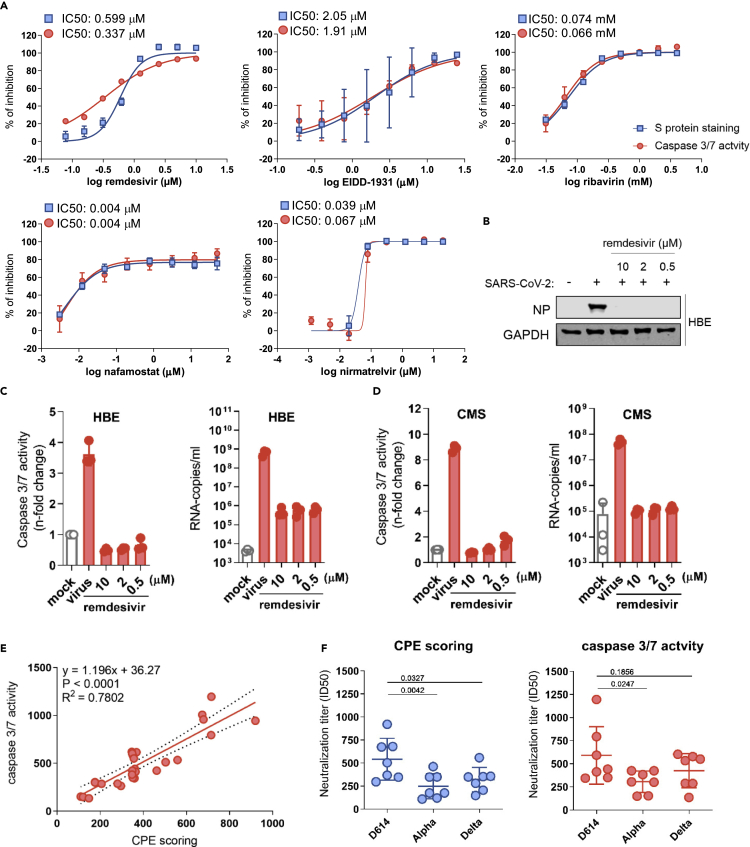
Next, we compared caspase 3/7 activity and S protein staining for the detection of the antiviral activity of drugs with known efficacy against SARS-CoV-2, including remdesivir (RNA-dependent RNA polymerase [RdRp] inhibitor), EIDD-1931 (active form of molnupiravir that induces “error catastrophe” in newly produced SARS-CoV-2 genomes), ribavirin (broad-spectrum antiviral drug), nirmatrelvir (3C-like protease/main protease inhibitor), and nafamostat (TMPRSS2 inhibitor)51,52 in SARS-CoV-2 variant G614-infected Caco-2-F03 cells. Both detection methods resulted in very similar IC50s (concentrations that inhibit virus activity by 50%) (Figure 3A).
Figure 3.
Caspase 3/7 activity for the determination of the antiviral activity of anti-SARS-CoV-2 agents and neutralization assays
(A) Dose-response curves and concentrations that inhibit virus infection by 50% (IC50) of antiviral agents as determined by caspase 3/7 activity and immunostaining for the coronavirus S protein in G614 (MOI 0.01)-infected Caco-2-F03 cells 24 h postinfection.
(B) Effects of the approved anti-SARS-CoV-2 drug remdesivir on cellular levels of the viral NP protein in G614 (MOI 1)-infected air liquid interface (ALI) cultures of primary human bronchial epithelial (HBE) cells 120 h postinfection.
(C) Effects of remdesivir on caspase 3/7 activity and virus titers (genomic RNA copy numbers determined by PCR) in G614 (MOI 1)-infected ALI HBE cultures 120 h postinfection.
(D) Effects of remdesivir on caspase 3/7 activity and virus titers in G614 (MOI 1)-infected primary human cardiomyocytes (CMS) 48 h postinfection.
(E) Correlation of the neutralization capacity of sera derived from seven donors two weeks after their second dose of the mRNA-1273 vaccine determined by caspase 3/7 activity or cytopathogenic effect (CPE) scoring in D614-, Alpha-, and Delta-infected Caco-2-F03 cells 48 h postinfection. Correlations were determined using the Pearson correlation coefficient.
(F) Determination of neutralization titers by caspase 3/7 activity or CPE scoring using sera derived from seven donors two weeks after their second dose of the mRNA-1273 vaccine in Caco-2-F03 cells infected with D614, Alpha, and Delta isolates 72 h postinfection. p values were calculated using paired t-test. P-values were determined by ANOVA.
Results are expressed as the mean ± standard deviation.
Caspase 3/7 activity also enabled the monitoring of the antiviral activity of remdesivir in SARS-CoV-2-infected primary ALI HBE cell cultures that do not display a recognizable CPE. The validity of the results obtained by caspase 3/7 assay was confirmed by determining virus titers and Western Blot analysis of SARS-CoV-2 N protein levels (Figures 3B and 3C). Finally, caspase 3/7 activity reflected the effect of remdesivir on SARS-CoV-2 replication in CMS (Figure 3D).
Taken together, these findings demonstrate that caspase 3/7 activity enables the monitoring of the antiviral drug response in a broad range of cell culture models.
Caspase 3/7 activity for the determination of neutralizing antibody titers
Caspase 3/7 activity also enabled the determination of neutralizing antibody titers in sera derived from seven donors two weeks after their second dose of the mRNA-1273 vaccine, as indicated by a close correlation with results obtained by CPE scoring in Caco-2-F03 cells infected with different SARS-CoV-2 variants (Figures 3E and 3F). The neutralization capacity of the sera was higher against the early SARS-CoV-2 strain FFM3 (G614) than against Alpha (B1.1.7) and Gamma (P.2) variant isolates (Figure 3F), which is in line with the immune evasion properties documented for these variants.53
Caspase 3/7 activity for detection of SARS-CoV-2 resistance
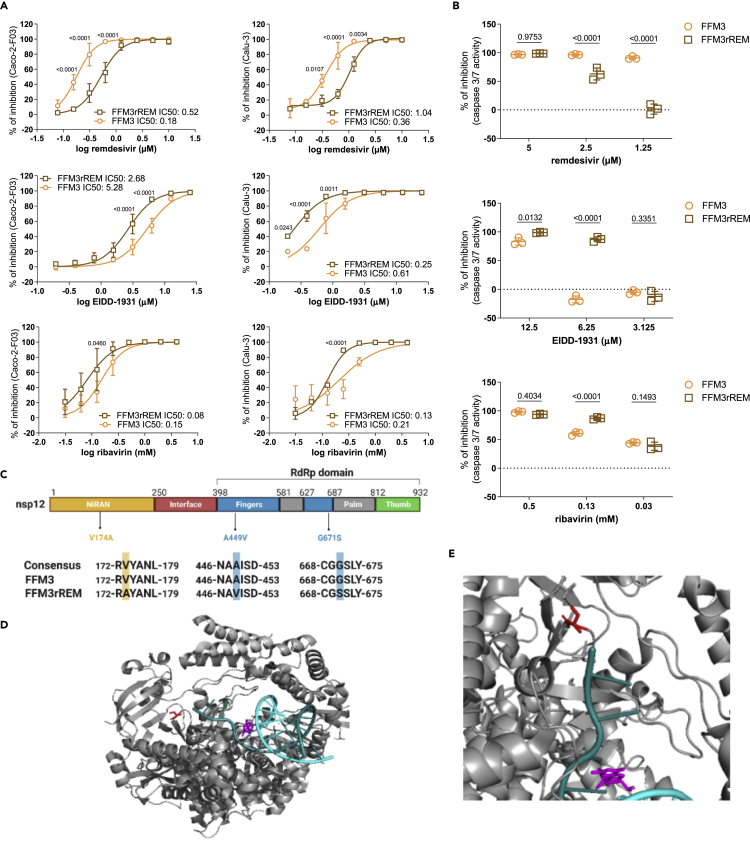
Next, we tested whether the caspase 3/7 activity assay may be used for phenotypic screens identifying resistant virus strains. To establish a drug-resistant strain, SARS-CoV-2 strain FFM3 (G614) was passaged in the presence of increasing remdesivir concentrations starting with 0.5 μM (the IC50 concentration) until it could be cultivated in the presence of remdesivir 2 μM (FFM3rREM). FFM3rREM displayed a significantly reduced sensitivity to remdesivir, as indicated by determination of cellular S levels in Caco-2-F03 and Calu-3 cells (Figure 4A) and by caspase 3/7 activity in Caco-2-F03 cells (Figure 4B). Interestingly, this remdesivir-resistant strain displayed increased sensitivity to EIDD-1931 and ribavirin relative to the parental strain (Figures 4A and 4B).
Figure 4.
Caspase 3/7 activity for the phenotypic resistance testing of SARS-CoV-2 strains
(A) Drug dose-response curves and concentrations that reduce cellular levels of the SARS-CoV-2 S protein by 50% (IC50) in Caco-2-F03 and Calu-3 cells as determined by immunostaining 24 h (Caco-2-F03) or 48 h (Calu-3) postinfection with the SARS-CoV-2 strain FFM3 or its remdesivir-adapted substrain FFM3rREM at MOI 0.01.
(B) Drug concentrations that reduce caspase 3/7 activity in FFM3 and FFM3rREM (MOI 0.01)-infected Caco-2-F03 cells 48 h postinfection. P-values were determined by Student’s t-test.
