Abstract
Under certain cellular conditions, transcription and mRNA translation in prokaryotes appear to be “coupled,” in which the formation of mRNA transcript and production of its associated protein are temporally correlated. Such transcription-translation coupling (TTC) has been evoked as a mechanism that speeds up the overall process, provides protection against premature termination, and/or regulates the timing of transcript and protein formation. What molecular mechanisms underlie ribosome-RNAP coupling and how they can perform these functions have not been explicitly modeled. We develop and analyze a continuous-time stochastic model that incorporates ribosome and RNAP elongation rates, initiation and termination rates, RNAP pausing, and direct ribosome and RNAP interactions (exclusion and binding). Our model predicts how distributions of delay times depend on these molecular features of transcription and translation. We also propose additional measures for TTC: a direct ribosome-RNAP binding probability and the fraction of time the translation-transcription process is “protected” from attack by transcription-terminating proteins. These metrics quantify different aspects of TTC and differentially depend on parameters of known molecular processes. We use our metrics to reveal how and when our model can exhibit either acceleration or deceleration of transcription, as well as protection from termination. Our detailed mechanistic model provides a basis for designing new experimental assays that can better elucidate the mechanisms of TTC.
Significance
Transcription-translation coupling (TTC) in prokaryotes is thought to control the timing of protein production relative to transcript formation. The marker for such coupling has typically been the measured time delay between the first completion of transcript and protein. We formulate a stochastic model for ribosome and RNAP elongation that also includes RNAP pausing and ribosome-RNAP binding. The model is able to predict how these processes control the distribution of delay times and the level of protection against premature termination. We find relative speed conditions under which ribosome-RNAP interactions can accelerate or decelerate transcription. Our analysis provides insight on the viability of potential TTC mechanisms under different conditions and suggests measurements that may be potentially informative.
Introduction
In prokaryotic cells, transcription and translation of the same genes are sometimes “coupled” in that the first mRNA transcript is detected coincidentally with the first protein associated with that transcript. This observation suggests the proximity of the ribosome to the RNA polymerase (RNAP), as was directly observed by Miller et al. (1). These findings contributed to the idea that the ribosome and RNAP may interact directly or indirectly. Ribosome-RNAP interactions in prokaryotes are thought to maintain the processivity of RNAP by physically pushing it out of the paused, backtracking state (2). Paused RNAP has also been associated with transcriptional error correction and premature mRNA cleavage (3,4,5,6); thus, ribosome-RNAP interactions that catalyze RNAP unstalling and speeding up the overall process might do so at the expense of more errors in the transcript. Transcription-translation coupling (TTC) may also play an important role in protecting mRNA from premature transcription termination (7,8,9). This protection might arise from steric shielding of the elongation complex by the leading ribosome, preventing attack by Rho (9,10).
Evidence for TTC has come from two types of experiments. The first is “time-of-flight” experiments that quantify the time delay between first detection of a complete transcript and a complete protein. For example, IPTG-induced LacZ completion experiments (LacZ assays) measure the mean time of mRNA completion and the mean time of protein completion by the leading ribosome , with the latter measured from the time of first RNAP engagement (11,12,13). Since the transcript length L is known, the effective velocities of the RNAP and ribosome over the entire transcript can be estimated by
| (1) |
These measurements are performed at the population level, averaging the time-dependent signal from many newly formed transcripts and corresponding proteins. Thus, the individual molecular coupling mechanisms between RNAP and ribosomes cannot be resolved by the time delay unless single-molecule time-of-flight experiments can be designed.
Another class of experiments uses a variety of in vitro and in vivo assays to probe direct and indirect molecular interactions between RNAPs and ribosomes (9,14,15,16). A recent structural study identified active transcription-translation complexes and confirmed an indirect molecular interaction in situ (17).
Two modes of coupling between the leading ribosome and the RNAP have been proposed. One mode of coupling occurs through a “collided expressome” in which the ribosome and RNAP are held in close proximity (9,14) by direct association, which may be mediated by entropic effects of the intervening mRNA (18). The second coupling mode occurs through a larger complex in which ribosome-RNAP interactions are mediated by the proteins NusG and NusA (15,16,17,18). There have been no reports that this mode alters elongation speeds or RNAP processivity, although both are possible. It has been shown that the NusG-coupled expressome can inhibit Rho-induced premature transcription termination (19).
Both potential coupling mechanisms require at least some moments of close proximity between the RNAP and the leading ribosome during the simultaneous transcription-translation process (see Fig. 1), followed by recruitment of NusG for the NusG-coupled expressome mechanism. The ribosome-RNAP proximity requirement can be met if the ribosome elongation speed is, on average, faster than that of the RNAP. Even if the ribosome is fast, proximity also depends on initial condition (ribosome initiation delay after RNAP initiation) and the length of the transcript L. Moreover, both RNAPs and ribosomes are known to experience, respectively, pausing through backtracking (6) and through “slow codons” for which the associated tRNA is scarce (20).
Figure 1.
Schematic of translation of a nascent mRNA transcript (polypeptide not shown). Ribosomes translocate at rate p while RNAPs elongate at rate q. (A) The transcript associated with the RNAP at position n along the gene is shown with a leading ribosome at position m along the mRNA. Ribosomes attach to open initiation sites at rate α. (B) A nearly complete transcript is shown. If the leading ribosome has caught up to the RNAP and m is close to n, the two may bind with rate to form a “coupled expressome.” Ribosome-RNAP complexes can spontaneously dissociate with rate . We assume that the leading ribosome “terminates” upon reaching the stop codon (not shown). Protein-mediated expressomes (not shown) form larger complexes that can accommodate longer mRNA segments within it.
A number of open questions remain. In the “strong coupling” picture, the ribosome and RNAP are nearly always in contact and the ribosome is thought to maintain RNAP processivity during mRNA transcription. Without the ribosome, mRNA transcription can be particularly slow. Therefore, the ribosome elongation rate dictates the RNAP elongation rate. Administration of antibiotics to slow down translation also slowed down transcription (11).
However, other experiments have shown that the distance between the ribosome and RNAP can be large most of the time, leading to a “weak coupling” picture (21,22). The biological role of weak coupling is unclear since any shielding provided by the ribosome would be limited and ribosome and RNAP speeds could be independently modulated. Even though direct ribosome-RNAP interactions may still arise after an RNAP has stalled for a sufficiently long time, any apparent ribosome-RNAP coordination would be largely coincidental.
