Abstract
Short-term or long-term exposure to fine particulate matter (PM2.5) is related to increased incidences of respiratory diseases. This study aimed to investigate the influences of omega-3 polyunsaturated fatty acids (ω-3 PUFAs) supplementation on oxidative stress, inflammation, lung metabolic profile, and gut microbiota in PM2.5-induced lung injury mice. Mice were divided into four groups (n = 15, per group): two unsupplemented groups, control group and PM2.5 group, and two supplemented groups with ω-3 PUFAs, ω-3 PUFAs group, and ω-3 PUFAs + PM2.5 group. Mice in the supplemented groups were placed on an ω-3 PUFAs-enriched diet (ω-3 PUFAs, 21 g/kg). During the 5th to 6th week of dietary supplementation, mice were exposed to PM2.5 by intra-tracheal instillation. ω-3 PUFAs ameliorate lung histopathological injury, reduce inflammatory responses and oxidative stress, affect lung metabolite profile, and modulate gut microbiota in PM2.5-induced lung injury mice. Thus, supplementary ω-3 PUFAs showed effectiveness in attenuation of PM2.5-induced lung injury, indicating that the interventions exhibited preventive and therapeutic potential.
Supplementary information
The online version contains supplementary material available at 10.1007/s11356-022-25111-0.
Keywords: Fine particulate matter, Omega-3 polyunsaturated fatty acids, Oxidative stress, Metabolic profile, Gut microbiota, Lung injury
Introduction
Fine particulate matter (PM2.5) is one of the major global air pollutants and has increasingly become a great threat to public health in recent years (Manisalidis et al. 2020). Some studies have reported that PM2.5 can easily reach the end of the respiratory tract and deposit in the alveoli, inducing adverse lung injury (Kirrane et al. 2019). Compared to other systems, the respiratory system is more susceptible to being the primary target of the toxicological effect of PM2.5 (Liu et al. 2021). Numerous epidemiological investigations have indicated that short-term or long-term exposure to PM2.5 is related to increased incidences of respiratory diseases, such as chronic obstructive pulmonary disease (COPD) (Hart et al. 2018), asthma (Williams et al. 2019), and even lung cancer (Wei et al. 2020). Although the potential biological mechanisms related to PM2.5 have not been completely elucidated, inflammatory responses and oxidative stress are considered to be crucial for PM2.5-induced lung injury (Feng et al. 2016).
Omega-3 polyunsaturated fatty acids (PUFAs) found in fish oil are beneficial to human health (Itariu et al. 2012). Eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), as the potent components of ω-3 PUFAs, play significant roles in anti-inflammatory, antioxidant, and immune-modulation, which are extensively utilized as nutritional supplements (Lobo et al. 2016). A recent study has also shown that ω-3 PUFAs administered parenterally considerably alleviate PM2.5-induced lung injury by reducing inflammation and anti-oxidative activity (Li et al. 2019). However, the exact mechanisms for ω-3 PUFAs inhibition of PM2.5-induced lung injury have not been fully understood.
More than 1014 bacterial species make up the gut microbiota, which is involved in homeostatic functions such as digestion, nutrition absorption, immune system activation, and development (Palm et al. 2015; Parekh et al. 2015). Changes in the structure of the gut microbiota, particularly change in microbial diversity and abundance, have a significant impact on human health and disease. Environmental exposures and dietary structure are considered to be important determinants of the composition of the gut microbiota, which in turn can impact health (Palmer et al. 2007; Wu et al. 2011). Studies demonstrated that PM2.5 exposure could alter gut microbial composition and function through either promoting or inhibiting the growth of certain microorganisms (Gao et al. 2017). The gastrointestinal tract is readily exposed to air pollutants through inhalation, especially smaller particles such as PM2.5. Following inhalation, PM2.5 is deposited in the bronchiolar and alveolar spaces, where they are engulfed by alveolar macrophages. Particles are cleared from the lungs by mucociliary transport toward the oropharynx and are subsequently swallowed (Bailey et al. 2020). Furthermore, ω-3 PUFAs have been reported to alter the composition of the gut microbiota (Watson et al. 2018; Gui et al. 2019), which restore the gut microecosystem and reduce inflammation (Caesar et al. 2015; Zhuang et al. 2018). On the one hand, increasing evidence indicates that there is a vital interaction between gut microbiota and the lung, which is called the “gut-lung axis.” The gut microbiota may influence the lung's immunological status through the gut-lung axis (Keely et al. 2012), while lung inflammation may alter the gut microbiota via the lung-gut axis (Dumas et al. 2018). On the other hand, the diet-gut-physiology axis is of increasing interest to researchers. Changing diet structure to shape the gut microbiota has emerged as a potential treatment for illness alleviation (McKenzie et al. 2017).
Up to now, few studies have investigated the influences of ω-3 PUFAs supplementation on oxidative stress, inflammation, lung metabolic profile, and gut microbiota in PM2.5-induced lung injury mice. We assumed that ω-3 PUFAs supplementation may show protective effects against PM2.5 exposure induced lung injury in mice by impacting the gut microbiota and lung metabolic profile. Therefore, the primary goal of this study is to assess the effectiveness of ω-3 PUFAs supplementation on PM2.5-induced lung injury. The secondary aim was to investigate the effects of ω-3 PUFAs supplementation on the gut microbiome and the changes in the lung metabolic profile by the untargeted LC–MS metabolomics and 16S rRNA gene sequencing method, which provide the potential pharmacological mechanism and a novel strategy to ameliorate lung injury caused by PM2.5 exposure.
Methods
Animals and diets
Before the experiments, male C57BL/6 N mice (Shanghai SLAC Laboratory Animal Co., Ltd) group-housed (five mice/cage) with temperature (22–23 °C) and humidity controlled (60 ± 5%), in a 12-h light/dark cycle (light from 6:00 a.m.) with ad libitum access to drinking water and standard chow diet (American Institute of Nutrition (AIN)-93G diets) for a 1-week acclimatization period. All animal procedures performed were approved by the Animal Ethical Committee of Shaoxing People’s Hospital and followed the Guide for the Care and Use of Laboratory Animals of National Administration Regulations on Laboratory Animals of China. All attempts were made to minimize animal suffering and the number of animals required for the required studies.
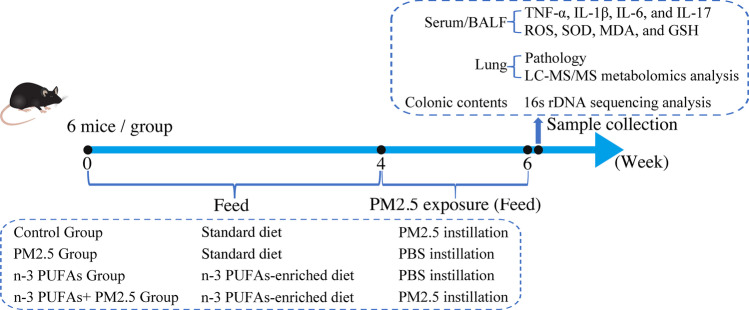
The sample size was determined based on previous experiments and the literature. Mice were randomly divided into four groups (n = 15, per group): two unsupplemented groups, control group and PM2.5 group, and two supplemented groups with ω-3 PUFAs, ω-3 PUFAs group, and ω-3 PUFAs + PM2.5 group. Mice in the two unsupplemented groups continued to be fed the standard chow diet, whereas the supplemented groups were placed on an ω-3 PUFAs-enriched diet for 6 weeks. The dietary intervention duration was chosen based on our preliminary experiment and the published literatures (Li et al. 2019; Huang et al. 2020). The two diets were only differed only in oil composition, whereas other macronutrients and total energy content were kept the same (Table 1). Briefly, the ω-3 PUFAs-enriched diet (ω-3 PUFAs, 21 g/kg) was made with fish oil containing EPA/DHA (3:2 ratio) instead of soybean oil. To avoid fatty acid oxidation, both diets were kept at − 20 °C in sealed plastic bags filled with nitrogen. A fresh aliquot of the diet was supplied every day, and feeds offered and refused were daily recorded, and then the data were used for calculations of feed consumption. Recorded data indicated that there were no significant differences in food intake were noted between the diet groups. The dietary intervention was carried out for 6 weeks and body weight was measured weekly.
Table 1.
Composition of experimental diets
| Ingredient (g/kg) | Control diet | ω-3 PUFA-supplemented diet |
|---|---|---|
| Casein | 200 | 200 |
| L-Cystine | 3 | 3 |
| Corn Starch | 397 | 397 |
| Maltodextrin | 132 | 132 |
| Sucrose | 100 | 100 |
| Cellulose | 50 | 50 |
| Soybean Oil | 70 | 0 |
| Fish Oil | 0 | 70 |
| t-Butylhydroquinone | 0.014 | 0.014 |
| Mineral Mix | 35 | 35 |
| Vitamin Mix | 10 | 10 |
| Choline Bitartrate | 2.5 | 2.5 |
PM2.5 exposure
PM2.5 used in the mice instillation experiment was purchased from the Standard Reference Materials and was certified by the National Institute of Standards and Technology (SRM 1649b; Urban Dust; National Institute of Standards and Technology; U.S. Department of Commerce: Gaithersburg, MD). The detailed particulate material composition is listed in Supplementary Table S1. Prior to instillation, the PM2.5 powder was dissolved in PBS to make solutions of 5.4 mg/cm3.