(C) Sequence variants in FFM3rREM compared to FFM3.
(D) The polymerase complex with nsp7 and nsp8 and a template-primer RNA (cyan and deep teal) and remdesivir (magenta) bound. Gly671Ser is shown in red (as serine).
(E) The SARS-CoV-2 polymerase Gly671Ser sequence variant. Residue 671 is shown in red as serine, which would be able to form a hydrogen bond with Thr402 which would not be present as Gly671. All p values were calculated by two-way ANOVA.
Results are expressed as the mean ± standard deviation.
Sequencing of FFM3rREM identified a 154452G>A mutation (present in >90% of alleles) in the coding region of the RNA-dependent RNA polymerase, which results in a change from glycine to serine in position 671(Gly671Ser) (Figure 4C). Gly671Ser is located in the polymerase domain of the RNA-dependent RNA polymerase in close vicinity to where RNA leaves (or enters) the active site (Figure 4D). Gly671Ser could have an effect on the protein structure as it is located in a bend between two beta sheets, where glycine often has an important role. In addition, Gly671Ser introduces a side chain capable of forming a hydrogen bond with Thr402 on an adjacent loop (Figure 4E), which could have an effect on the flexibility and conformation of the protein. Therefore, Gly671Ser seems likely to either reduce the binding affinity for remdesivir or enable the polymerase to overcome the effect of the drug.
Although our structural analysis plausibly explains why Gly671Ser in the RNA-dependent RNA polymerase is likely to mediate remdesivir resistance, it would have been impossible to determine this as a resistance variant without the prior knowledge that the change had happened in response to SARS-CoV-2 adaptation to remdesivir. Hence, this finding emphasizes the relevance of phenotypic assays for the identification of resistant strains that cannot be identified by the analysis of viral genomic information and the subsequent elucidation of the underlying resistance mechanisms. Notably, the caspase 3/7 activity assay also provides an easy-to-use readout for such phenotypic virus resistance testing approaches.
Comparison of Caco-2F03 with other cell line candidates for the identification of anti-SARS-CoV-2 drug candidates in screening assays
Next, we directly compared Caco-2F03 to other SARS-CoV-2 cultivation models that could be used for the identification of anti-SARS-CoV-2 drug candidates in screening assays. We focused on permanent cell lines that are easy to cultivate and maintain.
Suitable cell line candidates should be highly permissive for a broad spectrum of SARS-CoV-2 variants and display high caspase 3/7 activity upon infection. We had already shown that Caco-2-F03 cells display high susceptibility to a broad range of SARS-CoV-2 isolates (Figure 2). Here, we directly compared the susceptibility of A549-ACE2, Calu-3, Vero, and Caco-2-F03 cells to D614, G614, Alpha, Beta, and Delta isolates. S immunostaining and caspase 3/7 activity showed that Caco-2-F03 displayed the most pronounced broad-spectrum permissiveness to all tested SARS-CoV-2 isolates (Figures S5A and S5B).
Recently, drug-induced phospholipidosis was demonstrated to affect antiviral screens by causing false-positive hits due to unspecific effects that do not translate into the clinical setting.6 In particular, cationic amphiphilic drugs such as hydroxychloroquine were found to induce phospholipidosis.6 Hence, SARS-CoV-2 culture systems for phenotypic antiviral screening would ideally avoid false positives due to phospholipidosis.
Treatment with hydroxychloroquine resulted in considerable phospholipidosis and inhibited a Delta isolate in A549-ACE2 and Vero cells but not in Calu-3 or Caco-2-F03 cells (Figures S5C and S5D). Owing to the susceptibility to the broadest range of SARS-CoV-2 isolates and the insensitivity to drug-induced phospholipidosis, we selected Caco-2-F03 cells for a proof-of-concept screen for anti-SARS-CoV-2 drug candidates. Notably, Caco-2-F03 cells and the other clonal Caco-2 sublines presented in this manuscript are available to other researchers via the Resistant Cancer Cell Line (RCCL) Collection (https://research.kent.ac.uk/industrial-biotechnology-centre/the-resistant-cancer-cell-line-rccl-collection/).
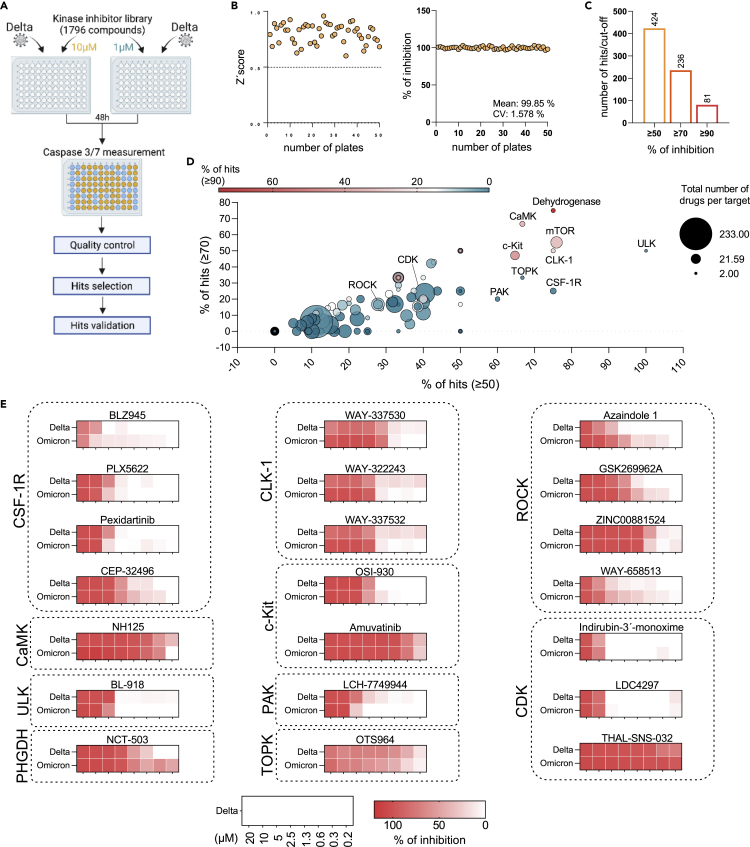
Proof-of-concept kinase inhibitor screen for drug candidates that inhibit SARS-CoV-2 replication
Next, we used the caspase 3/7 activity assay in Caco-2F03 cells to screen the Kinase Inhibitor Library (96-well)-L1200 (Selleck) for anti-SARS-CoV-2 drug candidates (Figure 5A). All compounds were tested at a concentration of 10 μM and 1 μM (Table S2). Z’scores were determined as quality controls on all plates as previously described,54 and only plates with a Z'score ≥0.5 were further analyzed (Figure 5B). Moreover, remdesivir (10 μM) was used as positive control on each plate and produced consistent results (Figure 5B).
Figure 5.
Proof-of-concept screen for anti-SARS-CoV-2 drug candidates using the Caco-2-F03 cell line platform and caspase 3/7 activity as readout method
(A) Overview of the proof-of-concept screen for anti-SARS-CoV-2 compounds using the Kinase inhibitor library L-1200 (Selleckchem, Germany) containing 1,796 compounds (Selleckchem, Germany) in Delta (MOI 0.01)-infected Caco-2-F03 cells using caspase 3/7 activity as readout 48 h postinfection. For the screen, every compound was tested at a concentration of 10 and 1 μM. 21 selected hits were then confirmed by determining drug-response curves.
(B) Quality controls, Z’scores served as quality controls (left). Only plates with a Z'score ≥0.5 were further analyzed. Remdesivir (10 μM) was used as positive control on each plate and produced consistent results (right).
(C) Number of hits at different inhibition cutoffs.
(D) Visualization of the distribution of hits according to their targets. Targets for which inhibitors were selected for confirmation are indicated.
(E) Heatmaps of the anti-SARS-CoV-2 activity of 21 hits by the determination of dose-response in Delta and Omicron (MOI 0.01)-infected Caco-2-F03 cells using immunostaining for the viral S protein as readout 24 h postinfection.
Using ≥90% inhibition of caspase 3/7 activity at 10 μM as a cutoff, our screen resulted in 81 hits (Figure 5C, Table S2). Moreover, compounds may exert antiviral effects at 1 μM but toxic effects at 10 μM, which would be expected to result in higher caspase 3/7 inhibition at 1 μM than at 10 μM. Hence, we additionally analyzed our screening data for compounds that displayed >90% caspase 3/7 inhibition at 1 μM and at least 30% higher caspase 3/7 inhibition at 1 μM than at 10 μM. However, this did not result in additional hits (Table S2).
Most hits were identified among inhibitors that target dehydrogenases, CaMK (calcium/calmodulin-dependent protein kinase), mTOR (mechanistic target of rapamycin kinase), ULK (Unc-51 like autophagy activating kinase), CLK-1, TOPK (lymphokine-activated killer T-cell-originated protein kinase, also known as PBK, PDZ binding kinase), CSF-1R (colony stimulating factor 1 receptor), and PAK (P21 [RAC1] activated kinase) (Figure 5D). CaMK, mTOR, ULK, TOPK, and PAK had already been proposed as antiviral drug targets for SARS-CoV-2.55,56,57,58,59 However, we could not find any information on potential anti-SARS-CoV-2 effects caused by CLK-1 or CSF-1R inhibition. We also included the PHGDH inhibitor NCT-50360,61 in our confirmation experiments. Although dehydrogenases had been known to contribute to SARS-CoV-2 replication,57 PHGDH had not previously been shown to be involved.