Besides the strong and weak coupling dichotomy, another unresolved question is whether or not TTC can slow down transcription. Traditionally, TTC has been invoked as a mechanism for maintaining RNAP processivity by rescuing RNAP from paused states. However, in vivo experiments by Kohler et al. (9) reported that, when translation is inhibited, the mutant in which RNAP does not associate with ribosome exhibited faster proliferation than that of wild-type RNAP that can associate with ribosomes. Coupled transcription through ribosome-RNAP association may give rise to slower transcription. Although in situ cryo-EM has also shown protein-mediated complexes of ribosome and RNAP (23), additional studies should be devised to confirm and characterize ribosome-RNAP interactions in vivo. Nonetheless, our mathematical model can provide possible mechanistic insights into TTC.
To help resolve the puzzles discussed above, provide a quantitative way to explore different molecular mechanisms that may contribute to TTC, and generate predictions that can be compared with experimental observations, we formulate a stochastic model that combines a number of known molecular mechanisms from transcription, keeping track of ribosome and RNAP states and positions along the gene. While an earlier model combined transcription and translation in prokaryotes (24), it did not explicitly incorporate mechanisms of direct transcription and translation coupling and only assumed simple volume exclusion between the RNAP and the leading ribosome.
Here, we explicitly allow for RNAP pausing and formal association and dissociation of the ribosome-RNAP complex. Here, “association” will be generally used to denote specific or nonspecific, direct or indirect molecular interactions. The typical assay used to probe TTC involves measuring the time delay between the completion of mRNA and its associated protein. Although time delays can be used as a metric for defining TTC, absence of delay is a necessary but not sufficient condition for direct ribosome-RNAP coupling. Even a direct observation of an interacting expressome does not imply functional consequences of TTC. After formulating our model, we construct additional metrics that better define and reflect possible attributes of TTC. However, since the time delay is the most experimentally measurable quantity, we will still derive and compute the full probability density of delay times .
Model and methods
Based on existing structural and interaction information, we formulate a continuous-time Markov chain to model ribosome and RNAP kinetics. As shown in Fig. 1, we describe the position of the head of the leading ribosome along the nascent transcript by , where 0 denotes a ribosome-free transcript. We also track the length of the nascent mRNA transcript that has cleared the exit channel of the RNAP through the discrete variable . The positions are described in terms of triplets of nucleotides corresponding to codons, the fundamental step size during ribosome elongation.
Here, L is the length of the coding region of the transcript, typically about codons. Since we are interested in the elongation phase of coupled transcription and translation, the length L is measured from the ribosome binding site immediately upstream of the start codon to the stop codon. We carefully choose the definition of m and n so that ribosome and RNAP sizes are irrelevant and that the difference precisely describes the length of the free intervening mRNA between them. Effects of TTC on transcription termination have been previously discussed in (25,26). Therefore, , , and is interpreted as a completed mRNA while indicates a completed polypeptide. Thus triangular state-space defined by often arises in related stochastic models of interacting particles in one dimension (6,27,28). Thus far, we assumed monocistronic mRNAs. Polycistronic mRNA with multiple internal ribosome entry sites can be modeled as successive processes with shared RNAP coordinates, independent ribosomes, and L being the respective cistron length.
Within each positional state , the leading ribosome and RNAP can exist in different internal configurations describing their molecular states. The RNAP at site n can switch between two states, a processive state and a paused state. In the processive state, the RNAP can move forward by one codon at rate or it can transition to a paused or “backtracking” state with stalling rate . The RNAP elongation rate can also depend on its position n through different abundances of corresponding nucleotides. For simplicity, we assume that RNAPs in the backtracking state are fixed and do not elongate () but may transition back to the processive state with unstalling rate . The waiting time distributions in the processive and paused states are exponential with mean and , respectively. The leading ribosome at site m will be assumed to always be in a processive state with forward hopping rate if and only if the next site is empty (not occupied by the downstream RNAP). In general, the ribosome translation rate can depend on the position m through the codon usage at that site.
When the distance between the leading ribosome and the RNAP is within an interaction range , (), they may associate or “bind” with rate to form a coupled expressome and dissociate with rate (Eqs. 6 and 7). Note that simply describes proximity and not necessarily molecular coupling. In cases where ribosome-RNAP complex formation is mediated by other proteins and cofactors, such as NusG, etc., would represent an effective complex formation rate that could depend on NusG availability. To enumerate coupled and paused-RNAP internal states, we define such that refers to an associated, or “bound” ribosome-RNAP complex, and refers to an RNAP in a backtracking, or a “paused” or “stalled” state. When , the ribosome is not bound to the RNAP, and when , the RNAP is in the processive state. The state space of our discrete stochastic model is given by , with representing ribosome-free configurations. By appropriately choosing , , and , our model can effectively describe both the collided expressome and the NusG-mediated expressome. Experimentally, the collided expressome is typically observed in a stalled configuration (9,17,18,29). This observation is consistent with a small and a large , which suggests that the coupled, processive state a is transient. We will define which is not restricted to positive values.
Other than steric exclusion (which constrains ) and ribosome-RNAP association and dissociation, we incorporate a contact-based RNAP “pushing” mechanism. The processing ribosome can directly push (powerstroke) against a stalled RNAP and/or reduce the entropy of a backtracking RNAP to bias it toward a processive state. A similar mechanism arises in RNAP-RNAP interactions, as discussed in (6). To quantify this pushing mechanism, we simply modify the paused-to-processive RNAP () transition rate from to whenever the ribosome abuts the RNAP (). The enhanced rate arises from a reduction in the total transition free energy barrier provided by the adjacent ribosome. Typical model parameters relevant to prokaryotic transcription and translation are listed in Table 1.
Table 1.