During the 5th to 6th week of dietary supplementation, mice were exposed to PBS or PM2.5 by intra-tracheal instillation on day 1, day 5, day 9, and day 13 (Fig. 1). We selected this protocol because it has been shown to induce lung histopathological injury, cause inflammatory responses, and perturb lung metabolic profiles in mice in our previous study (Li et al. 2020a). The PM2.5 instillation dose was 5.4 mg/kg, which was determined by our previous research (Li et al. 2020a). To reduce agglomeration, the PM2.5 suspension was sonicated for 10 min prior to instillation. The remaining control groups received an equivalent volume of PBS.
Fig. 1.
Schematic diagram of experimental design
Sample preparation
Following completion of exposure to PM2.5 for two weeks as described above, the mice were sacrificed at 24 h after the final intra-tracheal instillation of PM2.5 suspension.
The trachea was cannulated and the lungs were lavaged with phosphate-buffered saline (6 ml, 2 ml per time). The recovery BALFs were nearly 80% of the volume instilled. BALFs were pooled and centrifuged for 20 min at 1500 rpm at 4 °C. The supernatant was collected in a new Eppendorf tube and frozen at − 80 °C for cytokine measurement later.
The blood of mice was taken and centrifuged for 20 min at 1500 rpm at 4 °C to extract serum. The lung tissues of non-lavaged mice were harvested using sterile instruments and cleaned with phosphate-buffered saline at 4 °C, blood removed, and the tissues dried with filter paper. A portion of the lung was immediately fixed in 4% formaldehyde for further histological analyses. Serum and other lung tissue samples were kept at − 80 °C until follow-up analysis.
In addition, colon content samples (0.5 g weight per mouse) were also collected and snap-frozen in liquid nitrogen, and then stored at − 80 °C for later analysis of gut microbiome composition in feces.
Histological analysis
The lung tissue samples were fixed in 4% formaldehyde and embedded in paraffin before being sliced into 5 µm thick slices. Hematoxylin and eosin (H&E) were used to stain the tissue slices, and morphological alterations were assessed.
Enzyme-linked immunosorbent assay for inflammatory factors
TNF-α, IL-1β, IL-6, and IL-17 levels in the BALF and serum were determined using commercially available ELISA kits (Nanjing Jiancheng Biotechnology Institute, Nanjing, China) according to the manufacturer’s instruction.
Determination of oxidative stress
The activity of reactive oxygen species (ROS), total-superoxide dismutase (T-SOD), as well as malondialdehyde (MDA) and glutathione (GSH) levels were detected using commercial kits following manufacturer’s instructions (Nanjing Jiancheng Biotechnology Institute, Nanjing, China).
LC–MS/MS metabolomics analysis of lung tissue
The lung tissues were thawed on ice. Homogenize it (Jingxin, Shanghai) with 500 µL of ice-cold methanol/water (70%, v/v, Merck, USA) for 30 Hz for 2 min. Centrifuge the mixture at 12,000 rpm at 4 ℃ for 10 min (Eppendorf, Germany) and suck supernatant 400 µL into another new EP tube. Add 500 µL of ethyl acetate/methanol (1:3, v/v, Thermo Fisher, USA) into the original EP tube and centrifuge the mixture again. Merge the two supernatants and concentrate them. Then add 100 µL of 70% methanol–water (Merck, USA) into the dried product and perform ultrasonic treatment for 3 min. Finally, centrifuge it again, and suck 60 µL of supernatant for LC–MS/MS analysis.
All sample extracts were analyzed by the LC–MS/MS system (1290 Infinity ultra-high performance liquid chromatography, QTOF/MS-6545, Agilent, USA) followed machine orders. The analytical conditions were as follows, UPLC: column, Waters ACQUITY UPLC HSS T3 C18 (1.8 µm, 2.1 mm*100 mm); solvent system, water (0.1% formic acid): acetonitrile (0.1% formic acid); column temperature, 40 ℃; flow rate, 0.4 mL/min; injection volume, 2 μL; gradient program, 95:5 V/V at 0 min, 10:90 V/V at 11.0 min, 10:90 V/V at 12.0 min, 95:5 V/V at 12.1 min, 95:5 V/V at 14.0 min. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap mass spectrometer. Linear ion trap and triple quadrupole scans were acquired on the mass spectrometer system, equipped with an ESI Turbo Ion-Spray port, operating in positive ion and negative ion mode and controlled by Analyst 1.6.3 software (Sciex, USA). The major ESI source operation settings were as follows: voltage 250 and 1500 V (for positive and negative ion modes, respectively), gas temperature 325 °C, drying gas flow 8 L/min, sheath gas temperature 325 °C, sheath gas flow 11 L/min, nebulizer pressure gas 40 psi.
The original data file obtained by LC–MS analysis is converted into mzML format by ProteoWizard software (NET Framework 4.7.2, USA). Metabolic identification information was obtained by searching the laboratory’s self-built database (from Metware Biotechnology Co., Ltd., Wuhan, China) and integrating the public database and metDNA. Statistical analysis was carried out by the R program. Statistical analysis includes univariate analysis and multivariate analysis. Univariate statistical analysis includes Student’s t-test and variance multiple analysis. Multivariate statistical analysis includes principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA). Pathway enrichment analysis of differential metabolites was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg) database and MetaboAnalyst 5.0 (Montre al, QC, Canada), and significance of which was determined by the p-value.
16 s rDNA sequencing analysis of gut microbiota
Total genomic DNA was extracted from colonic contents (200 mg) by QIAamp DNA Stool Mini Kit (QIAGEN, German). Detailed information on DNA extraction, library preparation, and sequencing was previously described(Li et al. 2020a). Samples were sequenced on an Illumina NovaSeq platform according to the manufacturer’s recommendations (Illumina, USA).
Raw sequencing data were subjected to sequence processing and analysis using FLASH (V1.2.7), QIIME software (V1.9.1), UCHIME algorithm, and Uparse software (V7.0.1001). Sequences with > 97% similarity were assigned to the same operational taxonomic units (OTUs). Representative sequences were chosen for each OTU, and taxonomic data were then annotated for each representative sequence using the Silva Database (V123).
Alpha diversity is applied in analyzing the complexity of species diversity for a sample through 5 indices, including Chao1, ACE, Shannon, and Simpson, and all indices in our samples were calculated with QIIME. Beta diversity analysis was used to evaluate the differences of samples in species complexity, calculated by principle coordinates analysis (PCoA) and cluster analysis by QIIME. Linear discriminant analysis (LDA) effect size (LEfSe) analysis was applied to evaluate differential taxa between groups.
Statistical analysis
All the data were presented as the means ± SD. The GraphPad Prism (V8.0, La Jolla, CA, USA) was used for statistical analysis. Variables were checked for normality assumptions using Q–Q plot, skewness, kurtosis, and normality tests. An unpaired Student’s t-test was used to analyze the statistical significance between two groups. Two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison post-test was used to evaluate the statistical significance among three or more groups. The correlation between lung metabolome and fecal microbiome was analyzed using the Spearman correlation tests. Graphs were generated using the R software (Version 3.3.1). P-value < 0.05 was considered statistically significant.
Results
The body weight changes are not affected by the diets or the PM2.5 exposure
At the beginning of the study, the average body weight was 21.98 ± 0.18 g for the control group, 21.34 ± 0.25 g for the PM2.5 group, 21.48 ± 0.15 g for the ω-3 PUFAs group, and 21.65 ± 0.23 g for the ω-3 PUFAs + PM2.5 groups. At the end of the study, the average body weight was calculated as 23.26 ± 0.36 g, 22.85 ± 0.28 g, 22.46 ± 0.27 g, and 22.56 ± 0.38 g, respectively, without any significant difference between the groups. The body weights of the mice in different groups and changes during the experiment were summarized in Supplementary Table S2.
ω-3 PUFAs ameliorate PM2.5-induced lung histopathological injury
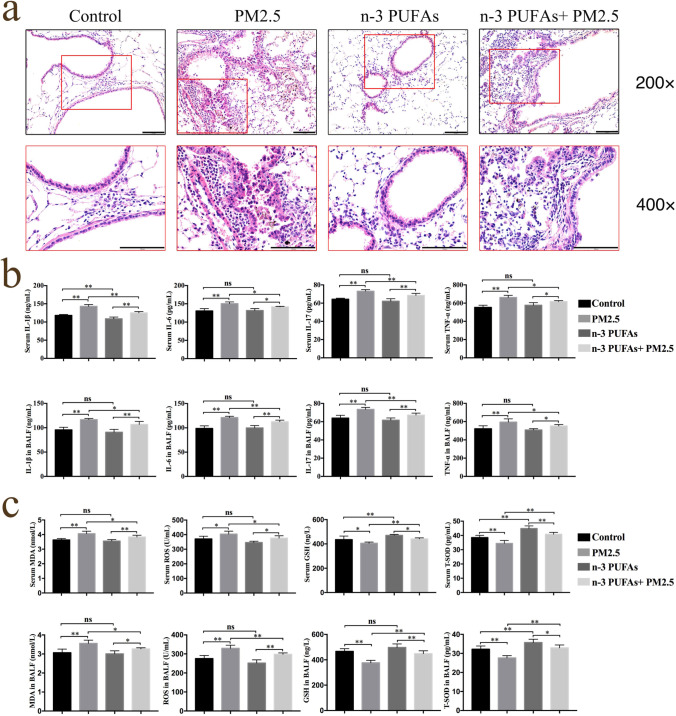
The alveolar size in the control and ω-3 PUFAs groups were generally normal, and capillaries in the lung tissue were not enlarged, according to light microscopy. Black foreign body particles were found in the alveolar cavity and bronchus, neutrophils were observed in the alveolar space, and the alveolar wall was thickened to some degree in the PM2.5 group. Some black particles in alveolar cavities and bronchi were seen and the alveolar structure was mostly complete in the ω-3 PUFAs + PM2.5 group. Compared with the PM2.5 group, the pathologic damage was milder in the ω-3 PUFAs + PM2.5 group, with less thickness of local alveolar septal and inflammatory cells. In summary, these results demonstrate that PM2.5 exposure induces significant inflammatory damage, which may be mitigated by ω-3 PUFA supplementation. (Fig. 2A).