In addition to inhibitors of the targets described above, ROCK (Rho-associated coiled-coil containing protein kinase) and CDK (cyclin-dependent kinase) inhibitors were also included in the confirmation experiments. ROCK was described to be involved in the SARS-CoV-2-induced suppression of the host cell interferon response.62 CDK inhibitors had previously been shown to inhibit SARS-CoV-2 replication.63,64 The determination of dose-response curves for all of the 21 inhibitors using immunostaining for the SARS-CoV-2 S protein confirmed the results of the screen (Figure 5E, Figure S6).
PHGDH inhibitor NCT-503 as anti-SARS-CoV-2 drug candidate
Because PHGDH is a new potential antiviral drug target for the treatment of SARS-CoV-2 infection, we further investigated NCT-503. To investigate whether PHGDH inhibition is critical for NCT-503-mediated SARS-CoV-2 inhibition, we compared its effects and those of a chemically closely related analogue, which does not inhibit PHGDH and is commonly used as inactive NCT-503 control (Figure S7A),60,65 for antiviral activity. Only NCT-503 but not the inactive control inhibited Delta- and Omicron-induced caspase 3/7 activation indicating that the antiviral effects of NCT-503 are indeed mediated by PHGDH inhibition (Figure S7B).
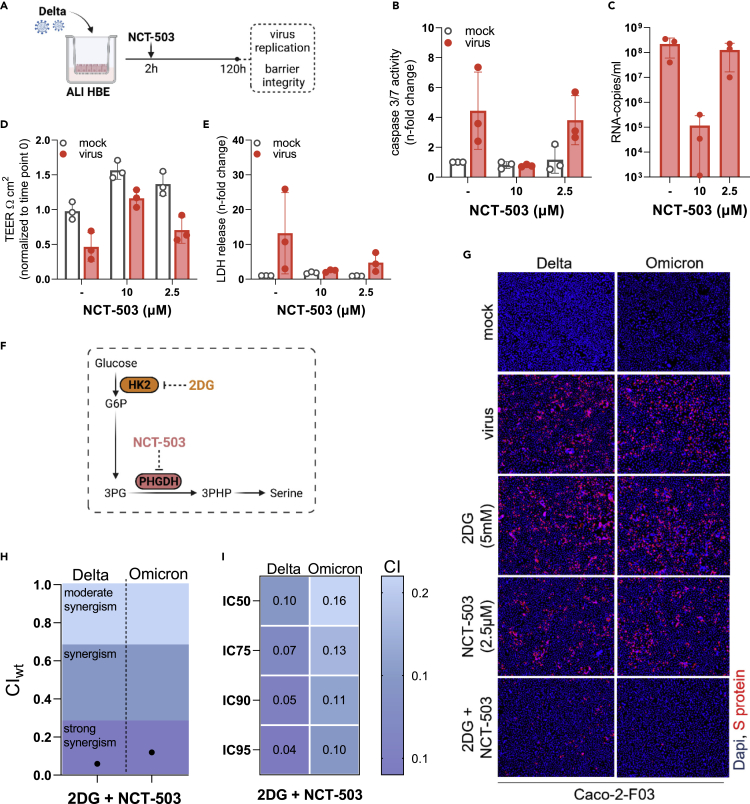
NCT-503 also inhibited SARS-CoV-2 replication in primary human bronchial epithelial cell ALI cultures (Figure 6A) as indicated by SARS-CoV-2-induced caspase 3 activity (Figure 6B), viral titers (determined as copy numbers of genomic RNA by PCR) (Figure 6C), cell layer integrity (Figure 6D), and lack of SARS-CoV-2-induced cytotoxicity (as indicated by LDH release) (Figure 6E).
Figure 6.
Investigation of the anti-SARS-CoV-2 effects of the PHGDH inhibitor NCT-503 alone or in in combination with 2-Deoxy-D-glucose (2DG)
(A–E) Scheme of the testing of NCT-503 for anti-SARS-CoV-2 activity in air liquid interface (ALI) cultures of primary human bronchial epithelial (HBE) cells. Effect of NCT-503 on (B) caspase 3/7 activity, (C) virus titers (determined as genomic RNA copy numbers by qPCR), (D) transepithelial electrical resistance (TEER), and (E) LDH release in ALI HBE cultures infected with Delta (MOI 1) 120 h postinfection.
(F) Anti-SARS-CoV-2 effects of NCT-503 in combination with 2-Deoxy-D-glucose (2DG). Illustration of how NCT-503 and 2DG can exert combined effects on a common metabolic pathway.
(G) Representative fluorescence images indicating the number of Delta- and Omicron (MOI 0.01)-infected cells in NCT503 and/or 2DG-treated Caco-2-F03 cultures 24 h postinfection.
(H) and I) Weighted combination indices (CIwt) determined by the method of Chou and Talalay66 indicating a strong synergism of NCT-503 and 2DG.
Results are expressed as the mean ± standard deviation.
Taken together, NCT-503 is a novel antiviral drug candidate for the treatment of SARS-CoV-2 infections that inhibits virus replication via PHGDH inhibition and is effective in different model systems including primary human bronchial epithelial cell ALI cultures, the system considered to be most physiologically relevant.67
NCT-503 in combination with 2-deoxy-D-glucose (2DG)
The discovery of PHGDH as novel antiviral drug target and of NCT-503 as antiviral drug candidate offers potential additional opportunities for combination therapies that display higher efficacy than either single treatment.
De novo serine synthesis is a side branch of glycolysis that includes the conversion of the glycolytic intermediate 3-phosphoglycerate (3 PG) into 3-phosphohydroxypyruvate (3PHP) by PHGDH (Figure 6F).68 The production of 3 PG in the glycolytic cycle depends on the phosphorylation of glucose into glucose-6-phosphate (G6P) by hexokinase II (HK2) as an initial step (Figure 6F).69 Notably, the HK2 inhibitor 2-Deoxy-D-glucose (2DG) has already been shown to inhibit SARS-CoV-2 replication.10,70 Hence, we hypothesized that the combined inhibition of de novo serine synthesis by 2DG and NCT-503 may result in further enhanced antiviral effects (Figure 6F).
Indeed, the combination of 2DG and NCT-503 resulted in stronger Delta and Omicron BA.1 inhibition than either drug alone (Figures 6G and S7C). The determination of combination indices (CIs) by the method of Chou and Talalay66 indicated a strong synergism of NCT-503 and 2DG against both SARS-CoV-2 isolates (Figures 6H and 6I).
Discussion
Here, we developed a novel screening assay for the identification of anti-SARS-CoV-2 compounds, based on using caspase 3/7 activity determined by the Caspase-Glo Assay System as readout indicating SARS-CoV-2 replication. SARS-COV-2 infection also activated the caspases 8 and 9, but the caspase 3/7 assay was superior to the detection of these other caspases as indicated by substantially higher signal-to-basal (S/B) ratios and Z′ scores. However, caspase 3/7 activation is not essentially involved in SARS-CoV-2 replication. The caspase inhibitor emricasan inhibited virus-induced caspase 3/7 activity but not SARS-CoV-2 replication. Hence, compounds that interfere with caspase 3/7 activity may produce false-positive hits. Therefore, hits identified by caspase 3/7 assay need to be confirmed by alternative assays (e.g., immunostaining for virus proteins and determination of virus titers) prior to further investigations.
In contrast to immunostaining approaches that require multiple handling steps, the Caspase-Glo 3/7 assay is a one-step readout assay that can be used by the large number of laboratories, which are equipped with the required plate readers that are in common use. Moreover, the assay is widely (potentially universally) applicable to different SARS-CoV-2 strains and clinical isolates as well as cell culture systems, as indicated by our wide range of pilot experiments. Moreover, our findings show that caspase 3/7 activity can also be used to determine SARS-CoV-2 replication in neutralization assays determining the antibody response in the plasma of individuals and for the phenotypic resistance testing of virus variants. Notably, the caspase 3/7 assay also detects SARS-CoV-2 replication in cultivation systems that do not develop a CPE and in which viability assays such as the MTT assay and the CellTiter Glo Assay did not reflect SARS-CoV-2 replication.
For our proof-of-concept experiment for phenotypic resistance testing, we established a remdesivir-resistant SARS-CoV-2 strain by adapting the SARS-CoV-2 strain FFM3 to replication in the presence of remdesivir. Our results confirmed previous observations9,26 demonstrating that SARS-CoV-2 resistance formation against clinically approved antiviral drugs poses a relevant risk. Notably, the genetic sequence of our remdesivir-adapted SARS-CoV-2 strain would not have enabled us to identify this as a resistant strain by a genotypic approach, which emphasizes the potential need for effective phenotypic resistance testing platforms in the future.