Model parameters
| Parameters | Description | Typical valuesa | Refs. |
|---|---|---|---|
| α | translation initiation rate | (25,30,31,32) b | |
| L | gene and transcript length | (33) | |
| m | ribosome position from mRNA 5′ | – | |
| n | RNAP position from mRNA 5′ | – | |
| p | free ribosome translocation rate | codons/s | (11,25,34,35) |
| q | free processing RNAP transcription rate | codons/s | (11,12,13,36) c |
| processive RNAP paused RNAP rate | (37) d | ||
| paused RNAP processive RNAP rate | (37) | ||
| paused RNAP processive RNAP rate (pushed) | estimated | ||
| ribosome-RNAP association, dissociation rates | , | (14) | |
| maximum mRNA length in bound complex | codons | (29) |
The unit of length assumed throughout the paper will be nucleotide triplets (codons).
Translation initiation is highly variable. In accordance with LacZ completion assays (25,30), we initially set in our simulations and later discuss the effects of varying translation initiation rates.
Typical noninteracting RNAP transcription rates are codons/s. Since typically , we use typical values codons/s for the unimpeded transcription rate of processing RNAP.
The pausing probability along an RNAP trajectory has been measured as per 100 nucleotides. By using the estimated mean RNAP velocity of codons/s, we convert this probability to a pausing rate /s.
The length may influence direct molecular coupling and stochastic dynamics of transcription. In vitro studies of ribosome and RNAP structure provide constraints on the configuration space accessible to coupled expressomes. Wang et al. (29) and Webster et al. (18) found that collided expressomes are stable only when the spacer mRNA between the ribosome and the RNAP is nucleotides ( codons). Because the intervening mRNA must be at least 12 nucleotides to extend beyond the RNA exit channel of the RNAP, the free intervening RNA within an intact collided expressome can vary between 0 and 12 nucleotides. In contrast, the NusG-mediated expressome can accommodate free mRNA nucleotides. RNA looping might allow for even longer spacer mRNA, but there has so far been no in vivo evidence that collided expressomes exist with mRNA loops.
Since mRNA is flexible, we can also assume that is constant for . The association rate may be dependent on the distance between the ribosome and the RNAP; for example, a distance-dependent association rate might take the form , where represents the effective volume fraction of the leading ribosome and ξ is the configuration flexibility of ribosome-RNAP binding when they are close. If we adopt such a distance-dependent , we would also have to let the ratio be dependent on in order to conserve free energy during approach and binding steps. To simplify matters, we will assume that and take to be a constant for and zero for .
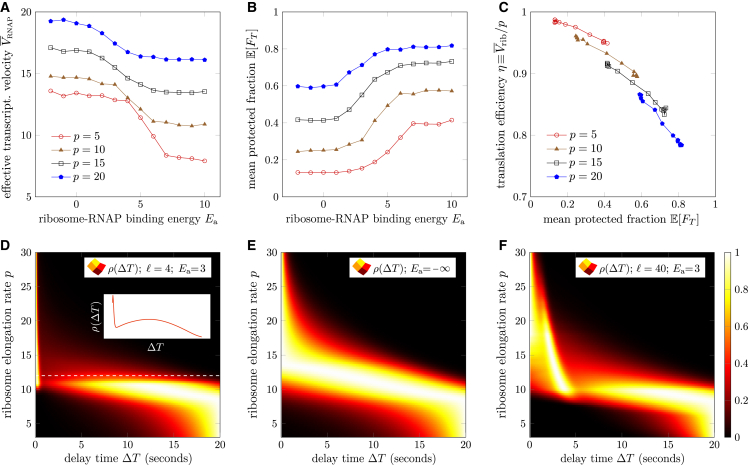
The overall kinetics of the internal states pictured in the insets of Fig. 2 can be explicitly summarized by considering the intervening mRNA length between RNAP and the ribosome. Fig. S1 in Appendix S1 explicitly depicts the transitions for different d. Since, in our model, the maximum length of mRNA that can fit within the complex is , a processing ribosome-bound RNAP at cannot advance to lengthen the already compressed transcript (see Fig. 2 E). The only way a coupled state with can reach any state where is for the ribosome and RNAP to first dissociate (we assume dissociation rates in all states remain constant at ). Molecular coupling effectively slows down transcription by preventing RNAP elongation in the state. Such ribosome-mediated slowing down of transcription has been proposed in previous studies (2,9).
Figure 2.
(A) State space of the stochastic model defined in terms of the leading ribosome and RNAP positions . The initial time is defined as the time RNAP first produces a ribosome initiation site, starting the system in . For , as the RNAP is elongating, the first ribosome binds at rate α. Here, a ribosome binds after the RNAP first reaches position . Red and blue trajectories indicate scenarios in which the RNAP is relatively fast and slow, respectively. Within each position exist internal molecular microstates. (B) In the “interior” states ( in this example), the ribosome and RNAP are too distant to be bound, and only stalled and processing RNAP states arise, with transition rates between them. (C) When , the ribosome and RNAP are adjacent without any intervening mRNA, allowing them to associate with rate . The RNAP can be in either stalled or processive states. In the stalled state, whether associated or not, the adjacent volume-excluding ribosome entropically “pushes” the stalled RNAP, catalyzing its transition to a processive state so that . (D) When , the ribosome and the RNAP are close enough to bind with rate . Here, the intervening mRNA dissipates the entropic pushing (so that the stalled RNAP processing RNAP transition rate is ) and also allows an RNAP in the processive state to elongate with rate q, regardless of whether it is bound to the ribosome. (E) Only when the ribosome and the RNAP are separated by is a bound RNAP prevented from processing as this would reel in more mRNA than can be fit inside the complex, either a collided or NusG-mediated expressome. Molecular binding prevents complexed ribosome and RNAP to be separated by more than mRNA codons.
We now list all allowed transitions in the state space of our continuous-time stochastic Markov model. The probability that an allowable transition from state ω to state occurs in time increment is , where the complete set of rates is given by
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
Using these rules, we performed event-based stochastic simulations (38,39) of the model as detailed in Appendix S1 of the supporting material. For completeness, the master equation associated with our model is also formally given in Appendix S2.
Construction of time-delay distribution
Our stochastic molecular model allows for the explicit calculation of the probability density of time delay between mRNA and polypeptide completion. To find , we first find the distribution of ribosome positions at the moment the RNAP first reaches site i. denotes the instant the mRNA is completed. The initial value is known because, immediately after initiation of RNAP at site , the ribosome is not yet present but is trying to bind at a rate of α. As detailed in Appendix S3, we can iteratively find the distribution of given that of . By the same method, the distribution of association values at the instant of RNAP completion can be computed. After constructing the probability distribution , we evolve it to find the distribution of the additional amount of time required for m to first reach L.