Fig. 2.
Effect of PM2.5 exposure and ω-3 PUFAs on lung histopathological changes, inflammatory response and oxidative stress in mice. (a) Representative H&E-stained lung tissues from mice in four different groups. 200 × magnification and amplification areas of 400 × magnification is shown for the indicated areas. Scale bars, 100 μm. (b) The cytokines levels of IL‐1β, IL‐6, IL‐17, and TNF‐α in the serum and the BALF. (c) The level of MDA and ROS, and the activities of GSH and SOD activities in the serum and the BALF. Data are shown as mean ± SD. n = 6 per group. *p < 0.05 and **p < 0.01. IL‐1β, interleukin 1β. IL‐6, interleukin 6. IL‐17, interleukin 17. TNF‐α, tumor necrosis factor alpha. MDA, malondialdehyde. ROS, reactive oxygen species. GSH, glutathione. T-SOD, total-superoxide dismutase
ω-3 PUFAs reduce lung and systemic inflammatory responses in PM2.5-induced lung injury
The pro-inflammatory cytokines, including TNF-α, IL-1β, IL-6, and IL-17 from the BALF and serum were detected by ELISA. As shown in Fig. 2B, PM2.5 instillation significantly increased the levels of TNF-α, IL-1β, IL-6, and IL-17 (p < 0.05), while ω-3 PUFAs supplementation could mitigate their secretion. No significant difference was observed between the control group and the ω-3 PUFAs group (p > 0.05). These results suggest that ω-3 PUFAs supplementation reduces inflammation levels of the BALF and serum in PM2.5-induced lung injury mice.
ω-3 PUFAs reduce systemic and lung oxidative stress in PM2.5-induced lung injury
In order to determine the potential role of ω-3 PUFAs in relieving oxidative stress, we examined the concentration of ROS, the oxidant, and MDA, the end product of lipid oxidation in serum and BALF. As shown in Fig. 2C, compared with the control group, the ROS and MDA concentration was remarkably increased by PM2.5 exposure (p < 0.05) but decreased in the ω-3 PUFAs + PM2.5 group (p < 0.05). Additionally, we detected the level of antioxidant GSH and antioxidant enzyme T-SOD in serum and BALF. The results showed that the content of GSH and T-SOD in mice exposed to PM2.5 were decreased compared to the control group (p < 0.05), and ω-3 PUFAs supplementation significantly increased the GSH and T-SOD levels compared to the PM2.5 group (p < 0.05). These results suggest that ω-3 PUFAs supplementation alleviates oxidative stress in PM2.5-induced lung injury mice.
ω-3 PUFAs affect lung metabolite profile in PM2.5-induced lung injury
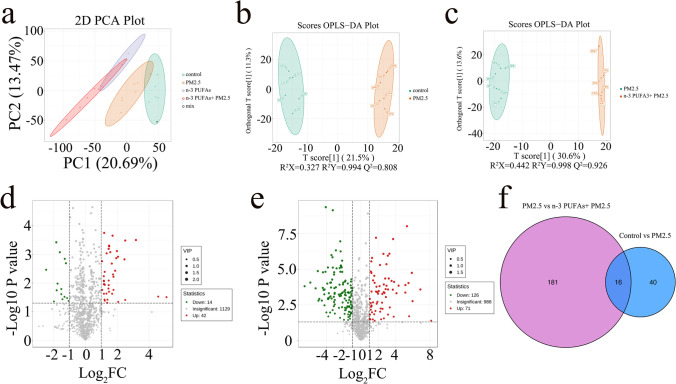
To comprehensively illustrate mechanisms underlying the amelioration effects of ω-3 PUFAs on PM2.5-induced lung injury, lung samples were analyzed using LC–MS/MS in the positive ion mode. To examine the systemic alterations of the metabolome in PM2.5-induced lung injury mice supplemented with ω-3 PUFAs, principal component analysis (PCA) was used to investigate the tendency of metabolic profiles between the groups. The PCA revealed that the major metabolic components were significantly different across the four groups, with 20.69% and 13.47% variation explained by the principle components PC1 and PC2, which suggested that both PM2.5 exposure and ω-3 PUFAs diet altered metabolic profiles (Fig. 3A). Therefore, we used orthogonal partial least squares discriminant analysis (OPLS-DA) to further visualize the metabolic alterations occurring between the control group and the PM2.5 group as well as between the PM2.5 group and the ω-3 PUFAs + PM2.5 group. The OPLS-DA score plot likewise exhibited a clear separation between the control and PM2.5 groups, and between the PM2.5 and ω-3 PUFAs + PM2.5 groups, which showed high goodness of fit and high prediction ability (R2X = 0.327, R2Y = 0.994, Q2 = 0.808; R2X = 0.442, R2Y = 0.998, Q2 = 0.826, respectively) (Fig. 3B and C). Volcano plots may be applied to rapidly screen differential metabolites based on the p-values < 0.05, and fold-change values ≥ 2 or ≤ 0.5. According to the volcano plots between the control group and the PM2.5 group, 42 metabolites were significantly upregulated, and 14 metabolites were significantly downregulated (Fig. 3D). In the PM2.5 and ω-3 PUFAs + PM2.5 groups, there were 197 metabolites with significant differences, with 71 upregulated metabolites and 126 down-regulated metabolites (Fig. 3E). By showing the relationship between the two groups of differential metabolites in the form of a difference Venn diagram, we found 16 metabolites with common differences (Fig. 3F).
Fig. 3.
Effect of PM2.5 exposure and ω-3 PUFAs on metabolite profiles in mice lungs. (a) Global metabolite analysis based on PCA score plots from the different groups. (b) OPLS-DA score plots of the control and PM2.5 groups. (c) OPLS-DA score plots of the PM2.5 and ω-3 PUFAs + PM2.5 groups. (d) Volcano plot for analyses of differential metabolites in lung between the control and PM2.5 groups. (e) Volcano plot for analyses of differential metabolites in lung between the PM2.5 and ω-3 PUFAs + PM2.5 groups. (f) Venn diagram of differential metabolites. Control group vs PM2.5 group (blue) and PM2.5 group vs ω-3 PUFAs + PM2.5 group (purple). Each dot represents a specific metabolite. Fold change ≥ 2 and p < 0.05 are marked in red, fold change ≤ 0.5 and p < 0.05 are marked in green, and the rest are in grey. PCA, principal component analysis. OPLS-DA, orthogonal partial least squares discriminant analysis
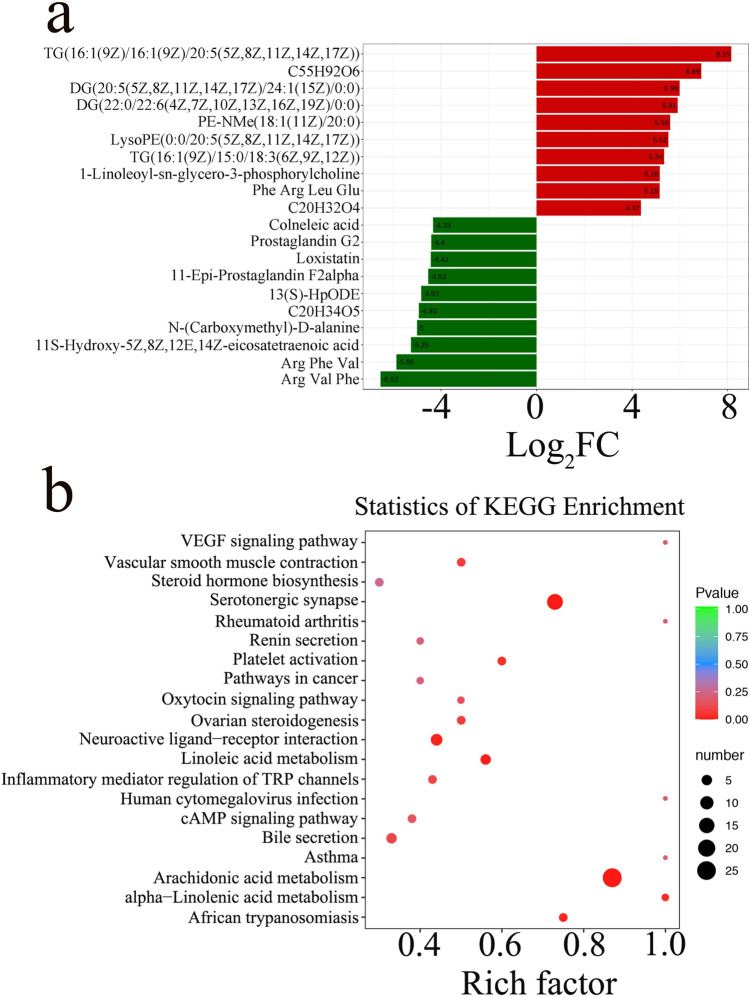
In addition, linear discriminate analysis effect size (LEfSe) was performed to determine the differential metabolites; the threshold of the LDA score was 4. The results with the changed metabolites between the PM2.5 and ω-3 PUFAs + PM2.5 groups were visualized by the metabolite difference bar plot (Fig. 4A). Key metabolites with significant differences between the PM2.5 and ω-3 PUFAs + PM2.5 groups were imported into the MetaboAnalyst 5.0 based on KEGG to determine the meaningful metabolic pathways. As shown in Fig. 4B, the enrichment analysis of metabolic pathways of the differential metabolites indicated that VEGF signaling pathway, asthma, arachidonic acid metabolism, and alpha − Linolenic acid metabolism were the main metabolic pathways altered by ω-3 PUFAs supplementation when compared against the PM2.5 group. These results suggested that ω-3 PUFAs did cause an effect on the lung metabolism in mice with PM2.5-induced lung injury, and these metabolic pathways might be related to the potential mechanism of ω-3 PUFAs in the treatment of PM2.5-induced lung injury.