In addition to identifying an easy-to-handle readout assay for anti-SARS-CoV-2 agent screens, we were also interested in identifying a well-suited cell culture platform. We considered permanent cell lines to be the most promising candidates because they are readily available and require a minimum of handling. A number of continuous cell lines (e.g., A549-ACE, Calu-3, Vero, and Caco-2) had already been used in different phenotypic screening approaches for the identification of antiviral drug candidates against SARS-CoV-2.3,11,12,13,14,15,16 Based on our comparison of different candidate cell lines, however, we identified Caco-2-F03 as the best platform as it displayed susceptibility to the widest range of SARS-CoV-2 strains and isolates and was not affected by drug-induced phospholipidosis that has been shown to result in false-positive hits during the testing of anti-SARS-CoV-2 drug candidates.6
Notably, Caco-2 cells were (in contrast to Calu-3, A549, or Vero cells) shown to be highly susceptible to seasonal coronaviruses such as HCoV-229E or HCoV-OC43.29,71,72,73,74 In this context, we found here that the caspase 3/7 assay also enabled the monitoring of HCoV-229E replication in Caco-2-F03 cells, indicating that this may also serve as a unique broad-spectrum drug screening platform for (seasonal) coronaviruses. Moreover, Caco-2 cells have been described to be susceptible to further viruses, including enteroviruses (coxsackie B1-B6, poliovirus types 1–3, most echoviruses, and coxsackie A viruses), adenovirus, herpes simplex virus, measles morbillivirus, respiratory syncytial virus (RSV), and parainfluenza type 2 virus.28 Hence, Caco-2-F03 cells may have potential for identification of agents with activity against further viruses.
Our study also provided an explanation for the contradictory findings on the SARS-CoV-2 susceptibility of Caco-2 cells reported in previous studies.10,14,35,36,37,38,39,40,41,42,43,44 When we investigated newly acquired Caco-2 cell lines from different providers (DSMZ, CLS, Sigma) for SARS-CoV-2 susceptibility, they did indeed not present the level of SARS-CoV-2 permissiveness that we find in our Caco-2-F03 cell line. The subsequent analysis of 21 clonal sublines of the newly purchased lowly SARS-CoV-2-susceptible Caco-2 cell line from DSMZ (Caco-2A) resulted in a broad range of susceptibility phenotypes, suggesting that a highly SARS-CoV-2-susceptible subpopulation has inadvertently become the dominant population in our Caco-2-F03 cell line. Notably, the susceptibility phenotype of Caco-2-F03 appears to be stable as we have used this cell line for the cultivation of SARS-CoV and SARS-CoV-2 since 2003.32,33 Moreover, the SARS-CoV-2 susceptibility phenotype of Caco-2-F03 was maintained for a further 30 passages within the current study. Notably, such phenotypic differences between samples of the same cell line obtained from different sources are not surprising and have been described for different cell lines.75,76,77
Next, we used the caspase 3/7 activity assay in Caco-2-F03 cells to screen the Kinase Inhibitor Library (96-well)-L1200 (Selleck) for anti-SARS-CoV-2 drug candidates, which resulted in 81 hits that reduced SARS-CoV-2-induced caspase 3/7 activity by ≥ 90% at a concentration of 10 μM. These hits included inhibitors of known potential anti-SARS-CoV-2 drug targets (CaMK, mTOR, ULK, TOPK, PAK, ROCK, and CDK)14,55,56,57,58,59,62 and those that interfere with drug targets that had not previously been identified to be relevant during SARS-CoV-2 replication (CLK-1 and CSF-1R). We determined dose-response curves for 21 of the hit compounds using immunostaining for the viral S protein, which confirmed their anti-SARS-CoV-2 activities.
Among these hits, we further investigated the PHGDH inhibitor NCT-50360,61 as it interferes with a dehydrogenase that had not previously been shown to be involved in SARS-CoV-2 replication. In addition to NCT-503, we tested a structurally closely related analogue that does not inhibit PHGDH.60,65 This inactive NCT-503 analogue did not affect SARS-CoV-2 replication, indicating that the anti-SARS-CoV-2 effects of NCT-503 are caused by its effect on PHGDH.
PHGDH activity is critically involved in de novo serine synthesis,68 a pathway downstream of the glycolytic cycle that depends on the phosphorylation of glucose into G6P by HK2 as an initial step.69 Because the HK2 inhibitor 2DG has already been shown to inhibit SARS-CoV-2 replication,10,70 we tested whether the combined interference with this pathway using NCT-503 and 2DG resulted in further increased antiviral effects. Indeed, the combination resulted in strongly synergistic anti-SARS-CoV-2 activity. Such antiviral combination therapies have been suggested to be of critical importance for the sustained control of virus outbreaks as they are not only more effective but also anticipated to reduce and, ideally, prevent resistance formation.78 Future studies will have to show, whether NCT-503 also inhibits SARS-CoV-2 in vivo, alone and in combination with 2DG.
In conclusion, we here present a novel phenotypic screening platform for the identification of drug candidates with activity against SARS-CoV-2 and other coronaviruses based on the determination of caspase 3/7 activity using the one-step Caspase-Glo 3/7 Assay System as readout. Caspase 3/7 activity is also a suitable readout for neutralization assays and phenotypic resistance testing. The Caco-2-F03 cell line was identified as the best-suited cell culture platform. It is susceptible to a particularly broad range of SARS-CoV-2 isolates, and its susceptibility phenotype remains stable over many passages. Moreover, Caco-2-F03 is not affected by phospholipidosis, which is known to cause false-positive hits during the testing of potential anti-SARS-CoV-2 agents.6 Hence, the determination of caspase 3/7 activity in SARS-CoV-2-infected Caco-2-F03 cells represents a newly established screening platform that is easy to use also for groups without experience in drug-discovery projects. A proof-of-concept screen of a kinase inhibitor library containing 1,796 compounds resulted in known and novel anti-SARS-CoV-2 drug targets. The PHGDH inhibitor NCT-503 was identified as novel antiviral drug candidate, whose activity was further increased by 2DG (an inhibitor of the PHGDH upstream HK2), which is under clinical development for the treatment of COVID-19 treatment.79
Limitations of the study
Although we tested a wide range of SARS-CoV-2 isolates and other coronaviruses and performed a proof-of-concept drug screen, such a study can never cover all eventualities. Hence, future research will have to provide further evidence on the prospects and limitations of determining caspase 3/7 activity in Caco-2-F03 cells infected with SARS-CoV-2 and other coronaviruses as platform for the discovery of antiviral drugs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS-CoV-2 spike protein antibody | Sino Biological | Cat: 40592-R190 |
| peroxidase-conjugated anti-rabbit secondary antibody | Dianova | SKU: 111-035-045 |
| Alexa Fluor 488 anti-rabbit secondary antibody | Invitrogen | Cat # A32731; RRID: AB_2633280 |
| GAPDH | Trevigen | #2275-PC-100; RRID: AB_2107456 |
| SARS-CoV-2 NP | Sino Biological | #40143-R019; RRID: AB_2827973 |
| ACE2 | Abcam | #ab15348; RRID: AB_301861 |
| TMPRSS2 | Abcam | #ab92323; RRID: AB_10585592 |
| IRDye®800CW Goat anti-Rabbit | LI-COR Biotechnology | 926–32211; RRID: AB_621843 |
| Virus strains | ||
| D614 | Goethe University | SARS-CoV-2/FFM1, GenBank: MT358638 |
| G614 | Goethe University | SARS-CoV-2/FFM7, GenBank: MT358643 |
| Alpha | Goethe University | SARS-CoV-2/FFM-UK7931/2021, Gen Bank: MZ427280 |
| Beta | Goethe University | SARS-CoV-2/FFM-ZAF1/2021, GenBank: MW822592 |
| Delta | Goethe University | SARS-CoV-2/FFM-IND8424/2021, GenBank: MZ315141 |
| Zeta | Goethe University | SARS-CoV-2/FFMBRA1/2021, GenBank: MW822593 |
| Omicron | Goethe University | SARS-CoV-2/FFM-SIM0550/2021, GenBank: OL800702 |
| SARS-CoV | Goethe University | Cinatl et al., 2004 |
| MERS-CoV | BEI | EMC/2012, NR-44260 |
| HCoV-229E | ATCC | CCL-137 |
| Biological samples | ||
| Primary bronchial epithelial cells | Dr. Ruth Olmer | van Wetering et al., 2000 |
| Human induced pluripotent stem cell-derived cardiomyocytes | Prof. Stefanie Dimmeler | Breckwoldt et al., 2017 |
| Primary human hepatocytes | Dr. Florian Vondran | Vondran et al., 2008 |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Thermo Scientific | D1306 |
| Alexa FluorTM 647 Phalloidin | Invitrogen | #A22287 |
| Kinase inhibitor library L-1200 | Selleckchem | Catalog No.L1200 |
| Zosuquidar | MedChemExpress | Cat. No.: HY-15255 |
| Remdesivir | MedChemExpress | Cat. No.: HY-104077 |
| NBD-PE | ThermoFisher | N360 |
| Critical commercial assays | ||
| Caspase-Glo® 3/7 Assay Systems | Promega | G8093 |
| Caspase-Glo® 8 Assay Systems | Promega | G8201 |
| Caspase-Glo® 9 Assay Systems | Promega | G8211 |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | G7571 |
| QIAamp Viral RNA Kit | QIAGEN | Cat. No. / ID: 52962 |
| Experimental models: Cell lines | ||
| Caco-2A | DSMZ | ACC 169 |
| Caco-2B | Sigma | 86010202-1VL |
| Caco-2C | CLS | 300137 |
| Caco-2-F03 | RCCL | Available upon request |
| Caco-2 clones | RCCL | Available upon request |
| A549-ACE2 | Invivogen | a549-hace2 |
| Vero | DSMZ | ACC 33 |
| Calu-3 | ATCC | ATCC HTB-55 |
| Oligonucleotides | ||
| RdRP_SARSr-F2 (GTG ARA TGG TCA TGT GTG GCG G) | Synthesized by ThermoFisher | NA |
| RdRP_SARSr-R1 (CAR ATG TTA AAS ACA CTA TTA GCA TA) | Synthesized by ThermoFisher | NA |
| Recombinant DNA | ||
| pEX-A128-RdRP | Dr. Marek Widera | NA |
| Software and algorithms | ||
| Image J | open source software | https://imagej.nih.gov/ij/download.html |
| CalcuSyn | Biosoft | http://www.biosoft.com/w/calcusyn.htm |
| GraphPad Prism 9 | Graphstats Technologies | https://www.graphstats.net/graphpad-prism |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jindrich Cinatl (cinatl@em.uni-frankfurt.de).