Although we are able to construct the whole distribution of delay times that might provide a more resolved metric, especially if single-molecule or single-cell assays can be developed, a short time delay is a necessary but not sufficient condition for TTC. To provide direct information on molecular ribosome-RNAP interactions, we construct additional metrics.
Analytic approximation
Although our stochastic model contains a complex state space with many parameters, simple physical limits are immediately apparent. If the free ribosome translocation rate p is much greater than the free RNAP transcription rate q and , the ribosome, for much of the time, abuts the RNAP, inducing it to transcribe at an average rate . Here, we predict an expected delay , , and . If the ribosome is slow and , the ribosome and RNAP are nearly always free, , , and . However, when p is intermediate, more intricate kinetics can arise, including tethered elongation and transcription slowdown.
To gain insight into this intermediate case, we first assume the large system limit and divide the states into those with and those with . Within each metastate, the behavior of the ribosome and RNAP is relatively homogeneous. States with are completely decoupled, while the proximal states can include associated or unassociated ribosome/RNAP substates.
To characterize the transition between the two metastates, we define the mean dwell time in metastate by and the mean dwell time in the metastate by . The variability in the state dwell time arises mainly from RNAP stalling and unstalling. When translation is slow, the RNAP may undergo multiple stalling-unstalling cycles before leaving the metastate. For faster translation, one stalling event will typically be enough to leave the metastate. Let be the probability of leaving following one RNAP stalling event. With details provided in Appendix S4 of the supporting material, we find analytic approximations to and in the and small limit:
| (11) |
| (12) |
As , the ribosome and RNAP are less likely to be proximal and . Once , the RNAP and the ribosome remain separated. On the other hand, as , , , and is always monotonically increasing with p. Kinetics between these two metastates qualitatively capture the behavior of the model in the translation-invariant, infinite length limit. The approximations, and will used in some of our subsequent results.
Metrics of TTC
We now define additional metrics that help characterize TTC. To more explicitly quantify direct molecular coupling, we define the coupling coefficient C by
| (13) |
the probability that the ribosome is associated with the RNAP () at the moment the mRNA transcript is completed. The coupling parameter C provides a more direct measure of molecular coupling and further resolves configurations that have short or negligible delays. While delay time distributions do not directly quantify ribosome-RNAP contact, the coupling coefficient C does not directly probe the trajectories or history of ribosome-RNAP dynamics.
To also characterize the history of ribosome-RNAP interactions, we quantify TTC by the fraction of time that the ribosome “protects” the RNAP across the entire transcription process. There are different ways of defining how the transcript is protected. While both modes of TTC are proposed to shield the mRNA from premature termination, neither has been directly observed in vivo. We assume that a termination protein has size and that, if the ribosome and RNAP are closer than , the termination factor is excluded. Thus, we define the protected time as the total time that codons, divided by the time to complete transcription:
| (14) |
Since the transcription-termination protein Rho has an mRNA footprint of about 80 nt, codons (40). A high reflects transcription that has been protected against exposure to Rho-mediated termination. The fraction of transcription events that is prematurely terminated is then proportional to . Exploration of this fraction under different conditions might be a way to experimentally estimate .
Using these metrics, including the effective velocities and , we will explore properties and predictions of our model.
Results and discussion
Here, we present analyses of solutions to our model obtained from numerical recursion and Gillespie-type kinetic Monte Carlo simulations detailed in Appendices S1, S2, and S3 of the supporting material. Predictions derived from using different parameter sets are compared, and mechanistic interpretations are provided.
Comparison of TTC metrics
We evaluate our stochastic model to provide quantitative predictions for the coupling indices, , C, and . The results are summarized in Fig. 3.
Figure 3.
Comparison of different TTC indices. Common parameters for all these plots are /s, , , , , , and /s, unless stated otherwise. (A) The delay-time density plotted for codons/s and codons/s. Densities for /s, /s (rarely pausing RNAPs, green curves), and /s, /s (frequently pausing RNAPs, red curves) are shown. Within these cases, strong-binding (, /s, ) and no-binding (, ) subcases are indicated by solid and dashed curves, respectively. (B) Mean delay as a function of p and q. (C) The direct coupling coefficient C as a function of the relative velocity . Each point represents C evaluated at specific values of (p,q), each chosen from all integers between 3 and 27 codons/s. The dashed curve represents the analytic approximation given by Eq. 15. (D) Heatmap of . (E) Values of , each derived from 1000 kinetic Monte-Carlo (kMC) trajectories, plotted against . The analytic approximation given by Eq. 16 is shown by the dashed curve. (F) The heatmap of .
Limitations of mean delay times
Fig. 3A shows delay-time distributions for various parameter sets and reveals subtle differences in the kinetic consequences of coupling. Without molecular coupling , the distribution has a single peak around the mean delay time. With molecular coupling, the distribution can exhibit two peaks with one at . This short-time peak reflects trajectories that terminate as a bound ribosome-RNAP complex. These finer structures in cannot be resolved by evaluating only the mean delay time. Fig. 3 B plots the mean delay as a function of p and q. For our chosen parameters, in particular /s and , we see that is rather featureless, with a significant delay arising only for small p. Thus, the mean delay time provides little information about the details of TTC.
Coupling coefficient
The dimensionless ratio is a key indicator of the overall level of coupling possible. If , the speed of the ribosome exceeds the average speed of the RNAP, allowing them to approach each other and potentially form a coupled expressome. If , the ribosome speed is slower than the average RNAP speed and the system can at most be only transiently coupled. It turns out that the coupling coefficient C is mostly determined by alone, particularly if the effects of translation initiation vanishes for large L (see “effects of translation initiation rates” section later). Essentially, the transition to a coupled system (large C) is predicted when . In Fig. 3 C, we find the values of C for multiple values of p and q (each dot corresponds to each pair), and plot them as a function of , with . The mean values of C as a function of p and q are plotted in Fig. 3 D and are qualitatively distinct from the mean times shown in (B). Given Eqs. 11 and 12, the steady-state coupling coefficient C for can be approximated as
| (15) |
which qualitatively agrees with numerical results as shown in Fig. 3 C.