Fig. 4.
Pathway analysis of differential metabolites. (a) LDA effect size analysis of top 20 differential metabolites between the PM2.5 and ω-3 PUFAs + PM2.5 groups. Significant increased metabolites are highlighted in red and decreased metabolites are in green (p < 0.05). (b) Enrichment analysis of metabolic pathways between the PM2.5 and ω-3 PUFAs + PM2.5 groups. The number of metabolites enriched in KEGG terms, p value, and rich factor are shown in scatterplot. LDA, Linear discriminate analysis. C55H92O6, [(2S)-3-[(Z)-hexadec-9-enoyl]oxy-2-[(6Z,9Z,12Z)-octadeca-6,9,12-trienoyl]oxypropyl](6Z,9Z,12Z)-octadeca-6,9,12-trienoate. C20H32O4, 8(R)-HPETE;(5Z,9E,11Z,14Z)-(8R)-8-Hydroperoxyeicosa-5,9,11,14-tetraenoate;(5Z,9E,11Z,14Z)-(8R)-8-Hydroperoxyicosa-5,9,11,14-tetraenoate. C20H34O5, (Z)-7-[(2R,3S)-3,5-dihydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]cyclopentyl]hept-5-enoic acid. KEGG, Kyoto encyclopedia of genes and genomes
ω-3 PUFAs modulate gut microbiota in PM2.5-induced lung injury
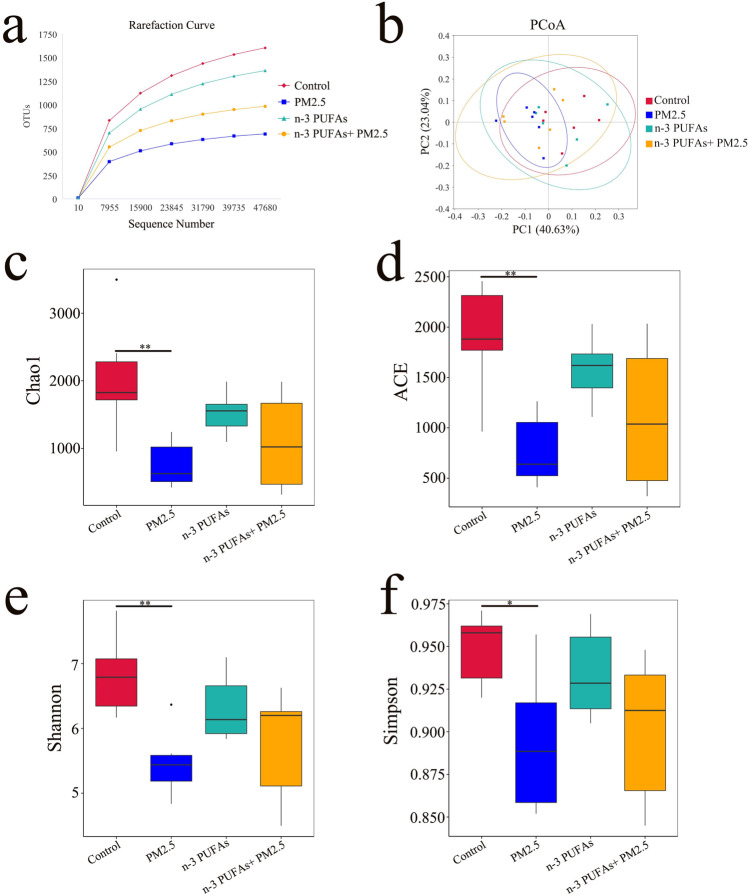
16S rRNA sequencing analysis was performed to explore PM2.5 exposure-associated differences in the microbiota community of the colonic contents and to investigate the effect of ω-3 PUFAs supplementation on the gut microbial ecology. The multiple sample rarefaction curves tended to be flat when the sequence number increased to 47,000, which indicated that the sequencing depth of each sample in this study is reasonable (Fig. 5A). The principal co-ordinates analysis (PCoA) with weighted UniFrac was performed to measure the phylogenetic similarities between microbial communities among different groups. The PCoA revealed that there is no significant separation in the gut microbiota community structure of the four mice groups, with PC1 explained by 40.63% and PC2 explained by 23.04%, respectively (Fig. 5B). Compared with the control group, the α-diversity indexes (including the Chao1, ACE, Shannon, and Simpson indexes) of the PM2.5 group were significantly decreased (p < 0.05 or < 0.01). The α-diversity indexes in the ω-3 PUFAs + PM2.5 group had an increasing tendency compared with the PM2.5 group, but no statistical difference (p > 0.05), which indicates that ω-3 PUFAs supplementation has no obvious effect on the richness and diversity of gut microbiota (Fig. 5C-F).
Fig. 5.
Effect of PM2.5 exposure and ω-3 PUFAs on the structure and the diversity of the gut microbiota. (a) Bacterial rarefaction curves based on observed species were used to assess the depth of coverage for all groups. (b) PCoA plot based on the relative abundance of phylum using UniFrac distance. Each point in the plot represented the gut microbiota of one mouse. (c-f) Alpha diversity indicating species abundance and diversity of microbiota among the different groups. Alpha diversity indexes including Chao1, ACE, Shannon, and Simpson. Data are shown as mean ± SD, analyzed by Wilcoxon rank sum test, n = 6 per group, *p < 0.05 and **p < 0.01. PCoA, Principal coordinate analysis
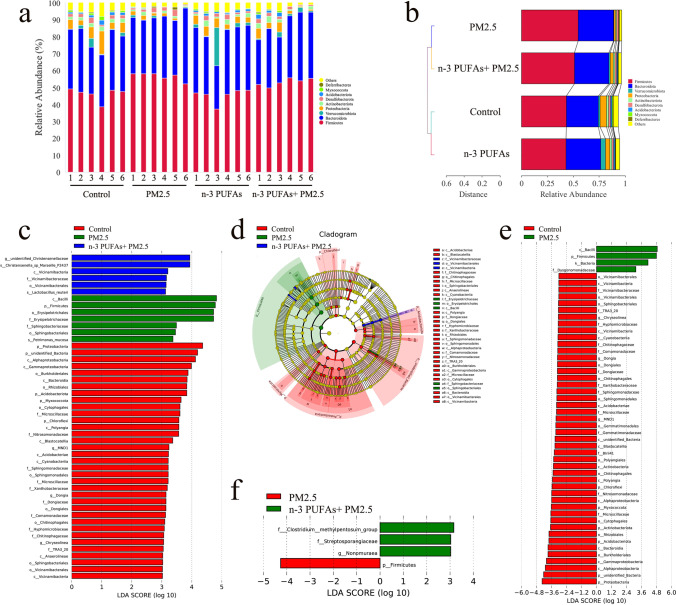
The degree of bacterial taxonomic similarity at the phylum level was analyzed to compare the overall composition of gut microbiota across the four groups. Firmicutes and Bacteroidota were the dominant phyla among these groups, with a total abundance of nearly 80% (Fig. 6A). PM2.5 exposure increased the relative abundance of Firmicutes, and Bacteroidota, and decreased the relative abundance of Verrucomicrobiota, Proteobacteria, Actinobacteriota, and Acidobacteriota. However, the changes in the fraction of Firmicutes, Bacteroidota, Verrucomicrobiota, Proteobacteria, Actinobacteriota, and Acidobacteriota were attenuated by ω-3 PUFAs supplementation (Fig. 6B). The linear discriminant analysis effect size (LEfSe) analysis, which highlights the statistical significance and biological correlation, was also conducted to explore the different bacteria among all the groups, with an LDA Score log 10 > 3. A total of 48 genera were in the control, PM2.5, ω-3 PUFAs, and ω-3 PUFAs + PM2.5 groups (Fig. 6C and D). The composition of gut bacteria showed significant changes in the control, PM2.5, and ω-3 PUFAs + PM2.5 groups, but not the ω-3 PUFAs group. In detail, p_Proteobacteria, c_Alphaproteobacteria, and c_Gammaproteobacteria were the main functional microbiota in the control group, while c_Bacilli, p_Firmicutes, o_Erysipelotrichales, and f_Erysipelotrichaceae were the main functional microbiota in the PM2.5 group. Notably, the bacteria associated with the ω-3 PUFAs + PM2.5 group included g_unidentified_Christensenellaceae and s_Christensenella_sp_Marseille_P2437. There were 50 significantly changed bacteria between the control group and PM2.5 groups (Fig. 6E). Differential microbial lineages of the PM2.5 group included c_Bacilli, p_Firmicutes, k_Bacteria, and f_Dysgonomonadaceae. The following bacteria were the most abundant in the control group: p_Proteobacteria, c_Alphaproteobacteria, and c_Gammaproteobacteria. In addition, a total of 4 bacteria changed significantly between the PM2.5 group and ω-3 PUFAs + PM2.5 groups (Fig. 6F). The microorganisms with statistical biomarkers of the PM2.5 group was p_Firmicutes, while the bacteria associated with the ω-3 PUFAs + PM2.5 group were f_Clostridium_methylpentosum_group, f_Streptosporangiaceae, and g_Nonomuraea. Collectively, these results demonstrated that ω-3 PUFAs supplementation modulates the gut microbiota of mice with PM2.5-induced lung injury.