Materials availability
Caco-2-F03 and clonal Caco-2-F03 sublines are available from the Resistant Cancer Cell Line (RCCL) collection (https://research.kent.ac.uk/industrial-biotechnology-centre/the-resistant-cancer-cell-line-rccl-collection/) with Martin Michaelis (M.Michaelis@kent.ac.uk) being the main contact.
Experimental models and subject details
Cell culture
Caco-2A (DSMZ), Caco-2B (Sigma), Caco-2C (CLS), Vero (DSMZ), Calu-3 (ATCC), and Caco-2-F03 (Resistant Cancer Cell Line collection, https://research.kent.ac.uk/industrial-biotechnology-centre/the-resistant-cancer-cell-line-rccl-collection/) were grown at 37 °C in minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 μg/mL streptomycin. All culture reagents were purchased from Sigma. A549-ACE2 (Invivogen) was grown in DMEM supplemented with 10% FBS, 2% L-glutamine, 100 μg/ml normocin, 0.5 μg/mL puromycin, 100 IU/mL penicillin, and 100 μg/mL of streptomycin. All cell lines were regularly authenticated by short tandem repeat (STR) analysis and tested for mycoplasma contamination.
Primary bronchial epithelial cells were isolated from the lung explant tissue of a patient with lung emphysema as previously described.80 For differentiation into air-liquid interface (ALI) cultures, cells were resuscitated, passaged once in PneumaCult-Ex Medium (StemCell technologies), and seeded on transwell inserts (12-well plate, Sarstedt) at 4x104 cells/insert. After reaching confluence, medium on the apical side of the transwell insert was removed and medium in the basal chamber was replaced with PneumaCult ALI Maintenance Medium (StemCell Technologies) including Antibiotic/Antimycotic solution (Sigma Aldrich) and MycoZap Plus PR (Lonza). Criteria for successful differentiation were the development of ciliary movement, an increase in transepithelial electric resistance, and mucus production.
Human induced pluripotent stem cell-derived cardiomyocytes (hiPS-CMs) of two donors were obtained with an embryoid body-based protocol as previously described.81 hiPS-CMs were cultured in RPMI/B27 medium at 37 °C and 5% CO2 for 4 to 5 days prior to viral infection.
Primary human hepatocytes (PHHs) were isolated as previously described82 and were maintained in William’s Medium E (PAN Biotech, Aidenbach, Germany) containing 10% fetal bovine serum (Biochrom, Cambridge, UK) and 10,000 U penicillin/streptomycin, 1% l-glutamine, 1% non-essential amino acids, 5 mmol/L HEPES (Thermo Fisher Scientific, Schwerte, Germany), 2% dimethyl sulfoxide (DMSO, Roth, Karlsruhe, Germany), 5 μg/mL insulin, and 0.05 mmol/L hydrocortisone (Sigma Aldrich, Munich, Germany).
Virus preparation and infection of different cell types
Caco-2-F03 cells were used for the isolation SARS-CoV-2 variants applied in this study. Information on the following isolates is available from GenBank: D614 (SARS-CoV-2/FFM1, GenBank: MT358638), G614 (SARS-CoV-2/FFM7, GenBank: MT358643), Alpha (SARS-CoV-2/FFM-UK793½021, GenBank: MZ427280), Beta (SARS-CoV-2/FFM-ZAF1/2021, GenBank: MW822592), Delta (SARS-CoV-2/FFM-IND8424/2021, GenBank: MZ315141), Zeta (SARS-CoV-2/FFMBRA1/2021, GenBank: MW822593), Omicron (SARS-CoV-2/FFM-SIM0550/2021, GenBank: OL800702). Additional isolates were not further characterized. SARS-CoV-2 stocks were cultivated for a maximum of three passages in Caco-2-F03 cells and stored at −80°C. SARS-CoV stocks were prepared on Caco-2-F03 cells as previously described.33 MERS-CoV was obtained from BEI Resources (EMC/2012, NR-44260) and passaged once on Vero cells prior experiments. Viral stocks of HCoV-229E (ATCC no. CCL-137) were prepared using Caco-2-F03 cells. Virus titers were determined as TCID50/mL in confluent cells in 96-well microtiter plates.
Primary bronchial and nasal epithelial cells in ALI cultures were infected with SARS-CoV-2 from the apical site. The inoculum was incubated for 2 h, then removed and cells were washed three times with PBS. For testing of antiviral activity of drugs, the compounds were added after the infection period from both the apical and the basal site. The apical medium was removed after one day.
Method details
Caspase activity assay
Caspase 3/7, 8 and 9 activity was measured using the Caspase-Glo assay kit (Promega, Madison, WI, USA), according to the manufacturer’s instructions. Briefly, 100 μL of Caspase-Glo reagent were added to each well, mixed, and incubated at room temperature for 30 min. Luminescence intensity was measured using an Infinite M200 microplate reader (Tecan).
Viability assay
Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye reduction assay. 25 μL of MTT solution (2 mg/mL in PBS) were added per well, and the plates were incubated at 37 °C for 4 h. After this, the cells were lysed using 100 μL of a buffer containing 20% SDS and 50% N,N-dimethylformamide with the pH adjusted to 4.7 at 37 °C for 4 h. Absorbance was determined at 560 nm (reference wavelength 620 nm) using a Tecan infinite M200 microplate reader (TECAN).
Alternatively, cell viability was determined using the CellTiter-Glo (Promega), which measures ATP production, according to the manufacturer’s protocol. Luminescence was measured on a Tecan infinite M200 microplate reader (TECAN).
Immunocytochemistry of viral antigen
Cells were fixed with acetone:methanol (40:60) solution and immunostaining was performed using a monoclonal antibody directed against the spike (S) protein of SARS-CoV-2 (1:1500, Sinobiological), which was detected with a peroxidase-conjugated anti-rabbit secondary antibody (1:1,000, Dianova), followed by addition of AEC substrate. The S positive area was scanned and quantified by the Bioreader 7000-F-Z-I microplate reader (Biosys). The results are expressed as percentage of inhibition relative to virus control which received no drug.
Immunofluorescence labeling
Cells were fixed with 3% PFA permeabilized with 0.1% Triton X-100. Prior to primary antibody labeling, cells were blocked with 5% donkey serum in PBS or 1% BSA and 2% goat serum in PBS for 30 min at room temperature. Spike (S) protein was detected using a specific antibody (1:1500, Sinobiological) and an Alexa Fluor 488 anti-rabbit secondary antibody (1:200, Invitrogen). The nucleus was labeled using DAPI (1:1000, Thermo Scientific). Cardiomyocytes were counterstained with Alexa FluorTM 647 Phalloidin (1:100, #A22287, Invitrogen). Images were taken using Spark Mulitmode microplate reader (TECAN) at 10x magnification.
Immunoblot assay
Cells were lysed using Triton X-100 sample buffer (Sigma-Aldrich), and proteins were separated by SDS-PAGE. Detection occurred by using specific antibodies against GAPDH (1:1000 dilution, #2275-PC-100, Trevigen), SARS-CoV-2 NP (1:1000 dilution, #40143-R019, Sino Biological), ACE2 (1:500 dilution, #ab15348, Abcam), and TMPRSS2 (1:1000 dilution, Recombinant Anti-TMPRSS2 antibody [EPR3861], #ab92323, Abcam) followed by incubation with IRDye-labeled secondary antibodies (LI-COR Biotechnology, IRDye800CW Goat anti-Rabbit, 926-32211, 1:40,000) according to the manufacturer’s instructions. Protein bands were visualized by laser-induced fluorescence using an infrared scanner for protein quantification (Odyssey, Li-Cor Biosciences, Bad Homburg, Germany).
qRT-PCR
SARS-CoV-2 RNA from cell culture supernatant samples was isolated using AVL buffer and the QIAamp Viral RNA Kit (QIAGEN) according to the manufacturer’s instructions. Quantification of viral RNA was performed as previously described10,38 using primers targeting the RNA-dependent RNA polymerase (RdRp): RdRP_SARSr-F2 (GTG ARA TGG TCA TGT GTG GCG G) and RdRP_SARSr-R1 (CAR ATG TTA AAS ACA CTA TTA GCA TA). Standard curves were created using plasmid DNA (pEX-A128-RdRP) harboring the corresponding amplicon regions for RdRP target sequence according to GenBank Accession number NC_045512. All quantification experiments have been carried out with biological replicates.
Neutralization assay
Serum of double mRNA-1273-vaccinated individuals was serially diluted and pre-incubated with 4000 TCID50/mL of SARS-CoV-2 variants at 37°C for 1 h prior transfer to Caco-2-F03 monolayers in 96 well plate. The neutralization titer was determined either by visual scoring of CPE 72 h post infection or caspase 3/7 activity measurement.