Fraction of time protected
Each point in Fig. 3 E indicates the mean value , , for different values of p and q, arranged along values of . Each value was computed by averaging protected-time fractions (Eq. 14) from 1000 simulated trajectories. As expected, increases with ribosome translation rate p until saturation to above for codons/s.
Comparing C and from Fig. 3 D and F we find that C and are qualitatively similar across various values of p and q, although in general we find . The transition from low to high values occurs at lower values of p for since the condition for protection () is not as stringent as that for ( and binding). Thus, there can be value of for which is small but is close to one.
The similarity between and C is restricted to the dependence on p and q. The coupling and the protection time fraction may respond to changes in other parameters in drastically different ways. For example, C is nonzero only if molecular binding is present, rendering it sensitive to . However, directly measures the dynamics of TTC and does not depend on actual molecular coupling, so it will be relatively insensitive to , particularly when p is large. Thus, may be a better index if we wish to quantify functional consequences of TTC. The standard deviation of the simulated values is large and approximately (shown in Appendix S5 of the supporting material), limiting the suitability of the mean protected-time fraction as a robust metric.
As with the coupling coefficient C, can be estimated under our heuristic approximations:
| (16) |
From Eq. 16, we see that is comprised of two terms, the protection provided by coupling , and the protection provided by ribosome elongation when . Although Eq. 16 is valid mainly for positive and , the protection from physical proximity exists even if molecular association is completely absent. This formula confirms our observation that .
Binding-induced slowdown
We now use our model to reveal the major factors contributing to TTC-induced slowdown of transcription and discuss limits this slowdown.
Unstalling rate dictates ribosome efficiency
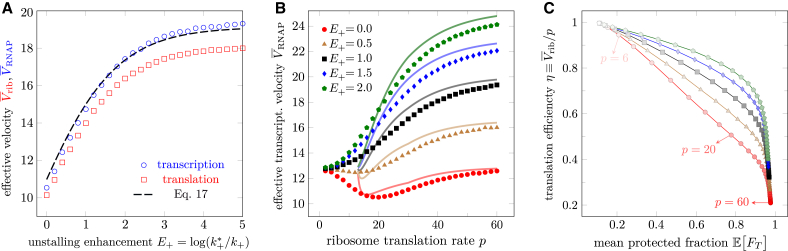
The principal factor that influences the overall velocity of a coupled expressome is the interplay between two antagonistic mechanisms: ribosome-mediated dislodging of an adjacent stalled RNAP and bound-ribosome deceleration of the RNAP. When the reduction in activation free energy of unstalling, , is large, the ribosome is less likely to be impeded by a stalled RNAP. Fig. 4 A plots the effective velocities and as a function of and illustrates the increases in overall speed when the ribosome is more effective at dislodging a stalled RNAP (higher ).
Figure 4.
Slowdown induced by molecular coupling. The common parameters used are the same as those in Fig. 3. (A) Effective velocities as a function activation energy reduction in ribosome-induced RNAP unstalling, codons/s and codons/s. (B) Effective velocity of RNAP as a function of the free ribosome translation rate p. (C) The trade-off between translation efficiency and mean fraction of time protected . The efficiency is defined by the ratio of the mean ribosome speed to the translation rate of an isolated ribosome. In (B) and (C), codons/s. The variances (not shown) for the plotted quantities are large, typically overlapping the mean-value curves in (A and B). The dashed curve in (A) and solid curves in (B) are the analytical predictions of the effective velocities using Eq. 17. To see this figure in color, go online.
The decrease in the velocity of a coupled processing RNAP is primarily determined by the ribosome translation speed p. For different values of , the dependence of on p can be quite different, as is shown in Fig. 4 B. For large , when the ribosome efficiently pushes stalled RNAPs, increasing p allows the ribosome to more frequently abut the RNAP and dislodge it, leading to faster overall transcription. However, for inefficient unstalling (small ), we see that faster ribosomes can decrease . This feature arises because for inefficient unstalling, a larger p increases the fraction of time the ribosome and RNAP are bound , allowing a binding-induced slowdown to arise more often. Besides , the emergence of a decreasing transcription velocity with increasing ribosome translation rate p depends intricately on factors, such as , , , and arises only if is sufficiently large and is not too large.
Although the decrease in is not large, it certainly suggests that increasing p under small is not advantageous. This observation motivates us to define a translation efficiency as the ratio of the effective ribosome speed to its unimpeded translation speed p: . The loss measures how much a ribosome is impeded due to its interactions with the RNAP. As p is increased, we find trajectories that display a trade-off between translation efficiency and protected time. Higher p leads to more proximal ribosomes and protected RNAP at the expense of translation efficiency η. Fig. 4 C shows that the decrease in unstalling activation energy affects this level of trade-off. For large , increasing p can speed up ribosomes beyond the velocity determined by so that η decreases more slowly than . At the same time, the system is only slightly less coupled, leading to a subtle decrease in . In the end, larger leads to a higher η versus curve.
Low η values are likely selected against since a cell would be expending more resources than necessary to maintain high levels of tRNA and other translation factors. A potentially optimal setting may be to maintain , which is the minimally sufficient velocity to keep the RNAP protected. This intermediate choice of p for the ribosome may explain the recent observations that slower ribosomes did not appreciably slow down transcription (22) or prevent folding of specific mRNA segments (21).
When and , the ribosome and the RNAP intermittently close on each other and share a common effective velocity. In states , the velocity is determined by the slower of the ribosome or the processive RNAP. While the system is in states , its velocity is given by , as detailed in Appendix S4. Finally, the overall effective velocity is estimated by
| (17) |
Eq. 17 agrees qualitatively well with simulation and reproduces the slowdown, as shown in Fig. 4 A and B.
Limits of binding-induced slowdown
When and , we explore the lower bound for the effective velocity of RNAP by examining . For sufficiently large and , the term is approximately , and lower bounds for are
| (18) |
The first equality holds when , and the second equality holds when . We conclude that the maximum slowdown induced by binding is essentially limited by the slowdown of RNAP due to transcriptional road blocks. The latter plays a fundamental role in the significantly slower rate of mRNA transcription nt/s relative to rRNA transcription nt/s.