Fig. 6.
Effect of PM2.5 exposure and ω-3 PUFAs on the composition of the gut microbiota. (a) Relative abundance of gut microbiota at the phylum level. (b) Weighted unifrac clustering analysis of the gut microbiota at the phylum level. (c) The LefSe was adopted to identify the bacterial groups that showed significant differences in abundance between the control, PM2.5, and ω-3 PUFAs + PM2.5 groups. (d) The cladogram of the gut microbiota of mice between the control, PM2.5, and ω-3 PUFAs + PM2.5 groups derived from LEfSe analysis. (e) Histogram of the LDA scores computed for features differentially abundant between the control and PM2.5 groups. (f) Histogram of the LDA scores computed for features differentially abundant between the PM2.5 and ω-3 PUFAs + PM2.5 groups. p, phylum; c, class; o, order; f, family; g, genus. The specific differential taxa that enriches in each group is characterized with different colors. LDA scores > 3 and significance of P < 0.05 as determined by Wilcoxon’s signed-rank test. LefSe, linear discriminant analysis effect size
Correlation analysis of gut microbiota and lung metabolites associated with ω-3 PUFAs supplementation in PM2.5-induced lung injury
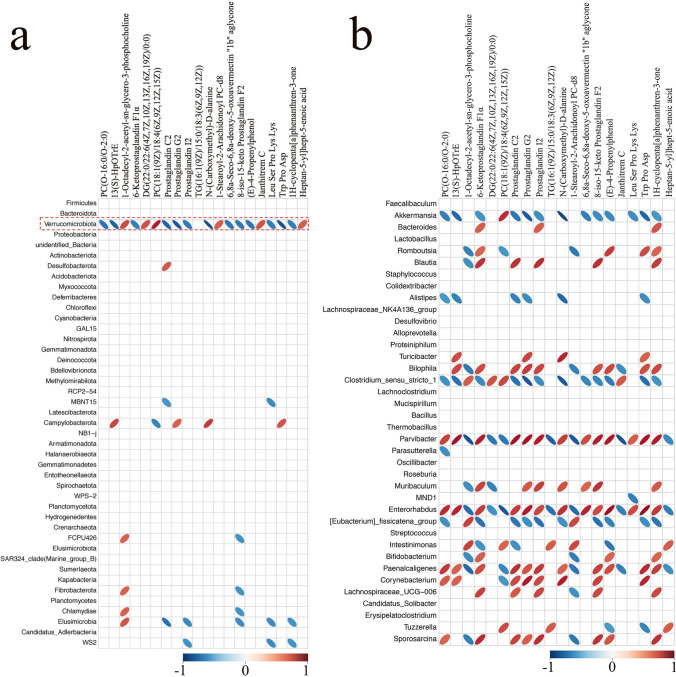
To comprehensively analyze the potential relationship between gut microbiota changes and lung metabolites, a correlation matrix was generated by calculating the Spearman’s rank correlation coefficient. As shown in Fig. 7A, there was a strong correlation between Verrucomicrobiota and significant difference metabolites. Verrucomicrobiota showed a positive correlation with PC(18:1(9Z)/18:4(6Z,9Z,12Z,15Z)) (r = 0.80, p < 0.01), 1-Octadecyl-2-acetyl-sn-glycero-3-phosphocholine (r = 0.65, p < 0.05), DG(22:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)/0:0) (r = 0.64, p < 0.05), and Janthitrem C (r = 0.64, p < 0.05), whereas Verrucomicrobiota was positively correlated with N-(Carboxymethyl)-D-alanine (r = − 0.91, p < 0.01), Prostaglandin G2 (r = − 0.85, p < 0.01), Trp Pro Asp (r = − 0.85, p < 0.01), 13(S)-HpOTrE (r = − 0.81, p < 0.01), and Prostaglandin C2 (r = − 0.80, p < 0.01). The correlation between gut microbiota changes at the genus level and the significant difference metabolites was further analyzed (Fig. 7B). Thus, these findings revealed the potential interactions between ω-3 PUFAs–modulated gut microbial species and lung metabolites.
Fig. 7.
Associations between altered gut microbial species and disturbed lung metabolites. The altered gut microbiota exhibit correlations with lung metabolic biomarkers (a, phylum; b, genus). Color red indicates that altered lung metabolites were positively correlated with perturbed gut microbiota. Color blue indicates that lung metabolites were negatively correlated with perturbed colon microbiota. The larger the absolute value of the correlation, the thinner the ellipse. Empty grids indicate significance P values greater than 0.05
Discussion
Epidemiological studies have shown that PM2.5 exposure is positively associated with an increased incidence of respiratory disease, and it plays a significant role in the occurrence and development of respiratory diseases (Luo et al. 2020). The gut-lung axis is a newly developed concept that involves bidirectional interactions and regulation between the two physiological compartments. Many evidences indicated that gut microbiota is related to human health and host immunity (Nie et al. 2019), whereas gut dysbiosis and its interactions with the gut-lung axis have a complicated impact on health and respiratory disorders (Saint-Criq et al. 2021). One of the potential mechanisms of adjunctive efficacy is the modulation of gut microbiota by specific dietary supplementation (Singh et al. 2017). ω-3 PUFAs supplementation has a therapeutic benefit in alleviating several lung disorders, including COPD (Wood 2015), asthma (Bisgaard et al. 2016), ARDS (Kohama et al. 2014), and even coronavirus disease COVID-19 (Safaei Ardestani & Rahideh 2022). However, the influence of ω-3 PUFAs supplementation on PM2.5 exposure induced lung injury remains unclear. Thus, the present study was designed to investigate whether ω-3 PUFAs supplementation could decrease PM2.5-induced lung injury. To our knowledge, this is the first study to explore the impact of ω-3 PUFAs supplementation on the interaction between gut microbiota and lung metabolic profile in mice with PM2.5-related lung injury using an integrated method of 16S rDNA gene sequencing and LC–MS/MS-based metabolomics.
Pro-inflammatory cytokines, which may drive macrophage migration, expand the inflammatory cascade, and aggravate lung damage, play an important role in the development of PM2.5-induced lung injury (Li et al. 2020b; Xu et al. 2020). In the present study, we found that PM2.5 exposure elevated the production of pro-inflammatory cytokines, such as IL-1β, IL-6, IL-17, and TNF-α in BALF and serum, indicating lung and systemic inflammation. These findings were consistent with previous studies. It has been reported that PM2.5 exposure promotes the production of pro-inflammatory cytokines such as IL- 1β, IL-6, IL-8, and TNF-α in airway epithelial cells, causing airway inflammation by stimulating lymphocytes, neutrophils, and antigen-presenting cells (Honda et al. 2021). Another study also reported that PM2.5 exposure up-regulated IL-17 expression, which aggravated neutrophil airway inflammation and mixed neutrophils/eosinophils inflammation in asthmatic mice (Yu et al. 2019). T helper 17 (Th17) cells, the third subgroup of Th cells, have been identified as the principal producers of IL-17, and Polycyclic aromatic hydrocarbons (PAHs) in PM2.5 may drive Th17-cell activation, which cause an increase in IL‐17 (Weaver et al. 2006; Schmidt-Weber et al. 2007; Veldhoen et al. 2008). In addition, the overproduction of ROS could induce the release of inflammatory cytokines, leading to lung damage, which is recognized as another key player in PM2.5-induced lung injury (Kim et al. 2017). In this study, exposure to PM2.5 increased the levels of MDA and ROS, and decreases the levels of GSH and SOD in the BALF and serum, indicating that PM2.5 exposure induces oxidative stress. The chemical components of PM2.5, such as metals, may play a role in the mechanisms underlying PM2.5-induced oxidative stress. Similar findings were observed that PM2.5 lowered the activity of antioxidant enzymes such as CAT and GSH-PX in alveolar macrophages while increasing the concentration of MDA, causing an imbalance in the pro-oxidant and antioxidant status (Liu et al. 2018). Therefore, PM2.5 may penetrate deep into the lower respiratory tract and adhere to lung epithelial cells or be phagocytosed by alveolar macrophages, inducing oxidative stress and inflammation, which are considered to be the key molecular mechanisms of PM2.5-induced lung injury.
Dietary ω-3 PUFAs are known to have anti-inflammatory and antioxidant capacities (Alnahdi and Sharaf 2019). Our results from this study showed that ω-3 PUFAs supplementation significantly alleviated inflammation and oxidative stress in PM2.5-induced lung injury mice. Consistent with this study, an in vitro experiment found that supplementing with ω-3 PUFAs reduce the inflammation and oxidative stress in vascular endothelial cells treated with PM2.5 (Bo et al. 2016). Previous researches also manifested that ω-3 PUFAs provide protection against lung injury induced by PM2.5 through improving antioxidant metabolism, reducing lipid peroxidation, and inhibition of pyrin domain-containing 3 (NLRP3) inflammasome activation (Samet et al. 2001; Li et al. 2019). The inflammasome could facilitate the maturation and release of some pro-inflammatory cytokines, while ROS is thought to be one of the main triggers of NLRP3 inflammasome activation (Tschopp and Schroder 2010). Thus, the protective effects of ω-3 PUFAs may be attributed to their capacity to scavenge ROS (Dasilva et al. 2017; Kucharská et al. 2021; Durán et al. 2022). Interestingly, we found that feeding with a high ω-3 PUFAs diet could reduce systemically IL-1β level and increase SOD and GSH levels in the ω-3 PUFAs groups compare to the control group. We argue that the administration of ω-3 PUFAs may have health benefits even in healthy individuals. These findings suggest a potential modulatory role of ω-3 PUFAs in protecting the lung from PM2.5 exposure. However, the specific mechanisms for ω-3 PUFAs inhibition of PM2.5-induced lung inflammation and oxidative stress have not been fully revealed, which require further in-depth study.