Selection of drug-resistant variant
SARS-CoV-2/FFM3 was serially passaged with increasing concentration (starting concentration - 500 nM) of remdesivir in Caco-2-F03. Viral replication was monitored by observation for any cytopathogenic effect present in the culture. Infected cultures were frozen at −80°C and thawed once prior a passaging. Virus was serially passaged by using 1 aliquot of viral stock from the preceding passage to infect fresh Caco-2-F03 cells (MOI of 0.1) in the presence of increasing concentrations of compound for a total of 30 passages, resulting in a strain that could be readily passaged in the presence of remdesivir 2 μM (FFM3rREM).
Sequencing
Extracted nucleic acid was DNase treated, reverse transcribed, and randomly amplified using a Sequence-Independent Single-Primer Amplification (SISPA) method described previously.83 Illumina sequencing used the Nextera XT protocol with 2 × 150-bp paired-end sequencing on a MiSeq.
Phospholipidosis quantification
Phospholipidosis was assessed as previously described.6 Cells were treated with hydroxychloroquine in the presence of 7.5 μM nitrobenzoxadiazole-conjugated phosphoethanolamine (NBD-PE) (ThermoFisher). Images were taken and the fluorescence was quantified using a Spark Mulitmode microplate reader (TECAN).
Screening assay
The Kinase inhibitor library L-1200 (Selleckchem) containing 1796 compounds was tested in a proof-of-concept screen in Delta-infected Caco-2-F03 cells for the identification of antivirally active agents. Caco-2-F03 cells were seeded into 96-well plates (50,000 cells/well) and incubated at 37°C for 4 days. After the cells reached confluence, the supernatant was replaced by 25 μL/well of medium containing the ABCB1 inhibitor zosuquidar (final concentration 1 μM), 25 μL/well of medium containing kinase inhibitors (final concentration 10 μM) in singlets, and 50 μL/well SARS-CoV-2 suspension (MOI 0.01). Remdesivir (10 μM) was used as positive control. Plates were incubated at 37°C for 48h prior to the measurement of caspase 3/7 activity as described above. For each plate the Z′ score, a measure of statistical effect size and an index for assay quality control, was calculated by: Z′ = 1 − (3∗SD.signal +3∗SD.basal)/(Meansignal − Meanbasal). Only plates with Z'score ≥0.5 were further analyzed.
Drug combination studies
To evaluate antiviral activity of drug combinations, drugs were tested alone or in fixed combinations at 1:2 dilutions using monolayers of Caco-2-F03 cells infected with the indicated SARS-CoV-2 isolates at MOI 1. Antiviral effects were detected 24 h post infection by immunofluorescence staining for S protein. The calculation of IC50, IC75, IC90 and IC95 for single drugs and their combinations as well as combination indices (CIs) was performed using the software CalcuSyn (Biosoft) based on the method of Chou and Talalay.66 The weighted average CI value (CIwt) was calculated according to the formula: CIwt [CI50 + 2CI75 + 3CI90 + 4CI95]/10. CIwt values were calculated for mutually exclusive interactions where CIwt < 1 indicates synergism, CIwt = 1 indicates additive effects, and CIwt ˃1 suggest antagonism.
Quantification and statistical analysis
The results are expressed as the mean ± SD of at least three experiments. The Student’s t test was used for comparing two groups. Three and more groups were compared by ANOVA. GraphPad Prism 9 was used to determine IC50 and CC50. Correlations were determined using the Pearson correlation coefficient. The statistical details of experiments can be found in the figures and their legends.
Acknowledgments
We thank Kerstin Euler, Sebastian Grothe, and Lena Stegman for their technical assistance. This work was supported by the Frankfurter Stiftung für krebskranke Kinder, the Goethe-Corona-Fonds, BMBF (COVID-Protect), the Corona Accelerated R&D in Europe (CARE) project from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 101005077, and the BBSRC (SoCoBio DTP training program).
Author contributions
D.B., M.M., and J.C. jr. conducted the study. D.B., P.R., L.P., M.B., T.R., J.D.K., A.P., J.U.G.W., M.S., T.T., F.R., R.Z., R.S., M.N.W., and J.C. jr. performed experiments. R.O., U.M., and F.W.R. contributed primary human bronchial epithelial cells. K.L.O., S.T.P., and M.W.C. sequenced virus genomes. D.B., S.D., S.C., M.N.W., M.M., and J.C. jr. supervised research. All authors analyzed data. M.M. wrote the manuscript draft. All authors revised and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.105944.
Contributor Information
Martin Michaelis, Email: m.michaelis@kent.ac.uk.
Jindrich Cinatl, Jr., Email: cinatl@em.uni-frankfurt.de.
Supplemental information
Data and code availability
-
•
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/yzbfm4v7y7.1.
-
•
This paper does not report original code. All data reported in this paper will be shared by the lead contact (Jindrich Cinatl, cinatl@em.uni-frankfurt.de) upon request. Any additional information required to reanalyze the data reported in this paper is also available from the lead contact upon request.
-
•
Data on the SARS-CoV-2 isolates is available from GenBank: D614 (SARS-CoV-2/FFM1, GenBank: MT358638), G614 (SARS-CoV-2/FFM7, GenBank: MT358643), Alpha (SARS-CoV-2/FFM-UK7931/2021, GenBank: MZ427280), Beta (SARS-CoV-2/FFM-ZAF1/2021, GenBank: MW822592), Delta (SARS-CoV-2/FFM-IND8424/2021, GenBank: MZ315141), Zeta (SARS-CoV-2/FFMBRA1/2021, GenBank: MW822593), Omicron (SARS-CoV-2/FFM-SIM0550/2021, GenBank: OL800702).
References
- 1.Gao J., Sun F. Drug discovery to treat COVID-19 two years after its outbreak. Drug Discov. Ther. 2021;15:281–288. doi: 10.5582/ddt.2021.01302. [DOI] [PubMed] [Google Scholar]
- 2.Gentile I., Schiano Moriello N. COVID-19 prophylaxis in immunosuppressed patients: beyond vaccination. PLoS Med. 2022;19:e1003917. doi: 10.1371/journal.pmed.1003917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dittmar M., Lee J.S., Whig K., Segrist E., Li M., Kamalia B., Castellana L., Ayyanathan K., Cardenas-Diaz F.L., Morrisey E.E., et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021;35:108959. doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bojkova D., Rothenburger T., Ciesek S., Wass M.N., Michaelis M., Cinatl J., Jr. SARS-CoV-2 Omicron variant virus isolates are highly sensitive to interferon treatment. Cell Discov. 2022;8:42. doi: 10.1038/s41421-022-00408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H., Lu L., Peng Z., Chen L.L., Meng X., Zhang C., Ip J.D., Chan W.M., Chu A.W.H., Chan K.H., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 2022;11:277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tummino T.A., Rezelj V.V., Fischer B., Fischer A., O'Meara M.J., Monel B., Vallet T., White K.M., Zhang Z., Alon A., et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021;373:541–547. doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Bárcena M., et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramirez S., Fernandez-Antunez C., Galli A., Underwood A., Pham L.V., Ryberg L.A., Feng S., Pedersen M.S., Mikkelsen L.S., Belouzard S., et al. Overcoming culture restriction for SARS-CoV-2 in human cells facilitates the screening of compounds inhibiting viral replication. Antimicrob. Agents Chemother. 2021;65:e0009721. doi: 10.1128/AAC.00097-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szemiel A.M., Merits A., Orton R.J., MacLean O.A., Pinto R.M., Wickenhagen A., Lieber G., Turnbull M.L., Wang S., Furnon W., et al. In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 2021;17:e1009929. doi: 10.1371/journal.ppat.1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Touret F., Gilles M., Barral K., Nougairède A., van Helden J., Decroly E., de Lamballerie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020;10:13093. doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z.R., Zhang Y.N., Li X.D., Zhang H.Q., Xiao S.Q., Deng F., Yuan Z.M., Ye H.Q., Zhang B. Zhang B. A cell-based large-scale screening of natural compounds for inhibitors of SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:218. doi: 10.1038/s41392-020-00343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellinger B., Bojkova D., Zaliani A., Cinatl J., Claussen C., Westhaus S., Keminer O., Reinshagen J., Kuzikov M., Wolf M., et al. A SARS-CoV-2 cytopathicity dataset generated by high-content screening of a large drug repurposing collection. Sci. Data. 2021;8:70. doi: 10.1038/s41597-021-00848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Damme E., De Meyer S., Bojkova D., Ciesek S., Cinatl J., De Jonghe S., Jochmans D., Leyssen P., Buyck C., Neyts J., Van Loock M. In vitro activity of itraconazole against SARS-CoV-2. J. Med. Virol. 