Testing the molecular coupling hypothesis
It is informative to use our proposed indices to compare scenarios that predict molecular coupling to those that do not. We now vary the binding energy for different velocity ratios . For , Fig. 5 A shows as a function of for various values of p. Although higher p leads to increased , for each value of p, increasing the binding energy increases coupling and leads to RNAP slowdown. Since the most significant slowdown arises when translation is slow, association-induced slowdown is indicative of strong coupling. Both p and increase , as shown in Fig. 5 B. The variation in and with also suggests that, within the context of our model, one can use experimental measurements of these indices to estimate . As is tuned in our model we also predict a trade-off between ribosome efficiency and protection, as shown in Fig. 5 A. Since our analytic approximation is most accurate in the , regime, deviations in and are mainly due to finite L effects when ; thus, we forgo plotting our analytic approximations in Fig. 5 A–C. In Appendix S6, we provide additional simulation results that confirm the dependence in Eq. 17 and in .
Figure 5.
Effects of molecular coupling. For those parameters not varied, we use the same values used to generate Figs. 3 and 4. (A) For , the effective transcription velocity as a function of binding-energy depth between the ribosome and RNAP . (B) Mean protected-time fraction as a function of binding energy depth (). (C) The trade-off between efficiency and protection for . (D) Rescaled heatmap of the delay-time distribution as a function of ribosome translocation rate p. The brightness indicates the relative probability, and the inset shows the probability distribution at codons/s indicated by the dashed white line. Here, the binding energy and . For codons/s, is bimodal in . (E) Delay-time distribution in the absence of ribosome-RNAP binding (). Here, the dependence disappears and is mono-modal. (F) Delay-time distribution for and . Bimodality arises in more than one regime of p. To see this figure in color, go online.
Additional parallel experiments may show consistent evidence of coupling; thus, we investigate the distribution of delay times as p is varied. Fig. 5 D shows rescaled so that the largest value is set to unity for easier visualization. We see that for intermediate values of (unit codons/s, omitted henceforth), can be bimodal. Fig. 5 E depicts a single peaked when coupling is completely turned off by setting . In this case, is irrelevant. Fig. 5 F shows the rescaled in the presence of coupling () for . Here, there are two regimes, and , that exhibit bimodality. The delay-time density would be ideally measured via single-molecule or single-cell experiments combined with proper fluorescent imaging. However, in Appendix S7 we also consider how improved bulk LacZ completion assays can be used to estimate features of . We find that the distribution of ribosome completion times is always mono-modal and that the bimodality of is mainly the result of bimodality in .
Genome-wide variability of coupling
We have so far assumed all parameters are homogeneous along the transcript and time independent. However, a cell is able to dynamically regulate the transcription and translation of different genes by exploiting the transcript sequence or other factors that mediate the process. Such regulation can be effectively described within our model by varying its parameters in the appropriate way.
Regulation of RNAP pausing
The RNAP pausing rate is one parameter that can be modulated by specific DNA sequences and other roadblocks along the gene (36,41,42,43). There is evidence that consensus pause sequences are enriched at the beginning of genes (44,45). In addition to leading ribosomes, a trailing RNAP can also push the leading RNAP out of a paused state by increasing , much like ribosomes (6,36). Even if and are varied in our model, the overall predicted performance regimes of the system are still delineated by values of , and the effective transcription velocity can still be predicted by Eq. 17.
Effects of translation initiation rates
Translation initiation is another process that can be altered by the cell through, e.g., initiation factors that modulate the initiation rate α (46). Genome-wide analysis reveals that translation initiation times in E. coli are highly variable, ranging from less than 1 s to more than 500 s (31,47).
As shown in Figs. S8 A–C of Appendix S8, varying the translation initiation rate α straightforwardly affects TTC. As indicated in (A), the peak in at /s persists across different values of p. Slower translation initiation results in larger initial separations , decreasing the overall fraction of protected times, as shown in (B) and (C). To mitigate large initial distances and lower likelihood coupling due to slow initiation, RNAP pausing occurs more often at the start of the gene to allow time for a slow-initiating ribosome to catch up. Thus, delayed ribosome initiation and early RNAP pausing are two “opposing” processes that can regulate coupling and efficiency, especially for short genes.
A simple analytic description of how the interplay between length L and transcription initiation rate α affects our TTC metrics could not be found. However, large/small α generally leads to higher/lower C and compared with those associated with the approximations (Eqs. 15 and 16), as shown in Fig. S9 in Appendix S8. For sufficiently small α, a rough estimate of the deviation from steady state is , the typical time needed for the ribosome to catch up to the RNAP divided by the time needed for the RNAP to finish the transcription.
Ribosome translocation rate profiles
Although we have thus far assumed uniform ribosome translocation rates, it is known that codon bias and tRNA/amino acid availability can locally affect ribosome translocation (20,48). Snapshots of ribosome positions along transcripts have been inferred from ribosome profiling experiments. After imposing a stochastic exclusion model (49), Khanh and coworkers (50) reconstructed position-dependent ribosome translocation rates . They found that hopping rates are larger near the 5′ end and decrease toward the 3′ end. Although they reconstructed the entire genome-wide profile, translocation rates are gene dependent, so we will propose and test simple profiles .
To qualitatively match the inferred profile (50), we define profile 1 by increasing p by 50% for the first 40 codons, and decreasing it by 50% for the second 40 codons. The rest of the transcript retains the constant baseline value of p. Profile 2 is similarly defined except that, instead of being the second group of 40 codons, the speed across the last 40 codons is decreased. We compared the performance of the three different profiles in Fig. S8 D–F as a function of the mean translation rate . We conclude that the differences in the effective velocities and protection fraction are subtle, suggesting that these performance statistics are relatively insensitive to different speed profiles.
Conclusions
We have presented a detailed stochastic model of TTC. The continuous-time discrete-state model tracks the distance between the leading ribosome and the RNAP and assumes they sterically exclude each other along the nascent mRNA transcript. All current experimental understanding of interactions between RNAP and ribosome, including ribosome initiation, RNAP pausing, and direct ribosome-RNAP association, have also been incorporated. Our model exhibits a number of rich features that depend on the interplay of these intermediate mechanisms.