Metabolomics has made significant progress in helping to understand the pathogenesis of many diseases, which has demonstrated the usefulness of metabolite profiling (Johnson et al. 2016). In this study, alterations in endogenous small molecule metabolites were revealed in the UPLC-MS spectra of lung samples, which may provide light on the mechanism by which ω-3 PUFAs intervention ameliorate PM2.5-induced lung injury. Oral supplementary of ω-3 PUFAs obviously reduced lung levels of colneleic acid, prostagland G2, loxistatin, N-(carboxymethyl)-D-alanine, etc., and significantly increased lung levels of Phe Arg Leu Glu, 1-linoleoyl-sn-glycero-3-phosphorylcholine, etc. in PM2.5-induced lung injury mice. The metabolic pathway enrichment analysis of the differential lung metabolites indicated that ω-3 PUFAs supplementation alleviated PM2.5-induced lung injury by regulating pathways involved in VEGF signaling pathway, asthma, arachidonic acid metabolism, and alpha − Linolenic acid metabolism. Lin et al. found that PAHs-rich air pollution particles induced pro-inflammatory responses in arachidonic acid metabolism (Lin et al. 2022). It was observed that fish oil supplementation modulated genes involved in arachidonic acid metabolism (Lu et al. 2011; Magbanua et al. 2011). Additionally, EPA and DHA have been demonstrated to competitively inhibit arachidonic acid metabolism by cyclooxygenase and 5-lipoxygenase, which produce prostaglandins and leukotrienes implicated in inflammation (Calder 2006, 2020). These findings suggest that ω-3 PUFAs may exert antioxidant effects in PM2.5-induced lung injury mice by regulating arachidonic acid metabolism. Alpha-linolenic acid, as a precursor to ω-3 PUFAs, has been associated with a decrease in the production of pro-inflammatory cytokines (Monk et al. 2016). It has been reported that Alpha-linolenic acid exhibited a protective effect on lipopolysaccharide-induced acute lung injury through inhibiting NF-κB activation (Zhu et al. 2020). However, no study has yet reported the role of Alpha-linolenic acid in PM2.5-induced lung injury. Overall, these results might be helpful in future studies looking into the possible mechanisms of ω-3 PUFAs in PM2.5-induced lung injury.
Dysbiosis of the gut microbiota is believed to be critical for the pathophysiology of PM2.5-induced lung injury (Fouladi et al. 2020). Dietary intervention to alter gut microecology is a hot topic in science right now. In this study, we identified how dietary ω-3 PUFAs could modulate gut microbiota in PM2.5-induced lung injury mice. In general, a decrease in gut microbiota diversity is regarded as a less healthy condition, as it reflects a disruption of gut homeostasis, which is especially evident when external agents are taken, such as antibiotics and medications (Dudek-Wicher et al. 2018). In this study, analyses of the mice colon content microbiota α-diversity indicated that PM2.5 exposure induced a significant decrease in the gut microbiota diversity and richness, while the intervention effects of ω-3 PUFAs were not significant. The absence of a substantial change in α -diversity after ω-3 PUFA intervention is consistent with findings from another study (Robertson et al. 2017). However, Zhu et al. reported that the ω-3 PUFAs administration significantly increased the gut microbiota diversity and richness (Zhu et al. 2021). The reason for the inconsistency between these two results may be the regulation of ω-3 PUFAs on microbiota α-diversity needs to reach a certain dose. Dietary interventions have been seen as a critical element in modifying the structure of the gut microbiota (Bai et al. 2018). However, the results of PCoA in this study showed that the gut microbiota structure of the ω-3 PUFAs group and the ω-3 PUFAs + PM2.5 group was more similar to that in the control group, revealing that the alleviation effect was not due to a drastic shift in the gut microbiota structure. We proceeded to analyze the effects of ω-3 PUFAs on the relative abundance of gut bacteria. Firmicutes and Bacteroidota dominated at the phylum level in all four groups in this study, which was consistent with earlier research (Qin et al. 2010). The compositions of gut microbiota were found to be different significantly between the control and PM2.5 groups, and most of these differences were diminished by ω-3 PUFAs supplementation. At the phylum level, ω-3 PUFAs restore gut dysbiosis by upregulating Proteobacteria and Actinobacteriota, and by downregulating Firmicutes and Bacteroidota, revealing that dietary ω-3 PUFAs may attenuate gut dysbiosis presumably by modulating the gut microbiota composition. This finding is consistent with previous research that found a greater of Proteobacteria in mice given a high fish oil diet, indicating that the antioxidation of ω-3 PUFAs in fish oil might cause the increased abundance of Proteobacteria (Li et al. 2017). Moreover, our results are in agreement with a previous study on early-life stress, which reported that high-dose supplementation of ω-3 PUFAs elevated the levels of several members Actinobacteria (Pusceddu et al. 2015). Liu et al. indicated that ω-3 PUFAs increased the proportion of species of the Bacteroidetes phylum (Liu et al. 2012). However, it has been shown that ω-3 PUFAs derived from flaxseed seem to reduce the proportion of Bacteroidetes, which is consistent with our results. In addition, following treatment with ω-3 PUFAs, the decrease in the abundance of the Bacteroides was also shown to decrease in the IBD patients (Santoru et al. 2017). Furthermore, Yu et al. also indicated that ω-3 PUFAs from fish oil lower the population of Firmicutes in mice (Yu et al. 2014). We also performed a LEfSe analysis to detect the effect of ω-3 PUFAs on the gut microbiome was assessed by comparing specific bacteria. Our results showed that gut microbiota of the ω-3 PUFAs + PM2.5 group was mainly associated with increased levels of short-chain fatty acids (SCFAs) producing bacteria compared with the PM2.5 group, including f_Clostridium_methylpentosum_group, f_Streptosporangiaceae, and g_Nonomuraea. It is worth mentioning that SCFAs, as metabolites released by bacterial components of the gut microbiota, have been reported to possess anti-inflammatory activity (Akhtar et al. 2022). Together, our data support the expected beneficial effects of ω-3 PUFAs on restoring the composition of the gut microbiota of mice in PM2.5 exposure induced lung injury.
It is reported that the gut microbiota plays an important role in regulating the host’s metabolic pathways and in a variety of disease phenotypes (van Soest et al. 2020). In the present study, ω-3 PUFAs supplementation increased the abundance of Verrucomicrobiota in PM2.5-induced lung injury mice, which was associated with an increase in the levels of PC(18:1(9Z)/18:4(6Z,9Z,12Z,15Z)), 1-Octadecyl-2-acetyl-sn-glycero-3-phosphocholine, DG(22:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)/0:0), and Janthitrem C, and with a decrease in the levels of N-(Carboxymethyl)-D-alanine, Prostaglandin G2, Trp Pro Asp, 13(S)-HpOTrE, and Prostaglandin C2. Given the potential health advantages of ω-3 PUFAs, we proposed that the protective effect of ω-3 PUFAs on PM2.5-induced lung injury is also related to the modulation of the interaction between gut microbiota and these major differential metabolites. However, more evidence is required to understand better the possible molecular involvement of ω-3 PUFAs in PM2.5-induced lung injury.
Limitations should be also noted in our study. To begin with, the present research did not explore the potential antioxidant and anti-inflammatory effects of soybean polyphenols, nor did it investigate the components of ambient PM2.5 that caused these observed differences. Secondly, intratracheal instillation for PM2.5 exposure differs significantly from inhalation exposure in terms of toxicokinetics and toxicodynamics. We acknowledge that inhalation exposure may better reflect human exposure than intratracheal instillation. Thirdly, further investigation concerning the gut bacterial community should be conducted through shotgun metagenomics by providing more accurate composition, as well as functional information. Fourth, the pathway analysis was exploratory, with its inherent restriction of not considering the effect direction. Last but not least, the relationship between the key microbiota and differential metabolites in response to ω-3 PUFAs intervention needs further verification.
Conclusions
In summary, the present study suggested that supplementary ω-3 PUFAs showed effectiveness in attenuation of PM2.5-induced lung injury, indicating that the interventions exhibited preventive and therapeutic potential. This study further revealed that the protective effect of ω-3 PUFAs is associated with ameliorated inflammation and oxidative stress, restored lung metabolite profile, and attenuated gut dysbiosis. Our findings not only extend the understanding of PM2.5-induced lung injury but also provide a potential therapeutic strategy for PM2.5-induced lung injury.
Supplementary information
Below is the link to the electronic supplementary material.
Author contribution
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Jingli Li, Yang Chen, and Qiangqiang Shi. The first draft of the manuscript was written by Jingli Li. Previous versions of the manuscript were commented by Jian Sun and Chunyi Zhang. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Medical Science and Technology Project of Zhejiang Province (2022RC272, 2023RC289); Key Project of Young Research Fund of Shaoxing People's Hospital (2021YA08).