2021;93:4454–4460. doi: 10.1002/jmv.26917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan K., Rawle D.J., Le T.T., Suhrbier A. Simple rapid in vitro screening method for SARS-CoV-2 anti-virals that identifies potential cytomorbidity-associated false positives. Virol. J. 2021;18:123. doi: 10.1186/s12985-021-01587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caccuri F., Zani A., Messali S., Giovanetti M., Bugatti A., Campisi G., Filippini F., Scaltriti E., Ciccozzi M., Fiorentini S., Caruso A. A persistently replicating SARS-CoV-2 variant derived from an asymptomatic individual. J. Transl. Med. 2020;18:362. doi: 10.1186/s12967-020-02535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Y., Li X., Mou T., Zhou X., Li D., Wang L., Zhang Y., Dong X., Zheng H., Guo L., et al. Distinct infection process of SARS-CoV-2 in human bronchial epithelial cell lines. J. Med. Virol. 2020;92:2830–2838. doi: 10.1002/jmv.26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bielarz V., Willemart K., Avalosse N., De Swert K., Lotfi R., Lejeune N., Poulain F., Ninanne N., Gilloteaux J., Gillet N., Nicaise C. Susceptibility of neuroblastoma and glioblastoma cell lines to SARS-CoV-2 infection. Brain Res. 2021;1758:147344. doi: 10.1016/j.brainres.2021.147344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wurtz N., Penant G., Jardot P., Duclos N., La Scola B. Culture of SARS-CoV-2 in a panel of laboratory cell lines, permissivity, and differences in growth profile. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:477–484. doi: 10.1007/s10096-020-04106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia G., Jr., Sharma A., Ramaiah A., Sen C., Purkayastha A., Kohn D.B., Parcells M.S., Beck S., Kim H., Bakowski M.A., et al. Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication. Cell Rep. 2021;35:108940. doi: 10.1016/j.celrep.2021.108940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thi Nhu Thao T., Labroussaa F., Ebert N., V'kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582:561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- 23.Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X., Quan S., Xu M., Rodriguez S., Goh S.L., Wei J., Fridman A., Koeplinger K.A., Carroll S.S., Grobler J.A., et al. Generation of SARS-CoV-2 reporter replicon for high-throughput antiviral screening and testing. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025866118. e2025866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiscox J.A., Khoo S.H., Stewart J.P., Owen A. Shutting the gate before the horse has bolted: is it time for a conversation about SARS-CoV-2 and antiviral drug resistance? J. Antimicrob. Chemother. 2021;76:2230–2233. doi: 10.1093/jac/dkab189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang K.S., Leeuwon S.Z., Xu S., Liu W.R. Evolutionary and structural insights about potential SARS-CoV-2 evasion of nirmatrelvir. J. Med. Chem. 2022;65:8686–8698. doi: 10.1021/acs.jmedchem.2c00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fogh J., Fogh J.M., Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J. Natl. Cancer Inst. 1977;59:221–226. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- 28.Reigel F. Isolation of human pathogenic viruses from clinical material on CaCo2 cells. J. Virol. Methods. 1985;12:323–327. doi: 10.1016/0166-0934(85)90143-0. [DOI] [PubMed] [Google Scholar]
- 29.Collins A.R. Comparison of the replication of distinct strains of human coronavirus OC43 in organotypic human colon cells (Caco-2) and mouse intestine. Adv. Exp. Med. Biol. 1990;276:497–503. doi: 10.1007/978-1-4684-5823-7_69. [DOI] [PubMed] [Google Scholar]
- 30.Chan J.F.W., Chan K.H., Choi G.K.Y., To K.K.W., Tse H., Cai J.P., Yeung M.L., Cheng V.C.C., Chen H., Che X.Y., et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013;207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan K.H., Yan M.K., To K.K.W., Lau S.K., Woo P.C., Cheng V.C.C., Li W.S., Chan J.F.W., Tse H., Yuen K.Y. Use of the human colorectal adenocarcinoma (Caco-2) cell line for isolating respiratory viruses from nasopharyngeal aspirates. J. Med. Virol. 2013;85:874–879. doi: 10.1002/jmv.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cinatl J., Jr., Hoever G., Morgenstern B., Preiser W., Vogel J.U., Hofmann W.K., Bauer G., Michaelis M., Rabenau H.F., Doerr H.W. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell. Mol. Life Sci. 2004;61:2100–2112. doi: 10.1007/s00018-004-4222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bojkova D., Wagner J.U.G., Shumliakivska M., Aslan G.S., Saleem U., Hansen A., Luxán G., Günther S., Pham M.D., Krishnan J., et al. SARS-CoV-2 infects and induces cytotoxic effects in human cardiomyocytes. Cardiovasc. Res. 2020;116:2207–2215. doi: 10.1093/cvr/cvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bojkova D., Bechtel M., McLaughlin K.M., McGreig J.E., Klann K., Bellinghausen C., Rohde G., Jonigk D., Braubach P., Ciesek S., et al. Aprotinin inhibits SARS-CoV-2 replication. Cells. 2020;9:2377. doi: 10.3390/cells9112377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D., Behrens P., Böddinghaus B., Götsch U., Naujoks F., et al. Evidence of SARS-CoV-2 infection in returning travelers from wuhan, China. N. Engl. J. Med. 2020;382:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klann K., Bojkova D., Tascher G., Ciesek S., Münch C., Cinatl J. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol. Cell. 2020;80:164–174.e4. doi: 10.1016/j.molcel.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toptan T., Hoehl S., Westhaus S., Bojkova D., Berger A., Rotter B., Hoffmeier K., Cinatl J., Jr., Ciesek S., Widera M. Optimized qRT-PCR approach for the detection of intra- and extra-cellular SARS-CoV-2 RNAs. Int. J. Mol. Sci. 2020;21:4396. doi: 10.3390/ijms21124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bojkova D., McGreig J.E., McLaughlin K.M., Masterson S.G., Antczak M., Widera M., Krähling V., Ciesek S., Wass M.N., Michaelis M., Cinatl J. Differentially conserved amino acid positions may reflect differences in SARS-CoV-2 and SARS-CoV behaviour. Bioinformatics. 2021;37:2282–2288. doi: 10.1093/bioinformatics/btab094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gower G., Picazo P.I., Fumagalli M., Racimo F. Detecting adaptive introgression in human evolution using convolutional neural networks. Elife. 2021;10:e64669. doi: 10.7554/eLife.64669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widera M., Mühlemann B., Corman V.M., Toptan T., Beheim-Schwarzbach J., Kohmer N., Schneider J., Berger A., Veith T., Pallas C., et al. Surveillance of SARS-CoV-2 in frankfurt am main from October to December 2020 reveals high viral diversity including spike mutation N501Y In B.1.1.70 and B.1.1.7. Microorganisms. 2021;9:748. doi: 10.3390/microorganisms9040748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu H., Chan J.F.W., Yuen T.T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C.Y., Tsang J.O.L., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet. Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S., Yoon G.Y., Myoung J., Kim S.J., Ahn D.G. Robust and persistent SARS-CoV-2 infection in the human intestinal brush border expressing cells. Emerg. Microbes Infect. 2020;9:2169–2179. doi: 10.1080/22221751.2020.1827985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung M.L., Teng J.L.L., Jia L., Zhang C., Huang C., Cai J.P., Zhou R., Chan K.H., Zhao H., Zhu L., et al. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell. 2021;184:2212–2228.e12. doi: 10.1016/j.cell.2021.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Connolly P.F., Fearnhead H.O. Viral hijacking of host caspases: an emerging category of pathogen-host interactions. Cell Death Differ. 2017;24:1401–1410. doi: 10.1038/cdd.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S., Zhang Y., Guan Z., Li H., Ye M., Chen X., Shen J., Zhou Y., Shi Z.L., Zhou P., Peng K. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 2020;5:235. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 50.Shiffman M.L., Pockros P., McHutchison J.G., Schiff E.R., Morris M., Burgess G. Clinical trial: the efficacy and safety of oral PF-03491390, a pancaspase inhibitor - a randomized placebo-controlled study in patients with chronic hepatitis C. Aliment. Pharmacol. Ther. 2010;31:969–978. doi: 10.1111/j.1365-2036.2010.04264.x. [DOI] [PubMed] [Google Scholar]
- 51.Apaydın Ç.B., Çınar G., Cihan-Üstündağ G. Small-molecule antiviral agents in ongoing clinical trials for COVID-19. Curr. Drug Targets. 2021;22:1986–2005. doi: 10.2174/1389450122666210215112150. [DOI] [PubMed] [Google Scholar]
- 52.Simonis A., Theobald S.J., Fätkenheuer G., Rybniker J., Malin J.J. A comparative analysis of remdesivir and other repurposed antivirals against SARS-CoV-2. EMBO Mol. Med. 2021;13:e13105. doi: 10.15252/emmm.202013105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Banga Ndzouboukou J.L., Gan M., Lin X., Fan X. Immune evasive effects of SARS-CoV-2 variants to COVID-19 emergency used vaccines. Front. Immunol. 2021;12:771242. doi: 10.3389/fimmu.2021.771242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X.P., Gan H.Y., Li F.X., Tian Q., Zhang J., Liang R.L., Li M., Yang X.X., Wu Y.S. A method to quantify cell-free fetal DNA fraction in maternal plasma using next generation sequencing: its application in non-invasive prenatal chromosomal aneuploidy detection. PLoS One. 2016;11:e0146997. doi: 10.1371/journal.pone.0146997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shahinozzaman M., Basak B., Emran R., Rozario P., Obanda D.N. Artepillin C: a comprehensive review of its chemistry, bioavailability, and pharmacological properties. Fitoterapia. 2020;147:104775. doi: 10.1016/j.fitote.2020.104775. [DOI] [PubMed] [Google Scholar]