To quantitatively investigate the predictions of our model, we constructed three different metrics to quantify TTC, the delay-time probability distribution , the probability C that the ribosome and the RNAP are in a bound state at termination, and the fraction of time that the ribosome and the RNAP are proximal over the entire process. is a measure of protection against binding of termination proteins. These metrics were computed or simulated under different model parameters. Specifically, since a bound RNAP at distance from the trailing ribosome needs to first detach before can be increased, the states shown in Figs. 2 and S1 form an effective attractive well that tethers RNAP to ribosome. By allowing direct ribosome-RNAP binding, we find that this effective attraction zone can allow a slower ribosome to dynamically hold back bound RNAP, leading to decreased . Indirect coupling mediated by Nus proteins provides a longer interaction range, which may improve translation efficiency η compared with direct coupling where codons.
Qualitatively, our model predicts two different regimes of TTC that appear to be consistent with observations. One limit can arise when is large, resulting in close proximity and strong molecular coupling that may lead to slowdown of RNAP, while the other arises when is small leading to intermittent contacts and perhaps modest speed up of pausing RNAPs. Besides , our model suggests that , α, and also control which type of TTC arises. Across different genes, and are expected to be unchanged, but variations in (and to some degree α) can affect the balance between these qualitative models of TTC. Our analysis suggests that the unstalling enhancement is a key factor determining how TTC is manifested. For inefficient unstalling, increased coupling slows down the whole expressome, which might be disadvantageous under growth conditions. Gene-dependent kinetic parameters and signaling pathways involving ppGpp (12,22) and other factors can also be incorporated into our model to provide a more complete picture of how TTC influences cellular regulation.
Our model reveals that a bimodal time-delay distribution when is a hallmark of molecular association. We provide a discussion of possible measurements of this distribution through traditional bulk assays in the supporting material. By quantitatively controlling ribosome speed p and measuring the corresponding effective velocity measured in the LacZ-completion assay or other single-molecule assays. such as DNA curtains, important kinetic information associated with the delay-time density may be revealed by fitting our model to data.
Measurement of important parameters and indices, such as and , could be obtained from quantitative analysis of previous experiments (2,22). Cryo-EM approaches used in (17) might also be used to obtain a genome-wide estimate of the coupling coefficient C. Finally, FRET experiments or super-resolution imaging may shed light on macromolecular-level ribosome and RNAP dynamics (51). Our model can guide how in vitro measurements can be designed and used to inform delay-time distributions , coupling coefficients C, protected-time fractions , and efficiencies η.
Modeling of transcription and translation coupling may also inform the stochastic modeling of gene transcription dynamics. Simple kinetic models of transcription that incorporate the active and inactive states of the gene, the transcription initiation, elongation, pausing, premature termination, and the degradation of mRNA products have been developed to understand cell-to-cell variability of gene-specific mRNA levels (52,53,54). Such models could be merged with our modeling approach to accommodate TTC.
Author contributions
X.L. and T.C. devised and analyzed the model and wrote the paper. X.L. developed the computational algorithms and performed the numerical calculations and kinetic Monte Carlo simulations.
Acknowledgments
This work was supported by grants from the NIH through grant R01HL146552 and the NSF through grant DMS-1814364 (to T.C.).
Declaration of interests
The authors declare no competing interests.
Editor: Jason Kahn.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.09.041.
Supporting material
References
- 1.Miller O.L., Hamkalo B.A., Thomas C.A. Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- 2.Stevenson-Jones F., Woodgate J., et al. Zenkin N. Ribosome reactivates transcription by physically pushing RNA polymerase out of transcription arrest. Proc. Natl. Acad. Sci. USA. 2020;117:8462–8467. doi: 10.1073/pnas.1919985117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sydow J.F., Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr. Opin. Struct. Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. Catalysis and regulation/Proteins. [DOI] [PubMed] [Google Scholar]
- 4.Knippa K., Peterson D.O. Fidelity of RNA polymerase II transcription: role of Rbp9 in error detection and proofreading. Biochemistry. 2013;52:7807–7817. doi: 10.1021/bi4009566. [DOI] [PubMed] [Google Scholar]
- 5.Sahoo M., Klumpp S. Backtracking dynamics of RNA polymerase: pausing and error correction. J. Phys. Condens. Matter. 2013;25:374104. doi: 10.1088/0953-8984/25/37/374104. [DOI] [PubMed] [Google Scholar]
- 6.Zuo X., Chou T. Density- and elongation speed-dependent error correction in RNA polymerization. Phys. Biol. 2022;19:026001. doi: 10.1088/1478-3975/ac45e2. [DOI] [PubMed] [Google Scholar]
- 7.Chalissery J., Muteeb G., et al. Sen R. Interaction surface of the transcription terminator Rho required to form a complex with the C-terminal domain of the antiterminator NusG. J. Mol. Biol. 2011;405:49–64. doi: 10.1016/j.jmb.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 8.Lawson M.R., Ma W., et al. Berger J.M. Mechanism for the regulated control of bacterial transcription termination by a universal adaptor protein. Mol. Cell. 2018;71:911–922.e4. doi: 10.1016/j.molcel.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohler R., Mooney R.A., et al. Cramer P. Architecture of a transcribing-translating expressome. Science. 2017;356:194–197. doi: 10.1126/science.aal3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma C., Mobli M., et al. Lewis P.J. RNA polymerase-induced remodelling of NusA produces a pause enhancement complex. Nucleic Acids Res. 2015;43:2829–2840. doi: 10.1093/nar/gkv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proshkin S., Rahmouni A.R., et al. Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer S., Le D., Park B.R., Kim M. Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli. Nat. Microbiol. 2018;3:741–748. doi: 10.1038/s41564-018-0161-3. [DOI] [PubMed] [Google Scholar]
- 13.Vogel U., Jensen K.F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan H., Conn A.B., et al. Blaha G.M. Transcription–translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Res. 2017;45:11043–11055. doi: 10.1093/nar/gkx719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooney R.A., Davis S.E., et al. Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxena S., Myka K.K., et al. Gottesman M.E. Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol. Microbiol. 2018;108:495–504. doi: 10.1111/mmi.13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Reilly F.J., Xue L., Graziadei A., Sinn L., Lenz S., Tegunov D., Blötz C., Singh N., Hagen W.J.H., Cramer P., Stülke J., Mahamid J., Rappsilber J. In-cell architecture of an actively transcribing-translating expressome. Science. 2020;369:554–557. doi: 10.1126/science.abb3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webster M.W., Takacs M., et al. Weixlbaumer A. Structural basis of transcription-translation coupling and collision in bacteria. Science. 2020;369:1355–1359. doi: 10.1126/science.abb5036. [DOI] [PubMed] [Google Scholar]