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Research involving human participants and/or animals
All animal procedures performed were approved by the Animal Ethical Committee of Shaoxing People’s Hospital and followed the Guide for the Care and Use of Laboratory Animals of National Administration Regulations on Laboratory Animals of China.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Akhtar M, Chen Y, Ma Z, Zhang X, Shi D, Khan JA, Liu H. Gut microbiota-derived short chain fatty acids are potential mediators in gut inflammation. Anim Nutr. 2022;8:350–360. doi: 10.1016/j.aninu.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnahdi HS, Sharaf IA. Possible prophylactic effect of omega-3 fatty acids on cadmium-induced neurotoxicity in rats' brains. Environ Sci Pollut Res Int. 2019;26:31254–31262. doi: 10.1007/s11356-019-06259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Zhu Y, Dong Y. Modulation of gut microbiota and gut-generated metabolites by bitter melon results in improvement in the metabolic status in high fat diet-induced obese rats. J Funct Foods. 2018;41:127–134. doi: 10.1016/j.jff.2017.12.050. [DOI] [Google Scholar]
- Bailey MJ, Naik NN, Wild LE, Patterson WB, Alderete TL. Exposure to air pollutants and the gut microbiota: a potential link between exposure, obesity, and type 2 diabetes. Gut Microbes. 2020;11:1188–1202. doi: 10.1080/19490976.2020.1749754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard H, Stokholm J, Chawes BL, Vissing NH, Bjarnadóttir E, Schoos AM, Wolsk HM, Pedersen TM, Vinding RK, Thorsteinsdóttir S, Følsgaard NV, Fink NR, Thorsen J, Pedersen AG, Waage J, Rasmussen MA, Stark KD, Olsen SF, Bønnelykke K. Fish oil-derived fatty acids in pregnancy and wheeze and asthma in Offspring. N Engl J Med. 2016;375:2530–2539. doi: 10.1056/NEJMoa1503734. [DOI] [PubMed] [Google Scholar]
- Bo L, Jiang S, Xie Y, Kan H, Song W, Zhao J. Effect of vitamin E and omega-3 fatty acids on protecting ambient PM2.5-induced inflammatory response and oxidative stress in vascular endothelial cells. PLoS One. 2016;11:e0152216. doi: 10.1371/journal.pone.0152216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR Signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder PC. Use of fish oil in parenteral nutrition: rationale and reality. Proc Nutr Soc. 2006;65:264–277. doi: 10.1079/pns2006500. [DOI] [PubMed] [Google Scholar]
- Calder PC (2020): N-3 PUFA and inflammation: from membrane to nucleus and from bench to bedside. Proc Nutr Soc, 1-13. 10.1017/s0029665120007077 [DOI] [PubMed]
- Dasilva G, Pazos M, García-Egido E, Gallardo JM, Ramos-Romero S, Torres JL, Romeu M, Nogués MR, Medina I. A lipidomic study on the regulation of inflammation and oxidative stress targeted by marine ω-3 PUFA and polyphenols in high-fat high-sucrose diets. J Nutr Biochem. 2017;43:53–67. doi: 10.1016/j.jnutbio.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Dudek-Wicher RK, Junka A, Bartoszewicz M. The influence of antibiotics and dietary components on gut microbiota. Prz Gastroenterol. 2018;13:85–92. doi: 10.5114/pg.2018.76005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas A, Bernard L, Poquet Y, Lugo-Villarino G, Neyrolles O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol. 2018;20:e12966. doi: 10.1111/cmi.12966. [DOI] [PubMed] [Google Scholar]
- Durán AM, Beeson WL, Firek A, Cordero-MacIntyre Z, De León M. Dietary omega-3 polyunsaturated fatty-acid supplementation upregulates protective cellular pathways in patients with type 2 diabetes exhibiting improvement in painful diabetic neuropathy. Nutrients. 2022;14:761. doi: 10.3390/nu14040761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Gao D, Liao F, Zhou F, Wang X. The health effects of ambient PM2.5 and potential mechanisms. Ecotoxicol Environ Saf. 2016;128:67–74. doi: 10.1016/j.ecoenv.2016.01.030. [DOI] [PubMed] [Google Scholar]
- Fouladi F, Bailey MJ, Patterson WB, Sioda M, Blakley IC, Fodor AA, Jones RB, Chen Z, Kim JS, Lurmann F, Martino C, Knight R, Gilliland FD, Alderete TL. Air pollution exposure is associated with the gut microbiome as revealed by shotgun metagenomic sequencing. Environ Int. 2020;138:105604. doi: 10.1016/j.envint.2020.105604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Chi L, Mahbub R, Bian X, Tu P, Ru H, Lu K. Multi-Omics Reveals that Lead Exposure Disturbs Gut Microbiome Development, Key Metabolites, and Metabolic Pathways. Chem Res Toxicol. 2017;30:996–1005. doi: 10.1021/acs.chemrestox.6b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L, Chen S, Wang H, Ruan M, Liu Y, Li N, Zhang H, Liu Z. ω-3 PUFAs alleviate high-fat diet-induced circadian intestinal microbes dysbiosis. Mol Nutr Food Res. 2019;63:e1900492. doi: 10.1002/mnfr.201900492. [DOI] [PubMed] [Google Scholar]
- Hart JE, Grady ST, Laden F, Coull BA, Koutrakis P, Schwartz JD, Moy ML, Garshick E. Effects of indoor and ambient black carbon and PM2.5 on pulmonary function among individuals with COPD. Environ Health Perspect. 2018;126:127008. doi: 10.1289/ehp3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Okuda T, Nagao M, Miyasaka N, Tanaka M, Takano H. PM2.5 collected using cyclonic separation causes stronger biological responses than that collected using a conventional filtration method. Environ Res. 2021;198:110490. doi: 10.1016/j.envres.2020.110490. [DOI] [PubMed] [Google Scholar]
- Huang LM, Hu Q, Huang X, Qian Y, Lai XH. Preconditioning rats with three lipid emulsions prior to acute lung injury affects cytokine production and cell apoptosis in the lung and liver. Lipids Health Dis. 2020;19:19. doi: 10.1186/s12944-019-1137-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itariu BK, Zeyda M, Hochbrugger EE, Neuhofer A, Prager G, Schindler K, Bohdjalian A, Mascher D, Vangala S, Schranz M, Krebs M, Bischof MG, Stulnig TM. Long-chain n-3 PUFAs reduce adipose tissue and systemic inflammation in severely obese nondiabetic patients: a randomized controlled trial. Am J Clin Nutr. 2012;96:1137–1149. doi: 10.3945/ajcn.112.037432. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely S, Talley NJ, Hansbro PM. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. 2012;5:7–18. doi: 10.1038/mi.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Choi MG, Park MK, Seo YR. Predictive and prognostic biomarkers of respiratory diseases due to particulate matter exposure. J Cancer Prev. 2017;22:6–15. doi: 10.15430/jcp.2017.22.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirrane EF, Luben TJ, Benson A, Owens EO, Sacks JD, Dutton SJ, Madden M, Nichols JL. A systematic review of cardiovascular responses associated with ambient black carbon and fine particulate matter. Environ Int. 2019;127:305–316. doi: 10.1016/j.envint.2019.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohama K, Nakao A, Terashima M, Aoyama-Ishikawa M, Shimizu T, Harada D, Nakayama M, Yamashita H, Fujiwara M, Kotani J. Supplementation of parenteral nutrition with fish oil attenuates acute lung injury in a rat model. J Clin Biochem Nutr. 2014;54:116–121. doi: 10.3164/jcbn.13-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharská J, Poništ S, Vančová O, Gvozdjáková A, Uličná O, Slovák L, Taghdisiesfejir M, Bauerová K. Treatment with coenzyme Q10, omega-3-polyunsaturated fatty acids and their combination improved bioenergetics and levels of coenzyme Q9 and Q10 in skeletal muscle mitochondria in experimental model of arthritis. Physiol Res. 2021;70:723–733. doi: 10.33549/physiolres.934664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhu Y, Zhao F, Song S, Li Y, Xu X, Zhou G, Li C. Fish oil, lard and soybean oil differentially shape gut microbiota of middle-aged rats. Sci Rep. 2017;7:826. doi: 10.1038/s41598-017-00969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li H, Li H, Guo W, An Z, Zeng X, Li W, Li H, Song J, Wu W. Amelioration of PM2.5-induced lung toxicity in rats by nutritional supplementation with fish oil and Vitamin E. Respir Res. 2019;20:76. doi: 10.1186/s12931-019-1045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hu Y, Liu L, Wang Q, Zeng J, Chen C. PM2.5 exposure perturbs lung microbiome and its metabolic profile in mice. Sci Total Environ. 2020;721:137432. doi: 10.1016/j.scitotenv.2020a.137432. [DOI] [PubMed] [Google Scholar]
- Li T, Wu YN, Wang H, Ma JY, Zhai SS, Duan J. Dapk1 improves inflammation, oxidative stress and autophagy in LPS-induced acute lung injury via p38MAPK/NF-κB signaling pathway. Mol Immunol. 2020;120:13–22. doi: 10.1016/j.molimm.2020.01.014. [DOI] [PubMed] [Google Scholar]