- 56.Jamaly S., Tsokos M.G., Bhargava R., Brook O.R., Hecht J.L., Abdi R., Moulton V.R., Satyam A., Tsokos G.C. Complement activation and increased expression of Syk, mucin-1 and CaMK4 in kidneys of patients with COVID-19. Clin. Immunol. 2021;229:108795. doi: 10.1016/j.clim.2021.108795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shang C., Zhuang X., Zhang H., Li Y., Zhu Y., Lu J., Ge C., Cong J., Li T., Li N., et al. Inhibition of autophagy suppresses SARS-CoV-2 replication and ameliorates pneumonia in hACE2 transgenic mice and xenografted human lung tissues. J. Virol. 2021;95:e0153721. doi: 10.1128/JVI.01537-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agrawal P., Sambaturu N., Olgun G., Hannenhalli S. A path-based analysis of infected cell line and COVID-19 patient transcriptome reveals novel potential targets and drugs against SARS-CoV-2. Front. Immunol. 2022;13:918817. doi: 10.3389/fimmu.2022.918817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basile M.S., Cavalli E., McCubrey J., Hernández-Bello J., Muñoz-Valle J.F., Fagone P., Nicoletti F. The PI3K/Akt/mTOR pathway: a potential pharmacological target in COVID-19. Drug Discov. Today. 2022;27:848–856. doi: 10.1016/j.drudis.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pacold M.E., Brimacombe K.R., Chan S.H., Rohde J.M., Lewis C.A., Swier L.J.Y.M., Possemato R., Chen W.W., Sullivan L.B., Fiske B.P., et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamanaka R.B., Nigdelioglu R., Meliton A.Y., Tian Y., Witt L.J., O'Leary E., Sun K.A., Woods P.S., Wu D., Ansbro B., et al. Inhibition of phosphoglycerate dehydrogenase attenuates bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2018;58:585–593. doi: 10.1165/rcmb.2017-0186OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang C., Li W., Lei X., Xie Z., Qi L., Wang H., Xiao X., Xiao J., Zheng Y., Dong C., et al. Targeting lysophospholipid acid receptor 1 and ROCK kinases promotes antiviral innate immunity. Sci. Adv. 2021;7:eabb5933. doi: 10.1126/sciadv.abb5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gutierrez-Chamorro L., Felip E., Ezeonwumelu I.J., Margelí M., Ballana E. Cyclin-dependent kinases as emerging targets for developing novel antiviral therapeutics. Trends Microbiol. 2021;29:836–848. doi: 10.1016/j.tim.2021.01.014. [DOI] [PubMed] [Google Scholar]
- 64.Hahn F., Hamilton S.T., Wangen C., Wild M., Kicuntod J., Brückner N., Follett J.E.L., Herrmann L., Kheimar A., Kaufer B.B., et al. Development of a PROTAC-based targeting strategy provides a mechanistically unique mode of anti-cytomegalovirus activity. Int. J. Mol. Sci. 2021;22:12858. doi: 10.3390/ijms222312858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arlt B., Mastrobuoni G., Wuenschel J., Astrahantseff K., Eggert A., Kempa S., Deubzer H.E. Inhibiting PHGDH with NCT-503 reroutes glucose-derived carbons into the TCA cycle, independently of its on-target effect. J. Enzyme Inhib. Med. Chem. 2021;36:1282–1289. doi: 10.1080/14756366.2021.1935917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 67.Mulay A., Konda B., Garcia G., Jr., Yao C., Beil S., Villalba J.M., Koziol C., Sen C., Purkayastha A., Kolls J.K., et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021;35:109055. doi: 10.1016/j.celrep.2021.109055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geeraerts S.L., Heylen E., De Keersmaecker K., Kampen K.R. The ins and outs of serine and glycine metabolism in cancer. Nat. Metab. 2021;3:131–141. doi: 10.1038/s42255-020-00329-9. [DOI] [PubMed] [Google Scholar]
- 69.Pajak B., Siwiak E., Sołtyka M., Priebe A., Zieliński R., Fokt I., Ziemniak M., Jaśkiewicz A., Borowski R., Domoradzki T., Priebe W. 2-Deoxy-d-Glucose and its analogs: from diagnostic to therapeutic agents. Int. J. Mol. Sci. 2019;21:234. doi: 10.3390/ijms21010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bojkova D., Costa R., Reus P., Bechtel M., Jaboreck M.C., Olmer R., Martin U., Ciesek S., Michaelis M., Cinatl J., Jr. Targeting the pentose phosphate pathway for SARS-CoV-2 therapy. Metabolites. 2021;11:699. doi: 10.3390/metabo11100699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang B.S.F., Chan K.H., Cheng V.C.C., Woo P.C.Y., Lau S.K.P., Lam C.C.K., Chan T.L., Wu A.K.L., Hung I.F.N., Leung S.Y., Yuen K.Y. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 2005;79:6180–6193. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshikawa T., Hill T.E., Yoshikawa N., Popov V.L., Galindo C.L., Garner H.R., Peters C.J., Tseng C.T.K. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One. 2010;5:e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Michaelis M., Doerr H.W., Cinatl J., Jr. Investigation of the influence of EPs® 7630, a herbal drug preparation from Pelargonium sidoides, on replication of a broad panel of respiratory viruses. Phytomedicine. 2011;18:384–386. doi: 10.1016/j.phymed.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramani R., Laplante J.M., Church T.M., Farrell G.M., Lamson D.M., St George K. CACO-2 cells: a continuous cell line with sensitive and broad-spectrum utility for respiratory virus culture. J. Virol. Methods. 2021;293:114120. doi: 10.1016/j.jviromet.2021.114120. [DOI] [PubMed] [Google Scholar]
- 75.Feichtinger J., Hernández I., Fischer C., Hanscho M., Auer N., Hackl M., Jadhav V., Baumann M., Krempl P.M., Schmidl C., et al. Comprehensive genome and epigenome characterization of CHO cells in response to evolutionary pressures and over time. Biotechnol. Bioeng. 2016;113:2241–2253. doi: 10.1002/bit.25990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ben-David U., Siranosian B., Ha G., Tang H., Oren Y., Hinohara K., Strathdee C.A., Dempster J., Lyons N.J., Burns R., et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y., Mi Y., Mueller T., Kreibich S., Williams E.G., Van Drogen A., Borel C., Frank M., Germain P.L., Bludau I., et al. Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat. Biotechnol. 2019;37:314–322. doi: 10.1038/s41587-019-0037-y. [DOI] [PubMed] [Google Scholar]
- 78.White J.M., Schiffer J.T., Bender Ignacio R.A., Xu S., Kainov D., Ianevski A., Aittokallio T., Frieman M., Olinger G.G., Polyak S.J. Drug combinations as a first line of defense against coronaviruses and other emerging viruses. mBio. 2021;12:e0334721. doi: 10.1128/mbio.03347-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sahu K.K., Kumar R. Role of 2-Deoxy-D-Glucose (2-DG) in COVID-19 disease: a potential game-changer. J. Family Med. Prim. Care. 2021;10:3548–3552. doi: 10.4103/jfmpc.jfmpc_1338_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Wetering S., van der Linden A.C., van Sterkenburg M.A., de Boer W.I., Kuijpers A.L., Schalkwijk J., Hiemstra P.S. Regulation of SLPI and elafin release from bronchial epithelial cells by neutrophil defensins. Am. J. Physiol. Lung Cell Mol. Physiol. 2000;278:L51–L58. doi: 10.1152/ajplung.2000.278.1.L51. [DOI] [PubMed] [Google Scholar]
- 81.Breckwoldt K., Letuffe-Brenière D., Mannhardt I., Schulze T., Ulmer B., Werner T., Benzin A., Klampe B., Reinsch M.C., Laufer S., et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017;12:1177–1197. doi: 10.1038/nprot.2017.033. [DOI] [PubMed] [Google Scholar]
- 82.Kleine M., Riemer M., Krech T., DeTemple D., Jäger M.D., Lehner F., Manns M.P., Klempnauer J., Borlak J., Bektas H., Vondran F.W.R. Explanted diseased livers - a possible source of metabolic competent primary human hepatocytes. PLoS One. 2014;9:e101386. doi: 10.1371/journal.pone.0101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lewandowski K., Xu Y., Pullan S.T., Lumley S.F., Foster D., Sanderson N., Vaughan A., Morgan M., Bright N., Kavanagh J., et al. Metagenomic nanopore sequencing of influenza virus direct from clinical respiratory samples. J. Clin. Microbiol. 2019;58 doi: 10.1128/JCM.00963-19. e00963-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/yzbfm4v7y7.1.
-
•
This paper does not report original code. All data reported in this paper will be shared by the lead contact (Jindrich Cinatl, cinatl@em.uni-frankfurt.de) upon request. Any additional information required to reanalyze the data reported in this paper is also available from the lead contact upon request.
-
•
Data on the SARS-CoV-2 isolates is available from GenBank: D614 (SARS-CoV-2/FFM1, GenBank: MT358638), G614 (SARS-CoV-2/FFM7, GenBank: MT358643), Alpha (SARS-CoV-2/FFM-UK7931/2021, GenBank: MZ427280), Beta (SARS-CoV-2/FFM-ZAF1/2021, GenBank: MW822592), Delta (SARS-CoV-2/FFM-IND8424/2021, GenBank: MZ315141), Zeta (SARS-CoV-2/FFMBRA1/2021, GenBank: MW822593), Omicron (SARS-CoV-2/FFM-SIM0550/2021, GenBank: OL800702).