- 19.Burmann B.M., Schweimer K., et al. Rösch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 20.Chou T., Lakatos G. Clustered bottlenecks in mRNA translation and protein synthesis. Phys. Rev. Lett. 2004;93:198101. doi: 10.1103/PhysRevLett.93.198101. [DOI] [PubMed] [Google Scholar]
- 21.Chen M., Fredrick K. Measures of single- versus multiple-round translation argue against a mechanism to ensure coupling of transcription and translation. Proc. Natl. Acad. Sci. USA. 2018;115:10774–10779. doi: 10.1073/pnas.1812940115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu M., Mori M., et al. Dai X. Disruption of transcription–translation coordination in Escherichia coli leads to premature transcriptional termination. Nat. Microbiol. 2019;4:2347–2356. doi: 10.1038/s41564-019-0543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washburn R.S., Zuber P.K., et al. Frank J. Escherichia coli NusG links the lead ribosome with the transcription elongation complex. iScience. 2020;23:101352. doi: 10.1016/j.isci.2020.101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mäkelä J., Lloyd-Price J., et al. Ribeiro A.S. Stochastic sequence-level model of coupled transcription and translation in prokaryotes. BMC Bioinf. 2011;12:121–213. doi: 10.1186/1471-2105-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson G.E., Lalanne J.-B., et al. Li G.-W. Functionally uncoupled transcription–translation in Bacillus subtilis. Nature. 2020;585:124–128. doi: 10.1038/s41586-020-2638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R., Zhang Q., et al. Shi H. Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the Rho-independent terminator. Nucleic Acids Res. 2016;44:2554–2563. doi: 10.1093/nar/gkv1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou T. Peeling and sliding in nucleosome repositioning. Phys. Rev. Lett. 2007;99:058105. doi: 10.1103/PhysRevLett.99.058105. [DOI] [PubMed] [Google Scholar]
- 28.Teimouri H., Spaulding C., Kolomeisky A.B. Optimal pathways control fixation of multiple mutations during cancer initiation. Biophys. J. 2022;121:3698–3705. doi: 10.1016/j.bpj.2022.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C., Molodtsov V., et al. Ebright R.H. Structural basis of transcription-translation coupling. Science. 2020;369:1359–1365. doi: 10.1126/science.abb5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai X., Zhu M., Warren M., et al. Hwa T. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2016;2:16231. doi: 10.1038/nmicrobiol.2016.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaham G., Tuller T. Genome scale analysis of Escherichia coli with a comprehensive prokaryotic sequence-based biophysical model of translation initiation and elongation. DNA Res. 2018;25:195–205. doi: 10.1093/dnares/dsx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennell D., Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J. Mol. Biol. 1977;114:1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- 33.Xu L., Chen H., et al. Luo Z.W. Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between the two kingdoms. Mol. Biol. Evol. 2006;23:1107–1108. doi: 10.1093/molbev/msk019. [DOI] [PubMed] [Google Scholar]
- 34.Young R., Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem. J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu M., Dai X., Wang Y.-P. Real time determination of bacterial in vivo ribosome translation elongation speed based on LacZα complementation system. Nucleic Acids Res. 2016;44:e155. doi: 10.1093/nar/gkw698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epshtein V., Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 37.Neuman K.C., Abbondanzieri E.A., et al. Block S.M. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 38.Bortz A.B., Kalos M.H., Lebowitz J.L. A new algorithm for Monte Carlo simulation of Ising spin systems. J. Comput. Phys. 1975;17:10–18. [Google Scholar]
- 39.Gillespie D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. A. 1977;81:2340–2361. [Google Scholar]
- 40.Koslover D.J., Fazal F.M., et al. Block S.M. Binding and translocation of termination factor Rho studied at the single-molecule level. J. Mol. Biol. 2012;423:664–676. doi: 10.1016/j.jmb.2012.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komissarova N., Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 42.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davenport R.J., Wuite G.J., et al. Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 44.Hatoum A., Roberts J. Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol. Microbiol. 2008;68:17–28. doi: 10.1111/j.1365-2958.2008.06138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larson M.H., Mooney R.A., et al. Weissman J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou T. Ribosome recycling, diffusion, and mRNA loop formation in translational regulation. Biophys. J. 2003;85:755–773. doi: 10.1016/S0006-3495(03)74518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siwiak M., Zielenkiewicz P. Transimulation - protein biosynthesis web service. PLoS One. 2013;8:e73943. doi: 10.1371/journal.pone.0073943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klumpp S., Dong J., Hwa T. On ribosome load, codon bias and protein abundance. PLoS One. 2012;7:e48542. doi: 10.1371/journal.pone.0048542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacDonald C.T., Gibbs J.H. Concerning the kinetics of polypeptide synthesis on polyribosomes. Biopolymers. Biopolymers. 1969;7:707–725. [Google Scholar]
- 50.Dao Duc K., Song Y.S. The impact of ribosomal interference, codon usage, and exit tunnel interactions on translation elongation rate variation. PLoS Genet. 2018;14:e1007166. doi: 10.1371/journal.pgen.1007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morisaki T., Lyon K., et al. Stasevich T.J. Real-time quantification of single RNA translation dynamics in living cells. Science. 2016;352:1425–1429. doi: 10.1126/science.aaf0899. [DOI] [PubMed] [Google Scholar]
- 52.Choubey S., Kondev J., Sanchez A. Deciphering transcriptional dynamics in vivo by counting nascent RNA molecules. PLoS Comput. Biol. 2015;11:e1004345. doi: 10.1371/journal.pcbi.1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu H., Skinner S.O., et al. Golding I. Stochastic kinetics of nascent RNA. Phys. Rev. Lett. 2016;117:128101. doi: 10.1103/PhysRevLett.117.128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filatova T., Popovic N., Grima R. Statistics of nascent and mature RNA fluctuations in a stochastic model of transcriptional initiation, elongation, pausing, and termination. Bull. Math. Biol. 2020;83:3. doi: 10.1007/s11538-020-00827-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.