- Lin Y, Lu X, Qiu X, Yin F, Faull KF, Tseng CH, Zhang JJ, Fiehn O, Zhu T, Araujo JA, Zhu Y. Arachidonic acid metabolism and inflammatory biomarkers associated with exposure to polycyclic aromatic hydrocarbons. Environ Res. 2022;212:113498. doi: 10.1016/j.envres.2022.113498. [DOI] [PubMed] [Google Scholar]
- Liu T, Hougen H, Vollmer AC, Hiebert SM. Gut bacteria profiles of Mus musculus at the phylum and family levels are influenced by saturation of dietary fatty acids. Anaerobe. 2012;18:331–337. doi: 10.1016/j.anaerobe.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Liu CW, Lee TL, Chen YC, Liang CJ, Wang SH, Lue JH, Tsai JS, Lee SW, Chen SH, Yang YF, Chuang TY, Chen YL. PM2.5-induced oxidative stress increases intercellular adhesion molecule-1 expression in lung epithelial cells through the IL-6/AKT/STAT3/NF-κB-dependent pathway. Part Fibre Toxicol. 2018;15:4. doi: 10.1186/s12989-018-0240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang T, Si B, Du H, Liu Y, Waqas A, Huang S, Zhao G, Chen S, Xu A. Intratracheally instillated diesel PM2.5 significantly altered the structure and composition of indigenous murine gut microbiota. Ecotoxicol Environ Saf. 2021;210:111903. doi: 10.1016/j.ecoenv.2021.111903. [DOI] [PubMed] [Google Scholar]
- Lobo BW, Lima CK, Teixeira MS, Silva NL, Takiya CM, Ramos MF, Miranda AL, Dellamora-Ortiz GM. Fish oil attenuates persistent inflammatory pain in rats through modulation of TNF-α and resolvins. Life Sci. 2016;152:30–37. doi: 10.1016/j.lfs.2016.03.034. [DOI] [PubMed] [Google Scholar]
- Lu Y, Boekschoten MV, Wopereis S, Müller M, Kersten S. Comparative transcriptomic and metabolomic analysis of fenofibrate and fish oil treatments in mice. Physiol Genomics. 2011;43:1307–1318. doi: 10.1152/physiolgenomics.00100.2011. [DOI] [PubMed] [Google Scholar]
- Luo G, Zhang L, Hu X, Qiu R. Quantifying public health benefits of PM2.5 reduction and spatial distribution analysis in China. Sci Total Environ. 2020;719:137445. doi: 10.1016/j.scitotenv.2020.137445. [DOI] [PubMed] [Google Scholar]
- Magbanua MJ, Roy R, Sosa EV, Weinberg V, Federman S, Mattie MD, Hughes-Fulford M, Simko J, Shinohara K, Haqq CM, Carroll PR, Chan JM. Gene expression and biological pathways in tissue of men with prostate cancer in a randomized clinical trial of lycopene and fish oil supplementation. PLoS ONE. 2011;6:e24004. doi: 10.1371/journal.pone.0024004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manisalidis I, Stavropoulou E, Stavropoulos A, Bezirtzoglou E. Environmental and Health Impacts of Air Pollution: A Review. Front Public Health. 2020;8:14. doi: 10.3389/fpubh.2020.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie C, Tan J, Macia L, Mackay CR. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol Rev. 2017;278:277–295. doi: 10.1111/imr.12556. [DOI] [PubMed] [Google Scholar]
- Monk JM, Liddle DM, Cohen DJ, Tsang DH, Hillyer LM, Abdelmagid SA, Nakamura MT, Power KA, Ma DW, Robinson LE. The delta 6 desaturase knock out mouse reveals that immunomodulatory effects of essential n-6 and n-3 polyunsaturated fatty acids are both independent of and dependent upon conversion. J Nutr Biochem. 2016;32:29–38. doi: 10.1016/j.jnutbio.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Nie P, Li Z, Wang Y, Zhang Y, Zhao M, Luo J, Du S, Deng Z, Chen J, Wang Y, Chen S, Wang L. Gut microbiome interventions in human health and diseases. Med Res Rev. 2019;39:2286–2313. doi: 10.1002/med.21584. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015;159:122–127. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh PJ, Balart LA, Johnson DA. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin Transl Gastroenterol. 2015;6:e91. doi: 10.1038/ctg.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusceddu MM, El Aidy S, Crispie F, O'Sullivan O, Cotter P, Stanton C, Kelly P, Cryan JF, Dinan TG. N-3 Polyunsaturated Fatty Acids (PUFAs) Reverse the Impact of Early-Life Stress on the Gut Microbiota. PLoS ONE. 2015;10:e0139721. doi: 10.1371/journal.pone.0139721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson RC, Seira Oriach C, Murphy K, Moloney GM, Cryan JF, Dinan TG, Paul Ross R, Stanton C. Omega-3 polyunsaturated fatty acids critically regulate behaviour and gut microbiota development in adolescence and adulthood. Brain Behav Immun. 2017;59:21–37. doi: 10.1016/j.bbi.2016.07.145. [DOI] [PubMed] [Google Scholar]
- Safaei Ardestani SZ, Rahideh ST. The effect of omega-3 fatty acid supplementation on clinical and biochemical parameters of critically ill patients with COVID-19: a randomized clinical trial. J Transl Med. 2022;20:32. doi: 10.1186/s12967-022-03237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Criq V, Lugo-Villarino G, Thomas M. Dysbiosis, malnutrition and enhanced gut-lung axis contribute to age-related respiratory diseases. Ageing Res Rev. 2021;66:101235. doi: 10.1016/j.arr.2020.101235. [DOI] [PubMed] [Google Scholar]
- Samet JM, Hatch GE, Horstman D, Steck-Scott S, Arab L, Bromberg PA, Levine M, McDonnell WF, Devlin RB. Effect of antioxidant supplementation on ozone-induced lung injury in human subjects. Am J Respir Crit Care Med. 2001;164:819–825. doi: 10.1164/ajrccm.164.5.2008003. [DOI] [PubMed] [Google Scholar]
- Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S, Ibba I, Lai MA, Orrù S, Blois S, Loizedda AL, Griffin JL, Usai P, Caboni P, Atzori L, Manzin A. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. 2017;7:9523. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Weber CB, Akdis M, Akdis CA. TH17 cells in the big picture of immunology. J Allergy Clin Immunol. 2007;120:247–254. doi: 10.1016/j.jaci.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- van Soest APM, Hermes GDA, Berendsen AAM, van de Rest O, Zoetendal EG, Fuentes S, Santoro A, Franceschi C, de Groot L, de Vos WM (2020): Associations between Pro- and Anti-Inflammatory Gastro-Intestinal Microbiota, Diet, and Cognitive Functioning in Dutch Healthy Older Adults: The NU-AGE Study. Nutrients 12. 10.3390/nu12113471 [DOI] [PMC free article] [PubMed]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Watson H, Mitra S, Croden FC, Taylor M, Wood HM, Perry SL, Spencer JA, Quirke P, Toogood GJ, Lawton CL, Dye L, Loadman PM, Hull MA. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut. 2018;67:1974–1983. doi: 10.1136/gutjnl-2017-314968. [DOI] [PubMed] [Google Scholar]
- Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Wei FR, Xiong LL, Li W, Wang XR, Hong X, Chen BA. Relationship between Short-term Exposure to PM2.5 and Daily Lung Cancer Mortality in Nanjing. Biomed Environ Sci. 2020;33:547–551. doi: 10.3967/bes2020.072. [DOI] [PubMed] [Google Scholar]
- Williams AM, Phaneuf DJ, Barrett MA, Su JG. Short-term impact of PM2.5 on contemporaneous asthma medication use: Behavior and the value of pollution reductions. Proc Natl Acad Sci U S A. 2019;116:5246–5253. doi: 10.1073/pnas.1805647115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LG. Omega-3 polyunsaturated fatty acids and chronic obstructive pulmonary disease. Curr Opin Clin Nutr Metab Care. 2015;18:128–132. doi: 10.1097/mco.0000000000000142. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Gan CX, Chen SS, Li JQ, Liu MZ, Guo GH. BMSC-derived exosomes alleviate smoke inhalation lung injury through blockade of the HMGB1/NF-κB pathway. Life Sci. 2020;257:118042. doi: 10.1016/j.lfs.2020.118042. [DOI] [PubMed] [Google Scholar]
- Yu HN, Zhu J, Pan WS, Shen SR, Shan WG, Das UN. Effects of fish oil with a high content of n-3 polyunsaturated fatty acids on mouse gut microbiota. Arch Med Res. 2014;45:195–202. doi: 10.1016/j.arcmed.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Yu PF, Pang LL, Mao QS, Zou SC, Shi Y, Lin DJ. Dose dependency PM2.5 aggravated airway inflammation in asthmatic mice via down-regulating expression of ITGB4. Eur Rev Med Pharmacol Sci. 2019;23:1688–1697. doi: 10.26355/eurrev_201902_17131. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wang B, Zhang X, Chen X, Zhu J, Zou Y, Li J. Alpha-linolenic acid protects against lipopolysaccharide-induced acute lung injury through anti-inflammatory and anti-oxidative pathways. Microb Pathog. 2020;142:104077. doi: 10.1016/j.micpath.2020.104077. [DOI] [PubMed] [Google Scholar]
- Zhu X, Bi Z, Yang C, Guo Y, Yuan J, Li L, Guo Y (2021): Effects of different doses of omega-3 polyunsaturated fatty acids on gut microbiota and immunity. Food Nutr Res 65 10.29219/fnr.v65.6263 [DOI] [PMC free article] [PubMed]
- Zhuang P, Shou Q, Wang W, He L, Wang J, Chen J, Zhang Y, Jiao J. Essential Fatty Acids Linoleic Acid and α-Linolenic Acid Sex-Dependently Regulate Glucose Homeostasis in Obesity. Mol Nutr Food Res. 2018;62:e1800448. doi: 10.1002/mnfr.201800448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.